Abstract
Rationale
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is a lethal developmental disorder of lung morphogenesis caused by insufficiency of FOXF1 (forkhead box F1) transcription factor function. The cellular and transcriptional mechanisms by which FOXF1 deficiency disrupts human lung formation are unknown.
Objectives
To identify cell types, gene networks, and cell–cell interactions underlying the pathogenesis of ACDMPV.
Methods
We used single-nucleus RNA and assay for transposase-accessible chromatin sequencing, immunofluorescence confocal microscopy, and RNA in situ hybridization to identify cell types and molecular networks influenced by FOXF1 in ACDMPV lungs.
Measurements and Main Results
Pathogenic single-nucleotide variants and copy-number variant deletions involving the FOXF1 gene locus in all subjects with ACDMPV (n = 6) were accompanied by marked changes in lung structure, including deficient alveolar development and a paucity of pulmonary microvasculature. Single-nucleus RNA and assay for transposase-accessible chromatin sequencing identified alterations in cell number and gene expression in endothelial cells (ECs), pericytes, fibroblasts, and epithelial cells in ACDMPV lungs. Distinct cell-autonomous roles for FOXF1 in capillary ECs and pericytes were identified. Pathogenic variants involving the FOXF1 gene locus disrupt gene expression in EC progenitors, inhibiting the differentiation or survival of capillary 2 ECs and cell–cell interactions necessary for both pulmonary vasculogenesis and alveolar type 1 cell differentiation. Loss of the pulmonary microvasculature was associated with increased VEGFA (vascular endothelial growth factor A) signaling and marked expansion of systemic bronchial ECs expressing COL15A1 (collagen type XV α 1 chain).
Conclusions
Distinct FOXF1 gene regulatory networks were identified in subsets of pulmonary endothelial and fibroblast progenitors, providing both cellular and molecular targets for the development of therapies for ACDMPV and other diffuse lung diseases of infancy.
Keywords: alveolar capillary dysplasia, FOXF1, pulmonary microvasculature
At a Glance Commentary
Scientific Knowledge on the Subject
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is a lethal developmental disorder of lung morphogenesis causing diffuse lung disease and respiratory failure in infancy. Most cases are caused by pathogenic variants involving the FOXF1 (forkhead box F1) gene locus. The cellular and transcriptional mechanisms whereby FOXF1 deficiency disrupts human lung formation are unknown.
What This Study Adds to the Field
This study integrated pulmonary histopathology, immunofluorescence microscopy, and single-nucleus RNA and assay for transposase-accessible chromatin sequencing data obtained from lung tissue from infants with ACDMPV. Alterations in cellular composition, cell type–specific gene expression patterns and networks, and cell–cell interactions underlying the disruption of lung morphogenesis and respiratory failure in infants with ACDMPV were identified. Pathogenic variants involving the FOXF1 gene locus perturbed gene expression in endothelial cell progenitors, inhibiting the differentiation or survival of capillary 2 endothelial cells and cell–cell interactions necessary for normal pulmonary vasculogenesis and alveolar epithelial cell differentiation, while demonstrating marked expansion of the systemic bronchial circulation. The cell type–selective transcriptional targets of FOXF1 and associated signaling networks identified in this study provide insights into the mechanisms of lung morphogenesis and a framework for the development of therapies for disorders of pulmonary vascular formation and function.
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV; Mendelian Inheritance in Man #265380) is one of several developmental disorders of lung morphogenesis causing diffuse lung disease and respiratory failure in infancy (1–3). Most ACDMPV cases are caused by de novo single-nucleotide variants (SNVs) or copy-number variant (CNV) deletions involving the FOXF1 (forkhead box F1) gene locus, altering the expression or activity of FOXF1; however, familial transmission also occurs. To date, more than 100 pathogenic SNVs, including missense, nonsense, and frameshift mutations and indels, as well as 70 CNV deletions involving FOXF1 have been reported (4). Most infants with ACDMPV present immediately after birth with unremitting cyanosis, pulmonary hypertension, and respiratory failure despite ventilatory support, vasodilators, or extracorporeal membrane oxygenation. Affected infants die in the first days or months after birth. Less common are infants who survive into childhood with pulmonary hypertension and respiratory insufficiency but often require lung transplantation early in life for survival (2, 5, 6). Histopathologic findings in ACDMPV include deficient alveolar development resulting in lobular simplification, thickened alveolar septa containing a paucity of pulmonary capillaries, arterial hypertensive changes, and “mislocalization” of pulmonary veins in bronchovascular bundles rather than being restricted to the pleural and interlobular septa as in the normal lung (1). In addition, vascular perfusion studies have demonstrated prominent right-to-left intrapulmonary vascular shunt pathways in ACDMPV, linking systemic and pulmonary circulations that bypass the alveolar capillary bed (7, 8).
Studies of mice in which Foxf1 was deleted or in which an ACDMPV human pathogenic variant was introduced into the mouse Foxf1 locus have recapitulated some, but not all, of the findings seen in the human disorder. A majority of these mice die of respiratory failure at birth, associated with pulmonary hypertension and abnormalities in the alveolar microvasculature (9–11). In the mouse, Foxf1 is expressed throughout the embryonic septum transversum and splanchnic mesenchyme, where it influences pulmonary, cardiac, and gastrointestinal tissue morphogenesis, consistent with the multiorgan malformations that can be seen in human infants with ACDMPV (10, 12).
During lung morphogenesis, FOXF1 is expressed throughout the splanchnic mesenchyme, including subsets of fibroblast and endothelial progenitors, as well as their derivatives (9, 13). FOXF1 functions in gene regulatory networks (GRNs) controlling epithelial–mesenchymal interactions required for lung growth and patterning (14). Pathogenic variants in FOXF1 and related GRNs disrupt pulmonary morphogenesis, causing deficient lung development in mice and infants with ACDMPV and other forms of diffuse lung growth abnormalities (15, 16). In the present study, we integrated histopathology, immunofluorescence microscopy, and single-nucleus RNA sequencing (snRNA-seq) and assay for transposase-accessible chromatin (ATAC) sequencing data to identify cells and cell type–specific GRNs and to predict cell–cell interactions underlying the disruption of lung morphogenesis and respiratory failure in infants with ACDMPV.
Methods
Peripheral lung tissue samples from deidentified subjects with ACDMPV were obtained from the Cincinnati Children’s Hospital Medical Center and the Washington University School of Medicine. Large airways or large bronchovascular bundles were not included in the study. Combinatorial barcoding single-nucleus ATAC sequencing (snATAC-seq) (17) and 10X Chromium (10x Genomics) snRNA-seq were performed on three frozen lung tissue samples (one biopsy specimen, two explants) from subjects with ACDMPV (ACD1–ACD3). 10X Multiome (10x Genomics) single-nucleus ATAC and gene expression sequencing was performed on autopsy lung tissue samples from subjects ACD4 and ACD5. For control lung samples, we obtained publicly available snATAC-seq and snRNA-seq data from three 3-year-old control donors and three preterm neonate lungs (17) and single-cell RNA sequencing (scRNA-seq) data (two biological replicates; depleted of CD45 [cluster of differentiation 45] positive cells by bead selection) from a 2-month-old normal lung (18). Single-cell RNA and ATAC analyses are described in the online supplement. Immunofluorescence confocal microscopy and RNA in situ hybridization were used for cross-validation experiments.
Results
Single-Nucleus Transcriptional Profiles and Chromatin Accessibility in Human ACDMPV Lung Tissues with Varying Disease Severity
Peripheral lung samples were obtained from six infants with histopathologic diagnoses of ACDMPV associated with SNVs or CNV deletions involving the FOXF1 gene locus. Five subjects were prioritized for snRNA-seq and snATAC-seq (Table 1; see Table E1 in the online supplement). Three of the subjects died at 2–5 weeks of age with histopathologic diagnoses confirmed by lung biopsy (ACD1) or autopsy (ACD4 and ACD5). The remaining two subjects had less severe lung phenotypes, resulting in prolonged survival and allowing lung transplantation at 9 months (ACD3) and 3.5 years (ACD2) of age, with ACDMPV histopathology confirmed in lung explants. Lung histology was diagnostic of ACDMPV in all subjects, with a paucity of alveolar septal capillaries, arterial hypertensive changes, and atypical placement of pulmonary veins within the bronchovascular structures (Figures 1 and E1). ACDMPV histopathologic features were diffusely present within the lungs of infants dying within 2–5 weeks of birth (ACD1, ACD4, and ACD5). In contrast, lung explants from the subjects surviving to lung transplantation at 3.5 years (ACD2) and 9 months (ACD3) of age had nonuniform ACDMPV features with patchy areas of deficient alveolar development interspersed with areas of normally formed alveoli characterized by thin alveolar septa containing appropriately formed capillary networks correctly positioned subjacent to differentiated alveolar type 1 (AT1) and alveolar type 2 (AT2) cells (Figure 1). To profile the wide spectrum of ACDMPV phenotypes, lung samples from neonates ACD1 (biopsy), ACD4 (autopsy), and ACD5 (autopsy) were selected to represent the most severe phenotypes, with lung explantation samples from 9-month-old (ACD3) and 3.5-year-old (ACD2) subjects chosen to represent a gradient of less severe phenotypes. Data obtained from snRNA-seq/snATAC-seq of these five ACDMPV lung tissues were compared with nondiseased lung published data sets (17) (see Table E1) from neonates (n = 3; preterm [29–31 wk of gestational age] and 1–4 d of age) and control donor lungs (n = 3; 3 yr old) to identify cell types expressing the FOXF1 gene and to impute gene regulatory pathways influenced by FOXF1.
Table 1.
Characteristics of Subjects with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins
| Subject ID | Sex | Age | Race/Ethnicity | FOXF1 Variant |
|---|---|---|---|---|
| ACD1* | Male | 2 wk | White/non-Hispanic | c.249del;p.Phe84Serfs*74 (heterozygous) |
| ACD2* | Male | 3.5 yr | Other/non-Hispanic | c.175A>G;p.Met59Val (heterozygous) |
| ACD3* | Male | 9 mo | White/Hispanic | c.144G>C;p.Lys48Asn (heterozygous) |
| ACD4* | Male | 5 wk | White | 1.3-Mb deletion at 16q24.1 (hg19, chr16:85,038,804–86,353,606) |
| ACD5* | Male | 1 mo | White | 1.48-Mb deletion at 16q24.1 (hg19, chr16:84,738,447–86,218,175) |
| ACD6 | Female | 2 h | White | 1.7-Mb deletion at 16q24.1 (hg19, Chr16:85,063,102–86,721,852) |
Definition of abbreviations: FOXF1 = forkhead box F1; ID = identifier.
Lung tissue samples were used for single-nucleus RNA and assay for transposase-accessible chromatin sequencing.
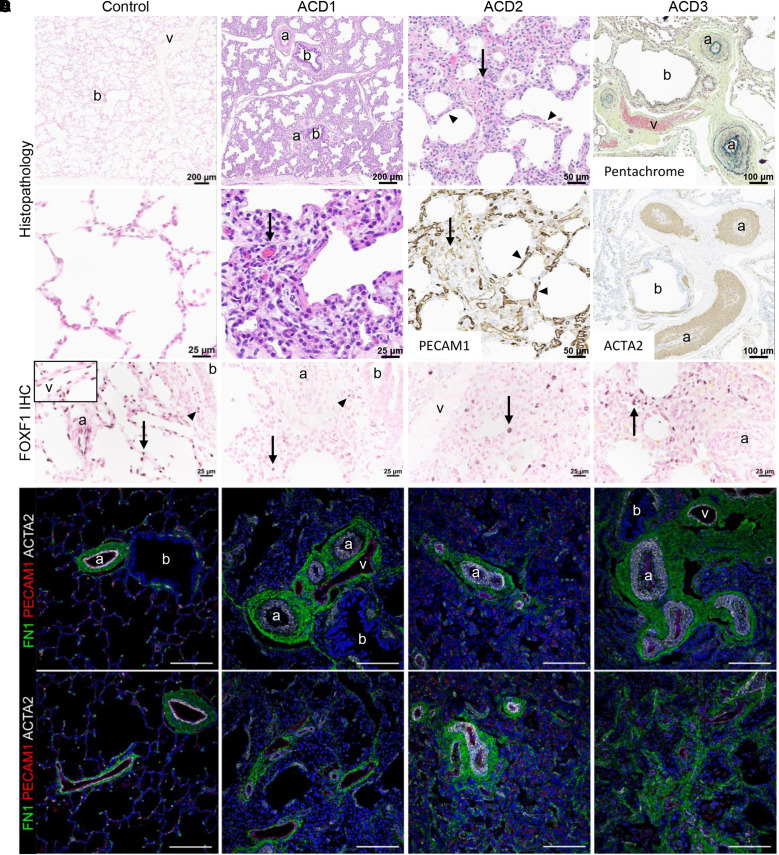
Figure 1.
Human lungs with alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) with a spectrum of phenotype severity used for single-nucleus RNA and assay for transposase-accessible chromatin sequencing. (A) Lung histology of patient biopsy (ACD1) and explants (ACD2 and ACD3) was diagnostic of ACDMPV in all subjects and markedly differed from control lungs. ACDMPV lungs had deficient alveolar development with thickened alveolar septa containing a paucity of abnormally patterned capillaries (arrows) highlighted by PECAM1 (platelet and EC adhesion molecule 1) immunohistochemistry (IHC). Marked arterial hypertensive changes were present, with medial smooth muscle thickening of the small arteries (a) highlighted by ACTA2 (actin alpha 2, smooth muscle) IHC and pentachrome stains. ACDMPV lungs also had atypical placement of dilated veins (v) adjacent to arteries and bronchioles (b), as opposed to the veins’ being restricted to the pleura and interlobular septa in control lung (6 months of age). Explanted lungs from ACD2 and ACD3, who underwent transplantation at 3.5 years and 9 months of age, respectively, contained patchy ACDMPV features (ACD2, arrow) interspersed with more normally developed alveoli separated by thin septa containing capillaries immediately subjacent to the alveolar epithelium as seen in control lungs and highlighted by PECAM1 IHC (ACD2, arrowheads). Histology of ACD4 and ACD5 ACDMPV lungs at autopsy is shown in Figure E1. (B) FOXF1 (forkhead box F1) IHC showing diffuse alveolar septal (arrow), airway smooth muscle cell (arrowhead), arterial (a) and venous (v, inset) endothelial cell (EC) staining in control lung (6 months of age). FOXF1 expression was absent in arterial and venous ECs in ACDMPV lungs, with patchy alveolar septal (arrows) and airway smooth muscle cell staining (arrowhead). (C) Immunofluorescence staining for FN1 (fibronectin 1), PECAM1, and ACTA2 showing bronchovascular structures in control lung and misalignment of dilated veins (v) adjacent to bronchioles (b) and arteries (a), with hypertensive changes including thickened mural smooth muscle in ACDMPV lungs. Scale bars, 100 μm. (D) Deficient alveolar development and a paucity of alveolar septal capillaries in ACDMPV lungs compared with control lungs are highlighted by PECAM1 staining with diffuse deficiency of the capillary network in subject ACD1, with the most severe phenotype, and patchy areas with a more extensive capillary network in subject ACD2, with the least severe phenotype. Scale bars, 100 μm.
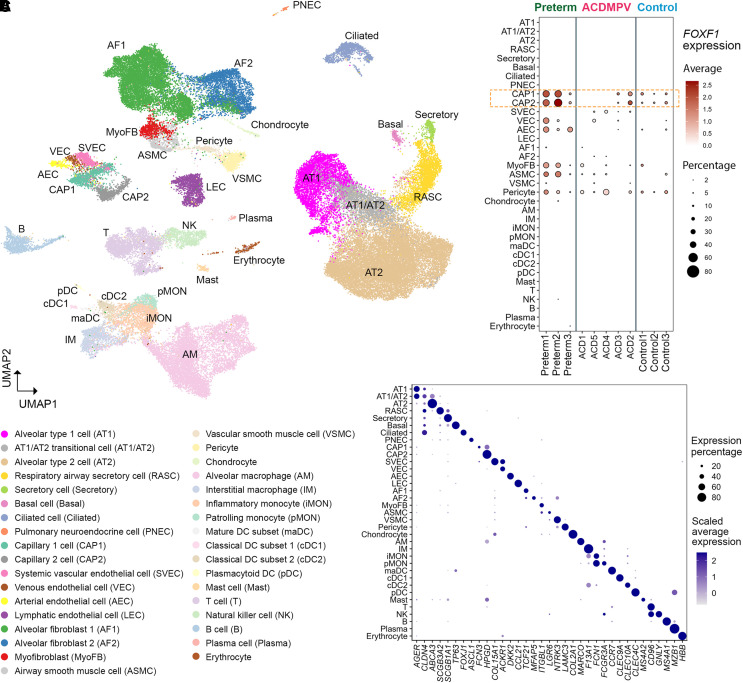
Cell types in snRNA-seq analysis were identified using a combination of three approaches: 1) iterative unbiased clustering analysis, 2) automated cell type annotation using cell identities in the published control data (17) and the recently released LungMAP Human Lung CellRef (19), and 3) and selective expression of cell type markers in LungMAP CellRef (19) and CellCards (20). Manual curation of the results from these three approaches was performed to identify consensus cell types at the cluster level (see the online supplement and Figures E2 and E3). In total, 35 distinct cell types were identified using snRNA profiling of ACDMPV (32,300), preterm neonate (15,817), and control infant lung (17,692) nuclei. Eight epithelial cell types (AT1, AT2, AT1/AT2 transitional [AT1/AT2], respiratory airway secretory, secretory, ciliated, basal, and pulmonary neuroendocrine cells), 6 endothelial cell (EC) types (capillary 1 [CAP1] cells [also known as general capillary cells] [21], capillary 2 [CAP2] cells [also known as aerocytes or Car4+ ECs] [21–23], systemic vascular ECs [SVECs; also known as peribronchial vascular ECs (24) or bronchial vessels (17), coexpressing COL15A1 (collagen type XV α 1 chain) and PECAM1 (platelet and endothelial cell adhesion molecule 1) (24) as well as ABCB1 (ATP binding cassette subfamily B member 1) (17)], venous ECs [coexpressing ACKR1 (atypical chemokine receptor 1) and CPE (carboxypeptidase E) (25) but not COL15A1 (26)], arterial ECs, and lymphatic ECs), 7 mesenchymal (alveolar fibroblast 1 [AF1] and alveolar fibroblast 2 [AF2] cells, myofibroblasts, airway/vascular smooth muscle cells, pericytes, and chondrocytes), and 14 immune cell types were identified (Figure 2). Expression of the proliferation marker MKI67 (marker of proliferation Ki-67) was low in the snRNA-seq data (see Figure E3G). Using unbiased clustering of chromatin accessibility patterns, we identified major cell populations and EC types from snATAC profiling of ACDMPV (36,335 nuclei), preterm (26,074 nuclei), and control (31,507 nuclei) lungs, consistent with cell identities identified in the snRNA-seq analysis (see Figure E4).
Figure 2.
Single-nucleus transcriptome atlas of alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) and control human lungs. (A) UMAP embedding of snRNA-seq of nuclei (n = 65,809) from ACDMPV (n = 5), preterm neonates (n = 3; 1–4 d of age and 29–31 wk of gestational age), and control lungs (n = 3; 3 yr old). (B) Dot plot of average expression amounts and expression percentage of FOXF1 RNA in each cell type in each lung. Expression with percentage ⩾2% is shown. (C) Dot plot of RNA expression of marker genes in single-nucleus RNA–identified cell types in A. Gene expression with percentage ⩾5% is shown. ABCA3 = ATP binding cassette subfamily A member 3; ACKR1 = atypical chemokine receptor 1; AEC = arterial endothelial cell; AF1 = alveolar fibroblast 1; AF2 = alveolar fibroblast 2; AGER = advanced glycosylation end-product specific receptor; AM = alveolar macrophage; ASCL1 = achaete-scute family BHLH transcription factor 1; ASMC = airway smooth muscle cell; AT1 = alveolar type 1 cell; AT2 = alveolar type 2 cell; CAP1 = capillary 1 cell; CAP2 = capillary 2 cell; CCL21 = C-C motif chemokine ligand 21; CCR7 = C-C motif chemokine receptor 7; CD96 = cluster of differentiation 96; cDC1 = classical dendritic cell subset 1; cDC2 = classical dendritic cell subset 2; CLDN4 = claudin 4; CLEC = C-type lectin domain containing; COL15A1 = collagen type XV α 1 chain; COL2A1 = collagen type II α 1 chain; DC = dendritic cell; DKK2 = Dickkopf WNT signaling pathway inhibitor 2; F13A1 = coagulation factor XIII A chain; FCGR3A = Fc gamma receptor IIIa; FCN = ficolin; FOXF1 = forkhead box F1; FOXJ1 = forkhead box J1; GNLY = granulysin; HBB = hemoglobin subunit β; HPGD = 15-hydroxyprostaglandin dehydrogenase; IM = interstitial macrophage; iMON = inflammatory monocyte; ITGBL1 = integrin subunit β like 1; LAMC3 = laminin subunit gamma 3; LEC = lymphatic endothelial cell; LGR6 = leucine rich repeat–containing G 55protein–coupled receptor 6; maDC = mature dendritic cell subset; MARCO = macrophage receptor with collagenous structure; MFAP5 = microfibril associated protein 5; MS4 = membrane spanning 4-domains; MyoFB = myofibroblast; MZB1 = marginal zone B and B1 cell-specific protein; NK = natural killer; NTRK3 = neurotrophic receptor tyrosine kinase 3; pDC = plasmacytoid dendritic cell; pMON = patrolling monocyte; PNEC = pulmonary neuroendocrine cell; RASC = respiratory airway secretory cell; SCGB3A = secretoglobin family 3A; snRNA-seq = single-nucleus RNA sequencing; SVEC = systemic vascular endothelial cell; TCF21 = transcription factor 21; TP53 = tumor protein P53; UMAP = uniform manifold approximation and projection; VEC = venous endothelial cell; VSMC = vascular smooth muscle cell.
FOXF1 RNA was expressed in endothelial and mesenchymal cells (CAP1 and CAP2 cells, pericytes, venous ECs, arterial ECs, and smooth muscle cells) in snRNA-seq of preterm neonates and healthy control infants (Figure 2B). We downloaded and reannotated recently published scRNA-seq (two biological replicates, 16,424 cells) of a 2-month-old normal infant lung and found that major cell types expressing FOXF1 RNA were consistent with those in the snRNA-seq data (see Figures E5A and E5B). Publicly available RNA expression data from human fetal lung (27) was also queried and demonstrated FOXF1 expression in capillary ECs and a subset of mesenchymal cells (see Figure E6). In ACDMPV lungs, FOXF1 RNA was absent in capillary ECs (CAP1 and CAP2 cells) from three of five subjects (ages 2–5 wk), despite its expression in other mesenchymal cell types. FOXF1 RNA was expressed in capillary cells from the two subjects with ACDMPV with less severe histopathologic changes and prolonged survival (ACD2 [3.5 yr old] and ACD3 [9 months old]). Consistent with the snRNA-seq results, FOXF1 protein was readily detected in all control lung tissues examined from donors at 1 day to 13 months of age (n = 5) localizing to mesenchymal cells within the alveolar septa, airway smooth muscle cells, and ECs lining the pulmonary arteries and veins (Figure 1). Remarkably, FOXF1 protein was not seen in venous or arterial ECs in ACDMPV lungs, despite being detected in mesenchymal cells within the alveolar septa and in airway smooth muscle cells (Figure 1). Patchy areas with more prominent FOXF1 protein expression within alveolar septal mesenchymal cells were seen in the ACDMPV explanted lungs (ACD2 and ACD3), consistent with the nonuniform histopathology and the less severe phenotypes with prolonged survival of these two subjects with ACDMPV.
Loss of Pulmonary Capillary and Expansion of Systemic ECs in ACDMPV Lungs
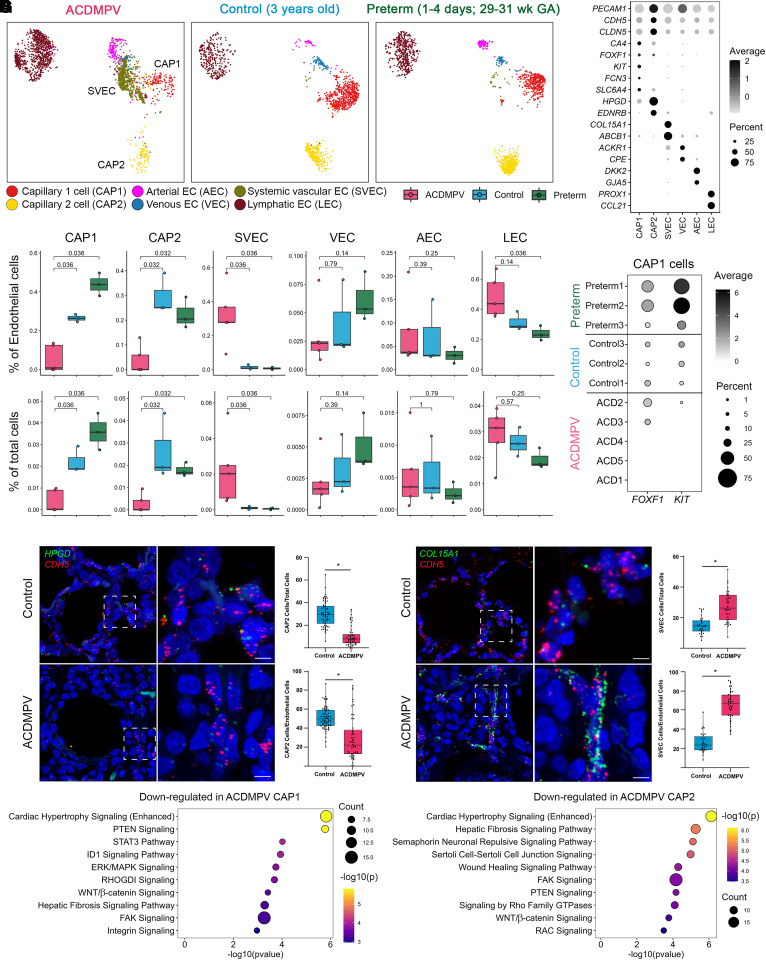
Significant differences in EC fractions were found between ACDMPV and control donor lungs (Figures 3A–3C, E5C, and E5D). CAP2 cells were markedly reduced or absent in all subjects with ACDMPV. CAP1 cells, considered CAP2 cell progenitors (21), were significantly reduced in ACDMPV lungs. A relatively higher proportion of CAP1 cells was identified in the two subjects with ACDMPV with prolonged survival (ACD2 [3.5 yr old] and ACD3 [9 months old]), likely reflecting patchy or nonuniform ACDMPV malformations within their lung explants and less severe phenotypes (5, 6). Accordingly, FOXF1 RNA expression amounts and frequencies varied in proportion to capillary cell frequencies in snRNA-seq of ACDMPV lungs (see Figure E7). Despite the expression of FOXF1 RNA in the retained CAP1 cells in ACD2 and ACD3, the expression of KIT (KIT proto-oncogene, receptor tyrosine kinase) was markedly reduced or absent in those retained CAP1 cells (Figures 3D and E5C), suggesting that FOXF1+/KIT+ CAP1 cells, an EC progenitor cell population that differentiates into arterial, venous, and CAP2 cells via FOXF1-mediated signaling in the mouse (28–31), were lost in ACDMPV lungs. In sharp contrast to the loss of pulmonary capillary ECs, SVECs, coexpressing COL15A1 and PECAM1 (24, 26) as well as ABCB1 (17), were markedly increased in ACDMPV lungs, indicating relative expansion of the systemic circulation in ACDMPV lungs. Previous studies using scRNA-seq and immunohistology analyses reported that COL15A1/PECAM1-expressing ECs were localized to systemically supplied vessels of the bronchial vascular plexus or visceral pleura in normal human lung (24, 26) and were increased in areas of bronchiolization and fibrosis in lung from patients with idiopathic pulmonary fibrosis (24). The reduction of CAP1 and CAP2 ECs and the increase of SVECs in ACDMPV were consistent compared with lungs from both control donors and preterm neonates, providing evidence that the alterations were disease associated. ATAC analysis of CAP1 and CAP2 cell and SVEC frequency changes in ACDMPV was consistent with the RNA analyses (see Figure E4).
Figure 3.
Cell and gene expression changes in lung endothelial cells (ECs) in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). (A) Uniform manifold approximation and projection embedding of ECs from snRNA-seq of ACDMPV (n = 5), control (n = 3; 3 yr old), and preterm neonate (n = 3; 1–4 d of age and 29–31 wk of GA) lungs. Cells are colored by the predicted EC types. (B) Dot plot of expression of marker genes for EC types. Gene expression with percentage ⩾5% is shown. (C) Changes in EC proportions in snRNA-seq of ACDMPV compared with control infant or preterm neonate lungs. P value represents the significance of the difference in cell proportions using a two-tailed Wilcoxon rank sum test. (D) Expression of FOXF1 and KIT RNA in capillary 1 (CAP1) cells of individual donors. Gene expression with percentage ⩾1% is shown. (E and F) RNAscope analysis validating the decrease in HPGD/cadherin 5 (CDH5) coexpressing capillary 2 (CAP2) cells in ACDMPV (n = 6) (ACD1 shown) versus control (n = 7) (1-day-old shown) lungs (E) and increase in COL15A1/CDH5 SVECs (F) in ACDMPV (n = 5) (ACD3 shown) versus control (n = 4) (13-month-old shown) lungs. Scale bars, 5 μm. (G) Ingenuity Pathway Analysis identified pathways significantly associated with genes downregulated in CAP1 (left) or in CAP2 (right) cells in snRNA-seq of ACDMPV versus control lungs. Shown are the top 10 signaling pathways ranked by P value. Downregulated genes satisfied the following criteria: differentially expressed in ACDMPV versus control snRNA cells (P < 0.05, fold change ⩾ 1.5, and expression percentage ⩾ 20%), selectively expressed in the selected cell type in either ACDMPV or control lung snRNA-seq data (P < 0.05, fold change ⩾ 1.2, and expression percentage ⩾ 10%), and not a mitochondrial gene. The two-tailed Wilcoxon rank sum test was used for differential expression analysis. Boxplots represent 25%, 50%, and 75% quantiles. ABCB1 = ATP binding cassette subfamily B member 1; ACKR1 = atypical chemokine receptor 1; AEC = arterial endothelial cell; CA4 = carbonic anhydrase 4; CCL21 = C-C motif chemokine ligand 21; CLDN5 = claudin 5; COL15A1 = collagen type XV α 1 chain; CPE = carboxypeptidase E; DKK2 = Dickkopf WNT signaling pathway inhibitor 2; EDNRB = endothelin receptor type B; ERK = extracellular signal-related kinase; FAK = focal adhesion kinase; FCN3 = ficolin 3; FOXF1 = forkhead box F1; GA = gestational age; GJA5 = gap junction protein α 5; GTPase = guanosine triphosphatase; HPGD = 15-hydroxyprostaglandin dehydrogenase; ID1 = inhibitor of DNA binding 1; KIT = KIT proto-oncogene, receptor tyrosine kinase; LEC = lymphatic endothelial cell; MAPK = mitogen-activated protein kinase; PECAM1 = platelet and endothelial cell adhesion molecule 1; PROX1 = Prospero homeobox 1; PTEN = phosphatase and tensin homolog; snRNA-seq = single-nucleus RNA sequencing; SLC6A4 = solute carrier family 6 member 4; STAT3 = signal transducer and activator of transcription 3; SVEC = systemic vascular endothelial cell; VEC = venous endothelial cell.
Immunofluorescence confocal microscopy and RNAscope (Advanced Cell Diagnostics) were performed to orthogonally validate the EC fraction changes in ACDMPV, demonstrating significantly decreased numbers of HPGD (15-hydroxyprostaglandin dehydrogenase)/CDH5 (cadherin 5)–coexpressing CAP2 cells and increased numbers of COL15A1/CDH5-coexpressing SVECs in ACDMPV compared with control lungs (Figures 3E and 3F). A direct correlation was noted between the number of CAP2 ECs and disease severity, with markedly decreased CAP2 ECs in ACDMPV lungs from subjects who died at 2–5 weeks of age (ACD1, ACD4, and ACD5), intermittent numbers of CAP2 ECs in the subject transplanted at 9 months of age (ACD3), and a less notable CAP2 EC decrease in ACD2, with the least severe phenotype, surviving to lung transplantation at 3.5 years of age, providing further evidence that the decrease in capillary ECs was disease associated (see Figure E8). Ingenuity Pathways Analysis (QIAGEN) identified pathways associated with gene expression downregulated in the retained capillary ECs in ACDMPV lungs (Figure 3G). Reductions of PTEN (phosphatase and tensin homolog), ERK (extracellular signal-related kinase)/MAPK (mitogen-activated protein kinase), STAT3 (signal transducer and activator of transcription 3), FAK (focal adhesion kinase), integrin, WNT/β-catenin, and ID1 (inhibitor of DNA binding 1) signaling were demonstrated in CAP1 cells, and loss of pathways regulating cell migration and extension, for example, semaphorin signaling, FAK, and Rho family of guanosine triphosphatases, was demonstrated in the remaining CAP2 cells.
FOXF1 Influences Pericyte and Fibroblast Cell Function and Gene Expression in ACDMPV Lungs
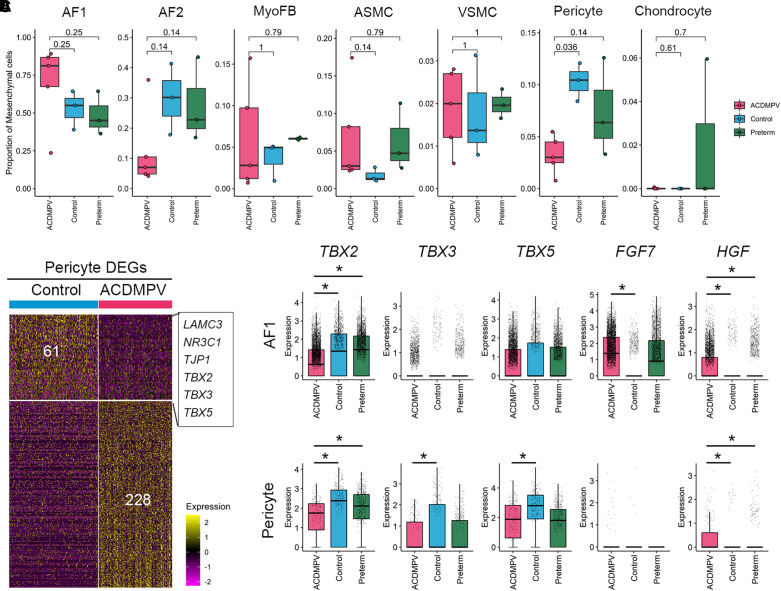
Consistent with the expression of FOXF1 RNA in normal pulmonary mesenchymal progenitors and derivatives, marked changes in fibroblast cell types and pericytes were observed on snRNA-seq. Within mesenchymal cells, pericyte proportions were significantly reduced in ACDMPV, whereas AF1 cells were increased and AF2 cells were decreased in four of five subjects with ACDMPV (Figures 4A and E5E). Differential expression analysis was performed to identify gene expression changes in mesenchymal cells in ACDMPV compared with control lungs (see Table E2). Genes (n = 61) downregulated in ACDMPV pericytes included LAMC3 (laminin subunit gamma 3; a pericyte marker gene) (20), NR3C1 (nuclear receptor subfamily 3 group C member 1; a glucocorticoid receptor regulating signaling required for normal perinatal lung morphogenesis and function), TJP1 (tight junction protein ZO-1), and multiple T-box transcription factors (TFs; TBX2 [T-box TF 2], TBX3, and TBX5) (Figure 4B; see Table E2). Recent studies using chromatin immunoprecipitation sequencing (ChIP-seq) identified TBX2 and TBX4 binding sites near and within FOXF1 and FENDRR (FOXF1 adjacent noncoding developmental regulatory RNA), suggesting potential interactions between TBX4–FGF10 (fibroblast growth factor 10) and SHH (Sonic Hedgehog signaling molecule)–FOXF1 signaling (15, 32). Using snRNA-seq, we identified T-box TFs and growth factors that were differentially expressed in ACDMPV mesenchymal cells. Expression of T-box TFs (TBX2/3/5) was reduced in AF1 cells and/or pericytes in ACDMPV lungs, whereas the expression of growth factors (FGF7 and HGF [hepatocyte growth factor]) was increased in AF1 cells and/or pericytes in ACDMPV lungs (Figure 4C; see Table E2), likely indicating a compensatory response to the absence of FOXF1.
Figure 4.
Cell and gene expression changes in lung mesenchymal cells in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). (A) Changes in mesenchymal cell type proportions using snRNA-seq of ACDMPV (n = 5), control (n = 3; 3 yr old), and preterm neonate (n = 3; 1–4 d of age and 29–31 wk of gestational age) lungs. P value represents the significance of the difference in cell proportions using a two-tailed Wilcoxon rank sum test. (B) Gene expression changes between ACDMPV and control lungs in pericytes in snRNA-seq. The criteria were as follows: differentially expressed in ACDMPV (P < 0.05, fold change ⩾ 1.5, and expression percentage ⩾ 20%) and selectively expressed in pericytes in either ACDMPV or control lungs (P < 0.05, fold change ⩾ 1.2, and expression percentage ⩾ 10%). A two-tailed Wilcoxon rank sum test was used. (C) Differential expression of FGF7, HGF, and T-box transcription factors in ACDMPV AF1 cells and pericytes compared with control or preterm neonate lungs. The asterisk denotes gene expression changes satisfying the following criteria: Bonferroni-adjusted P < 0.1, expression percentage ⩾ 20%, and fold change of average expression or expression percentage ⩾ 1.5. Boxplots represent 25%, 50%, and 75% quantiles. AF1 = alveolar fibroblast 1; AF2 = alveolar fibroblast 2; ASMC = airway smooth muscle cell; DEG = differentially expressed gene; FGF7 = fibroblast growth factor 7; HGF = hepatocyte growth factor; LAMC3 = laminin subunit gamma 3; MyoFB = myofibroblast; NR3C1 = nuclear receptor subfamily 3 group C member 1; snRNA-seq = single-nucleus RNA sequencing; TBX = T-box transcription factor; TJP1 = tight junction protein ZO-1; VSMC = vascular smooth muscle cell.
Impaired Differentiation of Alveolar Epithelial Cells in ACDMPV Lungs
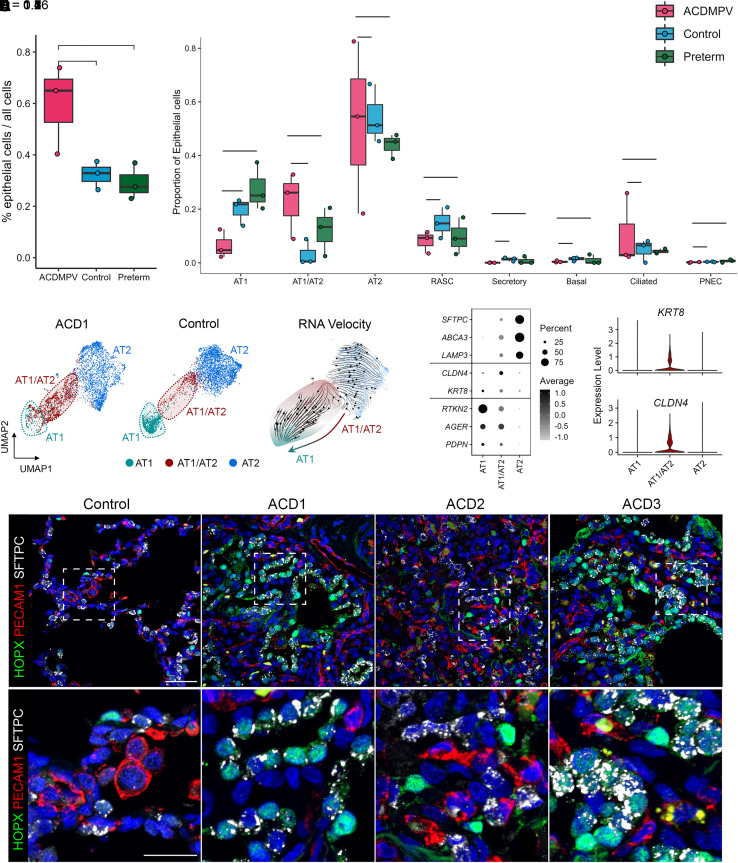
Although FOXF1 was not expressed in epithelial cell lineages, we identified striking changes in epithelial cell populations in ACDMPV lungs. Epithelial cell numbers were increased in ACDMPV compared with control and preterm lungs, attributed primarily to increases in AT1/AT2 cells and a loss of AT1 cells in the ACDMPV epithelium (Figures 5A, 5B, E5F, E5G, and E9). RNA velocity analysis (33) predicted the transitional state of AT1/AT2 cells and its differentiation potential to AT1 cells (Figure 5C). snRNA-seq analysis demonstrated that AT1/AT2 cells expressed marker genes of AT1 and AT2 cells, as well as KRT8 (keratin 8) and CLDN4 (claudin 4), genes associated with damage-associated transient progenitors (34) (Figure 5D), a cell type recently identified as a transitional cell detected in various disease settings and during alveolar regeneration (34–36). Cell type compositional analysis together with the RNA velocity analyses predicted the arrest of AT1/AT2 cell differentiation to mature AT1 cells, consistent with the deficient alveolar development in ACDMPV lungs. Immunofluorescence analysis demonstrated a marked increase in HOPX (HOP homeobox)/SFTPC (surfactant protein C)–coexpressing AT1/AT2 cells and loss of mature AT1 cells in ACDMPV compared with control lungs (Figure 5E).
Figure 5.
Impaired distal epithelial cell differentiation in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) lungs. (A) Increased overall epithelial cell proportions in ACDMPV lungs. (B) Comparison of epithelial cell proportions in ACDMPV (ACD1, ACD4, and ACD5), control (n = 3; 3 yr old), and preterm neonate (n = 3; 1–4 d of age and 29–31 wk of gestational age) lungs. Cell proportions were calculated using the snRNA-seq data. P values represent the significance of the difference using a two-tailed Wilcoxon rank sum test. Figure E9 shows comparisons using snRNA-seq from all five ACDMPV lungs. Boxplots represent 25%, 50%, and 75% quantiles. (C) RNA velocity analysis of differentiation relationships among alveolar epithelial cells from snRNA-seq of ACD1 lung. (D) Left: dot plot of expression of alveolar type 1 (AT1), alveolar type 2 (AT2), and damage-associated transient progenitor cell markers in AT1, AT2, and AT1/AT2 transitional (AT1/AT2) cells from snRNA-seq of ACDMPV lungs (n = 5). Gene expression with percentage ⩾5% is shown. Right: violin plot of KRT8 and CLDN4 in AT1, AT2, and AT1/AT2 cells from snRNA-seq of ACDMPV lungs (n = 5). (E) Immunofluorescence confocal microscopy of control lung (6-mo-old) and ACDMPV lungs demonstrating loss of mature AT1 and increased HOPX/SFTPC coexpressing AT1/AT2 cells in ACDMPV. Lungs from subjects with the most severe phenotype showed diffuse loss of mature AT1 cells and increased coexpressing AT1/AT2 cells (ACD1). Lungs from subjects with less severe phenotypes had decreased mature AT1 cells with markedly increased AT1/AT2 cells in regions with deficient microvasculature (ACD3) alternating with rare AT1/AT2 cells in regions with preserved capillary development in the subject with the least severe phenotype (ACD2), who received a lung transplant at 3.5 years of age. Scale bars, 40 μm (upper panel) and 20 μm (lower panel). ABCA3 = ATP binding cassette subfamily A member 3; AGER = advanced glycosylation end-product specific receptor; CLDN4 = claudin 4; HOPX = HOP homeobox; KRT8 = keratin 8; LAMP3 = lysosomal associated membrane protein 3; PDPN = podoplanin; PECAM1 = platelet and endothelial cell adhesion molecule 1; PNEC = pulmonary neuroendocrine cell; RASC = respiratory airway secretory cell; RTKN2 = rhotekin 2; SFTPC = surfactant protein C; snRNA-seq = single-nucleus RNA sequencing; UMAP = uniform manifold approximation and projection.
FOXF1 Regulatory Targets and Alterations in Capillary and Pericyte Cells in ACDMPV Lungs
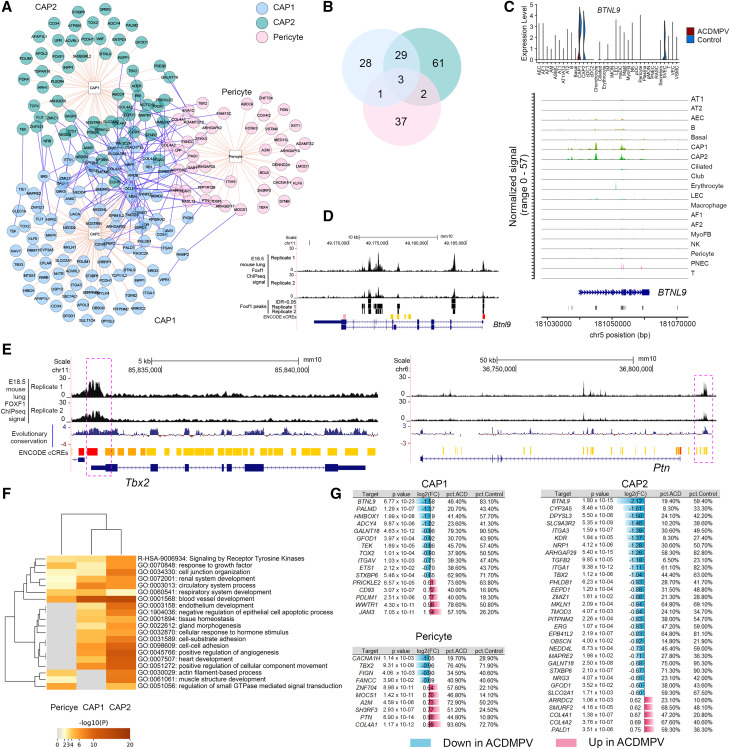
Pericytes and CAP1 and CAP2 ECs, which express FOXF1 in normal lung, were significantly reduced in ACDMPV, suggesting a primary regulatory role for FOXF1 in these cell types. To identify cell-specific regulatory networks mediated by FOXF1, we used PECA2 (37) to integrate snATAC and snRNA data from the 3-year-old control donor lungs to infer regulatory targets of FOXF1 in CAP1 and CAP2 cells and pericytes (see the online supplement; Figure 6). In total, 161 targets were predicted, and each cell type had shared and unique targets (Figures 6A and 6B). To assess the accuracy of the predictions, we processed and analyzed publicly available Embryonic Day 18.5 wild-type mouse lung FOXF1 ChIP-seq data (38) and identified reproducible FOXF1 binding sites in mouse lung (irreproducible discovery rate < 0.05) (see Figure E10). Of the human FOXF1 targets predicted by the multiomic analysis, 86.3% (139 of 161) of the predicted targets shared FOXF1 ChIP-seq binding sites near their mouse orthologs (see Table E3). We also performed the prediction using DIRECT-NET (39), a recently published single-cell GRN inference method. As the snRNA-seq/snATAC-seq of 3-year-old control lung data was not derived from paired profiling, we performed the DIRECT-NET predictions using the snATAC-seq data (see the online supplement). On average, 58.8% of the PECA2-predicted FOXF1 targets were supported by the DIRECT-NET prediction (see Table E3). Figures 6C and 6D show BTNL9 (butyrophilin like 9) as a FOXF1 target gene predicted by the PECA2 multiomic analysis (see Table E3), which 1) was selectively expressed in CAP1 and CAP2 ECs in snRNA-seq of control lungs; 2) had open chromatin accessibility in CAP1 and CAP2 ECs in snATAC-seq of control lungs; and 3) had mouse lung FOXF1 ChIP-seq peaks near its mouse ortholog (Btnl9) gene promoter region. Predicted targets, including LIFR (LIF receptor subunit α), TBX2, TBX3, CD93, PTN (pleiotrophin), NFIB (nuclear factor I B), and COL4A1/2 (collagen type IV α 1/2 chain), had nearby mouse lung FOXF1 ChIP-seq binding orthologous sites that overlap with the evolutionarily conserved regions (Figures 6E and E11). Functional enrichment analysis using Metascape (40) showed common and unique biological processes associated with the predicted targets in each cell type, indicating common and distinct cell-autonomous roles for FOXF1 in ECs and pericytes (Figure 6F). FOXF1 targets were closely shared by CAP1 and CAP2 cells but were less concordant with pericytes, likely consistent with their divergent lineages. Differential expression analysis identified cell type–specific RNA expression changes of the PECA2-predicted FOXF1 regulatory targets in ACDMPV lungs (Figure 6G).
Figure 6.
Multiomic prediction of cell type–specific FOXF1 regulatory targets. (A) Predicted FOXF1 regulatory targets in capillary 1 (CAP1) cells, capillary 2 (CAP2) cells, and pericytes in normal human lung. The prediction was performed using PECA2, integrating cell type–specific snRNA-seq and snATAC-seq data from control lungs (n = 3; 3 yr old). Shown are targets that satisfied the following criteria: false discovery rate < 0.001 of P value of regulatory potential in at least two snRNA–snATAC sample pairs and selectively expressed (P < 0.05, fold change (FC) ⩾ 1.2, and percentage ⩾ 10%) in the corresponding cell type in snRNA-seq of control lungs. Visualization was generated using the “Interaction Network” function of ToppCluster (https://toppcluster.cchmc.org) using the predicted targets as input gene sets. (B) The predicted regulatory targets overlapped among the three cell types. (C) Expression of BTNL9 (butyrophilin like 9) in snRNA-seq of alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) versus control lung (top) and chromatin accessibility near BTNL9 in CAP1 and CAP2 cells in snATAC-seq of control lungs (bottom). (D) Chromatin immunoprecipitation sequencing (ChIP-seq) analysis demonstrated reproducible FOXF1 binding sites (IDR < 0.05) near Btnl9 in E18.5 wild-type mouse lung. (E) Predicted FOXF1 regulatory targets that have nearby mouse lung FOXF1 ChIP-seq binding sites that overlapped with evolutionarily conserved regions (placental mammal base-wise conservation by phyloP). Positive scores (blue) denote sites predicted to be conserved, and negative scores (red) denote sites predicted to be fast evolving. Shown are TBX2 and PTN. An extended list is shown in Figure E11. (F) Metascape analysis of biological processes and pathways significantly associated with the predicted FOXF1 regulatory targets. Shown are the top 20 functional annotation clusters identified. (G) Differential expression of FOXF1 regulatory targets in CAP1 and CAP2 cells and pericytes in ACDMPV versus control lungs. The criteria were as follows: P value of two-tailed Wilcoxon rank sum test < 0.05, FC ⩾ 1.5, and expression percentage ⩾ 20%. A2M = alpha-2-macroglobulin; ADCY4 = adenylate cyclase 4; AEC = arterial endothelial cell; AF1 = alveolar fibroblast 1; AF2 = alveolar fibroblast 1; AM = alveolar macrophage; ARHGAP29 = Rho GTPase activating protein 29; ARRDC2 = arrestin domain containing 2; ASMC = airway smooth muscle cell; AT1 = alveolar type 1 cell; AT1/AT2 = AT1/AT2 transitional cell; AT2 = alveolar type 2 cell; CACNA1H = calcium voltage-gated channel subunit alpha1 H; cCRE = candidate cis-regulatory element; CD93 = cluster of differentiation 93; cDC1 = classical dendritic cell subset 1; cDC2 = classical dendritic cell subset 2; chr = chromosome; COL4A = collagen type IV α; CYP3A5 = cytochrome P450 family 3 subfamily A member 5; DPYSL3 = dihydropyrimidinase like 3; E18.5 = Embryonic Day 18.5; EEPD1 = endonuclease/exonuclease/phosphatase family domain containing 1; ENCODE = Encyclopedia of DNA Elements; EPB41L2 = erythrocyte membrane protein band 4.1 like 2; ERG = ETS transcription factor ERG; ETS1 = ETS proto-oncogene 1, transcription factor; FANCC = FA complementation group C; FIGN = fidgetin, microtubule severing factor; GALNT16 = polypeptide N-acetylgalactosaminyltransferase; GFOD1 = glucose-fructose oxidoreductase domain containing 1; GO = Gene Ontology; GTPase = guanosine triphosphatase; HMBOX1 = homeobox containing 1; IDR = irreproducible discovery rate; IM = interstitial macrophage; ITGA = integrin subunit α; JAM3 = junctional adhesion molecule 3; KDR = kinase insert domain receptor; LEC = lymphatic endothelial cell; MAPRE2 = microtubule associated protein RP/EB family member 2; MKLN1 = muskelin 1; MOCS1 = molybdenum cofactor synthesis 1; MyoFB = myofibroblast; NEDD4L = NEDD4 like E3 ubiquitin protein ligase; NK = natural killer; NRG3 = neuregulin 3; NRP1 = neuropilin 1; OBSCN = obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF; PALD1 = phosphatase domain containing paladin 1; PALMD = palmdelphin; pct = percentage; pDC = plasmacytoid dendritic cell; PDLIM1 = PDZ and LIM domain 1; PHLDB1 = pleckstrin homology like domain family B member 1; PITPNM2 = phosphatidylinositol transfer protein membrane associated 2; PNEC = pulmonary neuroendocrine cell; PRICKLE2 = prickle planar cell polarity protein 2; PTN = pleiotrophin; SH3RF3 = SH3 domain containing ring finger 3; SLCO2A1 = solute carrier organic anion transporter family member 2A1; SMURF2 = SMAD specific E3 ubiquitin protein ligase 2; snATAC-seq = single-nucleus assay for transposase-accessible chromatin sequencing; snRNA-seq = single-nucleus RNA sequencing; STXBP6 = syntaxin binding protein 6; SVEC = systemic vascular endothelial cell; TBX2 = T-box transcription factor 2; TEK = TEK receptor tyrosine kinase; TGFB2 = transforming growth factor β 2; TMOD3 = tropomodulin 3; TOX2 = TOX high mobility group box family member 2; VEC = vascular endothelial cell; VSMC = vascular smooth muscle cell; WWTR1 = WW domain containing transcription regulator 1; ZMIZ1 = zinc finger MIZ-type containing 1; ZNF704 = zinc finger protein 704.
Recent single-cell transcriptomic analysis of an ACDMPV mouse model identified genes (n = 142 [93 downregulated, 49 upregulated]) significantly changed in CAP1 cells in Foxf1WT/S52F mouse lung compared with wild-type mouse lung at Embryonic Day 18.5 (29). None of the 49 upregulated genes were predicted as CAP1-specific FOXF1 targets by either PECA2 or DIRECT-NET (see Table E4). Among the 93 downregulated genes in the mouse CAP1 cells, 15 were predicted as FOXF1 targets in human lung CAP1 cells and have nearby FOXF1 ChIP-seq binding sites in mouse lung, including ACVRL1 (activin A receptor like type 1), FOXF1, AQP1 (aquaporin 1), BCAM (basal cell adhesion molecule), CASZ1 (castor zinc finger 1), CDH5, CPNE8 (copine 8), DAAM1 (disheveled associated activator of morphogenesis 1), DLL4 (delta-like canonical Notch ligand 4), ID1, PODXL (podocalyxin like), PRX (periaxin), PTPRM (protein tyrosine phosphatase receptor type M), TFPI (tissue factor pathway inhibitor), and TSPAN18 (tetraspanin 18), indicating that they were potential direct targets of FOXF1 in CAP1 cells in ACDMPV (see Table E4). Using the mouse model, Wang and colleagues (29) showed that FOXF1 synergizes with FLI1 (Fli-1 proto-oncogene, ETS transcription factor) to stimulate ACVRL1 gene transcription and that the S52F Foxf1 mutation disrupts BMP9 (bone morphogenetic protein 9)/ACVRL1 signaling in pulmonary endothelial progenitor cells, likely decreasing neonatal lung angiogenesis and alveolarization in Foxf1WT/S52F mice. Our integrative analysis of multiomics data from human and mouse lung validated these FOXF1 targets and predicted new cell-specific targets for further studies.
Alterations in Alveolar Niche Signaling in ACDMPV Lungs
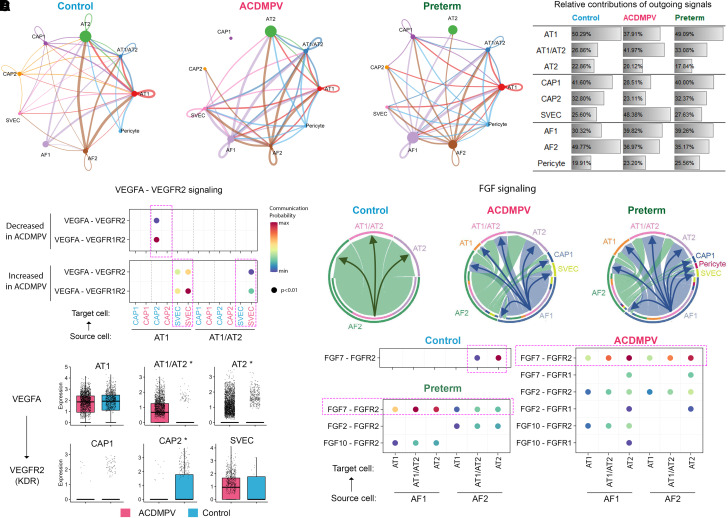
We applied CellChat (41) analysis on the snRNA-seq data to decipher cell–cell communication patterns on the basis of cell type–selective expression of ligands and receptors. CellChat analysis identified marked changes in ligand receptor–based communication patterns among alveolar cell types in ACDMPV compared with both 2-month-old and 3-year-old control infant or preterm neonate lungs (Figures 7A and E5H). In lungs from control infants and preterm neonates, AT1 and CAP1 cells were predicted to be primary signaling hubs in alveolar epithelium and endothelium, respectively (Figure 7B). In ACDMPV lungs, signaling from CAP1 and CAP2 cells to other cell types was largely disrupted, whereas signaling between epithelial cells (AT1 and AT1/AT2 cells) and SVECs was increased. In mesenchyme, AF2 cells play a signaling hub role in control infant lungs, whereas the signaling from AF1 cells was significantly increased in lungs from ACDMPV and preterm neonates (Figure 7B). Major signaling pathways known to pattern murine and rhesus alveolar development (22, 42) were identified by CellChat. For example, in control infant lung, VEGFA (vascular endothelial growth factor A) was expressed primarily in AT1 cells, and its receptor, KDR (kinase insert domain receptor) (VEGFR2 [vascular endothelial growth factor receptor 2]), was expressed in CAP2 cells, the latter located in close proximity to AT1 cells, likely contributing to the patterning of the alveolar gas exchange surface (21, 22, 42). This signaling pattern was disrupted in ACDMPV lungs (Figure 7C), wherein VEGFA–KDR signaling was absent from AT1 to CAP2 cells, likely related to the marked loss of CAP2 cells. In sharp contrast, VEGFA–KDR signaling was increased from the AT1/AT2 cells to SVECs (Figure 7C), likely contributing to the abnormal expansion of the systemic ECs in ACDMPV lungs. FGF signaling from mesenchymal to epithelial cells is important for normal alveolar development and maturation. In ACDMPV lungs, expression of FGF7 ligand and communication probabilities of FGF signaling was largely increased from AF1 cells to AT1/AT2 and AT2 cells, a pattern that was also identified in lungs from preterm neonates (Figures 4C and 7D). Expression of HGF, THBS (thrombospondin), and PTN ligands, all active in fetal lung morphogenesis, was increased in ACDMPV alveolar fibroblasts and/or pericytes (Figures 4C and E12; see Table E2). We compared the mesenchymal signaling, which was increased in ACDMPV lungs, with that increased in preterm neonate lungs. Sixty percent of the predicted interactions increased in alveolar mesenchymal–epithelial/endothelial cell–cell communications in ACDMPV lungs were also increased in preterm human lungs, wherein strong WNT2/FGF/HGF signaling is predicted from mesenchymal to epithelial cells (see Figure E12). The present findings support the concept that cell–cell communications activated in ACDMPV lungs represent signaling that is more active in earlier stages of lung morphogenesis.
Figure 7.
Alterations in alveolar niche cell–cell communications in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) lungs. (A) CellChat analysis predicted alterations in ligand receptor–based cell–cell communications among alveolar cell types in ACDMPV (middle) compared with control (left; 3 yr old) and preterm neonate (right; 1–4 d of age and 29–31 wk of gestational age) lungs. Edge thickness is proportional to the number of CellChat-inferred ligand–receptor interactions for each cell–cell pair. Shown are edges with the top 50% number of interactions. Edge color represents the cell type expressing the ligands. Node size is proportional to the number of cells in a cell type. (B) Relative contributions of outgoing signaling by each cell type in ACDMPV, control, and preterm neonate lungs. The relative contribution of a cell type was calculated as the total number of outgoing signals from the cell type divided by the total number of outgoing signals from the cell types of the same lineage. (C) Top: CellChat analysis predicted the loss of VEGFA (vascular endothelial growth factor A)–VEGFR2 (vascular endothelial growth factor receptor 2) signaling from alveolar type 1 (AT1) to capillary 2 (CAP2) cells and an increase in VEGFA–VEGFR2 signaling from AT1 and AT1/AT2 to SVECs in ACDMPV lungs. Node color represents normalized communication probability calculated using CellChat. Label colors represent cell types in ACDMPV (red) or in control (blue) lung. Bottom: differential expression of VEGFA in epithelial cells and KDR in endothelial cells in snRNA-seq of ACDMPV versus control lungs. The asterisk represents gene expression changes satisfying the following criteria: P value of two-tailed Wilcoxon rank sum test < 0.05, expression percentage ⩾ 20%, and fold change of average expression or expression percentage ⩾ 1.5. (D) CellChat analysis predicted an increase in FGF (fibroblast growth factor) signaling from AF1 cells to alveolar epithelial cells in ACDMPV lungs. Top: FGF signaling among alveolar cell types in the control, ACDMPV, and preterm lungs. Edge thickness is proportional to the normalized aggregated communication probabilities of significant FGF ligand–receptor interactions for a cell–cell pair. Edges are from source to target cells. Bottom: significant (P < 0.01) FGF ligand–receptor interactions inferred by CellChat using snRNA-seq of control, ACDMPV, and preterm lungs. Node color represents normalized communication probability. AF1 = alveolar fibroblast 1; AF2 = alveolar fibroblast 2; AT1/AT2 = AT1/AT2 transitional cell; AT2 = alveolar type 2 cell; CAP1 = capillary 1 cell; FGFR=fibroblast growth factor receptor; KDR = kinase insert domain receptor; snRNA-seq = single-nucleus RNA sequencing; SVEC = systemic vascular endothelial cell.
Discussion
Single-cell multiomics, histopathology, and confocal microscopy were used to identify cell types and cell type–specific gene expression patterns and to predict alterations in cell–cell interactions caused by pathogenic variants (SNVs and CNV deletions) involving the FOXF1 gene locus in human ACDMPV lungs. Changes in gene expression were consistent with the critical role of FOXF1 and the orchestration of distinct GRNs in both endothelial and fibroblast progenitors required for the formation of alveolar capillary microvasculature and the growth and differentiation of mesenchymal fibroblast progenitors required for branching morphogenesis and alveolarization. Our data show that FOXF1 is required for production or survival of alveolar endothelial CAP2 cells from endothelial CAP1 progenitors, supporting the concept that the loss of paracrine signaling from the CAP2 cells impairs AT1 cell differentiation from AT1/AT2 progenitors and inhibits the formation of the alveolar gas exchange region in ACDMPV lungs. Pericytes, which are required for precise control of VEGFA signaling for normal vasculogenesis (43), were decreased in ACDMPV lungs. The present findings support distinct, cell-autonomous roles for FOXF1 in ECs, fibroblasts, and pericyte progenitors. The FOXF1 TF orchestrates cell–cell signaling among pulmonary mesenchymal, alveolar epithelial, and capillary ECs in the developing lung tubules to control diverse cellular processes underlying ACDMPV pathogenesis.
The Role of FOXF1 in the Formation of the Alveolar Microvasculature
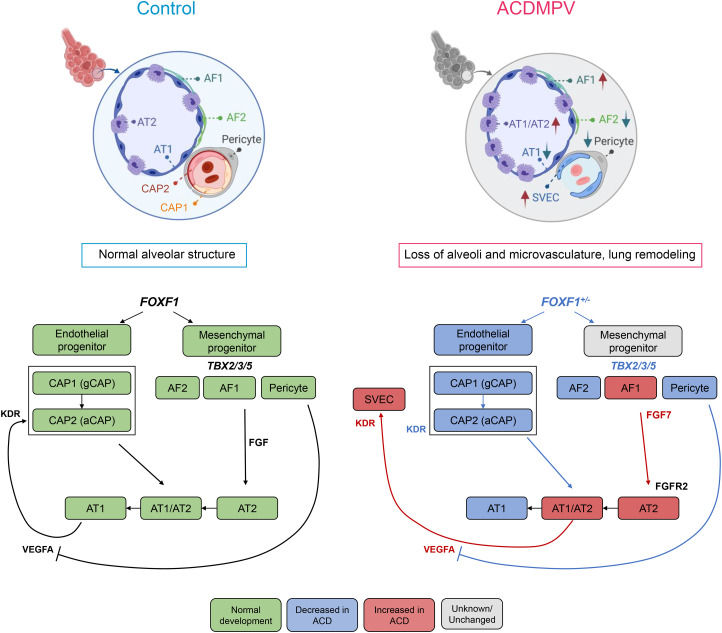
Pulmonary hypertension and unrelenting hypoxemia are clinical hallmarks of ACDMPV and are consistent with the present observations demonstrating a paucity of pulmonary microvasculature and a loss of capillary ECs (CAP1 and CAP2 cells). CAP2 cells, also termed aerocytes (21), form in close apposition to alveolar epithelial AT1 cells to create the alveolar gas exchange interface. We found that CAP2 cells were remarkably reduced or absent in ACDMPV lungs. Our data support the contention that FOXF1 is required for the maintenance of cKIT+, FOXF1+, and CAP1 cell populations, which serve as progenitors of CAP2 ECs in ACDMPV lungs. In contrast to the loss of CAP1 and CAP2 cells, COL15A1-positive SVECs, characteristic of bronchial vessels (17, 24–26), were markedly increased, consistent with previous anatomic studies demonstrating intrapulmonary shunts from the systemic circulation that bypass the pulmonary capillary bed (7, 8, 44). The loss of FOXF1-expressing pericytes in ACDMPV lungs also likely influences the formation and stability of the microvasculature. Pericytes express PDGFRB (platelet-derived growth factor receptor β), required for the precise control of VEGFA signaling needed for normal vessel sprouting and stability (43). VEGFA RNA was found to be highly expressed in AT1/AT2 epithelial cells and markedly increased in ACDMPV lungs; thus, uncontrolled VEGFA signaling may contribute to the disruption of the alveolar microvasculature formation in ACDMPV lungs (Figure 8). The present findings support a cell-autonomous role for FOXF1 in capillary endothelial progenitors required for the formation of the pulmonary microvasculature. The remarkable expansion of the systemic vasculature may compensate for the lack of pulmonary arterial perfusion during lung morphogenesis in ACDMPV lungs. Nanoparticle delivery of FOXF1 or STAT3 to ECs in newborn mice rescues pulmonary vasculogenesis, consistent with a cell-autonomous role for FOXF1 in endothelial progenitors (9, 11, 45). The present integration of ATAC and RNA sequencing data identified transcriptional targets of FOXF1 in diverse pulmonary mesenchymal cells, several of which have been validated in mouse and human ACDMPV lungs (46).
Figure 8.
Schematic showing alveolar structure and cell–cell communications in normal human lung and alterations in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). FOXF1 (forkhead box F1) is expressed in pulmonary mesenchymal cells, including fibroblasts, pericytes, and endothelial cell (EC) progenitors. FOXF1 was required for the differentiation or survival of endothelial and fibroblast progenitors, which in turn influence the growth and differentiation of the pulmonary epithelial progenitors during the formation of the peripheral lung. FOXF1 deficiency in ACDMPV disrupted gene expression in EC progenitors, preventing differentiation or survival of CAP2 ECs, critical for the formation of the alveolar gas exchange, resulting in hypoxemia at birth. Increased expression of VEGFA by atypical alveolar epithelial progenitors (AT1/AT2) is likely to enhance the proliferation and migration of the systemic vasculature (SVECs), a characteristic of ACDMPV. aCAP = aerocyte capillary cell; ACD = alveolar capillary dysplasia; AF1 = alveolar fibroblast 1; AF2 = alveolar fibroblast 2; AT1 = alveolar type 1 cell; AT1/AT2 = AT1/AT2 transitional cell; AT2 = alveolar type 2 cell; CAP1 = capillary 1; CAP2 = capillary 2; FGFR = fibroblast growth factor receptor; gCAP = general capillary; KDR = kinase insert domain receptor; SVEC = systemic vascular endothelial cell; TBX = T-box transcription factor; VEGFA = vascular endothelial growth factor A.
Confocal microscopy and transcriptomic studies demonstrated a remarkable loss of AT1 cells in ACDMPV lungs, which are located in close apposition to microvasculature ECs in control lungs. The indeterminate differentiation of AT1/AT2 progenitors and the lack of AT1 cell differentiation resulted in a cuboidal epithelium lining the malformed immature distal airspaces in ACDMPV lungs, likely contributing to the loss of normal cell–cell communications among CAP1 progenitors and alveolar epithelial and endothelial CAP2 cells that, in turn, disrupt the alveolar epithelial–capillary interface, resulting in pulmonary hypertension and cyanosis after birth. Results of analysis of the GRNs lost in ACDMPV lungs support the concept that CAP1 endothelial progenitors fail to generate CAP2 cells; alternatively, FOXF1 is needed for survival of CAP2 via STAT3-, PTEN-, or ID1-mediated pathways, consistent with the failure of FOXF1+/cKIT+ endothelial progenitors to produce or maintain the pulmonary microvasculature in FOXF1-deficient mice (29–31). The present single-cell data support a role for a PTEN signaling deficiency in CAP1 progenitors, consistent with murine studies wherein the loss of PTEN in the pulmonary mesenchyme caused an ACDMPV-like phenotype (47). Thus, the CAP1 progenitors represent important cellular targets for genetic or pharmacological therapies for ACDMPV or other lung disorders, for example, bronchopulmonary dysplasia and small patellar syndrome (also known as ischiocoxopodopatellar syndrome) caused by TBX4 variants, affecting the pulmonary microvasculature. The present transcriptomic analyses support a cell-autonomous role for FOXF1 in the differentiation or survival of lung mesenchymal cells. Non–cell-autonomous roles for FOXF1 controlling cell–cell interactions were demonstrated by single-cell RNA analyses using CellChat, supporting the important role of FOXF1 in regulating alveolar fibroblast–AT2 and CAP2–AT1 interactions, predicted to be mediated in part by FGF and VEGFA signaling (Figure 8), and supporting recent observations from bulk RNA expression profiles obtained from ACDMPV lung tissues (46).
The present studies are limited to lung tissue samples from subjects who were previously treated for respiratory distress and thus may be influenced by the intensive care (mechanical ventilation, oxygen, extracorporeal membrane oxygenation, pulmonary vasodilators) needed to sustain critically ill infants with ACDMPV after birth. Although previous RNA studies of lung tissues from patients with ACDMPV demonstrated increased inflammation (48), the present single-cell RNA data and histopathology identified relatively few inflammatory cells, especially in subjects who died soon after birth. Nevertheless, the expression studies are likely to have been influenced by the diverse clinical conditions and heterogeneity of FOXF1 pathogenic variants in these subjects. Likewise, differences in age and the severity of pulmonary disease between the ACDMPV and control lung tissues may have influenced the present findings.
Conclusions
Single-cell multiomics and confocal microscopy were utilized to identify cell types and cell type gene expression patterns in lungs from infants with variants involving the FOXF1 gene locus, the cause of ACDMPV. The critical role of FOXF1 and its transcriptional targets in the regulation of endothelial, mesenchymal, and epithelial cell growth and differentiation were identified, providing insights into the genetic networks mediating human lung morphogenesis.
Acknowledgments
Acknowledgment
The authors thank the staff of the Pathology Research Core and the Single Cell Genomics Core at Cincinnati Children’s Hospital Medical Center; the Center for Epigenomics at the University of California (UC), San Diego; and the LungMAP Human Tissue Core at the University of Rochester Medical Center for assistance with this study. Work at the Center for Epigenomics was supported in part by the UC San Diego School of Medicine. The authors thank the QB3 Macrolab at UC Berkeley for purification of the Tn5 transposase. This publication includes data generated at the UC San Diego IGM Genomics Center using an Illumina NovaSeq 6000 that was purchased with funding from an NIH Shared Instrumentation Grant (S10 OD026929).
Footnotes
Supported by NHLBI grants R01HL164414 (J.A. Whitsett), U01HL122642 and U01HL148856 (J.A. Whitsett and Y.X.), U01HL134745 (J.A. Whitsett and Y.X.), R01HL153045 (Y.X.), R01HL141174 (V.V.K), R01HL149631 (V.V.K), R01HL152973 (V.V.K), U01HL148867 (X.S.), and R01HL166283 (M. Gu and M. Guo); National Institute of Diabetes and Digestive and Kidney Diseases grant P30DK117467 (J.A. Whitsett, K.A.W.-B., and Y.X.); and LAM Foundation grant LAM0150C01-22 (M. Guo).
Author Contributions: J. A. Whitsett and M. Guo conceived and designed the experiments and wrote the manuscript. M. Guo, C.J., K.A.W.-B., Y.X., and G.W. analyzed the data. K.A.W.-B. performed pathologic analysis and identified and selected samples of alveolar capillary dysplasia with misalignment of pulmonary veins for single-nucleus analysis. K.A.W.-B. and J.A. Kitzmiller conducted sample preparation, histopathology, immunofluorescence, RNAscope, and confocal microscopy experiments. A.W., S.P., X.H., and J.B. performed combinatorial barcoding single-nucleus assay for transposase-accessible chromatin sequencing and 10X single nucleus RNA sequencing data generation. Y.M. refined the protocol for the 10X Multiome data generation. P.S. performed FOXF1 genetic analysis. J.A. Wambach provided samples along with FOXF1 alterations and clinical diagnosis. K.A.W.-B., J.A. Kitzmiller, V.V.K., P.S., J.A. Wambach, Y.X., J.A. Karolak, Y.M., M. Gu, D.B.F., W.J.Z., S.P., and X.S. provided scientific insights, edited the manuscript, and revised the manuscript critically for important intellectual content.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202210-2015OC on July 18, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Vincent M, Karolak JA, Deutsch G, Gambin T, Popek E, Isidor B, et al. Clinical, histopathological, and molecular diagnostics in lethal lung developmental disorders. Am J Respir Crit Care Med . 2019;200:1093–1101. doi: 10.1164/rccm.201903-0495TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. Am J Respir Crit Care Med . 2011;184:172–179. doi: 10.1164/rccm.201010-1697CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kurland G, Deterding RR, Hagood JS, Young LR, Brody AS, Castile RG, et al. American Thoracic Society Committee on Childhood Interstitial Lung Disease (chILD) and the chILD Research Network An official American Thoracic Society clinical practice guideline: classification, evaluation, and management of childhood interstitial lung disease in infancy. Am J Respir Crit Care Med . 2013;188:376–394. doi: 10.1164/rccm.201305-0923ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szafranski P, Gambin T, Dharmadhikari AV, Akdemir KC, Jhangiani SN, Schuette J, et al. Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins. Hum Genet . 2016;135:569–586. doi: 10.1007/s00439-016-1655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Towe CT, White FV, Grady RM, Sweet SC, Eghtesady P, Wegner DJ, et al. Infants with atypical presentations of alveolar capillary dysplasia with misalignment of the pulmonary veins who underwent bilateral lung transplantation. J Pediatr . 2018;194:158–164.e1. doi: 10.1016/j.jpeds.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edwards JJ, Murali C, Pogoriler J, Frank DB, Handler SS, Deardorff MA, et al. Histopathologic and genetic features of alveolar capillary dysplasia with atypical late presentation and prolonged survival. J Pediatr . 2019;210:214–219.e2. doi: 10.1016/j.jpeds.2019.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galambos C, Sims-Lucas S, Abman SH. Three-dimensional reconstruction identifies misaligned pulmonary veins as intrapulmonary shunt vessels in alveolar capillary dysplasia. J Pediatr . 2014;164:192–195. doi: 10.1016/j.jpeds.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galambos C, Sims-Lucas S, Ali N, Gien J, Dishop MK, Abman SH. Intrapulmonary vascular shunt pathways in alveolar capillary dysplasia with misalignment of pulmonary veins. Thorax . 2015;70:84–85. doi: 10.1136/thoraxjnl-2014-205851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pradhan A, Dunn A, Ustiyan V, Bolte C, Wang G, Whitsett JA, et al. The S52F FOXF1 mutation inhibits STAT3 signaling and causes alveolar capillary dysplasia. Am J Respir Crit Care Med . 2019;200:1045–1056. doi: 10.1164/rccm.201810-1897OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalinichenko VV, Gusarova GA, Kim IM, Shin B, Yoder HM, Clark J, et al. Foxf1 haploinsufficiency reduces Notch-2 signaling during mouse lung development. Am J Physiol Lung Cell Mol Physiol . 2004;286:L521–L530. doi: 10.1152/ajplung.00212.2003. [DOI] [PubMed] [Google Scholar]
- 11. Sun F, Wang G, Pradhan A, Xu K, Gomez-Arroyo J, Zhang Y, et al. Nanoparticle delivery of STAT3 alleviates pulmonary hypertension in a mouse model of alveolar capillary dysplasia. Circulation . 2021;144:539–555. doi: 10.1161/CIRCULATIONAHA.121.053980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns . 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 13. Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res . 2014;115:709–720. doi: 10.1161/CIRCRESAHA.115.304382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones MR, Chong L, Bellusci S. Fgf10/Fgfr2b signaling orchestrates the symphony of molecular, cellular, and physical processes required for harmonious airway branching morphogenesis. Front Cell Dev Biol . 2021;8:620667. doi: 10.3389/fcell.2020.620667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karolak JA, Vincent M, Deutsch G, Gambin T, Cogné B, Pichon O, et al. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am J Hum Genet . 2019;104:213–228. doi: 10.1016/j.ajhg.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ustiyan V, Bolte C, Zhang Y, Han L, Xu Y, Yutzey KE, et al. FOXF1 transcription factor promotes lung morphogenesis by inducing cellular proliferation in fetal lung mesenchyme. Dev Biol . 2018;443:50–63. doi: 10.1016/j.ydbio.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang A, Chiou J, Poirion OB, Buchanan J, Valdez MJ, Verheyden JM, et al. NHLBI LungMap Consortium Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife . 2020;9:e62522. doi: 10.7554/eLife.62522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zepp JA, Morley MP, Loebel C, Kremp MM, Chaudhry FN, Basil MC, et al. Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science . 2021;371:eabc3172. doi: 10.1126/science.abc3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo M, Morley MP, Wu Y, Du Y, Zhao S, Wagner A, et al. Guided construction of single cell reference for human and mouse lung. 2023;14:4566. doi: 10.1038/s41467-023-40173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun X, Perl AK, Li R, Bell SM, Sajti E, Kalinichenko VV, et al. NHLBI LungMAP Consortium A census of the lung: CellCards from LungMAP. Dev Cell . 2022;57:112–145.e2. doi: 10.1016/j.devcel.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, et al. Capillary cell-type specialization in the alveolus. Nature . 2020;586:785–789. doi: 10.1038/s41586-020-2822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, et al. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell . 2020;52:617–630.e6. doi: 10.1016/j.devcel.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife . 2020;9:e53072. doi: 10.7554/eLife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv . 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature . 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schupp JC, Adams TS, Cosme C, Jr, Raredon MSB, Yuan Y, Omote N, et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation . 2021;144:286–302. doi: 10.1161/CIRCULATIONAHA.120.052318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao J, O’Day DR, Pliner HA, Kingsley PD, Deng M, Daza RM, et al. A human cell atlas of fetal gene expression. Science . 2020;370:eaba7721. doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang G, Wen B, Ren X, Li E, Zhang Y, Guo M, et al. Generation of pulmonary endothelial progenitor cells for cell-based therapy using interspecies mouse-rat chimeras. Am J Respir Crit Care Med . 2021;204:326–338. doi: 10.1164/rccm.202003-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang G, Wen B, Deng Z, Zhang Y, Kolesnichenko OA, Ustiyan V, et al. Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling. Nat Commun . 2022;13:2080. doi: 10.1038/s41467-022-29746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wen B, Li E, Ustiyan V, Wang G, Guo M, Na CL, et al. In vivo generation of lung and thyroid tissues from embryonic stem cells using blastocyst complementation. Am J Respir Crit Care Med . 2021;203:471–483. doi: 10.1164/rccm.201909-1836OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren X, Ustiyan V, Guo M, Wang G, Bolte C, Zhang Y, et al. Postnatal alveologenesis depends on FOXF1 signaling in c-KIT+ endothelial progenitor cells. Am J Respir Crit Care Med . 2019;200:1164–1176. doi: 10.1164/rccm.201812-2312OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karolak JA, Gambin T, Szafranski P, Stankiewicz P. Potential interactions between the TBX4-FGF10 and SHH-FOXF1 signaling during human lung development revealed using ChIP-seq. Respir Res . 2021;22:26. doi: 10.1186/s12931-021-01617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol . 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 34. Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell . 2020;27:366–382.e7. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobayashi Y, Tata A, Konkimalla A, Katsura H, Lee RF, Ou J, et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol . 2020;22:934–946. doi: 10.1038/s41556-020-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun . 2020;11:3559. doi: 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duren Z, Chen X, Xin J, Wang Y, Wong WH. Time course regulatory analysis based on paired expression and chromatin accessibility data. Genome Res . 2020;30:622–634. doi: 10.1101/gr.257063.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dharmadhikari AV, Sun JJ, Gogolewski K, Carofino BL, Ustiyan V, Hill M, et al. Lethal lung hypoplasia and vascular defects in mice with conditional Foxf1 overexpression. Biol Open. 2016;5:1595–1606. doi: 10.1242/bio.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang L, Zhang J, Nie Q. DIRECT-NET: an efficient method to discover cis-regulatory elements and construct regulatory networks from single-cell multiomics data. Sci Adv . 2022;8:eabl7393. doi: 10.1126/sciadv.abl7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun . 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell–cell communication using CellChat. Nat Commun . 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toth A, Steinmeyer S, Kannan P, Gray J, Jackson CM, Mukherjee S, et al. Inflammatory blockade prevents injury to the developing pulmonary gas exchange surface in preterm primates. Sci Transl Med . 2022;14:eabl8574. doi: 10.1126/scitranslmed.abl8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eilken HM, Diéguez-Hurtado R, Schmidt I, Nakayama M, Jeong HW, Arf H, et al. Pericytes regulate VEGF-induced endothelial sprouting through VEGFR1. Nat Commun . 2017;8:1574. doi: 10.1038/s41467-017-01738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chambers BR, Norris JW. Carotid bruit controversy. Arch Neurol . 1985;42:423. doi: 10.1001/archneur.1985.04060050017002. [DOI] [PubMed] [Google Scholar]
- 45. Bolte C, Ustiyan V, Ren X, Dunn AW, Pradhan A, Wang G, et al. Nanoparticle delivery of proangiogenic transcription factors into the neonatal circulation inhibits alveolar simplification caused by hyperoxia. Am J Respir Crit Care Med . 2020;202:100–111. doi: 10.1164/rccm.201906-1232OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karolak JA, Gambin T, Szafranski P, Maywald RL, Popek E, Heaney JD, et al. Perturbation of semaphorin and VEGF signaling in ACDMPV lungs due to FOXF1 deficiency. Respir Res . 2021;22:212. doi: 10.1186/s12931-021-01797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tiozzo C, Carraro G, Al Alam D, Baptista S, Danopoulos S, Li A, et al. Mesodermal Pten inactivation leads to alveolar capillary dysplasia- like phenotype. J Clin Invest . 2012;122:3862–3872. doi: 10.1172/JCI61334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sen P, Dharmadhikari AV, Majewski T, Mohammad MA, Kalin TV, Zabielska J, et al. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PLoS One . 2014;9:e94390. doi: 10.1371/journal.pone.0094390. [DOI] [PMC free article] [PubMed] [Google Scholar]