Abstract
Vitamin D deficiency is a common deficiency worldwide, particularly among women of reproductive age. During pregnancy, it increases the risk of immune-related diseases in offspring later in life. However, exactly how the body remembers exposure to an adverse environment during development is poorly understood. Herein, we explore the effects of prenatal vitamin D deficiency on immune cell proportions in offspring using vitamin D deficient mice established by dietary manipulation. We show that prenatal vitamin D deficiency alters immune cell proportions in offspring by changing the transcriptional properties of genes downstream of vitamin D receptor signaling in hematopoietic stem and progenitor cells of both the fetus and adults. Further investigations of the associations between maternal vitamin D levels and cord blood immune cell profiles from 75 healthy pregnant women and their term babies also confirm that maternal vitamin D levels significantly affect immune cell proportions in the babies. Thus, lack of prenatal vitamin D, particularly at the time of hematopoietic stem cell migration from the liver to the bone marrow, has long-lasting effects on immune cell proportions. This highlights the importance of providing vitamin D supplementation at specific stages of pregnancy.
INTRODUCTION
Vitamin D, a micronutrient/hormone, regulates transcription by binding to its nuclear receptor, the vitamin D receptor (VDR). Humans can obtain vitamin D through photosynthesis on the skin and food intake. Approximately 50% to 90% of vitamin D is synthesized on the skin via sunlight exposure, while the remainder comes from the diet. Cutaneous vitamin D synthesis depends on environmental factors (geographic latitude, season, and amount of air pollution), skin type, clothing habitats, and lifestyle1. Despite country-level vitamin D fortification programs in many countries, the prevalence rate of vitamin D deficiency, especially in reproductive-age women2–7, is still high worldwide8–14. The most severe consequence of vitamin D deficiency is rickets or impaired bone formation (reviewed by Holick15). Besides its importance in bone formation, VDR signaling regulates gene transcription in nearly every tissue in our bodies, including the brain, heart, muscle, kidney, and immune system (reviewed in Pike et.al16). During development, the micronutrient status of offspring is entirely dependent on the status of the mother; therefore, developing embryos are vulnerable to adverse micronutrient conditions of deficient mothers17–21. To study the adverse consequences of prenatal vitamin D exposure, animal models with maternal dietary manipulations22–35 and knockout mouse model studies18,36–42 have been utilized. Studies using knockout mice models have shown that depletion of VDR causes a rickets-like phenotype after weaning. Both maternal dietary manipulation and knockout mouse models revealed that prenatal vitamin D deficiency can lead to immune defects in offspring. Epidemiological studies in humans have demonstrated that prenatal vitamin D deficiency has been associated with susceptibility to a number of immune-related diseases affecting their children, including asthma43,44, multiple sclerosis45, and type I diabetes46–48. These findings indicate that prenatal vitamin D deficiency disturbs immune cell development. In addition, it is possible that the hematopoietic system poses a long-term memory of exposure to vitamin D deficiency during development. However, this memory mechanism has not been extensively studied.
In this work, we show that prenatal vitamin D deficiency alters immune cell proportions of offspring during adulthood, attributed to cell fate decisions influenced by transcriptional alterations of VDR signaling pathway genes. Our data also supports that changing the cell fate of stem cells is the key component of the long-term effects of maternal vitamin D deficiency on the hematopoietic system of offspring.
RESULTS:
Lack of impact of maternal vitamin D deficient diet feeding on offspring bone development
Daily food intake was comparable between the vitamin D-sufficient diet (VDsuf, 2.39 g/day/mouse) and the vitamin D-deficient diet (VDdef, 2.38 g/day/mouse). After five weeks of feeding VDdef, serum vitamin D (25-hydroxyvitamin D3, 25(OH)D3) concentrations of the females reached the vitamin D deficient threshold (5.1±3.8 ng/mL), which is about 6.5 times lower than that of the VDsuf-fed females (33.0±5.44 ng/mL) (Supplementary Fig. 1A). While the number of offspring per dam was smaller in VDdef-fed females (mean = 3.82, standard deviation (sd) = 2.8, n=16) than in VDsuf-fed females (mean=4.7, sd=1.7, n=10), the difference was not significant (p=0.32). We did not observe any obvious adverse effects on these female mice. The offspring of VDdef-fed females (VDD) and VDsuf-fed females (VDS) were further investigated. The growth of VDD and VDS based on their body weight were comparable for all of the time points we measured (postnatal day 1, 5 weeks, 9 weeks, 13 weeks) (Supplementary Fig. 1B and 1C). The serum vitamin D concentrations of VDD were lower than the detection limit (0.6 ng/ml, n=3 per group) at postnatal day 1 (Supplementary Fig. 1A). While the concentration of VDD was still significantly lower than that of VDS (n=5 per group), it was not deficient anymore (Supplementary Fig. 1A). We collected tissues and weighted liver, lung, kidney, thymus, spleen, and heart from the VDD and VDS at the young adult stage (15–17 weeks of age) (n=4–6 for each group). The average weight of each tissue was not significantly different between the groups, except the heart was larger in male VDD compared to male VDS (p=0.041) (Supplementary Fig. 1D). We also assessed the bone and tissue mineral density of the humerus of the left arm of the offspring using an X-ray CT system (n=5–6 for each group). Bone volume, bone mineral content, bone mineral density, tissue mineral content, tissue mineral density, and bone volume fraction were assessed, and no significant alterations were observed (Supplementary Fig. 1E).
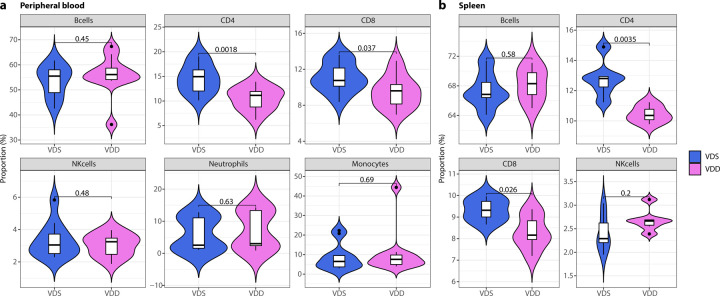
Prenatal vitamin D deficiency decreases CD4+ and CD8+ T cell proportions of the offspring in peripheral blood and spleen
We collected peripheral blood from VDD and VDS and compared the immune cell profiles (n=11 per group) at 16 weeks of age. We observed a significant reduction of CD4+ T cells (28.2% decrease at p=0.0018) and CD8+ T cells (14.7% decrease at p=0.037) in VDD males compared to VDS males (Fig. 1a). This reduction was not observed in females (Supplementary Fig. 2). We also compared the immune cell profiles of spleen (n=6 per group) and observed the significant reduction of CD4+ T cells (18.4% decrease at p=0.0035) and CD8+ T (11.1% decrease at p=0.026) cells in VDD (Fig. 1b). Representative flow cytometry traces are shown in Supplementary Fig. 3.
Fig. 1: Prenatal vitamin D deficiency reduces CD4+ and CD8+ T cell proportions at the adult stage.
a The violin plots illustrate the proportions of immune cells in the peripheral blood, specifically showing that the alteration is only present in T cell proportions (n=11 per group). b The violin plots depicting the proportions of immune cells in the spleen also show that the decrease in cell proportion is specifically limited to T cells (n=6 per group). The white box shows the range between the first and third quartiles. The upper and lower whiskers represent the 1.5x inter-quantile range, while the black bars show the median. The values in the plot are p-values (Student’s t-test).
The reduced proportion of lymphocytes in the periphery reflects the cellular composition changes in the bone marrow.
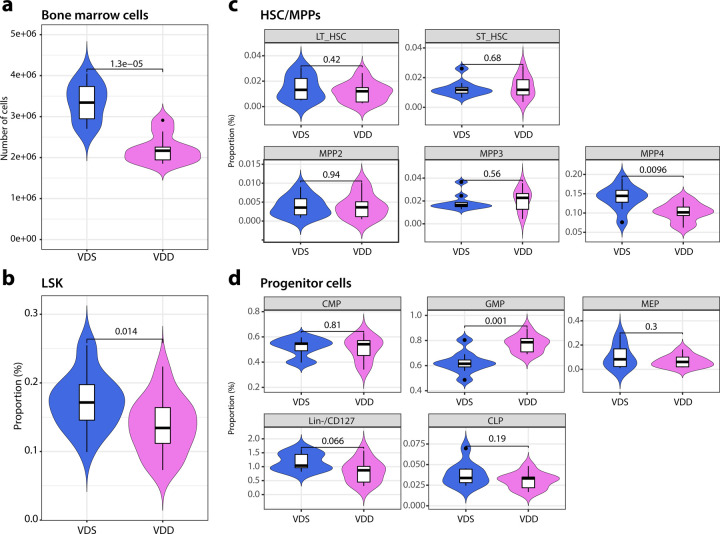
During hematopoiesis, hematopoietic stem cells (HSCs) undergo differentiation into three multipotent progenitor cells (MPPs) in the bone marrow49. MPP2 and MPP3 are further differentiated primarily into the myeloid lineage, while MPP4 is lymphoid. MPP2 is biased toward the production of megakaryocytes and erythrocytes, and MPP3 towards granulocytes and macrophages49. To test if the reductions in the periphery are reflected in the alterations in bone marrow, we collected mononucleated cells from bone marrow from the offspring (n=9–10 per group). The total number of bone marrow mononucleated cells in VDD was 34% decreased compared to VDS (p=0.000013, Fig. 2a). We, then analyzed the proportions of HSCs, MPPs, and hematopoietic progenitor cells using the previously reported definitions49,50; lineage negative (Lin−,CD3−/CD4−/CD8−/B220−/Ter119−/CD11b−/Gr-1−/CD127−), Lin−/Sca-1+/c-Kit+ (LSK), long-term hematopoietic stem cell (LT-HSC, LSK/Flk2−/CD150+/CD48−), short-term hematopoietic stem cell (ST-HSC, LSK/Flk2−/CD150−/CD48−), multipotent progenitor 2 (MPP2, LSK/Flk2−/CD150+/CD48+), multipotent progenitor 3 (MPP3, LSK/Flk2−/CD150−/CD48+), multipotent progenitor 4 (MPP4, LSK/Flk2+), common myeloid progenitor cells (CMP, Lin−/c-Kit+/CD34+/CD16/32−), common lymphoid progenitor cells (CLP, Lin−/c-Kit+/Flk2+/CD127+), (GMP, Lin−/c-Kit+/CD34+/CD16/32+), (MEP, Lin−/c-Kit+/CD34−/CD16/32−), Lin−/CD127+ (early T cell progenitor). We observed a significant reduction of LSK (57.6% decrease, p=0.014, Fig. 2b) and MPP4 (61.7% decrease, p=0.0096, Fig. 2c), and an increase of GMP (50.1% increase, p=0.001) in VDD compared to VDS (Fig. 2d). Representative flow cytometry traces are shown in Supplementary Fig. 4.
Fig. 2: Prenatal vitamin D deficiency alters hematopoietic cell cellular compositions of bone marrow at the adult stage.
a The violin plots show that prenatal vitamin D deficiency reduces the total number of bone marrow cells (n=9 (VDD) and n=10 (VDS)). b The proportion of LSK fraction in bone marrow cells is reduced in VDD mice (n=9 per group). c The differences between the proportions of total HSC, long-term and short-term HSC, and three MPPs in the bone marrow of VDD and VDS mice are displayed in the violin plots (n=9 (VDD) and n=10 (VDS)). d The violin plots illustrate the proportions of hematopoietic progenitor cells in the bone marrow (n=9 per group). The white box shows the range between the first and third quartiles. The upper and lower whiskers represent the 1.5x inter-quantile range, while the black bars show the median. The values in the plot are p-values (Student’s t-test).
Prenatal vitamin D deficiency alters gene expression profiles of MPP4 cells and postnatal day 1 liver
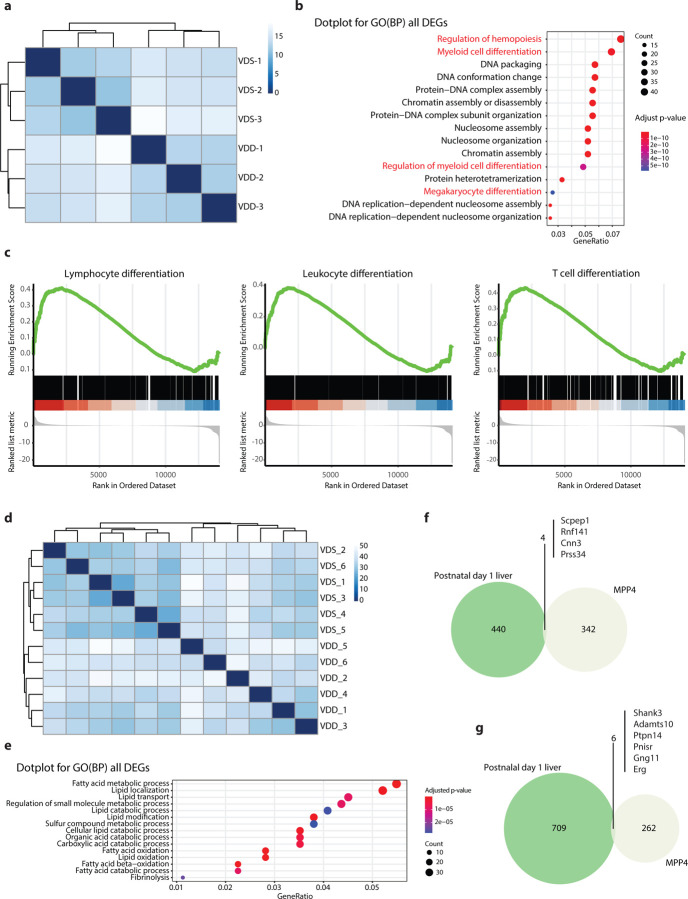
We performed transcriptome analysis on bone marrow MPP4 from VDD and VDS (n=3 per group) to see the transcriptional alterations at 16 weeks of age. The sequencing status and quality of each sample are summarized in Supplementary Data 1. All samples have passed primary quality checks. A hierarchical clustering analysis showed a clear dissociation, suggesting genome-wide transcriptional alterations in VDD (Fig. 3a). We identified 612 differentially expressed genes (DEGs) with at least 1.2-fold change and a false discovery rate adjusted p-value (FDR-adj p-value) <0.05 (Supplementary Data 2). A Gene Ontology (GO) enrichment analysis showed genes of regulation of hemopoiesis (q=1.18×10−13), and myeloid differentiation (q=2.50×10−11) were the top 2 enriched GO biological processes terms in DEGs (Fig. 3b, Supplementary Data 3). Gene Set Enrichment Analysis (GSEA) showed leukocyte differentiation (GO:0002521, enrichment score = 0.385, q-value= 0.010), lymphocyte differentiation (GO:0030098, enrichment score = 0.412, q-value= 0.026), T cell differentiation (GO:0030217, enrichment score =0.435, q-value=0.023) were enriched (Fig. 3c, Supplementary Data 4). We then tested the transcriptional profile of postnatal day 1 liver of VDS and VDD (n=6 per group) using bulk RNA-seq. The sequencing status and quality of each sample are summarized in Supplementary Data 5. All samples have passed primary quality checks. In mice, while the transition of the hematopoietic stem cell production site from the fetal liver to bone marrow starts after embryonic day 16 (E16), the transition from fetal to adult HSC properties occurs 3–4 weeks after birth51; thus, postnatal day 1 liver still contains embryonic HSCs. A hierarchical analysis showed that VDS and VDD have distinct gene expression profiles (Fig. 3d). We identified 1159 DEGs (>1.2-fold change and FDR-adj p-value<0.05) (Supplementary Data 6). Interestingly, 107 out of 444 up-regulated genes overlapped with reported embryonic day 18.5 mouse livers lacking hepatocyte nuclear factor 4alpha (HNF4alpha) up-regulated genes52 (Odds ratio= 5.92, adjusted p-value = 1.49 ×10−37), and 146 were overlapped with reported RXR, the dimerization partner of VDR, binding site harboring genes53 (Supplementary Data 7). The GO enrichment analyses showed that these DEGs are enriched in metabolic processes, specifically lipid metabolism (Fig. 3e). We searched overlapped DEGs between MPP4 and postnatal day 1 liver, showing the same directional changes. We identified four common up-regulated genes (Fig. 3f) and six down-regulated genes (Fig. 3g). This result suggests that prenatal vitamin D deficiency induces long-term transcriptional alterations in hematopoietic stem cells that persist into adulthood.
Fig. 3: Bulk RNA-seq analyses reveal the transcriptional alterations in MPP4 and postnatal day 1 liver.
a The heatmap shows a distinct difference in the transcriptional profiles of MPP4 between VDD and VDS (n=3 per group). b The GO enrichment analysis reveals that the DEGs between VDD and VDS MPP4 are enriched in hematopoiesis and myeloid differentiation-related pathways. c The GSE analysis shows that downregulated genes in MPP4 from VDD mice are enriched in lymphocyte, leukocyte, and T cell differentiation pathways. d The heatmap shows a distinct difference in postnatal day 1 liver transcriptional profiles between VDD and VDS (n=6 per group). e The GO enrichment analysis reveals that the DEGs between postnatal day 1 liver of VDD and VDS mice are enriched in lipid metabolism-related pathways. Four downregulated (f) and six upregulated (g) genes in VDD MPP4 are also differentially expressed in postnatal day 1 liver of VDD in the same direction.
Prenatal vitamin D deficiency alters cellular compositions of the E14.5 embryonic liver, suggesting immune cell proportion changes start during development.
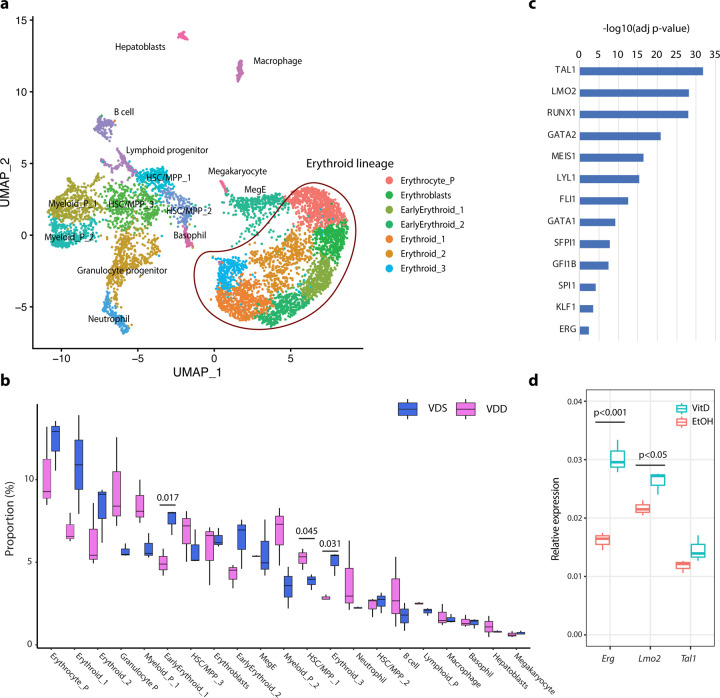
To assess if the cell composition alteration started at the embryonic stage, we performed single-cell RNA-seq (scRNA-seq) on the E14.5 fetal liver of both VDS and VDD embryos (n=3 per group) using the 10x Genomics Chromium platform. Multiplexing with cell hash antibodies was used to reduce the technical batch effect. The obtained sequences were aligned by the 10x Genomics software, CellRanger, and the matrix was demultiplexed and analyzed by Seurat, an R-package 54–56. After eliminating low-quality cells (<1000 genes/cell, <5000 reads/cell, and >10% mitochondrial reads/cell) and cells without cell hashing information, we have identified 21 different cell clusters in a total of 6947 cells. We identified the cell types of each cell cluster based on the marker gene expression status (Fig. 4a, Supplementary Data 8). We detected three HSC/MPP populations, lineage-specific progenitor cells, erythroid lineage cells, and hepatoblast cells in the dataset. Concordant with the previous report57, more than half of the cells were identified as erythroid lineage cells. The cellular composition analysis showed that one of the HSC/MPP populations (HSC/MPP 1) was significantly increased in VDD embryos, and two erythroid lineage cells (Early Erythroid 1 and Erythroid 3) were decreased (Fig. 4b), suggesting the cell composition changes started at least at the embryonic stage E14.5. A pseudo bulk RNA-seq analysis on the scRNAseq datasets showed that down-regulated genes were enriched in the genes regulated by hematopoietic system transcription factors (Fig. 4c, Supplementary Data 9-11). Among these transcription factors, Tal1, Lmo2, and Erg were identified as VDD downregulated genes in MPP4 at the adult stage (Supplementary Data 2). To test whether vitamin D alters the expression of Tal1, Lmo2, and Erg, we measured their expression by quantitative RT-PCR on an embryonic stem cell-derived hematopoietic progenitor cell line, HPC-758, after treating 1-alpha-25-dihydroxyvitamin D3, a ligand of VDR, or ethanol (solvent) for 24 hours by quantitative RT-PCR (Fig. 4d). We observed a significant increase of Lmo2 (p=0.034) and Erg (p=0.005) expression. This result suggests that the gene expression alterations observed at E14.5 were attributed to the dysregulation of hematopoietic transcription factors, such as Lmo2 and Erg, regulated by VDR signaling pathways.
Fig. 4: Prenatal vitamin D deficiency alters cellular compositions of the embryonic liver, suggesting immune cell proportion changes start during development.
a UMAP representation of single-cell RNA-seq gene expression data and cellular lineage identification of E14.5 fetal liver (n=3 per group). b The boxplots indicate prenatal vitamin D deficiency alters cellular compositions of E14.5 fetal liver. c Genes downregulated in VDD E14.5 fetal liver are enriched in the genes regulated by hematopoietic transcription factors. d Treating HPC7 cells with 1-alpha-25-dihydroxyvitamin D3 significantly increases gene expression levels of Erg and Lmo2, suggesting these genes are regulated by VDR (n=3 per treatment group).
Maternal serum vitamin D status in the second trimester is positively associated with the CD8+ T cell proportion in the cord blood.
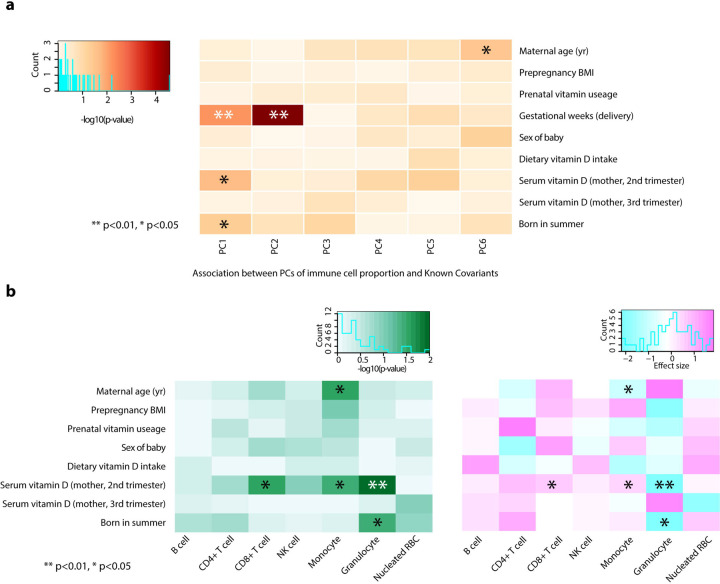
To test the associations between maternal vitamin D levels and immune cell proportions of offspring, we assessed 75 pregnant Japanese women who were recruited as participants of The Birth Cohort Gene and Environment Interaction Study of TMDU (BC-GENIST) project at the Tokyo Medical and Dental University, Bunkyo, Tokyo, Japan59,60. We measured the concentration of serum 25(OH)D3 at two-time points: time point 1 (T1), intended to represent the second trimester from week 10 to week 29, and time point 2 (T2), to represent the third trimester from week 33 to week 40 of gestational age. The immune cell proportions were estimated from the bulk DNA methylation profiles of the cord blood of the fetus using a Bioconductor package FlowSorted.CordBloodCombined.450k61–64. We excluded one participant who did not have T1 serum 25(OH)D3 status from the analysis. The demographic, clinical, and phenotypic information for the study participants is provided in Table 1. All participants are healthy pregnant Japanese women without smoking or drinking during their pregnancy. No participants had hypertension. The average maternal age at delivery was 34.2 (sd 4.0) years old, with pre-pregnancy BMI 17.1–29.2 (average 20.73, sd 2.6). 70.3% of women took prenatal vitamin supplements, and the average estimated daily dietary vitamin D intake was 5.1 (sd 4.4) µg/day65,66. 34 offspring were males (45.9%), and the average gestational week of the delivery was 39.1 (sd 1.2). The average concentration of serum 25(OH)D3 was 25.2 (sd 11.8) ng/ml in the second trimester (average gestation week 19.0 (sd 4.5), T1) and 28.0 ng/ml (sd 14.4) in the third trimester (average gestation week 35.9 (sd 0.9), T2). 30 participants were vitamin D deficient (serum 25(OH)D3 <20 ng/ml) at T1 (40.5%), and 23 were vitamin D deficient at T2 (31.8%). Of those, eight participants were vitamin D deficient at both T1 and T2. While we observed significant associations between the serum 25(OH)D3 levels and sampling season (Summer and Winter) (p=0.047 and p<0.001, T1 and T2, respectively), no significant associations were observed in maternal age, pre-pregnancy BMI, sex of fetuses, prenatal vitamin supplements usage and dietary vitamin D intake. The results of the univariate model fitting are shown in Supplementary Data 12. To analyze the associations between maternal serum 25(OH)D3 levels and immune cell profiles of the cord blood, we calculated principal components (PCs) of immune cell profiles and assessed the contributions of each covariate. Gestation weeks at delivery showed the most significant contributions to PC1 (p= 0.0063 and adjusted r2=0.087) and PC2 (p= 0.000027 and adjusted r2=0.208), followed by maternal serum 25(OH)D3 at T1 to PC1 (p= 0.021 and adjusted r2=0.059) and being born in summer (p= 0.049 and adjusted r2=0.04) (Fig. 5a). We then assessed the associations of covariates to each cell type. We observed that the gestational weeks at delivery were negatively associated with the proportions of CD4 T cells (estimate - 0.027 and p=0.00006) and B cells (estimate −1.12 and p=0.00002) and positively associated with proportions of granulocytes (estimate 0.028 and p=0.0014). Maternal serum 25(OH)D3 at T1 was positively associated with proportions of CD8 T cells (estimate 0.0007 and p=0.0216) and monocytes (estimate 0.0005 and p=0.0349) and negatively associated with proportions of granulocytes (estimate - 0.0024 and p=0.0365) (Supplementary Fig. 5). These associations were still significant after adjusting for the sex of the fetus and gestational weeks at delivery, season of T1, and the gestational week at T1 (Fig. 5b, Supplementary Data 13). These findings indicate that maternal vitamin D levels, especially in the second trimester, affect the immune cell proportions in the babies at birth.
Table 1:
Demographic information of BC-GENIST cohort
| Total N (%) | 74 (100.0) | |
|
| ||
| Maternal age (yr) | Mean (SD) | 34.2 (4.0) |
| Pre-pregnancy BMI | Mean (SD) | 20.7 (2.6) |
| Prenatal vitamin usage, N (%) | Yes | 52 (70.3) |
| Fetal sex, N (%) | Female | 40 (54.1) |
| Gestational age at delivery (weeks) | Mean (SD) | 39.1 (1.2) |
| Gestational age at T1 | Mean (SD) | 19.0 (4.5) |
| Serum vitamin D concentration (ng/ml) at T1 | Mean (SD) | 25.2 (11.8) |
| Gestational age at T2 | Mean (SD) | 35.9 (0.9) |
| Serum vitamin D concentration (ng/ml) at T2 | Mean (SD) | 28.0 (14.4) |
| Estimated dietary vitamin D intake per day | Mean (SD) | 5.1 (4.4) |
| Maternal serum vitamin D deficiency levels, N(%) ☨ | Deficient | 8 (10.8) |
| Insufficient | 20 (27.0) | |
| Sufficient | 46 (62.2) | |
| Estimated immune proportions from cord blood DNA methylation profiles | ||
|
| ||
| CD8 T cell | Mean (SD) | 5.23(3.35) |
| CD4 T cell | Mean (SD) | 19.25(7.15) |
| NK cell | Mean (SD) | 0.71(1.24) |
| B cell | Mean (SD) | 6.67(2.84) |
| Monocyte | Mean (SD) | 3.15(2.27) |
| Granulocyte | Mean (SD) | 61.46(9.2) |
| Nucleated red blood cell (nRBC) | Mean (SD) | 4.09(3.26) |
Deficient, both T1 and T2 serum vitamin D concentrations were less than 20 ng/ml; insufficient, one of the measurements was less than 20 ng/ml and the other was less than 30 ng/ml
Fig. 5: Maternal serum vitamin D status in the second trimester is positively associated with the CD8+ T cell proportion in the cord blood.
a The heatmap shows that the gestational week at the delivery has the strongest associations with immune cell composition variations assessed by the principal component (PC), followed by maternal serum vitamin D (2nd trimester) and being born in the summer season. b After adjusting for the sex of the fetus, gestational age, the season of T1, and the gestational week at T1, maternal serum vitamin D (2nd trimester) maintains significant associations with immune cell composition, specifically positive association with proportions of CD8+ T cell and monocytes, and negative association with granulocytes. Asterisks indicate the significance (** p<0.01 and * p<0.05, Student’s t-test). The left panel shows −log10(p-value), and the right panel shows the direction of the associations.
DISCUSSION:
In this study, we found that prenatal vitamin D deficiency reduced peripheral T-cell and bone marrow MPP4 proportions in adults. Bone marrow MPP4 is a lymphoid-committed multipotent progenitor49, suggesting that the bone marrow MPP proportions might reflect decreased peripheral lymphocyte proportion. The transcriptome analysis revealed that Erg, an essential transcription factor for definitive hematopoiesis67 and is required for self-renewal of HSCs68, expression status significantly reduced in both postnatal day 1 liver and MPP4 of VDD offspring. It has been reported that a reduction in the self-renewal activity of HSC leads to a myeloid-biased output in regenerative conditions49. In the progenitor proportion analysis, we observed increased GMP and reduced early T cell progenitor proportions, suggesting VDD offspring might have a reduction in the self-renewal activity of HSCs. A study using ERG mutant and chimeric embryos reported that Erg is critical for HSC maintenance during embryonic development, and without Erg leads to the rapid exhaustion of definitive hematopoiesis69. The dosage of Erg is associated with the survival of hematopoietic stem and progenitor cells70. We were unable to detect significant differences between VDD and VDS in the expression of Erg in the E14.5 fetal liver scRNA-seq analysis, as its expression was weak in all samples. However, the transcription factor network analysis on e14.5 fetal liver scRNA-seq revealed that Tal1, Lmo2, Runx1, Gata2, and Erg downstream genes were significantly enriched in VDD downregulated genes. In MPP4 of VDD offspring, we observed a significant downregulation of Erg, Tal1, and Lmo2. Intriguingly, the ERG mutant and chimeric embryo study mentioned above reported that ERG is a direct upstream regulator of Runx1 and Gata2 in fetal livers69. In addition, we found that the expression of Erg and Lom2 are regulated by VDR, as the treatment with 1-alpha-25-dihydroxyvitamin D3 (1,25(OH)2D3) led to an increase in Erg expression in HPC7 cells. These suggest that prenatal vitamin D deficiency exposure perturbs the transcription profiles of HSC and MPP during development via modulating hematopoiesis transcription factors regulated by VDR and causes long-term changes in immune cell compositions.
Besides insufficient nutritional intake or photosynthesis on the skin, genetic mutations significantly contribute to vitamin D deficiency. In humans, mutations in genes of the vitamin D biosynthetic pathway are the most common cause of heritable vitamin D-dependent rickets (VDDR), which can be treated with active form vitamin D supplementation, whereas VDR mutation results in hereditary vitamin D-resistant rickets (HVDR), which could partially be ameliorated with Ca2+ supplementation71,72. Animal models with gene knockout have been studied to understand the functions of genes and diseases associated with them. A VDR Knockout mouse model study revealed that VDR null mice are developed without any significant defects before birth; however, they developed a rickets-like phenotype after weaning, and most of the animals died within 15 weeks after birth40, suggesting VDR is critical in the growth and bone formation in the post-weaning stage. Subsequent studies using the same VDR knockout mouse line feeding normal chow diet identified that while VDR knockout mice showed immune defects, no significant differences in the numbers or percentages of red and white cells compared to their wildtype littermates were observed36,37. Interestingly, correcting hypocalcemia by feeding a lactose-rich or polyunsaturated fat-rich diet was able to restore the immune abnormalities observed in VDR knockout mice37. Another research group independently developed another VDR knockout mouse line in C57BL/6 background and reported similar phenotypes, vitamin D-dependent rickets type II with alopecia after weaning42. Studies using this mouse line revealed that this line also developed immune abnormalities39,73. Additionally, the nonobese diabetic (NOD) mouse line with a VDR knockout showed that although diabetes onset is not accelerated by VDR deletion, NOD VDR knockout mice develop rickets and have lower numbers of natural killer T-cells and CD4+CD25+ T-cells41. These findings from independent knockout mouse studies indicate that VDR-depleted animals are phenotypically normal at birth but develop hypocalcemia within the first month of life and immune abnormalities. The active form 1-α−25-dihydroxyvitamin D [1,25(OH)2D] is synthesized from its precursor 25 hydroxyvitamin D [25(OH)D] via catalytic action of the 25(OH)D-1alpha-hydroxylase [1α(OH)ase] enzyme, encoded by Cyp27b1. Mice deficient in 1α(OH)ase developed hypocalcemia, skeletal abnormalities characteristics of rickets, female infertile, and a reduction in CD4 and CD8 peripheral T lymphocytes after weaning18. A zebrafish study showed that loss of Cyp27b1-mediated biosynthesis by gene knockdown significantly reduced runx1 expression and hematopoietic stem and progenitor cell productions71. In this study, we found that prenatal vitamin D deficiency-exposed offspring at the adult stage did not show significant alterations in bone morphology, suggesting that vitamin D depletion during development does not affect embryonic bone development and bone morphology later in life. On the other hand, the immune phenotype of prenatal vitamin D deficiency-exposed offspring we observed was much more severe than those of VDR knockout models and similar to the Cyp27b1 deletion models. Since we observed this phenotype in offspring at 16 weeks of age, while VDR knockout mice typically do not survive beyond 15 weeks40, age might be another factor in developing this phenotype. However, our findings suggest that the immune phenotypes in offspring exposed to prenatal vitamin D deficiency may be caused by a lack of ligands rather than receptor malfunctions during the developmental phase.
A large biological difference between our study and the Cyp27b1 deletion models is 1,25(OH)2D availability after weaning. All offspring were fed VDS diets after weaning, and the serum vitamin D status of the VDD offspring became normal at five weeks of age. Therefore, the alterations we observed at 16 weeks of age were not associated with the vitamin D deficiency status at the time of measurement. But it does indicate that prenatal vitamin D deficiency irreversibly alters bone marrow development in the offspring. This aligns with a previous study conducted on rats74. The study found that the rats with prenatal vitamin D deficiency by maternal dietary manipulation showed significant reductions in the total number of nucleated bone marrow cells and in the colony-forming unit (CFU) content with a corresponding increase in cell cycle rate74. The authors measured the CFU content by transplanting nucleated bone marrow cells to AJ mice purchased from Jackson Laboratory (vitamin D-sufficient diet-fed mice) and found that while 1,25(OH)2D3- or 24,25(OH)D3-treatment on vitamin D deficient rats in postnatal life corrected the serum calcium and phosphate, the treatments did not reverse the CFU content alterations. Their findings clearly demonstrate that prenatal and early-life vitamin D deficiency could cause irreversible effects on HSC development. However, we cannot fully rule out the possibility that alterations in hematopoietic stem cell niches might also be irreversibly altered. HSC niches in different developmental stages play critical roles in the maintenance and differentiation of HSCs (reviewed by Gao et al.75). It would be beneficial to explore this possibility further in upcoming research.
Our study also revealed that higher maternal vitamin D levels during the second trimester were associated with increased proportions of CD8+ T cells and decreased granulocytes in cord blood in humans. The associations were maintained after adjusting for the season of the second trimester, the gestational week at the maternal blood draw, the sex of the fetus, and the gestational week at birth. Hematopoietic development consists of primitive and definitive hematopoiesis76. In mice, the definitive hematopoiesis starts in the aorta-gonad mesonephros (AGM) region at approximately E10.5, then moves to the liver around E12.5 and the bone marrow around E17.5 77,78. After birth, the bone marrow is the only site where HSCs are maintained and expanded. In human development, the transition of the hematopoietic stem cell production site from the fetal liver to bone marrow occurs in the second trimester78,79 when we observed significant associations. Therefore, our findings suggest that the vitamin D levels during the transition period of fetal liver to bone marrow are crucial for the development of immune cells. Recently, Elgormus et al. reported that newborn serum vitamin D levels were negatively correlated with neutrophil-to-lymphocyte ratios (NLR) in newborn babies80. The most common white blood cell is the granulocyte, composed of three distinct types: neutrophils, eosinophils, and basophils. Among them, the neutrophil is the most abundant type of the three. Our results and their finding concordantly indicate that the immune cell compositions, especially granulocyte (neutrophil) and lymphocyte, of the babies are influenced by vitamin D levels in early life. Of note, cord blood NLR has been proposed as an indicator for the diagnosis of early neonatal sepsis combined with other laboratory tests and clinical manifestations81–83. The associations between neonatal sepsis and cord blood vitamin D levels have been well reported. A meta-analysis of 18 studies revealed that low maternal and cord blood vitamin D levels were significantly associated with the incidence of neonatal sepsis84. Additionally, a study on 4,340 neonates appropriate for gestational age found a negative correlation between cord blood NLR and fetal malnutrition. This indicates that cord blood NLR could be utilized as a marker for fetal malnutrition85. Supplementing with Vitamin D during pregnancy may lower the risks of neonatal sepsis and other adverse outcomes.
In conclusion, this study demonstrates that prenatal vitamin D deficiency reduces the number and the proportion of MPP4 cells in the bone marrow, changing the expression status of the genes that may be directly and indirectly regulated by VDR, and this alteration results in reduced T cell proportions in peripheral blood and spleens of the offspring. The association between T cell proportion and maternal vitamin D status was also observed in our BC-GENIST mother-baby cohorts. We and others have reported that micronutrient deficiency changed the cellular compositions of adult mature organs and may contribute to the phenotypes using animal dietary manipulation models21,35,86. These findings indicate that the cell fate decision alteration of stem cells could be the key component of the long-term memory of prenatal micronutrient deficiency, which links to disease risks later in life. Nevertheless, this study has several limitations. A study conducted in Greece on mother-infant pairs revealed that insufficient levels of vitamin D (<50 nmol/L) in pregnant mothers were linked to an increased occurrence of micronucleated cells in binucleated T-cells87. Micronuclei are early indicators of genetic effects used to test the relationship between exposure to genotoxic substances and cancer. However, our study did not explore whether maternal VDD increases DNA damage risks in offspring. Sexual dimorphism is another covariate we must explore. Since female mice did not show significant alterations in immune cell proportions in adults, we only performed transcriptional analysis on male mice. Both males and females may have prenatal and neonatal immune cell proportional alterations. Furthermore, there could be transcriptional changes in certain genes in adult females, but these changes may not affect cell fate decisions. These possibilities should be further investigated in future studies.
METHODS:
Maternal vitamin D deficiency mouse model
We performed dietary manipulations on C57BL/6J female mice. Five weeks old female mice (F0) were fed vitamin D deficient (VDdef) or sufficient (VDsuf) diets for five weeks before mating to the control diet-fed male mice and throughout the subsequent pregnancy. VDdef (0.0 IU/g vitamin D) and nutrient-matched VDsuf (1.0 IU/g vitamin D) diets were obtained from Research Diets Inc. (10 kcal%fat, 20 kcal%Protein and 70 kcal%Carbonate). The detailed ingredients of each diet are listed in Supplementary Data 14. VDdef-fed females were supplemented with 1.5% calcium gluconate water for drinking water. After delivery, all F0 mice were fed VDsuf diets. The offspring were fed VDsuf diet after weaning and maintained the diet until the sampling. All animal studies were approved by the Institutional Animal Care and Use Committee at the Albert Einstein College of Medicine.
Serum vitamin D level of mouse serum samples
Serum vitamin D (25(OH)D) levels of mouse serum samples were assessed by commercially available ELISA kits (Eagle Biosciences, Inc. Amherst NH, or Abcam, Cambridge, MA) according to the manufacturer’s instructions. The signal was detected with BioTek Synergy 4 Microplate reader (Agilent Technologies), and the results were analyzed using a 4-parameter logistic regression algorithm (http://www.elisaanalysis.com/app). The measurements were performed as duplicates.
Immune cell profiling on peripheral blood and spleen
Immune cell profiling analysis was performed using flow cytometry (FACS Aria2, BD Biosciences) after fluorescent dye conjugate antibodies staining. The obtained data were analyzed with FlowJo_10.6.1_CL (https://www.flowjo.com/). Peripheral blood samples were collected using submandibular vein bleeding methods. The spleen was obtained after euthanizing the animals with carbon dioxide inhalation. The spleen samples were dissociated on 70 µm filters. The obtained single-cell suspensions were stained with antibodies of immune cell surface marker proteins to identify cell types. The antibodies used in this study are listed in Supplementary Data 15.
Isolating multipotent progenitor cells from bone marrow
Bone marrow cells were collected by crushing bones (tibia, Femur, iliac, sternum, and vertebrae). The red cells were lysed with lysis buffer (150 mM NH4Cl, 1 mM KHCO3, and 0.1 mM EDTA). HSC/MPP fractions were defined by the previously reported definition49. MPP4, lymphoid-primed hematopoietic multipotent progenitor cells were isolated for gene expression analysis. Representative flow cytometry traces are shown in Supplementary Fig. 4.
Cell culture
Hematopoietic progenitor cell line HPC-7 was kindly gifted by Dr. Britta Will at Albert Einstein College of Medicine. HPC-7 cells were maintained at density of 1–10 ×105/ml in Iscove’s modified Dulbecco’s medium (Invitrogen) supplemented with 50–100 ng/ml of mouse stem cell factor (Gemini Bio-Products), 1 mM Sodium Pyruvate, 6.9 ng/mL α-Monothioglycerol (SIGMA-Aldrich), 5% of bovine calf serum and Penicillin-Streptomycin. To examine the effect of vitamin D treatment on gene expression levels, the HPC-7 cells were treated with 0.1 µM of 1alpha,25-Dihydroxyvitamin D3 (SIGMA-Aldrich) or 0.1% (vol/vol) ethanol (solvent) for 24 hours.
RNA-seq library construction and sequencing
We performed RNA-seq on FACS isolate MPP4 cells (lymphoid primed-multipotent progenitor cells) and postnatal day 1 neonate liver. Total RNA was isolated with AllPrep DNA/RNA micro kit (QIAGEN). After we depleted ribosomal RNAs from total RNA, we generated the RNA-seq libraries using KAPA RNA HyperPrep with RiboErase kit (Roche). The generated libraries were sequenced on an Illumina NOVA-seq sequencer (Novogene Co., Ltd., USA).
RNA-seq alignment
After checking the quality of the sequencing files using FastQC88 and trimming low-quality reads and adapter sequences using Cutadapt89, the obtained sequences were aligned to the mouse mm10 reference genome with the gencode M15 gene annotation using STAR aligner90. The quality of the library was assessed with RSeQC91. The obtained transcript counts were analyzed with DESeq2. We identified significant differentially expressed genes (DEGs) that showed two times higher or lower expression with a false discovery rate-adjusted p-value less than 0.05. The detailed sequencing and alignment status are listed in Supplementary Data 1 and 5.
Enrichment analysis for Gene Ontology (GO)
We used the Gene Ontology (GO) enrichment analyses and Gene Set Enrichment Analyses (GSEA) of a Bioconductor package ClusterProfiler92 to see the functional enrichment of DEGs. Q-values <0.05 were considered significant.
Quantitative RT-PCR
Total RNA samples were isolated with AllPrep DNA/RNA micro kit, and cDNA libraries were synthesized using SuperScript III transcriptase (Invitrogen) with random hexamer. The real-time PCR was performed with Roche LightCycler 480 SYBR GREEN I Master mix on a LightCycler 480 system (Roche). Relative gene expression abundance between samples was calculated with the CT method using Gapdh as an internal control. The sequences of primers used in this study are listed in Supplementary Data 16.
Single-cell RNA sequencing library preparation
We used e14.5 male mouse liver (n=3 per group) to study transcriptional alteration at single-cell resolution. The e14.5 mouse liver samples were dissociated on 70 µm filters, then each sample was stained with unique cell hashing antibodies (BioLegend). 3,000–4,000 cells per sample were targeted on the 10x Genomics Chromium platform. Single-cell mRNA libraries were built using the Chromium Next GEM Single Cell 3’ Library Construction V3 Kit, libraries sequenced on an Illumina NOVA-seq. Sequencing data were aligned to mm10 mouse reference using the Cell Ranger 3.0.2 pipeline (10x Genomics). Counting cell hashing tag were performed using CITE-seq Count version 1.3.493.
scRNA-seq data processing, batch correction, clustering, cell-type labeling, and data visualization
All scRNA-seq analysis and data visualization were performed using an R package, Seurat54–56. After demultiplexing based on the cell hashing tag information, low-quality cells (<1000 genes/cell, <5000 reads/cell, and >10% mitochondrial reads/cell) were eliminated from the further analyses. Data integration and identifying cell clusters were carried out after performing SCTransform56. Cell types of each cell cluster were identified based on the expression of the marker genes94. Proportions of assigned cell types were analyzed using Student’s t-test. P-values <0.05 were considered significant. Differentially expressed genes of each cell type between VDD and VDS were identified as at least 25% of cells expressed the gene, the log fold change greater than 0.5, and the FDR adjusted p-value less than 0.05. GO enrichment analysis was performed using a Bioconductor package ClusterProfiler92 with q-values <0.05 considered significant. The enrichment status of the transcription factors was assessed using CHEA transcription factor targets dataset95 provided by Enrichr (http://amp.pharm.mssm.edu/Enrichr)96.
Measurement of bone mineral density
Bone mineral density was measured humerus of the offspring at dissection. After the dissection, the left arms (n=5–6 animals for each group) were scanned using an X-ray CT system, Inveon Multimodality scanner (Siemens). The CT x-rays were generated by an 80kV peak voltage difference between the cathode and tungsten target at 0.5 mA current and 200 millisecond exposure time. The arm samples were placed on the 38mm width bed tandemly. The CT field of view was 5.5 cm by 8.5 cm with an overall resolution without magnification of 60 microns. A Scout View was performed before the start of the CT Acquisition to ensure the correct positioning of the subject in the field of view. Image analysis was performed using MicroView (https://microview.parallax-innovations.com/).
Serum vitamin D level of serum samples from pregnant women
Healthy pregnant women were recruited as participants in The Birth Cohort Gene and Environment Interaction Study of TMDU (BC-GENIST) project at the Tokyo Medical and Dental University, Bunkyo, Tokyo, Japan59,60. Written informed consent was obtained from the participants, and the study was approved by the Institutional Review Board of Tokyo Medical and Dental University (No. G2000–181, 29 July 2014). In this study, all participants were healthy Japanese females aged 27–42 years old without smoking or drinking alcohol during their pregnancy. We used the following demographic information of the participants in this study; maternal age at delivery, prepregnant body mass index, prenatal vitamin supplements usage, sex of the fetus, gestation weeks, and estimated daily vitamin D intake. The daily vitamin D intake was estimated 3-day food record questionnaire. Maternal serum samples were collected twice, around 20 and 36 gestational weeks, and aliquoted samples were stored at −150 °C until use. Serum 25(OH)D3 levels were measured using a modified LC-APCI-MS/MS method97. As previously described, this method involves the use of deuterated 25(OH)D3 (d6-25(OH)D3) as an internal standard compound and the selection of a precursor and product ion with an MS/MS multiple reaction monitoring (MRM) method. The internal standard d6-25(OH)D3 (0.5 ng/10 μL) was added to serum (40 μL) and precipitated with acetonitrile (200 μL). After evaporation of the supernatant, the precipitant was dissolved with ethyl acetate (400 μL) and distilled water (200 μL) with vigorous shaking. The ethyl acetate phase was removed and evaporated. Extracted vitamin D metabolites from serum were derivatized by 4-[2-(6,7-dimethoxy-4-methyl-3-oxo-3,4-dihydroquinoxalyl)ethyl]-1,2,4-triazoline-3,5-dione (DMEQ-TAD) to obtain high sensitivity by increasing ionization efficiency 98. Separation was carried out using a reverse-phase C18 analytical column (CAPCELL PAK C18 UG120, 5 μm; (4.6 I.D. × 250 mm) (SHISEIDO, Tokyo, Japan) with a solvent system consisting of (A) acetonitrile, (B) distilled water (0–5 min A = 30%, 5–34 min (A) = 30 → 70%, and 34–37 min (A) = 70 → 100%) as the mobile phase and a flow rate of 1.0 mL/min. All MS data were collected in the positive ion mode, and quantitative analysis was carried out using MS/MS-MRM of the precursor/product ion for DMED-TAQ-25(OH)D3 (m/z; 746.5/468.1) and DMED-TAQ-d6-25(OH)D3 (m/z; 752.5/468.1) with a dwell time of 200 ms (AB Sciex LLC., Framingham, MA, USA).
Estimation of immune cell profiles of human cord blood
We used a modified deconvolution approach to estimate the immune cell profiles of human cord blood from the bulk DNA methylation profiles99. Bulk DNA methylation profiles of cord blood were accessed using Infinium HumanMethylation450 BeadChip, and the estimation was performed using a Bioconductor package FlowSorted.CordBloodCombined.450k61.
Statistical analysis
Mouse phenotype results were analyzed using Student’s t-test. The associations between maternal serum vitamin D levels and immune cell profiles in human cord blood were tested using one-way ANOVA. P-values <0.05 were considered significant. Differences in gene expression between VDD-F1 and VDS-F1 in RNA-seq were analyzed using DESeq2 with FDR-adjused p-value <0.05. R v4.0 (https://www.r-project.org/) was used for most of the analyses.
Supplementary Material
ACKNOWLEDGMENTS:
The authors thank Dr. Britta Will at Albert Einstein College of Medicine for generously providing the HPC7 cells. Additionally, we would like to acknowledge the MicroPET Facility, supported by The M. Donald Blaufox Laboratory for Molecular Imaging and NIH (1S10RR029545” MicroPET/SPECT/CT Animal Imaging Device”), the Flow Cytometry Core Facility, and the Genomic Core Facility at Albert Einstein College of Medicine.
Funding Statement
This work was supported by the Human Genomic Pilot Grant; Department of Genetics, Albert Einstein College of Medicine (M.S.), internal Texas A&M AgriLife Research (M.S.), the National Institutes of Health under award number R01HL145302 (M.S.), Nanken-Kyoten, Tokyo Medical and Dental University, under award number 2021-kokusai 02 (M.S.), and Mishima Kaiun Memorial Fund (M.S.). This work was also supported by Jane A. and Myles P. Dempsey and by NIH grant R35CA253127 (to U.G.S.). U.G.S. holds the Edward P. Evans Endowed Professorship in Myelodysplastic Syndromes at Albert Einstein College of Medicine. The Endowed Professorship was supported by a grant from the Edward P. Evans Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DATA AVAILABILITY:
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files, except for human cord blood analyses, which may contain sensitive information. All sequencing data, RNA-seq and scRNA-seq of this study, is deposited in NCBI’s Gene Expression Omnibus GEO database under the accession number GSE242043, and processed data and code used in this study are available upon request.
ETHICS DECLARATIONS:
Competing interests
The authors declare no competing interests.
REFERENCE
- 1.Mostafa W. Z. & Hegazy R. A. Vitamin D and the skin: Focus on a complex relationship: A review. J. Advanc. Res. 6, 793–804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodnar L. M. et al. Maternal vitamin D deficiency increases the risk of preeclampsia. J. Clin. Endocrinol. Metab. 92, 3517–3522 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Pligt P. et al. Associations of Maternal Vitamin D Deficiency with Pregnancy and Neonatal Complications in Developing Countries: A Systematic Review. Nutrients 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zosky G. R. et al. Vitamin D deficiency at 16 to 20 weeks’ gestation is associated with impaired lung function and asthma at 6 years of age. Ann. Am. Thorac. Soc. 11, 571–577 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Weinert L. S. & Silveiro S. P. Maternal-fetal impact of vitamin D deficiency: a critical review. Matern. Child Health J. 19, 94–101 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Hart P. H. et al. Vitamin D in fetal development: findings from a birth cohort study. Pediatrics 135, e167–73 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Gilani S. & Janssen P. Maternal Vitamin D Levels During Pregnancy and Their Effects on Maternal-Fetal Outcomes: A Systematic Review. J. Obstet. Gynaecol. Can. (2019) doi: 10.1016/j.jogc.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Cashman K. D. Vitamin D Requirements for the Future-Lessons Learned and Charting a Path Forward. Nutrients 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth D. E. et al. Vitamin D supplementation in pregnancy and lactation and infant growth. N. Engl. J. Med. 379, 535–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashman K. D. Vitamin D deficiency: defining, prevalence, causes, and strategies of addressing. Calcif. Tissue Int. 106, 14–29 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Aspelund T. et al. Effect of Genetically Low 25-Hydroxyvitamin D on Mortality Risk: Mendelian Randomization Analysis in 3 Large European Cohorts. Nutrients 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest K. Y. Z. & Stuhldreher W. L. Prevalence and correlates of vitamin D deficiency in US adults. Nutr. Res. 31, 48–54 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Liu X., Baylin A. & Levy P. D. Vitamin D deficiency and insufficiency among US adults: prevalence, predictors and clinical implications. Br. J. Nutr. 119, 928–936 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Lips P. Worldwide status of vitamin D nutrition. J. Steroid Biochem. Mol. Biol. 121, 297–300 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Holick M. F. Resurrection of vitamin D deficiency and rickets. J. Clin. Invest. 116, 2062–2072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pike J. W., Meyer M. B., Lee S.-M., Onal M. & Benkusky N. A. The vitamin D receptor: contemporary genomic approaches reveal new basic and translational insights. J. Clin. Invest. 127, 1146–1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yetgin S. & Ozsoylu S. Myeloid metaplasia in vitamin D deficiency rickets. Scand. J. Haematol. 28, 180–185 (1982). [DOI] [PubMed] [Google Scholar]
- 18.Panda D. K. et al. Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: evidence for skeletal, reproductive, and immune dysfunction. Proc Natl Acad Sci USA 98, 7498–7503 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maka N. et al. Vitamin D deficiency during pregnancy and lactation stimulates nephrogenesis in rat offspring. Pediatr. Nephrol. 23, 55–61 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Gezmish O. & Black M. J. Vitamin D deficiency in early life and the potential programming of cardiovascular disease in adulthood. J. Cardiovasc. Transl. Res. 6, 588–603 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Foong R. E. et al. The effects of in utero vitamin D deficiency on airway smooth muscle mass and lung function. Am. J. Respir. Cell Mol. Biol. 53, 664–675 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hawes J. E. et al. Maternal vitamin D deficiency alters fetal brain development in the BALB/c mouse. Behav. Brain Res. 286, 192–200 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Wu J., Zhong Y., Shen X., Yang K. & Cai W. Maternal and early-life vitamin D deficiency enhances allergic reaction in an ovalbumin-sensitized BALB/c mouse model. Food Nutr. Res. 62, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belenchia A. M., Johnson S. A., Ellersieck M. R., Rosenfeld C. S. & Peterson C. A. In utero vitamin D deficiency predisposes offspring to long-term adverse adipose tissue effects. J. Endocrinol. 234, 301–313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belenchia A. M. et al. Maternal vitamin D deficiency during pregnancy affects expression of adipogenic-regulating genes peroxisome proliferator-activated receptor gamma (PPARγ) and vitamin D receptor (VDR) in lean male mice offspring. Eur. J. Nutr. 57, 723–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichetzeder C. et al. Maternal vitamin D deficiency and fetal programming--lessons learned from humans and mice. Kidney Blood Press. Res. 39, 315–329 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Nicholas C. et al. Maternal vitamin D deficiency programs reproductive dysfunction in female mice offspring through adverse effects on the neuroendocrine axis. Endocrinology 157, 1535–1545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N. Q. et al. Dietary vitamin D restriction in pregnant female mice is associated with maternal hypertension and altered placental and fetal development. Endocrinology 154, 2270–2280 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Xue J., Schoenrock S. A., Valdar W., Tarantino L. M. & Ideraabdullah F. Y. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin. Epigenetics 8, 107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandes de Abreu D. A. et al. Prenatal vitamin D deficiency induces an early and more severe experimental autoimmune encephalomyelitis in the second generation. Int. J. Mol. Sci. 13, 10911–10919 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maia-Ceciliano T. C. et al. Maternal vitamin D-restricted diet has consequences in the formation of pancreatic islet/insulin-signaling in the adult offspring of mice. Endocrine 54, 60–69 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Nascimento F. A. M., Ceciliano T. C., Aguila M. B. & Mandarim-de-Lacerda C. A. Maternal vitamin D deficiency delays glomerular maturity in F1 and F2 offspring. PLoS ONE 7, e41740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento F. A. M., Ceciliano T. C., Aguila M. B. & Mandarim-de-Lacerda C. A. Transgenerational effects on the liver and pancreas resulting from maternal vitamin D restriction in mice. J. Nutr. Sci. Vitaminol. 59, 367–374 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Liang Y. et al. Vitamin D deficiency worsens maternal diabetes induced neurodevelopmental disorder by potentiating hyperglycemia-mediated epigenetic changes. Ann. N. Y. Acad. Sci. (2020) doi: 10.1111/nyas.14535. [DOI] [PubMed] [Google Scholar]
- 35.Lundy K. et al. Vitamin D deficiency during development permanently alters liver cell composition and function. Front Endocrinol (Lausanne) 13, 860286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Kelly J. et al. Normal myelopoiesis but abnormal T lymphocyte responses in vitamin D receptor knockout mice. J. Clin. Invest. 109, 1091–1099 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieu C. et al. In vitro and in vivo analysis of the immune system of vitamin D receptor knockout mice. J. Bone Miner. Res. 16, 2057–2065 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Yu S. & Cantorna M. T. Epigenetic reduction in invariant NKT cells following in utero vitamin D deficiency in mice. J. Immunol. 186, 1384–1390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu S. & Cantorna M. T. The vitamin D receptor is required for iNKT cell development. Proc Natl Acad Sci USA 105, 5207–5212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshizawa T. et al. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat. Genet. 16, 391–396 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Gysemans C. et al. Unaltered diabetes presentation in NOD mice lacking the vitamin D receptor. Diabetes 57, 269–275 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Li Y. C. et al. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci USA 94, 9831–9835 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camargo C. A. et al. Maternal intake of vitamin D during pregnancy and risk of recurrent wheeze in children at 3 y of age. Am. J. Clin. Nutr. 85, 788–795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brehm J. M. et al. Serum vitamin D levels and severe asthma exacerbations in the Childhood Asthma Management Program study. J. Allergy Clin. Immunol. 126, 52–8.e5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirzaei F. et al. Gestational vitamin D and the risk of multiple sclerosis in offspring. Ann. Neurol. 70, 30–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulligan M. L., Felton S. K., Riek A. E. & Bernal-Mizrachi C. Implications of vitamin D deficiency in pregnancy and lactation. Am. J. Obstet. Gynecol. 202, 429.e1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stene L. C., Ulriksen J., Magnus P. & Joner G. Use of cod liver oil during pregnancy associated with lower risk of Type I diabetes in the offspring. Diabetologia 43, 1093–1098 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Vitamin D supplement in early childhood and risk for Type I (insulin-dependent) diabetes mellitus. The EURODIAB Substudy 2 Study Group. Diabetologia 42, 51–54 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Pietras E. M. et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Challen G. A., Boles N., Lin K. K.-Y. & Goodell M. A. Mouse hematopoietic stem cell identification and analysis. Cytometry A 75, 14–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luis T. C., Killmann N. M. B. & Staal F. J. T. Signal transduction pathways regulating hematopoietic stem cell biology: introduction to a series of Spotlight Reviews. Leukemia 26, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Battle M. A. et al. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA 103, 8419–8424 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boergesen M. et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32, 852–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satija R., Farrell J. A., Gennert D., Schier A. F. & Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A., Hoffman P., Smibert P., Papalexi E. & Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafemeister C. & Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X. et al. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res. 30, 1109–1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinto do O P., Kolterud A. & Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17, 5744–5756 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavethynath S. et al. Metabolic and Immunological Shifts during Mid-to-Late Gestation Influence Maternal Blood Methylation of CPT1A and SREBF1. Int. J. Mol. Sci. 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato N. et al. Placenta mediates the effect of maternal hypertension polygenic score on offspring birth weight: a study of birth cohort with fetal growth velocity data. BMC Med. 19, 260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucas A., K. G., Meaghan C.,. Kelly M.,. Devin C.,. John K.,. Karl T.,. Robert Lyle, Brock C.,. Janine Felix. FlowSorted.CordBloodCombined.450k. Bioconductor (2019) doi: 10.18129/b9.bioc.flowsorted.cordbloodcombined.450k. [DOI] [Google Scholar]
- 62.Gervin K. et al. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin. Epigenetics 11, 125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salas L. A. et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 19, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koestler D. C. et al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL). BMC Bioinformatics 17, 120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imai C. et al. Diet Quality and Its Relationship with Weight Characteristics in Pregnant Japanese Women: A Single-Center Birth Cohort Study. Nutrients 15, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imai C. et al. Application of the Nutrient-Rich Food Index 9.3 and the Dietary Inflammatory Index for Assessing Maternal Dietary Quality in Japan: A Single-Center Birth Cohort Study. Nutrients 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loughran S. J. et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 9, 810–819 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Ng A. P. et al. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood 118, 2454–2461 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Taoudi S. et al. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 25, 251–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Y. et al. Reduced erg dosage impairs survival of hematopoietic stem and progenitor cells. Stem Cells 35, 1773–1785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cortes M. et al. Developmental vitamin D availability impacts hematopoietic stem cell production. Cell Rep. 17, 458–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fraser D. et al. Pathogenesis of hereditary vitamin-D-dependent rickets. An inborn error of vitamin D metabolism involving defective conversion of 25-hydroxyvitamin D to 1 alpha,25-dihydroxyvitamin D. N. Engl. J. Med. 289, 817–822 (1973). [DOI] [PubMed] [Google Scholar]
- 73.Yu S., Bruce D., Froicu M., Weaver V. & Cantorna M. T. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc Natl Acad Sci USA 105, 20834–20839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wientroub S., Hagan M. P. & Reddi A. H. Reduction of hematopoietic stem cells and adaptive increase in cell cycle rate in rickets. Am. J. Physiol. 243, C303–6 (1982). [DOI] [PubMed] [Google Scholar]
- 75.Gao X., Xu C., Asada N. & Frenette P. S. The hematopoietic stem cell niche: from embryo to adult. Development 145, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orkin S. H. & Zon L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baron M. H., Isern J. & Fraser S. T. The embryonic origins of erythropoiesis in mammals. Blood 119, 4828–4837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soares-da-Silva F., Peixoto M., Cumano A. & Pinto-do-Ó P. Crosstalk between the hepatic and hematopoietic systems during embryonic development. Front. Cell Dev. Biol. 8, 612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tavian M. & Péault B. Embryonic development of the human hematopoietic system. Int. J. Dev. Biol. 49, 243–250 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Elgormus Y., Okuyan O. & Uzun H. The relationship between hematological indices as indicators of inflammation and 25-hydroxyvitamin D3 status in newborns. BMC Pediatr. 23, 83 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xin Y. et al. Accuracy of the neutrophil-to-lymphocyte ratio for the diagnosis of neonatal sepsis: a systematic review and meta-analysis. BMJ Open 12, e060391 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sumitro K. R., Utomo M. T. & Widodo A. D. W. Neutrophil-to-Lymphocyte Ratio as an Alternative Marker of Neonatal Sepsis in Developing Countries. Oman Med. J. 36, e214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Panda S. K., Nayak M. K., Rath S. & Das P. The Utility of the Neutrophil-Lymphocyte Ratio as an Early Diagnostic Marker in Neonatal Sepsis. Cureus 13, e12891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Workneh Bitew Z., Worku T. & Alemu A. Effects of vitamin D on neonatal sepsis: A systematic review and meta-analysis. Food Sci. Nutr. 9, 375–388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Can E. & Can C. The value of neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) parameters in analysis with fetal malnutrition neonates. J. Perinat. Med. 47, 775–779 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Chen F. et al. Prenatal retinoid deficiency leads to airway hyperresponsiveness in adult mice. J. Clin. Invest. 124, 801–811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Callaghan-Gordo C. et al. Vitamin D insufficient levels during pregnancy and micronuclei frequency in peripheral blood T lymphocytes mothers and newborns (Rhea cohort, Crete). Clin. Nutr. 36, 1029–1035 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Andrews/Babraham Institute, S. FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010). [Google Scholar]
- 89.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, 10 (2011). [Google Scholar]
- 90.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L., Wang S. & Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 92.Yu G., Wang L.-G., Han Y. & He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stoeckius M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao S. et al. Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res. 32, 38–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lachmann A. et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nishikawa M. et al. Generation of 1,25-dihydroxyvitamin D3 in Cyp27b1 knockout mice by treatment with 25-hydroxyvitamin D3 rescued their rachitic phenotypes. J. Steroid Biochem. Mol. Biol. 185, 71–79 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Higashi T., Awada D. & Shimada K. Simultaneous determination of 25-hydroxyvitamin D2 and 25-hydroxyvitamin D3 in human plasma by liquid chromatography-tandem mass spectrometry employing derivatization with a Cookson-type reagent. Biol. Pharm. Bull. 24, 738–743 (2001). [DOI] [PubMed] [Google Scholar]
- 99.Houseman E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.