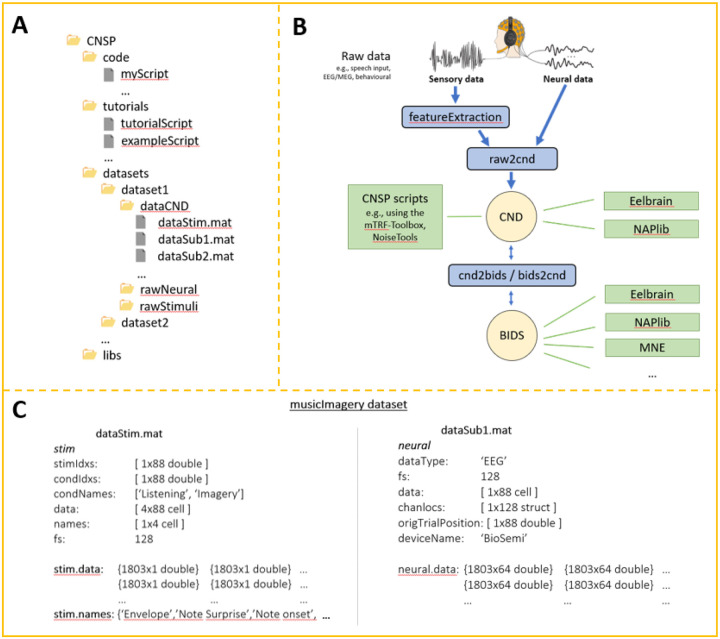
Figure 5. The Continuous-event Neural Data (CND) structure.
(A) CND folder structure. ‘dataStim.mat’ refers to the stimulus features, while ‘dataSubX.mat’ refers to the neural data. In the example, all participants were presented with the same stimuli. It is also possible to associate a ‘dataStim’ file to each participant if needed e.g., dataStim1.mat, dataStim2.mat. (B) Conceptual data preprocessing and analysis pipeline. In addition to the CNSP resources, Eelbrain-toolkit and NAPLib can also process CND data structures. It is also possible to export the CND data structure into Brain Imaging Data Structure (BIDS), enabling the use of general-purpose toolboxes such as MNE-Python. (C) Example of dataStim and dataSub structures. dataStim contains the stimulus data matrices (stim.data) for each of the four stimulus features and for each of the 88 experimental trials. ‘names’ specified what the four stimulus features are i.e., envelope, note expectation, note onset, metronome. ‘stimIdxs’ indicate the stimulus index of each trial. The ‘rawStimuli’ folder should include documentation regarding what stimulus file corresponds to each index. Here, indices 1–4 refer to pieces named ‘chor-096’, ‘chor-038’, ‘chor-101’, and ‘chor-019’ respectively. ‘condIdxs’ refers to the task that was carried out in each trial i.e., music listening or imagery. The EEG data structure is straightforward in that it simply contains basic meta-data, such as the ‘deviceName’, and a data matrix (neural.data), with a [samples × channel] data matrix per trial.