Abstract
Although the relationship between gut microbiota and flavan-3-ol metabolism differs greatly between individuals, the specific metabolic profiles, known as metabotypes, have not yet been clearly defined. In this study, fecal batch fermentations of 34 healthy donors inoculated with (−)-epicatechin were stratified into groups based on their conversion rate of (−)-epicatechin and their quali–quantitative metabolic profile. Fast and slow converters of (−)-epicatechin, high producers of 1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol (3-HPP-2-ol) and 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone (3,4-DHPVL) were identified. Fecal microbiota analysis revealed that fast conversion of (−)-epicatechin was associated with short-chain fatty acid (SCFA)-producing bacteria, such as Faecalibacterium spp. and Bacteroides spp., and higher levels of acetate, propionate, butyrate, and valerate were observed for fast converters. Other bacteria were associated with the conversion of 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol into 3-HPP-2-ol (Lachnospiraceae UCG-010 spp.) and 3,4-DHPVL (Adlercreutzia equolifaciens). Such stratification sheds light on the mechanisms of action underlying the high interindividual variability associated with the health benefits of flavan-3-ols.
Keywords: metabotype, phenyl-γ-valerolactone, fecal batch fermentation, gut microbiota, metabolic profile, conversion rate
1. Introduction
Flavan-3-ols are the principal contributors to dietary (poly)phenols intake, owing to consumption of tea, red wine, cocoa products, fruits, and nuts.1−3 These molecules have positive effects on health, notably in the prevention of noncommunicable diseases such as obesity, diabetes, cancer, and neurodegenerative and cardiovascular diseases.4−9 It is worth noting that a recent dietary bioactive guideline recommended the daily intake of 400–600 mg flavan-3-ols to benefit from a cardiometabolic protective effect.10
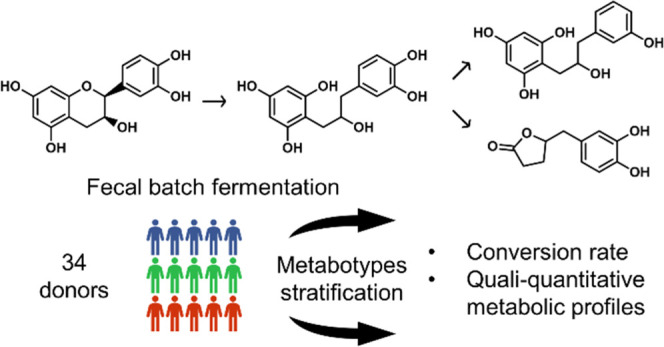
Gut microbiota is known to drive the health effects of flavan-3-ols.11−13 In fact, only a small fraction of monomeric flavan-3-ols is absorbed by the small intestine, while the majority of the ingested flavan-3-ols reach the colon and undergo gut microbiota degradation. This leads to the formation of bioavailable metabolites such as phenyl-γ-valerolactones (PVLs) and phenylvaleric acids (PVAs) with potential bioactivity as shown in Figure 1.14−16 Another aspect to consider in the flavan-3-ol metabolism is the large interindividual variability in the capacity of the gut microbiota to produce these metabolites across the population.16 To better understand this variability, the concept of metabotypes has been introduced to stratify individuals into groups with similar metabolic capacity.
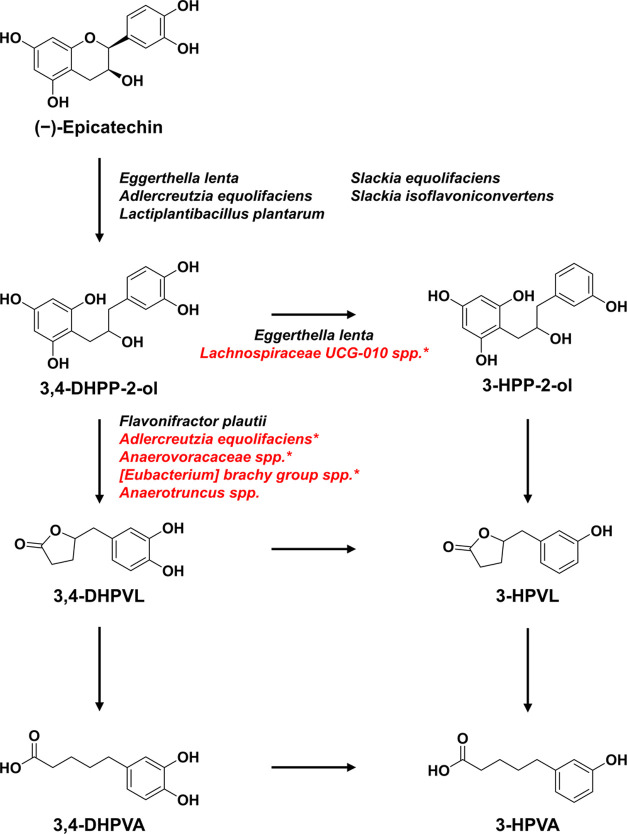
Figure 1.
Proposed metabolic pathways for the metabolism of (−)-epicatechin by gut microbiota. The pathway is based on the microbial metabolites formed during the fermentation and on the literature.16 Bacteria previously involved in (−)-epicatechin metabolism are noted in black,23−27 while bacterial species associated with specific transformation in this study are written in red and annotated with an asterisk.
Metabotypes have been defined by the presence or absence of specific metabolites, since they have previously been described as metabolic phenotypes defined by the production of metabolites resulting from the degradation of specific (poly)phenols by the gut microbiota and by the ecology of the latter in terms of composition and activity.13 For example, Tomás-Barberán et al. proposed three metabotypes associated with the metabolism of ellagic acids and ellagitannins according to the qualitative (absence vs presence) excretion of urolithin A, isourolithin A, and urolithin B in urine and feces.17 However, unlike ellagitannins metabotypes, there is yet no clear and robust definition of flavan-3-ol metabotypes and they cannot be defined using this strict qualitative criterion. For instance, Mena et al. attempted to define flavan-3-ol metabotypes based on a quali–quantitative criterion (low vs high producers) according to the urinary excretion of PVLs and 3-(hydroxyphenyl)propanoic acids following green tea18 and cranberry19 supplementation. In contrast, Cortés-Martín et al. did not identify any metabotypes related to flavan-3-ols gut microbial metabolism, but observed high interindividual variability in the urinary excretion of PVLs and phenylpropanoic acids.20 Therefore, there’s still no definitive agreement regarding the presence of genuine flavan-3-ol metabotypes.
In vitro fecal batch fermentations have been used to investigate the interindividual variability associated with the gut metabolism of (−)-epicatechin21 and (+)-catechin.22 The first study with (−)-epicatechin provided preliminary insights on the bacterial phyla associated with the production of specific metabolites and attempted to cluster the 24 subjects included in the study into metabotypes according to the concentration of metabolites in the fermentation after 2 h.21 So far, bacterial species involved in the metabolism of (−)-epicatechin, such as Eggerthella lenta, Adlercreutzia equolifaciens and Flavonifractor plautii, have been identified using simple fermentations (Figure 1).23−27 Li et al. introduced a new aspect in the characterization of the interindividual variability by evaluating the conversion rate of (+)-catechin by the gut microbiota and reported large differences between donors.22 They also provided preliminary findings on the bacterial species associated with the conversion rate (fast vs slow converters), but these results need to be validated in larger cohorts since only 12 subjects were included in this work.
Hence, the objective of the present study is to characterize the interindividual variability associated with the metabolism of (−)-epicatechin by the gut microbiota. The 34 subjects included in this study were stratified into groups based on their conversion rate of (−)-epicatechin and their quali–quantitative metabolic profile (production of microbial metabolites such as PVLs and PVAs by the gut microbiota) using in vitro fecal batch fermentations. The fecal microbiota of each donor was analyzed through 16S rRNA sequencing, and short-chain fatty acids (SCFA) were quantified in feces to characterize the proposed groups in terms of fecal microbiota composition and function.
2. Material and Methods
2.1. Chemicals
The following products were purchased from Fisher Scientific (Ottawa, Canada): arabinogalactan, xylan, pectin, glucose, proteose peptone, l-cysteine, K2HPO4, KH2PO4, SCFA standards, sterile airtight containers, anaerobic sachets and LC/MS-grade acetonitrile, methanol, and formic acid. (−)-Epicatechin, sodium thioglycolate, porcine mucin, potato starch, Na2HPO4, lysozyme and mutanolysine were acquired from Sigma-Aldrich (Oakville, Canada). Yeast extract was bought from BioBasic (Markham, Canada). 5-(3′-Hydroxyphenyl)valeric acid (3-HPVA), 5-(3′-hydroxyphenyl)-γ-valerolactone (3-HPVL) and 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone (3,4-DHPVL) were obtained from Enamine (Monmouth Jct., NJ), while 5-(3′,4′-dihydroxyphenyl)valeric acid (3,4-DHPVA) was purchased from Toronto Research Chemicals (Toronto, Canada). Deuterated internal standards for the quantification of (−)-epicatechin and its microbial metabolites were acquired from C/D/N Isotopes (Pointe-Claire, Canada). Leucine-enkephaline was purchased from Waters (Milford, MA). Ultrapure water (18.2 MΩ·cm, TOC ≤ 3 ppb) was obtained from a Millipore Milli-Q water purification system (Oakville, Ontario). The kit for DNA extraction (Quick-DNA Fecal/Soil Microbe MiniPrep Kit) was purchased from Zymo Research (Irvine, California).
2.2. Recruitment of Fecal Donors
To inoculate the fecal batch fermentation, 34 healthy donors were recruited from INAF’s list of volunteers and came from the greater Quebec City community area. In total, 14 men and 20 women aged between 23 and 63 years (36 years old on average) with a mean BMI of 23.4 kg/m2 participated in the study. Participants did not use antibiotic and/or probiotic 3 months prior to the study and were asked to strictly avoid consumption of food or beverage containing flavan-3-ols (see Table S1 for the complete list) for 1 week prior to the donation. The day of the donation, fresh fecal materials were collected in sterile airtight containers including an anaerobic sachet to maintain anoxic conditions until processing. Informed consent was obtained from all human subjects and the study was approved by the ethics committee for research involving human beings of Laval University (Québec, Canada) under Registration Number 2019-312.
2.3. Fecal Batch Fermentation
(−)-Epicatechin underwent an upper predigestion using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME, Prodigest, Ghent, Belgium) to mimic the stomach and small intestine passage with a dynamic delivery of secretion (acid, pancreatic/bile juice).28 (−)-Epicatechin was added to the single reactor to obtain a final concentration of 250 μM. Immediately after the predigestion, aliquots of 10 mL were stored at −80 °C until batch fermentation.
The day of the fecal fermentation, predigested (−)-epicatechin aliquots were thawed and 20% (w/V) fecal slurry was prepared as described by Roussel et al.29 Briefly, 100 mL of anaerobic phosphate buffer (8.8 g/L K2HPO4, 6.8 g/L KH2PO4, 1.0 g/L sodium thioglycolate) was added to 20 g of fresh fecal materials before being homogenized using a Stomacher lab blender (Seward, Bohemia, New York). The fecal slurry was then centrifuged at 500g for 2 min to remove larger solid particles. Aliquots of 500 μL were kept at −80 °C until DNA extraction. The rest of the fecal donation was transferred to 5 mL tubes and stored at −80 °C till SCFA analysis.
Fecal batch fermentation was performed in 50 mL penicillin bottles by adding 10 mL of sterile nutritive medium (1.2 g/L arabinogalactan, 0.5 g/L xylan, 2 g/L porcine mucin, 2.0 g/L pectin, 0.5 g/L glucose, 3.0 g/L yeast extract, 1.0 g/L proteose peptone, 4.0 g/L potato starch, 0.5 g/L l-cysteine) adjusted to pH = 6.4 with KH2PO4 (4.76 g/L) and Na2HPO4 (2.66 g/L) to 5 mL of 20% (w/V) fecal slurry and 10 mL of (−)-epicatechin predigested to attain a final fecal concentration of 4% (w/V) and a final (−)-epicatechin concentration of 100 μM. The final concentration of (−)-epicatechin was chosen to be comparable to the study performed by Liu et al.21 To ensure anaerobic conditions, the bottles were sealed and flushed with N2 gas. Incubations were performed at 37 °C and 80 rpm with an orbital shaker for only 24 h to avoid bacterial growth inhibition. Three replicates were performed for each donor.
Aliquots of 500 μL were collected after 0, 1, 2, 3, 4, 5, 6, 7, 8 and 24 h of incubation and immediately mixed with 500 μL of ice-cold methanol to stop enzymatic activity. The resulting solution was centrifuged at 21 500g and 4 °C for 8 min. The supernatant was transferred to a clean 1.5 mL tube and kept at −80 °C till analysis.
2.4. Quantification of (−)-Epicatechin and Its Microbial Metabolites in the Fecal Fermentation by Ultra-Performance Liquid Chromatography Coupled with Quadrupole Time of Flight (UPLC-QToF)
(−)-Epicatechin and its microbial metabolites in the fecal fermentation were quantified by UPLC-QToF using an Acquity I-Class UPLC coupled with a Synapt G2-Si QToF (Waters, Milford, Massachusetts). Before UPLC-QToF analysis, 20 μL of 50% aqueous methanol (v/V) spiked with internal standards (see Table S2 for concentrations) was added to 200 μL of the sample before being filtered using a 0.22 μm water wettable polytetrafluoroethylene filter plate at 1500g and 4 °C for 5 min. For chromatographic separation, 2 μL of the sample, kept at 6 °C in the autosampler, was injected onto an ACQUITY Premier HSS T3 column (2.1 mm × 100 mm, 1.8 μm) (Waters, Milford, MA) protected with an ACQUITY Premier HSS T3 VanGuard FIT precolumn (2.1 mm × 5 mm, 1.8 μm) (Waters, Milford, MA) heated to 40 °C. The column was eluted isocratically at a flow rate of 0.4 mL/min with 98% mobile phase A (0.01% formic acid in water) for 0.4 min, followed by a linear gradient to 45% mobile phase B (0.01% formic acid in acetonitrile) over 6.35 min. Then, the proportion of mobile phase B was rapidly increased to 95% to wash the column for 4.5 min. Finally, the column was reequilibrated with initial conditions for 3.6 min.
MS data were acquired in negative electrospray ionization and sensitivity mode (resolution ≈ 15 000) with the following source parameters: capillary voltage, −0.8 kV; cone voltage, 40 V; source temperature, 150 °C; desolvation temperature, 500 °C; cone gas flow, 50 L/h; desolvation gas flow, 1000 L/h. MSE acquisition was performed with a scan time of 0.2 s over a range (m/z) of 50–1200. In the high energy function, a collision energy ramp from 5 to 20 V was applied. To perform internal mass correction, leucine-enkephaline (200 pg/μL) was infused with a flow rate of 10 μL/min. Data were processed using Skyline 21.1.30
(−)-Epicatechin and its microbial metabolites were quantified using their authentic analytical standards with the calibration range from 0.1 to 100 μM. Since analytical standards for 1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol (3-HPP-2-ol) and 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol (3,4-DHPP-2-ol) are not commercially available, the two metabolites were quantified with the calibration curve obtained from (−)-epicatechin. Standard solutions used for calibration curves were prepared in methanol and were diluted in blank fermentation solution (without feces and (−)-epicatechin) to take into account the matrix effect. As reported by Li et al. with (+)-catechin,22 the concentration of (−)-epicatechin in the fecal fermentation was not constant due to the limited solubility of (−)-epicatechin in the predigestate. Hence, instead of presenting results as concentration, they were expressed as molar mass recovery to take into account this variability. The results obtained from triplicates of each subject were averaged in order to perform statistical analysis at the subject level. Finally, first-order elimination rate constant was calculated as reported by Li et al.22 using the following equation
| 1 |
where C0 was the initial concentration of (−)-epicatechin, Ct represented the remaining concentration of (−)-epicatechin at t, t was the sampling time studied (h), and t0 was the initial time (0 h).
2.5. Quantification of SCFA in Feces by Gas Chromatography Coupled with Flame Ionization Detector (GC-FID)
To extract SCFA, 0.5 g of feces was suspended in 5 mL of ultrapure water and homogenized with a Bead Ruptor (Omni, Kennesaw, Georgia) at 4.0 m/s for 2 min. Then, fecal suspension was centrifuged at 5500g and 4 °C for 30 min and 500 μL of the supernatant was transferred to a clean tube and extracted as thoroughly described elsewhere.31 Acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, isovaleric acid, and hexanoic acid were quantified by GC-FID (Shimadzu, Kyoto, Japan). SCFA analysis was carried out in duplicate. The results were averaged and expressed as μmol per gram of dry feces.
2.6. DNA Extraction and 16S Sequencing
DNA was extracted from pellets obtained after centrifugation of 500 μL of 20% (w/V) fecal slurry using the kit Zymo Research according to the manufacturer’s protocol. Enzymatic lysis of DNA was completed using lysozyme (20 mg) and mutanolysine (10 KU). DNA extracts were eluted in 1× TE buffer (Tris and EDTA) and stored at −20 °C until sequencing. The quality of DNA was analyzed by gel electrophoresis (1.2% w/V agarose) (Life Technologies, Madrid, Spain). Concentrations were measured by Qubit (Thermo Fisher Scientific, Waltham) and the DNA was stored at −20 °C until 16S rRNA library preparation.
The V3–V4 hypervariable region of 16S rRNA was amplified using primer pairs F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and R (5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) (341F–805R). According to the Qiaseq 16S Region panel protocol (Qiagen, Hilden, Germany), the QIAseq 16S/ITS 384-Index I kit (Qiagen) was applied for the amplicon library preparation. The 16S metagenomic libraries were qualified by an Agilent High Sensitivity DNA Kit (Agilent, Palo Alto) using a Bioanalyser to verify the amplicon size and quantified with both a Quant-iT PicoGreen dsDNA Kit (Thermo Fisher Scientific, Waltham), and a Qubit (Thermo Fisher Scientific, Waltham). The final PCR products were pooled, followed by paired-end sequencing using the MiSeq 600 cycles Reagent Kit V3 by an Illumina MiSeq System (Illumina, San Diego).
Demultiplexed raw data files covering all of the samples were imported into R Studio 2022.12.0 environment with R 4.1.3. Amplicon sequence variants (ASV) were inferred using the DADA2 R package (1.20.0), applying the recommended workflow.32 Briefly, sequence reads were first filtered and trimmed with the following parameters: truncQ = 2, truncLen = c(250, 215), maxEE = c(2,2). Filtered reads were denoised using the DADA2 algorithm, which infers the sequencing errors. After removing chimeras, ASVs sequences were subsequently merged and classified using the SILVA database SSU Ref NR 99 release 138 using default parameters.33 Unassigned taxa and singletons were removed. To deal with differences in the sampling depth, the data were rescaled to proportions for further analysis.
Raw 16S rRNA gene amplicon sequencing data were deposited under the BioProject accession number PRJNA955174.
2.7. Statistical Analysis
Statistical analysis was performed with R 4.1.3 using R Studio 2022.12.0. The package FactoMineR (2.7) was used to perform principal component analysis (PCA) and the results were represented graphically using the packages ggplot2 (3.4.0) and ggrepel (0.9.3). Clustering by k-means algorithm was carried out with the package stats (4.1.0), while hierarchical clustering was done with the package cluster (2.1.4) using Euclidean distance and Ward clustering. Kruskal–Wallis rank sum test and Wilcoxon rank sum test with Benjamini & Hochberg correction were performed using the package rstatix (0.7.2). Boxplots and lineplots were obtained with the package ggpubr (0.5.0). Microbiome analyses were performed with phyloseq (1.36.0) for exploring microbiome profiles, vegan (2.6–4) for computing α-diversity indexes, and DESeq2 (1.32.0) for differential analysis of normalized counts data between conditions.
3. Results
3.1. Analysis of (−)-Epicatechin and Its Microbial Metabolites in Fecal Batch Fermentation
In this study, (−)-epicatechin and four of its microbial metabolites (PVLs and PVAs) were identified and quantified with their authentic analytical standard, while 3,4-DHPP-2-ol and 3-HPP-2-ol were identified based on their exact mass and their fragmentation pattern (match using in silico MS2 fragmentation and the literature22) and quantified as epicatechin equivalent (Table 1). All of these molecules were detected in the fecal batch fermentations. The major metabolites formed from the metabolism of (−)-epicatechin by the fecal microbiota, as a proxy for gut microbiota, were 3,4-DHPP-2-ol, 3-HPP-2-ol, and 3,4-DHPVL (Figure S1). 3,4-DHPP-2-ol was formed via C-ring cleavage of (−)-epicatechin at the beginning of the fermentation (Figure 1). Then, 3,4-DHPP-2-ol was dehydroxylated into 3-HPP-2-ol, and 3,4-DHPVL was formed from the degradation of the A-ring of 3,4-DHPP-2-ol later during the fermentation (Figure 1). 3-HPVL and PVAs derivatives were only detected in limited concentrations and only trace amounts of (−)-epicatechin were detected after 24 h of fermentation (Figure S1). Molar mass recoveries over 100% were obtained since 3,4-DHPP-2-ol and 3-HPP-2-ol were not quantified with their authentic standard. Hence, their respective concentrations were probably overestimated using the calibration curve obtained from (−)-epicatechin, resulting in higher molar mass recoveries (Figure S1).
Table 1. Identification of (−)-Epicatechin and Its Microbial Metabolites in Fecal Batch Fermentation by UPLC-QToF.
| class | metabolitea | abbreviation | RT (min) | [M–H]–, m/z detected | mass accuracy (ppm) | MS2 fragments, m/z (relative intensity) | ID levelb |
|---|---|---|---|---|---|---|---|
| epicatechin | (−)-epicatechin | 3,79 | 289,0713 | 0,3 | 245,0818 (100); 203,0714 (54); 125,0242 (42); 109,0291 (38); 151,0394 (27) | 1 | |
| PP-2-ols | 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol | 3,4-DHPP-2-ol | 3,87 | 291,0870 | 0,3 | 123,0437 (100); 247,0966 (75); 167,0341 (69); 135,0438 (40); 205,0854 (32); 151,0390 (27); 189,0548 (17) | 2 |
| 1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol | 3-HPP-2-ol | 4,61 | 275,0923 | 1,5 | 107,0491 (100); 231,1025 (73); 191,0709 (71); 125,0236 (65); 167,0346 (54); 217,0504 (44) | 2 | |
| PVLs | 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone | 3,4-DHPVL | 4,19 | 207,0659 | 1,0 | 163,0761 (100); 122,0368 (28) | 1 |
| 5-(3′-hydroxyphenyl)-γ-valerolactone | 3-HPVL | 5,21 | 191,0706 | –1,0 | 147,0810 (100); 106,0417 (42) | 1 | |
| PVAs | 3-(3′,4′-dihydroxyphenyl)valeric acid | 3,4-DHPVA | 4,99 | 209,0813 | –0,5 | 135,0449 (100); 191,0707 (96); 122,0371 (68); 147,0810 (34); 165,0914 (30) | 1 |
| 5-(3′-hydroxyphenyl)valeric acid | 3-HPVA | 5,99 | 193,0869 | 2,1 | 175,0765 (100); 149,0968 (40); 106,0418 (24) | 1 |
Metabolites were named according to the standardized nomenclature proposed by Kay et al.46
Identification levels were established according to Sumner et al.47 Level 1 identifications were validated by comparing the retention time (RT) and MS2 fragmentation spectra an authentic standard, while level 2 identifications were proposed according to exact mass, MS2 fragmentation spectra, and UV absorption spectra compared to the literature.22
Incubation of (−)-epicatechin in the absence of fecal microbiota was also performed to assess the stability of (−)-epicatechin and ensure that microbial metabolites production observed in fecal batch fermentation was solely due to fecal microbiota. (−)-Epicatechin was stable during the 24 h of incubation (Figure S2) and no microbial metabolite was detected.
3.2. Stratification Based on the Conversion Rate of (−)-Epicatechin
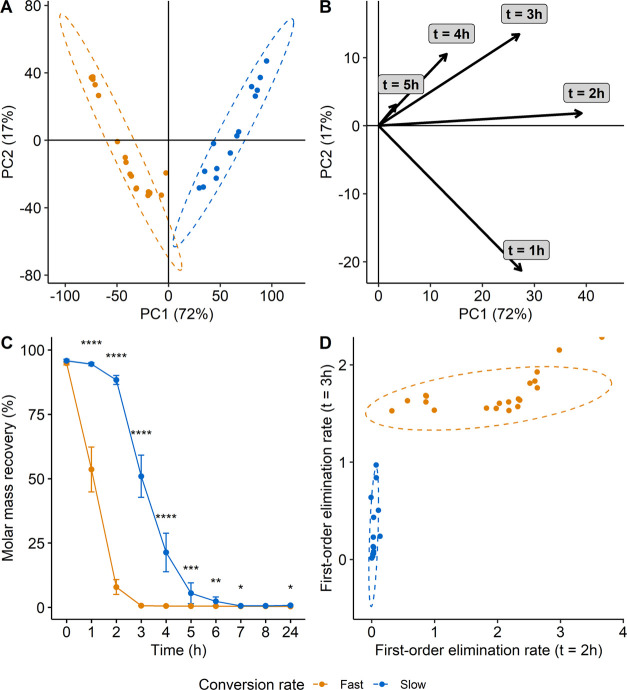
The first step of the study was to stratify the fecal donors into fast and slow converters of (−)-epicatechin. PCA was carried out using the concentration of (−)-epicatechin at each time point (0, 1, 2, 3, 4, 5, 6, 7, 8 and 24 h) as variables. Two distinct groups were obtained either by k-means or by hierarchical clustering and separated by the first principal component (PC) explaining over 72% of the total variation on the score plot (Figure 2A). The partition was mainly guided by the concentration of (−)-epicatechin after 1, 2 and 3 h of fermentation and, to a lesser extent, by the concentration after 4 and 5 h as shown by the projection of the variables in the loading plot (Figure 2B). Fast converters completely metabolized (−)-epicatechin after 3 h of fermentation, while slow converters only started to degrade (−)-epicatechin in the same timeframe and needed more than 7 h to entirely transform the substrate into metabolites (Figures 2C and S3). Using the first-order elimination rate constant after 2 and 3 h of fermentation to estimate the conversion rate, the two groups were clearly separated (Figure 2D). Only a negligible proportion of (−)-epicatechin was metabolized after 2 h of fermentation by the slow converters (first-order conversion rate constant close to 0) in contrast to fast converters that all displayed a first-order elimination rate constant higher than 0, indicating (−)-epicatechin metabolization. After 3 h of fermentation, only a few slow converters started to metabolize (−)-epicatechin, while the conversion was completed by the fast ones. In total, out of the 34 subjects, 14 were slow converters and 20 were fast converters.
Figure 2.
Stratification of the subjects into groups based on the conversion rate of (−)-epicatechin. Score (A) and loading (B) plot of the PCA performed with concentration of (−)-epicatechin in fecal fermentation after 0, 1, 2, 3, 4, 5, 6, 7, 8 and 24 h as variables. Data were not scaled prior to PCA. % of the total variability explained by each PC is reported between parenthesis in axis titles. (C) Concentration of (−)-epicatechin in the fecal fermentation at each sampling point for the two groups. Data are expressed as mean ± standard error of mean. Statistical significance at each sampling point was assessed with Wilcoxon test adjusted for multiple comparisons using the Benjamini and Hochberg method and the results were represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). (D) Conversion rate of (−)-epicatechin after 2 and 3 h of fermentation represented using the first-order elimination rate constant. The point at the top right of the figure is cut into half because this subject completely metabolized (−)-epicatechin after 3 h, resulting in a first-order elimination rate at t = 3 h value equal to infinity.
3.3. Stratification Based on Quali–Quantitative Metabolic Profiles
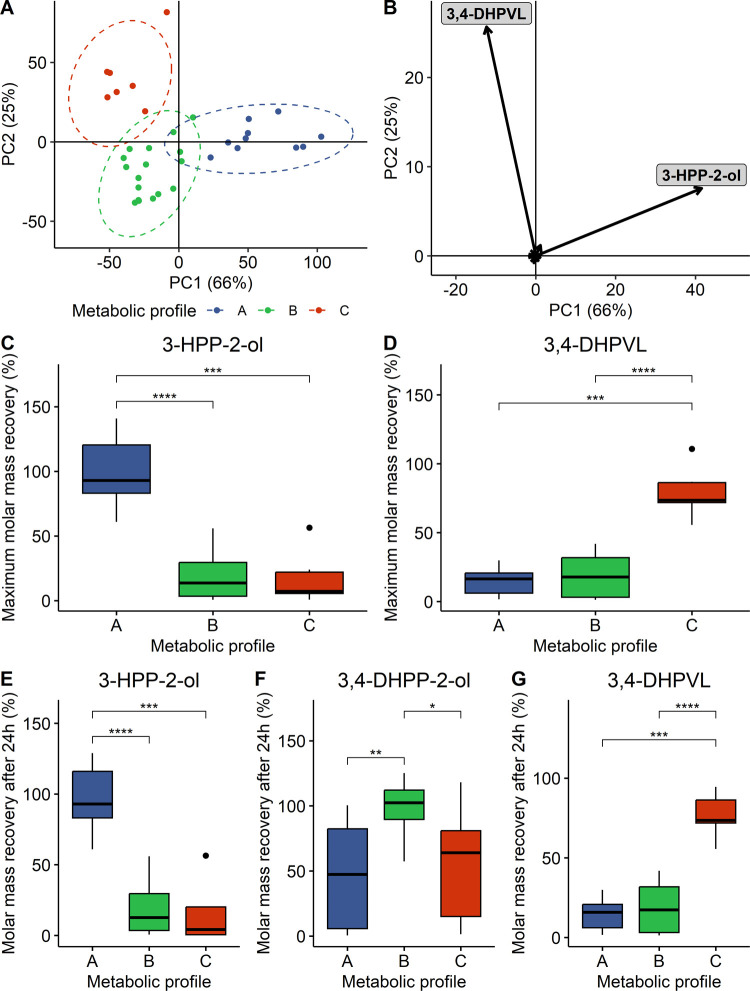
As previously proposed by Mena et al.,18,19 a quali–quantitative criterion was used to stratify the subjects included in the study. The goal was to regroup the fecal donors based on their capacity to produce specific metabolites at different concentrations (low vs high producers). Hence, PCA was performed on the maximal concentration reached during the fecal batch fermentation of each metabolite. Three well-defined groups were obtained either by k-means or by hierarchical clustering (Figure 3A). The first PC, explaining 66% of the total variability, separated metabolic profile A from metabolic profiles B and C, while the second PC, explaining 25% of the total variability, discriminated metabolic profile B from metabolic profile C (Figure 3A). In total, almost all of the variability was explained by the first two PC (91%). Two metabolites were driving the separation of the three metabolic profile, namely, 3-HPP-2-ol and 3,4-DHPVL, which were respectively associated with PC1 and PC2 (Figure 2B). Hence, metabolic profile A was associated with a significant (adjusted p-values ≤ 0.001) higher production of 3-HPP-2-ol (Figure 3C,E), while subjects within metabolic profile C were high producers of 3,4-DHPVL (adjusted p-values ≤ 0.001, Figure 3D,G). Since PCA was carried out using maximal concentration of each metabolite, 3,4-DHPP-2-ol was not discriminant, considering all of the subjects were able to convert (−)-epicatechin to 3,4-DHPP-2-ol via C-ring cleavage (Figure 1). However, metabolic profile B was defined by its limited capacity to transform 3,4-DHPP-2-ol into other metabolites (adjusted p-values ≤ 0.05), while metabolic profiles A and C predominantly converted 3,4-DHPP-2-ol into 3-HPP-2-ol and 3,4-DHPVL, respectively (Figures 3F, S4, and S5). Half of the subjects were included in metabolic profile B, while metabolic profiles A and C were composed of 10 and 7 subjects, respectively.
Figure 3.
Stratification of the subjects into metabolic profiles based on their quali–quantitative differences. Score (A) and loading (B) plot of the PCA carried out with maximal concentration of each metabolite detected during the fecal batch fermentation. Data were not scaled prior to PCA. % of the total variability explained by each PC is reported between parenthesis in axis titles. Maximal molar mass recoveries of 3-HPP-2-ol (C) and 3,4-DHPVL (D) and molar mass recoveries after 24 h of fecal batch fermentation of 3-HPP-2-ol (E), 3,4-DHPP-2-ol (F), and 3,4-DHPVL (G) were represented according to the metabolic profile stratification. Statistical significance was assessed with Kruskal–Wallis test followed by Wilcoxon test adjusted for multiple comparisons using the Benjamini and Hochberg method and the results were represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
3.4. Microbial Characterization of the Groups Based on the Differential Conversion Rate of (−)-Epicatechin
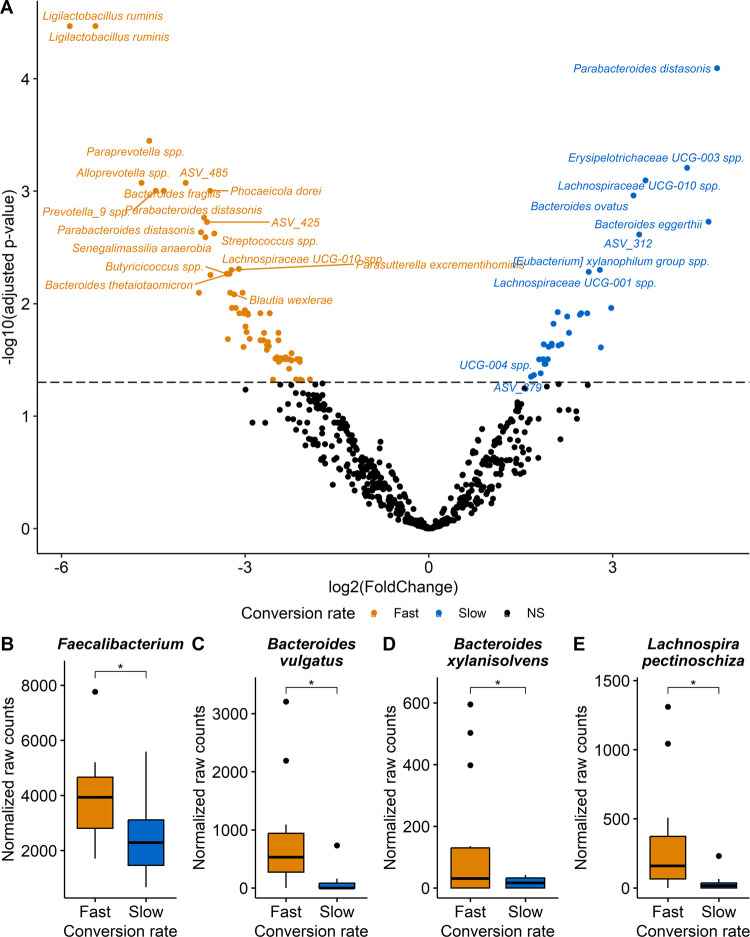
DESeq2 analysis was performed to assess quantitative differences in fecal microbiota composition between the two distinct groups, qualified as fast and slow converters. Several bacterial genera and species were significantly discriminant (adjusted p-value ≤ 0.05) between the fast and slow converters (Figures 4A and S6A, Tables S3 and S4). Among the 17 genera and 100 species emerging as significant (Figures S6B–D and S7), most of them were caused by the very large abundance of these bacteria in only a few individuals. However, we consider as more biologically pertinent to discuss the differences observed in most subjects rather than those driven by only a few individuals, even if statistically significant. Hence, these differences are considered meaningful and are thus the only ones discussed further. Therefore, Faecalibacterium, Bacteroides vulgatus (ASV_24), Bacteroides xylanisolvens (ASV_141) and Lachnospira pectinoschiza (ASV_46) were significantly (adjusted p-values ≤ 0.05) more abundant in fast converters than in slow converters (Figure 4B–E). α diversity (Shannon, Simpson, and Fisher index) and richness (Chao1 index) analysis were conducted, but no significant differences were found between fast and slow converters, with a persisting interindividual variability intragroup (Figure S8).
Figure 4.
Bacteria discriminating fast and slow converters of (−)-epicatechin assessed by DESeq2 analysis. (A) Volcano plot highlighting significant ASV (p-value < 0.05) obtained with Wald test adjusted for multiple comparisons using the Benjamini and Hochberg method. Dotted line was added at adjusted p-value = 0.05 to indicate statistical significance. Boxplots of selected meaningful significant (adjusted p-value < 0.05) genus (B) and species (C–E). Statistical significance, as determined on the volcano plot, was represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
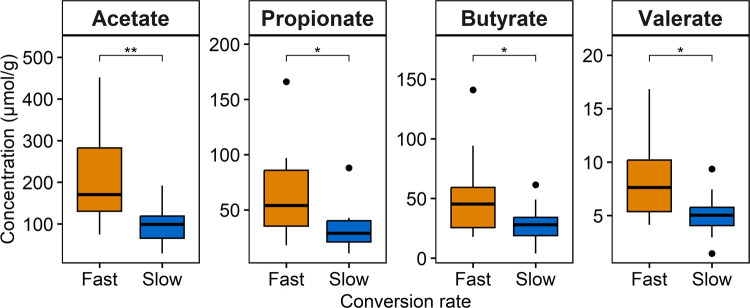
SCFA were quantified in the feces, since these key metabolites generated by the microbiota were previously associated with the conversion rate of (+)-catechin in the in vitro fecal batch fermentations.22,34 Feces of fast converters of (−)-epicatechin contained significantly more SCFA, namely, acetate (adjusted p-value ≤ 0.01), propionate (adjusted p-value ≤ 0.05), butyrate (adjusted p-value ≤ 0.05) and valerate (adjusted p-value ≤ 0.05), than slow converters (Figure 5). No significant difference was found for isobutyrate, isovalerate and hexanoate (data not shown).
Figure 5.
Concentration of SCFA in donor feces according to the groups based on the conversion rate of (−)-epicatechin. SCFA were measured in fresh feces and the results were expressed as μmol per gram of dried feces to normalize the concentration. Statistical significance was assessed with Wilcoxon test adjusted for multiple comparisons using the Benjamini and Hochberg method and the results were represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
3.5. Microbial Characterization of the Metabolic Profiles Based on Quali–Quantitative Differences
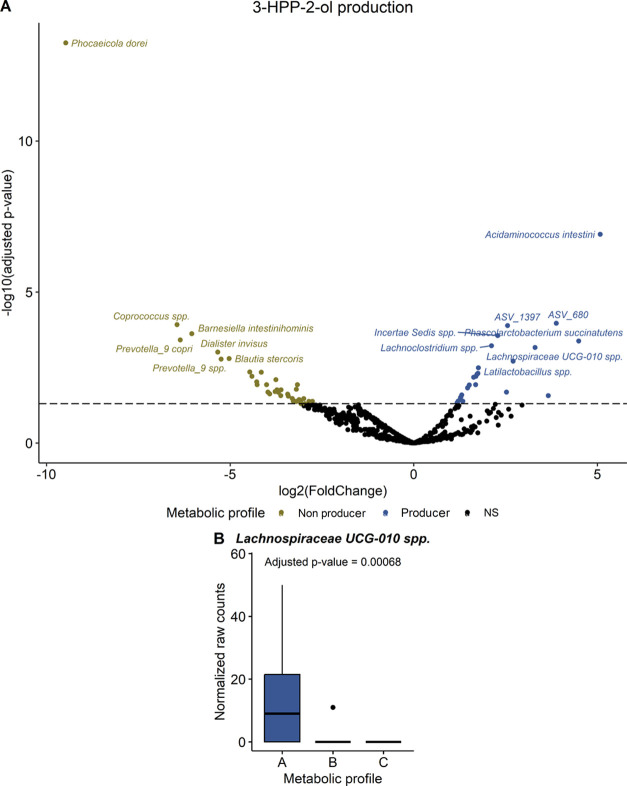
DESeq2 analysis was also applied to investigate the differences in fecal microbiota composition between the metabolic profiles based on the quali–quantitative production of specific metabolites. The statistical approach was slightly different than the one used with groups based on the conversion rate of (−)-epicatechin. Instead of comparing the three metabolic profiles simultaneously, since metabolic profile A was defined by its capacity to dehydroxylate 3,4-DHPP-2-ol into 3-HPP-2-ol and metabolic profile C was associated with the ability to transform 3,4-DHPP-2-ol into 3,4-DHPVL, metabolic profile A was contrasted with pooled metabolic profiles B and C and metabolic profile C was opposed to pooled metabolic profiles A and B. Such comparisons provided more statistical power to identify bacterial genera or species potentially responsible for the dehydroxylation of 3,4-DHPP-2-ol into 3-HPP-2-ol and the production of 3,4-DHPVL from 3,4-DHPP-2-ol, respectively. In total, three genera were significantly associated (adjusted p-value ≤ 0.05) with the production of 3-HPP-2-ol. However, these genera were not considered as meaningfully different because they were driven by extreme values in only a few subjects (Figure S9 and Table S5). In fact, this was the case for all of the following DESeq analyses and only the meaningful differences are discussed. At the species level, 28 ASV were more abundant in metabolic profile A and only Lachnospiraceae UCG-010 spp. (ASV_846) was meaningfully linked to 3-HPP-2-ol production (Figures 6, S10, and Table S6).
Figure 6.
Bacterial species associated with the conversion of 3,4-DHPP-2-ol into 3-HPP-2-ol assessed by DESeq2 analysis. (A) Volcano plot highlighting significant ASV (adjusted p-value < 0.05) obtained with the Wald test adjusted for multiple comparisons using the Benjamini and Hochberg method. Dotted line was added at adjusted p-value = 0.05 to indicate statistical significance. (B) Boxplot of the meaningful significant (adjusted p-value < 0.05) species. Statistical significance, as determined on the volcano plot, was represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
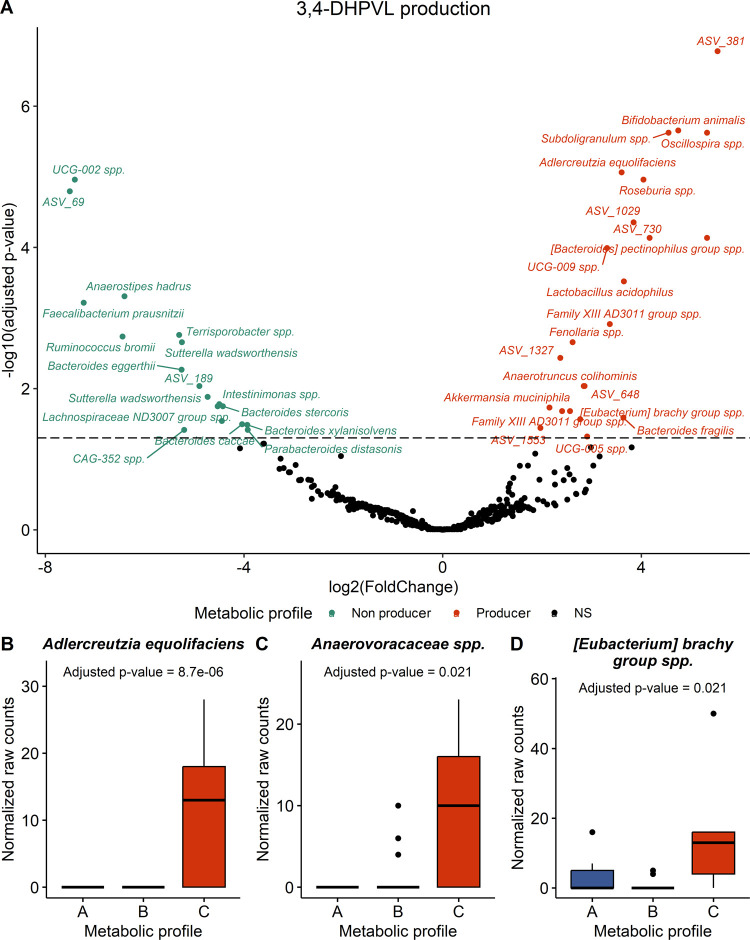
The production of 3,4-DHPVL was significantly associated with three genera, especially Anaerotruncus (Figure S11 and Table S7) and with 23 species (Figures 7A and S12, and Table S8). Among these significant species, Adlercreutiza equolifaciens (ASV_1134), a species from Anaerovoracaceae family (ASV_765), more specifically of family XIII AD2011 group, and [Eubacterium] brachy group spp. (ASV_499) were meaningfully more abundant in subjects from metabolic profile C (Figure 7B–D).
Figure 7.
Bacterial species associated with the conversion of 3,4-DHPP-2-ol into 3,4-DHPVL assessed by DESeq2 analysis. (A) Volcano plot highlighting significant ASV (adjusted p-value < 0.05) obtained with the Wald test adjusted for multiple comparisons using the Benjamini and Hochberg method. Dotted line was added at adjusted p-value = 0.05 to indicate statistical significance. Boxplots of the meaningful significant (adjusted p-value < 0.05) species (B–D). Statistical significance, as determined on the volcano plot, was represented with asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
Finally, in this study, 3-HPP-2-ol and 3,4-DHPVL production from 3,4-DHPP-2-ol were not significantly linked with species previously reported to be able to perform these transformations, namely, E. lenta (ASV_1015) and F. plautii (ASV_349), respectively (Figures 1 and S13). Moreover, there was no significant difference between the three metabolic profiles based on quali–quantitative criteria in terms of α diversity and richness (Figure S14) nor in the SCFA quantified in feces (Figure S15).
4. Discussion
In this study, the interindividual variability associated with the metabolism of (−)-epicatechin by the gut microbiota was investigated by in vitro fecal batch fermentations inoculated with feces from 34 donors. To the best of our knowledge, this is the first time that such a large number of subjects were clustered based on both their conversion rate of (−)-epicatechin and their levels of production of specific metabolites (low vs high producers) using short-term in vitro fecal fermentations. During the fermentations, (−)-epicatechin was converted to 3,4-DHPP-2-ol via C-ring cleavage. Then, 3,4-DHPP-2-ol was mainly metabolized into 3-HPP-2-ol by dehydroxylation and into 3,4-DHPVL by A-ring degradation. Only limited amounts of 3-HPVL, 3,4-DHPVA, and 3-HPVA were detected after 24 h of fermentation. These results are coherent with those reported by Li et al., assessing the conversion of (+)-catechin by 12 donors in fecal batch fermentation.22 However, they are contrasting with the report of Liu et al.,21 where the main metabolites observed after 6 h of fecal fermentation were 3,4-DHPVL and 3-HPVL and almost only 3-HPVA was detected after 24 h. These discrepancies could be explained by the different culture medium used for the fecal batch fermentation. In fact, we used a very similar culture medium to Li et al., containing a mixture of nutriments essential for bacterial growth and survival, while Liu et al. performed fermentations solely in phosphate buffer saline. Hence, bacteria went into metabolic starvation and rapidly consumed their sole available carbon source, (−)-epicatechin.28 This could explain why 3,4-DHPVL and 3-HPVL were metabolized into 3-HPVA between 6 and 24 h of fermentation. However, this result is not representative of the colonic fermentation of flavan-3-ols in humans.
In previous clinical trials assessing the supplementation of flavan-3-ols from green tea,16 cranberry19 and apple,35 PVLs and PVAs were the main metabolites excreted in urine, while excretion of 3,4-DHPP-2-ol and 3-HPP-2-ol was not reported. Since the average residence time of (poly)phenols in the colon is 35 h,36 it is probable that 3,4-DHPP-2-ol and 3-HPP-2-ol could be further metabolized into PVLs and, then, into PVAs during this time period. In fact, PVAs were mainly excreted after 48 h of fecal fermentation with (+)-catechin.22 In addition, excretion of 3-HPP-2-ol and 3,4-DHPP-2-ol in urine has never, to our knowledge, been reported and are probably poorly absorbed in the colon and are therefore excreted in feces. Hence, our results obtained from fecal batch fermentations are coherent with these previous clinical trials performed in human subjects.
In our study, a strong interindividual variability in the conversion rate of (−)-epicatechin was observed between the 34 donors, as reported in previous studies.21,22,37 To cope with such variability, the subjects were stratified into two groups, namely, fast and slow converters. Close repeated sample collections enabled us to conclude that the separation between fast and slow converters was mainly driven by the concentration of (−)-epicatechin between 1 and 3 h of fermentation. This result was not observed by Li et al., since they measured (+)-catechin concentration only after 4 h of fermentation.22 Interestingly, the fast converters were associated with higher abundance of Faecalibacterium, B. vulgatus (ASV_24), B. xylanisolvens (ASV_141) and L. pectinoschiza (ASV_46), which are, so far, not known to be involved in the metabolism of (−)-epicatechin, but rather in the transformation of dietary fibers into SCFA.38,39 In fact, fast converters excreted significantly more SCFA in their feces, particularly, acetate, propionate, butyrate, as well as some minor SCFA, namely, valerate, compared to slow converters. Although no significant differences were found in fecal fermentation of (+)-catechin for SCFA production, the same genera were more abundant in fast converters than slow converters in a previous study.22 Hence, we surmise that individuals frequently consuming flavan-3-ols-rich foods, such as green tea, cocoa products, fruits and nuts,1−3 are fast converters of (−)-epicatechin due to habituation of the gut microbiota to metabolize this compound. In fact, higher levels of SCFA-producing bacteria, such as Faecalibacterium, in fast converters could be associated with the consumption of flavan-3-ols-rich foods, since it has been demonstrated that epigallocatechin gallate (abundant in green tea) and red wine can stimulate the growth of these bacteria.40−42
A quali–quantitative criterion was also used in our study to stratify the 34 donors into metabolic profiles based on their capacity to produce specific metabolites at different concentrations (low vs high producers). Remarkably, three distinct metabolic profiles were obtained depending on the ability of the fecal microbiota to convert 3,4-DHPP-2-ol into higher concentrations of 3-HPP-2-ol (metabolic profile A) and 3,4-DHPVL (metabolic profile C), while metabolic profile B was defined by its limited metabolization of 3,4-DHPP-2-ol. In human clinical trials, three metabotypes were proposed using this criterion following green tea18 and cranberry19 supplementations. In the green tea study, the first metabotype was associated with high excretion of 5-(3′,4′,5′-trihydroxyphenyl)-γ-valerolactone and 3,4-DHPVL, the second metabotype was linked to high production of 3,4-DHPVL, and the third metabotype was defined by high excretion of 3-(hydroxyphenyl)propanoic acids.18 In the cranberry study, which does not contain trihydroxylated flavan-3-ols, the subjects were classified as high 5-(hydroxyphenyl)-γ-valerolactones (both 3′ and 4′ derivatives) and 3-(hydroxyphenyl)propanoic acid excreters, high 3,4-DHPVL producers, or low excreters of PVLs and 3-(hydroxyphenyl)propanoic acids.19 Since green tea extracts have high amounts of trihydroxylated flavan-3-ols such as epigallocatechin, it is hard to compare these results with ours. However, the metabolic profiles obtained in our study are coherent with those reported after cranberry consumption.19 In fact, with slightly longer fermentation time, it is probable that donors from metabolic profile A would convert 3-HPP-2-ol into 3-HPVL and, eventually, into 3-(3′-hydroxyphenyl)propanoic acid. Also, metabolic profile C would be high excreters of 3,4-DHPVL and subjects from metabolic profile B would be low excreters of PVLs. Moreover, we confirmed that flavan-3-ol metabotypes cannot be defined using a qualitative criterion (absence vs presence of specific metabolites), since all subjects were able to produce each metabolite, but at different concentrations.
Altogether, the stratification into metabolic profiles in this study using a quali–quantitative criterion allowed to determine which bacteria are potentially responsible for the transformation of 3,4-DHPP-2-ol into 3-HPP-2-ol and 3,4-DHPVL. Previous studies reported that E. lenta was able to dehydroxylate 3,4-DHPP-2-ol into 3-HPP-2-ol and that F. plautii could convert 3,4-DHPP-2-ol into 3,4-DHPVL,23−25 but these species were not associated with these transformations in our study. Interestingly, we observed that 3,4-DHPP-2-ol conversion to 3-HPP-2-ol was linked to Lachnospiraceae UCG-010 spp (ASV_846), while A. equolifaciens (ASV_1134), a specie from Anaerovoracaceae family (ASV_765), [Eubacterium] brachy group spp. (ASV_499), and Anaerotruncus spp. were associated with the production of 3,4-DHPVL. Among these four bacteria, only A. equolifaciens has been reported to convert (−)-epicatechin into 3,4-DHPP-2-ol via C-ring cleavage.23,26 Our result thus confirmed the crucial role of this bacterium on (−)-epicatechin metabolism in the colon. Finally, since all of the subjects were able to convert (−)-epicatechin into 3,4-DHPP-2-ol, it was not possible to associate bacteria to this C-ring cleavage transformation in our study.
Recently, Iglesias-Aguirre et al. reported two co-cultures able to convert ellagic acids into urolithins reproducing urolithin metabotypes A and B using only two gut bacteria per co-culture and the administration of these bacterial consortia successfully replicated urolithin metabotypes in rat.43,44 Hence, new probiotic combinations could be formulated to engineer the gut microbiota of individuals unable to produce bioactive urolithins (urolithins metabotype 0) into urolithins metabotype A or B. Similar studies should be conducted to identify a bacterial consortium able to convert (−)-epicatechin, or more generally flavan-3-ols, into 3,4-DHPVL. This co-culture could enable subjects from metabolic profile B to further metabolize 3,4-DHPP-2-ol into bioavailable and bioactive metabolites and potentially benefit more from the positive health effects of flavan-3-ols. A. equolifaciens is an ideal candidate for this action since it is potentially able to cleave (−)-epicatechin C-ring and further degrade 3,4-DHPP-2-ol into 3,4-DHPVL. Moreover, since the conversion rate of (−)-epicatechin is associated with SCFA-producing bacteria, dietary fibers should be added to these probiotic formulations to promote fast (−)-epicatechin metabolism.
In conclusion, the 34 subjects included in this study were stratified according to their conversion rate of (−)-epicatechin and their quali–quantitative metabolic profiles, separately. These two criteria are not mutually exclusive and are intermingled in vivo, since both can influence the concentration of the different metabolites formed and then absorbed in the colon, reaching the systemic circulation where they can exhibit beneficial effects. In fact, we hypothesize that fast converters of (−)-epicatechin have more chances of producing metabolites further down in the pathway, such as PVLs and PVAs, since they can achieve more transformation than slow converters in the same time span. Despite our relatively small sample size, we were able to observe that a greater proportion of slow converters belonged to metabolic profile B (64.3%) in contrast to fast converters (40%) (Figure S16). Moreover, 60% of the fast converters were able to transform high amounts of 3,4-DHPP-2-ol into 3-HPP-2-ol and 3,4-DHPVL, while only 35.7% of slow converters belonged to metabolic profiles A and C. Larger cohorts will be needed to confirm the link between the conversion rate of (−)-epicatechin and the quali–quantitative metabolic profile. Also, metabolic profiles obtained from in vitro fecal batch fermentation should be confirmed in clinical trials following flavan-3-ols supplementation to robustly define flavan-3-ol metabotypes. In vitro experiments offer valuable perspectives on the individual differences linked to the metabolism of flavan-3-ols. However, these models have certain drawbacks, including the lack of metabolite absorption and the reliance on fecal microbiota as opposed to gut microbiota. The link between flavan-3-ol metabotypes and their health effects should be appraised. In addition to quantifying PVLs and PVAs in urine and plasma, these studies should assess the bioavailability of 3,4-DHPP-2-ol and 3-HPP-2-ol. Also, to further characterize flavan-3-ol metabotypes, metatranscriptomics analysis should be performed to identify which gene expression related to gut microbiota discriminates the different metabotypes. Finally, the classification of the population into metabotypes is a necessary step to fully understand the mechanisms of action of flavan-3-ols and their health benefits. This stratification is of utmost importance in personalized nutrition45 and could ameliorate the outcome of clinical trials investigating the health effects of flavan-3-ols by characterizing the important interindividual variability associated with their metabolism by the gut microbiota.
Acknowledgments
This research was funded by the NSERC–Diana Food Industrial Chair on prebiotic effects of fruits and vegetables. Financial support to Jacob Lessard-Lord was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC). The authors thank Pamela Généreux and Valentina Cattero for their help with the fecal batch fermentation, Ana-Sofia Medina-Larqué for her support with the recruitment of fecal donors, and Joseph Lupien-Meilleur for helpful discussion on the manuscript. The authors also acknowledge INAF platforms for providing access to the analytical instruments used in this work, especially Roxanne Nolet and Perrine Feutry for their help with SCFA analysis.
Glossary
Abbreviations
- PVLs
phenyl-γ-valerolactones
- PVAs
phenylvaleric acids
- SCFA
short-chain fatty acids
- 3-HPVA
5-(3′-hydroxyphenyl)valeric acid
- 3,4-DHPVA
5-(3′,4′-dihydroxyphenyl)valeric acid
- 3-HPVL
5-(3′-hydroxyphenyl)-γ-valerolactone
- 3,4-DHPVL
5-(3′,4′-dihydroxyphenyl)-γ-valerolactone
- 3-HPP-2-ol
1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol
- 3,4-DHPP-2-ol
1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol
- UPLC-QtoF
ultra-performance liquid chromatography coupled with quadrupole time of flight
- GC-FID
gas chromatography coupled with flame ionization detector
- ASV
amplicon sequence variant
- PCA
principal component analysis
- PC
principal component
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.3c05491.
Food and beverage restriction 1 week prior to the donation (Table S1); concentration of the internal standards (Table S2); statistics obtained from DESeq2 analysis (Tables S3–S8); microbial metabolites formed during fecal batch fermentations (Figure S1); stability of (−)-epicatechin in incubation without fecal microbiota (Figure S2); interindividual variability of the conversion rate of (−)-epicatechin among the 34 donors included in the study (Figure S3); kinetic profile of microbial metabolites produced during fecal batch fermentations depending on the metabolic profile (Figure S4); interindividual variability of the production of microbial metabolites among the 34 donors included in the study (Figure S5); bacterial genera discriminating fast and slow converters of (−)-epicatechin assessed by DESeq2 analysis (Figure S6); significant bacterial species discriminating fast and slow converters influenced by extreme values (Figure S7); α diversity according to the groups based on the conversion rate of (−)-epicatechin (Figure S8); bacterial genera associated with the conversion of 3,4-DHPP-2-ol into 3-HPP-2-ol assessed by DESeq2 analysis (Figure S9); significant bacterial species associated with the conversion of 3,4-DHPP-2-ol into 3-HPP-2-ol influenced by extreme values (Figure S10); bacterial genera associated with the conversion of 3,4-DHPP-2-ol into 3,4-DHPVL assessed by DESeq2 analysis (Figure S11); significant bacterial species associated with the conversion of 3,4-DHPP-2-ol into 3,4-DHPVL influenced by extreme values (Figure S12); abundance of bacteria known to be involved in 3,4-DHPP-2-ol metabolism (Figure S13); α diversity according to the metabolic profiles based on quali–quantitative differences (Figure S14); concentration of SCFA in donor feces according to the metabolic profiles based on quali–quantitative differences (Figure S15); and proportion of each metabolic profile based on quali–quantitative differences within fast and slow converters (Figure S16) (PDF)
The authors declare no competing financial interest.
Notes
Yves Desjardins holds an NSERC–Diana Food Industrial Chair on prebiotic effects of fruit and vegetable polyphenols.
Supplementary Material
References
- Ziauddeen N.; Rosi A.; Del Rio D.; Amoutzopoulos B.; Nicholson S.; Page P.; Scazzina F.; Brighenti F.; Ray S.; Mena P. Dietary intake of (poly)phenols in children and adults: cross-sectional analysis of UK National Diet and Nutrition Survey Rolling Programme (2008–2014). Eur. J. Nutr. 2019, 58, 3183–3198. 10.1007/s00394-018-1862-3. [DOI] [PubMed] [Google Scholar]
- Castro-Barquero S.; Tresserra-Rimbau A.; Vitelli-Storelli F.; Doménech M.; Salas-Salvadó J.; Martín-Sánchez V.; Rubín-García M.; Buil-Cosiales P.; Corella D.; Fitó M.; Romaguera D.; Vioque J.; Alonso-Gómez Á. M.; Wärnberg J.; Martínez J. A.; Serra-Majem L.; Tinahones F. J.; Lapetra J.; Pintó X.; Tur J. A.; Garcia-Rios A.; García-Molina L.; Delgado-Rodriguez M.; Matía-Martín P.; Daimiel L.; Vidal J.; Vázquez C.; Cofán M.; Romanos-Nanclares A.; Becerra-Tomas N.; Barragan R.; Castañer O.; Konieczna J.; González-Palacios S.; Sorto-Sánchez C.; Pérez-López J.; Zulet M. A.; Bautista-Castaño I.; Casas R.; Gómez-Perez A. M.; Santos-Lozano J. M.; Rodríguez-Sanchez M. Á.; Julibert A.; Martín-Calvo N.; Hernández-Alonso P.; Sorlí J. V.; Sanllorente A.; Galmés-Panadés A. M.; Cases-Pérez E.; Goicolea-Güemez L.; Ruiz-Canela M.; Babio N.; Hernáez Á.; Lamuela-Raventós R. M.; Estruch R. Dietary Polyphenol Intake is Associated with HDL-Cholesterol and A Better Profile of other Components of the Metabolic Syndrome: A PREDIMED-Plus Sub-Study. Nutrients 2020, 12, 689. 10.3390/nu12030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Ros R.; Sacerdote C.; Ricceri F.; Weiderpass E.; Roswall N.; Buckland G.; St-Jules D. E.; Overvad K.; Kyrø C.; Fagherazzi G.; Kvaskoff M.; Severi G.; Chang-Claude J.; Kaaks R.; Nöthlings U.; Trichopoulou A.; Naska A.; Trichopoulos D.; Palli D.; Grioni S.; Mattiello A.; Tumino R.; Gram I. T.; Engeset D.; Huerta J. M.; Molina-Montes E.; Argüelles M.; Amiano P.; Ardanaz E.; Ericson U.; Lindkvist B.; Nilsson L. M.; Kiemeney L. A.; Ros M.; Bueno-de-Mesquita H. B.; Peeters P. H. M.; Khaw K. T.; Wareham N. J.; Knaze V.; Romieu I.; Scalbert A.; Brennan P.; Wark P.; Vineis P.; Riboli E.; González C. A. Flavonoid and lignan intake in relation to bladder cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Br. J. Cancer 2014, 111, 1870–1880. 10.1038/bjc.2014.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter H.; Heiss C.; Spencer J. P.; Keen C. L.; Lupton J. R.; Schmitz H. H. Recommending flavanols and procyanidins for cardiovascular health: Current knowledge and future needs. Mol. Aspects Med. 2010, 31, 546–557. 10.1016/j.mam.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Curtis P. J.; Sampson M.; Potter J.; Dhatariya K.; Kroon P. A.; Cassidy A. Chronic Ingestion of Flavan-3-ols and Isoflavones Improves Insulin Sensitivity and Lipoprotein Status and Attenuates Estimated 10-Year CVD Risk in Medicated Postmenopausal Women With Type 2 Diabetes: A 1-year, double-blind, randomized, controlled trial. Diabetes Care 2012, 35, 226–232. 10.2337/dc11-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio D.; Rodriguez-Mateos A.; Spencer J. P. E.; Tognolini M.; Borges G.; Crozier A. Dietary (Poly)phenolics in Human Health: Structures, Bioavailability, and Evidence of Protective Effects Against Chronic Diseases. Antioxid. Redox Signaling 2013, 18, 1818–1892. 10.1089/ars.2012.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mateos A.; Vauzour D.; Krueger C. G.; Shanmuganayagam D.; Reed J.; Calani L.; Mena P.; Del Rio D.; Crozier A. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: an update. Arch. Toxicol. 2014, 88, 1803–1853. 10.1007/s00204-014-1330-7. [DOI] [PubMed] [Google Scholar]
- Sloan R. P.; Wall M.; Yeung L.-K.; Feng T.; Feng X.; Provenzano F.; Schroeter H.; Lauriola V.; Brickman A. M.; Small S. A. Insights into the role of diet and dietary flavanols in cognitive aging: results of a randomized controlled trial. Sci. Rep. 2021, 11, 3837 10.1038/s41598-021-83370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani J. I.; Heiss C.; Spencer J. P. E.; Kelm M.; Schroeter H. Recommending flavanols and procyanidins for cardiovascular health: Revisited. Mol. Aspects Med. 2018, 61, 63–75. 10.1016/j.mam.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Crowe-White K. M.; Evans L. W.; Kuhnle G. G. C.; Milenkovic D.; Stote K.; Wallace T.; Handu D.; Senkus K. E. Flavan-3-ols and Cardiometabolic Health: First Ever Dietary Bioactive Guideline. Adv. Nutr. 2022, 13, 2070–2083. 10.1093/advances/nmac105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Van de Wiele T. Gut microbiota as a driver of the interindividual variability of cardiometabolic effects from tea polyphenols. Crit. Rev. Food Sci. Nutr. 2023, 63, 1500–1526. 10.1080/10408398.2021.1965536. [DOI] [PubMed] [Google Scholar]
- Narduzzi L.; Agulló V.; Favari C.; Tosi N.; Mignogna C.; Crozier A.; Del Rio D.; Mena P. (Poly)phenolic compounds and gut microbiome: new opportunities for personalized nutrition. Microbiome Res. Rep. 2022, 1, 16. 10.20517/mrr.2022.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín J. C.; González-Sarrías A.; Tomás-Barberán F. A. The gut microbiota: A key factor in the therapeutic effects of (poly)phenols. Biochem. Pharmacol. 2017, 139, 82–93. 10.1016/j.bcp.2017.04.033. [DOI] [PubMed] [Google Scholar]
- Ottaviani J. I.; Momma T. Y.; Heiss C.; Kwik-Uribe C.; Schroeter H.; Keen C. L. The stereochemical configuration of flavanols influences the level and metabolism of flavanols in humans and their biological activity in vivo. Free Radical Biol. Med. 2011, 50, 237–244. 10.1016/j.freeradbiomed.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Monagas M.; Urpi-Sarda M.; Sánchez-Patán F.; Llorach R.; Garrido I.; Gómez-Cordovés C.; Andres-Lacueva C.; Bartolomé B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- Mena P.; Bresciani L.; Brindani N.; Ludwig I. A.; Pereira-Caro G.; Angelino D.; Llorach R.; Calani L.; Brighenti F.; Clifford M. N.; Gill C. I. R.; Crozier A.; Curti C.; Del Rio D. Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019, 36, 714–752. 10.1039/C8NP00062J. [DOI] [PubMed] [Google Scholar]
- Tomás-Barberán F. A.; García-Villalba R.; González-Sarrías A.; Selma M. V.; Espín J. C. Ellagic Acid Metabolism by Human Gut Microbiota: Consistent Observation of Three Urolithin Phenotypes in Intervention Trials, Independent of Food Source, Age, and Health Status. J. Agric. Food Chem. 2014, 62, 6535–6538. 10.1021/jf5024615. [DOI] [PubMed] [Google Scholar]
- Mena P.; Ludwig I. A.; Tomatis V. B.; Acharjee A.; Calani L.; Rosi A.; Brighenti F.; Ray S.; Griffin J. L.; Bluck L. J.; Del Rio D. Inter-individual variability in the production of flavan-3-ol colonic metabolites: preliminary elucidation of urinary metabotypes. Eur. J. Nutr. 2019, 58, 1529–1543. 10.1007/s00394-018-1683-4. [DOI] [PubMed] [Google Scholar]
- Mena P.; Favari C.; Acharjee A.; Chernbumroong S.; Bresciani L.; Curti C.; Brighenti F.; Heiss C.; Rodriguez-Mateos A.; Del Rio D. Metabotypes of flavan-3-ol colonic metabolites after cranberry intake: elucidation and statistical approaches. Eur. J. Nutr. 2022, 61, 1299–1317. 10.1007/s00394-021-02692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés-Martín A.; Selma M. V.; Espín J. C.; García-Villalba R. The Human Metabolism of Nuts Proanthocyanidins does not Reveal Urinary Metabolites Consistent with Distinctive Gut Microbiota Metabotypes. Mol. Nutr. Food Res. 2019, 63, 1800819 10.1002/mnfr.201800819. [DOI] [PubMed] [Google Scholar]
- Liu C.; Vervoort J.; Beekmann K.; Baccaro M.; Kamelia L.; Wesseling S.; Rietjens I. M. C. M. Interindividual Differences in Human Intestinal Microbial Conversion of (−)-Epicatechin to Bioactive Phenolic Compounds. J. Agric. Food Chem. 2020, 68, 14168–14181. 10.1021/acs.jafc.0c05890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Van Herreweghen F.; Onyango S. O.; De Mey M.; Van de Wiele T. In Vitro Microbial Metabolism of (+)-Catechin Reveals Fast and Slow Converters with Individual-Specific Microbial and Metabolite Markers. J. Agric. Food Chem. 2022, 70, 10405–10416. 10.1021/acs.jafc.2c00551. [DOI] [PubMed] [Google Scholar]
- Takagaki A.; Nanjo F. Bioconversion of (−)-Epicatechin, (+)-Epicatechin, (−)-Catechin, and (+)-Catechin by (−)-Epigallocatechin-Metabolizing Bacteria. Biol. Pharm. Bull. 2015, 38, 789–794. 10.1248/bpb.b14-00813. [DOI] [PubMed] [Google Scholar]
- Jin J.-S.; Hattori M. Isolation and Characterization of a Human Intestinal Bacterium Eggerthella sp. CAT-1 Capable of Cleaving the C-Ring of (+)-Catechin and (−)-Epicatechin, Followed by p-Dehydroxylation of the B-Ring. Biol. Pharm. Bull. 2012, 35, 2252–2256. 10.1248/bpb.b12-00726. [DOI] [PubMed] [Google Scholar]
- Kutschera M.; Engst W.; Blaut M.; Braune A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011, 111, 165–175. 10.1111/j.1365-2672.2011.05025.x. [DOI] [PubMed] [Google Scholar]
- Takagaki A.; Nanjo F. Biotransformation of (−)-epicatechin, (+)-epicatechin, (−)-catechin, and (+)-catechin by intestinal bacteria involved in isoflavone metabolism. Biosci., Biotechnol., Biochem. 2016, 80, 199–202. 10.1080/09168451.2015.1079480. [DOI] [PubMed] [Google Scholar]
- Sánchez-Patán F.; Tabasco R.; Monagas M.; Requena T.; Peláez C.; Moreno-Arribas M. V.; Bartolomé B. Capability of Lactobacillus plantarum IFPL935 To Catabolize Flavan-3-ol Compounds and Complex Phenolic Extracts. J. Agric. Food Chem. 2012, 60, 7142–7151. 10.1021/jf3006867. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T.; Van den Abbeele P.; Ossieur W.; Possemiers S.; Marzorati M.. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx K.; Cotter P.; López-Expósito I.; Kleiveland C.; Lea T.; Mackie A.; Requena T.; Swiatecka D.; Wichers H., Eds.; Springer International Publishing: Cham, 2015; pp 305–317. [PubMed] [Google Scholar]
- Roussel C.; De Paepe K.; Galia W.; De Bodt J.; Chalancon S.; Leriche F.; Ballet N.; Denis S.; Alric M.; Van de Wiele T.; Blanquet-Diot S. Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 2020, 18, 141. 10.1186/s12915-020-00860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. J.; Pratt B.; Bose N.; Dubois L. G.; St John-Williams L.; Perrott K. M.; Ky K.; Kapahi P.; Sharma V.; MacCoss M. J.; Moseley M. A.; Colton C. A.; MacLean B. X.; Schilling B.; Thompson J. W. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel C.; Chabaud S.; Lessard-Lord J.; Cattero V.; Pellerin F.-A.; Feutry P.; Bochard V.; Bolduc S.; Desjardins Y. UPEC Colonic-Virulence and Urovirulence Are Blunted by Proanthocyanidins-Rich Cranberry Extract Microbial Metabolites in a Gut Model and a 3D Tissue-Engineered Urothelium. Microbiol. Spectrum 2022, 10, e02432–21. 10.1128/spectrum.02432-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B. J.; McMurdie P. J.; Rosen M. J.; Han A. W.; Johnson A. J. A.; Holmes S. P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren M. R.; Callahan B. J.. Silva 138.1 prokaryotic SSU taxonomic training data formatted for DADA2 Zenodo 2021.
- Staley C.; Weingarden A. R.; Khoruts A.; Sadowsky M. J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesi A.; Mena P.; Bub A.; Ulaszewska M.; Del Rio D.; Kulling S. E.; Mattivi F. Quantification of Urinary Phenyl-γ-Valerolactones and Related Valeric Acids in Human Urine on Consumption of Apples. Metabolites 2019, 9, 254. 10.3390/metabo9110254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf A. M.; Phillips S. F.; Zinsmeister A. R.; MacCarty R. L.; Beart R. W.; Wolff B. G. Simplified assessment of segmental colonic transit. Gastroenterology 1987, 92, 40–47. 10.1016/0016-5085(87)90837-7. [DOI] [PubMed] [Google Scholar]
- van Velzen E. J. J.; Westerhuis J. A.; van Duynhoven J. P. M.; van Dorsten F. A.; Grün C. H.; Jacobs D. M.; Duchateau G. S. M. J. E.; Vis D. J.; Smilde A. K. Phenotyping Tea Consumers by Nutrikinetic Analysis of Polyphenolic End-Metabolites. J. Proteome Res. 2009, 8, 3317–3330. 10.1021/pr801071p. [DOI] [PubMed] [Google Scholar]
- Nogal A.; Valdes A. M.; Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212 10.1080/19490976.2021.1897212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M.; Chen Y.; Ma Z.; Zhang X.; Shi D.; Khan J. A.; Liu H. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim. Nutr. 2022, 8, 350–360. 10.1016/j.aninu.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; de Bruijn W. J. C.; Bruins M. E.; Vincken J.-P. Microbial Metabolism of Theaflavin-3,3′-digallate and Its Gut Microbiota Composition Modulatory Effects. J. Agric. Food Chem. 2021, 69, 232–245. 10.1021/acs.jafc.0c06622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Indias I.; Sánchez-Alcoholado L.; Pérez-Martínez P.; Andrés-Lacueva C.; Cardona F.; Tinahones F.; Queipo-Ortuño M. I. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. 2016, 7, 1775–1787. 10.1039/C5FO00886G. [DOI] [PubMed] [Google Scholar]
- Queipo-Ortuño M. I.; Boto-Ordóñez M.; Murri M.; Gomez-Zumaquero J. M.; Clemente-Postigo M.; Estruch R.; Cardona Diaz F.; Andrés-Lacueva C.; Tinahones F. J. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012, 95, 1323–1334. 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- Iglesias-Aguirre C. E.; García-Villalba R.; Beltrán D.; Frutos-Lisón M. D.; Espín J. C.; Tomás-Barberán F. A.; Selma M. V. Gut Bacteria Involved in Ellagic Acid Metabolism To Yield Human Urolithin Metabotypes Revealed. J. Agric. Food Chem. 2023, 71, 4029–4035. 10.1021/acs.jafc.2c08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Aguirre C. E.; González-Sarrías A.; Cortés-Martín A.; Romo-Vaquero M.; Osuna-Galisteo L.; Cerón J. J.; Espín J. C.; Selma M. V. In vivo administration of gut bacterial consortia replicates urolithin metabotypes A and B in a non-urolithin-producing rat model. Food Funct. 2023, 14, 2657–2667. 10.1039/D2FO03957E. [DOI] [PubMed] [Google Scholar]
- Cortés-Martín A.; Selma M. V.; Tomás-Barberán F. A.; González-Sarrías A.; Espín J. C. Where to Look into the Puzzle of Polyphenols and Health? The Postbiotics and Gut Microbiota Associated with Human Metabotypes. Mol. Nutr. Food Res. 2020, 64, 1900952 10.1002/mnfr.201900952. [DOI] [PubMed] [Google Scholar]
- Kay C. D.; Clifford M. N.; Mena P.; McDougall G. J.; Andres-Lacueva C.; Cassidy A.; Del Rio D.; Kuhnert N.; Manach C.; Pereira-Caro G.; Rodriguez-Mateos A.; Scalbert A.; Tomás-Barberán F.; Williamson G.; Wishart D. S.; Crozier A. Recommendations for standardizing nomenclature for dietary (poly)phenol catabolites. Am. J. Clin. Nutr. 2020, 112, 1051–1068. 10.1093/ajcn/nqaa204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L. W.; Amberg A.; Barrett D.; Beale M. H.; Beger R.; Daykin C. A.; Fan T. W. M.; Fiehn O.; Goodacre R.; Griffin J. L.; Hankemeier T.; Hardy N.; Harnly J.; Higashi R.; Kopka J.; Lane A. N.; Lindon J. C.; Marriott P.; Nicholls A. W.; Reily M. D.; Thaden J. J.; Viant M. R. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.