Abstract
In this report, we present details of two rapid molecular detection techniques based on 16S and 23S rRNA sequence data to identify and differentiate Yersinia species from clinical and environmental sources. Near-full-length 16S rRNA gene (rDNA) sequences for three different Yersinia species and partial 23S rDNA sequences for three Y. pestis and three Y. pseudotuberculosis strains were determined. While 16S rDNA sequences of Y. pestis and Y. pseudotuberculosis were found to be identical, one base difference was identified within a highly variable region of 23S rDNA. The rDNA sequences were used to develop primers and fluorescently tagged oligonucleotide probes suitable for differential detection of Yersinia species by PCR and in situ hybridization, respectively. As few as 102 Yersinia cells per ml could be detected by PCR with a seminested approach. Amplification with a subgenus-specific primer pair followed by a second PCR allowed differentiation of Y. enterocolitica biogroup 1B from biogroups 2 to 5 or from other pathogenic Yersinia species. Moreover, a set of oligonucleotide probes suitable for rapid (3-h) in situ detection and differentiation of the three pathogenic Yersinia species (in particular Y. pestis and Y. pseudotuberculosis) was developed. The applicability of this technique was demonstrated by detection of Y. pestis and Y. pseudotuberculosis in spiked throat and stool samples, respectively. These probes were also capable of identifying Y. enterocolitica within cryosections of experimentally infected mouse tissue by the use of confocal laser scanning microscopy.
The genus Yersinia comprises 11 species, of which Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica possess the potential to be pathogenic in humans and animals. The pathogenicity of these three species is controlled by the common 64- to 75-kb virulence plasmid pYV (or pCD1 for Y. pestis) (5). In contrast to the enteropathogenic yersiniae (Y. enterocolitica and Y. pseudotuberculosis), the plague bacillus (Y. pestis) usually harbors two additional virulence plasmids (pCP1 and pMT1).
Y. enterocolitica can be divided into six biogroups (biogroups 1A, 1B, and 2 to 5) and more than 50 serovars (8, 35). Y. enterocolitica strains belonging to biogroup 1B are commonly isolated in the United States, whereas strains of other biogroups are ubiquitously distributed. Those isolates formerly called Y. enterocolitica-like isolates were reclassified and assigned to eight different species (Y. frederiksenii, Y. intermedia, Y. kristensenii, Y. mollaretii, Y. bercovieri, Y. aldovae, Y. rhodei, and Y. ruckeri) (1).
Unfortunately, the established cultivation and serological techniques are not sufficient for the diagnosis of all Yersinia species (for reviews, see references 8 and 28). There is accumulating evidence that Y. enterocolitica may be difficult to recover in chronic infections by using standard cultivation techniques, although indirect immunofluorescence allows detection of the organism within clinical specimens (15, 18). Furthermore, some of the eight nonpathogenic Yersinia species share surface antigens with serotypes of Y. enterocolitica (8) that are pathogenic in humans, leading to false identification.
Rapid identification of Y. pestis is important in the monitoring of enzootic plague and during outbreaks of human plague. Cultivation of Y. pestis from clinical specimens requires approximately 2 days; this is followed by biotyping and detection of, e.g., fraction 1 antigen. Several reports have identified unusual Y. pestis strains, isolated from patients or rodents, which lack plasmid pCP1 or production of F1 antigen (for a review, see reference 28). Moreover, the pigmentation phenotype characteristic for Y. pestis has also been observed with freshly isolated Y. pseudotuberculosis strains (10).
Thus, rapid and reliable procedures for the direct detection and differentiation of yersiniae in clinical samples may prove helpful to both clinicians and public health authorities (8, 28).
Therefore, a 16S rRNA-based detection approach was developed, since this molecule has been used extensively to elucidate phylogenetic relationships of bacteria at intra- and intergeneric levels and it is also an excellent target for diagnostic PCR and fluorescent in situ hybridization assays (4, 30). Near-full-length 16S rRNA gene (rDNA) sequences for the three pathogenic Yersinia species were determined. A portion of the 23S rRNA gene was also analyzed for Y. pestis and Y. pseudotuberculosis. These sequence data were used to develop primer sets and fluorescently labelled oligonucleotide probes suitable for group- and subspecies-specific rDNA amplification reactions (PCRs) and for in situ hybridization of pathogenic Yersinia species within clinical specimens, respectively.
MATERIALS AND METHODS
Preparation of samples for in situ hybridization and PCR.
All bacterial strains used in this study are listed in Table 1. They were grown aerobically in Luria-Bertani (LB) broth at 26°C. Bacterial cells were harvested while in exponential growth phase, centrifuged, washed in 1 M NaCl, resuspended in TE buffer (10 mM Tris, 1 mM EDTA [pH 8]), and diluted to an optical density of 1.0 at 600 nm. One microliter of each cell suspension was used in PCR assays. For in situ hybridization, harvested cells were processed and fixed with paraformaldehyde as previously described (3).
TABLE 1.
Reference organisms, sources, and results of whole-cell hybridizations
| Organism and straina | Serotype(s)/biotypeb | Hybridization with probe
|
|||||
|---|---|---|---|---|---|---|---|
| Accession no. | Y.16S-69 | Y.ent.16S-184 | Y.p.16S-997 | Y.pseu.23S-1526 | E.car.16S-636 | ||
| Yersinia enterocolitica | |||||||
| IP 132c | O1, O2, O3/3 | + | + | − | − | − | |
| IP 1154c | O2, O3/3 | + | + | − | − | − | |
| Y 108-ce,n | O3*/4 | Z75316 | + | + | − | − | − |
| MYOk | O3/4 | + | + | − | + | − | |
| NF-0e | O5/1A | + | + | − | − | − | |
| Y-5,27f | O5, O27/2 | + | + | − | + | − | |
| 0310/90f | O6, O30/1A | + | + | − | − | − | |
| MY 79k | O9/2 | + | + | − | − | − | |
| WA-314d | O8/1B | Z75318,oZ75324p | + | + | − | − | − |
| 8081c | O8/1B | + | + | − | − | − | |
| 1209-79d | O13/1B | + | + | − | − | − | |
| 1223-75-1d | O20/1B | + | + | − | − | − | |
| Yersinia pestis | |||||||
| Kumag | —/antiqua | + | − | + | − | − | |
| Yokohamag | —/antiqua | + | − | + | − | − | |
| EV 76g,n | —/orientalis | Z75317 | + | − | + | − | − |
| A1122g | —/orientalis | + | − | + | − | − | |
| 6/69h | —/orientalis | + | − | + | − | − | |
| MP6g | —/orientalis | + | − | + | − | − | |
| M23g | —/orientalis | + | − | + | − | − | |
| G32g | —/orientalis | + | − | + | − | − | |
| KIMg | —/mediaevalis | + | − | + | − | − | |
| Yersinia pseudotuberculosis | |||||||
| H346.36/88f | O1 | + | − | + | + | − | |
| 2020j | O1 | + | − | + | + | − | |
| 444aj | O1 | + | − | + | + | − | |
| 2019j | O1 | + | − | + | + | − | |
| H 191/91k | O1A | + | − | + | + | − | |
| H 47/91k | O1A | + | − | + | + | − | |
| PB1g | O1A | + | − | + | + | − | |
| 123i | O1A | + | − | + | + | − | |
| 1468i | O1A | + | − | + | + | − | |
| 4587i | O1A | + | − | + | + | − | |
| H 268/91k | O1B | + | − | + | + | − | |
| H 260/91k | O1B | + | − | + | + | − | |
| O16i | O1B | + | − | + | + | − | |
| O6i | O1B | + | − | + | + | − | |
| 541i | O1B | + | − | + | + | − | |
| 487i | O1B | + | − | + | + | − | |
| Y-p-Tl | O3 | + | − | + | + | − | |
| YPIIIg,n | O3 | Z21939 | + | − | + | + | − |
| H 267/91k | O3 | + | − | + | + | − | |
| Yersinia frederikseniic | O3 | Z75319,oZ75325p | − | − | − | + | NDm |
| Yersinia mollaretiic | O3 | Z75322,oZ75328p | − | − | − | + | ND |
| Yersinia intermediac | O3 | Z75320,oZ75326p | + | − | − | − | ND |
| Yersinia kristenseniic | O3 | Z75321,oZ75327p | − | − | − | − | ND |
| Y. ruckerik | Z75323,oZ75329p | − | − | − | − | ND | |
| Hafnia alvei DSM 30163 | − | + | − | − | ND | ||
| Serratia marcescens ATCC 9141 | − | − | − | + | ND | ||
| Serratia fonticola DSM 4576 | − | + | − | − | ND | ||
| Rahnella aquatilis DSM 4594 | − | − | − | − | ND | ||
| Erwinia carotovora ATCC 43762 | − | − | + | − | + | ||
| Erwinia rhapontici LMG 2462 | − | − | − | + | − | ||
| Proteus mirabilisf | − | − | − | − | ND | ||
| Escherichia coli XL-1-Blue | − | − | − | − | ND | ||
| Klebsiella oxytocaf | − | − | − | − | ND | ||
| Citrobacter freundiif | − | − | − | − | ND | ||
| Salmonella enteritidisf | − | − | − | − | ND | ||
| Vibrio parahaemolyticus DSM 10027 | − | − | − | − | ND | ||
| Aeromonas hydrophila DSM 6173 | − | − | − | − | ND | ||
| Pasteurella multocida ATCC 12945 | − | − | − | − | ND | ||
DSM, Deutsche Stammsammlung von Mikroorganismen und Zellkulturen, Brunswick, Germany; ATCC, American Type Culture Collection, Rockville, Md.; LMG, Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium.
—, no serotype detected.
Strain as described in reference 11.
Strain as described in reference 22.
Strain as described in reference 13.
Strain obtained from Institut für Hygiene und Mikrobiologie, Würzburg, Germany.
Strain as described in reference 31.
Strain as described in reference 29.
Strain obtained from I. Semenova, St. Petersburg, Russia.
Strain obtained from J. Lomov, Rostov, Russia.
Strain obtained from Hygiene Institut, Hamburg, Germany.
Strain as described in reference 32.
ND, not determined.
Complete 16S rRNA sequence was determined.
V1 and V2 regions (positions 56 to 197) were sequenced.
V3 region (positions 420 to 650) was sequenced.
Female BALB/c mice (6 to 8 weeks old), purchased from Charles River Wiga, Sulzfeld, Germany, were inoculated intravenously with 3 × 105 bacterial cells (Y. enterocolitica WA-314) and sacrificed 4 days postexposure. The spleen, liver, and lung were aseptically removed from each mouse and cut into small pieces. The tissue pieces were immediately immersed in freshly prepared, cold 3% paraformaldehyde and refrigerated at 4°C for 24 h, to allow complete penetration of the fixative. The fixed tissue was washed in phosphate-buffered saline (PBS) for 2 h, mounted in O.C.T. Tissuetek (Miles Laboratories Inc., Elkart, Ind.), and snap-frozen in liquid nitrogen. Frozen tissue blocks were cut in 5-μm-thick sections with a cryostat and stored at −70°C.
Tissue samples from livers of sterile BALB/c mice were homogenized and spiked with different numbers of Y. enterocolitica WA-314. These samples were prepared for PCR analysis by using the QIAGEN (Hilden, Germany) tissue kit as recommended by the manufacturer. Five microliters of each of the resulting preparations was analyzed to evaluate the sensitivity of the different PCR approaches.
Three random stool samples and three random throat swabs submitted to the diagnostic laboratory of the Max von Pettenkofer Institut were collected and prepared for in situ hybridization as follows: 1 g of each stool specimen was resuspended in 9 ml of sterile PBS and processed further as described by Langendijk et al. (25). The throat swabs were placed in 500 μl of sterile PBS in a sterile 1.5-ml Eppendorf tube, and the remaining fluid was expressed from the swab by pressing it against the wall of the tube. The resulting suspension was centrifuged (6,000 × g, 10 min) and washed once in sterile PBS. The cell pellet was resuspended in 100 μl of a 1:1 mixture of PBS and 96% ethanol. An aliquot of the fixed specimens was spiked with ≥106 Y. pseudotuberculosis 487 and Y. pestis A1122, respectively. Ten microliters of each sample was analyzed by in situ hybridization.
PCR amplification and sequencing of rDNA.
Amplification and sequencing with universal primers were performed as described by Lane (24). Partial-length 16S and 23S rDNAs were amplified with primer pairs 27f-1525r and 1104f-1608r, respectively; one of each primer was biotinylated in each of two reciprocal reactions. Single-stranded DNA was obtained for direct sequencing by using the streptavidin-coated magnetic bead separation technique (17). Single-stranded DNAs were sequenced with multiple internal primers by the Taq cycle DyeDeoxy terminator method, combined with an ABI PRISM 373A automatic sequencer (PE Applied Biosystems, Weiterstadt, Germany). Sequences for both rDNA strands were determined. The nucleotide sequence data reported here have been deposited in the EMBL sequence database (Table 1).
PCR with Yersinia-specific primers (Table 2 and Fig. 1) was performed with all isolates specified in Table 1, each in a 50-μl reaction mixture containing 20 pmol of each primer, 1 μl of the bacterial cell suspension, 0.20 mM each dATP, dCTP, dGTP, and dTTP (Pharmacia LKB Biotechnology, Freiburg, Germany), 4 mM MgCl2, and 2.5 U of Taq DNA polymerase (AmpliTaq; PE Applied Biosystems) in a buffer with 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 0.001% (wt/vol) gelatin (PE Applied Biosystems). For each amplification reaction, negative controls containing water, instead of template DNA, were run in parallel. After the initial denaturation (80°C, 5 min), 30 cycles of amplification were carried out in a GeneAmp 2400 thermal cycler (PE Applied Biosystems). Each cycle consisted of 1 min of denaturation at 94°C, 1 min of annealing at 63°C (the exception is given in Table 2), and 2 min of extension at 72°C. Five microliters of each PCR product was analyzed by electrophoresis on 1.5% agarose gels containing 0.5 μg of ethidium bromide per ml.
TABLE 2.
PCR primers and hybridization probes
| Primer or probe | Sequence (5′-3′) | Target group | Position (nt)a | Reference |
|---|---|---|---|---|
| PCR primers | ||||
| Y.16S-86fb | GCGGCAGCGGGAAGTAGTTTA | Subgenus | 66–86 | PSc |
| Y.e.eur.16S-455r | CAATCACAAAGGTTATTAACCTTTATG | Y. enterocolitica European serotypes | 455–481 | PS |
| Y.e.ame.16S-455r | CAATCCAACAACGTATTAAGTTATTGG | Y. enterocolitica American serotypes | 455–481 | PS |
| Y.p.16S-455r | CAATGATTGAGCGTATTAAACTCAACC | Y. pestis and Y. pseudotuberculosis | 455–481 | PS |
| B.16S-794r | TACAGCGTGGACTACCAGGGT | Kingdom Bacteria | 794–814 | 36 |
| Probes | ||||
| Y.16S-69 | TAAACTACTTCCCGCTGC | Subgenus | 69–85 | PS |
| Y.ent.16S-184 | CCCACTTTGGTCCGAAGA | Y. enterocoliticad | 184–202 | PS |
| Y.p.16S-997 | CTCTGCCAAATTCTGTGG | Y. pestis and Y. pseudotuberculosisd | 997–1013 | PS |
| Y.pseu.23S-1526 | CTGCACCGTAGTGCATCG | Y. pseudotuberculosisd | 1526–1549 | PS |
| B.16S-338 | GCTGCCTCCCGTAGGAGT | Kingdom Bacteria | 338–355 | 3 |
| cB.16S-338 | CGACGGAGGGCATCCTCA | Negative control | c338–335 | 3 |
Numbering of the target positions corresponds to the E. coli numbering described in reference 9.
The annealing temperature for PCR with primer Y.16S-86f was 53°C.
PS, present study.
Exceptions are mentioned in the text.
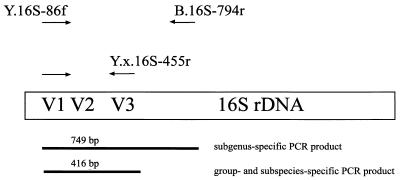
FIG. 1.
Positions and specificities of PCR primers and lengths of the amplification products generated with different PCR primers.
Development of PCR primers and hybridization probes.
An alignment of 29 partial Yersinia 16S rRNA sequences was used for the design of PCR primers and hybridization probes. Primer and probe designations, sequences, positions, and references are listed in Table 2.
The oligonucleotides for in situ hybridization were synthesized with a C6-trifluoroacetyl amino-linker [6-(trifluoroacetylamino)-hexyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidit] (MWG, Ebersberg, Germany). Labelling with 5 (and 6)-carboxytetramethylrhodamine, succinimidyl-ester [5(6)-TAMRA,SE] (CT; Molecular Probes, Eugene, Oreg.), 5-(-6-)carboxyfluorescein-N-hydroxysuccinimide-ester (FLUOS; Boehringer GmbH, Mannheim, Germany), and Cy5 (Biological Detection Systems, Pittsburgh, Pa.) was performed as described previously (3, 34).
Whole-cell hybridization.
In situ hybridization on glass slides was performed as described by Amann et al. (3). For the detection of pathogenic Yersinia species, two different probe combinations were hybridized simultaneously to the reference cells and to the clinical samples. Probes Y.16S-69-FLUOS and Y.ent.184-CT were used together for the detection of Y. enterocolitica, while probes Y.p.16S-997-FLUOS and Y.pseu.23S-1526-CT were applied simultaneously for the identification and differentiation of Y. pestis and Y. pseudotuberculosis. The addition of 30% formamide to the hybridization buffer resulted in a specific hybridization of the oligonucleotides to their respective target organisms. To reduce the amount of toxic waste, formamide was not used in the washing buffer in hybridization reactions. According to the formula of Lathe et al. (26), the NaCl concentration was instead decreased in the washing buffer to obtain the same stringency as that of the hybridization buffer.
Probe Y.-16S-69-CT was used to detect Yersinia cells within tissue sections. Probe cB.16S-338-CT, complementary to universal probe B.16S-338, was also hybridized to these samples to monitor nonspecific binding of labelled probes to bacterial and human cells. Citifluor (Citifluor Ltd., London, United Kingdom) was used as a mounting medium on hybridized slides, and the slides were examined with a Leica (Heerbrugg, Switzerland) TCS NT scanning confocal microscope equipped with a standard filter set. For probe excitation, an argon-krypton laser was used. Three different fluorochromes (CT, FLUOS, and Cy5) could be detected simultaneously with three different photomultipliers and represented the green (FLUOS), red (CT), and blue (Cy5) channels of the Leica software package. For the tissue sections, optical sectioning (0.5- to 1.0-μm width) was performed to reveal the three-dimensional localization of the probe-conferred fluorescence within the samples. The standard software delivered by the manufacturer was used to further process the digitized images.
RESULTS
Sequence analysis and phylogeny.
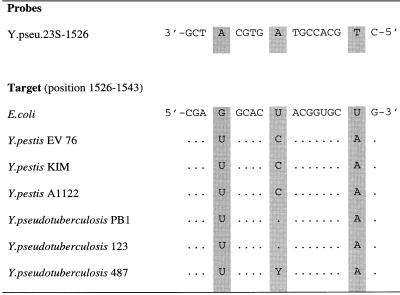
The PCR primers 27f and 1525r directed the synthesis of a 1,535-bp 16S rDNA fragment. Almost complete double-stranded sequences of these amplicons were determined for the three strains Y-108-c (Y. enterocolitica O:3), EV 76 (Y. pestis biovar orientalis), and YPIII (Y. pseudotuberculosis O:3). Only one region of intergeneric variability, corresponding to region V1, and two of intrageneric variability, corresponding to regions V2 and V3, could be detected (27). These regions were also sequenced for seven other Yersinia strains. In contrast to the work of Ibrahim et al. (20), the present analysis of 16S rDNA sequences revealed no differences between Y. pestis and Y. pseudotuberculosis. Therefore, a variable portion of the 23S rDNA of three Y. pestis and three Y. pseudotuberculosis strains encompassing the region corresponding to Escherichia coli positions 1104 to 1608 (9) was sequenced. The sequences of the six strains were identical except for position 1534 (Fig. 2). Direct partial sequencing of the 23S rRNA of Y. pseudotuberculosis 487 revealed two different possible bases (C or U) in position 1534.
FIG. 2.
Alignment of the target region for probe Y.pseu.23S-1526. Numbering of the target positions corresponds to the E. coli numbering described in reference 9. Identical nucleotides are represented by dots.
Sensitivity and specificity of PCR.
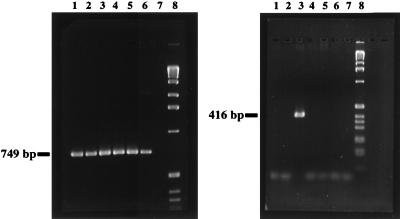
A subgenus-specific primer set (genus Yersinia excluding Y. frederiksenii, Y. mollaretii, and some strains of Y. kristensenii) was developed. Also, for the Y. pestis-Y. pseudotuberculosis group and for the two subspecies of Y. enterocolitica, specific primers were developed. The specificity of the different amplification reactions was evaluated by PCR by using genomic DNA preparations of closely related bacteria (Table 1) and human DNA. The PCR approach described above generated no amplification products with any of these DNA preparations. Figure 3 shows the results of a PCR assay using the subgenus-specific primer pair Y.16S-86f and B.16S-794r followed by a seminested amplification with primers Y.16S-86f and Y.e.eur.16S-455r. The sensitivity of this seminested PCR approach was compared to previously described alternative PCR systems based on amplification of the ail (23) and the yst (19) genes. Since no data about sensitivity were given by the authors of these earlier reports, we tested these two PCR approaches in comparison to our seminested PCR. A total of 5 × 103 cells per ml could be detected by amplification of the ail and the yst genes, whereas the seminested-PCR approach presented in this study generated a visible amplicon with 1 × 102 cells per ml.
FIG. 3.
Result of a seminested-PCR amplification of Yersinia rDNA by using primer pair Y.16S-86f–B.16S-794r for the first PCR (left panel) and primer pair Y.16S-86f–Y.e.eur.16S-455r for the second PCR (right panel) analyzed with a 1.5% agarose gel. One microliter of the first PCR mixture served as a template for the second PCR mixture. Lane 1, Y. pestis EV 76; 2, Y. pseudotuberculosis YPIII; 3, Y. enterocolitica Y-108-c; 4, Y. enterocolitica WA-314; 5, Y. intermedia; 6, Y. ruckeri; 7, H. alvei; 8, 1-kb ladder (Bethesda Research Laboratories, Eggenstein, Germany).
Sensitivity and specificity of in situ hybridization.
Probe Y.16S-69 is targeted to essentially the same rRNA region as primer Y.16S-86f; the theoretical specificity of this probe is therefore the same as that described for primer Y.16S-86f. Primers Y.e.ame.16S-455r and Y.p.16S-455r have also been labelled and tested as hybridization probes. Since these probes failed to hybridize to their respective target organisms, we designed probes Y.ent.16S-184 and Y.p.16S-997 for the detection of Y. enterocolitica and the Y. pestis-Y. pseudotuberculosis group, respectively.
Probe Y.ent.16S-184 is complementary to all Y. enterocolitica sequences but also to some phytopathogenic Erwinia species, Xenorhabdus beddingii, and Hafnia alvei as revealed by a gapped BLAST search (2). The probe target region for Y.p.16S-997 was found in all known Y. pestis and Y. pseudotuberculosis strains and also in 16S rDNA sequences of two Erwinia carotovora strains. For probe Y.pseu.23S-1526–CT, no BLAST matches were identified, although specificity testing showed that all Y. pseudotuberculosis strains, some European serotypes of Y. enterocolitica, Y. mollaretii, Y. frederiksenii, and Serratia fonticola also hybridized with this probe.
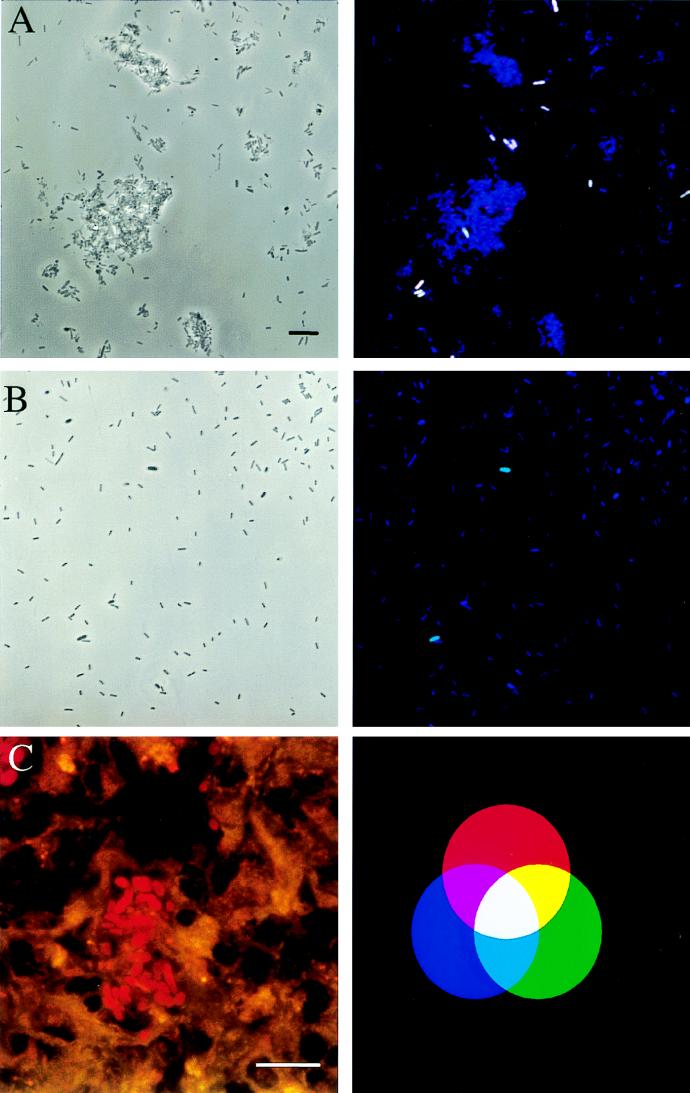
Whereas none of the regions sequenced is suitable for unequivocal identification of the three potentially pathogenic Yersinia species, a combination of two different rRNA regions unique for each of these yersiniae could be found. The presence of Y. enterocolitica could therefore be unequivocally detected by the combined application of Y.16S-69-FLUOS and Y.ent.16S-184-CT, and Y. pestis could be differentiated from Y. pseudotuberculosis and other Yersinia species by simultaneously hybridizing probes Y.pseu.23S-1526-CT and Y.p.16S-997-FLUOS to the samples. This probe combination was hybridized to three stool samples: no signal was obtained with any of the three samples, although more than 95% of the cells present hybridized with bacterial probe B.16S-338-Cy5. Within the spiked stool samples, Y. pseudotuberculosis cells could be easily identified (Fig. 4A). The same probe combination hybridized to specimens prepared from the three throat swabs also unequivocally detected Y. pestis within the spiked samples but not within the original samples (Fig. 4B). Probe E.car.16S-636-FLUOS, developed to monitor the presence of E. carotovora within the investigated samples, did hybridize to E. carotovora but not to any of the Yersinia species.
FIG. 4.
Detection of Yersinia species by in situ hybridization. Bars, 10 μm. Binding of at least two differently labelled probes results in distinct mixed colors, as shown in the additive-color illustration (C, right panel). Dual combinations of the red, green, and blue colors result in yellow (green and red), purple (red and blue), and turquoise (green and blue). White is a result of a combination of all three colors. (A and B) The same microscopic fields were viewed by phase-contrast microscopy (left panels) and by epifluorescence microscopy (right panels). Oligonucleotides Y.pseu.23S-1526-CT, Y.p.997-FLUOS, and B.16S-338-Cy5 were simultaneously applied to spiked stool (A) and throat (B) samples. As indicated by the white color, Y. pseudotuberculosis hybridized to all three labeled probes, whereas the turquoise color of the Y. pestis cells in the throat swab specimen is a result of the simultaneous binding of probes B.16S-338-Cy5 and Y.p.16S-997-FLUOS. (C) In situ detection of Y. enterocolitica in spleen sections of an infected mouse (left panel). Tissue sections were hybridized with probe Y.16S-69-CT and detected with the double-exposure option for the green and the red fluorescence of the Leica software package. Single bacterial cells are clearly visible within the spleen sections.
In situ detection of Yersinia cells in tissue samples.
Spleen and liver cryosections from mice experimentally infected with Y. enterocolitica 108-c were hybridized with probe Y.16S-69-CT. All tissue sections showed moderate to strong autofluorescence. Despite such interference, the bacterial cells could be easily detected in all tissues by using confocal laser scanning microscopy, whereas no bacterial cells could be detected in tissue samples hybridized with the negative control probe cB.16S-338-CT. Application of the dual-detector option of the Leica confocal laser scanning microscope (Fig. 4C; left panel) further improved image quality.
DISCUSSION
We evaluated the potential of rRNA-targeted PCR and fluorescent in situ hybridization for detection and differentiation of Yersinia species with respect to diagnosis of yersiniosis and plague, respectively.
The present analysis of complete and partial 16S rDNA sequences generally confirmed the results of a previous study (20). In contrast to this study was the finding of identical, near-full-length, 16S rDNAs in Y. pseudotuberculosis and Y. pestis. This is in good concordance with a previous study showing a close relationship between these bacterial species (6) and encouraged us to further investigate a portion of the 23S rRNA which contains the largest variable rRNA region encompassing helices 54 to 59 (16) and a smaller variable region (33) around helix 45. The 23S rRNA sequence data corroborate our 16S rRNA sequences, with the finding of only one base difference between Y. pestis and Y. pseudotuberculosis. For Y. pseudotuberculosis 487, sequencing reactions in both directions indicate that two different bases are present at position 1534. The existence of different rRNA operons within Y. pseudotuberculosis 487 is a reasonable explanation for this ambiguity. However, the in situ hybridization results indicate that this sequence ambiguity did not influence the reliable identification of this Y. pseudotuberculosis strain by in situ hybridization.
The sequence data obtained from rDNA sequences could be successfully used for the construction of both PCR primers and hybridization probes, allowing a rapid genotype-based detection of Yersinia species on different taxonomic levels. Labelled primers Y.e.ame.16S-455r and Y.p.16S-455r did not, however, hybridize to their target cells in situ. Since these primers perform well in PCR, noncomplementarity of probes and target could be ruled out. The most probable explanation is that RNA-RNA or RNA-protein interactions within 16S rRNA prevent hybridization of the probes. The influence of rRNA higher-order structure on probe-conferred fluorescence after in situ hybridization has long been proposed (4) and has been recently demonstrated (12).
A unique 16S rRNA target region suitable for the detection of Y. enterocolitica by in situ hybridization could not be identified. Therefore, an identification approach was developed by employing two different probes with broader specificities. The combined use of probes Y.ent.16S-184 and Y.16S-69 allowed an unequivocal identification of Y. enterocolitica. Furthermore, the simultaneous application of probes Y.p.16S-997-FLUOS and Y.pseu.23S-1526-CT proved to be a suitable tool for the differentiation of Y. pestis and Y. pseudotuberculosis. Nine Y. pestis and 19 Y. pseudotuberculosis strains were correctly identified by this approach, whereas no false-positive signal was observed with 31 other variably related bacterial species. The only exceptions were some E. carotovora strains, which, to our knowledge, have never been isolated from clinical sources. Nonetheless, probe E.car.16S-636 was developed to rule out the presence of this organism within a particular clinical sample. The specificity of this approach was further demonstrated by the successful detection of Y. pseudotuberculosis and Y. pestis in spiked clinical specimens, since presumably more than 400 bacterial species can be found in stool samples and samples from the oral cavity harbor up to 200 different bacterial species (7).
The potential to detect and identify the closely related yersiniae within 3 h without extensive preparation of nucleic acids from clinical samples is intriguing and may prove useful considering that misidentification of Y. pestis as Y. pseudotuberculosis and vice versa may occur when identification is based on the detection of capsular antigen F1 or pigmentation (10, 28). Moreover, this investigation showed that Yersinia cells infecting different mouse tissues carry enough ribosomes to be detected with fluorochrome-labelled oligonucleotides. The probes readily penetrate tissue samples and bacterial cell walls. This technique is well suited to detect the location of a pathogen within the body, particularly in combination with confocal laser scanning microscopy.
Future studies will evaluate whether in situ hybridization techniques are sensitive enough to detect dormant or metabolically inactive Yersinia cells within tissues, where they probably survive intracellularly (18). The importance of this issue stems from the observation that yersiniae associated with chronic infection are often noncultivable (15, 18). However, the sensitivity of in situ hybridization is comparatively low (4). In samples containing fewer than 105 cells per ml, more sensitive techniques must be applied. The use of sensitive PCR approaches has been described for the detection of Yersinia species (14, 21). Compared to these procedures, the present seminested-PCR assay covers the entire spectrum of pathogenic yersiniae and differentiates Y. enterocolitica biotype 1B from other, less pathogenic, Y. enterocolitica biotypes with high sensitivity, although rDNA-based PCRs do not discriminate between pathogenic and nonpathogenic Y. enterocolitica strains. For this purpose, other published target sequences such as the chromosomal gene for Y. enterocolitica heat-stable enterotoxin (yst gene) or genes of the virulence plasmid pYV have to be employed in the PCR (19, 21, 23). The yst-based PCR, however, showed reduced sensitivity compared to that of our approach, and the latter approaches could lead to false-negative results, since target plasmids can be lost during cultivation.
Both of the molecular methods investigated in this study offer alternatives to more traditional diagnostic methods for detection of yersiniosis. In particular, whole-cell hybridization holds great promise as a rapid, cultivation-independent method for detection of bacterial pathogens within clinical samples.
ACKNOWLEDGMENTS
This study was supported by a grant from the Friedrich Bauer Stiftung.
We thank Thomas Fritsche for critically reading the manuscript and Kristin Adler for excellent technical assistance.
REFERENCES
- 1.Aleksic S, Bockemühl J. Proposed revision of the Wauters et al. antigenic scheme for serotyping of Yersinia enterocolitica. J Clin Microbiol. 1984;20:99–102. doi: 10.1128/jcm.20.1.99-102.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. doi: 10.1016/0147-619x(81)90019-6. [DOI] [PubMed] [Google Scholar]
- 6.Bercovier H, Mollaret H H, Alonso J M, Brault J, Fanning G R, Steigerwalt A G, Brenner D J. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis. Curr Microbiol. 1980;4:225–229. [Google Scholar]
- 7.Berg R D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 8.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Burrow T W. Observation on the pigmentation of Yersinia pseudotuberculosis. In: Winblaad S, editor. Contributions to microbiology and immunology. Vol. 2. Basel, Switzerland: Karger; 1973. pp. 184–189. [Google Scholar]
- 11.Caugant D A, Aleksic S, Mollaret H H, Selander R K, Kapperud G. Clonal diversity and relationships among strains of Yersinia enterocolitica. J Clin Microbiol. 1989;27:2678–2683. doi: 10.1128/jcm.27.12.2678-2683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 13.Heesemann J, Algermissen B, Laufs R. Genetically manipulated virulence of Yersinia enterocolitica. Infect Immun. 1984;46:105–110. doi: 10.1128/iai.46.1.105-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch J, Schwan T G. New method for plaque surveillance using polymerase chain reaction to detect Yersinia pestis in fleas. J Clin Microbiol. 1993;31:1511–1514. doi: 10.1128/jcm.31.6.1511-1514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoogkamp-Korstanje J A, de-Koning J, Heesemann J. Persistence of Yersinia enterocolitica in man. Infection. 1988;16:81–85. doi: 10.1007/BF01644307. [DOI] [PubMed] [Google Scholar]
- 16.Höpfl P, Ludwig W, Schleifer K-H, Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989;185:355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- 17.Hultman T, Ståhl S, Hornes E, Uhlén M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppertz H-I, Heesemann J. Experimental Yersinia infection of human synovial cells: persistence of living bacteria and generation of bacterial antigen deposits including “ghosts,” nucleic acid free bacterial rods. Infect Immun. 1996;64:1484–1487. doi: 10.1128/iai.64.4.1484-1487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim A, Liesack W, Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992;30:1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim A, Goebel B M, Liesack W, Griffith M, Stackebrandt E. The phylogeny of the genus Yersinia based on 16S rRNA sequences. FEMS Microbiol Lett. 1993;114:173–178. doi: 10.1111/j.1574-6968.1993.tb06569.x. [DOI] [PubMed] [Google Scholar]
- 21.Kapperud G, Varund T, Skjerve E, Hornes E, Michaelsen T E. Detection of pathogenic Yersinia enterocolitica in food and water by immunomagnetic separation, nested polymerase chain reactions, and colorimetric detection of amplified DNA. Appl Environ Microbiol. 1993;59:2938–2944. doi: 10.1128/aem.59.9.2938-2944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay B A, Wachsmuth K, Gemski P. New virulence-associated plasmid in Yersinia enterocolitica. J Clin Microbiol. 1982;15:1161–1163. doi: 10.1128/jcm.15.6.1161-1163.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwaga J, Iversen J O, Misra V. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J Clin Microbiol. 1992;30:2668–2673. doi: 10.1128/jcm.30.10.2668-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 25.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183:11–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 27.Neefs J-M, van de Peer Y, Hendriks L, de Wachter R. Compilation of small ribosomal subunit RNA sequence. Nucleic Acids Res. 1990;18:2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry R D, Fetherstone J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt T M, Relman D A. Phylogenetic identification of uncultured pathogenes using ribosomal RNA sequences. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 31.Sikkema D J, Brubaker R R. Resistance to pesticin, storage of iron and invasion of HeLa cells by yersiniae. Infect Immun. 1987;55:572–578. doi: 10.1128/iai.55.3.572-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tertti R, Granfors K, Lehtonen O P, Mertsola J, Mäkelä A L, Välimäki I, Hänninen P, Toivanen A. An outbreak of Yersinia pseudotuberculosis infection. J Infect Dis. 1984;149:245–250. doi: 10.1093/infdis/149.2.245. [DOI] [PubMed] [Google Scholar]
- 33.Van Camp G, Chapell S, de Wachter R. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr Microbiol. 1993;27:147–151. doi: 10.1007/BF01576012. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. In situ identification of ammonia oxidizing bacteria. Syst. Appl. Microbiol. 18:251–264.
- 35.Wauters G, Kandolo K, Janssen M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987;9:14–21. [PubMed] [Google Scholar]
- 36.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]