Abstract
CRISPR-Cas transcriptional tools have been widely applied for programmable regulation of complex biological networks. In comparison to eukaryotic systems, bacterial CRISPR activation (CRISPRa) has stringent target site requirements for effective gene activation. While genes may not always have an NGG protospacer adjacent motif (PAM) at the appropriate position, PAM-flexible dCas9 variants can expand the range of targetable sites. Here we systematically evaluate a panel of PAM-flexible dCas9 variants for their ability to activate bacterial genes. We observe that dxCas9-NG provides a high dynamic range of gene activation for sites with NGN PAMs while dSpRY permits modest activity across almost any PAM. Similar trends were observed for heterologous and endogenous promoters. For all variants tested, improved PAM-flexibility comes with the tradeoff that CRISPRi-mediated gene repression becomes less effective. Weaker CRISPR interference (CRISPRi) gene repression can be partially rescued by expressing multiple sgRNAs to target many sites in the gene of interest. Our work provides a framework to choose the most effective dCas9 variant for a given set of gene targets, which will further expand the utility of CRISPRa/i gene regulation in bacterial systems.
Keywords: Engineered Cas9, PAM, CRISPRi, CRISPRa
Graphical Abstract
1. Introduction
CRISPR-Cas transcriptional regulation enables programmable control over gene activation and repression in bacteria.1–3 These tools can be used to regulate endogenous gene networks both to probe biological function and to engineer new behaviors. However, effective CRISPR activation (CRISPRa) is limited by complex and stringent target site requirements.4–7 Previous work has demonstrated a sharply periodic pattern of effective target sites occurring every 10 bp within a 60–100 bp region upstream of the TSS.2,4,8,9 Targeting these sites with S. pyogenes dCas9 requires a compatible protospacer adjacent motif (PAM) at the right position, and endogenous genes that lack an appropriate PAM may be incompatible for activation via Sp-Cas9.
We previously demonstrated that one of the first expanded PAM dCas9 variants, dxCas9(3.7),10 could activate some genes that lacked an appropriately-positioned NGG PAM.4 Specifically, dxCas9(3.7) produced at least two-fold increases in gene expression at three out of seven endogenous promoters tested.4 Although these results were encouraging, all seven of the candidate promoters were predicted to encode PAMs compatible with dxCas9(3.7) and all seven were expected to be activated. Since our initial work with dxCas9(3.7), several new expanded PAM dCas9 variants have been described,11–13 raising the possibility that improved variants could further expand the number of targetable genes for CRISPRa in bacteria.
In this work, we demonstrate that PAM-flexible dCas9 variants can improve transcriptional activation at endogenous genes compared to both dCas9 and dxCas9(3.7). We observe activation at previously inaccessible gene targets, and we observe a tradeoff between fold-activation and PAM flexibility. We also demonstrate that expanded PAM dCas9 variants are partially impaired for CRISPR interference (CRISPRi) gene repression. This effect can be mitigated by targeting multiple CRISPR complexes to the desired gene. By systematically characterizing the properties of PAM-flexible dCas9 variants in bacterial CRISPRa/i, we provide a framework to choose the most effective variant for a given gene target or set of targets. This toolbox of dCas9 variants will further expand the utility of CRISPRa/i in bacterial systems for a broad range of applications including metabolic engineering and genome-wide functional screens.
2. Results
2.1. In-silico PAM availability analysis to predict effective CRISPRa sites
In bacterial systems, effective CRISPRa requires a target site that is precisely positioned upstream of the gene of interest.4 Consequently, the ability to activate an arbitrary gene is dependent on an appropriately positioned PAM at the desired target site. The widely-used S. pyogenes Cas9 (Sp-Cas9) is generally limited to NGG PAMs, but several engineered Cas9 variants have been developed with more flexibility to accommodate various PAMs. We examined two groups of engineered Cas9 variants (Figure 1A). The first group includes xCas9-NG, a variant that exhibits high nuclease efficiency and CRISPRa/i performance at NGN PAMs in mammalian systems.10–12 The second group includes SpG and SpRY.12–16 The SpRY variant was engineered to be near-PAMless with some preference for NRN PAMs.
Figure 1: Engineered PAM-flexible Cas9 variants and PAM availability analysis.
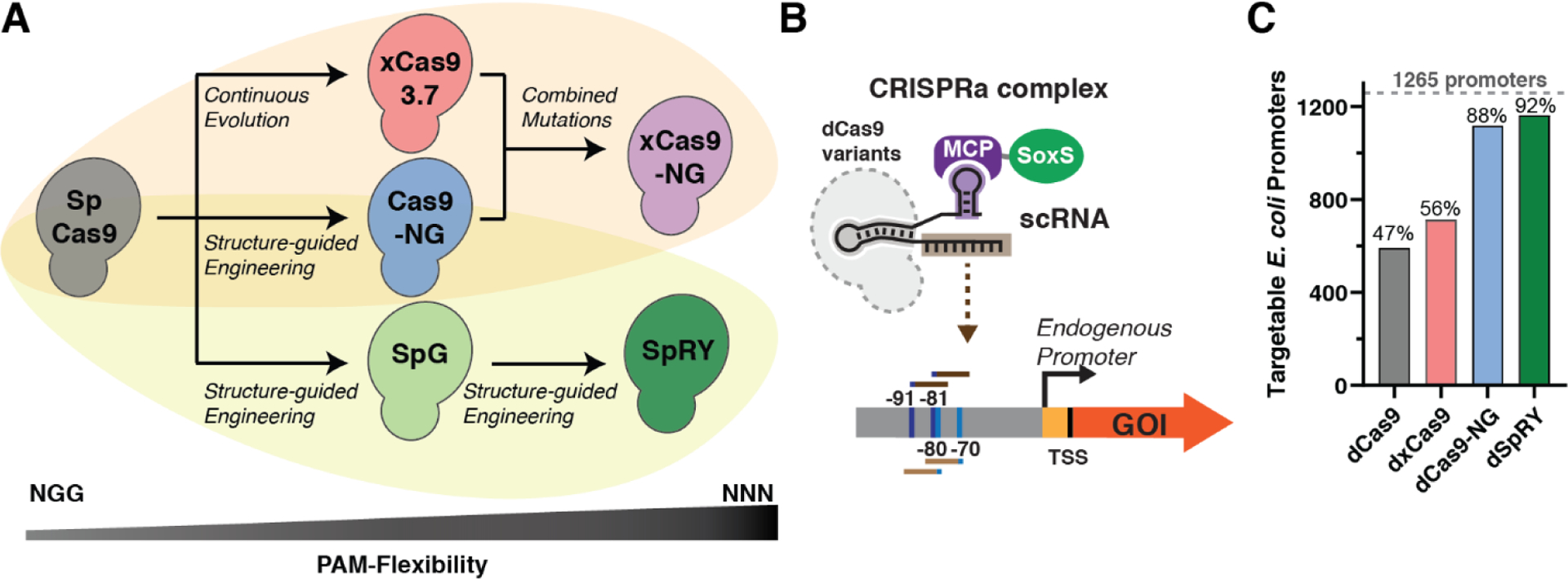
(A) PAM-flexible S. pyogenes Cas9 (Sp-Cas9) have been engineered using various rational design, screening, and directed evolution methodologies (e.g. phage-assisted continuous evolution (PACE), structure-guided engineering monitored via HT-PAMDA).10–16 (B) The CRISPRa complex consists of dCas9, an scRNA, and the MCP-SoxS activator.2 Previous work suggests that four precisely-positioned sites upstream of the TSS are most effective for CRISPRa (−70 and −80 for the template strand and −81 and −91 for non-template strand relative to the TSS).4 (C) 1265 E. coli endogenous promoters were analyzed for target site availability with different PAM-flexible dCas9 variants. Promoters were identified as targetable if they have at least one effective target site with a compatible PAM. For each variant, PAM preferences were obtained from previously-reported nuclease screening experiments (see Methods; PAMs with at least 20% of Sp-Cas9 activity at NGG PAMs were considered compatible).
We previously identified a set of four precisely-positioned sites upstream of the TSS that are the most effective for CRISPRa with the SoxS activator (positions −70 and −80 for the template strand and −81 and −91 for non-template strand) (Figure 1B).4 We performed an in-silico analysis to identify promoters with accessible PAMs at one or more of the optimal upstream positions in Escherichia coli and Pseudomonas putida. We restricted the search to promoters with high-confidence TSS positions17 and with sigma factors previously identified to be effective for CRISPRa with the SoxS activator.4 Together these criteria were met for 1265 out of 4042 promoters in E. coli. PAM compatibility for each Cas9 variant was predicted based on the reported nuclease activity with variable PAM targets (See Methods and Supplementary Methods).11,13,18 We performed the analysis with 4 deactivated Cas9 variants (dCas9, dxCas9(3.7), dCas9-NG, and dSpRY) of different PAM-flexibility (Table S4). We expect the predictions for dCas9-NG to be representative for dxCas9-NG. Consistent with this expectation, we found that xCas9-NG and Cas9-NG exhibit comparable levels of PAM flexibility in mammalian cell assays for nuclease activity and CRISPRa (Figure S2). The number of promoters with at least one PAM at a suitable position was 47% for dCas9, and increased to almost all promoters for the engineered variants (89% and 93% for dCas9-NG and dSpRY, respectively) (Figure 1C). A similar trend was observed in P. putida (Figure S3). It is important to note that these predictions are based on nuclease activity in mammalian cells and may not accurately predict bacterial CRISPRa activity, since binding and cleavage determinants may differ.
2.2. CRISPRa on non-NGG PAM is improved with engineered dCas9 variants
To test whether PAM-flexible Cas9 variants can increase the number of available CRISPRa target sites, we constructed expression cassettes for the catalytically-inactive versions of each Cas9 variant (dCas9, dxCas9(3.7), dCas9-NG, dxCas9-NG, dSpG, and dSpRY). Our bacterial CRISPRa system uses dCas9 and a modified guide RNA (termed scaffold RNA, scRNA) with an MS2 hairpin to recruit the MCP-SoxS activator (Figure 2A).2,4 We first determined whether each dCas9 variant was effective for CRISPRa at a canonical AGG PAM target site. We used a previously-described reporter gene (J3-BBa_J23117-mRFP) with an AGG PAM positioned at −81 relative to the TSS (Fontana et al., 2020). All tested dCas9 variants exhibited similar activation (~40–50-fold) with the canonical AGG PAM (Figure 2). We also evaluated the growth burden associated with each dCas9 variant because dCas9 expression can cause growth defects,19,20 and PAM-flexible variants can potentially bind the genome non-specifically at many more sites than the parent dCas9,21,22 For each dCas9 variant, we observed similar growth profiles and similar effects on expression capacity (Figure S4), suggesting that PAM flexibility does not produce additional growth burden effects.
Figure 2: CRISPRa with PAM-flexible dCas9 variants.
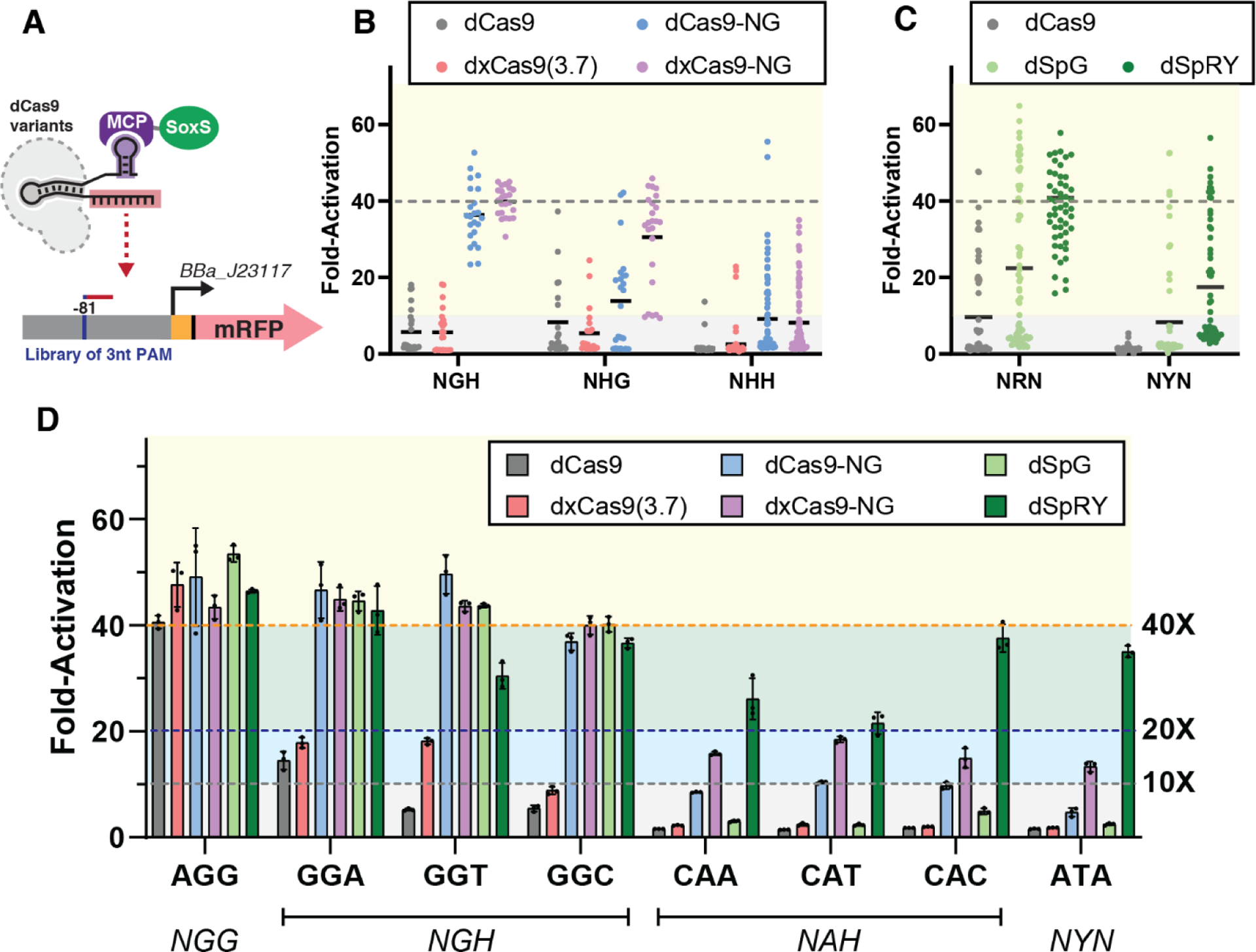
(A) CRISPRa for PAM-flexible dCas9 variants was tested on an mRFP reporter gene with libraries of alternative 3 nucleotide PAMs at the −81 target site. (B) Reporter gene expression with the dxCas9-NG group (Figure 1A) at NGH, NHG, and NHH PAM libraries. dxCas9-NG outperforms other variants in every PAM library. (C) Reporter gene expression with the dSpRY group (Figure 1A) at NRN and NYN PAM libraries. dSpRY exhibited the highest CRISPRa efficiency. (D) Direct comparisons of reporter gene expression with dCas9, dxCas9(3.7), dCas9-NG, dxCas9-NG, dSpG, and dSpRY at a representative set of PAMs. dCas9-NG, dxCas9-NG, and dSpRY performed best at NGN PAMs while dSpRY outperformed other variants at NAN and NYN PAMs. An scRNA targeting the J306 sequence was used for all library screens and individual PAM assays. To calculate fold-activation, we used an off-target scRNA (hAAVS1) with an AGG PAM reporter to define the basal expression level. Basal reporter expression levels vary <1.5-fold with different dCas9 variants or PAMs (Figure S5). Values in panel D represent the mean ± standard deviation calculated from n = 3.
To evaluate the effectiveness of each dCas9 variant for recognizing non-canonical PAM sites (non-NGG), we constructed libraries of reporters with varied PAM sequences. The first group of dCas9 variants (dxCas9(3.7), dCas9-NG, and dxCas9-NG) has a preference for NGN PAMs, so we screened three PAM libraries (NGH, NHG, and NHH, where H is not G). This approach follows the strategy previously used to characterize several Cas9 variants in mammalian systems.11 Taken together, the data indicate that dxCas9-NG provided the highest fold-activation across the largest number of alternative PAM reporters (Figure 2B). dxCas9-NG was effective at all NGH reporters (23 out of 23, or 100% had >10-fold activation) and most NHG reporters (21 out of 24, or 88% had >10-fold activation), but displayed substantially diminished effectiveness at NHH reporters (19 out of 71, or 27% had >10-fold activation). dCas9-NG performed similarly to dxCas9-NG at NGH and NHH PAMs, but notably weaker at NHG PAMs (NGH: 23 out of 23, or 100% had >10-fold activation; NHH: 22 out of 72, or 31% had >10-fold activation; NHG: 13 out of 24, or 54% had >10-fold activation). dCas9 and dxCas9(3.7) led to consistently reduced fold-activation and activated fewer reporters than dxCas9-NG across all three PAM libraries. The broad effectiveness of dxCas9-NG at NGH/NHG PAMs, along with its ability to activate some NHH PAMs, is consistent with prior reports from mammalian systems.11
To evaluate the effectiveness of the dSpRY group of dCas9 variants (Figure 1A), we screened NRN and NYN PAM libraries (R = A or G; Y = C or T) following the previously-determined PAM preferences for dSpRY in mammalian cells.13,16 As expected, we observed the strongest performance across the broadest range of PAM reporters with dSpRY. This variant produced >10-fold CRISPRa at all tested reporters in the NRN library and 29 out of 64 (45%) of the reporters in the NYN library. In both libraries, dSpRY consistently outperformed both dSpG and dCas9 (Figure 2C).
We proceeded to directly compare the CRISPRa efficiency of dxCas9-NG and dSpRY at specific non-NGG PAMs. We tested reporters with NGH (GGA, GGT, GGC), NAN (CAA, CAT, CAC), and NYN (ATA) PAMs (Figure 2D). We found that dxCas9-NG and dSpRY outperformed both dCas9 and dxCas9(3.7) at all of the non-NGG PAMs. dxCas9-NG exhibited the highest activity at NGH (≥40-fold activation) and moderately weaker activity at NAN and NYN (between 10-fold to 20-fold activation). Compared to dxCas9-NG, dSpRY produced similar or stronger activation at NAN and NYN PAMs (>20-fold activation). These data indicate that different PAM-flexible variants have distinct patterns of optimal PAMs. Together with the PAM library screens, these results suggest a framework for choosing an effective dCas9 variant. For a given target gene of interest, PAMs at the appropriate target site positions should be identified (−70 and −80 for the template strand and −81 and −91 for non-template strand upstream of the TSS). dCas9 should be used for NGG, either dCas9-NG, dxCas9-NG or dSpG should be used for NGH, dxCas9-NG should be used for NHG, and dSpRY should be used for NHH PAMs. When the four potential target sites include multiple alternative PAMs, they should be prioritized in the order NGG, NGH or NHG, and NHH. Following these guidelines, for subsequent experiments we prioritized dxCas9-NG for NGH/NHG PAMs and dSpRY for NHH PAMs.
2.3. CRISPRi efficiency is impaired with PAM-flexible dCas9 variants
CRISPRi acts by physically blocking transcription, and effective repression in bacteria can generally be obtained by targeting within the promoter or near the beginning of the gene on the non-template strand.3,23 CRISPRi is not subject to the same stringent target site requirements as CRISPRa, and there are often many canonical NGG PAMs available in the promoter or at the beginning of the gene. However, because we desire to express a single dCas9 protein to simultaneously target multiple genes for CRISPRa or CRISPRi, it is important to evaluate the performance of expanded PAM variants for CRISPRi. Previous work in bacterial and eukaryotic systems suggests that expanded PAM variants exhibit impaired CRISPRi function.11,24 We proceeded to evaluate the PAM-flexible variants dxCas9(3.7), dxCas9-NG, and dSpRY for CRISPRi gene repression. We tested multiple distinct sgRNA target sites, with one site in the promoter region and two sites within the ORF, all with NGG PAMs (Figure 3A). At each of these sites, dCas9 produces ~95-fold repression, while the PAM-flexible variants exhibit varying degrees of impaired repression (Figure 3B and Figure S6). For target sites within the ORF, the PAM-flexible dCas9 variants repress gene expression by 5 to 30-fold; these effects are significant but substantially weaker than the ~95-fold repression obtained with dCas9 at these sites. For the target site at the promoter, the dxCas9(3.7) performs similarly to dCas9, while dxCas9-NG and dSpRY produce weaker repression effects (28-fold and 14-fold, respectively).
Figure 3: CRISPRi with PAM-flexible dCas9 variants.
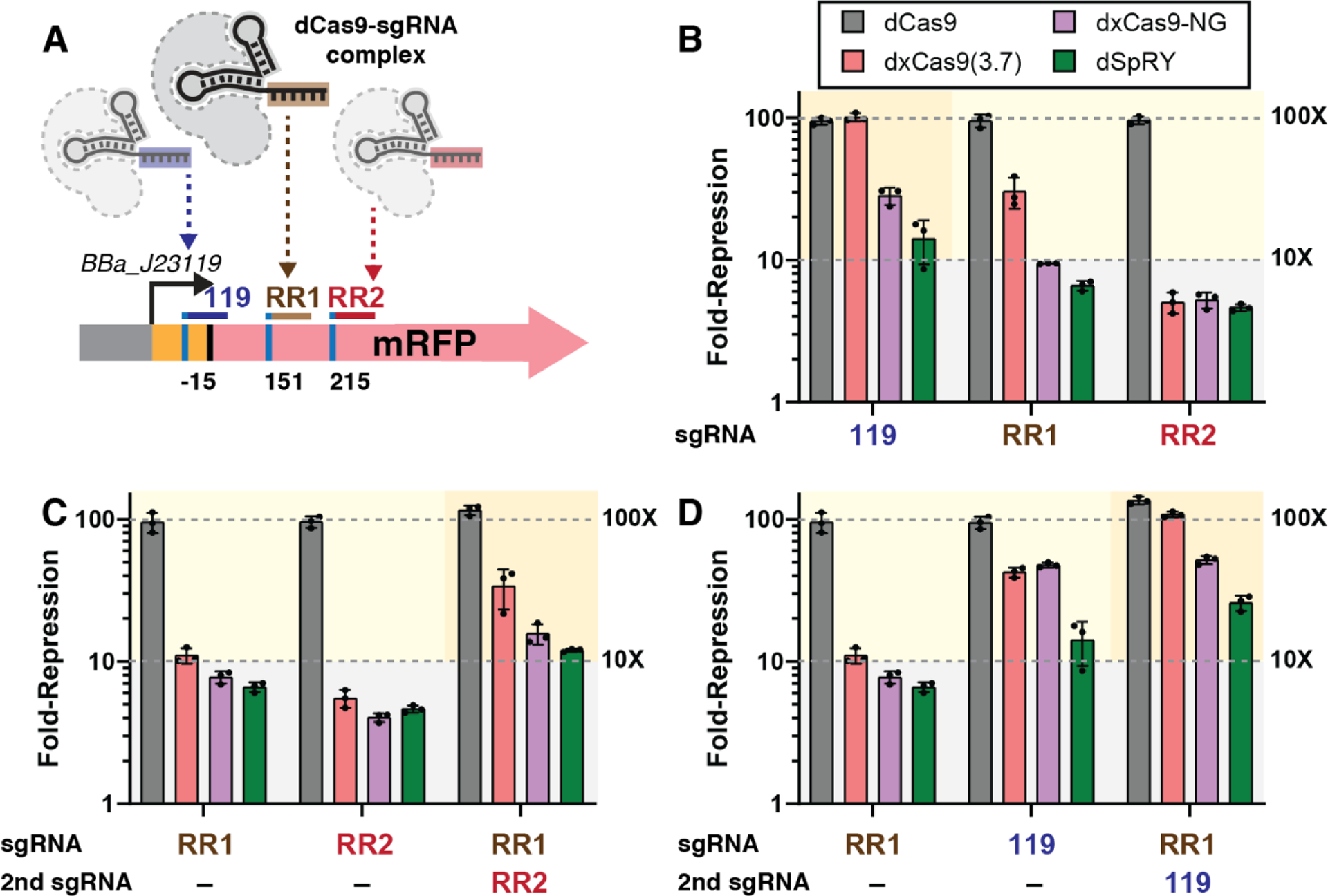
(A) CRISPRi for PAM-flexible dCas9 variants was tested on an mRFP reporter gene with sgRNAs targeting the promoter (119) or coding sequence (RR1 and RR2), where each target site encodes an NGG PAM. (B) Fold-repression of the mRFP reporter gene with a single expressed sgRNA. (C & D) Comparison of fold-repression with one or two sgRNAs expressed. Fold-repression consistently increases when two sgRNAs are expressed. See Figure S6 for a comparison of all dCas9 variants, including dCas9-NG and dSpG, with single and multiple sgRNAs. Values in panel B, C, and D represent the mean ± standard deviation calculated from n = 3.
CRISPRi repression with dCas9 can be improved by targeting multiple sgRNAs to the same gene.3 We therefore tested whether pairs of sgRNAs could be used to improve CRISPRi repression with PAM-flexible dCas9 variants. In each case, we observed improved repression (Figure 3B and 3C). Most notably, dSpRY repression can be improved from 14-fold to 26-fold. The dSpRY variants has one of the broadest targeting ranges due to its wide tolerance of PAMs for CRISPRa (Figure 2D), and the ability to improve its CRISPRi function via gRNA multiplexing suggests that dSpRY can be used for multi-gene CRISPRa/i programs with a large dynamic range of activation and repression.
2.4. PAM-flexible dCas9 variants improve CRISPRa at endogenous promoters
We previously demonstrated that the expanded PAM variant dxCas9(3.7) enables activation of some endogenous genes that are inaccessible to dCas9.4 To determine if dxCas9-NG and dSpRY further increase the pool of activatable endogenous genes, we examined four endogenous promoters previously tested with dxCas9(3.7): yajGp, uxuRp, araEp, and ppiDp2. For each endogenous promoter, we tested one NGG and one non-NGG PAM at appropriate positions upstream of the TSS at −70 or −80 (template strand) or at −81 or −91 (non-template strand), following the targeting rules defined previously.4 For two promoters previously activated by dxCas9(3.7), yajGp and uxuRp, dxCas9-NG and dSpRY produced comparable or improved activation compared to dxCas9(3.7) (Figure 4B). We also observed that two promoters that could not be activated by dxCas9(3.7) were slightly activated with dxCas9-NG and dSpRY: araEp was activated by 1.3-fold by dxCas9-NG while ppiDp2 was activated by 3.1-fold by dSpRY (Figure S7C).
Figure 4: CRISPRa at endogenous promoters is enhanced with PAM-flexible dCas9 variants.
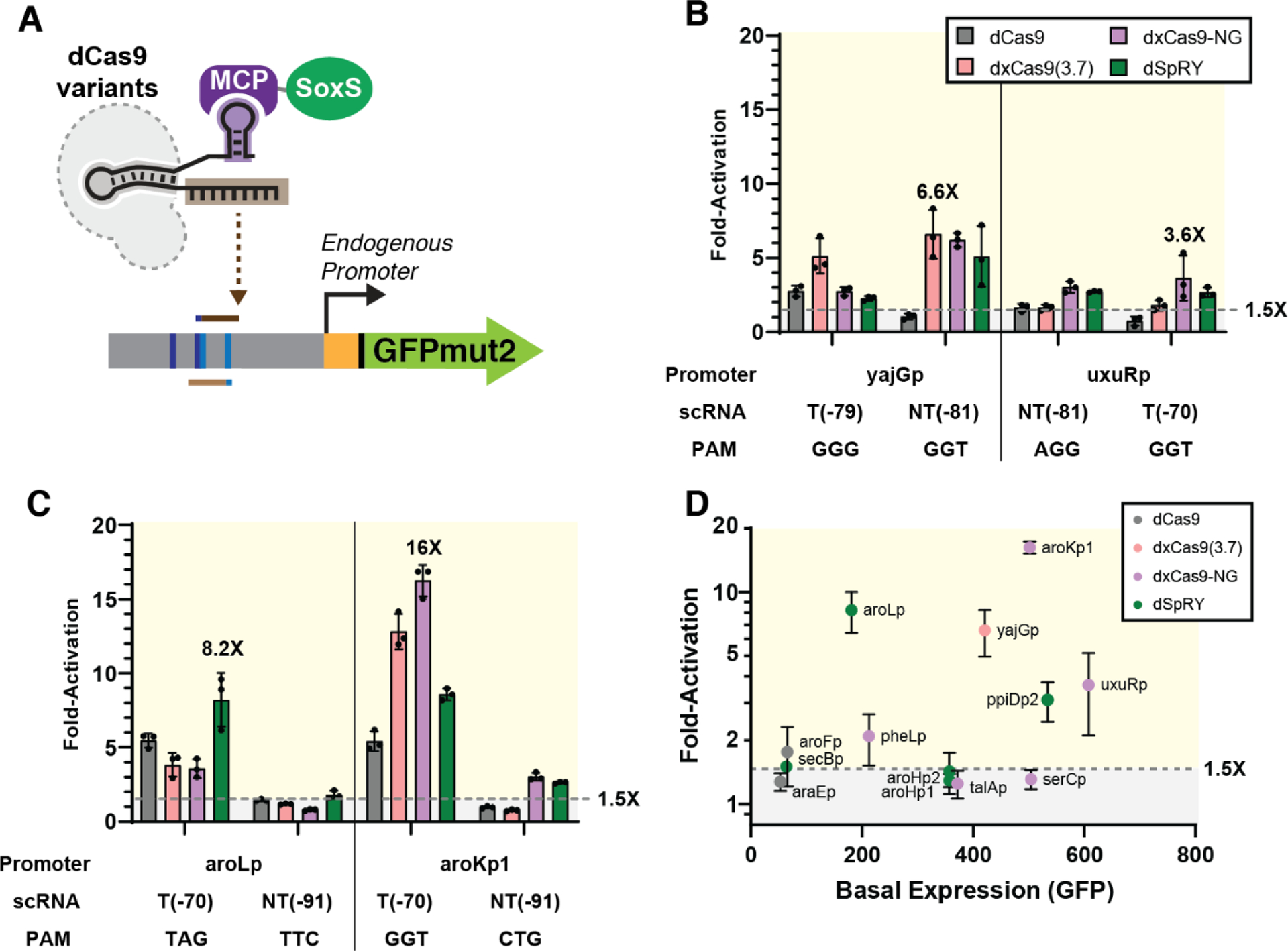
(A) CRISPRa at endogenous promoters of E. coli was tested using two of the four optimal scRNA positions for each promoter (see Supplemental Methods). (B) CRISPRa at endogenous promoters previously tested with dxCas9(3.7) (yajGp and uxuRp) using NGG or non-NGG PAMs. See Figure S7C for araEp and ppiDp2 promoters. (C) CRISPRa at endogenous promoters involved in aromatic amino acid biosynthesis using non-NGG PAMs. See Figure S7D for additional promoters (D) Plot of fold-activation versus basal expression level for all endogenous promoters tested in (B) & (C). See Table S5 for additional details. Data were collected by flow cytometry and fold-activation was calculated relative to a strain expressing the corresponding dCas9 variant and an off-target hAAVS1 scRNA. Values in panel B, C, and D represent the mean ± standard deviation calculated from n = 3.
We also tested nine new weakly-expressed endogenous promoters chosen from metabolic pathways related to aromatic amino acid biosynthesis (See Supplementary Methods).25 None of these promoters have NGG PAMs at the ideal distances relative to the TSS for CRISPR activation. For each promoter, we identified two candidate non-NGG targets that we expected to be compatible with dxCas9-NG or dSpRY (see Table S3). These sites were selected with the target site position rules described above.4 Out of nine new promoters tested, five can be activated by more than 1.5-fold. Activation of aroKp1 by dxCas9-NG and activation of aroLp by dSpRY demonstrated the largest fold-activation values (16-fold and 8-fold, respectively) (Figure 4C). We observed modest, ~2-fold activation for pheLp, secBp, and aroFp, and no activation (<1.5-fold) for aroHp1, aroHp2, talAp, and serCp (Figure S7D). For all promoters that were activated >1.5-fold, we observed distinct behaviors with different PAM-flexible variants. For aroKp1, dxCas9-NG significantly outperforms other variants at the GGT PAM. For aroLp, dSpRY outperforms other variants at the TAG PAM. These behaviors are consistent with our results from the mRFP reporter assay, which suggested the use of dxCas9-NG at NGN PAMs and dSpRY at NHH PAMs (Figure 2D). Taken together, these results suggest that the newer PAM-flexible variants dxCas9-NG and dSpRY can outperform dCas9 and dxCas9(3.7) for bacterial CRISPRa.
Out of the 13 total endogenous promoters tested, we found that five could not be activated above the 1.5-fold threshold (Figure 4B&C, S6C&D). One possible explanation for the failure of some target promoters to activate is that their basal expression levels are too high or too low. We previously observed that bacterial CRISPRa is sensitive to basal promoter strength, with no activation at the weakest promoters, effective activation at moderately weak promoters, and progressively smaller increases in gene expression as basal expression levels increased.4 Promoters with innately high expression levels may be inaccessible to high activation (>1.5-fold) due to the metabolic burden associated with increasing protein concentration.26,27 However, none of the five inaccessible promoters have unusually high basal expression levels (Figure 4D). Four of the promoters with <1.5 fold activation (aroHp1, aroHp2, talAp, and serCp) fall in a basal expression range that is higher than aroLp and lower than aroKp1, the two most highly activated promoters. Other factors beyond basal expression levels may be responsible for the failure of these four promoters to activate. The remaining inaccessible promoter, araEp, has the lowest basal expression level of all promoters tested and may be too weak to be activated with our current CRISPRa system.
3. Discussion
In this work, we have demonstrated that PAM-flexible dCas9 variants can improve bacterial CRISPRa in both synthetic and endogenous promoter contexts. Target distance from the TSS has been previously shown to be an important factor for effective bacterial CRISPRa.4–7 PAM-flexible variants expand the scope of accessible PAM sites and therefore enable targeting at precise, optimal positions upstream of the TSS.
Although PAM-flexible dCas9 variants enable CRISPRa at a majority of previously inaccessible gene targets, not all endogenous promoters with target sites at the appropriate position were able to be activated (Figure 4). Additional native regulatory machinery at endogenous gene targets may be responsible for preventing activation in these cases. We have previously shown that transcription factor binding can interfere with bacterial CRISPRa, presumably by physically obstructing binding of the CRISPRa complex or blocking the SoxS effector protein from engaging with RNA polymerase.4 We hypothesize that cryptic and unannotated transcription factor binding sites could be responsible for preventing activation at the endogenous genes targeted with PAM-flexible dCas9 variants in this work. Alternatively, we note that our guidelines for effective CRISPRa/i are based on experiments with fluorescent reporter genes, and while many endogenous targets appear to behave according to these rules, some gene sequences could have unexpected, context-specific effects on transcription or translation.
Improved bacterial CRISPRa with PAM-flexible variants comes with tradeoffs. We found that PAM-flexible dCas9 variants exhibited weaker CRISPRi-based gene repression compared to dCas9 (Figure 3B). One possible explanation for this behavior follows from the observation that increasing PAM promiscuity reduces affinity towards NGG PAMs.10–13,28 This reduced PAM-binding affinity could allow RNA polymerase to more readily displace the CRISPRi complex. Alternatively, a near-PAMless dCas9 variant would be expected to interrogate almost every DNA sequence in the bacterial genome21,22 and increase the time needed to find the correct target site. During every cell division, the CRISPRi complex is displaced from the genome,3 and increased time will be needed to bind the target site,29 which could allow for increased leaky expression and consequently weaker CRISPRi compared to the parent dCas9.
To implement complex, programmable genetic regulatory networks, both upregulation and downregulation controls with broad dynamic ranges are desirable.30–32 Thus, identifying systems that improve CRISPRa while maintaining effective CRISPRi is crucial. Multiplexing the CRISPRi targets with additional sgRNAs led to significantly higher fold-repression, although still weaker than CRISPRi with the parent dCas9 (FIgure 3C–3D). It remains to be seen whether these improvements are sufficient for genetic circuit applications. If stronger repression is needed, an alternative approach could be to distribute each engineering task to a different subpool of Cas9 proteins.22 In this application, a PAM-flexible variant could be used for CRISPRa and an orthogonal Cas protein could be used for CRISPRi.
An additional challenge for multi-gene CRISPRa/i regulation is the possibility that a CRISPRa target site ~80–90 bases upstream of a target gene could have undesired CRISPRi effects on the gene immediately upstream. The average intergenic region upstream of an E. coli gene is ~140 bases,33 so CRISPRa target sites could overlap with promoters or the 3’ end of an ORF, depending on the orientation of the preceding gene, and either situation could produce CRISPRi repression.3 The use of the PAM-flexible dCas9 variants described here has no impact on this challenge, and any DNA-targeting transcriptional activation system in bacteria is likely to face the same issue. Alternative gene regulation methods, including mRNA-targeting CRISPR translational activation systems,34 could be useful in this scenario.
While this manuscript was under revision, another manuscript was published that reported broadly consistent findings with the dSpRY variant.35 Our work includes additional comparisons with multiple PAM-flexible dCas9 variants and identifies situations where some of these variants (notably dxCas9-NG) are more effective than dSpRY. We also observed tradeoffs with CRISPRi efficiency that have not been previously described. Overall, the independently-obtained results support the broader idea that PAM-flexible Cas9 variants provide an effective means to overcome challenges associated with bacterial CRISPRa/i gene regulation.
These improvements to bacterial CRISPRa bring us closer to the long-standing goal of performing CRISPRa gain-of-function screens for basic discovery and engineering applications. Previously, some successes have been reported for activating natural product biosynthesis pathways36,37 but the ability to perform genome-wide CRISPRa screens in bacteria lags far behind eukaryotic systems.38–41 By unlocking these capabilities in bacteria, CRISPRa screens could enable rapid functional annotation of uncharacterized genes. For bioindustrial applications, we envision identifying genes that can overcome bottlenecks in routing metabolic flux or that confer robustness in harsh, non-native growth conditions. In the long term, combined CRISPRi and CRISPRa screens could provide even more information to map biological functions and deconvolute regulatory networks. In mammalian cells, combining information from single gene CRISPRa/i screens enabled improved chemical genetic profiling to identify drug targets,42 and dual-gene activation/repression screens have identified genetic interactions and functional relationships between genes.43,44 Combined CRISPRa/i screens should be possible in bacteria, as CRISPRi loss-of-function screens have been broadly applied,45 and it is straightforward to target multiple genes with multiple gRNAs for activation and repression.2,9,46 Some of these goals are plausibly within reach in bacteria using current CRISPRa tools, and further improvements in CRISPRa systems at endogenous gene targets will enable rapid progress in basic research and bioindustrial applications.
Materials and methods
Bacterial strains and plasmid constructs
E. coli K-12 substrain MG1655 was used for all CRISPRa tests unless specified. CD38 with highly expressed mRFP was used for CRISPRi experiments (Supplementary Table S1). Plasmid constructs were cloned using standard molecular biology methods.4,9 All PCR fragments were amplified with Phusion DNA Polymerase (Thermo-Fisher Scientific) for Infusion Cloning (Takara Bio). Plasmids were transformed into chemically competent NEB Turbo E. coli (New England Biolabs) cells, plated on LB-agar, and cultured in LB media supplied with the appropriate antibiotics used in the following concentrations: 100 μg/mL Carbenicillin, 25 μg/mL Chloramphenicol, 30 μg/mL Kanamycin. All plasmid constructs were confirmed by Sanger sequencing (GENEWIZ). Selected plasmids will be available upon request on Addgene (https://www.addgene.org/Jesse_Zalatan/).
PAM-flexible dCas9 variants were cloned from existing dCas9 and dxCas9(3.7) plasmids (pCD442 and pCD564).4 dxCas9-NG was cloned by replacing the C-terminus of dxCas9(3.7) with dCas9-NG mutations ordered as a gBlock (IDT). dCas9-NG was cloned by fusing the N-terminus of dCas9 and C-terminus of dxCas9-NG together. dSpG and dSpRY sequences were cloned into a bacterial codon optimized vector (Addgene #101199) and then subcloned into the pCD442 vector. Complete sequences are provided in the supplementary information.
Each dCas9 variant (dCas9, dxCas9, dCas9-NG, dxCas9-NG, dSpG, and dSpRY) was expressed from the endogenous Sp.pCas9 promoter in a p15A vector (Supplementary Table S2). MCP-SoxS (R93A, S101A) (abbreviated MCP-SoxS) was expressed from the BBa_J23107 promoter (http://parts.igem.org) in the same plasmid with dCas9. The single guide RNAs (sgRNA) or modified scaffold RNAs b2.1xMS2 (scRNAs) were expressed from the strong BBa_J23119 promoter, either in the same plasmid with the dCas9-carrying plasmid or in a separate ColE1 plasmid.2,4 All 20 bp scRNA/sgRNA target sequences are provided in Supplementary Table S3. The mRFP reporter was expressed from the weak J3-BBa_J23117 promoter on a pSC101** plasmid (Supplementary Table S2). For CRISPRi experiments, a construct expressing mRFP from the strong BBa_J23119 promoter (strain CD38, Supplementary Table S1) was integrated into the E. coli genome using a previously-described lambda red system.2,47 For endogenous promoter CRISPRa experiments, we used GFPmut2 reporters on pSC101** vectors as described previously.25 Reporters were purchased from Horizon Discovery or constructed with the same methodology.25
Plate reader experiments
Single colonies from LB plates were inoculated in 400 μL of EZ-RDM (Teknova) supplemented with the appropriate antibiotics and grown in 96-deep-well plates at 37 °C with shaking overnight 900 RPM on a Heidolph titramax 1000. 150 μL of the overnight culture were transferred into flat, clear-bottomed black 96-well plates (Corning) and the OD600 and fluorescence were measured in a Biotek Synergy HTX plate reader. Data were analyzed using the BioTek Gen5 2.07.17 software. For mRFP1 detection, the excitation wavelength was 540 nm and emission wavelength was 600 nm. For GFPmut2 detection, the excitation wavelength was 485 nm and emission wavelength was 528 nm.
Flow cytometry
Single colonies from LB plates were inoculated in 400 μL EZ-RDM (Teknova) supplemented with appropriate antibiotics and grown in 96-deep-well plates at 37 °C, 900 RPM on a Heidolph titramax 1000. Cultures were grown overnight and then diluted in 1:100 in Dulbecco’s phosphate-buffered saline (PBS) and analyzed on a MACSQuant VYB flow cytometer with the MACSQuantify 2.8 software (Miltenyi Biotec). To select single cells, we used a previously-described gating procedure.2 A side scatter threshold trigger (SSC-H) was applied to select for single cells until 10000 events were collected. FlowJo 10.0.7 software was used to apply a narrow gate along the diagonal line on the SSC-H vs SSC-A plot to exclude the events where multiple cells were grouped together. Within the selected population, events that appeared on the edges of the FSC-A vs. SSC-A plot and the fluorescence histogram were excluded.
E. coli growth profiles and expression capacity
To evaluate E. coli expression capacity, we incorporated an sfGFP capacity monitor expressed from a constitutive, medium-strength promoter (BBa_J23110) into the mRFP CRISPRa reporter plasmid. This method follows previously-described approaches to evaluate expression capacity.48 For measurement of expression burden, CRISPRa plasmids with different dCas9 variants and an scRNA (J306 for CRISPRa and hAAVS1 for an off-target control) were co-transformed with the reporter plasmids (pJF143.J3 or pCK760). Single colonies were inoculated in 400 μL EZ-RDM with appropriate antibiotics and endpoint fluorescent protein levels were measured after 18 hours with a plate reader as described above.
Growth profile time courses (OD600 vs. time) were obtained with strains inoculated from overnight stationary cultures with a 1:100 dilution. Growth profiles were also initiated from exponentially-growing cultures by first subculturing overnight cultures 1:100 into 2 mL EZ-RDM in 14 mL culture tubes for 2 hours, then diluting all cultures back to OD600 = 0.1. 200 μL of subcultures were transferred into flat, clear-bottomed black 96-well plates (Corning). OD600 values were measured in a plate reader for 16 hours at 37 °C.
Pooled PAM library construction and screening
To generate the pooled reporter library with different PAMs at the target site −81 bp from the TSS, we used pJF143.J3 (Supplementary Table S2) as a PCR template with oCK679_NNN (5’-CTCGTCTCCTCACTTTNNNACGGAGCGTTCTGGACACAACG-3’) as a forward primer and oCK680 (5’-AAGTGAGGAGACGAGCGAACGC-3’) as a reverse primer. Amplified linear fragments were treated with DpnI to remove the parental vector and circularized with Infusion. oCK679_NNN oligos with NGH, NHG, NHH, NRN, and NYN were used to construct each corresponding PAM library. For each screened variant, the number of colonies picked was 2X the number of sequence variants. For example, we picked 24 colonies for NGH (12 possible sequences) and 64 colonies for NRN (32 possible sequences). Fold activation was calculated relative to a strain with pJF143.J3 and an off-target scRNA.
CRISPRa at endogenous promoters
CRISPRa at endogenous promoters was performed with a three plasmid system — dCas9 plasmid, scRNA plasmid, and reporter plasmid. The reporter plasmids were adapted from the E. coli promoter collection (Dharmacon), a commercially available library of promoter-GFPmut2 fusions.25 We have previously confirmed that CRISPRa effects on these fluorescent protein reporters are also observed by RT-qPCR of the endogenous genomic transcript.4 Four promoters tested here (yajGp, uxuRp, araEp, and ppiDp2) were evaluated previously with dxCas9(3.7).4 These promoters were chosen based on the criteria: 1) genes should not be highly expressed, and 2) genes should be regulated by the sigma70 family.4 A second set of endogenous promoters (aroKp1, aroLp, pheLp, secBp, aroFp, aroHp1, aroHp2, talAp, and serCp) were selected from genes involved in aromatic amino acid biosynthesis pathways that meet the same criteria described above. scRNAs for each promoter were designed to target the optimal positions (−70 and −80 for the template strand and −81 and −91 for non-template strand relative to the TSS). One scRNA from each strand was chosen, based on which PAM was predicted to be accessible to the highest number of dCas9 variants. Accessibility was assessed based on the moderate performance threshold cutoff (see Supplemental Table S4 and Supplementary Methods).
Bioinformatic analysis of targetable genes
Previously reported data for the activity of PAM-flexible dCas9 variants were used to predict the targetable genes for each dCas9 variant. To identify the compatible PAMs for each engineered dCas9 variant, we used data from Cas9 nuclease assays for each variant11,13,18. Predicted compatible PAMs for each variant are provided in Supplementary Table S4 (see Supplementary Methods for further details). We then examined E. coli and P. putida genome sequences for PAM availability. For E. coli, 1265 omoters with strong confidence in TSS were retrieved from RegulonDB.17 For P. putida, 1104 experimentally-confirmed primary transcriptional units were used for the analysis.49 The PAMs at optimal target site positions were retrieved: −70 and −80 for the template strand and −81 and −91 for non-template strands relative to the TSS. The promoters with at least one compatible PAM out of four target sites were considered targetable. Further information can be found in Figure S3.
Data analysis
Flow cytometry analysis was conducted on FlowJo 10.0.7 software or Python FlowCytometryTools on Jupyter Notebooks and then further processed with Microsoft Excel and Graphpad Prism. Data represents the average and standard deviation of at least three biologically-independent replicates, unless specified. Fold activation and fold repression were calculated by comparing sample fluorescence with a strain expressing off-target scRNA/sgRNA.
Supplementary Material
Acknowledgements
We thank Benjamin I. Tickman, Ian D. Faulkner, Diego Alba Burbano, Ryan Cardiff, and members of Carothers and Zalatan laboratories for thorough discussion during the experimental planning and data collection/analysis strategy. We also thank Madelynn N. Whittaker for cloning the original bacterial codon optimized dSpG and dSpRY constructs. This work was supported by NSF Award 1817623 (J.M.C, J.G.Z.), DOE Award DE-EE0008927 (J.M.C, J.G.Z.), and NIH/NHGRI DP2HG010099 (N.E.S.). B.P.K. acknowledges funding from an MGH ECOR Howard M. Goodman Award. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-2140004 (A.V.K.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Disclosure and competing interests statement
B.P.K. is an inventor on patents and/or patent applications filed by Mass General Brigham that describe genome engineering technologies, including for the development of SpRY. B.P.K. is a consultant for EcoR1 capital and ElevateBio, and is an advisor to Acrigen Biosciences, Life Edit Therapeutics, and Prime Medicine. N.E.S. is an advisor to Vertex and Qiagen. J.M.C. and J.G.Z. are advisors to Wayfinder Biosciences.
Supporting Information
he Supporting information is available free of charge online.
Schematic of mutations in PAM-flexible dCas9 variants; CRISPRko and CRISPRa screens in mammalian systems; PAM availability and distribution in E. coli and P. putida promoters; growth burden comparisons; effect of dCas9 variants on basal reporter gene expression; CRISPRi with PAM-flexible dCas9 variants; CRISPRa screening at endogenous promoters; tables of bacterial strains, plasmids, and gRNA target sites; DNA sequences of reporter genes, proteins, and bacterial promoters, and supplementary methods (PDF).
Data availability
Screen data generated during this study will be deposited to GEO.
References
- (1).Bikard D; Jiang W; Samai P; Hochschild A; Zhang F; Marraffini LA Programmable Repression and Activation of Bacterial Gene Expression Using an Engineered CRISPR-Cas System. Nucleic Acids Res 2013, 41 (15), 7429–7437. 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dong C; Fontana J; Patel A; Carothers JM; Zalatan JG Synthetic CRISPR-Cas Gene Activators for Transcriptional Reprogramming in Bacteria. Nat Commun 2018, 9 (1), 2489. 10.1038/s41467-018-04901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Qi LS; Larson MH; Gilbert LA; Doudna JA; Weissman JS; Arkin AP; Lim WA Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152 (5), 1173–1183. 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fontana J; Dong C; Kiattisewee C; Chavali VP; Tickman BI; Carothers JM; Zalatan JG Effective CRISPRa-Mediated Control of Gene Expression in Bacteria Must Overcome Strict Target Site Requirements. Nat. Commun 2020, 11 (1), 1618. 10.1038/s41467-020-15454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ho H-I; Fang JR; Cheung J; Wang HH Programmable CRISPR-Cas Transcriptional Activation in Bacteria. Mol. Syst. Biol 2020, 16 (7), e9427. 10.15252/msb.20199427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Liu Y; Wan X; Wang B Engineered CRISPRa Enables Programmable Eukaryote-like Gene Activation in Bacteria. Nat. Commun 2019, 10 (1), 3693. 10.1038/s41467-019-11479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Villegas Kcam MC; Tsong AJ; Chappell J Rational Engineering of a Modular Bacterial CRISPR–Cas Activation Platform with Expanded Target Range. Nucleic Acids Res. 2021, 49 (8), 4793–4802. 10.1093/nar/gkab211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fontana J; Sparkman-Yager D; Zalatan JG; Carothers JM Challenges and Opportunities with CRISPR Activation in Bacteria for Data-Driven Metabolic Engineering. Curr. Opin. Biotechnol 2020, 64, 190–198. 10.1016/j.copbio.2020.04.005. [DOI] [PubMed] [Google Scholar]
- (9).Kiattisewee C; Dong C; Fontana J; Sugianto W; Peralta-Yahya P; Carothers JM; Zalatan JG Portable Bacterial CRISPR Transcriptional Activation Enables Metabolic Engineering in Pseudomonas Putida. Metab. Eng 2021, 66, 283–295. 10.1016/j.ymben.2021.04.002. [DOI] [PubMed] [Google Scholar]
- (10).Hu JH; Miller SM; Geurts MH; Tang W; Chen L; Sun N; Zeina CM; Gao X; Rees HA; Lin Z; Liu DR Evolved Cas9 Variants with Broad PAM Compatibility and High DNA Specificity. Nature 2018, 556 (7699), 57–63. 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Legut M; Daniloski Z; Xue X; McKenzie D; Guo X; Wessels H-H; Sanjana NE High-Throughput Screens of PAM-Flexible Cas9 Variants for Gene Knockout and Transcriptional Modulation. Cell Rep. 2020, 30 (9), 2859–2868.e5. 10.1016/j.celrep.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Nishimasu H; Shi X; Ishiguro S; Gao L; Hirano S; Okazaki S; Noda T; Abudayyeh OO; Gootenberg JS; Mori H; Oura S; Holmes B; Tanaka M; Seki M; Hirano H; Aburatani H; Ishitani R; Ikawa M; Yachie N; Zhang F; Nureki O Engineered CRISPR-Cas9 Nuclease with Expanded Targeting Space. Science 2018, 361 (6408), 1259–1262. 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Walton RT; Christie KA; Whittaker MN; Kleinstiver BP Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 2020, eaba8853. 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kleinstiver BP; Pattanayak V; Prew MS; Tsai SQ; Nguyen NT; Zheng Z; Joung JK High-Fidelity CRISPR–Cas9 Nucleases with No Detectable Genome-Wide off-Target Effects. Nature 2016, 529 (7587), 490–495. 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kleinstiver BP; Prew MS; Tsai SQ; Topkar VV; Nguyen NT; Zheng Z; Gonzales APW; Li Z; Peterson RT; Yeh J-RJ; Aryee MJ; Joung JK Engineered CRISPR-Cas9 Nucleases with Altered PAM Specificities. Nature 2015, 523 (7561), 481–485. 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Walton RT; Hsu JY; Joung JK; Kleinstiver BP Scalable Characterization of the PAM Requirements of CRISPR–Cas Enzymes Using HT-PAMDA. Nat. Protoc 2021, 16 (3), 1511–1547. 10.1038/s41596-020-00465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Santos-Zavaleta A; Salgado H; Gama-Castro S; Sánchez-Pérez M; Gómez-Romero L; Ledezma-Tejeida D; García-Sotelo JS; Alquicira-Hernández K; Muñiz-Rascado LJ; Peña-Loredo P; Ishida-Gutiérrez C; Velázquez-Ramírez DA; Del Moral-Chávez V; Bonavides-Martínez C; Méndez-Cruz C-F; Galagan J; Collado-Vides J RegulonDB v 10.5: Tackling Challenges to Unify Classic and High Throughput Knowledge of Gene Regulation in E. Coli K-12. Nucleic Acids Res. 2019, 47 (D1), D212–D220. 10.1093/nar/gky1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kim N; Kim HK; Lee S; Seo JH; Choi JW; Park J; Min S; Yoon S; Cho S-R; Kim HH Prediction of the Sequence-Specific Cleavage Activity of Cas9 Variants. Nat. Biotechnol 2020. 10.1038/s41587-020-0537-9. [DOI] [PubMed] [Google Scholar]
- (19).Cho S; Choe D; Lee E; Kim SC; Palsson B; Cho B-K High-Level DCas9 Expression Induces Abnormal Cell Morphology in Escherichia Coli. ACS Synth. Biol 2018, 7 (4), 1085–1094. 10.1021/acssynbio.7b00462. [DOI] [PubMed] [Google Scholar]
- (20).Zhang S; Voigt CA Engineered DCas9 with Reduced Toxicity in Bacteria: Implications for Genetic Circuit Design. Nucleic Acids Res. 2018, 46 (20), 11115–11125. 10.1093/nar/gky884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Anders C; Niewoehner O; Duerst A; Jinek M Structural Basis of PAM-Dependent Target DNA Recognition by the Cas9 Endonuclease. Nature 2014, 513 (7519), 569–573. 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Collias D; Beisel CL CRISPR Technologies and the Search for the PAM-Free Nuclease. Nat. Commun 2021, 12 (1), 555. 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Peters JM; Colavin A; Shi H; Czarny TL; Larson MH; Wong S; Hawkins JS; Lu CHS; Koo B-M; Marta E; Shiver AL; Whitehead EH; Weissman JS; Brown ED; Qi LS; Huang KC; Gross CA A Comprehensive, CRISPR-Based Functional Analysis of Essential Genes in Bacteria. Cell 2016, 165 (6), 1493–1506. 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang J; Teng Y; Zhang R; Wu Y; Lou L; Zou Y; Li M; Xie Z-R; Yan Y Engineering a PAM-Flexible SpdCas9 Variant as a Universal Gene Repressor. Nat. Commun 2021, 12 (1), 6916. 10.1038/s41467-021-27290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zaslaver A; Bren A; Ronen M; Itzkovitz S; Kikoin I; Shavit S; Liebermeister W; Surette MG; Alon U A Comprehensive Library of Fluorescent Transcriptional Reporters for Escherichia Coli. Nat. Methods 2006, 3 (8), 623–628. 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- (26).Gyorgy A; Jiménez JI; Yazbek J; Huang H-H; Chung H; Weiss R; Del Vecchio D Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J 2015, 109 (3), 639–646. 10.1016/j.bpj.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wu G; Yan Q; Jones JA; Tang YJ; Fong SS; Koffas MAG Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol. 2016, 34 (8), 652–664. 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- (28).Corsi GI; Qu K; Alkan F; Pan X; Luo Y; Gorodkin J CRISPR/Cas9 GRNA Activity Depends on Free Energy Changes and on the Target PAM Context. Nat. Commun 2022, 13 (1), 3006. 10.1038/s41467-022-30515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jones DL; Leroy P; Unoson C; Fange D; Ćurić V; Lawson MJ; Elf J Kinetics of DCas9 Target Search in Escherichia Coli. Science, 2017, 357, 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Alon U Network Motifs: Theory and Experimental Approaches. Nat. Rev. Genet 2007, 8 (6), 450–461. 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- (31).Brophy JAN; Voigt CA Principles of Genetic Circuit Design. Nat. Methods 2014, 11 (5), 508–520. 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tickman BI; Burbano DA; Chavali VP; Kiattisewee C; Fontana J; Khakimzhan A; Noireaux V; Zalatan JG; Carothers JM Multi-Layer CRISPRa/i Circuits for Dynamic Genetic Programs in Cell-Free and Bacterial Systems. Cell Syst. 2021, S2405471221004191. 10.1016/j.cels.2021.10.008. [DOI] [PubMed] [Google Scholar]
- (33).Fontana J; Voje WE; Zalatan JG; Carothers JM Prospects for Engineering Dynamic CRISPR-Cas Transcriptional Circuits to Improve Bioproduction. J Ind Microbiol Biotechnol 2018. 10.1007/s10295-018-2039-z. [DOI] [PubMed] [Google Scholar]
- (34).Otoupal PB; Cress BF; Doudna JA; Schoeniger JS CRISPR-RNAa: Targeted Activation of Translation Using DCas13 Fusions to Translation Initiation Factors. Nucleic Acids Res. 2022, 50 (15), 8986–8998. 10.1093/nar/gkac680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Klanschnig M; Cserjan-Puschmann M; Striedner G; Grabherr R CRISPRactivation-SMS, a Message for PAM Sequence Independent Gene up-Regulation in Escherichia Coli. Nucleic Acids Res. 2022, gkac804. 10.1093/nar/gkac804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ameruoso A; Villegas Kcam MC; Cohen KP; Chappell J Activating Natural Product Synthesis Using CRISPR Interference and Activation Systems in Streptomyces. Nucleic Acids Research, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ke J; Robinson D; Wu Z-Y; Kuftin A; Louie K; Kosina S; Northen T; Cheng J-F; Yoshikuni Y CRAGE-CRISPR Facilitates Rapid Activation of Secondary Metabolite Biosynthetic Gene Clusters in Bacteria. Cell Chem. Biol 2022, 29 (4), 696–710.e4. 10.1016/j.chembiol.2021.08.009. [DOI] [PubMed] [Google Scholar]
- (38).Joung J; Engreitz JM; Konermann S; Abudayyeh OO; Verdine VK; Aguet F; Gootenberg JS; Sanjana NE; Wright JB; Fulco CP; Tseng Y-Y; Yoon CH; Boehm JS; Lander ES; Zhang F Genome-Scale Activation Screen Identifies a LncRNA Locus Regulating a Gene Neighbourhood. Nature 2017, 548 (7667), 343–346. 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kampmann M CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol 2018, 13 (2), 406–416. 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sanson KR; Hanna RE; Hegde M; Donovan KF; Strand C; Sullender ME; Vaimberg EW; Goodale A; Root DE; Piccioni F; Doench JG Optimized Libraries for CRISPR-Cas9 Genetic Screens with Multiple Modalities. Nat. Commun 2018, 9 (1), 5416. 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Schmidt R; Steinhart Z; Layeghi M; Freimer JW; Bueno R; Nguyen VQ; Blaeschke F; Ye CJ; Marson A CRISPR Activation and Interference Screens Decode Stimulation Responses in Primary Human T Cells. Science, 2022, 375, eabj4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Jost M; Chen Y; Gilbert LA; Horlbeck MA; Krenning L; Menchon G; Rai A; Cho MY; Stern JJ; Prota AE; Kampmann M; Akhmanova A; Steinmetz MO; Tanenbaum ME; Weissman JS Combined CRISPRi/a-Based Chemical Genetic Screens Reveal That Rigosertib Is a Microtubule-Destabilizing Agent. Mol. Cell 2017, 68 (1), 210–223.e6. 10.1016/j.molcel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Boettcher M; Tian R; Blau JA; Markegard E; Wagner RT; Wu D; Mo X; Biton A; Zaitlen N; Fu H; McCormick F; Kampmann M; McManus MT Dual Gene Activation and Knockout Screen Reveals Directional Dependencies in Genetic Networks. Nat. Biotechnol 2018, 36 (2), 170–178. 10.1038/nbt.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Najm FJ; Strand C; Donovan KF; Hegde M; Sanson KR; Vaimberg EW; Sullender ME; Hartenian E; Kalani Z; Fusi N; Listgarten J; Younger ST; Bernstein BE; Root DE; Doench JG Orthologous CRISPR–Cas9 Enzymes for Combinatorial Genetic Screens. Nat. Biotechnol 2018, 36 (2), 179–189. 10.1038/nbt.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Todor H; Silvis MR; Osadnik H; Gross CA Bacterial CRISPR Screens for Gene Function. Host-Microbe Interact. Bact. Viruses 2021, 59, 102–109. 10.1016/j.mib.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wu Y; Liu Y; Lv X; Li J; Du G; Liu L CAMERS-B: CRISPR/Cpf1 Assisted Multiple-Genes Editing and Regulation System for Bacillus Subtilis. Biotechnol Bioeng 2020, 117 (6), 1817–1825. 10.1002/bit.27322. [DOI] [PubMed] [Google Scholar]
- (47).Fontana J; Dong C; Ham JY; Zalatan JG; Carothers JM Regulated Expression of SgRNAs Tunes CRISPRi in E. Coli. Biotechnol J 2018, 13 (9), e1800069. 10.1002/biot.201800069. [DOI] [PubMed] [Google Scholar]
- (48).Ceroni F; Algar R; Stan G-B; Ellis T Quantifying Cellular Capacity Identifies Gene Expression Designs with Reduced Burden. Nat. Methods 2015, 12 (5), 415–418. 10.1038/nmeth.3339. [DOI] [PubMed] [Google Scholar]
- (49).D’Arrigo I; Bojanovič K; Yang X; Holm Rau M; Long KS Genome-Wide Mapping of Transcription Start Sites Yields Novel Insights into the Primary Transcriptome of Pseudomonas Putida. Environ. Microbiol 2016, 18 (10), 3466–3481. 10.1111/1462-2920.13326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Screen data generated during this study will be deposited to GEO.


