Abstract
Thymic activation improves the outcome of COVID-19 patients with severe pneumonia. The rs2204985 genetic polymorphism within the TCRA-TCRD locus, which affects thymic output in healthy individuals, was found here to modify SARS-CoV-2–specific immunity and disease severity in COVID-19 patients with severe pneumonia. Forty patients with severe COVID-19 pneumonia were investigated. The GG genotype at the rs2204985 locus was associated, independently of age and sex, with stronger and long-lasting anti–SARS-CoV-2 helper and cytotoxic T cell responses 6 months after recovery. The GG genotype was also associated with less severe lung involvement, higher thymic production, and higher counts of blood naïve T lymphocytes, including recent thymic emigrants, and a larger population of activated stem cell memory CD4+ T cells. Overall, GG patients developed a more robust and sustained immunity to SARS-CoV-2. Polymorphism at rs2204985 locus should be considered as an additional predictive marker of anti–SARS-CoV-2 immune response.
A genetic polymorphism linked to thymopoiesis correlates with anti-SARS-CoV-2 immunity and clinical outcome in severe COVID-19.
INTRODUCTION
At least 700 to 800 million patients have been infected worldwide by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), most of them recovering after a mild to moderate disease. However, a minority of patients developed severe multi-organ failure and acute respiratory distress syndrome, leading to about 7 million deaths. Although in most cases the fatal outcome has been associated with old age and comorbidities (1, 2), it can also occur in younger individuals without overt ongoing pathological conditions. In this context, it would be extremely important to identify markers that can help to determine which patients with coronavirus disease 2019 (COVID-19) are at high risk of complications and death.
The role of the host genetic background on susceptibility and outcome has been previously reported in multiple infectious diseases, and several relevant genetic variants, which contribute to the inter-individual variability of COVID-19 severity, have been identified. For example, rare loss-of-function variants of Toll-like receptor 3 and interferon (IFN) regulatory factor 7 genes, which affect the production of type I IFNs, were found enriched in patients with life-threatening COVID-19 pneumonia (3). Three missense single-nucleotide polymorphisms (SNPs) in the Rab46 gene (EFCAB4B), potentially involved in the thrombotic and inflammatory events, were recently reported to be associated with COVID-19 fatality independently of risk factors (4). Moreover, genome-wide and transcriptome-wide association studies discovered significant genetic association between life-threatening COVID-19 and genes coding for proteins playing a key role in host antiviral defense and inflammatory response: antiviral restriction enzyme activators 2′-5′-oligoadenylate synthetase 1 and 3, tyrosine kinase 2, dipeptidyl peptidase 9, interferon receptor, C-C chemokine receptor type 2 (5).
We recently found that among severely ill COVID-19 patients with pulmonary involvement, thymic reactivation, evidenced by enhanced thymic production of new lymphocytes, with or without thymus enlargement, was associated with a more favorable outcome (6), an observation that was confirmed by subsequent studies (7, 8). The origin of the enhanced thymic function observed in this subgroup of patients with COVID-19 remained unexplored. Four SNPs (rs8013419, rs10873018, rs12147006, and rs2204985), located in the intergenic T cell receptor (TCR) D2 and D3 segments within the TCRA-TCRD locus, were identified in a general population cohort as associated with different levels of thymopoïesis (9). Among them, rs2204985, located in an open-chromatin region (10) close to a binding site of the ubiquitous Zn-finger protein CTCF (CCCTC-binding factor) involved in chromatin looping and in chromatin recombination, showed a predominant phenotypic effect (11). The rs2204985 GG genotype was associated with the highest level of thymic production and T cell diversity, the AA genotype with the lowest, the GA genotype being intermediate (9). The pathophysiological relevance of the rs2204985 SNP has recently been examined in the context of unrelated hematopoietic cell stem transplantation in patients with hematologic malignancies. Donor AA genotype in single human leukocyte antigen mismatched cases was associated with significantly lower, overall and disease-free survival (12). However, there are no studies so far that have evaluated the impact of rs2204985 SNP on the immune response in the context of severe infection.
Here, we investigated whether the rs2204985 polymorphism might constitute a genetic marker associated with the level of disease severity in hospitalized patients with severe COVID-19 and explored its impact on anti–SARS-CoV-2 immunity. We found that GG patients developed a more robust and sustained immunity to SARS-CoV-2 than individuals with the other genotypes.
RESULTS
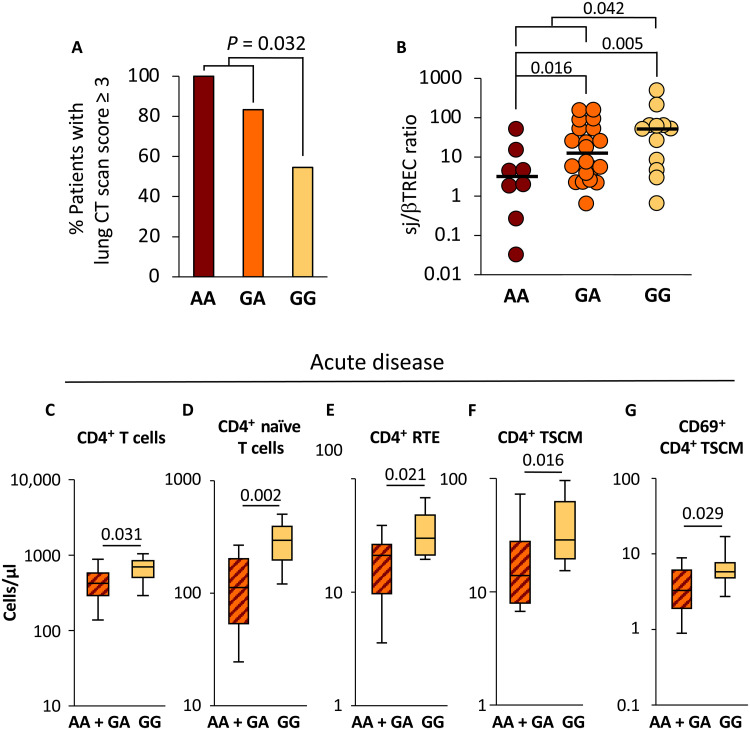
Forty consecutive patients hospitalized in the same intensive care unit (ICU) for severe COVID-19 with pulmonary involvement, who gave written consent for TCRA-TCRD locus DNA sequencing, were included in the study. The sequence of the rs2204985-containing region in patients with COVID-19 identified 8 AA, 20 GA, and 12 GG genotypes, close to the expected allelic distribution (https://gnomad.broadinstitute.org). The GG genotype was associated with less severe lung involvement: only 54.5% of GG patients showed a computerized tomograpgy (CT) scan score of 3 or higher, compared to 83.3 and 100% in GA and AA patients, respectively. A logistic regression model showed a significant difference between AA + GA and GG patients (odds ratio = 0.16, 95% confidence interval: [0.03–0.8], P = 0.032; Fig. 1A). Clinical characteristics of patients are shown in Table 1. Age and sex, two main factors associated with COVID-19 severity, were comparable in GG and AA + GA groups. Among comorbidities commonly related to the severity of COVID-19, none was significantly associated with either GG or AA + GA genotype. However, the percentage of patients with hypertension or diabetes was higher in AA + GA than in GG patients (57% versus 25% and 25% versus 8%, respectively). We then performed a multivariate analysis with hypertension, body mass index (BMI), diabetes, cancer, chronic obstructive pulmonary disease (COPB), and renal failure as covariables. Using multiple logistic regression in this setting, the GG genotype appeared as an independent factor affecting the lung CT scan score (table S1). These data are consistent with the hypothesis that the rs2204985 GG genotype somehow protects patients with COVID-19 against the most severe forms of lung involvement.
Fig. 1. Patients with GG genotype displayed less severe pulmonary involvement and higher thymic output.
Patients with COVID-19 were classified according to their genotype at the rs2204985 (n = 8 AA, n = 20 GA, and n = 12 GG) locus and plotted as a function of pulmonary involvement severity (A) and thymic function (sj/βTREC ratio; B). Total CD4+ T cells, naïve (CD45RA+, CD95−, CD27+, CD28+) CD4+ T cells, CD4+ RTE (CD45RA+, CD95−, CD27+, CD28+, CD31Hi), CD4+ TSCM (CD45RA+, CD95+, CD27+, CD28+), and activated (CD69+) CD4+ TSCM were quantified by fluorescence-activated cell sorting in peripheral blood mononuclear cells (PBMCs) from patients with COVID-19 sampled during the acute phase of the disease (C to G). Statistical significance of the differences between groups is shown on top [logistic regression model for (A) and Mann-Whitney test for (B) to (G)]. Patients with AA and GA genotypes were analyzed in the same group. The gating strategy for T cell subsets is shown in fig. S1.
Table 1. Patients’ characteristics.
Continuous variables were compared using Mann-Whitney nonparametric test for groups with less than 30 patients. Categorical variables were analyzed using Fisher exact test. CRP, C-reactive protein.
| Rs2204985 genotype | GA or AA | GG | |
|---|---|---|---|
| Number | 28 (8AA, 20GA) | 12 | |
| Age (years) | 66.5 | 67 | NS |
| Sex ratio (F/H) | 8/20 | 3/9 | NS |
| BMI (kg/m2) | 29.4 [20.4–43.3] | 26.2 [24.5–40.3] | NS |
| Hypertension (N) | 16 (57%) | 3 (25%) | NS |
| Diabetes (N) | 7 (25%) | 1 (8%) | NS |
| COPB (N) | 1 (3.5%) | 0 (0%) | NS |
| Smoking (N) | 2 (7%) | 3 (25%) | NS |
| Renal failure (N) | 2 (7%) | 0 (0%) | NS |
| Cancer (N) | 2 (7%) | 1 (8%) | NS |
| Hemoglobin (g/dl) | 12.0 [8.4–14.7] | 11.8 [9.4–15.8] | NS |
| Leukocytes (G/liter) | 7.4 [3.6–15.2] | 8 [4.2–13] | NS |
| Lymphocytes (G/liter) | 1.2 [0.3–3.3] | 1.5 [0.6–2.2] | NS |
| Monocytes (G/ml) | 0.6 [0.1–1.8] | 0.6 [0.4–1] | NS |
| Platelets (×103/mm3) | 305 [105–623] | 410 [203–564] | 0.024 |
| CRP (mg/liter) | 18 [<1–395] | 24 [14–153] | NS |
| Steroids (N) | 17 | 7 | NS |
| Death (N) | 2 | 0 | NS |
We next investigated whether the rs2204985 genotype would be associated with different levels of thymopoïesis in patients with COVID-19, estimated through the quantification of T cell receptor excision circles (TRECs). The extent of intrathymic precursor T cell proliferation, reflected by the signal-joint TREC/DJbeta TREC (sj/βTREC) ratio in circulating lymphocytes (see Materials and Methods), is proportional to thymic export of new T cells into the blood (13, 14). Thymic output was correlated with a higher number of circulating T cells and greater TCR repertoire polyclonality after stem cell transplantation (15). The sj/βTREC ratio was reported to decrease with age both in healthy populations (9, 13) and in patients with COVID-19 (6). Despite a comparable age distribution in the three groups of patients with COVID-19 [median age (range): AA:59y (32 to 76); GA:67y (29 to 84); GG: 67y (32 to 77)], the median sj/βTREC ratio was higher in rs2204985 GG than in GA and AA individuals (51.9, 12.51, and 3.17, respectively) and significantly different in the GG group, compared to the combined AA + GA group (P = 0.042; Fig. 1B). Moreover, GG patients also displayed higher counts of total and naïve blood CD4+ T lymphocytes, compared to AA + GA patients (P = 0.031 and P = 0.002, respectively; Fig. 1, C and D). A similar difference was observed for recent thymic emigrants (RTEs), the subgroup of naïve CD4+ T cells having recently left the thymus, which still express high levels of the CD31 antigen (P = 0.021; Fig. 1E). In contrast, central memory and effector memory T cells were not affected by the genotype at the rs2204985 polymorphism (fig. S2). Patients with the GG genotype also displayed higher counts of stem cell memory CD4+ T cells (TSCM; P = 0.016; Fig. 1F), a larger proportion of them expressing CD69, a marker of recent activation (P = 0.03; Fig. 1G). These findings support the hypothesis that individuals with rs2204985 GG genotype developed a more efficient adaptive immune response against SARS-CoV-2 during acute infection.
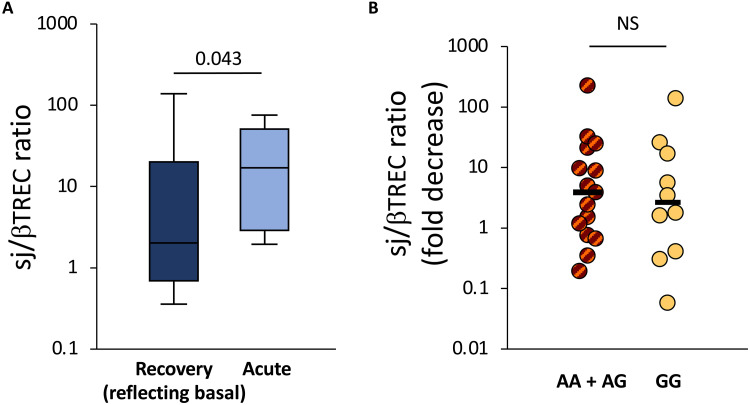
We next examined to what extent the rs2204985 polymorphism might, in addition, be associated with a stimulating impact of SARS-CoV-2 infection on thymic function. Assuming that the sj/βTREC ratio measured at distance of the acute infection would somewhat reflect basal thymic production, a subgroup of 25 patients who fully recovered from COVID-19 and accepted a second investigation (4 AA, 11 GA, and 10 GG) were tested again 6 months after the first assessment. Consistent with a SARS-CoV-2 stimulating effect, the sj/βTREC ratios were globally higher during the acute phase than during the recovery phase (P = 0.043; Fig. 2A). However, the fold increase in thymic production during acute SARS-CoV-2 infection was not different between the two groups of patients (2.63- and 3.87-fold in the GG and the AA + GA groups, respectively; P = 0.38; Fig. 2B). These data indicate that the fold increase in thymic function consecutive to viral infection is independent of the rs2204985 genotype. Notably, the enrichment in the naive, RTE, and TSCM subpopulations observed in GG patients during the acute disease tended to persist for at least 6 months and remained significant for the subset of activated (CD69+) TSCM (P = 0.04; fig. S3). In contrast, AA + GA and GG patients displayed very similar numbers of circulating CD8+ T cell subsets at both time points (fig. S4).
Fig. 2. Thymic function during acute disease and after recovery from COVID-19.
Thymic function was assessed through the quantification of the sj/βTREC ratio in patients with COVID-19 during their first hospitalization in ICU and 6 months after recovery are shown (A). The fold decrease in sj/βTREC ratio between samplings for AA + GA and GG patients is shown (B). Horizontal bars represent medians in each patient group. Statistical differences between AA + GA and GG patients at each time points (Mann-Whitney test for two independent samples). The recovery phase is expected to reflect some features present during the basal conditions. NS, not significant.
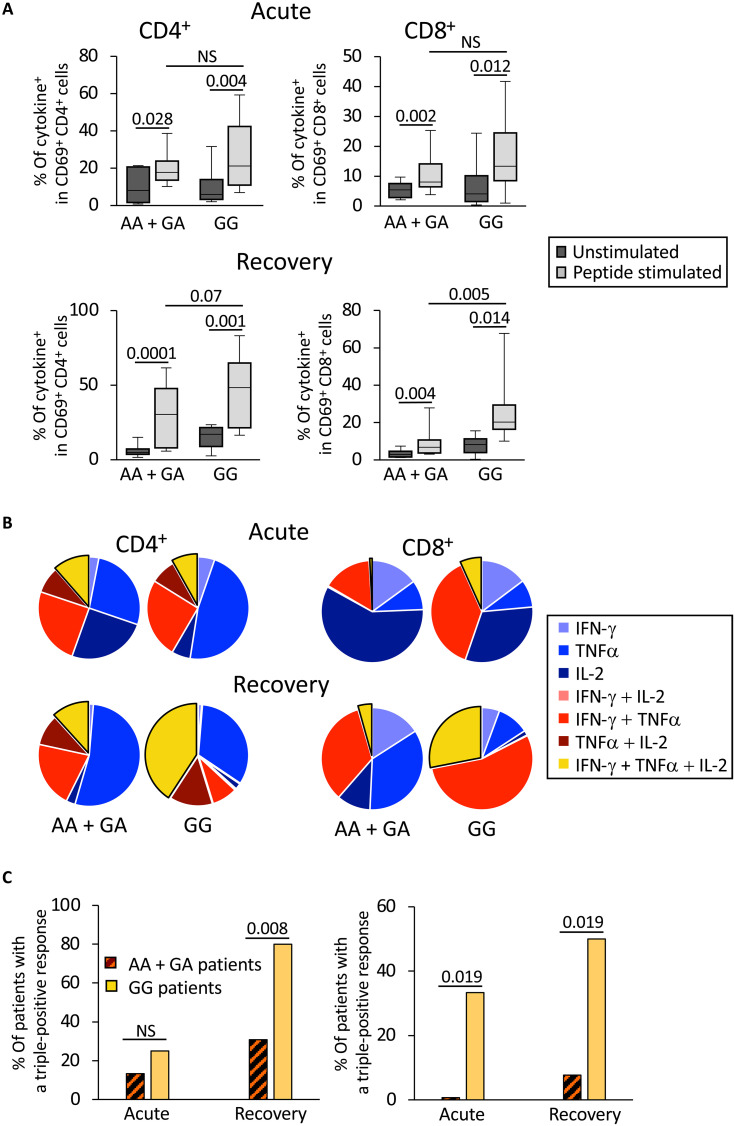
To estimate the impact of the higher thymopoïesis observed in GG patients on their ability to mount a more effective and sustained immune response against SARS-CoV-2, we compared virus-specific T cell responses in GG and AA + GA patients during acute disease and 6 months after recovery. Peripheral blood cells of patients were stimulated in vitro with a pool of 88 major histocompatibility complex (MHC) class I– and class II–restricted peptides derived from the whole proteome of SARS-CoV-2. Staining for intracellular IFN-γ, tumor necrosis factor (TNF-α), and interleukin-2 (IL-2) was performed to identify responder cells: The frequency of SARS-CoV-2–specific cells among activated (CD69+) CD3+CD4+ and CD3+CD8+ cells was assessed for each patient under peptide-stimulated and nonstimulated conditions (fig. S5). During both the acute and recovery phases of the infection, peptide-specific CD4 and CD8 T cell responses, identified by the expression of any combination of the tested cytokines upon peptide stimulation, were observed in both patient groups (Fig. 3A). During the recovery phase, although the median frequency of cytokine+ CD4+CD69+ cells was not significantly different between the GG and the AA + GA groups (46.6% versus 29.7%, P = 0.07), that of cytokine+ CD8+CD69+ cells was higher in GG patients (19.55% versus 6.3%, P = 0.005).
Fig. 3. SARS-CoV-2–specific T cell responses are stronger in patients with GG genotype at the rs2204985 locus than in AA + GA patients.
SARS-CoV-2– specific T cells in PBMCs were identified by cytokine expression after in vitro stimulation by a selected pool of viral peptides. (A) The frequency of cytokine-positive cells in in vitro–stimulated PBMCs sampled during acute disease (top) and after recovery (bottom), in either CD69+CD4+ (left) and CD69+CD8+ (right) is shown for both groups of patients. Statistical significance of the differences between peptide stimulated and unstimulated conditions as well as between the two groups of patients is shown on top (Mann-Whitney test). (B) Simple (IFN-γ, IL-2, or TNFα; blue sectors), double (any pair of cytokines; red sectors), and triple (all three cytokines; yellow sectors) expression of cytokines by CD69+CD4+ (left) and CD69+CD8+ right) T cells stimulated in vitro with SARS-CoV-2 peptides is represented as a pie chart. During the acute phase of the infection (top), the frequency of polyfunctional CD8+ T cells was significantly larger in GG patients than in AA + GA patients (P = 0.027). During the recovery phase (bottom), the proportion of both CD4+ and CD8+ polyfunctional T cells was significantly larger in GG patients (P = 0.011 and 0.003; respectively). The frequency of CD69+CD8+ cells expressing both IFN-γ and TNFα was significantly higher in GG patients at the second time point (P = 0.033). (C) The frequency of patients showing more than 5% of triple-positive (i.e., expressing IFN-γ, IL-2, and TNFα) cells among CD69+CD4+ (left) and CD69+CD8+ (right) T cells is shown for both groups (GG and AA + GA) during the acute and recovery phases. Statistical differences between AA + GA and GG patients at each time point are shown (Student’s t test).
The effect of GG genotype on the development of anti–SARS-CoV-2 T cell responses was further analyzed through the quantification of peptide-specific T cells expressing the various combinations of cytokines. Polyfunctional T cells expressing all three cytokines after peptide stimulation were significantly overrepresented in GG patients, during the acute phase in CD8+ T cells, and after recovery for both CD4+ and CD8+ responses (Fig. 3B and fig. S6). The overrepresentation of polyfunctional T cells in GG individuals was corroborated by the proportion of patients displaying ≥5% of CD4+ and CD8+ polyfunctional SARS-CoV-2–responding cells, in particular during the recovery phase (Fig. 3C; P = 0.008 for the CD4+ T cell response after recovery; P = 0.019 for CD8+ T cell response at both time points). Together, these data indicate that anti–SARS-CoV-2 blood helper and cytotoxic memory T cells are more effective in GG patients than in the other group, this advantage being already detectable for cytotoxic T cells during the acute phase.
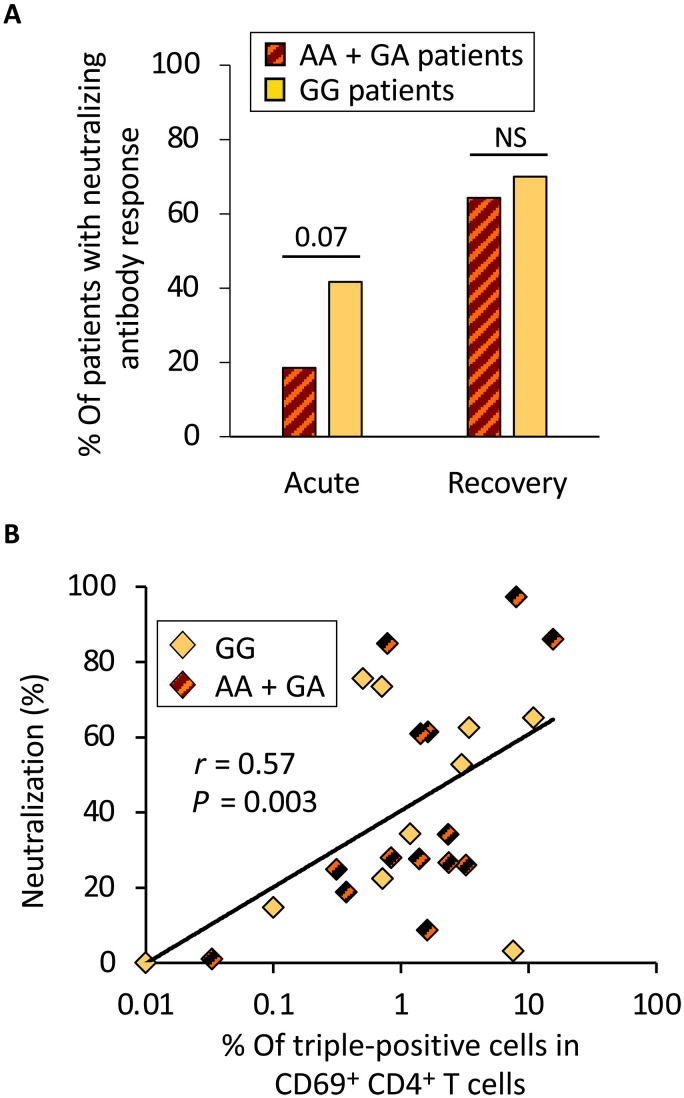
We next determined the SARS-CoV-2 neutralizing capacity of patient blood immunoglobulins (Ig) during acute COVID-19 and after recovery, by measuring their inhibitory effect on the interaction between the virus spike protein and its receptor ACE2 (Fig. 4). During acute disease, although this difference was not statistically significant, the percentage of patients of the GG group displaying neutralizing Ig was higher than in the other group (41.7% as compared to 18.5% In GG and AA + GA patients, respectively; P = 0.07; Fig. 4A). Consistent with the requirement of an appropriate helper T cell response for the development of neutralizing antibodies, the percentage of virus neutralization during acute disease was directly correlated with polyfunctional anti–SARS-CoV-2 CD4+ T cell response (Fig. 4B). Comparable frequencies were observed after recovery, likely because most patients received the first injection of mRNA vaccine, due to the initiation of the national anti–SARS-CoV-2 vaccination campaign between the two sampling dates (Fig. 4A).
Fig. 4. Neutralizing antibody response is stronger in patients with a GG genotype than in AA + GA patients.
(A) Antibody neutralizing activity was assessed by measuring the capacity of plasma to inhibit SARS-CoV-2 spike protein binding to its receptor (ACE2). The frequency of patients displaying a neutralizing antibody response (>50% neutralization at 1/80 dilution) is shown for both groups of patients (GG and AA + GA genotypes), at both time points. Statistical significance of the differences between the two groups of patients is shown on top (Mann-Whitney test). (B) The percentage of neutralization by patients’ plasma at the acute phase of the infection correlates with the frequency of triple-positive CD69+CD4+ T cells after in vitro stimulation with a selected pool of SARS-CoV-2 peptides. The correlation coefficient (Spearman’s r) and the associated probability (P) are shown.
Severe COVID-19 patients with pneumonia often experience systemic inflammation, caused by the release of numerous proinflammatory cytokines, chemokines, and IFNs, partly due to the viral activation of innate immunity defense mechanisms (16). We then investigated whether the rs2204985 polymorphism would affect blood cytokine levels in the context of SARS-CoV-2 infection. Fifteen of 31 tested cytokines and factors were significantly overexpressed in patients with COVID-19, as compared to ICU-hospitalized control individuals (Mann-Whitney test; Table 2). Ten of them were differentially more expressed in GG patients than in controls, including six (IL-15, IL17E, IL-21, IL-33, CCL3, and CCL20) that were not significantly different in AA + GA patients compared to controls. These data suggest that the global difference documented between patients with COVID-19 and controls is at least in part attributable to the enhanced expression in GG patients.
Table 2. Panel of cytokines, chemokines, and growth factors in patients with COVID-19 and controls.
The indicated cytokines, chemokines, and growth factors were measured in the serum of patients and controls during the initial hospitalization. Mean values are indicated. The comparisons between COVID patients and controls were performed using Mann-Whitney test. The comparisons between COVID patients (GG or GA + AA) and controls were performed using Kruskal-Wallis test and Dunn’s posttest.
| Cytokines, chemokines, and growth factors | Median value (pg/ml) | Mann-Whitney | Krustal-Wallis and Dunn’s posttest | |||||
|---|---|---|---|---|---|---|---|---|
| Control | COVID-19 | GG | AA + GA | COVID-19 versus Ctrls | AA + GA versus Ctrls | GG versus Ctrls | GG versus AA + GA | |
| IL-15 | 4.26 | 5.24 | 6.97 | 4.18 | 1.65 × 10−02 | NS | 3.7 × 10−04* | 6.1 × 10−03 |
| IL-17E | 0.00 | 0.47 | 1.47 | 0.00 | 6.3 × 10−04* | 1.6 × 10−02 | 1.2 × 10−04* | 4.9 × 10−02 |
| IL-21 | 0.12 | 0.57 | 0.88 | 0.49 | 1.1 × 10−03* | 2.6 × 10−02 | 2.7 × 10−04* | 1.5 × 10−02 |
| IL-33 | 0.13 | 0.57 | 1.59 | 0.48 | 4.1 × 10−06* | 7.3 × 10−03 | 4.8 × 10−06* | 1.3 × 10−02 |
| MIP-1α (CCL3) | 20.86 | 36.78 | 55.24 | 35.31 | 2.8 × 10−07* | 1.9 × 10−03 | 1.4 × 10−06* | 1.6 × 10−02 |
| MIP-3α (CCL20) | 9.9 | 18.5 | 34.7 | 16.8 | 6.6 × 10−04* | 1.6 × 10−02 | 6.4 × 10−06* | 8.8 × 10−03 |
| IL-7 | 0.40 | 3.43 | 6.36 | 2.78 | 1.3 × 10−09* | 1.6 × 10−05* | 2.9 × 10−06* | NS |
| IFN-α | 0.03 | 0.09 | 0.15 | 0.08 | 1.3 × 10−07* | 3.1 × 10−05* | 3.7 × 10−06* | NS |
| IFN-γ | 1.29 | 7.22 | 7.69 | 7.22 | 2.9 × 10−06* | 1.4 × 10−05* | 8.9 × 10−05* | NS |
| VEGF | 23.1 | 73.8 | 92.5 | 67.8 | 7.7 × 10−06* | 1.2 × 10−04* | 3.6 × 10−04* | NS |
| IL-10 | 0.26 | 0.62 | 0.64 | 0.58 | 9.7 × 10−05* | 5.9 × 10−04* | 3.8 × 10−03 | NS |
| GMCSF | 0.00 | 0.02 | 0.03 | 0.01 | 1.8 × 10−03* | 1.3 × 10−02 | 2.8 × 10−03 | NS |
| TSLP | 1.87 | 3.16 | 2.62 | 3.41 | 2.8 × 10−03* | 1.2 × 10−03 | NS | NS |
| TNF-α | 1.83 | 2.90 | 2.88 | 2.90 | 6.8 × 10−04* | 6.2 × 10−03 | 9.4 × 10−03 | NS |
| IL-8 | 2.64 | 6.14 | 6.89 | 5.89 | 5.9 × 10−04* | 5.6 × 10−03 | 6.6 × 10−03 | NS |
| MIP-1β (CCL4) | 54.0 | 77.4 | 86.6 | 70.2 | 1.2 × 10−03* | 4.1 × 10−02 | 7.6 × 10−03 | NS |
| IL-17F | 0.88 | 3.34 | 3.79 | 2.78 | 7.4 × 10−03 | 2.5 × 10−02 | 9.9 × 10−03 | NS |
| IL-23 | 0.06 | 0.68 | 0.54 | 0.71 | 6.8 × 10−03 | 2.7 × 10−02 | 4.8 × 10−02 | NS |
| IL-22 | 0.26 | 0.47 | 0.41 | 0.49 | 1.0 × 10−02 | 1.1 × 10−02 | NS | NS |
| IFN-β | 0.00 | 0.30 | 0.63 | 0.30 | 2.3 × 10−02 | 2.6 × 10−02 | NS | NS |
| SDF-1 (CXCL12) | 894 | 1085 | 1083 | 1085 | 1.2 × 10−02 | 1.3 × 10−02 | NS | NS |
| IL-1β | 0.06 | 0.10 | 0.06 | 0.14 | NS | NS | NS | NS |
| IL-4 | 0.00 | 0.00 | 0.01 | 0.00 | NS | NS | 8.2 × 10−03 | NS |
| IL-6 | 3.31 | 5.26 | 5.05 | 5.26 | NS | NS | NS | NS |
| IL-12 | 0.03 | 0.05 | 0.05 | 0.04 | NS | NS | NS | NS |
| IL-17A | 0.72 | 0.73 | 0.82 | 0.63 | NS | NS | NS | NS |
| IL-27 | 233 | 196 | 179 | 197 | NS | NS | NS | NS |
| MDC | 804 | 778 | 1001 | 761 | NS | NS | NS | NS |
| TGF-β1 | 4706 | 2917 | 5268 | 2327 | NS | NS | NS | NS |
| TGF-β2 | 4.89 | 7.38 | 10.65 | 4.99 | NS | NS | NS | NS |
| TGF-β3 | 0.15 | 0.20 | 0.27 | 0.16 | NS | NS | 2.5 × 10−02 | NS |
*Statistical significance after Holm’s correction is shown.
DISCUSSION
We investigated the functional significance of the rs2204985 polymorphism within the TCRA-TCRD locus in ICU patients with SARS-CoV-2–associated severe pneumonia. This polymorphism was previously reported to affect basal thymopoïesis levels in the general healthy population, independently of age and sex (9). Here, we demonstrate that it markedly impacts on thymic T cell production in the pathological context of SARS-CoV-2 infection, likely affecting the immunological parameters and the clinical presentation of the disease. During the acute phase, patients with the GG genotype showed a less severe pulmonary involvement; they also displayed enhanced release of new naïve CD4+ T lymphocytes into the blood, including CD4+ RTEs, compared to individuals harboring other genotypes (AA + GA). Patients with a GG genotype also displayed a larger population of CD4+ TSCM with an activated phenotype during the acute phase of the disease, which persisted 6 months later during the recovery phase.
Since the enhanced thymopoïesis observed in GG patients is a consequence of enhanced intrathymic T cell proliferation, it is expected to be associated with a larger T cell repertoire, as reported in human and mouse models (9, 12). Such a larger T cell repertoire should facilitate the selection of virus-specific T cell clones with enhanced affinity for viral peptides and thus allow the development of a more efficient specific immune response. Consistent with this hypothesis, we observed a larger expansion of CD4+ stem cell memory T cells harboring an activated phenotype during the acute phase in GG patients. Moreover, while a virus-specific CD4+ T cell response was detectable in all patients, it was globally stronger in GG individuals than in AA + GA patients, as shown by the higher proportion of patients displaying polyfunctional CD4+ and CD8+ T cell responses upon peptide stimulation 6 months after recovery (fig. S7, A and B). In contrast, polyfunctional T cell responses were not different between vaccinated and nonvaccinated individuals (fig. S7, C and D).
During the acute phase, there was a trend for a higher proportion of individuals exhibiting a neutralizing anti–SARS-CoV-2 humoral response among GG patients, the strength of the neutralizing antibody response being proportional to the anti–SARS-CoV-2–specific CD4+ T cell response. This difference was blunted during the recovery phase as a likely consequence of the anti–SARS-CoV-2 vaccination received by most patients during the 6-month interval, which boosted the neutralizing antibody response, as reported previously (17, 18). Whereas vaccinated individuals demonstrated a stronger neutralizing activity than nonvaccinated patients during the recovery phase, the percentage of neutralization was comparable in GG and AA + GA patients (fig. S7, E and F).
Since anti–SARS-Cov-2 vaccination with mRNA vaccines only marginally enhances virus-specific CD4+ T cell immunity in recovered patients (18, 19), the stronger capacity of GG patient to mount an efficient helper T cell response against SARS-Cov-2 likely reflects an intrinsic quality of GG individuals.
Although there was no significant difference between the two groups in circulating CD69+ stem cell memory CD8+ T cell counts, GG patients displayed polyfunctional virus-specific CD8+ T cells more often, at both acute and recovery phases. This observation indicates, as for the CD4+ response, that a stronger basal thymic function in GG patients facilitates the selection of virus-specific CD8+ T cell clones with enhanced affinity for viral peptides. Polyfunctionality of CD8+ T cells was reported to be proportional to the affinity of their T cell receptors for MHC-peptide complexes (20). Because of the higher thymic production of naïve T cells with a more diversified repertoire in GG patients, the probability of selecting CD8+ T cell clones with high affinity for SARS-CoV-2 is likely enhanced.
Consistent with most previous observations (21, 22), blood concentration of 15 cytokines, chemokines, and growth factors was increased in recently infected COVID-19 patients, compared to uninfected ICU-hospitalized controls. Among the cytokines overexpressed in GG patients, IL-17E, IL-21, IL-33, and CCL3, expressed by activated T cells, are known to participate in the development of antigen-specific T cell responses and might contribute to the robustness of anti–SARS-CoV-2 T cell responses in GG individuals. IL-15, specifically enhanced in the GG group, plays an essential homeostatic role for both naïve and memory T cells by providing proliferative and anti-apoptotic signals.
The relatively small size of our study group (40 individuals) is explained by the sudden drop in ICU-hospitalized COVID-19 patients with severe pulmonary involvement during the recruitment period. The systematic vaccination of all French population, which modified the “naïve” immunological status against SARS-CoV-2, likely plays an essential role in this event. However, our patients were all hospitalized in the same ICU, for severe COVID with pulmonary involvement, during the same period of time, contributing to limit heterogeneity.
The precise mechanisms leading to increased intrathymic precursor T cell proliferation in individuals with GG genotype have not been elucidated yet. The rs2204985 SNP is localized very close to the recombination site used for the excision of the TCRD locus leading to the generation of the sjTREC, in the vicinity of binding sites for transcription factors, which regulate thymocyte division. For example, the highly conserved 11-zinc finger protein CTCF has multiple binding sites near the TCRA segments (23, 24) In mice, CTCF is involved in V(D)J recombination and cell cycle progression of αβ T cells in the thymus and in the TCR up-regulation in double-positive thymocytes. Some CTCF-binding elements, localized in the close vicinity of the rs2204985, were shown to regulate TCRA repertoire diversity, probably as an indirect consequence of their impact on TCRD rearrangement (25). In the context of the lymphopenia caused by SARS-CoV-2 infection and of IL-7–stimulated thymopoïesis, the presence of a G at this particular position might allow immature thymocytes to divide more times between the recombination at the TCRB locus and the excision of the TCRD locus, leading to a more diversified T cell repertoire, as reported by Clave et al. (9).
In conclusion, we report the first demonstration that rs2204985 polymorphism can affect the clinical and immunological responses to severe viral infections, by affecting both the baseline production of T cells by the thymus and its infection-induced enhancement. Patients with the GG genotype, who display a greater basal thymic activity and a larger naïve T cell pool, were able to mount stronger and more efficient T cell responses against SARS-CoV-2. The analysis of the rs2204985 sequence variants might be included among the predictive tests in critically ill COVID-19 patients and more generally in severe infections and other clinical situations where stimulated thymopoïesis has mechanistic and/or prognostic importance.
MATERIALS AND METHODS
Patients
This observational study was conducted in 40 consecutive adult COVID-19 patients and 41 controls with COVID-19–unrelated disease, hospitalized in the ICU of the Clinique Ambroise Paré (Neuilly, France) from the beginning of March 2020 to the end of February 2022 (Table 1). A second sampling was gathered from a subgroup of 25 patients with COVID-19 who accepted a second investigation 6 months after recovery. Control individuals were admitted in the same ICU during the same period, for scheduled hospitalization in medical cardiology (n = 13), cardiac surgery (n = 24), spinal surgery (n = 4) and presented postoperative biological inflammatory syndrome. During the time period, this ICU, which, under normal circumstances, principally treats patients admitted for cardiovascular diseases, contributed to the national French program aimed at enhancing the hospitalization capacity for severely affected SARS-CoV-2–infected patients. The present study was approved by the French ethical committee (Comité de protection des personnes Est II; #20.11.06.73935); written informed consent was obtained from all patients upon admission.
Thoracic CT scans
Thirty-seven COVID+ and all COVID− patients had CT scans at admission. Thoracic CT scans were performed for the initial evaluation of pulmonary status upon admission. All images were independently reviewed and classified by two radiologists. In case of disagreement, they reviewed the scans together and were asked to give a final common classification, as described (6).
Cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from total blood on Ficoll gradient (Sigma-Aldrich) by centrifugation at 1000g for 20 min at room temperature. Cells were frozen in fetal calf serum 10% dimethyl sulfoxide and preserved in liquid nitrogen until use.
TREC quantification
TRECs were quantified by nested multiplex quantitative polymerase chain reaction (qPCR), adapted from (14). Briefly, 3.106 PBMCs were resuspended in a lysis buffer [tris-HCl (pH 8.0), 20 mM (Sigma-Aldrich); 0.1% NP-40; 0.1% Tween 20 (Sigma Aldrich); proteinase K (200 μg/ml; Eurobio)] and incubated for 30 min at 56°C. Proteinase K was heat-inactivated for 15 min at 95°C. Multiplex PCR amplification was performed for sjTREC together with the CD3γ chain, in a final volume of 100 μl (10-min initial denaturation at 95°C, then 22 cycles of 30 s at 95°C, 30 s at 60°C, and 2 min at 72°C) using outer 3′/5′ primer pairs described in (14). The qPCR conditions in LightCycler experiments, using inner primer pairs described in (14) and performed on 1/100th of the initial PCR products, were 1-min initial denaturation at 95°C, then 40 cycles of 1 s at 95°C, 10 s at 60°C, and 15 s at 72°C. Measurements of the fluorescent signals were performed at the end of elongation steps. sjTREC and CD3γ LightCycler quantifications were performed in independent experiments, using the same first-round serial dilution standard curve. Similarly, DJβ1TRECs (DJβ1.1-1.6) and DJβ2TRECs (DJβ2.1-2.7) were quantified in multiplex quantitative PCR assays, using the primers described in (14). This highly sensitive nested quantitative PCR assay made it possible to detect one TREC molecule in 105 cells for any excision circle. The sj/βTREC ratio was calculated according to the formula below, described in (13)
SNP sequencing
Because of the presence of numerous repeated sequences nearby, the region containing the rs2204985 SNP was amplified using nested PCR. A first round of 35 cycles PCR [95°C 10 min; (95°C 30 s; 60°C 30 s; 72°C 30s) *35] was performed using outer primers (rs2204985-Out5 5′-CAGGAAACAGCTATGACCGCTTGAACCCAAGGGGCTAT-3′ and rs2204985-Out3 5′-TAATACGACTCACTATAGGGGGTTTCCCACTGAGGAGTTT-3′). A second round PCR was performed on 1/10th of the initial PCR products with inner primers (rs2204985-In5 5′-CAGGAAACAGCTATGACCATATGGGCTACTGGCCTGAA-3′ and rs2204985-In3 5′-TAATACGACTCACTATAGGGATGCTGTGGGTTAGGCAGAA-3′) according to the same cycling protocol. Electrophoresis was performed in a 2% agarose gel, and amplicons were gel-purified using the Nucleospin gel and the PCR clean-up kit (Macherey Nagel) according to the manufacturer’s protocol. Samples were sent to the Eurofins Genomics platform for Sanger sequencing using T7 and M13 sequencing primers. Analysis was performed using the Geneious R7 software.
Flow cytometry phenotyping
Cells were stained for viability using the Zombie NIR fixable viability kit (BioLegend) according to the manufacturer’s protocol and then washed in Dulbecco's phosphate-buffered saline (D-PBS) without Ca2+ and Mg2+ (Gibco) by centrifugation 5 min at 600g at 4°C. Antibodies (anti-CD3:OKT3, anti-CD4:OKT4, anti-CD8:SK1, anti-CD45RA:Hl100, anti-CD45RO:UCHL1, anti-CD95:DX2, anti-CD27:M-T271, anti-CD28:CD28.2, and anti-CD31:WM59, all supplied by BioLegend) were then added and incubated 30 min at 4°C. Cells were washed twice in D-PBS without Ca2+ and Mg2+ and fixed in 2% paraformaldehyde. Spectral acquisition was performed on an Aurora flow cytometer (Cytek), and data were analyzed using the FlowJo 10 software.
Quantification of anti–SARS-CoV-2 T cell responses
Anti–SARS-CoV-2–specific T cell responses were quantified using the Miltenyi Biotec SARS-CoV-2 Select T Cell Analysis Kit (PBMC), in triplicate experiments, as recommended by the provider. Briefly, PBMCs were thawed and let to rest overnight at 37°C, 5%CO2 before use. Peptide stimulation was performed on 106 cells with the pool of 88 peptides defined by the provider on the entire proteome of SARS-CoV-2. Cells were then cultured for 6 hours, brefeldin A being added to the culture for the last 4 hours. Extra- and intracellular staining was lastly performed according to the recommended protocol with anti-CD3, anti-CD4, anti-CD8, anti-CD14, anti-CD20, anti-CD69, anti-CD154, anti–IFN-γ, anti-TNF-α, and anti–IL-2 antibodies and Viobility 405/452 Fixable Dye. Data were collected on an AURORA spectral cytometer and analyzed with FlowJo software (fig. S2).
Quantification of neutralizing antibodies
Neutralizing antibodies were quantified for their ability to inhibit the binding of the SARS-CoV-2 spike protein to its receptor Angiotensin-converting enzyme 2 (ACE2) using the SARS-CoV-2 Neutralizing Antibody enzyme-linked immunosorbent assay Kit (CD Creative Diagnostics), in duplicate, as recommended by the provider.
Statistical analyses
Descriptive statistics were used to present baseline characteristics for the entire population and for the various groups. Continuous variables are presented as mean ± SD or median (25 to 75% interquartile range), according to their distribution, and categorical variables are presented as the number of patients in each category and the corresponding percentages. No imputation was applied for missing values; patients were excluded from analysis.
Categorial ordinal variable were ranked in two groups to define a binary variable and then analyzed using logistic regression. On the basis of this analysis, we calculated the odds ratio (95% confidence interval).
Continuous variables were analyzed either in two or three groups, using Mann-Whitney nonparametric test, analysis of variance (ANOVA), or Kruskal-Wallis and Dunn’s tests in the eventuality of unequal variances. VassarStats website (Vassar University), Real Statistics 8.4 (Charles Zaiontz), Graphpad Prism version 9.3.1, and R/RStudio version 1.4.1106 for macOS software were used for the above-described statistical analyses. P < 0.05 was considered to be statistically significant.
Acknowledgments
We greatly acknowledge Genom’IC and Cybio core facilities at Institut Cochin. We thank both healthy volunteers and patients for participation in the study.
Funding: This work was supported by Inserm, CNRS, Université Paris Cité, and the ANRS/MIE (grant ANRS0289) from a charity fund of GMF.
Author contributions: Conceptualization: S.M. and R.C. Investigation: H.R., A.M., B.C.-D., S.F.-M., V.A.-F., P.C., and P.S. Management of the cohort of patients: P.S., C.N., F.B., and G.G. Supervision: R.C., A.C.-C., J.D., and P.S. Writing—original draft: S.M. and R.C. Writing—review and editing: S.M., R.C., A.M., A.C.-C., and J.D.
Competing interests: V.A.-F. received honoraria as member of a board for Gilead and Viiv. All other authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Table S1
Figs. S1 to S7
REFERENCES AND NOTES
- 1.F. Zhou, T. Yu, R. Du, G. Fan, Y. Liu, Z. Liu, J. Xiang, Y. Wang, B. Song, X. Gu, L. Guan, Y. Wei, H. Li, X. Wu, J. Xu, S. Tu, Y. Zhang, H. Chen, B. Cao, Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 395, 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.T. Chen, D. Wu, H. Chen, W. Yan, D. Yang, G. Chen, K. Ma, D. Xu, H. Yu, H. Wang, T. Wang, W. Guo, J. Chen, C. Ding, X. Zhang, J. Huang, M. Han, S. Li, X. Luo, J. Zhao, Q. Ning, Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ 368, m1091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Q. Zhang, P. Bastard, Z. Liu, J. Le Pen, M. Moncada-Velez, J. Chen, M. Ogishi, I. K. D. Sabli, S. Hodeib, C. Korol, J. Rosain, K. Bilguvar, J. Ye, A. Bolze, B. Bigio, R. Yang, A. A. Arias, Q. Zhou, Y. Zhang, F. Onodi, S. Korniotis, L. Karpf, Q. Philippot, M. Chbihi, L. Bonnet-Madin, K. Dorgham, N. Smith, W. M. Schneider, B. S. Razooky, H.-H. Hoffmann, E. Michailidis, L. Moens, J. E. Han, L. Lorenzo, L. Bizien, P. Meade, A.-L. Neehus, A. C. Ugurbil, A. Corneau, G. Kerner, P. Zhang, F. Rapaport, Y. Seeleuthner, J. Manry, C. Masson, Y. Schmitt, A. Schluter, T. Le Voyer, T. Khan, J. Li, J. Fellay, L. Roussel, M. Shahrooei, M. F. Alosaimi, D. Mansouri, H. Al-Saud, F. Al-Mulla, F. Almourfi, S. Z. Al-Muhsen, F. Alsohime, S. Al Turki, R. Hasanato, D. van de Beek, A. Biondi, L. R. Bettini, M. D'Angio, P. Bonfanti, L. Imberti, A. Sottini, S. Paghera, E. Quiros-Roldan, C. Rossi, A. J. Oler, M. F. Tompkins, C. Alba, I. Vandernoot, J.-C. Goffard, G. Smits, I. Migeotte, F. Haerynck, P. Soler-Palacin, A. Martin-Nalda, R. Colobran, P.-E. Morange, S. Keles, F. Colkesen, T. Ozcelik, K. K. Yasar, S. Senoglu, S. N. Karabela, C. Rodriguez-Gallego, G. Novelli, S. Hraiech, Y. Tandjaoui-Lambiotte, X. Duval, C. Laouenan; COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group, A. L. Snow, C. L. Dalgard, J. D. Milner, D. C. Vinh, T. H. Mogensen, N. Marr, A. N. Spaan, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, M. J. Ciancanelli, I. Meyts, T. Maniatis, V. Soumelis, A. Amara, M. Nussenzweig, A. Garcia-Sastre, F. Krammer, A. Pujol, D. Duffy, R. P. Lifton, S.-Y. Zhang, G. Gorochov, V. Beziat, E. Jouanguy, V. Sancho-Shimizu, C. M. Rice, L. Abel, L. D. Notarangelo, A. Cobat, H. C. Su, J.-L. Casanova, Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Wang, S. D. Wiktor, C. W. Cheng, K. J. Simmons, A. Money, L. Pedicini, A. Carlton, A. L. Breeze, L. McKeown, EFCAB4B (CRACR2A) genetic variants associated with COVID-19 fatality. medRxiv 01.17.22269412 (2022).
- 5.E. Pairo-Castineira, S. Clohisey, L. Klaric, A. D. Bretherick, K. Rawlik, D. Pasko, S. Walker, N. Parkinson, M. H. Fourman, C. D. Russell, J. Furniss, A. Richmond, E. Gountouna, N. Wrobel, D. Harrison, B. Wang, Y. Wu, A. Meynert, F. Griffiths, W. Oosthuyzen, A. Kousathanas, L. Moutsianas, Z. Yang, R. Zhai, C. Zheng, G. Grimes, R. Beale, J. Millar, B. Shih, S. Keating, M. Zechner, C. Haley, D. J. Porteous, C. Hayward, J. Yang, J. Knight, C. Summers, M. Shankar-Hari, P. Klenerman, L. Turtle, A. Ho, S. C. Moore, C. Hinds, P. Horby, A. Nichol, D. Maslove, L. Ling, D. McAuley, H. Montgomery, T. Walsh, A. C. Pereira, A. Renieri; GenOMICC Investigators; ISARIC4C Investigators; COVID-19 Human Genetics Initiative; 23andMe Investigators; BRACOVID Investigators; Gen-COVID Investigators, X. Shen, C. P. Ponting, A. Fawkes, A. Tenesa, M. Caulfield, R. Scott, K. Rowan, L. Murphy, P. J. M. Openshaw, M. G. Semple, A. Law, V. Vitart, J. F. Wilson, J. K. Baillie, Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98 (2021). [DOI] [PubMed] [Google Scholar]
- 6.P. Cuvelier, H. Roux, A. Couedel-Courteille, J. Dutrieux, C. Naudin, B. Charmeteau de Muylder, R. Cheynier, P. Squara, S. Marullo, Protective reactive thymus hyperplasia in COVID-19 acute respiratory distress syndrome. Crit. Care 25, 4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. An, X. Li, P. Qin, Y. Ye, J. Zhang, H. Guo, P. Duan, Z. He, P. Song, M. Li, J. Wang, Y. Hu, G. Feng, Y. Lin, Predicting model of mild and severe types of COVID-19 patients using Thymus CT radiomics model: A preliminary study. Math. Biosci. Eng. 20, 6612–6629 (2023). [DOI] [PubMed] [Google Scholar]
- 8.O. Berkan, I. Kiziloğlu, E. Keles, L. Duman, M. Bozkurt, Z. Adibelli, G. Oncel, N. Berkan, Y. Ekemen Keles, J. H. Jones, A. H. Inan, C. Solak, M. Emiroğlu, M. Yildirim, A. Dursun, E. Ilhan, A. Camyar, O. Inceer, A. Nart, M. B. Yilmaz, Does the Thymus Index Predict COVID-19 Severity? J. Comput. Assist. Tomogr. 47, 236–243 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E. Clave, I. L. Araujo, C. Alanio, E. Patin, J. Bergstedt, A. Urrutia, S. Lopez-Lastra, Y. Li, B. Charbit, C. R. MacPherson, M. Hasan, B. L. Melo-Lima, C. Douay, N. Saut, M. Germain, D. A. Tregouet, P.-E. Morange, M. Fontes, D. Duffy, J. P. Di Santo, L. Quintana-Murci, M. L. Albert, A. Toubert; Milieu Intérieur Consortium , Human thymopoiesis is influenced by a common genetic variant within the TCRA-TCRD locus. Sci. Transl. Med. 10, eaao2966 (2018). [DOI] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Z. Carico, M. S. Krangel, Chromatin Dynamics and the Development of the TCRα and TCRδ Repertoires. Adv. Immunol. 128, 307–361 (2015). [DOI] [PubMed] [Google Scholar]
- 12.C. Tsamadou, S. Gowdavally, U. Platzbecker, E. Sala, T. Valerius, E. Wagner-Drouet, G. Wulf, N. Kroger, N. Murawski, H. Einsele, K. Schaefer-Eckart, S. Freitag, J. Casper, M. Kaufmann, M. Durholt, B. Hertenstein, S. Klein, M. Ringhoffer, S. Frank, C. Neuchel, I. Rode, H. Schrezenmeier, J. Mytilineos, D. Fuerst, Donor genetic determinant of thymopoiesis rs2204985 impacts clinical outcome after single HLA mismatched hematopoietic stem cell transplantation. Bone Marrow Transplant. 57, 1539–1547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. L. Dion, J. F. Poulin, R. Bordi, M. Sylvestre, R. Corsini, N. Kettaf, A. Dalloul, M. R. Boulassel, P. Debre, J. P. Routy, Z. Grossman, R. P. Sekaly, R. Cheynier, HIV infection rapidly induces and maintains a substantial suppression of thymocyte proliferation. Immunity 21, 757–768 (2004). [DOI] [PubMed] [Google Scholar]
- 14.M. L. Dion, R. P. Sekaly, R. Cheynier, Estimating thymic function through quantification of T-cell receptor excision circles. Methods Mol. Biol. 380, 197–213 (2007). [DOI] [PubMed] [Google Scholar]
- 15.K. Talvensaari, E. Clave, C. Douay, C. Rabian, L. Garderet, M. Busson, F. Garnier, D. Douek, E. Gluckman, D. Charron, A. Toubert, A broad T-cell repertoire diversity and an efficient thymic function indicate a favorable long-term immune reconstitution after cord blood stem cell transplantation. Blood 99, 1458–1464 (2002). [DOI] [PubMed] [Google Scholar]
- 16.M. S. Diamond, T. D. Kanneganti, Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 23, 165–176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Sokal, G. Barba-Spaeth, I. Fernandez, M. Broketa, I. Azzaoui, A. de La Selle, A. Vandenberghe, S. Fourati, A. Roeser, A. Meola, M. Bouvier-Alias, E. Crickx, L. Languille, M. Michel, B. Godeau, S. Gallien, G. Melica, Y. Nguyen, V. Zarrouk, F. Canoui-Poitrine, F. Pirenne, J. Megret, J. M. Pawlotsky, S. Fillatreau, P. Bruhns, F. A. Rey, J.-C. Weill, C.-A. Reynaud, P. Chappert, M. Mahévas, mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants. Immunity 54, 2893–2907.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Lozano-Rodriguez, J. Valentin-Quiroga, J. Avendano-Ortiz, A. Martin-Quiros, A. Pascual-Iglesias, V. Terron-Arcos, K. Montalban-Hernandez, J. C. Casalvilla-Duenas, M. Bergon-Gutierrez, J. Alcami, J. Garcia-Perez, A. Cascajero, M. A. Garcia-Garrido, A. D. Balzo-Castillo, M. Peinado, L. Gomez, I. Llorente-Fernandez, G. Martin-Miguel, C. Herrero-Benito, J. M. Benito, N. Rallon, C. Vela-Olmo, L. Lopez-Morejon, C. Cubillos-Zapata, L. A. Aguirre, C. D. Fresno, E. Lopez-Collazo, Cellular and humoral functional responses after BNT162b2 mRNA vaccination differ longitudinally between naive and subjects recovered from COVID-19. Cell Rep. 38, 110235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. I. Samanovic, A. R. Cornelius, S. L. Gray-Gaillard, J. R. Allen, T. Karmacharya, J. P. Wilson, S. W. Hyman, M. Tuen, S. B. Koralov, M. J. Mulligan, R. S. Herati, Robust immune responses are observed after one dose of BNT162b2 mRNA vaccine dose in SARS-CoV-2-experienced individuals. Sci. Transl. Med. 14, eabi8961 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. P. Tan, A. B. Gerry, J. E. Brewer, L. Melchiori, J. S. Bridgeman, A. D. Bennett, N. J. Pumphrey, B. K. Jakobsen, D. A. Price, K. Ladell, A. K. Sewell, T cell receptor binding affinity governs the functional profile of cancer-specific CD8+ T cells. Clin. Exp. Immunol. 180, 255–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Wang, M. Jiang, X. Chen, L. J. Montaner, Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 108, 17–41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. Binayke, A. Zaheer, J. Dandotiya, S. K. Gupta, S. Mani, M. R. Tripathy, U. Madan, T. Shrivastava, Y. Kumar, A. K. Pandey, D. K. Rathore, A. Awasthi, Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19. Vaccines (Basel) 10, 1762 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. M. Choi, A. J. Feeney, CTCF and ncRNA Regulate the Three-Dimensional Structure of Antigen Receptor Loci to Facilitate V(D)J Recombination. Front. Immunol. 5, 49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. Heath, C. Ribeiro de Almeida, F. Sleutels, G. Dingjan, S. van de Nobelen, I. Jonkers, K.-W. Ling, J. Gribnau, R. Renkawitz, F. Grosveld, R. W. Hendriks, N. Galjart, CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 27, 2839–2850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. Chen, Z. Carico, H. Y. Shih, M. S. Krangel, A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat. Immunol. 16, 1085–1093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figs. S1 to S7