Abstract
Pathology studies of SARS-CoV-2 Omicron variants of concern (VOC) are challenged by the lack of pathogenic animal models. While Omicron BA.1 and BA.2 replicate in K18-hACE2 transgenic mice, they cause minimal to negligible morbidity and mortality, and less is known about more recent Omicron VOC. Here, we show that in contrast to Omicron BA.1, BA.5-infected mice exhibited high levels of morbidity and mortality, correlating with higher early viral loads. Neither Omicron BA.1 nor BA.5 replicated in brains, unlike most prior VOC. Only Omicron BA.5–infected mice exhibited substantial weight loss, high pathology scores in lungs, and high levels of inflammatory cells and cytokines in bronchoalveolar lavage fluid, and 5- to 8-month-old mice exhibited 100% fatality. These results identify a rodent model for pathogenesis or antiviral countermeasure studies for circulating SARS-CoV-2 Omicron BA.5. Further, differences in morbidity and mortality between Omicron BA.1 and BA.5 provide a model for understanding viral determinants of pathogenicity.
Infection with Omicron BA.5, but not BA.1, leads to severe morbidity and mortality in partially aged K18 hACE-2 mice.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). Since the emergence of SARS-CoV-2 in November 2019, several new variants of concern (VOC) have emerged and spread globally, including Alpha, Beta, Gamma, Delta, and Omicron. At this time, Omicron has replaced Delta as the major circulating VOC; it first emerged at the end of November 2021 as what is now known as the Omicron BA.1 subvariant and has continued to evolve. The BA.1 lineage was replaced by the BA.2 and BA.4/5 lineages, with Omicron BA.5 now one of the predominant sublineages globally with its continued evolution likely along with second-generation BA.2 lineages, simple and complex recombination, antigenic drift, and convergent evolution (1). Hence, Omicron lineages have remarkable diversity, being roughly as distinct from each other as Alpha, Beta, Gamma, and Delta are to one another (2, 3). Especially concerning are the remarkable differences in the spike (S) protein of Omicron lineages, with more diversity in antigenic regions than previous VOC. The receptor binding domain (RBD) of Omicron S protein has multiple substitutions that alter species tropism, particularly to mouse angiotensin-converting enzyme 2 (ACE2) (4, 5). Wild-type mice have been shown to be readily infected by Omicron BA.1 and BA.2, unlike other variants, although not resulting in pathogenic phenotypes (5–8). SARS-CoV-2 S protein has been the major target for vaccines and monoclonal antibody treatments (9, 10). With more than 30 mutations in the S protein, vaccines and monoclonal antibody treatments have shown reduced efficacy in mice and humans against Omicron variants, especially the BA.4 and BA.5 lineages (11–13).
Pathology studies and preclinical development of antiviral countermeasures against SARS-CoV-2 Omicron VOC have been hindered by the lack of pathological small animal models (7, 14). First, such models would be highly useful to understand the drivers of pathology based on both intrinsic differences among non-Omicron and Omicron SARS-CoV-2 VOC, as well as their corresponding host immune responses. Further, the development of vaccines and antivirals, either prophylactic or therapeutic, would greatly benefit from such pathological rodent models of SARS-CoV-2 Omicron infection. Our study addresses these needs, providing evidence for host determinants important in Omicron pathogenesis.
Inflammatory cells and their cytokines have been implicated in progression of severe COVID-19 disease, which can result in cytokine release syndrome (CRS), an extreme inflammatory response leading to acute respiratory distress syndrome (ARDS) (15). Some key cytokines in this process include interleukin-6 (IL-6), tumor necrosis factor–α (TNF-α), IL-1, IL-2, and interferon-γ (IFN-γ), although more than 38 cytokines have been associated with increased disease severity (16). Beyond the lung, CRS may contribute to breakdown of the blood-brain barrier, leading to disruption of the central nervous system (CNS) and neurological symptoms, with or without direct brain infection (17), although brain infection has been observed in humans (17, 18). Direct brain infection of K18-hACE2 mice has been observed with Alpha, Beta, and Delta VOC, although brain infection has not yet been seen with Omicron (19).
Multiple SARS-CoV-2 VOC, including Alpha, Beta, Gamma, and Delta, have shown severe and lethal infection in K18-hACE2 mice (20). However, Omicron lineage viruses BA.1 and BA.2 have resulted in minimal to negligible pathogenesis, despite establishing infections (5–8, 14). More recent lineages, such as Omicron BA.5, remain minimally tested in mouse models. Here, we compare infection of Omicron BA.1 and BA.5 in K18-hACE2 mice, chosen for their high expression of hACE2, driven by the cytokeratin-18 gene promoter (21). We report severe disease and lethality of the Omicron BA.5 lineage, but in agreement with previous reports, not with Omicron BA.1. Further, we confirm that Omicron BA.5 causes more robust lung inflammation, immune infiltrates, and proinflammatory cytokine levels, yet at similar viral burden to Omicron BA.1. Furthermore, we observe an effect of age on disease outcome for SARS-CoV-2 Omicron BA.5 in this pathological rodent model.
RESULTS
SARS-CoV-2 VOC Omicron BA.5 induces lethal disease in K18-hACE2 transgenic mice
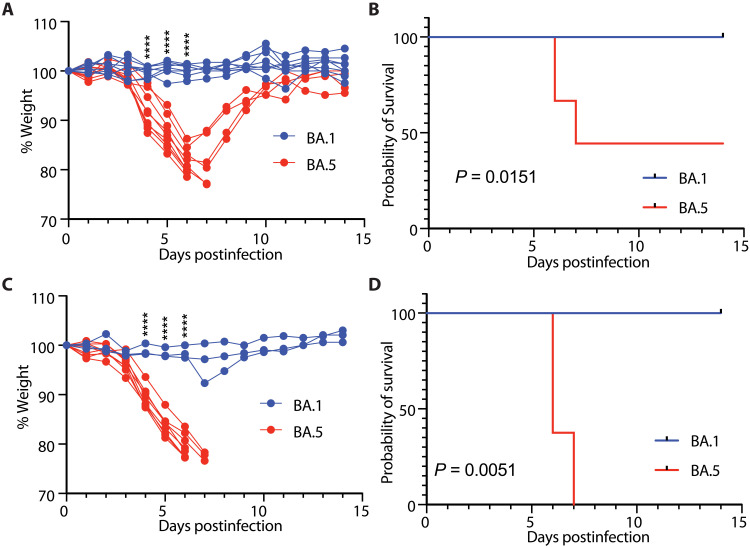
We tested whether the new circulating SARS-CoV-2 VOC Omicron BA.5–induced disease in K18-hACE2 transgenic mice, compared to Omicron BA.1 (22–24). First, 3-month-old and 5- to 8-month-old mice were intranasally infected with 3.25 × 104 plaque-forming units (PFU) of Omicron BA.1 or BA.5, and surviving mice were weighed and monitored through 14 days postinfection (dpi). Similar to prior reports, Omicron BA.1 mice did not lose significant weight following infection (Fig. 1A) (14). However, 3-month-old Omicron BA.5–infected mice started displaying significant weight loss 4 dpi compared to Omicron BA.1–infected mice (Fig. 1A). By 6 to 7 dpi, five of nine of the Omicron BA.5–infected mice had reached humane standards for euthanasia (<80% of their original weight) (Fig. 1, A and B). The surviving Omicron BA.5–infected mice regained weight, and on 14 dpi, their weights were comparable to Omicron BA.1–infected mice (Fig. 1A). All mice infected with Omicron BA.1 lost little to no weight and survived the infection to endpoint (Fig. 1B). We then tested the effect of host age on this response. The 5- to 8-month-old mice infected with Omicron BA.1 did not lose weight following infection (Fig. 1C). However, Omicron BA.5 infection caused significant weight loss starting on 3 to 4 dpi and required euthanasia as early as 6 dpi, reaching 100% mortality (Fig. 1, C and D). Together, while Omicron BA.1 infection did not result in significant weight loss or mortality in K18-hACE2 mice, Omicron BA.5 infection resulted in significant weight loss, frequently leading to lethality, particularly in older mice (Fig. 1, B and D).
Fig. 1. SARS-CoV-2 Omicron BA.5–induced lethality in K18-hACE2 transgenic mice is exacerbated with age.
(A) Three-month-old K18-hACE2 mice (BA.1 n = 8, BA.5 n = 9) were infected with 3.25 × 104 PFU SARS-CoV-2 Omicron BA.1 or BA.5, and weight was monitored daily. (B) Percent survival following SARS-CoV-2 Omicron BA.1 or BA.5 infection. (C) Five- to 8-month old K18-hACE2 mice (BA.1 n = 3, BA.5 n = 8) were infected with 3.25 × 104 PFU SARS-CoV-2 Omicron BA.1 or BA.5, and weight was monitored daily. (D) Percent survival following SARS-CoV-2 Omicron BA.1 or BA.5 infection. Two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test (A and C). ****P < 0.0001. Log-rank (Mantel-Cox) test (B and D).
Similar virus titers but distinct pathology follow SARS-CoV-2 Omicron BA.1 versus BA.5 infection in K18-hACE2 mice
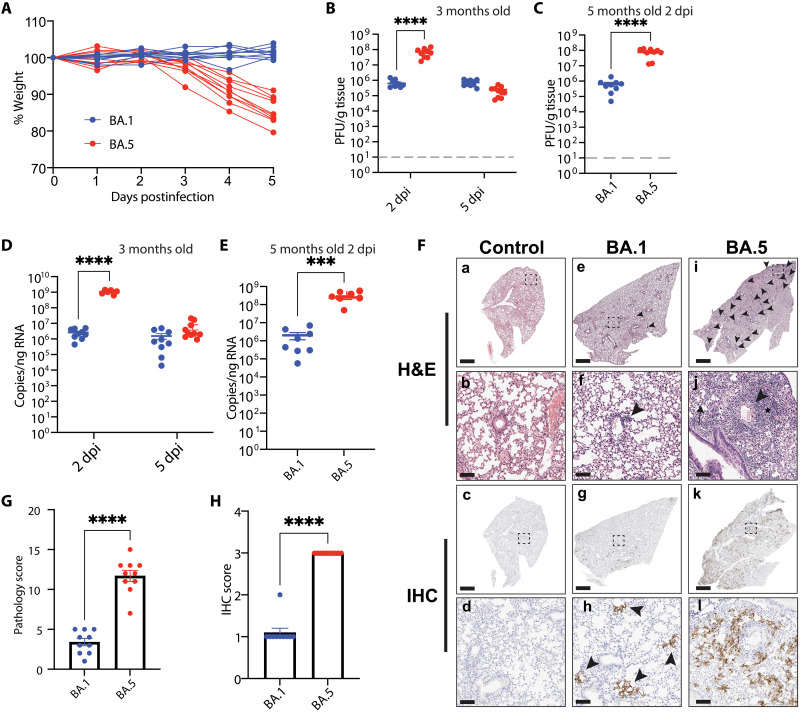
To gain additional insight into the Omicron BA.5–induced weight loss and mortality, lungs and brains were collected from Omicron BA.1– or BA.5–infected mice at their humane endpoint or at 14 dpi if they survived infection, to analyze the pathology and virus dispersion in the tissue at the time of euthanasia. Hematoxylin and eosin (H&E) staining of lung sections showed an increase in interstitial and perivascular infiltrates, accompanied with increase alveolar cells in the airways in Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (fig. S1A). Higher histopathological scores were also observed in Omicron BA.5–infected mice as compared to Omicron BA.1 (fig. S1B), consistent with over half of the Omicron BA.5–infected animals succumbing to severe disease. Immunolabeling of the SARS-CoV-2 nucleocapsid (N) protein showed an increase in positively labeled cells in Omicron BA.5 as compared to Omicron BA.1–infected mice (fig. S1A). While all Omicron BA.5–infected mice euthanized 6 and 7 dpi (triangles) were virus positive by immunolabeling, those that survived until 14 dpi (circles) were negative (fig. S1C). Five- to 8-month-old mice displayed similar trends for lung pathology scores and immunohistochemistry (IHC) quantification (fig. S1, D to F). To further confirm the presence of virus following infection, we quantified both infectious viral particles via plaque assays and viral RNA via quantitative polymerase chain reaction (qPCR) from lungs and brains. Infectious virus was not detected in mice euthanized at 14 dpi (circle), but 3 of 5 3-month-old Omicron BA.5–infected mice that reached endpoint before 14 dpi were positive (fig. S1G). Viral RNA in the lung was detected in 2/8 of the Omicron BA.1–infected mice and 2/4 of the Omicron BA.5–infected mice from 14 dpi (fig. S1H). The majority of Omicron BA.5–infected mice which were euthanized before 14 dpi were positive for viral RNA in the lung (fig. S1H).
In the older mice, while virus was not detected in Omicron BA.1–infected mice which survived to 14 dpi, virus was detected in Omicron BA.5–infected mice which reached endpoint at either 6 or 7 dpi (fig. S1I). Viral RNA was detected in 1/3 of the Omicron BA.1–infected mice and in all the Omicron BA.5–infected mice (fig. S1J). In contrast to our prior findings for the original WA-1 or Delta strains, in which viral infection was detected in the brain (25), little to no infectious virus, nor viral RNA, was detected in the brain of mice of any age (fig. S1, K and L). Largely, Omicron BA.5–infected mice had higher levels of virus and pathology at endpoint, although this difference was skewed as the majority of the Omicron BA.5–infected animals were euthanized at an early humane endpoint. Thus, experiments where animals were all euthanized at the same time point postinfection were needed to make fair comparisons and were subsequently conducted.
Virus load and histopathology at 5 dpi differentiate SARS-CoV-2 Omicron BA.5 from BA.1 infection in K18-hACE2 mice
To more closely compare the dynamics of Omicron BA.1 and BA.5, we infected 3.5- and 5-month-old mice with 3.25 × 104 PFU. Oral swabs were collected 24 and 48 hours postinfections to monitor early viral shedding in the oral cavity. In addition, we collected lungs and brains to assess viral load, H&E, and IHC at 2 and 5 dpi. Bronchoalveolar lavage fluid (BALF) and lymph nodes were also harvested for cytokine profiling and immune cell analysis. Consistent with our initial observations (Fig. 1, A and C), Omicron BA.1–infected mice did not lose weight following infection (Fig. 2A); however, Omicron BA.5–infected mice lost on average 15% of their original weight by 5 dpi (Fig. 2A). Although not significant, more viral RNA was detected in the oral cavity of 3-month-old mice infected with Omicron BA.5 than with Omicron BA.1 at 24 and 48 hours postinfection (fig. S2, A and B). Significantly more viral RNA was detected in the oral cavity of 5-month-old Omicron BA.5–infected mice than Omicron BA.1–infected mice, both at 24 and 48 hours postinfection (fig. S2, A and B). Higher titers of infectious virus were observed in the lungs of 3-month-old Omicron BA.5–infected mice at 2 dpi compared to Omicron BA.1–infected mice (Fig. 2B). Infectious titers were comparable in both Omicron BA.1– and Omicron BA.5–infected mice at 5 dpi (Fig. 2B). Further analysis of the viral plaques revealed that Omicron BA.5 plaques were significantly larger than Omicron BA.1 plaques, potentially linked to the difference of pathogenicity observed (fig. S2, C and D) (26, 27). Consistent with the observation in 3-month-old mice, 5-month-old Omicron BA.5–infected mice had significantly higher titers of virus in the lungs than Omicron BA.1–infected mice (Fig. 2C). In accordance with infectious titers, higher viral RNA was detected in the Omicron BA.5–infected mice at 2 dpi than in the Omicron BA.1–infected mice (Fig. 2D). At 5 dpi, the levels were similar in both Omicron BA.5– and BA.1–infected mice (Fig. 2E). Analogously, viral RNA was significantly higher in 5-month-old Omicron BA.5–infected mice than Omicron BA.1–infected mice at 2 dpi (Fig. 2E). Correlating with the weight changes, we observed significantly more pathology in the lungs of Omicron BA.5–infected mice compared to Omicron BA.1–infected mice 5 dpi, characterized by interstitial and perivascular infiltrates, increased alveolar cells in the airways and alveolar edema as determined by histopathology (Fig. 2, F and G). Immunolabeling for SARS-CoV-2 nucleocapsid was more widespread in the lungs of mice infected with Omicron BA.5 compared to Omicron BA.1 (Fig. 2, F and H).
Fig. 2. SARS-CoV-2 Omicron BA.5 induces increased pathology at 5 dpi.
(A) Three-month-old K18-hACE2 mice (n = 10 per Omicron variant) were infected with 3.25 × 104 PFU of SARS-CoV-2 Omicron BA.1 or BA.5. Weights were monitored daily, and mice were euthanized 5 dpi. Two-way ANOVA with Sidak’s multiple comparisons test. (B) Lung viral titers. Tissues were collected from 3-month-old infected mice 2 and 5 dpi and homogenized, and virus was collected for plaque assay. Two-way ANOVA with Tukey’s multiple comparisons test. Data are means ± SEM. (C) Lung viral titers in the lung of 5-month-old mice 2 dpi. Two-tailed Mann-Whitney test. Data are means ± SEM. (D) Viral RNA in the lungs of 3-month-old mice at 2 and 5 dpi by qRT-PCR. Two-way ANOVA with Tukey’s multiple comparisons test. Data are means ± SEM. (E) Viral RNA in the lungs of 5-month-old mice 2 dpi. Two-tailed Mann-Whitney test. Data are means ± SEM. (F) Representative H&E staining (a, b, e, f, i, and j) of lung histopathology or IHC staining with an anti–SARS-CoV-2 N protein antibody (c, d, g, h, k, and l) (one section analyzed per mouse) at death or study endpoint in control (a to d) or mice infected with Omicron BA.1 (e to h) or Omicron BA.5 (i to l). Perivascular infiltrates (arrowheads) were more numerous in mice infected with Omicron BA.5 compared to Omicron BA.1. Mice infected with Omicron BA.5 developed interstitial infiltrates (asterisk) and inflammatory cells in alveoli (arrow). Scale bars, 1 mm (a, c, e, g, i, and k) and 100 μm (b, d, f, h, j, and l). For histopathology and IHC analyses, representative images were selected on the basis of mean pathology score. (G) Quantification of the histopathology in the lung. Two-tailed Mann-Whitney test. Data are means ± SEM. (H) Quantification of virus nucleoprotein positive lung sections. Two-tailed Mann-Whitney test. Data are means ± SEM. ***P < 0.001, ****P < 0.0001.
Earlier SARS-CoV-2 VOC Alpha, Beta, and Delta have been shown to result in productive infection in the brain of mice, albeit at varying levels (22, 24, 28–30). However, we observed no infectious virus, viral RNA, pathology, or SARS-CoV-2 N protein immunolabeling in the brains of Omicron BA.1– or Omicron BA.5–infected mice at 5 dpi, further confirming the absence of viral infection in the brain (fig. S2, A, B, and D). Overall, we found that at 5 dpi, both Omicron BA.1 and BA.5 mice were infected; however, only Omicron BA.5 induced significant weight loss and pathology. Omicron BA.5 established a more robust infection than Omicron BA.1 in the lungs early during infection, but viral titers were comparable by 5 dpi, at which point morbidity and pathology were drastically different. In addition, no brain infection was observed at 5 dpi, further differentiating Omicron BA.1 and BA.5 as likely non-neurotropic viruses compared to our own and other groups’ prior SARS-CoV-2 VOC studies (22, 24, 25, 28, 29, 31).
Cellular immune profiling at 5 dpi differentiates inflammation induced by SARS-CoV-2 Omicron BA.5 versus BA.1
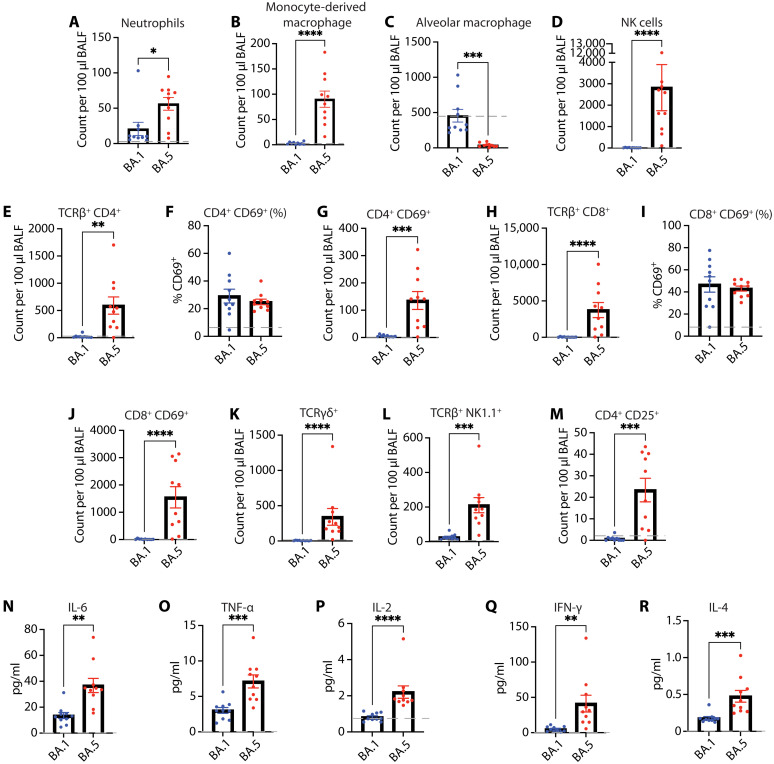
Immune responses to SARS-CoV-2 infection are vital for efficient virus clearance (32), but hyperactivation and dysregulation of the immune response have been associated with severe disease outcomes in both humans and animal models (33–35). Both innate and adaptive immune responses have been implicated in progression of severe disease phenotypes resulting in development of CRS or ARDS (36, 37). Therefore, we characterized the immune cell profile of K18-hACE2 mice at 5 dpi with Omicron BA.1 or BA.5 to better understand the underlying responses (fig. S3). High neutrophil counts and excessive monocyte/macrophage activation have been correlated with severe cases of COVID-19 (38, 39). We observed increased neutrophils (Fig. 3A) and monocyte-derived macrophages (Fig. 3B) in the BALF of Omicron BA.5–infected mice compared to Omicron BA.1. By contrast, Omicron BA.5–infected mice had significantly less alveolar macrophages than Omicron BA.1 or mock-infected (gray line) mice (Fig. 3C). This could potentially be attributed to cell death or pyroptosis that could be resultant of direct infection or Fc receptor–mediated entry (40, 41). We also found significantly higher numbers of natural killer (NK) cells in the BALF of Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3D).
Fig. 3. SARS-CoV-2 Omicron BA.5 infection induces increased cellular immune response at 5 dpi.
K18-hACE2 mice were infected with SARS CoV-2 Omicron BA.1 or BA.5 (n =10 per Omicron variant). At 5 dpi, BALF were collected, and cells were characterized by flow cytometry. Gray dashed line represents average values for mock-infected mice. (A) Number of neutrophils. (B) Number of monocyte-derived macrophages 5 dpi. (C) Alveolar macrophages. (D) NK cells. (E) Number of CD4+ T cells. (F) Percent CD4+ CD69+ T cells. (G) Number of CD4+ CD69+ T cells. (H) Number of CD8 + T cells. (I) Percent CD8+ CD69+ T cells. (J) Number of CD8+ CD69+ T cells. (K) Number of γδ T cells. (L) Number of NK1.1+ TCRβ+ NK T cells. (M) Number of CD4+ CD127Lo CD25Hi Tregs. (N to R) Cytokine expression in BALF 5 dpi. Levels of expression of (N) IL-6, (O) TNF-⍺, (P) IL-2, (Q) IFN-γ, and (R) IL-4. A two-tailed unpaired t test used to analyze data that followed a normal distribution and Mann-Whitney test for data not normally distributed. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, we broadly investigated the T cell compartment, due to their known involvement in severe COVID-19 disease progression (42, 43). We observed significantly more CD4+ T cells in the BALF of Omicron BA.5–infected mice as compared to Omicron BA.1–infected mice (Fig. 3E). Total CD4+ T cell increase in the lung was mirrored in the mediastinal lymph node (fig. S4A). Further characterization of these cells uncovered no differences in the percent of CD69+-activated CD4+ T cells between the groups, both in the BALF and lymph node (Fig. 3F and fig. S4B). However, Omicron BA.5–infected mice had significantly higher numbers of activated CD4+ T cells compared to Omicron BA.1–infected mice (Fig. 3G). In the CD8+ T cell subset, we saw high numbers of total CD8+ T cells in the BALF of Omicron BA.5–infected mice compared to BA.1 (Fig. 3H), which were also mirrored in the mediastinal lymph node (fig. S4C). No differences in percent-activated T cells were observed between the two groups both in BALF and mediastinal lymph node (Fig. 3I and fig. S4D). While the percent of CD69+-activated CD8+ T cells was not different, analogous to the CD4+ T cells, we saw a significant increase in the number of activated CD8+ T cells in the BALF of Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3J). We also investigated the levels of γδ T cells and found that their numbers too were elevated in the BALF of Omicron BA.5–infected mice compared to BA.1, but similar levels were detected in the lymph node (Fig. 3K and fig. S4E). NK T cells (NK1.1+ TCRαβ+), which have been associated with both positive and negative outcomes following SARS-CoV-2 infection in humans (43–46), were increased in Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3L). We also analyzed the number of B cells in the lymph node, and no differences were noted between groups at this stage of infection (fig. S4F), which was not unexpected as B cell responses typically take more time to amplify. Last, we looked at the number of regulatory T cells (CD4+ CD25Hi Tregs) which can express IL-10, an important cytokine in preventing excessive inflammation during virus infection (47), but which have been associated with severe disease in COVID-19 (48, 49). The number of Tregs was significantly up-regulated in Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3M).
To gain insights into the cytokine profile in Omicron BA.1– and Omicron BA.5–infected mice, we evaluated secreted cytokines in the BALF at 5 dpi. The levels of proinflammatory cytokines IL-6 and TNF-α were significantly higher in Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3, N and O). The levels of the T helper cell 1 (TH1) cytokines (50), IL-2 and IFN-γ, were also elevated in Omicron BA.5–infected mice compared to Omicron BA.1–infected mice (Fig. 3, P and Q). Last, we found that the levels of the TH2 cytokine IL-4 was higher in Omicron BA.5–infected mice compared to Omicron BA.1 (Fig. 3R).
Overall, we observed an increase in immune infiltrates and proinflammatory cytokines in the lungs of Omicron BA.5–infected mice compared to Omicron BA.1–infected mice at 5 dpi. The extent to which they contribute to the pathology and mortality observed in the Omicron BA.5–infected mice is an area that requires further investigation, but reports with prior VOC have correlated an increase in immune cells and proinflammatory cytokines with increased pathology and disease (34, 35).
Whole genome sequence comparison of SARS-CoV-2 Omicron BA.1 and BA.5 evaluates differences between the VOC
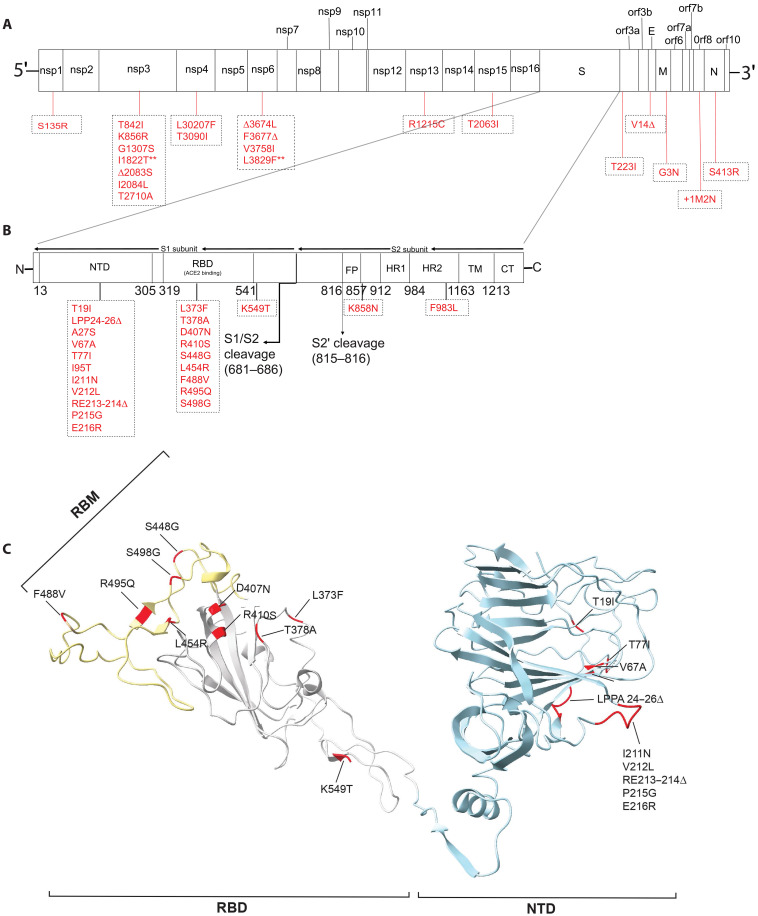
To better understand the different phenotypes following SARS-CoV-2 Omicron BA.1 and BA.5 infections, we sequenced the Omicron variants used in the aforementioned infections to determine whether there were unique mutations that might explain the Omicron BA.5–induced pathogenesis. The percentage nucleotide similarity between our Omicron BA.1 (OP984771) and BA.5 (OP984772) sequences with the representative sequences of each group (BA.1 OL672836.1 and BA.5 ON249995.1) was 99.9 and 99.8%, respectively, with only one unique amino acid mutation (nsp6 L3829F, summarized in Fig. 4A). As expected, the majority of the differences between our Omicron BA.1 and BA.5 isolates were localized in the S protein (Fig. 4B). These were mostly in the N-terminal domain (NTD) and the RBD of the S1 subunit. Several NTD mutations clustered in short flexible loops that likely represent antigenic sites (51), with RBD mutations generally outside of the ACE-2–interacting residues (Fig. 4C and fig. S5) (52). Only two mutations were observed in the S2 subunit, K858N upstream of the HR1 domain (just outside of the fusion peptide) and F983L in the HR2 domain. The next set of mutations were in ORF1a and included mutations in nsp1, nsp3, nsp4, and nsp6 and one unique substitution in nsp6 (L3826F), (Fig. 4A). nsp3, nsp4, and nsp6, which displayed the most variability between Omicron BA.5 and BA.1, have been shown to be important for SARS-CoV-1 replication, particularly in the formation of double-membrane vesicles (53–55). In ORF1b, mutations were observed in nsp13 and nsp15, a T223I substitution in ORF3a, a deletion was found in the envelope (E) protein V14D, with a G3N substitution in the matrix (M) protein, two amino acid insertions +1M and +2N in ORF8, which has been shown to dampen CD8+ T cell response by down-regulation major histocompatibility complex I (MHC-I) (56), and a S413R substitution in the N protein (Fig. 4A).
Fig. 4. Comparison of SARS-CoV-2 Omicron BA.1 and BA.5 genome sequences.
(A) Full genome comparison of Omicron BA.5 to BA.1. In red are amino acid differences, ** represent mutations unique to our viral stock, and Δ represent a deletion. (B) S protein comparison between Omicron BA.5 and BA.1. (C) Omicron BA.1 S protein [structure source Protein Data Bank (pdb): 7tgw]. Blue, NTD; grey, RBD; yellow, Receptor-binding motif; red, mutated residues in Omicron BA.5.
Collectively, Omicron BA.5 is distinct from Omicron BA.1, with most of the mutations in the S protein and ORF1a. A comparison to Wuhan-Hu-1 S is also shown in fig. S5. In comparing SARS-CoV-2 WA1/USA to Omicron BA.1, Liu et al. (4) showed that a region between nsp1 and nsp12 is most likely important for the difference in pathogenicity. Further studies are required to better understand if Omicron BA.5 has regained these mutations and if they are driving the pathology observed.
DISCUSSION
SARS-CoV-2 Omicron variants BA.1 and BA.2 result in attenuated disease and low mortality in mouse and hamster models of disease (14, 57). The inability to cause significant morbidity and mortality has limited the testing of vaccines and antiviral therapies in small animal models, as well as our understanding of viral drivers of pathogenesis. Here, we report that Omicron BA.5 causes significant disease and mortality in K18-hACE2 mice, which was age dependent, with 5- to 8-month-old mice displaying increased morbidity and mortality compared to 3-month-old animals. Our results agree with previous reports showing the importance of age in SARS CoV-2 pathogenicity (58–61). It has been shown that the initial activation and priming of both the innate and adaptive immune response can be delayed or dysregulated with aging (38, 62–65). This dysregulation can lead to reduced effectiveness while being accompanied by heightened inflammation (38). In addition, factors such as ineffective T cell priming, a decrease in the naïve T cell repertoire, and slower antibody maturation can result in inadequate adaptive immune responses to SARS-CoV-2 and correlate with older age (65–67). These aspects of immunosenescence may contribute to increased pathogenicity in older mice and humans while also leading to exacerbated inflammation.
We also report that Omicron BA.5 establishes infection faster than Omicron BA.1, with significantly higher viral levels observed in the lungs of Omicron BA.5–infected mice than Omicron BA.1–infected animals particularly early postinfection. It is possible that this rapid replication both in the oral cavity and the lungs of Omicron BA.5 mice triggers severe lung injury leading to the increased pathogenicity observed. Although the levels of infectious virus are comparable between Omicron BA.1– and Omicron BA.5–infected mice at 5 dpi, IHC data show higher levels of viral N protein in the lungs, alluding to higher amounts of viral antigen in the lungs of Omicron BA.5–infected mice. These findings suggest that Omicron BA.5 established a robust infection faster than Omicron BA.1 (68). In addition, that K18-hACE2 mice could be used as a model for pathological, vaccination, and antiviral studies of the VOC Omicron BA.5.
Studies using SARS-CoV-2 Omicron VOC have produced variable outcomes in the ability to induce morbidity and mortality in K18-hACE2 mice (3, 4, 14, 57, 69). Shuai et al. (69) observed weight loss and lethality in 47% of K18-hACE2 mice infected with SARS-CoV-2 B.1.1.529 Omicron. In contrast, another study observed no weight loss or mortality following infection with SARS-CoV-2 B.1.1.529 Omicron (14). One recent study reported that Omicron BA.5 did not cause disease or lethality in K18-hACE2 mice (70). However, the latter study used only younger female mice, while our study used older mice of both genders. It is well documented that both age and gender are significant factors in SARS-CoV-2 pathogenesis, with males displaying more severe disease than females. Thus, distinctions between our study and prior studies could be a result of several factors, including but not limited to animal age and gender, differences in the animal microbiome, or specific mutations between the viral isolates used for infection. It is well recognized that microbiomes can vary from one facility to another, mouse to mouse or human to human. These differences can differentially affect immune responses to therapy, vaccination, and infection dynamics leading to variable outcomes (71–75). Virus propagation can also give rise to mutations that can either drive or attenuate pathogenesis (76–79). We found two mutations in ORF1a (L3515P and L3826F) and a valine deletion at position 14 in the E protein between our Omicron BA.5 sequence and that used by Uraki et al (70). It is possible that these mutations contributed to the virulence differences observed. Overall, it is clear that multiple groups have observed differences in pathogenicity of Omicron variants, and further studies are required to investigate the effects of these variables and to better define the viral determinants of pathogenesis we and others observe for SARS-CoV-2 Omicron BA.5.
An important observation with the early SARS-CoV-2 WA1 strain was the infection of the respiratory and olfactory epithelia leading to brain infection that resulted in symptoms such as anosmia and frequently death (31, 80). VOC Alpha, Beta, and Delta have all been shown to cause significant brain infection in K18-hACE2 mice (22, 24, 25, 28–30). It has even been further suggested that brain infection is necessary for these early VOC to cause weight loss and mortality, as aerosolized inoculation methods in mice resulted in similar lung infection to intranasal infection, yet no brain infection nor mortality (81). However, despite causing significant weight loss and mortality, infection with Omicron BA.5 did not lead to neurological symptoms. We did not detect infectious viral particles or significant levels of viral RNA in the brains of Omicron BA.1– or BA.5–infected mice, a unique finding as compared with previous work on earlier VOC. As ACE2 levels in the CNS are very low, it is thought that other host factors, such as neuropilin-1 (NRP1), may serve to permit neurotropism (18, 82). Our data align with recent studies with cells in which NRP1 is the dominant receptor for SARS-CoV-2, such as using human brain organoids and astrocytes, where SARS-CoV-2 Omicron showed minimal infection compared to SARS-CoV-2 WA-1 and Delta strains (82). It is probable that as SARS-CoV-2 evolved and new VOC emerged, there was an increase in hACE-2 binding, leading to enhanced viral spread. However, concurrently, this could have resulted in decreased binding to NRP-1, potentially leading to reduced brain infections. However, further studies are needed to rule out infections in other organs and to determine whether and how much they contribute to disease and mortality.
Among the unique observations made in the study was that despite having similar levels of virus in the lung at 5 dpi, Omicron BA.5–infected mice displayed more severe disease and pathology compared to Omicron BA.1–infected mice. It is possible that the severe disease and pathology are driven by the more rapid establishment of infection by Omicron BA.5 compared to BA.1, which could cause immune hyperactivation leading to CRS and ARDS that has been associated with severe SARS-CoV-2 outcomes in patients (34, 83). To better understand the role of the immune response in the observed pathology, we characterized the immune phenotypes of Omicron BA.1– and BA.5–infected animals at 5 dpi. Inflammatory cells such as neutrophils, monocyte-derived macrophages, NK cells, and activated T cells were increased in BALF of Omicron BA.5–infected animals. While these cells are important for virus clearance, they have also been shown to contribute to increased pathology and disease when overactivated (33, 34, 36, 84). For instance, NK cells have been shown to be important for early antiviral responses, but in patients with severe COVID-19, they are hyperactivated, characterized by increased CD57 and NKG2C expression (85, 86). Notably, a reduction in alveolar macrophages was observed in Omicron BA.5–infected animals. A decrease in alveolar macrophages has been observed in severe cases of COVID-19 in patients (36, 87, 88). Several studies have investigated potential mechanisms underlying this reduction in alveolar macrophages following SARS-CoV-2 infection. Among the proposed mechanisms is the internalization of the virus by macrophages and monocytes through an FcγR-mediated pathway, triggering inflammasome activation and subsequent pyroptosis (41, 89). Another hypothesis suggests that SARS-CoV-2 infection and the subsequent loss of alveolar type 2 cells contribute to a decline in GM-CSF, a critical cytokine involved in the self-renewal of alveolar macrophages (87, 90). Further work would need to be performed to determine whether these cells die because of infection, a decrease in survival factors, or both. In addition to proinflammatory cells, we also observed an increase in CD4+ CD25Hi Tregs in the Omicron BA.5–infected mice. While these cells are classically associated with suppressive functions that are important in resolving inflammation (91), recent work with SARS-CoV-2 has shown an increase of Tregs in patients with severe COVID-19 (49), and while these cells were found to express suppressive factors, they can also express proinflammatory cytokines that can exacerbate inflammation, thus pointing to a multifactorial role for Tregs in SARS-CoV-2 (49). In addition, high levels of IL-10 have been found in patients with severe COVID-19 (48, 92), further highlighting potential roles for this cytokine that may extend beyond suppression of inflammatory responses. Concurrently, IL-6, TNF-α, the TH1 cytokines IL-2 and IFN-γ, and IL-4, which can activate B cells, were all elevated in the BALF. It is possible that these cytokines not only help mount an antiviral immune response in BA.5-infected mice but also could exacerbate inflammation and thus lead to CRS. Whereas we focused on immunological characterization at 5 dpi, it may be valuable to study the immune response at earlier time points. BA.1 and BA.5 viral kinetics show viral load differences at 1 and 2 dpi, and we posit that the early differences in viral load may account for later differences in immune responses.
Several mutations have emerged in Omicron BA.5, and it is possible that such mutations could play an important role in the differences in pathogenicity between Omicron BA.1 and BA.5. The majority of the differences between the two Omicron variants are in the S protein. While the S protein is critical in infection, particularly cellular entry, it is partially implicated in pathogenesis (26, 76), along with other viral proteins that have been shown to play a role (56, 93). Multiple proteins of SARS-CoV-2 are involved in the modulation of the immune response such as inhibition of IFN-β, which include nsp1, nsp3, nsp6, nsp12, nsp13, nsp14, ORF3, ORF6, and the M protein (94–96), several of which were mutated between Omicron BA.1 and BA.5. The ORF8 in our Omicron BA.5 isolate carries two additional amino acids, +1M and +2N, at the N terminus of the protein. Zhang et al. (56) have shown that SARS-CoV-2 ORF8 protein is important for immune invasion by down-regulating MHC-I proteins, leading to evasion of cell lysis of infected cells by cytotoxic T cells (CD8+ T cells). In addition to their role in agonizing the immune response to infection, nsp2-16 forms replication-transcription complex, which is important for viral replication (97). Of important interest are nsp3, nsp4, and nsp6, which displayed the highest mutations differences outside the S protein between Omicron BA.1 and BA.5 and are critical for the formation of double-membrane vesicles, critical compartments for SARS-CoV-1 and SARS-CoV-2 viral replication (53–55, 98). Given the rapid establishment of infection by Omicron BA.5, it is possible that the mutations in nsp3, nsp4, and nsp6 enhance the formation of double-membrane vesicles by Omicron BA.5, leading to increase virus production. This may in turn hyperstimulate the immune response leading to CRS, increased pathology, and ultimately death. These changes between Omicron BA.1 and BA.5 may result in enhanced immune invasion and replication by Omicron BA.5, perhaps leading to increased pathogenicity.
Compared to earlier variants, the Omicron variants have been characterized as less pathogenic especially in animal models of disease. Initial in vitro studies attributed the attenuation of the Omicron B.1.1.529 to inefficient use of the transmembrane serine protease 2 (TMPRSS2) and to decreased spike protein cleavage (69, 99). However, while most initial studies focused on the role of the S protein on this attenuation, Liu et al. (4) showed that the S protein was likely not a major determinant of the attenuation observed in animal models since a recombinant Omicron BA.1 carrying the SARS-CoV-2 WA-1 S did not cause significantly more disease or mortality compared to a the equivalent virus carrying the Omicron-S. Rather, attenuation was attributed to the 5′ untranslated region to NSP12 regions (4). A recent study showed that the combination of the Omicron S protein and NSP6, in which we observed several differences between Omicron BA.1 and BA.5, played a role in the attenuation of SARS-CoV-2 WA-1 (93). However, the study did not investigate changes induced by NSP6 alone. Further, gain of pathogenicity studies have not been performed, particularly among circulating Omicron strains. We found that Omicron BA.5 is more pathogenic than Omicron BA.1, which makes such pathogenicity studies possible. Although at this point the viral proteins driving pathogenicity in Omicron BA.5 are unknown, we have established an excellent model to further elucidate the role of the S and the nonstructural proteins in driving pathogenicity in Omicron BA.5.
In summary, we uncovered age-dependent severe disease in K18-hACE2 mice following Omicron BA.5, but not Omicron BA.1 infection, despite active viral replication in the lungs of both groups. This pathology correlated with increases in immune cell infiltrates and proinflammatory cytokines in the lungs of Omicron BA.5–infected mice. Overall, we establish K18-hACE2 mice as a lethal model to understand age-dependent SARS-CoV-2 Omicron disease and pathological differences between SARS-CoV-2 Omicron strains, with high potential for utility in pathogenicity studies as well as vaccine and antiviral development.
MATERIALS AND METHODS
Cell and virus expansion
Vero cells expressing hACE2 and TMPRSS2 [Vero E6-TMPRSS2-T2A-ACE2; Biodefense and Emerging Infections Research Resources Repository (BEI): NR-54970] were cultured in Dulbecco’s modified Eagle’s medium (DMEM) solution with 10% fetal bovine serum (FBS), penicillin-streptomycin (100 IU/ml), and puromycin (10 μg/ml). Cells were cultivated in a T-75 flask with 5% CO2 at 37°C and cell density between 0.25 and 2 million cells/ml. SARS-CoV-2 Omicron sublineages BA.1(BEI: NR-56461) and BA.5 (BEI: NR-58620) were obtained from BEI Resources. The viruses were propagated in Vero AT cells, supplemented with 2% FBS and penicillin-streptomycin (100 IU/ml) for 2 days.
Whole genome sequencing
Whole genome sequence of viral stocks Omicron BA.1 (OP984771) and Omicron BA.5 (OP984772) was performed using a tailed amplicon method using the primers and protocol previously described (100). Following each multiplex PCR, libraries were constructed using the Twist Library Preparation EF Kit 2.0 and the Twist CD Index Adapter set 1-96 (Twist Bioscience). The sequencing was carried out in the iSeq platform using an iSeq 100 flow cell and cartridge (Illumina). Resulting reads were trimmed using Trimmomatic (101) and mapped using BWA-MEM2 (102) against a reference genome of SARS-CoV2 (accession number OP295757) in The Galaxy platform (103).
SARS-CoV-2 infection in K18-hACE2 mice
In vivo studies were conducted in compliance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. All experiments were carried out under Biosafety Level 3 (BSL-3) conditions and were authorized by Cornell University's Institutional Animal Care and Use Committee (mouse protocol no. 2017-0108 and BSL-3 Institutional Biosafety Committee no. MUA-16371-1). For this study, 3-month-old and 5- to 8-month-old heterozygous K18-hACE2 c57BL/6J mice [strain: 2B6.Cg-Tg(K18-hACE2)2Prlmn/J] (21, 22, 80) were used (The Jackson Laboratory, Bar Harbor, ME). Female and male mice were housed separately in groups of five per cage and provided a regular chow diet. Intranasal SARS CoV-2 Omicron BA.1 or BA.5 virus inoculation was conducted with 3.25 × 104 PFU per mouse under isoflurane anesthesia. The weight of the mice was monitored and recorded daily for any weight loss after infection. Mice were euthanized at day 2 and 5 postinfection or at humane endpoint according to specified protocols to reduce distress and were euthanized when weight loss reaches 20% of the original weight from the day of challenge. Oral swabs (24 and 48 hours postinfection) and organ samples from the lung, brain, lymph node, and BALF were collected immediately after euthanasia.
Real-time PCR
Oral swabs, lung, and brain tissues were homogenized in 1 ml of TRIzol LS (Invitrogen). According to the manufacturer’s protocol, total RNA extractions were carried out using chloroform. The aqueous phase was collected, mixed with isopropanol, and washed with 70% ethanol after isopropanol removal. The RNA pellet was resuspended in the ribonuclease (RNase) deoxyribonuclease (DNase)–free water, and the RNA quality and genomic DNA contamination were validated according to the previous reports (104). Using the iTag Universal Probes One-Step Kit (Bio-Rad), qRT PCR was performed as described by the manufacturer’s protocol. Primers and probe (Integrated DNA Technologies DNA: TGGCCGCAAATTGCACAATT;TGTAGGTCAACCACGTTCCC;/56FAM/CGCATTGGCATGGAAGTCAC/3BHQ_1/) (105) were individually resuspended to 20 μM stock solutions in RNase DNase-free water. qPCRs were conducted in 384-well plates and read by the ViiA 7 Real-Time PCR System (Thermo Fisher Scientific).
Plaque assay
Oral swabs, lung, and brain organs were homogenized in DMEM supplemented with 2% FBS, and the supernatant was used for plaque assay. Vero AT cells were cultured at 5 × 105 cells per well density in 12-well plates and incubated overnight at 37°C before infection. Serial dilutions of the virus in DMEM with 2% FBS were prepared and, after washing cells with serum free DMEM, distributed on cells for 1 hour in 37°C and gently mixed every 10 min. Cells were overlaid with 1 ml per well of 0.3% agarose-containing 2% FBS medium and incubated at 37°C for 3 days. To visualize and count the plaques, plates were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet in 30% methanol (25). PFUs were calculated according to the number of plaques, and the size of the plaques was quantified using ImageJ software (Fuji).
Flow cytometry
Lymphocytes were harvested from mediastinal lymph nodes and BALF immediately following euthanasia. The lymph node organ was mashed through a 70-μm filter, and BALF lymphocytes were separated from the supernatant after centrifugation. Cells were stained for three separate panels (table S1). Stained cells were run on the Attune NxT (Thermo Fisher Scientific) and analyzed using FlowJo Software 10 (BD Biosciences).
Multiplex assay
BALF was assayed for IL-2, IL-4, IL-6, TNF-α, and IFN-γ using the Procartaplex Mouse High Sensitivity Panel 5-plex (Invitrogen). Briefly, samples were thawed on ice and prepared as described by the manufacturer’s protocol, and plates were read in the MAGPIX (Luminex), quantified with xPONENT Software (Luminex), and graphed per analyte.
Histopathology
Following euthanasia, organ samples including lung and brain were collected for histologic examinations and assessed as previously described (25). Briefly, the tissues were preserved in 10% formalin for more than 72 hours before being embedded in paraffin. Tissue sections (4 μm) were stained with H&E, examined, and scored in a blinded manner by an anatomic pathologist. For the lung, the following criteria were used to assign scores based on the percentage of various tissue types (alveolus, vessels, etc.) affected: normal (0); less than 10% (1); between 10 and 25% (2); 26 to 50% (3); and higher than 50% (4) (106). Lung pathology of Omicron BA.1– and BA.5–infected K18-hACE2 mice was assessed using lung sections stained with hematoxylin. Histologic grading of perivascular inflammation was performed on brains using the most seriously afflicted vessel and the following criteria: (0) no perivascular inflammation; (1) incomplete cuff one cell layer thick; (2) complete cuff one cell layer thick; (3) complete cuff two to three cells thick; and (4) complete cuff four or more cells thick. The most seriously affected region and the following criteria were used to assess necrotic cells in the neuroparenchyma per 0.237 mm2 field: (0) no necrotic cells; (1) rare individual necrotic cells; (2) less than 10 necrotic cells; (3) 11 to 25 necrotic cells; (4) 26 to 50 necrotic cells; and (5) higher than 50 necrotic cells. Sections were stained with anti–SARS-CoV-2 N protein rabbit immunoglobulin G monoclonal antibody (GeneTex; GTX635679) at a dilution of 1:5000 to identify viral antigen. The stained sections were processed using a Leica Bond Max automated IHC stainer. Leica Bond Polymer Refine Detection (Leica; DS9800) with 3,3'-Diaminobenzidine was used as the chromogen. Images were acquired using a Roche Ventana DP200 slide scanner, and immunolabeling was scored by a blinded anatomic pathologist using the following criteria based on the tissue affection: (0) none; (1) less than 10%; (2) 10 to 25%; (3) 26 to 50%; and (4) higher than 50%.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism v9.0 with two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons, two-tailed unpaired test, two-tailed Mann-Whitney test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were statistically significant.
Acknowledgments
We thank A. Redko and the Cornell CARE staff for mouse colony maintenance and members of the Cornell BSL-3, ABSL-3, and Cornell Biosafety staff including but not limited to P. Jenettee, N. Kushner, J. Turse, S. Mattoon, D. Miller, H. B. Roman, B. Singh, D.G. Collin, and T.L. Van Deusen. We thank the Cornell Flow Cytometry Core, the Cornell Animal Health and Diagnostic Center Anatomic Pathology lab, and D. Gludish for comments and feedback. We also thank C. Florence at the NIH/NIAID for sharing the SARS-CoV-2 Omicron variants.
Funding: This work was supported by the National Institutes of Health grants R01AI138570, R25GM125597, and T32EB023860 to A.A.; R01AI109022 to H.C.A.; and 75N93021C00014, R01AI161175, R21 AI173816, and R01AI161363 to L.M.-S.
Author contributions: B.I., S.E., D.W.B., J.S., A.A., and H.C.A initialized and designed the mouse studies. B.I., S.E., and D.W.B. performed most of the mouse and in vitro studies. C.Y. and L.M.-S. provided the viruses used in the studies. B.I., S.E., D.W.B.., J.S., C.Y., X.A.O.-C., A.Z., C.K., K.O., A.C., L.G., G.R.W., L.M.-S., A.A., and H.C.A. performed data analysis. K.O., R.A.A., and S.K. performed in vitro experiments. B.I., S.E., D.W.B., J.S., and H.C.A. wrote the initial draft of the manuscript. All authors provided feedback and edits to the manuscript.
Competing interests: A.A. receives research support from 3M Company. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The sequence of Omicron BA.1 and BA.5 can be found as GenBank accession numbers OP984771 and OP984772, respectively (https://ncbi.nlm.nih.gov/genbank/).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.R. Cornelius, H. Ryan, F. Nicholas, S. Hitoshi, G. Federico, P. P. Thomas, SARS-CoV-2 evolution, post-Omicron. (2022).
- 2.L. B. Shrestha, C. Foster, W. Rawlinson, N. Tedla, R. A. Bull, Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 32, e2381 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. Tarrés-Freixas, B. Trinité, A. Pons-Grífols, M. Romero-Durana, E. Riveira-Muñoz, C. Ávila-Nieto, M. Pérez, E. Garcia-Vidal, D. Perez-Zsolt, J. Muñoz-Basagoiti, D. Raïch-Regué, N. Izquierdo-Useros, C. Andrés, A. Antón, T. Pumarola, I. Blanco, M. Noguera-Julián, V. Guallar, R. Lepore, A. Valencia, V. Urrea, J. Vergara-Alert, B. Clotet, E. Ballana, J. Carrillo, J. Segalés, J. Blanco, Heterogeneous infectivity and pathogenesis of SARS-CoV-2 variants beta, delta and omicron in transgenic K18-hACE2 and wildtype mice. Front. Microbiol. 13, 840757 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. F. Liu, P. Selvaraj, K. Sangare, B. Q. Luan, T. T. Wang, Spike protein-independent attenuation of SARS-CoV-2 Omicron variant in laboratory mice. Cell Rep. 40, 111359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. Zhang, K. Shi, Q. Geng, G. Ye, H. Aihara, F. Li, Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant. Proc. Natl. Acad. Sci. U.S.A. 119, e2206509119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Y. N. Zhang, Z. R. Zhang, H. Q. Zhang, N. Li, Q. Y. Zhang, X. D. Li, C. L. Deng, F. Deng, S. Shen, B. Zhu, B. Zhang, Different pathogenesis of SARS-CoV-2 Omicron variant in wild-type laboratory mice and hamsters. Signal Transduct. Target. Ther. 7, 62 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. A. A. Mahdy, W. Younis, Z. Ewaida, An overview of SARS-CoV-2 and animal infection. Front. Vet. Sci. 7, 596391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Kawaoka, R. Uraki, M. Kiso, S. Iida, M. Imai, E. Takashita, M. Kuroda, P. Halfmann, S. Loeber, T. Maemura, S. Yamayoshi, S. Fujisaki, Z. Wang, M. Ito, M. Ujie, K. Iwatsuki-Horimoto, Y. Furusawa, R. Wright, Z. Chong, S. Ozono, A. Yasuhara, H. Ueki, Y. Sakai, R. Li, Y. Liu, D. Larson, M. Koga, T. Tsutsumi, E. Adachi, M. Saito, S. Yamamoto, S. Matsubara, M. Hagihara, K. Mitamura, T. Sato, M. Hojo, S.-I. Hattori, K. Maeda, M. Okuda, J. Murakami, C. Duong, S. Godbole, D. Douek, S. Watanabe, N. Ohmagari, H. Yotsuyanagi, M. Diamond, H. Hasegawa, H. Mitsuya, T. Suzuki, Y. Kawaoka; IASO study team , Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Nature 607, 119–127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Yuan, H. J. Liu, N. C. Wu, C. C. D. Lee, X. Y. Zhu, F. Z. Zhao, D. L. Huang, W. L. Yu, Y. Z. Hua, H. Tien, T. F. Rogers, E. Landais, D. Sok, J. G. Jardine, D. R. Burton, I. A. Wilson, Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.W. Dejnirattisai, D. M. Zhou, H. M. Ginn, H. M. E. Duyvesteyn, P. Supasa, J. B. Case, Y. G. Zhao, T. S. Walter, A. J. Mentzer, C. Liu, B. B. Wang, G. C. Paesen, J. Slon-Campos, C. Lopez-Camacho, N. M. Kafai, A. L. Bailey, R. E. Chen, B. L. Ying, C. Thompson, J. Bolton, A. Fyfe, S. Gupta, T. K. Tan, J. Gilbert-Jaramillo, W. James, M. Knight, M. W. Carroll, D. Skelly, C. Dold, Y. C. Peng, R. Levin, T. Dong, A. J. Pollard, J. C. Knight, P. Klenerman, N. Temperton, D. R. Hall, M. A. Williams, N. G. Paterson, F. K. R. Bertram, C. A. Siebert, D. K. Clare, A. Howe, J. Radecke, Y. Song, A. R. Townsend, K. Y. A. Huang, E. E. Fry, J. Mongkolsapaya, M. S. Diamond, J. S. Ren, D. I. Stuart, G. R. Screaton, The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183–2200.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Tuekprakhon, R. Nutalai, A. Dijokaite-Guraliuc, D. M. Zhou, H. M. Ginn, M. Selvaraj, C. Liu, A. J. Mentzer, P. Supasa, H. M. E. Duyvesteyn, R. Das, D. Skelly, T. G. Ritter, A. Amini, S. Bibi, S. Adele, S. A. Johnson, B. Constantinides, H. Webster, N. Temperton, P. Klenerman, E. Barnes, S. J. Dunachie, D. Crook, A. J. Pollard, T. Lambe, P. Goulder, N. G. Paterson, M. A. Williams, D. R. Hall, E. E. Fry, J. D. Huo, J. Mongkolsapaya, J. S. Ren, D. I. Stuart, G. R. Screaton; OPTIC Consortium; ISARIC4C Consortium , Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 185, 2422–2433.e13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D. Yamasoba, Y. Kosugi, I. Kimura, S. Fujita, K. Uriu, J. Ito, K. Sato; Genotype to Phenotype Japan (G2P-Japan) Consortium , Genotype Phenotype Japan, Neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 22, 942–943 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Y. L. Cao, A. Yisimayi, F. C. Jian, W. L. Song, T. H. Xiao, L. Wang, S. Du, J. Wang, Q. Q. Li, X. S. Chen, Y. L. Yu, P. Wang, Z. Y. Zhang, P. L. Liu, R. An, X. H. Hao, Y. Wang, R. Feng, H. Y. Sun, L. J. Zhao, W. Zhang, D. Zhao, J. Zheng, L. L. Yu, C. Li, N. Zhang, R. Wang, X. Niu, S. J. Yang, X. T. Song, Y. Y. Chai, Y. Hu, Y. S. Shi, L. L. Zheng, Z. Q. Li, Q. Q. Gu, F. Shao, W. J. Huang, R. H. Jin, Z. Y. Shen, Y. C. Wang, X. X. Wang, J. Y. Xiao, X. S. Xie, BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.P. J. Halfmann, S. Iida, K. Iwatsuki-Horimoto, T. Maemura, M. Kiso, S. M. Scheaffer, T. L. Darling, A. Joshi, S. Loeber, G. Singh, S. L. Foster, B. L. Ying, J. B. Case, Z. L. Chong, B. Whitener, J. Moliva, K. Floyd, M. Ujie, N. Nakajima, M. Ito, R. Wright, R. Uraki, P. Warang, M. Gagne, R. Li, Y. Sakai-Tagawa, Y. A. Liu, D. Larson, J. E. Osorio, J. P. Hernandez-Ortiz, A. R. Henry, K. Ciuoderis, K. R. Florek, M. Patel, A. Odle, L. Y. R. Wong, A. C. Bateman, Z. D. Wang, V. V. Edara, J. Franks, T. Jeevan, T. Fabrizio, J. DeBeauchamp, L. Kercher, P. Seiler, A. S. Gonzalez-Reiche, E. M. Sordillo, L. A. Chang, H. van Baker, V. Simon, D. C. Douek, N. J. Sullivan, L. B. Thackray, H. Ueki, S. Yamayoshi, M. Imai, S. Perlman, R. J. Webby, R. A. Seder, M. S. Suthar, A. Garcia-Sastre, M. Schotsaert, T. Suzuki, A. C. M. Boon, M. S. Diamond, Y. Kawaoka, S., Consortium Mt Sinai Pathogen, SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou, J. He, L. Liu, H. Shan, C. Lei, D. S. C. Hui, B. Du, L. Li, G. Zeng, K. Y. Yuen, R. Chen, C. Tang, T. Wang, P. Chen, J. Xiang, S. Li, J. L. Wang, Z. Liang, Y. Peng, L. Wei, Y. Liu, Y. H. Hu, P. Peng, J. M. Wang, J. Liu, Z. Chen, G. Li, Z. Zheng, S. Qiu, J. Luo, C. Ye, S. Zhu, N. Zhong; China Medical Treatment Expert Group for Covid-19 , China med treatment expert, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y. X. Liu, C. Zhang, F. M. Huang, Y. Yang, F. X. Wang, J. Yuan, Z. Zhang, Y. H. Qin, X. Y. Li, D. D. Zhao, S. W. Li, S. G. Tan, Z. Q. Wang, J. X. Li, C. G. Shen, J. M. Li, L. Peng, W. B. Wu, M. L. Cao, L. Xing, Z. X. Xu, L. Chen, C. Z. Zhou, W. J. Liu, L. Liu, C. Y. Jiang, Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 7, 1003–1011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.G. Aghagoli, B. G. Marin, N. J. Katchur, F. Chaves-Sell, W. F. Asaad, S. A. Murphy, Neurological involvement in COVID-19 and potential mechanisms: A review. Neurocritical Care 34, 1062–1071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.F. Crunfli, V. C. Carregari, F. P. Veras, L. S. Silva, M. H. Nogueira, A. Antunes, P. H. Vendramini, A. G. F. Valenca, C. Brandao-Teles, G. D. Zuccoli, G. Reis-de-Oliveira, L. C. Silva-Costa, V. M. Saia-Cereda, B. J. Smith, A. C. Codo, G. F. de Souza, S. P. Muraro, P. L. Parise, D. A. Toledo-Teixeira, I. M. S. de Castro, B. M. Melo, G. M. Almeida, E. M. S. Firmino, I. M. Paiva, B. M. S. Silva, R. M. Guimaraes, N. D. Mendes, R. L. Ludwig, G. P. Ruiz, T. L. Knittel, G. G. Davanzo, J. A. Gerhardt, P. B. Rodrigues, J. Forato, M. R. Amorim, N. S. Brunetti, M. C. Martini, M. N. Benatti, S. S. Batah, S. Y. Li, R. B. Joao, I. K. Aventurato, M. R. de Brito, M. J. Mendes, B. A. da Costa, M. K. M. Alvim, J. R. da Silva, L. L. Damiao, I. M. P. de Sousa, E. D. da Rocha, S. M. Goncalves, L. H. L. da Silva, V. Bettini, B. M. Campos, G. Ludwig, L. A. Tavares, M. C. Pontelli, R. M. M. Viana, R. B. Martins, A. S. Vieira, J. C. Alves, E. Arruda, G. G. Podolsky-Gondim, M. V. Santos, L. Neder, A. Damasio, S. Rehen, M. A. R. Vinolo, C. D. Munhoz, P. Louzada, R. D. Oliveira, F. Q. Cunha, H. I. Nakaya, T. Mauad, A. N. Duarte-Neto, L. F. F. da Silva, M. Dolhnikoff, P. H. N. Saldiva, A. S. Farias, F. Cendes, P. M. M. Moraes-Vieira, A. T. Fabro, A. Sebollela, J. L. Proenca-Modena, C. L. Yasuda, M. A. Mori, T. M. Cunha, D. Martins-de-Souza, Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 119, e2200960119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.F. Seehusen, J. J. Clark, P. Sharma, E. G. Bentley, A. Kirby, K. Subramaniam, S. Wunderlin-Giuliani, G. L. Hughes, E. I. Patterson, B. D. Michael, A. Owen, J. A. Hiscox, J. P. Stewart, A. Kipar, Neuroinvasion and Neurotropism by SARS-CoV-2 Variants in the K18-hACE2 Mouse. Viruses 14, 1020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.H. Y. Huang, H. Y. Liao, X. R. Chen, S. W. Wang, C. W. Cheng, M. Shahed-Al-Mahmud, Y. M. Liu, A. Mohapatra, T. H. Chen, J. M. Lo, Y. M. Wu, H. H. Ma, Y. H. Chang, H. Y. Tsai, Y. C. Chou, Y. P. Hsueh, C. Y. Tsai, P. Y. Huang, S. Y. Chang, T. L. Chao, H. C. Kao, Y. M. Tsai, Y. H. Chen, C. Y. Wu, J. T. Jan, T. J. R. Cheng, K. I. Lin, C. Ma, C. H. Wong, Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci. Transl. Med. 14, eabm0899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.P. B. McCray, L. Pewe, C. Wohlford-Lenane, M. Hickey, L. Manzel, L. Shi, J. Netland, H. P. Jia, C. Halabi, C. D. Sigmund, D. K. Meyerholz, P. Kirby, D. C. Look, S. Perlman, Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.F. S. Oladunni, J. G. Park, P. A. Pino, O. Gonzalez, A. Akhter, A. Allue-Guardia, A. Olmo-Fontanez, S. Gautam, A. Garcia-Vilanova, C. J. Ye, K. Chiem, C. Headley, V. Dwivedi, L. M. Parodi, K. J. Alfson, H. M. Staples, A. Schami, J. I. Garcia, A. Whigham, R. N. Platt, M. Gazi, J. Martinez, C. Chuba, S. Earley, O. H. Rodriguez, S. D. Mdaki, K. N. Kavelish, R. Escalona, C. R. A. Hallam, C. Christie, J. L. Patterson, T. J. C. Anderson, R. Carrion Jr., E. J. Dick Jr., S. Hall-Ursone, L. S. Schlesinger, X. Alvarez, D. Kaushal, L. D. Giavedoni, J. Turner, L. Martinez-Sobrido, J. B. Torrelles, Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 11, 6122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. Radvak, H. J. Kwon, M. Kosikova, U. Ortega-Rodriguez, R. X. Xiang, J. N. Phue, R. F. Shen, J. Rozzelle, N. Kapoor, T. Rabara, J. Fairman, H. Xie, SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 12, 6559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.E. S. Winkler, A. L. Bailey, N. M. Kafai, S. Nair, B. T. McCune, J. S. Yu, J. M. Fox, R. T. E. Chen, J. T. Earnest, S. P. Keeler, J. H. Ritter, L. I. Kang, S. Dort, A. Robichaud, R. Head, M. J. Holtzman, M. S. Diamond, SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.T. Shapira, I. A. Monreal, S. P. Dion, D. W. Buchholz, B. Imbiakha, A. D. Olmstead, M. Jager, A. Desilets, G. Gao, M. Martins, T. Vandal, C. A. H. Thompson, A. Chin, W. D. Rees, T. Steiner, I. R. Nabi, E. Marsault, J. Sahler, D. G. Diel, G. R. Van de Walle, A. August, G. R. Whittaker, P. L. Boudreault, R. Leduc, H. C. Aguilar, F. Jean, A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605, 340–348 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. Suzuki, D. Yamasoba, I. Kimura, L. Wang, M. Kishimoto, J. Ito, Y. Morioka, N. Nao, H. Nasser, K. Uriu, Y. Kosugi, M. Tsuda, Y. Orba, M. Sasaki, R. Shimizu, R. Kawabata, K. Yoshimatsu, H. Asakura, M. Nagashima, K. Sadamasu, K. Yoshimura, H. Sawa, T. Ikeda, T. Irie, K. Matsuno, S. Tanaka, T. Fukuhara, K. Sato; The Genotype to Phenotype Japan (G2P-Japan) Consortium , Genotype Phenotype Japan, Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603, 700–705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.I. Kimura, D. Yamasoba, T. Tamura, N. Nao, T. Suzuki, Y. Oda, S. Mitoma, J. Ito, H. Nasser, J. Zahradnik, K. Uriu, S. Fujita, Y. Kosugi, L. Wang, M. Tsuda, M. Kishimoto, H. Ito, R. Suzuki, R. Shimizu, M. M. Begum, K. Yoshimatsu, K. T. Kimura, J. Sasaki, K. Sasaki-Tabata, Y. Yamamoto, T. Nagamoto, J. Kanamune, K. Kobiyama, H. Asakura, M. Nagashima, K. Sadamasu, K. Yoshimura, K. Shirakawa, A. Takaori-Kondo, J. Kuramochi, G. Schreiber, K. J. Ishii, T. Hashiguchi, T. Ikeda, A. Saito, T. Fukuhara, S. Tanaka, K. Matsuno, K. Sato; Genotype to Phenotype Japan (G2P-Japan) Consortium , Genotype Phenotype Japan, Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 185, 3992–4007.e16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.I. Ullah, J. Prevost, M. S. Ladinsky, H. Stone, M. L. Lu, S. P. Anand, G. Beaudoin-Bussieres, K. Symmes, M. Benlarbi, S. L. Ding, R. Gasser, C. Fink, Y. Z. Chen, A. Tauzin, G. Goyette, C. Bourassa, H. Medjahed, M. Mack, K. O. Chung, C. B. Wilen, G. A. Dekaban, J. D. Dikeakos, E. A. Bruce, D. E. Kaufmann, L. Stamatatos, A. T. McGuire, J. Richard, M. Pazgier, P. J. Bjorkman, W. Mothes, A. Finzi, P. Kumar, P. D. Uchil, Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity 54, 2143–2158.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Kumari, H. A. Rothan, J. P. Natekar, S. Stone, H. Pathak, P. G. Strate, K. Arora, M. A. Brinton, M. Kumar, Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses-Basel 13, 132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Lee, G. Y. Yoon, S. J. Lee, Y. C. Kwon, H. W. Moon, Y. J. Kim, H. Kim, W. Lee, G. U. Jeong, C. Kim, K. D. Kim, S. J. Kim, D. G. Ahn, Immunological and pathological peculiarity of severe acute respiratory syndrome coronavirus 2 beta variant. Microbiol. Spectr. 10, e0237122 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. T. Tang, D. W. Buchholz, K. M. Szigety, B. Imbiakha, S. Gao, M. Frankfurter, M. Wang, J. Yang, P. Hewins, P. Mericko-Ishizuka, N. A. Leu, S. Sterling, I. A. Monreal, J. Sahler, A. August, X. Zhu, K. A. Jurado, M. Xu, E. E. Morrisey, S. E. Millar, H. C. Aguilar, M. L. Kahn, Cell-autonomous requirement for ACE2 across organs in lethal mouse SARS-CoV-2 infection. PLOS Biol. 21, e3001989 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.B. Israelow, T. Y. Mao, J. Klein, E. Song, B. Menasche, S. B. Omer, A. Iwasaki, Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 6, eabl4509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Schulte-Schrepping, N. Reusch, D. Paclik, K. Bassler, S. Schlickeiser, B. W. Zhang, B. Kramer, T. Krammer, S. Brumhard, L. Bonaguro, E. De Domenico, D. Wendisch, M. Grasshoff, T. S. Kapellos, M. Beckstette, T. Pecht, A. Saglam, O. Dietrich, H. E. Mei, A. R. Schulz, C. Conrad, D. Kunkel, E. Vafadarnejad, C. J. Xu, A. Horne, M. Herbert, A. Drews, C. Thibeault, M. Pfeiffer, S. Hippenstiel, A. Hocke, H. Muller-Redetzky, K. M. Heim, F. Machleidt, A. Uhrig, L. B. de Jarcy, L. Jurgens, M. Stegemann, C. R. Glosenkamp, H. D. Volk, C. Goffinet, M. Landthaler, E. Wyler, P. Georg, M. Schneider, C. Dang-Heine, N. Neuwinger, K. Kappert, R. Tauber, V. Corman, J. Raabe, K. M. Kaiser, M. T. Vinh, G. Rieke, C. Meisel, T. Ulas, M. Becker, R. Geffers, M. Witzenrath, C. Drosten, N. Suttorp, C. von Kalle, F. Kurth, K. Handler, J. L. Schultze, A. C. Aschenbrenner, Y. Li, J. Nattermann, B. Sawitzki, A. E. Saliba, L. E. Sander; Deutsche COVID-19 OMICS Initiative (DeCOI) , Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. Mathew, J. R. Giles, A. E. Baxter, D. A. Oldridge, A. R. Greenplate, J. E. Wu, C. Alanio, L. Kuri-Cervantes, M. B. Pampena, K. D'Andrea, S. Manne, Z. Y. Chen, Y. J. Huang, J. P. Reilly, A. R. Weisman, C. A. G. Ittner, O. Kuthuru, J. Dougherty, K. Nzingha, N. Han, J. Kim, A. Pattekar, E. C. Goodwin, E. M. Anderson, M. E. Weirick, S. Gouma, C. P. Arevalo, M. J. Bolton, F. Chen, S. F. Lacey, H. Ramage, S. Cherry, S. E. Hensley, S. A. Apostolidis, A. C. Huang, L. A. Vella, M. R. Betts, N. J. Meyer, E. J. Wherry; UPenn COVID Processing Unit; Michael R Betts , Unit, Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Lucas, P. Wong, J. Klein, T. B. R. Castro, J. Silva, M. Sundaram, M. K. Ellingson, T. Y. Mao, J. E. Oh, B. Israelow, T. Takahashi, M. Tokuyama, P. W. Lu, A. Venkataraman, A. Park, S. Mohanty, H. W. Wang, A. L. Wyllie, C. B. F. Vogels, R. Earnest, S. Lapidus, I. M. Ott, A. J. Moore, M. C. Muenker, J. B. Fournier, M. Campbell, C. D. Odio, A. Casanovas-Massana, R. Herbst, A. C. Shaw, R. Medzhitov, W. L. Schulz, N. D. Grubaugh, C. Dela Cruz, S. Farhadian, A. I. Ko, S. B. Omer, A. Iwasaki, I. T. Yale, Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–+ (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. T. Chen, M. D. Park, D. M. Del Valle, M. Buckup, A. Tabachnikova, R. C. Thompson, N. W. Simons, K. Mouskas, B. Lee, D. Geanon, D. D'Souza, T. Dawson, R. Marvin, K. Nie, Z. Zhao, J. LeBerichel, C. Chang, H. Jamal, G. Akturk, U. Chaddha, K. Mathews, S. Acquah, S.-A. Brown, M. Reiss, T. Harkin, M. Feldmann, C. A. Powell, J. L. Hook, S. Kim-Schulze, A. H. Rahman, B. D. Brown, N. D. Beckmann, S. Gnjatic, E. Kenigsberg, A. W. Charney, M. Merad; Mount Sinai COVID-19 Biobank Team , A shift in lung macrophage composition is associated with COVID-19 severity and recovery. Sci. Transl. Med. 14, eabn5168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Merad, C. A. Blish, F. Sallusto, A. Iwasaki, The immunology and immunopathology of COVID-19. Science 375, 1122–1127 (2022). [DOI] [PubMed] [Google Scholar]
- 38.C. Qin, L. Q. Zhou, Z. W. Hu, S. Q. Zhang, S. Yang, Y. Tao, C. H. Xie, K. Ma, K. Shang, W. Wang, D. S. Tian, Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 71, 762–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Gomez-Rial, I. Rivero-Calle, A. Salas, F. Martinon-Torres, Role of monocytes/macrophages in Covid-19 pathogenesis: Implications for therapy. Infect. Drug Resist. 13, 2485–2493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. D. Lv, Z. F. Wang, Y. J. Qu, H. Zhu, Q. Q. Zhu, W. Tong, L. L. Bao, Q. Lv, J. Cong, D. Li, W. Deng, P. Yu, J. P. Song, W. M. Tong, J. N. Liu, Y. Y. Liu, C. Qin, B. Huang, Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 7, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.C. Junqueira, A. Crespo, S. Ranjbar, L. B. de Lacerda, M. Lewandrowski, J. Ingber, B. Parry, S. Ravid, S. Clark, M. R. Schrimpf, F. L. Ho, C. Beakes, J. Margolin, N. Russell, K. Kays, J. Boucau, U. Das Adhikari, S. M. Vora, V. Leger, L. Gehrke, L. A. Henderson, E. Janssen, D. Kwon, C. Sander, J. Abraham, M. B. Goldberg, H. Wu, G. Mehta, S. Bell, A. E. Goldfeld, M. R. Filbin, J. Lieberman, FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 606, 576–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T. Carvalho, F. Krammer, A. Iwasaki, The first 12 months of COVID-19: A timeline of immunological insights. Nat. Rev. Immunol. 21, 245–256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.S. Kreutmair, S. Unger, N. G. Nunez, F. Ingelfinger, C. Alberti, D. De Feo, S. Krishnarajah, M. Kauffmann, E. Friebel, S. Babaei, B. Gaborit, M. Lutz, N. P. Jurado, N. P. Malek, S. Goepel, P. Rosenberger, H. A. Haberle, I. Ayoub, S. Al-Hajj, J. Nilsson, M. Claassen, R. Liblau, G. Martin-Blondel, M. Bitzer, A. Roquilly, B. Becher, Distinct immunological signatures discriminate severe COVID-19 from non-SARS-CoV-2-driven critical pneumonia. Immunity 54, 1578–1593.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. A. Zingaropoli, V. Perri, P. Pasculli, F. C. Dezza, P. Nijhawan, G. Savelloni, G. La Torre, C. D'Agostino, F. Mengoni, M. Lichtner, M. R. Ciardi, C. M. Mastroianni, Major reduction of NKT cells in patients with severe COVID-19 pneumonia. Clin. Immunol. 222, 108630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Y. Zhang, X. M. Wang, X. D. Xing, Z. Xu, C. Zhang, J. W. Song, X. Fan, P. Xia, J. L. Fu, S. Y. Wang, R. N. Xu, X. P. Dai, L. Shi, L. Huang, T. J. Jiang, M. Shi, Y. X. Zhang, A. Zumla, M. Maeurer, F. Bai, F. S. Wang, Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 21, 1107–1118 (2020). [DOI] [PubMed] [Google Scholar]
- 46.A. Mazzoni, L. Salvati, L. Maggi, M. Capone, A. Vanni, M. Spinicci, J. Mencarini, R. Caporale, B. Peruzzi, A. Antonelli, M. Trotta, L. Zammarchi, L. Ciani, L. Gori, C. Lazzeri, A. Matucci, A. Vultaggio, O. Rossi, F. Almerigogna, P. Parronchi, P. Fontanari, F. Lavorini, A. Peris, G. M. Rossolini, A. Bartoloni, S. Romagnani, F. Liotta, F. Annunziato, L. Cosmi, Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 130, 4694–4703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Sun, R. Madan, C. L. Karp, T. J. Braciale, Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15, 277–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H. Han, Q. F. Ma, C. Li, R. Liu, L. Zhao, W. Wang, P. G. Zhang, X. H. Liu, G. S. Gao, F. Liu, Y. G. Jiang, X. M. Cheng, C. L. Zhu, Y. C. Xia, Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 9, 1123–1130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. Galván-Peña, J. Leon, K. Chowdhary, D. A. Michelson, B. Vijaykumar, L. Yang, A. Magnuson, Z. Manickas-Hill, A. Piechocka-Trocha, D. P. Worrall, K. E. Hall, M. Ghebremichael, B. D. Walker, J. Z. Li, X. G. Yu, D. Mathis, C. Benoist; MGH COVID-19 Collection & Processing Team , Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. U.S.A. 118, e2111315118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.G. C. Zeng, G. L. Zhang, X. C. Chen, Th1 cytokines, true functional signatures for protective immunity against TB? Cell. Mol. Immunol. 15, 206–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Cantoni, M. J. Murray, M. D. Kalemera, S. J. Dicken, L. Stejskal, G. Brown, S. Lytras, J. D. Coey, J. McKenna, S. Bridgett, D. Simpson, D. Fairley, L. G. Thorne, A. K. Reuschl, C. Forrest, M. Ganeshalingham, L. Muir, M. Palor, L. Jarvis, B. Willett, U. F. Power, L. E. McCoy, C. Jolly, G. J. Towers, K. J. Doores, D. L. Robertson, A. J. Shepherd, M. B. Reeves, C. G. G. Bamford, J. Grove, Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep. 23, e54322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. Moulana, T. Dupic, A. M. Phillips, J. Chang, S. Nieves, A. A. Roffler, A. J. Greaney, T. N. Starr, J. D. Bloom, M. M. Desai, Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat. Commun. 13, 7011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. M. Angelini, M. Akhlaghpour, B. W. Neuman, M. J. Buchmeier, Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. Mbio 4, e00524-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T. Y. Taha, I. P. Chen, J. M. Hayashi, T. Tabata, K. Walcott, G. R. Kimmerly, A. M. Syed, A. Ciling, R. K. Suryawanshi, H. S. Martin, B. H. Bach, C.-L. Tsou, M. Montano, M. M. Khalid, B. K. Sreekumar, G. Renuka Kumar, S. Wyman, J. A. Doudna, M. Ott, Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6. Nat. Commun. 14, 2308–2308 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Ricciardi, A. M. Guarino, L. Giaquinto, E. V. Polishchuk, M. Santoro, G. Di Tullio, C. Wilson, F. Panariello, V. C. Soares, S. S. G. Dias, J. C. Santos, T. M. L. Souza, G. Fusco, M. Viscardi, S. Brandi, P. T. Bozza, R. S. Polishchuk, R. Venditti, M. A. De Matteis, The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 606, 761–768 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. W. Zhang, Y. S. Chen, Y. Z. Li, F. Huang, B. H. Luo, Y. C. Yuan, B. J. Xia, X. C. Ma, T. Yang, F. Yu, J. Liu, B. F. Liu, Z. Song, J. L. Chen, S. M. Yan, L. Y. Wu, T. Pan, X. Zhang, R. Li, W. J. Huang, X. He, F. Xiao, J. S. Zhang, H. Zhang, The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proc. Natl. Acad. Sci. U.S.A. 118, e2024202118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. Uraki, M. Kiso, S. Iida, M. Imai, E. Takashita, M. Kuroda, P. J. Halfmann, S. Loeber, T. Maemura, S. Yamayoshi, S. Fujisaki, Z. D. Wang, M. Ito, M. Ujie, K. Iwatsuki-Horimoto, Y. Furusawa, R. Wright, Z. L. Chong, S. Ozono, A. Yasuhara, H. Ueki, Y. Sakai-Tagawa, R. Li, Y. N. Liu, D. Larson, M. Koga, T. Tsutsumi, E. Adachi, M. Saito, S. Yamamoto, M. Hagihara, K. Mitamura, T. Sato, M. Hojo, S. I. Hattori, K. Maeda, R. Valdez, M. Okuda, J. Murakami, C. Duong, S. Godbole, D. C. Douek, S. Watanabe, A. Gordon, N. Ohmagari, H. Yotsuyanagi, M. S. Diamond, H. Hasegawa, H. Mitsuya, T. Suzuki, Y. Kawaoka; IASO study team , Characterization and antiviral susceptibility ofSARS-CoV-2 Omicron BA.2. Nature 607, 119–127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. R. Leist, K. H. Dinnon, A. Schafer, L. V. Tse, K. Okuda, Y. X. J. Hou, A. West, C. E. Edwards, W. Sanders, E. J. Fritch, K. L. Gully, T. Scobey, A. J. Brown, T. P. Sheahan, N. J. Moorman, R. C. Boucher, L. E. Gralinski, S. A. Montgomery, R. S. Baric, A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.K. H. Dinnon, S. R. Leist, A. Schafer, C. E. Edwards, D. R. Martinez, S. A. Montgomery, A. West, B. L. Yount, Y. X. J. Hou, L. E. Adams, K. L. Gully, A. J. Brown, E. Huang, M. D. Bryant, I. C. Choong, J. S. Glenn, L. E. Gralinski, T. P. Sheahan, R. S. Baric, A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.N. Osterrieder, L. D. Bertzbach, K. Dietert, A. Abdelgawad, D. Vladimirova, D. Kunec, D. Hoffmann, M. Beer, A. D. Gruber, J. Trimpert, Age-dependent progression of SARS-CoV-2 infection in syrian hamsters. Viruses 12, 779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.P. Yu, F. F. Qi, Y. F. Xu, F. D. Li, P. P. Liu, J. Y. Liu, L. L. Bao, W. Deng, H. Gao, Z. G. Xiang, C. Xiao, Q. Lv, S. R. Gong, J. N. Liu, Z. Q. Song, Y. J. Qu, J. Xue, Q. Wei, M. Y. Liu, G. P. Wang, S. Y. Wang, H. S. Yu, X. Liu, B. Y. Huang, W. L. Wang, L. Zhao, H. J. Wang, F. Ye, W. M. Zhou, W. Zhen, J. Han, G. Z. Wu, Q. Jin, J. W. Wang, W. J. Tan, C. Qin, Age-related rhesus macaque models of COVID-19. Animal Models Exp. Med. 3, 93–97 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. Agrawal, S. Agrawal, J. N. Cao, H. F. Su, K. Osann, S. Gupta, Altered innate immune functioning of dendritic cells in elderly humans: A role of phosphoinositide 3-kinase-signaling pathway. J. Immunol. 178, 6912–6922 (2007). [DOI] [PubMed] [Google Scholar]
- 63.S. Della Bella, L. Bierti, P. Presicce, R. Arienti, M. Valenti, M. Saresella, C. Vergani, M. L. Villa, Peripheral blood dendritic cells and monocytes are differently regulated in the elderly. Clin. Immunol. 122, 220–228 (2007). [DOI] [PubMed] [Google Scholar]
- 64.J. M. Bartleson, D. Radenkovic, A. J. Covarrubias, D. Furman, D. A. Winer, E. Verdin, SARS-CoV-2, COVID-19 and the aging immune system. Nat. Aging 1, 769–782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. R. Moderbacher, S. I. Ramirez, J. M. Dan, A. Grifoni, K. M. Hastie, D. Weiskopf, S. Belanger, R. K. Abbott, C. Kim, J. Choi, Y. Kato, E. G. Crotty, S. A. Rawlings, J. Mateus, L. P. V. Tse, A. Frazier, R. Baric, B. Peters, J. Greenbaum, E. O. Saphire, D. M. Smith, A. Sette, S. Crotty, Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.T. Takahashi, M. K. Ellingson, P. Wong, B. Israelow, C. Lucas, J. Klein, J. Silva, T. Y. Mao, J. E. Oh, M. Tokuyama, P. W. Lu, A. Venkataraman, A. Park, F. M. Liu, A. Meir, J. Sun, E. Y. Wang, A. Casanovas-Massana, A. L. Wyllie, C. B. F. Vogels, R. Earnest, S. Lapidus, I. M. Ott, A. J. Moore, A. Shaw, J. B. Fournier, C. D. Odio, F. Shelli, C. Dela Cruz, N. D. Grubaugh, W. L. Schulz, A. M. Ring, A. I. Ko, S. B. Omer, A. Iwasaki, I. R. T. Yale, Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 588, 315–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.E. J. Yager, M. Ahmed, K. Lanzer, T. D. Randall, D. L. Woodland, M. A. Blackman, Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J. Exp. Med. 205, 711–723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. Nchioua, F. Diofano, S. Noettger, P. von Maltitz, S. Stenger, F. Zech, J. Munch, K. M. J. Sparrer, S. Just, F. Kirchhoff, Strong attenuation of SARS-CoV-2 Omicron BA.1 and increased replication of the BA.5 subvariant in human cardiomyocytes. Signal Transduct. Target. Ther. 7, 395 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.H. P. Shuai, J. F. W. Chan, B. J. Hu, Y. Chai, T. T. T. Yuen, F. F. Yin, X. E. Huang, C. M. Yoon, J. C. Hu, H. Liu, J. L. Shi, Y. C. Liu, T. R. Z. Zhu, J. J. Zhang, Y. X. Hou, Y. X. Wang, L. Lu, J. P. Cai, A. J. X. Zhang, J. Zhou, S. F. Yuan, M. A. Brindley, B. Z. Zhang, J. D. Huang; K. K. W. To, K. Y. Yuen, H. Chu, Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699 (2022). [DOI] [PubMed] [Google Scholar]
- 70.R. Uraki, P. J. Halfmann, S. Iida, S. Yamayoshi, Y. Furusawa, M. Kiso, M. Ito, K. Iwatsuki-Horimoto, S. Mine, M. Kuroda, T. Maemura, Y. Sakai-Tagawa, H. Ueki, R. Li, Y. Liu, D. Larson, S. Fukushi, S. Watanabe, K. Maeda, A. Pekosz, A. Kandeil, R. J. Webby, Z. Wang, M. Imai, T. Suzuki, Y. Kawaoka, Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. Nature 612, 540–545 (2022). [DOI] [PubMed] [Google Scholar]
- 71.V. Gopalakrishnan, C. N. Spencer, L. Nezi, A. Reuben, M. C. Andrews, T. V. Karpinets, P. A. Prieto, D. Vicente, K. Hoffman, S. C. Wei, A. P. Cogdill, L. Zhao, C. W. Hudgens, D. S. Hutchinson, T. Manzo, M. P. de Macedo, T. Cotechini, T. Kumar, W. S. Chen, S. M. Reddy, R. S. Sloane, J. Galloway-Pena, H. Jiang, P. L. Chen, E. J. Shpall, K. Rezvani, A. M. Alousi, R. F. Chemaly, S. Shelburne, L. M. Vence, P. C. Okhuysen, V. B. Jensen, A. G. Swennes, F. McAllister, E. M. R. Sanchez, Y. Zhang, E. Le Chatelier, L. Zitvogel, N. Pons, J. L. Austin-Breneman, L. E. Haydu, E. M. Burton, J. M. Gardner, E. Sirmans, J. Hu, A. J. Lazar, T. Tsujikawa, A. Diab, H. Tawbi, I. C. Glitza, W. J. Hwu, S. P. Patel, S. E. Woodman, R. N. Amaria, M. A. Davies, J. E. Gershenwald, P. Hwu, J. E. Lee, J. Zhang, L. M. Coussens, Z. A. Cooper, P. A. Futreal, C. R. Daniel, N. J. Ajami, J. F. Petrosino, M. T. Tetzlaff, P. Sharma, J. P. Allison, R. R. Jenq, J. A. Wargo, Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.T. Hagan, M. Cortese, N. Rouphael, C. Boudreau, C. Linde, M. S. Maddur, J. Das, H. Wang, J. Guthmiller, N. Y. Zheng, M. Huang, A. A. Uphadhyay, L. Gardinassi, C. Petitdemange, M. P. McCullough, S. J. Johnson, K. Gill, B. Cervasi, J. Zou, A. Bretin, M. Hahn, A. T. Gewirtz, S. E. Bosinger, P. C. Wilson, S. Z. Li, G. Alter, S. Khurana, H. Golding, B. Pulendran, Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–1328.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R. Uchiyama, B. Chassaing, B. Y. Zhang, A. T. Gewirtz, Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 210, 171–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Z. D. Shi, J. Zou, Z. Zhang, X. Zhao, J. Noriega, B. Y. Zhang, C. Y. Zhao, H. Ingle, K. Bittinger, L. M. Mattei, A. J. Pruijssers, R. K. Plemper, T. J. Nice, M. T. Baldridge, T. S. Dermody, B. Chassaing, A. T. Gewirtz, Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 179, 644–658.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.T. Ichinohe, I. K. Pang, Y. Kumamoto, D. R. Peaper, J. H. Ho, T. S. Murray, A. Iwasaki, Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.B. A. Johnson, X. P. Xie, A. L. Bailey, B. Kalveram, K. G. Lokugamage, A. Muruato, J. Zou, X. W. Zhang, T. Juelich, J. K. Smith, L. H. Zhang, N. Bopp, C. Schindewolf, M. Vu, A. Vanderheiden, E. S. Winkler, D. Swetnam, J. A. Plante, P. Aguilar, K. S. Plante, V. Popov, B. Lee, S. C. Weaver, M. S. Suthar, A. L. Routh, P. Ren, Z. Q. Ku, Z. Q. An, K. Debbink, M. S. Diamond, P. Y. Shi, A. N. Freiberg, V. D. Menachery, Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.J. J. Baczenas, H. Andersen, S. Rashid, D. Yarmosh, N. Puthuveetil, M. Parker, R. Bradford, C. Florence, K. J. Stemple, M. G. Lewis, S. L. O'Connor, Propagation of SARS-CoV-2 in calu-3 cells to eliminate mutations in the furin cleavage site of spike. Viruses 13, 2434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.P. Wang, S. Y. Lau, S. F. Deng, P. Chen, B. W. Y. Mok, A. J. Zhang, A. C. Y. Lee, K. H. Chan, R. C. Y. Tam, H. R. Xu, R. H. Zhou, W. J. Song, L. Liu; K. K. W. To, J. F. W. Chan, Z. W. Chen, K. Y. Yuen, H. L. Chen, Characterization of an attenuated SARS-CoV-2 variant with a deletion at the S1/S2 junction of the spike protein. Nat. Commun. 12, 2790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. G. P. Funnell, B. Afrough, J. J. Baczenas, N. Berry, K. R. Bewley, R. Bradford, C. Florence, Y. Le Duff, M. Lewis, R. V. Moriarty, S. L. O. Connor, K. L. Osman, S. Pullan, S. Rashid, K. S. Richards, K. J. Stemple, I. Knezevic, A cautionary perspective regarding the isolation and serial propagation of SARS-CoV-2 in Vero cells. Npj Vaccines 6, 83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.J. Zheng, L. Y. R. Wong, K. Li, A. K. Verma, M. E. Ortiz, C. Wohlford-Lenane, M. R. Leidinger, C. M. Knudson, D. K. Meyerholz, P. B. McCray, S. Perlman, COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.V. Fumagalli, M. Rava, D. Marotta, P. Di Lucia, C. Laura, E. Sala, M. Grillo, E. Bono, L. Giustini, C. Perucchini, M. Mainetti, A. Sessa, J. M. Garcia-Manteiga, L. Donnici, L. Manganaro, S. Delbue, V. Broccoli, R. De Francesco, P. D'Adamo, M. Kuka, L. G. Guidotti, M. Iannacone, Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 7, eabl9929 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.W. L. Kong, M. Montano, M. J. Corley, E. Helmy, H. Kobayashi, M. Kinisu, R. Suryawanshi, X. Y. Luo, L. A. Royer, N. R. Roan, M. Ott, L. C. Ndhlovu, W. C. Greene, Neuropilin-1 mediates SARS-CoV-2 infection of astrocytes in brain organoids, inducing inflammation leading to dysfunction and death of neurons. Mbio 13, e0230822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.D. M. Del Valle, S. Kim-Schulze, H. H. Huang, N. D. Beckmann, S. Nirenberg, B. Wang, Y. Lavin, T. H. Swartz, D. Madduri, A. Stock, T. U. Marron, H. Xie, M. Patel, K. Tuballes, O. Van Oekelen, A. Rahman, P. Kovatch, J. A. Aberg, E. Schadt, S. Jagannath, M. Mazumdar, A. W. Charney, A. Firpo-Betancourt, D. R. Mendu, J. Jhang, D. Reich, K. Sigel, C. Cordon-Cardo, M. Feldmann, S. Parekh, M. Merad, S. Gnjatic, An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.A. J. Wilk, A. Rustagi, N. Q. Zhao, J. Roque, G. J. Martinez-Colon, J. L. McKechnie, G. T. Ivison, T. Ranganath, R. Vergara, T. Hollis, L. J. Simpson, P. Grant, A. Subramanian, A. J. Rogers, C. A. Blish, A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.C. Maucourant, I. Filipovic, A. Ponzetta, S. Aleman, M. Cornillet, L. Hertwig, B. Strunz, A. Lentini, B. Reinius, D. Brownlie, A. Cuapio, E. H. Ask, R. M. Hull, A. Haroun-Izquierdo, M. Schaffer, J. Klingstrom, E. Folkesson, M. Buggert, J. K. Sandberg, L. I. Eriksson, O. Rooyackers, H. G. Ljunggren, K. J. Malmberg, J. Michaelsson, N. Marquardt, Q. Hammer, K. Stralin, N. K. Bjorkstrom, C.-S. G. Karolinska, Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 5, eabd6832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.N. K. Bjorkstrom, A. Ponzetta, Natural killer cells and unconventional T cells in COVID-19. Curr. Opin. Virol. 49, 176–182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.C. Bosteels, K. F. A. Van Damme, E. De Leeuw, J. Declercq, B. Maes, V. Bosteels, L. Hoste, L. Naesens, N. Debeuf, J. Deckers, B. Cole, M. Pardons, D. Weiskopf, A. Sette, Y. Vande Weygaerde, T. Malfait, S. J. Vandecasteele, I. K. Demedts, H. Slabbynck, S. Allard, P. Depuydt, E. Van Braeckel, J. De Clercq, L. Martens, S. Dupont, R. Seurinck, N. Vandamme, F. Haerynck, D. F. Roychowdhury, L. Vandekerckhove, M. Guilliams, S. J. Tavernier, B. N. Lambrecht, Loss of GM-CSF-dependent instruction of alveolar macrophages in COVID-19 provides a rationale for inhaled GM-CSF treatment. Cell Rep. Med. 3, 100833 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.M. F. Liao, Y. Liu, J. Yuan, Y. L. Wen, G. Xu, J. J. Zhao, L. Cheng, J. X. Li, X. Wang, F. X. Wang, L. Liu, I. Amit, S. Y. Zhang, Z. Zhang, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 89.E. Sefik, R. H. Qu, C. Junqueira, E. Kaffe, H. Mirza, J. Zhao, J. R. Brewer, A. L. Han, H. R. Steach, B. Israelow, H. N. Blackburn, S. E. Velazquez, Y. G. Chen, S. Halene, A. Iwasaki, E. Meffre, M. Nussenzweig, J. Lieberman, C. B. Wilen, Y. Kluger, R. A. Flavell, Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.J. Gschwend, S. P. M. Sherman, F. Ridder, X. G. Feng, H. E. Liang, R. M. Locksley, B. Becher, C. Schneider, Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth. J. Exp. Med. 218, e20210745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. Chaudhry, R. M. Samstein, P. Treuting, Y. Q. Liang, M. C. Pils, J. M. Heinrich, R. S. Jack, F. T. Wunderlich, J. C. Bruning, W. Muller, A. Y. Rudensky, Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34, 566–578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.S. K. Dhar, K. Vishnupriyan, S. Damodar, S. Gujar, M. Das, IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: Results from meta-analysis and regression. Heliyon 7, e06155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.D.-Y. Chen, C. V. Chin, D. Kenney, A. H. Tavares, N. Khan, H. L. Conway, G. Liu, M. C. Choudhary, H. P. Gertje, A. K. O'Connell, S. Adams, D. N. Kotton, A. Herrmann, A. Ensser, J. H. Connor, M. Bosmann, J. Z. Li, M. U. Gack, S. C. Baker, R. N. Kirchdoerfer, Y. Kataria, N. A. Crossland, F. Douam, M. Saeed, Spike and nsp6 are key determinants of SARS-CoV-2 Omicron BA.1 attenuation. Nature 615, 143–150 (2023). [DOI] [PubMed] [Google Scholar]
- 94.M. Thoms, R. Buschauer, M. Ameismeier, L. Koepke, T. Denk, M. Hirschenberger, H. Kratzat, M. Hayn, T. Mackens-Kiani, J. D. Cheng, J. H. Straub, C. M. Sturzel, T. Frohlich, O. Berninghausen, T. Becker, F. Kirchhoff, K. M. J. Sparrer, R. Beckmann, Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369, 1249–1255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.X. B. Lei, X. J. Dong, R. Y. Ma, W. J. Wang, X. Xiao, Z. Q. Tian, C. H. Wang, Y. Wang, L. Li, L. L. Ren, F. Guo, Z. D. Zhao, Z. Zhou, Z. C. Xiang, J. W. Wang, Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.H. J. Xia, Z. G. Cao, X. P. Xie, X. W. Zhang, J. Y. C. Chen, H. L. Wang, V. D. Menachery, R. Rajsbaum, P. Y. Shi, Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.B. Malone, N. Urakova, E. J. Snijder, E. A. Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 23, 21–39 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.G. Wolff, R. Limpens, J. C. Zevenhoven-Dobbe, U. Laugks, S. Zheng, A. W. M. de Jong, R. I. Koning, D. A. Agard, K. Grunewald, A. J. Koster, E. J. Snijder, M. Barcena, A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.B. Meng, A. Abdullahi, I. Ferreira, N. Goonawardane, A. Saito, I. Kimura, D. Yamasoba, P. P. Gerber, S. Fatihi, S. Rathore, S. K. Zepeda, G. Papa, S. A. Kemp, T. Ikeda, M. Toyoda, T. S. Tan, J. Kuramochi, S. Mitsunaga, T. Ueno, K. Shirakawa, A. Takaori-Kondo, T. Brevini, D. L. Mallery, O. J. Charles, J. E. Bowen, A. Joshi, A. C. Walls, L. Jackson, D. Martin, K. G. C. Smith, J. Bradley, J. A. G. Briggs, J. Choi, E. Madissoon, K. B. Meyer, P. Mlcochova, L. Ceron-Gutierrez, R. Doffinger, S. A. Teichmann, A. J. Fisher, M. S. Pizzuto, A. de Marco, D. Corti, M. Hosmillo, J. H. Lee, L. C. James, L. Thukral, D. Veesler, A. Sigal, F. Sampaziotis, I. G. Goodfellow, N. J. Matheson, K. Sato, R. K. Gupta, Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.M. Martins, P. M. Boggiatto, A. Buckley, E. D. Cassmann, S. Falkenberg, L. C. Caserta, M. H. V. Fernandes, C. Kanipe, K. Lager, M. V. Palmer, D. G. Diel, From deer-to-deer: SARS-CoV-2 is efficiently transmitted and presents broad tissue tropism and replication sites in white-tailed deer. PlOS Pathogens 18, e1010197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.A. M. Bolger, M. Lohse, B. Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.L. Heng, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. (2013); https://api.semanticscholar.org/CorpusID:14669139.
- 103.E. Afgan, D. Baker, B. Batut, M. van den Beek, D. Bouvier, M. Cech, J. Chilton, D. Clements, N. Coraor, B. A. Gruning, A. Guerler, J. Hillman-Jackson, S. Hiltemann, V. Jalili, H. Rasche, N. Soranzo, J. Goecks, J. Taylor, A. Nekrutenko, D. Blankenberg, The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 Update. Nucleic Acid. Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.J. P. Brosseau, J. F. Lucier, E. Lapointe, M. Durand, D. Gendron, J. Gervais-Bird, K. Tremblay, J. P. Perreault, S. Abou Elela, High-throughput quantification of splicing isoforms. RNA 16, 442–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.T. Toptan, S. Hoehl, S. Westhaus, D. Bojkova, A. Berger, B. Rotter, K. Hoffmeier, J. Cinatl, S. Ciesek, M. Widera, Optimized qRT-PCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci. 21, 4396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D. K. Meyerholz, A. P. Beck, Histopathologic Evaluation and Scoring of Viral Lung Infection, in Mers Coronavirus: Methods and Protocols, R. Vijay, Ed. (Methods in Molecular Biology, Humana Press Inc, Totowa, 2020), vol. 2099, pp. 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1