Abstract
Non-Hodgkin lymphoma (NHL) is a heterogeneous disease, encompassing a wide variety of individually distinct neoplastic entities of mature B-, T-, and NK-cells. While they constitute a broad category together they are the most common hematologic malignancies in the world. The distinction between different neoplastic entities requires a multi-modal approach including flow cytometric immunophenotyping which can both exclude a neoplastic proliferation and help narrow the differential diagnosis. This article describes a flow cytometric test developed at Memorial Sloan Kettering Cancer Center that can assess B-, T-, an NK-cells in a single tube, 21 antibody, 19-color assay. The assay can identify most B- and T-cell NHLs with high specificity and sensitivity and significantly narrow the differential when a specific diagnosis cannot be made. The Basic Protocol provides a detailed operational procedure for processing, staining, and cytometric acquisition of samples. The Support Protocol provides typical steps and caveats for data analysis in lymphoproliferative disorders, and in distinguishing a variety of specific disease entities from each other as well as normal lymphoid populations.
Basic Protocol 1:
Processing, staining, and cytometric analysis of samples for B- and T-cell assessment
Support Protocol 1:
Analysis and interpretation of the B- and T-cell lymphocyte assay
Keywords: Non-Hodgkin lymphoma, B-cells, T-cells, Lymphoproliferative disorder, Lymphoid leukemia
INTRODUCTION
Non-Hodgkin lymphoma (NHL) is a heterogeneous disease, encompassing a wide variety of individually distinct neoplastic disease entities of B-, T-, and NK-cells. It is the most common hematologic malignancy and accounts for 3-4% of all new cancers and cancer deaths (Siegel et al., 2022; Thandra et al., 2021). Diagnosis and subtyping of NHLs require a multi-modal approach including morphology, immunohistochemistry (IHC), and flow cytometry (FC) (Cheson et al., 2014; Horwitz et al., 2022).
Adequate immunophenotyping is essential for the diagnosis and subclassification of NHLs, and FC is a robust and rapid method (Cheson et al., 2014; Horwitz et al., 2022). Its potential for fast turn-around time can guide further work-up including IHC staining. This method also benefits from high sensitivity and its ability to utilize specimens from a variety of sampling methods (Cozzolino et al., 2016; Yu et al., 2014).
In this article, we describe an assay for combined assessment of B-, T-, and NK-cells developed at Memorial Sloan Kettering Cancer Center, which can identify abnormal lymphoid populations, subtype different disease categories, and guide further work-up. The use of 21 antibodies and assessment of both B- and T-cells in a single assay reduces technologist labor, equipment costs, and cytometer machine time. The method is also modular, allowing for analysis of only B- or T-cells depending on the clinical context, opening channels for addition of different antibodies as work-up requires.
The Basic Protocol describes the procedure for processing, staining, and cytometric analysis of cells from virtually any sample including bone marrow (BM), peripheral blood (PB), lymph node (LN) and other tissue specimens, fine needle aspirates (FNAs), and fluid samples. Within this article, “fluid” samples refer to cerebral spinal fluid (CSF), ascites, or pleural fluid samples. The Support Protocol discusses approaches used for the interpretation of resulting FC data. Samples were obtained with patient consent under protocol reviewed and approved by an institutional review board.
STRATEGIC PLANNING
This method requires an instrument capable of analyzing a minimum of 19 colors simultaneously. The assay was specifically developed for use on an LSR-Fortessa X20 equipped with four lasers and a FACS-Symphony A3 equipped with five. Instrument settings should be configured for maximal sensitivity while allowing separation between spectrally overlapping fluorochromes. Target setting examples for instrument configurations and relevant photomultiplier tubes (PMTs) have been published previously (Gao & Roshal, 2022). The use of multiple instruments requires standardization of the instruments; details of setup and standardization are beyond the scope of this article (Selliah et al., 2019). The procedure as described uses a variety of samples including EDTA-anticoagulated PB and BM aspirates, tissue, FNA, and fluid samples. Samples may be analyzed up to 48 hours post-collection if they are stored and/or shipped refrigerated; frozen samples or tissue samples stored in formalin (or any other fixatives) are unacceptable. Irreplaceable samples may be processed up to 72 hours post-collection.
Antibodies must be titered for saturation prior to use, using the same staining procedure including staining volume, as the procedure used for the test. The manufacturer’s recommended volume per test can be used as a starting point but titers must be verified in the laboratory. Titration procedures are again beyond the scope of this article and are described elsewhere (Maciorowski et al., 2017).
To avoid errors when adding the large number of antibodies required to a single sample, we advise creating three separate cocktails containing groups of antibodies for staining both B- and T-cells (in combination) or separate B- or T- cell compartments (Table 1) with additional brilliant violet buffer, to make 100μL/sample cocktails. Two separate cocktails (B- or T-cell cocktails) add modularity, saving materials when evaluation of only one cell type is required, such as in follow-up samples. Additionally, drop-in antibodies can also be added as needed with more opened channels and therefore increase the versatility of the tube and reduce the number of reflex tests. Cocktails must undergo quality control testing using a sample showing positive expression of all antigens tested. Once made, cocktails can be stored refrigerated and protected from light for up to 2 weeks. Cocktail stability can be validated by following the CLSI guideline (CLSI, 2021).
Table 1.
BT, B, T Panel Surface Antibody Combinations
| BT panel | B panel | T panel | Fluorochrome | Clone | Vendor | Catalog no. |
|---|---|---|---|---|---|---|
| Kappa | Kappa | n/a | FITC | polytypic | Agilent | F043401-1 |
| CD8 | n/a | CD8 | FITC | REA734 | Miltenyi | 130-110-815 |
| Lambda | Lambda | n/a | PE | polytypic | Agilent | R043701-1 |
| CD56 | n/a | CD56 | PE | N901 | Beckman Coulter | IM2073U |
| CD25 | CD25 | CD25 | PE-Dazzle 594 | BC96 | BioLegend | 302646 |
| CD22 | CD22 | n/a | PC5.5 | SJ10.1H11 | Beckman Coulter | A80712 |
| CD19 | CD19 | n/a | PC7 | J3-119 | Beckman Coulter | IM3628U |
| CD10 | CD10 | CD10 | APC | ALB1 | Beckman Coulter | IM3633 |
| CD7 | n/a | CD7 | APC-Alexa700 | 8H8.1 | Beckman Coulter | A70201 |
| CD45 | CD45 | CD45 | APC-H7 | 2D1 | BD Horizon | 641408 |
| CD5 | CD5 | CD5 | BV421 | L17F122 | BioLegend | L17F12 |
| CD38 | CD38 | CD38 | BV480 | HIT2 | BD Horizon | 566137 |
| CD279 (PD1) | CD279 (PD1) | CD279 (PD1) | BV605 | EH12.2H7 | BioLegend | 329924 |
| CD20 | CD20 | n/a | BV650 | 2H7 | BD Optibuild | 563780 |
| TCRγδ | n/a | TCRγδ | BV711 | 11F2 | BD Optibuild | 624148 |
| CD2 | n/a | CD2 | BV786 | RPA-2.10 | BD Optibuild | 740960 |
| CD26 | n/a | CD26 | BUV395 | M-A261 | BD Optibuild | 744454 |
| CD4 | n/a | CD4 | BUV563 | SK3 | BD Horizon | 612912 |
| CD3 | n/a | CD3 | BUV737 | UCHT1 | BD Horizon | 612750 |
| CD14 | CD14 | CD14 | BUV805 | M5E2 | BD Horizon | 612903 |
| TCRCβ1 | n/a | TCRCβ1 | BB700 | JOVI.1 | BD Optibuild | 7497974 |
| Brilliant Violet Buffer Plusa | N/A | N/A | BD Horizon | 566385 |
Brilliant violet buffer must be used to mitigate staining artifacts when multiple BD Horizon Brilliant dye-conjugated reagents are used.
Panel selection (BT, B, or T tubes) is done for every sample received in the laboratory upon a thorough history review. In general, BT tube is performed on all patients that are newly admitted to our institution, regardless of the sample type. For follow-up specimens, B, T, or BT tubes may be selected accordingly based on the prior diagnosis or flow cytometric findings. BT, the combinational tube, is used on all new tissue/FNA specimens regardless of the disease status, as both B- or T-cell lymphoma can be seen simultaneously in these samples. DAPI, the viability dye, is added to tissue specimens to help remove dead cells/debris.
BASIC PROTOCOL 1
PROCESSING, STAINING, AND CYTOMETRIC ANALYSIS OF SAMPLES FOR B- AND T-CELL ASSESSMENT
The following protocol is designed for efficient processing, staining, and cytometric analysis of tissue (including lymph node tissue), bone marrow, peripheral blood, and other fluid samples. The procedure aims to ensure that the proper number of cells are available for analysis. Steps are taken to minimize absolute cell loss and to preserve the original cell proportions for proper cell quantitation. The processing is also optimized to reduce processing-induced antigenic and scatter changes and to minimize background due to unbound residual antibodies. Finally, instrument acquisition is aimed at minimizing carryover that may confound data interpretation. If the procedure is performed correctly with adequate samples, up to 500,000 cells will be analyzed and available for data interpretation.
NOTE: All protocols involving human samples should first be reviewed and approved by an institutional review board and or local ethics committee.
Materials
EDTA-anticoagulated PB or BM aspirate, or tissue sample in RPMI-1640 (Corning, cat. no. 10-041-CV)
Phosphate-buffered saline (PBS; see recipe in Reagents and Solutions)
ViaStain AOPI (Nexcelom Bioscience, cat. no. CS2-0106-5ML)
4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; see recipe in Reagents and Solutions)
BT antibody cocktail (Table 1)
B-cell antibody cocktail (Table 1)
T-cell antibody cocktail (Table 1)
Lyse-fix reagent (see recipe in Reagents and Solutions)
PBA medium (see recipe in Reagents and Solutions) or RPMI-1640 medium (see recipe in Reagents and Solutions)
70-μm cell strainers (Miltenyi Biotec, cat. no. 130-098-462)
15-mL conical tubes
Vortex
Cellometer Auto 2000 (Nexcelom Bioscience)
10–1000 μL single-channel pipets and pipet tips
1-10 mL serological pipets
96-well microtiter plate (Fisher Scientific, cat. no. 07-200-127)
Cellometer cell counting chambers SD-100 (Nexcelom Bioscience, cat. no. CHT4-SD100-014)
5 mL (12 × 75-mm) polystyrene tubes (Fisher, cat. no. 149595)
Flow tube caps
Serofuge (Clay Adams) or equivalent
BD LSR Fortessa X-20 Flow Cytometer (BD Biosciences)
BD FACSymphony A3 Flow Cytometer (BD Biosciences)
Gauze or Kimwipes
Stainless steel surgical scalpels
Plastic petri dishes
18-gauge needles
Protocol steps with step annotations
Sample preparation
All specimens are processed in the biosafety cabinet following our institutional guidelines. Individual institutional guidelines regarding sample handling may differ and should be followed accordingly.
PB/BM samples
Upon receipt, examine the sample for proper collection, delivery, and labeling.
- Filter BM samples into a pre-labeled 15 mL conical tube using a 70 μm cell strainer (this step can be skipped for peripheral blood samples).The filtration step removes large particulate matter that may clog cytometer fluidics during acquisition. No filtration is required for peripheral blood.
Mix the sample thoroughly by vortexing on a low setting for 5s or inverting the closed tube 10 times.
- Obtain a WBC count and viability using the Cellometer Auto 2000 automatic cell counter.The nucleated count can also be obtained using another instrument such as a small CBC analyzer. Make sure that any alternative method can count nucleated cells in the presence of non-nucleated RBCs and platelets. Viability determination is helpful to assess sample quality but is not absolutely required for the method. In general, blood and marrow samples should have viability >90%. This method as described below uses a nucleic acid cell-permanent dye (acridine orange; AO) to obtain nucleated cell count. The ratio of damaged cells staining positively with cell-impermeant nucleic acid dye (propidium iodide) to AO-positive cells determines the reported viability. The calculation is performed automatically. Refer to instrument user manual for limitations and troubleshooting.
Dilute the sample at a 1:10 ratio by adding 10 μL of sample to 90 μL of PBS in a well of a 96 well microtiter plate. Mix the diluted sample thoroughly with the pipet tip by swirling and pipetting up and down in the well.
Remove plastic coverings (front and back) from one SD-100 slide. Label one side of the counting chamber with two patient identifiers. Set slide aside for later use.
Add 20 μL of ViaStain AOPI to another well of the microtiter plate.
Transfer 20 μL of the diluted sample to the ViaStain AOPI and mix thoroughly with the pipet tip by swirling and pipetting up and down. Aliquot 20 μL of this mixture into the loading chamber of the labeled SD-100 slide from step 6.
Wait until the mixture is completely loaded into the reading chamber without air bubbles. Repeat prior step if air bubbles are observed.
Insert slide into cell counter. Select the “Diluted” setting, click “count”, and record the result.
- For high cell count above the linear range of the instrument, dilute according to previously published guidelines and repeat steps 5-10 (Gao & Roshal, 2022).The maximum of analytical measurement range (AMR) of the Cellometer is 108 cells per ml (accounting for dilution), and thus highly concentrated samples must be diluted to produce precise counts. Each laboratory should verify the AMR based on instrumentation, sample types, and dilution procedures.
Tissue/core biopsy samples
-
12.Upon receipt, examine the sample for freezing, collection medium, as well as proper labeling.Tissue/core biopsy samples are acceptable in RPMI-1640 (preferred) or saline.
-
13.
Pour the contents of the patient sample from the collection tube into a petri dish.
-
14.
Rinse the conical tube 2x with RPMI-1640 medium and transfer it to the same petri dish.
-
15.Tease the samples apart with two 18-gauge needles.If there are large pieces of tissue/core biopsy, the samples should be minced with scalpels into smaller pieces before teasing.
-
16.Use the thumb press on a syringe plunger to crush samples in a circular motion.This step is to ensure all possible cells can be squeezed out of the solid samples.
-
17.
Transfer all the cell suspension from the petri dish using a transfer pipette and filter the suspension into a labeled 50 mL conical tube with a 70 μm cell strainer.
-
18.
Rinse the petri dish with 1-2 mL of RPMI-1640 medium (see recipe in Reagents and Solutions) and repeat the step 18 three times.
-
19.
Rinse the cell strainer with 1-2 mL of RPMI-1640 medium two times.
-
20.
Cap the conical tube and spin it down for 10 minutes at 500 x g at room temperature.
-
21.
Aspirate the supernatant without touching the cell pellet.
-
22.
Add approximately 25 mL of RPMI-1640 medium solution and repeat steps 20 to 21.
-
23.
Repeat step 22.
-
24.Resuspend the cell pellet in 300 μL of RPMI-1640 medium.The amount of resuspension solution should be adjusted based on the pellet size. Commonly, if a sizable excisional biopsy is processed, the cell pellet can be resuspended into 1 to 5 mL of solution.
-
25.Perform the cell count following steps 4 to 11.Note that if no RBC contamination is observed, the tissue/core specimens can be counted directly without performing 1:10 dilution.
CSF/body fluid samples
-
26.Upon receipt, examine the sample for freezing, collection medium, as well as proper labeling.CSF or other body fluids are preferred to be collected in a sterile container. CSF should be delivered to the laboratory immediately after the sample collection to preserve cell viability. If the fluid appears to be bloody (contaminated with red blood cells), a collection within anticoagulated tubes is preferred.
-
27.
Filter the specimen into a 15 mL or 50 mL conical tube using a 70 μm cell strainer (CSF does not need to be filtered).
-
28.
Fill the tube with RPMI-1640 medium (up to 25 mL in a 50 mL conical tube).
-
29.
Follow steps 20-21 and repeat the washing 2X.
-
30.Resuspend the cell pellet following step 24 and perform another cell count following steps 4-11.Note that we usually skip cell counting on CSF samples since the cell number is often below the instrument’s lower AMR (105 for the Cellometer).
Sample staining
-
31.Define the volume of sample needed for staining using approximately 1 million cells per tube, cap the maximum volume at 200 μl per tube for staining.For example, if the cell concentration is 10 million/mL, the required sample staining amount is 100 μL. If the concentration is below AMR, default to use 200 μL per sample. For tissue specimens, we use 50 μL per tube for an easier operational approach if cell concentration is low. CSF may be split equally based on the number of tubes required for the evaluation. However, the laboratory can adjust the minimum of cell number to be aliquoted based on the cell recovery of the staining procedure. Since this assay is not used for MRD purposes, a lower number of input cells may be used.
-
32.
Aliquot the appropriate volume of cells into a pre-labeled 5 mL polystyrene tube (flow tube), Follow steps 34-39 for PB/BM samples; go to step 40 for tissue/CSF/other fluids.
-
33.
Wash the cells with 3 mL of PBA.
-
34.
Centrifuge the tube for 5 minutes at 500 x g at room temperature.
-
35.
Aspirate the supernatant (as low as possible) without touching the pellet.
-
36.
Repeat the wash with PBA and aspirate the supernatant (steps 33-35).
-
37.
Wash the cells with 3 mL of RPMI-1640 medium.
-
38.
Centrifuge the tube for 5 minutes at 500 x g at room temperature.
-
39.
Aspirate the supernatant and resuspend the cell pellet in 50 to 100 μL of RPMI-1640 medium. Mix the pellet thoroughly by pipetting up and down 10 times.
-
40.Add up to 100 μL of the selected cocktail to the bottom of the tube. Mix sample with cocktail by vortexing on low for 5s or gently pipetting up and down.The cocktail volume should be equal to the sample volume per tube if the sample volume used is below 100 μL. E.g., 50 μL of sample requires 50 μL of the cocktail. When more than 200 μL of sample is used, add 100 μL cocktail per tube.
-
41.
Incubate the sample/cocktail mixture in the dark for 15 minutes at room temperature.
-
42.Mix cells by vortexing the tube on low for 5 seconds and add lyse-fix reagent.The lyse-fix reagent volume is added according to the aliquoted sample volume.1.5 ml of lyse-fix for sample volumes of 100 μL.3.0 ml of lyse-fix for sample volumes of 200 μL.
-
43.
Incubate in the dark for 15 minutes at room temperature.
-
44.
Centrifuge the tube for 5 minutes at 500 x g at room temperature.
-
45.
Decant and vortex on low to resuspend the cell pellet.
-
46.
Add 3 mL of PBA to the tube.
-
47.
Centrifuge the tube for 5 minutes at 500 x g at room temperature.
-
48.
Decant and vortex to resuspend the cell pellet.
-
49.Add 100 μL of PBA to the bottom of the tubeThis sample is ready for acquisition.
-
50.Add 1 μL of diluted DAPI and incubate the tube in the dark for 10 minutes at room temperature.This step only applies to tissue samples. Go to step 50 for PB/BM/Fluids.
-
51.Place the tube on ice and protect it from light until acquisition on flow cytometer.Acquisition on a flow cytometer should be performed within 1 hour.
Sample acquisition
-
52.Create and label the experiment with appropriate sample ID on the flow cytometer. Match patient identifiers from the tube label with the experiment name before acquisition.Follow cytometer software instructions to accomplish this step.
-
53.Clean the sample injection tube (SIT) and flush the system before acquiring the sample.
- Spray reagent-grade water and wipe SIT with gauze or Kimwipe after the acquisition of each tube.
- Acquire at least two reagent-grade water tubes filled with 1 mL of water on the flow cytometer after each patient tube, for at least 1 minute at a high flow rate.
A small number of cells from prior acquisitions may remain in the sample probe or instrument tubing. This step prevents carry-over from previously acquired samples. If the instrument is capable of self-cleaning the probe automatically after acquisition (ex. SIT flush capability), step 53a may be skipped. Number of water tubes required between samples may vary. Cleaning procedure should be verified and/or validated. -
54.Immediately prior to acquisition, vortex tube containing sample on low setting and place specimen on SIT to acquire. Check forward (FSC) and side scatter (SSC) to make sure all populations are on scale and adjust as needed. Standardization of FSC/SSC parameters is discussed in a previous protocol (Gao & Roshal, 2022).We find it helpful to standardize FSC and SSC parameters for all samples stained with the same methodology, which prevents unnecessary loss of sample and extra time for readjustment of scatter parameters. A lymphocyte bounding gate may be drawn to ensure that the lymphoid population falls within the expected range.
-
55.
Acquire up to 500,000 events or drain the tube – whichever happens first.
-
56.
Export the fcs or lmd files for analysis.
SUPPORT PROTOCOL 1
ANALYSIS AND INTERPRETATION OF B- AND T-CELL LYMPHOCYTE ASSAY
This Support Protocol provides a guide to the analysis and interpretation of data obtained from the performance of the Basic Protocol 1. The section details a gating strategy that the authors find optimal for isolating mature B-, T-, and NK-cells and identifying populations of interest. Normal cell populations expected to be identified in various sample types are described in some detail, to allow the reader to distinguish normal lymphoid populations from neoplastic and/or abnormal counterparts. The context of sample type is important, both for the normal populations expected and the differential diagnosis of abnormal populations. Typical immunophenotypes of several neoplastic disease categories are described. Comprehensive immunophenotyping of all possible lymphomas is beyond the scope of this article, but there are many other papers that describe their phenotypes (Craig & Dorfman, 2017; Debord et al., 2020; Glynn et al., 2019; Jevremovic & Olteanu, 2019; Kroft & Harrington, 2017, 2022). Supplemental Table 1 describes additional markers that can be tested for further subclassification when specific entities are suspected. While this assay can screen for and roughly categorize most lymphomas and lymphoid leukemias, detection of populations suspicious for some diagnostic entities should prompt further work-up with more specific flow assays.
Materials
Woodlist software (generous gift from Dr. Brent L. Wood) or other FC visualization software.
While the examples in this protocol use Woodlist (a custom software not available for purchase), the instructions are mostly software-neutral and can be used in any FC visualization package that allows for sequential gating of populations (e.g. FCS Express (De Novo Software), FlowJo (BD Biosciences), Infinicyt (Cytogonos), and others).
Time gate
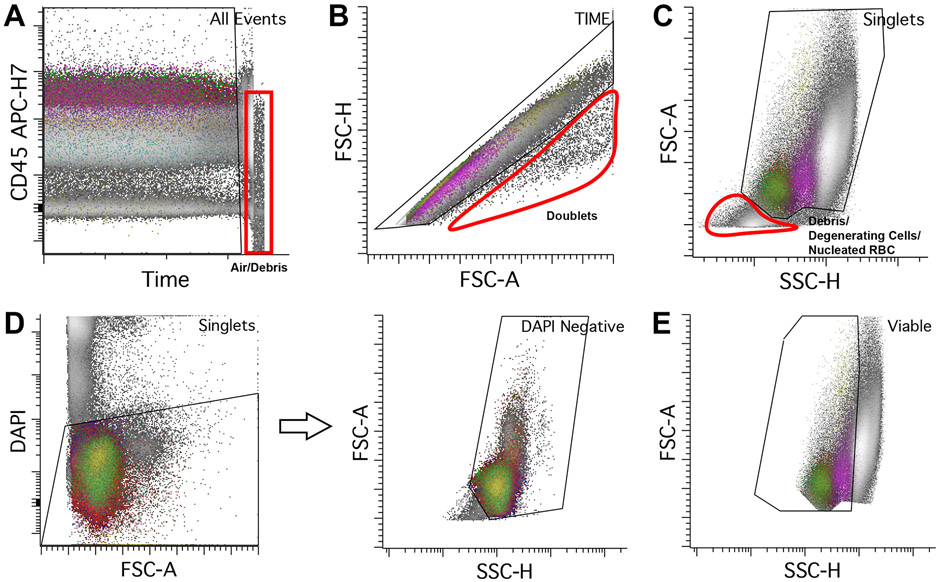
Gating of Time versus one of the fluorochromes is used to identify certain instrumental stability issues that may affect time delay, including sample clotting and fluid/pressure instability. Occasionally, acquisition of excessive air/debris due to sample exhaustion may be identified and should be excluded from the Time gate (Figure 1A). It is best to examine the channel with a fluorochrome that shows variable intensities in the stained populations. The fluorochrome must be excited by a laser other than the one used for measurement of SSC (i.e., 488-nm blue laser) to examine time delay. In this assay, CD45 APC-H7 is used to check the acquisition stability (Figure 1A).
Figure 1.
Initial gating from all events to mononuclear cells. A. CD45 APC-H7 versus time. Stable acquisition has all populations showing consistent intensities of a single selected fluorochrome (in this case CD45 APC-H7) over time. Towards the end of the acquisition, there is an abrupt drop in CD45 intensity (red box) from acquisition of air or debris due to sample exhaustion. It is recommended to remove these events from downstream analysis. B. Doublets (circled in red) should be excluded from analysis. An individual single cell has defined area, height, and width. To gate for singlets, events should be plotted using FSC height versus area, using a roughly diagonal singlet gate as shown. Cell aggregates falling outside of this linear zone are aggregates which should be excluded from downstream analysis. C. Viable cells can be gated using FSC (linear scale) versus SSC (log scale) to exclude degenerated cells, nucleated red cells, and other debris (circled in red). D. DAPI can also be used to exclude non-viable cell material; all cells that are negative for DAPI are gated which should yield similar results compared to the method in C; we use DAPI as a viability gating tool in all samples from solid tissue. E. Gating of mononuclear cells using FSC (linear scale) versus SSC (log scale). Include lymphocytes (lowest FSC and SSC) and monocytes (intermediate FSC and SSC), and exclude granulocytes (highest FSC and SSC). Note that immature B-cells (hematogones) usually have lower FSC and SSC than mature lymphocytes. Other cells that are captured within the mononuclear gate include basophils, blasts, dendritic cells, and plasma cells.
Doublets
A singlet gate using scatter parameters should be used to exclude cell aggregates or doublets to reduce coincident events that may be confused with abnormal cells (Figure 1B). Doublet exclusion is typically done by using a combination of area, height, and/or width.
The area scaling must be properly set up for accurate discrimination.
Viability
A viable gate is used to inspect the sample staining quality and exclude unwanted artifactual events, such as degenerated cells and lysed nucleated red cells. It is often examined by using FSC and SSC (Figure 1C). Degenerating cells typically show reduced FSC and increased SSC, while cell debris and nucleated red cells (depending on the sample type) usually shows reduction of both scatter parameters. In tissue samples (either FNAs or biopsy specimens), DAPI can be added to exclude dead or poorly viable cells. DAPI is a nucleic acid dye that binds to the DNA of cells with compromised cell membranes; DAPI positive populations should be excluded from the viable gate (Figure 1D)
It is important to set up an appropriate threshold during sample acquisition to minimize cell debris and lysed nucleated red cells (depending on sample type) during analysis. Additionally, the amount of DAPI for sample staining should be evaluated, as too much DAPI may increase the background with the dead cells presenting out of the maximum detectable scale; or the resolution between the live and dead cells may be poor if too little DAPI is added.
Normal B-cells in the context of the B-/T-tube
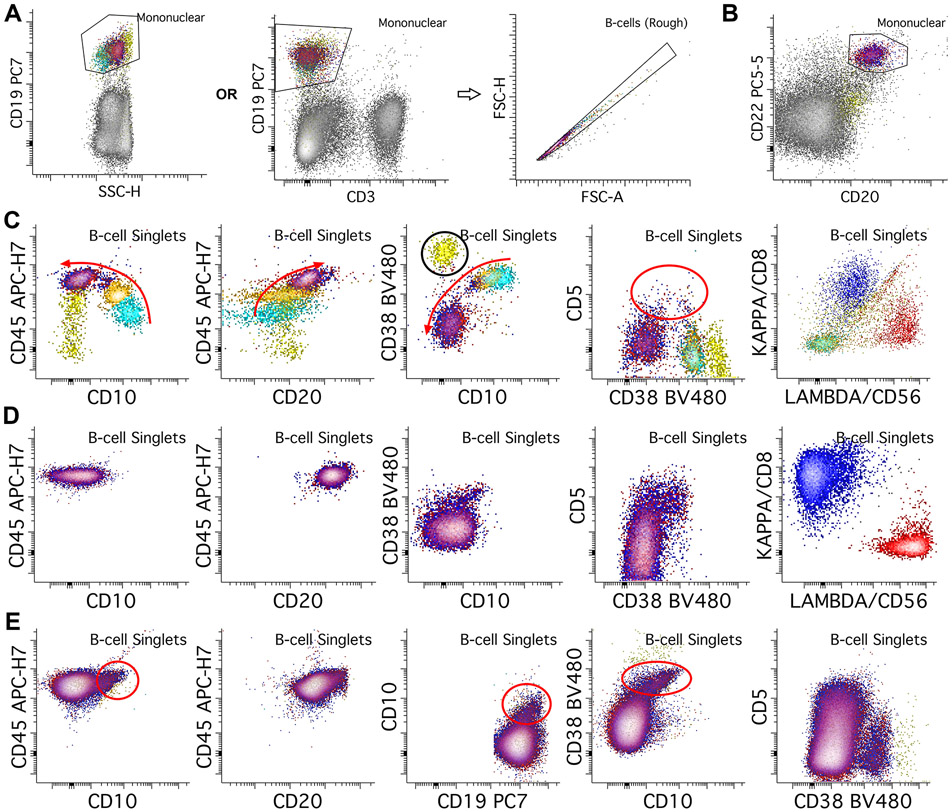
B-cell composition varies between sample types. B-cell development starts in the BM; these samples normally contain both immature B-cells (hematogones) and mature B-cells. In contrast, other sample types (PB, LN, extranodal tissue, fluids) contain mostly mature B-cells, though PB and LN samples may contain small populations of transitional B-cells. The primary B-cell gating can be done using CD19 and SSC on mononuclear cells, though alternative gates are useful in some situations (Figure 2A). Normal B-cells (both mature and immature) and some plasma cells are positive for CD19 and negative for CD3. While we can remove most doublets using the previous singlet gate, B-cells may aggregate due to CD19 engagement, and may not be cleared from the initial singlet gate because of variable cell sizes and granularities (Dezorella et al., 2016). Therefore, we recommend an additional, B-cell specific singlet gate to further reduce aggregated cells (Figure 2A).
Figure 2.
B-cell gating and normal patterns. A. B-cells are initially gated using CD19 versus SSC (log scale) plot, or alternatively with CD19 versus a T-cell marker such as CD3 to include B-cells and exclude T-cells from further B-cell assessment. We then place a second, B-cell singlet gate to further reduce B-cell aggregates. B. In some situations B-cells may lose CD19 expression or the antibody may not bind to B-cells due to interference from therapeutic agents; in these cases, if only mature B-cells assessment is being performed, alternative markers such as CD20 and/or CD22 can be used to gate for B-cells. C. In the bone marrow, the most immature B-cells (hematogones, light blue population) express bright CD10 and CD38, without CD20 or light chains, and with dim CD45. As the B-cells mature through intermediate (orange population) and mature stages (red/blue overlayed), they lose CD10 and CD38, while gaining CD20, CD45, and either kappa or lambda surface light chains (red arrows show changes with maturity). A transitional B-cell component can also express CD5 (red circle). Note also the plasma cells with brightest CD38 (yellow population, black circle). D. In normal peripheral blood, only mature B-cell populations (red and blue populations) should be present, with small amounts of transitional cells which can express CD5 and CD10. E. In benign lymph node specimens, normal B-cells typically show a germinal center component that expresses dim CD10 and bright CD38. Transitional B-cell populations can also be seen like those in peripheral blood.
Note that using CD19 as the sole B-cell gating agent poses several issues. CD19 may be dim or negative in some types of mature B-cell lymphoma (e.g., follicular lymphoma and large B cell lymphoma). Expression or staining of CD19 may be lost after targeted therapies. In addition, CD19 can bind to a subset of NK cells that may be mistaken for neoplastic B cells (Supplemental Figure 1). We recommend including at least one or more B-cell lineage markers to confirm the B-lineage. For example, we included both CD20 and CD22 as mature B cell-lineage markers in our tube (Figure 2B). Alternatively, surface light chain expression usually also helps confirm the B-cell identity.
In the BM, B-cell subsets include immature, maturing, and mature B-cells (Figure 2C). CD19 and CD22 are expressed on all normal maturing B-cells. CD20 is not expressed on the most immature B-cells and increases in expression as B-cells mature. CD45 is dimly expressed on immature B-cells and increases as they mature. CD38 is expressed by immature B-cells and becomes dim to absent with maturity. Plasma cells (PCs) show the brightest CD38 expression, and they are typically seen in the bone marrow in low proportions. CD10 is expressed by immature B-cells and lost as they mature. Some transitional B-cells may express CD5 (Palanichamy et al., 2009). Surface light chains are not expressed on immature B-cells; polytypic light chains are gained as the B-cells mature.
In PB samples, B-cells are largely mature, except for small populations of transitional B-cells (Figure 2D). The B-cells should express CD19, CD20, CD22, CD45, and either kappa or lambda surface light chains. CD38 expression is variable in the normal B-cell subsets in the peripheral blood. CD10 and CD5 are usually negative in mature B-cells of the PB, though they can be positive in PB transitional B-cell populations.
Lymph nodes contain of several B-cell subsets, including germinal center B-cells (GCBs) and marginal zone B-cells. Essentially all B-cell subsets in normal lymph nodes are mature and express CD19, CD20, CD22, CD45, and either kappa or lambda surface light chains. GCBs also express CD10 without CD5, with intermediate to bright CD38 expression (Figure 2E). Marginal zone B-cells are negative for both CD5 and CD10 and show dim to negative CD38 expression. Transitional B-cells with CD5 expression may be present. PCs again show the brightest CD38 expression.
Other tissue types typically show relatively low B-cell content but contain the same subsets as normal LNs. Fluid specimens including CSF typically contain low B-cell content, but the few B-cells present are from similar subsets as those seen in PB. In BM, tissue, and fluid specimens, estimation of granulocyte proportion to assess for evidence of PB contamination manifesting as an increased proportion of granulocytes, with reporting adjusted accordingly.
Normal T-/NK-cells in the context of the B-/T-cell tube
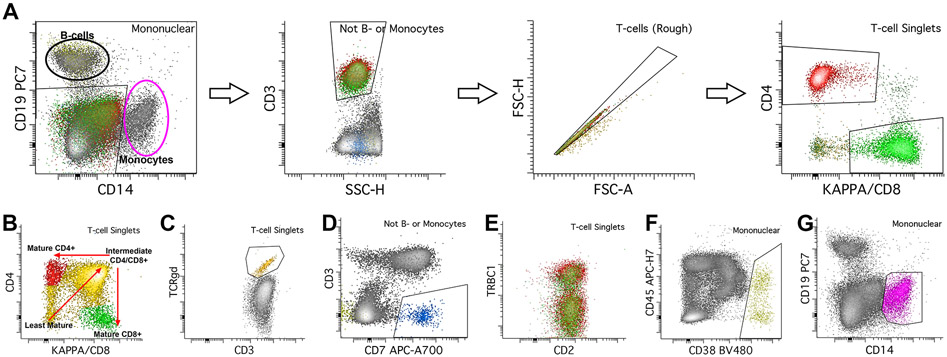
The primary T-cell gating is performed in two stages. First, B-cells and monocytes are gated within the mononuclear cell gate, using a combination of CD19 for B-cells and CD14 for monocytes (Figure 3A). Using everything outside of this gate (e.g., cells that are neither B-cells nor monocytes), T-cells are gated using CD3 and SSC. Like B-cells, we recommend an additional T-cell specific singlet gate to further reduce aggregated cells (Figure 3A). While normal T-cells express surface CD3, some abnormal T-cells can express diminished or negative CD3, and alternative antigens such as CD2, CD5, and CD7 can be used to gate T-cells. T-cells are then further sub-gated into CD4 and CD8 positive subsets (Figure 3A).
Figure 3.
Gating of T-cells, NK cells, plasma cells, and monocytes. A. Before gating T-cells, first gate B-cells and monocytes using a gate that capture CD19 positive B-cells and CD14 positive monocytes. Everything outside of this gate is neither B-cells nor monocytes. Using this inverted gate, gate T-cells using CD3 and SSC. We use a second, T-cell singlet gate to further reduce aggregates. Lastly, gate CD4 and CD8 positive subsets of T-cells. In blood, bone marrow, most tissue, and fluid specimens, there are mostly CD4 or CD8 positive T-cell subsets, with very small numbers of double positive or double negative specimens. B. In the normal thymus, there are maturing T-cell populations which start as CD4/CD8 double negative, then become CD4/CD8 double positive, and then finally become either CD4 or CD8 positive (red arrows show maturation pattern). C. TCRgd is used to gate for gamma-delta T-cell populations. D. NK cells are gated using CD3 and CD7; NK-cells are CD7 positive while CD3 negative, and should also express CD2, while negative for CD5 (not shown). E. TRBC1 can be used as a surrogate for clonality; mature T-cells express either TRBC1 or TRBC2, so polyclonal T-cell populations should show a mixture of TRBC1 negative and positive cells. F. Plasma cells are gated using CD38 and CD45; plasma cells show the brightest CD38 expression of any normal cells. G. Lastly, monocytes are gated using CD14.
T-cell maturation occurs in the thymus rather than the BM. While this tube is not designed for full review of T-cell maturation, it may be important to distinguish the normal thymic T cells from neoplastic mature T cells or abnormal immature T cells for screening purposes (Figure 3B). Thymic T-cell subsets include the most immature CD4/CD8 double negative populations, intermediate CD4/CD8 double positive populations, and mature CD4 and CD8 positive populations. Mature T-cells express pan-T-cell surface antigens CD2, CD3, CD5, and CD7.
Other sample types contain mostly mature T-cell subsets, typically at a CD4:CD8 ratio of 1:1 to 2:1. Minute subsets of CD4 and CD8 double negative and/or double positive T-cells are present in some normal samples. Increased proportions of CD4/CD8 double positive T-cells can be seen in LNs in the context of nodular lymphocyte predominant Hodgkin lymphoma (Chen et al., 2021; Rahemtullah et al., 2006).
Small subsets of T-large granular lymphocytes (LGL) can be seen in most specimens, though these populations are most pronounced in the BM and PB. These populations have reduced CD5 and CD7 expression compared to other T-cells. Importantly, this reduction is characterized by the variable distribution of CD5 and CD7 expression, rather than discrete CD5 and/or CD7 negative populations. The majority of LGLs express CD8 rather than CD4. Lymph nodes also contain T-follicular helper (TFH) cells, which co-express CD4 and bright CD279/PD-1 and can also show the expression of CD10. The antigens CD25 and CD26 are variably expressed on subsets of activated T-cells seen in normal samples.
Gamma-delta T-cells can also be detected using this assay via TCRgd antibody; minute populations (usually <1% of total WBCs) of gamma-delta T-cells can be detected in most normal specimens, with slightly larger proportions detected in BM, PB, and intestinal mucosal specimens (Figure 3C). NK cells show expression of CD7, express CD2, CD8 (subset), and CD56, and are negative for CD3, CD4, and CD5 (Figure 3D). Occasionally a minute CD5 positive NK subset maybe occasionally noted in reactive conditions (Supplemental Figure 1). Most normal specimens contain small numbers of NK cells, which may be higher in BM and PB samples.
TCRCβ1 (TRBC1) is used as evidence of clonality (Figure 3E). Normal mature T-cells express either TCRCβ1 or TCRCβ2; non-clonal populations have an approximately equal distribution of negatively and positively staining cells. Restricted positive or negative TRBC1 expression is used as evidence of clonality (Horna et al., 2021).
Gating of other cell populations
Additional cell populations that are important to gate include plasma cells and monocytes. Plasma cell populations are mononuclear cells with bright CD38 expression (Figure 3F). Mature monocyte populations are identified by CD14 expression on mononuclear cells (Figure 3G). Additionally, the expression of CD4 without CD3 can help identify monocytic cells.
Abnormal B-cell populations
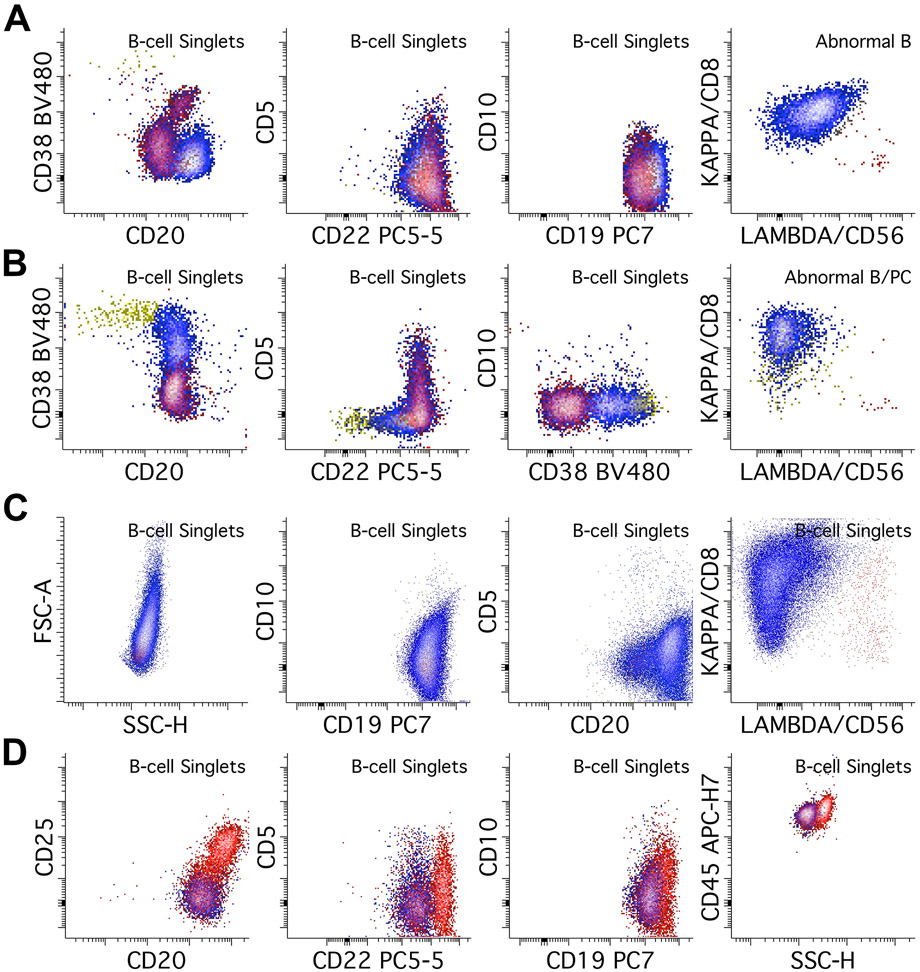
Neoplastic B-cell populations can be seen in any sample type; major hallmarks of abnormal B-cells are clonality and aberrant phenotype. Clonality in B-cells is assessed through surface light chain expression. Mature B-cells express either Kappa or Lambda light chain; light chain restriction or absence is used as evidence of clonality with this assay. Aberrancy is also required, and its identification requires knowledge of expected normal populations in a sample to identify antigens expressed at levels that are too bright, dim, uniform, and/or other aberrancies. Large B-cell lymphomas often have increased FSC and SSC, they should not be excluded from early gates, and evidence of large cell size should be annotated during reporting and prompt further work-up for aggressive lymphomas. Abnormal B-cells can be either dim or negative for CD19, particularly after CD19 targeted therapy. This may require an alternative gating strategy using CD20 and/or CD22 (Figure 2B). Neoplastic B-cells can be broadly categorized into CD5 positive, CD10 positive, or negative for both.
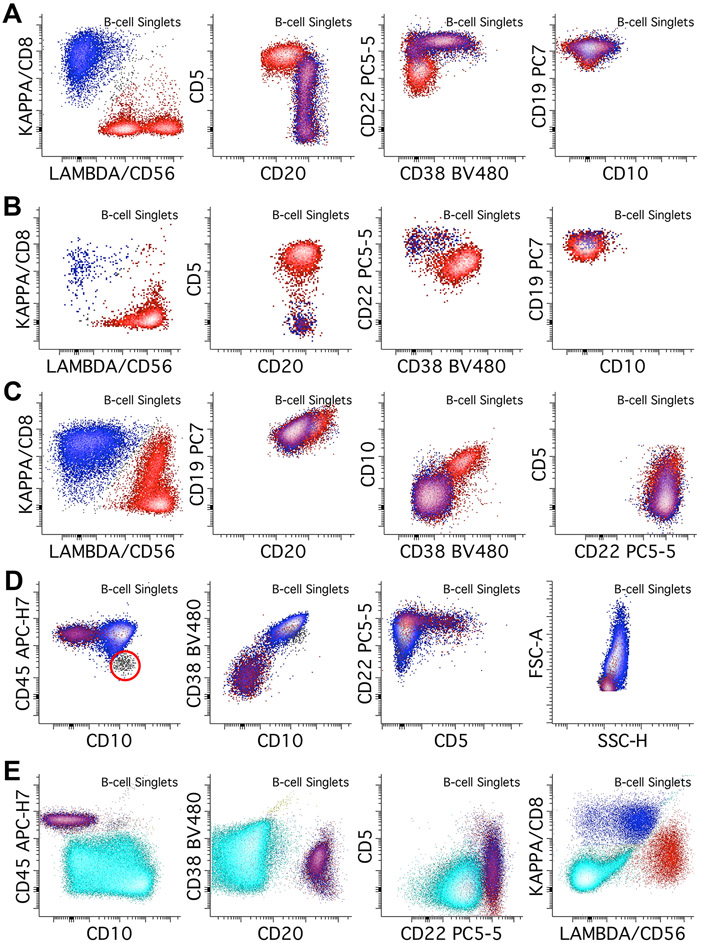
The main differential diagnosis for CD5-positive B-cell populations include chronic lymphocytic leukemia / small lymphocytic lymphoma (CLL/SLL), mantle cell lymphoma (MCL), and normal B-cells. Other B-cell processes that can express CD5 include subsets of marginal zone lymphoma (MZL), lymphoplasmacytic lymphoma (LPL), diffuse large B-cell lymphoma (DLBCL), and B-prolymphocytic leukemia/lymphoma. Rare cases of CD5-positive hairy cell leukemia (HCL) (Jain et al., 2016) and follicular lymphoma (FCL) (Li et al., 2015) are described. The classic CLL/SLL immunophenotype is CD5 positive, CD10 negative, CD20 dim to negative, CD22 dim to negative, with dim monotypic expression of either Kappa or Lambda light chain (Figure 4A). CD38 is usually dim to negative. Of note light chain expression may be negative and several clonal populations with different light chain expression may co-exist (oligoclonal/biclonal pattern). Small populations with a CLL/SLL-like phenotype are often seen, particularly in BM and PB specimens, often representing a monoclonal B-cell lymphocytosis (MBL) rather than involvement by a true CLL/SLL. Typical MCL phenotype is CD5 positive and CD10 negative, with normal expression of CD20 and CD22, and with usually brighter expression of surface light chain compared to CLL/SLL (Figure 4B). While the classic examples of CLL/SLL and MCL can often be differentiated by their intensity differences in CD20, CD22, CD38, and light chain expression, some cases may show unusual phenotypes. It is therefore recommended to perform further workups with markers that can help further distinguish these two neoplasms, such as CD23, CD79b, CD81, and CD200. In our lab, we perform a separate 14-antibody CLL tube to establish the full phenotype for follow-up (Gao & Roshal, 2022).
Figure 4.
Abnormal B-cells with CD5 or CD10 expression. Kappa positive B-cells are colored blue while lambda positive B-cells are colored red. A. An example of CLL/SLL, which shows dim/weak light chain restriction (in this case lambda restriction), with CD20 and CD22 dim, CD5 positive, CD38 negative, and CD10 negative abnormal B-cells. B. In contrast, this example of mantle cell lymphoma shows relatively bright light chain restriction (in this case lambda), with normal expression of CD20. CD22 may be dim in MCL. It also shows expression of CD5, dim CD38, and is negative for CD10. C. This lambda restricted follicular lymphoma typically shows normal CD19, though CD19 expression is often dim in follicular lymphoma. It also shows dim expression of CD10, bright CD20 expression, normal CD22 expression, without CD5, and positive CD38, though CD38 can be variable. D. Large B-cell lymphomas such as DLBCL and Burkitt lymphoma can also express CD10. In this case of a kappa-restricted Burkitt lymphoma, the population shows CD10 expression at the level of immature B-cells (gray population, red circle), CD45 expression at the level of background mature B-cells, bright CD38 expression, dim CD22 expression, and absence of CD5. FSC and SSC plots show increased scatter characteristics, consistent with large cell size. E. Lastly, B-lymphoblastic leukemia/lymphoma can be detected by this assay, though it is not designed for extensive immunophenotyping in B-ALL. Abnormal immature B-cells (cyan population) show dim/absent CD45 and CD20 expression, with variable CD10 expression, consistent with immature B-cells. CD38 is dim, CD5 is negative, and CD22 is dim. The blasts show no surface light chain expression.
The major differential diagnosis for CD10-positive abnormal B-cell populations includes FCL, GCB-type of DLBCL, Burkitt lymphoma, B-lymphoblastic leukemia/lymphoma (B-ALL), and normal B-cells (typically immature B-cells in BM or mature GCBs in LN samples). Other neoplastic B-cell processes should also be considered; CD10-positive HCL constitutes approximately 15% of HCL cases (Wang et al., 2016) and some MCLs (Xu et al., 2018) are described. FCL is typically CD10 positive, CD19 dim, and CD20 bright. CD38 is often dim or negative but can vary substantially (Figure 4C) (Aren et al., 2022). GCB-type DLBCL and Burkitt lymphoma in this assay are commonly CD10 and CD38 bright (may be brighter than GCBs), may show increased FSC/SSC, though their phenotypes may otherwise vary substantially (Figure 4D). While this assay is not designed for a complete assessment of B-ALL, abnormal B-lymphoblasts are usually CD19 positive, CD20 negative, CD45 dim populations, often with bright CD10 and dim CD38, and without light chain expression, though this may vary substantially (Figure 4E) (Cherian & Soma, 2021). If B-ALL is suspected, further tests designed for a more thorough assessment of immature B-cells must be performed; in our laboratory we perform a 14-color assay for B-ALL assessment (Gao et al., 2023).
The main differential for CD5/CD10 negative neoplastic B-cell populations includes MZL, LPL, HCL, and non-GCB type DLBCLs. The most common MZL phenotype is CD19 and CD20 bright, CD5 negative, and CD10 negative (Figure 5A). They can show evidence of plasmacytic or PC differentiation with increased expression of CD38, sometimes up to the level of plasma cells. Further assessment of plasma cells with a plasma cell-specific assay is also recommended in cases where PC differentiation is suspected. LPL can show a similar immunophenotype to MZL (Figure 5B). Non-GCB type DLBCLs are usually CD5 negative and by definition CD10 negative, usually with increased FSC/SSC (Figure 5C). HCL’s immunophenotype in this assay is typically CD5 negative, CD10 negative, CD19 bright, CD20 bright, CD22 bright, CD25 positive, and with increased SSC (Figure 5D). Identification of populations suspicious of HCL should be followed by testing for additional markers such as CD103, CD11c, and CD200.
Figure 5.
Abnormal B-cell populations without CD5 or CD10 expression. Kappa positive B-cells are colored blue while lambda positive B-cells are colored red. A. This case of a kappa restricted marginal zone lymphoma shows bright expression of CD20, dim CD22, and bright CD19, without CD5, CD10, or CD38 expression. B. This kappa restricted lymphoplasmacytic lymphoma shows no CD5 or CD10 expression, and shows variable CD38, increasing up to the level of plasma cells (yellow population). Further assessment of plasma cells using a plasma cell specific assay showed additional kappa restricted plasma cell population with an otherwise normal immunophenotype (not pictured). C. This case of a kappa restricted diffuse large B-cell lymphoma shows increased FSC/SSC consistent with larger cell size. It is negative for CD5 and CD10. D. Hairy cell leukemia can be detected by this assay; this lambda restricted example shows bright expression of CD20, CD22, CD19, and CD45, with increased side scatter, aberrant expression of CD25, and absence of CD5 and CD10. Additional testing with CD11c, CD103, and CD123 showed positive expression of those markers, consistent with HCL (not shown).
Abnormal T-/NK-cell assessment
Like B-cells, the FC hallmarks of T-cell neoplasia are both clonality and aberrancy. TCRCβ1 is a useful screening marker for clonality assessment. Normal mature T-cells express either TCRCβ1 or TCRCβ2; non-clonal populations have a variable expression of TCRCβ1 with subsets of negative and positive cells. Restricted positive or negative TCRCβ1 expression is used as evidence of clonality. It is important to remember that TCRCβ1 surface expression requires expression of both surface CD3 and alpha-beta T cell receptor (non-gamma-delta); TCRCβ1 loses its utility when abnormal T-cell populations are negative for CD3, a relatively common aberrancy (Horna et al., 2021).
Immunophenotypic aberrancy in T-cells is characterized by abnormal levels of expression of pan T-cell antigens such as CD2, CD3, CD5, CD7, as well as CD4 and/or CD8. Abnormally dim or negative surface CD3 expression can result in an abnormal population being outside of the traditional CD3-based T-cell gate, requiring alternative T-cell gating. Like B-cell neoplasms, T-cell lymphoma cells can show increased size evidenced by increased FSC/SSC parameters.
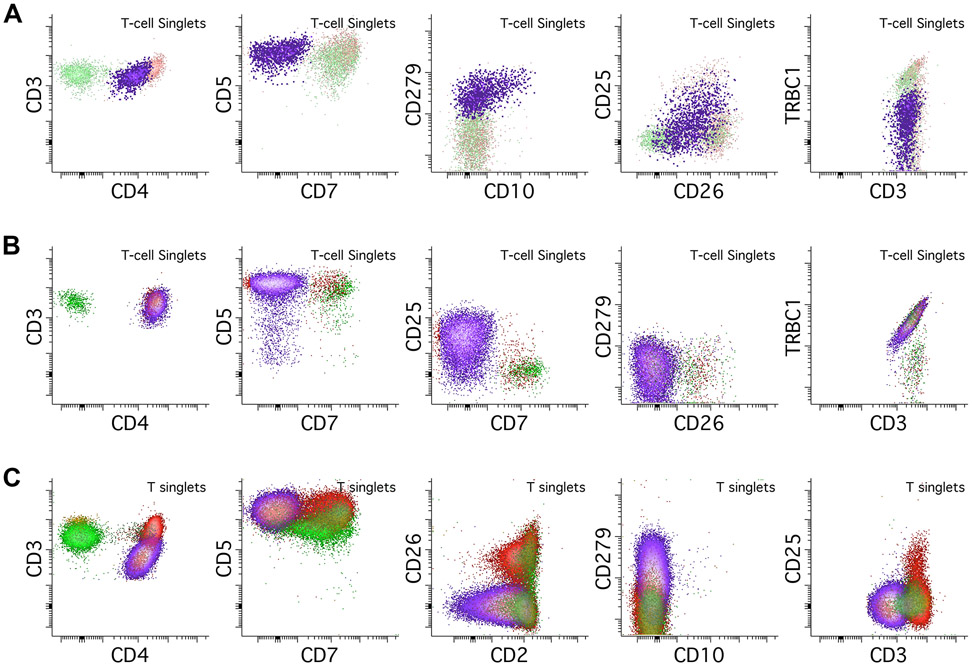
The immunophenotype of a T-cell lymphoma can significantly narrow the differential and in some cases be outright diagnostic. Expression of bright CD279/PD-1 is seen in T-cell neoplasms with TFH phenotypes such as angioimmunoblastic T-cell lymphoma (AITL), which is also classically surface CD3 negative, CD4 positive, CD7 negative, and occasionally CD10 positive, though normal CD3 expression is not infrequent (Figure 6A) (Jevremovic & Olteanu, 2019). Involvement of blood and marrow are nearly universal at diagnosis of AITL (Yabe et al., 2020). A combination of dim CD3, CD4 positivity, and CD7 negativity in the context of CD25 expression is relatively specific for adult T-cell leukemia/lymphoma (ATLL, Figure 6B) (Dahmoush et al., 2002). Cutaneous T-cell lymphomas such as mycosis fungoides (MF) and its circulating counterpart Sezary syndrome are usually CD4 positive, CD7 negative, CD26 negative, and often express CD279/PD-1 (Figure 6C) (Lewis et al., 2022).
Figure 6.
Abnormal T-cell populations. In all examples abnormal populations are colored purple, background CD4 positive T-cell are colored red, and background CD8 positive T-cells are colored green. A. This example of angioimmunoblastic T-cell lymphoma (AITL, purple population) shows a CD4 positive T-cell population with abnormal loss of CD7. It also shows expression of uniform bright PD-1/CD279 and partial co-expression of CD10. While there is some CD25 expression, it is below the level of the normal background T-cells. The population shows monotypic negative TRBC1 expression, a surrogate for clonality. B. This example of adult T-cell leukemia/lymphoma (ATLL, purple population) shows a CD4 positive T-cell population with abnormal loss of CD7 and partial loss of CD5. There is relatively bright expression of CD25 when compared to the background T-cells. There is no CD26 or PD-1/CD279 expression, and the population shows monotypic TRBC1 positivity, consistent with clonality. C. This example of a cutaneous T-cell lymphoma (CTCL) circulating in the peripheral blood shows abnormally dim expression of CD3 in a CD4 positive population. There is abnormal loss of CD7 and CD2, with CD26 negativity. There is also PD-1/CD279 expression, but no evidence of CD10 expression.
T-prolymphocytic leukemia (T-PLL) shows a brighter than normal expression of CD5 and CD7 with no expression of PD-1 and can express CD4, CD8, neither, or both (Figure 7A) (Jevremovic & Olteanu, 2019). T-large granular lymphocytic leukemia (T-LGLL) is defined by unexplained persistently increased numbers of LGLs in the PB associated with cytopenias; their typical immunophenotype is CD5 dim/negative, CD7 dim/negative, CD8 positive, and CD56 negative with restricted TRBC1 (Figure 7B) (Jevremovic & Olteanu, 2019). Detection of populations suspicious for T-PLL or T-LGLL can be followed by more specific assays; for T-PLL we add additional antibodies against cytoplasmic TCL1, and for T-LGLL we add on CD57 and sometimes the cytotoxic markers TIA1 and Granzyme B.
Figure 7.
Abnormal T-cell populations, continued. All abnormal populations are colored purple, background CD4 positive T-cells are red, and background CD8 positive T-cells are green. A. An example of T-prolymphocytic leukemia/lymphoma (T-PLL), showing abnormal absence of surface CD3 with bright CD7, dim CD2, bright CD5 and CD4 expression. T-PLL can be double negative or double positive for both CD4 and CD8; it rarely expresses CD8 alone. There is no expression of CD279/PD-1 or CD10, in contrast to angioimmunoblastic T-cell lymphoma (AITL) Additional testing showed positive expression of CD52 and TCL1 (not pictured). B. An example of a clonal T-large granular lymphocyte (T-LGL) population, which shows CD8 expression, dim CD5 and CD7 expression, and partial CD56 expression. TRBC1 shows monotypic positive expression consistent with clonality. C. An example of an anaplastic large cell lymphoma (ALCL), ALK negative. This case shows retained expression of CD3, CD2, and variable CD5 expression, with loss of CD7. It expresses CD4 but can be CD8 positive as well. CD25 can also be expressed, often brighter than background T-cells, which can also raise a possibility of adult T-cell leukemia/lymphoma. FSC and SSC parameters show marked increases suggestive of large size and high complexity. Addition of a CD30 antibody is strongly recommended in cases like this one.
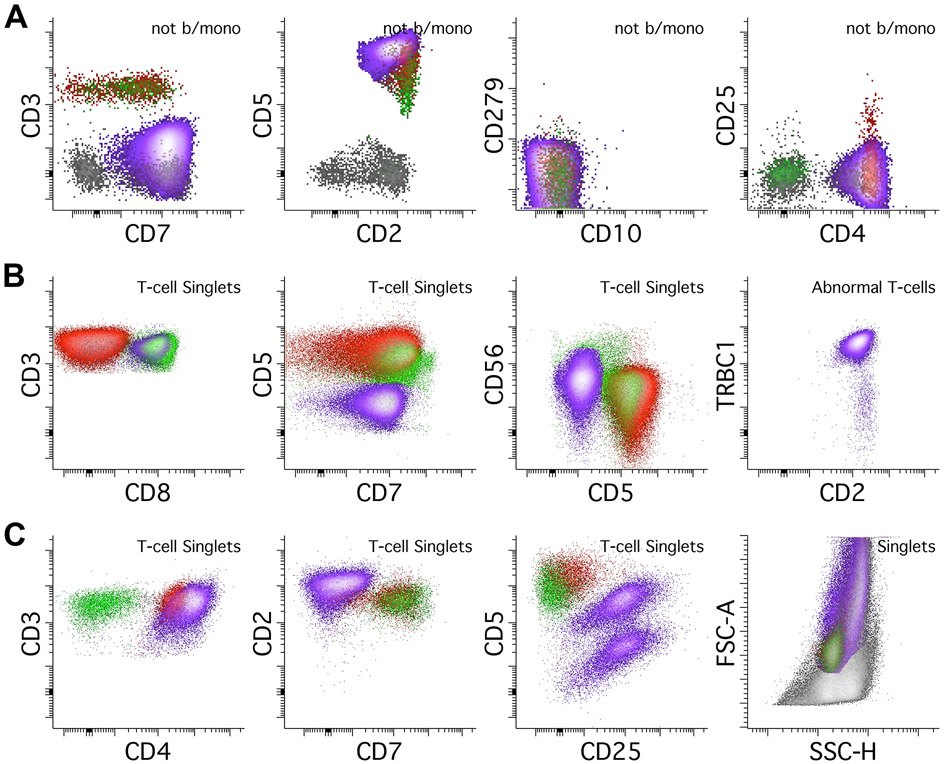
Anaplastic large cell lymphoma (ALCL) typically shows loss of T-cell antigens and occasionally can be negative for all pan-T-cell antigens (a so-called “null” phenotype). The most commonly retained markers are CD2, CD4, and CD5 (Swerdlow et al., 2017). In addition, ALCL shows markedly increased FSC/SSC parameters, often overlapping with monocytes and granulocytes (Figure 7C). If ALCL is suspected, addition of a CD30 antibody is strongly recommended (Supplemental Figure 2). Hepatosplenic T-cell lymphoma (HSTCL) is a rare aggressive type of T-cell lymphoma with a cytotoxic phenotype and often expressing gamma-delta T-cell receptors. It is often CD5 negative, CD4/8 double negative, and can express CD56. It is usually T-cell receptor gamma-delta positive and T-cell receptor alpha-beta negative, though occasionally can express alpha-beta (Kapur et al., 2014). Additional testing with cytotoxic markers is useful if HSTCL is suspected.
T-lymphoblastic leukemia/lymphoma (T-ALL) can also be detected using this method, though it is not designed for an extensive evaluation of T-ALL (Figure 8A). Immature T-cells are typically negative for surface CD3 negative, CD7 bright, CD38 bright, and CD45 dim to negative with or without CD10, often with CD4 and CD8 double negativity or positivity. In samples where T-ALL is suspected, further testing is vital, and antibodies for CD1a, CD34, CD48, CD99, TdT may be particularly useful for assessing T-cell immaturity (Supplemental Figure 2).
Figure 8.
Abnormal T-cells and NK cells, continued. All abnormal populations are colored purple. A. This assay can also detect T-lymphoblastic leukemia/lymphoma (T-ALL) but is not designed for deep immunophenotyping of T-ALL. T-ALL shows absence of surface CD3 and bright CD7 expression. The expression of CD10 on CD45 dim T-cells should prompt a thorough assessment for T-ALL. In contrast to AITL, T-ALL does not express CD279/PD-1. B. NK cell lymphomas can also be detected by this assay. This case of an extranodal NK/T-cell lymphoma shows CD2 expression in the absence of surface CD3 and CD5 expression. CD7 is mostly negative, which is an aberrant finding in NK-cells. The NK-cells express neither CD4 nor CD8 and are brightly positive for CD56. Additional testing in this case showed expression of CD94, CD335, and cytoplasmic cytotoxic markers TIA1 and Granzyme B (not shown).
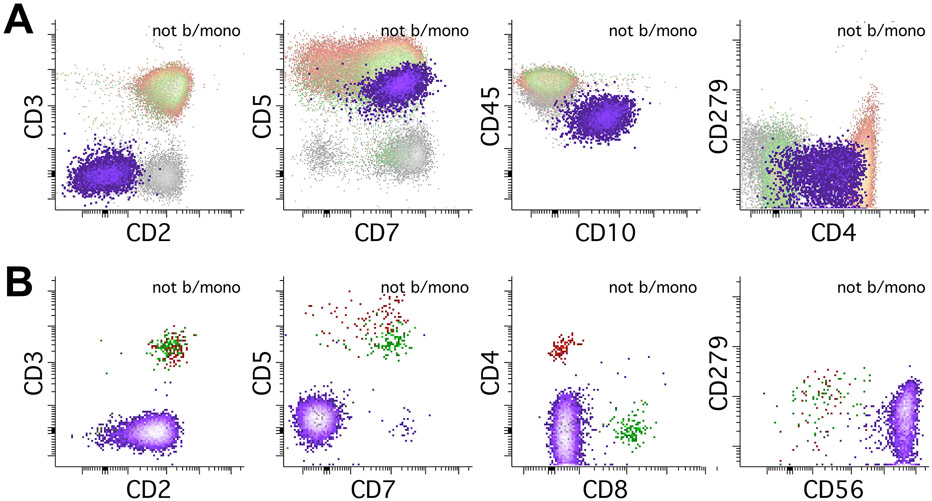
Lastly, abnormal NK-cells show nonspecific immunophenotypes that can overlap with abnormal T-cells using this assay. They can show aberrancies such as loss of CD2, CD7, and loss of CD56 (Figure 8B) (Swerdlow et al., 2017). As this test is not designed for extensive phenotyping of NK-cells, testing to confirm NK-cell lineage with markers such as CD94 or CD335, and/or cytoplasmic cytotoxic markers such as TIA1 or Granzyme B is recommended when abnormal NK-cell populations are suspected (Supplemental Figure 2).
An important pitfall to remember is that conventional dendritic cells (cDCs) may be sometimes identified as neoplastic T cells as they may show CD4, CD2, CD5, expression without CD3 or PD-1 (Supplemental Figure 3). Awareness of potential existence of this normal subset is important to avoid mistaking it for neoplastic T cells. Some helpful tips to distinguish the cDCs is that they usually show a slightly increased SSC (next to monocytes) without increasing FSC compared to the normal T cells and are always negative for CD279/PD-1. A negative cytoplasmic CD3 staining is helpful to rule out the T-lineage, and additional markers to confirm dendritic cell lineage can also be used.
Gating abnormal populations
When gating any abnormal populations, it is recommended to gate using multiple abnormal antigens when possible. Multiple sequential gates to refine a population can be used but this should be minimized if possible. Gating using markers of clonality such as Kappa, Lambda, or TRBC1 is not advised, as gating on these markers results in an artificially clonal-appearing population. When considering the number of cells in an abnormal population, there is no agreed upon standard for minimum population size; the assay has no formally established limit of detection. The size of a detectable population depends on a combination of the disease type, desired sensitivity limits based on both clinical and technical considerations, desired level of precision, and the number of cells acquired in a single test. In routine practice, we require a minimum of 20 events that cluster together, except on few occasions with clearly abnormal but minute populations, where we may require only 10 events to label something a “population” (Roshal, 2020).
REAGENTS AND SOLUTIONS
Lyse-fix reagent
Add 80.2 g of ammonium chloride (NH4Cl), 8.4 g of sodium bicarbonate (NaHCO3), and 3.7 g of EDTA to a 1 L volumetric flask. Fill flask to 1 L with reagent-grade water. Mix thoroughly using a stir bar or magnetic stir plate. Measure pH and adjust using sodium hydroxide (NaOH) and/or hydrochloric acid (HCl) to 7.2-7.4. Filter through a 0.45 mm Millipore filter. This is a 10x stock solution that can be stored at 2°-8° degrees C for 3 months. Add 50 mL of 10x stock and 12.5 mL of 10% formaldehyde (methanol-free, Polysciences, cat. No. 040181) to 500mL volumetric flask. Fill flask to 500 mL with reagent grade water and transfer to a light protected container.
PBA
Add one packet of phosphate-buffered saline powder (Sigma-Aldrich, cat. no. P3813-5 x10PAK), 10 mL of 30% bovine serum albumin (Sigma-Aldrich, cat. no. A7284), and 20 mL of 5% sodium azide (Ricca Chemical Company, cat. no. 7144.8-16) to a 1 L volumetric flask. Fill flask to 1 L with reagent grade water. Mix thoroughly by using a stir bar or magnetic stir plate and filter through a 0.45 mm Millipore filter. PBA can be stored at 2°-8° C for 1 month.
PBS
Add one packet of phosphate-buffered saline powder (Sigma-Aldrich, cat. no. P3813-5 x10PAK) to a 1 L volumetric flask. Fill flask to 1 L with reagent grade water. Mix thoroughly by using a stir bar or magnetic stir plate and filter through a 0.45mm Millipore filter.
RPMI-1640 medium
Add 50 mL of pre-thawed room temperature newborn calf serum (Life Technologies, cat. no. 26010-074) and 4 mL of pre-thawed room-temperature 100x penicillin-streptomycin (Gibco, cat. no. 15140-122) to a 500mL bottle of RPMI-1640 (Corning, cat. no. 10-041-CV). Mix thoroughly. RPMI-1640 medium can be stored at 2°-8°C for 2 weeks. Aliquot a small portion for daily use.
DAPI
Aliquot 5 μL of the DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) stock solution (1.0 mg/ml, BD Pharmingen, cat. no. 564907) into each of the microcentrifuge tubes upon reception of the reagent and label the tubes properly. Store the stock aliquots in −20°C freezer. When needed, obtain a vial of the DAPI stock solution, and thaw the vial at room temperature (protected from the light). Add 495 μL of Reagent Grade Water to the thawed DAPI stock and make a solution with 1:100 dilution (10 μg/mL). Store the vial at 2-8°C in the dark if not in use. The diluted DAPI solution can be stored up to 1 month from the day of dilution preparation.
COMMENTARY
Background Information
The assay described in this article provides a platform for detection and rough categorization of virtually all B- and T-cell non-Hodgkin lymphomas. The assay can guide further morphologic and IHC assessment for complete diagnosis and subclassification. While it is not designed for assessment of immature lymphoid neoplasms, it can screen for and detect them.
Separate cocktails for B- and T-cell assessment imparts a degree of modularity. While combined B- and T-cell assessment is vital in initial assessment for lymphoma, this is not always required in follow-up samples. Using a single cocktail for isolated B- or T-cell analysis also allows the use of additional drop-in antibodies that would be otherwise required in a separate reflex test.
The combined assessment of B- and T-cell populations in a single tube reduces use of materials, instrument time, and technologists’ labor. Prior to development of this protocol, new lymphoma assessments in our laboratory required separate assays for B- and T-cells. In our validation study, this assay resulted in a 52% reduction in the number of tubes stained compared to the previous method, improving workflow efficiency while maintaining test efficacy.
Critical Parameters
Reproducible analysis requires standardization of each step involved. Light exposure of the antibodies and antibody-stained samples should be minimized to prevent variability induced by light degradation of tandem fluorochromes. Cocktail stability should be validated. Instruments must be checked for optical and fluidic stability; multiple instruments should be standardized with respect to each other. Description of instrument setup, verification, standardization, and spill-over calculation is beyond the scope of this article (CLSI, 2021). However, successful analysis requires analyses on different instruments and days to be indistinguishable.
Analysis of abnormal B- and T-cells in various sample types requires understanding of the normal populations expected in each sample type. Normal samples of each possible sample type must be tested and analyzed during validation to verify detection of normal lymphoid populations. Critical aspects of the assay must be validated within each laboratory. The investigators are referred to excellent validation resources by CLSI (CLSI, 2021).
Troubleshooting
As with any clinical-grade assay, unexpected results should be treated with caution. Verification of sample identity is paramount, and verification that each step of the protocol was followed according to the written standard operating procedure. Expiration dates, storage, and handling conditions of each reagent should be established and adhered to.
Occasional samples will show nonspecific antibody binding or aggregation. This rarely interferes with analysis as aggregates can be ignored or gated out of downstream analysis. However, if nonspecific reactivity is extensive, additional washing with PBS-BSA may be considered prior to re-staining.
Time Considerations
The Basic Protocol 1 takes 1-1.5 hours to perform.
The Support Protocol 1 takes 5-15 minutes to perform.
Supplementary Material
ACKNOWLEDGEMENT:
This study was funded by the Center for Hematologic Malignancies at Memorial Sloan Kettering Cancer Center and in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Footnotes
CONFLICT OF INTEREST STATEMENT:
The authors have no relevant conflicts of interest to disclose.
Non-relevant disclosures:
Mikhail Roshal: Auron Pharma (advisory role); Roche (research funding).
DATA AVAILABILITY STATEMENT:
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Literature Cited
- Aren M, Marce S, Jurado R, Tapia G, Puigdefabregues L, Raya M, … Sorigue M (2022). Flow cytometry to detect bone marrow involvement by follicular lymphoma. Cytometry B Clin Cytom, 102(6), 427–439. doi: 10.1002/cyto.b.22098 [DOI] [PubMed] [Google Scholar]
- Chen ZW, Wizniak J, Shang C, & Lai R (2021). Flow Cytometric Detection of the Double-Positive (CD4+CD8+)/PD-1bright T-Cell Subset Is Useful in Diagnosing Nodular Lymphocyte-Predominant Hodgkin Lymphoma. Arch Pathol Lab Med. doi: 10.5858/arpa.2020-0726-OA [DOI] [PubMed] [Google Scholar]
- Cherian S, & Soma LA (2021). How I Diagnose Minimal/Measurable Residual Disease in B Lymphoblastic Leukemia/Lymphoma by Flow Cytometry. Am J Clin Pathol, 155(1), 38–54. doi: 10.1093/ajcp/aqaa242 [DOI] [PubMed] [Google Scholar]
- Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, … United Kingdom National Cancer Research, I. (2014). Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol, 32(27), 3059–3068. doi: 10.1200/JCO.2013.54.8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. (2021). Validation of assays performed by flow cytometry (1st ed). In CLSI guideline H62. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cozzolino I, Rocco M, Villani G, & Picardi M (2016). Lymph Node Fine-Needle Cytology of Non-Hodgkin Lymphoma: Diagnosis and Classification by Flow Cytometry. Acta Cytol, 60(4), 302–314. doi: 10.1159/000448389 [DOI] [PubMed] [Google Scholar]
- Craig JW, & Dorfman DM (2017). Flow Cytometry of T cells and T-cell Neoplasms. Clin Lab Med, 37(4), 725–751. doi: 10.1016/j.cll.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Dahmoush L, Hijazi Y, Barnes E, Stetler-Stevenson M, & Abati A (2002). Adult T-cell leukemia/lymphoma: a cytopathologic, immunocytochemical, and flow cytometric study. Cancer, 96(2), 110–116. doi: 10.1002/cncr.10480 [DOI] [PubMed] [Google Scholar]
- Debord C, Wuilleme S, Eveillard M, Theisen O, Godon C, Le Bris Y, & Bene MC (2020). Flow cytometry in the diagnosis of mature B-cell lymphoproliferative disorders. Int J Lab Hematol, 42 Suppl 1, 113–120. doi: 10.1111/ijlh.13170 [DOI] [PubMed] [Google Scholar]
- Dezorella N, Kay S, Baron S, Shapiro M, Porat Z, Deutsch V, … Katz BZ (2016). Measurement of lymphocyte aggregation by flow cytometry-physiological implications in chronic lymphocytic leukemia. Cytometry B Clin Cytom, 90(3), 257–266. doi: 10.1002/cyto.b.21263 [DOI] [PubMed] [Google Scholar]
- Gao Q, Liu Y, Aypar U, Baik J, Londono D, Sun X, … Roshal M (2023). Highly sensitive single tube B-lymphoblastic leukemia/lymphoma minimal/measurable residual disease test robust to surface antigen directed therapy. Cytometry B Clin Cytom. doi: 10.1002/cyto.b.22120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, & Roshal M (2022). Minimal/Measurable Disease Analysis in Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma by Flow Cytometry. Curr Protoc, 2(8), e503. doi: 10.1002/cpz1.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E, Soma L, Wu D, Wood BL, & Fromm JR (2019). Flow Cytometry for Non-Hodgkin and Hodgkin Lymphomas. In Kuppers R (Ed.), Lymphoma: Methods and Protocols (2nd ed.). New York, NY, USA: Springer Nature. [DOI] [PubMed] [Google Scholar]
- Horna P, Shi M, Olteanu H, & Johansson U (2021). Emerging Role of T-cell Receptor Constant beta Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies. Int J Mol Sci, 22(4). doi: 10.3390/ijms22041817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz SM, Ansell S, Ai WZ, Barnes J, Barta SK, Brammer J, … Sundar H (2022). T-Cell Lymphomas, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw, 20(3), 285–308. doi: 10.6004/jnccn.2022.0015 [DOI] [PubMed] [Google Scholar]
- Jain D, Dorwal P, Gajendra S, Pande A, Mehra S, & Sachdev R (2016). CD5 positive hairy cell leukemia: A rare case report with brief review of literature. Cytometry B Clin Cytom, 90(5), 467–472. doi: 10.1002/cyto.b.21365 [DOI] [PubMed] [Google Scholar]
- Jevremovic D, & Olteanu H (2019). Flow Cytometry Applications in the Diagnosis of T/NK-Cell Lymphoproliferative Disorders. Cytometry B Clin Cytom, 96(2), 99–115. doi: 10.1002/cyto.b.21768 [DOI] [PubMed] [Google Scholar]
- Kapur LH, Khaled Y, Solh M, Ward D, & Chang CC (2014). De novo CD3 negative hepatosplenic T-cell lymphoma: diagnostic challenges and pitfalls. Arch Pathol Lab Med, 138(7), 969–973. doi: 10.5858/arpa.2013-0074-CR [DOI] [PubMed] [Google Scholar]
- Kroft SH, & Harrington AM (2017). Flow Cytometry of B-Cell Neoplasms. Clin Lab Med, 37(4), 697–723. doi: 10.1016/j.cll.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Kroft SH, & Harrington AM (2022). How I Diagnose Mature T-Cell Proliferations by Flow Cytometry. Am J Clin Pathol, 158(4), 456–471. doi: 10.1093/ajcp/aqac079 [DOI] [PubMed] [Google Scholar]
- Lewis NE, Gao Q, Petrova-Drus K, Pulitzer M, Sigler A, Baik J, … Roshal M (2022). PD-1 improves accurate detection of Sezary cells by flow cytometry in peripheral blood in mycosis fungoides/Sezary syndrome. Cytometry B Clin Cytom, 102(3), 189–198. doi: 10.1002/cyto.b.22070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu S, Zuo Z, Hong M, Lin P, Li S, … Yin CC (2015). CD5-positive follicular lymphoma: clinicopathologic correlations and outcome in 88 cases. Mod Pathol, 28(6), 787–798. doi: 10.1038/modpathol.2015.42 [DOI] [PubMed] [Google Scholar]
- Maciorowski Z, Chattopadhyay PK, & Jain P (2017). Basic Multicolor Flow Cytometry. Curr Protoc Immunol, 117, 5 4 1–5 4 38. doi: 10.1002/cpim.26 [DOI] [PubMed] [Google Scholar]
- Palanichamy A, Barnard J, Zheng B, Owen T, Quach T, Wei C, … Anolik JH (2009). Novel human transitional B cell populations revealed by B cell depletion therapy. J Immunol, 182(10), 5982–5993. doi: 10.4049/jimmunol.0801859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahemtullah A, Reichard KK, Preffer FI, Harris NL, & Hasserjian RP (2006). A Double-Positive CD4+CD8+ T-Cell Population Is Commonly Found in Nodular Lymphocyte Predominant Hodgkin Lymphoma. American Journal of Clinical Pathology, 126(5), 805–814. doi: 10.1309/y8kd32qgryfn1xqx [DOI] [PubMed] [Google Scholar]
- Roshal M. (2020). Measurable disease evaluation in patients with myeloma. Best Pract Res Clin Haematol, 33(1), 101154. doi: 10.1016/j.beha.2020.101154 [DOI] [PubMed] [Google Scholar]
- Selliah N, Eck S, Green C, Oldaker T, Stewart J, Vitaliti A, & Litwin V (2019). Flow Cytometry Method Validation Protocols. Curr Protoc Cytom, 87(1), e53. doi: 10.1002/cpcy.53 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Fuchs HE, & Jemal A (2022). Cancer statistics, 2022. CA Cancer J Clin, 72(1), 7–33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe E, Pileri SA, Stein H, … Siebert R (2017). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (4th ed. Vol. 2). Lyon, France: International Agency for Research on Cancer Press. [Google Scholar]
- Thandra KC, Barsouk A, Saginala K, Padala SA, Barsouk A, & Rawla P (2021). Epidemiology of Non-Hodgkin’s Lymphoma. Med Sci (Basel), 9(1). doi: 10.3390/medsci9010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tadros AS, Hoh CK, & Wang HY (2016). CD10-Positive Hairy Cell Leukemia Involving Multiple Deep Lymph Nodes. Clin Lymphoma Myeloma Leuk, 16(5), e51–53. doi: 10.1016/j.clml.2016.02.041 [DOI] [PubMed] [Google Scholar]
- Xu J, Medeiros LJ, Saksena A, Wang M, Zhou J, Li J, … Li S (2018). CD10-positive mantle cell lymphoma: clinicopathologic and prognostic study of 30 cases. Oncotarget, 9, 11441–11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe M, Gao Q, Ozkaya N, Huet S, Lewis N, Pichardo JD, … Roshal M (2020). Bright PD-1 expression by flow cytometry is a powerful tool for diagnosis and monitoring of angioimmunoblastic T-cell lymphoma. Blood Cancer J, 10(3), 32. doi: 10.1038/s41408-020-0301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GH, Vergara N, Moore EM, & King RL (2014). Use of flow cytometry in the diagnosis of lymphoproliferative disorders in fluid specimens. Diagn Cytopathol, 42(8), 664–670. doi: 10.1002/dc.23106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.