Abstract
Amplified-fragment length polymorphism (AFLP) analysis is the name given to a genotypic technique in which adapter oligonucleotides are ligated to restriction enzyme fragments and then used as target sites for primers in a PCR amplification process. The amplified fragments are electrophoretically separated to give strain-specific band profiles. We have developed a single-enzyme approach that did not require costly equipment or reagents for the fingerprinting of strains of Helicobacter pylori. The method was assessed with 46 isolates of H. pylori from 28 patients, and the results were compared with those from other genotypic tests. The AFLP profiles derived from HindIII fragments differentiated strains of H. pylori from unrelated individuals and confirmed the common origin of strains in some family members. AFLP analysis was also applied to investigate persistent infection following antibiotic therapy. Overall, the modified technique was relatively rapid and technically simple yet gave reproducible and discriminatory results. AFLP analysis samples variation throughout the genome and is a valuable addition to the existing genotypic fingerprinting methods for H. pylori.
In recent years the range of molecular techniques available for epidemiological fingerprinting has expanded, and there are now many genotypic methods that allow high levels of discrimination between bacterial strains. In this study we have modified and evaluated a PCR-based technique, amplified-fragment length polymorphism (AFLP) analysis, for use in fingerprinting strains of Helicobacter pylori. The method was originally developed for the typing of crop plants (European patent application 0534858A1 [14]) and has been applied to the typing of plant, animal, and prokaryotic DNAs (12). It has proved to be useful for differentiating between strains of Legionella pneumophila (11).
A number of different AFLP-based techniques have been described (3, 4, 11, 12). There are four steps to the AFLP procedure used here: (i) digestion of the extracted DNA with a single enzyme, (ii) ligation of an adapter, designed to disrupt the enzyme restriction site, to each sticky end of the digestion fragments, (iii) PCR amplification of the adapter-tagged fragments with a single primer which is complementary to the adapter sequence, and (iv) electrophoretic separation and ethidium bromide detection of the amplified fragments in an agarose gel. Not all adapter-tagged fragments are amplified because the primer sequence extends one nucleotide, at the 3′ end, beyond the adapter and into the DNA fragment and thereby confers selectivity on the amplification process.
H. pylori infects approximately 40% of individuals in the United Kingdom, but the exact routes and modes of transmission of the organism remain unclear. Accurate fingerprinting techniques are required to address such questions and to help determine if the most common cause of treatment failure is due to the acquisition of antibiotic resistance by the organism or through reinfection with a new strain. In this study a single-enzyme approach to AFLP analysis was developed as a new method for typing strains of H. pylori. AFLP analysis was performed with 46 strains of H. pylori that were characterized by at least one other molecular method (urease genotyping or ribotyping). These included isolates that were epidemiologically unrelated, strains isolated from the same patients before and after antibiotic treatment, and strains from family groups. The AFLP analysis results were assessed for reproducibility and were analyzed in light of the other genotypic and phenotypic data available for the isolates examined.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The 46 isolates of H. pylori used in this study were from gastric biopsy specimens from 28 patients and included 3 strains obtained from the National Collection of Type Cultures (NCTC), 9 further strains from unrelated subjects, 10 strains from individuals belonging to four different families in Belfast, Northern Ireland, and 24 strains from six unrelated individuals who each provided isolates from two pretreatment and two posttreatment biopsy specimens. Details of the isolates are listed in Tables 1 to 3. The strains of H. pylori were preserved in 10% (vol/vol) glycerol in Nutrient Broth (Oxoid) over liquid nitrogen or at −80°C and were cultured on blood agar (Columbia agar base; Oxoid) with 10% (vol/vol) defibrinated horse blood at 37°C under microaerobic conditions (4% O2, 5% CO2, 3% H2, and 88% N2) in a variable atmosphere incubator (Don Whitley Scientific Ltd., Yorkshire, United Kingdom).
TABLE 1.
Details for isolates of H. pylori from unrelated individuals examined by AFLP analysisa
| Strain | Urease AB genotypeb |
|---|---|
| NCTC 11637 | U-2 |
| NCTC 11638 | U-10 |
| NCTC 12455 | U-3 |
| A222/89 | U-28 |
| A650/91 | U-6 |
| A657/91 | U-34 |
| A610/92 | U-7 |
| 10C | U-22 |
| 20A | U-3 |
| 41A | U-46 |
| 46C | U-1 |
| 86C | U-15 and U-5c |
All isolates had unique AFLP profiles.
Urease AB genotype according to the new classification of Owen (5).
Mixed bacterial culture in terms of urease genotype.
TABLE 3.
Details for multiple isolates of H. pylori obtained before and after antibiotic treatment from single patients
| Patient | Biopsy specimena | Antibiotic susceptibilityb
|
Urease AB typec | AFLP typed | |
|---|---|---|---|---|---|
| Cla | Mz | ||||
| 48 | Pre-A | S | R | U-3 | D |
| Pre-C | S | R | U-3 | D | |
| Post-A | R | R | U-3 | D | |
| Post-C | R | R | U-3 | D | |
| 205 | Pre-A | S | R | U-2 | E |
| Pre-C | S | R | U-2 | E | |
| Post-A | R | R | U-2 | E | |
| Post-C | R | R | U-2 | E | |
| 13 | Pre-A | S | S | U-2 | F |
| Pre-C | S | S | U-2 | F | |
| Post-A | R | S | U-71 | G | |
| Post-C | R | S | U-71 | G | |
| 67 | Pre-A | S | S | U-43 | H1 |
| Pre-C | S | S | U-43 | H1 | |
| Post-A | R | R | U-43 | H2 | |
| Post-C | R | R | U-43 | H1 | |
| 217 | Pre-A | S | S | U-29 | I1 |
| Pre-C | S | S | U-29 | I2 | |
| Post-A | S | S | U-29 | I1 | |
| Post-C | S | S | U-29 | I3 | |
| Ae | Pre-A | ND | ND | U-105 | J |
| Pre-A | ND | ND | U-105 | J | |
| Post-A | ND | ND | U-3 | K | |
| Post-A | ND | ND | U-3 | K | |
Pre-, pretreatment biopsy specimen; Post-, posttreatment biopsy specimens; A antral biopsy specimen; C, corpus biopsy specimen.
Cla, clarithromycin; Mz, metronidazole; S, sensitive; R, resistant; ND, not determined.
Urease AB type according to the new classification of Owen (5).
The AFLP type was arbitrarily designated when visual comparison indicated a high degree of overall similarity in the band patterns. Numbers indicate pattern variations.
Ribotypes were previously determined for these strains (6). The two pretreatment strains were ribotype 1, and the two posttreatment strains were ribotype 4.
DNA extraction, restriction endonuclease digestion, and ligation of adapters.
DNA was extracted either by the cetyltrimethylammonium bromide method by the DNA miniprep protocol of Wilson (13) or by using a nucleic acid extraction kit (IsoQuick; Orca Research Inc., Bothell, Wash.). The precipitated DNA was dissolved in 50 to 100 μl of distilled water, and the concentration and purity of the samples were determined by obtaining absorbance readings at 230, 260, and 280 nm. An aliquot containing 10 μg of DNA was digested overnight (16 h) at 37°C with 24 U of HindIII (NBL Gene Sciences, Northumberland, United Kingdom) in the buffer provided with the enzyme (50 mM Tris-HCl [pH 8.3], 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol) with 5 mM spermidine trihydrochloride (Sigma, Poole, United Kingdom) added in a final volume of 20 μl. A 5-μl aliquot containing 2.5 μg of digested DNA was used in a ligation reaction containing 0.2 μg of each adapter oligonucleotide (detailed below), 1 U of T4 DNA ligase (Boehringer Mannheim, East Sussex, United Kingdom), and single-strength ligase buffer (66 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP) in a final volume of 20 μl held at 37°C for 3 to 4 h. Combined restriction-ligation reactions involved the digestion of 2.5 μg of DNA with 24 U of HindIII in the ligase buffer and reaction mixture as described above for the ligation reaction only.
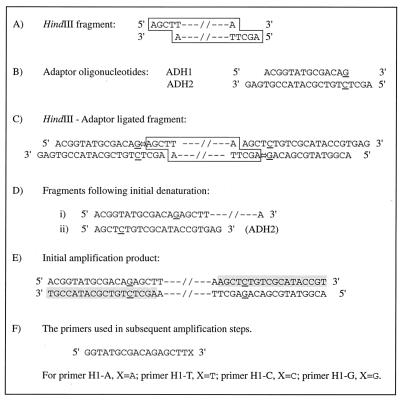
The complementary oligonucleotide sequences used for the adapter are shown in Fig. 1 and were synthesized by PE-Applied Biosystems, Warrington, United Kingdom. These oligonucleotides incorporated an additional base pair in the restriction site (11) in order to eliminate it after ligation of the adapter to the restricted fragment, as illustrated in Fig. 1. This allows the restriction enzyme to be present in the ligation mixture to prevent rejoining of the DNA fragments. The adapter oligonucleotides were not phosphorylated, and because T4 ligase requires a 5′ phosphate as well as a 3′ hydroxyl group to catalyze the linkage of two DNA strands, only one of the two oligonucleotides comprising the adapter was ligated. In our experiments, the 5′ phosphate of the restricted DNA fragment was used to ligate the shorter oligonucleotide, ADH1.
FIG. 1.
Diagram illustrating the AFLP technique used in the study. (A) The double-stranded fragment produced from digestion with HindIII (recognition site A↓AGCTT). (B) Sequences of the two complementary oligonucleotides forming the adapter which is ligated to each end of the HindIII restriction fragment. Nucleotides which have been inserted into the adapter sequence to eliminate the restriction site after ligation are underlined. (C) Fragment formed following ligation to the adapter. The double-headed arrow marks the point where ligation occurs. (D) Fragments present after initial denaturation at 94°C. (E) Initial amplification product. Newly synthesized DNA is shaded. (F) Primers used in subsequent amplification reactions. These are based on the adapter sequence. Selectivity is conferred by the final 3′ base of each primer.
PCR template preparation for AFLP analysis.
The ligated DNA was precipitated with ammonium acetate (final concentration, 2.5 M in a total volume of 100 μl) and 200 μl of chilled absolute ethanol. The samples were held at −20°C for 30 min and were then centrifuged at 12,000 × g for 15 min at 4°C. The precipitated DNA was washed in 70% (vol/vol) ethanol and resuspended in 100 μl of distilled water. A 5-μl aliquot was used as template for PCR.
For experiments in which the precipitation step was omitted, a ligated DNA sample was diluted (1/5 or 1/10) in distilled water and was then heated to 80°C for 10 min to inactivate the T4 ligase. A 5-μl aliquot was used as template for PCR.
PCR primers and PCR for AFLP analysis.
The four primers used in the PCR are shown in Fig. 1. They were synthesized by PE-Applied Biosystems. Amplification reactions were performed in a total volume of 50 μl containing 5 μl of template DNA, 2.5 mM MgCl2, 150 ng of a single primer, and 0.2 μl (1 U) of Taq DNA polymerase (Gibco BRL) in 1× PCR buffer provided by the manufacturer. The amplification cycles were an initial denaturing step of 94°C for 4 min, followed by 33 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2.5 min. The PCR primer used had the same sequence as ADH1 and required the complementary sequence, that of primer ADH2, for annealing. However, ADH2 was not ligated to the DNA fragment and so ADH2 dissociated from the DNA fragment during the initial denaturation steps in the PCR. As a result, in the first stages of the reaction the Taq polymerase filled in the overhang area left by the departure of ADH2 (Fig. 1). Amplified fragments were separated by electrophoresis in a 1.5% (wt/vol) agarose gel (Ultrapure Agarose; Gibco BRL) in TBE buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA) and were stained with ethidium bromide (0.5 μg/ml).
Genotyping by ureA and ureB PCR-restriction fragment length polymorphism (RFLP) analysis.
PCR amplification in reaction mixtures containing 100 ng of template DNA was undertaken as described previously (8). A 2,410-bp fragment spanning nucleotides 2648 and 5057 of the urease A and B genes (ureA and ureB) was amplified with the primers 5′-AGGAGAATGAGATGA-3′ and 5′-ACTTTATTGGCTGGT-3′ of Foxall et al. (1). The amplification cycles were initial denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min and a final extension step of 72°C for 5 min.
Restriction digestion analysis was carried out as described previously (9) with HaeIII to digest 8 to 16 μl of PCR product from the ureA and ureB genes. Fragments were separated on a 3% (wt/vol) agarose gel (UltraPure, Gibco BRL), stained with ethidium bromide, and viewed under UV light. Profiles with different patterns were assigned arbitrary numbers (5).
Antibiotic sensitivity tests.
A suspension of the test organism was prepared to a density equivalent to that of a McFarland no. 4 standard in maximum recovery diluent (Oxoid, Basingstoke, United Kingdom). Swabs were used to innoculate blood agar plates (Columbia agar base; Oxoid) containing 10% (vol/vol) defibrinated horse blood with the suspensions. Epsilometer (E-test) strips (Cambridge Diagnostic Services, Cambridge, United Kingdom) or 5-μg metronidazole discs (Oxoid) were placed on the surface of each plate, which was incubated under microaerobic conditions at 37°C for 2 to 3 days. A breakpoint MIC of <2 mg/liter was used to indicate resistance to clarithromycin, and a growth inhibition zone of <20 mm in diameter was interpreted as resistance to metronidazole.
RESULTS
Effects of different primers on AFLP profiles.
Four primers with sequences complementary to the adapter sequence but extending one base into the fragment DNA (i.e., adapter sequence with a final 3′ base of an A or a T or a G or a C) were tested. Although the annealing temperature for the PCR (60°C) was above the estimated melting temperatures of the primers (50.1 to 56°C), the specificities of the primers were reflected in their very different PCR product banding patterns (Fig. 2A and B). The utility of each primer for the fingerprinting of H. pylori was assessed on the basis of the results for the three NCTC strains. Primer HI-A (Fig. 2B) was judged to produce the most satisfactory results for ease of visual analysis, primer HI-G (Fig. 2A) gave a more complex banding pattern than that given by primer HI-A, primer HI-T (Fig. 2A) produced numerous products that ran as a smear on the gel, and primer HI-C (Fig. 2A) produced too few bands (fewer than nine) for comparative analysis. Primer HI-A was subsequently used to type all strains.
FIG. 2.
(A) AFLP banding patterns produced for strains of H. pylori with three different selective primers: H1-C (lanes 1 to 3), H1-T (lanes 4 to 6), and H1-G (lanes 7 to 9). Template DNA was amplified from strains NCTC 11637 (lanes 1, 4, and 7), NCTC 11638 (lanes 2, 5, and 8), and NCTC 12455 (lanes 3, 6, and 9). (B) AFLP banding patterns obtained with primer H1-A from PCR template DNA digested and ligated in T4 ligase buffer (lanes 1, 3, and 5) compared with those from template DNA which was digested in restriction enzyme buffer in the presence of 5 mM spermidine trihydrochloride prior to ligation of the adapters. Lanes 1 and 2, NCTC 11637; lanes 3 and 4, NCTC 11638; lanes 5 and 6, NCTC 12455. DNA size standards (123-bp marker; Gibco BRL) are shown in lanes marked m.
Effects of different DNA template preparations on AFLP profiles.
For two of the three NCTC strains tested, the AFLP pattern obtained when the DNA was digested in the presence of sperimidine prior to ligation was different from the pattern obtained when digestion took place in combination with the ligation reaction in T4 ligase buffer without added spermidine (Fig. 2B). The predigested DNA produced fewer amplified fragments, indicating a more complete digestion. In separate experiments we showed that the levels of spermidine (none, 2 mM, or 5 mM) in the restriction digests modified the AFLP pattern (data not shown). Although transfer of digested DNA to the ligation mixture resulted in a final concentration of 1.25 mM spermidine in the ligation buffer, there was no evidence that this level inhibited the T4 ligase.
Most of the results presented here were obtained from a PCR with template DNA precipitated following the adapter-ligation step. We also conducted experiments in which the time-consuming precipitation step was omitted. Identical AFLP patterns (primer HI-A) were obtained for the six strains (strains NCTC 11637, NCTC 11638, NCTC 12455, A610/92, H224, and H263) tested when template DNA was taken directly from the ligation mixture, diluted 1/5 or 1/10, and heated to 80°C for 10 min to inactivate the T4 ligase.
AFLP profiles for strains from unrelated individuals.
We observed diversity in the AFLP profiles of strains from the unrelated individuals with H. pylori infection listed in Table 1 (Fig. 3A), although it was noted that up to 55% of the profile bands for isolates from unrelated individuals were the same size. The urease genotyping results for these strains are presented in Table 1. The initial urease results for strain 86C indicated that the strain was actually a mixture of strains, and on further investigation, strains with two different urease types (types 15 and 5) were identified. The AFLP pattern for strain 86C did not have more bands than the number of bands in AFLP patterns for most other isolates examined, even though it had a mixed urease genotype.
FIG. 3.
(A) AFLP profiles (primer H1-A) for nine strains of H. pylori (see Table 1) other than the NCTC strains from unrelated individuals. The profile for strain 86C, referred to in the text, is shown in lane 9. (B) AFLP banding patterns obtained for strains isolated from family members. Lanes are marked according to strain origin: m, mother; f, father; s, son; d, daughter. The different families (families K, M, H, and B) are indicated by the braces under the lanes. DNA size standards (123-bp marker; Gibco BRL) are shown in the first lanes of panels A and B.
AFLP profiles for isolates from family members.
The AFLP profiles for isolates from family members are shown in Fig. 3B, and the urease gene RFLP analysis and ribotyping results are presented in Table 2.
TABLE 2.
Details of isolates of H. pylori from family sets
| Family | Relation, age (yr) | Ribotypea | Urease genotypea | AFLP analysis genotypeb |
|---|---|---|---|---|
| K | Son, 11 | K | 2 | A1 |
| Father, 40 | K | 2 | A2 | |
| Mother, 35 | K | 2 | A3 | |
| M | Son, 13 | Unique | NDc | Unique |
| Father, 43 | M | 8 | B1 | |
| Mother, 43 | M | 8 | B2 | |
| H | Son, 10 | Ha | 2 | C1 |
| Father, 34 | Hb | 2 | C2 | |
| B | Daughter, 10 | Unique | 5 | Unique |
| Mother, 37 | Unique | 2 | Unique |
Ribotype and urease type are as designated previously (2).
The AFLP type was arbitrarily designated when visual comparison indicated a high degree of overall similarity in the band patterns. Numbers indicate pattern variations.
ND, not determined.
(i) Family K.
The AFLP profiles for the strains from the father and son in family K were identical except for one band difference and showed a high degree of similarity to the AFLP profile of the strain from the mother. The urease genotyping and ribotyping results for this group of strains also indicated identity between them.
(ii) Family M.
Isolates from the father and mother in family M had similar AFLP profiles (two minor band differences), the same ribotype, and the same urease genotype, indicating common infection with the same strain. The results obtained for the strain from the son showed that it had a different AFLP pattern (although 50% of the bands were of a similar size) as well as a unique ribotype, which indicated that the parents were not the source of his infection.
(iii) Family H.
The strains derived from the father and son of family H had AFLP profiles that differed by just two bands, had similar ribotypes, and had the same urease genotype.
(iv) Family B.
The AFLP profiles for the isolates from the mother and daughter of family B showed an overall similarity in band grouping, although fewer than 60% of the bands were the same size. The ribotypes and urease genotypes were different and indicated infection with different strains.
AFLP profiles of paired pre- and posttreatment isolates.
Twenty-four strains of H. pylori from six patients, each of whom provided isolates from two pretreatment and two posttreatment biopsy specimens, were examined. The AFLP profiles of these strains are shown in Fig. 4, and the clarithromycin and metronidazole sensitivity patterns together with the urease gene RFLP results are presented in Table 3. All four isolates from two of the six patients (patients 48 and 205) had identical AFLP profiles, although the organisms from both patients had acquired resistance to clarithromycin during treatment. The AFLP analysis results for these strains and the urease genotyping data indicated that the patients each remained infected with their original strain. Likewise, the AFLP patterns of the isolates from two further patients (patients 67 and 217) indicated that the infecting strains were the same after and before antibiotic treatment, although minor differences in band presence or absence or band position within the profiles were observed. The same pattern variations were present on repeat testing of the samples. The AFLP analysis results and the urease gene RFLP analysis data confirmed that the original infecting strain in patient 67 had acquired resistance to both antibiotics during treatment. The AFLP patterns for strains isolated from the final two patients (patient 13 and patient A) showed that for each patient the two pretreatment strains were identical but were different from the two posttreatment strains, which were also identical. The urease gene RFLP analysis results for the pre- and posttreatment isolates from patient 13 differed, and the pre- and posttreatment isolates from patient A had different ribotypes and urease genotypes.
FIG. 4.
AFLP profiles of isolates of H. pylori from biopsy specimens obtained from six patients before and after antibiotic treatment. Patient identity is indicated by the braces below the lanes. The first two lanes for each patient are the profiles for pretreatment isolates; the second two lanes in each set are the profiles for posttreatment isolates. DNA size standards (123-bp marker; Gibco BRL) are in the lanes marked m.
DISCUSSION
AFLP analysis is a technique in which adapter molecules are ligated to restriction enzyme fragments and are subsequently used as target sites for primers in a PCR amplification process (3, 4, 11, 12). It can be applied to the fingerprinting of a wide variety of microbial species, and in this study we modified the single-restriction-enzyme approach described by Valsangiacomo and colleagues (11) in order to type strains of H. pylori. In the original protocol (11), the ligated DNA was precipitated by a procedure designed to remove adapter oligonucleotides that may act as primers in the subsequent PCR amplification. The design of our adapters allowed the precipitation step to be omitted without altering the results that we obtained. Unligated ADH1 oligonucleotides present in the PCR mixture were unlikely to serve as primers because the estimated melting temperature of ADH1 was 40.3°C, well below the annealing temperature of 60°C of the PCR. The second oligonucleotide in the adapter, ADH2, did not prime the amplification because the 3′ end pointed outward from the adapter-linked fragments.
In theory, AFLP analysis provides a means of examining DNA segments distributed over the entire genome of an organism, and it offers this advantage over methods that examine restriction site changes in single genes, for example, PCR-RFLP techniques. Here, we examined both the utility and the limitations of AFLP analysis when it was applied to the differentiation of strains of H. pylori. The reproducibility of the results was evident from the patterns obtained for multiple isolates cultured from biopsy specimens from a single patient, taken at different times and from separate locations in the stomach, even when template preparation and PCR were performed on different days. However, we have shown that the conditions governing the restriction digestion reaction require strict definition, and this would be an important parameter for the interlaboratory comparison of results. In other experiments (data not shown), we found that repeated freezing-thawing of the ligated template DNA led to a decreased amount of product. This was most likely due to the fragility of the single-strand linkage between the adapter and the fragment.
All strains of H. pylori examined in the study described here were fingerprinted by using HindIII to digest the DNA, indicating a high level of typeability with this enzyme. Previous studies have shown that up to 25% of strains of H. pylori are resistant to digestion with other commonly used restriction enzymes such as HaeIII (7, 8). The variation in the profiles of strains from unrelated individuals indicated the discriminatory power of the technique, even though there could be up to 55% similarity in band sizes. AFLP analysis is not as rapid as some other PCR-based assays such as randomly amplified polymorphic DNA analysis, but when compared to ribotyping performance of AFLP analysis is less labor-intensive and, because the profiles contain more bands, the results are more informative.
Although pulsed-field gel electrophoresis is an established typing technique for examination of the entire genome, it has not been widely used to type strains of H. pylori. This may result from problems with in situ extraction of DNA from the organisms and the occurrence of resistance to digestion by the commonly used rarely cutting enzymes. However, whereas interpretive criteria have been proposed for pulsed-field gel electrophoresis patterns (10), there are underlying problems with the interpretation of the AFLP profiles. In the absence of Southern blotting and hybridization experiments, it is not possible to determine if similarly sized fragments within the range of 50 to 2,500 bp obtained by AFLP analysis are derived from the same part of the genome, and the insertion or deletion of DNA in an existing fragment would not be readily identified. Also, because only a small proportion of fragments are selectively amplified, it is generally not possible to predict whether the creation of a restriction site following a point mutation in an existing band would lead to the appearance of one or two smaller bands or simply to the loss of that band. There are limitations to the size of fragment that Taq polymerase can reliably amplify (<5 kbp), and the factors governing restriction enzyme activity, including those that lead to some sites being more refractory to digestion than others, are not fully understood. In addition, the general genomic stability of H. pylori in vivo is unknown, and together, these factors may limit the interpretation of the profiles in terms of definable genetic events. As a consequence of these problems, the AFLP patterns cannot be readily used to identify mixed bacterial cultures. Although the results obtained in this study do illustrate some problems with data interpretation, they also point to the utility of the AFLP technique for providing additional information. For example, the AFLP profiles for the strains from the mother and daughter in family B showed similarities in overall AFLP patterns but differences in ribotyping and urease genotyping results. In this case the AFLP profiles provided useful information concerning the likely common origin of the strains, but the results are not conclusive and require confirmation with further genomic data.
In conclusion, we find that the AFLP technique described here can be applied to H. pylori without the need for expensive equipment or reagents, and if the DNA precipitation step is omitted, its performance is relatively rapid and technically simple. The profiles obtained were reproducible, and on visual analysis, they differentiated strains of H. pylori from unrelated individuals, confirmed the common origin of strains, and demonstrated continuing infection with the same strain following antibiotic treatment. Because AFLP analysis samples the variation throughout the genome, it is a valuable addition to the existing genotypic fingerprinting methods available for H. pylori.
ACKNOWLEDGMENTS
We thank Mathew Williams, Royal Free Hospital, London, United Kingdom, and Kathleen Bamford, Queens University, Belfast, Northern Ireland, for providing some of the strains used in this study. We also thank Henry Smith, Laboratory of Enteric Pathogens, and Norman Fry, Respiratory and Systemic Infection Laboratory, for technical discussion.
REFERENCES
- 1.Foxall P A, Hu L, Mobley H L T. Use of polymerase chain reaction amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739–741. doi: 10.1128/jcm.30.3.739-741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurtado A, Owen R J. Urease gene polymorphisms in Helicobacter pylori from family members. Med Microbiol Lett. 1993;2:386–393. [Google Scholar]
- 3.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 4.Mazurak G H, Reddy V, Marston B J, Haas W A, Crawford J T. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996;34:2386–2390. doi: 10.1128/jcm.34.10.2386-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen, R. J. 1998. Unpublished data.
- 6.Owen R J, Bickley J, Hurtado A, Fraser A, Pounder R E. Comparison of PCR-based restriction length polymorphism analysis of urease genes with rRNA gene profiling for monitoring Helicobacter pylori infections in patients on triple therapy. J Clin Microbiol. 1994;32:1203–1210. doi: 10.1128/jcm.32.5.1203-1210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen R J, Hunton C, Bickley J, Moreno M, Linton D. Ribosomal RNA gene restriction patterns of Helicobacter pylori: analysis and appraisal of HaeIII digests as a molecular typing system. Epidemiol Infect. 1992;109:35–47. [PMC free article] [PubMed] [Google Scholar]
- 8.Owen R J, Fraser J, Costas M, Morgan D, Morgan D R. Signature patterns of DNA restriction fragments of Helicobacter pylori before and after treatment. J Clin Pathol. 1990;43:646–649. doi: 10.1136/jcp.43.8.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero-Lopez C, Owen R J, Desai M. Differentiation between isolates of Helicobacter pylori by PCR-RFLP analysis of urease A and B genes and comparison with ribosomal RNA gene patterns. FEMS Microbiol Lett. 1993;110:37–44. doi: 10.1111/j.1574-6968.1993.tb06292.x. [DOI] [PubMed] [Google Scholar]
- 10.Tenover F C, Arbeit R D, Goering R V, Mickleson P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valsangiacomo C, Baggi F, Gaia V, Balmelli T, Peduzzi R, Piffaretti J. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J Clin Microbiol. 1995;33:1716–1719. doi: 10.1128/jcm.33.7.1716-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 2.4.1–2.4.2. [Google Scholar]
- 14.Zabeau M, Vos P. Selective restriction fragment amplification: a general method for DNA fingerprinting. Publication 0534858A1. Munich, Germany: European Patent Office; 1993. [Google Scholar]