Abstract
Histone deacetylase 6 (HDAC6) mediates DNA damage signaling by regulating the mismatch repair and nucleotide excision repair pathways. Whether HDAC6 also mediates DNA double-strand break (DSB) repair is unclear. Here, we report that HDAC6 negatively regulates DSB repair in an enzyme activity-independent manner. In unstressed cells, HDAC6 interacts with H2A/H2A.X to prevent its interaction with the E3 ligase RNF168. Upon sensing DSBs, RNF168 rapidly ubiquitinates HDAC6 at lysine 116, leading to HDAC6 proteasomal degradation and a restored interaction between RNF168 and H2A/H2A.X. H2A/H2A.X is ubiquitinated by RNF168, precipitating the recruitment of DSB repair factors (including 53BP1 and BRCA1) to chromatin and subsequent DNA repair. These findings reveal novel regulatory machinery based on an HDAC6–RNF168 axis that regulates the H2A/H2A.X ubiquitination status. Interfering with this axis might be leveraged to disrupt a key mechanism of cancer cell resistance to genotoxic damage and form a potential therapeutic strategy for cancer.
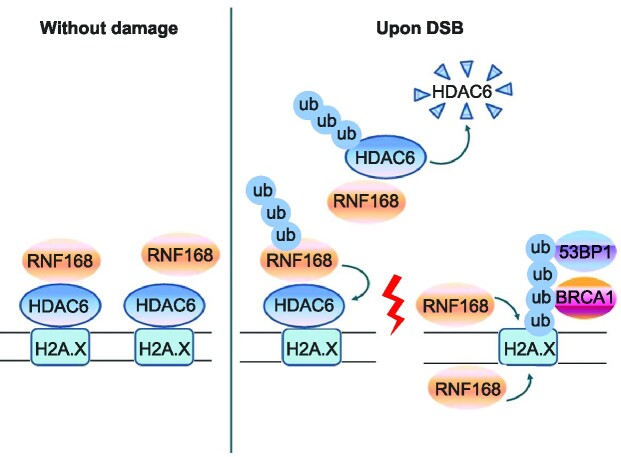
Graphical Abstract
Graphical Abstract.
INTRODUCTION
DNA is constantly under assault by various endogenous and environmental agents that can induce DNA lesions (1). The most cytotoxic and deleterious of these lesions are DNA double-strand breaks (DSBs), in which the phosphate backbones of the two complementary DNA strands are broken simultaneously (2,3). If left unrepaired or mis-repaired, DSB lesions can lead to genome mutations and wider-scale structural rearrangements, which can ultimately cause a variety of diseases, including cancers and neurodegenerative disorders, et al. (4,5). In response, mammals have evolved complex cellular pathways—namely homologous recombination (HR) and non-homologous DNA end-joining (NHEJ) pathways (6,7)—to ensure DSB repair and genome stability. Ionizing radiation (IR) and anticancer therapeutic drugs, such as cisplatin, temozolomide and etoposide (VP16), are well known exogenous agents that can cause DSBs damage (8). In addition, paused or blocked replication forks, which can be induced by a variety natural impediments, can also lead to DSBs (8,9). Thus, to gain a deeper understanding of DSB repair, a greater and more detailed understanding of the underlying mechanisms is necessary.
Histone deacetylase 6 (HDAC6) is a widely expressed, unique isoenzyme belonging to the HDAC family; it contains two deacetylation functional catalytic (DAC) domains and a zinc finger (ZnF) motif that assist in ubiquitin binding (10). HDAC6 and its role in DNA damage signaling is involved in two pathways – mismatch repair (MMR) and nucleotide excision repair (NER), in which it serves as deacetylase and ubiquitase. For example, the nuclear-HDAC6 DAC2 domain blocks the assembly process of the MMR MutSα–MutLα complex by deacetylating MLH1 and MSH2, which in turn facilitates the tolerance of DNA damage repair (11,12). HDAC6 also deacetylates replication protein A (RPA1) in the nuclei in unstimulated status, disrupting the interaction between RPA1 and the key NER protein XPA, resulting in compromised NER (13,14). In addition, the DAC1 domain of HDAC6 has been reported to function as an E3 ligase, ubiquitinating MSH2 in response to genotoxic stress (11). It is clear therefore that HDAC6 impairs DNA damage repair in a deacetylase activity-dependent manner. Whether and how HDAC6 might be involved in DSB repair, however, is unknown.
RNF168 is a nuclear E3 ubiquitin ligase that regulates various ubiquitin signaling pathways, such as mono-ubiquitination (15) and the K48-linked (16) and K63-linked ubiquitin chains (17,18), which are required during DNA damage repair. RNF168 is a rate-limiting mediator of DNA damage repair factor recruitment, as set out below. Once a DSB occurs, the serine/threonine protein kinase ATM phosphorylates the histone protein H2A.X at Ser 139 (γH2A.X), which is read by mediator of checkpoint 1 (MDC1) (19,20). Recruited MDC1 is also phosphorylated by ATM, recognized by the E3 ubiquitin ligase RNF8, and subsequently further recruits the key E3 ligase, RNF168, to DSB regions (21–24). RNF168 then in turn ubiquitylates the K13 and K15 residues of histone H2A/H2A.X, which is a prerequisite for repair factors (e.g. 53BP1 and BRCA1) to localize at DNA damage sites (15,25–27). Thus, RNF168-regulated H2A/H2A.X ubiquitination is a tightly controlled, essential process driving the DSB response.
Any factors that regulate RNF168 function itself could affect H2A/H2A.X ubiquitination status and thus DSB repair efficiency. Identifying other factors that also precisely regulate H2A/H2AX to safeguard genome stability is another important and active area of research. Given that the modification of H2A/H2A.X is integral to DSB repair, we focused our research efforts on whether HDAC6 mediates H2A/H2A.X ubiquitination, and if the underlying mechanisms involve RNF168. In this study, we investigated whether HDAC6 might play a role in the repair response to DSBs and, if so, its significance in cancer treatment. We provide evidence to support the hypothesis that chromatin-associated HDAC6 functions as a barrier to antagonize the inappropriate H2A/H2AX ubiquitination by RNF168 at unstress status in a deacetylase-independent manner. Our delineation of the mechanisms underlying this process provides a basis for the further understanding of an enzyme-independent biological mechanism of a deacetylase in genotoxic treatment for cancer.
MATERIALS AND METHODS
Cell culture
HCT116, HeLa, EJ5-GFP U2OS and DR-GFP U2OS cells were cultured in McCoy's 5A medium or Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin. These cell lines were maintained in a humidified incubator with 5% CO2 atmosphere at 37°C.
Antibodies and reagents
The antibodies used in this study included: anti-HDAC6 (Proteintech, 12834-1-AP), anti-H3 (Abcam, ab1791), anti-phospho-H2A.X (S139, Cell Signaling, 9718), anti-Lamin-B1 (Proteintech, 12987-1-AP), anti-β-actin (Santa Cruz, sc-47778), anti-p53 (Cell Signaling Technology, 9282S), anti-p62 (Cell Signaling Technology, 23214), anti-FLAG (Sigma, F1804), anti-HA (MBL, M180-3), anti-FK2 (Millipore), anti-RNF168 (Proteintech, 21393-1-AP), anti-RNF8 (Santa Cruz, sc-271462), anti-UHRF1 (Millipore, ABE551), anti-α-tubulin (Santa Cruz, sc-398103), anti-His (MBL, PM032), anti-Myc (MBL, M047-3), anti-BRCA1(Cell Signaling Technology, 9010S), anti-53BP1 (Novus, NB100-304), anti-RAD51 (Abcam, ab133534), anti-NBS1 (Cell Signaling Technology, 14956S), anti-RAD50 (Abcam, 3427S), anti-ATM (Abcam, GTX70103), anti-Ku86 (Santa Cruz, sc-56135), anti-Ku70 (Abcam, ab92450), anti-PARP1 (proteintech, 13371-1-AP), anti-Ac-α-tubulin (Cell Signaling Technology, 5335S), anti-HDAC1 (Santa Cruz, sc-6298), anti-HDAC2 (Cell Signaling, 2545), anti-H2A.X (Cell Signaling, 2595), anti-H2A (ubiquityl K119) (Abcam, ab193203), anti-GST (Abcam, 10000-0-AP), anti-K48 and anti-K63-linkage-specific polyubiquitin (Cell Signaling Technology, 8081S and 5621S). Etoposide, adriamycin, geneticin, puromycin, neocarzinostatin, and trichostatin A were purchased from Sigma-Aldrich, and MG132 was purchased from Selleckchem, USA.
Plasmids and siRNA
All plasmids and small interfering RNAs (siRNAs) were transfected with Lipofectamine 2000 (Life Technologies-Invitrogen, USA), according to the manufacturer's instructions. HDAC6 and RNF168 cDNAs were amplified from a human cell line and cloned into the p3 × FLAG-CMV10, pEGFP-C2, pcDNA 3.1, pGEX4T-3, and pET28a vectors (Addgene, USA). Various fragments of HDAC6 (DAC1 domain, 1–440 aa; DAC2 domain, 441–835 aa; ZnF domain, 836–1215 aa) were cloned into the p3 × FLAG-CMV10 or pET28a vector (Addgene, USA). Various fragments of RNF168 (RING domain, 1–58 aa; UDM1 and UDM2 domain, 59–571 aa; RING and UDM1 domain, 1–297 aa; UDM2 domain, 298–571 aa) were cloned into the pGEX4T-3 vector (Addgene, USA). RNF168 domain deletion fragments were cloned into a pGEX-6P1 vector. Site-specific mutations of HDAC6 (Y386F/Y782F, K116R) were generated using a site-directed mutagenesis kit (Vazyme, China). All RNAi oligonucleotides were purchased from Shanghai GenePharma Company. The following siRNAs were used to silence target genes:
HDAC6: 5′-GCAUUAUCCUUAUCCUAGA-3′
HDAC6: 5′-CACUGAUCAGGCCAUAUUU-3′
RNF168: 5′-GGCGAAGAGCGAUGGAGGATT-3′
RNF168: 5′-GACACUUUCUCCACAGAUATT-3′
nonspecific siRNA: 5′-UUCUCCGAACGUGUCACGUTT-3′
nonspecific siRNA: 5′-ACGUGACACGUUCGGAGAATT-3′.
CRISPR-cas9
HDAC6 and RNF168 single guide RNA (sgRNA) sequences were constructed in PX459/Puro vector (Addgene). Cells were transfected with Lipofectamine 2000 and subjected to selection with puromycin. The effect of knockout was confirmed by immunoblotting. PCR and sequencing were then used to confirm homozygous editing of the gene loci. The following sgRNA sequences were used:
HDAC6: 5′-CTCTATCCCCAATCTAGCGGAGG-3′
HDAC6: 5′-AACTATGACCTCAACCGGCCAGG-3′
RNF168: 5′-TCGCCTTTTCGACGGTCGACTGG-3′
RNF168: 5′-CTTCCAGTCGACCGTCGAAAAGG-3′
GST pull-down
His-tagged or GST-tagged plasmids were transformed into Escherichia coli BL21 cells (TransGen, China), incubated with 0.1 mM IPTG (Sigma, USA) overnight at 16–24°C, and then purified with glutathione-Sepharose 4B beads (GE Healthcare, USA) or Ni-IDA Sepharose gel (Selleck, USA). Equal amounts of His-tagged proteins in the eluate were incubated with GST-tagged protein fusion beads in TEN buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 100 mM NaCl) for 4 h at 4°C under rotation. The beads were washed three times with TEN buffer, and the precipitated components were analyzed by immunoblotting.
In vitro ubiquitination assay
His-tagged and GST-tagged proteins were purified from E. coli BL21 cells (TransGen, China). For HDAC6 ubiquitination, assays were set up in a total volume of 25 μl of ubiquitination assay reaction buffer (50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 μM ZnCl2, 3 mM ATP, 1 mM TCEP, 0.2 μM UBE1, 0.5 μM E2 (UbcH5c)) containing 5 μM Flag-ubiquitin, 0.25 μM His-HDAC6, and 1–4 μM GST-RNF168. For H2A ubiquitination, assays were set up in a total volume of 25 μl of ubiquitination assay reaction buffer, 5 μM ubiquitin (Sigma), 1 μM E3 (GST-RNF168), and 0.25 μM substrates H2A/H2B dimer or supplemented with 5 μM HDAC6 or HDAC6 domain or not. All reaction mixtures were incubated at 32°C for 3 h.
Comet assay
A comet assay was performed as previously described (28). Briefly, cells were treated to induced DNA damage and harvested after recovery. The cells were then mixed with 37°C molten low-melting point agarose at a ratio of 1:10 (vol/vol) and the cell suspensions were transferred to prewarmed comet slides. The slides were kept at 4°C in the dark for 30 min and immersed in prechilled lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris·HCl, 1% N-lauroylsarcosine sodium, and 1% Triton X-100) for 1 h at 4°C followed by further immersion in freshly prepared alkaline buffer for 30 min. Then, the slides were washed twice with 1 × TBE buffer (90 mM Tris, 90 mM boric acid, and 3 mM EDTA) and subjected to TBE electrophoresis at 1.0 V/cm for 20 min. The slides were fixed in 100% ethanol for 5 min, air dried, and stained with 5 μg/ml propidium iodide at room temperature in the dark for 10 min. Images were captured and quantified by ImageJ software with the OpenComet plugin.
Colony formation assay
After DNA DSB treatment and release for 1 h, cells were counted and re-cultured in fresh medium for 14 days. Then, cells were stained with crystal violet and the number of colonies consisting of >50 cells were counted.
Protein extraction
For whole cell lysate extraction, cells were counted, harvested, and washed twice with ice-cold PBS, then re-suspended in 40 μl PBS (containing cocktail protease inhibitor) per 106 cells. The re-suspended cells were added to an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer (containing 5% 2-mercaptoethanol) and then the samples were boiled for 10 min with a pulse vortex every 5 min and stored at –20°C.
For histone acid extraction, cells were harvested and lysed in 1 ml of hypotonic lysis buffer (10 mM Tris–HCl pH 8.0, 1 mM KCl, 1.5 mM MgCl2) supplemented with 1 mM DTT, 0.2 M sulfuric acid, and protease inhibitor cocktail (Roche, Switzerland). The samples were incubated at 4°C for at least 30 min and then the supernatant was collected by centrifugation for 10 min at 12 000 g. The supernatant was added dropwise to trichloroacetic acid (TCA) to a final concentration of 33% and incubated on ice for 30 min. The histone pellets were collected by centrifuging at 12 000 g for 10 min at 4°C, then washing twice with ice-cold acetone. After removing the excess residue, the histone pellets were dissolved in ddH2O and stored at –20°C.
For cytoplasmic and nuclear extraction, after harvesting, cells were suspended in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) supplemented with 1 mM DTT, 0.15% NP-40, and a protease inhibitor cocktail (Roche, Switzerland) and lysed on ice for 10 min. The lysate was centrifuged at 12 000 g for 1 min, and the supernatant was collected as cytoplasmic proteins. The pellet was washed twice with buffer A and then suspended in buffer B (20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA) supplemented with 1 mM DTT, 0.5% NP-40, and protease inhibitor cocktail (Roche, Switzerland). The lysate was incubated on ice for 15 min, with a pulse vortex for 30 s every 3 min, and then centrifuged at 12 000 g for 1 min at 4°C. The supernatant was collected as nuclear proteins and stored at –20°C.
For chromatin fractionation, cells were suspended in buffer I (50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM EDTA) supplemented with 0.1% Triton X-100 and protease inhibitor cocktail (Roche, Switzerland) and then lysed on ice for 3 min. The lysate was centrifuged at 12 000 g for 3 min, and the supernatant was discarded. The pellet was dissolved in buffer I supplemented with 200 μg/ml RNase A and protease inhibitor cocktail and incubated at room temperature for 30 min. The supernatant was removed after centrifugation; the pellet was then collected by chromatin fractionation and stored at –20°C.
Western blotting
Western blotting was used to estimate protein levels according to the protocol supplied by Cell Signaling Technology. Briefly, equal amounts of proteins were resolved by 6–15% SDS-PAGE and transferred to nitrocellulose or PVDF membranes. After blocking with 5% non-fat milk or bovine serum albumin, the blots were probed with primary antibodies (1:500–1:10 000). The bound antibodies were detected with an HRP-conjugated secondary antibody (1:2000–1:10 000) and visualized using a Tanon 5200 imaging system using medium-sensitive ECL substrates (4A-Biotech).
Co-immunoprecipitation (co-IP) assay
For the whole cell lysate co-IP assay, after treatment, cells were harvested and incubated in NP-40 lysis buffer (20 mM Tris–HCl pH 8.0, 137 mM NaCl, 10% glycerol), supplemented with 1% NP-40 and protease inhibitor cocktail (Roche, Switzerland), on ice for 10 min. The mixtures were sonicated at 30% intensity (10 times, on ice, each for 2 s). The supernatants were collected and incubated with applicable beads at 4°C for 2–3 h, then washed three times with ice-cold PBS. Finally, the samples were boiled in SDS loading buffer and analyzed by western blotting.
The nuclear fraction co-IP assay was performed as previous described (29). Briefly, after harvesting, cells were incubated with buffer A supplemented with 0.15% NP-40 and a protease inhibitor cocktail (Roche, Switzerland), then lysed on ice for 10 min. The pellet was collected and suspended in buffer B supplemented with 0.5% NP-40 and protease inhibitor cocktail. The lysate was incubated on ice for 15 min and sonicated at 30% intensity (10 times, on ice, each for 2 s). The supernatants were collected and incubated with applicable beads at 4°C for 2–3 h, then washed three times with ice-cold PBS. The samples were boiled in SDS loading buffer and analyzed by western blotting.
Laser micro-irradiation
Cells were cultured in a glass-bottomed dish and irradiated with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 40% output) generated using a Micropoint System (Andor).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 for 15 min, incubated with 1% bovine serum albumin for 1 h, then incubated with the indicated primary antibody (1:100–1:500) overnight at 4°C. After washing three times with PBS, each sample was incubated with secondary antibodies conjugated to Alexa Fluor 488 or 594 dye (1:500–1:1000) for 1 h and then counterstained with DAPI for 3 min. After washing three times with PBS, immunofluorescent images were captured under an Andor confocal microscope.
NHEJ and HR assays
DR-U2OS (HR) and EJ5-U2OS (NHEJ) cells, containing a single copy integrated into the genome of a DR-GFP or EJ5-GFP reporter gene, respectively, were used for DSB repair assays. The assays were performed as previously described (30). After transfected with indicated siRNA or plasmids for 24 h and then infected with retrovirus expressing I-SceI. After 48 h, each sample was analyzed by fluorescence-activated cell sorting (FACS).
Chromosomal abnormalities assay
After DNA DSB treatment and release for 8 h, cells were treated with colcemid (0.03 μg/ml) for another 6 h and then harvested hypotonic swelled with 0.8% sodium citrate (31 mM) at 37°C. After 15 min, the samples were fixed using methanol and acetic acid (3:1) and spread on glass slides. All samples were stained with DAPI and analyzed using an Andor confocal microscope.
Statistics
Comparisons between two groups were analyzed using the Student's t-test. A P-value <0.05 was considered statistically significant (ns, no significance, P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001). At least three independent experiments were performed in all cases.
RESULTS
HDAC6 is involved in DSB repair
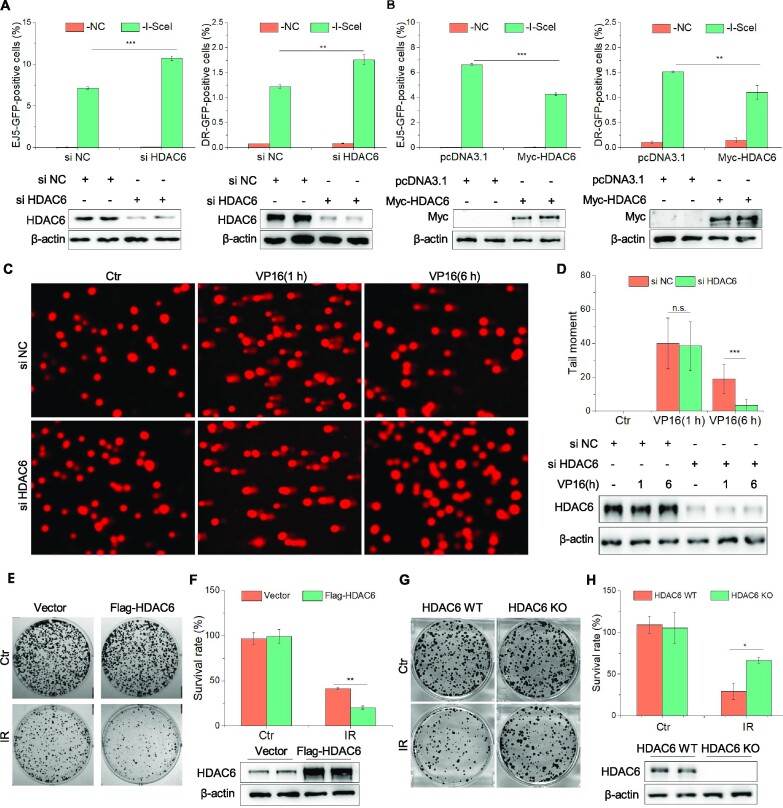
HDAC6 is involved in the DNA damage response via the MMR and NER pathways (11,14), but its role in DSB repair is unknown. Here, we first tested if HDAC6 can help repair DSBs via either the HR or NHEJ pathways using the DR-GFP and pEJ5-GFP U2OS cell reporter systems, respectively. We transfected both U2OS cell models with negative control (NC) or HDAC6 siRNAs and then subjected them to NHEJ and HR assays. Knockdown of HDAC6 increased both NHEJ and HR repair (Figure 1A), while overexpression of HDAC6 had the opposite effect (Figure 1B). These findings suggest that HDAC6 is a negative regulator in DNA DSB repair.
Figure 1.
HDAC6 is involved in DSB repair. (A) pEJ5-GFP U2OS or DR-U2OS cells were transfected with HDAC6 or negative control (NC) siRNAs and subjected to NHEJ and HR assays, separately. (B) pEJ5-GFP U2OS or DR-U2OS cells were transfected with Myc-HDAC6 or empty vector and subjected to NHEJ and HR assays, separately. (C, D) HeLa cells were transfected with HDAC6 or NC siRNAs for 48 h, treated with 10 μM VP16 for 2 h, and washed three times with fresh medium before recovery for the indicated times. All samples were then subjected to a comet assay. Representative images (C) and statistical analyses (D) are shown. (E, F) HeLa cells were transfected with Flag-HDAC6 plasmids for 48 h. After 3 Gy irradiation, the cells were counted and seeded for the colony formation assays. For the control (Ctr) group, 500 cells were seeded in each plate. For the IR-treated group, 5000 cells were seeded in each plate. Representative images (E) and statistical analyses (F) are shown. (G, H) HDAC6 WT or HDAC6 KO HCT116 cells underwent 3 Gy irradiation (IR) and were counted and seeded for colony formation assays. For the control (Ctr) group, 500 cells were seeded in each plate. For the IR-treated group, 5000 cells were seeded in each plate. Representative images (G) and statistical analyses (H) are shown. All data represent the means ± SD (n = 3, *P< 0.05, **P< 0.01, ***P< 0.001).
We confirmed a role for HDAC6 in regulating DSB repair using a comet assay. We first induced DSBs with the DNA-damaging agent etoposide (VP16, 10 μM for 2 h followed by recovery for various times). Compared with the siHDAC6 HTC116 cells, we saw a higher proportion of control cells containing residual DSB lesions, as indicated by the longer tail moment at 6 h (Figure 1C, D). Consistently, HDAC6 overexpression resulted in a longer tail moment than seen in HDAC6 wild-type (WT) cells at 6 h after inducing DSBs with irradiation (IR; 10 Gy) (Supplementary Figure S1A, B).
Finally, we assessed the role of HDAC6 in cell survival after DSBs by performing colony formation assays. HDAC6-overexpressing HeLa cells exhibited hypersensitivity to DSBs, as evidenced by an approximately 21% decrease in colony numbers compared with HDAC6 WT cells (Figure 1E, F and Supplementary Figure S1C, D). We next generated HDAC6 knockout (KO) HCT116 cells using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology (Supplementary S1E). In contrast with the HDAC6 WT cells, HDAC6-KO HCT116 cells exhibited markedly increased survival, as evidenced by an approximately 37% increase in colony numbers (Figure 1G, H and Supplementary Figure S1F, G). These data indicate that HDAC6 is involved in DSB repair.
HDAC6 interacts with H2A/H2A.X in response to DSBs, inhibiting H2A/H2A.X ubiquitination
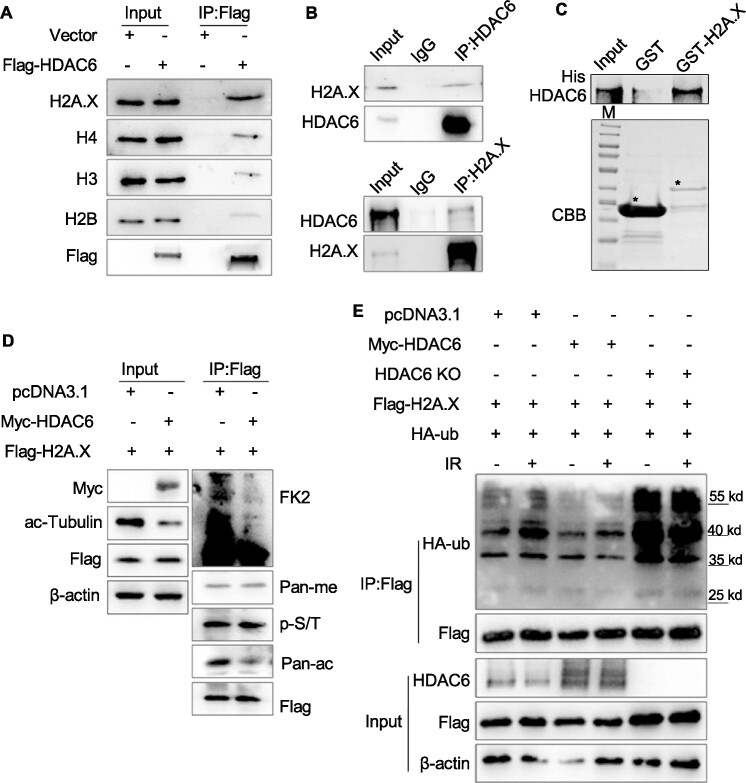
Considering that HDAC6 is a member of the class IIb histone deacetylases, we first examined the interaction of HDAC6 with histone or histone variants by co-immunoprecipitation (co-IP) using Flag-HDAC6 or empty plasmid-transfected HeLa cells. Interestingly, we found that exogenous HDAC6 interacted strongly with both H2A.X and H2A, but only weakly with H3 or H4 (Figure 2A and Supplementary Figure S2A). We then confirmed this endogenous interaction between HDAC6 and H2A.X (Figure 2B). To determine whether the interaction was direct, we carried out an in vitro GST pull-down assay. After incubating the GST-tagged H2A.X with His-tagged HDAC6, we observed that HDAC6 directly interacted with H2A.X (Figure 2C). To further map the regions of HDAC6 responsible for the interaction with H2A.X, we constructed various HDAC6 fragments and purified fragment proteins. Using a GST pull-down assay, we found that full-length HDAC6 and the DAC1 and DAC2 domains interacted with H2A.X, but the ZnF domain did not (Supplementary Figure S2B, C), further suggesting that HDAC6 directly interacts with H2A.X at the DAC1/2 regions.
Figure 2.
HDAC6 interacts with H2A/H2A.X in response to DSBs, inhibiting H2A/H2A.X ubiquitination. (A) HeLa cells were transfected with Flag-HDAC6 or an empty plasmid for 48 h. Chromatin fractions were subjected to immunoprecipitation with an anti-Flag antibody. Western blots of indicated proteins are shown. (B) Whole cell lysates from HeLa cells were extracted and immunoprecipitated using an anti-HDAC6 (up) or an anti-H2A.X (down) antibody. Rabbit IgG was used as a negative control. Western blotting was performed with the indicated antibodies. (C) GST or GST-H2A.X fusion proteins were expressed in bacteria, purified, and then incubated with His-HDAC6 protein. Western blotting was performed to detect HDAC6 protein levels and Coomassie Brilliant Blue (CBB) staining was performed to detect GST or GST-H2A.X levels. The asterisk indicates the corresponding protein bands. (D) HCT116 cells were co-transfected with Flag-H2A.X, Myc-HDAC6, or an empty plasmid for 48 h. Flag-H2A.X was immunoprecipitated from whole cell lysates and eluted for western blotting with the indicated antibodies (FK2: anti-ubiquitin conjugate antibody, Pan-me: anti-pan methyl Lysine antibody, p-S/T: anti-Phospho-(Ser/Thr) antibody, Pan-ac: anti-acetyl lysine antibody) to detect post-translational modifications. (E) HDAC6 WT or HDAC6 KO HCT116 cells were transfected with Flag-H2A.X and HA-ub, with or without Myc-HDAC6 for 48 h, before they were exposed to IR (10 Gy) and released for 1 h. Flag-H2A.X proteins were immunoprecipitated from the whole cell lysates and eluted for western blotting to detect changes in ubiquitination status.
We next explored the relationship between HDAC6 and H2A.X. To do so, we co-transfected Flag-H2A.X with Myc-HDAC6 or an empty plasmid into HeLa cells and subjected whole cell lysates to co-IP. Overexpression of HDAC6 decreased H2A.X ubiquitination and acetylation levels (Figure 2D). We corroborated this finding using HDAC6 KO cells, in which H2A.X ubiquitination levels were increased (Supplementary Figure S2D).
The function of H2A and H2A.X in the DNA damage response mainly depends on their phosphorylation and ubiquitination (31), so we examined whether HDAC6 is involved in DSB repair by regulating H2A.X and H2A ubiquitination. To do so, we co-transfected HDAC6 (WT) or HDAC6 (KO) HCT116 cells with Flag-H2A.X/H2A and HA-ub, with or without Myc-HDAC6, and exposed the cells to IR (10 Gy) before releasing them for 1 h. We then subjected the whole cell lysates to co-IP and observed that IR-induced H2A/H2A.X ubiquitination was enhanced in HDAC6 KO cells and decreased in Myc-HDAC6 overexpressing cells (Figure 2E and Supplementary Figure S2E).
In response to DSBs, RNF168 mediates the ubiquitination of H2A/H2A.X at lysine 13/15 and BMI1 mediates ubiquitination at lysine 119 (32,33). Therefore, we decided to co-transfect HeLa cells with Flag-HDAC6 or empty plasmid and again exposed the cells to IR (10 Gy). After a release for 1 h, we isolated the histone fractions and incubated them with an anti-ub-K119-specific antibody. HDAC6-regulated ubiquitination of H2A did not occur at the K119 residue (Supplementary Figure S2F). These data suggest that HDAC6 interacts with H2A/H2A.X in response to DSBs and decreases its ubiquitination levels.
HDAC6 is associated with the H2A/H2A.X ubiquitination signaling cascade in DSB repair
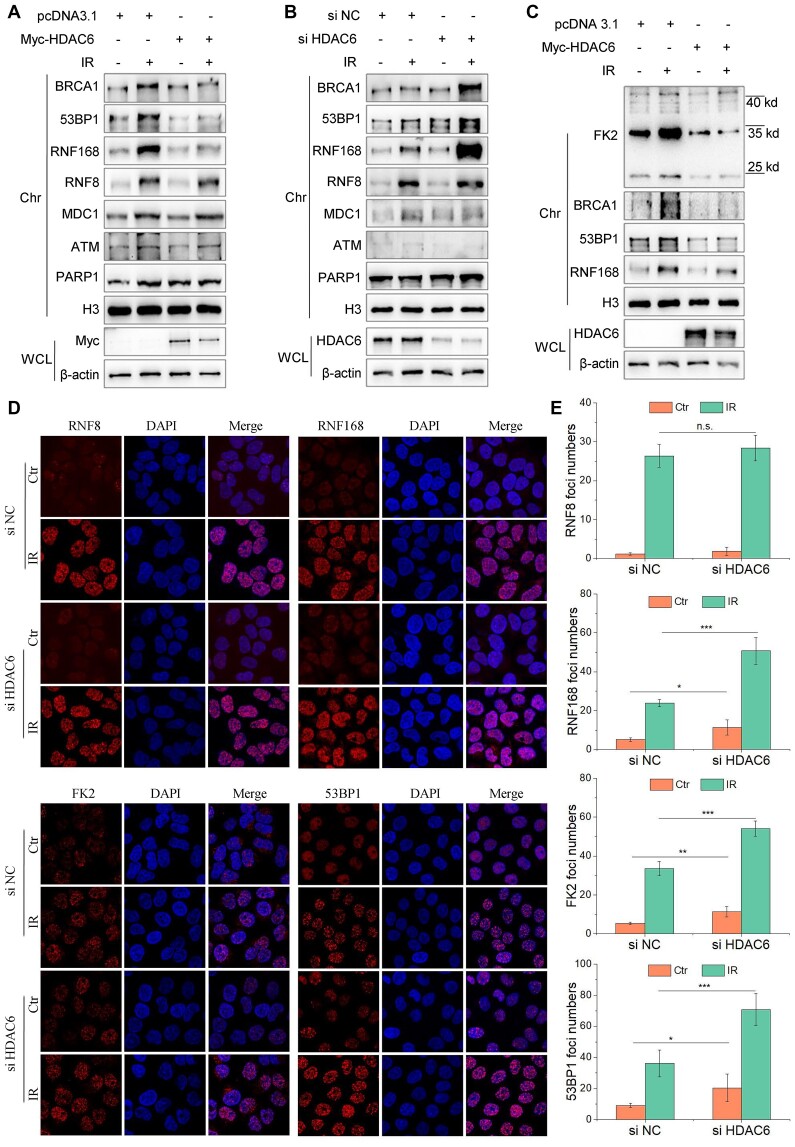
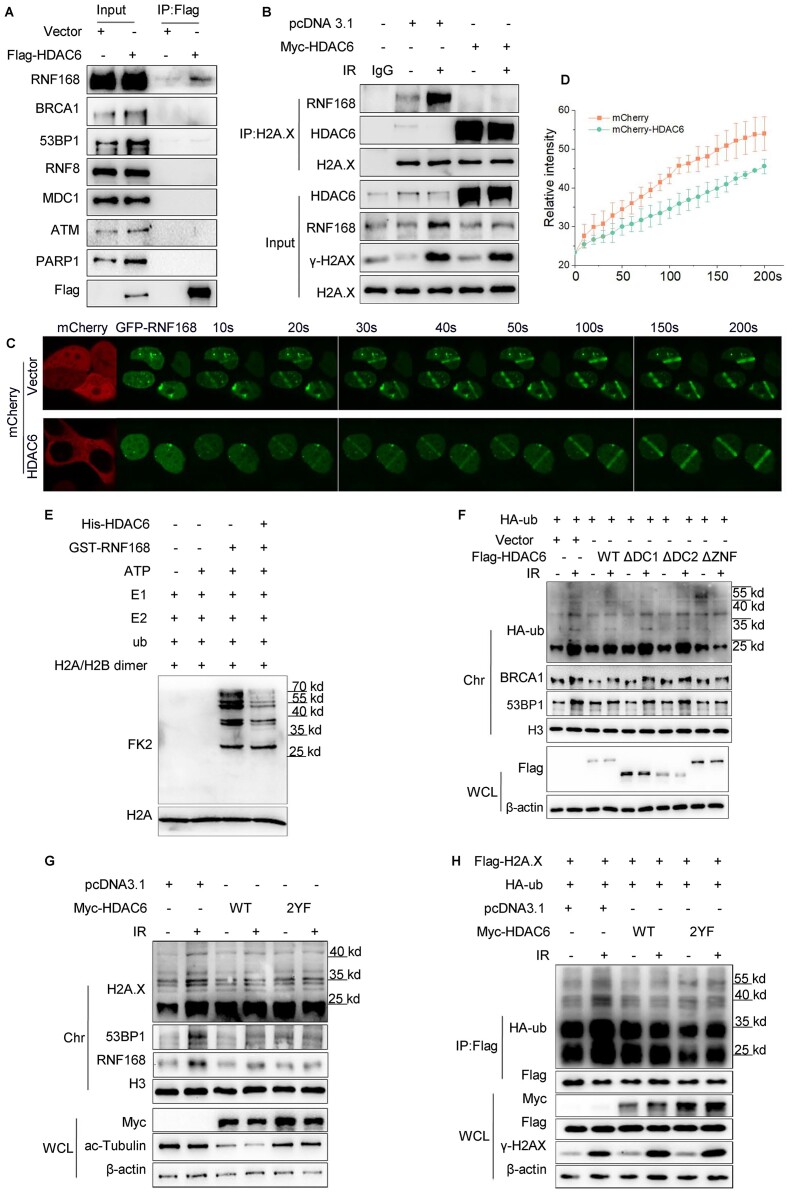
The H2A/H2A.X ubiquitination signaling cascade serves as a recruitment scaffold for DSB repair factors at damaged chromatin (34). Having observed so far that HDAC6 can interact with H2A/H2A.X and affect its ubiquitination status, we next investigated the function of HDAC6-mediated H2A/H2A.X ubiquitination in DSB repair. To do so, we first explored whether HDAC6 favors for the recruitment of DSB repair factors that are involved in H2A/H2A.X ubiquitination signaling cascade. We transfected HeLa cells with Myc-tagged HDAC6 or an empty plasmid for 48 h and then exposed the cells to IR (10 Gy) and released them for 1 h or treated them with etoposide (VP16, 20 μM) for 2 h. HDAC6 overexpression attenuated the loading of RNF168, 53BP1, and BRCA1 to chromatin, but not PARP1, ATM, MDC1, or RNF8, in response to either IR or VP16 treatment (Figure 3A and Supplementary Figure S3A). Conversely, HDAC6 knockdown using siRNA resulted in enhanced recruitment of 53BP1, BRCA1, and RNF168 to chromatin after IR (Figure 3B). Overexpressing HDAC6 in these HDAC6 KO cells restored the normal recruitment of these factors to chromatin after inducing DSBs with irradiation (10 Gy) (Figure 3C).
Figure 3.
HDAC6 is associated with the H2A/H2A.X ubiquitination signaling cascade in DSB repair. (A–E) All cells were irradiated with 10 Gy followed by release for 1 h, after transfection with the indicated plasmids or siRNAs. (A) HeLa cells were transfected with Myc-HDAC6 or an empty plasmid for 48 h. Whole cell lysate and chromatin fractions were subjected to western blotting. (B) HeLa cells were transfected with HDAC6 or negative control (NC) siRNAs for 48 h. Whole cell lysate and chromatin fractions were subjected to western blotting. (C) HDAC6 KO HCT116 cells were transfected with Myc-HDAC6 or empty plasmid for 48 h. Whole cell lysate and chromatin fractions were subjected to immunoblotting. (D, E) HeLa cells were transfected with HDAC6 or negative control (NC) siRNAs for 48 h. The cells were then fixed, and the samples were labeled with the indicated antibodies. Representative images (D) and statistical analyses (E) are shown. The data represent the means ± SD (n = 5, *P< 0.05, **P< 0.01, ***P< 0.001).
We also performed immunostaining to visualize the foci of factors involved in the H2A/H2A.X ubiquitination signaling cascade. After labeling, we saw that RNF168, 53BP1, and FK2 foci were significantly increased in HDAC6 knockdown cells compared with HDAC6 WT cells (Figure 3D, E). The converse was true in HDAC6 overexpressing cells (Supplementary Figure S3B, C). However, MDC1 and RNF8 foci were not significantly changed (Figure 3D, E and Supplementary Figure S3B-C). These data indicate that HDAC6 is associated with the H2A/H2A.X ubiquitination signaling cascade during DSB repair.
HDAC6 regulates the H2A/H2A.X ubiquitination signaling cascade in an RNF168-dependent manner during DSB repair
The H2A/H2A.X ubiquitination signaling cascade factors are sequentially sequestered to DSB sites (25). Our next aim was to identify which step of the H2A/H2A.X ubiquitination signaling cascade is regulated by HDAC6 during DSB repair. By overexpressing Flag-HDAC6 in HeLa cells, we observed that HDAC6 interacted with RNF168 but not with other related factors, such as RNF8 or MDC1 (Figure 4A).
Figure 4.
HDAC6 regulates the H2A/H2A.X ubiquitination signaling cascade in an RNF168-dependent manner during DSB repair. (A) HeLa cells were transfected with Flag-HDAC6 or an empty plasmid for 48 h. Whole cell lysates were subjected to immunoprecipitation with an anti-Flag antibody before western blotting. (B) HeLa cells were transfected with Myc-HDAC6 or empty plasmid for 48 h and exposed to 10 Gy irradiation (IR) and released for 1 h. Chromatin fractions were subjected to immunoprecipitation with an anti-H2A.X antibody followed by western blotting. Rabbit IgG was used as a negative control. (C, D) HeLa cells were co-transfected with GFP-RNF168 and mCherry-HDAC6 or an empty plasmid for 48 h and then subjected to laser micro-IR. Images were captured every 10 s for 200 s (C), and the IR path signal intensity was quantified (D). (E) Recombinant His-HDAC6 and GST-RNF168 were subjected to in vitro ubiquitination assays in the presence of ATP, E1 (UBE1), E2 (UbcH5c), ub (ubiquitin) and H2A/H2B dimer as indicated. Western blotting was performed with the indicated antibodies. (F, G) HeLa cells were transfected with the indicated plasmids for 48 h and then exposed to 10 Gy irradiation and released for 1 h. Whole cell lysate and chromatin fractions were analyzed by western blotting with the indicated antibodies. WT: wild-type; ΔDC1: DAC1 domain deleted; ΔDC2: DAC2 domain deleted; ΔZnF: ZnF-UBP domain deleted (F); HDAC6 catalytic mutant (Y386F/Y782F) (G). (H) HeLa cells were co-transfected with Flag-H2A.X, HA-ub, with Myc-HDAC6 WT or Myc-HDAC6 2YF (catalytic mutant) for 48 h and then exposed to 10 Gy irradiation and released for 1 h. The Flag-H2A.X proteins were immunoprecipitated from the whole cell lysates and eluted to detect the ubiquitination changes by western blotting with the indicated antibodies. All data represent the means ± SD.
As RNF168 is known to polyubiquitylate H2A/H2A.X at K13-15, we investigated whether HDAC6 inhibits RNF168 function in DSB repair. We overexpressed a Myc-tagged HDAC6 plasmid in HeLa cells; we then exposed them to IR (10 Gy) and released them for 1 h. We next subjected the chromatin fraction to co-IP. The interaction between H2A/H2A.X and RNF168 was reduced in HDAC6-overexpressing cells compared with control (empty vector) cells after IR treatment (Figure 4B and Supplementary Figure S4A). Next, we used live-cell imaging of the signal intensity across a laser micro-irradiation path and observed that GFP-RNF168 but not GFP-RNF8 was delayed in its recruitment to DSBs in HDAC6-overexpressing cells; such retarded recruitment was reversed in HDAC6 siRNA knockdown cells (Figure 4C-D and Supplementary Figure S4B–E).
Now knowing that HDAC6 reduced RNF168 interaction with H2A.X and impaired RNF168 recruitment to DSB sites, we next performed an in vitro ubiquitination assay to confirm whether HDAC6 directly affects RNF168-catalyzed H2A ubiquitination in vitro. The presence of HDAC6 decreased the capacity of RNF168 to ubiquitinate H2A/H2B dimer (Figure 4E). The HDAC6 ZnF-UBP domain can recognize and bind to ubiquitin (35,36); therefore, we investigated whether HDAC6-mediated chromatin ubiquitination is independent of the ZnF-UBP domain. To do so, we overexpressed wild-type HDAC6 (WT), DAC1 domain-deleted (ΔDC1), DAC2 domain-deleted (ΔDC2), or ZnF-UBP domain-deleted (ΔZnF) plasmids in HeLa cells and then subjected them to IR (10 Gy). After releasing the cells for 1 h, we saw that ΔZnF did not rescue the HDAC6-mediated inhibitory effect on chromatin ubiquitination in DSB repair (Figure 4F). However, both ΔDC1 and ΔDC2 abrogated this effect (Figure 4F). Data from in vitro ubiquitination assays also confirmed that wild-type full-length HDAC6, as well wild-type DC1 and DC2 fragments, but not the ZnF-UBP domain, impeded RNF168-induced H2A ubiquitination (Supplementary Figure S4F).
Finally, because HDAC6 is a class IIb histone deacetylase, we considered whether its enzymatic activity is required for suppressing the RNF168-mediated H2A/H2A.X ubiquitination signaling cascade in DSB repair. We transfected HeLa cells with wild-type HDAC6 (HDAC6-WT) or a HDAC6 catalytic mutant (HDAC6-2YF:Y386F/Y782F) and exposed the cells to IR (10 Gy) followed by release for 1 h. The HDAC6 2YF mutant failed to rescue the inhibition seen upon H2A/H2A.X ubiquitination (Figure 4G, H). Taken together, these data indicate that HDAC6 regulates the H2A/H2A.X ubiquitination signaling cascade by restraining RNF168 activity during DSB repair. This effect of HDAC6 is independent of its enzymatic activity and ubiquitin-binding domain.
Nuclear HDAC6 is released from chromatin and degraded via the proteasome in response to DSBs
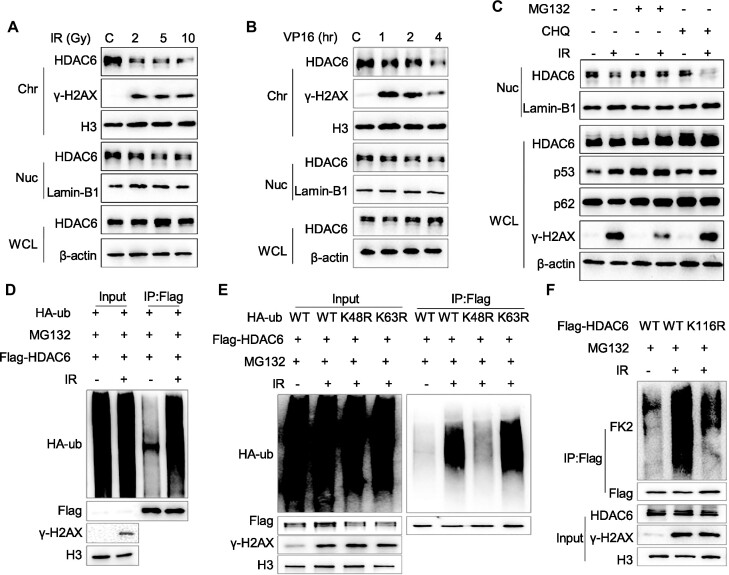
Based on our findings thus far, we proposed that chromatin-bound HDAC6 affects the RNF168-regulated H2A/H2A.X ubiquitination signaling cascade during DSB repair. This mechanism implies that HDAC6 needs to be dynamically regulated to maintain normal DSB repair. Thus, we examined the dynamic changes in HDAC6 after exposing cells to various DSB-inducing stimuli. We treated HCT116 or HeLa cells with different doses of IR and released them for different times or treated these cells with different doses of VP16 for different time periods. Interestingly, the results showed that both nuclear and chromatin HDAC6 levels were decreased, while total HDAC6 showed no obvious change after the induction of DSBs (Figure 5A, B and Supplementary Figure S5A, B). Meanwhile, IR did not result in a decrease in HDAC1 or HDAC3 chromatin levels (Supplementary Figure S5C). We further observed HDAC6 at the laser-induced DSB stripe by staining the endogenous HDAC6. The results indicated that HDAC6 dissociated from chromatin upon laser micro-irradiation at the γH2AX-positive region, which was used to indicate the DSB stripe (Supplementary Figure S5D, E).
Figure 5.
Nuclear HDAC6 is displaced from chromatin and degraded via the proteasome in response to DSBs. (A, B) HeLa cells were exposed to an increasing dose of irradiation (IR) and released after 1 h or treated with 10 μM etoposide (VP16) for the indicated time. Then, whole cell lysate (β-actin), nuclear (Lamin-B1), and chromatin (H3) fractions were isolated and analyzed by western blotting. (C) HeLa cells were treated with 10 μM MG132 for 3 h or 50 μM CHQ for 12 h, then exposed to 10 Gy IR and released for 1 h before analysis of the whole cell lysate and nuclear fractions by western blotting. (D–F) HeLa cells were co-transfected with Flag-HDAC6 and HA-ub (D), Flag-HDAC6 and different HA-ubiquitin constructs (WT: wild type ubiquitin, K48R: K48-mutant ubiquitin, K63R: K63-mutant ubiquitin) (E) or Flag-HDAC6 wild-type (WT) or mutant (K116R) (F) for 48 h. Then, the cells were treated with 10 μM MG132 for 2 h, exposed to 10 Gy IR, and released for 1 h. The nuclear fractions were then subjected to immunoprecipitation with an anti-Flag antibody and analyzed by western blotting.
To determine whether the decrease in nuclear and chromatin HDAC6 levels in response to DSBs was due to protein instability or a change in mRNA expression, we isolated the mRNA after exposure to 10 Gy IR for different times and monitored the RNA levels by quantitative PCR. HDAC6 mRNA levels were not downregulated following IR, indicating that the decrease in HDAC6 protein levels may be due to protein degradation (Supplementary Figure S5F).
We next treated these cells with a proteasome inhibitor (MG132, 10 μM for 3 h) or a lysosome inhibitor (chloroquine, CHQ, 50 μM for 12 h) and again exposed the cells to IR (10 Gy). Here, MG132, but not CHQ, inhibited IR-induced nuclear HDAC6 decrease, implying that proteasome-mediated degradation is the underlying mechanism for nuclear HDAC6 degradation in response to DSBs (Figure 5C). We confirmed this finding by co-transfecting Flag-tagged HDAC6 with an HA-tagged ubiquitin (HA-ub) plasmid in HeLa or HCT116 cells and exposed them to IR (10 Gy with 1 h release) or VP16 (10 μM for different time periods). After isolating the nuclear fractions and performing a co-IP assay, we observed that ubiquitinated nuclear HDAC6 levels were increased compared with those seen in untreated (IR or VP16) cells (Figure 5D and Supplementary Figure S5G).
We also co-expressed HA-tagged K48R and K63R together with Flag-HDAC6 and found that nuclear HDAC6 was predominantly ubiquitylated by K48-linked, but not by K63-linked, polyubiquitin chains in response to DSBs (Figure 5E). These findings are consistent with the notion that K48-linked polyubiquitin chains are usually involved in proteasomal degradation (37). As HDAC6 lysine 116 (K116) is a single ubiquitination site (38), we assessed whether HDAC6 K116 is responsible for nuclear HDAC6 ubiquitination during DSB repair. We transfected HeLa cells with wild-type (WT) Flag-HDAC6 or K116R-mutated (K116R) plasmids and then exposed the cells to IR (10 Gy with 1 h release) or VP16 (10 μM for 2 h). The co-IP results showed that K116R, but not WT-HDAC6, failed to promote nuclear HDAC6 ubiquitylation during the DSB response (Figure 5F and Supplementary Figure S5H), suggesting that HDAC6 K116 is indeed ubiquitinated in response to DSBs. These data indicate that upon sensing DSBs, nuclear HDAC6 is ubiquitinated at K116, which mediates its degradation via the proteasome.
RNF168 directly interacts with HDAC6 and mediates DSB-induced nuclear HDAC6 degradation
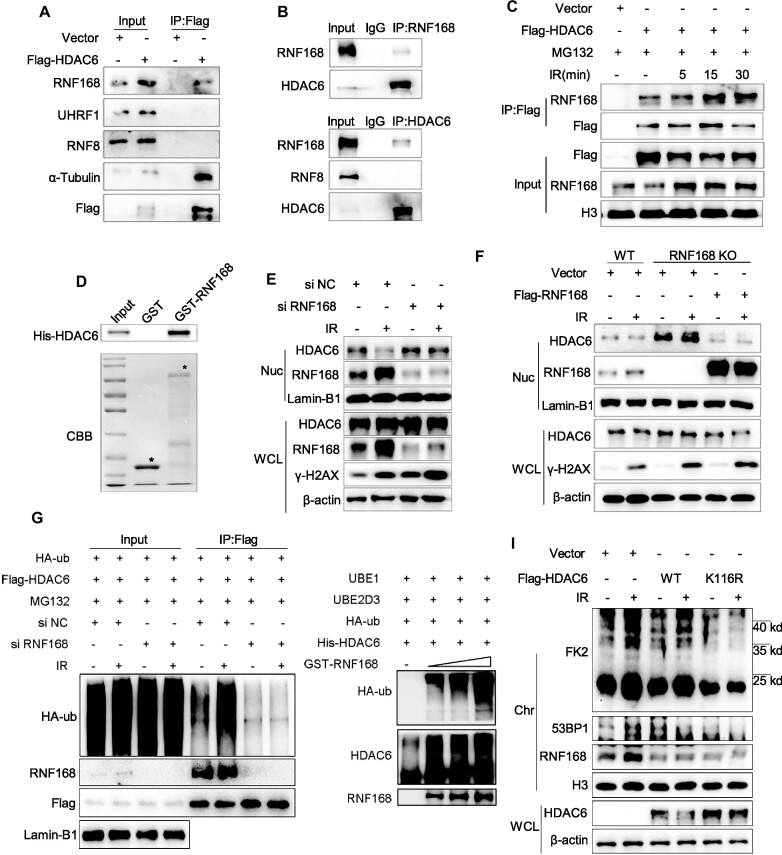
We also sought to identify the E3 ligases that can specifically target nuclear HDAC6 for degradation. To do so, we transfected Flag-HDAC6 or an empty plasmid into HeLa cells and analyzed the proteins extracted from whole cell lysate by co-IP assay. Flag-HDAC6 interacted with the E3 ligase RNF168, but not RNF8 or UHRF1 (Figure 6A). We also confirmed the endogenous interaction between HDAC6 and RNF168 in the nuclear fraction of HeLa cells (Figure 6B).
Figure 6.
RNF168 directly interacts with HDAC6 and mediates DSB-induced nuclear HDAC6 degradation. (A) HeLa cells were transfected with Flag-HDAC6 or an empty plasmid for 48 h. Whole cell lysates were subjected to immunoprecipitation with an anti-Flag antibody and analyzed by western blotting. (B) Whole cell lysates from HeLa cells were extracted and subjected to immunoprecipitation using an anti-RNF168 (up) or an anti-HDAC6 (down) antibody and subsequent analysis by western blotting. Rabbit IgG served as a negative control. (C) HeLa cells were transfected with Flag-HDAC6 or an empty plasmid for 48 h. Then, the cells were treated with 10 μM MG132 for 2 h, exposed to 10 Gy irradiation (IR), and released for the indicated times. The nuclear fractions were subjected to immunoprecipitation with an anti-Flag antibody and analyzed by western blotting. (D) GST or GST-RNF168 fusion proteins were incubated with His-HDAC6 protein. Western blotting was performed to detect HDAC6 protein levels, and Coomassie Brilliant Blue (CBB) staining was performed to detect GST or GST-RNF168 levels (indicated by the asterisk). (E) HeLa cells were transfected with RNF168 or negative control (NC) siRNAs for 48 h then exposed to 10 Gy IR and released for 1 h before the whole cell lysate and nuclear fractions were analyzed by western blotting. (F) RNF168-WT or RNF168 KO HCT116 cells were transfected with Flag-RNF168 or empty plasmids for 48 h, then exposed to 10 Gy IR and released after 1 h before the whole cell lysate and nuclear fractions were analyzed by western blotting. (G) HCT116 cells were transfected with RNF168 or negative control (NC) siRNAs for 12 h, then co-transfected with the indicated plasmids for 48 h. Then, the cells were treated with 10 μM MG132 for 2 h and exposed to 10 Gy IR and released for 1 h before the nuclear fractions were subjected to immunoprecipitation with an anti-Flag antibody and analyzed by western blotting. (H) Recombinant His-HDAC6 and GST-RNF168 were subjected to in vitro ubiquitination assays performed in a reactive system containing E1 (UBE1), E2 (UBE2D3), HDAC6 (substrate), and ubiquitin with increasing RNF168 (E3 ligase). The reactions were analyzed by western blotting. (I) HDAC6 KO cells were transfected with Flag-HDAC6 wild-type (WT) or mutant (K116R) for 48 h, then exposed to 10 Gy IR and released after 1 h before the whole cell lysate and chromatin fractions were analyzed by western blotting.
Next, we transfected HeLa cells with Flag-HDAC6 or an empty plasmid and treated them with IR (10 Gy). The interaction between HDAC6 and RNF168 was increased following IR (Figure 6C). We then carried out an in vitro GST pull-down assay by incubating the GST-tagged RNF168 with His-tagged HDAC6 and found that His-HDAC6 was precipitated by GST beads, indicating a direct interaction between HDAC6 and RNF168 in vitro (Figure 6D).
To map the regions of HDAC6 responsible for the interaction with RNF168, we constructed various RNF168 and HDAC6 fragments (Supplementary Figure S6A and C) and purified the protein fragments. According to the GST pull-down assay, full-length RNF168 and the ubiquitin-dependent DSB recruitment module 1 (UDM1) and UDM2 domains, but not the RING domain, interacted with HDAC6 (Supplementary Figure S6B). Reciprocally, purified His-tagged HDAC6 fragments interacted with GST-RNF168 via the deacetylase domain 1 (DC1) and DC2 domains (Supplementary Figure S6D). Together, these data indicate that HDAC6 directly interacts with RNF168.
The above observations promoted us to investigate whether RNF168 regulates the degradation of nuclear HDAC6. We knocked down RNF168 using siRNA and evaluated HDAC6 levels in HeLa cells after inducing DSBs by IR (10 Gy with 1 h release) or VP16 (10 μM for 2 h). Notably, RNF168 knockdown markedly alleviated the downregulation of nuclear HDAC6 in response to DSBs (Figure 6E and Supplementary Figure S6E). We next generated RNF168 knockout (KO) HCT116 cells using CRISPR-Cas9 technology (Supplementary Figure S6F). As before, IR-induced nuclear HDAC6 degradation was abolished in these KO cells, but this effect was reversed by overexpressing RNF168 (Figure 6F).
To confirm whether RNF168 is required for nuclear HDAC6 degradation, HeLa cells were transfected with RNF168 or negative control siRNAs for 12 h and co-transfected with Flag-HDAC6 and HA-ub plasmids for 48 h. After IR (10 Gy) treatment and release for 1 h, the nuclear fractions were subjected to co-IP. We observed that RNF168 knockdown markedly prevented nuclear HDAC6 ubiquitination in response to IR-induced DNA damage (Figure 6G). Co-expression of RNF168 with HDAC6 in the presence of HA-ub led to an increase in HDAC6 ubiquitination at K48 but not at the K63 ubiquitin chain (Supplementary Figure S6G).
To check whether HDAC6 is ubiquitinated by RNF168 directly, we performed an in vitro ubiquitination assay with bacterially purified HDAC6 and RNF168. HDAC6 was efficiently ubiquitinated by RNF168 in a dose-dependent manner (Figure 6H). In addition, overexpression of a Flag-HDAC6 mutant (K116R) more strongly inhibited than that of Flag-HDAC6 WT in 53BP1 recruitment and ubiquitination of chromatin (Figure 6I). Collectively, these results suggest that RNF168-mediated HDAC6 ubiquitination at lysine 116 leads to nuclear HDAC6 degradation in response to DSBs.
RNF168-mediated nuclear HDAC6 degradation is beneficial for DSB repair and cell survival
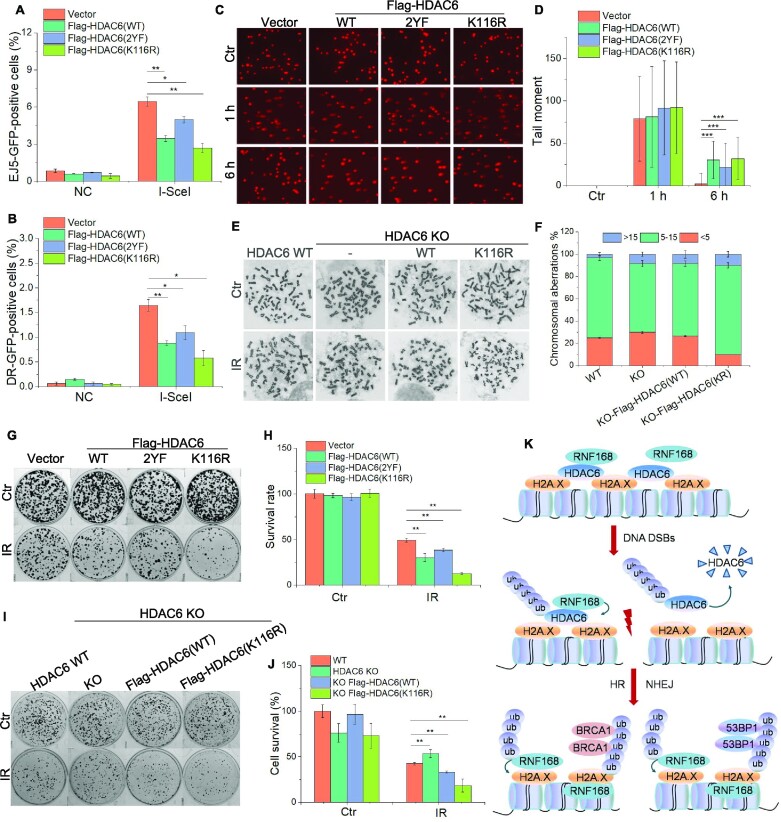
In our final analyses, we explored the biological and functional relevance of the HDAC6-mediated H2A/H2A.X ubiquitination signaling cascade. We first tested the involvement of HDAC6 in DSB repair by using the DR-GFP and pEJ5-GFP reporter systems. Overexpression of Flag-HDAC6 WT or HDAC6 K116R and 2YF mutants inhibited both the NHEJ and HR repair pathways (Figure 7A, B). Inhibition of HDAC6 enzymatic activity by a specific HDAC6 inhibitor (ricolinostat) also affected the efficiency of DNA damage repair (Supplementary Figure S7A, B). Next, we performed comet assays in HCT116 cells transfected with Flag-HDAC6 WT or Flag-HDAC6 mutants. Upon IR treatment (10 Gy), we observed a significant increase in comet tails in the context of both HDAC6 WT and mutant overexpression compared with the empty plasmid control cells (Figure 7C, D). We also observed an elevated frequency of chromosomal abnormalities in Flag-HADC6 K116R stably expression cells (Figure 7E, F).
Figure 7.
RNF168-mediated nuclear HDAC6 degradation is beneficial for DSB repair and cell survival. (A, B) pEJ5-GFP U2OS (A) and DR-U2OS (B) cells were transfected with Flag-HDAC6 wild-type (WT) or mutants (K116R or 2YF) for 48 h and subjected to NHEJ (A) and HR (B) assays, respectively. (C, D) HeLa cells were transfected with Flag-HDAC6 wild-type (WT) or mutants (K116R or 2YF) for 48 h, exposed to 10 Gy irradiation (IR), and then allowed to recover under normal conditions for the indicated times. All samples were then analyzed by comet assay. Representative images (C) and data analysis (D) are shown. (E, F) HDAC6-WT, HDAC6 KO, HDAC6 KO re-transfected Flag-HDAC6(WT), and Flag-HDAC6(K116R) stably expressing cells were exposed to 3 Gy IR and released for 8 h. The cells were then treated with colcemid (0.03 μg/ml) for an additional 6 h before chromosomal abnormality analysis. Representative images (E) and data analysis (F) are shown. (G, H) HeLa cells were transfected with Flag-HDAC6 wild-type (WT) or mutants (K116R or 2YF) for 48 h and then exposed to 3 Gy IR. The cells were counted and seeded for colony formation assays. For the control (Ctr) group, 500 cells were seeded in each plate, while for the IR-treated group, 5000 cells were seeded in each plate. (I, J) HDAC6-WT, HDAC6 KO, HDAC6 KO re-transfected Flag-HDAC6 (WT), and HDAC6 KO re-transfected Flag-HDAC6 (K116R) stably expressing cells were exposed to 3 Gy IR before being counted and seeded for colony formation assays. For the control (Ctr) group, 500 cells were seeded in each plate, while for the IR-treated group, 5000 cells were seeded in each plate. (K) A model of the role played by HDAC6 in DSB repair. All data represent the means ± SD (n = 60 for E–F, I–J; n = 3 for A–D, G–H; *P< 0.05, **P< 0.01, ***P< 0.001).
We next examined the biological relevance of HDAC6 in cancer cell resistance to DNA-damaging treatment. Here, we expressed Flag-HDAC6 wild-type (WT) or HDAC6 mutant (2YF) and (K116R) plasmids in HeLa cells and exposed them to IR (3 Gy, release for 8 h). Colony formation assays showed that WT and K116R-HDAC6-expressing cells exhibited greater hypersensitivity to IR treatment compared with the empty plasmid transfected cells, as evidenced by an approximately 30% decrease in colony numbers between the two groups (Figure 7G, H). However, in 2YF-HDAC6 cells, the survival rate was decreased by approximately 11%. Next, we compared clonogenic survival following the rescue of HDAC6-deficient cells by overexpressing HDAC6 WT or ubiquitin-defective HDAC6 K116R. Both WT and K116R abrogated the cell survival induced by an HDAC6-deficiency in response to IR (Figure 7I, J). Taken together, these results indicate that the degradation of nuclear HDAC6 promotes DSB repair and confers cancer cell resistance to DNA-damaging treatments.
DISCUSSION
Despite the modification of H2A/H2A.X being integral to DSB repair, precisely how H2A/H2AX signaling is regulated under physiological conditions remain poorly understood. In this study, we aimed to understand the role HDAC6 in the regulation of the H2A/H2AX signaling cascade during DSB repair. After performing a series of experiments, we propose a working model whereby under normal conditions, HDAC6 antagonizes H2A/H2A.X ubiquitination by preventing RNF168 loading to H2A/H2A.X. Following DSBs, RNF168 directly interacts with HDAC6 and mediates DSB-induced degradation of nuclear HDAC6. This effect allows RNF168 to bind and ubiquitylate H2A/H2A.X, thus creating a recruitment scaffold for DSB repair factors (Figure 7K).
HDAC6 overexpression or nuclear localization is frequently described in various cancers (10). It seems that nuclear HDAC6 decreases DNA MMR or NER efficiency via a mechanism that is dependent on its deacetylase and ubiquitase activities (11,13,14). Here, we show that HDAC6 is also involved in DNA DSB repair but in a deacetylase-independent manner. To the best of our knowledge, this is the first report to uncover an enzymatic-independent function of nuclear HDAC6 in DSB repair. Despite possessing both deacetylase and ubiquitin-binding activity, through our experiments we observed that HDAC6 serves as a physical barrier by interacting with H2A/H2A.X and thus preventing an RNF168–H2A/H2A.X interaction. Both RNF168 and H2A.X interact with the DAC1 and DAC2 domains of HDAC6. Under physiological conditions, HDAC6 interacts with H2A.X, which prevents RNF168 binding and excessively amplifies the H2A/H2A.X ubiquitination signaling cascade. Upon DNA DSBs, there is massive recruitment of RNF168, which in turn disrupts the balance of HDAC6, H2A/H2A.X and RNF168 by interacting and regulating HDAC6 degradation, thus weakening the HDAC6-H2A.X interaction. This physiological role for nuclear HDAC6 during DSB stimulation seems to be specific to the type of DNA damage induced. Indeed, similar phenomena have been described for the SIRT6 deacetylase. SIRT6 deacetylates H3K56 to regulate recruitment of the ISWI-chromatin remodeler SNF2H during DSB repair (39), but as a ribosylase, SIRT6 mono-ADP ribosylates PARP1 at the K521 residue during base excision repair (40). In previous work, we also demonstrated that SIRT6 interacts with and recruits CHD4 in an enzymatic activity-independent manner and promotes chromatin relaxation for HR repair (41). Thus, a ubiquitin-binding and deacetylase-independent role for HDAC6 in DSB repair seems reasonable. It will be interesting to further explore the interplay between the enzymatic and nonenzymatic roles of HDAC6 in DNA damage response regulation.
It has been reported that UV exposure promotes the rapid translocation of nuclear HDAC6 to the cytoplasm, thus preventing HDAC6–RPA1 interaction and promoting NER (13). Once in the cytoplasm, HDAC6 is degraded by the ubiquitin E3 ligases TRIM28 (42) and VHL (43). Here, we show that HDAC6 is also degraded in the nucleus via E3 ligase-mediated ubiquitination, in this case by RNF168. RNF168 is widely involved in the ubiquitination-mediated DNA damage response. Besides H2A/H2A.X K15 ubiquitination and non-degradable K63 or K27-linked poly-ubiquitination (15,18,44), RNF168 also catalyzes K48-linked poly-ubiquitination of non-histone proteins for chromatin eviction or degradation. For example, the H4K20me2-binding protein JMJD2A is ubiquitinated by RNF8 and RNF168 and degraded by proteasome, allowing efficient 53BP1 chromatin recruitment (16). Here, we demonstrated for the first time that HDAC6 is a substrate of RNF168-mediated K48-linked poly-ubiquitination.
Although RNF8 often exhibits similar roles to RNF168 in response to DNA damage-induced ubiquitination (22,23,45), we have excluded a role for RNF8 in ubiquitinating HDAC6, as no interaction between HDAC6 and RNF8 was detected in our system. This finding might be because HDAC6 interacts directly with the RNF168 UDM domain, which is a domain that RNF8 lacks. In our previous study, we also showed a similar case where RNF8, but not RNF168, ubiquitinates lysine methyltransferase 5A (KMT5A) in response to DNA damage (29). Based on our observations in the current study, together with our previous findings, we therefore put forward a differential regulation mechanism based on HDAC6 stability and a novel role for both HDAC6 and RNF168 in DSB repair.
The ubiquitination of H2A and its variant H2A.X enables the creation of a platform that can recruit major DNA damage repair factors to DNA damage sites, and this process is critical for the DNA damage response (46–49). Specifically, RNF168-mediated H2A/H2A.X ubiquitination at K13–K15 permits the recruitment of repair factors such as 53BP1 and BRCA1 to DSB sites (31). RNF2-regulated H2A ubiquitination at K119 is important for ATM recruitment to the damage site (50). Here, we confirm that HDAC6-mediated ubiquitination of H2A/H2A.X is dependent on RNF168. RNF168 is the priming ligase for the ubiquitination of H2A/H2A.X at K13–K15 (15). Thus, the dynamic change of RNF168 requires strict controls to safeguard its appropriate function. It has been reported that RNF8-mediated L3MBTL2 ubiquitination is required for RNF168 recruitment in DNA DSB repair (24). In addition, the E3 ligases, TRIP12 and UBR5, are responsible for RNF168 degradation to prevent excessive RNF168 accumulation (51). However, under physiological conditions, RNF168 also functions as an E3 ligase involved in genomic stability. For example, RNF168 directly ubiquitylates DHX9 to facilitate its recruitment to R-loop-prone genomic loci (52). Here, we further observed that RNF168 is present in chromatin under normal conditions, which is consistent with the findings of our previous study (29). Most importantly, we show that HDAC6 deficiency results in an inappropriate increase in H2A/H2A.X ubiquitination even in the absence of stress. Based on this observation, we confirm that HDAC6 is a key inhibitory protein to prevent RNF168 activity and H2A/H2A.X ubiquitination under physiological conditions.
DNA damage repair is one of the major barriers to effective genotoxic treatment, and disrupting the key signaling pathways underlying the DNA damage repair process would represent a potential therapeutic strategy for cancer (53–55). For example, the small molecular antagonists MM-102 and OICR-9429, which target the MLL1/WDR5 protein–protein interaction, efficiently inhibit the catalytic activity of MLL1 and suppress H3K4 methylation, which in turn enhances apoptosis and chemosensitivity to cisplatin (56–58). We showed here that RNF168 interacts with HDAC6 and promotes HDAC6 degradation—a key step in initiating DNA DSB repair. Importantly, blocking RNF168-mediated HDAC6 degradation impairs effective DNA damage repair. In this study, we observed that although inhibition of HDAC6 enzymatic activity can affect the efficiency of DNA damage repair, the effect on DNA damage efficiency is much lower than that treated by HDAC6 RNAi. Because ACY-1215 is a selective inhibitor of HDAC6 by disrupting the DAC1/2 domains’ interaction with downstream substrates to inhibit HDAC6 enzymatic activity, it is undoubtedly the case that HDAC6 inhibitors also affect the efficiency of DSB repair (HDAC6 interacts with RNF168 at the DAC1/2 domains). This phenomenon provides insights into a new approach to improve the sensitivity of radio/chemotherapies in cancer therapy based on disrupting the RNF168/HDAC6 protein–protein interaction. This approach offers the potential for the development of new cancer therapies.
In conclusion, our findings provide novel insights into the role of HDCA6 in negatively regulating RNF168-mediated H2A/H2AX ubiquitination and DSB repair. This may indicate the possible relevance of HDAC6 in clinical research into cancer radio/chemotherapies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Jessica Tamanini of ETediting, UK for English language editing prior to submission.
Author contributions: Conceptualization, L.Y.Q., Y.C.C., W.-G.Z., J.Z.; Methodology, L.Y.Q., X.P. L., Q.Z., Y. T., J.Z.; Investigation, L.Y.Q., W.C.X., F.C., J.Z.; Writing–Original Draft, L.Y.Q., J.Z.; Writing–Review & Editing, L.Y.Q., J.Z.; W.-G.Z.; Funding Acquisition, W.-G.Z., J.Z.; Resources, X.Y.L., Y.Q.W., X.-H.P., X.Z.X.; Supervision, W.-G.Z., J.Z.
Contributor Information
Lingyu Qiu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Wenchao Xu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Xiaopeng Lu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Feng Chen, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Yongcan Chen, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Yuan Tian, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Qian Zhu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Xiangyu Liu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Yongqing Wang, Division of Rheumatology and Immunology, University of Toledo Medical Center, 3120 Glendale Avenue, Toledo 43614, OH, USA.
Xin-Hai Pei, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Anatomy and Histology, Shenzhen University Medical School, Shenzhen 518055, China.
Xingzhi Xu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Cell Biology and Medical Genetics, Shenzhen University Medical School, Shenzhen 518055, China.
Jun Zhang, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China.
Wei-Guo Zhu, International Cancer Center, Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Marshall Laboratory of Biomedical Engineering, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen 518055, China; School of Basic Medical Sciences, Wannan Medical College, Wuhu, Anhui 241002, China; Department of Biochemistry and Biophysics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [32090030, 81720108027, 81530074, 82002986]; Science and Technology Program of Guangdong Province in China [2017B030301016]; Guangdong Basic and Applied Basic Research Foundation [2021A1515011126]; Shenzhen Municipal Commission of Science and Technology Innovation [JCYJ20200109114214463, JCYJ20220818100015032, RCYX20210706092040047]; Shenzhen University 2035 Program for Excellent Research. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Panier S., Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 2013; 14:661–672. [DOI] [PubMed] [Google Scholar]
- 4. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang K., Ning Y., Kong F., Chen X., Cai Y.. Genome instability in pathogenesis of tuberculosis. Genome Instab. Dis. 2021; 2:331–338. [Google Scholar]
- 6. Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S.. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004; 73:39–85. [DOI] [PubMed] [Google Scholar]
- 7. Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roos W.P., Thomas A.D., Kaina B.. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016; 16:20–33. [DOI] [PubMed] [Google Scholar]
- 9. Zhou B.B., Elledge S.J.. The DNA damage response: putting checkpoints in perspective. Nature. 2000; 408:433–439. [DOI] [PubMed] [Google Scholar]
- 10. Li T., Zhang C., Hassan S., Liu X., Song F., Chen K., Zhang W., Yang J.. Histone deacetylase 6 in cancer. J. Hematol. Oncol. 2018; 11:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang M., Xiang S., Joo H.Y., Wang L., Williams K.A., Liu W., Hu C., Tong D., Haakenson J., Wang C.et al.. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol. Cell. 2014; 55:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang M., Hu C., Moses N., Haakenson J., Xiang S., Quan D., Fang B., Yang Z., Bai W., Bepler G.et al.. HDAC6 regulates DNA damage response via deacetylating MLH1. J. Biol. Chem. 2019; 294:5813–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He H., Wang J., Liu T.. UV-induced RPA1 acetylation promotes nucleotide excision repair. Cell Rep. 2017; 20:2010–2025. [DOI] [PubMed] [Google Scholar]
- 14. Zhao M., Geng R., Guo X., Yuan R., Zhou X., Zhong Y., Huo Y., Zhou M., Shen Q., Li Y.et al.. PCAF/GCN5-mediated acetylation of RPA1 promotes nucleotide excision repair. Cell Rep. 2017; 20:1997–2009. [DOI] [PubMed] [Google Scholar]
- 15. Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K.. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 16. Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S.. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012; 31:1865–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guturi K.K.N., Bohgaki M., Bohgaki T., Srikumar T., Ng D., Kumareswaran R., El Ghamrasni S., Jeon J., Patel P., Eldin M.S.et al.. RNF168 and USP10 regulate topoisomerase iialpha function via opposing effects on its ubiquitylation. Nat. Commun. 2016; 7:12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bohgaki M., Bohgaki T., El Ghamrasni S., Srikumar T., Maire G., Panier S., Fradet-Turcotte A., Stewart G.S., Raught B., Hakem A.et al.. RNF168 ubiquitylates 53BP1 and controls Its response to DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2013; 110:20982–20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P.. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005; 123:1213–1226. [DOI] [PubMed] [Google Scholar]
- 20. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 21. Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T.et al.. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006; 21:187–200. [DOI] [PubMed] [Google Scholar]
- 22. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J.. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. [DOI] [PubMed] [Google Scholar]
- 23. Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J.. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007; 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nowsheen S., Aziz K., Aziz A., Deng M., Qin B., Luo K., Jeganathan K.B., Zhang H., Liu T., Yu J.et al.. L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat. Cell Biol. 2018; 20:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu J., Qin B., Lou Z.. Ubiquitin and ubiquitin-like molecules in DNA double strand break repair. Cell & Bioscience. 2020; 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fradet-Turcotte A., Canny M.D., Escribano-Diaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F.et al.. 53BP1 is a reader of the DNA-damage-induced H2A lys 15 ubiquitin mark. Nature. 2013; 499:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu J., Lu L.Y., Yu X.. The role of BRCA1 in DNA damage response. Protein Cell. 2010; 1:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao W., McNutt M.A., Zhu W.G.. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods. 2009; 48:46–53. [DOI] [PubMed] [Google Scholar]
- 29. Lu X.P., Xu M., Zhu Q., Zhang J., Liu G., Bao Y.T., Gu L., Tian Y., Wen H., Zhu W.G.. RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. FASEB J. 2021; 35:e21326. [DOI] [PubMed] [Google Scholar]
- 30. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Attikum H., Gasser S.M.. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009; 19:207–217. [DOI] [PubMed] [Google Scholar]
- 32. Cao R., Tsukada Y., Zhang Y.. Role of Bmi-1 and Ring1A in H2A ubiquitylation and hox gene silencing. Mol. Cell. 2005; 20:845–854. [DOI] [PubMed] [Google Scholar]
- 33. Ulrich H.D., Walden H.. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010; 11:479–489. [DOI] [PubMed] [Google Scholar]
- 34. Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M.et al.. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009; 136:420–434. [DOI] [PubMed] [Google Scholar]
- 35. Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Muller C.W., Khochbin S.. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006; 25:3357–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hook S.S., Orian A., Cowley S.M., Eisenman R.N.. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2002; 99:13425–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kolla S., Ye M., Mark K.G., Rape M.. Assembly and function of branched ubiquitin chains. Trends Biochem. Sci. 2022; 47:759–771. [DOI] [PubMed] [Google Scholar]
- 38. Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J.et al.. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011; 44:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S.et al.. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013; 51:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V.. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011; 332:1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hou T., Cao Z., Zhang J., Tang M., Tian Y., Li Y., Lu X., Chen Y., Wang H., Wei F.Z.et al.. SIRT6 coordinates with CHD4 to promote chromatin relaxation and DNA repair. Nucleic Acids Res. 2020; 48:2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Z., Lu X., Liu Y., Zhao J., Ma S., Yin H., Huang S., Zhao Y., He X.. Gain of LINC00624 enhances liver cancer progression by disrupting the histone deacetylase 6/tripartite motif containing 28/zinc finger protein 354C corepressor complex. Hepatology. 2021; 73:1764–1782. [DOI] [PubMed] [Google Scholar]
- 43. Ran J., Liu M., Feng J., Li H., Ma H., Song T., Cao Y., Zhou P., Wu Y., Yang Y.et al.. ASK1-Mediated phosphorylation blocks HDAC6 ubiquitination and degradation to drive the disassembly of photoreceptor connecting cilia. Dev. Cell. 2020; 53:287–299. [DOI] [PubMed] [Google Scholar]
- 44. Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M.G., Aebersold R., Penengo L.. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015; 10:226–238. [DOI] [PubMed] [Google Scholar]
- 45. Ma T., Keller J.A., Yu X.. RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta Biochim. Biophys. Sin. 2011; 43:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uckelmann M., Sixma T.K.. Histone ubiquitination in the DNA damage response. DNA Repair (Amst.). 2017; 56:92–101. [DOI] [PubMed] [Google Scholar]
- 47. Schmid J.A., Berti M., Walser F., Raso M.C., Schmid F., Krietsch J., Stoy H., Zwicky K., Ursich S., Freire R.et al.. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S phase. Mol. Cell. 2018; 71:897–910. [DOI] [PubMed] [Google Scholar]
- 48. Zhang J., Lu X.P., MoghaddamKohi S., Shi L., Xu X.Z., Zhu W.G.. Histone lysine modifying enzymes and their critical roles in DNA double-strand break repair. DNA Repair (Amst.). 2021; 107:103206. [DOI] [PubMed] [Google Scholar]
- 49. Nowsheen S., Deng M., Lou Z.. Ubiquitin and the DNA double-strand break repair pathway. Genome Instab. Dis. 2020; 1:69–80. [Google Scholar]
- 50. Pan M.R., Peng G., Hung W.C., Lin S.Y.. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 2011; 286:28599–28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S.et al.. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012; 150:697–709. [DOI] [PubMed] [Google Scholar]
- 52. Patel P.S., Abraham K.J., Guturi K.K.N., Halaby M.J., Khan Z., Palomero L., Ho B., Duan S., St-Germain J., Algouneh A.et al.. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J. Clin. Invest. 2021; 131:e140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu X., Nowsheen S., Deng M.. Exploring the DNA damage response pathway for synthetic lethality. Genome Instab. Dis. 2023; 4:98–120. [Google Scholar]
- 54. Li G., Tian Y., Zhu W.G.. The roles of histone deacetylases and their inhibitors in cancer therapy. Front. Cell Dev. Biol. 2020; 8:576946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang J., Zhang J., Xu W., Wu Q., Zeng R., Liu Z., Tao W., Chen Q., Wang Y., Zhu W.-G.. Structure-Based Discovery of Selective Histone Deacetylase 8 Degraders with Potent Anticancer Activity. J. Med. Chem. 2022; 66:1186–1209. [DOI] [PubMed] [Google Scholar]
- 56. Karatas H., Townsend E.C., Cao F., Chen Y., Bernard D., Liu L., Lei M., Dou Y., Wang S.. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 2013; 135:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J., Zhou Q., Xie K., Cheng L., Peng S., Xie R., Liu L., Zhang Y., Dong W., Han J.et al.. Targeting WD repeat domain 5 enhances chemosensitivity and inhibits proliferation and programmed death-ligand 1 expression in bladder cancer. J. Exp. Clin. Cancer Res. 2021; 40:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou Q., Chen X., He H., Peng S., Zhang Y., Zhang J., Cheng L., Liu S., Huang M., Xie R.et al.. WD repeat domain 5 promotes chemoresistance and Programmed Death-Ligand 1 expression in prostate cancer. Theranostics. 2021; 11:4809–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.