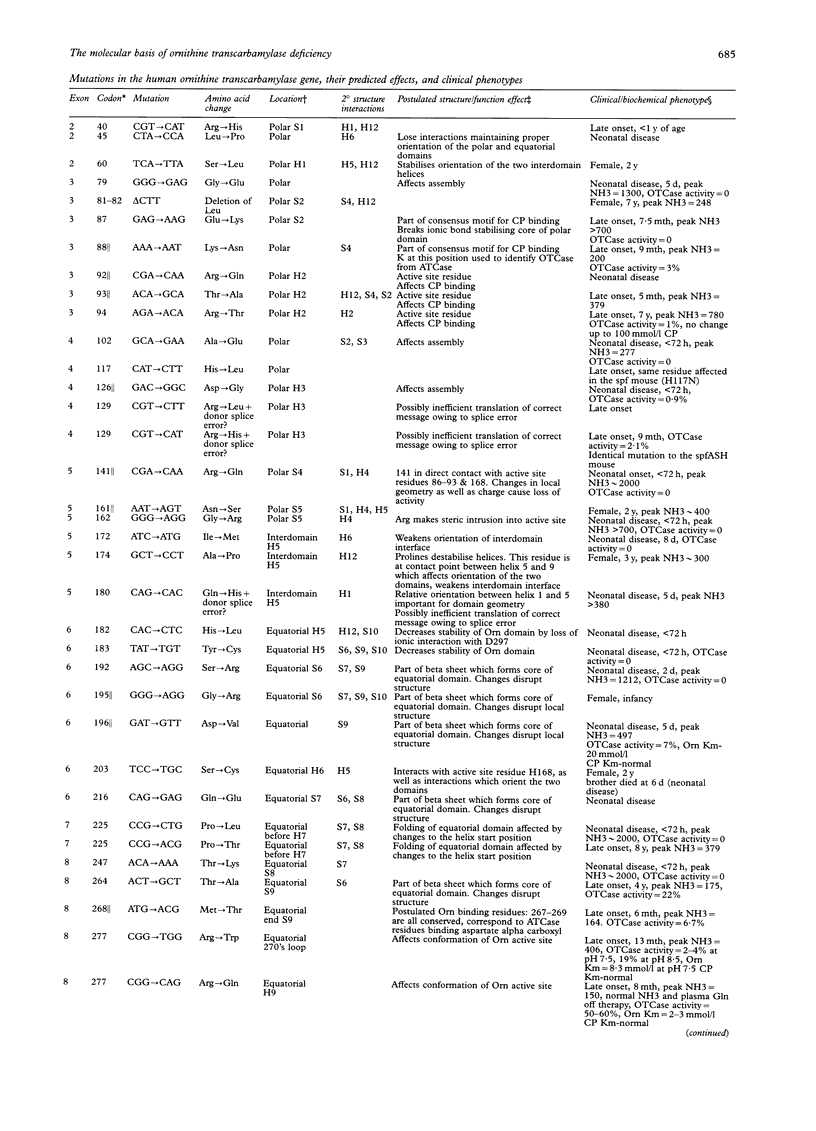
Abstract
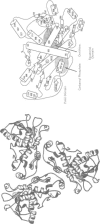
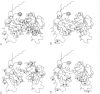
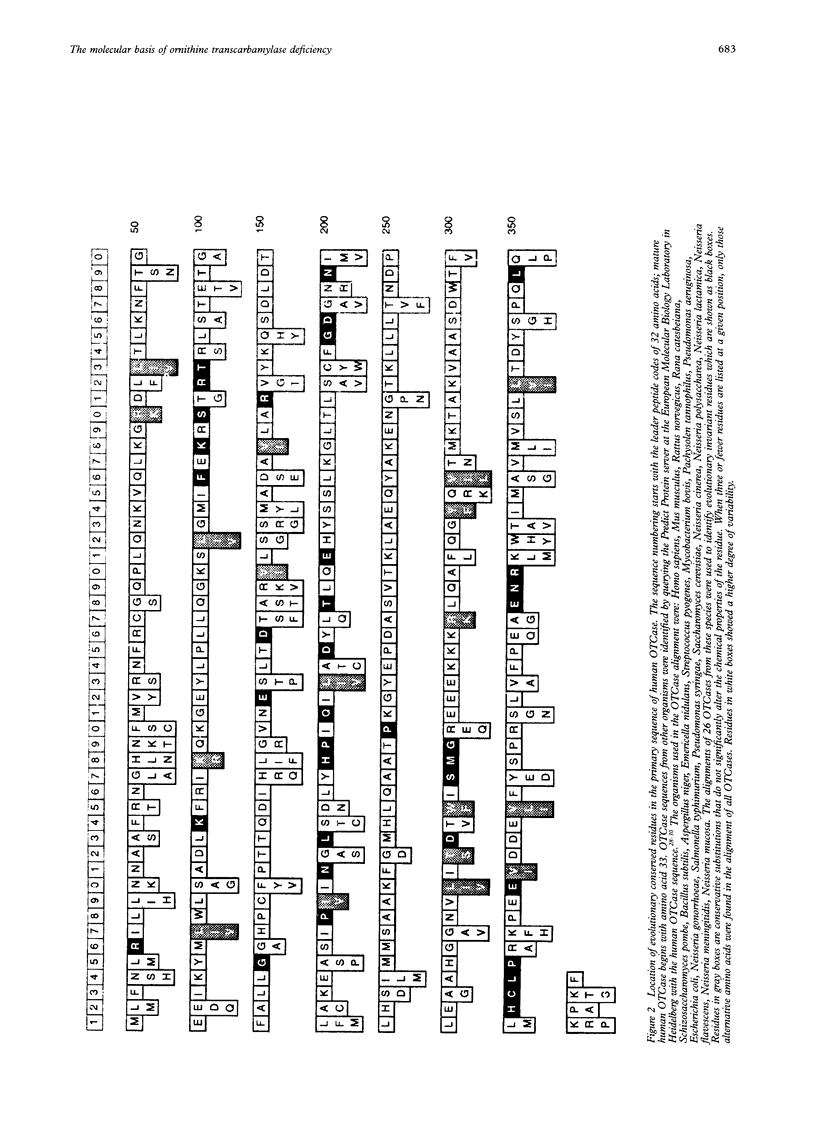
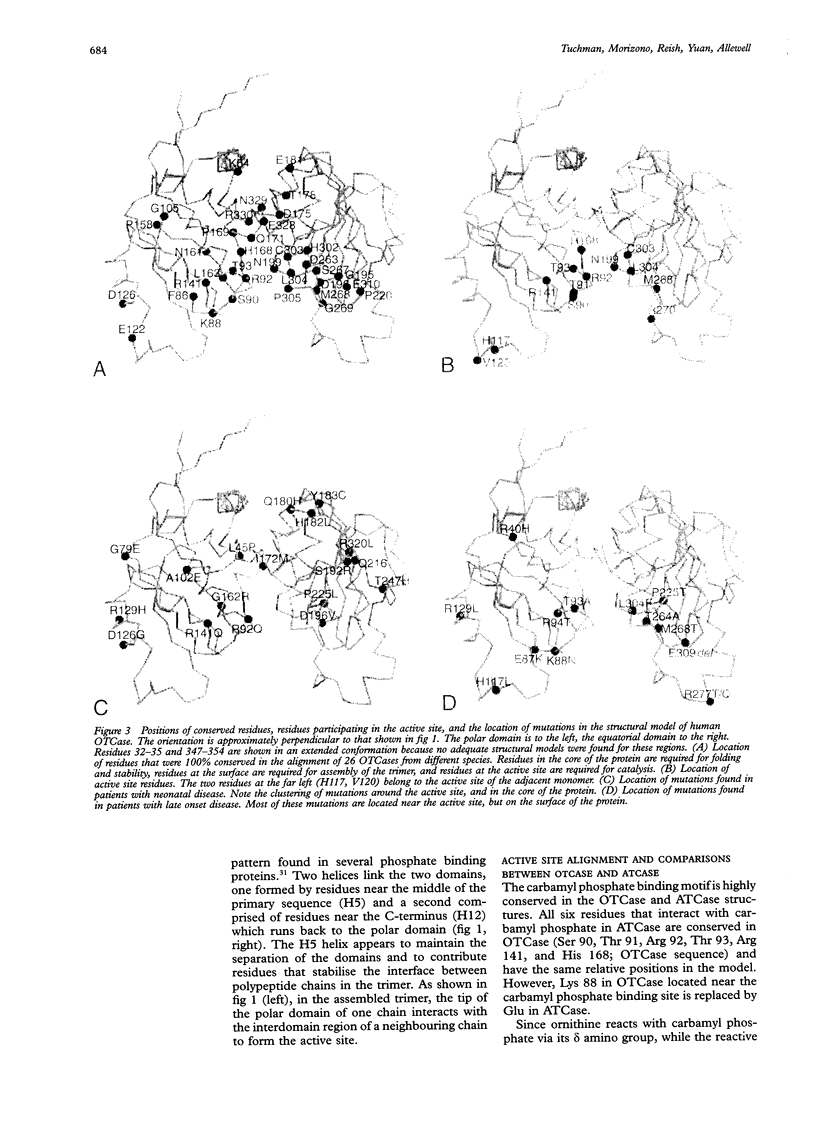
Human ornithine transcarbamylase is a trimer with 46% amino acid sequence homology to the catalytic subunit of E coli aspartate transcarbamylase. Secondary structure predictions, distributions of hydrophilic and hydrophobic regions, and the pattern of conserved residues suggest that the three dimensional structures of the two proteins are likely to be similar. A three dimensional model of ornithine transcarbamylase was generated from the crystal structure of the catalytic subunit of E coli aspartate transcarbamylase in the holoenzyme, by aligning the sequences, building in gaps, and minimising the energy. The binding sites for carbamyl phosphate in both enzymes are similar and the ornithine binding site in ornithine transcarbamylase appears to be in the same location as the L-aspartate binding site in aspartate transcarbamylase, with negatively charged side chains replaced by positively charged residues. Mutations in the ornithine transcarbamylase gene found in patients with hyperammonaemia of the "neonatal type" are clustered in important structural or functional domains, either in the interior of the protein, at the active site, or at the interchain interface, while mutations found in patients with milder "late onset" disease are located primarily on the surface of the protein. The predicted effects of all known missense mutations and in frame deletions in the ornithine transcarbamylase gene on the structure and function of the mature enzyme are described.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allewell N. M. Escherichia coli aspartate transcarbamoylase: structure, energetics, and catalytic and regulatory mechanisms. Annu Rev Biophys Biophys Chem. 1989;18:71–92. doi: 10.1146/annurev.bb.18.060189.000443. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cathelineau L., Briand P., Petit F., Nuyts J. P., Farriaux J. P., Kamoun P. P. Kinetic analysis of a new human ornithine carbamoyltransferase variant. Biochim Biophys Acta. 1980 Jul 10;614(1):40–45. doi: 10.1016/0005-2744(80)90165-5. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Allosteric interactions in aspartate transcarbamylase. II. Evidence for different conformational states of the protein in the presence and absence of specific ligands. Biochemistry. 1968 Feb;7(2):538–552. doi: 10.1021/bi00842a600. [DOI] [PubMed] [Google Scholar]
- Hata A., Tsuzuki T., Shimada K., Takiguchi M., Mori M., Matsuda I. Structure of the human ornithine transcarbamylase gene. J Biochem. 1988 Feb;103(2):302–308. doi: 10.1093/oxfordjournals.jbchem.a122265. [DOI] [PubMed] [Google Scholar]
- Horwich A. L., Fenton W. A., Williams K. R., Kalousek F., Kraus J. P., Doolittle R. F., Konigsberg W., Rosenberg L. E. Structure and expression of a complementary DNA for the nuclear coded precursor of human mitochondrial ornithine transcarbamylase. Science. 1984 Jun 8;224(4653):1068–1074. doi: 10.1126/science.6372096. [DOI] [PubMed] [Google Scholar]
- Houghton J. E., Bencini D. A., O'Donovan G. A., Wild J. R. Protein differentiation: a comparison of aspartate transcarbamoylase and ornithine transcarbamoylase from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4864–4868. doi: 10.1073/pnas.81.15.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton J. E., O'Donovan G. A., Wild J. R. Reconstruction of an enzyme by domain substitution effectively switches substrate specificity. Nature. 1989 Mar 9;338(6211):172–174. doi: 10.1038/338172a0. [DOI] [PubMed] [Google Scholar]
- Kalousek F., François B., Rosenberg L. E. Isolation and characterization of ornithine transcarbamylase from normal human liver. J Biol Chem. 1978 Jun 10;253(11):3939–3944. [PubMed] [Google Scholar]
- Kosman R. P., Gouaux J. E., Lipscomb W. N. Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity. Proteins. 1993 Feb;15(2):147–176. doi: 10.1002/prot.340150206. [DOI] [PubMed] [Google Scholar]
- Kuo L. C., Caron C., Lee S., Herzberg W. Zn2+ regulation of ornithine transcarbamoylase. II. Metal binding site. J Mol Biol. 1990 Jan 5;211(1):271–280. doi: 10.1016/0022-2836(90)90026-I. [DOI] [PubMed] [Google Scholar]
- Kuo L. C., Lipscomb W. N., Kantrowitz E. R. Zn(II)-induced cooperativity of Escherichia coli ornithine transcarbamoylase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2250–2254. doi: 10.1073/pnas.79.7.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L. C., Seaton B. A. X-ray diffraction analysis on single crystals of recombinant Escherichia coli ornithine transcarbamoylase. J Biol Chem. 1989 Sep 25;264(27):16246–16248. [PubMed] [Google Scholar]
- Lee S., Shen W. H., Miller A. W., Kuo L. C. Zn2+ regulation of ornithine transcarbamoylase. I. Mechanism of action. J Mol Biol. 1990 Jan 5;211(1):255–269. doi: 10.1016/0022-2836(90)90025-H. [DOI] [PubMed] [Google Scholar]
- Legrain C., Stalon V. Ornithine carbamoyltransferase from Escherichia coli W. Purification, structure and steady-state kinetic analysis. Eur J Biochem. 1976 Mar 16;63(1):289–301. doi: 10.1111/j.1432-1033.1976.tb10230.x. [DOI] [PubMed] [Google Scholar]
- Lindgren V., de Martinville B., Horwich A. L., Rosenberg L. E., Francke U. Human ornithine transcarbamylase locus mapped to band Xp21.1 near the Duchenne muscular dystrophy locus. Science. 1984 Nov 9;226(4675):698–700. doi: 10.1126/science.6494904. [DOI] [PubMed] [Google Scholar]
- Miller A. W., Kuo L. C. Ligand-induced isomerizations of Escherichia coli ornithine transcarbamoylase. An ultraviolet difference analysis. J Biol Chem. 1990 Sep 5;265(25):15023–15027. [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Newton C. J., Kantrowitz E. R. Importance of domain closure for homotropic cooperativity in Escherichia coli aspartate transcarbamylase. Biochemistry. 1990 Feb 13;29(6):1444–1451. doi: 10.1021/bi00458a015. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C., Schneider R. PHD--an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994 Feb;10(1):53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum Mutat. 1993;2(3):174–178. doi: 10.1002/humu.1380020304. [DOI] [PubMed] [Google Scholar]
- Tuchman M., Plante R. J. Mutations and polymorphisms in the human ornithine transcarbamylase gene: mutation update addendum. Hum Mutat. 1995;5(4):293–295. doi: 10.1002/humu.1380050404. [DOI] [PubMed] [Google Scholar]
- Wild J. R., Wales M. E. Molecular evolution and genetic engineering of protein domains involving aspartate transcarbamoylase. Annu Rev Microbiol. 1990;44:193–218. doi: 10.1146/annurev.mi.44.100190.001205. [DOI] [PubMed] [Google Scholar]