Abstract
Acclimation and adaptation of metabolism to a changing environment are key processes for plant survival and reproductive success. In the present study, 241 natural accessions of Arabidopsis (Arabidopsis thaliana) were grown under two different temperature regimes, 16 °C and 6 °C, and growth parameters were recorded, together with metabolite profiles, to investigate the natural genome × environment effects on metabolome variation. The plasticity of metabolism, which was captured by metabolic distance measures, varied considerably between accessions. Both relative growth rates and metabolic distances were predictable by the underlying natural genetic variation of accessions. Applying machine learning methods, climatic variables of the original growth habitats were tested for their predictive power of natural metabolic variation among accessions. We found specifically habitat temperature during the first quarter of the year to be the best predictor of the plasticity of primary metabolism, indicating habitat temperature as the causal driver of evolutionary cold adaptation processes. Analyses of epigenome- and genome-wide associations revealed accession-specific differential DNA-methylation levels as potentially linked to the metabolome and identified FUMARASE2 as strongly associated with cold adaptation in Arabidopsis accessions. These findings were supported by calculations of the biochemical Jacobian matrix based on variance and covariance of metabolomics data, which revealed that growth under low temperatures most substantially affects the accession-specific plasticity of fumarate and sugar metabolism. Our findings indicate that the plasticity of metabolic regulation is predictable from the genome and epigenome and driven evolutionarily by Arabidopsis growth habitats.
Plasticity of primary metabolism is connected to the geographical origin of natural Arabidopsis accessions.
Introduction
Acclimation and adaptation of metabolism to changing environments are key processes for plant survival and reproductive success. The multitude of different abiotic and biotic stressors requires plant metabolism to be highly flexible, as the degree of reprograming of metabolism depends on the type and strength of stress to which plants are exposed (Caldana et al. 2011; Krasensky and Jonak 2012). The metabolic response to changing environmental factors differs substantially between plant species (Weston et al. 2011), as well as among ecotypes or cultivars of the same species (Barah et al. 2013; Shankar et al. 2016; Demirel et al. 2020). Temperature affects plant development and has been shown to be an important determinant for the geographical distribution range of many temperate plant species, e.g. Arabidopsis (Arabidopsis thaliana) (Hoffmann 2002). Considering that only 5% of the land mass worldwide is free of freezing events (Hurry 2017) and that low-temperature damage leads to significant losses in agricultural yield (Larcher 1981; Thakur et al. 2010), the investigation of plant cold response holds a large potential in establishing a sustainable supply of food for a growing world population (Mahajan and Tuteja 2005).
Exposure to low temperature immediately affects plant metabolism by reducing enzymatic reaction rates, which has a substantial effect on biosynthesis, degradation, and transport processes (see, e.g. Patzke et al. 2019). Within a process termed cold acclimation, metabolism is adjusted to low temperature, which, in many temperate plant species, results in increased freezing tolerance (Levitt 1980). Cold acclimation is a multigenic process involving hundreds of genes, numerous signaling cascades, and metabolic pathways to stabilize the photosynthetic capacity and plant performance (Herrmann et al. 2019). Cold exposure typically results in a rapid increase of the C-repeat binding factor (CBF) transcription factors (TFs) that regulate >100 genes, the so-called CBF regulon that plays a dominant role in cold acclimation (Thomashow 2010). Comparing Italian and Swedish A. thaliana accessions revealed a lower induction of the CBF regulon in the Italian accessions, which contributed to lower freezing tolerance compared to the Swedish accessions (Gehan et al. 2015). Although CBF TFs rapidly increase after cold exposure, comparison of time-resolved cold response of A. thaliana revealed an even faster metabolic response when compared to the transcriptional response (Caldana et al. 2011). This finding indicates a complex mode of regulation, which, in addition to transcription, also includes translational, posttranslational, and metabolic regulation (Ding et al. 2020).
A. thaliana inhabits a large latitudinal range (Koornneef et al. 2004) and is therefore confronted with a wide range of climatic conditions. This widespread distribution and the predominantly selfing reproduction type have led to the development of a large number of genetically distinct (homozygous) inbred lines, called accessions, which are well adapted to the respectively prevailing microclimate (Nordborg et al. 2005; Mitchell-Olds and Schmitt 2006; Kleessen et al. 2012). Accessions feature large variances in cold and freezing resistance, acquired after cold acclimation, relative to naïve states, i.e. without cold acclimation. These adaptions were shown to be connected to the mean minimum temperature at the site of origin, indicating a selective pressure on the ability to adapt to low temperatures (Hannah et al. 2006; Horton et al. 2016). The variance in freezing tolerance along geographical clines of origin was found correlated to several differences in the accumulation of sugars and the expression of a number of CBF-regulated genes, after an acclimation phase at low, nonfreezing temperatures (Zuther et al. 2012). Another example of the adaptation of A. thaliana to local climates was recently given, by showing a strong connection of climate of origin and the life-history strategy, i.e. the prevalence of winter or summer annuality (Martínez-Berdeja et al. 2020).
It has been reported earlier that habitat temperature of natural A. thaliana accessions determines the response of physiological parameters, such as photosynthesis and transpiration, to growth temperature (Adams et al. 2016). Although it is known that photosynthesis needs to be tightly linked to carbohydrates and primary metabolism to sustain growth and development under low temperature, it remains unclear how plasticity of metabolism relates to natural genetic variation and geographical origin of plant species. In this study, the natural variation of dynamics of primary metabolites under moderate (16 °C) and low (6 °C) temperature was monitored together with growth rates of 241 natural A. thaliana accessions originating from a wide geographical range across Europe and Asia. Genomic prediction was applied together with epigenome-wide association studies (EWAS) and mathematical modeling to reveal how pathway regulation in primary metabolism is related to observed metabolome plasticity.
Results
Natural allelic variation allows for genomic prediction of metabolome plasticity and metabolic distance between 6 °C and 16 °C growth conditions
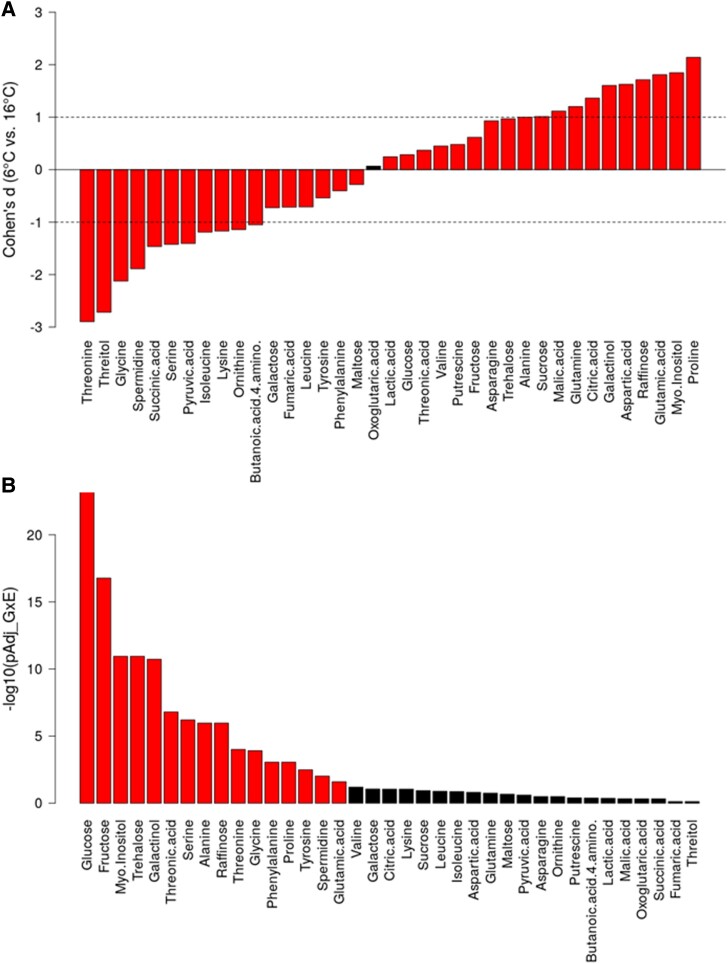
The natural metabolic variation in 241 Arabidopsis accessions was quantified by a metabolomics platform. To provide a comparable data set and for all further statistics, we calculated absolute metabolite concentrations (with additionally applied normalization steps, see Materials and Methods) for a set of 37 central primary metabolites of which 22 changed with a Cohen's d effect size greater than one (between the two different growth temperature regimes, i.e. 16 °C and 6 °C, and averaged over all 241 accessions [Fig. 1A]). Plants grown at 6 °C showed a strong (absolute Cohen's d > 1) increase of proline, myo-inositol, glutamic acid, raffinose, aspartic acid, galactinol, citric acid, glutamine, malic acid, sucrose, and alanine (in descending order of |Cohen's d|), whereas under the 16 °C growth condition, threonine, threitol, glycine, spermidine, succinic acid, serine, pyruvic acid, isoleucine, lysine, ornithine, and 4-amino-butanoic acid accumulated relative to the respective alternative, lower growth temperature condition. For 16 of the 37 metabolites, a strong G × E interaction effect was observed (two-way ANOVA with interaction term; Fig. 1B), revealing a conserved role of soluble carbohydrate metabolism, and particularly of hexoses, in different environments.
Figure 1.
Effects of growth temperature on primary plant metabolism. A) Differential metabolite levels captured by effect size (Cohen's d) for the 37 metabolites at 6 °C and 16 °C, respectively, considered in this study. Metabolite levels were taken as normalized absolute levels as explained in Materials and Methods. Based on t-tests, all but one (oxoglutarate) differences were significant (PFDR < 0.05). Note: Cohen's d can be roughly interpreted as the difference of the mean values (levels at 6 °C minus levels at 16 °C) normalized by the average of the respective standard deviations. Absolute Cohen's d values exceeding “1” (dashed lines) can be regarded as large effects. B) Interaction effects, accessions (genotype, G) × temperature (=environment, E) from a two-way ANOVA analysis with an interaction term. Color indicates significance, red PFDR < 0.05, black otherwise. FDR-adjusted P values are displayed as their -log_base10 transformed values. Note, except 4-amino-butanoic acid, all metabolites were found significantly different across the 241 different accessions (ANOVA PFDR for accession, i.e. genotype effect alone). Statistical tests were performed using all individual samples (Ntot = 1,624 with N16 °C = 817 and N6 °C = 807), i.e. normalized, but not averaged per accession.
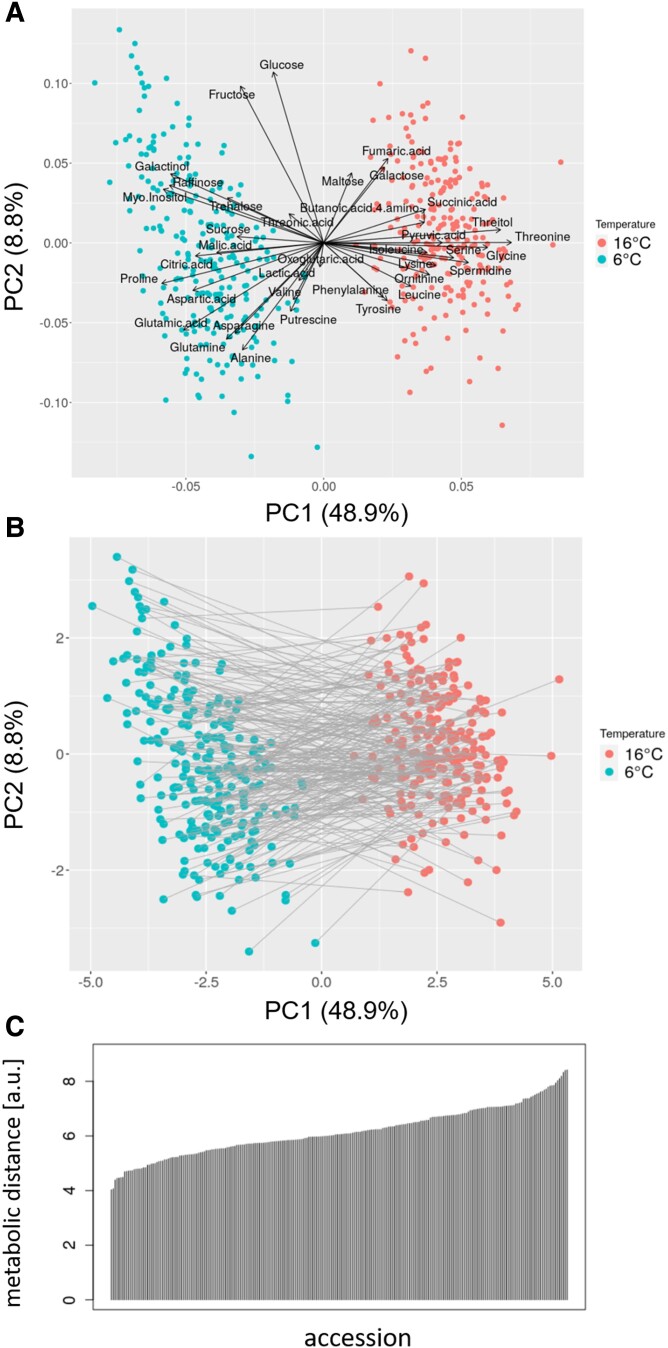
The two different temperature regimes were a stronger influencing factor on metabolite levels than genotypic variation. Principal component 1 of a performed principal component analysis (PCA) perfectly separated the two growth conditions, capturing 49% of the global variance (Fig. 2A). By contrast, PC2, found to largely capture genotypic variation, accounted for 9% only. Respective loadings of the 37 metabolites onto PC1 were in line with the changes reported above. Interestingly, loadings of the sugars fructose and glucose were found to be aligned almost perpendicularly to PC1 and to capture variance associated with the 241 accessions, rather than temperature.
Figure 2.
PCA and illustration of MDs. A) Scaled PCA biplot of the first two principal components (PC 1 and 2), with points corresponding to the scores of the 241 accessions exposed to the two growth temperature conditions and arrows signifying the respective 37 metabolite loadings. Here, every combination of accession and treatment corresponds to the profile of mean normalized metabolite levels, i.e. repeat-averaged samples. Percentages in parentheses denote the variance captured by the respective PC. B) PCA score plot with gray line segments and their respective lengths denoting the MDs calculated as the Euclidean distance of metabolite profiles between samples of the same accession exposed to the two different growth temperatures (Eq. 1). Note, unlike in Fig. 1A, scores are not scaled and MDs are computed in this unscaled value range respectively, i.e. the score plot corresponds to a rotation in the 37-dimensional Euclidean space to align with PC 1 and 2. C) Overview of the distribution and range of MD values across all 241 accessions.
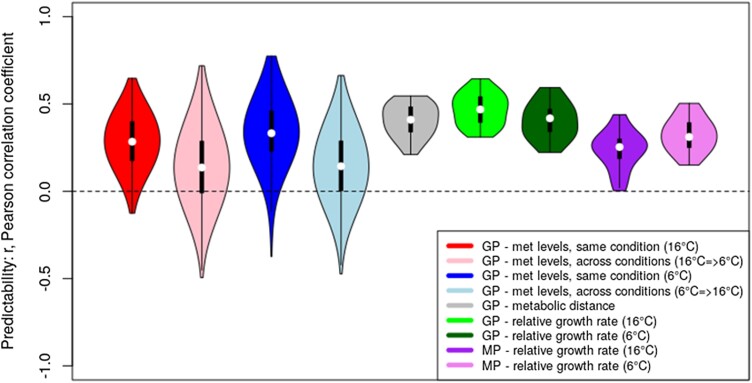
To further investigate the natural variation of metabolic responses to different growth temperatures, we calculated “metabolic distances” (MDs) for all accessions to investigate the natural variation of metabolic responses to the cold growth condition (Eq. 1; Fig. 2B). For a given accession, its MD value corresponds to the Euclidean distance of the normalized metabolite profiles measured at the 2 different growth temperatures. Hence, MDs represent a measure of how strongly the metabolism differed between the plants grown at 6 °C and 16 °C, respectively (for details, please refer to the Materials and Methods section). The computed MDs effectively capture the integral information about all quantified metabolites and, therefore, allowed to glean insights into the amplitude of changes of a large part of primary metabolism between different growth temperature conditions (Fig. 2B). MDs showed a large variation within the set of natural accessions (spread of 71% relative to its mean, Fig. 2C), indicative of a genotype-specific magnitude (plasticity) of response to changing temperatures. To assess the extent to which the metabolic phenotypes of the 241 accessions and the detected metabolic plasticity as measured by MDs is determined genetically, we performed genomic predictions by applying the ridge regression best linear unbiased predication (rrBLUP) methodology (Endelman 2011) that revealed a strong predictability of metabolite profiles (Fig. 3). For within-condition prediction, the predictability, scored by the Pearson correlation coefficient, r, of observed versus predicted values was similar for both temperatures (median of r = 0.28 for 16 °C and r = 0.33 for 6 °C, respectively). As expected, cross-condition prediction accuracy was lower (median of r = 0.13 for 16 °C → 6 °C and r = 0.14 for 6 °C → 16 °C, respectively), albeit significantly above zero (Fig. 3).
Figure 3.
Predictability of metabolite levels, MDs, and RGRs as measured by the Pearson correlation coefficient, r, of predicted versus observed values in a cross-validation setting and performing rrBLUP predictions. GP, genomic prediction, i.e. predictor variables = SNP-matrix (16,544 unique SNPs); MP, metabolic prediction, i.e. 37 input metabolite levels. With regard to metabolite (met) levels, all 37 measured metabolites were considered target variables predicted by rrBLUP using a regression model derived from a training set of 180 randomly chosen accessions and applied to the remaining 61 accessions; likewise, MD, and growth rate (at T = 6 °C and 16 °C) were taken as target variables as well. “Same condition” refers to the situation of training a regression model at a given temperature and applying it to the test set samples of the same conditions, whereas “across conditions” tests for the predictability of metabolite levels of the respective other temperatures regime. Distributions were obtained from 25 repeat runs of random 180-training/61-test set accessions. Note that for metabolites, this was done separately for each metabolite and condensed into one distribution for all 37 metabolites per test condition (four “GP- met levels” distributions). Violin plots display the characteristics of the respective distributions: median (open circle), 25 to 75 percentile range (black rectangle) and range excluding outliers (vertical line).
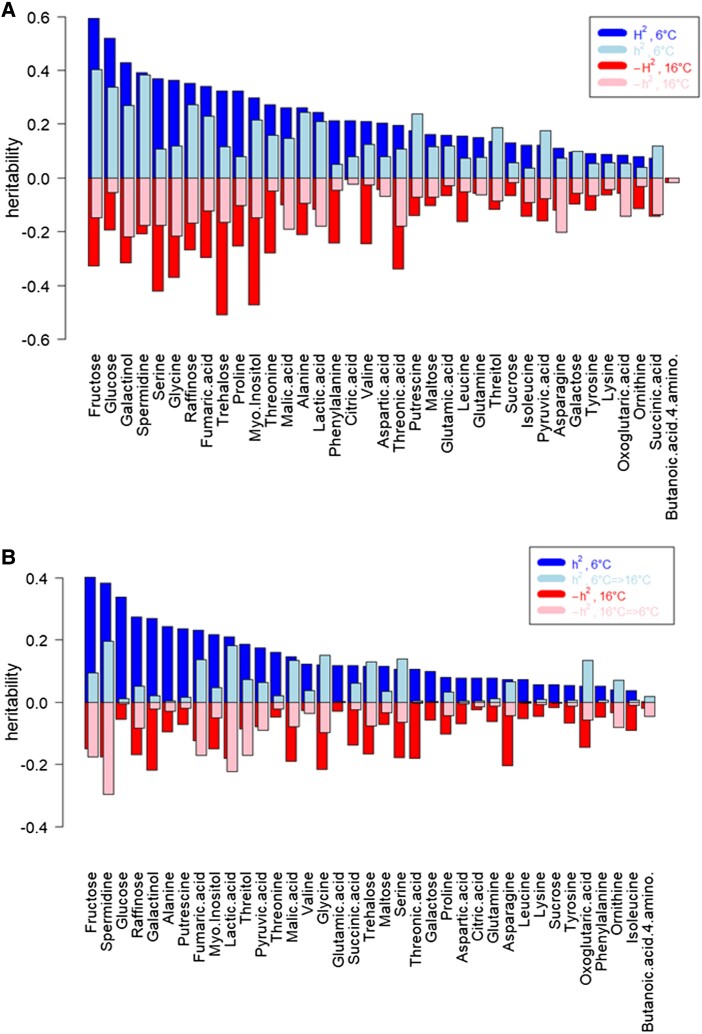
To further test to what extent genetic variance was explaining the observed variation in metabolite levels, broad-sense heritabilities, H2, were calculated for each metabolite at both temperatures. We also computed estimates of narrow-sense heritabilities, h2, taken as the coefficient of determination of metabolite levels in an rrBLUP cross-validation setting (r2, see Materials and Methods) (Fig. 4A). In self-test settings, these r2 values were observed to be highly correlated with h2 values obtained using the function “marker_h2” (R package “heritability”, Supplemental Fig. S1). The different heritability scales H2 and h2 were observed to be highly correlated (Pearson correlation coefficient r = 0.77 [P = 2.1E−8] for T = 6 °C, and r = 0.55 [P = 4.01E−4] for T = 16 °C, respectively), albeit with noticeable differences. At T = 6 °C, greatest heritabilities were found for fructose (with respect to H2 and h2), while at T = 16 °C, trehalose we found with greatest H2 heritability, and galactinol with greatest h2 heritabililty. Further, following the approach to estimate h2 values using rrBLUP coefficients of determination, we inspected the heritability of cross-temperature metabolite levels. Predictabilities/heritabilities were lower in the cross-conditions (trans) setting than in the same-condition (cis) setting (Fig. 4B) and were found only moderately similar (cis/trans Pearson correlation coefficients of h2 values, T = 6 °C: r = 0.34 [P = 0.04], and for T = 16 °C: r = 0.45 [P = 0.004]).
Figure 4.
Temperature effects on heritabilities of metabolite levels. A) Broad-sense heritability, H2, as estimated for the two different temperatures and narrow-sense heritability, h2, taken as the coefficient of determination in cross-validation (average over 25 repeat runs, see Materials and Methods for details on H2 and h2 estimates). H2 values for T = 6 °C and T = 16 °C are correlated at a level of Pearson correlation coefficients r = 0.65 (P = 1.3E−5), and for h2: r = 0.39 (P = 0.017). The different heritability scales H2 and h2 are correlated at r = 0.77 (P = 2.1E−8) for T = 6 °C, and r = 0.55 (P = 4.01E−4) for T = 16 °C, respectively. B)h2 estimates based in cis (same-temperature) and trans (across different temperatures) settings, i.e. cross-validated rrBLUP estimates of coefficients of determinations, taken as h2, trained under a chosen temperature and applied either in cis (same temperatures) or trans (respective alternative temperature). Cis/trans correlations of values, T = 6 °C: r = 0.34 (P = 0.04), and for T = 16 °C: r = 0.45 (P = 0.005). Note that the absolute values of H2 and h2 should not be directly compared, as they are computed using very different procedures. Negative h2-value estimates were replaced by 0 before averaging. For graphical compactness, values have been multiplied by −1 for T = 16 °C. In both plots, metabolites were sorted according to their T = 6 °C values.
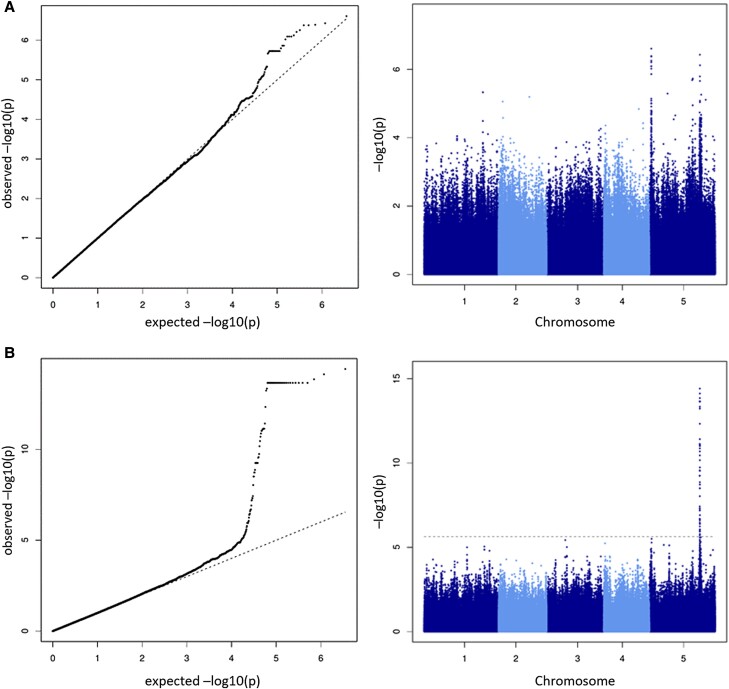
Our analyses show that there is a genetic basis for both the variation of certain metabolites at specific temperatures and for the response to changing temperatures, with noticeable interaction effects between genotypes (accessions) and environments (temperatures). To further investigate the genetic background of differences in metabolism and to identify candidate genes associated with metabolite levels, a genome-wide association study (GWAS) was conducted using the metabolite levels in both conditions. The most significant association was found for single-nucleotide polymorphisms (SNPs) in the promotor region of FUMARASE2 (FUM2, AT5g50950, q = 1.39E−9) under the 6 °C condition, which highlights the influence of genetic variation in the regulation of fumarate metabolism under the applied growth condition (Fig. 5). This association was not detected as significant at T = 16 °C. Additional GWAS candidate SNPs at q-value levels < 0.05 were found for trehalose (q = 0.004, chromosome 4, position ∼7.3 Mb), lactic acid (0.03, chr1, 4.03 Mb; 0.04, chr3, 9.06 Mb) at T = 6 °C and for spermidine (0.04, chr2, 10.2 Mb) and glycine (0.04, chr1, 29.7 Mb) at T = 16 °C (see Supplemental Data Set 1 for a complete listing and more details, including information on the closest gene found at the respective SNP position and Supplemental Data Set 2 for all qq- and Manhattan plots).
Figure 5.
Metabolic GWAS of fumarate levels. A) Growth temperature 16 °C, and B) growth temperature 6 °C. Left panels: qq-plot, right panels: Manhattan plot, where the dashed horizontal line indicates FDR threshold of FDR = 0.05, the peak (T = 6 °C) on chromosome 5 corresponds to the locus of the FUM2 gene. Please note the difference in y value range between the two graphs. qq- and Manhattan plots of all other metabolites under both temperatures are provided in the Supplemental Data Set 2.
After first having focused on genomic determination of individual metabolite levels, we then inspected the predictability of integrative phenotypic parameters, specifically, MDs and measured relative growth rates (RGRs) of accessions. As shown in Fig. 3, MD was predicted with a slightly better correlation coefficient (median r = 0.46) than the average of individual metabolites in the metabolite profile, indicating that the amplitude of change to environmental perturbation is closely related to genome variation. Applied to RGRs as the considered target variables, rrBLUP predictabilities of median r = 0.42 (6 °C) and median r = 0.47 (16 °C) were obtained and, thus, better than for the metabolite levels and comparable to MDs (Fig. 3). When using metabolite levels as predictor variables, growth rates were found predictable at median levels of r = 0.31 and r = 0.25 for T = 6 °C and 16 °C, respectively (“metabolic prediction” of growth rates) and, thus, at noticeably lower levels than achieved for using genotype (SNP) information as input (Fig. 3), likely explained by the substantially larger number of available predictor variables (∼16K SNPs vs. 37 metabolites). Note, as prediction was done in a cross-validation setting, the effect of difference in the number of predictor variables is not a sign of overfitting. As expected, RGRs were lower at T = 6 °C (0.05 ± 0.004 [mean ± Sd]) than at T = 16 °C (0.17 ± 0.014), with RGR in units of 1/d.
Q1 temperature at the natural origin of Arabidopsis accessions is linked to MDs between cold and warm growth conditions
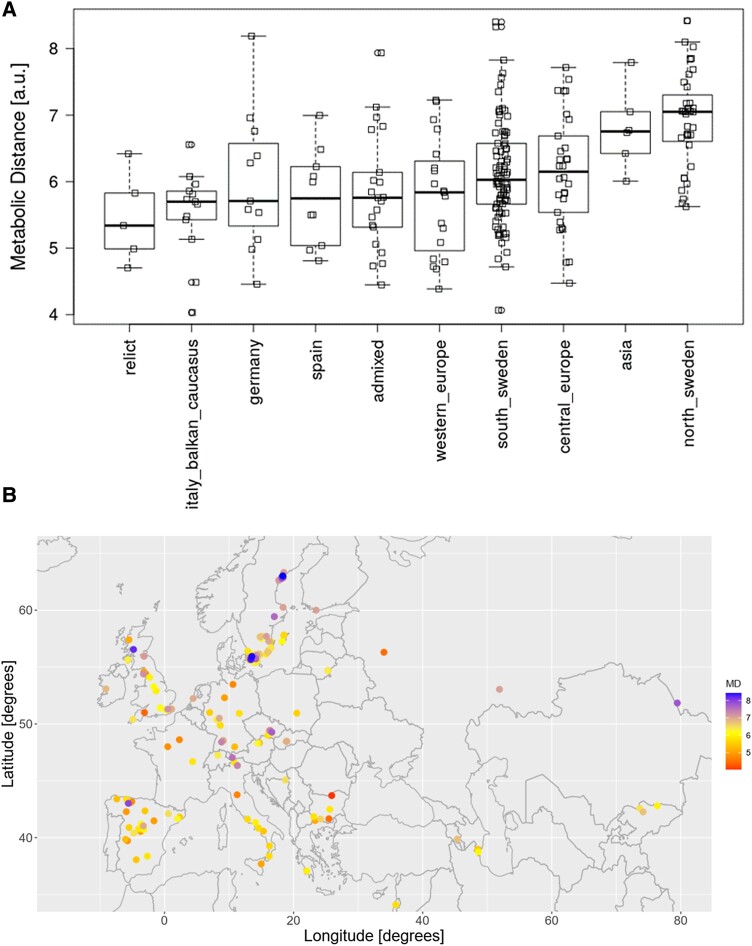
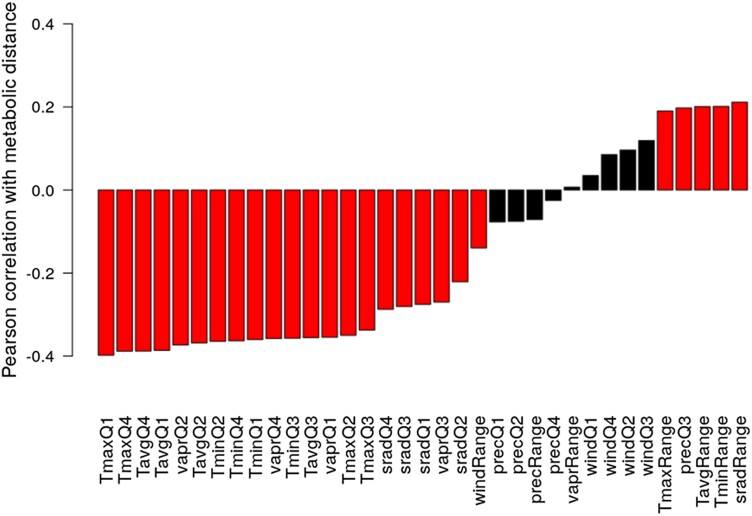
Accessions showed a large diversity in metabolic adjustments to the different growth temperature conditions, reflected by a large range of MDs (Fig. 2C). Every accession was assigned to a genetic admixture group (The 1001 Genomes Consortium 2016) and a one-way ANOVA revealed significant differences in MD between the groups (Fig. 6A, one-way ANOVA P = 1.7E−08). A positive correlation of MD and latitude of origin of accessions was revealed (r = 0.34, P = 4.2E−8, Supplemental Fig. S2), indicating a directed influence of genetic and geographic origin on the metabolic response to cold growth conditions. For Southern accessions, relatively small MDs were observed, while Northern accessions showed relatively large MDs (Figs. 6B and S2). As the MDs roughly correlate with a north–south gradient, a set of available climatic variables were tested for correlations between specific climate parameters of geographic location of origin for the analyzed accessions and their metabolic response. To investigate the relationship of metabolic response to changing growth temperatures and climate of origin, Pearson correlation coefficients between the MDs and environmental variables at the respective geographic location of accessions and averaged by quarter (three-month intervals), as well as their annual ranges, comprising temperature, solar radiation, water vapor pressure, precipitation, and wind speed, were calculated. The highest absolute correlation coefficients with MD were observed for temperature variables characteristic for January to March, i.e. the first quarter of the year (Q1) and October to December (Q4), with local habitat temperatures being negatively correlated with MDs (Fig. 7). In addition to temperature, water vapor pressure was found strongly negatively correlated with MDs as well. However, it has to be borne in mind that some climate parameters are strongly correlated (such as water vapor pressure and temperature, Supplemental Fig. S3) and can therefore not clearly be separated with regard to significance. Wind had a negligible correlation with MD, indicating a low impact of this factor on regulation of metabolic reactions of plants to the applied cold growth conditions. Precipitation during Q3 and, in addition, annual ranges of temperature and radiation were found to be significantly positively correlated with MD (Fig. 7).
Figure 6.
Geographic association of MDs. A) MDs of Arabidopsis natural accessions, grouped by genetic admixture group. Boxplots with median (horizontal lines), 25 to 75 percentile range (rectangles), and range excluding outliers (vertical dashed lines). Squares correspond to the individual accessions. B) Geographic location of accessions used in this study and their corresponding color-coded MDs (i.e. Euclidean distances of between metabolite profiles [37 metabolites] measured at two different temperatures [6 °C/16 °C]).
Figure 7.
Correlation of MDs of accessions with climate variables (averaged by quarter, Q) at their respective geographic location of origin. Red bars indicate significance (PFDR < 0.05); black otherwise. “Range” refers to the annual range of the respective variable (max–min of quarterly values). T, temperature (maximal, average, and minimal), vapr, water vapor pressure; srad, solar radiation; prec, precipitation; wind, wind.
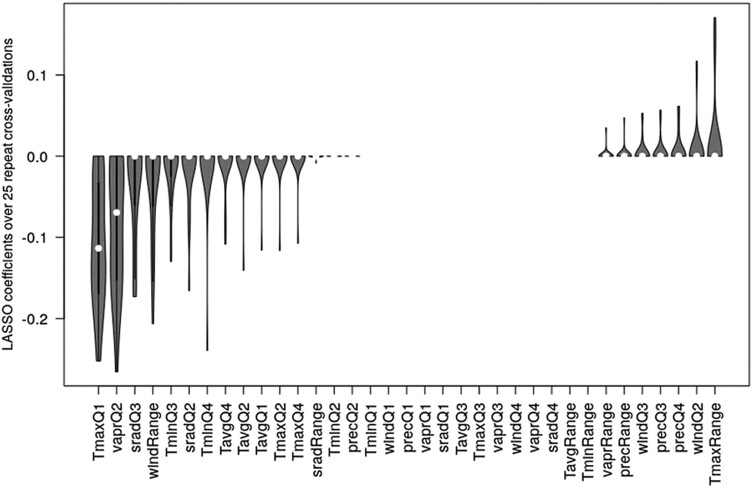
To identify the most predictive (regarding MD) climate variables and combinations thereof, we performed Lasso regressions in a cross-validation setting (25 repeats). Figure 8 shows the distribution of Lasso coefficients (as climate variables were scaled, they can be directly compared). Only two parameters were appreciably different from zero, Tmax, Q1, and with a wider spread, water vapor pressure Q2. On average, the Lasso models predicted MDs with Pearson correlation coefficient between actual and predicted MD in cross-validation of r = 0.3 (Sd = 0.04).
Figure 8.
Lasso regression coefficients with MD as the target variable and scaled climate parameters as independent predictor variables. Violin plots over 25 cross-validation runs (180 training, 59 testing, randomly selected in every run). Note, for two accessions, no climate parameters were available. The regularization parameter lambda was determined in each of the repeats in a separate cross-validation using the respective training accessions only and taken as the value associated with the minimum error. “Range” refers to the annual range of the respective variable (max–min of quarterly values). T, temperature (maximal, average, and minimal), vapr, water vapor pressure; srad, solar radiation; prec, precipitation; wind, wind; Q, quarter. Violin plots display the characteristics of the respective distributions: median (open circle), 25 to 75 percentile range (black rectangle), and range excluding outliers (vertical line).
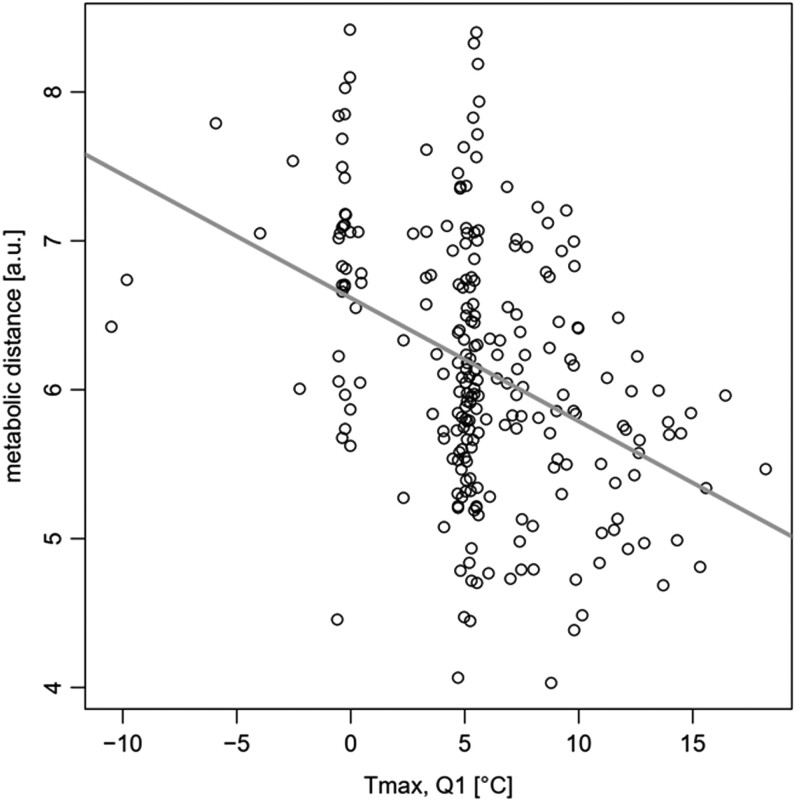
With the maximal temperature during Q1 at the geographic location of origin determined as the climate parameter most strongly correlated with MDs (i.e. metabolic plasticity of accessions in response to changing temperatures), Fig. 9 displays the scatter plot of both variables for all accessions. Despite substantial scatter, the significant negative correlation between both variables is evident (r = −0.4, P = 1.69E−10).
Figure 9.
Relationship of maximum Q1 temperature (January to March, Tmax, and Q1) and MD (n = 239). Note, for two accessions, no climate parameters were available. Pearson correlation coefficient r = −0.4 (P = 1.69E−10). The gray line corresponds to the best linear fit to the data.
In summary, accessions from locations with low temperatures, and in particular during Q1, exhibit larger metabolic plasticities in response to changing temperatures than those originating from warmer climates.
Accessions from cold and warm climates have specific metabolite profiles under cold growth conditions
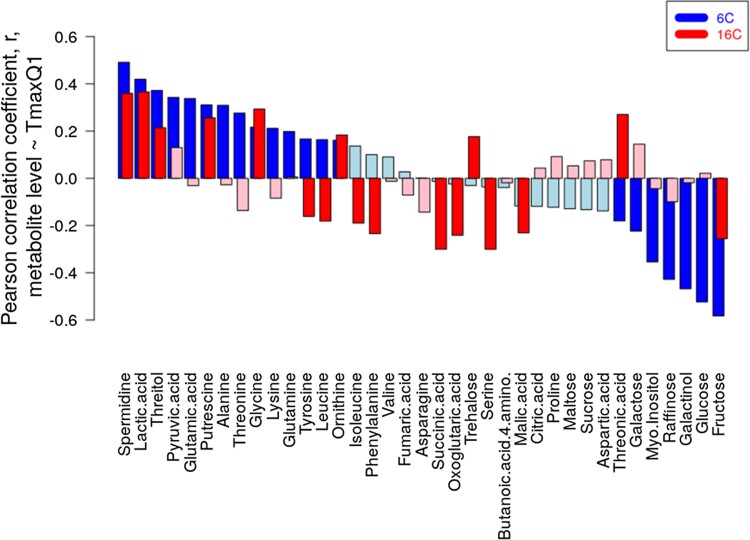
We observed that the magnitude of metabolic response to changing growth temperatures, as measured by MD, of accessions correlated with the maximal temperature during the first quarter of the year (TmaxQ1) at the geographic site of origin (Fig. 9). We now examined the association of all 37 investigated metabolites across all accessions to their respective TmaxQ1 and separately for both growth temperatures (Fig. 10). Significant associations (positive and negative correlations of metabolite level to TmaxQ1) were detected under both growth temperatures. However, only a moderate concordance between the two temperatures regimes was observed. The correlation of the two association profiles was determined at r = 0.3 (P = 0.057), where association profile is defined as the vector of 37 correlation coefficients metabolite level ∼TmaxQ1 at either T = 6 °C or T = 16 °C. At T = 6 °C (in descending order of absolute correlation levels and reporting significant correlations [PFDR < 0.05] only), spermidine, lactic acid, threitol, pyruvic acid, glutamic acid, putrescine, alanine, threonine, glycine, lysine, glutamate, tyrosine, leucine, and ornithine were found positively correlated with TmaxQ1, while fructose, glucose, galactinol, raffinose, myo-inositol, galactose, and threonic acid were observed to be negatively correlated with TmaxQ1. By comparison, at T = 16 °C (asterisks indicate sign-flip compared to T = 6 °C), lactic acid, spermidine, glycine, threonic acid*, putrescine, threitol, ornithine, and trehalose* were positively correlated, and serine, succinic acid, fructose, glutaric acid, phenylalanine*, malic acid, isoleucine*, leucine*, and tyrosine* were observed to be negatively correlated with TmaxQ1.
Figure 10.
Correlation of metabolite levels (growth condition T = 6 °C [blue] T = 16 °C [red]) with maximum temperature in the first quarter of the year, TmaxQ1, at the geographic site of origin of accessions. Bar color indicates significance, with red/blue: PFDR < 0.05, pale red/blue otherwise. Both profiles are correlated at Pearson r = 0.31, P = 0.057.
Biochemical Jacobian reveals checkpoints of cold stress adaptation
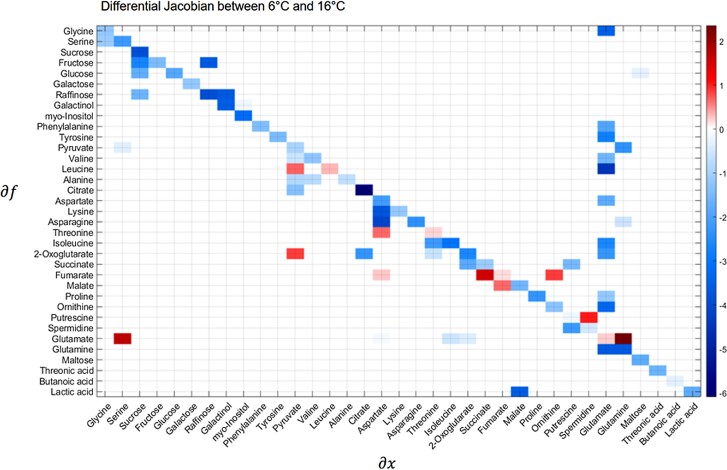
To investigate the regulation of metabolism, Jacobian matrices were calculated based on a method that connects metabolite variance information with a metabolic network (Nägele et al. 2014; Weckwerth 2019; Wilson et al. 2020). Metabolite data of the two growth conditions, i.e. 6 °C versus 16 °C were used for calculating a differential biochemical Jacobian. Individual entries of the differential Jacobian indicate the sensitivities of an enzymatic reaction rate to the change of a specific metabolite in the underlying biochemical network (Weckwerth 2019). Accordingly, the changes or ratios of the differential Jacobian entries point to perturbations in the biochemical network regulation between 6 °C and 16 °C growth conditions. Increasing ratios between 6 °C and 16 °C suggested a stronger impact, e.g. due to limiting concentration, of a metabolite (denominator) on a metabolic function (nominator) under 6 °C. Consequently, decreasing ratios indicated less regulatory impact of a metabolite on metabolism at 6 °C than at 16 °C. Most pronounced dynamics in Jacobian entries were observed for citrate, fumarate, glutamate, raffinose, and leucine metabolism (Fig. 11). Jacobian matrix entries related to fumarate metabolism had a strong influence on the separation of the two growth conditions, pointing to a differential regulation of fumarate metabolism under low temperature, which is highlighted further by the strong correlation of SNPs in the promotor region of FUM2 with fumarate levels in the 6 °C condition and not in 16 °C condition (see Fig. 5).
Figure 11.
Calculation of the differential biochemical Jacobian according to Nägele et al. (2014). Shown is a heatmap of Jacobian matrix entries and their respective change from 6 °C growth conditions to 16 °C growth conditions. Color bar: ratios of Jacobian entries 6 °C/16 °C. Ordinate: affected metabolic functions. Abscissa: affecting metabolites. Strong perturbations in biochemical reaction rates in response to changes in metabolite concentrations are marked by dark red and dark blue color values in the heatmap. Fumarate shows substantial changes with respect to aspartate, succinate, and ornithine. Malate shows substantial changes with respect to fumarate. This is in accordance with GWAS analysis (see Fig. 5). In GWAS, we found a SNP marker close to gene FUM2, whose gene product fumarase catalyzes the interconversion between fumarate and malate, to be strongly associated to fumarate levels revealing the overall high variance of fumarate metabolism as a cold response effect in different Arabidopsis ecotypes.
Although the Jacobian matrix differed with regard to substrate/product relation of fumarate biosynthesis (df(fumarate)/d(succinate)), which is catalyzed by succinate dehydrogenase and not fumarase, this finding suggests a substantial impact of temperature on fumarate metabolism. Simultaneously, plasticity of citrate consumption, i.e. df(citrate)/d(citrate), was decreased at 6 °C indicting a shift of metabolic regulation from the early to the late reactions of the TCA cycle. A cold-induced increase of Jacobian entries related to glutamate/glutamine metabolism was observed, suggesting differential regulation of nitrogen assimilation rates at 6 °C. Maybe as a consequence, also leucine metabolism, and particularly its pyruvate-dependent biosynthesis (df(Leucine)/d(Pyruvate)), becomes more limiting under 6 °C than under 16 °C (Fig. 11). Finally, metabolic functions related to raffinose metabolism including sucrose, galactinol, and myo-inositol dependencies were found to be perturbed under 6 °C.
EWAS reveal potential linkage of methylation levels and metabolite pools
In addition to assessing the genetic influence on accession-specific metabolite profiles and plasticities, we tested for the role of epigenetic factors in determining metabolite levels. Specifically, we tested for an association of regions showing differential methylation, so-called differentially methylated regions (DMRs) and metabolite levels at different temperatures. We performed EWAS for each combination of DMR context, temperature, and metabolite level. Based on available public domain methylation data, we were able to test 138 of the 241 accessions used in this study for a significant association of DMRs and metabolite level (see Materials and Methods). When using kinship matrices based on both DMRs and SNPs, the respective quantile–quantile (QQ) plots in all cases followed the expected generally uniform distribution of P values indicating a correct accounting for population structure. While we analyzed DMRs in all contexts, we considered DMRs in the CpG context most reliable, since those methylations are stably inherited, while CHH/CHG can change substantially during abiotic stress, leading to potential difference between the methylome of the 1001 Epigenomes Project and this experiment (Hofmeister et al. 2017; Liu and He 2020). Significant DMR-metabolite associations in all contexts were obtained (Tables 1 and 2; see Supplemental Data Set 3 for qq- and Manhattan plots for all metabolites and methylation contexts).
Table 1.
Significant associations of DMRs and metabolite levels, sorted by DMR-type (context)at T = 6 °C
| Chromo some | Position | Metabolite | DMR | Genes in <2 kb vicinity | Q values (FDR) | |
|---|---|---|---|---|---|---|
| SNPkin | DMRkin | |||||
| 1 | 19402179 | Leucine | C_DMR | AT1G52130 (mannose-binding lectin superfamily protein) | 0.03 | 0.03 |
| 5 | 7870808 | Spermidine | C_DMR | AT5G23390 (polygalacturonase inhibitor), AT5G23380 (hypthetical protein) | 0.035 | 0.035 |
| 5 | 19474491 | Glycine | C_DMR | AT5G48060 (C2 calcium/lipid-binding plant phosphoribosyltransferase family protein), AT5G48050 (TE) | 0.24 | 0.049 |
| 3 | 11657590 | Glutamic acid | C_DMR | AT3G29776 (TE), AT3G29777 (TE) | 0.04 | 0.74 |
| 5 | 16142038 | Butanoic acid | CG_DMR | AT5G40348 (natural antisense transcript) | 0.028 | 0.017 |
| 1 | 11776549 | Fructose | CG_DMR | AT1G32570 (hypothetical protein) | 0.033 | 0.027 |
| 1 | 19402179 | Leucine | CG_DMR | AT1G52130 (mannose-binding lectin superfamily protein) | 0.025 | 0.025 |
| 3 | 2422836 | Leucine | CG_DMR | AT3G07580 (hypothetical protein), AT3G07590 (SMD1A) | 0.025 | 0.025 |
| 3 | 19138953 | Lysine | CG_DMR | AT3G51610 (NPU), AT3G51600 (LTP5) | 0.005 | 0.005 |
| 4 | 9144364 | Myo Inositol | CG_DMR | AT4G16146(cAMP-regulated phosphoprotein 19-related protein) | 0.037 | 0.031 |
| 5 | 18201876 | Succininc acid | CG_DMR | AT5G45085 (TE), AT5G45090 (phloem protein, ATPP2-A7) | 0.018 | 0.017 |
“SNPkin” and “DMRkin” q values refer to significance levels achieved using either genomic SNP-based or DMR-based kinship matrices. FDR, false discovery rate (q values, applied per metabolite and condition [T and DMR context]); TE, transposable element gene.
Table 2.
Significant associations of DMRs and metabolite levels, sorted by DMR-type (context) at T = 16 °C
| Chromo some | Position | Metabolite | DMR | Genes in <2 kb vicinity | Q values (FDR) | |
|---|---|---|---|---|---|---|
| SNkin | DMRkin | |||||
| 4 | 5038897 | Alanine | C_DMR | AT4G08108 (TE) | 0.033 | 0.032 |
| 4 | 8189392 | Glutamine | C_DMR | AT4G14200 (pentatricopeptide repeat superfamily protein), AT4G14210 (phytoene dehydrogenase) | 0.004 | 0.003 |
| 4 | 2170845 | Malic acid | CG_DMR | AT4G04405 (TE) | 0.016 | 0.021 |
| 5 | 12392130 | Malic acid | CG_DMR | AT5G32690 (pseudogene), AT5G32975 (TE) | 0.016 | 0.021 |
| 4 | 7830616 | Threonine | CH_DMR | AT4G13470 (TE) | 0.024 | 0.018 |
“SNPkin” and “DMRkin” q values refer to significance levels achieved using either genomic SNP-based or DMR-based kinship matrices. FDR, false discovery rate (q values, applied per metabolite and condition [T and DMR context]); TE, transposable element gene.
At T = 6 °C, more candidate DMR-metabolite associations were found than at T = 16 °C (11 (metabolites: butanoic acid, fructose, glutamic acid, glycine, leucine, lysine, myo-inositol, spermidine, and succinic acid) vs. five (metabolites: alanine, glutamine, threonine, and malic acid), Tables 1 and 2). While there are several significant associations for the CG/C context, only one significant candidate association at for the CHH context (T = 16 °C) was identified. As expected, many DMRs appear to be associated with transposable element (TE) genes. However, a few DMRs were found positioned close to protein coding genes of seemingly relevant functions. A C-DMR was found significantly related to spermidine, which has been connected to growth rate in cold conditions (Meyer et al. 2007). The gene close to this DMR is a polygalacturonase inhibitor (AT5G23390). Polygalacturonase is involved in determining the softness of the cell wall and has been connected to cold acclimation (Takahashi et al. 2019). Spermidine and other polyamines are known to accumulate at the cell wall during biotic stress (Hura et al. 2015). Lysine was found associated with a CG-DMR close to the gene LIPID TRANSFER PROTEIN 5 (LTP5). LTP proteins have been associated with diverse roles, including stress response (Chae et al. 2010).
In general, plant defense response appears to be a common function of genes near candidate DMR regions, such as AT1G52130 (MANNOSE-BINDING LECTIN SUPERFAMILY PROTEIN), AT4G16146 (cAMP-REGULATED PHOSPHOPROTEIN 19-related protein), and AT5G45090 (PHLOEM PROTEIN 2-A7), all three of which are annotated with being involved in “defense response.”
When comparing results between the different kinship matrices used in the computations (DMR-call-based or SNP-based), it is evident that they are highly similar (Tables 1 and 2). In cases where only analysis with one kinship matrix has a significant hit, the respective DMR in the other analysis was among the hits with the highest significance, albeit below the significance threshold.
Inspecting EWAS candidate regions for overlaps with GWAS-SNPs (q < 0.05) and considering as overlaps segments of length 10 kb, no overlaps were found.
RGR is connected to different metabolite pools depending on temperature
To investigate correlations between metabolite levels and overall RGRs in both growth conditions, stepwise forward linear regression modeling (under Akaike information criterion [AIC] termination criterion) was used to find the strongest connection between metabolism and growth. For the 6 °C growth condition, the resulting model contained 11 metabolites (in decreasing order of their absolute coefficients (using scaled metabolite level data) and with added signs to signify positive or negative correlation, respectively): +spermidine, +alanine, +trehalose, −serine, −galactinol, −sucrose, +phenylalanine, −lysine, −proline, −galactose, and −threitol (Pearson correlation of predicted vs. observed RGRs: r = 0.63, P = 1.03E−27). Under the 16 °C condition, the AIC-terminated stepwise forward linear model contained 15 metabolites: −serine, −asparagine, +trehalose, +spermidine, +glutamine, −myo-inositol, +galactose, −lysine, +glycine, −lactate, −threitol, −pyruvate, +proline, +threonine, and −fructose (r = 0.74, P = 1.4E−43). Comparing the two models, 7 metabolites (spermidine, trehalose, threitol, serine, lysine, proline, and galactose) correlated with the RGR in both conditions, indicating a general connection of growth with these metabolites—albeit proline and galactose with opposite signs, while galactinol, alanine, sucrose, and phenylalanine occurred only in the model for 6 °C, and glycine, asparagine, myo-inositol, pyruvate, glutamine, threonine, lactate, and fructose were only contained in the model for growth in the 16 °C condition. When performed in a cross-validation setting (25 repeats of random 180 training vs. 61 test set splits of accessions), RGRs were predictable under both temperature regimes but with higher accuracy at T = 16 °C compared to T = 6 °C (Pearson correlation coefficient between predicted and actual values of r = 0.46 ± 0.07 [Sd] [mean p-value = 0.00138] at T = 6 °C and r = 0.62 ± 0.06 [Sd] [mean p-value = 2.649E-6] at T = 16°C).
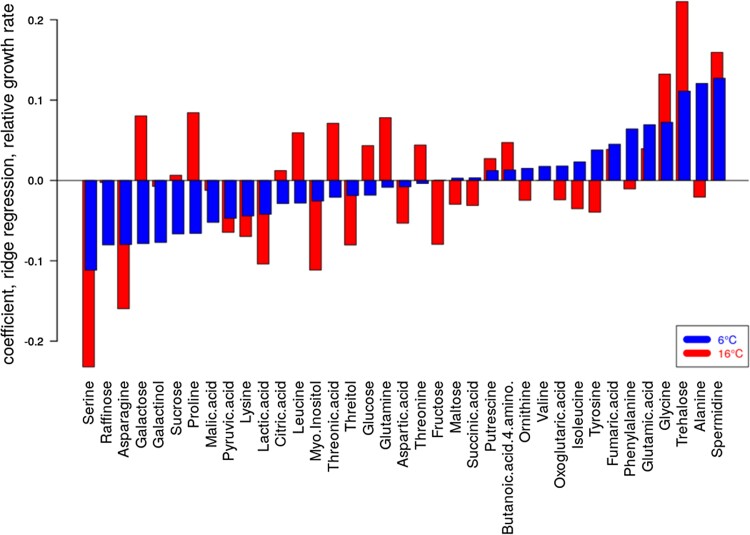
In addition to stepwise forward modeling, we performed ridge regression predictions of RGRs, which handle correlated variables better (assignment of significance). Figure 12 shows the obtained coefficients for both temperatures. As described above for the stepwise forward regression modeling, RGRs were associated with related metabolite profiles (correlation of coefficients for the two temperatures r = 0.52 [P = 0.0009]), albeit pronounced differences were also apparent; e.g. galactose and proline had opposite effects with being positively associated with RGR under 16 °C, while negatively correlated at T = 6 °C.
Figure 12.
Ridge regression of RGR using scaled metabolite level data as predictor variables for the 241 accessions and at the two different growth temperatures examined in this study. Coefficients of the 37 metabolites, signifying their importance, at the T = 16 °C and T = 6 °C were correlated at r = 0.52 (P = 0.0012). Metabolites are sorted in ascending order of coefficients for T = 6 °C. The regularization parameter lambda was determined in cross-validation and taken as the value associated with the observed minimum error in cross-validation.
Discussion
Natural habitat temperature in the first quarter of the year predicts the response of primary metabolism in cold grown plants
It has been described earlier that freezing tolerance of cold acclimated plants significantly correlates with latitude of geographical origin of natural Arabidopsis accessions (Zhen and Ungerer 2008; Zuther et al. 2012). In the present study, we have shown that the extent of primary metabolism response to growth at 6 °C separates Northern from Southern accessions and that it is strongly connected to local habitat temperatures, in particular temperatures between January to March (Q1) (see Figs. 6 and 7). While the general direction of separation of metabolic profiles between the growth temperature conditions was similar for all included accessions (see Fig. 2B), the magnitude of the metabolic response to the 6 °C relative to the 16 °C condition was stronger in plants originating from colder climates (Fig. 9). This finding suggests that accessions from colder regions can respond more flexibly in their metabolism to low temperature than accessions originating from warmer regions. The reason for this observation remains speculative here, but it may result from a differential degree of substrate saturation of metabolic enzymes due to a cold-induced increase of protein amounts. For Col-0, it has been shown earlier that cold acclimation results in significantly increased protein amounts of enzymes catalyzing limiting steps of carbohydrate metabolism (Strand et al. 2003). Comparing two natural Arabidopsis accessions with distinct cold acclimation capacities further revealed higher activity of sucrose phosphate synthase enzymes in a more freezing tolerant accession to prevent limitation of sucrose biosynthesis and photosynthesis during cold exposure (Nägele et al. 2012).
Climatic range boundaries of A. thaliana distribution were shown to be determined by a combination of temperature and precipitation (Hoffmann 2002), which can explain the connection of temperature in Q1, and metabolic reaction to cold. In the present study, however, no significant correlation of metabolic distance and simultaneously temperature and precipitation in the same quarter of the year was detected at the original habitat (Fig. 7). This finding suggests that even though both temperature and precipitation limit the distribution range, the metabolic response to these factors is generally uncoupled from each other. Additionally, the correlation of metabolic distance with original habitat temperature was less significant during the warmer part of the year, i.e. Q2/3 (Figs. 7 and 8). This allows us to speculate that success of germination, flowering, and seed production is determined within the first quarter of the year although no clear association exists between flowering habit and geographic distribution (Pigliucci 1998).
Soluble carbohydrates belonging to the raffinose family oligosaccharide (RFO) biosynthesis pathway, i.e. sucrose, galactinol, myo-inositol, and raffinose, significantly accumulated at low temperature but were negatively correlated to RGR (Figs. 1A and 12). This indicates a trade-off between metabolic acclimation and biomass accumulation. Previous studies have demonstrated that soluble sugars positively correlate with the capability of natural accessions to acclimate to low temperature and to increase freezing tolerance; see e.g. Hannah et al. (2006). However, plant development and growth under low temperature result in a different metabolic constitution than observed for plants that were shifted from ambient to low temperature in mature stage for cold acclimation (Strand et al. 1997). In particular, Arabidopsis leaves developing at 5 °C accumulated relatively high amounts of soluble sugars but, in contrast to cold shifted plants, released suppression of photosynthetic genes, which the authors discussed to be essential for development of full freezing tolerance (Strand et al. 1997). Although CO2 assimilation has not been measured in the present study, evaluating the correlation of metabolites with growth rates at 16 °C and 6 °C revealed a significantly positive correlation with spermidine and trehalose while phenylalanine only correlated positively under 6 °C (Fig. 12). Interestingly, spermidine and trehalose have previously been found to correlate positively with growth under 20 °C, while phenylalanine correlated negatively under these conditions (Meyer et al. 2007). Following the discussion of Meyer et al., who interpreted growth-correlated metabolites as positive (or negative) signals, this would indicate that also under low-temperature spermidine and trehalose represent growth-related metabolite pools. Consequently, due to its negative correlation under ambient temperature (Meyer et al. 2007) and an absent correlation with growth rate at 16 °C, phenylalanine would represent a metabolite which correlates with cold-specific growth dynamics. Phenylalanine represents a central metabolic precursor for numerous secondary metabolites, e.g. flavonoids and lignin (Vanholme et al. 2010; Schulz et al. 2016). Hence, phenylalanine might even play a more complex role than currently assumed and might serve as a central metabolic integrator for growth, development, and stress protection under low temperature.
The extent of the metabolic response to changing temperatures, and thus plasticity, was predictable from genetic variation among the investigated genotypes (Fig. 3). Interestingly, the predictability increased from the 16 °C to the 6 °C growth condition and showed the best predictability for metabolic distance and growth rate (Fig. 3). Metabolic distance comprises the sum of all metabolite perturbations from reference growth (16 °C) to stressed condition (6 °C). Accordingly, it is the most condensed, comprehensive, and synergetic parameter for the description of the cold stress response in relation to the reference metabolome and, thus, corresponds even more to genetic variation than individual metabolite concentrations. This indicates the necessity to resolve and quantify temperature-induced dynamics of metabolism, e.g. by calculating metabolic distances, to reveal a genotype × phenotype relationship.
In contrast to the relatively high predictability of metabolite levels using genomic SNP information and using a ridge regression approach that (potentially) employs all markers (Fig. 3) and the detected relatively high heritabilities of metabolites (Fig. 4), trying to associate polymorphisms (SNPs or DMRs) to metabolite levels via GWAS did not, with the exception of a strong association for fumarate at 6 °C (Fig. 5) and several candidate regions of borderline statistical significance, yield strong associations of genomic alterations and changed metabolites levels. This discrepancy may have two explanations. (i) Metabolite levels may be determined in a polygenic fashion, such that a combined influence of many regions/genes may exert their effects, which are discernable via genomic prediction but not GWAS. (ii) Testing ∼1.7 million SNPs in GWAS and the associated multiple testing correction may wipe out many true associations. It is a principal “dilemma” of GWAS that aiming for dense marker coverage to increase resolution inevitably lessens the chance to detect true associations as studies become underpowered. Here, recently introduced approaches to focus on genes, rather than SNPs via transcriptome-wide association studies (TWAS) may be a promising avenue for further pursuits (Gamazon et al. 2015; Lan et al. 2021; Li and Ritchie 2021).
Metabolite covariance connects natural genetic variation with metabolic regulation
As a consequence of differential metabolite covariances, the calculated Jacobian matrices revealed strong differences between the two growth conditions (Fig. 11). In general, Jacobian matrices allow the investigation of causal relations between metabolites and point to differences in metabolic regulation (Nägele et al. 2014; Weckwerth 2019; Wilson et al. 2020). Most pronounced differences in Jacobian entries were found in fumarate, glutamate, leucine, spermidine, sucrose, and raffinose metabolism. Under low temperature, fumarate serves as a carbon sink in leaf metabolism of A. thaliana, aiding in the acclimatization of photosynthesis to a new homeostasis (Dyson et al. 2016). The differential accumulation of fumarate under stress in A. thaliana accessions with different cold acclimation potential could already be shown in an earlier study (Weiszmann et al. 2018) and FUM2 was described to have a strong effect on carbon partition and growth rates in A. thaliana accessions (Dyson et al. 2016; Riewe et al. 2016; Saunders et al. 2022). The mitochondrial fumarase, FUM1, has previously been shown to have a negligible effect on fumarate accumulation across a large temperature range (Saunders et al. 2022), which could be due to an immediate metabolization within the TCA cycle. Hence, fumarate accumulation in Arabidopsis might require cytosolic biosynthesis by FUM2 because it enables direct transport into the vacuole. Finally, organic acids such as fumarate play an important role in regulating the accumulation of solutes within the vacuole by controlling cold induced acidification (Schulze et al. 2012), which makes them a good target for metabolic regulation, as indicated by the differences in Jacobian matrices.
The score for the influence of ornithine on the metabolic function of fumarate additionally points to the plastidial conversion of ornithine to arginine via ornithine carbamoyltransferase (OTC), argininosuccinate synthase (ASSY), and argininosuccinate lyase (ASL). This transformation is an important part of nitrogen cycling and homeostasis (Witte 2011; Planchais et al. 2014). The connection of natural variation along the gradient of Q1 temperature of origin and this pathway is supported by the prevalence of a SNP in the L-aspartase-like family protein (ASL) gene (At5g10920), which is highly correlated with temperature of origin in >1,000 Arabidopsis accessions (Ferrero-serrano and Assmann 2019). This points to differences in the extent of regulation of amino acid metabolism caused by low temperature between accessions originating from colder and accessions originating from warmer climate. It has been shown that amino acid metabolism and nitrogen usage have to be heavily adjusted under stress conditions to allow survival in plants (Guy et al. 2008; Less and Galili 2008; Espinoza et al. 2010).
Sucrose and raffinose metabolism play a central role in plant development, stress response, and growth regulation. Its cyclic metabolism, composed of invertase-driven cleavage and cytosolic resynthesis, represents an important buffer mechanism against environmental fluctuation (Geigenberger and Stitt 1991; Weiszmann et al. 2018; Wienkoop 2008). Hence, the observed differences in reaction elasticities connected to sucrose metabolism point to differences in regulation in this pathway between plants from warmer and colder habitats. Futile cycling of sucrose might stabilize the cellular energy status by providing electron acceptor molecules for photosynthesis and serve as an efficient mechanism to control carbon partitioning (Ruan 2014).
EWAS reveals potential targets for epigenetic regulation
EWAS for different DMR—metabolite associations revealed potential targets of epigenetic regulation, specifically, for several amino/organic acids, fructose, myo-inositol, and the polyamine spermidine (Tables 1 and 2). However, further investigation is needed. With regard to associating methylation patterns to metabolite levels, we had to resort to published data taken as part of different experiments and conditions than tested here. For a subset of 138 accessions only, comparable methylome data were available. Furthermore, Kawakatsu et al. grew accessions at 22 °C, while metabolite data in this study were measured at 16 °C and 6 °C. As methylation is dynamic and potentially condition specific, the EWAS results have to be interpreted with caution. However, methylation patterns have also been reported to be stable. Previously, it was reported that 99.998% of methylated regions are stably inherited across a generation (Hofmeister et al. 2017). Abiotic stress response-induced methylation patterns have been reported to be stably transferred between generations, acting as a transgenerational memory (Liu and He 2020). Furthermore, a recent study shows that using genome-wide single cytosine methylation levels, Arabidopsis plants clustered according to their accession, irrespective of stress condition, specifically in the CG and CHG contexts (Lin et al. 2022). Therefore, it seems reasonable to assume that two plants from the same accession (ecotype) share a very similar methylation pattern, which in turn influences gene expression/stress response compared to plants from a different ecotype.
Conclusions
Changes in ambient temperatures have different ecological implications in the different growth habitats of Arabidopsis. We could show that within a large group of accessions from diverse growth habitats, there exists a significant connection of the temperature at the geographic site of origin, especially from January to March, and metabolic reaction to cold. This metabolic reaction correlates with genomic and epigenomic information indicating that genotype × environment × phenotype information might be traceable and predictable by plasticity information about metabolism rather than absolute metabolite amounts. Finally, the presented data indicate a trade-off between metabolic acclimation and biomass accumulation, which might result in or be induced by differential accumulation of sugars, organic acids, and amino acids.
Materials and methods
Experimental design and plant growth
As described before (Clauw et al. 2022), seeds of natural accessions (metabolite profiling was successfully conducted on 241 of the initial 249 accessions; Supplemental Data Set 4) of Arabidopsis (A. thaliana) described in the 1001 Genomes Project (The 1001 Genomes Consortium 2016) were sown on sieved (6 mm) substrate (Einheitserde ED63). Pots were filled with 71.5 g ± 1.5 g of soil to assure homogenous packing. The prepared pots were all covered with blue mats (Junker et al. 2015) to enable a robust performance of the high-throughput image analysis algorithm. Stratification was done for 4 d at 4 °C in the dark, upon which the seeds were germinated and seedlings established at 21 °C for 14 d (relative humidity: 55%; light intensity: 160 µmol m−2 s−1; 14-h light). The temperature treatments were started by transferring the seedlings to either 6 °C or 16 °C. To simulate natural conditions temperatures fluctuated diurnally between 16 °C to 21 °C, 0.5 °C to 6 °C, and 8 °C to 16 °C for respectively the 21 °C initial growth conditions and the 6 °C and 16 °C treatments. Relative humidity (55% to 95%) and light intensity (160 µmol m−2 s−1) were kept the same for all experiments. Daylength was 9 h during the 16 °C and 6 °C treatments and 14 h during the 21 °C initial growth conditions. Each temperature treatment was repeated 3 times. Five replicate plants were grown for every genotype per experiment. Plants were randomly distributed across the growth chamber with an independent randomization pattern for each experiment. During the temperature treatments (14 d after sawing [DAS]—35 DAS), plants were photographed twice a day (1 h, after/before lights switched on/off), using an RGB camera (IDS uEye UI-548xRE-C; 5MP) mounted to a robotic arm. At 35 DAS, whole rosettes were harvested, immediately frozen in liquid nitrogen and stored at −80 °C until further analysis. Rosette areas were extracted from the plant images using Lemnatec OS (LemnaTec GmbH, Aachen, Germany) software.
RGRs (units: 1/d) were computed as described before (Clauw et al. 2022) from which all generated phenotyping data are filed under https://doi.org/10.5281/zenodo.6076948.
Metabolite quantification and profiling via GC-MS
Frozen leaf material of 3 to 4 replicate plants for each accession and treatment was homogenized in a ball mill (Retsch GmbH, Haan, Germany). Polar metabolites were extracted and measured on gas chromatography coupled to mass spectrometry (GC-MS) as previously described (Weiszmann et al. 2018), with slight modifications. In brief, homogenized plant material was extracted with a methanol–chloroform–water (MCW) mixture (2.5/1/0.5, v/v/v) on ice for 15 min, which was split in a polar and an apolar fraction by addition of water after extraction. Subsequently, pellets were reextracted twice using 80% (v/v) ethanol at 80 °C for 30 min. Ethanol extracts were combined with the polar phase of the MCW extraction and dried in a vacuum concentrator (ScanVac, LaboGene, Allerød, Denmark). To compensate technical variance in the measurements, 2 internal standards, i.e. pentaerythritol and phenyl β-D-glucopyranoside (both Sigma-Aldrich), were spiked to the extracts before drying. Dry extracts were derivatized by methoximation for 90 min at 30 °C with methoxylamine hydrochloride (Merck KGaA, Darmstadt, Germany) in pyridine and silylation for 30 min at 37 °C with N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA, Macherey–Nagel, Düren, Germany). Measurements were conducted on a LECO Pegasus GCxGC-TOF mass spectrometer (LECO Corporation, St. Joseph, USA) coupled to an Agilent 6890 gas chromatograph (Agilent Technologies, Santa Clara, USA) using an Agilent HP-5Ms column (length: 30 m; inner diameter: 0.25 mm; film: 0.25 mm). For targeted analysis, baseline correction, chromatogram deconvolution, peak finding, retention index calculation, and peak area extraction were done in the software LECO Chromatof. Retention index calculation was conducted by measuring a mixture of linear alkanes (C12 to C40) with every measurement batch. All metabolites included in the targeted analysis were identified and quantified by measuring a mixture of pure standard compounds in different concentrations in every measurement batch. Areas of each metabolite were normalized to the internal standard with a minimum distance of retention time to the metabolite. Internal standard normalized areas were then normalized to the slope of peak areas of the corresponding externally measured standard row and to sample fresh weight yielding the absolute amount of metabolites (µmol gFW−1). The data table containing all metabolite quantifications can be found in the supplement (raw absolute and normalized values, Supplemental Data Set 5). Original files are available at MetaboLights, a database for metabolomics experiments and derived information (https://www.ebi.ac.uk/metabolights/; study identifier MTBLS4655).
Metabolite level data normalization
Metabolite level data were further normalized to render their distribution Gaussian and to reduce batch and repeat effects. Raw absolute metabolite levels of every of the 37 annotated metabolites across all 1,624 samples (241 accessions, 2 temperature conditions, and 3 to 5 repeats) were first rendered “normal” by applying the function “orderNorm” of the package R-package “bestNormalize.” Metabolite profile data of every sample was then median-centered, and every metabolite was standardized (mean = 0, variance = 1). To lessen replicate and batch effects, the function “ComBat” from the R-package “sva” was applied in succession. Afterwards, repeat data were aggregated (mean) across all repeats, yielding for each of the 37 metabolites a single value for all 482 average sample data sets (241 accessions and two temperatures).
Absolute and normalized metabolite level data (per sample and aggregated per accession) are available in Supplemental Data Set 5.
MDs
The magnitude of change of metabolite profiles of every accession was quantified by their metabolic distance (Houshyani et al. 2012). The metabolic distance (MD) of a given accession, a, is defined as the Euclidean distance between its normalized and averaged (over repeats) metabolite profile at T6 = 6 °C vs. T16 = 16 °C (Eq. 1):
| (1) |
where a is accession, m metabolite, n is the number of metabolites (n = 37), and L refers to metabolite level (value after preprocessing and averaging). MD data along with more relevant accession-related information (RGR and climate data) are available in Supplemental Data Set 4. Figure 2B illustrates all MD values (gray lines) projected onto the first two principal components (score plot of samples).
Climate data
Climate data (“5-min” resolution, ∼85 km2) were taken from the WorldClim Database (Fick and Hijmans 2017), which was further used to calculate summary variables as three-month means. The worldClim2 data were linked to each natural Arabidopsis accession based on longitude and latitude of their origin. Two accessions had no valid climate data and were not considered in the respective analyses (accession 7471, RLD-1, had no geographic location information; accession 9416, Kru-3, had no climate information for that location).
Statistical analyses
Statistical analyses were conducted within the free statistical software environment R (R Core Team 2019). Data manipulation, summarization, and plotting were conducted using the R package “tidyverse” (Wickham et al. 2019).
Stepwise forward linear regression was performed using R under AIC-constraint. Ridge and Lasso regression was performed using the R package “glmnet” (Friedman et al. 2010) with penalty parameter λ (lambda) determined in cross-validation runs (no use of the ultimate test data sets).
Genetic heritabilities
Broad-sense heritability, H2, was estimated by applying a random-effects model (R package “lmer”) with accessions (genotypes) considered the random effect and with H2 = Vg/(Vg + Ve), where Vg is the variance associated with genotype and (Vg + Ve) estimates the phenotypic variance as the sum of genotypic and residual error variance. Metabolite levels were taken as their normalized levels (see above) and considering all biological repeats as separate samples (i.e. levels were not averaged per accession).
Narrow-sense heritability, h2, was estimated using the function marker_h2 (R package “heritability”) using metabolite levels and the computed additive relationship matrix (R package “rrBLUP”) as input. The function marker_h2 estimates genetic and residual variances using an AI-REML algorithm (Gilmour et al. 1995) and reports as an estimator of narrow-sense heritability a value introduced as hr2 in Kruijer et al. (2015). Metabolite levels were taken as their preprocessed (see above) level values averaged over repeat samples, i.e. one value per metabolite, accession, and temperature. As expected and shown in this study (Supplemental Fig. S1), h2 correlates with the coefficient of determination (r2 with r being the Pearson correlation coefficient of rrBLUP predicted and actual metabolite levels). We used r2 obtained in a cross-validation setting (180 randomly chosen accessions and predicted metabolite levels in the 61 hold-out test set) as a surrogate measure of h2. This approach has the advantage of reducing overfitting during rrBLUP-based r2 estimates in a self-test (training = test data set). Occasionally, occurring negative r values (correlation between predicted and actual metabolite levels) were set to 0 to indicate no predictability, which, upon inspection, did not introduce any artifacts (Supplemental Fig. S1).
Data-driven inverse modeling
Calculation of Jacobian matrices was conducted as previously described to reveal whether metabolic covariance information reveals conserved metabolic pathway regulation comparing 16 °C and 6 °C growth conditions (Nägele 2014; Nägele et al. 2014). In brief, covariance data, calculated directly from the metabolite concentrations in the 6 °C and the 16 °C condition, were connected with biochemical network information and used for an inverse approximation of biochemical Jacobian matrices. The inverse calculation of Jacobian matrices was conducted 1 × 104 times, and the resulting median was normalized to the inverse variance of all calculations. The calculations were done using scripts of the toolbox COVAIN (Sun and Weckwerth 2012) in the numerical software environment MATLAB (R2019b).
Metabolic GWAS and genomic prediction
SNP information was obtained from the Arabidopsis 1001 Genomes Project information portal (SNPEFF file, version 3.1, fully imputed set). Requiring the minor allele to be present in at least 13 accessions (minor allele frequency [MAF] > 5% of all 241 accessions) yielded 1,756,214 biallelic SNPs and thus a high-resolution set of ∼one SNP per 77 bp used for the GWAS computations. For genomic prediction purposes, where SNP coverage is of lesser concern, we rendered the SNP data set unique (no repeating SNP profiles across the 241 accessions [vectors of length 241 with elements “1” and “−1”]) and selected from the original data set only those that required at most one missing allele to be imputed (high confidence set), yielding 16,544 biallelic SNPs (one SNP per ∼8 kb). Given a genome size of ∼135 Mb and considering an average linkage disequilibrium (LD) distance of 10 kb (Kim et al. 2007; Perlaza-Jiménez and Walther 2018), SNP coverage was deemed adequate for both GWAS and genomic prediction purposes. Of note also, while more predictor variables (SNPs) may be desirable, here, more SNPs would mean to rely more and more on imputed (“guessed”) information (most SNPs have missing allele calls, i.e. no allele call in certain accessions), possibly leading to false conclusions. In addition, we tested the prediction performance using sets with no imputation (∼4K unique SNPs), at most one missing allele (∼16K), and the fully imputed SNP set (∼860K unique SNPs) and observed no relevant differences in metabolite level predictions (Supplemental Fig. S4). Alleles were encoded as −1 and 1 to reflect the two different diploid homozygous genotypes.
Genomic prediction was performed applying the best linear unbiased predication (BLUP) methodology as implemented in the R-package “rrBLUP” (Endelman 2011). Cross-validation (split of accessions into training and test population) was performed on 180 (training)/61 (test) random accession splits. As a metric of predictability, Pearson correlation coefficients of predicted versus observed considered target variable (metabolite levels, MD, and growth rate) were computed.
GWAS analysis was performed using the GWAS function of the R-package “rrBLUP” (Endelman 2011), with a SNP-based additive relationship matrix to correct for population structure as a confounding factor, computed using the A.mat function of rrBLUP. q values to estimate false discovery rates (FDRs) were computed on the returned GWAS scores, transformed back to P values (P = 10−score), using the R-package “qvalue.”
EWAS
Methylation data for 138 accessions were obtained from Kawakatsu et al. (2016), GEO accession number GSE43857. DMR calls were performed using the methylpy pipeline (Schultz et al. 2015). The protocol of Kawakatsu et al. was followed. Separate DMR calls for methylation in a CpG context, termed CG-DMRs, CHH/CHG context, termed CH-DMRs, and both contexts, termed C-DMRs, were performed. This resulted in 67,224, 43,773, and 123,881 unique DMRs, respectively. The R-package “rrBLUP” (Endelman 2011) was utilized for the association study and applied as described above for GWAS. Input was a matrix with columns representing DMR positions and rows representing accessions. If an accession at a DMR position was significantly hypermethylated, it was represented as “1”; if it was significantly hypomethylated, the cell contained “−1,” all other accessions were represented by “0.” DMR and genomic SNP (described above) data were used to estimate the respective additive relationship matrix to correct for population structure as a confounding factor. While testing both (DMR-based and SNP-based kinship), the DMR-based approach was found to be a better control for the rate of false positives than a kinship matrix based on SNP data. A similar approach has been applied earlier (Gugger et al. 2016).
Accession numbers
Sequence data from this article can be obtained from the Arabidopsis 1001 Genomes Project information portal (https://1001genomes.org/).
Supplementary Material
Acknowledgments
We would like to thank Klara Wuketich and Anneliese Auer for the technical support during the phenotyping experiments. Further, we thank Matthias Nagler for assisting with sample preparation, Lena Fragner and Martin Brenner for technical assistance, and Lisa Fürtauer for critical discussion. The Vienna BioCenter Core Facilities GmbH (VBCF) Plant Sciences Facility acknowledges funding from the Austrian Federal Ministry of Education, Science and Research and the City of Vienna.
Contributor Information
Jakob Weiszmann, Molecular Systems Biology (MOSYS), Department of Functional and Evolutionary Ecology, Faculty of Life Sciences, University of Vienna, 1030 Vienna, Austria; Vienna Metabolomics Center (VIME), University of Vienna, 1030 Vienna, Austria.
Dirk Walther, Bioinformatics, Max-Planck-Institute of Molecular Plant Physiology, 14476 Potsdam, Germany.
Pieter Clauw, Austrian Academy of Sciences, Gregor Mendel Institute (GMI), 1030 Vienna, Austria.
Georg Back, Bioinformatics, Max-Planck-Institute of Molecular Plant Physiology, 14476 Potsdam, Germany.
Joanna Gunis, Austrian Academy of Sciences, Gregor Mendel Institute (GMI), 1030 Vienna, Austria.
Ilka Reichardt, Genome Engineering Facility, Max Planck Institute of Molecular Cell Biology and Genetics, 01307 Dresden, Germany.
Stefanie Koemeda, Plant Sciences Facility, Vienna BioCenter Core Facilities GmbH (VBCF), 1030 Vienna, Austria.
Jakub Jez, Plant Sciences Facility, Vienna BioCenter Core Facilities GmbH (VBCF), 1030 Vienna, Austria.
Magnus Nordborg, Austrian Academy of Sciences, Gregor Mendel Institute (GMI), 1030 Vienna, Austria.
Jana Schwarzerova, Molecular Systems Biology (MOSYS), Department of Functional and Evolutionary Ecology, Faculty of Life Sciences, University of Vienna, 1030 Vienna, Austria; Brno University of Technology, Faculty of Electrical Engineering and Communication, Department of Biomedical Engineering, Technická 12, 616 00 Brno, Czech Republic.
Iro Pierides, Molecular Systems Biology (MOSYS), Department of Functional and Evolutionary Ecology, Faculty of Life Sciences, University of Vienna, 1030 Vienna, Austria.
Thomas Nägele, LMU Munich, Faculty of Biology, Plant Evolutionary Cell Biology, 82152 Planegg, Germany.
Wolfram Weckwerth, Molecular Systems Biology (MOSYS), Department of Functional and Evolutionary Ecology, Faculty of Life Sciences, University of Vienna, 1030 Vienna, Austria; Vienna Metabolomics Center (VIME), University of Vienna, 1030 Vienna, Austria.
Author contributions
J.W.: GC-MS analysis, first submission: statistics, programming, and wrote the paper. D.W.: for all parts (except Jacobian analysis): analyses, statistics, programming, and wrote the paper. P.C., J.G., I.R., S.K., J.J., and M.N.: growth analysis and phenotyping. P.C.: GWAS and heritability analysis. G.B. and J.S.: EWAS analysis and genomic prediction. I.P.: Jacobian analysis. T.N. and W.W.: conceived the study, supervision of GC-MS analysis, statistics, and wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Relationship of h2 heritabilty of metabolite levels taken as the molecular phenotype and rrBLUP predictability assessed by the squared Pearson correlation coefficient, r2 (coefficient of determination) of predicted vs. actual metabolite level.
Supplemental Figure S2 . MD of accessions as a function of geographic latitude of their location of origin.
Supplemental Figure S3 . Similarity of quarterly climate parameters.
Supplemental Figure S4 . Predictability of metabolite level in cross-validation as judged by the correlation coefficient between observed and rrBLUP-predicted levels of all 37 metabolites.
Supplemental Data Set 1 . Metabolic phenotype associations.
Supplemental Data Set 2 . Results of GWAS.
Supplemental Data Set 3 . Results of EWAS.
Supplemental Data Set 4 . Natural accessions.
Supplemental Data Set 5 . Metabolite information.
Funding
This work was supported by the University of Vienna (ViME) at the University of Vienna, and by TRR175/D03 and NA 1545/5-1 both funded by the Deutsche Forschungsgemeinschaft (DFG) as well as DOC 111 doc.funds and I 5234 by the Austrian Science Fund (FWF).
Data availability
All data needed to evaluate the presented conclusions of this study are provided in the paper, supplement, or cited repositories.
References
- Adams WW, Stewart JJ, Cohu CM, Muller O, Demmig-Adams B. Habitat temperature and precipitation of Arabidopsis thaliana ecotypes determine the response of foliar vasculature, photosynthesis, and transpiration to growth temperature. Front Plant Sci. 2016:7(1026). 10.3389/fpls.2016.01026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P, Jayavelu ND, Rasmussen S, Nielsen HB, Mundy J, Bones AM. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 2013:14(1):722. 10.1186/1471-2164-14-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011:67(5):869–884. 10.1111/j.1365-313X.2011.04640.x [DOI] [PubMed] [Google Scholar]
- Chae K, Gonong BJ, Kim SC, Kieslich CA, Morikis D, Balasubramanian S, Lord EM. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J Exp Bot. 2010:61(15):4277–4290. 10.1093/jxb/erq228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P, Kerdaffrec E, Gunis J, Reichardt-Gomez I, Nizhynska V, Koemeda S, Jez J, Nordborg M. Locally adaptive temperature response of vegetative growth in Arabidopsis thaliana. eLife. 2022:11:e77913. 10.7554/eLife.77913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel U, Morris WL, Ducreux LJM, Yavuz C, Asim A, Tindas I, Campbell R, Morris JA, Verrall SR, Hedley PE, et al. Physiological, biochemical, and transcriptional responses to single and combined abiotic stress in stress-tolerant and stress-sensitive potato genotypes. Front Plant Sci. 2020:11(169). 10.3389/fpls.2020.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S. Molecular regulation of plant responses to environmental temperatures. Mol Plant. 2020:13(4):544–564. 10.1016/j.molp.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Dyson BC, Miller MAE, Feil R, Rattray N, Bowsher CG, Goodacre R, Lunn JE, Johnson GN. FUM2, A cytosolic fumarase, is essential for acclimation to low temperature in Arabidopsis thaliana. Plant Physiol. 2016:172(1):118–127. 10.1104/pp.16.00852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman JB. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome. 2011:4(3):250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One 2010:5(11):e14101. 10.1371/journal.pone.0014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero-serrano Á, Assmann SM. Phenotypic and genome-wide association of natural variation with the local environment of Arabidopsis. Nat Ecol Evol. 2019:3(2):1–41. 10.1038/s41559-018-0754-5 [DOI] [PubMed] [Google Scholar]
- Fick SE, Hijmans RJ. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017:37(12):4302–4315. 10.1002/joc.5086 [DOI] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010:33(1):1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, GTEx Consortium, Nicolae DL, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015:47(9):1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehan MA, Park S, Gilmour SJ, An C, Lee CM, Thomashow MF. Natural variation in the C-repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J. 2015:84(4):682–693. 10.1111/tpj.13027 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. A futile cycle of sucrose synthesis and degradation is involved regulating partitioning between sucrose starch and respiration in cotyledons of germinating Ricinus communis L. seedlings when phloem transport is inhibited. Planta 1991:185(1):81–90. 10.1007/BF00194518 [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Thompson R, Cullis BR. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 1995:51(4):1440–1450. 10.2307/2533274 [DOI] [Google Scholar]
- Gugger PF, Fitz-Gibbon S, PellEgrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol. 2016:25(8):1665–1680. 10.1111/mec.13563 [DOI] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiol Plant. 2008:132(2):220–235. 10.1111/j.1399-3054.2007.00999.x [DOI] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006:142(1):98–112. 10.1104/pp.106.081141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann HA, Schwartz J-M, Johnson GN. Metabolic acclimation—a key to enhancing photosynthesis in changing environments? J Exp Bot. 2019:70(12):3043–3056. 10.1093/jxb/erz157 [DOI] [PubMed] [Google Scholar]
- Hoffmann MH. Biogeography of Arabidopsis thaliana (L.). J Biogeogr. 2002:29(1):125–134. 10.1046/j.1365-2699.2002.00647.x [DOI] [Google Scholar]
- Hofmeister BT, Lee K, Rohr NA, Hall DW, Schmitz RJ. Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 2017:18(1):155. 10.1186/s13059-017-1288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Willems G, Sasaki E, Koornneef M, Nordborg M. The genetic architecture of freezing tolerance varies across the range of Arabidopsis thaliana. Plant Cell Environ. 2016:39(11):2570–2579. 10.1111/pce.12812 [DOI] [PubMed] [Google Scholar]
- Houshyani B, Kabouw P, Muth D, de Vos RCH, Bino RJ, Bouwmeester HJ. Characterization of the natural variation in Arabidopsis thaliana metabolome by the analysis of metabolic distance. Metabolomics 2012:8(S1):131–145. 10.1007/s11306-011-0375-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura T, Dziurka M, Hura K, Ostrowska A, Dziurka K. Free and cell wall-bound polyamines under long-term water stress applied at different growth stages of xTriticosecale wittm. PLoS One 2015:10(8):e0135002. 10.1371/journal.pone.0135002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurry V. Metabolic reprogramming in response to cold stress is like real estate, it's all about location. Plant Cell Environ. 2017:40(5):599–601. 10.1111/pce.12923 [DOI] [PubMed] [Google Scholar]
- Junker A, Muraya MM, Weigelt-Fischer K, Arana-Ceballos F, Klukas C, Melchinger AE, Meyer RC, Riewe D, Altmann T.. Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Front Plant Sci. 2015:5:52. 10.3389/fpls.2014.00770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SS, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016:166(2):492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007:39(9):1151–1155. 10.1038/ng2115 [DOI] [PubMed] [Google Scholar]
- Kleessen S, Antonio C, Sulpice R, Laitinen R, Fernie AR, Stitt M, Nikoloski Z. Structured patterns in geographic variability of metabolic phenotypes in Arabidopsis thaliana. Nat Commun. 2012:3(1):1317–1319. 10.1038/ncomms2333 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004:55(1):141–172. 10.1146/annurev.arplant.55.031903.141605 [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012:63(4):1593–1608. 10.1093/jxb/err460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJB, van Eeuwijk FA. Marker-based estimation of heritability in immortal populations. Genetics 2015:199(2):379–398. 10.1534/genetics.114.167916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Sun R, Ouyang J, Ding W, Kim MJ, Wu J, Li Y, Shi T. AtMAD: Arabidopsis thaliana multi-omics association database. Nucleic Acids Res. 2021:49(D1):D1445–D1451. 10.1093/nar/gkaa1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher W. Effects of low temperature stress and frost injury on plant productivity. In: Johnson CB, editors. Physiological processes limiting plant productivity. London: Butterworth; 1981. p. 253–269. [Google Scholar]
- Less H, Galili G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008:147(1):316–330. 10.1104/pp.108.115733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stresses. New York: Academic Press, INC; 1980. [Google Scholar]
- Li B, Ritchie MD. From GWAS to gene: transcriptome-wide association studies and other methods to functionally understand GWAS discoveries. Front Genet. 2021:12:713230. 10.3389/fgene.2021.713230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Zhou M, Yao J, Li QQ, Zhang YY. Phenotypic and methylome responses to salt stress in Arabidopsis thaliana natural accessions. Front Plant Sci. 2022:13:841154. 10.3389/fpls.2022.841154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front Plant Sci. 2020:11:595603. 10.3389/fpls.2020.595603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005:444(2):139–158. 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Martínez-Berdeja A, Stitzer MC, Taylor MA, Okada M, Ezcurra E, Runcie DE, Schmitt J. Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2020:117(5):2526–2534. 10.1073/pnas.1912451117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007:104(11):4759–4764. 10.1073/pnas.0609709104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 2006:441(7096):947–952. 10.1038/nature04878 [DOI] [PubMed] [Google Scholar]
- Nägele T. Linking metabolomics data to underlying metabolic regulation. Front Mol Biosci. 2014:1:1–6. 10.3389/fmolb.2014.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägele T, Mair A, Sun X, Fragner L, Teige M, Weckwerth W. Solving the differential biochemical Jacobian from metabolomics covariance data. PLoS One 2014:9(4):e92299. 10.1371/journal.pone.0092299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägele T, Stutz S, Hörmiller II, Heyer AG. Identification of a metabolic bottleneck for cold acclimation in Arabidopsis thaliana. Plant J. 2012:72(1):102–114. 10.1111/j.1365-313X.2012.05064.x [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005:3(7):e196. 10.1371/journal.pbio.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke K, Prananingrum P, Klemens PAW, Trentmann O, Rodrigues CM, Keller I, Fernie AR, Geigenberger P, Bölter B, Lehmann M, et al. The plastidic sugar transporter pSuT influences flowering and affects cold responses. Plant Physiol. 2019:179(2):569–587. 10.1104/pp.18.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlaza-Jiménez L, Walther D. A genome-wide scan for correlated mutations detects macromolecular and chromatin interactions in Arabidopsis thaliana. Nucleic Acids Res. 2018:46(16):8114–8132. 10.1093/nar/gky576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. Ecological and evolutionary genetics of Arabidopsis. Trends Plant Sci. 1998:3(12):485–489. 10.1016/S1360-1385(98)01343-0 [DOI] [Google Scholar]
- Planchais S, Cabassa C, Toka I, Justin AM, Renou JP, Savoure A, Carol P. BASIC AMINO ACID CARRIER 2 gene expression modulates arginine and urea content and stress recovery in Arabidopsis leaves. Front Plant Sci. 2014:5:330. 10.3389/fpls.2014.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- Riewe D, Jeon HJ, Lisec J, Heuermann MC, Schmeichel J, Seyfarth M, Meyer RC, Willmitzer L, Altmann T. A naturally occurring promoter polymorphism of the Arabidopsis FUM2 gene causes expression variation, and is associated with metabolic and growth traits. Plant J. 2016:88(5):826–838. 10.1111/tpj.13303 [DOI] [PubMed] [Google Scholar]
- Ruan Y-L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014:65(1):33–67. 10.1146/annurev-arplant-050213-040251 [DOI] [PubMed] [Google Scholar]
- Saunders HA, Calzadilla PI, Schwartz J-M, Johnson GN. Cytosolic fumarase acts as a metabolic fail-safe for both high and low temperature acclimation of Arabidopsis thaliana. J Exp Bot. 2022:73(7):2112–2124. 10.1093/jxb/erab560 [DOI] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015:523(7559):212–216. 10.1038/nature14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci Rep. 2016:6(1):34027. 10.1038/srep34027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WX, Schneider T, Starck S, Martinoia E, Trentmann O. Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 2012:69(3):529–541. 10.1111/j.1365-313X.2011.04812.x [DOI] [PubMed] [Google Scholar]
- Shankar R, Bhattacharjee A, Jain M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci Rep. 2016:6(1):23719. 10.1038/srep23719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Foyer CH, Gustafsson P, Gardeström P, Hurry V. Altering flux through the sucrose biosynthesis pathway in transgenic Arabidopsis thaliana modifies photosynthetic acclimation at low temperatures and the development of freezing tolerance. Plant Cell Environ. 2003:26(4):523–535. 10.1046/j.1365-3040.2003.00983.x [DOI] [Google Scholar]
- Strand A, Hurry V, Gustafsson P, Gardeström P. Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J. 1997:12(3):605–614. 10.1111/j.0960-7412.1997.00605.x [DOI] [PubMed] [Google Scholar]
- Sun XL, Weckwerth W. COVAIN: a toolbox for uni- and multivariate statistics, time-series and correlation network analysis and inverse estimation of the differential Jacobian from metabolomics covariance data. Metabolomics 2012:8(S1):81–93. 10.1007/s11306-012-0399-3 [DOI] [Google Scholar]
- Takahashi D, Gorka M, Erban A, Graf A, Kopka J, Zuther E, Hincha DK. Both cold and sub-zero acclimation induce cell wall modification and changes in the extracellular proteome in Arabidopsis thaliana. Sci Rep. 2019:9(1):2289. 10.1038/s41598-019-38688-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H. Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot. 2010:67(3):429–443. 10.1016/j.envexpbot.2009.09.004 [DOI] [Google Scholar]
- The 1001 Genomes Consortium . 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016:166(2):481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010:154(2):571–577. 10.1104/pp.110.161794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010:153(3):895–905. 10.1104/pp.110.155119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W. Toward a unification of system-theoretical principles in biology and ecology—the stochastic lyapunov matrix equation and its inverse application. Front Appl Math Stat. 2019:5:29. 10.3389/fams.2019.00029 [DOI] [Google Scholar]
- Weiszmann J, Fürtauer L, Weckwerth W, Nägele T. Vacuolar sucrose cleavage prevents limitation of cytosolic carbohydrate metabolism and stabilizes photosynthesis under abiotic stress. FEBS J. 2018:285(21):4082–4098. 10.1111/febs.14656 [DOI] [PubMed] [Google Scholar]
- Weston DJ, Karve AA, Gunter LE, Jawdy SS, Yang X, Allen SM, Wullschleger SD. Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max. Plant Cell Environ. 2011:34(9):1488–1506. 10.1111/j.1365-3040.2011.02347.x [DOI] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan LDA, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. Welcome to the Tidyverse. J Open Source Software. 2019:4(43):1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- Wienkoop S, Morgenthal K, Wolschin F, Scholz M, Selbig J, Weckwerth W.. Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol Cell Proteomics. 2008:7(9):1725–1736. 10.1074/mcp.M700273-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JL, Nägele T, Linke M, Demel F, Fritsch SD, Mayr HK, Cai Z, Katholnig K, Sun X, Fragner L, et al. Inverse data-driven modeling and multiomics analysis reveals Phgdh as a metabolic checkpoint of macrophage polarization and proliferation. Cell Rep. 2020:30(5):1542–1552.e1547. 10.1016/j.celrep.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP. Urea metabolism in plants. Plant Sci. 2011:180(3):431–438. 10.1016/j.plantsci.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Zhen Y, Ungerer MC. Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytologist. 2008:177(2):419–427. 10.1111/j.1469-8137.2007.02262.x [DOI] [PubMed] [Google Scholar]
- Zuther E, Schulz E, Childs LH, Hincha DK. Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 2012:35(10):1860–1878. 10.1111/j.1365-3040.2012.02522.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the presented conclusions of this study are provided in the paper, supplement, or cited repositories.