Abstract
Saccharomyces boulardii (nom. inval.) has been used for the treatment of several types of diarrhea. Recent studies have confirmed that S. boulardii is effective in the treatment of diarrhea, in particular chronic or recurrent diarrhea, and furthermore that it is a safe and well-tolerated treatment. The aim of the present study was to identify strains of S. boulardii to the species level and assess their virulence in established murine models. Three strains of S. boulardii were obtained from commercially available products in France and Italy. The three S. boulardii strains did not form spores upon repeated testing. Therefore, classical methods used for the identification of Saccharomyces spp. could not be undertaken. Typing by using the restriction fragment length polymorphisms (RFLPs) of the PCR-amplified intergenic transcribed spacer regions (including the 5.8S ribosomal DNA) showed that the three isolates of S. boulardii were not separable from authentic isolates of Saccharomyces cerevisiae with any of the 10 restriction endonucleases assessed, whereas 9 of the 10 recognized species of Saccharomyces could be differentiated. RFLP analysis of cellular DNA with EcoRI showed that all three strains of S. boulardii had identical patterns and were similar to other authentic S. cerevisiae isolates tested. Therefore, the commercial strains of S. boulardii available to us cannot be genotypically distinguished from S. cerevisiae. Two S. boulardii strains were tested in CD-1 and DBA/2N mouse models of systemic disease and showed intermediate virulence compared with virulent and avirulent strains of S. cerevisiae. The results of the present study show that these S. boulardii strains are asporogenous strains of the species S. cerevisiae, not representatives of a distinct and separate species, and possess moderate virulence in murine models of systemic infection. Therefore, caution should be advised in the clinical use of these strains in immunocompromised patients until further study is undertaken.
Saccharomyces boulardii (nom. inval. [19]) has been widely used in Europe to treat diarrhea. This organism was first isolated from litchi fruit in Indochina (33, 49). A lyophilized form has been studied for oral administration in the United States by Biocodex Inc. (Seattle, Wash.), and the patent strain is held in the American Type Culture Collection (ATCC).
S. boulardii has been used in Europe and has been proposed for use in the United States for the treatment of several types of diarrhea, either as a preventative agent for antibiotic-associated diarrhea or as a treatment for diarrhea in adults and children infected with Clostridium difficile, for diarrhea in human immunodeficiency virus-infected patients, and for acute diarrhea in children and adults. The efficacy of these treatments has been previously reviewed (33). Several recent studies have confirmed that S. boulardii is effective in the treatment of diarrhea, in particular chronic or recurrent diarrhea (17, 25, 34, 35), and in the prevention of antibiotic-associated diarrhea (51). Furthermore, a recent study, which assessed the effect of 3 weeks of oral administration of S. boulardii in humans, showed that there was an increase in the brush border enzyme activity of the duodenal mucosa as well as a positive effect on the maturation of enterocytes (22). Several studies have shown that S. boulardii is a safe and well-tolerated treatment (25, 35, 39). However, there are also several documented reports of S. boulardii fungemia and septicemia resulting from the oral administration of this organism in immunocompromised and immunocompetent patients (46, 55, 57).
The taxonomy of the genus Saccharomyces has undergone significant changes recently, with more importance being placed on genotypic rather than phenotypic methods for the identification of isolates to the species level. This has resulted in a change in the number of species of Saccharomyces from 41 (27) to 7 (3) or 10 (4, 42). Recent and ongoing research in our laboratories has investigated the genetics and virulence of clinical and wild-type isolates of Saccharomyces cerevisiae (8, 13–16, 30, 31). The aim of the present study was to attempt to identify isolates of S. boulardii to the species level and assess their virulence in established murine models.
MATERIALS AND METHODS
Isolates.
Three isolates of S. boulardii were obtained from commercially available products in France (Ultralevure; two different batches, no. 48 and 49 [Sb 48 and Sb 49]) and Italy (Codex; Sb It). The company distributing the product in France is the parent company of the U.S. subsidiary clinically studying the product. The isolate of S. boulardii deposited at the ATCC (no. 74012) was requested; however, access was denied by the company (Biocodex Inc.) holding the patent on this isolate. Other isolates of Saccharomyces used (8, 14) are listed in Table 1 and the legend for Fig. 1.
TABLE 1.
Recovery of S. boulardii and S. cerevisiae from the brain in murine models of systemic infection
| Isolatea | Log10 geometric mean CFU/brain (95% CIb) for murine model:
|
|
|---|---|---|
| CD-1 | DBA/2N | |
| YJM145 | 4.51 (4.4–4.6) | 4.73 (4.5–4.9) |
| Sb 48 | 3.73 (3.6–3.9) | 4.42 (4.3–4.6) |
| Sb 49 | 3.22 (3.1–3.3) | 3.57 (3.5–3.6) |
| Y55 | 1.64 (1.3–2.0) | 4.51 (4.4–4.6) |
Sb 48 and Sb 49 are commercial isolates of S. boulardii; the others are S. cerevisiae isolates (14).
CI, confidence interval.
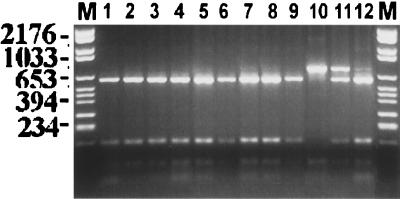
FIG. 1.
Photograph of the ethidium bromide-stained, UV-transilluminated, MaeI-digested PCR products after electrophoresis within a 3% agarose gel. The DNA from the PCR had been first purified by the Wizard PCR Preps purification system prior to overnight digestion with the 10 U of restriction endonuclease MaeI. Molecular size markers are in lanes M, and their corresponding sizes (in base pairs) are given on the left of the figure. Lanes: 1, S. boulardii Sb 48; 2, S. boulardii Sb 49; 3, S. boulardii Sb It; 4, S. cerevisiae ATCC 52530; 5, S. cerevisiae ATCC 26108; 6, S. cerevisiae YJM128, a clinical isolate (14, 30); 7, S. cerevisiae ItB9, a clinical isolate (28); 8, S. cerevisiae ItB8, a clinical isolate (28); 9, S. bayanus ATCC 76515; 10, S. paradoxus ATCC 76856; 11, S. paradoxus/S. cerevisiae hybrid YJM508 (29); 12, S. bayanus/S. cerevisiae hybrid YJM334 (29).
Genetic tests and phenotyping.
Standard yeast genetic techniques were attempted (50) to assess spore viability and to test for species (40). These included an assay of S. boulardii isolates for the ability to mate with known mating-competent S. cerevisiae laboratory strains. A complementation test was used to assess the mating competence of S. boulardii. Ethidium bromide-induced petite (mitochondrial respiration deficient) derivatives of S. boulardii were made (50) and were mixed with mating-competent MATa Lys2 and MATα Lys2 S. cerevisiae strains on a YEPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose, and 2% [wt/vol] agar) plate. The mixed strains were grown overnight and then replica plated to minimal medium containing ethanol and glycerol as the sole carbon sources, on which neither the petite S. boulardii strains nor the auxotrophic S. cerevisiae strains can grow. If the S. boulardii strains were able to mate, the S. boulardii/S. cerevisiae hybrids would grow on the minimal medium containing ethanol and glycerol.
The S. boulardii isolates were tested for their abilities to ferment galactose, maltose, and raffinose and for their abilities to grow on minimal medium with dextrose (50). Sugar fermentation was tested by replicate plating from a YEPD plate to plates with galactose, maltose, or raffinose as the carbon source and with antimycin A to block respiration. An isolate showing abundant growth is fermenting and would be designated “+” (e.g., Gal+ equals galactose fermenting).
Genotyping.
Cellular DNA was isolated by previously described methods (16, 45, 48).
Intergenic transcribed spacer region PCR (ITS-PCR) ribotyping is a method that utilizes restriction fragments of PCR products derived from the 5.8S ribosomal DNA (rDNA) and associated intergenic spacer regions (ITS1 and ITS2). Primers for the amplification of the 5.8S rRNA genes were ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′). DNA was amplified, and the amplicons were purified as described previously (29). For the restriction fragment length polymorphism (RFLP) analysis of the amplicons, the purified PCR products were digested with one of the following restriction enzymes: MaeI, HaeIII, CfoI, DdeI, BglII, BamHI, HindIII, EcoRI, SmaI, or PstI (Boehringer Mannheim, Indianapolis, Ind.) and the PCR products, with and without endonuclease digestion, were then analyzed by electrophoresis and visualized by staining and transillumination (29).
For RFLP analysis of the cellular DNA, approximately 3 μg of the cellular DNA was digested with 20 U of restriction endonuclease EcoRI for 6 h at 37°C. The DNA fragments were separated through a 0.7% (wt/vol) agarose gel in TAE buffer (40 mM Tris-acetate, 0.2 mM EDTA [pH 8.3]) for 20 h at 2 V/cm and visualized by UV transillumination at 302 nm after ethidium bromide staining.
In vivo studies.
For the in vivo study of virulence, two experimental murine models of systemic infection were studied to assess the relative virulence of S. boulardii, as described previously (8, 14). S. cerevisiae YJM145 and Y55 (14, 30) and two of our isolates of S. boulardii, randomly selected, Sb 48 and Sb 49, were grown overnight at ambient temperature in YPD broth on a gyratory shaker. In the nonimmunocompromised model, 6-week-old male CD-1 mice (Charles River Laboratories, Hollister, Calif.) were used. In the immunocompromised model, DBA/2N mice (Taconic, Germantown, N.Y.) were used; however, these were 6-week-old mice instead of the previously studied 4-week-old animals (8). Infection was initiated for groups of 10 mice by intravenous inoculation with 2 × 107 CFU of yeast. Two weeks postinfection, all mice were euthanized, and the number of viable yeast cells in the brain of each animal was determined by quantitative plating of serially diluted organ homogenates (14). The mean burdens of fungi were compared by using the nonparametric Mann-Whitney U test (14).
RESULTS
Genetics, genotyping, and phenotyping.
None of the three strains of S. boulardii produced any spores upon repeated testing at 16, 20, 24, and 30°C. The S. boulardii strains did not mate with known mating-competent S. cerevisiae strains. Therefore, the classical genetic methods for identification to the species level (40) could not be undertaken for these strains. All three isolates were Gal+ Mal+ Raf+. All three grew on minimal medium, showing that they do not have an auxotrophic requirement for any amino acid or base.
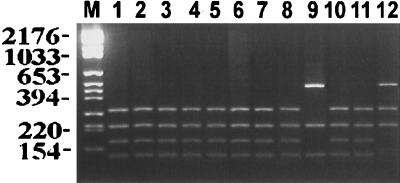
Typing using RFLPs of the PCR-amplified intergenic transcribed spacer regions (including the 5.8S rDNA) showed that the three isolates of S. boulardii were not separable from the well-authenticated strains of S. cerevisiae with any of the 10 restriction endonucleases assessed, whereas 9 of the 10 recognized species of Saccharomyces could be differentiated. Saccharomyces bayanus and Saccharomyces pastorianus could not be differentiated; these two species were previously postulated to be conspecific (29). Figures 1 and 2 show representative examples of the two most discriminatory restriction endonuclease (MaeI [Fig. 1] and HaeIII [Fig. 2]) digests of the PCR products. In each instance, the PCR product size was approximately 1,000 bp and the product gave a single amplicon, which was subsequently digested into smaller fragments. The interspecific hybrid species gave a PCR product that was the same size as that of the parental species. However, restriction of that PCR product revealed two separate amplicons. Thus, in Fig. 1 (lane 11) and Fig. 2 (lane 12), the hybrids share the ITS-PCR ribotyping pattern of the parental species.
FIG. 2.
Photograph of the ethidium bromide-stained, UV-transilluminated, HaeIII-digested PCR products after electrophoresis within a 3% agarose gel. The DNA from the PCR had been first purified by the Wizard PCR Preps purification system prior to overnight digestion with the 10 U of restriction endonuclease HaeIII. Molecular size markers are in lane M, and their corresponding sizes (in base pairs) are given on the left of the figure. Lane assignments are the same as those for Fig. 1.
RFLP analysis of cellular DNA with EcoRI showed that all three isolates of S. boulardii had identical patterns. These RFLP patterns were similar to those of other genotype A (16) S. cerevisiae isolates and strains used (Fig. 3).
FIG. 3.
Representative photograph of UV-transilluminated, ethidium bromide-stained agarose gel. RFLPs generated by EcoRI digestion of S. cerevisiae DNA are shown. Molecular size markers are in lanes M, and their corresponding sizes (in kilobases) are given on the left of the figure. Lane assignments are the same as those for Fig. 1.
In vivo studies.
The results of the in vivo testing of S. boulardii isolates for virulence are presented in Table 1. In CD-1 mice, comparison with strains YJM145 and Y55 of S. cerevisiae showed that both isolates of S. boulardii were significantly (P < 0.001) more virulent than Y55 but were significantly less virulent than YJM145 (P < 0.001) (Table 1). In comparison with each other, Sb 48 was more virulent than Sb 49 (P < 0.001). In CD-1 mice, YJM145 is intermediate (and above the median) in virulence and Y55 is avirulent (14). Thus, in this model the rank order of virulence was YJM145 > Sb 48 > Sb 49 > Y55. In comparison with previous work (14), the S. boulardii isolates would also be ranked more virulent than some clinical and nonclinical isolates. In the DBA/2N mice the S. boulardii isolates proved to be different from each other (Table 1). YJM145 was again significantly more virulent than the other strains (P < 0.01 to 0.001). Y55 was equivalent to Sb 48 in virulence (P > 0.05), and both were significantly more virulent than Sb 49 (P < 0.001). Differences in relative virulence between the two models for individual isolates have been previously described (8). In DBA/2N mice, YJM145 has intermediate virulence and Y55 is less virulent (but is not avirulent) (8). Overall, the two isolates of S. boulardii showed some capacity for virulence in these models and are considered to be of intermediate virulence.
DISCUSSION
The commercial strains of S. boulardii available to us could not be distinguished by the typing methods used in this study. The results of the present study show that these strains are asporogenous subtypes within the circumscription of S. cerevisiae and are not representatives of a separate species.
The classical genetic methods for species assignation cannot be utilized for these organisms as they do not produce spores. The method of ITS-PCR ribotyping used here for species designation is a relatively new technique. This method assesses the DNA that encodes the rRNA genes (rDNA) and has been utilized by many investigators for the determination of species of a wide variety of fungi (1, 5, 18, 20, 24, 26, 43, 56). This methodology has been applied also to Saccharomyces species for the authentication of strains in the S. cerevisiae “complex” (21). Messner and Prillinger (36) used a similar, yet more thorough method for the differentiation of 10 genotypically distinct Saccharomyces species. A recent investigation showed that ITS-PCR ribotyping is a simple method that can distinguish all Saccharomyces species, not just S. bayanus and S. pastorianus (29). Previous research has shown the latter two species to be very closely related (2, 21, 37, 44, 53). Furthermore, the findings of this recent study showed that interspecific hybrids resulting from the mating of two closely related yet distinct species of Saccharomyces shared the ITS-PCR ribotyping pattern of both parental species (29). This research (29) therefore allowed for the differentiation of isolates for which differentiation was previously not possible (40). It was anticipated that this technique would be able to differentiate the commercial strains of S. boulardii from other species of Saccharomyces and would therefore confirm previous reports that these strains belonged to a separate species. However, the findings of the present study showed that the commercial strains designated S. boulardii are not representatives of a separate species and should be reidentified as isolates of S. cerevisiae.
The results of the EcoRI-generated RFLPs of cellular DNA showed that these three strains, obtained from two regions of Europe, are genotypically indistinguishable. These results also support the above contention that these strains, purportedly S. boulardii, do not belong to a separate species as the RFLP patterns appear similar to those of the S. cerevisiae isolates studied here. The RFLP pattern of the S. boulardii isolates appears similar to those of over 80 S. cerevisiae isolates we have studied in previous research (16, 28).
A recent communication (32) from the company which holds the patent on this strain emphatically states that “S. boulardii is a completely different species of yeast from baker’s, brewer’s or wine yeast,” implying that the commercial isolates (and that deposited at ATCC) are derived directly from the originally described (49) isolate. The author supports the statement about species with seven pieces of evidence, which are analyzed as follows. (i) The strain has a separate designation by the ATCC. This designation would have been done only at the request of the patent-holding supplier; it does not confer species status on the organism, and the strain has not been made available for study. (ii) The quoted reference for phenotypic data which reportedly can separate this strain from S. cerevisiae is an unpublished personal communication and cannot therefore be assessed. Furthermore, the author states that S. boulardii does not use galactose as a carbon source as do wild-type S. cerevisiae strains. This is an unacceptable marker for species identification as the utilization of galactose has been shown to be variable among S. cerevisiae strains (3); indeed, sugar utilization is not useful for species delimitation or as a marker for DNA homology (41). Moreover, many S. cerevisiae isolates are Gal− in our hands and in the literature; these include well-authenticated laboratory isolates related to strain S288c (38) and strain Y55 and clinical isolates YJM436 and YJM522 (14). In addition, many clinical isolates are heterozygous for mutations affecting galactose fermentation ability (30). At least one group of S. cerevisiae is entirely or predominantly Gal− (41). Fermentation tests for galactose for all three of the S. boulardii strains in the present study were positive. This may be explained in part by differences in the methodology used for this test; the previous communication (32) gave no indication as to how the author arrived at the conclusion that S. boulardii is unable to utilize galactose as a carbon source. (iii) Three separate genetic studies which reportedly can distinguish this organism from S. cerevisiae are quoted; however, all of these are again unpublished data and cannot therefore be assessed. (iv) The final two references which are cited to support the distinct status of S. boulardii are the most interesting as they are published and can be studied. One (20) assessed the four sibling species of the genus Saccharomyces (S. bayanus, S. cerevisiae, S. paradoxus, and S. pastorianus) for variation by restriction analysis of mitochondrial DNA. Isolates of S. boulardii were not studied, nor was this organism mentioned in that article. The last study quoted is that by Cardinali and Martini (9), who investigated the electrophoretic karotypes of authentic strains representative of the genus Saccharomyces sensu stricto. This careful study used a principal-component analysis of the molecular weights of the bands of the electrophoretic karyotypes of 32 certified authentic strains to develop a matrix in which strains of the four species studied were clustered. This established a reference for assessing the localization of electrophoretic karyotype profiles obtained from unknown, noncertified yeast. Utilizing this model these investigators assessed the statistical distance (residual standard deviation [RSD]) of an isolate of S. boulardii (isolate DBVPG 6699; see Table 3 in reference 9) as 0.3730 compared to that of a standard S. cerevisiae isolate. The reference RSD value for S. cerevisiae in this model was given as 0.3952, and according to this analysis, an unknown strain belongs to a given species when its RSD value falls below the calculated reference RSD for the universe representing that species (9). Therefore, the only possible conclusion is that S. boulardii is a subtype of the species S. cerevisiae, a conclusion supported by our findings.
A recent study using the random amplified polymorphic DNA analysis methodology also concluded that S. boulardii is a member of the genotypic species S. cerevisiae (37). DNA reassociation data have recently shown 95% homology of S. boulardii and S. cerevisiae (54). Strains with >80% of their base sequences in common have been considered to belong to the same species (3, 52).
The name S. boulardii is invalid per the International Code of Botanical Nomenclature (ICBN) (19), because it was not effectively published in a readily available forum and lacks a description in Latin and the designation of accessible dried or lyophilized material (ICBN Articles 29.1, 32.1, 36, and 37). Taxonomic manuals and reviews (3, 4, 54) consider S. boulardii an abolished term and synonymous with S. cerevisiae. On the basis of this review of the literature and the results of the current investigation, it is not possible to support the previously made contention that the strain previously designated S. boulardii is a separate species from S. cerevisiae. Moreover, Biodiphar, the Biocodex subsidiary marketing the product in Belgium, also describes S. boulardii as S. cerevisiae (11). If indeed, as indicated previously (32, 33), the commercial isolates are direct descendants of the originally described isolate (49), then the latter may also be correctly described as S. cerevisiae, a conclusion that may be supported by the work of others (9, 37, 54).
There have been an increasing number of reports of human infections due to S. cerevisiae. For example, McCusker et al. (31) cited 12 such publications in 1994, 11 of these dealing with systemic infections, and that was not an exhaustive bibliography at the time. Many previously unpublished descriptions of clinical isolates appear in a study by Clemons et al. (14). Although immunocompromised patients are at risk, infection in healthy hosts also occurs (14, 31). Systemic infection can occur after oral ingestion (23). To assess the pathogenic potential of S. boulardii, two of the strains were used as the infecting organisms in murine models. Our previous work (8, 14) with these models has demonstrated that isolates of S. cerevisiae display a continuum of virulence potential and that virulence, as previously defined, in mice (14) was significantly associated with clinical isolates. In comparison with virulent and avirulent isolates of S. cerevisiae (8, 14), the two isolates of S. boulardii proved to have an intermediate degree of virulence. Mortality in the DBA/2N model used in this study was not seen, likely due to the increased resistance to fungal infection of (even slightly) older mice (7). While the level of virulence of S. boulardii is modest, it raises the question of the wisdom of using a potential pathogen as a therapeutic agent, especially in immunocompromised patients, in light of the demonstration that uptake of this organism from the gut lumen and translocation across the mucosa may occur (10) and reports of fungemia caused by S. boulardii administered orally (46, 55, 57). In addition, two isolates from different S. boulardii batches significantly differed in virulence in the same direction in two different models. This suggests that different batches of the preparation may not be uniform and may differ in a gene or genes that determine virulence. Possibly also relevant to strain variation is that the initial publication (49) described a sporulating yeast, whereas our isolates are asporogenous, as shown by repeated testing.
It would appear then that this strain of S. cerevisiae, which has been previously arbitrarily designated S. boulardii, has been shown to be useful in a range of clinical situations, particularly in the preventive treatment of diarrhea. It is likely that some other strains of S. cerevisiae also have these properties. In fact, animal model studies (6) and at least two clinical studies (12, 47) have shown that other S. cerevisiae strains do have such therapeutic properties.
ACKNOWLEDGMENTS
This research was funded in part by a Fellowship from the Commonwealth AIDS Research Grants Committee of the National Health and Medical Research Council of the Australian Federal Government.
We thank Richard C. Summerbell, Ministry of Health, Toronto, Canada; D. Yarrow, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; June Kwon-Chung, National Institutes of Health, Bethesda, Md.; and Michael McGinnis, University of Texas, Galveston, for helpful discussions with respect to taxonomic issues.
REFERENCES
- 1.Baleiras Couto M M, Vogels J T, Hofstra H, Huis in’t Veld J H, Vossen J M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food-borne yeasts. J Appl Bacteriol. 1995;79:525–535. doi: 10.1111/j.1365-2672.1995.tb03173.x. [DOI] [PubMed] [Google Scholar]
- 2.Banno I, Kaneko Y. A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast. 1989;5:S373–S377. [PubMed] [Google Scholar]
- 3.Barnett J A. The taxonomy of the genus Saccharomyces Meyen ex Rees: a short review for the non-taxonomists. Yeast. 1992;8:1–23. [Google Scholar]
- 4.Barnett J A, Payne R W, Yarrow D. Yeast characteristics and identification. Cambridge, England: Cambridge University Press; 1990. [Google Scholar]
- 5.Boekhout T, Kurtzman C P, O’Donnell K, Smith M T. Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26S ribosomal DNA nucleotide sequences. Int J Syst Bacteriol. 1994;44:781–786. doi: 10.1099/00207713-44-4-781. [DOI] [PubMed] [Google Scholar]
- 6.Brandao R L, Castro I M, Bambirra E A, Amaral S C, Fietto L G, Tropia M J M, Neves M J, Dos Santos R G, Gomes N C M, Nicoli J R. Intracellular signal triggered by cholera toxin in Saccharomyces boulardii and Saccharomyces cerevisiae. Appl Environ Microbiol. 1998;64:564–568. doi: 10.1128/aem.64.2.564-568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brass C, Stevens D A. Maturity as a critical determinant of resistance to fungal infections: studies in murine blastomycosis. Infect Immun. 1982;36:387–395. doi: 10.1128/iai.36.1.387-395.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byron J K, Clemons K V, McCusker J H, Davis R W, Stevens D A. Pathogenicity of Saccharomyces cerevisiae in complement factor five-deficient mice. Infect Immun. 1995;63:478–485. doi: 10.1128/iai.63.2.478-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardinali G, Martini A. Electrophoretic karyotypes of authentic strains of the sensu stricto group of the genus Saccharomyces. Int J Syst Bacteriol. 1994;44:791–797. doi: 10.1099/00207713-44-4-791. [DOI] [PubMed] [Google Scholar]
- 10.Cartwright-Shamoon J, Dickson G R, Dodge J, Carr K E. Uptake of yeast (Saccharomyces boulardii) in normal and rotavirus treated intestine. Gut. 1996;39:204–209. doi: 10.1136/gut.39.2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centre Belge d’Information Pharmacotherapeutique. Repertoire commente des medicaments. Brussels, Belgium: Ministry of Public Health, General Inspection of Pharmacy; 1997. p. 74. [Google Scholar]
- 12.Chia J K S, Chan S M, Goldstein H. Baker’s yeast as adjunctive therapy for relapses of Clostridium difficile diarrhea. Clin Infect Dis. 1995;20:1581. doi: 10.1093/clinids/20.6.1581. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 13.Clemons K V, Hanson L C, Stevens D A. Colony phenotype switching in clinical and non-clinical isolates of Saccharomyces cerevisiae. J Med Vet Mycol. 1996;34:259–264. doi: 10.1080/02681219680000441. [DOI] [PubMed] [Google Scholar]
- 14.Clemons K V, McCusker J H, Davis R W, Stevens D A. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis. 1994;169:859–867. doi: 10.1093/infdis/169.4.859. [DOI] [PubMed] [Google Scholar]
- 15.Clemons K V, McCusker J H, Davis R W, Stevens D A. Saccharomyces cerevisiae and the host-fungus interplay. In: Vanden Bossche H, Stevens D A, Odds F C, editors. Host-fungal interplay. Bethesda, Md: National Foundation for Infectious Diseases; 1997. pp. 193–198. [Google Scholar]
- 16.Clemons K V, Park P, McCusker J H, McCullough M J, Davis R W, Stevens D A. Application of DNA typing methods and genetic analysis to epidemiology and taxonomy of Saccharomyces isolates. J Clin Microbiol. 1997;35:1822–1828. doi: 10.1128/jcm.35.7.1822-1828.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elmer G W, Surawicz C M, McFarland L V. Biotherapeutic agents. A neglected modality for the treatment and prevention of selected intestinal and vaginal infections. JAMA. 1996;275:870–876. doi: 10.1001/jama.275.11.870. [DOI] [PubMed] [Google Scholar]
- 18.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greuter W, Barrie F R, Burdet H M, Chaloner W C, Demoulin V, Hawksworth D L, Jorgenson P M, Nicholson D H, Silva P C, Trehane P, McNeill J. International code of botanical nomenclature (Tokyo Code). Konigstein, Germany: Koeltz Scientific Books; 1994. [Google Scholar]
- 20.Guillamon J M, Barrio E, Huerta T, Querol A. Rapid characterization of four species of the Saccharomyces sensu stricto complex according to mitochondrial DNA patterns. Int J Syst Bacteriol. 1994;44:708–714. doi: 10.1099/00207713-44-4-708. [DOI] [PubMed] [Google Scholar]
- 21.Huffman J L, Molina F I, Jong S C. Authentication of ATCC strains in the Saccharomyces cerevisiae complex by PCR fingerprinting. Exp Mycol. 1992;16:316–319. [Google Scholar]
- 22.Jahn H U, Ullrich R, Schneider T, Liehr R M, Schieferdecker H L, Holst H, Zeitz M. Immunological and trophical effects of Saccharomyces boulardii on the small intestine in healthy human volunteers. Digestion. 1996;57:95–104. doi: 10.1159/000201320. [DOI] [PubMed] [Google Scholar]
- 23.Jensen D P, Smith D L. Fever of unknown origin secondary to brewer’s yeast ingestion. Arch Intern Med. 1976;136:332–333. [PubMed] [Google Scholar]
- 24.Johnston C G, Aust S D. Detection of Phanerochaete chrysosporium in soil by PCR and restriction enzyme analysis. Appl Environ Microbiol. 1994;60:2350–2354. doi: 10.1128/aem.60.7.2350-2354.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchelle A, Fruhwein N, Toburen D. Treatment of persistent diarrhea with S. boulardii in returning travelers. Results of a prospective study. Fortschr Med. 1996;114:136–140. . (In German.) [PubMed] [Google Scholar]
- 26.Kumeda Y, Asao T. Single-strand conformation polymorphism analysis of PCR-amplified ribosomal DNA internal transcribed spacers to differentiate species of Aspergillus Section Flavi. Appl Environ Microbiol. 1996;62:2947–2952. doi: 10.1128/aem.62.8.2947-2952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodder J, Kreger-van Rij N J W. The yeasts: a taxonomic study. Amsterdam, The Netherlands: North Holland Publishing Company; 1952. [Google Scholar]
- 28.McCullough M J, Clemons K V, Farina C, McCusker J H, Stevens D A. Epidemiological investigation of vaginal Saccharomyces cerevisiae isolates by a genotypic method. J Clin Microbiol. 1998;36:557–562. doi: 10.1128/jcm.36.2.557-562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCullough M J, Clemons K V, McCusker J H, Stevens D A. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J Clin Microbiol. 1998;36:1035–1038. doi: 10.1128/jcm.36.4.1035-1038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCusker J H, Clemons K V, Stevens D A, Davis R W. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCusker J H, Clemons K V, Stevens D A, Davis R W. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFarland L V. Saccharomyces boulardii is not Saccharomyces cerevisiae. Clin Infect Dis. 1996;22:200–201. doi: 10.1093/clinids/22.1.200. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 33.McFarland L V, Bernasconi P. Saccharomyces boulardii: a review of an innovative biotheraputic agent. Microb Ecol. 1993;6:157–171. [Google Scholar]
- 34.McFarland L V, Surawicz C M, Greenberg R N, Elmer G W, Moyer K A, Melcher S A, Bowen K E, Cox J L. Prevention of beta-lactam-associated diarrhea by Saccharomyces boulardii compared with placebo. Am J Gastroenterol. 1995;90:439–448. [PubMed] [Google Scholar]
- 35.McFarland L V, Surawicz C M, Greenberg R N, Fekety R, Elmer G W, Moyer K A, Melcher S A, Bowen K E, Cox J L, Noorani Z. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA. 1994;271:1913–1918. [PubMed] [Google Scholar]
- 36.Messner R, Prillinger H. Saccharomyces species assignment by long range ribotyping. Antonie Leeuwenhoek. 1995;67:363–370. doi: 10.1007/BF00872936. [DOI] [PubMed] [Google Scholar]
- 37.Molnar O, Messner R, Prillinger H, Stahl U, Slavikova E. Genotypic identification of Saccharomyces species using random amplified polymorphic DNA analysis. Syst Appl Microbiol. 1995;18:136–145. [Google Scholar]
- 38.Mortimer R T, Johnston J R. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller J, Remus N, Harms K H. Mycoserological study of the treatment of paediatric cystic fibrosis patients with Saccharomyces boulardii (Saccharomyces cerevisiae Hansen CBS 5926) Mycoses. 1995;38:119–123. doi: 10.1111/j.1439-0507.1995.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 40.Naumov G I. Genetic basis for classification and identification of the ascomycetous yeast. Stud Mycol. 1987;30:469–475. [Google Scholar]
- 41.Naumov G I. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol. 1996;17:295–302. [Google Scholar]
- 42.Naumov G I, Naumov E S, Gailadrin C. Genetic and karyotypic identification of wine Saccharomyces bayanus yeast isolated in France and Italy. Syst Appl Microbiol. 1993;16:274–279. [Google Scholar]
- 43.O’Donnell K, Gray L E. Phylogenetic relationships of the soybean sudden death syndrome pathogen Fusarium solani f. sp. phaseoli inferred from rDNA sequence data and PCR primers for its identification. Mol Plant-Microbe Interact. 1995;8:709–716. doi: 10.1094/mpmi-8-0709. [DOI] [PubMed] [Google Scholar]
- 44.Peterson S W, Kurtzman C P. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst Appl Microbiol. 1991;14:124–129. [Google Scholar]
- 45.Philippsen P, Stotz A, Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 46.Pletincx M, Legein J, Vandenplas Y. Fungemia with Saccharomyces boulardii in a 1-year-old girl with protracted diarrhea. J Pediatr Gastroenterol Nutr. 1995;21:113–115. doi: 10.1097/00005176-199507000-00022. [DOI] [PubMed] [Google Scholar]
- 47.Schellenberg D A, Bonington A, Champion C M, Lancaster R, Webb S, Main J. Treatment of Clostridium difficile diarrhea with brewer’s yeast. Lancet. 1994;343:171–172. doi: 10.1016/s0140-6736(94)90960-1. [DOI] [PubMed] [Google Scholar]
- 48.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seguela J P, Bastide J M, Massot J. Abstracts of the VIth International Symposium on Yeasts. 1984. Saccharomyces boulardii: criteres d’identification; pp. XIV–II-P. [Google Scholar]
- 50.Sherman F, Fink G R, Lawrence C W. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 51.Surawicz C M, Elmer G W, Speelman P, McFarland L V, Chinn J, Van Belle G. Prevention of antibiotic-associated diarrhea by Saccharomyces boulardii: a prospective study. Gastroenterology. 1989;96:981–988. doi: 10.1016/0016-5085(89)91613-2. [DOI] [PubMed] [Google Scholar]
- 52.van der Walt J P. The typological yeast species, and its delimitation. In: Rose A H, Harrison J S, editors. The yeasts. 2nd ed. Vol. 1. London, United Kingdom: Academic Press; 1987. pp. 95–121. [Google Scholar]
- 53.Vaughn-Martini A, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 54.Vaughn-Martini A, Martini A. Saccharomyces Meyen ex Rees. In: Kurtzman C P, Fell J W, editors. The yeasts: a taxonomic study. Amsterdam, The Netherlands: Elsevier; 1998. pp. 358–371. [Google Scholar]
- 55.Viggiano M, Badetti C, Bernini V, Garabedian M, Manelli J C. Saccharomyces boulardii fungemia in a patient with severe burns. Ann Fr Anesth Reanim. 1995;14:356–358. doi: 10.1016/s0750-7658(05)80603-3. . (In French.) [DOI] [PubMed] [Google Scholar]
- 56.Williams D W, Wilson M J, Lewis M A, Potts A J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zunic P, Lacotte J, Pegoix M, Buteux G, Leroy G, Mosquet B, Moulin M. Saccharomyces boulardii fungemia. Apropos of a case. Therapie. 1991;46:498–499. . (Letter.) (In French.) [PubMed] [Google Scholar]