Abstract
Background
Evidence regarding the effects of infant feeding type (exclusive breastfeeding compared with exclusive formula feeding) on the gut microbiota and how it impacts infant growth status is limited.
Objectives
The primary objective was to compare gut microbiota by feeding type and characterize the associations between gut microbiota and infant growth status.
Methods
Stool samples from healthy, full-term infants (4–5 mo-old) who were either exclusively breastfed (BF) or exclusively formula-fed (FF) in Denver, CO, United States were collected, and fecal 16S ribosomal ribonucleic acid gene-based profiling was conducted. Length and weight were measured at the time of stool collection. Length-for-age z-score, weight-for-age z-scores (WAZ), and weight-for-length z-scores were calculated based on the World Health Organization standards. Associations between gut microbial taxa and anthropometric z-scores were assessed by Spearman’s rank correlation test.
Results
A total of 115 infants (BF n = 54; FF n = 61) were included in this study. Feeding type (BF compared with FF) was the most significant tested variable on gut microbiota composition (P < 1 × 10-⁶), followed by mode of delivery and race. Significant differences were observed in α-diversity, β-diversity, and relative abundances of individual taxa between BF and FF. BF infants had lower α-diversity than FF infants. Abundances of Bifidobacterium and Lactobacillus were greater in the breastfeeding group. FF infants had a higher relative abundance of unclassified Ruminococcaceae (P < 0.001), which was associated with a higher WAZ (P < 0.001) and length-for-age z-score (P < 0.01). Lactobacillus was inversely associated with WAZ (P < 0.05).
Conclusions
Feeding type is the main driver of gut microbiota differences in young infants. The gut microbiota differences based on feeding type (exclusive breast- or formula feeding) were associated with observed differences in growth status.
This trial was registered at clinicaltrials.gov as NCT02142647, NCT01693406, and NCT04137445.
Keywords: mode of feeding, gut microbiota, growth status, breastfeeding, infant formula
Introduction
Before solid foods are introduced, the liquid diet (breastmilk compared with infant formula) accounts for virtually all of the infant’s dietary intake, which affects the gut microbiota [1]. The gut microbiota has been shown to associate with human health indicators and disease development, such as allergies, autoimmune diseases, and obesity [[2], [3], [4]]. During infancy and early childhood, the gut microbiota also plays a role in developing certain health conditions [5]. More importantly, underdeveloped or nonage-appropriate gut microbiota has been shown to negatively impact infant growth in low-source settings [6,7]. Thus, a thorough understanding of the gut microbiota and the role of feeding type in modulating the gut microbiota during infancy is critical [1].
Research to date has produced inconsistent findings regarding the effects of feeding mode on infant gut microbiota. For example, although the majority of the literature showed that breastfed (BF) infants tend to have a higher abundance of Bifidobacterium [8] and lower overall α-diversity [9], some reported significantly higher Bifidobacterium in mixed-fed infants, defined as infants fed a mixture of both breastmilk and formula, compared with exclusively BF infants [10]. A meta-analysis concluded that exclusive breastfeeding is strongly associated with a greater abundance of the family Bifidobacteriaceae and lower α-diversity before 6 mo compared with nonexclusive breastfeeding [11]. One limitation of the current literature is that most findings were based on breastfeeding exclusivity, namely comparing exclusively BF with nonexclusively BF infants. Nonexclusive breastfeeding includes exclusive formula feeding and feeding both infant formula and breastmilk, adding noise to the comparison and making it difficult to interpret the results. Emerging research also suggests that gut microbiota may modulate growth and weight gain early in life [12]. One animal study [13] found that gut microbiota drives bone growth in juvenile mice. A cohort study [14] identified bacterial species, such as Ruminococcus gnavus, whose proportional representation defined healthy and mature gut microbiota during the first year of life in Malawian infants. It remains unknown to what extent, if any, these types of growth-discriminatory taxa might have in a high-resourced Westernized setting and/or in healthy, well-nourished populations.
In the present study, the gut microbiota of exclusively BF infants and exclusively formula-fed (FF) infants at 4–5 mo of life from Denver, CO, were assessed. The 2 objectives were as follows: 1) to compare gut microbial profiles by feeding type and 2) to examine the potential association between gut microbial composition and infant growth status. We hypothesized that 1) the gut microbiota composition would differ by feeding type, with BF having higher abundances of potential commensals, such as Bifidobacterium, and with FF being more diverse and having taxa representing more “mature” gut microbiota; and 2) gut microbial composition would associate with infant linear and ponderal growth status (eg, z-scores).
Methods
Participants
This secondary analysis uses fecal samples collected from 3 healthy, full-term (gestational age ≥37 wk) infant cohorts in Denver, CO. Healthy was defined as not having significant congenital anomalies or known chronic conditions affecting feeding, growth, or developmental potential. Two cohorts had BF infants exclusively (no formula exposure since birth), and 1 had FF infants exclusively (breastmilk exposure <2 wk). Fecal samples were collected from all 3 infant cohorts at 4–5 mo of age, which was the baseline time point of 1 completed, 1 ongoing infant dietary intervention study, and the end point of a completed observational study [15]. Thus, none of the participants received any intervention at the time of stool sample collection (4–5 mo). The Colorado Multiple Institutional Review Board approved all 3 studies. Written informed consent was obtained from the parents or legal guardians of the infants. These studies were registered at clinicaltrials.gov (NCT02142647, NCT01693406, and NCT04137445).
Stool sample collection
To collect stool samples, sterile diaper liners, gloves, and prelabeled ziplock bags were given to caregivers to place in the infant’s diaper. The liner was biodegradable, which effectively collected stool, but allowed passage of urine. Caregivers were given instructions on how to place and remove the liner from the diaper once stool was produced. The soiled liner was then placed by the caregiver in zip lock bags with written collection time and date. The caregiver placed the bagged samples in home freezers (–20°C) and notified the study coordinator immediately. The study coordinator collected the samples within 24 h and transferred them to the University of Colorado Anschutz Medical Campus on ice packs. Stool samples were then collected by research personnel from the liner in a laminar flow hood and stored in sterile vials in –80°C freezers until analysis. All 3 infant cohorts followed the same sample collection procedures, and samples were sequenced in 1 batch. One cohort of BF infants used RNAlater (Qiagen) in the sterile vials to store the stool samples, whereas the other 2 cohorts used sterile vials only. Comparison of sequencing data and other variables between the 2 BF cohorts that samples were stored with or without RNAlater showed no differences, and the 2 BF cohorts were combined.
Weight and length measurements
Infant weight and length were measured in all participants by trained pediatric research personnel at the Colorado Clinical and Translational Research Center at Children’s Hospital Colorado. All measurements were performed in triplicate. The length was measured in a recumbent position using an infant stadiometer accurate to 0.1 cm (Holtain Ltd.). An electronic digital balance (Sartorius Corporation) was used to obtain naked infant weight. Growth z-scores (the number of SDs above or below the population median) were calculated based on WHO/CDC BF infant growth standards [16]. In brief, the weight-for-age z-score (WAZ, weight parameter) and length-for-age z-score (LAZ, length parameter) are based on weight and length using the WHO/CDC growth standards controlled for age and sex [16]. Weight-for-length z-score (WLZ) is derived from WAZ and LAZ.
Microbiota analysis
16S amplicon library construction
Bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes following our previously described methods [17,18]. In brief, DNA was extracted from 25 to 50 mg of stool using the QIAamp PowerFecal DNA kit (Qiagen Inc.), which employs chemical and mechanical biomass disruption. Samples were bead-beaten using a MagNA Lyser (Roche Inc.) at 10,000 × g; 60 s. PCR amplicons were generated using barcoded [19] primers that target ∼450 basepairs of the V3V4 variable region of the 16S rRNA gene (338F: 5’ACTCCTACGGGAGGCAGCAG and 806R: 5’ GGACTACHVGGGTWTCTAAT) [20,21]. PCR products were normalized using a SequalPrep kit (Invitrogen) and then pooled. The amplicon pool was partially lyophilized to reduce its volume and then purified and concentrated using a DNA Clean and Concentrator Kit (Zymo). Pooled amplicons were quantified using a Qubit Fluorometer 2.0 (Invitrogen). Illumina paired-end sequencing was performed following the manufacturer’s protocol on the MiSeq platform using a 600-cycle version 3 reagent kit and version 2.4 of the MiSeq Control Software. All samples were sequenced in a single batch.
Analysis of illumina paired-end reads
Illumina MiSeq paired-end reads were aligned to human reference genome hg19 with bowtie2 and matching sequences discarded [22,23]. As previously described [17,18], demultiplexed paired reads were assembled using phrap [24,25], and pairs that did not assemble were discarded. Assembled sequences were trimmed over a moving window of 5 nucleotides until the mean quality met or exceeded 20. Trimmed sequences with >1 ambiguity or <350 nt were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) [26] using the Schloss [27] Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.3.0-r23838) [28] using the 418,497 bacterial sequences in Silva 115NR99 [29] as reference configured to yield the Silva taxonomy. Taxonomic assignment by SINA used the lowest common ancestor approach with default parameters. Operational taxonomic units were produced by binning sequences with identical taxonomic assignments. A single sample generated <5000 sequences and was excluded from further analysis. The remaining 114 sequence libraries had a median of 90,914 sequences/sample (interquartile range: 73,820–106,109), and all libraries had Good’s coverage values >99%. The software package Explicet (version 2.10.5) [30] was used to calculate α-diversity indices through 1000 replicate resamplings.
Statistical and data analyses
Values are presented as mean ± SD for continuous variables. P values of ≤0.05 were considered to be significant. The software packages R (version 4.1.0) [31] and Explicet (version 2.10.5) [30] were used to analyze and visualize data. Independent Student’s t-test was used to compare differences in growth and other demographic data between groups. For the gut microbiota analysis, differences in overall composition (ie, β-diversity) were assessed through permutational ANOVA (PERMANOVA) with the Aitchison dissimilarity index [32,33]. PERMANOVA P values were inferred through 106 label permutations. For 2- and 3-factor PERMANOVA tests, margin P values are reported for each covariate. Principal coordinates analysis was carried out using Aitchison dissimilarities and the vegan wcmdscale function. Alpha-diversity indices (ie, Chao1, Shannon H, and Shannon H/Hmax) were assessed by ANOVA. Individual taxa differing between treatment groups were identified using the ANOVA-like differential expression (ALDEx2) R package [34,35]. The distribution of taxa in each sequence library was estimated through 1000 Dirichlet Monte Carlo resamplings of sequence count data. To account for the compositional nature of microbiome data, [36] sequence count data were subjected to a centered log-ratio (CLR) transformation with all features used as the denominator. Either nominal or false discovery rate (FDR)-corrected [37] P values are reported, as indicated in the text and figures. Effect size plots are derived from the outputs of ALDEx2 and represent the median effect sizes, calculated as the median between-group difference in CLR values between groups divided by the largest within-group difference in CLR values [34,35]. Associations between anthropometric measures and CLR-transformed microbiota count data were assessed by nonparametric Kendall rank correlation tests.
Results
Participants
A total of 115 infants, aged 4–5 mo, were initially included in the study; 54 infants were exclusively BF, and 61 infants were exclusively FF at the time of stool collection. The BF group was 87% White (n = 47), 13% non-White (n = 7), 50% male (n = 27), and 50% female (n = 27). The FF cohort was 69% White (n = 42), 31% non-White (n = 19), 52.5% male (n = 32), and 47.5% female (n = 29). For maternal education, 73% and 74% had an associate degree or higher in BF and FF groups, without differences between groups, respectively. Three participants had a gross family income <$18,000/y (n = 1 in FF; n = 2 in BF); 80% of families’ income was >$50,000/y for both BF and FF. Table 1 depicts infant demographics, maternal BMI (kg/m2), maternal height, delivery mode, antibiotic use, and growth z-scores. There was a significant difference between BF and FF infants in both WAZ and WLZ. No significant difference in LAZ was observed between groups.
TABLE 1.
Characteristics of study participants
| BF (n = 54) | FF (n = 61) | |
|---|---|---|
| Race1 | White n = 47 (87%) | White n = 42 (69%) |
| Sex | Male n = 27 (50%) | Male n = 32 (52%) |
| Maternal BMI1 (kg/m2) | 25.6 ± 5.5 | 28.0 ± 6.5 |
| Maternal height (cm) | 164 ± 7 | 166 ± 7 |
| Mode of delivery | C-section n = 15 (28%) | C-section n = 16 (26%) |
| Antibiotic use (n) | 1 (2%) | 2 (3%) |
| LAZ | –0.386 ± 0.942 | –0.197 ± 0.945 |
| WAZ1 | –0.510 ± 0.923 | –0.088 ± 0.774 |
| WLZ1 | –0.296 ± 0.945 | 0.125 ± 0.796 |
Abbreviations: BF, breastfed infants; BMI, body mass index; FF, formula-fed infants; LAZ, length-for-age z-score; WAZ, weight-for-age z-score; WLZ, weight-for-length z-score.
indicate variables that were statistically significant (P < 0.05) between groups.
Dietary intakes
At the time of stool sample collection, none of the BF infants had received complementary formula or foods. Likewise, none of the FF infants had consumed human milk for >2 wk (cumulatively) since birth or any complementary foods by caregivers’ reports. All BF infants took vitamin D supplements. FF infants consumed several types of infant formula, including Enfamil AR (n = 2), Enfamil Gentlease (n = 14), Enfamil Infant (n = 17), Gerber Goodstart Gentle (n = 3), Nutramigen (n = 1), Similac Advance (n = 16), or Similac Sensitive (n = 8). Supplementary Table 1 shows the macronutrient composition of each type of formula. No differences in the infant gut microbiota were found among formula types, degree of protein hydrolysis, or percentage of whey protein compared with casein or lactose content (PERMANOVA tests with no significant P values). Also, none of the formulas on the United States market at the time of sample collection (2013–2015) contained human milk oligosaccharides, which are prebiotics in human milk that could impact gut microbiota. Thus, all FF infants were combined as 1 group for analysis purposes and treated equally.
Type of feeding and infant gut microbiota
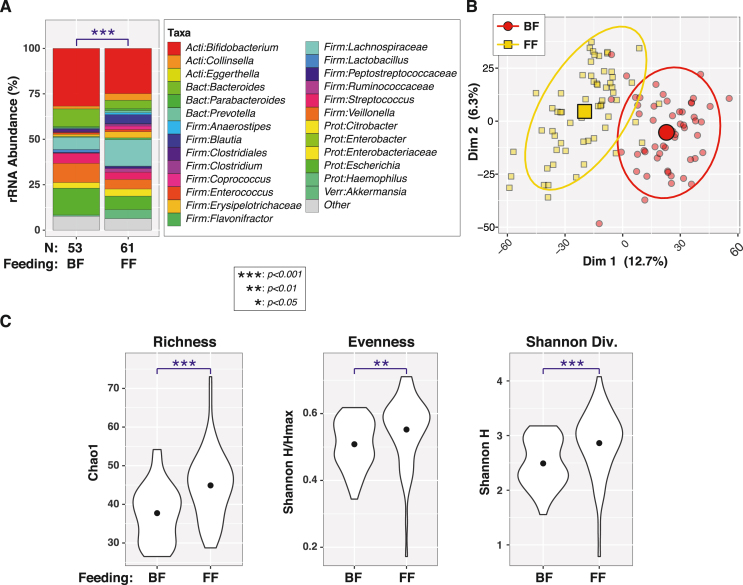
Bacterial profiles were generated for all 115 infant fecal samples by 16S rRNA gene sequencing; 1 infant sample was excluded from further analysis because of low sequence counts (<5000). Overall, β-diversity differed significantly between BF and FF infants (PERMANOVA P < 1 × 10-6; Figure 1A). A principal coordinates analysis plot showed clear clustering of participants by feeding type (Figure 1B). FF infants also have greater α-diversity than BF infants (Figure 1C), as measured by richness (Chao1; P = 6.6e-06), evenness (Shannon H/Hmax; P = 0.0095), and diversity (Shannon H; P = 0.00034).
FIGURE 1.
Variation in 4–5 mo-old breastfed (BF) and formula-fed (FF) infant microbiota between feeding types. (A) Bar charts with PERMANOVA results. Percent relative abundance (%RA) of genus-level taxa, stratified by feeding type. Taxa with %RA <1% were collapsed into the “Other” category. The results of PERMANOVA tests are summarized in the plot. (B) Principal coordinates plot. Individual subjects are indicated by smaller symbols (circles or squares), with group affiliations designated by symbol shapes and color coding. Mean PC values for each group along the x- and y-axes are indicated by larger shapes, whereas ellipses represent 95% CIs. (C) Alpha-diversity. Violin plots show distributions of α-diversity indices by feeding type. ANOVA test results are indicated above each plot. BF (n = 54), FF (n = 61). Taxa listed at the family level are those that cannot be identified at the genus level. ANOVA, analysis of variance; CI, confidence interval; PERMANOVA, permutational analysis of variance; rRNA, ribosomal ribonucleic acid; Dim 1 & Dim 2, Principal coordinates dimensions 1 and 2.
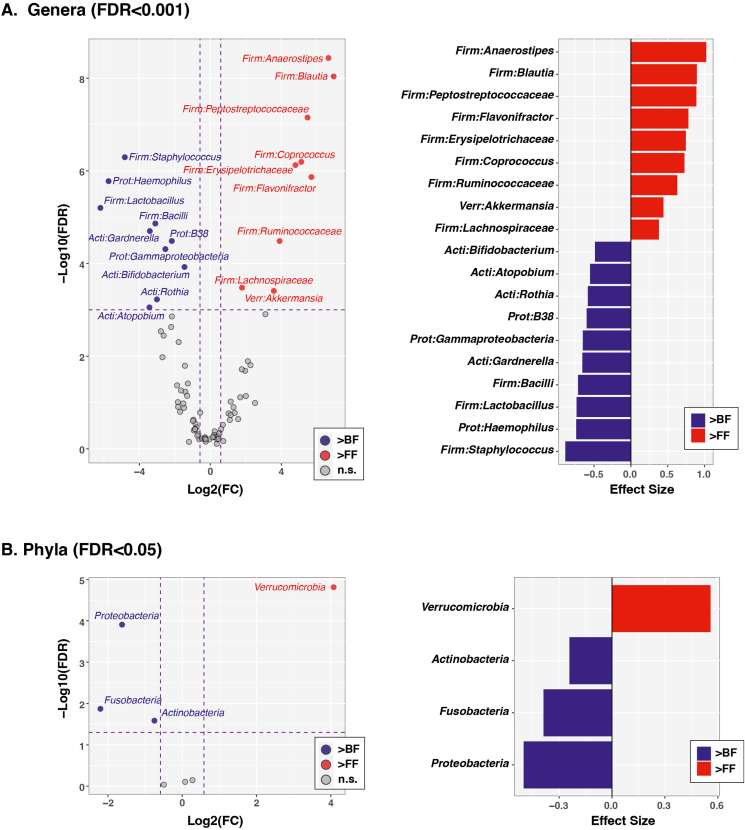
A few genera (Figure 2A) and phyla (Figure 2B) had significantly different abundances between feeding types based on a stringent cutoff of FDR-corrected P < 0.001. BF infants had higher relative abundances of Staphylococcus, Lactobacillus, Haemophilus, unclassified Bacilli, Gardeneralla, unclassified Gammaproteobacteria, B38, Rothia, Atopobium, Gemella, and Bifidobacterium. FF infants had higher relative abundances of Anaerostipes, unclassified Peptostreptococcaceae, Blautia, Flavonifractor, Erysipelotrichaceae, Coprococcus, unclassified Ruminococcaceae, Akkermansia, and unclassified Lachnospiraceae. Two phylum-level taxa (Proteobacteria and Verrucomicrobia) also had FDR-corrected P values of <0.001, with Proteobacteria higher in BF and Verrucomicrobia higher in FF. Actinobacteria and Fusobacteria were higher in BF infants at a less stringent cutoff (FDR < 0.05). Individual taxa relative abundances are in supplementary materials.
FIGURE 2.
Individual taxa varying by feeding type in 4–5 mo-old breastfed (BF) and formula-fed (FF) infants. Tests were conducted at the genus (A) and phylum (B) levels. The left column shows volcano plots of fold-change (FC; Log2 transformed) compared with FDR-corrected P values (–Log10 transformed) ascertained by ALDEx2 analysis. Vertical and horizontal dashed lines represent significance cutoffs of FC ≥2 and FDR-corrected P value ≤ 0.001, respectively. Taxa in the upper left quadrant and colored blue were enriched in BF relative to FF infants (“>BF”). Taxa in the upper right quadrant and colored red were enriched in FF relative to BF infants (“>FF”). Taxa that did not meet the significance cutoffs are colored gray (“NS”; ie, FC <2 or FDR-corrected P value >0.001). The right column shows plots of ALDEx2-calculated effect sizes of taxa meeting FC and FDR cutoffs. BF (n = 54), FF (n = 61). Taxa listed at the family level are those that cannot be identified at the genus level. ALDEx2, ANOVA-like differential expression; FDR, false discovery rate.
Other determinants of infant gut microbiota
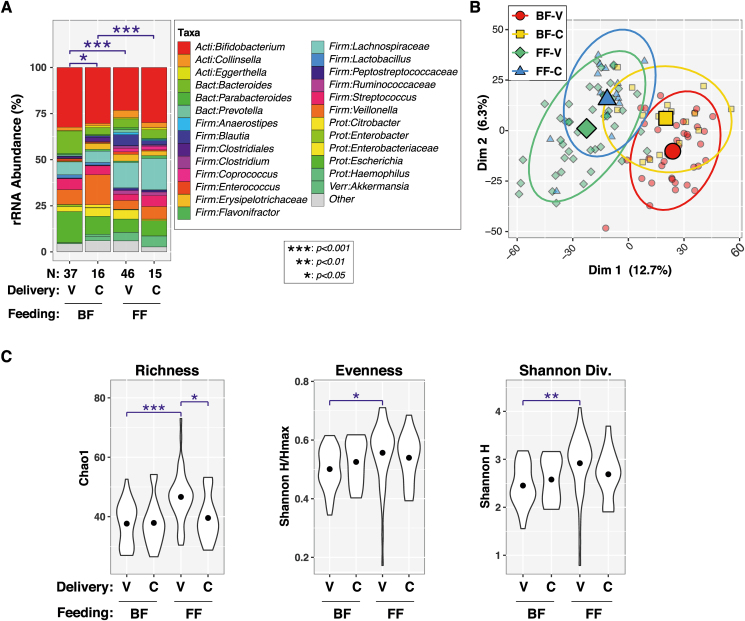
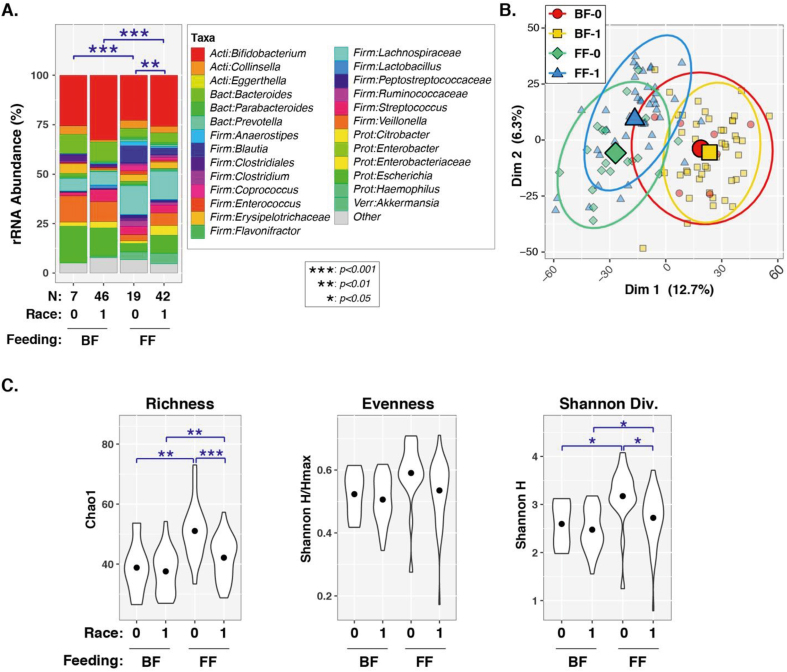
Besides feeding type, maternal and infant demographic/clinical variables were also examined. Univariable PERMANOVA tests (Table 2) found significant or marginally significant associations between overall microbiota composition (ie, β-diversity) and feeding type (P < 1 × 10-6), infant race (P = 0.001), delivery mode (P = 0.010), maternal weight (P = 0.0061), infant WAZ (P = 0.067), and infant WLZ (P = 0.079). On the contrary, infant sex, antibiotic use, maternal BMI, maternal height, or infant LAZ were not significantly associated with gut microbiota diversity (P > 0.05). Only race (P = 0.038) and delivery mode (P = 0.006) remained significant when feeding type was included as a covariate in bivariable PERMANOVA tests. A 3-variable PERMANOVA test showed that feeding type (P < 1e-06), race (P = 0.064), and delivery mode (P = 0.010) were all independently associated with β-diversity when adjusted for the other 2 variables (Table 2). Effects of delivery mode and race were further assessed within each feeding type (FIGURE 3, FIGURE 4). Significant differences in β-diversity were observed between vaginal delivery and cesarean delivery groups of BF infants (P = 0.025), whereas the difference was marginal for FF infants (P = 0.069). Vaginally delivered FF infants also had greater α-diversity (richness, evenness, and Shannon diversity) than the other infants (Figure 3C). Beta diversity was different between White and non-White FF infants (P = 0.0082), but not BF infants (P = 0.56). No differentially abundant taxa were identified for either delivery mode or race by FDR-corrected P values (Supplementary Figure 1).
TABLE 2.
Variables and their associations with the gut microbiota using 3 different models
| Variable | PERMANOVA P value1 |
||
|---|---|---|---|
| Univariable | Adjusted for feeding type | 3-variable model | |
| Feeding type | 1.00 × 10–6 | na | 1.00 × 10–6 |
| Race | 0.0011 | 0.038 | 0.064 |
| Sex | 0.290 | 0.157 | na |
| Delivery mode | 0.010 | 0.0060 | 0.010 |
| Antibiotics | 0.54 | 0.44 | na |
| Maternal BMI2 | 0.19 | 0.53 | na |
| Maternal Hgt | 0.51 | 0.39 | na |
| Maternal Wgt | 0.0061 | 0.28 | na |
| Infant LAZ | 0.40 | 0.52 | na |
| Infant WAZ | 0.067 | 0.79 | na |
| Infant WLZ | 0.079 | 0.61 | na |
Abbreviations: BMI, body mass index; LAZ, length-for-age z-score; na, not applicable; PERMANOVA, permutational analysis of variance; WAZ, weight-for-age z-score; WLZ, weight-for-length z-score.
Univariable PERMANOVA tests were used to test associations between overall microbiota composition (ie, β-diversity) and variables listed in the variable column–Aitchison dissimilarity. 106 permutations
Included underweight, normal weight, overweight, and obese categories.
FIGURE 3.
Effects of delivery mode and feeding type on α- and β-diversity in 4–5 mo-old breastfed (BF) and formula-fed (FF) infants. (A) Bar charts with PERMANOVA results. Percent relative abundance (%RA) of genus-level taxa, stratified by feeding type. Taxa with %RA <1% were collapsed into the “Other” category. The results of PERMANOVA tests are summarized above each plot for pairwise tests (blue lines/symbols). (B) Principal coordinates plot. Individual subjects are indicated by smaller symbols (circles/squares/diamonds/triangles), with group affiliations designated by symbol shapes and color coding. Mean PC values for each group along the x- and y-axes are indicated by larger shapes, whereas ellipses show 95% CI. (C) Alpha diversity. Violin plots show distributions of α-diversity indices by feeding type. Pairwise test results are indicated above each plot (blue lines/symbols). BF-V = BF infants born by vaginal delivery (n = 39), BF-C = BF infants born by cesarean delivery (n = 15), FF-V = FF infants born by vaginal delivery (n = 45), FF-C = FF infants born by cesarean delivery (n = 16). CI, confidence interval; PERMANOVA, permutational analysis of variance; rRNA, ribosomal ribonucleic acid; Dim 1 & Dim 2, Principal coordinates dimensions 1 and 2.
FIGURE 4.
Effects of race and feeding type on α- and β-diversity in 4–5 mo-old breastfed (BF) and formula-fed (FF) infants. (A) Bar charts with PERMANOVA results. Percent relative abundance (%RA) of genus-level taxa, stratified by feeding type. Taxa with %RA <1% were collapsed into the “Other” category. The results of PERMANOVA tests are summarized above each plot for pairwise tests (blue lines/symbols). (B) Principal coordinates plot. Individual subjects are indicated by smaller symbols (circles/squares/diamonds/triangles), with group affiliations designated by symbol shapes and color coding. Mean PC values for each group along the x- and y-axes are indicated by larger shapes, whereas ellipses show 95% CI. (C) Alpha diversity. Violin plots show distributions of α-diversity indices by feeding type. Pairwise test results are indicated above each plot (blue lines/symbols). BF-0 = BF infants of non-White race (n = 7), BF-1 = BF infants of White race (n = 47), FF-0 = FF infants of non-White race (n = 19), FF-1 = FF infants of White race (n = 42). CI, confidence interval; PERMANOVA, permutational analysis of variance; rRNA, ribosomal ribonucleic acid; Dim 1 & Dim 2, Principal coordinates dimensions 1 and 2.
Association of gut microbiota with infant growth parameters
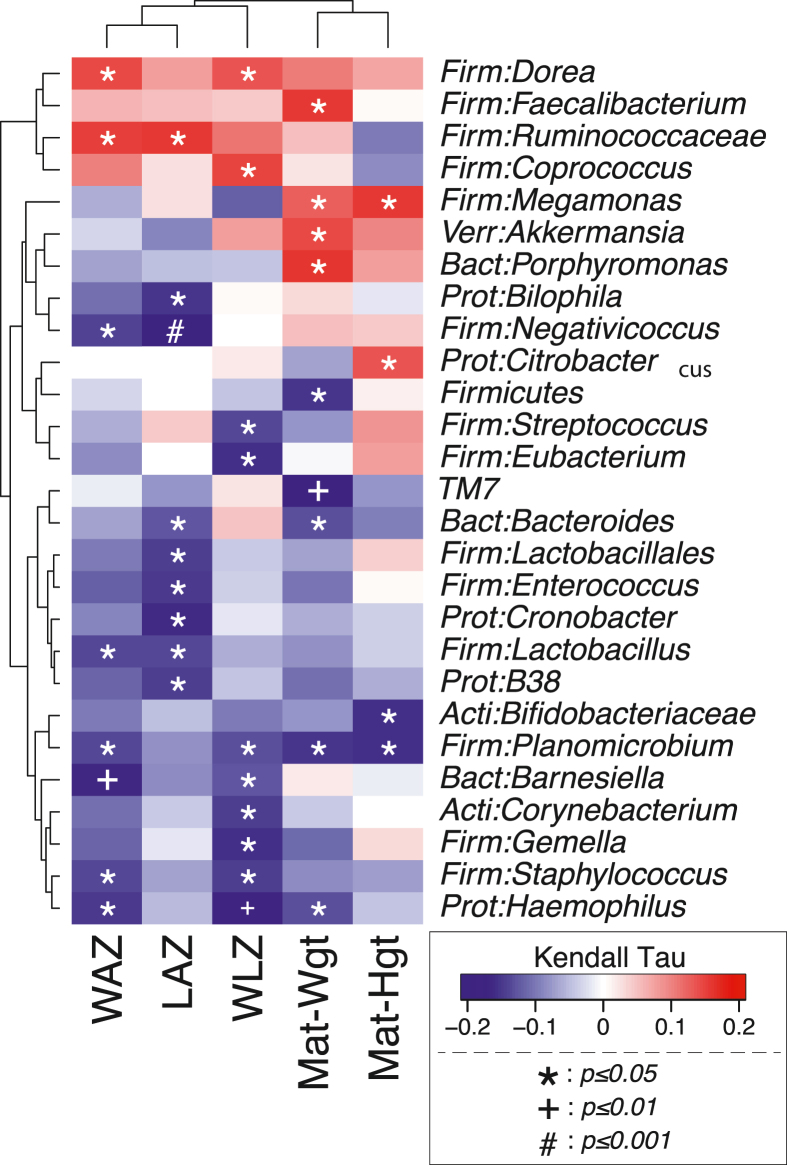
Associations between anthropometric measures and CLR-transformed microbiota count data were assessed by Pearson correlations. A heatmap of growth z-scores compared with genus-level infant gut microbiota is presented in Figure 5. There were several genera that were either positively or negatively associated with infant growth status and maternal height and weight. For example, unclassified Ruminococcaceae was positively associated with both WAZ (τ = 0.15; P < 0.05) and LAZ (τ = 0.16; P < 0.05). In contrast, Lactobacillus was negatively associated with WAZ (τ = –0.14; P < 0.05) and LAZ (τ = –0.14, P < 0.05). Maternal weight was positively associated with Akkermensia, and Barnesiella was negatively associated with WAZ WLZ.
FIGURE 5.
Associations between anthropometric z-scores and infant gut microbiota in 4–5 mo-old breastfed (BF) and formula-fed (FF) infants. Associations were assessed by Pearson correlations between anthropometric measures, and centered log-ratio (CLR) transformed microbiota count data. Darker blue indicates stronger negative Kendall rank correlation coefficients between taxa and growth parameters, whereas darker red indicates stronger positive Kendall rank correlation coefficients. LAZ = length-for-age z-score, WAZ = weight-for-age z-score, WLZ = weight-for-length z-score.
Discussion
Findings from the present study suggest that feeding type (exclusive breastfeeding compared with exclusive formula feeding) is a major determinant of the infant gut microbial composition and diversity in 4–5 mo-old United States infants. More importantly, the gut microbiota is associated with infant growth status. In the present study, Proteobacteria was higher in BF infants, although the actual abundance was relatively low (19% in BF and 12% in FF). Previous research [38] found lower Proteobacteria abundance in BF infants compared to non-BF infants. Proteobacteria have been shown to associate with adverse health conditions, such as necrotizing enterocolitis [39] and lower hematocrit [40] in preterm infants and slower growth trajectories and smaller brain volumes in mice [41]. The higher Proteobacteria abundance in BF did not appear to negatively affect infant growth status (Figure 5). Given that the participants were healthy, term United States infants and the overall Proteobacteria abundance was low; the observed higher Proteobacteria in BF did not appear to be concerning. Consistent with previous research [42,43], the present study also found a higher abundance of Bifidobacterium in BF infants. Although some research showed that the differences in Bifidobacterium by feeding type disappeared at 3 mo [1], the present study found that Bifidobacterium was still more abundant in BF at 4–5 mo of age, right before solid foods are normally introduced, indicating that the impact by feeding type could be robust and persistent in young infants. Although the present study did not find associations between infant growth status and Bifidobacterium, emerging research suggests a positive association between Bifidobacterium and weight gain in preterm infants [44,45]. One recent study showed that Bifidobacterium species B. infantis treatment/supplementation promoted weight gain in malnourished Bangladeshi infants [46]. It appears that in undesirable health conditions (preterm and malnutrition), Bifidobacterium could promote weight gain. However, its role in growth still needs to be explored in full-term, healthy infants.
Genus Ruminococcus is known as 1 of the SCFA producers, which plays a critical role in energy harvesting and could affect both linear growth and body weight [47]. Emerging research also has suggested that Ruminococcus may directly impact infant linear growth (eg, LAZ) as a “growth-discriminating” strain that is absent or low in growth-impaired infants or immature gut microbiota [48]. In a recent study of 118 Bangladeshi children, those who received a microbiota-directed complementary food prototype were linked to 21 bacterial taxa, including Ruminococcus, that were positively correlated with WLZ [14]. Although the present study did not identify Ruminococcus at the genus level to associate with growth z-scores, the family level Ruminococcaceae was positively associated with both LAZ and WAZ.
In general, during the first year of life, BF infants are leaner than FF infants [49]. In the present study, there was a higher relative abundance of Lactobacillus in BF compared with FF, which was negatively associated with both WAZ and LAZ. Similar to Bifidobacterium, Lactobacillus also promotes weight gain in preterm infants [50]. Thus, the effects of these bacteria need to be considered in the context of the study population (ie, preterm compared with healthy full-term infants). Another taxon Barnesiella was negatively associated with both WAZ and WLZ. Human milk oligosaccharides promote the growth of Barnesiella, which reduces infections in animal models [51]. Although there was no statistical difference in Barnesiella abundance between BF and FF, it could still partially explain the lower WAZ and WLZ in BF.
The present study showed that BF infants had lower gut microbiota α-diversity. Although most studies in developed settings [52,53] reported lower α-diversity with breastfeeding, studies of African infants [54] and Chinese infants [55] found that feeding type did not impact gut microbiota diversity, possibly because of potential environmental impact. As illustrated in Figure 3C, the higher α-diversity in FF appeared to be driven by vaginally delivered infants, and gut microbiota richness was higher in vaginally delivered infants. These findings were consistent with previous research [56].
Although recent studies suggest that varying formula sources may have differing effects on the infant microbiota [57], the present study showed no difference between formula components (protein size or percentage of whey, casein, or lactose) and the infant gut microbiota composition. This might be because of the limited sample size in some formula groups in our study (Table 2). Additionally, all infant formulas were cow milk-based instead of soy compared with cow milk-based, which might account for the limited difference in gut microbiota among FF infants [52].
One of the strengths of the present study was the inclusion of exclusively BF and FF infants. The formula feeding group was considered exclusive, although some infants received breastmilk (<2 wk within the first month of life). This allowed us to avoid possible influences of complicated mixed liquid diets and/or complementary feeding as potential confounders. Another strength is the inclusion of growth status assessment and their associations with the gut microbiota. Some limitations of the study include the homogeneity of the study population [58]. The small sample size of each of the various infant formula subtypes was a limitation and restricted our ability to robustly characterize the impact of formula type on the gut microbiota of exclusively FF infants.
In conclusion, feeding type is a major driver of the observed gut microbiota composition in 4–5 mo-old, healthy, full-term infants. Several genera were associated with infant growth status (growth z-scores), including some previously known taxa that promote weight gain in preterm infants, although their roles in infant growth appeared different in the current cohort. Gut microbiota could be a potential target of interest in directing future feeding guidelines during infancy.
Author contributions
The authors’ responsibilities were as follows – MT, NFK, and BEY: designed the study; MT, NFK, BEY, KND, and LMB: conducted the study; DNF, CER, and JMK: analyzed the samples; DNF, MT, and EO: analyzed the data. All authors wrote the paper, and all authors: read and approved the final manuscript.
Conflict of interest
The authors report no conflicts of interest.
Funding
This study was supported by the NIDDK 1K01DK111665, NIH (T32 DK007658, DK067009), NIH/National Center for Advancing Translational Sciences (NCATS) Colorado Clinical and Translational Science Awards (CTSA) grant number UL1 TR002535, NICHD F32-HD0978068, AHA, National Pork Board, Beef Checkoff, Foundation for Meat and Poultry Research and Education.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tjnut.2023.07.009.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Martin R., Makino H., Cetinyurek Yavuz A., Ben-Amor K., Roelofs M., Ishikawa E., et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLOS ONE. 2016;11(6) doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuoka K., Kanai T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015;37(1):47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018;9(1):141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Indias I., Cardona F., Tinahones F.J., Queipo-Ortuño M.I. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cukrowska B., Bierła J.B., Zakrzewska M., Klukowski M., Maciorkowska E. The relationship between the infant gut microbiota and allergy. The role of Bifidobacterium breve and prebiotic oligosaccharides in the activation of anti-allergic mechanisms in early life. Nutrients. 2020;12(4) doi: 10.3390/nu12040946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkar A., Yoo J.Y., Valeria Ozorio Dutra S., Morgan K.H., Groer M. The association between early-life gut microbiota and long-term health and diseases. J. Clin. Med. 2021;10(3) doi: 10.3390/jcm10030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penders J., Thijs C., van den Brandt P.A., Kummeling I., Snijders B., Stelma F., et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallani M., Young D., Scott J., Norin E., Amarri S., Adam R., et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010;51(1):77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 9.Borewicz K., Suarez-Diez M., Hechler C., Beijers R., de Weerth C., Arts I., et al. The effect of prebiotic fortified Infant Formulas on microbiota composition and dynamics in early life. Sci. Rep. 2019;9(1):2434. doi: 10.1038/s41598-018-38268-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagpal R., Kurakawa T., Tsuji H., Takahashi T., Kawashima K., Nagata S., et al. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: a quantitative assessment. Sci. Rep. 2017;7(1):10097. doi: 10.1038/s41598-017-10711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho N.T., Li F., Lee-Sarwar K.A., Tun H.M., Brown B.P., Pannaraj P.S., et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018;9(1):4169. doi: 10.1038/s41467-018-06473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nash M.J., Frank D.N., Friedman J.E. Early microbes modify immune system development and metabolic homeostasis-the ”restaurant” hypothesis revisited. Front Endocrinol. (Lausanne). 2017;8:349. doi: 10.3389/fendo.2017.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351(6275):854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 14.Chen R.Y., Mostafa I., Hibberd M.C., Das S., Mahfuz M., Naila N.N., et al. A microbiota-directed food intervention for undernourished children. N. Engl. J. Med. 2021;384(16):1517–1528. doi: 10.1056/NEJMoa2023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young B.E., Patinkin Z.W., Pyle L., de la Houssaye B., Davidson B.S., Geraghty S., et al. Markers of oxidative stress in human milk do not differ by maternal BMI but are related to infant growth trajectories. Matern. Child Health J. 2017;21(6):1367–1376. doi: 10.1007/s10995-016-2243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grummer-Strawn L.M., Reinold C., Krebs N.F. Centers for Disease Control and Prevention (CDC). Use of World Health Organization and CDC growth charts for children aged 0-59 months in the United States. MMWR Recomm. Rep. 2010;59(RR-9):1–15. [PubMed] [Google Scholar]
- 17.Frank D.N., Qiu Y., Cao Y., Zhang S., Lu L., Kofonow J.M., et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene. 2022;41(9):1269–1280. doi: 10.1038/s41388-021-02137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang M., Frank D.N., Hendricks A.E., Ir D., Esamai F., Liechty E., et al. Iron in micronutrient powder promotes an unfavorable gut microbiota in Kenyan infants. Nutrients. 2017;9(7) doi: 10.3390/nu9070776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank D.N. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D.J., Pace B., Olsen G.J., Stahl D.A., Sogin M.L., Pace N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U S A. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homo Sapiens UCSC Hg19 Human Genome Sequence From iGenome: Illumina. 2009. http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn Available from. [Google Scholar]
- 23.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing B., Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome. Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 25.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome. Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 26.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss P.D., Westcott S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011;77(10):3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruesse E., Peplies J., Glöckner F.O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic. Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson C.E., Harris J.K., Wagner B.D., Granger D., Browne K., Tatem B., et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29(23):3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R.C. Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A language and environment for statistical computing. Vienna, Austria. [Google Scholar]
- 32.Anderson M.J., Crist T.O., Chase J.M., Vellend M., Inouye B.D., Freestone A.L., et al. Navigating the multiple meanings of beta diversity: a roadmap for the practicing ecologist. Ecol. Lett. 2011;14(1):19–28. doi: 10.1111/j.1461-0248.2010.01552.x. [DOI] [PubMed] [Google Scholar]
- 33.Oksanen J., Blanchet G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. 2019. Vegan: Community Ecology Package.http://vegan.r-forge.r-project.org R Package version 2.5-7. Available from: [Google Scholar]
- 34.Fernandes A.D., Macklaim J.M., Linn T.G., Reid G., Gloor G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes A.D., Reid J.N., Macklaim J.M., McMurrough T.A., Edgell D.R., Gloor G.B. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57(1):289–300. [Google Scholar]
- 38.Lee S.A., Lim J.Y., Kim B.S., Cho S.J., Kim N.Y., Kim O.B., et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using Pyrosequencing. Nutr. Res. Pract. 2015;9(3):242–248. doi: 10.4162/nrp.2015.9.3.242. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pammi M., Cope J., Tarr P.I., Warner B.B., Morrow A.L., Mai V., et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5(1):31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho T.T.B., Kumar A., Louis-Jacques A.F., Dishaw L.J., Yee A.L., Groer M.W. The development of intestinal dysbiosis in anemic preterm infants. J. Perinatol. 2020;40(7):1066–1074. doi: 10.1038/s41372-020-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rendina D.N., Lubach G.R., Lyte M., Phillips G.J., Gosain A., Pierre J.F., et al. Proteobacteria abundance during nursing predicts physical growth and brain volume at one year of age in young rhesus monkeys. FASEB J. 2021;35(6) doi: 10.1096/fj.202002162R. [DOI] [PubMed] [Google Scholar]
- 42.Forbes J.D., Azad M.B., Vehling L., Tun H.M., Konya T.B., Guttman D.S., et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172(7) doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J., Li Z., Zhang W., Zhang C., Zhang Y., Mei H., et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci. Rep. 2020;10(1):15792. doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Younge N.E., Newgard C.B., Cotten C.M., Goldberg R.N., Muehlbauer M.J., Bain J.R., et al. Disrupted maturation of the microbiota and metabolome among extremely preterm infants with postnatal growth failure. Sci. Rep. 2019;9(1):8167. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Underwood M.A., Salzman N.H., Bennett S.H., Barman M., Mills D.A., Marcobal A., et al. A randomized placebo-controlled comparison of 2 prebiotic/probiotic combinations in preterm infants: impact on weight gain, intestinal microbiota, and fecal short-chain fatty acids. J. Pediatr. Gastroenterol. Nutr. 2009;48(2):216–225. doi: 10.1097/MPG.0b013e31818de195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barratt M.J., Nuzhat S., Ahsan K., Frese S.A., Arzamasov A.A., Sarker S.A., et al. Bifidobacterium infantis treatment promotes weight gain in Bangladeshi infants with severe acute malnutrition. Sci. Transl. Med. 2022;14(640) doi: 10.1126/scitranslmed.abk1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baxter N.T., Schmidt A.W., Venkataraman A., Kim K.S., Waldron C., Schmidt T.M. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1) doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351(6275) doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dewey K.G., Heinig M.J., Nommsen L.A., Peerson J.M., Lönnerdal B. Breast-fed infants are leaner than formula-fed infants at 1 y of age: the DARLING study. Am. J. Clin. Nutr. 1993;57(2):140–145. doi: 10.1093/ajcn/57.2.140. [DOI] [PubMed] [Google Scholar]
- 50.Spreckels J.E., Wejryd E., Marchini G., Jonsson B., de Vries D.H., Jenmalm M.C., et al. Lactobacillus reuteri colonisation of extremely preterm infants in a randomised placebo-controlled trial. Microorganisms. 2021;9(5) doi: 10.3390/microorganisms9050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss G.A., Chassard C., Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br. J. Nutr. 2014;111(9):1602–1610. doi: 10.1017/S0007114513004200. [DOI] [PubMed] [Google Scholar]
- 52.Brink L.R., Mercer K.E., Piccolo B.D., Chintapalli S.V., Elolimy A., Bowlin A.K., et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am. J. Clin. Nutr. 2020;111(6):1190–1202. doi: 10.1093/ajcn/nqaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugino K.Y., Ma T., Kerver J.M., Paneth N., Comstock S.S. Human milk feeding patterns at 6 months of age are a major determinant of fecal bacterial diversity in infants. J. Hum. Lact. 2021;37(4):703–713. doi: 10.1177/0890334420957571. [DOI] [PubMed] [Google Scholar]
- 54.Wood L.F., Brown B.P., Lennard K., Karaoz U., Havyarimana E., Passmore J.S., et al. Feeding-related gut microbial composition associates with peripheral T-cell activation and mucosal gene expression in African infants. Clin. Infect. Dis. 2018;67(8):1237–1246. doi: 10.1093/cid/ciy265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang R., Gao R., Cui S., Zhong H., Zhang X., Chen Y., et al. Dynamic signatures of gut microbiota and influences of delivery and feeding modes during the first 6 months of life. Physiol Genomics. 2019;51(8):368–378. doi: 10.1152/physiolgenomics.00026.2019. [DOI] [PubMed] [Google Scholar]
- 56.Rutayisire E., Huang K., Liu Y., Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 2016;16(1):86. doi: 10.1186/s12876-016-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabiano V., Indrio F., Verduci E., Calcaterra V., Pop T.L., Mari A., et al. Term Infant Formulas influencing gut microbiota: an overview. Nutrients. 2021;13(12) doi: 10.3390/nu13124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li R., Perrine C.G., Anstey E.H., Chen J., MacGowan C.A., Elam-Evans L.D. Breastfeeding trends by race/ethnicity among US children born from 2009 to 2015. JAMA Pediatr. 2019;173(12) doi: 10.1001/jamapediatrics.2019.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.