Abstract
Terpenes are volatile compounds responsible for aroma and the postharvest quality of commercially important xiangfei (Torreya grandis) nuts, and there is interest in understanding the regulation of their biosynthesis. Here, a transcriptomics analysis of xiangfei nuts after harvest identified 156 genes associated with the terpenoid metabolic pathway. A geranyl diphosphate (GPP) synthase (TgGPPS) involved in production of the monoterpene precursor GPP was targeted for functional characterization, and its transcript levels positively correlated with terpene levels. Furthermore, transient overexpression of TgGPPS in tobacco (Nicotiana tabacum) leaves or tomato (Solanum lycopersicum) fruit led to monoterpene accumulation. Analysis of differentially expressed transcription factors identified one basic helix-loop-helix protein (TgbHLH95) and one basic leucine zipper protein (TgbZIP44) as potential TgGPPS regulators. TgbHLH95 showed significant transactivation of the TgGPPS promoter, and its transient overexpression in tobacco leaves led to monoterpene accumulation, whereas TgbZIP44 directly bound to an ACGT-containing element in the TgGPPS promoter, as determined by yeast 1-hybrid test and electrophoretic mobility shift assay. Bimolecular fluorescence complementation, firefly luciferase complementation imaging, co-immunoprecipitation, and GST pull-down assays confirmed a direct protein–protein interaction between TgbHLH95 and TgbZIP44 in vivo and in vitro, and in combination these proteins induced the TgGPPS promoter up to 4.7-fold in transactivation assays. These results indicate that a TgbHLH95/TgbZIP44 complex activates the TgGPPS promoter and upregulates terpene biosynthesis in xiangfei nuts after harvest, thereby contributing to its aroma.
A bHLH-bZIP transcription complex regulates terpene biosynthesis by activating TgGPPS expression in Torreya grandis nuts during the postharvest ripening stage
Introduction
Xiangfei (Torreya grandis) is an evergreen coniferous tree belonging to the yew family (Taxaceae), subgenus Torreya. Xiangfei nuts have high nutritional value and potential health benefits attributed to a myriad of bioactive compounds, including sciadonic acid, tocopherol, squalene, and β-sitosterol (Suo et al. 2019). Roasted xiangfei nuts are valued by consumers for their unique flavor and aroma (Hu et al. 2022), with the latter being an important index for xiangfei nut quality evaluation. The desirable aroma results from a series of coordinated biosynthetic processes that occur under appropriate temperature and humidity conditions during postharvest ripening over a period of 10 to 15 days, resulting in a reduction in astringency, nutrient conversion, and aroma formation (Zhang et al. 2020b; Hu et al. 2022; Song et al. 2022). However, with the rapid development of the xiangfei industry and the associated prioritization of output, postharvest quality has declined, notably a reduction in the unique nut aroma (Li et al. 2005). To address this problem, there is interest in understanding the biosynthetic and regulatory processes that govern aroma formation during the ripening process.
Aroma is a major factor in the sensory quality of fruit and influences food flavor, thereby strongly affecting consumer preferences (El Hadi et al. 2013). Common aroma components in fruits and seeds belong to several chemical groups, including terpenes, esters, alcohols, aldehydes, ketones, and sulfur compounds, the profiles of which vary depending on the variety, developmental stage, postharvest management, and storage conditions (Schwab et al. 2008). The corresponding biosynthetic pathways include terpenoids, fatty acids, amino acids, and carbohydrates (Aragüez et al. 2013). Most studies of the regulation of aroma compounds in fruits have focused on horticulturally important fruits, and there are far fewer equivalent studies of aroma substances in woody nuts. Indeed, those that have been reported for nuts have mainly focused on differences in substance composition and content, and there are only a few reports describing the gene regulation related to nut aroma (Valdés García et al. 2021). In our previous research, 5 major types of aromatic components were identified in xiangfei nuts after harvest: terpenes, aldehydes, alcohols, alkanes, and esters. Among these, terpenes constituted the main aroma compounds during the postharvest ripening stage, accounting for 56% to 87% (Hu et al. 2022).
Volatile terpenoids, which include isoprene (C5), monoterpenes (C10), and sesquiterpenes (C15), are the most numerous and diverse class of secondary metabolites (Nagegowda 2010). In plants, these C5-isoprene building units are generated through either the cytoplasmic mevalonate (MVA) pathway or the plastidial 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway. Volatile sesquiterpenes are produced by the MVA pathway, whereas volatile hemiterpenes, monoterpenes, and diterpenes are produced by the MEP pathway (Vranová et al. 2013). Numerous plants have been identified that contain the genes for these 2 pathways’ enzymes, e.g. Arabidopsis (Arabidopsis thaliana) (Hong et al. 2012), tomato (Solanum lycopersicum; Zhou and Pichersky 2020), wormwood (Artemisia annua; Lv et al. 2016), grape (Vitis vinifera; Duchêne et al. 2009), and apple (Malus × domestica; Nieuwenhuizen et al. 2013). Interestingly, the identified key terpene biosynthetic genes are different in different species. For example, the identified key genes for the biosynthesis of grape terpenes encode a 1-deoxy-D-xylulose-5-phosphate synthase (DXS; VviDXS) and a terpene synthase (TPS; VviTPS52) (Battilana et al. 2009; Martin et al. 2010), while in apple peel, the identified α-farnesene biosynthesis genes are a 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR; MdHMGR) and an α-farnesene synthase (AFS; MdAFS) (Pechous and Whitaker 2004). Furthermore, biosynthesis of terpenes is affected by many environmental factors, and the biosynthetic genes can respond differently to different environmental factors. For example, UV-B can influence the production of α-farnesene, linalool, and nerolidol in peach (Prunus persica) fruit by regulating the terpene biosynthesis genes PpTPS1/2/3, respectively (Liu et al. 2017). Notably, the biosynthetic genes involved in terpene biosynthesis in postharvested xiangfei nuts remain to be identified.
The expression of multiple genes in metabolic pathways is regulated by transcription factors (TFs), which makes them suitable candidates for pathway engineering (Iwase et al. 2009). The synthesis of plant terpenes is known to be regulated by many TFs including members of the APETALA2/ethylene-responsive factor (AP2/ERF), basic-helix-loop-helix (bHLH), v-myb avian myeloblastosis viral oncogene homolog (MYB), NAM, ATAF1/2, CUC1/2 (NAC), WRKY, and basic leucine zipper (bZIP) families (Supplemental Table S1; Xu et al. 2019). For example, CitAP2.10 transcriptionally activates the CsTPS1 gene and regulates the biosynthesis of the sesquiterpene valencene in sweet orange (Citrus sinensis; Shen et al. 2016). The A. thaliana bHLH TF MYC2 interacts with DELLA (Asp-Glu-Leu-Leu-Ala motif containing) proteins and regulates the expression of sesquiterpene synthase TPS21/11 and mediate jasmonic acid-induced sesquiterpene (E)-β-caryophyllene production (Hong et al. 2012). As another example, AaNAC1 can bind to the AaADS promoter and upregulate its expression to increase artemisinin content and enhance drought stress tolerance (Lv et al. 2016).
In our previous report, terpenes were identified as the main aroma compounds in xiangfei nuts after harvest, and their content was greatly affected by temperature and humidity (Hu et al. 2022). However, biosynthetic genes and regulatory mechanisms underlying their generation during the postharvest ripening stage remain unclear. This study focused on the biosynthesis of terpenes after harvest with 2 goals: (i) screening and functional validation of key genes for terpene biosynthesis and (ii) analysis of transcriptional regulatory mechanisms of the related biosynthetic genes. We present insights into aroma production to be employed for improving the ripening quality of xiangfei and other woody nuts.
Results
TgGPPS expression positively correlates with terpene contents in harvested xiangfei (T. grandis) nuts
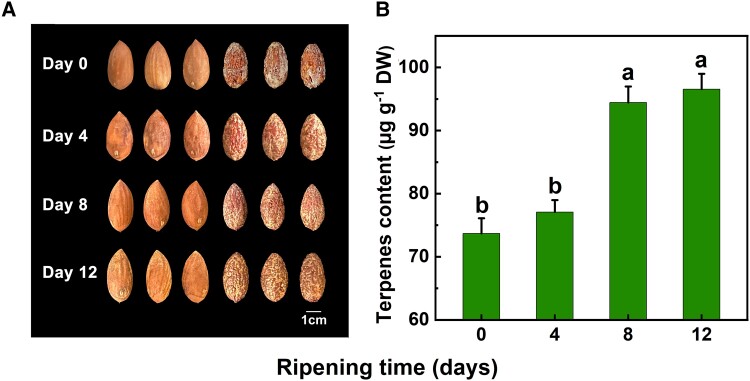
Terpenes were found to be the predominant aroma compounds in our previous study that evaluated the aroma components in raw xiangfei nuts throughout the postharvest ripening stage, and their levels significantly increased at high humidity (Hu et al. 2022). In this current study, the aroma composition of xiangfei nuts changed at various postharvest ripening stages, and 56 volatiles were found using gas chromatography-MS (GC-MS) analysis (Fig. 1A and Supplemental Fig. S1). As expected, terpenes were the main aroma compounds, and the concentrations were low from days 0 to 4, and increased significantly from days 4 to 8, then maintained stable until day 12, reaching a maximum of 96.53 µg/g on day 12 after harvest (Fig. 1B).
Figure 1.
Changes in terpene production in xiangfei (Torreya grandis) nuts during the postharvest ripening stage. A) Nuts were collected at different postharvest ripening times. Images were digitally extracted for comparison. Scale bars = 1 cm. B) Total terpene content of xiangfei nuts during the postharvest ripening stage. Error bars represent Se based on 3 biological replicates. The different letters indicate significant differences (P < 0.05) during the postharvest ripening stage by Duncan's multiple range test. DW, dry weight.
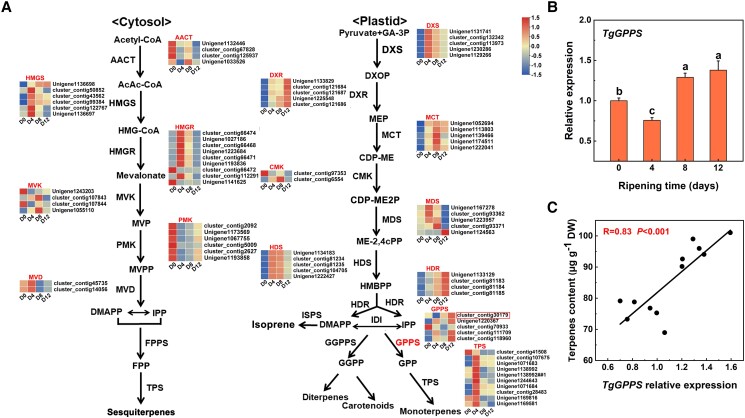
As shown in Fig. 2A, putative genes involved in postharvest terpene biosynthesis were identified by combining transcriptome data with reverse transcription quantitative PCR (RT-qPCR) data from days 4 and 8 (Fig. 2A). Transcript analysis revealed the upregulation of biosynthetic genes from the MVA pathway (AACT, HMGS, HMGR, MVK, PMK, and MVD), among which 8 genes were annotated to AACT, 11 genes annotated to HMGS, 27 genes annotated to the HMGR, 5 genes annotated to MVK, 7 genes annotated to PMK, and 3 genes annotated to MVD. DXS, DXR, MCT, CMK, MDS, HDS, HDR, GPPS, and TPS are the key biosynthetic genes in the MEP pathway. Here, we found 13 genes annotated to DXS, 8 genes annotated to DXR, 5 genes annotated to MCT, 3 genes annotated to CMK, 12 genes annotated to MDS, 5 genes annotated to HDS, 5 genes annotated to HDR, 8 genes annotated to GPPS, and 36 genes annotated to TPS (Supplemental Table S2). Highly expressed terpene biosynthetic genes were identified based on the fragments per kilobase of exon per million mapped fragments (FPKM) values, and a heatmap was created (Fig. 2A). A correlation analysis between the FPKM value and the total terpene levels revealed that a geranyl diphosphate synthase (GPPS) unigene (cluster_contig30179), subsequently called TgGPPS, was highly and positively correlated with the total terpene content (Fig. 2A). To confirm the transcriptome analysis, RT-qPCR analysis was performed and confirmed that the expression level of TgGPPS was significantly higher during days 4 to 8 after harvest (Fig. 2B), which was consistent with the total terpene content (R = 0.83, P < 0.001; Fig. 2C), consistent with a role for TgGPPS in terpene biosynthesis during the postharvest ripening stage.
Figure 2.
Gene expression and FPKM analysis of terpene biosynthetic genes in xiangfei nuts during the postharvest ripening stage. A) Biosynthesis of terpenoid volatile organic compounds and the FPKM values for terpenoid biosynthetic genes. The cytosolic MVA pathway produces sesquiterpenes. The plastidial MEP pathway produces monoterpenes, diterpenes, and volatile carotenoid derivatives. B) The TgGPPS expression pattern in xiangfei nuts during the postharvest ripening stage. Error bars represent Se based on 3 biological replicates. The different letters indicate significant differences (P < 0.05) during the postharvest ripening stage by Duncan's multiple range test. C) Correlation analysis between the total terpene content and TgGPPS expression. IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; AACT, acetyl-CoA acetyltransferase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase; MVK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD, mevalonate diphosphate decarboxylase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 50 -diphospho)-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4- cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase; GGPPS, geranylgeranyl pyrophosphate synthase; FPPS, farnesyl pyrophosphate synthase; DW, dry weight.
Effect of overexpressing TgGPPS on terpene production in tobacco (Nicotiana tabacum) leaves and tomato (S. lycopersicum) fruit
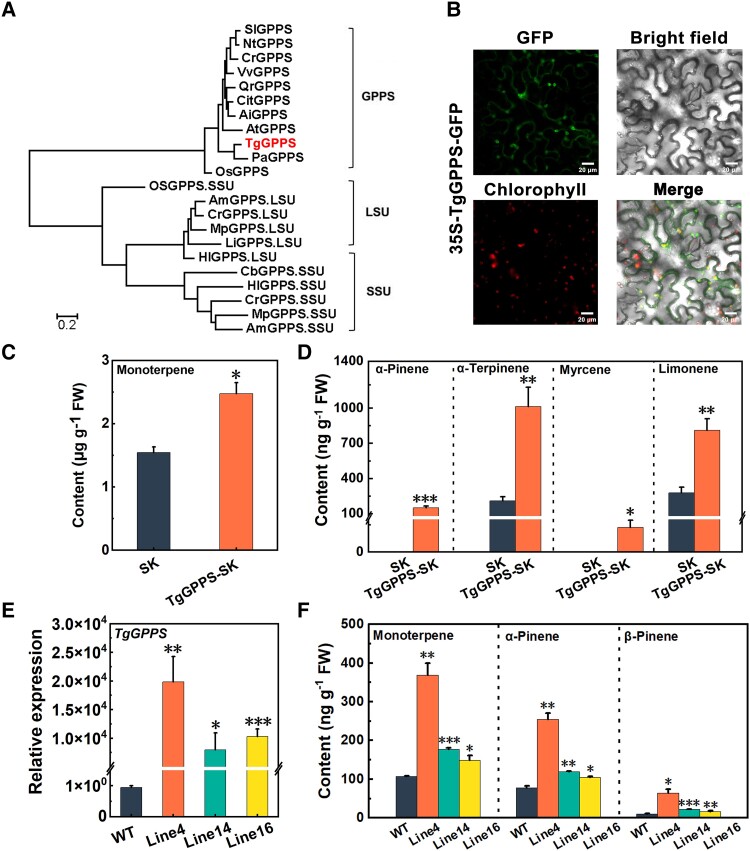
A comparison of several GPPS sequences revealed that TgGPPS included 2 important aspartate-rich (Asp-rich) motifs of the consensus sequence DD(X)2-4D (where X may be any amino acid) (Supplemental Fig. S2; Wang and Ohnuma 2000). A phylogenetic tree of TgGPPS and 21 other plant GPPS proteins from selected gymnosperm and angiosperm species (Fig. 3A) showed that TgGPPS clustered with the gymnosperm Picea abies PaGPPS, which has been described as a chloroplast-localized regulator of terpene synthesis (Fig. 3A; Schmidt and Gershenzon 2007, 2008). A subcellular localization assay was performed in N. benthamiana leaves, which indicated that TgGPPS was also located in the chloroplasts (Fig. 3B), further suggesting a role for TgGPPS in terpene biosynthesis.
Figure 3.
Functional characterization of TgGPPS. A) Phylogenetic analysis of TgGPPS and GPPS proteins from other species. All the sequences were obtained from the NCBI database. Accession numbers for these GPPSs are given in Materials and methods section. B) Subcellular localization analysis of TgGPPS. The GFP fluorescence indicates TgGPPS localization and the red fluorescence indicates chloroplast fluorescent protein (CFP). Scale bars = 20 µm. C) Transient overexpression of TgGPPS increases total monoterpene content in tobacco leaves. GC-MS identified 10 monoterpene components in tobacco leaves: α-pinene, β-pinene, myrcene, limonene, γ-terpinene, α-terpinene, terpinolene, linalool, geraniol, and menthol. D) Levels of α-pinene, α-terpinene, myrcene, and limonene in tobacco leaves transiently overexpressing TgGPPS or in leaves transformed with the empty vector (SK) as a control. The relative content was calculated according to the peak area of the internal standard (2-octanol). E) Relative expression of TgGPPS in transgenic tomato fruit. F) Overexpressing TgGPPS induces monoterpene synthesis in tomato. GC-MS detected 4 monoterpene components in tomato fruit: α-pinene, β-pinene, limonene, and linalool. SEs are all obtained from 3 biological replicates, and the significant differences are indicated with asterisks using Students's test (*P < 0.05, **P < 0.01 and ***P < 0.001). FW, fresh weight.
To confirm TgGPPS function, we used a highly efficient tobacco leaf transient expression assay. Agrobacterium tumefaciens containing TgGPPS-SK or empty SK vector was injected into tobacco (N. tabacum) leaves, and volatile substances were detected by GC-MS 1 wk post transfection. Compared with the negative control, TgGPPS overexpressing tobacco leaves exhibited 1.6-fold higher monoterpene levels (Fig. 3C and Supplemental Fig. S3). Major monoterpenes showing an increase included α-pinene and myrcene present at trace levels in the control (Fig. 3D). Moreover, α-terpinene and limonene showed 4.8- and 4.4-fold increases in overexpressing leaves, respectively (Fig. 3D).
We also performed a heterologous stable transgenic experiment in tomato (cv. MicroTom) plants. Five independent stable transgenic lines were obtained, of which the 3 showing the highest TgGPPS expression levels were selected for volatile analysis and RT-qPCR verification. Gene expression analysis confirmed that TgGPPS was overexpressed in the fruit (Fig. 3E), and compared with the wild type (WT), the monoterpene levels were 1.3 to 3.3 times higher in 3 independent transgenic plants. The abundance of α-pinene and β-pinene showed particularly significant increases (Fig. 3F and Supplemental Fig. S4). These results confirmed that TgGPPS is involved in terpene biosynthesis in xiangfei nuts during the postharvest ripening stage (Fig. 3).
TgbHLH95 functions as a positive indirect regulator in terpene biosynthesis
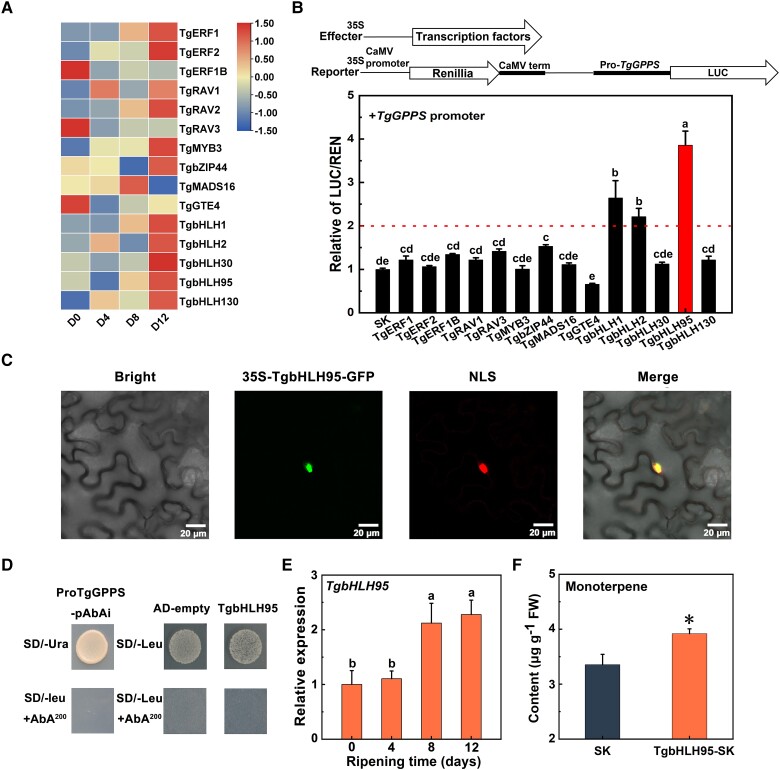
Following the confirmation of TgGPPS as a key gene in terpene biosynthesis, we next investigated mechanisms of its regulation. Differentially expressed TFs were identified using the RNA-seq database to further explore transcriptional regulatory mechanisms, and 15 TFs were identified with an expression pattern highly similar to that of TgGPPS (correlation index > 0.6 on days 4 to 8 of postharvest ripening) (Fig. 4A). The 15 TF sequences, which collectively were annotated as belonging to the AP2/ERF, MYB, bHLH, bZIP, MADS, and GTE families, were individually inserted into the pGreenII0029 62-SK vector, while the TgGPPS promoter sequence was cloned into the pGreen II 0800-LUC vector. The results of a dual-luciferase assay in N. benthamiana leaves (threshold was set as 2) suggested that 3 TFs activated the TgGPPS promoter: TgbHLH1, TgbHLH2, and TgbHLH95, with TgbHLH95 displaying the strongest activation effect, of approximately 3.9-fold (Fig. 4B).
Figure 4.
Functional characterization of TgbHLH95. A) Heatmap analysis of differentially expressed TFs. B) The regulatory effect of TFs on the TgGPPS promoter, the LUC/REN value of the empty SK vector on the TgGPPS promoter was set to 1.0, and Se values were calculated using 3 biological replicates. Different letters indicate significant differences (P < 0.05) during the postharvest ripening stage by Duncan's multiple range test. C) Subcellular localization analysis of TgbHLH95. The fluorescent signals were detected in tobacco cells cotransfected with TgbHLH95-GFP and the nuclear RFP-NLS marker. Scale bar = 20 μm. D) Yeast 1-hybrid analysis of TgbHLH95 binding capacity to the TgGPPS promoter, with the empty vector pGADT7 (AD) used as a negative control. E) The TgbHLH95 expression pattern in xiangfei nuts during the postharvest ripening stage. Error bars represent Se based on 3 biological replicates. The different letters indicate significant differences (P < 0.05) during the postharvest ripening stage by Duncan's multiple range test. F) Transient overexpression of TgbHLH95 increases total monoterpene content in tobacco leaves. Error bars represent Se based on 3 biological replicates. Significant differences are indicated with asterisks using Students's test (*P < 0.05).
TgbHLH95, which showed the strongest activation effect, was selected as a candidate to investigate terpene biosynthesis regulation. First, subcellular localization analysis suggested that TgbHLH95 was a nucleus-localized protein (Fig. 4C) consistent with the proposed TF function. Besides, a yeast 1-hybrid test was used to analyze the interactions between TgbHLH95 and the TgGPPS promoter. The growth of yeast containing TgbHLH95 was inhibited as was the empty vector, suggesting that TgbHLH95 binds indirectly to the TgGPPS promoter (Fig. 4D). Next, RT-qPCR data indicated that TgbHLH95 expression pattern was similar to that of TgGPPS, as both were significantly upregulated on days 4 to 8 after harvest (Fig. 4E). By transiently overexpressing TgbHLH95 in tobacco leaves, the monoterpene content greatly increased, from 3.35 µg/g toward 3.92 µg/g (Fig. 4F). Collectively, these results revealed that TgbHLH95 functioned as a positive indirect regulator activating TgGPPS expression to promote terpene production.
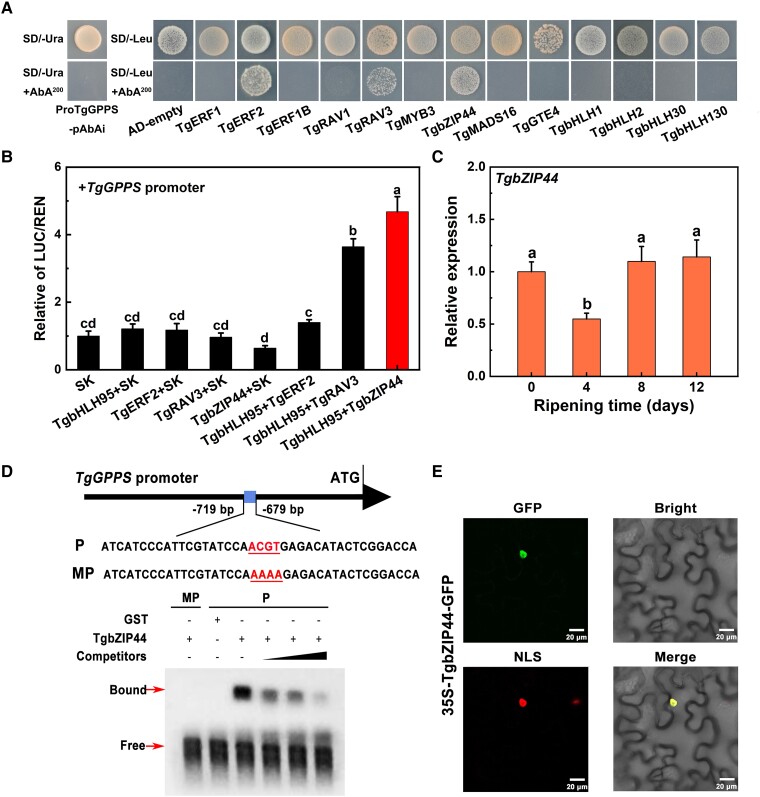
TgbZIP44 acts as a direct regulator in terpene biosynthesis
The gene expression results, the dual-luciferase assay, and the transient overexpression validation in tobacco leaves indicated that TgbHLH95 transactivated the TgGPPS promoter, but did not directly bind to it. Therefore, it is essential to discover other TFs that can interact with TgbHLH95, which may serve as a bridge in connecting TgbHLH95 and TgGPPS. A yeast 1-hybrid assay was used to analyze the interactions between other differentially expressed TFs and TgGPPS promoter. The results revealed that TgERF2, TgRAV3, and TgbZIP44 showed normal growth under 200 ng ml−1 aureobasidin A (AbA) compared with the empty vector (Fig. 5A). This indicated that TgERF2, TgRAV3, and TgbZIP44 could directly bind to the TgGPPS promoter, suggesting that the activation by TgbHLH95 might occur through a transcriptional complex with TgERF2, TgRAV3, and TgbZIP44. Accordingly, we next investigated whether TgbHLH95 and TgERF2, TgRAV3 and TgbZIP44 are synergistically involved in terpene biosynthesis. In a dual-luciferase assay, TgbHLH95 and TgbZIP44 in combination showed the strongest induction of the TgGPPS promoter, with a LUC/REN ratio of 4.68, compared with transfection of TgbHLH95 (1.21-fold activation) or TgbZIP44 (0.64-fold activation) in combination with the empty vector (Fig. 5B). The combination of TgbHLH95 and TgRAV3 showed a relatively lower additive effect (3.64-fold activation), and TgbHLH95 with TgERF2 showed no additive effect (Fig. 5B). Consequently, TgbZIP44 was chosen for further study. The expression pattern of TgbZIP44 via RT-qPCR analysis was similar to that of TgGPPS, which were significantly upregulated on days 4 to 8 (Fig. 5C). In vitro experiments using electrophoretic mobility shift assay (EMSA) revealed that TgbZIP44 could directly recognize and bind to the ACGT-containing elements in the TgGPPS promoter (Fig. 5D). When the specific ACGT element was mutated by AAAA sites, the putative TgbZIP44 protein binding was eliminated. Meantime, the binding affinity of the biotinylated probe was substantially reduced as the concentration of the competitor (cold probe) increased (Fig. 5D). In addition, subcellular localization of TgbZIP44 showed a strong fluorescent signal in the nucleus, indicating TgbZIP44 as a nucleus-localized protein (Fig. 5E). Their colocalization implied that TgbHLH95 and TgbZIP44 might form a protein–protein interaction in the nucleus. Collectively, the combination of TgbHLH95 and TgbZIP44 was chosen for further protein–protein interaction assays.
Figure 5.
Functional characterization of TgbZIP44. A) Yeast 1-hybrid analysis of other TFs binding capacity to the TgGPPS promoter, with the empty vector pGADT7 (AD) used as a negative control. B) Synergistic transactivation effect of different TFs on the TgGPPS promoter. The LUC/REN ratio of the empty vector (SK) plus the promoter was used as a calibrator (value set to 1.0). Error bars represent Se based on 3 biological replicates. The different letters indicate significant differences (P < 0.05) by Duncan's multiple range test. C) The TgbZIP44 expression pattern in xiangfei nuts during the postharvest ripening stage. Error bars represent Se based on 3 biological replicates. The different letters indicate significant differences (P < 0.05) during the postharvest ripening stage by Duncan's multiple range test. D) In vitro binding ability of TgbZIP44 to the TgGPPS promoter performed by EMSA. The presence (+) or absence (−) of specific probes is marked. The black arrows become larger from left to right indicating an increase in the concentration of competitive probes. GST protein was used as a negative control. E) Subcellular localization analysis of TgbZIP44. The fluorescent signals were detected in tobacco cells cotransfected with TgbZIP44-GFP and the nuclear RFP-NLS marker. Scale bar = 20 μm.
A TgbHLH95/TgbZIP44 complex activates the TgGPPS promoter
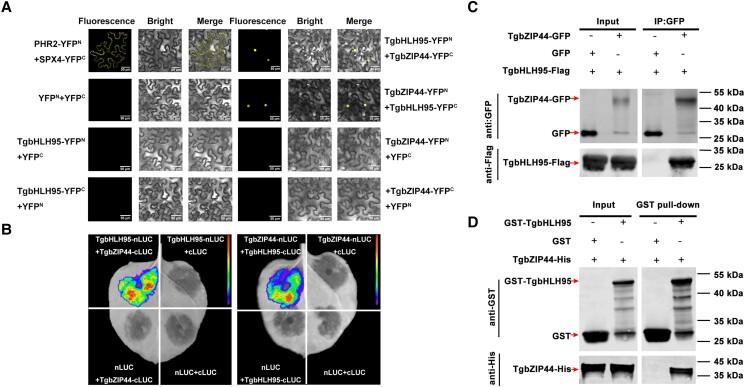
To test if TgbHLH95 interacts with TgbZIP44, transient bimolecular fluorescence complementation (BiFC) assays were conducted accordingly. N. benthamiana leaves cotransformed with PHR2-YC, and SPX4-YN (positive controls) showed a YFP fluorescent signal in the cytoplasm, in contrast to those expressing the combination of a single construct (i.e. TgbHLH95-YFPN + YFPC, TgbHLH95-YFPC + YFPN, TgbZIP44-YFPN + YFPC, and TgbZIP44-YFPC + YFPN) and the corresponding bidirectional empty vector (negative controls), which did not produce any YFP signal (Fig. 6A). TgbHLH95-YFPC cotransformed with TgbZIP44-YFPN or TgbHLH95-YFPN cotransformed with TgbZIP44-YFPC displayed a nuclear YFP fluorescent signal (Fig. 6A), indicating that TgbHLH95 could interact with TgbZIP44 in the nucleus. The interaction between TgbHLH95 and TgbZIP44 was also examined using a luciferase complementation imaging (LCI) assay to confirm the results obtained by BiFC. When nLUC-TgbHLH95 and cLUC-TgbZIP44 or nLUC-TgbZIP44 and cLUC-TgbHLH95 were coinfiltrated into N. benthamiana leaves, a high LUC luminescence signal showed (Fig. 6B), whereas the negative controls had no detectable LUC activity (the combinations of nLUC + cLUC, nLUC + cLUC-TgbHLH95, nLUC-TgbHLH95 + cLUC, nLUC + cLUC-TgbZIP44, nLUC-TgbZIP44 + cLUC) (Fig. 6B). A co-immunoprecipitation (Co-IP) test was also used to confirm the interaction between TgbHLH95 and TgbZIP44. N. benthamiana leaf protein samples were utilized for immunoprecipitation using an anti-GFP antibody. After that, anti-GFP and anti-Flag antibodies were used to immunoblot the eluted protein. The outcome demonstrated that the TgbZIP44-GFP fusion protein interacted with TgbHLH95 in N. benthamiana leaves in vivo, but not the GFP negative control (Fig. 6C). An in vitro GST pull-down test was utilized to further validate the interaction between the TgbHLH95 and TgbZIP44 proteins. The recombinant TgbZIP44-His protein was incubated in vitro with the recombinant TgbHLH95-GST protein and the GST control. Anti-GST and anti-His antibodies were used to immunoblot the protein after it had been eluted using GST-tag beads. The results showed that TgbHLH95-GST was pulled down, but GST alone was not. Therefore, TgbZIP44-His interacted directly with the TgbHLH95-GST protein (Fig. 6D). Collectively, BiFC, LCI, Co-IP, and GST pull-down assays all revealed that TgbHLH95 and TgbZIP44 formed a protein–protein complex in vivo and in vitro (Fig. 6).
Figure 6.
TgbHLH95 protein interacts with TgbZIP44. A) BiFC analysis of the interaction between TgbHLH95 and TgbZIP44. The pairs of fusion proteins tested were TgbHLH95-YFPN + TgbZIP44-YFPC and TgbZIP44-YFPN + TgbHLH95-YFPC. PHR2-YN and SPX4-YC were used as the positive control; YFPN and YFPC were used as negative controls. Nuclear location signal (NLS) indicates the position of the nucleus. Scale bars = 50 µm. B) LCI analysis of the interaction between TgbHLH95 and TgbZIP44. Tobacco leaves were infiltrated with Agrobacterium strain GV3101 harboring different constructs. The luminescence signals of the infiltrated areas were measured 48 h after infiltration. The experiment was repeated at least 3 times. C) Co-IP assay of TgbHLH95 and TgbZIP44. TgbHLH95-Flag plus TgbZIP44-GFP proteins were immunoprecipitated with an anti-GFP antibody and immunoblotted with anti-GFP and anti-Flag antibodies. D) Pull-down assay detection of the interaction of TgbHLH95 and TgbZIP44. The recombinant TgbHLH95-GST or GST was incubated with TgbZIP44-His. Blots were probed with anti-GST and anti-His antibody.
Discussion
TgGPPS is a key terpene biosynthetic gene in xiangfei (T. grandis) nuts after harvest
GPPS (EC 2.5.1.1) is the main enzyme responsible for the production of the monoterpene precursor GPP (Dudareva et al. 2006), and studies in several plant species have shown that GPPS can form homodimers or heterodimers (Rai et al. 2013). Heterodimeric GPPS has been identified in the leaves of the angiosperms peppermint (Mentha piperita), snapdragon (Antirrhinum majus), and hops (Humulus lupulus) (Bouvier et al. 2000; Schmidt and Gershenzon 2008; Wang and Dixon 2009) and consist of large subunits (LSUs) and small subunits (SSU). For example, LSU and SSU in heterogenous GPPS in peppermint have no catalytic activity alone, while the interaction between the 2 subunits leads to the formation of active GPPS. Structural studies also showed that LSU acts as a catalytic unit, while SSU acts as a regulatory unit (Chang et al. 2010). In gymnosperms, only homodimer GPPS complexes have been identified to be involved in terpene biosynthesis, such as PaGPPS in Picea abies (Schmidt and Gershenzon 2007). This current study showed a phylogenetic clustering of TgGPPS and PaGPPS, consistent with its involvement in terpene biosynthesis (Fig. 3A). GPPS has also been located in the nongreen plastids of the active MEP pathway and in chloroplasts of photosynthetic cells (Bouvier et al. 2000). Subcellular localization analysis indicated that TgGPPS was localized in chloroplasts (Fig. 3B), further revealing its role in terpene biosynthesis. In addition, the TgGPPS transcript levels were positively correlated with the total terpene content after harvest (Figs. 1 and 2). Furthermore, transient heterologous overexpression of TgGPPS in tobacco (N. tabacum) leaves or tomato (S. lycopersicum) fruit resulted in a significant increase in monoterpene levels (Fig. 3, C to F), indicating that TgGPPS contributes to terpene biosynthesis in nuts after harvest.
TgbHLH1, TgbHLH2, and TgbHLH95 are positive regulators of terpene biosynthesis in xiangfei nuts after harvest
There are many reports describing the transcriptional regulation of terpene biosynthesis, but the regulatory mechanisms acting on GPPS are less well studied. One such study, performed in spearmint (M. spicata), showed that MsMYB inhibits GPPS.LSU expression to reduce the terpene content (Reddy et al. 2017); however, the role of other TFs on GPPS expression remains to be elucidated. In the current study, 3 bHLH TFs were identified that exerted significant activation effects on the TgGPPS promoter. TgbHLH95 showed the highest activation effect, gene expression, subcellular localization analysis, and transient overexpression assays in tobacco indicated that it was indeed a positive regulator of terpene biosynthesis, although it did not bind directly to the TgGPPS promoter in Y1H assay (Fig. 4). The expression analysis also revealed that TgbHLH1 and TgbHLH2 expression increased significantly on days 4 to 8 after harvest (Supplemental Fig. S5). Furthermore, tobacco overexpression assays showed that overexpression of TgbHLH1 significantly increased terpene content, although overexpression of TgbHLH2 did not (Supplemental Fig. S6), indicating that TgbHLH1 also indirectly regulates TgGPPS, while TgbHLH2 is not involved in terpene biosynthesis. This study suggested that TgbHLH95 and TgbHLH1 activate TgGPPS expression to promote terpene production.
The proteins in the plant bHLH TF family, the second-largest TF family, have a conserved bHLH domain that has a length of about 60 amino acids (Feller et al. 2011). Members of the bHLH family have been identified and characterized in many species, such as A. thaliana and apple (Pires and Dolan 2010; Mao et al. 2017). These bHLH TFs play important roles in various metabolic pathways, including hormone synthesis (Qi et al. 2014), stress response (Fan et al. 2014), and metabolite (flavonoid, alkaloid, terpenoid) biosynthesis (Xie et al. 2017). In many plant models, the contribution of bHLH members to terpene biosynthesis has been demonstrated (Hong et al. 2012; Xu et al. 2018). In tomato, the bHLH TF SlMYC1 inhibits sesquiterpene production in stem trichomes while acting as a positive regulator of monoterpene biosynthesis in leaf and stem trichomes (Xu et al. 2018). In peach fruit, PpbHLH1 directly bound to the PpTPS3 promoter to regulate production of the key aroma compound linalool (Wei et al. 2021). In xiangfei nuts, TgbHLH1 and TgbHLH95 were shown to be involved in postharvest terpene biosynthesis through indirect regulation of the TgGPPS promoter. These studies suggested that bHLH family members play an important role in the regulation of terpene synthesis.
A TgbHLH95/TgbZIP44 transcription complex promotes terpene production via activation of the TgGPPS promoter
Terpene production has been linked to a number of TFs, but most of these TFs act alone on individual biosynthetic genes. For example, MdERF3 and MdMYC2 regulate α-farnesene synthesis through the activation of expression of the terminal enzyme MdAFS. Among these, MdERF3 can directly bind to the DRE element of the MdAFS promoter (Wang et al. 2020). Xu et al. (2017) reported that MdMYC2 can activate MdERF3 transcription by binding to the MdERF3 promoter, i.e. MdMYC2 can act as an upstream regulator of MdERF3 and coregulate it to induce α-farnesene synthesis in apple, further expanding this regulatory network. Less is known about the transcriptional complexes that regulate terpene production, although a MYB-bHLH protein complex has been identified (Yang et al. 2020). In flowers of Freesia hybrida, FhMYB21 substantially activated FhTPS1 expression, while FhMYC2 showed an inverse expression pattern compared with FhTPS1. Upon FhMYC2 coexpression with FhMYB21, FhMYC2 reduced the activation effect of FhMYB21 through interactions between the 2 TFs, demonstrating that the MYB-bHLH complex cooperated in the regulation of linalool synthesis. The mechanism of MYB-bHLH complexes in regulating terpene aromatics is conserved in plants such as A. thaliana (Yang et al. 2020). This report shows a bHLH-bZIP transcription complex regulating terpene synthase genes (Fig. 7). TgbHLH95 indirectly activates the TgGPPS promoter, while TgbZIP44 binds directly to it, with formation of a TgbHLH95/TgbZIP44 protein complex participating in terpene biosynthesis (Figs. 4 to 6). These findings add to our understanding of the regulatory mechanisms affecting terpene biosynthesis in nuts and provide a foundation for further research into terpene biosynthesis in other woody nuts.
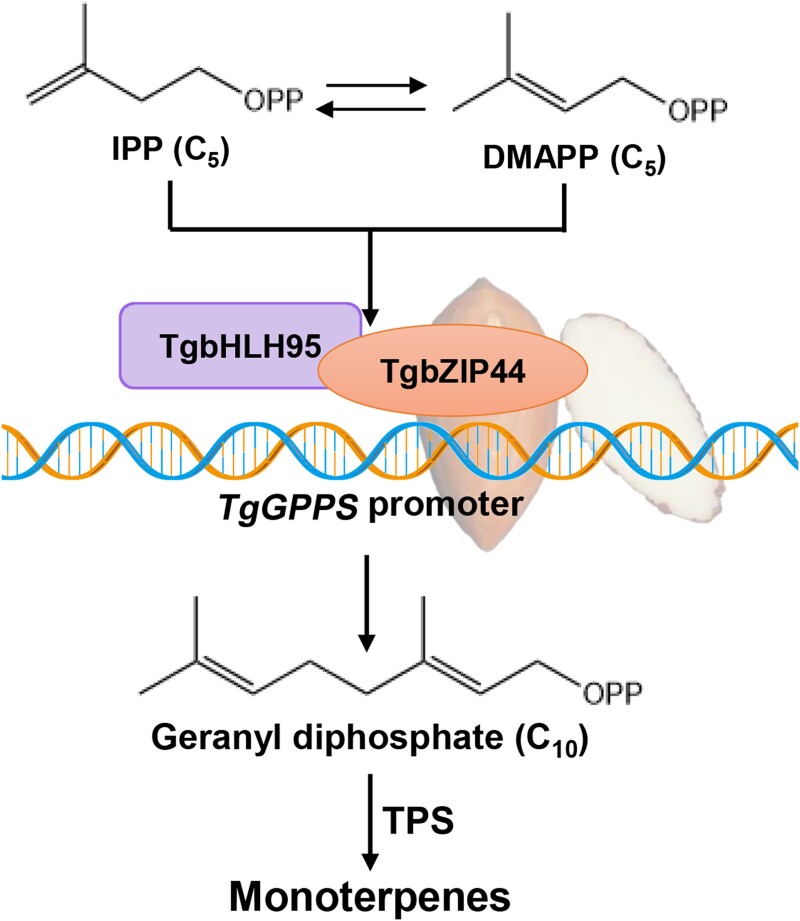
Figure 7.
A proposed regulatory model of the TgbHLH95-TgbZIP44 protein complex promoting terpene accumulation via activation of TgGPPS expression in xiangfei nuts during the postharvest ripening stage.
Conclusions
Accumulation of terpenes in xiangfei nuts after harvest is important for nut quality and consumption. Here, based on both in vitro and in vivo assays, a TgGPPS gene was cloned and characterized encoding a protein capable of promoting terpene production. TgbHLH95 activated TgGPPS transcription indirectly, and TgbHLH95 overexpression promoted the terpene content in tobacco leaves. Y1H and EMSA analysis showed that TgbZIP44 directly bound to the TgGPPS promoter, and protein binding assays further confirmed an interaction between TgbHLH95 and TgbZIP44, with a synergistic effect in combination on the TgGPPS promoter to regulate the synthesis of terpenes. These hypotheses, identified in xiangfei nuts, have yet to be examined in other tree nut species. Knowledge of the regulation of volatiles’ biosynthesis pathways can facilitate future efforts to enhance nut flavor quality and screen for in breeds or genetically modified plants showing the best aroma composition targeting certain terpenes.
Materials and methods
Plant materials and experimental treatments
Xiangfei (T. grandis cv. “Merrillii”) nuts were harvested 525 days after full bloom from an orchard in Xinchang, Zhejiang, China. On the day of harvest, the nuts were brought to the laboratory. The arils were manually removed, thoroughly cleaned with clear water, and then allowed to air dry until there was no more moisture present outside the shell. Nuts with uniform size and free of visible wounding were selected and divided into 4 groups. They were kept at controlled temperature conditions (20 °C ± 2 °C) and relative humidity (RH) levels (90% ± 2% RH), and 30 nuts were sampled on days 0, 4, 8, and 12 in 3 biological replicates. After removing the shell, the kernels were frozen in liquid nitrogen and stored at −80 °C for further studies.
Tobacco (N. tabacum) plants for transient overexpression, N. benthamiana plants for dual-luciferase assay, and tomato (S. lycopersicum, cv. MicroTom) plants for stable overexpression were grown in a greenhouse with a light/dark cycle of 16:8 h at 25 °C.
Analysis of volatiles by GC-MS
Volatiles were analyzed according to Zhang et al. (2010). Frozen xiangfei kernel tissue (1 g) and whole tomato fruit tissue (5 g) were ground into a powder in liquid nitrogen, which was then transferred to vials containing 5 mL of saturated NaCl solution. Frozen and ground N. tabacum leaves (1 g) were transferred to a vial containing 1.5 mL 200 mM EDTA and 1.5 mL 20% (w/v) CaCl2 solution. Subsequently, 2-octanol (0.8 mg/mL, 10 µL) was added as the internal standard, and then the vials were sealed. Agilent 7890A gas chromatograph and 5975C mass spectrometer linked to a solid-phase microextraction autosampler (Combi PAL, CTC Analytics, Agilent Technologies) homogenized and analyzed the mixture. A fiber coated with 65 μm of polydimethylsiloxane and divinylbenzene from the Supeclo Co. in Bellefonte, Pennsylvania, was used to capture volatiles. On a DB-WAX column (30 m × 0.25 mm, 0.25 μm phase film thickness, Agilent, USA), volatiles were separated. The flow rate of the helium carrier gas was 1 mL/min. The temperature program began at 40 °C and increased to 100 °C and 245 °C at rates of 3 °C/min and 5 °C/min, respectively. The column effluent was ionized at an energy of 70 eV with a source temperature of 230 °C and a transfer temperature of 250 °C. Volatiles were discovered by comparing electron ionization mass spectra and retention time data with the NIST Mass Spectral Library (NIST-08 and Flavor). The peak area of the internal standard was used as a base to calculate the amount of volatiles using the total ion chromatogram.
RNA-seq analysis
Total RNA was isolated from xiangfei kernel using the Hexadecyl trimethyl ammonium Bromide (CTAB) method (Chang et al. 1993). Then using agarose gel electrophoresis and a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, USA), the quantity and quality of the total RNA were evaluated. The cDNA libraries were created using a total of 2 g of RNA samples in accordance with the recommended procedure. The Illumina HiSeq 4,000 platform was used for the paired-end sequencing (Illumina Inc., California, USA). Using the Trinity software, the high-quality sequence reads were further combined to obtain unigenes after the low-quality reads, and adaptor sequences were filtered out (version 6.0). Six public databases, including the gene ontology, NCBI nonredundant protein sequences, NCBI nonredundant nucleotide sequences, SwissProt protein sequence database, and Clusters of Orthologous Groups of Proteins, were used to annotate putative terpene associated biosynthetic genes in xiangfei transcriptome database (Supplemental Table S2). Differentially expressed genes were those with a |log2FoldChange| > 1 and an adjusted P-value ≤ 0.05.
RNA isolation and RT-qPCR
Genome DNA in the total RNA was eliminated with the gDNA eraser. The PrimeScript RT regent Kit (Takara, Dalian, China) was used to create first-strand cDNA from 1 g of RNA after which it was diluted with water (1: 10). Utilizing the CFX96 equipment and the Ssofast Eva Green Supermix Kit, RT-qPCR was carried out (Bio-Rad, USA). Melting curves and product sequencing were used to confirm the specificity of primers before use. NCBI/Primer-BLAST was used out for primer design. The reaction mixture, which had a total volume of 20 µL, consisted of the following ingredients: 10 µL of SYBR PCR supermix (Bio-Rad, California, USA), 6 µL of diethylpyrocarbonate-treated water, 2 µL of cDNA template diluted 5 times, and 1 µL of each primer (10 µM) (Zhang et al. 202 °c). Data were processed, and relative expression levels were computed using the 2(−△△Ct) method to look at the differential expression of genes (Zhang et al. 2020b). The relative expression of control N. tabacum leaves expressing an empty vector (SK) was set as 1 for the transient overexpression assay. The relative expression in WT fruit was set as 1.0 for overexpression in tomato (cv. MicroTom). Supplemental Table S3 provides a list of the RT-qPCR primers.
Gene isolation, promoter cloning, and analysis
The coding sequence of the differentially expressed TFs and the promoter were cloned based on the RNA-seq data. The 1,426 bp-long TgGPPS promoter was amplified by PCR, sequenced, and subjected to an in-silico study starting from the start codon. The online tool http://bioinformatics.psb.ugent.be/webtools/plantcare/html/was used to analyze the TgGPPS promoter cis-element. Supplemental Table S4 contains a list of the primers.
Phylogenetic tree construction
The MEGA 7.0 program (Kumar et al. 2016) was used to align the full-length GPPS amino acid sequences and create a neighbor-joining tree. NCBI accession numbers are as follows: pedunculate oak QrGPPS (Quercus robur; XP_050287822.1), Norway spruce PaGPPS (Picea abies; ACA21459.1), grape VvGPPS (V. vinifera; XP_002268229.2), tomato SlGPPS (S. lycopersicum; NP_001234089.1), Clarkia brewer CbGPPS.SSU (AY534745), tobacco NtGPPS (N. tabacum; NP_001312253.1), satsuma mandarin CitGPPS (Citrus unshiu; AAN86061.1), neem AiGPPS (Azadirachta indica; AIG15448.1), peppermint MpGPPS.SSU (M. piperita; AAF08792.1), MpGPPS.LSU (AAF08793.1), hops HlGPPS.SSU (H. lupulus; ACQ90681), HlGPPS.LSU (ACQ90682), rice OsGPPS (Oryza sativa; XP_015644451.1), OsGPPS.SSU (EAY87007), Arabidopsis AtGPPS (A. thaliana; CAC16849.1), snapdragon AmGPPS.SSU (A. majus; AAS82859), AmGPPS.LSU (AAS82860.1), madagaskar periwinkle CrGPPS (Catharanthus roseus; AGL91647.1), CrGPPS.SSU (AGL91646.1), CrGPPS.LSU (AGL91645.1), and lavandin LiGPPS.LSU (Lavandula × intermedia; QHN60320.1).
Subcellular localization analysis
The full-length TgGPPS, TgERF2, TgRAV3, TgbZIP44, and TgbHLH95 coding sequences without stop codons were independently ligated into the pCAMBIA1302 vectors. Supplemental Table S4 provides a list of the primers used in vector construction. Following electroporation into A. tumefaciens GV3101, all of the recombinant vectors were transiently expressed in transgenic N. benthamiana leaves (Huang et al. 2013). The A. tumefaciens culture was brought to an OD600 of 0.75 using the infiltration buffer, which contains 10 mM MES, 10 mM MgCl2, and 150 mM acetosyringone at a pH of 5.6. Using a confocal laser scanning microscope (LSM510, Karl Zeiss, Germany), GFP fluorescence was measured and observed 2 days after infiltration (Zhang et al. 2020b). The excitation wavelength for GFP fluorescence was 488 nm, and fluorescence was detected at 490 to 520 nm. The excitation wavelength for NLS fluorescence was 594 nm, and fluorescence was detected at 612 to 635 nm.
Transient overexpression in N. tabacum leaves
Full-length cDNA of TgGPPS and TgbHLH95 sequences was separately cloned into the pGreenII0029 62-SK (SK) vector and transformed into Agrobacterium strain GV3101. Cultures were grown to OD600 = 0.75 with the same infiltration buffer as in subcellular localization analysis, and 1 mL was infiltrated into N. tabacum leaves until the whole leaf was infiltrated. Seven days after infiltration, volatile contents were evaluated. Negative control was empty vector infiltration (SK).
Stable overexpression in tomato fruit
The full-length TgGPPS cDNA was ligated into the pART-CAM vector (Xu et al. 2014). Supplemental Table S4 provides a list of the primers used in vector construction. Seeds were first placed in sterile distilled water for 30 min; sterilized with 75% (v/v) ethanol for 30 s, sodium hypochlorite solution (available chlorine is 2%) for 15 min, rinsed 4 times with sterile water, and inoculated on 1/2 MS solid medium. According to Wang et al., tomato (cv. MicroTom) transformation was accomplished using A. tumefaciens (Wang et al. 2005). The WT and transgenic T1 generations of tomato plants were grown in a greenhouse at 25 °C with 16 h of light and 8 h of darkness. Tomato fruits at the stage (breaker + 7 days) were frozen in liquid nitrogen and stored at −80 °C for later use. Three plants were selected from each tomato line to symbolize 3 biological replicates, each of which contained 10 fruits.
Dual-luciferase assay
The dual-luciferase assay was used to examine the transactivation capabilities of different TFs on target promoters. Full-length sequences of 14 TFs were amplified and inserted into the pGreen II 0029 62-SK (SK) vector, while the TgGPPS promoter sequence was recombined to the pGreen II 0800-LUC (LUC) vector (Zhang et al. 2020b). Supplemental Table S4 provides a list of the primers used in vector construction. GV3101 of the A. tumefaciens was electroporated with each of the SK and LUC vectors and then suspended in the same infiltration buffer as in the subcellular localization analysis, with an optimal density OD600 of 0.75. N. benthamiana leaves were infiltrated with A. tumefaciens combinations of promoters and TFs at a volume of 1:10 using needleless syringes. The LUC/REN fluorescence was assessed with the Dual Luciferase Reporter Assay System (Promega, Madison, USA) after 3 days of infiltration. Each TF–promoter interaction experiment consisted of 3 biological replicates.
Yeast 1-hybrid assay
The interaction of TFs with the TgGPPS promoter was tested using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, Mountain View, California). The pGADT7 vector was used to insert the full-length TF sequences, while the pAbAi vector was used to amplify and ligate the TgGPPS promoter sequence. Supplemental Table S4 provides a list of the primers used in vector construction. To test the promoter autoactivation, the Y1HGold yeast strain was transformed using the recombinant TgGPPS promoter-pAbAi vector after linearization in accordance with the system's instructions. The TF-pGADT7 plasmids and the empty vector (pGADT7) were individually transfected into the Y1HGold strain carrying the TgGPPS promoter.
Electrophoretic mobility shift assay
The full-length TgbZIP44 was inserted into pET32a vector resulting in His-tagged fusion proteins. The recombinant construct was purified and transformed into Escherichia coli BL21 (DE3). Recombinant TgbZIP44-His protein was expressed and purified as described by Zhang et al. (2020b). The Lightshift Chemiluminescent EMSA kit (ThermoFisher Scientific, Waltham, MA, USA) was used following the manufacturer's instructions. Sequences (40 bp) containing ACGT motif derived from the TgGPPS promoter (Supplemental Table S4) were labeled with 5′biotin by HuaGene. The identical DNA fragments without labels were used as competitors (cold probes). As mutant probes, DNA fragments with the ACGT motif were replaced with a polyA sequence.
Bimolecular fluorescence complementation
Using the methods described by Zhang et al. (2020a), the protein–protein interaction between TgbHLH95 and TgbZIP44 can be examined via the BiFC assay. Full-length TgbHLH95 and TgbZIP44 without termination codons were cloned into vectors containing the N-terminal and C-terminal sequences of yellow fluorescent protein (YFP), respectively. Supplemental Table S4 provides a list of the primers used in vector construction. All of the recombinant vectors were transiently expressed in N. benthamiana leaves by Agrobacterium transfection (GV3101). A confocal laser scanning microscope (LSM510, Karl Zeiss, Germany) was used to image the YFP fluorescence of infiltrated leaves after 2 days of infiltration. The fluorescence was measured at a wavelength of 520 to 540 nm. The excitation wavelength for YFP fluorescence was 488 nm, and fluorescence was detected at 490 to 520 nm. The excitation wavelength for NLS fluorescence was 594 nm, and fluorescence was detected at 612 to 635 nm.
Firefly LCI
Validation of the BiFC data was confirmed using the LCI assay. According to the methods described by Xiang et al. (2019), full-length TgbHLH95 and TgbZIP44 sequences were ligated into the pCAMBIA-nLUC vector and pCAMBIA-cLUC vectors, respectively. Supplemental Table S4 provides a list of the primers used in vector construction. By using Agrobacterium-mediated transformation, each recombinant vector was simultaneously introduced into N. benthamiana leaves and resuspended in infiltration buffer at a final density of OD600 = 1. Prior to infiltration, equal quantities of each Agrobacterium strain (i.e. nLUC + cLUC, TgbHLH95-nLUC + cLUC, TgbHLH95-cLUC + nLUC, TgbZIP44-nLUC + cLUC, TgbZIP44-cLUC + nLUC, TgbHLH95-nLUC + TgbZIP44-cLUC, TgbHLH95-cLUC + TgbZIP44-nLUC) were mixed and incubated at 25 °C for 3 h. The transformed tobacco leaves was used for LCI assays after 2 days of infiltration. After spraying luciferin (0.2 mM) onto the leaves’ infiltrated areas, they were left in the dark for 30 min. The luciferase signal was detected using a microplate luminometer (LSM510, Karl Zeiss).
Co-IP assay
The full-length of TgbHLH95 without stop codon was fused with 3× flag tags and cloned into pCAMBIA1302 vector, and TgbZIP44 was fused with GFP tag and cloned into pBinGFP2 vector. Following electroporation into A. tumefaciens GV3101, all of the recombinant vectors were transiently expressed in N. benthamiana leaves. Liquid nitrogen was used to grind the tobacco leaves, and an extraction buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, 1 mM PMSF, and 0.5 × proteinase inhibitor) was used to extract the proteins. After 15 min of centrifugation at 23,000 g, the extracts were incubated with anti-GFP antibody (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C for 3 h. The immunoprecipitated protein was boiled in 30 µL of protein loading solution for 5 min after being washed 4 times with dilution buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.5 mM EDTA). This protein was then examined by immunoblotting using an anti-GFP or anti-Flag antibody (Sigma-Aldrich, St. Louis, MO, USA).
GST pull-down assay
The full-length of TgbHLH95 was ligated into pGEX-6P-1 vector to produce GST-tagged fusion protein, while TgbZIP44 was ligated into pET32a vector to produce His-tagged fusion protein, respectively. According to the manufacturer's instructions, the pull-down assay was carried out using a Pierce Pull-Down PolyHis Protein: Protein Interaction Kit (ThermoFisher Scientific, Waltham, MA, USA). Western blotting was used to identify the eluted samples using anti-GST and anti-His antibodies (Sigma-Aldrich, St. Louis, MO, USA).
Statistical analysis
The Origin 8.0 was used for drawing the figures (Microcal Software). When comparing significant differences, SPSS Statistics 20.0 was used (SPSS Inc., Chicago, IL, USA). Additionally, a Student's t-test was applied to determine whether there were any significant differences between the 2 groups (*P < 0.05; **P < 0.01; ***P < 0.001).
Accession numbers
Sequence data from this article can be found in the Supplemental Table S5.
Supplementary Material
Acknowledgments
We would like to thank Professor Mohamed A. Farag (Cairo University) for comments and suggestions for the manuscript.
Contributor Information
Zuying Zhang, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China; Zhejiang Provincial Key Laboratory of Forest Aromatic Plants-based Healthcare Functions, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Liu Tao, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Lingling Gao, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Yadi Gao, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Jinwei Suo, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Weiyu Yu, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Yuanyuan Hu, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Chunyan Wei, Zhejiang Academy of Agricultural Sciences, Institute of Horticulture, Desheng Middle Road No. 298, Hangzhou, 310021 Zhejiang Province, China.
Mohamed A Farag, Pharmacognosy Department, College of Pharmacy, Cairo University, Kasr el Aini st., Cairo 11562, Egypt.
Jiasheng Wu, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Lili Song, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China; Zhejiang Provincial Key Laboratory of Forest Aromatic Plants-based Healthcare Functions, Zhejiang A&F University, Lin’an, 311300 Zhejiang Province, China.
Author contributions
L.S. and J.W. conceived the research plans and supervised the experiments; Z.Z. and L.T. carried out most of the experiments and analyzed the data; L.G., Y.G., J.S., W.Y., Y.H., and C.W. provided technical assistance to Z.Z.; Z.Z. and L.T. wrote and revised the article. M.A.F. took part in the discussion and revision of the manuscript; all authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gas chromatography-mass spectrometry (GC-MS) chromatogram of volatiles in xiangfei (Torreya grandis) nuts determined on different days during the postharvest ripening stage.
Supplemental Figure S2. Alignment of amino acid sequences of TgGPPS and GPPS proteins from other species.
Supplemental Figure S3. GC-MS chromatogram of volatiles determined 7 days after infiltration of tobacco (Nicotiana tabacum) leaves.
Supplemental Figure S4. GC-MS chromatogram of volatiles determined 7 days after the breaker stage of tomato (Solanum lycopersicum, cv. MicroTom) fruit.
Supplemental Figure S5. Relative expression of TgbHLH1 and TgbHLH2 on days 0 to 12 during the postharvest ripening stage of xiangfei nuts.
Supplemental Figure S6. Transient overexpression of TgbHLHs in tobacco (Nicotiana tabacum) leaves.
Supplemental Table S1. Isolated and identified transcription factors involved in regulating terpenes biosynthesis in different plant species.
Supplemental Table S2. The fragments per kilobase of exon per million mapped fragments (FPKM) value of all terpenoid synthesis genes from the 2-C-methyl-D-erythritol 4-phosphate (MEP) and mevalonate (MVA) pathways through annotation in Xiangfei transcriptome database.
Supplemental Table S3. Primers used for RT-qPCR.
Supplemental Table S4. Primers used for vector construction.
Supplemental Table S5. The full-length sequences of TFs and TgGPPS.
Funding
This work was financially supported by the National Natural Science Foundation of China (32101556), Zhejiang Forestry Science and Technology Project (2022B04), the Launching Funds for Zhejiang A&F University (2021FR019), Zhejiang Province Key R&D Project (2020C02019; 2021C02001), and the Zhejiang Provincial Cooperative Forestry Science and Technology Project (2019SY07; 2021SY01).
References
- Aragüez I, Osorio S, Hoffmann T, Rambla JL, Medina-Escobar N, Granell A, Botella MÁ, Schwab W, Valpuesta V. Eugenol production in achenes and receptacles of strawberry fruits is catalyzed by synthases exhibiting distinct kinetics. Plant Physiol. 2013:163(2):946–958. 10.1104/pp.113.224352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battilana J, Costantini L, Emanuelli F, Sevini F, Segala C, Moser S, Velasco R, Versini G, Grando MS. The 1-deoxy-D: -xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor Appl Genet. 2009:118(4):653–669. 10.1007/s00122-008-0927-8 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, d’Harlingue A, Backhaus RA, Camara B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 2000:24(2):241–252. 10.1046/j.1365-313x.2000.00875.x [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993:11(2):13–116. 10.1007/BF02670468 [DOI] [Google Scholar]
- Chang TH, Hsieh FL, Ko TP, Teng KH, Liang PH, Wang AH. Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. Plant Cell. 2010:22(2):454–467. 10.1105/tpc.109.071738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne E, Butterlin G, Claudel P, Dumas V, Jaegli N, Merdinoglu D. A grapevine (Vitis vinifera L.) deoxy-D: -xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor Appl Genet. 2009:118(3):541–552. 10.1007/s00122-008-0919-8 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Negre F, Nagegowda DA, Orlova I. Plant volatiles: recent advances and future perspectives. Crit Rev Plant Sci. 2006:25(5):417–440. 10.1080/07352680600899973 [DOI] [Google Scholar]
- El Hadi MA, Zhang FJ, Wu FF, Zhou CH, Tao J. Advances in fruit aroma volatile research. Molecules. 2013:18(7):8200–8229. 10.3390/molecules18078200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell. 2014:26(2):828–841. 10.1105/tpc.113.121111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011:66(1):94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012:24(6):2635–2648. 10.1105/tpc.112.098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhang Z, Hua B, Tao L, Chen W, Gao Y, Suo J, Yu W, Wu J, Song L. The interaction of temperature and relative humidity affects the main aromatic components in postharvest Torreya grandis nuts. Food Chem. 2022:368:130836. 10.1016/j.foodchem.2021.130836 [DOI] [PubMed] [Google Scholar]
- Huang C, Hu G, Li F, Li Y, Wu J, Zhou X. NbPHAN, a MYB transcriptional factor, regulates leaf development and affects drought tolerance in Nicotiana benthamiana. Physiol Plant. 2013:149(3):297–309. 10.1111/ppl.12031 [DOI] [PubMed] [Google Scholar]
- Iwase A, Matsui K, Ohme-Takagi M. Manipulation of plant metabolic pathways by transcription factors. Plant Biotechnol. 2009:26(1):29–38. 10.5511/plantbiotechnology.26.29 [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:33(7):1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZJ, Luo CF, Cheng XJ, Feng XJ, Yu WW. Component analysis and nutrition evaluation of seeds of Torreya grandis ‘Merrillii’. J Zhejiang A&F Univ. 2005:22(5):540–544. (in Chinese). [Google Scholar]
- Liu H, Cao X, Liu X, Xin R, Wang J, Gao J, Wu B, Gao L, Xu C, Zhang B, et al. UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 2017:40(10):2261–2275. 10.1111/pce.13029 [DOI] [PubMed] [Google Scholar]
- Lv Z, Wang S, Zhang F, Chen L, Hao X, Pan Q, Fu X, Li L, Sun X, Tang K. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016:57(9):1961–1971. 10.1093/pcp/pcw118 [DOI] [PubMed] [Google Scholar]
- Mao K, Dong Q, Li C, Liu C, Ma F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci. 2017:8:480. 10.3389/fpls.2017.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010:10(1):226. 10.1186/1471-2229-10-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda DA. Plant volatile terpenoid metabolism: biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010:584(14):2965–2973. 10.1016/j.febslet.2010.05.045 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Green SA, Chen X, Bailleul EJ, Matich AJ, Wang MY, Atkinson RG. Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 2013:161(2):787–804. 10.1104/pp.112.208249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous SW, Whitaker BD. Cloning and functional expression of an (E, E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta. 2004:219(1):84–94. 10.1007/s00425-003-1191-4 [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010:27(4):862–874. 10.1093/molbev/msp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell. 2014:26(3):1118–1133. 10.1105/tpc.113.121731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Smita SS, Singh AK, Shanker K, Nagegowda DA. Heteromeric and homomeric geranyl diphosphate synthases from Catharanthus roseus and their role in monoterpene indole alkaloid biosynthesis. Mol Plant. 2013:6(5):1531–1549. 10.1093/mp/sst058 [DOI] [PubMed] [Google Scholar]
- Reddy VA, Wang Q, Dhar N, Kumar N, Venkatesh PN, Rajan C, Panicker D, Sridhar V, Mao HZ, Sarojam R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol J. 2017:15(9):1105–1119. 10.1111/pbi.12701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J. Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation. Phytochemistry. 2007:68(21):2649–2659. 10.1016/j.phytochem.2007.05.037 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J. Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry. 2008:69(1):49–57. 10.1016/j.phytochem.2007.06.022 [DOI] [PubMed] [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E. Biosynthesis of plant-derived flavor compounds. Plant J. 2008:54(4):712–732. 10.1111/j.1365-313X.2008.03446.x [DOI] [PubMed] [Google Scholar]
- Shen SL, Yin XR, Zhang B, Xie XL, Jiang Q, Grierson D, Chen KS. CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J Exp Bot. 2016:67(14):4105–4115. 10.1093/jxb/erw189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Meng X, Yang L, Ma Z, Zhou M, Yu C, Zhang Z, Yu W, Wu J, Lou H. Identification of key genes and enzymes contributing to nutrition conversion of Torreya grandis nuts during post-ripening process. Food Chem. 2022:384:13245. 10.1016/j.foodchem.2022.132454 [DOI] [PubMed] [Google Scholar]
- Suo J, Tong K, Wu J, Ding M, Chen W, Yang Y, Lou H, Hu Y, Yu W, Song L. Comparative transcriptome analysis reveals key genes in the regulation of squalene and β-sitosterol biosynthesis in Torreya grandis. Ind Crop Prod. 2019:131:182–193. 10.1016/j.indcrop.2019.01.035 [DOI] [Google Scholar]
- Valdés García A, Sánchez Romero R, Juan Polo A, Prats Moya S, Maestre Pérez SE, Beltrán Sanahuja A. Volatile profile of nuts, key odorants and analytical methods for quantification. Foods. 2021:10(7):1611. 10.3390/foods10071611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013:64(1):665–700. 10.1146/annurev-arplant-050312-120116 [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci U S A. 2009:106(24):9914–9919. 10.1073/pnas.0904069106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005:17(10):2676–2692. 10.1105/tpc.105.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Ohnuma S. Isoprenyl diphosphate synthases. Biochim. Biophys Acta. 2000:1529(1–3):33–48. 10.1016/S1388-1981(00)00136-0 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu H, Zhang M, Liu S, Hao Y, Zhang Y. MdMYC2 and MdERF3 positively co-regulate α-farnesene biosynthesis in apple. Front Plant Sci. 2020:11:512844. 10.3389/fpls.2020.512844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Liu H, Cao X, Zhang M, Li X, Chen K, Zhang B. Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol J. 2021:19(10):2082–2096. 10.1111/pbi.13638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, Chen K. CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019:70(12):3111–3123. 10.1093/jxb/erz121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Zhao J, Hao YJ, Fang C, Wang Y. The ectopic expression of apple MYB1 and bHLH3 differentially activates anthocyanin biosynthesis in tobacco. Plant Cell. 2017:131(1):183–194. 10.1007/s11240-017-1275-7 [DOI] [Google Scholar]
- Xu J, van Herwijnen ZO, Drager DB, Sui C, Haring MA, Schuurink RC. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell. 2018:30(12):2988–3005. 10.1105/tpc.18.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang N, Jiao Y, Li R, Xiao D, Wang Z. The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol Biol Rep. 2014:41(8):5329–5342. 10.1007/s11033-014-3404-2 [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu C, Xu C, Sun J, Grierson D, Zhang B, Chen K. Integration of metabolite profiling and transcriptome analysis reveals genes related to volatile terpenoid metabolism in finger citron (C. medica var. sarcodactylis). Molecules. 2019:24(14):2564. 10.3390/molecules24142564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Liao YC, Lv FF, Zhang Z, Sun PW, Gao ZH, Hu KP, Sui C, Jin Y, Wei JH. Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) gilg. Plant Cell Physiol. 2017:58(11):1924–1933. 10.1093/pcp/pcx122 [DOI] [PubMed] [Google Scholar]
- Yang Z, Li Y, Gao F, Jin W, Li S, Kimani S, Yang S, Bao T, Gao X, Wang L. MYB21 Interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J Exp Bot. 2020:71(14):4140–4158. 10.1093/jxb/eraa184 [DOI] [PubMed] [Google Scholar]
- Zhang B, Shen JY, Wei WW, Xi W. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J Agric Food Chem. 2010:58(10):6157–6165. 10.1021/jf100172e [DOI] [PubMed] [Google Scholar]
- Zhang J, Yin XR, Li H, Xu M, Zhang MX, Li SJ, Liu XF, Shi YN, Grierson D, Chen KS. ETHYLENE RESPONSE FACTOR39-MYB8 complex regulates low-temperature-induced lignification of loquat fruit. J Exp Bot. 2020a:71(10):3172–3184. 10.1093/jxb/eraa085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jin H, Suo J, Yu W, Zhou M, Dai W, Song L, Hu Y, Wu J. Effect of temperature and humidity on oil quality of harvested Torreya grandis cv. Merrillii nuts during the after-ripening stage. Front Plant Sci. 2020c:11:573681. 10.3389/fpls.2020.573681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shi Y, Ma Y, Yang X, Yin X, Zhang Y, Xiao Y, Liu W, Li Y, Li S, et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol J. 2020b:18(11):2267–2279. 10.1111/pbi.13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Pichersky E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020:226(5):1341–1360. 10.1111/nph.16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.