Abstract
CDC group IVc-2 is a gram-negative, oxidase-positive, nonfermentative bacillus that has been implicated in human infections, including septicemia and peritonitis. Biochemically it most closely resembles Bordetella bronchiseptica and Alcaligenes sp. Results of cellular fatty acid (CFA) and 16S rRNA gene analysis were combined with biochemical data to assist in identification and classification. The predominant CFAs were hexadecanoic acid (16:0), cis-9-hexadecanoic acid (16:1ω7c), cis-11-octadecanoic acid (18:1ω7c), and Δ-cis-9,10-methylenehexadecanoic acid (17:0cyc). Small amounts (2 to 5%) of 3-hydroxytetradecanoic acid (3-OH-14:0), tetradecanoic acid (14:0), 2-hydroxyhexadecanoic acid (2-OH-16:0), and Δ-cis-11,12-methyleneoctadecanoic acid (19:0cyc) were also consistently present. The highest 16S rRNA gene similarity was with Ralstonia eutropha and Ralstonia solanacearum. The CFA and 16S rRNA gene sequence data support the inclusion of CDC group IVc-2 in the recently created genus Ralstonia, which includes R. eutropha, R. pickettii, and R. solanacearum.
CDC group IVc-2 is the vernacular name for a group of asaccharolytic, gram-negative, nonfermentative bacteria. The organism is found environmentally and is frequently associated with contaminated water sources (2, 3, 11, 13). Clinical cases of infection although rare, include 10 cases of bacteremia (1, 4, 5, 12, 17) and 2 cases of peritonitis (8, 25) in debilitated patients usually possessing indwelling catheters. Only a single case of infection in a patient lacking any predisposing pathologic condition has been reported (13).
Biochemically, CDC group IVc-2 shares many features with Bordetella avium, Bordetella bronchiseptica, Oligella ureolytica (18), and Ralstonia eutropha (formerly Alcaligenes eutrophus) (16). Cellular fatty acid (CFA) content, however, indicates a close association with the rRNA group II pseudomonads, which include Burkholderia cepacia, Ralstonia pickettii (formerly Burkholderia pickettii), and Ralstonia solanacearum (formerly Burkholderia solanacearum) (22). In order to facilitate identification and to further clarify the taxonomic status of this organism, we evaluated the results of biochemical, CFA, and 16S rRNA gene sequence analysis.
MATERIALS AND METHODS
Conventional identification.
One reference strain (CDC C6966) and four well-characterized stock clinical strains (JHH 1 to 4) were analyzed in this study. All isolates were initially evaluated by the following conventional tests: Gram stain, growth and morphologic characteristics on trypticase soy agar with 5% sheep blood and MacConkey agar, catalase, oxidase, triple sugar-iron agar reactions, motility, indole production, gelatin liquefaction, utilization of citrate, growth at 42°C, pigment production, oxidative-fermentative (OF) carbohydrate utilization (glucose, xylose, and maltose), utilization of sodium acetamide, decarboxylation of lysine, dihydrolase reaction of arginine, urease activity, susceptibility to 10 μg of colistin per ml, and hydrolysis of o-nitrophenyl-β-d-galactopyranoside (ONPG). Additional tests included acid production from 10% lactose; phenylalanine deamination; nitrate reduction and gas production; hydrolysis of esculin, starch, DNA, and phosphatidylcholine; allantoin utilization; growth in the presence of 6.5% NaCl; and staining of flagella. These tests were considered conventional identification methods, as described by others (7, 22). Antibiotic susceptibility was determined by the agar dilution method according to National Committee for Clinical Laboratory Standards guidelines (14).
Cellular fatty acid analysis.
Growth after 24 h from a plate containing trypticase soy agar with 5% sheep blood was processed for CFA analysis as previously described (15). Briefly, fatty acids liberated from the saponified cells were derivatized to form the corresponding fatty acid methyl esters (FAMEs) and then extracted into a hexane-ether mixture. FAMEs were analyzed with a Hewlett-Packard (Wilmington, Del.) 5890A gas chromatograph equipped with a flame ionization detector, automatic sampler, integrator, and computer. Separation of the FAMEs was achieved with a fused-silica capillary column (25m by 0.2 mm) with cross-linked 5% phenylmethyl silicone. The specific operating parameters for the instrument were controlled and set automatically by the computer software.
Identification and quantitation of the FAME were performed by the Microbial Identification System (MIS) software (Microbial ID, Newark, Del.). Subsequent organism identification was based on computer comparison of the unknown organism’s FAME profile with that of predetermined organism library profiles in the commercial database (version 3.9). Library organism profiles were established on the basis of a multivariate Gaussian model of at least 10 strains of each species when possible. In addition to American Type Culture Collection strains, well-characterized reference strains from a variety of environmental sources are utilized to account for possible heterogeneity in the CFA content.
The correlation of an unknown organism’s profile with a library entry is expressed as a similarity index (SI) on a numeric scale of 0 to 1.0. SI values of >0.6 are considered excellent matches, with a value of 0.5 representing approximately 3 standard deviations from the library profile mean.
Nucleic acid preparation.
Genomic DNA was Chelex extracted (Sigma, St. Louis, Mo.) from several well-isolated bacterial colonies by standard methods (20). The 16S rRNA gene was PCR amplified with two universal primers, 5F and 1540R (PE Applied Biosystems, Inc. [ABD], Foster City, Calif.), which are complementary to the phylogenetically conserved regions of the 5′ and 3′ ends of the 16S rRNA gene, respectively. Bacterial DNA (100 ng) was amplified in 100 μl of master mix from the MicroSeq 16S rRNA gene kit (ABD, Foster City, Calif.) and was amplified according to the manufacturer’s recommendations. The samples were subjected to 30 amplification cycles consisting of 30 s of denaturation at 95°C, 30 s of annealing at 60°C, and 45 s of extension at 72°C. An additional elongation cycle of 10 min at 72°C allowed for complete, final product extension. PCR product was confirmed by electrophoresis of 10-μl reaction volumes in a 2% agarose gel containing ethidium bromide. The remaining PCR products were purified by filtration with Microcon 100 (Amicon, Beverly, Mass.) spin columns. Cycle sequencing was subsequently performed with the PCR products with the sequencing module from the MicroSeq 16S rRNA gene kit, which contains six forward and six reverse sequencing primers (ABD). The sequencing mixture was purified of excess primers and unincorporated nucleotides by using Centri-Sep (Princeton Separations, Adelphia, N.J.) columns. The purified reaction product was dried under vacuum and resuspended in deionized formamide–50 mM EDTA. Fluorescently labeled sequencing products were separated on an ABI PRISM 377 DNA sequencer (ABD) fitted with a 5% polyacrylamide gel.
Nucleotide identification and signal processing were performed by the Sequencing Analysis program, version 3.0. A consensus sequence generated with the AutoAssembler program was submitted to the National Center for Biotechnology Information (NCBI) GenBank for sequence alignment and to the Ribosome Database Project (RDP) for similarity ranking (10). Identification with the MicroSeq 16S rDNA database (ABD) was also evaluated. The MicroSeq 16S rDNA database was established from nearly complete 16S rRNA gene sequence data derived from the analysis of individual American Type Culture Collection type strains. For most species, the amplicon size is 1,527 bp. Currently >1,100 eubacterial species are represented. The unknown sequence was subjected to an initial BLAST search to limit the number of sequences utilized in the more extensive full-length alignment library search. Similarity comparison results are generated rapidly. Values are expressed as percent sequence difference from type strains.
A phylogenetic tree incorporating sequence data from the MicroSeq 16S rDNA database was constructed by the neighbor-joining algorithm (19) with the sequence data from B. cepacia being used as the outgroup. Additional features of the MicroSeq identification and analysis software package enabled pairwise alignment of all the group IVc-2 strains and determination of sequence similarity. In particular, the concise alignment function depicts positions of nucleotide differences of unknown strains with library entries.
Nucleotide sequence accession number.
The rRNA gene sequence has been registered with the GenBank database under accession no. AF067657.
RESULTS
All of the CDC group IVc-2 strains grew well aerobically after 24 h on MacConkey agar and Trypticase soy agar with 5% sheep blood at 37°C. By Gram staining and flagellar staining, the bacteria were short gram-negative rods possessing peritrichous flagella, respectively. The isolates were positive only for citrate utilization, oxidase, catalase, and rapid urease production. In contrast to phenotypically similar R. eutropha and B. bronchiseptica, they failed to reduce nitrate or nitrite. Additionally they were negative for indole production, lysine decarboxylase, arginine dihydrolase, phenylalanine deamination, and the ability to utilize allantoin or acetamide. They did not hydrolyze esculin, gelatin, starch, DNA, ONPG, or phosphatidylcholine. No acid production was detected from OF glucose, xylose, maltose, or 10% lactose. All reactions were held at least 14 days before being assessed as negative.
Mean CFA values for the five strains are summarized in Table 1 and compared to reference profiles of R. eutropha, B. cepacia, B. bronchiseptica, and R. pickettii. All of these organisms share features either biochemically or in their CFA content with group IVc-2. The CFA profile of group IVc-2 is notable for possessing 16:1w7c and 16:0 as major acids; moderate amounts of 3-OH-14:0, 17:0cyc, and 18:1w7c; and trace to small amounts of 2-OH-16:1, 2-OH-18:1, 2-OH-14:0, 2-OH-16:0, and 19:0cyc11-12. The complete profile is consistent with published results (22).
TABLE 1.
CFA composition of CDC group IVc-2 and related organisms
| Fatty acida | % of fatty acid in organism:
|
||||
|---|---|---|---|---|---|
| Group IVc-2 | R. eutrophab | B. bronchi- septicac | B. cepaciac | R. pickettiic | |
| 12:0 | —d | — | 2 | — | — |
| 2-OH-12:0 | — | — | 4 | — | — |
| 14:0 | 4 | 4 | 6 | 4 | 5 |
| 15:0 | — | — | — | — | 1 |
| 2-OH-14:0 | — | 2 | — | — | 1 |
| 3-OH-14:0 | 7 | 9 | 5 | 5 | 7 |
| 16:1ω7c | 16 | 32 | 27 | 9 | 26 |
| 16:0 | 31 | 31 | 35 | 24 | 28 |
| 17:0cyc | 15 | 3 | 14 | 14 | 4 |
| 17:0 | — | — | — | — | 1 |
| 2-OH-16:1 | 1 | — | — | 1 | 2 |
| 2-OH-16:0 | 3 | — | — | 2 | 1 |
| 3-OH-16:0 | — | — | — | 4 | — |
| 18:1ω7c | 15 | 17 | 2 | 20 | 19 |
| 18:0 | 1 | — | 2 | 1 | 1 |
| 19:0cyc11–12 | 3 | — | — | 10 | 1 |
| 2-OH-18:1 | 1 | — | — | 2 | 2 |
| 2-OH-19:0cyc | — | — | — | 2 | — |
The number before the colon indicates the number of carbon atoms, and the number after is the number of double bonds. c, cis isomer; cyc, cyclopropane ring structure.
Data from MIS database.
Data from reference 22.
—, trace levels.
The overlap of the CFA profiles of Ralstonia, Burkholderia, and group IVc-2 is easily demonstrated compared to the MIS library database, since group IVc-2 was identified with a low confidence value (SI) as either B. cepacia, R. pickettii, or CDC group IVc-2. Additionally, it is common to have enteric negative rods such as Enterobacter aerogenes listed as possible choices. Strain C6966 was identified as B. cepacia (SI, 0.440), followed by Pantoea agglomerans (SI, 0.290) as the second choice. The remaining four strains were all identified as B. cepacia (SI, <0.500), followed by either CDC group IVc-2, Enterobacter cloacae (SI, <0.500), or R. pickettii (SI, <0.500) as the second choice.
All strains were resistant to ampicillin (>16 μg/ml), cefazolin (>16 μg/ml), gentamicin (>8 μg/ml), tobramycin (>8 μg/ml), and amikacin (>32 μg/ml). They were susceptible to tetracycline (2 μg/ml), ticarcillin (16 μg/ml), piperacillin (8 μg/ml), cefuroxime (8 μg/ml), cefotaxime (2 μg/ml), ceftazidime (8 μg/ml), trimethoprim-sulfamethoxazole (0.5/9.5 μg/ml), and ciprofloxacin (0.5 μg/ml). Similar results have been reported for other strains of group IVc-2 (1, 17, 25).
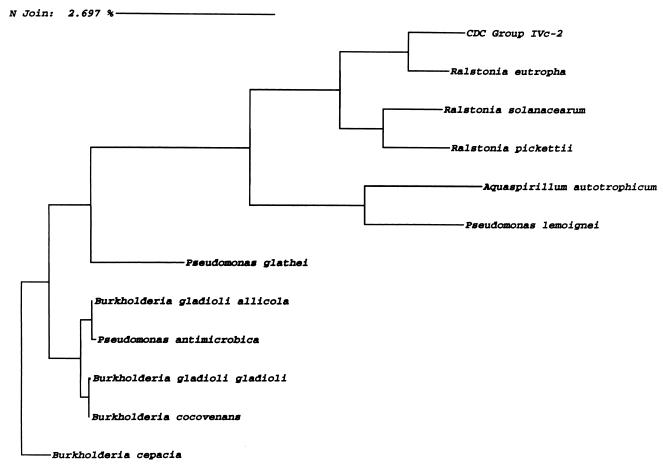
Comparison of the consensus sequence for strain C6966 with sequences in reference databases (RDP and GenBank) and the commercial database (MicroSeq) is summarized in Tables 2 to 4. Comparable results were achieved with the four clinical isolates. For each strain, R. eutropha or Alcaligenes sp. strain M91-3 was the closest match, followed by R. solanacearum and R. pickettii in each of the databases. The phylogenetic relationship between group IVc-2, R. eutropha, and R. solanacearum is further illustrated in the evolutionary tree (Fig. 1). Group IVc-2 was most closely linked to R. eutropha, and as shown in Fig. 1, it exhibited approximately 98% sequence similarity.
TABLE 2.
Search results for strain C6966 (1,527 bp) from public rRNA RDP databasea
| Name | Sequence length (no. of oligomers)b | Similarity valuec |
|---|---|---|
| Ralstonia eutropha 3CB-1 | 541 | 0.898 |
| Ralstonia eutropha 335 (R. Y. Stanier) | 1,407 | 0.896 |
| Alcaligenes sp. strain M91-3 | 1,363 | 0.894 |
| Ralstonia eutropha | 149 | 0.852 |
| Ralstonia solanacearum | 1,354 | 0.837 |
| R233 | 1,362 | 0.836 |
| Ralstonia solanacearum R780 | 1,340 | 0.835 |
| R506 | 1,342 | 0.833 |
| Pseudomonas syzygii R001 | 1,354 | 0.830 |
| Pseudomonas syzygii R058 | 1,354 | 0.830 |
Only the top 10 choices are listed.
Number of uniquely occurring oligomers within the given sequence. One oligomer = 7-mer.
Similarity value is the number of unique oligomers shared between an unknown sequence and a given RDP sequence divided by the lowest number of unique oligomers in either of the two.
TABLE 4.
Search results for strain C6966 (1,527 bp) from commercial MicroSeq databasea
| Name | Sequence length (bp) | % Differenceb |
|---|---|---|
| Ralstonia eutropha | 1,527 | 1.64 |
| Ralstonia solanacearum | 1,531 | 3.89 |
| Ralstonia pickettii | 1,533 | 4.19 |
| Burkholderia gladioli subsp. allicola | 1,528 | 7.59 |
| Aquaspirillum autotrophicum | 1,528 | 7.65 |
| Pseudomonas antimicrobica | 1,528 | 7.65 |
| Burkholderia gladioli subsp. gladioli | 1,528 | 7.72 |
| Pseudomonas lemoignei | 1,528 | 7.72 |
| Burkholderia cocovenans | 1,528 | 7.75 |
| Pseudomonas glathei | 1,526 | 7.79 |
Only the top 10 choices are listed.
Percent sequence difference of the unknown strain versus the type strains in the database.
FIG. 1.
Phylogenetic tree derived from neighbor-joining (N Join) analysis. B. cepacia was used as an outgroup. The distance between two species is obtained by adding the lengths of the connecting horizontal branches. Bar, 2.7% sequence difference.
Alignment of the 16S rRNA gene sequence for all group IVc-2 strains revealed >99.5% sequence similarity, while all strains yielded a >1.6% difference from the type strain of R. eutropha.
DISCUSSION
Recognition of CDC group IVc-2 in the clinical laboratory is important, since it can cause serious infections and may be refractory to standard antibiotic regimens. This is particularly evident when peritonitis develops as a complication of chronic ambulatory peritoneal dialysis. Typical treatment consists of vancomycin and tobramycin, and yet group IVc-2 isolates are resistant to both antibiotics. Prompt identification would facilitate administration of the appropriate therapy.
Among the nonfermentative gram-negative rods that do not oxidize glucose, are oxidase positive, and are capable of growth on MacConkey agar, group IVc-2 shares the most features with R. eutropha, B. bronchiseptica, and O. ureolytica. Nitrate reduction is an important differentiation characteristic, since group IVc-2 is the only one of the organisms described above that is usually negative for nitrate reductase. Only 11% of the strains have been reported in the literature to be capable of reducing nitrate (22). Like R. eutropha, B. bronchiseptica, and O. ureolytica, group IVc-2 is conspicuous for rapid hydrolysis of urea, while B. avium is not. The lack of definitive phenotypic tests complicates speciation when only physiological and biochemical features are considered.
Identification of microorganisms by analysis of CFA composition has become a routine method in many laboratories. Automation has enabled reproducible results to be generated rapidly, provided the strains are grown under specified standardized conditions. The CFA content of group IVc-2 is most similar to those of B. cepacia and R. pickettii, as reflected in the MIS identifications. For all of the strains tested, B. cepacia was the first choice, but at a low confidence value (SI, <0.500). The low SI value and the presence of multiple choices reflect the inability of the MIS system to definitively identify group IVc-2. However, the distinctive pattern of rRNA group II pseudomonads together with enteric negative rods necessitates the consideration of group IVc-2 as a probable identification, even if group IVc-2 is not listed as a possibility.
Even though it is biochemically distinct, R. pickettii possesses the closest overall composition, containing 16:1w7c, 16:0, and 18:1w7c as major acids, although with smaller amounts of 17:0cyc. The 17:0cyc content of group IVc-2 approaches that of the rRNA group II organisms such as B. cepacia but differs quantitatively in the 16:1w7c and 19:0cyc content and qualitatively by lacking 3-OH-16:0 and 2-OH-19:0cyc. Manual inspection of the profile reveals both qualitative and quantitative differences. Of the biochemically similar organisms, only B. bronchiseptica and R. eutropha are remotely related by CFAs. In addition to quantitative differences, both lack 19:0cyc11-12 and the 2-OH-16:0 carbon acids characteristic of group IVc-2, B. cepacia, and R. pickettii.
The combination of CFA and biochemical data is generally sufficient for accurate identification of group IVc-2. CFA analysis can be utilized as a rapid screening method followed by select biochemical tests to confirm the identification. When phenotypic methods prove inconclusive, confirmatory genotypic techniques such as 16S rRNA gene sequence analysis are beneficial.
Each of the databases searched in this study indicated that the group IVc-2 sequences showed the highest similarity to that of R. eutropha. However, the relatively low scores suggested the existence of a distinct species. Typically, two organisms are assumed to belong to the same species if they possess less than 5 to 15 nucleotide differences in their 16S rRNA gene sequences (6). The MicroSeq 16S rDNA database clearly depicts the close, yet distinct, relatedness to R. eutropha (1.6%), because a total of 25 bp differences are present (Table 5).
TABLE 5.
Nucleotide position differences among CDC group IVc-2 strains and the type strain of R. eutropha, ATCC 17697
| Nucleotide position | Nucleotide of strain:
|
|||||
|---|---|---|---|---|---|---|
| C6966 | JHH 1 | JHH 2 | JHH 3 | JHH 4 | ATCC 17697 | |
| 70 | G | G | G | G | G | A |
| 74 | A | A | A | A | A | G |
| 81 | T | T | T | T | T | C |
| 86 | C | C | C | C | C | T |
| 124 | A | A | A | A | A | G |
| 131 | T | T | T | T | T | A |
| 198 | G | G | A | G | G | G |
| 199 | A | A | A | C | C | C |
| 212 | A | A | A | A | A | T |
| 213 | G | G | G | G | G | A |
| 255 | G | G | G | G | G | A |
| 259 | C | C | C | C | C | T |
| 445 | T | T | T | T | T | G |
| 453 | C | C | C | C | C | T |
| 461 | T | T | T | T | T | C |
| 467 | A | A | A | A | A | C |
| 606 | G | G | G | G | G | A |
| 614 | C | C | C | C | C | T |
| 622 | T | T | T | T | T | G |
| 648 | G | G | G | G | G | A |
| 649 | C | C | C | C | C | T |
| 738 | G | G | G | G | G | C |
| 837 | T | T | T | T | T | C |
| 838 | C | C | C | C | C | T |
| 1125 | C | C | C | C | C | A |
| 1129 | A | A | A | A | A | G |
Among the available methods for evaluating phylogenetic relationships, chemotaxonomic and genotypic techniques are being utilized with increasing frequency (21). Two groups of macromolecules that have recently been made amenable to simplified analysis are CFA and rRNAs. Analysis of the 16S rRNA gene sequence has proven to be most useful in molecular systematics, since it is highly conserved, universally distributed, and contains diagnostically significant variable regions.
Polyphasic studies combining phenotypic characteristics, DNA-DNA and DNA-rRNA hybridization, lipid composition, and 16S rRNA gene sequence have been used to redefine the rRNA group II pseudomonads (23). The genetic bifurcation in this group has been further established (9), with the genus Ralstonia being proposed to accommodate one subdivision (24). In addition to R. pickettii and R. solanacearum, phenotypically dissimilar R. eutropha has been included because of the high levels of 16S rRNA gene sequence similarities.
CDC group IVc-2 is an organism phenotypically similar to R. eutropha. The results of this study indicate a high level of genotypic similarity as well and warrant the inclusion of this organism in the genus Ralstonia. Since the RDP Similarity Ranking and NCBI GenBank BLAST searches provide only reasonable approximations of phylogenetic analysis, further tests, such as cellular lipid analysis, DNA-DNA hybridization, and determination of the mole percent G+C, would be useful for confirmation. Further clarification of the taxonomic status would also be helpful in recognizing this organism in the clinical laboratory, since it can be a source of serious infection in immunocompromised patients. Traditionally, the technical complexity and laborious nature of 16S rRNA gene sequence analysis have limited its applications as a routine diagnostic tool. However, approaches to automate and standardize this methodology should facilitate its implementation as a common laboratory protocol.
TABLE 3.
Search results for strain C6966 (1,527 bp) from public rRNA database NCBI GenBank BLAST search
| Name (GenBank accession no.)a | Sequence length (bp) | No. of residues aligned/no. in sequence (%) |
|---|---|---|
| Ralstonia eutropha (AF027407) | 1,504 | 1,479/1,497 (98) |
| Alcaligenes eutrophus (M32021) | 1,511 | 1,466/1,506 (97) |
| Pseudomonas solanacearum (X67035) | 1,490 | 1,320/1,375 (96) |
| Pseudomonas solanacearum (X67036) | 1,470 | 1,319/1,375 (95) |
| Pseudomonas solanacearum (X67041) | 1,474 | 1,304/1,354 (96) |
| Pseudomonas solanacearum (X67040) | 1,470 | 1,303/1,354 (96) |
| Beta-proteobacterium species (X97093) | 1,233 | 1,217/1,233 (98) |
| Pseudomonas pickettii (L37367) | 1,359 | 1,236/1,287 (96) |
| Ralstonia sp. (D88001) | 1,463 | 948/969 (97) |
| Unidentified bacterium (Z99998) | 1,538 | 964/1,042 (92) |
Only the top 10 choices are listed.
ACKNOWLEDGMENTS
We thank the following staff of PE/Applied Biosystems and MIDI Labs for training and technical support: Nicole Ellis, Deborah Dodge, Scott Anderson, Doug Smith, Stacey Montgomery, Mike Waddington, John Bartel, Maggie Riehman, and Myron Sasser.
We give special thanks to PE/Applied Biosystems for equipment and support material.
REFERENCES
- 1.Anderson R R, Warnick P, Schreckenberger P C. Recurrent CDC group IVc-2 bacteremia in a human with AIDS. J Clin Microbiol. 1997;35:780–782. doi: 10.1128/jcm.35.3.780-782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspinall S T, Graham R. Two sources of contamination of a hydrotherapy pool by environmental organisms. J Hosp Infect. 1989;14:285–292. doi: 10.1016/0195-6701(89)90068-6. [DOI] [PubMed] [Google Scholar]
- 3.Clark W A, Hollis D G, Weaver R E, Riley P. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria. Atlanta, Ga: Centers for Disease Control; 1984. [Google Scholar]
- 4.Crowe H M, Brecher S M. Nosocomial septicemia with CDC group IV c-2, an unusual gram-negative bacillus. J Clin Microbiol. 1987;25:2225–2226. doi: 10.1128/jcm.25.11.2225-2226.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dan M, Berger S A, Aderka D, Levo Y. Septicemia caused by the gram-negative bacterium CDC IV c-2 in an immunocompromised human. J Clin Microbiol. 1986;23:803. doi: 10.1128/jcm.23.4.803-.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 7.Gilardi G L, editor. Nonfermentative gram-negative rods. New York, N.Y: Marcel Dekker, Inc.; 1985. pp. 17–84. [Google Scholar]
- 8.Hansen W, Glupczynski Y. Group IVc-2-associated peritonitis. Clin Microbiol Newsl. 1985;7:43–44. [Google Scholar]
- 9.Li X, Dorsch M, Del Dot T, Sly L I, Stackebrandt E, Hayward A C. Phylogenetic studies of the rRNA group II pseudomonads based on 16S rRNA gene sequences. J Appl Bacteriol. 1993;74:324–329. [Google Scholar]
- 10.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mania C M, Nunes O C, Morais P V, da Costa M S. Heterotrophic plate counts and the isolation of bacteria from mineral waters on selective and enrichment media. J Appl Bacteriol. 1990;69:871–876. doi: 10.1111/j.1365-2672.1990.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 12.Moissenet D, Tabone M-D, Girardet J-P, Leverger G, Garbarg-Chenon A, Vu-Thien H. Nosocomial CDC group IV c-2 bacteremia: epidemiological investigation by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1996;34:1264–1266. doi: 10.1128/jcm.34.5.1264-1266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musso D, Drancourt M, Bardot J, Legré R. Human infection due to the CDC group IVc-2 bacterium: case report and review. Clin Infect Dis. 1994;18:482–484. doi: 10.1093/clinids/18.3.482-a. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 15.Osterhout G J, Shull V H, Dick J D. Identification of clinical isolates of gram-negative nonfermentative bacteria by an automated cellular fatty acid identification system. J Clin Microbiol. 1991;29:1822–1830. doi: 10.1128/jcm.29.9.1822-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickett M J, Greenwood J R. Identification of oxidase-positive, glucose-negative, motile species of nonfermentative bacilli. J Clin Microbiol. 1986;23:920–923. doi: 10.1128/jcm.23.5.920-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos J M, Soriano F, Bernacer M, Esteban J, Zapardiel J. Infection caused by the nonfermentative gram-negative bacillus CDC group IVc-2: case report and literature review. Eur J Clin Microbiol Infect Dis. 1993;12:456–458. doi: 10.1007/BF01967442. [DOI] [PubMed] [Google Scholar]
- 18.Rossau R, Kersters K, Falsen E, Jantzen E, Segers P, Union A, Nehis L, Deley J. Oligella, a new genus including Oligella urethralis comb. nov. (formerly Moraxella urethralis) and Oligella ureolytica sp. nov. (formerly CDC group IVe): relationship to Taylorella equigenitalis and related taxa. Int J Syst Bacteriol. 1987;37:198–210. [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Singer-Sam J, Tanguay R L, Riggs A D. Use of Chelex to improve the PCR signal from a small number of cells. Amplifications, no. 3. Norwalk, Conn: Perkin-Elmer Cetus; 1989. [Google Scholar]
- 21.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jorda J G, Cook E C, Daneshvar M I. Identification of unusual pathogenic Gram-negative aerobic and facultatively anaerobic bacteria. 2nd ed. Baltimore, Md: Williams & Wilkins; 1996. [Google Scholar]
- 23.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 24.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 25.Zapardiel J, Blum G, Caramelo C, Fernandez-Roblas R, Rodriguez-Tudela J L, Soriano F. Peritonitis with CDC group IVc-2 bacteria in a patient on continuous ambulatory peritoneal dialysis. Eur J Clin Microbiol Infect Dis. 1991;10:509–511. doi: 10.1007/BF01963939. [DOI] [PubMed] [Google Scholar]