Abstract
Background
Our previous work shows that increased matrix stiffness not only alters malignant characteristics of hepatocellular carcinoma (HCC) cells, but also attenuates metformin efficacy in treating HCC cells. Here, we identified differential membrane proteins related to matrix stiffness-mediated metformin resistance for better understand therapeutic resistance of metformin in HCC.
Methods
Differential membrane proteins in HCC cells grown on different stiffness substrates before and after metformin intervention were screened and identified using isobaric tags for relative and absolute quantification (iTRAQ) labeling coupled with the liquid chromatography-tandem mass spectrometry (LC–MS/MS), then bioinformatic analysis were applied to determine candidate membrane protein and their possible signaling pathway.
Results
A total of 5159 proteins were identified and 354 differential membrane proteins and membrane associated proteins, which might be associated with matrix stiffness-mediated metformin resistance were discovered. Then 94 candidate membrane proteins including 21 up-regulated protein molecules and 73 down-regulated protein molecules were further obtained. Some of them such as Annexin A2 (ANXA2), Filamin-A (FLNA), Moesin (MSN), Myosin-9 (MYH9), Elongation factor 2 (eEF2), and Tax1 binding Protein 3 (TAX1BP3) were selected for further validation. Their expressions were all downregulated in HCC cells grown on different stiffness substrates after metformin intervention. More importantly, the degree of decrease was obviously weakened on the higher stiffness substrate compared with that on the lower stiffness substrate, indicating that these candidate membrane proteins might contribute to matrix stiffness-mediated metformin resistance in HCC.
Conclusions
There was an obvious change in membrane proteins in matrix stiffness-mediated metformin resistance in HCC cells. Six candidate membrane proteins may reflect the response of HCC cells under high stiffness stimulation to metformin intervention, which deserve to be investigated in the future.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12953-023-00216-7.
Keywords: Hepatocellular carcinoma, Metformin, Membrane proteins, Matrix stiffness, Drug resistance, iTRAQ
Background
Increasing evidence has showed that diabetes is closely related to the occurrence and development of tumors [1–3]. The incidence of multiple tumors in patients with diabetes is higher than that in the non-diabetic population, and the risk of the incidence of liver cancer is the highest (RR ≈ 2.5) [4]. Metformin, as a first-line hypoglycemic agent for the treatment of type 2 diabetes, has recently exhibited obvious anticancer effect in a variety of solid tumors including HCC [5–8]. However, the mechanism of metformin tolerance and resistance in solid tumor remains largely unclear. Past studies on the underlying mechanism of drug resistance mainly focus on biochemical and metabolic signals within tumor microenvironment, such as DNA mutations and metabolic reprogramming, etc. [9–12]. The effect of biomechanics signals within tumor microenvironment on therapeutic resistance has been rarely mentioned. Matrix stiffening, caused by massive deposition and cross-linking of matrix proteins, is the most frequent biomechanical characteristics of solid tumor, influences and drives the development of various cancers [13–15]. HCC almost exclusively develops in the condition of chronic liver diseases such as hepatitis, liver fibrosis, cirrhosis, etc. [16, 17], and HCC patients with severe cirrhosis usually present an unfavorable prognosis [18]. In clinic, liver stiffness has gradually served as a useful indicator to predict the development and progression of HCC [19, 20]. In vitro experiments reveal that increased matrix stiffness significantly strengthens the malignant properties of HCC cells including proliferation [21], invasion and metastasis [22, 23], aerobic glycolysis [24], stemness [25], epithelial-mesenchymal transition (EMT) [26] and pre-metastatic niche formation [27]. Besides, increased matrix stiffness also enhances resistance to chemotherapeutic agents remarkably like paclitaxel, 5-FU, cisplatin and oxaliplatin, etc. [25, 28, 29]. Similarly, our latest research also supports that increased matrix stiffness attenuates metformin efficacy in treating HCC cells, disclosing a significant linkage between matrix stiffness and therapeutic resistance in HCC [30]. Cells communicate with the surrounding microenvironment via cell membrane. Membrane proteins and their associated proteins including adhesion molecules, membrane surface receptors, ion channel proteins, etc., almost contribute to all kinds of physiological and pathological activities of cells. Composition and expression of membrane proteins usually determines the morphology of cells and their migration abilities. Membrane protrusion formation and cell morphological changes are the initial steps of invasion and migration of cancer cells, and the membrane proteins in HCC cells closely correlate with their malignant characteristics of liver cancer such as proliferation, invasion, and metastasis [31–33]. Increased matrix stiffness can also obviously alter the morphology of tumor cells [34, 35], improve their migration abilities [26], affect membrane proteins’ expression and distribution [36, 37]. However, little is known about the changes and functions of membrane proteins in matrix stiffness-mediated metformin resistance in HCC cells.
Here, we used isobaric tags for relative and absolute quantification (iTRAQ) labeling coupled with LC–MS/MS to identify differential membrane proteins related to matrix stiffness-mediated metformin resistance in HCC cells for better understand the mechanism of matrix stiffness-caused metformin resistance.
Methods
Preparation of gel substrates with stiffness 6,10,16 kPa in vitro
An in vitro gel-based culture system with tunable stiffness was established as the method described previously except for the coated matrix protein [21]. The established gel substrates with a diameter of 6 cm (stiffness 6,10 and16 kPa) were coated with 320 µl fibronectin (FN) solution (0.17 mg/ml). For further details, please refer to supplementary files attached (Additional file 1: Table S1).
Cell culture and metformin intervention
MHCC97H cells, a type of human HCC cell lines with high metastatic potential, was established at the Liver Cancer Institute of Fudan University. MHCC97H cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Biowest, South America Origin) and 1% penicillin/streptomycin (Gibco, USA). Approximately 1 × 106 cells in 0.3 ml of culture medium were placed evenly onto the surface of FN-coated gel substrates (diameter in 6 cm) and cultured for 2 h at room temperature. Subsequently, 10 ml culture medium was carefully added into culture dish. The cells were transferred to the incubator(37 °C, 5% CO2)for 48 h culture. The cells grown on different stiffness substrates were collected by a cell scraper.
1 mol/L of metformin (Sigma-Aldrich, St. Louis, MO, USA) was prepared as a stock solution for storage. Intervention concentration of metformin solution was determined based on a dose–response curve of metformin in HCC cells [30], that is 27 mM in MHCC97H cells.
Membrane protein extraction
Membrane proteins were extracted from HCC cells using Membrane Protein Extraction Kit (Thermo Fisher, USA) according to the manufacturer’s protocol. In briefly, the cells were collected and permeated with Permeabilization Buffer, a mild detergent, to release soluble cytoplasmic proteins. A second detergent Solubilization Buffer was used to solubilize and extract membrane and membrane-associated proteins. Protease and phosphatase inhibitors (Thermo Fisher, USA) were added into the Permeabilization and Solubilization Buffers to avoid protein degradation. The concentration of the extracted membrane proteins was measured using BCA Protein Assay Kit (Beyotime, China).
iTRAQ labeling coupled with LC–MS/MS detection
The quality and concentration of the extracted membrane proteins were confirmed to meet the requirement of subsequent experiment. Briefly, 100 µg membrane protein samples of each group were taken into new Eppendorf (EP) tubes, which is relatively small centrifuge tubes, and the volume was supplemented to a total volume of 150 µl with lysate, respectively. Tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher, USA) was added at final concentration of 10 mM and reacted for 60 min at 37 °C. Then we added 40 mM iodoacetamide (Sigma, USA) to each of the tubes and allowed to react for 40 min at room temperature, which also need to be protected from light. Pre-cooled acetone (CHINA SINOPHARM INTERNATIONAL (SHANGHAI) CO., LTD., China) was added to each tube at a ratio of 6:1 acetone: sample volume, and precipitated at -20 °C for 4 h. Centrifuged at 10,000 × g for 20 min and discarded the supernatant to remove the pellet. The precipitated sample was fully dissolved with 100 µl 100 mM tetraethyl ammonium bromide (TEAB) (Santa Cruz, USA). In the process above, the extracted protein completed reduction and alkylation. Then Trypsin (Hualishi Technology, China) was added to with ratio of enzyme to protein, 1:50 to enzymolyze the equal amounts of protein overnight at 37 °C. Following Trypsin digestion, peptides were evacuated by vacuum pump and then reconstituted with TEAB (0.4 M). The iTRAQ reagent (AB Sciex, USA) stored at -20 °C was brought to room temperature, centrifuged and isopropanol was added, followed by vortex centrifugation, then 1 tube of iTRAQ reagent was added to every 100 µg of peptide and incubated at room temperature for 2 h. And peptides were labeled with iTRAQ reagent as follows: NC-L, iTRAQ 113; NC-M, iTRAQ 114; NC-H, iTRAQ 115; Met-L, iTRAQ 116; Met-M, iTRAQ 117; Met-H, iTRAQ 118. Then, added 50 µl ultrapure water to each of the above mixture solution and placed at room temperature for 30 min. Next, The labeled peptides were mixed equally into a new tube, and dried by vacuum concentrator. Peptide samples reconstituted with Ultra-High Performance Liquid Chromatography (UPLC) loading buffer were subjected to high pH liquid phase separation using a reversed-phase C18 column (Waters, USA). UPLC loading buffer is consisted of A and B. Buffer A is 2% acetonitrile (adjusted to pH 10 with liquid ammonia) and buffer B is 80% acetonitrile (adjusted to pH 10 with liquid ammonia). UV detection wavelength was 214 nm. Flow rate was 200 µl/min. Gradient was 50 min. A total of 36 fractions were collected according to peak pattern and time, combined into 12 fractions, concentrated by vacuum centrifugation, dissolved with mass spectrometry loading buffer, and then analyzed on LC-MS/MS system (Thermo, USA). Full scan resolution was 70,000. MS/MS resolution was 17,500. Parent ion scanning range was 300–1800 m/z. The standardized collision energy value was 30 eV. The total running time was 120 min.
Database searching and bioinformatics
Data were acquired automatically on LC-MS/MS system by data dependent acquisition (DDA) mode. Swiss Prot-Human database was selected because HCC cells were human-derived. The raw file was submitted to the proteome discoverer software (version 1.4) during the database searching, selected the established database, and then performed the database search. False Discovery Rate (FDR) ≤ 0.01. Ratios of the 116/113, 117/114 and 118/115 tags from the iTRAQ-labeled peptides were calculated using MATLAB (version 2019b, MathWorks, USA). Fold changes more than 1.5 or less than 0.67 were set as cutoff values to screen significant differences in protein expression. P < 0.05 in three replicates was considered to be statistically significant for protein quantification.
The screened data were analyzed using bioinformatics. We further predicted the protein-protein interactions (PPI) network and signaling pathways which the candidate differentially expressed membrane proteins might be mediated or included using Ingenuity Pathway Analysis (IPA) software. R-Package clusterProfiler was used to analyze the target genes for K-means and Gene Ontology (GO) enrichment analysis, which was also used to map the volcano plots.
Coomassie brilliant blue staining and western blot
For Coomassie brilliant blue staining, approximately 20 µg proteins were loaded and separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), then placed the gel in an appropriate amount of Coomassie blue stain to ensure that the stain adequately covered the gel. Next, stained at room temperature for 1 h with gentle shaking on a shaker. Discarded the dye solution, and added an appropriate amount of Coomassie brilliant blue destaining solution, ensured that the destaining solution covered the gel, shaked slowly on a shaker at room temperature until the blue background was completely removed and the stained protein bands were clear. Finally, stained protein bands were photographed. Similarly, when performing western blot, approximately 20 µg proteins were loaded and separated by SDS-PAGE, and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, USA). Subsequently, the PVDF membrane was blocked in 1×Tris buffered saline (TBS)-Tween ( Sangon Biotech, China) containing 5% fat-free milk, then incubated overnight at 4 °C with the following primary antibodies against human β-Tubulin (1:5000, Affinity), Na-K ATPase (1:1000, Abcam), Filamin-A (1:10000, Abcam), Myosin IIA (1:1000, Abcam), EEF2 (1:10000, Abcam), Moesin (1:20000, Abcam), ANXA2 (1:1000, Abcam), TIP-1 (1:1000, Abcam). Afterwards, the membrane was incubated with Horseradish Peroxidase (HRP)-conjugated secondary antibody (1:5000, Proteintech) on a shaker at room temperature for 1 h. Finally, the target protein bands were visualized by an electrochemiluminescence kit (Tanon, China) and detected using Molecular Imager (Bio-Rad, Hercules, CA).
Statistical analysis
Data analysis was performed using SPSS 20.0 statistical software (SPSS Inc., Chicago, IL, USA). Quantitative variables were expressed as mean ± standard deviation (SD) and statistical analysis was using Student’s t-test. A P < 0.05 (two-tailed) was considered statistical significance.
Results
Identification and categorization of differential membrane proteins
Increased matrix stiffness not only alters obviously morphology and spreading area of HCC cells [21, 35], but also attenuates antitumor effects of therapeutic drugs including metformin [28–30]. These findings led us to speculate that the expression and distribution of membrane proteins might participate in matrix stiffness-mediated metformin resistance. Using same intervention system as the method reported previously [30], we developed in vitro matrix stiffness-mediated metformin resistance cell models for differential membrane proteomics analysis. A detailed flow chart of differential membrane proteomics analysis was shown in Fig. 1. According to the presence or absence of metformin, we divided HCC cells grown on low (6 kPa), medium (10 kPa) and high (16 kPa) stiffness substrates into NC group and Met group. Firstly, we evaluate the quality of the extracted membrane proteins and cytoplasmic protein by polyacrylamide gel electrophoresis staining analysis and internal reference protein analysis. Molecular weight distribution of the extracted membrane proteins in Coomassie brilliant blue staining from HCC cells was consistent with the pattern of membrane protein. Additionally, the extracted membrane proteins from HCC cells had the expression of Na-K ATPase, but almost no expressions in β-tubulin. On the other hand, the extracted cytoplasmic proteins exhibited high expression of β-tubulin, but no expression in Na-K ATPase. These above results suggested that the extracted membrane proteins and membrane junction proteins were qualified for subsequent differential membrane protein analysis (Additional file 2: Figure S1).
Fig. 1.
The workflow chart based on cell culture substrates with tunable stiffness in vitro. L: low stiffness; M: medium stiffness; H: high stiffness. NC refers to non-metformin intervention; Met refers to metformin intervention. iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively
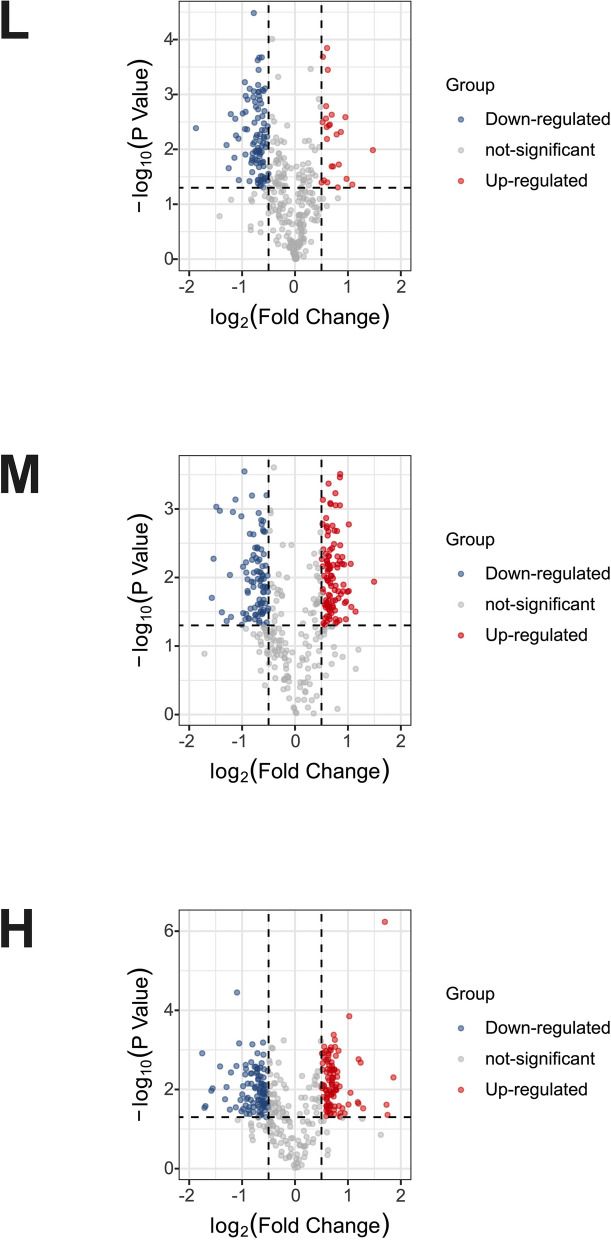
Subsequently, we used a quantitative proteomics technology iTRAQ labeling coupled with LC-MS/MS to screen differential membrane proteins related to matrix stiffness-mediated metformin resistance in HCC cells. A total of 5159 membrane proteins and membrane associated proteins were identified (Additional file 3: Table S2), and among them, 354 differential proteins, which might be associated with matrix stiffness-mediated metformin resistance, were discovered by defining fold-change > 1.5 or < 0.67 and P < 0.05 as a threshold (Additional file 4: Table S3). The volcano plot of differential membrane proteins and membrane associated proteins from HCC cells grown on 6 kPa (L), 10 kPa (M), and 16 kPa (H) substrates after metformin intervention (Fig. 2) showed that 20 upregulated proteins and 86 downregulated proteins in group L, 89 upregulated proteins and 84 downregulated proteins in group M, and 91 upregulated proteins and 93 downregulated proteins in group H. Based on the expression pattern of membrane proteins grown on different stiffness substrates before and after metformin intervention, we further clustered these differential membrane proteins and membrane associated proteins into 6 types of expression patterns using K-means cluster analysis (Fig. 3), and found that twelve typical expression patterns (Types I to VI of up-regulation group;Types A to F of down-regulation group) could reflect the response of HCC cells grown on different stiffness substrates to metformin intervention. We present the raw data for this section in Additional file 5: Figure S2 and Additional file 6: Figure S3. Importantly, the expression level changes of 94 differential membrane proteins (Types II, V, VI in Figure S2 and Types A, C, D in Figure S3) in HCC cells grown on the higher-stiffness substrate after metformin intervention were obviously weaker than those on the lower-stiffness substrate (Table 1), indicating that there exists a close linkage between these candidate membrane proteins and matrix stiffness-mediated metformin resistance.
Fig. 2.
The volcano plot of differentially expressed membrane proteins. Each point in the volcano map represents one protein molecule. Red dots indicate the upregulated proteins after metformin intervention, blue dots represent the downregulated proteins, and the black dots represent nondifferential expressed proteins. The abscissa represents the logarithmic value of the fold expression of a certain protein in the two samples before and after metformin intervention
Fig. 3.
K-means clustering analysis of 6 expression patterns with different trend plots according to the relative expression levels of proteins at different groups. "1" refers to Met-L/NC-L; "2" refers to Met-M/NC-M; and "3" refers to Met-H/NC-H. Maximum Normalization was performed
Table 1.
Typical membrane proteins screened based on resistance to the membrane inhibitory effect induced by high matrix stiffness
| ProteinName | GeneName | 116/113 | 117/114 | 118/115 |
|---|---|---|---|---|
| Prelamin-A/C | LMNA | 1.44066 | 1.503553 | 1.085137 |
| Small nuclear ribonucleoprotein E | SNRPE | 1.442228 | 1.540465 | 1.354483 |
| Insulin-like growth factor-binding protein 1 | IGFBP1 | 1.934701 | 2.022608 | 1.862529 |
| Cytochrome c | CYCS | 1.055029 | 2.020912 | 1.032612 |
| Rho GTPase-activating protein 17 | ARHGAP17 | 1.460284 | 1.885713 | 1.442544 |
| Calcium/calmodulin-dependent protein kinase type II subunit delta | CAMK2D | 1.380145 | 1.73068 | 1.372204 |
| Bcl-2-binding component 3 | BBC3 | 1.753446 | 1.876528 | 1.271211 |
| Lamina-associated polypeptide 2, isoform alpha | TMPO | 1.521199 | 1.408843 | 1.319708 |
| Keratin, type I cytoskeletal 17 | KRT17 | 1.823483 | 1.285828 | 1.145455 |
| Importin subunit alpha-5 | KPNA1 | 1.500398 | 1.471094 | 1.256237 |
| Nuclear mitotic apparatus protein 1 | NUMA1 | 1.964777 | 1.070844 | 1.026603 |
| Programmed cell death protein 4 | PDCD4 | 1.558858 | 1.062319 | 1.008753 |
| Zinc finger HIT domain-containing protein 2 | ZNHIT2 | 2.118072 | 1.276212 | 0.873924 |
| Lysine-specific demethylase 2A | KDM2A | 1.645833 | 1.185506 | 1.064857 |
| Chromodomain-helicase-DNA-binding protein 4 | CHD4 | 1.616529 | 1.310324 | 1.129761 |
| Ubiquitin-like domain-containing CTD phosphatase 1 | UBLCP1 | 1.514853 | 1.189058 | 1.150809 |
| Importin subunit alpha-3 | KPNA4 | 1.578245 | 1.115025 | 1.123152 |
| Complement decay-accelerating factor | CD55 | 1.724473 | 1.438625 | 1.576691 |
| Sodium/myo-inositol cotransporter | SLC5A3 | 1.615669 | 1.228817 | 1.454788 |
| TFIIH basal transcription factor complex helicase XPB subunit | ERCC3 | 1.526915 | 1.146046 | 1.165517 |
| High affinity cationic amino acid transporter 1 | SLC7A1 | 1.774459 | 1.184333 | 1.540541 |
| Annexin A2 | ANXA2 | 0.61652 | 0.899812 | 0.86238 |
| Pyruvate kinase PKM | PKM | 0.58267 | 0.953339 | 0.9183 |
| Moesin | MSN | 0.592999 | 0.965812 | 0.718781 |
| Peptidyl-prolyl cis–trans isomerase A | PPIA | 0.62565 | 0.790193 | 0.735385 |
| Na( +)/H( +) exchange regulatory cofactor NHE-RF1 | SLC9A3R1 | 0.620426 | 0.888193 | 0.711522 |
| 40S ribosomal protein SA | RPSA | 0.587755 | 0.767195 | 0.699355 |
| Ubiquitin-40S ribosomal protein S27a | RPS27A | 0.651907 | 0.823816 | 0.771223 |
| Destrin | DSTN | 0.534129 | 0.845319 | 0.834041 |
| Cofilin-1 | CFL1 | 0.430929 | 1.067309 | 0.864193 |
| LIM and SH3 domain protein 1 | LASP1 | 0.630703 | 0.832025 | 0.796359 |
| Fascin | FSCN1 | 0.665624 | 0.875154 | 0.806212 |
| General vesicular transport factor p115 | USO1 | 0.633543 | 1.006682 | 0.777284 |
| Cysteine and glycine-rich protein 1 | CSRP1 | 0.630921 | 1.09839 | 0.787614 |
| Calponin-2 | CNN2 | 0.651804 | 0.87804 | 0.813255 |
| Plasminogen activator inhibitor 1 RNA-binding protein | SERBP1 | 0.622013 | 0.906532 | 0.760247 |
| Protein S100-A6 | S100A6 | 0.607469 | 1.085248 | 0.689778 |
| 40S ribosomal protein S20 | RPS20 | 0.648542 | 1.040342 | 0.885948 |
| Protein S100-A10 | S100A10 | 0.53718 | 1.349493 | 0.94674 |
| Fatty acid-binding protein, epidermal | FABP5 | 0.552082 | 0.764246 | 0.680111 |
| Cdc42 effector protein 4 | CDC42EP4 | 0.569404 | 0.9141 | 0.787523 |
| Methionine aminopeptidase 2 | METAP2 | 0.523168 | 1.185742 | 0.885365 |
| Clustered mitochondria protein homolog | CLUH | 0.638349 | 0.760548 | 0.749095 |
| Programmed cell death protein 5 | PDCD5 | 0.561837 | 0.804717 | 0.652728 |
| Tax1-binding protein 3 | TAX1BP3 | 0.637478 | 1.147379 | 0.826159 |
| 28S ribosomal protein S21, mitochondrial | MRPS21 | 0.586635 | 0.711005 | 0.609355 |
| Apoptosis-associated speck-like protein containing a CARD | PYCARD | 0.604483 | 0.765976 | 0.701165 |
| Early endosome antigen 1 | EEA1 | 0.572098 | 0.869641 | 0.785589 |
| Tetratricopeptide repeat protein 17 | TTC17 | 0.629422 | 0.783653 | 0.776334 |
| Exocyst complex component 5 | EXOC5 | 0.628536 | 0.772019 | 0.733332 |
| Thioredoxin domain-containing protein 17 | TXNDC17 | 0.550895 | 0.739947 | 0.711344 |
| Malignant T-cell-amplified sequence 1 | MCTS1 | 0.593491 | 1.018201 | 0.726474 |
| Keratin, type II cuticular Hb6 | KRT86 | 0.569451 | 0.607246 | 0.570316 |
| Myotrophin | MTPN | 0.642847 | 1.312341 | 0.948826 |
| Thymosin beta-10 | TMSB10 | 0.570746 | 1.245873 | 0.678481 |
| Interleukin-1 alpha | IL1A | 0.615046 | 0.695885 | 0.661868 |
| Sulfiredoxin-1 | SRXN1 | 0.58674 | 0.637876 | 0.590507 |
| SH3 domain-binding glutamic acid-rich-like protein 3 | SH3BGRL3 | 0.476877 | 0.627616 | 0.619659 |
| Filamin-A | FLNA | 0.624602 | 0.843308 | 0.97049 |
| Glucose-6-phosphate 1-dehydrogenase | G6PD | 0.616705 | 0.800959 | 0.938911 |
| Myosin-9 | MYH9 | 0.523293 | 0.747035 | 0.853047 |
| Fatty acid synthase | FASN | 0.582805 | 0.659283 | 0.722716 |
| Elongation factor 2 | EEF2 | 0.599412 | 0.870375 | 0.918953 |
| Talin-1 | TLN1 | 0.600162 | 0.809829 | 0.902139 |
| 40S ribosomal protein S21 | RPS21 | 0.603461 | 0.922122 | 0.928002 |
| Transgelin-2 | TAGLN2 | 0.461376 | 0.84302 | 0.886943 |
| Cystatin-B | CSTB | 0.478538 | 0.692575 | 0.769809 |
| Flavin reductase (NADPH) | BLVRB | 0.65842 | 0.72311 | 0.734823 |
| Nucleosome assembly protein 1-like 1 | NAP1L1 | 0.457118 | 0.811807 | 0.891115 |
| Golgin subfamily A member 1 | GOLGA1 | 0.620774 | 0.686481 | 0.714003 |
| Programmed cell death protein 6 | PDCD6 | 0.646062 | 0.745869 | 0.84697 |
| Hepatocyte growth factor-regulated tyrosine kinase substrate | HGS | 0.633505 | 0.901127 | 1.041291 |
| Ubiquitin-conjugating enzyme E2 variant 1 | UBE2V1 | 0.615346 | 0.864989 | 0.933444 |
| Exocyst complex component 6 | EXOC6 | 0.652659 | 0.671117 | 0.719448 |
| Macrophage migration inhibitory factor | MIF | 0.6369 | 0.926661 | 0.927719 |
| Exocyst complex component 2 | EXOC2 | 0.522173 | 0.639032 | 0.69052 |
| cAMP-dependent protein kinase type I-alpha regulatory subunit | PRKAR1A | 0.62013 | 0.626398 | 0.744376 |
| Exocyst complex component 1 | EXOC1 | 0.577106 | 0.684352 | 0.72695 |
| Nucleosome assembly protein 1-like 4 | NAP1L4 | 0.613517 | 0.673389 | 0.839352 |
| Alanine–tRNA ligase, cytoplasmic | AARS | 0.563899 | 0.720974 | 0.853792 |
| E3 ubiquitin-protein ligase MARCH6 | 6-Mar | 0.637299 | 0.650759 | 0.83165 |
| Histone H3.1 | HIST1H3A | 0.663002 | 0.851717 | 1.037266 |
| Sequestosome-1 | SQSTM1 | 0.573854 | 0.57978 | 0.588272 |
| Peptidyl-prolyl cis–trans isomerase FKBP1A | FKBP1A | 0.419217 | 0.426265 | 0.572027 |
| Tubulin alpha chain-like 3 | TUBAL3 | 0.75047 | 0.593122 | 0.753641 |
| 28S ribosomal protein S11, mitochondrial | MRPS11 | 0.647251 | 0.637203 | 0.669923 |
| 39S ribosomal protein L14, mitochondrial | MRPL14 | 0.724559 | 0.615447 | 0.740257 |
| Protein phosphatase PTC7 homolog | PPTC7 | 0.760775 | 0.642389 | 0.774724 |
| Toll-interacting protein | TOLLIP | 0.66985 | 0.656658 | 0.762507 |
| Thrombospondin-1 | THBS1 | 0.432559 | 0.40842 | 0.580485 |
| Spondin-1 | SPON1 | 0.607419 | 0.383333 | 0.834185 |
| Midkine | MDK | 0.624462 | 0.609669 | 0.855459 |
| Tumor necrosis factor receptor superfamily member 1A | TNFRSF1A | 0.691871 | 0.624775 | 0.729306 |
| Serpin B7 | SERPINB7 | 0.407513 | 0.343795 | 0.460286 |
iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively
Validation of differential expression patterns of candidate membrane proteins
Due to their impacts on the malignant characteristics of HCC cells, some of candidate membrane proteins mentioned above like Annexin A2 (ANXA2), Filamin-A (FLNA), Moesin (MSN), Myosin-9 (MYH9), Elongation factor 2 (eEF2) and Tax1-binding Protein 3 (TAX1BP3) were further selected as the potential target proteins for future function analysis. As shown in Fig. 4, the expressions of the above membrane proteins were all downregulated in HCC cells grown on different stiffness substrates after metformin intervention. On the other hand, the degree of decrease was obviously weakened as matrix stiffness was increased. These results were in agreement with the results of iTRAQ analysis, indicating that these candidate membrane proteins might contribute to matrix stiffness-mediated metformin resistance in HCC.
Fig. 4.
Validation of differential expression patterns of selected membrane proteins. The expressions of FLNA, MYH9, EEF2, Moesin, ANXA2 and TAX1BP3 were all downregulated in HCC cells grown on different stiffness substrates after metformin intervention, While the degree of decrease was weakened as matrix stiffness was increased, consistent with the iTRAQ results
Prediction of signaling pathways in which the validated proteins above may be participated
The expression pattern of membrane proteins like ANXA2, FLNA, MSN, MYH9, eEF2 and TAX1BP3 indicated that they might be all involved in matrix stiffness-mediated metformin resistance in HCC. We further predicted the protein-protein interactions (PPI) network and signaling pathways which the above membrane proteins might be mediated or included using Ingenuity Pathway Analysis (IPA) software. R-Package clusterProfiler was used to analyze the target genes for Gene ontology (GO) enrichment analysis [38]. Coincidentally, the candidate membrane proteins mentioned above were all belonged to the cluster 1 in Fig. 3. Here, we analyzed them for GO enrichment analysis, and some potential pathways were also shown in Fig. 5. As showed in Fig. 5A, we found that cytoplasmic translation, regulation of actin filament-based process, actin filament organization, platelet aggregation, actin polymerization or depolymerization, homotypic cell-cell adhesion, regulation of actin cytoskeleton organization, actin filament severing, regulation of actin filament organization and regulation of cell morphogenesis were enriched in differential expressed proteins of the BP terms. Of the MF terms, cadherin binding, actin binding, actin filament binding, protein-membrane adaptor activity, calcium-dependent protein binding, structural constituent of ribosome, protein-disulfide reductase (NAD(P)) activity, virion binding, oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor and S100 protein binding were enriched (Fig. 5B). Of the CC terms, cytosolic small ribosomal subunit, secretory granule lumen, cytoplasmic vesicle lumen, vesicle lumen, ruffle, small ribosomal subunit, focal adhesion, cell-substrate junction, cytosolic ribosome and cell cortex were enriched (Fig. 5C). Besides, PPI network of the six candidate membrane proteins involved in important pathways were illustrated in Fig. 5D. Whether these predicted signal pathways are involved in matrix stiffness-mediated metformin resistance in HCC needs to be confirmed in our future work.
Fig. 5.
Gene Ontology (GO) enrichment analysis of genes in cluster 1 and protein–protein interaction (PPI) network construction of the six candidate membrane proteins. A The top 10 GO (biological process, BP) enrichment terms of the cluster 1 proteins; B The top 10 GO (molecular function, MF) enrichment terms of the cluster 1 proteins; C The top 10 GO (cellular component, CC) enrichment terms of the cluster 1 proteins; D PPI network of the six candidate membrane proteins involved in predicted pathways
Discussion
Membrane proteins and their associated proteins almost participate in all physiological and pathological activities of the cells such as cell adhesion, receptor signal transduction, metabolic exchange, ion transport, protein/drug macromolecular transport, immune response, etc. Additionally, they also serve as biomarkers for early diagnosis or prognosis of various cancers, or as the drug targets for evaluating pharmaceutical responses in human malignancies. Increased matrix stiffness can obviously alter the morphology of tumor cells [34, 35] and strengthen their malignant properties [26, 39], indicating that there exists a significant correlation among matrix stiffness, membrane associated protein and malignant behaviors. However, little is known about whether changes and remodeling of matrix stiffness-induced membrane proteins modulates the malignant characteristics of cancer cells. HCC ranks sixth in incidence and fourth in mortality among malignant tumors [40]. The main treatment for HCC is surgery, supplemented by radiotherapy, chemotherapy and transcatheter arterial chemoembolization (TACE). Typical chemotherapeutic drugs such as cisplatin, oxaliplatin, 5-fluorouracil (5-FU), and PD-1/PDL1 monoclonal antibody play an important role in inhibiting cancer progression and prolonging survival. As traditional hypoglycemic agent for type 2 diabetes, metformin alone or combined other drugs, recently exhibits obvious antitumor effect in HCC [41–43]. Biochemical or metabolic factors-mediated drug resistance have been well documented in tumor [44, 45]. However, biomechanical signal-caused therapeutic resistance remains largely unexplored. Recently, our work suggests that higher matrix stiffness significantly attenuates the inhibitory effect of metformin on HCC invasion and metastasis, and PTEN/PI3K/Akt/MMPs pathway contributes to matrix stiffness-mediated metformin resistance [30], highlighting a significant role of biomechanical signal in metformin intervention resistance in HCC. Membrane proteins deliver outside stimulating signals into cells, subsequently influence their biological function. Integrin β1 acts as a “bridge molecule” to transduce extracellular mechanical signal into the cell [21, 46]. Membrane proteins are also involved in stiffness-regulated biological behaviors such as tumor cell proliferation and differentiation, migration and invasion [37, 47, 48]. On the other hand, changes in the cell membrane composition of HCC cells could induce chemoresistance and affect tumor chemosensitivity. Transmembrane glycoprotein FAS ligand (FASLG) is negatively targeted by micoRNA-21-5p, and high expression of micoRNA-21-5p induces cisplatin resistance in HCC cells by inhibiting FASLG expression levels [49]. ABC transporters, also known as ATP-binding cassette proteins, is one of the conventional mechanisms of multidrug resistance (MDR), and effective MDR modulators are regarded as the key for enhancing tumor chemosensitivity. Quercetin inhibits ABCB1, ABCC1, and ABCC2 protein expression through the Wnt/β-catenin pathway to improve the chemosensitivity of HCC cells [50].
In addition, metformin, through the AMPK-CEBPβ pathway, targets and inhibits the expression of the HCC cell surface membrane protein CD133, suggesting that metformin regulates the expression of membrane proteins and plays a cancer-suppressing role [51]. Metformin combined with epigallocatechin gallate (EGCG) significantly reduced the expression level of the membrane interstitial protein phosphatidylinositol proteoglycan-3 (GPC3) in HCC cells and inhibited their proliferation [52]. These studies suggest that membrane protein molecules of HCC cells might be the targets of chemotherapeutic drugs including metformin, further play a role in tumor suppression.
In order to explore the changes of membrane proteins in matrix stiffness-mediated metformin resistance in HCC cells, here, we comparatively analyzed the changes of membrane proteins in HCC cells grown on variable stiffness substrates before and after metformin intervention. By defining fold-change > 1.5 or < 0.67 and P < 0.05 as a threshold, we first screened 354 differential expressed membrane proteins and membrane associated proteins among the total 5159 proteins in HCC cells before and after metformin intervention grown on different stiffness substrates. According to their expression patterns of group L, M and H, we further obtained 94 candidate membrane proteins related to matrix stiffness-mediated metformin resistance. Bioinformatic analysis showed that the expressions of 94 candidate membrane proteins were attenuated in HCC cells grown on high stiffness substrate compared with the control cells on low stiffness substrate. Among the 94 proteins mentioned above, we need to select out representative molecules for further validation. Representative proteins tended to been previously reported to regulate the malignant biological behaviors or chemosensitivity of cancer. Thus, six of these membrane proteins including Annexin A2 (ANXA2), Filamin-A (FLNA), Moesin (MSN), Myosin-9 (MYH9), Eukaryotic binding Elongation factor 2 (eEF2), Tax1-Protein 3 (TAX1BP3; TIP-1) were selected to validate their expression pattern. Since the expression patterns of the above six membrane protein molecules were consistent with the proteomics analysis, these candidate membrane proteins might contribute to matrix stiffness-mediated metformin resistance in HCC.
These six membrane proteins have been previously documented to be associated with malignant tumor progression and chemosensitivity. ANXA2, which is highly expressed in HCC tissues, interacts with ELMO1 to promote HCC cell chemotaxis and metastasis [53]. ANXA can be used as a supplementary serological marker for early diagnosis of HCC, and ANXA combining with AFP achieve a sensitivity of 87.4% for screening of early HCC [54]. Besides, ANXA2 contributes to therapeutic resistance in several tumors [55, 56]. Analysis of 113 patients with HCC after resection indicate a correlation between FLNA and HCC recurrence rate. FLNA expression could predict the early recurrence of HCC after hepatectomy and contribute to postoperative follow-up [57]. Additionally, a comparative proteomics analysis suggests that FLNA can be used as a potential marker for HCC progression [58]. Moreover, FLNA also serves as a predictor of chemoresistance in some cancers such as colorectal cancer, cervical cancer, etc. [59, 60]. Moesin, a member of the ezrin-radixin-moesin (ERM) family, is involved in the regulation of cell adhesion, polarity, and migration through cross-linking between plasma membrane proteins and the actin cytoskeleton, and has been found to play a key role in hepatic stellate cell activation and liver fibrogenesis [61]. Overexpression of miR-200c negatively regulates Moesin expression to inhibit the proliferation and invasion of glioma cells, Moesin is highly expressed in glioma specimens, and Moesin promotes glioma cell development [62]. Moesin participates in regulation of breast cancer therapeutic resistance [63, 64]. A proteomic study on colorectal liver metastases demonstrates that MYH9 overexpression was associated with shorter overall survival and disease-free survival, indicating that MYH9 has potential predictive value for colorectal liver metastases [65]. Other researches also shows that MYH9 was involved in cancer cells death resistance and promotes metastasis [66, 67]. eEF2 and phosphorylated eEF2 are prognostic indicators of HCC patient survival [68], and that eEF2 kinase promotes HCC angiogenesis and tumor progression through SP1/KLF5-mediated VEGF expression [69]. Modulation of eEF2 and its kinases is therefore a potential drug target for cancer therapy. Alkaloids in Coptidis rhizome suppresses eEF2 activity, and then inhibits tumor growth and angiogenesis in animal experiments, suggesting anti-hepatoma efficacy [70]. In addition, eEF2 was also related to chemoresistance in malignant tumors [71, 72]. Tax1 binding protein 3, also known as TIP-1, is widely involved in biological processes through selective protein interactions. TAX1BP3 expression levels are increased in human invasive breast cancer, and contribute to cell adhesion to extracellular matrix, invasion and lung metastasis [73]. TAX1BP3 expression could facilitate angiogenesis and tumor formation of human glioblastoma cells and were closely correlated with the prognosis of glioblastoma patients [74], suggesting that TAX1BP3 can be regarded as a prognostic marker for human glioblastoma. Additionally, TAX1BP3 can be a therapy target to regulate chemosensitivity of gastric cancer cells to 5-FU [75]. Accordingly, it is reasonable to assume that these membrane proteins might be involved in matrix stiffness-mediated metformin resistance in HCC. However, few studies related to the six membrane proteins above were focused on the biomechanics impact within tumor microenvironment on therapeutic resistance. Thus, little is known about whether changes and remodeling of matrix stiffness-induced membrane proteins modulates the malignant characteristics of HCC cells. The signaling pathways which the above membrane proteins might be mediated or participated predicted by GO enrichment analysis were also seldomly reported in cancer chemoresistance previously, which deserves further study. To search for the membrane proteins related to matrix stiffness-induced therapeutic resistance and explore the potential mechanisms, which is the purpose of our study.
Conclusions
In summary, we identified some differential membrane proteins and membrane associated proteins related to matrix stiffness-mediated metformin resistance in HCC cells in this study. Six candidate membrane proteins can reflect the response of HCC cells under high stiffness stimulation to metformin intervention, which deserve to be investigated in the future.
Supplementary Information
Additional file 1: Table S1. Ingredients of polyacrylamide gel substrates with variable stiffness.
Additional file 2: Figure S1. Efficiency and quality identification of membrane protein extraction. (A) The approximate location and range of membrane protein molecules performed by polyacrylamide gel electrophoresis with Coomassie brilliant blue staining. (B)(i, ii) Western blot were performed on the six groups of membrane proteins and cytoplasmic proteins.
Additional file 3: Table S2. A list of all identified proteins.
Additional file 4: Table S3. Differential membrane proteins and membrane associated proteins.
Additional file 5: Figure S2. Six typical expression patterns reflect the increase degree of membrane proteins. iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively.
Additional file 6: Figure S3. Six typical expression patterns reflect the decrease degree of membrane proteins. iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively.
Acknowledgements
All authors are very grateful to Dr Huanhuan Zhao from Institute of Biomedical Science, Fudan University for her technical help in iTRAQ.
Abbreviations
- HCC
Hepatocellular carcinoma
- iTRAQ
Isobaric tags for relative and absolute quantification
- LC–MS/MS
Liquid chromatography-tandem mass spectrometry
- ANXA2
Annexin A2
- FLNA
Filamin-A
- MSN
Moesin
- MYH9
Myosin-9
- eEF2
Eukaryotic Elongation Factor 2
- TAX1BP3
Tax1-binding protein 3
- EMT
Epithelial-mesenchymal transition
- FBS
Fetal bovine serum
- SDS-PAGE
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- PVDF
Polyvinylidene difluoride
- PPI
Protein–protein interactions
- IPA
Ingenuity Pathway Analysis
- GO
Gene Ontology
Authors’ contributions
YHY and JFC conceived and supervised the whole study; XYG performed the most experiments and wrote the manuscript text; JLQ helped to collect and analyze the data. YZ did the main data statistics and bioinformatics analysis work, prepared Figs. 2, 3 and 5. HMW helped to modify and polish the manuscript grammatically and linguistically. All authors reviewed and approved the final manuscript.
Funding
This study was supported by National Key R&D Program of China (2016YFC1305105) and National Natural Science Foundation of China (NO.81670751, 81573019, 81970711).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiefeng Cui, Email: cui.jiefeng@zs-hospital.sh.cn.
Yehong Yang, Email: yehongyang@fudan.edu.cn.
References
- 1.Umetsu S, Mizukami H, Saito T, Uchida C, Igawa A, Kudo K, Itabashi C, Osonoi S, Danyang G, Sasaki T, et al. Diabetes, an independent poor prognostic factor of non-B non-C hepatocellular carcinoma, correlates with dihydropyrimidinase-like 3 promoter methylation. Sci Rep. 2020;10(1):1156. doi: 10.1038/s41598-020-57883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suh S, Kim KW. Diabetes and Cancer: Cancer should be screened in Routine Diabetes Assessment. Diabetes Metab J. 2019;43(6):733–743. doi: 10.4093/dmj.2019.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mekala MR, Bangi BB, Lebaka NJ, Nadendla RR, Ginjupally LK. Association of diabetes with oral Cancer- an enigmatic correlation. Asian Pac J Cancer Prev. 2020;21(3):809–814. doi: 10.31557/APJCP.2020.21.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigneri P, Frasca F, Sciacca L, Pandini G, Vigneri R. Diabetes and cancer. Endocr Relat Cancer. 2009;16(4):1103–1123. doi: 10.1677/ERC-09-0087. [DOI] [PubMed] [Google Scholar]
- 5.Vacante F, Senesi P, Montesano A, Paini S, Luzi L, Terruzzi I. Metformin Counteracts HCC Progression and Metastasis Enhancing KLF6/p21 Expression and Downregulating the IGF Axis. International journal of endocrinology 2019, 2019:7570146. [DOI] [PMC free article] [PubMed]
- 6.Hu, Zeng Z, Xia Q, Liu Z, Feng X, Chen J, Huang M, Chen L, Fang Z, Liu Q et al. Metformin attenuates hepatoma cell proliferation by decreasing glycolytic flux through the HIF-1alpha/PFKFB3/PFK1 pathway. Life Sci 2019:116966. [DOI] [PubMed]
- 7.Klubo-Gwiezdzinska J, Jensen K, Costello J, Patel A, Hoperia V, Bauer A, Burman KD, Wartofsky L, Vasko V. Metformin inhibits growth and decreases resistance to anoikis in medullary thyroid cancer cells. Endocr Relat Cancer. 2012;19(3):447–456. doi: 10.1530/ERC-12-0046. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Tong D, Liu G, Xu J, Do K, Geary K, Zhang D, Zhang J, Zhang Y, Li Y, et al. Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-beta1/STAT3 axis-regulated EMT. Cell Death Dis. 2017;8(8):e3007. doi: 10.1038/cddis.2017.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magri A, Novara L et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science (New York, NY) 2019. [DOI] [PubMed]
- 10.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim et Biophys acta Reviews cancer. 2017;1868(1):1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Hirpara J, Eu JQ, Tan JKM, Wong AL, Clement MV, Kong LR, Ohi N, Tsunoda T, Qu J, Goh BC, et al. Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance. Redox Biol. 2019;25:101076. doi: 10.1016/j.redox.2018.101076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katara GK, Kulshrestha A, Mao L, Wang X, Sahoo M, Ibrahim S, Pamarthy S, Suzue K, Shekhawat GS, Gilman-Sachs A, et al. Mammary epithelium-specific inactivation of V-ATPase reduces stiffness of extracellular matrix and enhances metastasis of breast cancer. Mol Oncol. 2018;12(2):208–223. doi: 10.1002/1878-0261.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty S, Hong W. Linking Extracellular Matrix Agrin to the Hippo Pathway in Liver Cancer and Beyond. Cancers. 2018;10(2):45. doi: 10.3390/cancers10020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med. 2014;20(4):360–367. doi: 10.1038/nm.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan RL, Baack B, Smith BD, Yartel A, Pitasi M, Falck-Ytter Y. Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma: a meta-analysis of observational studies. Ann Intern Med. 2013;158(5 Pt 1):329–337. doi: 10.7326/0003-4819-158-5-201303050-00005. [DOI] [PubMed] [Google Scholar]
- 17.Affo S, Yu LX, Schwabe RF. The role of Cancer-Associated fibroblasts and fibrosis in Liver Cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127(5 Suppl 1):35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Masuzaki R, Tateishi R, Yoshida H, Yoshida H, Sato S, Kato N, Kanai F, Sugioka Y, Ikeda H, Shiina S, et al. Risk assessment of hepatocellular carcinoma in chronic hepatitis C patients by transient elastography. J Clin Gastroenterol. 2008;42(7):839–843. doi: 10.1097/mcg.0b013e318050074f. [DOI] [PubMed] [Google Scholar]
- 20.Masuzaki R, Tateishi R, Yoshida H, Goto E, Sato T, Ohki T, Imamura J, Goto T, Kanai F, Kato N, et al. Prospective risk assessment for hepatocellular carcinoma development in patients with chronic hepatitis C by transient elastography. Hepatology (Baltimore, MD) 2009;49(6):1954–1961. doi: 10.1002/hep.22870. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y, Xie X, Wang Z, Hu C, Zheng Q, Wang Y, Chen R, Xue T, Chen J, Gao D, et al. Increasing matrix stiffness upregulates vascular endothelial growth factor expression in hepatocellular carcinoma cells mediated by integrin beta1. Biochem Biophys Res Commun. 2014;444(3):427–432. doi: 10.1016/j.bbrc.2014.01.079. [DOI] [PubMed] [Google Scholar]
- 22.You Y, Zheng Q, Dong Y, Wang Y, Zhang L, Xue T, Xie X, Hu C, Wang Z, Chen R, et al. Higher Matrix Stiffness Upregulates Osteopontin expression in Hepatocellular Carcinoma cells mediated by integrin beta1/GSK3beta/beta-Catenin signaling pathway. PLoS ONE. 2015;10(8):e0134243. doi: 10.1371/journal.pone.0134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, Cui J, Qin Q, Zhang J, Liu L, Deng S, Wu C, Yang M, Li S, Wang C. Mechanical stiffness of liver tissues in relation to integrin beta1 expression may influence the development of hepatic cirrhosis and hepatocellular carcinoma. J Surg Oncol. 2010;102(5):482–489. doi: 10.1002/jso.21613. [DOI] [PubMed] [Google Scholar]
- 24.Liu QP, Luo Q, Deng B, Ju Y, Song GB. Stiffer Matrix accelerates Migration of Hepatocellular Carcinoma cells through enhanced Aerobic Glycolysis Via the MAPK-YAP signaling. Cancers. 2020;12(2):490. doi: 10.3390/cancers12020490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You Y, Zheng Q, Dong Y, Xie X, Wang Y, Wu S, Zhang L, Wang Y, Xue T, Wang Z, et al. Matrix stiffness-mediated effects on stemness characteristics occurring in HCC cells. Oncotarget. 2016;7(22):32221–32231. doi: 10.18632/oncotarget.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Y, Zheng Q, Wang Z, Lin X, You Y, Wu S, Wang Y, Hu C, Xie X, Chen J, et al. Higher matrix stiffness as an independent initiator triggers epithelial-mesenchymal transition and facilitates HCC metastasis. J Hematol Oncol. 2019;12(1):112. doi: 10.1186/s13045-019-0795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, Zheng Q, Xing X, Dong Y, Wang Y, You Y, Chen R, Hu C, Chen J, Gao D, et al. Matrix stiffness-upregulated LOXL2 promotes fibronectin production, MMP9 and CXCL12 expression and BMDCs recruitment to assist pre-metastatic niche formation. J Experimental Clin Cancer Res CR. 2018;37(1):99. doi: 10.1186/s13046-018-0761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Liu Y, Xie HG, Zhao S, Xu XX, Fan LX, Guo X, Lu T, Sun GW, Ma XJ. Role of three-dimensional matrix stiffness in regulating the chemoresistance of hepatocellular carcinoma cells. Biotechnol Appl Chem. 2015;62(4):556–562. doi: 10.1002/bab.1302. [DOI] [PubMed] [Google Scholar]
- 29.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology (Baltimore, MD) 2011;53(4):1192–1205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X, Qiao X, Xing X, Huang J, Qian J, Wang Y, Zhang Y, Zhang X, Li M, Cui J, et al. Matrix stiffness-upregulated MicroRNA-17-5p attenuates the intervention Effects of Metformin on HCC Invasion and Metastasis by targeting the PTEN/PI3K/Akt pathway. Front Oncol. 2020;10:1563. doi: 10.3389/fonc.2020.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha Y, Fang Y, Romecin Duran PA, Tolosa EJ, Moser CD, Fernandez-Zapico ME, Roberts LR. Induction of Lysosome‐associated protein transmembrane 4 Beta via sulfatase 2 enhances autophagic flux in Liver Cancer cells. Hepatol Commun. 2019;3(11):1520–1543. doi: 10.1002/hep4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietrich P, Hellerbrand C, Bosserhoff A. The Delta Subunit of Rod-Specific photoreceptor cGMP phosphodiesterase (PDE6D) contributes to Hepatocellular Carcinoma Progression. Cancers. 2019;11(3):398. doi: 10.3390/cancers11030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson MM, Jennings LK, Zhang XA. Tetraspanins and tumor progression. Clin Exp Metastasis. 2011;28(3):261–270. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- 34.Chiang MY, Yangben Y, Lin NJ, Zhong JL, Yang L. Relationships among cell morphology, intrinsic cell stiffness and cell-substrate interactions. Biomaterials. 2013;34(38):9754–9762. doi: 10.1016/j.biomaterials.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Chou SY, Cheng CM, LeDuc PR. Composite polymer systems with control of local substrate elasticity and their effect on cytoskeletal and morphological characteristics of adherent cells. Biomaterials. 2009;30(18):3136–3142. doi: 10.1016/j.biomaterials.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 36.Gopal S, Multhaupt HAB, Pocock R, Couchman JR. Cell-extracellular matrix and cell-cell adhesion are linked by syndecan-4. Matrix Biology. 2017;60–61:57–69. doi: 10.1016/j.matbio.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Wanggou S, Bodalia A, Zhu M, Dong W, Fan JJ, Yin WC, Min HK, Hu M, Draghici D, et al. A feedforward mechanism mediated by Mechanosensitive Ion Channel PIEZO1 and tissue mechanics promotes Glioma Aggression. Neuron. 2018;100(4):799–815e797. doi: 10.1016/j.neuron.2018.09.046. [DOI] [PubMed] [Google Scholar]
- 38.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Ma M, Dong G, Yao RR, Li JH, Zheng QD, Dong YY, Ma H, Gao DM, Cui JF, et al. Increased matrix stiffness promotes tumor progression of residual hepatocellular carcinoma after insufficient heat treatment. Cancer Sci. 2017;108(9):1778–1786. doi: 10.1111/cas.13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 41.Zhang HH, Zhang Y, Cheng YN, Gong FL, Cao ZQ, Yu LG, Guo XL. Metformin incombination with curcumin inhibits the growth, metastasis, and angiogenesis of hepatocellular carcinoma in vitro and in vivo. Mol Carcinog. 2018;57(1):44–56. doi: 10.1002/mc.22718. [DOI] [PubMed] [Google Scholar]
- 42.Abdelmonsif DA, Sultan AS, El-Hadidy WF, Abdallah DM. Targeting AMPK, mTOR and beta-catenin by combined metformin and aspirin therapy in HCC: an Appraisal in Egyptian HCC Patients. Mol Diagn Ther. 2018;22(1):115–127. doi: 10.1007/s40291-017-0307-7. [DOI] [PubMed] [Google Scholar]
- 43.Murff HJ, Roumie CL, Greevy RA, Hackstadt AJ, McGowan LEDA, Hung AM, Grijalva CG, Griffin MR. Metformin use and incidence cancer risk: evidence for a selective protective effect against liver cancer. Cancer Causes Control. 2018;29(9):823–832. doi: 10.1007/s10552-018-1058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montero J, Gstalder C, Kim DJ, Sadowicz D, Miles W, Manos M, Cidado JR, Paul Secrist J, Tron AE, Flaherty K, et al. Destabilization of NOXA mRNA as a common resistance mechanism to targeted therapies. Nat Commun. 2019;10(1):5157. doi: 10.1038/s41467-019-12477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R, Bi J, Ampah KK, Zhang C, Li Z, Jiao Y, Wang X, Ba X, Zeng X. Lipid raft regulates the initial spreading of melanoma A375 cells by modulating β1 integrin clustering. Int J Biochem Cell Biol. 2013;45(8):1679–1689. doi: 10.1016/j.biocel.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 48.Yeh YC, Ling JY, Chen WC, Lin HH, Tang MJ. Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: reciprocal regulation of caveolin-1 and β1 integrin. Sci Rep. 2017;7(1):15008. doi: 10.1038/s41598-017-14932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, Yang C, Sun C, Sun Y, Yang Z, Cheng S, Zhuge B. Mir-21-5p suppressed the sensitivity of Hepatocellular Carcinoma cells to cisplatin by Targeting FASLG. DNA Cell Biol. 2019;38(8):865–873. doi: 10.1089/dna.2018.4529. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z, Huang C, Ma T, Jiang L, Tang L, Shi T, Zhang S, Zhang L, Zhu P, Li J, et al. Reversal effect of quercetin on multidrug resistance via FZD7/beta-catenin pathway in hepatocellular carcinoma cells. Phytomedicine. 2018;43:37–45. doi: 10.1016/j.phymed.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 51.Maehara O, Ohnishi S, Asano A, Suda G, Natsuizaka M, Nakagawa K, Kobayashi M, Sakamoto N, Takeda H. Metformin regulates the expression of CD133 through the AMPK-CEBPβ Pathway in Hepatocellular Carcinoma Cell Lines. Neoplasia. 2019;21(6):545–556. doi: 10.1016/j.neo.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabry D, Abdelaleem OO, El Amin Ali AM, Mohammed RA, Abdel-Hameed ND, Hassouna A, Khalifa WA. Anti-proliferative and anti-apoptotic potential effects of epigallocatechin-3-gallate and/or metformin on hepatocellular carcinoma cells: in vitro study. Mol Biol Rep. 2019;46(2):2039–2047. doi: 10.1007/s11033-019-04653-6. [DOI] [PubMed] [Google Scholar]
- 53.Hongyan L, ⁎, Wanga Y. Yinying Lub,⁎⁎, Fei Lia,⁎⁎ Annexin A2 interacting with ELMO1 regulates HCC chemotaxis and metastasis. Life sciences 2019. [DOI] [PubMed]
- 54.Sun Y, Gao G, Cai J, Wang Y, Qu X, He L, Liu F, Zhang Y, Lin K, Ma S, et al. Annexin A2 is a discriminative serological candidate in early hepatocellular carcinoma. Carcinogenesis. 2013;34(3):595–604. doi: 10.1093/carcin/bgs372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan Y, Si W, Ji W, Wang Z, Gao Z, Tian R, Song W, Zhang H, Niu R, Zhang F. Rack1 mediates tyrosine phosphorylation of Anxa2 by src and promotes invasion and metastasis in drug-resistant breast cancer cells. Breast Cancer Res BCR. 2019;21(1):66. doi: 10.1186/s13058-019-1147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng X, Liu H, Zhang Z, Gu Y, Qiu H, He Z. Annexin A2 contributes to cisplatin resistance by activation of JNK-p53 pathway in non-small cell lung cancer cells. J Experimental Clin Cancer Res: CR. 2017;36(1):123. doi: 10.1186/s13046-017-0594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donadon M, Di Tommaso L, Soldani C, Franceschini B, Terrone A, Mimmo A, Vitali E, Roncalli M, Lania A, Torzilli G. Filamin A expression predicts early recurrence of hepatocellular carcinoma after hepatectomy. Liver Int. 2018;38(2):303–311. doi: 10.1111/liv.13522. [DOI] [PubMed] [Google Scholar]
- 58.Jianzhong Ai HH, Lv X, Tang Z, Chen M, Chen T, Duan W, Sun H, Li Q, Tan R, Liu Y, Duan J, Yang Y, Wei Y, Li Y, Qin Zhou FLNA and PGK1 are two potential markers for progression in Hepatocellular Carcinoma. Cell Physiol Biochemistry: Int J Experimental Cell Physiol Biochem Pharmacol. 2011;27:207–216. doi: 10.1159/000327946. [DOI] [PubMed] [Google Scholar]
- 59.Cheng M, Jiang Y, Yang H, Zhao D, Li L, Liu X. FLNA promotes chemoresistance of colorectal cancer through inducing epithelial-mesenchymal transition and smad2 signaling pathway. Am J Cancer Res. 2020;10(2):403–423. [PMC free article] [PubMed] [Google Scholar]
- 60.Jin YZ, Pei CZ, Wen LY. FLNA is a predictor of chemoresistance and poor survival in cervical cancer. Biomark Med. 2016;10(7):711–719. doi: 10.2217/bmm-2016-0056. [DOI] [PubMed] [Google Scholar]
- 61.Karvar S, Ansa-Addo EA, Suda J, Singh S, Zhu L, Li Z, Rockey DC. Moesin, an ERM family member, regulates hepatic fibrosis. Hepatology (Baltimore MD) 2019. [DOI] [PMC free article] [PubMed]
- 62.Qin Y, Chen W, Liu B, Zhou L, Deng L, Niu W, Bao D, Cheng C, Li D, Liu S, et al. MiR-200c inhibits the Tumor Progression of Glioma via Targeting Moesin. Theranostics. 2017;7(6):1663–1673. doi: 10.7150/thno.17886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yano K, Okabe C, Fujii K, Kato Y, Ogihara T. Regulation of breast cancer resistance protein and P-glycoprotein by ezrin, radixin and moesin in lung, intestinal and renal cancer cell lines. J Pharm Pharmacol. 2020;72(4):575–582. doi: 10.1111/jphp.13225. [DOI] [PubMed] [Google Scholar]
- 64.Alam F, Mezhal F, El Hasasna H, Nair VA, Aravind SR, Saber Ayad M, El-Serafi A, Abdel-Rahman WM. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumour Biology. 2017;39(9):1010428317714634. doi: 10.1177/1010428317714634. [DOI] [PubMed] [Google Scholar]
- 65.Ma YS, Huang T, Zhong XM, Zhang HW, Cong XL, Xu H, Lu GX, Yu F, Xue SB, Lv ZW, et al. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol Cancer. 2018;17(1):139. doi: 10.1186/s12943-018-0890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye G, Yang Q, Lei X, Zhu X, Li F, He J, Chen H, Ling R, Zhang H, Lin T, et al. Nuclear MYH9-induced CTNNB1 transcription, targeted by staurosporin, promotes gastric cancer cell anoikis resistance and metastasis. Theranostics. 2020;10(17):7545–7560. doi: 10.7150/thno.46001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Jiang Q, Liu X, Lin X, Tang Z, Liu C, Zhou J, Zhao M, Li X, Cheng Z, et al. Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3β/β-catenin signaling pathway. EBioMedicine. 2019;48:386–404. doi: 10.1016/j.ebiom.2019.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leona L, Pott SH, Reis H, Lorenz K, Bracht T, Herold T, Skryabin BV, Dominik A, Megger. Julia Kälsch, Frank Weber, Barbara Sitek, Hideo. Baba: eukaryotic elongation factor 2 is a prognostic marker and its kinase a potential therapeutic target in HCC. Oncotarget. 2017;8(7):11950–62. [DOI] [PMC free article] [PubMed]
- 69.Zhou Y, Li Y, Xu S, Lu J, Zhu Z, Chen S, Tan Y, He P, Xu J, Proud CG, et al. Eukaryotic elongation factor 2 kinase promotes angiogenesis in hepatocellular carcinoma via PI3K/Akt and STAT3. Int J Cancer. 2020;146(5):1383–1395. doi: 10.1002/ijc.32560. [DOI] [PubMed] [Google Scholar]
- 70.Tan HY, Wang N, Tsao SW, Zhang Z, Feng Y. Suppression of vascular endothelial growth factor via inactivation of eukaryotic elongation factor 2 by alkaloids in Coptidis rhizome in hepatocellular carcinoma. Integr Cancer Ther. 2014;13(5):425–434. doi: 10.1177/1534735413513635. [DOI] [PubMed] [Google Scholar]
- 71.Wang RX, Xu XE, Huang L, Chen S, Shao ZM. eEF2 kinase mediated autophagy as a potential therapeutic target for paclitaxel-resistant triple-negative breast cancer. Ann Translational Med. 2019;7(23):783. doi: 10.21037/atm.2019.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Zhang Y, Liu XY, Qin ZH, Yang JM. Expression of elongation factor-2 kinase contributes to anoikis resistance and invasion of human glioma cells. Acta Pharmacol Sin. 2011;32(3):361–367. doi: 10.1038/aps.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han M, Wang H, Zhang HT, Han Z. The PDZ protein TIP-1 facilitates cell migration and pulmonary metastasis of human invasive breast cancer cells in athymic mice. Biochem Biophys Res Commun. 2012;422(1):139–145. doi: 10.1016/j.bbrc.2012.04.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han M, Wang H, Zhang HT, Han Z. Expression of tax-interacting protein 1 (TIP-1) facilitates angiogenesis and tumor formation of human glioblastoma cells in nude mice. Cancer Lett. 2013;328(1):55–64. doi: 10.1016/j.canlet.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi H, Lee SK. TAX1BP1 downregulation by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer cells to 5-FU. Arch Virol. 2017;162(2):369–377. doi: 10.1007/s00705-016-3109-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Ingredients of polyacrylamide gel substrates with variable stiffness.
Additional file 2: Figure S1. Efficiency and quality identification of membrane protein extraction. (A) The approximate location and range of membrane protein molecules performed by polyacrylamide gel electrophoresis with Coomassie brilliant blue staining. (B)(i, ii) Western blot were performed on the six groups of membrane proteins and cytoplasmic proteins.
Additional file 3: Table S2. A list of all identified proteins.
Additional file 4: Table S3. Differential membrane proteins and membrane associated proteins.
Additional file 5: Figure S2. Six typical expression patterns reflect the increase degree of membrane proteins. iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively.
Additional file 6: Figure S3. Six typical expression patterns reflect the decrease degree of membrane proteins. iTRAQ tags 113, 114, 115, 116, 117 and 118 represent NC-L, NC-M, NC-H, Met-L, Met-M and Met-H, respectively.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.