Abstract
Background:
Sperm-associated antigen 5 (SPAG5) has been identified as a novel driver oncogene involved in multiple cancers; however, its role in lung adenocarcinoma (LUAD) needs further investigation. Our study aims to elucidate the potential significance of SPAG5 in LUAD prognosis and its implications for the efficacy of immunotherapy.
Methods:
In this study, we used bioinformatics analysis and tissue microarray (TMA) staining to examine the potential role of SPAG5 in LUAD survival and response to immunotherapy. We used the Oncomine, TIMER2.0, Gene Expression Profiling Interactive Analysis (GEPIA), Sangerbox, PredicScan, and Kaplan-Meier Plotter databases to examine the expression and prognostic role of SPAG5 in the LUAD of The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and other databases. We also used Cancer Single-cell State Atlas (CancerSEA) and Tumor Immune Estimation Resource (TIMER2.0) to analyze the association of SPAG5 with malignant phenotype and tumor immune microenvironment. Furthermore, Immune Cell Abundance Identifier (ImmuCellAI) analysis of TCGA sequencing data was used to predict the role of SPAG5 in determining the response to immune checkpoint blockade (ICB) treatment in LUAD. Co-expression analysis of programmed death-ligand 1 (PD-L1) and SPAG5 was performed using LUAD TMA immunohistochemistry (IHC) analysis.
Results:
Our findings indicate that SPAG5 is overexpressed in LUAD and is positively correlated with advanced clinical stage, poor overall survival, relapse-free survival, and progression-free survival outcomes. SPAG5 may be involved in regulating the cell cycle, proliferation, invasion, DNA damage and repair, and tumor immunosuppression. Furthermore, TMA IHC analysis showed a positive correlation between PD-L1 expression in LUAD and SPAG5 which suggests that SPAG5 may serve as a potential predictor of response to ICB therapy in LUAD.
Conclusions:
Our results highlight the role of SAPG5 in promoting a tumor malignancy phenotype and immunosuppression in LUAD and suggest that SPAG5 may serve as a potential response marker for ICB therapy.
Keywords: Sperm-associated antigen 5, prognosis, immune infiltration, lung adenocarcinoma, immune checkpoint blockade treatment
Introduction
Lung cancer is a malignant disease with the highest incidence and mortality globally. 1 Lung adenocarcinoma (LUAD) comprises more than 40% of all lung cancer cases and poses a systemic threat due to higher tendency of developing distant metastases and a mean overall survival (OS) of less than 5 years.2,3 Although immunotherapy such as immune checkpoint programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1) blockade therapy has made notable advances in treating various cancers including LUAD, there are still shortcomings such as low overall response rates, immune-related toxicities, and a lack of efficacy predictors.4-9 Thus far, PD-L1, tumor mutation burden (TMB), and microsatellite instability (MSI) have been approved as simultaneous diagnostic markers to guide immune checkpoint inhibitor therapy. 10 Nevertheless, accumulating evidence strongly suggests that using PD-L1 expression, MSI, or TMB solely as a single indicator is not comprehensive enough and cannot accurately predict the use of immunotherapy. 10 Therefore, it is essential to not only urgently identify the mechanisms of resistance to immunotherapy in LUAD but also screen for current predictive and prognostic markers to better improve the immune response.
Sperm-associated antigen 5 (SPAG5) is a mitotic spindle protein that has been observed to be highly expressed in various cancers and is indicated as a novel driver oncogene and a therapeutic target. 11 Studies have shown that elevated SPAG5 levels are associated with neoplastic growth, metastasis, chemoresistance, and shorter survival across a broad spectrum of cancers.12-16 Further research indicates that SPAG5 acts as an oncogene by modulating various signaling pathways to regulate tumorigenesis and progression, although counteracting the effects of various chemotherapies. For instance, SPAG5 involves AKT/mammalian target of rapamycin (mTOR), WNT/β-catenin, and PI3K/AKT signaling pathways to regulate tumor growth, apoptosis, and metastasis14,15,17 Although there is limited evidence, research suggests a strong link between SPAG5 and immune cell infiltration in cancer. A previous study has shown that SPAG5 is an alternative cancer vaccine target in several cancers, as it is positively associated with CD8 T cell infiltration based on a comprehensive analysis of tumor immunity programs. 18 In addition, there is a significant correlation between SPAG5 and CD8+ T cell infiltration in breast cancer. 12 In LUAD, SPAG5 has been found to promote tumor proliferation and motility and can be used as a prognostic marker.19-22 Despite existing studies that suggest SPAG5 is an oncogene in LUAD, a more comprehensive assessment of its role in LUAD and its relation to immune cell infiltration is needed.
In this study, we assessed the prognostic role of SPAG5 in LUAD using tissue microarray (TMA) staining. We also analyzed its relationship with tumor progression, clinical outcome, immune infiltration, and response to immunotherapy using bioinformatics analysis. Our findings can improve our understanding of the role of SPAG5 in LUAD and its relation to immune cell infiltration.
Material and Methods
Tissue microarray and immunohistochemistry staining
In this study, 2 TMAs were used, 1 containing 180 specimens (Cat no: ZL-LUC1801, WellBio, Shanghai), including 90 LUAD tissues and 90 adjacent non-cancerous tissues, and the other containing 140 specimens from 70 lung cancer patients (Cat no: LC1401, Alenabio, Xiena), each containing 2 duplicates of tumor tissues and PD-L1 expression information detected by 2 Food and Drug Administration (FDA)-proved antibodies (SP142 and SP263). Among them, the staining results for SP142 were classified into 3 categories: tumor cell ⩾50%, tumor cell <50%, and immune cell <10%, as well as negative. The staining results for SP163 were divided into 2 categories: tumor cell ⩾25% and negative. Therefore, in the correlation analysis, all patients who showed PD-L1 expression were considered as PD-L1 positive. Both TMAs were purchased from Yunnan WoSai Biology Science and Technology Co., Ltd. (Kunming, China). Therefore, LC1401 TMA was used to analyze the correlation between SPAG5 and PD-L1 expression. Moreover, after excluding all patients with incomplete clinical data in ZL-LUC1801 TMA, there were still tissues available from 75 patients for survival analysis and compare the expression difference of SPAG5 between tumor and normal tissue. The statistics of clinical features are shown in Supplementary Tables 1 and 2.
The immunohistochemistry (IHC) staining procedure can be summarized as follows: the TMAs were deparaffinized in xylene for 10 min, repeated twice, followed by rehydration using a graded ethanol series (100%, 95%, and 70% ethanol) for 3 min each. Antigen retrieval was performed by heating the sections in a microwave using citrate buffer (pH 6.0) until boiling. After thorough rinsing of the TMAs with phosphate-buffered saline (PBS), 0.3% hydrogen peroxide solution was applied to block endogenous peroxidase activity for 10 min at room temperature. Subsequently, serum blocking was carried out. The primary antibody against SPAG5 (HPA022008, Sigma-Aldrich, St. Louis, MO, USA) was incubated with the sections overnight at 4°C to ensure efficient binding. Following the overnight incubation, the TMAs were washed with PBS, and the secondary antibody was incubated with the sections for 1 h at room temperature. Finally, 3,3'-diaminobenzidine (DAB) staining was performed to visualize the targeted antigen, and the sections were mounted for capturing the whole stained area using a slide scanner. The SPAG5 level of each sample was calculated based on the staining scope and intensity as follows: H-score = (percentage of weak intensity × 1) + (percentage of moderate intensity × 2) + (percentage of strong intensity × 3).23-25 The correlation between SPAG5 expression and clinicopathologic characteristics was analyzed by the chi-square test, and P < .05 is considered statistically significant.
Oncomine database analysis
Oncomine database (http://www.oncomine.org) is a public platform for supplying microarray data download and mining for most tumors. 26 Here, the expression levels of SPAG5 in pan-cancer were determined by analysis of the Oncomine database. The cut-off value is set as fold change (FC) > 1.5 and P < .05.
Tumor Immune Estimation Resource database analysis
Tumor Immune Estimation Resource (TIMER) is a web platform that incorporated 10 009 samples across 23 cancer types from The Cancer Genome Atlas (TCGA). TIMER2.0 (http://timer.cistrome.org/) can be used to estimate immune infiltration levels for TCGA or customer’s RNA-seq data and explore the relationship between immune infiltration and other factors, such as genomics and transcriptome variation and clinical outcomes. 27 Here, we explore the expression levels of SPAG5 in various cancer types. In addition, a correlation analysis was performed between SPAG5 expression and infiltrating immune cells, including B cells, CD8+ T cells, Treg cells, macrophages (Mφ), neutrophils, and myeloid dendritic cells (MDCs). The gene expression levels are shown with log2TPM. The correlation between immune checkpoints and immune cells is determined based on TIMER2.0.
Functional state analysis
Cancer Single-cell State Atlas (CancerSEA; http://biocc.hrbmu.edu.cn/CancerSEA) is a specific database used to synthetically explore cancer cell function at the single-cell level across 41 900 cancer single cells from 25 cancer types. Cancer single-cell functional states of CancerSEA are involved in tumorigenesis, progression, and aggressive metastasis. 28 Here, the average correlation between SPAG5 and functional states in different cancers is shown, and the bar plot shows the number of data sets for which SPAG5 is significantly correlated with the corresponding state. And functional states are significantly correlated with SPAG5 in LUAD (ExpID, EXP0066), filter by P < .05.
The Cancer Cell Line Encyclopedia (CCLE, https://sites.broadinstitute.org/ccle/tools) is a publicly available database that provides molecular and phenotypic data for cancer cell lines. It serves as a valuable resource for investigating the molecular mechanisms and drug treatment strategies associated with tumors.29,30 In this study, we obtained the gene expression matrix for 70 LUAD cell lines from CCLE. Using the single-sample gene set enrichment analysis (ssGSEA) algorithm in the GSVA R package, we calculated the enrichment scores of each cell line for signatures derived from CancerSEA. We then performed the Spearman correlation analysis between the enrichment scores and the expression levels of the SPAG5 gene.
Connectivity Map (CMap, https://clue.io/) is a database that contains gene expression and drug response data. It is a useful tool for studying the relationship between small molecule compounds and cellular biological processes. In our analysis, we employed the DESeq2 algorithm to identify the differentially expressed genes between the high and low expression groups of SPAG5 in TCGA LUAD data. These differentially expressed genes were further used for CMap analysis to screen the top 10 small molecule drugs that exhibited positive and negative correlations with the differential gene expression.
Gene Expression Profiling Interactive Analysis database analysis
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) is a web tool that processes RNA-seq data based on TCGA and Genotype-Tissue Expression (GTEx) projects to comprehensively analyze gene expression at multiple levels. 31 Here, GEPIA validated the association between SPAG5 and clinical staging.
Immunotherapy response prediction
The LUAD transcriptome sequencing data were downloaded by the TCGA data download module of Sangerbox (http://vip.sangerbox.com/tool.html) and then uploaded to The Immune Cell Abundance Identifier (ImmuCellAI, 32 http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/analysis) website in accordance with the required format for immune checkpoint blockade (ICB) prediction. GraphPad Prism 8.0 software was used to draw the violin plots. Statistical significance was established using the Mann-Whitney test.
Survival analyses
In this study, Sangerbox, GEPIA, PrognoScan 33 database (http://www.abren.net/PrognoScan/) and Kaplan-Meier Plotter 34 database (http://kmplot.com/analysis/) were performed to detect the correlation between SPAG5 expression and survival. PrognoScan database is a public platform for cancer microarray data sets, which was used to explore the correlation between gene expression and patient prognosis, including OS and relapse-free survival (RFS). The Kaplan-Meier Plotter database is an online tool for exploring the relationship between gene expression and patient outcomes across a broad range of cancer types, based on RNA-seq data, microarray data, and clinical data from the Gene Expression Omnibus (GEO), European Genome-Phenome Archive (EGA), TCGA, and PubMed repositories. The Kaplan-Meier method was used to analyze OS and progression-free (first progression) survival (PFS). The threshold of SPAG5 is defined using the H-score median value.
DNA methylation and microRNA analysis
The Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/) is a publicly accessible database that allows for querying and downloading diverse types of cancer genomic data, epigenetic data, and clinical data. In this study, we downloaded gene expression, DNA methylation, and microRNA (miRNA) expression data from the GDC website for subsequent analysis using R software. The Spearman correlation analysis was employed to examine the relationship between SPAG5 expression and DNA promoter methylation.
The miRTarBase database (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php) and the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/) are comprehensive repositories of miRNA-target gene interactions.
In our study, we calculated the Spearman correlation between SPAG5 and miRNAs. To refine the results, we filtered for miRNA-mRNA interactions based on correlation coefficients (R < −0.15) and statistically significant P values (<.05), using both miRTarBase and miRWalk databases. The interactions between miRNAs and their target SPAG5 were visualized as a network diagram using Cytoscape v3.10.0 software.
Results
Increased sperm-associated antigen 5 expression was associated with unfavorable clinical staging in lung adenocarcinoma
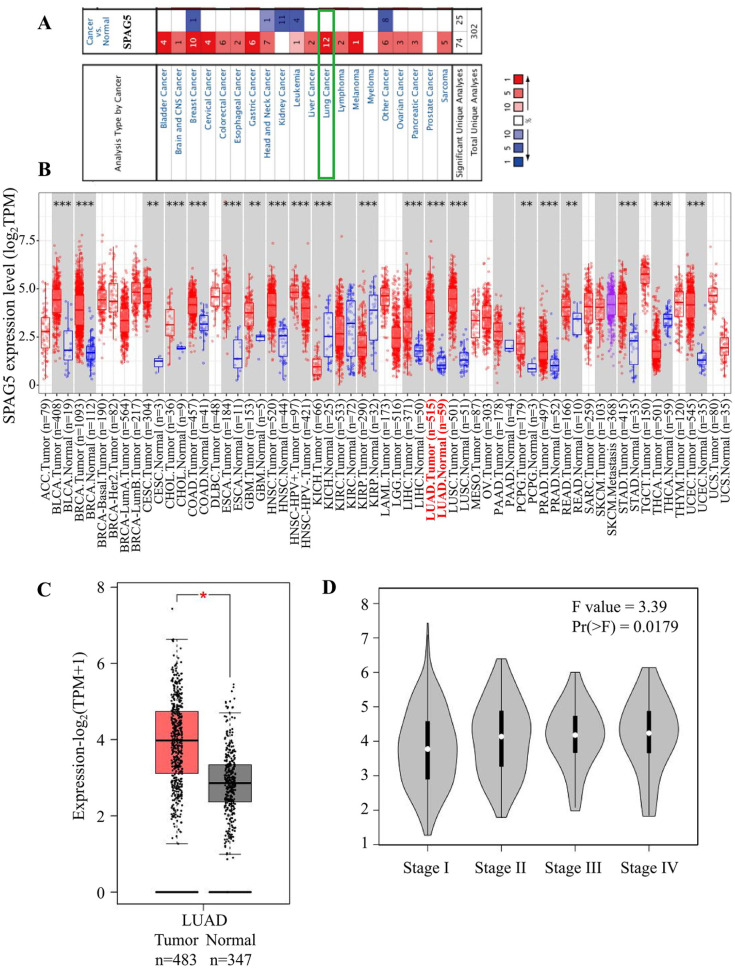
To investigate SPAG5 expression across various cancer types, we conducted analysis using both Oncomine and TCGA databases. Our findings indicate that SPAG5 is highly expressed in most tumors when compared with the corresponding normal tissues. In breast cancer, head and neck cancer, as well as several other unidentified cancers, SPAG5 was found to be generally highly expressed in tumors, albeit with a few exceptions. Conversely, we observed a low expression trend of SPAG5 in renal cancer and leukemia, particularly in cases of in renal cancer without exception. Notably, we observed that SPGA5 was highly expressed across all lung cancer types studied in up to 12 studies, indicating that SPAG5 may play a definite tumor-promoting role in lung cancer (Figure 1A). Consistent with this observation, the TCGA data analyzed on the Time2.0 website revealed higher expression of SPAG5 in most tumors compared with normal tissues (Figure 1B), including head and neck cancer and LUAD. Similar to the Oncomine data, SPAG5 was underexpressed in TCGA renal carcinoma samples, including kidney chromophobe (KICH) and kidney renal papillary cell carcinoma (KIRP) (Figure 1B). Moreover, the combined analysis of GTEx and TCGA data further confirmed the high expression of SPAG5 in LUAD tumor tissues (Figure 1C) and subsequent analysis showed a correlation between increased SPAG5 expression and unfavorable clinical staging in LUAD (Figure 1D). Overall, these findings suggest that SPAG5 may act as an oncogene in LUAD.
Figure 1.
Expression profile and clinical significance of SPAG5 in LUAD. (A) Expression profile of SPAG5 in various tumors and normal tissues derived from the Oncomine database. Upregulation is depicted in red, whereas downregulation is represented in blue. The green box emphasizes the high expression of SPAG5 across 12 lung cancer data sets. (B) Analysis of differential SPAG5 expression between tumors and normal tissues in TCGA pan-cancer was performed using the TIMER2.0 database. Notably, SPAG5 exhibits significantly increased expression in LUAD tumors (highlighted in red font). (C) GEPIA analysis revealed the expression difference of SPAG5 between LUAD tumor and normal tissues in the TCGA and GTEx databases. The box plot indicates a notable upregulation of SPAG5 in tumor tissues. (D) Correlation analysis conducted by GEPIA between TCGA-LUAD clinical staging and SPAG5 expression reveals a positive association between elevated SPAG5 expression levels and advanced tumor staging. CNS indicates central nervous system; GEPIA, Gene Expression Profiling Interactive Analysis; GTEx, genotype-tissue expression; KICH, kidney chromophobe; KIRP, kidney renal papillary cell carcinoma; LUAD, lung adenocarcinoma; SPAG5, sperm-associated antigen 5; TCGA, The Cancer Genome Atlas; TIMER, tumor immune estimation resource.
Statistical significance is denoted as *P < .05; **P < .01; ***P < .001.
DNA methylation and microRNAs may potentially regulate sperm-associated antigen 5
We initially evaluated the methylation status of SPAG5 promoter and its corresponding mRNA expression levels and observed that promoter methylation of SPAG5 was negatively correlated with mRNA expression (R = −0.25, P = 1.2e−07) (Supplemental Figure S1a). In addition, the use of correlation analysis indicated that hsa-miR-221-3p (R = −0.16, P = 2.280119e−04) and hsa-miR-877-3p (R = −0.21, P = 2.346523e−06) be involved in targeting regulation of SPAG5 (Supplemental Figure S1b).
The expression of sperm-associated antigen 5 in lung adenocarcinoma predicts a poor prognosis
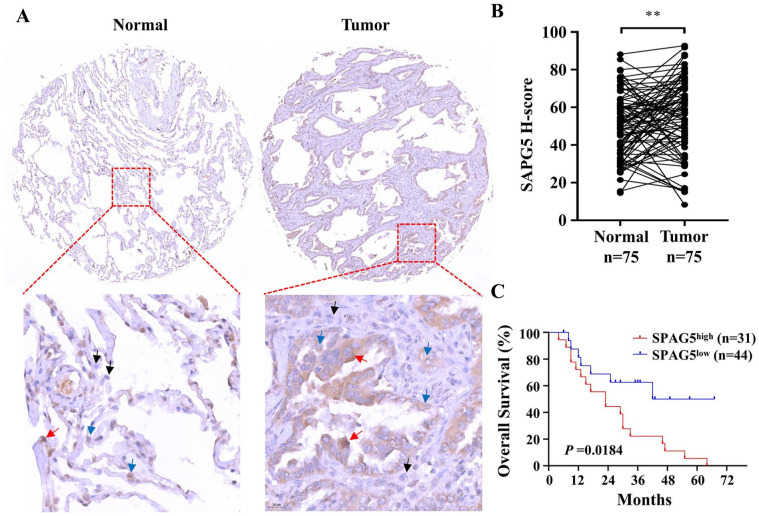
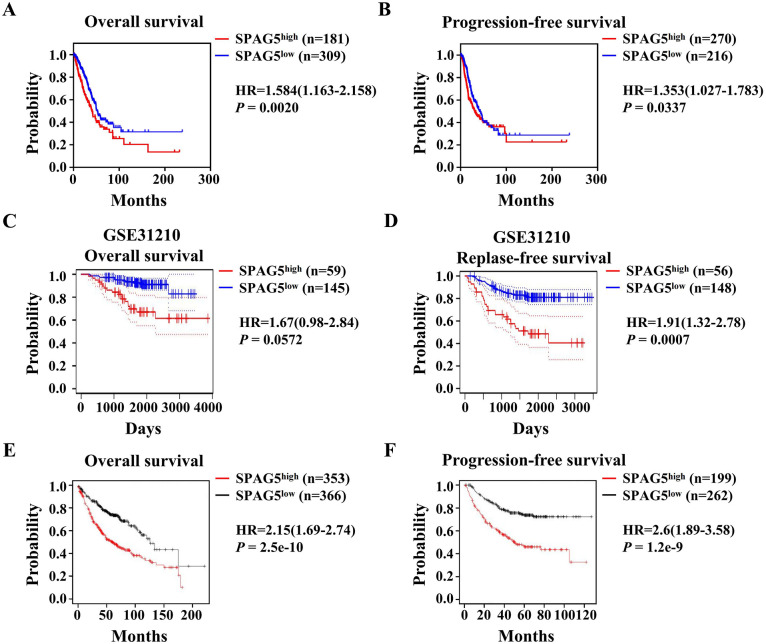
Following bioinformatics analysis of mRNA expression, we performed IHC staining of SPAG5 with TMA to further determine its protein expression and prognostic role in LUAD. The results indicated that SPAG5 was mainly expressed in the cytoplasm, and its expression was significantly higher in cancer cells than in normal lung epithelial cells (Figure 2A and B). Furthermore, the Kaplan-Meier survival curve analysis and log-rank tests demonstrated a significant correlation between elevated expression of SPAG5 and worse OS in LUAD (P = .0008) (Figure 2C). To support these findings, we analyzed the prognostic role of SPAG5 using multiple LUAD public data. Minimum P value analysis of TCGA-LUAD data via Sangerbox 3.0 software revealed that SPAG5 was associated with poor prognosis for OS (hazad ratio [HR] = 1.584, P = .0020) (Figure 3A) and PFS (HR = 1.353, P = .0337) (Figure 3B). Additional analysis of the GSE31210 LUAD data set demonstrated that strong SPAG5 expression predicted poor relapse-free survival (RFS) (HR = 1.91, P = .0007) and a trend toward worse OS (HR = 1.67, P = .0572), which did not reach statistical significance (Figure 3C and D). Moreover, using the Kaplan-Meier Plotter database based on microarray data, we validated the prognostic potential of SPAG5. As shown in Figure 3E and F, high SPAG5 expression indicated unfavorable OS (HR = 2.15, P = 2.5e−10) and PFS (HR = 2.6, P = 1.2e−9) of LUAD patients. Collectively, our data and others suggest that SPAG5 is a risk factor for OS and disease progression in LUAD.
Figure 2.
TMA IHC staining reveals elevated expression of SPAG5 in LUAD tumor tissue, and its upregulation correlates with unfavorable overall survival. (A) Representative images of SPAG5 IHC staining in LUAD TMA are presented, with the lower panel displaying a magnified view of the region highlighted by the red dashed box in the upper panel. Black, blue, and red arrows indicate cells with negligible expression, moderate expression, and high expression of SPAG5, respectively. (B) H-score analysis of IHC staining demonstrates significantly higher SPAG5 expression in LUAD tumor tissue (n = 75) compared with normal tissue (n = 75). (C) The Kaplan-Meier survival curve, analyzed using the log-rank test, illustrates a significant association between elevated expression of SPAG5 and a notably poorer prognosis (P = .0184). IHC indicates immunohistochemistry; LUAD, lung adenocarcinoma; SPAG5, sperm-associated antigen 5; TMA, tissue microarray.
**P < .01.
Figure 3.
High SPAG5 expression in LUAD is associated with an unfavorable prognosis. (A and B) Survival analysis of the TCGA data set using the GEPIA database reveals that LUAD patients with high SPAG5 expression exhibit significantly worse OS (P = .0020) and progression-free survival (P = .0337). (C and D) Survival analysis of the GSE31210 data set using the PrognoScan website indicates that LUAD patients with high SPAG5 expression show significantly poorer RFS (P = .0007) and a tendency toward reduced OS (P = .0572). (E and F) Survival analysis of LUAD data in the Kaplan-Meier plotter database demonstrates a substantial decrease in OS (P = 2.5e−10) and PFS (P = 1.2e−9) among patients with high SPAG5 expression. GEPIA indicates Gene Expression Profiling Interactive Analysis; LUAD, lung adenocarcinoma; OS, overall survival; PFS, progression-free survival; SPAG5, sperm-associated antigen 5; TCGA, The Cancer Genome Atlas; RFS, relapse-free survival.
Upregulation of sperm-associated antigen 5 was associated with malignant phenotypes of lung adenocarcinoma
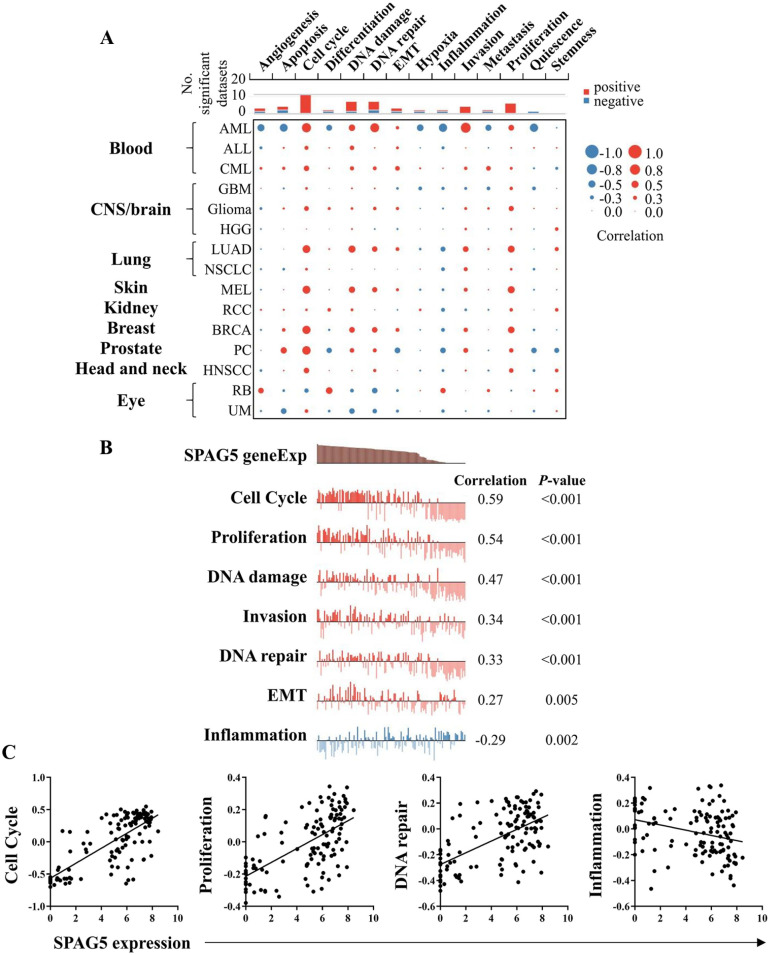
To elucidate the potential function of SPAG5 in LUAD, we employed the CancerSEA database to investigate the functional states of SPAG5. The functional states correlation analysis demonstrated that SPAG5 exhibited a positive correlation with cell cycle, DNA damage, DNA repair, invasion, proliferation, while being negatively correlated with angiogenesis, inflammation, and quiescence in most tumors (Figure 4A). Notably, in the case of LUAD, SPAG5 was found to be correlated with the malignant phenotypes, specifically displaying a positive correlation with cell cycle, proliferation, DNA damage, DNA repair, epithelial-mesenchymal transition (EMT), and a negative correlation with inflammation (Figure 4B and C). These findings suggest that SPAG5 may contribute to tumor progression by modulating multiple aspects of LUAD, including drug resistance, proliferation promotion, and immunosuppression.
Figure 4.
The CancerSEA analysis reveals a significant correlation between high SPAG5 expression and malignant tumor phenotypes. (A) Relevance of SPAG5 in 14 distinct functional states across various cancers. Red circles denote a positive correlation, whereas blue circles indicate a negative correlation. The size of the circles corresponds to the strength of the correlation. (B) The CancerSEA analysis was conducted on single-cell data sets (Kim KT, Genome Biology 2015), comprising 126 cells derived from patient-derived xenografts (PDX) of LUAD with the experimental ID EXP0066. Using a significance threshold of P < .05, it was observed that SPAG5 exhibited positive associations with functional states such as cell cycle, proliferation, DNA damage, invasion, DNA repair, and EMT, while demonstrating a negative correlation with the inflammation functional state. (C) The scatter plot illustrates the correlations between SPAG5 expression and selected functional states, including cell cycle, proliferation, DNA repair, and inflammation. CancerSEA indicates Cancer Single-cell State Atlas; CNS, central nervous system; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CML, chronic myelogenous leukemia; GBM, glioblastoma; HGG, high-grade glioma; MEL, melanoma; RCC, renal cell carcinoma; BRCA, breast cancer; PC, prostate cancer; HNSCC, head and neck cancer; EMT, epithelial-mesenchymal transition; LUAD, lung adenocarcinoma; NSCLC, non-small cell lung cancer; SPAG5, sperm-associated antigen 5.
Moreover, in the CCLE LUAD cell lines, we observed significant positive correlations between the expression of SPAG5 and scores of Cell Cycle, DNA repair, Proliferation, DNA damage, and EMT signatures (Supplemental Figure S2). These results align with the findings obtained from CancerSEA, further confirming the association between SPAG5 expression and the malignant phenotype of tumors.
Furthermore, based on the differentially expressed genes, we conducted a CMap analysis and identified 20 small molecule drugs that were related to SPAG5 expression (Supplementary Table 7). These drugs encompass purvalanol A, an inhibitor of CDK2/CDK1; TPCA-1, a direct dual inhibitor of STAT3 and NF-kappaB; JAK3-inhibitor-VI; PI-103, an inhibitor of PI3Kα/mTOR; and valproic acid, an inhibitor of HDAC2.
Functional annotation of sperm-associated antigen 5 co-expressed genes in lung adenocarcinoma
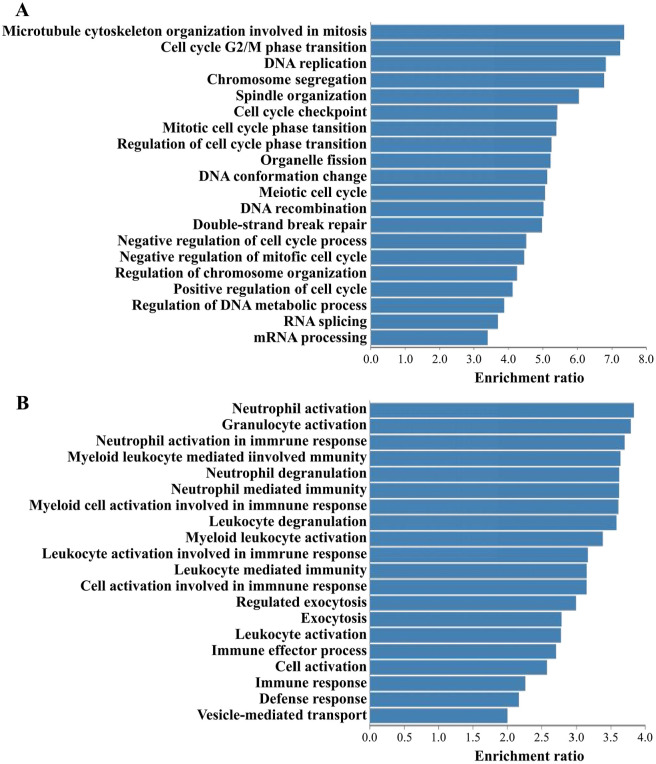
Subsequently, we filtered the genes co-expressed with SPAG5 based on TCGA-LUAD sequencing data, using an absolute value of the Spearman correlation coefficient >0.4 and P < .05 as filter criteria (Supplementary Table 3). An over-representation analysis (ORA) of the resulting co-expressed genes was conducted using the WebGestalt website 35 (http://www.webgestalt.org/#). The results showed that the positively correlated genes were primarily involved in the biological processes related to cell cycle and DNA repair, whereas the negatively correlated genes were mainly associated with immune responses, such as neutrophil activation, granulocyte activation, and myeloid cell activation (Figure 5A and B).
Figure 5.
Functional annotation of SPAG5 co-expressed genes in LUAD. Based on TCGA-LUAD sequencing data, over-representation analysis (ORA) of genes positively (A) or negatively (B) correlated with SPAG5 was performed on WebGestalt website. The top 20 functional terms with the highest enrichment ratios mediated by genes positively and negatively correlated with SPAG5 are shown in A and B, respectively. Co-expressed genes were filtered based on an absolute value of the Spearman correlation coefficient greater than 0.4 and a P value less than 0.05. LUAD indicates lung adenocarcinoma; SPAG5, sperm-associated antigen 5; TCGA, The Cancer Genome Atlas.
Sperm-associated antigen 5 promotes tumor immunosuppression and may be a predictor of response to lung adenocarcinoma immunotherapy
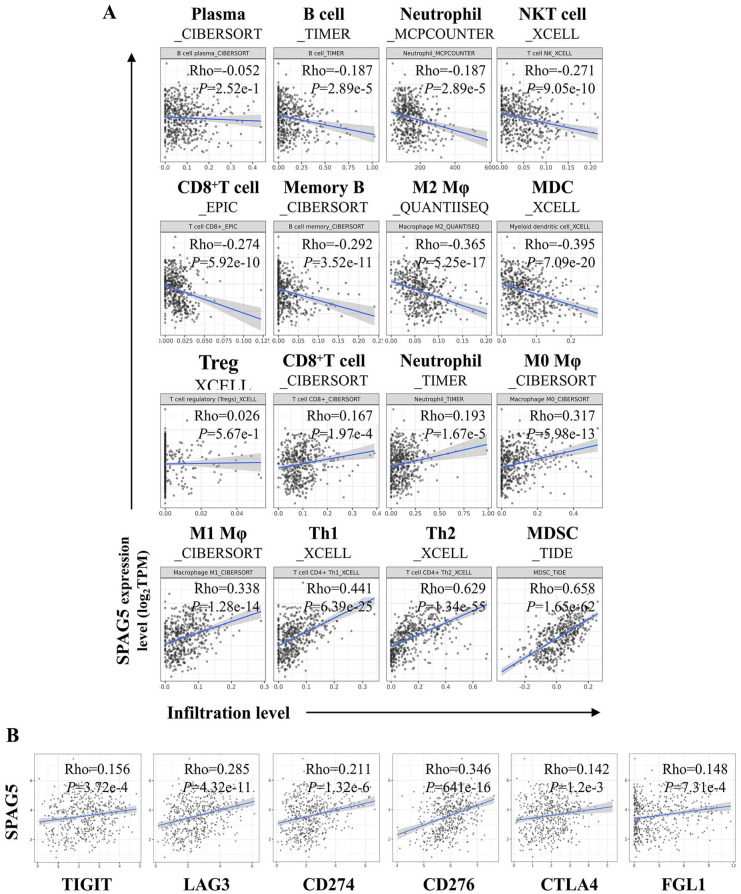
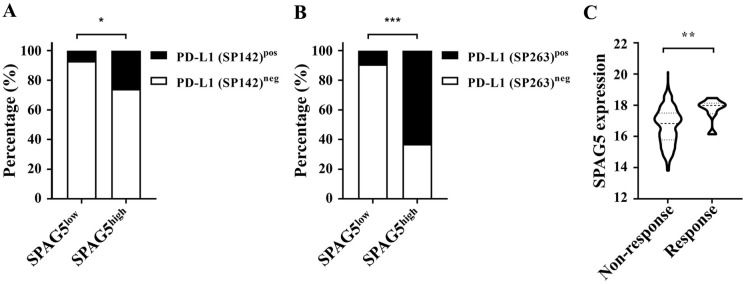
Sperm-associated antigen 5 has been identified to be associated with cell growth, invasion, and resistance to therapy in lung cancer. 36 However, its relationship with tumor immunity remains unclear. Here, the correlation between SPAG5 and tumor immune infiltration in LUAD was evaluated using the TIMER2.0 bioinformatics tool. Our findings indicated that SPAG5 was negatively correlated with the infiltration of several immune cells, including plasma cells, B cells, neutrophils, natural killer T (NKT) cells, CD8+ T cells, memory B cells, M2 Mφ, and MDCs. The immune cells that were positively correlated with SPAG5 expression were mainly CD8+ T cells, neutrophils, M0/M1 Mφ, Th-1, Th-2, and myeloid-derived suppressor cells (MDSCs), with no correlation observed with the infiltration of Tregs. Notably, the correlation of SPAG5 expression with some immune cells, such as CD8+ T cells and neutrophils, differed among different algorithm analyses. After removing these controversial cells, our results showed that SPAG5 expression was primarily negatively correlated with the invasion of anti-tumor immune cells and positively correlated with the infiltration of immunosuppressive cells, particularly Th-2 and MDSC cells, with a correlation of over 0.6 (Figure 6A). These findings suggest that the elevated SPAG5 expression is associated with an immunosuppressive microenvironment in LUAD. Therefore, we further analyzed the correlation between SPAG5 expression and immune checkpoints in LUAD. Our results revealed that SPAG5 was significantly positively associated with TIGIT, LAG3, CD274, CD276, CTLA4, and FGL1 inhibitory immune checkpoint molecules in LUAD (Figure 6B). To explore the correlation between SPAG5 and PD-L1, we also examined SPAG5 expression by IHC staining of TMA, which contained the expression information of PD-L1. Our chi-square test indicated that SPAG5 was positively correlated with PD-L1 expression detected by both FDA-approved antibodies (SP142 and SP263) 10 (Figure 7A and B and Supplementary Tables 1 and 4). Given the predictive role of PD-L1 in response to immunotherapy, we further investigated the predictive role of SPAG5 in response to immunotherapy in LUAD. Analysis of the LUAD sequencing data of TCGA by ImmuCellAI website showed that SPAG5 expression levels were significantly higher in the response group, indicating that SPAG5 expression could predict the response to ICB treatment (Figure 7C).
Figure 6.
Potential role of SPAG5 in tumor immunosuppression. Correlation between SPAG5 Expression and Tumor Immune Infiltration in LUAD. (A) The correlation between SPAG5 expression and tumor immune infiltration was systematically analyzed using the TIMER2.0 website. The scatter plots illustrate the correlation between SPAG5 expression and the infiltration levels of 16 distinct immune cell types in LUAD. The X-axis represents the level of immune infiltration, whereas the Y-axis denotes the expression level of SPAG5. (B) The TIMER2.0 website was employed to investigate the correlation between SPAG5 expression and the specific inhibitory immune checkpoint as indicated. LUAD indicates lung adenocarcinoma; SPAG5, sperm-associated antigen 5; TIMER, tumor immune estimation resource.
Figure 7.
SPAG5 may be a predictor of response to LUAD immunotherapy. (A and B) Based on the H-score evaluation of SPAG5 expression levels through IHC staining, a chi-square test was conducted to analyze the differential rates of PD-L1 positivity among LUAD samples with high and low SPAG5 expression. PD-L1 positivity was assessed using IHC staining with SP142 and SP263 antibodies in panels A and B, respectively. (C) Analysis of the LUAD sequencing data of TCGA by ImmuCellAI website showed that the expression of SPAG5 is positively correlated with good ICB therapy response. ICB indicates checkpoint blockade; IHC, immunohistochemistry; LUAD, lung adenocarcinoma; Mφ, macrophages; PD-L1, programmed death-ligand 1; SPAG5, sperm-associated antigen 5; TCGA, The Cancer Genome Atlas.
Discussion
Lung adenocarcinoma, the most common form of lung cancer, is prone to metastasis and recurrence, resulting in a low 5-year survival rate. In recent years, immunotherapy has emerged as a promising approach for LUAD treatment. However, the lack of predictive diagnostic markers hinders the identification of suitable individuals for LUAD immunotherapy. This study employs bioinformatics analysis to demonstrate the potential of SPAG5, a novel oncogene, as a predictive marker for immunotherapy in LUAD.
In recent years, bioinformatics analyses have contributed significantly to the identification of prognostic markers, therapeutic targets, and regulatory mechanisms for various types of tumors.37-41 Although the oncogenic role of SPAG5 has been established in several tumor types, its involvement in LUAD has primarily been inferred from bioinformatics analyses of RNA data. In this study, we have not only conducted a comprehensive bioinformatics analysis using multiple databases, but we have also analyzed the expression of SPAG5 at the protein level in both normal and tumor tissues of LUAD using IHC staining. Moreover, we have evaluated the prognostic significance of SPAG5 in LUAD for the first time. Our findings from both bioinformatics analysis and TMA staining confirm that SPAG5 is markedly overexpressed in LUAD and serves as an unfavorable prognostic indicator for OS and PFS. These results are in line with previous studies.
SPAG5 has been previously reported to promote the progression of prostate cancer by stimulating cell proliferation, migration, and invasion. 42 It has also been implicated in promoting lung cancer cell growth, migration, and invasion. 43 In this study, we conducted a comprehensive bioinformatics analysis of TCGA-LUAD data to thoroughly investigate the functional role of SPAG5 in LUAD. Our results reveal that SPAG5 is involved in the regulation of various important processes in LUAD, including the cell cycle, DNA damage/repair, EMT, invasion, proliferation, and inflammation. Furthermore, the over-representation analysis of the co-expressed genes of SPAG5 in LUAD demonstrates that the positively correlated genes are primarily associated with cell cycle–related biological processes, such as microtubule cytoskeleton organization involved in mitosis and DNA replication. On the contrary, the negatively correlated genes are mainly involved in the activation of immune cells, such as neutrophils, granulocytes, and other leukocytes involved in immune response. These findings suggest that SPAG5 plays a comprehensive role in the progression of LUAD, including tumor growth (proliferation), migration (EMT), chemotherapy resistance (DNA repair), and immune regulation (immune cell activation). Therefore, we speculate that SPAG5 may contribute to chemotherapeutic resistance and immunotherapy responses in LUAD. In addition, we also analyzed the functional roles of the co-expressed genes of SPAG5 in LUAD. The positively correlated genes are primarily associated with the cell cycle, DNA replication, repair, and other regulatory processes. Conversely, the negatively correlated genes are linked to the activation of immune cells, such as neutrophils and myeloid cells.
He et al 11 provided a comprehensive overview of the oncogenic role of SPAG5. They highlighted that SPAG5 exerts its influence by inhibiting mTORC1 activity, thereby regulating the cellular stress response. In addition, SPAG5 plays a role in inhibiting cell apoptosis through the p53-mediated DNA damage response pathway. This pathway governs the cellular response to DNA damage, redox stress, and abnormal oncogene amplification, ultimately promoting the expression of apoptotic genes. Furthermore, SPAG5 can upregulate WNT3 via the AKT/mTOR pathway. WNT3/β-catenin signaling pathway is involved in various biological processes, including the promotion of EMT and the development of chemoresistance in bladder cancer and human epidermal growth factor receptor 2 (HER2)-overexpressing breast cancer.14,44 In addition, SPAG5 enhances the transcriptional activity of c-Myc by increasing and interacting with c-Myc-binding protein (MYCBP). This interaction contributes to cellular mechanisms such as cell proliferation, apoptosis, and DNA repair. 12 Nonetheless, the precise functional mechanisms through which SPAG5 regulates LUAD remain unclear and require further investigation to elucidate.
Notably, some of the small molecule drugs predicted based on differential gene expression in the high-low SPAG5 expression groups have been reported to enhance the sensitivity of non-small cell lung cancer (NSCLC) drugs.45-49 Interestingly, 3 of the top 10 drugs with positive associations to differential gene expression are CDK inhibitors. Previous studies have demonstrated that CDK inhibitors significantly improve immunotherapy in recurrent breast cancer and chemotherapy response in recurrent small cell lung cancer.50,51 Therefore, it is worth further exploring the potential of CDK inhibitors as a treatment option for LUAD based on the expression levels of SPAG5.
Previous studies have established a positive correlation between SPAG5 expression in hepatocellular carcinoma and the infiltration of immune cells, including CD8+ T cells, Mφ, neutrophils B cells, and dendritic cells. 52 Zeng et al 53 identified that SPAG5 as one of the 15 key stemness-related genes (SRGs) in LUAD. Their findings revealed that the collective overexpression of these SRGs was associated with a decrease in immune invasion. However, the precise impact of SPAG5 on immune infiltration was not extensively explored in their study. Our study revealed a positive correlation between SPAG5 expression and the infiltration of anti-tumor immune cells, such as plasma cells and B cells. Conversely, SPAG5 exhibited a negative correlation with the infiltration of immunosuppressive Th-2 and MDSC cells, indicating its potential involvement in the formation of an immunosuppressive microenvironment. In addition, immune checkpoint correlation analysis showed a positive correlation between SPAG5 and the expression of various inhibitory immune checkpoint molecules, including TIGIT, LAG3, CD274 (coding PD-L1), CD276, CTLA4, and FGL1. The positive correlation between SPAG5 and PD-L1 was further validated at the protein level using TMA staining. Notably, we observed that the positive detection rates of the 2 PD-L1 antibodies, SP142 and SP263, were 14.3% (10/70) and 30% (21/70), respectively. Moreover, the consistency between the 2 antibodies was less than 80% (55/70) as shown in Supplementary Table 5. This inconsistency in detection rates could be attributed to the extensive glycosylation modification of PD-L1. The variation in detection rates among different antibodies highlights one of the limitations of PD-L1 as a predictor of immune efficacy. Therefore, it is advisable to supplement PD-L1 with additional indicators when used as a predictive diagnostic marker for immunotherapy. Sperm-associated antigen 5, being a protein that undergoes minimal modification by macromolecule glycosylation, presents itself as a promising candidate for analysis using the UniProt database (Supplementary Table 6). Consequently, in this study, we have also examined the predictive role of SPAG5 in response to ICB treatment. The findings revealed a positive association between elevated SPAG5 levels and a more favorable immune response to the treatment. This observation aligns with the positive correlation observed between SPAG5 and PD-L1, a well-known immune response molecule. Liu et al 54 have demonstrated both in vitro and in vivo the efficacy of SPAG5-targeted nano-siRNA drugs for the treatment of bladder cancer, highlighting the potential of SPAG5 as a therapeutic target. Furthermore, in a study by Li et al, 18 SPAG5 has also been proposed as an alternative target for cancer vaccine in various cancer types. These findings suggest the promising therapeutic potential of SPAG5. Our results indicate that upregulation of SPAG5 may serve as a predictive biomarker for the efficacy of immunotherapy. However, further in vitro and in vivo investigations are warranted to determine the suitability of targeting SPAG5 for combination immunotherapy.
Although we have thoroughly investigated the potential role of SPAG5 in LUAD and explored its potential as a predictor of immunotherapy response using bioinformatics and IHC analysis, our study still has limitations that should be acknowledged. First, the lack of validation using multi-center clinical samples undermines the generalizability of our findings. In addition, further mechanistic investigations are required to shed light on the intricate relationship between SPAG5 expression and the abundance, activity, and spatial distribution of various immune cells within LUAD tumor tissues. Such investigations hold the key to enhancing our comprehension of SPAG5’s effect on tumor immunity and its consequential impact on LUAD progression and immunotherapy outcomes. Addressing these limitations warrants future studies that leverage the advancements in single-cell sequencing and spatial transcriptomics.55,56 These innovative technologies provide an invaluable framework for unraveling the complex interplay between SPAG5 and immune cells within the dynamic tumor microenvironment. Therefore, capitalizing on the potential of single-cell omics technologies represents a promising avenue for elucidating the underlying mechanisms governing the actions of SPAG5.
Conclusions
In summary, our study highlights SPAG5’s role as a prognostic indicator for OS, RFS, and PFS in LUAD patients. Sperm-associated antigen 5 appears to be involved in regulating key processes including cell cycle control, proliferation, invasion, DNA damage and repair, and tumor immunosuppression. Furthermore, our findings suggest that SPAG5 holds promise as a potential predictor of immunotherapy response in LUAD.
Supplemental Material
Supplemental material, sj-docx-1-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-jpg-10-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-jpg-9-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-2-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-3-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-4-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-5-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-6-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-7-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-8-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Acknowledgments
The results shown here are in whole or part based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2020A1515011290) and the Guangzhou Municipal Science and Technology Project (202201010053).
Author contributions: Jianjiang Xie: designed the project.
Gang Xiao: performed the bioinformatic analysis and wrote the original manuscript and edited the final manuscript.
Xie Xu: performed the immunohistochemical staining experiment and conducted data analysis.
Zhibo Chen: performed the immunohistochemical staining experiment and conducted data analysis.
Jie Zeng: assisted with data analysis.
Gang Xiao: guided the experiments and the data analysis.
Jianjiang Xie: guided the experiments and the data analysis.
Availability of data and materials: The publicly available data sets are analyzed in this study. These data can be found here, Oncomine database (http://www.oncomine.org); TIMER2.0 database (http://timer.cistrome.org/); CancerSEA website (http://biocc.hrbmu.edu.cn/CancerSEA); GEPIA database (http://gepia.cancer-pku.cn/); PrognoScan database (http://www.abren.net/PrognoScan/); Kaplan-Meier Plotter database (http://kmplot.com/analysis/); Sangerbox website (http://vip.sangerbox.com/tool.html); ImmunCellAI website (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/analysis); WebGestalt database (http://www.webgestalt.org/#); UniProt database (https://www.uniprot.org/); GDC database (https://portal.gdc.cancer.gov/); miRTarBase database (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php); miRWalk database (http://mirwalk.umm.uni-heidelberg.de/); CCLE (https://sites.broadinstitute.org/ccle/tools); and CMap (https://clue.io/).
Consent for publication: Not applicable.
Ethics approval and consent to participate: Not applicable.
ORCID iD: Jianjiang Xie  https://orcid.org/0000-0002-8454-7753
https://orcid.org/0000-0002-8454-7753
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [DOI] [PubMed] [Google Scholar]
- 2. Shi J, Hua X, Zhu B, et al. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med. 2016;13:e1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steven A, Fisher SA, Robinson BW. Immunotherapy for lung cancer. Respirology. 2016;21:821-833. [DOI] [PubMed] [Google Scholar]
- 5. Tsiouprou I, Zaharias A, Spyratos D. The role of immunotherapy in extensive stage small-cell lung cancer: a review of the literature. Can Respir J. 2019;2019:6860432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alberti A, Mancin M, Cortinovis D, Bidoli P, Sala L. Disseminated intravascular coagulation in advanced lung adenocarcinoma during first-line pembrolizumab. Immunotherapy. 2020;12:629-633. [DOI] [PubMed] [Google Scholar]
- 7. Cui Y, Fang W, Li C, et al. Development and validation of a novel signature to predict overall survival in “driver gene-negative” lung adenocarcinoma (LUAD): results of a multicenter study. Clin Cancer Res. 2019;25:1546-1556. [DOI] [PubMed] [Google Scholar]
- 8. Bulat V, Likic R, Bradic L, Speeckaert R, Azdajic MD. Pembrolizumab-induced vitiligo in a patient with lung adenocarcinoma: a case report. Br J Clin Pharmacol. 2021;87:2614-2618. [DOI] [PubMed] [Google Scholar]
- 9. Xu-Monette ZY, Zhang M, Li J, Young KH. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response? Front Immunol. 2017;8:1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Tong Z, Zhang W, et al. FDA-approved and emerging next generation predictive biomarkers for immune checkpoint inhibitors in cancer patients. Front Oncol. 2021;11:683419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He J, Green AR, Li Y, Chan SYT, Liu DX. SPAG5: an emerging oncogene. Trends Cancer. 2020;6:543-547. [DOI] [PubMed] [Google Scholar]
- 12. Li M, Li A, Zhou S, Lv H, Yang W. SPAG5 upregulation contributes to enhanced c-MYC transcriptional activity via interaction with c-MYC binding protein in triple-negative breast cancer. J Hematol Oncol. 2019;12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z, Li H, Chen J, et al. SPAG5 promotes osteosarcoma metastasis via activation of FOXM1/MMP2 axis. Int J Biochem Cell Biol. 2020;126:105797. [DOI] [PubMed] [Google Scholar]
- 14. Liu JY, Zeng QH, Cao PG, et al. SPAG5 promotes proliferation and suppresses apoptosis in bladder urothelial carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway and predicts poorer survival. Oncogene. 2018;37:3937-3952. [DOI] [PubMed] [Google Scholar]
- 15. Yang YF, Zhang MF, Tian QH, et al. SPAG5 interacts with CEP55 and exerts oncogenic activities via PI3K/AKT pathway in hepatocellular carcinoma. Mol Cancer. 2018;17:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan LJ, Li JD, Zhang L, et al. SPAG5 upregulation predicts poor prognosis in cervical cancer patients and alters sensitivity to Taxol treatment via the mTOR signaling pathway. Cell Death Dis. 2015;6:e1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang J, Wang J, He X, et al. High expression of SPAG5 sustains the malignant growth and invasion of breast cancer cells through the activation of Wnt/beta-catenin signalling. Clin Exp Pharmacol Physiol. 2019;46:597-606. [DOI] [PubMed] [Google Scholar]
- 18. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang R, Li A. SPAG5 is associated with unfavorable prognosis in patients with lung adenocarcinoma and promotes proliferation, motility and autophagy in A549 cells. Exp Ther Med. 2020;20:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang T, Li K, Song H, et al. p53 suppression is essential for oncogenic SPAG5 upregulation in lung adenocarcinoma. Biochem Biophys Res Commun. 2019;513:319-325. [DOI] [PubMed] [Google Scholar]
- 21. Välk K, Vooder T, Kolde R, et al. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology. 2010;79:283-292. [DOI] [PubMed] [Google Scholar]
- 22. Liu P, Li H, Liao C, et al. Identification of key genes and biological pathways in Chinese lung cancer population using bioinformatics analysis. Peer J. 2022;10:e12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo R, Berry LD, Aisner DL, et al. MET IHC is a poor screen for MET amplification or MET Exon 14 mutations in lung adenocarcinomas: data from a tri-institutional cohort of the lung cancer mutation consortium. J Thorac Oncol. 2019;14:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paschalis A, Sheehan B, Riisnaes R, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol. 2019;76:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maclean A, Bunni E, Makrydima S, et al. Fallopian tube epithelial cells express androgen receptor and have a distinct hormonal responsiveness when compared with endometrial epithelium. Hum Reprod. 2020;35:2097-2106. [DOI] [PubMed] [Google Scholar]
- 26. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li T, Fu J, Zeng Z, et al.TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900-D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kao TJ, Wu CC, Phan NN, et al. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). 2021;13:17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh). 2020;7:1902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199-W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q, Wang Y, He J. MiR-133a-3p attenuates resistance of non-small cell lung cancer cells to gefitinib by targeting SPAG5. J Clin Lab Anal. 2021;35:e23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Demircioglu D, Cukuroglu E, Kindermans M, et al. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell. 2019;178:1465-1477.e17. [DOI] [PubMed] [Google Scholar]
- 38. Gong L, Zhang D, Dong Y, et al. Integrated bioinformatics analysis for identificating the therapeutic targets of aspirin in small cell lung cancer. J Biomed Inform. 2018;88:20-28. [DOI] [PubMed] [Google Scholar]
- 39. Holtstrater C, Schrors B, Bukur T, Lower M. Bioinformatics for cancer immunotherapy. Methods Mol Biol. 2020;2120:1-9. [DOI] [PubMed] [Google Scholar]
- 40. Liang W, Sun F. Identification of key genes of papillary thyroid cancer using integrated bioinformatics analysis. J Endocrinol Invest. 2018;41:1237-1245. [DOI] [PubMed] [Google Scholar]
- 41. Wang Q, Zhao S, Gan L, Zhuang Z. Bioinformatics analysis of prognostic value of PITX1 gene in breast cancer. Biosci Rep. 2020;40:BSR20202537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang H, Li S, Yang X, Qiao B, Zhang Z, Xu Y. miR-539 inhibits prostate cancer progression by directly targeting SPAG5. J Exp Clin Cancer Res. 2016;35:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song L, Dai Z, Zhang S, et al. MicroRNA-1179 suppresses cell growth and invasion by targeting sperm-associated antigen 5-mediated Akt signaling in human non-small cell lung cancer. Biochem Biophys Res Commun. 2018;504:164-170. [DOI] [PubMed] [Google Scholar]
- 44. Wu Y, Ginther C, Kim J, et al. Expression of Wnt3 activates Wnt/beta-catenin pathway and promotes EMT-like phenotype in trastuzumab-resistant HER2-overexpressing breast cancer cells. Mol Cancer Res. 2012;10:1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X, Liao Y, Long D, Yu T, Shen F, Lin X. The Cdc2/Cdk1 inhibitor, purvalanol A, enhances the cytotoxic effects of taxol through Op18/stathmin in non-small cell lung cancer cells in vitro. Int J Mol Med. 2017;40:235-242. [DOI] [PubMed] [Google Scholar]
- 46. Chen JH, Zheng YL, Xu CQ, et al. Valproic acid (VPA) enhances cisplatin sensitivity of non-small cell lung cancer cells via HDAC2 mediated down regulation of ABCA1. Biol Chem. 2017;398:785-792. [DOI] [PubMed] [Google Scholar]
- 47. Nishiya N, Sakamoto Y, Oku Y, Nonaka T, Uehara Y. JAK3 inhibitor VI is a mutant specific inhibitor for epidermal growth factor receptor with the gatekeeper mutation T790M. World J Biol Chem. 2015;6:409-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nan J, Du Y, Chen X, et al. TPCA-1 is a direct dual inhibitor of STAT3 and NF-kappaB and regresses mutant EGFR-associated human non-small cell lung cancers. Mol Cancer Ther. 2014;13:617-629. [DOI] [PubMed] [Google Scholar]
- 49. Zou ZQ, Zhang XH, Wang F, et al. A novel dual PI3Kalpha/mTOR inhibitor PI-103 with high antitumor activity in non-small cell lung cancer cells. Int J Mol Med. 2009;24:97-101. [DOI] [PubMed] [Google Scholar]
- 50. Uzhachenko RV, Bharti V, Ouyang Z, et al. Metabolic modulation by CDK4/6 inhibitor promotes chemokine-mediated recruitment of T cells into mammary tumors. Cell Rep. 2021;35:109271. [DOI] [PubMed] [Google Scholar]
- 51. Das M, Padda SK, Weiss J, Owonikoko TK. Advances in treatment of recurrent small cell lung cancer (SCLC): insights for optimizing patient outcomes from an expert roundtable discussion. Adv Ther. 2021;38:5431-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen W, Chen X, Li S, Ren B. Expression, immune infiltration and clinical significance of SPAG5 in hepatocellular carcinoma: a gene expression-based study. J Gene Med. 2020;22:e3155. [DOI] [PubMed] [Google Scholar]
- 53. Zeng H, Ji J, Song X, et al. Stemness related genes revealed by network analysis associated with tumor immune microenvironment and the clinical outcome in lung adenocarcinoma. Front Genet. 2020;11:549213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu J, Zhang Y, Zeng H, et al. Fe-doped chrysotile nanotubes containing siRNAs to silence SPAG5 to treat bladder cancer. J Nanobiotechnology. 2021;19:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang Z, Zhang T, Yang B, Su J, Song Q. spaCI: deciphering spatial cellular communications through adaptive graph model. Brief Bioinform. 2023;24:bbac563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Triozzi PL, Stirling ER, Song Q, et al. Circulating immune bioenergetic, metabolic, and genetic signatures predict melanoma patients’ response to anti-PD-1 immune checkpoint blockade. Clin Cancer Res. 2022;28:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-jpg-10-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-jpg-9-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-2-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-3-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-4-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-5-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-6-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-7-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology
Supplemental material, sj-xlsx-8-onc-10.1177_11795549231199915 for SPAG5 Expression Predicts Poor Prognosis and is Associated With Adverse Immune Infiltration in Lung Adenocarcinomas by Gang Xiao, Xie Xu, Zhibo Chen, Jie Zeng and Jianjiang Xie in Clinical Medicine Insights: Oncology