Abstract
The study presents the genome analysis of a new Pseudomonas sp. (SWRIQ11), which can alleviate salinity stress effects on growth of olive seedlings in greenhouse study. The strain SWRIQ11 can tolerate salinity up to 6%, produce siderophores, indole acetic acid (IAA), aminocyclopropane-1-carboxylate (ACC) deaminase, and has the phosphate-solubilizing capability. The SWRIQ11 genome contained an assembly size of 6,196,390 bp with a GC content of 60.1%. According to derived indices based on whole-genome sequences for species delineation, including tetra nucleotide usage patterns (TETRA), genome-to-genome distance (GGDC), and average nucleotide identity (ANI), Pseudomonas sp. SWRIQ11 can be considered a novel species candidate. The phylogenetic analysis revealed SWRIQ11 clusters with Pseudomonas tehranensis SWRI196 in the same clade. The SWRIQ11 genome was rich in genes related to stress sensing, signaling, and response, chaperones, motility, attachments, colonization, and enzymes for degrading plant-derived carbohydrates. Furthermore, the genes for production of exopolysaccharides, osmoprotectants, phytohormones, and ACC deaminase, ion homeostasis, nutrient acquisition, and antioxidant defenses were identified in the SWRIQ11 genome. The results of genome analysis (identification of more than 825 CDSs related to plant growth-promoting and stress-alleviating traits in the SWRIQ11 genome which is more than 15% of its total CDSs) are in accordance with laboratory and greenhouse experiments assigning the Pseudomonas sp. SWRIQ11 as a halotolerant plant growth-promoting bacterium (PGPB). This research highlights the potential safe application of this new PGPB species in agriculture as a potent biofertilizer.
Keywords: Plant growth-promoting rhizobacteria, Stress alleviation, Olive seedlings, Pseudomonas sp., Genome analysis, Genes and pathways
Introduction
Continuous world population growth in recent years and the expansion of soil salinity as one of the common abiotic stresses has led to the use of the plant growth-promoting rhizobacteria (PGPR) as a practical strategy for achieving sustainable agriculture (Balasubramanian et al. 2021; Alberton et al. 2020; Arora et al. 2020). PGPR provide cross-protection from several stresses and facilitate the growth of the associated plants (Leontidou et al. 2020). These bacteria alleviate salinity stress by various mechanisms such as modification of root morphology, nutrient acquisition, synthesis of exopolysaccharides, phytohormones, volatile compounds, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase, maintaining ion homeostasis, inducing accumulation of antioxidants and compatible solutes, induced systemic tolerance, and modulation of the stress-responsive genes (Mohammadipanah and Zamanzadeh 2019; Etesami 2020; Hoque et al. 2022)
Despite the extensive studies on PGPR as biofertilizers, the molecular pathways of PGPR function are not completely elucidated (Balasubramanian et al. 2021; Sekar et al. 2019; Hoque et al. 2022). Recently, next-generation sequencing, omics approaches, and computational tools have contributed to the molecular mechanisms of PGPR being demonstrated and discovery of novel genes and actions on plant–bacteria interactions (Meena et al. 2017; Shelake et al. 2019).
Pseudomonas spp. are Gram-negative rods, non-spore-forming, and motile bacteria belonging to the Gammaproteobacteria (Girard et al. 2021). They are ubiquitous bacteria that play crucial ecological roles in the environment (Girard et al. 2021; Nikolaidis et al. 2020). Pseudomonas spp.are prevalent plant-associated bacteria imposing multifaceted effects on plants (Gu et al. 2020). Most rhizospheric Pseudomonas spp. easily colonize various plant species and support plant growth, either directly by promoting plant growth or indirectly by protecting against stresses, including phytopathogens (Mavrodi et al. 2011; Zboralski et al. 2022). Pseudomonas, as the most complex genus, contains 312 validly published species and is presently the genus with the largest number of species among Gram-negative bacteria (Parte et al., accessed on 11 February, 2023).
In this study, the annotation and mining of the whole genome of a novel Pseudomonas sp. (SWRIQ11), capable of alleviating salinity stress in olive (Olea europaea L.) seedlings, were performed, and the corresponding genes and pathways to the beneficial activity of PGPR were characterized.
Material and methods
Bacterial strain
This strain was isolated from the olive rhizosphere in 2015, which was under salinity stress (12 dS/m) from the Qazvin province of Iran. The strain was identified as a Pseudomonas member. The strain was deposited as SWRIQ11 with CCSM-B00399 code in Culture Collection of Soil Microorganisms (CCSM) in Iran, and with CECT 30741 code in Spanish Type Culture Collection. Its 16S rRNA sequence was deposited in GenBank database, NCBI under accession number MH201206.
Morphological, biochemical, and physiological characterization of SWRIQ11
The Gram staining, oxidase, motility, catalase, and oxidative-fermentative tests were carried out according to Cappuccino and Sherman (2014). Furthermore, a fluorescence assay was performed by culturing strain on King B agar (Paez et al. 2005).
Plant growth-promoting assays
The siderophore production assay was performed using CAS (chrome-azurol sulfonate) agar (Senthilkumar et al. 2021). The production of indole-3-acetic acid (IAA) was assayed according to Bent and colleagues (2001). The ACC-deaminase production was evaluated according to Penrose and Glick (2003). The phosphate solubilization assay was carried out using Pikovskaya (PVK) liquid medium (Li et al. 2019).
Hypersensitive response (HR) assessment
Hypersensitive response (HR) can differentiate phytopathogenic bacteria from saprophytes. All plant pathogenic bacteria induce HR in the mesophyll tissue of the leaf of tobacco and geranium (Umesha et al. 2008). The HR test on tobacco is proper for detecting phytopathogenicity of Pseudomonas spp. (Scala et al. 2018). The HR test was performed based on the Klement method by injecting 200 µl suspension of SWRIQ11 at the concentration of 107 cfu/ml into the intercellular space of tobacco, geranium, and tomato leaves. Sterile water and Ralstonia solanacearoum suspension were injected as negative and positive controls, respectively. After 24–72 h of infiltration, leaves injected with phytopathogenic bacteria die off, but saprophytic bacteria do not produce necrosis in the injected leaves (Klement 1963; Umesha et al. 2008).
Hemolysis assay of SWRIQ11
Hemolysins are considered major virulence factors in animals and are commonly related to pathogenic bacteria (Mogrovejo et al. 2020). The hemolytic activity of SWRIQ11 was examined by culturing the strain on blood agar and incubating two series of cultures for 48h at 28 °C and 37 °C. Then, the existence of hemolysis zones surrounding the colonies was assayed (Cappuccino and Sherman 2014).
Evaluation of salinity tolerance of SWRIQ11
The salinity tolerance range of SWRIQ11 was assayed on nutrient agar (NA) with 2, 4, 6, and 8% w/v NaCl (Ramadoss et al. 2013).
Greenhouse assay of stress-alleviating activities
The effect of salinity stress (12 dS/m) and inoculation with SWRIQ11 suspension on the growth of six-month-old Koroneiki olive seedlings in pots with three replications at an average temperature of 23 °C were monitored after one year. Plastic pots (20 cm in diameter and volume of 2 kg) were filled with cocopeat and perlite (with a volume ratio of 2–1). Before transferring the seedlings to the pots, the roots of the seedlings were washed several times with water. 100 ml of SWRIQ11 suspension in saline solution (107 CFU/ml) was inoculated twice to pots, one month after planting the seedlings in pots and at the sixth month. In the fourth month, saline treatment was performed once a week for six months. The pots with salinity treatment and without bacterial inoculation (saline was added to pots) were considered as control. The treatment effect on seedlings growth was evaluated by measuring morphological parameters (seedling height, fresh and dry weight of shoot, number of lateral branches, and trunk diameter), and physiological parameters (photosynthetic pigments by UV–VIS Spectroscopy (Lichtenthaler and Buschmann 2001) after one year.
Genomic DNA preparation of SWRIQ11
The genomic DNA of Pseudomonas sp. SWRIQ11 was extracted utilizing the biotechrabbit (GenUP™ Bacteria gDNA Kit, Berlin, Germany) DNA purification kit. DNA quality and quantity were confirmed by NanoDrop™ spectrophotometer (Thermo Scientific NanoDrop 2000c) and agarose gel electrophoresis.
Genome sequencing and assembly
The whole genome sequencing was carried out by the Illumina HiSeq 4000 platform (150 bp paired-end reads) in Novogene company. FastQC v0.11.9 was applied for the quality control of raw sequences (Andrews et al. 2010), and then adapter and quality trimming was conducted using Trimmomatic 0.39 (Bolger et al. 2014). De novo assembly was carried out utilizing SPades v3.14.1, with default settings (Nurk et al. 2013). The assembly quality was assessed using the QUAST v2.3 software (Gurevich et al. 2013).
Annotation of the SWRIQ11 genome
The gene detection and genome annotation were conducted exploiting the Rapid Annotation using Subsystem Technology (RAST) server version 2.0 (Aziz et al. 2008), NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016), KEGG Automatic Annotation Server (KAAS) (Moriya et al. 2007), and UniProt database (MacDougall et al. 2020). Then, the annotated genes were assessed to assign the genes related to plant growth-promoting and stress-alleviating characteristics.
Gene network or pathway analysis
The presence of entire corresponding metabolic pathways to annotated genes related to plant growth-promoting characteristics was manually defined using comparisons to the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Kanehisa 2019). Gene clusters related to secondary metabolites biosynthesis were determined using antiSMASH bacterial version 6.6.1 (Blin et al. 2021).
Molecular identification of SWRIQ11
The full-length 16S rRNA sequence of the isolate was manually derived by integrating the sequence from 16S rRNA amplification and related contigs in the genome that were annotated as 16S rRNA (two sequences with the length of 1017 and 522 bp) using blastn. The 30 species of Pseudomonas spp. with the highest similarity and completeness in 16S rRNA sequence with strain SWRIQ11 were selected from the EzBioCloud database (Yoon et al. 2017a) for calculating ANI (average nucleotide identity) by the ANI calculator tool of EzBioCloud (Yoon et al. 2017b) and the JSpeciesWS web server based on BLAST (ANIb) and MUMmer (ANIm) (Richter et al. 2015), correlation indexes of their tetra-nucleotide signatures (Tetra) through the JSpeciesWS web server (Richter et al. 2015), and digital DNA–DNA hybridization (dDDH) (genome-to-genome distance) by Genome-to-Genome Distance Calculator (GGDC 3.0) (Meier-Kolthoff et al. 2022). Furthermore, the Tetra Correlation Search (TCS) based on tetra nucleotide frequencies and correlation coefficients against the entire genomes reference database (GenomesDB), was analyzed using the JSpeciesWS web server (Richter et al. 2015). In addition, the identification of SWRIQ11 was carried out using the Type Genome Server (TYGS) (Meier-Kolthoff and Goker 2019).
Phylogenetic analyses of SWRIQ11
The phylogenetic trees based on 16S rRNA sequences were constructed using MEGA version 10 with algorithms of Neighbor-Joining and Maximum-Likelihood (using the Tamura-Nei model), and with 10,000 and 1000 bootstrap replications, respectively (Tamura et al. 2021). The genome-based phylogenetic tree was constructed applying the Type (Strain) Genome Server (TYGS) on the basis of the Genome BLAST Distance Phylogeny (GBDP) method and with 100 bootstrap replications (Meier-Kolthoff and Goker 2019).
Comparative genome analysis
The genomic features of the SWRIQ11 strain were compared to genomes of closely related species (ANI > 83%) using the NCBI database and GenomesDB of JSpeciesWS. Some genomic characteristics, including the genome size, number of genes, the number of rRNAs and tRNAs, and the GC content of the species, were compared.
Results
Characterization of SWRIQ11
The strain SWRIQ11 is Gram-negative, rod-shaped, motile, fluorescent, oxidase-positive, catalase-positive, and non-fermentative. The evaluation of plant growth-promoting characteristics showed that the strain can produce siderophores, IAA, and ACC-deaminase and has phosphate-solubilizing activity. The strain could tolerate salinity up to 6% w/v NaCl. The strain inoculation did not induce necrosis in leaves of tobacco, geranium, and tomato after 48 h in the HR assay, and as a result, the strain is not considered phytopathogenic. No clear zones were not observed surrounding the colonies of SWRIQ11 on blood agar, and the strain had no hemolytic activity (gamma hemolysis).
Greenhouse assay of stress-alleviating effect of SWRIQ11
The results displayed that treatment with SWRIQ11 has a positive effect on the growth of olive seedlings under salinity stress. The inoculation with SWRIQ11 resulted in the increase of seedling height (20.69%), fresh weight of shoot (43.24%), dry weight of shoot (40%), number of lateral branches (66.7%), trunk diameter (28.57%), chlorophyll a + b (61.37%), and carotenoid (39.25%) amount compared to control seedlings (under salinity stress without bacteria inoculation).
Sequence statistics of strain SWRIQ11
Sequencing by the Illumina HiSeq 4000 platform led to nearly 185-fold coverage of the isolate genome. In total, 7,681,869 raw reads with a length of 150 bp were filtered for reads with > 10% Ns and 25–35 bases with a low-quality average (≤ Q20). Lastly, 7,564,065 (98.47%) clean reads were utilized for subsequent analyses and the de novo assembly using SPades v3.14.1 produced 101 scaffolds (≥ 200 bp).
Genomic features of strain SWRIQ11
The strain SWRIQ11 has a single circular chromosome of 6,196,390 bp with a genomic GC content of 60.1%, N50 157,109, and L50 14. The total numbers of 5566 genes were identified, from which 5420 were coding sequences (CDSs), 51 tRNA-coding genes, and 5 rRNA genes.
Taxonomic identification of strain SWRIQ11
According to the species definition, strains belonging to the same species typically show ≥ 95% ANI (Jain et al. 2018, Gomila et al. 2015), higher than 0.99 for the TETRA signature (Gomila et al. 2015), and > 70% of DNA–DNA hybridization (DDH) or GGDC. Accordingly, strain SWRIQ11 is a new species candidate in the pseudomonas genus (Table 1) which will be described in the follow-up study. Furthermore, according to the result of TCS (which compares the tetra nucleotide frequencies and correlation coefficients of the genome against GenomesDB), the highest Z-score (0.9988) belonged to Pseudomonas tehranensis SWRI196. Moreover, the highest records of ANI (EzBioCloud) (92.96%), ANIb (92.39%), ANIm (93.44%), and DDH (50.8%) belonged to P. tehranensis SWRI196 (GCA_014268615.1) (Table 1), which were less than the consensus threshold of species definition. In addition, according to identification using TYGS, the SWRIQ11 did not belong to any species found in databases.
Table 1.
The indices for identification of strain SWRIQ11 based on the 16S rRNA and genome analysis
| Species (Type strain) |
Similarity % (16S rRNA) | Completeness% (16S rRNA) | ANI% (EzBioCloud) | ANIb% (JSpeciesWS) | ANIm% (JSpeciesWS) | Tetra Correlation (Z-Score) | DDH or GGDC ( based on Formula 2 (Recommended)) |
|---|---|---|---|---|---|---|---|
| Pseudomonas tehranensis SWRI196 | 99.93 | 99 | 92.96 | 92.39 | 93.44 | 0.9988 |
Distance: 0.0697 DDH estimate: 50.80% |
| Pseudomonas piscicola P50 | 99.76 | 84.4 | 81.64 | 80.43 | 85.62 | 0.9669 |
Distance: 0.1694 DDH estimate: 25.70% |
| Pseudomonas bijieensis L22-9 | 99.66 | 100 | 88.04 | 87.42 | 88.97 | 0.99466 |
Distance: 0.1174 DDH estimate: 35.20% |
| Pseudomonas thivervalensis DSM 13194 | 99.52 | 100 | 87.76 | 87.28 | 88.93 | 0.98973 |
Distance: 0.1177 DDH estimate: 35.10% |
| Pseudomonas brassicacearum subsp. Brassicacearum DBK11 / CCUG:51,508 | 99.52 | 99.9 | 88.22 | 87.70 | 89.18 | 0.99399 |
Distance: 0.1157 DDH estimate: 35.60% |
| Pseudomonas corrugata ATCC 29736 / DSM:7228 | 99.44 | 98.6 | 86.08 | 85.41 | 87.66 | 0.99407 |
Distance: 0.1335 DDH estimate: 31.60% |
| Pseudomonas kilonensis DSM 13647 | 99.38 | 100 | 88.13 | 87.44 | 89.19 | 0.99298 |
Distance: 0.1154 DDH estimate: 35.70% |
| Pseudomonas viciae 11K1 | 99.31 | 100 | 88.30 | 87.69 | 89.22 | 0.99587 |
Distance: 0.1134 DDH estimate: 36.20% |
| Pseudomonas lini CFBP 5737 / DSM 16768 | 99.31 | 100 | 82.57 | 81.51 | 86.04 | 0.96318 |
Distance: 0.1627 DDH estimate: 26.60% |
| Pseudomonas chlororaphis subsp. Chlororaphis NBRC 3904 | 99.25 | 100 | 82.36 | 81.21 | 86.10 | 0.95255 |
Distance: 0.1651 DDH estimate: 26.30% |
| Pseudomonas mediterranea CFBP 5447 | 99.11 | 100 | 86.43 | 85.69 | 87.94 | 0.99189 |
Distance: 0.1313 DDH estimate: 32.10% |
| Pseudomonas mucoides P154a | 99.03 | 84.4 | 82.24 | 81.12 | 85.84 | 0.96132 |
Distance: 0.1656 DDH estimate: 26.20% |
| Pseudomonas fluorescens DSM50090 | 98.97 | 100 | 80.52 | 79.13 | 85.37 | 0.9568 |
Distance: 0.1794 DDH estimate: 24.30% [22 |
| Pseudomonas migulae NBRC 103157 | 98.97 | 100 | 82.47 | 81.33 | 86 | 0.96416 |
Distance: 0.1636 DDH estimate: 26.50% |
| Pseudomonas kielensis MBT-1 | 98.97 | 100 | 82.41 | 81.31 | 86.11 | 0.97128 |
Distance: 0.1627 DDH estimate: 26.60% |
|
Pseudomonas frederiksbergensis JAJ28 / SAMN04490185/ BS3655 LMG 19851 |
98.97 | 100 | 82.42 | 81.45 | 85.89 | 0.96646 |
Distance: 0.1636 DDH estimate: 26.50% |
| Pseudomonas taetrolens DSM 21104 | 98.9 | 100 | 78.82 | 77.32 | 84.75 | 0.90873 |
Distance: 0.1886 DDH estimate: 23.20% |
| Pseudomonas chlororaphis subsp. Aureofaciens NBRC 3521 | 98.9 | 100 | 82.34 | 81.25 | 86.18 | 0.95117 |
Distance: 0.1655 DDH estimate: 26.20% |
| Pseudomonas chlororaphis subsp. Aurantiaca DSM 19603 | 98.9 | 100 | 82.30 | 81.28 | 86.18 | 0.95328 |
Distance: 0.1643 DDH estimate: 26.40% |
| Pseudomonas chlororaphis subsp. Piscium DSM 21509 | 98.9 | 100 | 82.33 | 81.20 | 86.12 | 0.95268 |
Distance: 0.1654 DDH estimate: 26.20% |
| Pseudomonas arsenicoxydans CECT 7543 | 98.9 | 100 | 82.35 | 81.39 | 85.88 | 0.96866 |
Distance: 0.1647 DDH estimate: 26.30% |
| Pseudomonas salomonii CFBP 2022 / ICMP 14252 | 98.84 | 94.2 | 80.60 | 79.29 | 85.43 | 0.95021 |
Distance: 0.1780 DDH estimate: 24.50% |
| Pseudomonas prosekii LMG 26867 | 98.83 | 100 | 82.10 | 80.73 | 85.69 | 0.8877 |
Distance: 0.1686 DDH estimate: 25.80% |
| Pseudomonas haemolytica DSM 108987 | 98.83 | 100 | 80.55 | 79.07 | 85.50 | 0.96933 |
Distance: 0.1789 DDH estimate: 24.40% |
| Pseudomonas karstica HJ/4 / CCM 7891 | 98.76 | 99.3 | 80.65 | 79.18 | 85.41 | 0.97913 |
Distance: 0.1743 DDH estimate: 25.00% |
| Pseudomonas veronii DSM 11331 | 98.7 | 100 | 81.07 | 79.57 | 85.64 | 0.96096 |
Distance: 0.1753 DDH estimate: 24.80% |
| Pseudomonas caspiana FBF102 | 98.7 | 100 | 77.38 | 76.02 | 84.58 | 0.90133 |
Distance: 0.1957 DDH estimate: 22.40% |
| Pseudomonas spelaei SJ/9/1 / CCM 7893 | 98.69 | 99.3 | 80.77 | 79.36 | 85.48 | 0.97797 |
Distance: 0.1774 DDH estimate: 24.60% |
| Pseudomonas yamanorum 8H1 / LMG 27247 | 98.68 | 98.4 | 81.05 | 79.56 | 85.58 | 0.96659 |
Distance: 0.1749 DDH estimate: 24.90% |
| Pseudomonas marginalis ATCC 10844 / DSM 13124 | 98.67 | 92.7 | 80.96 | 79.72 | 85.64 | 0.96345 |
Distance: 0.1749 DDH estimate: 24.90% |
Comparative genome analysis
The genomic features of the strain SWRIQ11 and nine closely related genomes (ANI > 83%), including the sequence length, the number of genes (CDSs, tRNAs, and rRNAs), and GC content are presented in Table 2. The genome features of SWRIQ11 were similar to nine closely related species, and the most similarities were related to Pseudomonas tehranensis SWRI196, Pseudomonas brassicacearum subsp. Neoaurantiaca, and Pseudomonas corrugata which were located near SWRIQ11 based on phylogenetic analysis (Fig. 1 and Fig. 2).
Table 2.
Genome features of strain SWRIQ11 and closely related species
| Strains | Genome size (bp) | GC% | Scaffolds | No. of Genes | No. of rRNA | No. of tRNA | No. CDSs (with protein) | Bio-Project ID |
|---|---|---|---|---|---|---|---|---|
| Pseudomonas sp. SWRIQ11 | 6,196,390 | 60.1 | 101 | 5,566 | 5 | 51 | 5420 | PRJNA803818 |
| Pseudomonas tehranensis SWRI196 | 5,993,891 | 60.46 | 531 | 5,458 | 4 | 49 | 5,353 | PRJNA639797 |
| Pseudomonas brassicacearum subsp. Neoaurantiaca CDVBN10 | 6,172,913 | 60.8 | 68 | 5712 | 6 | 53 | 5467 | PRJNA546138 |
| Pseudomonas bijieensis L22-9 | 6,730,360 | 60.9 | 1 | 6,002 | 16 | 66 | 5,676 | PRJNA592828 |
| Pseudomonas thivervalensis DSM 13194 | 6,581,995 | 61.2 | 25 | Without published annotation | PRJNA290438 | |||
| Pseudomonas brassicacearum subsp. Brassicacearum DBK11 / CCUG:51,508 | 6,733,367 | 60.8 | 61 | 6307 | 13 | 60 | 6054 | PRJNA563568 |
| Pseudomonas viciae 11K1 | 6,704,877 | 60.3 | 2 | 5,894 | 16 | 64 | 5,741 | PRJNA514417 |
| Pseudomonas mediterranea CFBP 5447 | 6,319,692 | 61.1 | 32 | 5,545 | 2 | 53 | 5,355 | PRJNA210952 |
| Pseudomonas corrugata ATCC 29736 / DSM:7228 | 6,126,732 | 60.6 | 31 | Without published annotation | PRJNA290438 | |||
| Pseudomonas kilonensis DSM 13647 | 6,385,813 | 60.9 | 44 | Without published annotation | PRJNA290438 | |||
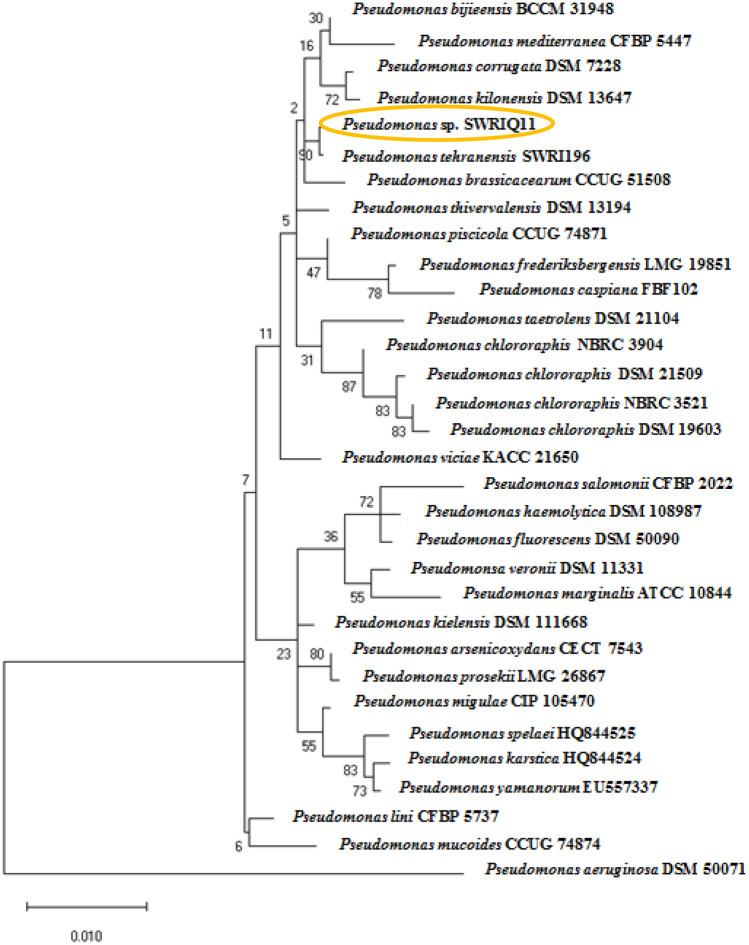
Fig. 1.
Phylogenetic tree of the Pseudomonas sp. SWRIQ11 and 30 closely related species based on 16S rRNA sequence analysis. Pseudomonas sp. SWRIQ11 clusters with Pseudomonas tehranensis SWRI196 in the same clade. Evolutionary analyses were conducted in MEGA 10 by the Maximum Likelihood method, the Tamura-Nei model, with 1000 bootstrap replications (Tamura et al. 2021). Pseudomonas aeruginosa DSM 50071 was applied as an outgroup
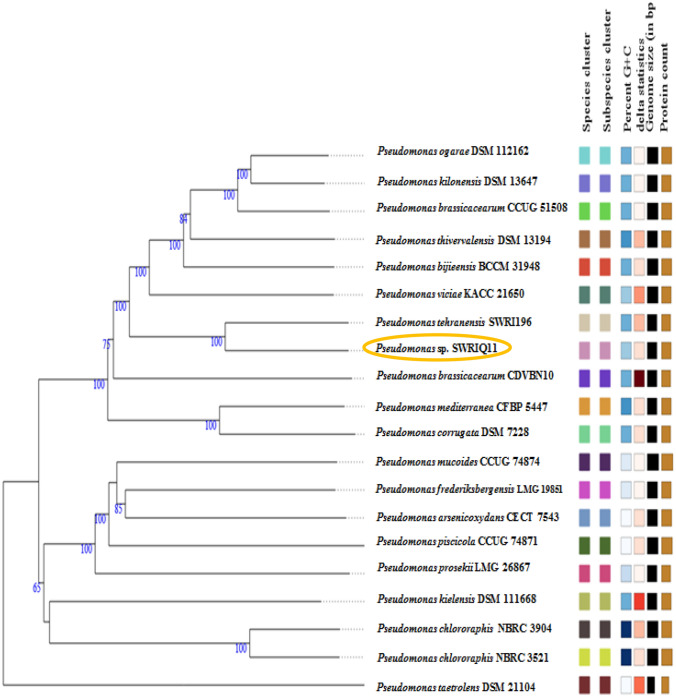
Fig. 2.
Phylogenetic tree of the Pseudomonas sp. SWRIQ11 and 20 closely related species based on whole genome sequence analysis. Pseudomonas sp. SWRIQ11 clusters with Pseudomonas tehranensis SWRI196 in the same clade. Evolutionary analysis was conducted by Type (Strain) Genome Server (TYGS) using the Genome BLAST Distance Phylogeny (GBDP) method with 100 bootstrap replications. Differences in genome size, GC percent, and protein count are indicated by color code (Meier-Kolthoff and Goker 2019)
Evolutionary relationships of strain SWRIQ11
The phylogenetic relations of strain SWRIQ11 and closely related species of Pseudomonas spp. based on 16S rRNA and whole genome sequences showed Pseudomonas sp. SWRIQ11 clusters with Pseudomonas tehranensis SWRI196 (GCA_014268615.1) in the same clade (Fig. S1, Figs. 1, and 2).
Genes related to plant growth promotion and stress alleviation characteristics in the genome of Pseudomonas sp. SWRIQ11
Genes and pathways related to plant growth-promotion and stress-alleviation characteristics, including stress response sigma factors, stress sensors, signaling or regulation proteins, chaperones, motility, attachments, colonization, enzymes for degrading plant-derived carbohydrates, exopolysaccharides, ion homeostasis, osmoprotectants, nutrient acquisition, phytohormones biosynthesis, ACC-deaminase, and antioxidants defenses were detected in the genome of strain SWRIQ11.
Genes related to stress sensing and halotolerance in the genome of SWRIQ11
Bacteria apply the cell surface extracytoplasmic function (ECF) sigma factors for sensing and responding to the surroundings. Moreover, ECF sigma factors can be a factor in establishing plants–bacteria interactions (Sheibani-Tezerji et al. 2015). The genome of SWRIQ11 contained at least 12 genes of the ECF sigma factors subfamily. In addition, there were several genes related to stress sensing, signaling, and response, and also other stress sigma like RNA polymerase sigma factors RpoE, RpoH, RpoS, serine phosphatase RsbU, outer membrane stress sensor protease DegS and DegQ (Table S1).
The adaptation mechanisms for tolerating salinity stress in SWRIQ11 can be attributed to the ion homeostasis, accumulation of osmolytes, and synthesis of universal proteins related to salt stress tolerance (Goyal et al. 2019). Transmembrane proteins acting as Na+/H+ antiporters participate significantly in conserving intracellular pH, cellular sodium amount, homeostasis, and cell volume (Goyal et al. 2019). Five types of Na+/H+ antiporters are known in prokaryotes, including NhaA, NhaB, NhaC, NhaD, and NapA (Kapoor and Kanwar 2019). Incitation of K+ uptake is the first quick reaction to an osmotic change by bacteria (Goyal et al. 2019). There are three K+ uptake systems, including Kup, Trk, and Kdp (Goyal et al. 2019). The genome of strain SWRIQ11 contained genes related to ion homeostasis, consisting of nhaA, nhaD, mrp, trkH, trkA, trkG, kup, kdpA, kdpB, kdpC, kdpD, and kdpE.
Osmolytes are accumulated either by intake from surroundings or by de novo synthesis (Mishra et al. 2018). Several genes potentially related to the importing systems of compatible solutes were detected in the genome of SWRIQ11, like genes of osmoprotectant ABC transporter, spermidine/putrescine import ABC transporter, ectoine/hydroxy ectoine ABC transporter, Omp family, EnvZ, NarL/FixJ family, choline-binding ABC transport, choline ABC transport system, high-affinity choline uptake, osmotically activated L-carnitine/choline ABC transporter, betaine ABC transporter, glycine betaine transporter, glycine betaine ABC transport system, glycine betaine/L-proline ABC transporter, L-proline/glycine betaine ABC transporter, glycerol ABC transporter, and glycerol uptake facilitator protein (Table S2).
Furthermore, the genes related to the synthesis of different osmolytes were present in the SWRIQ11 genome as well. Genes of proline synthesis in the SWRIQ11 genome included glutamate 5-kinase (proB), gamma-glutamyl phosphate reductase (proA), pyrroline-5-carboxylate reductase (proC), and pyrroline-5-carboxylate reductase ProG-like (proG). The genome of the strain contained genes of the choline and glycine betaine synthesis pathway, including the betC gene, which encodes a choline sulfatase that catalyzes the conversion of choline-O-sulfate into choline, the betA gene that encodes choline dehydrogenase that converts choline to betaine aldehyde, and the betB gene which encodes a betaine aldehyde dehydrogenase that participates in the production of the osmoprotectant glycine betaine through the irreversible oxidation of betaine aldehyde. SWRIQ11 contained a SAMDC1 gene, which encodes S-adenosylmethionine decarboxylase proenzyme, a crucial enzyme for the biosynthesis of polyamines. The genes of spermidine synthase (polyamine aminopropyl transferase) (speE), homospermidine synthase (hss), and gamma-butyrobetaine dioxygenase, which are related to the synthesis of spermidine, homospermidine, and carnitine, respectively, were present in the SWRIQ11 genome. The genes of the biosynthesis pathway of ectoine which is mediated by three enzymes, including N-alpha-acetyl-L-2,4-diamino butyrate deacetylase (ectA), L-2,4-diaminobutyric acid transaminase (ectB) and L-ectoine synthase (ectC) were identified in the genome of SWRIQ11. Accordingly, a gene cluster with a high similarity to the ectoine biosynthesis was observed during antiSMASH analysis (Fig. S2). Furthermore, the genes of arginine decarboxylase (speA), agmatine deiminase (aguA), and N-carbamoyl putrescine amidase (CPA) for putrescine biosynthesis via the agmatine pathway were identified in the SWRIQ11 genome. Five trehalose production pathways have been shown in bacteria consisting of TreS, OtsA/OtsB (Tps/Tpp), TreP, TreT, and TreY/TreZ (Liu et al. 2016; Nobre et al. 2008). The SWRIQ11 genome contained genes of trehalose synthesis pathways, including trehalose synthase (treS) from the TreS pathway, Malto-oligosyltrehalose synthase (treY), and malto-oligosyltrehalose trehalohydrolase (treZ) from the TreY/TreZ pathway.
In addition to genes related to the accumulation of osmoprotectants, the SWRIQ11 genome contained the aquaporin Z synthesis gene (aqpZ), a channel that adjusts the osmotically driven flow of water.
Heat-shock proteins (HSPs) or chaperones like DnaK, DnaJ, Clp family, GroES, GroEL, proteases, and sHSPs are upregulated upon osmotic stress. The SWRIQ11 genome contained genes of different chaperones like htrA, dnaJ, dnaK, grpE, groES, groEL, recN, djlA, cbp family, surA, clp family, hsc family, yidC, htp family, ccmE, mbtH, yegD, and mazG. Furthermore, the genome of strain SWRIQ11 contained several genes related to RNA chaperones (Hfl operon) that bind small regulatory RNA (sRNAs), mRNAs and with high specificity to tRNAs to assist mRNA translational regulation in reaction to environmental stress. (Rajkowitsch et al. 2007; Arce-Rodriguez et al. 2015). Moreover, the genes of the universal stress protein family in the SWRIQ11 genome, including uspA, uspB, uspC, uspD, uspE, uspF, and uspG were observed.
Genes related to motility, chemotaxis, and colonization in the SWRIQ11 genome
Bacterial root colonization starts when bacteria sense particular compounds in the root exudates. The influencing factors in the specificity of interactions between plants and bacteria are the quantity, and composition of root exudates and soil conditions (Etesami 2020).
The interaction of bacteria with plants is associated with a widespread set of genes related to chemotaxis, motility, adhesion, and colonization (Eida et al. 2020; Levy et al. 2018). A signal molecule, a chemoreceptor (such as the methyl-accepting chemotaxis protein [MCP]), a cytoplasmic signal transduction system, and a response regulator that regulates flagellar or pili activity involved in chemotaxis reaction (Levy et al. 2018). The SWRIQ11 genome contained chemotaxis genes (che and cet), flagellar gene operons (fla, flb, fli, flg, flh, fle, and mot), adhesion genes (oprQ and aidA), colonization genes (tad, cpa, and flp), and pili synthesis-related genes (pil, fim, cpa, and tad) (Table S3).
The genomes of many rhizobacteria encode enzymes for the degradation of plant-derived carbohydrates (Levy et al. 2018). The SWRIQ11 genome contained the genes related to cellulase M and xylanase.
Genes related to the nutrient acquisition in the SWRIQ11 genome
The genes related to nitrogen fixation (nif genes) were not identified in the genome of strain SWRIQ11. This strain contained genes of denitrification, including the nar gene cluster (nitrate reductase), nir gene cluster (nitrite reductase), nor gene cluster (nitric oxide reductase), and the nos gene cluster (nitrous oxide reductase). Furthermore, the genome contained the genes of dissimilatory nitrate reduction to ammonium (DNRA) process, which consists of two steps, nitrate is reduced to nitrite in the first step like denitrification (nar gene cluster), and then reduction of nitrite to ammonium by nitrite reductase, encoded by nrfA gene (Table S4) (Bu et al. 2017).
Siderophores are metal-chelating compounds produced by most PGPR with a vast chemical diversity. Bacterial siderophores consist of four main classes, including phenol catecholates, hydroxamates, carboxylate, and pyoverdines (Crowley 2006).
The antiSMASH analysis of the strain SWRIQ11 genome showed the existence of two gene clusters with high similarity to pyoverdine siderophore biosynthesis (pvd) (Figs. S3 and S4). Accordingly, the genes of pvd were identified in the genome of SWRIQ11 during annotation with different software and databases (Table S5). Achromobactin (ACR) is related to a group of non-peptide siderophores comprising a citrate core that is connected to diamino butyrate and ethanolamine, which are both condensed with α-ketoglutarate (Berti and Thomas 2009). The genes related to achromobactin siderophore synthesis were present in the genome of SWRIQ11 (acsABCDEF). Further, the strain genome contained the gene associated with ferrichrome (a hydroxamate-type siderophore)-iron receptor (fhuA).
Bacterial organic acids release phosphate. The most prevalent organic acids consist of gluconic acid (GA) and 2-keto gluconic acid. Glucose-1-dehydrogenase (gcd) synthesizes GA, and pyrrolo-quinolone quinine (PQQ) acts as its cofactor. Besides, GA dehydrogenase (gad) plays a role in GA synthesis and its transformation to 2-ketogluconate (Olenska et al. 2020). In addition, PGPR hydrolyze P-organic substrates enzymatically by non-specific acid phosphatases (NSAPs), including phytases, acid and alkaline phosphomonoesterases (phosphatases), phosphonatases, and C-P lyases (Olenska et al. 2020, Suarez et al. 2019). Pseudomonas sp. SWRIQ11 contained genes related to GA synthesis (gcd and pqqBCDEF) and different phosphatases (Table S6).
Genes related to phytohormones in the SWRIQ11 genome
Phytohormones support plants against abiotic stresses, and PGPR can modulate the level of endogenous phytohormones in plants by producing similar hormones (Mohammadipanah and Zamanzadeh 2019). PGPR hormones can trigger the division and growth of plant cells, alter root characteristics and play a significant role in organizing an array of genes, their regulators, and several signal transduction pathways when plants are under abiotic stresses conditions and make crops tolerant to the stresses (Etesami 2020). The main phytohormones produced by bacteria are abscisic acid (ABA), gibberellins (GA), auxins, ethylene, and cytokinins (Olenska et al. 2020).The amount of ABA increases under osmotic stress through elevated expression of multiple genes of ABA production, including genes of aldehyde oxidase, zeaxanthin epoxidase, 9-cis-epoxy carotenoid dioxygenase, and molybdenum cofactor sulturase (Khan et al. 2020). Genes related to different subunits of aldehyde oxidase were identified in the SWRIQ11 genome.
The pathways for auxin production in bacteria are categorized according to their intermediate, including indole-3-pyruvate (IPyA), indole-3-acetamide (IAM), indole-3-acetonitrile (IAN), tryptophan side-chain oxidase, tryptamine, and tryptophan independent (Gamalero and Glick 2011). IAM and IPyA are two main microbial pathways (Gupta et al. 2016), while most PGPR apply the IPyA pathway (Khatoon et al. 2020). The Pseudomonas sp. SWRIQ11 genome contained genes related to tryptophan synthesis as a precursor of auxin synthesis (anthranilate synthase, anthranilate phosphoribosyltransferase, phosphoribosyl anthranilate isomerase, different chains of tryptophan synthase, isochorismatase, indole-3-glycerol phosphate synthase, tryptophanyl-tRNA synthetase, phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase, para-aminobenzoate synthase, and aminodeoxychorismate lyase). The genes of the IPyA pathway, including the pyridoxal phosphate-dependent aminotransferase genes, which transforms tryptophan to IPyA, indole-3-pyruvate decarboxylase that changes IPyA to indole-3-acetaldehyde (IAAld), and indole-3-acetaldehyde dehydrogenase which synthesizes IAA, were identified in the SWRIQ11 genome. Furthermore, the SWRIQ11 genome contained the genes of the IAM pathway for IAA production, including the genes of tryptophan monooxygenase that transforms tryptophan to IAM and indole acetamide hydrolase (iaaH), which hydrolyzes IAM into IAA. Moreover, the SWRIQ11 genome contained the amidase gene, which contributes to the IAM pathway through the transformation of indole-3-acetamide to IAA.
Environmental stresses increase ethylene synthesis in the plant, which hampers the plants growth (Kumari et al. 2016). The ACCD-synthesizing bacteria can lessen the deleterious impact of the different stresses on plants by catabolizing ACC (precursor of ethylene) to α-ketobutyrate (precursor of leucine) and ammonia (Kumari et al. 2016, Jaya et al. 2019). ACCD is a pyridoxal phosphate-dependent enzyme encoded by the acdS gene (Kumari et al. 2016). acdS genes have been modulated through the leucine-responsive regulatory protein (LrP) and AcdB protein (Kumari et al. 2016). The SWRIQ11 genome contained the genes of ACCD (acdS), LrP (acdR), and several genes related to pyridoxal phosphate synthesis. In addition, the genes related to pyridoxal phosphate-dependent deaminase and D-cysteine desulfhydrase (dcyD) (homolog of ACCD) were identified in the SWRIQ11 genome.
Genes related to volatile organic compounds (VOCs) synthesis in the SWRIQ11 genome
Acetoin and 2,3-butanediol as VOCs are produced when two pyruvate molecules are compressed into acetolactate and transformed to acetoin via acetolactate decarboxylase, and lastly, acetoin reductase catalyzes 2,3-butanediol from acetoin (Liu et al. 2016; Suarez et al. 2019). Pseudomonas sp. SWRIQ11 genome contained the encoding gene of acetolactate synthase (ilv) but not genes related to acetolactate decarboxylase and acetoin reductase.
Genes related to antioxidant defense mechanism in the genome of strain SWRIQ11
Plants are equipped with antioxidant defense systems, including enzymatic and nonenzymatic mechanisms against the deleterious effects of reactive oxygen species (ROS) (Arora et al. 2020). There were genes related to antioxidant enzymes in the SWRIQ11 genome, including bifunctional enzyme catalase-peroxidase (katG), catalase (katE), glutathione reductase (gor), glutathione peroxidase (gpo), superoxide dismutase (sod), and alkyl hydroperoxide reductase subunit C-like protein (ahpC). Additionally, the SWRIQ11 genome contained genes related to nonenzymatic mechanisms (cysteine, glutathione, carotenoids, tocopherol, and ascorbate) (Table S7).
Genes related to exopolysaccharides (EPS) synthesis in the SWRIQ11 genome
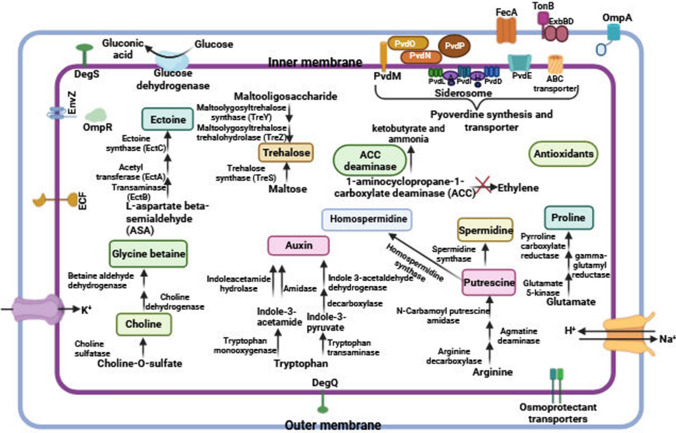
EPS production by PGPR forms hydrophilic biofilms conferring desiccation protection, regulates nutrients and water flow across plant roots, binds to Na+ and decreases the bioavailability of the ion, aggregates root-adhering soils (RAS), and stabilizes soil aggregates (Etesami 2020; Jiang et al. 2021). The SWRIQ11 genome contained the genes of the alginate production pathway, including algF, algJ, algX, algG, algK, algE, algP, algQ, algR, and algB. Moreover, the genes of capsular polysaccharide biosynthesis protein (capD), mannose-1-phosphate guanylyltransferase, mannose-6-phosphate isomerase, and glycosyl transferase from the exopolysaccharide biosynthesis pathway were detected in the SWRIQ11 genome. A schematic summary of metabolic pathways related to plant growth promotion and stress alleviation characteristics attributed to the SWRIQ11 genome has been shown in Fig. 3.
Fig. 3.
Schematic proteins and metabolic pathways involved in plant growth promotion and salinity stress alleviation identified in the genome of Pseudomonas sp. SWRIQ11. ECF Extracytoplasmic function (ECF) sigma factors, OmpA Outer membrane protein A
Existence of virulence genes in the genome of strain SWRIQ11
Previously, type III secretion systems (T3SSs) have been extensively investigated for their essential roles in bacterial pathogenesis in animals and plants. T3SSs have lately been known as a vital characteristic of an extensive range of symbioses in most Gram-negative bacteria that are closely associated with eukaryotes (Mavrodi et al. 2011; Zboralski et al. 2022). Genes of the T3SS cluster are conserved in both pathogenic and saprophytic species of pseudomonads (Mavrodi et al. 2011). The roles of T3SSs in plant-beneficial Pseudomonas spp., are not completely elucidated and seem as a determinant tool of rhizobacteria-plants interactions (Zboralski et al. 2022). According to RAST subsystem information, there is not any subsystem related to virulence and pathogenicity in the genome of strain SWRIQ11. Some genes of the hrp/hrc gene cluster, which encode T3SSs that are not complete gene cluster, were identified in the SWRIQ11 genome (Table S8). Several secreted proteins which cause disease in susceptible plants, including harpins and avirulence proteins (Avr) (Hueck 1998), were also not present in the genome of SWRIQ11.
Genes related to further functions in the SWRIQ11 genome
The Pseudomonas sp. SWRIQ11 genome contained the genes related to the synthesis of antibiotic compounds like phenazine and 4-hydroxybenzoate, which act as biocontrol agents and suppress plant pathogenic microorganisms. Furthermore, the gene cluster with similarity to the synthesis of herboxidiene was identified in the SWRIQ11 genome. Herboxidiene is a polyketide with herbicide and antitumor capabilities (Miller-Wideman et al. 1992; Hasegawa et al. 2011). Moreover, herboxidiene triggers the expression of genes related to responses to abiotic stress in plants (AlShareef et al. 2017). Furthermore, the SWRIQ11 genome contained the genes related to different pathways of aromatic compound degradation.
Discussion
The application of PGPR as an alternative solution for providing increasing nutrition needs and remediation of stress-affected soil is among the strategies triggered by the trend in climate change. For the use of PGPR as stress-alleviating biofertilizer, the molecular mechanisms of action in PGPR-plants interactions need to be revealed. Accordingly, the number of genomes of rhizobacteria assembled, bioprojects, and articles on this subject in PubMed have increased 16, 5, and 6 times in 2022 compared to 2010, respectively. However, because of the genetic and metabolic diversities of rhizobacteria and the complexity of their interactions, there are still unidentified gaps in molecular mechanisms of action by which PGPR protect the plants.
Pseudomonas spp. are one of the most ubiquitous plant-associated and metabolically versatile group of bacteria. Currently, the genome sequences of 348 species (13,521 strains) of the pseudomonas members have been deposited in the NCBI database upto September 2023. Eighty-five species (11,670 strains) are pathogenic, opportunistic pathogenic, and responsible for spoilage. Two hundred sixty-three species (1851 strains) have been isolated from the environment, from which, 87 species (1150 strains) have shown the potential for application as PGPR or for bioremediation purposes.
In this study, the genome of a new Pseudomonas sp. (SWRIQ11) that promote plant growth and alleviate salinity stress in olive seedlings, was sequenced and mined to reveal the molecular mechanisms of action and potential functional capabilities of this new strain. The genome data of SWRIQ11 supported the results of lab and greenhouse studies, indicating the PGP ability of this halotolerant strain. Through genome annotation and analysis, more than 825 CDSs related to plant growth-promoting and stress-alleviating traits were recognized in the genome of SWRIQ11. Among 30 species of Pseudomonas, which had more similarity with SWRIQ11 (according to EzBioCloud and JSpeciesWS servers), 10 species have records on plant growth promotion, and genes related to plant growth-promoting traits have been analyzed in 5 species, including P. corrugata (Zachow et al. 2017), P. chlororaphis subsp. Aurantiaca (Zhang et al. 2020), P. thivervalensis (Nascimento et al. 2021), P. fluorescens (Cho et al. 2015), and P. veronii (Montes et al. 2016).
Pseudomonas sp. SWRIQ11 was halotolerant (could grow in 6% NaCl), and the presence of several genes related to stress response sigma factors, ECF sigma factors, stress sensors, signaling and regulation proteins, chaperones, universal stress protein family, ion homeostasis including genes of Na+/H+ antiporters and K+ uptake systems, and the genes of osmoprotectants accumulation in the genome of SWRIQ11 confirmed the capability of this strain to tolerate the salinity. The genes related to osmoprotectants accumulation in the SWRIQ11 genome included different osmolytes import systems and de novo synthesis of proline, choline, glycine betaine, polyamines, spermidine, homospermidine, putrescine, carnitine, ectoine, and trehalose. Interestingly, Pseudomonas sp. SWRIQ11 contained two pathways (TreS and TreY/TreZ) for trehalose synthesis. The presence of several trehalose biosynthetic pathways can be due to the severe requirement to accumulate trehalose under stressful environmental conditions. Accordingly, the genes related to the synthesis of several osmoprotectants and two pathways of trehalose synthesis (TreS and TreY/TreZ) have been identified in the genomes of P. thivervalensis SC5 (Nascimento et al. 2021) and P. fluorescens PCL1751 (Cho et al. 2015), which were halotolerant species. In the SWRIQ11 genome, like many rhizobacteria, a high number of characterized CDSs were involved in the establishment of interaction with plants, including genes related to chemotaxis, motility, ECF sigma factors, adhesion, colonization, and enzymes for degrading plant-derived carbohydrates. The genomes of all of the five species of Pseudomonas (more similar to SWRIQ11) contained these indicated genes.
Genes related to ammonium production, including genes of dissimilatory nitrate reduction to ammonium (DNRA) process, were identified in the SWRIQ11 genome. For high efficiency iron uptake, this strain contained genes related to biosynthesis and transport of two siderophores, including pyoverdine and achromobactin, and a gene of the ferrichrome siderophore (fhuA) receptor for using xenoferrichrome. Pyoverdine biosynthesis genes have been the most predominant siderophore among the five species of Pseudomonas. The genes related to gluconic acid synthesis (gcd and pqq) as the most prevalent agent for releasing phosphate by phosphate-solubilizing bacteria and different phosphatases were found in the genome of SWRIQ11. The genomes of P. thivervalensis SC5 (Nascimento et al. 2021) and P. fluorescens PCL1751 (Cho et al. 2015), which could tolerate salinity stress, contain the genes gcd and pqq. The existence of genes associated with phosphate solubilization in these halotolerant Pseudomonas spp. can be correlated to the decrease in the bioavailability of phosphorus in saline soil (Xie et al. 2022).
The genes related to tryptophan synthesis as a precursor of auxin synthesis and two IAA synthesis pathways (IPyA and IAM pathways) were identified in the SWRIQ11 genome. PGPR support plants to moderate the abiotic stresses and prompt plant growth by supplying auxins which are synthesized and excreted by more than 80% of the rhizobacteria. However, the most common auxin (IAA) induces the expression of the ACC (precursor of ethylene) synthesizing enzyme gene at high concentrations. Therefore, IAA can enhance plant growth synergistically with ACC deaminase (ACCD). The ACCD gene that is key to the efficiency of PGPR in mitigating stress, its modulator, and the homolog of ACCD were present in the SWRIQ11 genome. All of the five mentioned species and several other assessed plant growth-promoting Pseudomonas spp. like P. putida LWPZF (Jin et al. 2022), P. chlororaphis GP72, P. fluorescens Pf-5, P. stutzeri A1501 (Shen et al. 2013), and P. aeruginosa FG106 (Ghadamgahi et al. 2022), harbor the acdS gene, which indicates the high prevalence of this pivotal gene in PGPR.
There were genes related to different antioxidant enzymes and nonenzymatic antioxidant mechanisms in the SWRIQ11 genome, which enabled the strain to tolerate different stresses. The SWRIQ11 genome included the genes of the alginate exopolysaccharide biosynthesis pathway. Interestingly, two halotolerant species (P. thivervalensis SC5 (Nascimento et al. 2021) and P. fluorescens PCL1751 (Cho et al. 2015) among five species, contained the genes associated with alginate EPS, which is efficient in salinity stress tolerance. However, further studies are needed to determine the expression and regulation of identified genes in the SWRIQ11 genome.
The absence of virulence genes and any subsystem related to virulence and pathogenicity in the SWRIQ11 genome was approved by the negative result in the HR biosafety assay and non-hemolytic activity of this strain. Some genes of the hrp/hrc gene cluster encode T3SSs (these protein appendages are found in many eukaryotes-associated Gram-negative bacteria), but no complete gene cluster was found in the SWRIQ11 genome. Several secreted proteins which cause disease in susceptible plants, including harpins and avirulence proteins (Avr), were not identified in the SWRIQ11 genome. Accordingly, Shariati et al. (2017) showed only the Pantoea agglomerans strain with pathogenic activity contained the complete hrc/hrp gene cluster. The genomic results of the assay of pathogenicity of SWRIQ11 are according to the negative result of the HR test Furthermore, the identification of genes related to the antibiotic, antitumor, and herbicide compounds biosynthesis and degradation of aromatic compounds in the SWRIQ11 genome shows more possible capabilities in this bacterium, which need to be experimentally proven.
By applying several methods and indices for identification of this strain because pseudomonas species are closely related, and regarding the threshold of species definitions (≥ 95% ANI, ≥ 0.99 TETRA signature, and > 70% of dDDH or GGDC), Pseudomonas sp. SWRIQ11 is a new species candidate clustered with Pseudomonas tehranensis SWRI196 in the same clade based on phylogenetic analysis.
Conclusions
Pseudomonas sp. SWRIQ11 is a PGPR that can alleviate salinity stress in olive seedlings. Comprehensive analysis of the Pseudomonas sp. SWRIQ11 genome confirmed observations of its PGPR characteristics in lab and greenhouse studies. SWRIQ11 is introduced as a new potent halotolerant plant growth-promoting and stress-alleviating rhizobacterium, which contained a high number of CDSs (more than 825) associated with tolerating salinity stress, interaction with plants, enhancing plant growth, and mitigating the salinity stress similar to assessed halotolerant plant growth-promoting Pseudomonas spp. However, further studies are needed to determine the expression and regulation of these genes. Regarding the salinity stress alleviation and the absence of any virulence genes in the SWRIQ11 genome, the strain can be applied as a potential safe plant stress protector. Furthermore, considering the identification of genes related to the antibiotic, antitumor, and herbicide compounds biosynthesis and degradation of aromatic compounds in the SWRIQ11 genome, further additional microbial, herbicidal or pesticidal activity may increase the competency of this strain as a biofertilizer.
Acknowledgements
We are grateful to Soil and Water Research Institute for providing Pseudomonas sp. SWRIQ11 and National Institute of Genetic Engineering and Biotechnology for conducting part of the experiments there.
Data availability
All analyzed data of this study are presented in the paper and its supplementary data files. The whole genome shotgun project was deposited at DDBJ/ENA/GenBank under the accession number JAKNQY000000000. Additionally, further information about the bacterial assembly is accessible in the NCBI database with BioProject ID: PRJNA803818 and BioSample ID: SAMN25688536.
Declarations
Ethical statements
The authors declare that they have no conflict of interest. This research contained no work on animal or tissue samples.
Contributor Information
Fatemeh Mohammadiapanah, Email: fmohammadipanah@ut.ac.ir.
Sajjad Sarikhan, Email: sarikhan@ibrc.ir.
References
- Alberton D, Valdameri G, Moure VR, Monteiro RA, Pedrosa FO, Muller-Santos M, Souza EM (2020) What did we learn from plant growth-promoting rhizobacteria (PGPR)-grass associations studies through proteomic and metabolomic approaches?. Front Sustain Food Syst 4:607343. 10.3389/fsufs.2020.607343.
- AlShareef S, Ling Y, Butt H, Mariappan KG, Benhamed M, Mahfouz MM. Herboxidiene triggers splicing repression and abiotic stress responses in plants. BMC Genom. 2017;18:260. doi: 10.1186/s12864-017-3656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Arce-Rodriguez A, Calles B, Nikel PI, de Lorenzo V. The RNA chaperone Hfq enables the environmental stress tolerance super-phenotype of Pseudomonas putida. Environ Microbiol. 2015;18(10):3309–3326. doi: 10.1111/1462-2920.13052. [DOI] [PubMed] [Google Scholar]
- Arora NK, Fatima T, Mishra J, Mishra I, Verma S, Verma R, Verma M, Bhattacharya A, Verma P, Mishra P, Bharti C. Halo-tolerant plant growth-promoting rhizobacteria for improving productivity and remediation of saline soils. J Adv Res. 2020;26:69–82. doi: 10.1016/j.jare.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian VK, Jansson C, Baker SE, Ahkami AH (2021) Molecular mechanisms of plant–microbe interactions in the rhizosphere as targets for improving plant productivity. In: Gupta VVSR, Sharma AK (eds) Rhizosphere biology: interactions between microbes and plants, Springer, Singapore, pp 295–338. 10.1007/978-981-15-6125-2_14.
- Bent E, Tuzun S, Chanway C, Enebak S. Alterations in plant growth and in root hormone levels of lodgepole pines inoculated with rhizobacteria. Can J Microbiol. 2001;47:793–800. doi: 10.1139/cjm-47-9-793. [DOI] [PubMed] [Google Scholar]
- Berti AD, Thomas MG. Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J Bacteriol Res. 2009;191(14):4594–4604. doi: 10.1128/JB.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Kautsar SA, Medema MH, Weber T (2021) The antiSMASH database version 3: increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res gkaa978. 10.1093/nar/gkaa978. [DOI] [PMC free article] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. J Bioinform btu170. 2014 doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque FG, Bertrand A, Claessens A. Alleviation of drought stress and metabolic changes in Timothy (Phleum pratense L.) colonized with Bacillus subtilis B26. Front Plant Sci. 2016 doi: 10.3389/fpls.2016.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Chakrabarty AM. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol. 1995;15(3):162–168. doi: 10.1007/BF01569821. [DOI] [PubMed] [Google Scholar]
- Bu C, Wang Y, Ge C, Ahmad HA, Gao B, Ni SQ. Dissimilatory nitrate reduction to ammonium in the yellow river estuary: rates, abundance, and community diversity. Sci Rep. 2017;7:6830. doi: 10.1038/s41598-017-06404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuccino JG, Sherman N (2014) Microbiology: a laboratory manual. Tenth edition. Pearson Education, United States of America.
- Catara V. Pseudomonas corrugata: plant pathogen and/or biological resource? Mol Plant Pathol. 2007;8(3):233–244. doi: 10.1111/J.1364-3703.2007.00391.X. [DOI] [PubMed] [Google Scholar]
- Cho ST, Chang HH, Egamberdieva D, Kamilova F, Lugtenberg B, Kuo CH. Genome analysis of Pseudomonas fluorescens PCL1751: a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS ONE. 2015;10(10):e0140231. doi: 10.1371/journal.pone.0140231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley DE (2006) Microbial siderophores in the plant rhizosphere. In: Barton LL, Abadia J (eds) Iron nutrition in plants and rhizospheric microorganisms. Springer, Dordrecht. 10.1007/1-4020-4743-6_8.
- Donate-Correa J, Leon-Barrios M, Perez-Galdona R. Screening for plant growth-promoting rhizobacteria in Chamaecytisus proliferus (tagasaste), a forage tree-shrub legume endemic to the Canary Islands. Plant Soil. 2004;266:261–272. doi: 10.1007/s11104-005-0754-5. [DOI] [Google Scholar]
- Eida AA, Bougouffa S, L’Haridon F, Alam I, Weisskopf L, Bajic VB, Saad MM, Hirt H. Genome insights of the plant-growth promoting bacterium Cronobacter muytjensii JZ38 with volatile-mediated antagonistic activity against Phytophthora infestans. Front Microbiol. 2020;11:369. doi: 10.3389/fmicb.2020.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etesami H (2020) Plant microbe interactions in plants and stress tolerance. In: Tripathi DK, Singh VP, Chauhan DK, Sharma S, Prasad SM, Dubey NK, Ramawat N (eds) Plant life under changing environment, Academic Press, Elsevier, London, pp 355–396. 10.1016/B978-0-12-818204-8.00018-7.
- Gamalero E, Glick BR (2011) Mechanisms used by plant growth-promoting bacteria. In: Maheshwari DK (ed) Bacteria in agrobiology: plant nutrient management, Springer-Verlag, Berlin Heidelberg, pp 17–46. 10.1007/978-3-642-21061-7_2.
- Garrido-Sanz D, Redondo-Nieto M, Martin M, Rivilla R (2021) Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. Nov Microb Genome 7:000593. 10.1099/mgen.0.000593. [DOI] [PMC free article] [PubMed]
- Ghadamgahi F, Tarighi S, Taheri P, Saripella GV, Anzalone A, Kalyandurg PB, Catara V, Ortiz R, Vetukuri RR. Plant growth-promoting activity of Pseudomonas aeruginosa FG106 and its ability to act as a biocontrol agent against potato, tomato and taro pathogens. Biology. 2022;11:140. doi: 10.3390/biology11010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Lood C, Hofte M, Vandamme P, Rokni-Zadeh H, Noort V, Lavigne R, De Mot R. The ever-expanding Pseudomonas genus: description of 43 new species and partition of the Pseudomonas putida Group. Microorganisms. 2021;9:1766. doi: 10.3390/microorganisms9081766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila M, Pena A, Mulet M, Lalucat J, Garcia-Valdes E. Phylogenomics and systematics in Pseudomonas. Front Microbiol. 2015;6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K, Kumar T, Sharma P, Rao M, Ahmed V, Chauhan NS (2019) Crop improvement through microbial biotechnology: a cross talk. In: Akhtar MS (ed) Salt stress, microbes, and plant interactions: mechanisms and molecular approaches, Springer, Singapore, pp 69–90. 10.1007/978-981-13-8805-7_4.
- Gu Y, Ma YN, Wang J, Xia Z, Wei HL. Genomic insights into a plant growth-promoting Pseudomonas koreensis strain with cyclic lipopeptide-mediated antifungal activity. Microbiologyopen. 2020;9:e1092. doi: 10.1002/mbo3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Seth R, Sharma A (2016) Plant growth-promoting rhizobacteria play a role as phytostimulators for sustainable agriculture. In: Choudhary DK, Varma A, Tuteja N (ed) Plant-microbe interaction: an approach to sustainable agriculture, Springer, Singapore, pp 475–495.10.1007/978-981-10-2854-0_22.
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Miura T, Kuzuya K, Inoue A, Won Ki S, Horinouchi S, Yoshida T, Kunoh T, Koseki K, Mino K, Sasaki R, Yoshida M, Mizukami T (2011) Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem Biol 18:6(3):229–33. 10.1021/cb100248e. [DOI] [PubMed]
- Hoque MN, Hannan A, Imran S, Paul NC, Mondal MF, Sadhin MMR. Plant growth-promoting rhizobacteria mediated adaptive responses. J Plant Growth Regul. 2022 doi: 10.1007/s00344-022-10633-1. [DOI] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62(2):379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EP, Christen R, Bizet C, Clermont D, Motreff L, Bouchier C, Zhukova NV, Crawford RJ, Kiprianova EA. Pseudomonas brassicacearum subsp. neoaurantiaca subsp. nov., orange-pigmented bacteria isolated from soil and the rhizosphere of agricultural plants. Int J Syst Evol Microbiol. 2009;59:2476–2481. doi: 10.1099/ijs.0.009654-0. [DOI] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya DK, Giyanto, Nurhidayat N, Antonius S (2019) Isolation, identification, and detection of ACC deaminase gene-encoding rhizobacteria from rhizosphere of stressed pineapple, Indones J Biotechnol 24(1): 17–25. 10.22146/ijbiotech.39018.
- Jiang LM, Lee YJ, Han HL, Lee MH, Jeong JC, Kim CY, Kim SW, Lee JY. Genome insights into the novel species Jejubacter calystegiae, a plant growth-promoting bacterium in saline conditions. Diversity. 2021;13:24. doi: 10.3390/d13010024. [DOI] [Google Scholar]
- Jin T, Ren J, Li Y, Bai B, Liu R, Wang Y. Plant growth-promoting effect and genomic analysis of the P. putida LWPZF isolated from C. japonicum rhizosphere. AMB Express. 2022;12:101. doi: 10.1186/s13568-022-01445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen K, Nielsen P. Diversity of Pseudomonas strains isolated with King's B and Gould's S1 agar determined by repetitive extragenic palindromic-polymerase chain reaction, 16S rDNA sequencing and Fourier transform infrared spectroscopy characterization. FEMS Microbiol Lett. 1999;173:155–162. doi: 10.1016/S0378-1097(99)00065-8. [DOI] [PubMed] [Google Scholar]
- Joshi A, Chitanand M. Complete genome sequence of plant growth promoting Pseudomonas aeruginosa AJ D 2 an isolate from monocropic cotton rhizosphere. Genomics. 2020;112(2):1318. doi: 10.1016/j.ygeno.2019.07.022. [DOI] [PubMed] [Google Scholar]
- Jousset A, Schuldes J, Keel C, Maurhofer M, Daniel R, Scheu S, Thuermer A (2014) Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc 2(2):e00322–14. 10.1128/genomeA.00322-14. [DOI] [PMC free article] [PubMed]
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M (2023) KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 51. 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed]
- Kapoor R, Kanwar SS. Genetic variations in salt tolerant and plant growth promoting rhizobacteria of the Western Himalayas. J Plant Biochem Biotechnol. 2019 doi: 10.1007/s13562-019-00489-0. [DOI] [Google Scholar]
- Khan N, Bano A, Ali S, Babar M. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020 doi: 10.1007/s10725-020-00571-x. [DOI] [Google Scholar]
- Khatoon Z, Huang S, Rafique M, Fakhar A, Kamran MA, Santoyo G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J Environ Manage. 2020;273:111–118. doi: 10.1016/j.jenvman.2020.111118. [DOI] [PubMed] [Google Scholar]
- Klement Z. Rapid detection of pathogenicity of phytopathogenic Pseudomonads. Nature. 1963;199:299–300. doi: 10.1038/199299b0. [DOI] [PubMed] [Google Scholar]
- Kumari S, Varma A, Tuteja N, Choudhary DK (2016) Bacterial ACC-deaminase: an eco-friendly strategy to cope abiotic stresses for sustainable agriculture. In: Choudhary DK, Varma A, Tuteja N (eds.) Plant-microbe interaction: an approach to sustainable agriculture, Springer, Singapore, pp 165–185. 10.1007/978-981-10-2854-0_8.
- Leontidou K, Genitsaris S, Papadopoulou A, Kamou N, Bosmali MT, Madesis P, Vokou D, Karamanoli K, Mellidou I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: genomic characterization and exploration of phyto-beneficial traits. Sci Rep. 2020;10:14857. doi: 10.1038/s41598-020-71652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Conway J, Dang JL, Woyke T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe. 2018 doi: 10.1016/j.chom.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Levy A, Gonzalez IS, Mittelviefhaus M, Clingenpeel S, Paredes SH, Miao J, Wang K, Devescovi G, Stillman K, Monteiro F, Alvarez BR. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50:138–150. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang J, Zhang J, Xu W, Mou Z. Characteristics of inorganic phosphate-solubilizing bacteria from the sediments of a eutrophic lake. Int J Environ Res. 2019;16:2141. doi: 10.3390/ijerph16122141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK and Buschmann C (2001) Chlorophylls and carotenoids—Measurement and characterisation by UV-VIS. Current Protocols in Food Analytical Chemistry (CPFA), (Supplement 1), F4.3.1–F 4.3.8. Wiley.New York. 10.1002/0471142913.faf0403s01
- Liu W, Wang Q, Hou J, Tu C, Luo Y, Christie P (2016) Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci Rep 6:26710. 10.1038/srep26710. [DOI] [PMC free article] [PubMed]
- Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LDH, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Paulsen IT. Comparative genomics of plant-associated Pseudomonas spp: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall A, Volynkin V, Saidi R, Poggioli D, Zellner H, Hatton-Ellis E, Joshi V, O’Donovan C, Orchard S, Auchincloss AH, Baratin D, Bolleman J, Coudert E, Castro E, Hulo C, Masson P, Pedruzzi I, Rivoire C, Arighi C, Wang Q, Chen C, Huang H, Garavelli J, Vinayaka CR, Yeh LS, Natale DA, Laiho K, Martin MJ, Renaux A, Pichler K. The UniProt Consortium, UniRule: a unified rule resource for automatic annotation in the UniProt Knowledgebase. Bioinformatics. 2020;36(17):4643–4648. doi: 10.1093/bioinformatics/btaa485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Joe A, Mavrodi OV, Hassan KA, Weller DM, Paulsen IT, Loper JE, Alfano JR, Thomashow LS. Structural and functional analysis of the type III secretion system from Pseudomonas fluorescens Q8r1–96. J Bacteriol. 2011;193(1):177–189. doi: 10.1128/JB.00895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena KK, Sorty AM, Bitla UM, Choudhary K, Gupta P, Pareek A, Singh DP, Prabha R, Sahu PK, Gupta VK, Singh HB, Krishanani KK, Minhas PS (2017) Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci 8:172. 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed]
- Meier-Kolthoff JP, Goker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Goker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022;50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Wideman M, Makkar N, Tran M, Isaac B, Biest N, Stonard R (1992) Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J Antibiot 45(6):914–21. 10.7164/antibiotics.45.914. [DOI] [PubMed]
- Mishra J, Fatima T, and Arora NK (2018) Role of secondary metabolites from plant growth-promoting rhizobacteria in combating salinity stress. In: Egamberdieva D, Ahmad P (eds) Plant microbiome: stress response, microorganisms for sustainability 5, Springer, Singapore, pp 127–163. 10.1007/978-981-10-5514-0_6.
- Mogrovejo DC, Perini L, Gostincar C, Sepcic K, Turk M, Ambrozic AJ, Brill FHH, Gunde-Cimerman N. Prevalence of antimicrobial resistance and hemolytic phenotypes in culturable arctic bacteria. Front Microbiol. 2020;11:570. doi: 10.3389/fmicb.2020.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadipanah F, Zamanzadeh M (2019) Bacterial mechanisms promoting the tolerance to drought stress in plants. In: Singh HB, Keswani C, Reddy MS, Sansinenea E, Garcia-Estrada C (eds) Secondary metabolites of plant growth promoting rhizomicroorganisms, Springer, Singapore, pp 185–224. 10.1007/978-981-13-5862-3_10.
- Montes C, Altimira F, Canchignia H, Castro A, Sanchez E, Miccono M, Tapia E, Sequeida VJ, Tapia P, Gonzalez C, Prieto H. A draft genome sequence of Pseudomonas veronii R4: a grapevine (Vitis vinifera L.) root-associated strain with high biocontrol potential. Stand in Genomic Sci. 2016;11:76. doi: 10.1186/s40793-016-0198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Bieber JE, Schmidt-Dannert MG, Nett RS, Peters RJ. A third class: functional gibberellin biosynthetic operon in beta-proteobacteria. Front Microbiol. 2018;9:2916. doi: 10.3389/fmicb.2018.02916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento FX, Uron P, Glick BR, Giachini A, Rossi MJ. Genomic analysis of the 1-aminocyclopropane-1-carboxylate deaminase-producing Pseudomonas thivervalensis SC5 reveals its multifaceted roles in soil and in beneficial interactions with plants. Front Microbiol. 2021;12:752288. doi: 10.3389/fmicb.2021.752288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis M, Mossialos D, Oliver SG, Amoutzias GD. Comparative analysis of the core proteomes among the Pseudomonas major evolutionary groups reveals species-specific adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity. 2020;12:289. doi: 10.3390/d12080289. [DOI] [Google Scholar]
- Nobre A, Alarico S, Fernandes Ch, Empadinhas N, Costa MS. A unique combination of genetic systems for the synthesis of trehalose in Rubrobacter xylanophilus: properties of a rare actinobacterial TreT. J Bacteriol. 2008 doi: 10.1128/JB.01055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA (2013). Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X (eds) Research in computational molecular biology. RECOMB. Lecture Notes in Computer Science 7821. Springer, Berlin, Heidelberg. 10.1007/978-3-642-37195-0_13.
- Olenska E, Malek W, Wojcik M, Swiecicka I, Thijs S, Vangronsveld J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: a methodical review. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140682. [DOI] [PubMed] [Google Scholar]
- Paez M, Martinez-Nieto P, Bernal-Castillo J. Siderophore producing Pseudomonas as pathogenic Rhisoctonia solani and Botrytis cinerea antagonists. Univ Sci. 2005;10(1):65–74. [Google Scholar]
- Parte AC, Carbasse JS, Meier-Kolthoff JP, Reimer LC, Goker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2022;70:5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth- promoting rhizobacteria. Physiol Plant. 2003;118(1):10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Poblete-Morales M, Plaza N, Almasia R, Corsini G, Silva-Moreno E. Draft genome sequence of Pseudomonas sp. strain M7D1, isolated from the rhizosphere of desert bloom plants. Microbiol Resour Announc. 2019;8:e00441–e519. doi: 10.1128/MRA.00441-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncheewin W, Diepeningen AD, Lee TAJ, Suarez-Diez M, Schaap PJ. Classification of the plant-associated lifestyle of Pseudomonas strains using genome properties and machine learning. Sci Rep. 2022;12:10857. doi: 10.1038/s41598-022-14913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA Chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4(3):118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Ramadoss D, Lakkineni VK, Bose P, Ali S, Annapurna K. Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. Springerplus. 2013;2:6. doi: 10.1186/2193-1801-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R, Glockner FO, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics btv681. 2015 doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala V, Pucci N, Loreti S. The diagnosis of plant pathogenic bacteria: a state of art. Front Biosci. 2018;10:449–460. doi: 10.2741/E832. [DOI] [PubMed] [Google Scholar]
- Sekar J, Saharan K, Raju K, Singh U, Vaiyapuri PR (2019) Consequences of bioinoculants and intercropping approach to alleviate plant drought and salinity stress for sustainable agriculture. In: Akhtar MS (ed) Salt stress, microbes, and plant interactions: mechanisms and molecular approaches, Springer, Singapore, pp 161–182. 10.1007/978-981-13-8805-7_8.
- Senthilkumar M, Amaresan N, Sankaranarayanan A. Plant-microbe interactions laboratory techniques. New York: Springer; 2021. pp. 177–181. [Google Scholar]
- Shariati V, Malboobi MA, Tabrizi Z, Tavakol E, Owlia P, Safari M (2017) Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci Rep. www.nature.com/scientificreports. [DOI] [PMC free article] [PubMed]
- Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B (2015) Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6(5):e00621–15. 10.1128/mBio.00621-15. [DOI] [PMC free article] [PubMed]
- Shelake RM, Pramanik D, Kim JY. Exploration of plant-microbe interactions for sustainable agriculture in CRISPR Era. Microorganisms. 2019;7:269. doi: 10.3390/microorganisms7080269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Hu H, Peng H, Wang W, Zhang X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genom. 2013;14:271. doi: 10.1186/1471-2164-14-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Singh RK, Guo DJ, Sharma A, Singh RN, Li DP, Malviya MK, Song XP, Lakshmanan P, Yang LT, Li YR. Whole genome analysis of sugarcane root-associated endophyte Pseudomonas aeruginosa B18—A plant growth-promoting bacterium with antagonistic potential against Sporisorium scitamineum. Front Microbiol. 2021;12:628376. doi: 10.3389/fmicb.2021.628376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Ratering S, Hain T, Fritzenwanker M, Goesmann A, Blom J, Chakraborty T, Bunk B, Sproer C , Overmann J, Schnell S (2019) Complete genome sequence of the plant growth-promoting bacterium Hartmannibacter diazotrophicus Strain E19. Int J Genomics 7586430. 10.1155/2019/7586430. [DOI] [PMC free article] [PubMed]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–24. 10.1093/nar/gkw569. PMID: 27342282. [DOI] [PMC free article] [PubMed]
- Trantas EA, Licciardello G, Almeida NF, Witek K, Strano CP, Duxbury Z, Ververidis F, Goumas DE, Jones JDG, Guttman DS, Catara V, Sarris PF. Comparative genomic analysis of multiple strains of two unusual plant pathogens: Pseudomonas corrugata and Pseudomonas mediterranea. Front Microbiol. 2015;6:811. doi: 10.3389/fmicb.2015.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesha S, Richardson PA, Kong P, Hong CX. A novel indicator plant to test the hypersensitivity of phytopathogenic bacteria. J Microbiol Methods. 2008;72:95–97. doi: 10.1016/j.mimet.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Vaishnav A, Varma A, Tuteja N, Choudhary DK (2016) PGPR-mediated amelioration of crops under salt stress. In: Choudhary DK, Varma A, Tuteja N (eds) Plant-microbe interaction: An approach to sustainable agriculture, Springer, Singapore, pp 205–226. 10.1007/978-981-10-2854-0_10.
- Xie W, Yang J, Gao S, Yao R, Wang X. The Effect and influence mechanism of soil salinity on phosphorus availability in coastal salt-affected soils. Water. 2022;14:2804. doi: 10.3390/w14182804. [DOI] [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. ANTON LEEUW INT J G. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- Zachow C, Muller H, Laireiter CM, Tilcher R, Berg G. Complete genome sequence of Pseudomonas corrugata strain RM1–1–4, a stress protecting agent from the rhizosphere of an oilseed rape bait plant. Stand. Genom. Sci. 2017;12:66. doi: 10.1186/s40793-017-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zboralski A, Biessy A, Filion M. Bridging the gap: type III secretion systems in plant-beneficial bacteria. Microorganisms. 2022;10:187. doi: 10.3390/microorganisms1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen W, Jiang Q, Fei Z, Xiao M (2020) Genome analysis of plant growth-promoting rhizobacterium Pseudomonas chlororaphis subsp. aurantiaca JD37 and insights from comparasion of genomics with three Pseudomonas strains. Microbiol Res 237:126483. 10.1016/j.micres.2020.126483. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All analyzed data of this study are presented in the paper and its supplementary data files. The whole genome shotgun project was deposited at DDBJ/ENA/GenBank under the accession number JAKNQY000000000. Additionally, further information about the bacterial assembly is accessible in the NCBI database with BioProject ID: PRJNA803818 and BioSample ID: SAMN25688536.