Summary
Background
Pseudomonas aeruginosa healthcare-associated infections are one of the top antimicrobial resistance threats world-wide. In order to analyze the current trends, we performed a Spanish nation-wide high-resolution analysis of the susceptibility profiles, the genomic epidemiology and the resistome of P. aeruginosa over a five-year time lapse.
Methods
A total of 3.180 nonduplicated P. aeruginosa clinical isolates from two Spanish nation-wide surveys performed in October 2017 and 2022 were analyzed. MICs of 13 antipseudomonals were determined by ISO-EUCAST. Multidrug resistance (MDR)/extensively drug resistance (XDR)/difficult to treat resistance (DTR)/pandrug resistance (PDR) profiles were defined following established criteria. All XDR/DTR isolates were subjected to whole genome sequencing (WGS).
Findings
A decrease in resistance to all tested antibiotics, including older and newer antimicrobials, was observed in 2022 vs 2017. Likewise, a major reduction of XDR (15.2% vs 5.9%) and DTR (4.2 vs 2.1%) profiles was evidenced, and even more patent among ICU isolates [XDR (26.0% vs 6.0%) and DTR (8.9% vs 2.6%)] (p < 0.001). The prevalence of Extended-spectrum β-lactamase/carbapenemase production was slightly lower in 2022 (2.1%. vs 3.1%, p = 0.064). However, there was a significant increase in the proportion of carbapenemase production among carbapenem-resistant strains (29.4% vs 18.1%, p = 0.0246). While ST175 was still the most frequent clone among XDR, a slight reduction in its prevalence was noted (35.9% vs 45.5%, p = 0.106) as opposed to ST235 which increased significantly (24.3% vs 12.3%, p = 0.0062).
Interpretation
While the generalized decrease in P. aeruginosa resistance, linked to a major reduction in the prevalence of XDR strains, is encouraging, the negative counterpart is the increase in the proportion of XDR strains producing carbapenemases, associated to the significant advance of the concerning world-wide disseminated hypervirulent high-risk clone ST235. Continued high-resolution surveillance, integrating phenotypic and genomic data, is necessary for understanding resistance trends and analyzing the impact of national plans on antimicrobial resistance.
Funding
MSD and the Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea—NextGenerationEU.
Keywords: Post-acute sequelae of SARS-CoV-2, PASC, COVID-19, SARS-CoV-2, Electronic health records
Research in context.
Evidence before this study
We searched PubMed, without language restrictions, for articles published before October 1st 2022 using the terms “Pseudomonas aeruginosa” and “surveillance” and “resistance” and “MDR”, “XDR” or “DTR”. We identified multiple global surveillance programs that analyzed P. aeruginosa resistance trends, including concerning MDR (multidrug resistance), XDR (extensively drug resistance), and/or DTR (difficult to treat resistance) phenotypes, and some of them used whole genome sequencing (WGS) to identify globally disseminated clones and associated resistance mechanisms. We identified also some national surveys (including Spain) that analyzed the prevalence of resistance that used as well WGS to decipher the involved mechanisms and to identify widespread MDR/XDR/DTR strains. All these studies have helped to settle the current knowledge on the global burden of P. aeruginosa antimicrobial resistance genomics and epidemiology, by integrating information from different time points and places. We did not identify however any study that analyzed at a nationwide scale how resistance phenotypes, mechanisms and epidemiology may change over a time lapse.
Added value of this study
Taking advantage of two large-scale surveys with identical methodology, we performed a Spanish nationwide high-resolution analysis of the trends in the susceptibility profiles, the genomic epidemiology and resistance mechanisms of P. aeruginosa isolates from hospital-acquired infections over a five-year time lapse. Positive findings included a sharp generalized decrease in resistance to older and newer antipseudomonals, as well as a dramatic decrease in the prevalence of XDR P. aeruginosa. The negative counterpart was a significant increase in the proportion of XDR and carbapenem resistant strains producing concerning horizontally-acquired carbapenemases, linked to a significant progression of the world-wide disseminated hypervirulent high-risk clone ST235.
Implications of all the available evidence
Continued high-resolution surveillance, integrating both, phenotypic and genomic data, is necessary for understanding resistance trends and analyzing the impact of national and global plans on antimicrobial resistance.
Introduction
Pseudomonas aeruginosa, is among the main causes of hospital-acquired and chronic infections and is associated with high antimicrobial resistance, morbidity and mortality.1 Indeed, antibiotic-resistant P. aeruginosa infections are estimated to be associated with over 300,000 annual deaths and are at the top of the WHO priority list for the need for research and development of new antibiotics.2,3 This growing threat results from the extraordinary capacity of this pathogen for developing resistance through chromosomal mutations and from the increasing prevalence of transferable resistance determinants, particularly those encoding carbapenemases or extended-spectrum β-lactamases (ESBLs).4,5 Combinations of such mechanisms lead to concerning and complex resistance profiles as defined by the European Centre for Disease Prevention Control (ECDC), [multidrug resistance (MDR), extensively drug resistance (XDR), and pandrug resistance (PDR)] or the infectious Diseases Society of America (IDSA) [difficult to treat resistance (DTR)].6,7 P. aeruginosa has a non-clonal epidemic population structure, composed of a limited number of widespread clones, which are selected from a background of a large quantity of rare and unrelated genotypes that are recombining at high frequency.8 Indeed, several surveys have provided evidence of the existence of XDR/DTR global clones, denominated high-risk clones, disseminated in hospitals worldwide.9, 10, 11 Moreover, beyond classical molecular epidemiology analysis and phenotypic assessment of resistance mechanisms, Whole Genome Sequencing (WGS) studies are providing relevant information for building up the complex and evolving resistome of MDR/XDR P. aeruginosa high-risk clones.12, 13, 14, 15
The recent introduction of newer β-lactam/β-lactamase inhibitors combinations (such as ceftolozane/tazobactam, ceftazidime/avibactam or imipenem/relebactam) has helped to mitigate, to some extent, the problem of XDR/DTR P. aeruginosa.16,17 Indeed, these agents show increased stability against the classic P. aeruginosa chromosomally-encoded β-lactam resistance mechanisms, such as the overexpression of the chromosomal β-lactamase AmpC and efflux pumps or OprD inactivation. However they are not exempt from resistance development through emerging mutational mechanisms, such as the modification of the catalytic center of AmpC or the modification of efflux pumps substrate specificity, evidenced right upon their introduction into clinical practice.18, 19, 20, 21 Likewise, they are not active against most potent transferable carbapenemases, particularly the metallo-β-lactamases (MBLs), and thus their use may lead to the selection of such concerning mechanisms.22 Therefore, emerging resistance to older and newer antibiotics is of particular concern and should be therefore closely monitored. Taking advantage of two large-scale (over 3.000 isolates from 66 hospitals) surveys, we performed a Spanish nation-wide high-resolution analysis of the trends in the susceptibility profiles, the molecular epidemiology and the resistome of P. aeruginosa over a five-year time lapse.
Methods
P. aeruginosa strains and susceptibility testing
A total of 1735 P. aeruginosa isolates were studied in this second Spanish nation-wide survey performed under the auspices of the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC) ensuring representation of all 17 Spanish regions. This collection included up to 30 consecutive healthcare-associated nonduplicated (one per patient) P. aeruginosa clinical isolates collected during October 2022 from each of the 66 participating hospitals (Figure S1). Species identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Bruker-Daltonics). Sample types [respiratory, urinary, bloodstream, skin soft tissue and osteoarticular (SST), others] and sources [Intensive care unit (ICU), medical ward, surgical ward, emergency room, others] were recorded for each isolate. MICs of piperacillin/tazobactam (4/4–256/4 mg/L), ceftazidime (1–64 mg/L), cefepime (1–64 mg/L), ceftolozane/tazobactam (0.5/4–32/4 mg/L), ceftazidime/avibactam (0.5/4–32/4 mg/L), aztreonam (2–128 mg/L), imipenem (0.5–64 mg/L), imipenem/relebactam (0.12/4–64/4 mg/L), meropenem (0.5–64 mg/L), ciprofloxacin (0.12–16 mg/L), tobramycin (0.25–32 mg/L), amikacin (2–128 mg/L), and colistin (0.5–16 mg/L) were determined by broth microdilution using Sensititre™ panels (Plate Code:FRCNRP2, Thermo Fisher Diagnostics, S.LU), except for imipenem/relebactam for which in house microdilution was performed according to ISO-EUCAST guidelines (http://www.eucast.org). EUCAST 2023 (v13.0) clinical breakpoints were used for interpretation of SIR categories. Results were compared with those previously obtained in the first Spanish nation-wide survey, which included 1445 isolates collected from 51 hospitals (all of them participating in the second survey) five years earlier (October 2017) with exactly the same criteria (up to 30 consecutive healthcare-associated nonduplicated isolates per hospital, same sample types and sources classification).15,23 Likewise, the same panel of antibiotics and concentrations were tested by the same reference laboratory (Microbiology Department, Hospital Son Espases, Palma de Mallorca) using the same protocols in both surveys. Moreover, to ensure interstudy reproducibility of susceptibility results, reference strains ATCC27853 and PAO1 were included as controls. Each of the reference strains was tested in at least 6 independent occasions during each of the studies, yielding no significant MIC differences between both periods, as shown in Table S1. To confirm reproducibility as well for testing resistant strains, 6 randomly selected XDR strains from the first (2017) study were retested in the second (2022) study yielding no significant MIC differences (Table S1). Finally, to compare SIR data for both studies, MICs of the first study were reinterpreted according to current EUCAST 2023 (v 13.0) clinical breakpoints.
According to established recommendations by ECDC,6 the MDR profile was defined as resistance to at least one agent in at least three of seven antibiotic classes including antipseudomonal penicillins + β-lactamase inhibitor combinations (piperacillin/tazobactam), antipseudomonal cephalosporins (ceftazidime and cefepime), monobactams (aztreonam), antipseudomonal carbapenems (imipenem and meropenem), fluoroquinolones (ciprofloxacin), aminoglycosides (tobramycin and amikacin), and polymyxins (colistin) and the XDR profile as resistance to at least one agent in all but one or two antibiotic classes. Likewise, PDR profile was defined as resistance to all agents in the seven antibiotic classes. The eight category (fosfonic acids, fosfomycin) included in the ECDC recommendations was not considered given the lack of current EUCAST clinical breakpoints. On the other hand, the DTR (Difficult to Treat Resistance) profile was defined according to IDSA recommendations as resistance to all first line (classical) agents: antipseudomonal penicillins + β-lactamase inhibitor combinations (piperacillin/tazobactam), antipseudomonal cephalosporins (ceftazidime and cefepime), monobactams (aztreonam), antipseudomonal carbapenems (imipenem and meropenem), and fluoroquinolones (ciprofloxacin).7 Thus, all DTR isolates meet the XDR criteria, since they are resistant to at least five of seven classes.
WGS
All XDR (and consequently all DTR) isolates, as well as those resistant to the any of the three newer β-lactam/β-lactamase inhibitor combinations herein evaluated (ceftolozane/tazobactam, ceftazidime/avibactam or imipenem/relebactam), and/or producing an acquired ESBL or carbapenemase from the 2022 study were fully sequenced (n = 138). Additionally, sequences from the 185 XDR isolates available from the 2017 study were included for comparison. It should be noted that a number of these isolates (n = 28) are now reclassified as MDR when applying current EUCAST breakpoints. Strains from the 2022 study that would have been classified as XDR in the 2017, but only as MDR with the current criteria (n = 19) were also sequenced to have the complete picture. Table S2 collects the information from the 342 strains sequenced from 2017 to 2022 studies.
Library preparation
Genomic DNA was obtained with a commercially available extraction kit (High Pure PCR Template Preparation Kit, Roche Diagnostics). Indexed paired-end libraries were generated by using the Illumina DNA Prep library preparation kit (Illumina Inc, USA) and then sequenced on either, an Illumina MiSeq® benchtop sequencer with MiSeq reagent kit v3 (600 cycles) or in an Illumina Novaseq 6000 system with NovaSeq 6000 SP Reagent Kit v1.5 (300 cycles).
Variant calling
Previously defined and validated protocols were used with slight modifications.24 The reads for each isolate were mapped against the genome of the P. aeruginosa reference strain PAO1 (RefSeq accession number NC_002516.2) using Bowtie 2 software, v2.2.6 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml).25 Pileups and raw files of the mapped reads were obtained by using SAMtools, v0.1.16 (https://sourceforge.net/projects/samtools/files/samtools/),26 and PicardTools, v1.140 (https://github.com/broadinstitute/picard). Read alignments surrounding all putative indels were realigned using the Genome Analysis Toolkit (GATK), v3.4-46 (https://www.broadinstitute.org/gatk/).27 The list of SNPs was compiled from the raw files that met the following criteria: a quality score of ≥50, a root mean square (RMS) mapping quality of ≥25 and a coverage depth of ≥3 reads. MicroIndels were extracted from the totalpileup files by use of the following criteria: a quality score of ≥500, a RMS mapping quality of ≥25 and support from at least one-fifth of the covering reads. SNPs and indels for each isolate were annotated by using SnpEff software v4.3 ().28 Finally, large chromosomal deletions were analyzed with Seqmonk v1.47.2 (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and R v4.2.3 within the RStudio v0.99.896 platform.29
De novo assembly
Reads were de novo assembled using SPAdes v3.15 with default options. De novo assemblies were used to define the sequence type (ST) by using MLST v2.23.0 according to PubMLST typing schemes (https://pubmlst.org/) developed by Keith Jolley.30 Additionally, assembled reads were used to study the structural integrity of the OprD porin. As different sequence variants of OprD have been described,31 the oprD gene was first classified according to their similarity to PAO1, LESB58, UCBP-PA14, MTB-1, FRD1 or F23197 reference sequences.
Analysis of the mutational resistome
A total of 48 genes involved in mutational resistance were selected according to findings of previous studies and analyzed.13,32,33 The complete list of the genes studied, indicating their role in antibiotic resistance, is shown in Table S3. Nucleotide sequence variants located within these genes were filtered by using a list of natural polymorphisms that have been previously defined by our group.34
Phylogenic analysis
With the aim to study the diversity of XDR ST175 and ST235, core genome phylogenetic reconstruction was performed with Parsnp from the Harvest Suite package v1.2 with default parameters,35 forcing the inclusion of all genomes (-c) and randomly selecting the reference genome (-r!). Additionally, a minimum-spanning tree (MST) for XDR and reference strains PAO1, PA14 and PA7 was inferred by using GrapeTree36 on the basis of cgMLST scheme for P. aeruginosa37 created using the open source ChewBBACCA algorithm.38
Acquired resistance determinants
To identify possible horizontally-acquired antimicrobial resistance genes, we used the online tool ResFinder v3.1.0. with default options (https://cge.cbs.dtu.dk//services/ResFinder/).39 Genomic information was complemented as needed by phenotypic (such as cloxacillin and EDTA tests) and molecular (PCR + sanger sequencing) methods for the detection of acquired β-lactamases.15
Statistical analysis
The data set analysed included a total of 3.180 nonduplicated P. aeruginosa clinical isolates from two Spanish nation-wide surveys performed in October 2017 (1.445 isolates) and 2022 (1.735 isolates), as well as the 342 genomes sequenced from 2017 (n = 185) and 2022 (n = 138) studies. The following variables were considered: Prevalence of resistance to each of 13 antipseudomonals agents for all isolates and for ICU isolates. Prevalence of MDR, XDR and DTR resistance profiles according to sample type [respiratory urinary, bloodstream, skin soft tissue and osteoarticular, others], hospital wards [ICU, medical, surgical, emergency room, others], production of ESBLs and carbapenemases and clonal types. We investigated how the different variables varied between the two periods (2017 and 2022) using Chi-square test. The Wilcoxon signed-rank test was used to compare the proportion of hospitals showing an increase or decrease of resistance for the different antibiotics tested in 2022 vs 2017. In all cases, a p value of <0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism 5 or IBM SPSS Statistics v22 software.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Data availability
Genomic sequences from the 2022 and 2017 studies have been deposited in the European Nucleotide Archive, under project numbers PRJEB61879 and PRJEB31047, respectively.
Results
Antimicrobial susceptibility and resistance profiles
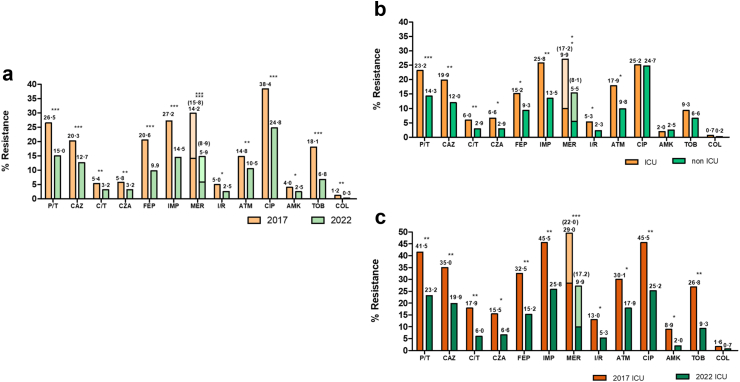
A comparative analysis of resistance rates for a panel of 13 antipseudomonal agents is shown in Fig. 1a. A statistically significant decrease in resistance for all antibiotics tested was documented in 2022 compared to 2017. In both periods, the antibiotic showing lowest resistance rates was colistin followed by amikacin, imipenem/relebactam, ceftolozane/tazobactam and ceftazidime/avibactam. In contrast, the highest resistance rates were documented for ciprofloxacin, followed by imipenem and piperacillin/tazobactam. Resistance rates were higher for all agents in the ICU (Fig. 1b), except for ciprofloxacin and amikacin, but again they were considerably lower in 2022 compared to 2017 (Fig. 1c).
Fig. 1.
a. Comparative analysis of the prevalence of resistance to 13 antipseudomonal agents in 2017 and 2022 Spanish nation-wide studies. b. Comparative analysis of the prevalence of resistance among ICU and nonICU isolates from the 2022 study. c. Comparative analysis of the prevalence of resistance in ICU isolates from 2017 to 2022 studies. In the case of meropenem, “I” (susceptible increased exposure) isolates were also represented (in a lighter tone and with numbers between brackets) in addition to resistant isolates for reference since these isolates show low level resistance. Statistical significance (Chi-square, X2) indicated (∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05).
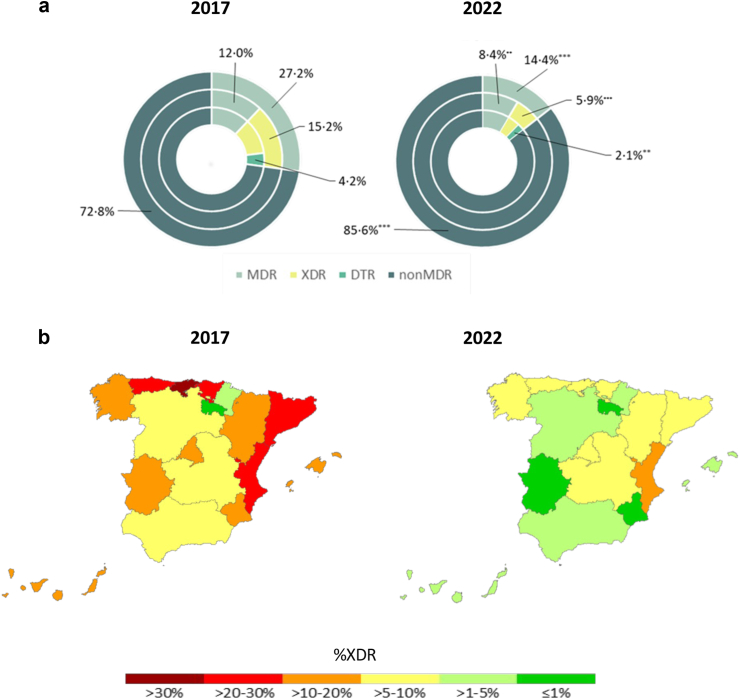
The distribution of resistance profiles is shown in Fig. 2a, revealing a major decrease as well in 2022 of MDR (27.2% vs 14.8%), XDR (15.2% vs 5.9%) and DTR (4.2% vs 2.1%) profiles. PDR profiles were not detected in either period. The generalized decrease in the prevalence of XDR profiles in nearly all Spanish regions is evidenced in Fig. 2b.
Fig. 2.
a. Comparative analysis of the distribution of MDR XDR and DTR profiles in 2017 and 2022 Spanish nation-wide studies. As described in the material and methods section, XDR isolates are a fraction of MDR isolates and DTR isolates a fraction of XDR isolates. Statistical significance (Chi-square, X2) indicated (∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05). b. Comparative analysis of the distribution of XDR profiles in the different Spanish region in 2017 and 2022 studies.
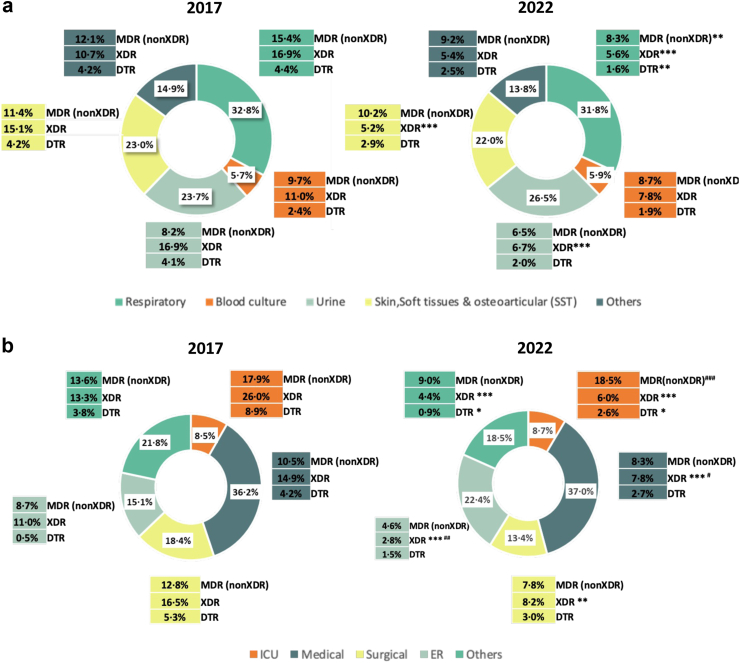
As shown in Fig. 3a the distribution of clinical sample types was nearly identical in both studied periods. Interestingly, while there was a generalized reduction in the prevalence of MDR/XDR/DTR phenotypes, the decrease was highest for respiratory samples and lowest for blood cultures. Likewise, the distribution of isolates according to the hospital ward of origin was very similar for both studies (Fig. 3b). Remarkably the highest decrease in the prevalence of XDR (26.0% vs 6.0%) and DTR (8.9% vs 2.6%) in 2022 vs 2017 was documented for ICU isolates. Moreover, contrasting dramatically with 2017 results, XDR and DTR profiles were not more frequent in the ICU than in other hospital wards in 2022. The prevalence of MDR (nonXDR) profiles were, however, very similar for ICU isolates in both study periods and much higher than those documented in other hospital wards.
Fig. 3.
Distribution of isolates and resistance profiles according to sample type (a) and hospital wards (b) in 2017 and 2022 Spanish nation-wide studies. ER, emergency room. Statistical significance (Chi-square, X2) comparing both studies (∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05) and between hospital wards in 2022 (###p < 0.0001; ##p < 0.01; #p < 0.05) are indicated.
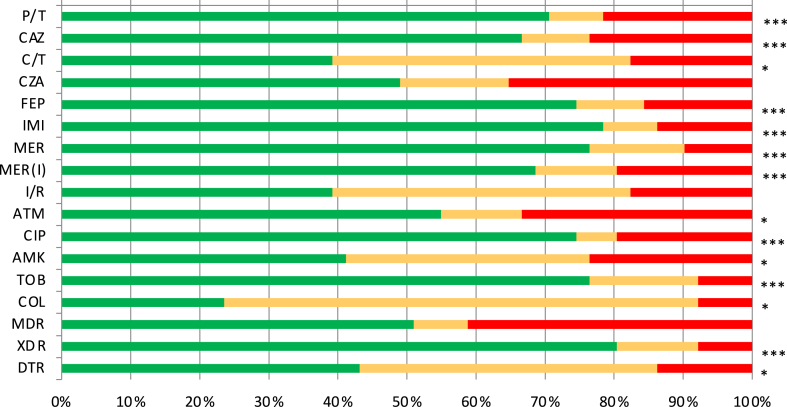
A paired analysis of the 51 hospitals participating in both studies further emphasizes the generalized decrease in resistance (Fig. 4). Particularly noteworthy, the percentage of XDR decreased in over 80% of the participating hospitals, while it increased in less than 10% of them. Among individual agents, the prevalence of resistance to imipenem, meropenem, tobramycin, ciprofloxacin, cefepime and piperacillin/tazobactam was decreased in >70% of the participating hospitals.
Fig. 4.
Percentage of hospitals showing lower (green), equal (orange) or higher (red) resistance rates in 2022 than in 2017. Only the 51 hospitals participating in both studies were included in the analysis. Statistical significance (Chi-square, X2) indicated (∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05).
Horizontally-acquired β-lactamases and distribution of high-risk clones
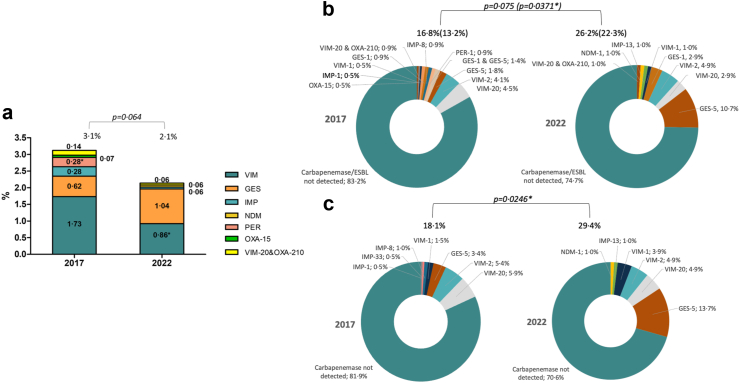
The nature and prevalence of horizontally-acquired ESBLs and carbapenemases is shown in Fig. 5a. The global prevalence of acquired enzymes tended to decrease from 3.1% in 2017 to 2.1% in 2022, but statistical significance was not reached (p = 0.064). Regarding the distribution of enzymes, a slight decrease in the prevalence of VIM MBLs was documented in 2022, while GES enzymes slightly increased. However, the proportion of carbapenemase producing isolates significantly increased among XDR isolates (13.2% vs 22.3%) (Fig. 5b) and among meropenem resistant isolates (18.1% vs 29.4%) (Fig. 5c) in 2022 compared to 2017.
Fig. 5.
a. Prevalence of ESBLs and carbapenemases for the complete collection of P. aeruginosa isolates from 2017 to 2022 studies. b. Prevalence of ESBLs and carbapenemases (carbapenemases specifically indicated in parenthesis) among XDR isolates. c. Prevalence of carbapenemases among meropenem resistant isolates. Statistical significance (Chi-square, X2) indicated (∗p < 0.05).
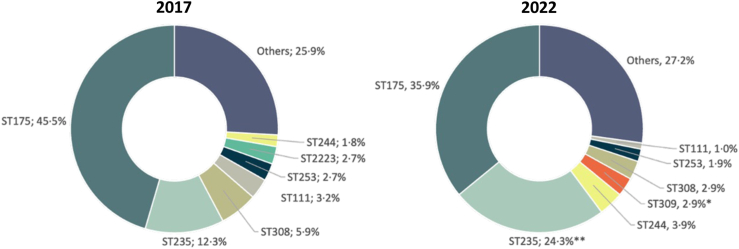
The distribution of clonal types among XDR isolates is shown in Fig. 6. While ST175 was still the most frequent clone in 2022 a slight reduction (not statistically significant) in its prevalence was noted when comparing 2017 (45.5%) with 2022 (35.9%). However, the most remarkable finding was the statistically significant increase in the prevalence of ST235 in 2022 (12.3% vs 24.3%).
Fig. 6.
Distribution of sequence types among XDR P. aeruginosa isolates recovered from the 2017 and 2022 studies. Statistical significance (Chi-square, X2) for the 2017 vs 2022 comparison indicated (∗∗p < 0.01; ∗p < 0.05). STs accounting for ≥2% of the XDR isolates in any of the two studies are shown individually, while all those representing <2% of isolates are included in “Others”.
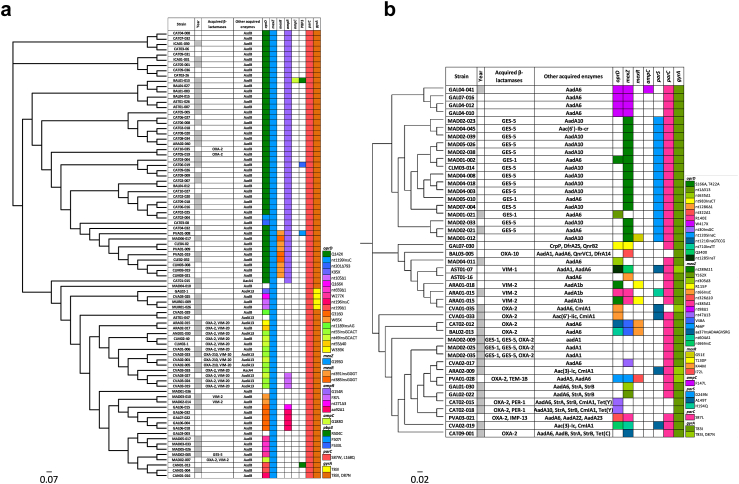
Phylogenetic reconstructions and association with horizontally acquired and mutational resistance are represented for XDR ST175 and ST235 clones in Fig. 7. As previously documented, XDR ST175 isolates were characterized by a strong mutational resistance genomic signature which included nearly uniform QRDR and mexZ mutations (Fig. 7a). Additionally, the most frequent cluster of ST175 isolates both in 2017 and 2022 showed the characteristic β-lactam mutational resistome described in isolates from nearly 15 years ago, including specific OprD (Q142∗) and AmpR (G154R) mutations.13 A second ST175 lineage, already documented in 2017, produced the VIM-20 variant but seemed not to have expanded in 2022. In sharp contrast, as shown in Fig. 7b, ST235 demonstrated an overwhelming association with the production of horizontally acquired resistance determinants, particularly noteworthy ESBLs and carbapenemases, and presented a more heterogeneous mutational resistome. Clonal expansion of a ST235 epidemic lineage in multiple hospitals from the Madrid region was particularly evident in 2022 and was associated with GES enzymes production and specific mexZ and parS mutations.
Fig. 7.
Core genome phylogenetic reconstruction of the 2017 and 2022 XDR ST175 (a) and ST235 (b)P. aeruginosa isolates. Year column labeled in gray and white corresponds with 2017 and 2022 isolates, respectively. Following columns correspond to β-lactamases and other acquired enzymes. Code description of changes in most commonly mutated genes (oprD, mexZ, mexR, ampR, ampC, parS, PBP3, parC and gyrA) is represented on hand-side. Each colour of each column corresponds to a single mutation.
Figure S2 shows the cgMLST analysis of the complete collection of XDR isolates from 2017 to 2022, further evidencing interregional transmission and persistence of several XDR clones, most noteworthy, but not only, ST175 and ST235. Increased association with acquired carbapenemases in 2022 was also documented for other less frequent high-risk clones. For example, ST308 was less frequent in 2022, but accounted for the single case of NDM production in the complete collection. Likewise, while the overall prevalence of ST253 had slightly decreased 2022, an increased association with MBL-production was observed including a clonal expansion of a VIM-1 lineage that was detected in a single hospital from Catalonia in 2017 but in three (including the first) from the same region in 2022 (Table S2). Moreover, an additional VIM-2-producing ST253 lineage was first detected in a different region in 2022. Globally expanding KPC enzymes were however not detected in any of the two periods. Finally, while the investigation of the transferable elements (plasmids and transposons) harboring the carbapenemase genes was beyond the scope of this work, the BLAST analysis of their genetic context confirmed that they were in all cases linked to class 1 integrons.
Mechanisms of resistance to newerβ-lactam β-lactamase inhibitors combinations
A specific analysis of β-lactam resistance mechanisms produced by strains resistant to the newer β-lactam/β-lactamase inhibitor combinations was performed for the 2022 study, and results are shown in Table 1. Production of horizontally acquired carbepenemases was most frequent among imipenem/relebactam resistant strains (74.4%), but less frequent among ceftazidime/avibactam resistant strains (25.0%). Particularly noteworthy, none of the ceftazidime/avibactam resistant strains produced a GES-5, in contrast to 33.3% of imipenem/relebactam resistant strains. Conversely, susceptibility rates among the 16 GES-5 producers was 93.8% for ceftazidime/avibactam, 68.8% for ceftolozane/tazobactam and 6.3% for imipenem/relebactam. As expected, susceptibility rates for MBL producers were below 10% for the three combinations. Regarding the mutational resistome, AmpC Ω-loop mutations were exclusively seen in ceftolozane/tazobactam (15.1%) and ceftazidime/avibactam (12.5%) resistant isolates, but not in those resistant to imipenem/relebactam. Moreover, ceftazidime/avibactam resistance was strongly associated with mutations in regulators of the expression of the efflux pump MexAB-OprM (55.4%).
Table 1.
Mechanisms detected among ceftolozane/tazobactam, ceftazidime/avibactam or imipenem/relebactam resistant strains from the 2022 study.
| Resistance mechanism/mutations | Ceftolozane/tazobactam resistant, n = 53 (%) | Ceftazidime/avibactam resistant, n = 56 (%) | Imipenem/relebactam resistant, n = 39 (%) |
|---|---|---|---|
| ESBL/Carbapenemases | 29 (54.7) | 16 (28.6) | 30 (76.9) |
| ESBL GES-1 | 4 (7.5) | 2 (3.6) | 1 (2.6) |
| Carbapenemases | 25 (47.2) | 14 (25.0) | 29 (74.4) |
| GES-5 | 9 (17.0) | 0 | 13 (33.3) |
| MBLs | 16 (30.2) | 14 (25.0) | 16 (41.0) |
| Narrow spectrum OXAs (only) | 3 (5.7) | 2 (3.6) | 0 |
| oprD | 23 (43.4) | 32 (57.1) | 18 (46.2) |
| ampC regulators (dacB, ampD, ampR) | 29 (54.7) | 29 (51.8) | 19 (48.7) |
| ampC (Ω-loop) | 8 (15.1) | 7 (12.5) | 0 |
| mexAB-OprM regulators (mexR, nalC, nalD) | 16 (30.2) | 31 (55.4) | 13 (33.3) |
| ftsI (PBP3) | 1 (1.9) | 4 (7.1) | 2 (5.1) |
| galU | 1 (1.9) | 3 (5.4) | 2 (5.1) |
Discussion
The May 2015 World Health Assembly (WHA) adopted a global action plan on antimicrobial resistance, after reaching the conclusion that it “threatens the very core of modern medicine and the sustainability of an effective, global public health response to the enduring threat from infectious diseases”. The five main objectives established included to (i) improve awareness and understanding of antimicrobial resistance (ii) to strengthen the knowledge through surveillance and research (iii) to reduce the incidence through prevention measures (iv) to optimize the use of antibiotics and (v) to develop the economic case for sustainable investment in new medicines, diagnostic tools, vaccines and other interventions. Some years before (November 17th 2011), the EU commission requested member states to develop national action plans on antimicrobial resistance, and in the case of Spain the first version was approved in 2014 with objectives similar to those adopted in the WHA (https://resistenciaantibioticos.es/es). Moreover, in 2018 the WHO published a priority list for the need for research and development of new antibiotics, in which carbapenem resistant P. aeruginosa, along with Enterobacterales and Acinetobacter baumannii were in top (critical) position.3
Within this scenario, in 2017 we performed a first nation-wide survey of P. aeruginosa antimicrobial susceptibility, resistance mechanisms and molecular epidemiology using whole genome sequencing tools.15 Overall resistance was found to be high, exceeding 20% for each available antipseudomonal except for toxic polymyxins and the newer β-lactam β-lactamase inhibitor combinations ceftolozane/tazobactam and ceftazidime/avibactam that were being introduced into clinical practice at that time. Moreover, MDR profiles were documented in nearly 30% of the isolates and XDR patterns in over 15%. The molecular epidemiology analysis performed revealed that close to half of the XDR isolates belonged to a single clone, ST175, that was associated with a specific resistome signature and that had been already detected for over one decade in Spanish hospitals, but that was quite uncommon in other countries besides France.13,40 Fortunately, the prevalence of the most concerning resistance mechanisms, inactivating even the newer β-lactam combinations, the horizontally-acquired carbapenemases, was relatively low even among XDR strains.
In this work, we decided to repeat the survey under the same conditions and using the same methodological approach in order to determine for the first time how the antimicrobial susceptibility, the molecular epidemiology and the resistance genomics had evolved at a nation-wide level during a five-year period. To our initial surprise we documented a major generalized reduction of antimicrobial resistance, affecting nearly all older as well as newer antipseudomonal agents. Particularly astonishing was the significant reduction of the prevalence of XDR strains from over 15% to below 6%. The reduction was patent in all regions and was even more evident in the ICUs with XDR profiles decreasing from 26% to 6%. A significant reduction of DTR profiles was as well documented in 2022, but should be noted that resistance to fluoroquinoles was the highest of all tested agents in both periods, supporting the concern on the use of these antibiotics as first line agents communicated by the European Medicines Agency (EUPAS37856 report).
While the factors underlying such a dramatic reduction are likely multifactorial and complex, the efforts of the Spanish national plan of antibiotic resistance (PRAN) should be acknowledged, including the implementation of country-wide infection control initiatives such as the Zero resistance in ICUs and antimicrobial stewardship programs.41,42 Another potential factor to consider is the role of the newer β-lactam β-lactamase inhibitor combinations that were active against the most prevalent XDR strains (ST175) and have been the first choice treatment for such infections in the last five years.43 The COVID-19 pandemic and associated measures, even if already highly relaxed by the last trimester of 2022, could have had an impact as well on resistance transmission.44
However, not all are positive findings in our survey, and at least two highly concerning and related facts need to be considered. The first is a significant increase in the proportion of carbapenemase-producing strains among XDR and carbapenem resistant isolates. The second is the documentation of a major progression of the word-wide disseminated high-risk clone ST235. Indeed, this concerning clone is strongly associated with epidemic settings and the acquisition of horizontally-acquired resistance determinants in hospitals world-wide, playing a determinant role in the global variation of the prevalence of carbapenemase-producing strains.45, 46, 47 Moreover, in addition to its strong association with epidemic dissemination and horizontally-acquired resistance, ST235 is associated with a higher virulence, due to the production of the ExoU toxin, unlike other MDR/XDR clones such as ST175.48, 49, 50 Therefore, findings of this study point towards a growing role of hypervirulent carbapenemase-producing ST235, and thus suggest that this clone should be subjected to an specific surveyance and that efforts should be made for the implementation of rapid diagnostic techniques for such infections in the national health system. Moreover, from the antimicrobial development perspective, efforts should likely be directed towards alternatives providing an optimal coverage of carbapenemase-producing P. aeruginosa.
The strengths of this work include the high number of hospitals participating in both surveys performed under the same experimental conditions and methodological approaches, providing robust data on how the antimicrobial susceptibility, the molecular epidemiology and the resistance genomics had evolved at a nation-wide level during a five-year period. However, the study has also some relevant limitations. First, the clinical characteristic of the P. aeruginosa infections were not analyzed, including risk factors, source of bacteremia, management and outcomes, and therefore conclusions related to these relevant aspects cannot be reached. Second, while the study revealed a major change in the epidemiology of P. aeruginosa antibiotic resistance at a nation-wide level, it was not designed to decipher the underlying causes of such findings that need to be analyzed in subsequent studies. Likewise, while the investigation of the transferable elements (plasmids and transposons) harboring the carbapenemase genes was beyond the scope of this work, it will be useful to characterize them in future studies to understand their potential dissemination across strains. Moreover, it will be of future interest to scale the findings from this single nation initiative to the European and/or global levels. Moreover, although the study includes the analysis of the resistance phenotypes and genotypes for the most relevant classical antipseudomonals, and the three most relevant newer β-lactam β-lactamase-inhibitors combinations already approved, the continuous introduction of new players such as the recently commercialized siderophore-cephalosporin cefiderocol or next generation of β-lactamase inhibitors under investigation, including those able to inhibit PBP2 (such as zidebactam) or MBLs (such as taniborbactam) need to be added in future surveillance initiatives. Finally, given the large number of statistical tests performed, some false positive results could have occurred with the 0.05 threshold, but the obtained p values, below 0.01 in most cases, support their true significance.
In summary, taking advantage of two large-scale surveys, we performed a Spanish nation-wide high resolution analysis of the trends in the susceptibility profiles, the molecular epidemiology and the resistome of P. aeruginosa over a five-year time lapse. Positive findings included a sharp generalized decrease in resistance to older and newer antipseudomonals, as well as a dramatic decrease in the prevalence of XDR P. aeruginosa. The negative counterpart was a significant increase in the proportion of XDR and carbapenem resistant strains producing concerning horizontally-acquired carbapenemases, linked to a significant progression of the major high-risk clone ST235. Continued high resolution surveillance, integrating phenotypic and genomic data, is necessary for understanding resistance trends and analyzing the impact of National plans on antimicrobial resistance.
Contributors
Members of GEMARA-SEIMC/CIBERINFEC Study Group carried out sample collection. SFMA, FMA, conducted laboratory assays with supervision and support from ZL and OA. GFMA, TB sequenced and analyzed the genomic data with supervision and support from LCC and OA. OA is the principal investigator and ASJ, MML, CR, LN, OIJ with OA were coordinating team members. All authors contributed important intellectual content during manuscript drafting or revision. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Data sharing statement
Genomic sequences from the 2022 and 2017 studies have been deposited in the European Nucleotide Archive, under project numbers PRJEB61879 and PRJEB31047, respectively.
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
This work was partially financed by a Grant from MSD to AO. The funders had no role in design, execution, analysis or reporting of the research. AO has participated in educational programs organized by MSD, Pfizer and Shionogi and had conducted research studies financed by MSD and Shionogi. RC has participated in educational programs organized by Menarini, MSD, Pfizer, Shionogi and had conducted research studies financed by MSD and Shionogi. NL has participated in educational programs organized by Menarini, MSD, Pfizer and Shionogi. LM has participated in educational programs organized by MSD, Pfizer and Shionogi and had conducted research studies financed by Janssen, MSD, Pfizer and Shionogi.
Acknowledgements
This study was made under the auspices of the Study group of mechanisms of antimicrobial action and resistance (GEMARA) from the Spanish society of clinical microbiology and infectious diseases (SEIMC) and the area of infectious diseases (CIBERINFEC) from the centers for network biomedical research (CIBER) of the Spanish national institute of health ISCIII. We are thankful to Dr Aina Millán from IdISBa for statistical support.
Funding: This work was supported by MSD and by the Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea—NextGenerationEU through grants PI21/00017 and Personalized and precision medicine grant (MePRAM Project, PMP22/00092).
Members of GEMARA-SEIMC/CIBERINFEC Pseudomonas study Group
Fátima Galán Sánchez (H. Universitario Puerta del Mar); Irene Gracia-Ahufinger y Luis Martínez Martínez (H. Universitario Reina Sofía); Carmen Liébana-Martos y Carolina Roldán (Complejo Hospitalario Ciudad de Jaén); Juan Manuel Sánchez-Calvo (H. Universitario de Jerez); Encarnación Clavijo y Laura Mora-Navas (H.Virgen de la Victoria); Javier Aznar, José Antonio Lepe y Angel Rodríguez-Villodres (Hospital universitario Virgen del Rocío); Esther Recacha (H. Virgen de la Macarena); Francisco Javier Casas-Círia y Carmen Martínez-Rubio (H. Universitario Puerto Real); Marco Antonio Sempere-Alcocer (HC Marbella Internacional); Lina Martin-Hita (H. Virgen de las Nieves); Cristina Seral (H. Clínico Universitario Lozano Blesa); Ana Isabel López-Calleja (H. Universitario Miguel Servet); Carmen Aspiroz (H. Royo Villanova); Marisa Monforte (H. Nuestra Señora de Gracia); Pedro de la Iglesia-Martínez (H. de Cabueñes); Gemma Jimenez-Guerra (H. Can Misses); Elena Riera-Pérez (H de Manacor); Carmen Collado y Carmen Gallegos (H. Son Llàtzer); Xavier Mulet, Almudena Fernández-Muñoz, Miquel Ángel Sastre-Femenía, María Antonia Gomis-Font, Laura Zamorano y Antonio Oliver (H. Son Espases); María Siller-Ruiz y Jorge Calvo (H. Universitario Marqués de Valdecilla); Dolores Quesada y Jun Hao Wang (H. Germans Tries i Pujol); Cristina Pitart y Francesc Marco (H Clínic de Barcelona); Nuria Prim, Juan Pablo Horcajada y Eduardo Padilla (H. del Mar); Ester del Barrio-Tofiño, Belen Viñado-Pérez y Nieves Larrosa (H. Vall d’Hebron); Fe Tubau (H. Universitario Bellvitge); Silvia Capilla y Antonio Casabella (Consorci Parc Taulí); Mar Olga Pérez-Moreno (H. Tortosa Verge de la Cinta); Emma Padilla y Mónica Ballestero (CatLab(Mutua Terrassa, H Terrassa y H de Martorell); Alba Rivera y Ferrán Navarro (H. de la Santa Creu i Sant Pau); Frederic Gómez-Bertomeu, Sergio Pardo-Granell, Ester Picó-Plana, Dolores Guerrero, Carolina Sarvisé-Buil (Hospital Universitari de Tarragona Joan XXIII); Alba Belles-Belles (H. Arnau de Vilanova de Lleida); Marta Fernández-Esgueva (H. Santos Reyes); María del Pilar Ortega-Lafont (Complejo Asistencial de Burgos); Inmaculada García (Complejo Asistencial Hospitalario de Salamanca); Noelia Arenal-Andrés, Susana Hernando-Real, Rosario Ibáñez, Jesús Martínez y Federico Becerra (H. General de Segovia); Carmen Aldea- Mansilla y Asmaa Alaoui-Sosse (Complejo Asistencial de Soria); José Carlos Gonzalez, Julia Guzman-Puche y Miguel Ángel Blázquez-Andrada (H. General de Ciudad Real); Nora Mariela Martínez-Ramírez (H.de Guadalajara); Alicia Beteta (H. General Nuestra Señora del Prado); Bárbara Gomila-Sard (H. General Universitario de Castellón); Salvador Giner Almaraz (H Universitario y Politécnico La Fe); Eugenio Garduño (Complejo Hospitalario de Badajoz); Pedro Miguel Juiz-González (H. Arquitecto Marcide); Jorge Arca-Suárez (Complejo Hospitalario A Coruña); Javier Alba, Pilar Alonso y Ana Isabel Rodríguez (H. Univeristario Lucus Augusti); María Isabel Paz-Vidal (Complexo Hospitalario Universitario de Orense); Marta García-Campello, Pablo Camacho y María de los Ángeles Pallarés (Complejo Hospitalario Pontevedra); María Luisa Pérez del Molino, Amparo Coira y Gema Barbeito (Complejo Hospitalario de Santiago); Anniris Rincón y Francisco José Vasallo-Vidal (H Álvaro Cunqueiro); Laura Alonso-Acero, Laura Iglesias-Llorente, Ana Bordes Benites y Laura Florén-Zabala (H. Dr Negrín); Jose Manuel Azcona, Carla Andrea Alonso y Yolanda Sáenz (H. San Pedro); Marta Lamata-Subero (H. Calahorra); David Molina y Ana González-Torralba (H. Getafe); Jennifer Villa y Esther Viedma (H. 12 de Octubre); Emilia Cercenado (H. Gregorio Marañón); Teresa Alarcón, Paula Vargas y María Diez (Hospital La Princesa); Rafael Cantón y Patricia Ruiz (H. Ramón y Cajal); María Isabel Sánchez-Romero (H. Puerta de Hierro); Felipe Pérez-García (H. Universitario Príncipe de Asturias); Genoveva Yagüe-Guirao (H. Virgen de la Arrixaca); Amaia Concepción Oteiza y José Leiva (Clínica Universidad de Navarra); María Eugenia Portillo (H. Universitario de Navarra); Andrés Canut-Blasco (H. Universitario de Álava); Matxalen Vidal (H. Universitario Basurto); Iker Alonso, Maider Zuriarrain y José Luis Barrios-Andrés (H. Universitario de Cruces).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanepe.2023.100736.
Contributor Information
Antonio Oliver, Email: antonio.oliver@ssib.es.
GEMARA-SEIMC/CIBERINFEC Pseudomonas study Group:
Fátima Galán-Sánchez, Irene Gracia-Ahufinger, Luis Martínez-Martínez, Carmen Liébana-Martos, Carolina Roldán, Juan Manuel Sánchez-Calvo, Encarnación Clavijo, Laura Mora-Navas, Javier Aznar, José Antonio Lepe, Ángel Rodríguez-Villodres, Esther Recacha, Francisco Javier Casas-Círia, Carmen Martínez-Rubio, Marco Antonio Sempere-Alcocer, Lina Martín-Hita, Cristina Seral, Ana Isabel López-Calleja, Carmen Aspiroz, Marisa Monforte, Pedro de la Iglesia-Martínez, Gemma Jimenez-Guerra, Elena Riera-Pérez, Carmen Collado, Carmen Gallegos, Xavier Mulet, Almudena Fernández-Muñoz, Miquel Àngel Sastre-Femenía, María Antonia Gomis-Font, Laura Zamorano, Antonio Oliver, María Siller-Ruiz, Jorge Calvo, Dolores Quesada, Jun Hao Wang, Cristina Pitart, Francesc Marco, Nuria Prim, Juan Pablo Horcajada, Eduardo Padilla, Ester Del Barrio-Tofiño, Belen Viñado-Pérez, Nieves Larrosa, Fe Tubau, Silvia Capilla, Antonio Casabella, Mar Olga Pérez-Moreno, Emma Padilla, Mónica Ballestero, Alba Rivera, Ferrán Navarro, Fréderic Gómez-Bertomeu, Sergio Pardo-Granell, Ester Picó-Plana, Dolores Guerrero, Carolina Sarvisé-Buil, Alba Belles-Belles, Marta Fernández-Esgueva, María del Pilar Ortega-Lafont, Inmaculada García, Noelia Arenal-Andrés, Susana Hernando-Real, Rosario Ibáñez, Jesús Martínez, Federico Becerra, Carmen Aldea-Mansilla, Asmaa Alaoui-Sosse, José Carlos González, Julia Guzman-Puche, Miguel Ángel Blázquez-Andrada, Nora Mariela Martínez-Ramírez, Alicia Beteta, Bárbara Gomila-Sard, Salvador Giner Almaraz, Eugenio Garduño, Pedro Miguel Juiz-González, Jorge Arca-Suárez, Javier Alba, Pilar Alonso, Ana Isabel Rodríguez, María Isabel Paz-Vidal, Marta García-Campello, Pablo Camacho, María de los Ángeles Pallarés, María Luisa Pérez del Molino, Amparo Coira, Gema Barbeito, Anniris Rincón, Francisco José Vasallo-Vidal, Laura Alonso-Acero, Laura Iglesias-Llorente, Ana Bordes-Benites, Laura Florén-Zabala, José Manuel Azcona, Carla Andrea Alonso, Yolanda Sáenz, Marta Lamata-Subero, David Molina, Ana González-Torralba, Jennifer Villa, Esther Viedma, Emilia Cercenado, Teresa Alarcón, Paula Vargas, María Díez, Rafael Cantón, Patricia Ruiz, María Isabel Sánchez-Romero, Felipe Pérez-García, Genoveva Yagüe-Guirao, Amaia Concepción Oteiza, José Leiva, María Eugenia Portillo, Andrés Canut-Blasco, Matxalen Vidal, Iker Alonso, Maider Zuriarrain, and José Luis Barrios-Andrés
Appendix A. Supplementary data
References
- 1.Horcajada J.P., Montero M., Oliver A., et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32:e000311. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.López-Causapé C., Cabot G., Del Barrio-Tofiño E., Oliver A. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol. 2018;9:685. doi: 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenover F.C., Nicolau D.P., Gill C.M. Carbapenemase-producing Pseudomonas aeruginosa an emerging challenge. Emerg Microbes Infect. 2022;11(1):811–814. doi: 10.1080/22221751.2022.2048972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magiorakos A.P., Srinivasan A., Carey R.B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 7.Kadri S.S., Adjemian J., Lai Y.L., et al. Difficult-to-Treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. (NIH–ARORI) Clin Infect Dis. 2018;67(12):1803–1814. doi: 10.1093/cid/ciy378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelegrin A.C., Palmieri M., Mirande C., et al. Pseudomonas aeruginosa: a clinical and genomics update. FEMS Microbiol Rev. 2021;45(6):fuab026. doi: 10.1093/femsre/fuab026. [DOI] [PubMed] [Google Scholar]
- 9.Oliver A., Mulet X., López-Causapé C., Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21-22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Del Barrio-Tofiño E., López-Causapé C., Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106196. [DOI] [PubMed] [Google Scholar]
- 11.Treepong P., Kos V.N., Guyeux C., et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Kos V.N., Déraspe M., McLaughlin R.E., et al. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother. 2015;59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabot G., López-Causapé C., Ocampo-Sosa A.A., et al. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother. 2016;60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaillard M., van Belkum A., Cady K.C., et al. Correlation between phenotypic antibiotic susceptibility and the resistome in Pseudomonas aeruginosa. Int J Antimicrob Agents. 2017;50:210–218. doi: 10.1016/j.ijantimicag.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Del Barrio-Tofiño E., Zamorano L., Cortes-Lara S., et al. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother. 2019;74(7):1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 16.Wright H., Bonomo R.A., Paterson D.L. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect. 2017;23:704–712. doi: 10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Yahav D., Giske C.G., Grāmatniece A., Abodakpi H., Tam V.H., Leibovici L. New β-Lactam-β-Lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34(1):e00115–e00120. doi: 10.1128/CMR.00115-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraile-Ribot P.A., Cabot G., Mulet X., et al. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother. 2018;73:658–663. doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 19.Gomis-Font M.A., Pitart C., Del Barrio-Tofiño E., et al. Emergence of Resistance to Novel Cephalosporin-β-lactamase Inhibitor Combinations through the Modification of the Pseudomonas aeruginosa MexCD-OprJ Efflux Pump. Antimicrob Agents Chemother. 2021;65(8) doi: 10.1128/AAC.00089-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields R.K., Stellfox M.E., Kline E.G., Samanta P., Van Tyne D. Evolution of Imipenem-Relebactam Resistance Following Treatment of Multidrug-Resistant Pseudomonas aeruginosa Pneumonia. Clin Infect Dis. 2022;75:710–714. doi: 10.1093/cid/ciac097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso-García I., Vázquez-Ucha J.C., Lasarte-Monterrubio C., et al. Simultaneous and divergent evolution of resistance to cephalosporin/β-lactamase inhibitor combinations and imipenem/relebactam following ceftazidime/avibactam treatment of MDR Pseudomonas aeruginosa Infections. J Antimicrob Chemother. 2023;78(Issue 5):1195–1200. doi: 10.1093/jac/dkad062. [DOI] [PubMed] [Google Scholar]
- 22.Ruedas-López A., Alonso-García I., Lasarte-Monterrubio C., et al. Selection of AmpC β-lactamase variants and metallo-β-lactamases leading to ceftolozane/tazobactam and ceftazidime/avibactam resistance during treatment of MDR/XDR Pseudomonas aeruginosa infections. Antimicrob Agents Chemother. 2022;66(2) doi: 10.1128/aac.02067-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraile-Ribot P.A., Zamorano L., Orellana R., et al. Activity of imipenem-relebactam against a large collection of Pseudomonas aeruginosa clinical isolates and isogenic β-lactam-resistant mutants. Antimicrob Agents Chemother. 2020;64(2):e021655–e021719. doi: 10.1128/AAC.02165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marvig R.L., Sommer L.M., Molin S., Krogh Johansen H. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 25.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Auwera G.A., Carneiro M.O., Hartl C., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110) doi: 10.1002/0471250953.bi1110s43. 11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cingolani P., Platts A., Wang le L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2023. R: A language and environment for statistical computing.https://www.R-project.org/ URL: [Google Scholar]
- 30.Jolley K.A., Maiden M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanbongi Y., Shimizu A., Suzuki T., et al. Classification of OprD sequence and correlation with antimicrobial activity of carbapenem agents in Pseudomonas aeruginosa clinical isolates collected in Japan. Microbiol Immunol. 2009;53(7):361–367. doi: 10.1111/j.1348-0421.2009.00137.x. [DOI] [PubMed] [Google Scholar]
- 32.del Barrio-Tofiño E., López-Causapé C., Cabot G., et al. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother. 2017;61(11):e015899–e015917. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Causapé C., Sommer L.M., Cabot G., et al. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep. 2017;7(1):5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortes-Lara S., del Barrio-Tofiño E., López-Causapé C., Oliver A., GEMARA-SEIMC/REIPI Pseudomonas study Group Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin Microbiol Infect. 2021;27(11):1631–1637. doi: 10.1016/j.cmi.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z., Alikhan N.F., Sergeant M.J., et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Sales R.O., Migliorini L.B., Puga R., Kocsis B., Severino P. A core genome multilocus sequence typing scheme for Pseudomonas aeruginosa. Front Microbiol. 2020;11:1049. doi: 10.3389/fmicb.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva M., Machado M.P., Silva D.N., et al. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4(3) doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabot G., Ocampo-Sosa A.A., Domínguez M.A., et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother. 2012;56(12):6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Álvarez-Lerma F., Catalán-González M., Álvarez J., et al. Impact of the "Zero Resistance" program on acquisition of multidrug-resistant bacteria in patients admitted to Intensive Care Units in Spain. A prospective, intervention, multimodal, multicenter study. Med Intensiva. 2023;47(4):193–202. doi: 10.1016/j.medine.2022.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Cercenado E., Rodríguez-Baño J., Alfonso J.L., et al. Antimicrobial stewardship in hospitals: expert recommendation guidance document for activities in specific populations, syndromes and other aspects (PROA-2) from SEIMC, SEFH, SEMPSPGS, SEMICYUC and SEIP. Enferm Infecc Microbiol Clín. 2023;41(4):238–242. doi: 10.1016/j.eimce.2022.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Mensa J., Barberán J., Soriano A., et al. Antibiotic selection in the treatment of acute invasive infections by Pseudomonas aeruginosa: guidelines by the Spanish society of Chemotherapy. Rev Esp Quimioter. 2018;31(1):78–100. [PMC free article] [PubMed] [Google Scholar]
- 44.Gaspari R., Spinazzola G., Teofili L., et al. Protective effect of SARS-CoV-2 preventive measures against ESKAPE and Escherichia coli infectionsEur. J Clin Invest. 2021;51(12) doi: 10.1111/eci.13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelstein M.V., Skleenova E.N., Shevchenko O.V., et al. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis. 2013;13(10):867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 46.Torrens G., van der Schalk T.E., Cortes-Lara S., et al. Susceptibility profiles and resistance genomics of Pseudomonas aeruginosa isolates from European ICUs participating in the ASPIRE-ICU trial. J Antimicrob Chemother. 2022;77(7):1862–1872. doi: 10.1093/jac/dkac122. [DOI] [PubMed] [Google Scholar]
- 47.Reyes J., Komarow L., Chen L., et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe. 2023;4(3):e159–e170. doi: 10.1016/S2666-5247(22)00329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peña C., Cabot G., Gómez-Zorrilla S., et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis. 2015;60(4):539–548. doi: 10.1093/cid/ciu866. [DOI] [PubMed] [Google Scholar]
- 49.Recio R., Sánchez-Diener I., Viedma E., et al. Pathogenic characteristics of Pseudomonas aeruginosa bacteraemia isolates in a high-endemicity setting for ST175 and ST235 high-risk clones. Eur J Clin Microbiol Infect Dis. 2020;39(4):671–678. doi: 10.1007/s10096-019-03780-z. [DOI] [PubMed] [Google Scholar]
- 50.Gómez-Zorrilla S., Juan C., Cabot G., et al. Impact of multidrug resistance on the pathogenicity of Pseudomonas aeruginosa: in vitro and in vivo studiesInt. J Antimicrob Agents. 2016;47(5):368–374. doi: 10.1016/j.ijantimicag.2016.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences from the 2022 and 2017 studies have been deposited in the European Nucleotide Archive, under project numbers PRJEB61879 and PRJEB31047, respectively.