Abstract
DNA methylation is critical for regulating gene expression, necessitating its accurate placement by enzymes such as the DNA methyltransferase DNMT3A. Dysregulation of this process is known to cause aberrant development and oncogenesis, yet how DNMT3A is regulated holistically by its three domains remains challenging to study. Here we integrate base editing with a DNA methylation reporter to perform in situ mutational scanning of DNMT3A in cells. We identify mutations throughout the protein that perturb function, including ones at an interdomain interface that block allosteric activation. Unexpectedly, we also find mutations in the PWWP domain, a histone reader, that modulate enzyme activity despite preserving histone recognition and protein stability. These effects arise from altered PWWP domain DNA affinity, which we show is a noncanonical function required for full activity in cells. Our findings highlight mechanisms of interdomain crosstalk and demonstrate a generalizable strategy to probe sequence-activity relationships of nonessential chromatin regulators.
Introduction
Chromatin regulation is essential for directing and safeguarding physiological processes. DNA methylation is a key chromatin modification that occurs throughout the human genome, primarily at CpG dinucleotide sites1,2. It has diverse functions, but canonically silences gene expression when present at promoter CpG islands2. DNA methylation is installed by the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B, which are essential for mammalian development3. Indeed, mutations in DNMT3A lead to developmental disorders, such as Tatton-Brown-Rahman Syndrome (TBRS)4 and Heyn-Sproul-Jackson Syndrome (HESJAS)5. Moreover, loss of DNMT3A in hematopoietic stem cells promotes self-renewal and leukemic transformation6,7, positioning DNMT3A mutations as important drivers of clonal hematopoiesis and acute myeloid leukemia (AML)8.
DNMT3A function is regulated by complex mechanisms involving its three domains. Beyond its direct role in catalysis, the methyltransferase (MTase) domain harbors interfaces enabling DNMT3A to complex with other copies of itself, DNMT3B, or the catalytically inactive homolog DNMT3L, thereby shaping its activity8-12. The two N-terminal domains, the ATRX-DNMT3-DNMT3L (ADD) and Pro-Trp-Trp-Pro (PWWP) domains, are readers of histone H3 unmethylated at lysine 4 (H3K4me0) and dimethylated at lysine 36 (H3K36me2), respectively13-15. H3K4me0 recognition activates DNMT3A by repositioning the ADD domain, which otherwise blocks the DNA substrate binding site13. Recent studies have also found that H3K36me2 stimulates DNMT3A activity selectively over H3K36me0 in vitro15,16, raising the question of whether crosstalk between the PWWP and MTase domains might exist.
Understanding the interplay between DNMT3A’s domains is critical to dissecting its role in disease and identifying novel therapeutics. Although structural approaches have yielded deep mechanistic insight into DNMT3A regulation12,13,17,18, to date no published structure has successfully resolved all three domains. A recent study evaluated how clinically observed mutations across DNMT3A’s full sequence impact function and stability19, demonstrating how large-scale mutational analysis can reveal novel insight. While this study used exogenously expressed DNMT3A, approaches such as CRISPR scanning could provide the opportunity to screen mutations introduced directly within the endogenous gene20-24. This approach has illuminated functional mechanisms of other chromatin regulators21-23, but is less easily applied to nonessential proteins like DNMT3A. This is because Cas9 predominately generates frameshift mutations that can obscure in-frame mutations of interest, unless actively removed from the gene pool due to a fitness disadvantage. CRISPR mutational scanning is greatly empowered by base editors, which avoid insertion-deletions (indels) and have more finely targeted and predictable mutational outcomes25. This technology, however, has so far been limited to viability-based screens26-33.
Here, we expand the scope of base editor screens using a genetically encoded methylation reporter to interrogate DNMT3A within its native state. Our study reveals novel insights into DNMT3A biochemistry and, more broadly, demonstrates a generalizable approach to studying chromatin regulators.
Results
A fluorescent reporter reads out endogenous DNMT3A activity
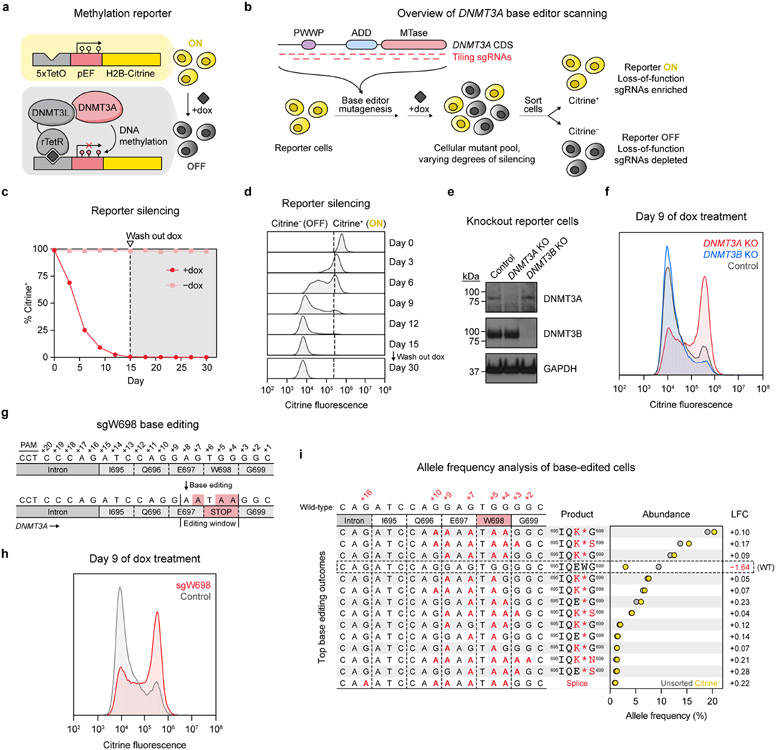
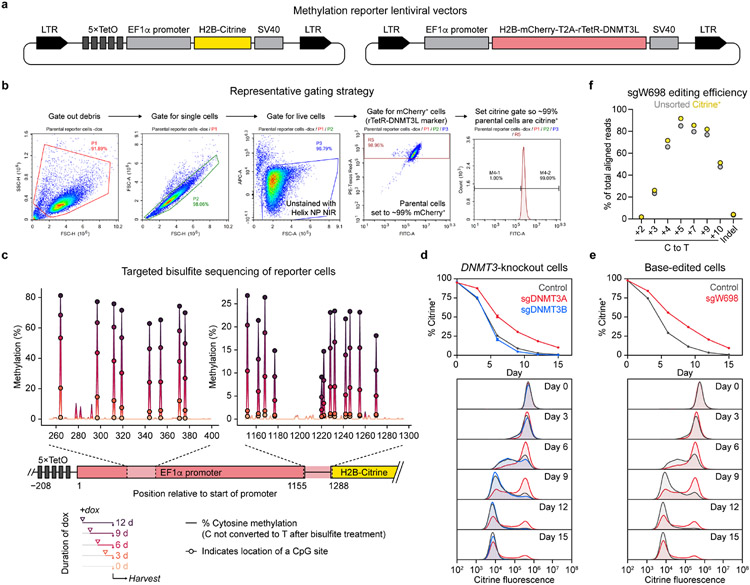
To enable readout of endogenous DNMT3A activity, we leveraged a chromatin regulation reporter providing a fluorescence-based readout of gene silencing activity34. We fused the accessory factor DNMT3L, which itself is catalytically inactive8,9, to the reverse Tet repressor (rTetR) to recruit endogenous DNMT3A to a genome-integrated reporter cassette in a doxycycline (dox)-inducible fashion (Fig. 1a and Extended Data Fig. 1a). Subsequent DNA methylation of the reporter silences citrine expression, providing a fluorescence-based readout of DNMT3A activity amenable for base editor scanning (Fig. 1b). Upon treating a clonal K562 reporter cell line with dox, we observed complete reporter silencing with clearly resolved silenced and unsilenced states (Fig. 1c,d and Extended Data Fig. 1b). Silenced cells did not revert after dox washout, consistent with stable gene repression due to DNA methylation34. Moreover, targeted bisulfite sequencing of the integrated reporter showed a time-dependent gain of promoter methylation in dox-treated cells, confirming that methylation accompanies silencing (Extended Data Fig. 1c).
Fig. 1 ∣. A live-cell reporter enables fluorescence-based readout of endogenous DNMT3A activity.
a. Schematic of the methylation reporter. TetO, tetracycline operator; pEF, EF1α promoter; rTetR, reverse tetracycline repressor. Red lollipops indicate DNA methylation.
b. Overview of DNMT3A base editor scanning experiment.
c. Timecourse of reporter silencing measured by flow cytometry. Dotted line indicates dox washout on day 15. Data are mean ± SD of n = 3 replicates.
d. Citrine fluorescence of dox-treated cells at indicated timepoints from c.
e. Immunoblot of DNMT3-knockout reporter cells. Control, sgRNA recognizing the luciferase coding sequence (nontargeting sgLucA). Uncropped images are provided as Source Data.
f. Citrine fluorescence of cells from e measured by flow cytometry after 9 d of dox treatment.
g. sgW698 target site in DNMT3A. Numbers indicate positions along the protospacer (antisense to the DNMT3A gene). Expected base editing mutations are highlighted in red (these appear as G to A because the protospacer is along the opposite strand).
h. Citrine fluorescence of cells treated with sgW698 or sgLucA control measured by flow cytometry after 9 d of dox treatment.
i. Allele frequencies in cells edited with sgW698 after 9 d of dox treatment, either sorted for citrine+ cells (yellow dots) or unsorted (gray dots). The wild-type allele is boxed, and only alleles with ≥1% allele frequency in at least one sample are shown. Protein product sequences are shown with nonsynonymous mutations in red. Splice, splice site mutation; LFC, log2(fold-change allele frequency in citrine+ versus unsorted cells). Genotyping was performed once (n = 1).
Histograms of citrine fluorescence in d, f, and h are representative of n = 3 replicates, and full results are shown in c and Extended Data Fig. 1d,e. Results in c–f and h are representative of two independent experiments. See also Extended Data Fig. 1.
We next evaluated reporter sensitivity toward DNMT3A loss-of-function, which should impair its silencing under dox treatment. Because mammalian cells possess a second de novo DNMT, DNMT3B, we first tested whether this might interfere with detection of DNMT3A-mediated silencing. CRISPR-Cas9 knockout of DNMT3A, but not DNMT3B, caused defective reporter silencing (Fig. 1e,f and Extended Data Fig. 1d), consistent with K562 cells predominantly expressing inactive isoforms of DNMT3B35. This showed that our reporter selectively detects loss of DNMT3A. To assess sensitivity toward inactivating C to T base editing mutations, we designed a single-guide RNA (sgRNA) with the goal of introducing a nonsense mutation at the W698 codon, named sgW698 (Fig. 1g). Reporter cells edited with sgW698 displayed a silencing defect similar to that of DNMT3A-knockout cells (Fig. 1h and Extended Data Fig. 1e). We next isolated cells remaining citrine+ after dox treatment using fluorescence-activated cell sorting (FACS) and deep sequenced the edited site. Relative to unsorted cells, citrine+ cells were markedly depleted of the wild-type allele and enriched with alleles containing the W698* mutation, while retaining low rates of indels (Fig. 1i and Extended Data Fig. 1f). Thus, loss-of-function mutations are indeed enriched in cells defective for silencing. These results demonstrate that our reporter can detect base editing mutations introduced in the endogenous DNMT3A gene, enabling mutational scanning of the native protein.
Base editor scanning charts mutations impacting DNMT3A
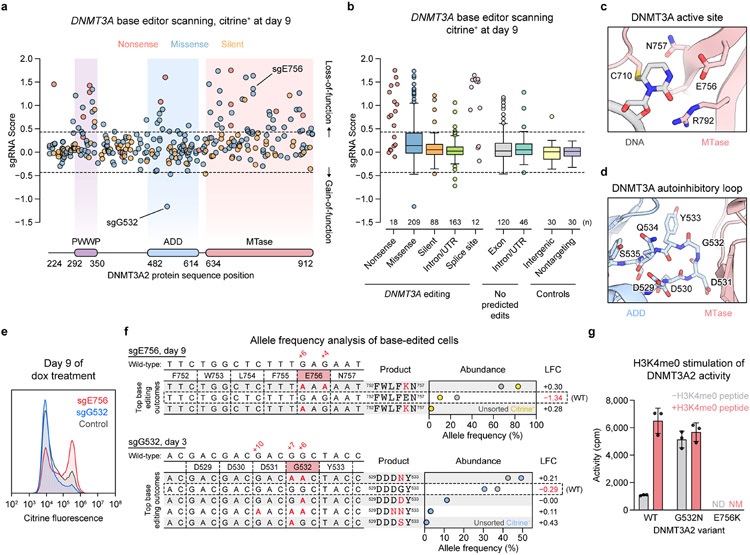
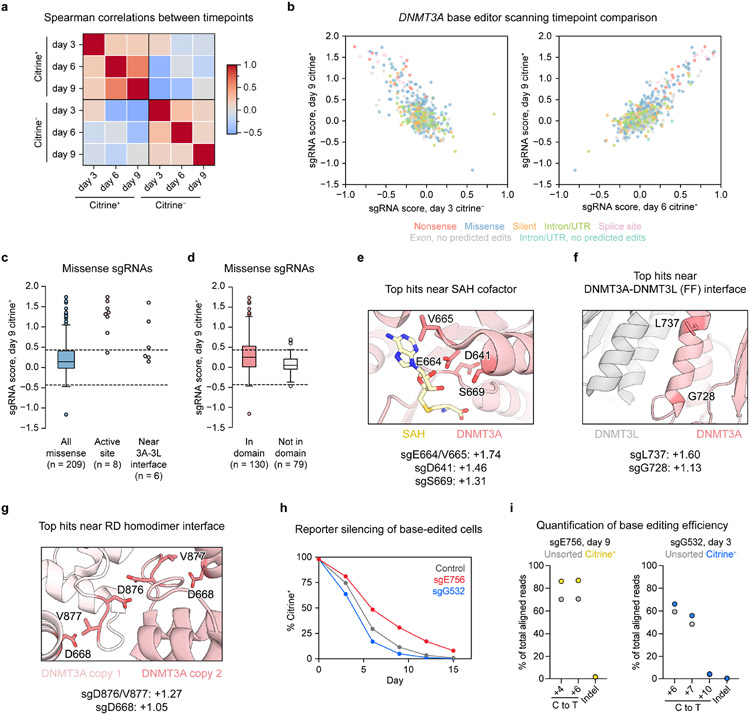
To carry out base editor scanning, we designed a library containing all sgRNAs targeting exons and flanking intronic sequences of DNMT3A isoform 2 (DNMT3A2), the predominant isoform expressed in K56235. We transduced this library into reporter cells, treated the cells with dox, and sorted them based on citrine fluorescence (Fig. 1b). Using deep sequencing, we quantified sgRNA abundances in citrine+ or citrine− fractions compared to in unsorted cells and calculated “sgRNA scores” representing whether each sgRNA was enriched or depleted (see Methods). As expected, sgRNA scores were inversely correlated between citrine+ and citrine− fractions (Extended Data Fig. 2a,b). Having confirmed these broad correlations, we focused our analysis on the day 9 citrine+ cells, which showed the greatest effect size.
We classified each sgRNA based on its predicted mutational outcome, assuming complete editing within the +4 to +8 editing window25. sgRNAs predicted to introduce nonsense or splice site-disrupting mutations (“nonsense sgRNAs” or “splice site sgRNAs,” respectively) were largely enriched, indicating loss-of-function, while most silent and intronic sgRNAs were unchanged in abundance (Fig. 2a,b). However, missense sgRNAs exhibited a wide range of scores, reflecting context-dependent effects. Consistent with expectations, those targeting residues at the active site or near the DNMT3A-DNMT3L interface were generally more enriched compared to all missense sgRNAs, as were those mapping within domains compared to those mapping outside of domains (Extended Data Fig. 2c,d). Residues targeted by the top MTase domain hits included E756, which participates in DNMT3A’s catalytic mechanism36; E664, which binds the SAH cofactor; G728 and L737, which reside in an α-helix at the DNMT3A-DNMT3L interface; and D876, which is known to play a critical role in stabilizing the RD homodimer interface10 (Fig. 2c and Extended Data Fig. 2e-g). Thus, our screen successfully recapitulated regions known to be vital for DNMT3A function.
Fig. 2 ∣. Base editor scanning charts mutations across DNMT3A that impact function.
(a–b) DNMT3A base editor scanning results for citrine+ cells at day 9. Dotted lines indicate ±2 SD of intergenic control sgRNAs, and data are the average of n = 3 replicates. Full screen data are provided in Supplementary Data 1-3.
a. Scatterplot of sgRNA scores for nonsense (red), missense (blue), and silent (orange) sgRNAs, plotted against the targeted site in the coding sequence.
b. Boxplot of sgRNA scores for sgRNAs classified by predicted editing outcome. Center line, median; box, interquartile range; whiskers, up to 1.5 × interquartile range per the Tukey method. The number of sgRNAs (n) in each category is printed on the plot above each label. Outliers and any categories with n < 20 are shown individually.
c. View of the DNMT3A active site (red) with the DNA substrate (gray) (PDB: 5YX2).
d. View of the ADD (blue)-MTase (red) autoinhibitory interface highlighting the autoinhibitory loop (PDB: 4U7P).
e. Citrine fluorescence of base editor-treated cells measured by flow cytometry after 9 d of dox treatment. Control, nontargeting sgLucA. Histograms are representative of n = 3 replicates and full results are shown in Extended Data Fig. 2e.
f. Allele frequencies in base-edited cells. Top, cells edited with sgE756 after 9 d of dox treatment, comparing citrine+ (yellow dots) to unsorted (gray dots) cells. Bottom, cells edited with sgG532 after 3 d of dox treatment, comparing citrine− cells (blue dots) to unsorted cells (gray dots). The wild-type allele is boxed, and only alleles with ≥1% allele frequency in at least one sample are shown. Protein product sequences are shown with nonsynonymous mutations in red. LFC, log2(fold-change allele frequency in sorted versus unsorted cells). Genotyping was performed once (n = 1).
g. Activity of purified DNMT3A2 in the presence (red) or absence (gray) of H3K4me0 peptide. Data are mean ± SD of n = 3 replicates. ND, not detected; NM, not measured.
Results in e and g are representative of two independent experiments. See also Extended Data Figs. 2-4.
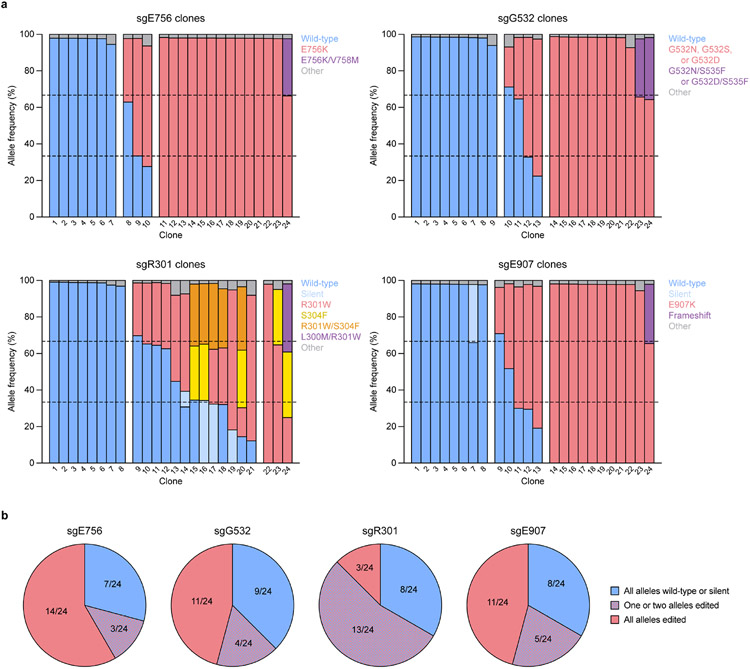
To further validate our screen, we picked two hits to study in depth, sgE756 and sgG532 (Fig. 2a). sgE756 was one of several highly enriched sgRNAs mutating E756 (Fig. 2c). Conversely, sgG532 was strongly depleted, implying a gain-of-function effect. Because sgG532 targets the ADD domain autoinhibitory loop (Fig. 2d), we hypothesized that it might disrupt autoinhibition. Testing of each sgRNA individually confirmed defective and enhanced silencing for sgE756- and sgG532-treated cells, respectively (Fig. 2e and Extended Data Fig. 2h). We genotyped cells to verify that the predicted mutations, E756K and G532N, were linked to these effects. Indeed, sgE756-treated cells remaining citrine+ at day 9 were enriched with the E756K allele and depleted of the wild-type allele (Fig. 2f and Extended Data Fig. 2i), consistent with E756K being loss-of-function. sgE756 displayed lower editing efficiency compared to sgW698, explaining its milder effect on reporter silencing. For sgG532, since gain-of-function mutations should be enriched in early silencing cells, we sorted citrine− cells at day 3, confirming these were enriched for the G532N allele. To determine how these alleles were distributed in individual cells, we genotyped clonal lines derived from sgRNA-treated cells (Extended Data Fig. 3a,b). Consistent with previous work29, most cells edited with sgE756 or sgG532 remained completely wild-type or contained edits in all alleles, with partial editing being uncommon.
To biochemically verify that E756K and G532N alter catalytic activity, we next purified recombinant DNMT3A2 (Extended Data Fig. 4a) and conducted enzyme activity assays. These confirmed that the E756K mutant is catalytically dead, while the G532N mutant is hyperactive relative to wild-type (Fig. 2g). We also tested the effect of adding H3K4me0 peptide, which promotes the release of DNMT3A autoinhibition13. Though wild-type DNMT3A2 was stimulated by H3K4me0 peptide, DNMT3A2 G532N could not be further stimulated, suggesting that its hyperactivity arises due to blocking the autoinhibited conformation. Taken together, these results demonstrate that our base editor scanning approach can accurately discern mutations that perturb DNMT3A function.
3D analysis highlights the ADD-MTase interface
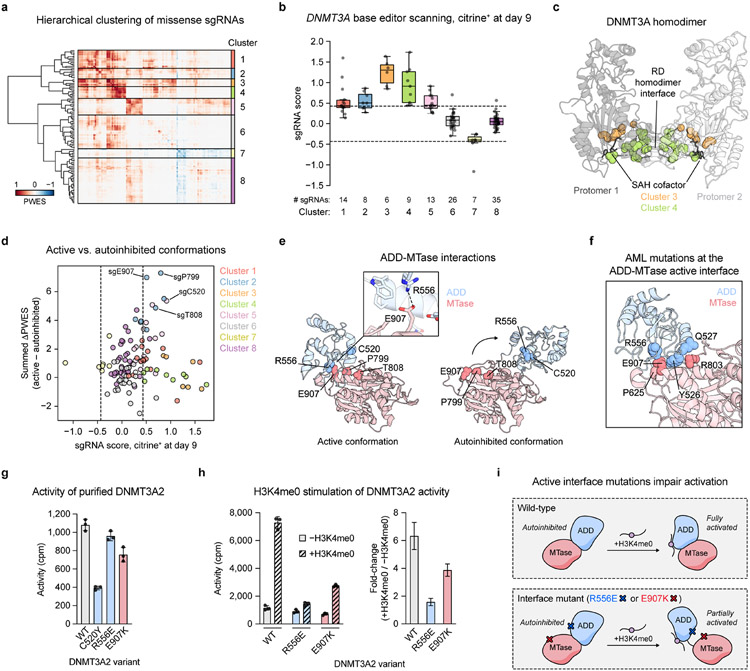
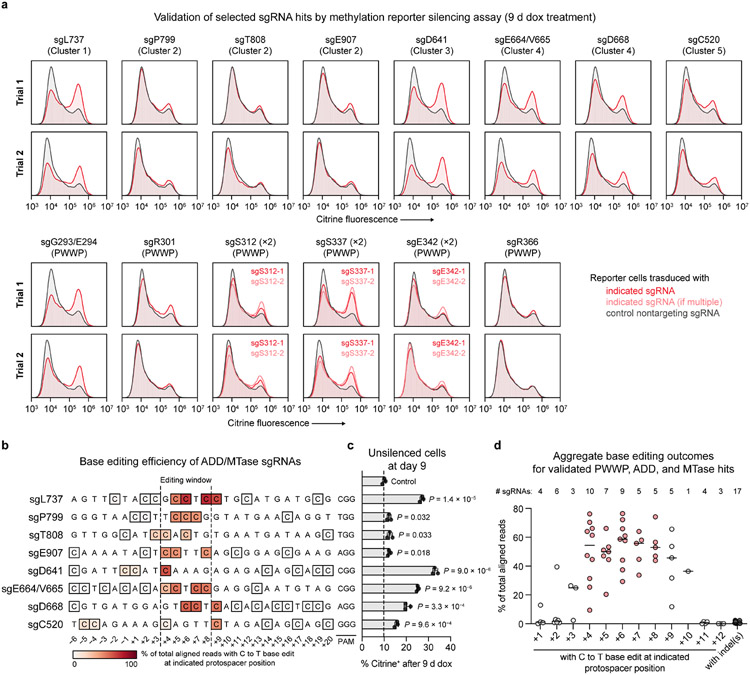
Since functional regions in proteins can comprise residues far apart in the linear sequence, we next considered our screen results in the context of 3D space. We mapped each missense sgRNA to its targeted residue in the active conformation of DNMT3A (ADD-MTase truncation)13, reasoning that this might highlight functionally critical areas. We calculated a proximity-weighted enrichment score (PWES)22 for each pair of sgRNAs describing their 3D proximity and combined sgRNA scores (see Methods). Hierarchical clustering of the resulting PWES matrix defined eight sgRNA clusters (Fig. 3a,b), with the two most highly enriched clusters, clusters 3 and 4, targeting known functional hotspots: the active site and RD homodimer interface (Fig. 3c). Clusters 3 and 4 contained most of the highest-scoring sgRNAs considered here, with the notable exception of sgL737 in cluster 1, which targets the DNMT3A-DNMT3L interface (Extended Data Fig. 2f). We validated a selection of these hits (sgD641, sgE664/V665, sgD668, and sgL737), confirming on-target base editing and impaired reporter silencing (Extended Data Figs. 2e-g and 5a-d).
Fig. 3 ∣. Functional hotspot analysis highlights an interdomain interface important for allosteric activation.
a. Heatmap depicting the PWES matrix for all pairs of missense sgRNAs (n = 118) mapping to resolved residues in the structure of active conformation DNMT3A (PDB: 4U7T). sgRNAs are ordered by hierarchical clustering.
b. Boxplot of sgRNA scores in citrine+ cells at day 9 of the base editor scanning experiment, with sgRNAs organized by clusters from a. Data are the average of n = 3 replicates. Dotted lines indicate ±2 SD of intergenic control sgRNAs. Center line, median; box, interquartile range; whiskers, up to 1.5 × interquartile range per the Tukey method. Individual data points are overlaid.
c. View of the DNMT3A homodimer with residues targeted by sgRNAs in clusters 3 and 4 highlighted (PDB: 4U7T).
d. Comparison of PWES values calculated using active conformation (PDB: 4U7T) 3D proximity versus those using autoinhibited conformation (PDB: 4U7P) 3D proximity, represented as the summed ΔPWES (see Methods for details). sgRNA scores, colors, and dotted lines correspond to those in b.
e. View of the ADD (blue)-MTase (red) interface in the structures of DNMT3A in active (left, PDB: 4U7T) or autoinhibited (right, PDB: 4U7P) conformations. Inset shows the ionic bond mediated by R556 and E907.
f. View of active conformation DNMT3A highlighting residues at the ADD-MTase interface that are mutated in AML (data from COSMIC) (PDB: 4U7T).
g. Activity of purified DNMT3A2. Data are mean ± SD of n = 3 replicates.
h. Stimulation of purified DNMT3A2 by H3K4me0 peptide. Right, fold-change in activity in the presence of H3K4me0 versus in the absence of H3K4me0. Data are mean ± SD of n = 3 replicates. Fold-change errors were propagated from the individual SDs. Results are representative of two independent experiments.
i. Cartoon depicting impaired H3K4me0 stimulation caused by mutations disrupting the active conformation ADD-MTase interface.
See also Extended Data Figs. 3-5 and Supplementary Data 4.
The ADD-MTase structure has been solved in both active and autoinhibited conformations13, and we hypothesized that comparing PWES values computed from the different structures might highlight residues involved in allostery. To investigate this, we calculated a “summed ΔPWES” for each sgRNA representing this comparison (see Methods). This measure was close to zero for most sgRNAs, but several sgRNAs in cluster 2 (sgP799, sgT808, and sgE907) and cluster 5 (sgC520) displayed large, positive summed ΔPWES values (Fig. 3d), indicating that the autoinhibited to active conformational switch strengthens the overall magnitude of their PWES correlations to other sgRNAs. These hits mapped to residues around the active conformation ADD-MTase interface (Fig. 3e). Interestingly, mutations at this interface have been identified in AML patients37 (Fig. 3f), suggesting this may be a clinically relevant functional hotspot. Individual validation of these sgRNAs showed on-target editing but subtle effects (sgP799, sgT808, and sgE907) on reporter silencing (Extended Data Fig. 5a-d). To test whether the allelic distribution of editing might help explain these subtle effects, we genotyped clonal lines of sgE907-treated cells. Although heterozygous editing was still uncommon for sgE907, it occurred at a higher rate than for the validated hit sgE756 (Extended Data Fig. 3a,b). Given lower editing, we reasoned that these ADD-MTase sgRNAs may have partial loss-of-function effects more difficult to detect in our reporter assay.
We therefore turned to biochemical assays to directly test the effect of mutations around this interface on catalytic activity. The C520Y mutant (predicted product of sgC520) was catalytically defective (Fig. 3g), although genotyping revealed that sgC520 also edited a splice site outside the editing window (+9 position) (Extended Data Fig. 5b), accounting in part for its effects. Focusing on residues directly at the ADD-MTase interface, we noted that E907 likely stabilizes the interface through an ionic bond with R556 (Fig. 3e). Surprisingly, both the E907K (product of sgE907) and R556E charge-reversal mutants showed comparable activity to wild-type, unlike the C520Y mutant (Fig. 3g). We considered whether these mutations might instead affect allosteric activation, as has been shown for the nearby Q527A and R803A mutations38. Indeed, both E907K and R556E mutants displayed a lower fold-change stimulation upon addition of peptide compared to wild-type (Fig. 3h), indicating that the mutations impair the release of autoinhibition by H3K4me0, and that the active ADD-MTase interface is necessary for full allosteric activation. Blocking the E907-R556 interaction may force the H3K4me0-bound ADD domain to adopt a distinct orientation, such as an MTase-unbound state, that is less amenable to catalysis (Fig. 3i); alternatively, H3K4me0 binding itself may be disfavored. Both E907 and R556 are mutated in AML37, suggesting that this loss-of-function mechanism is clinically relevant. Thus, our analysis shows that the active ADD-MTase interface is a functional hotspot, demonstrating the utility of 3D structural analysis to yield mechanistic insight.
PWWP domain mutations variably impact protein stability
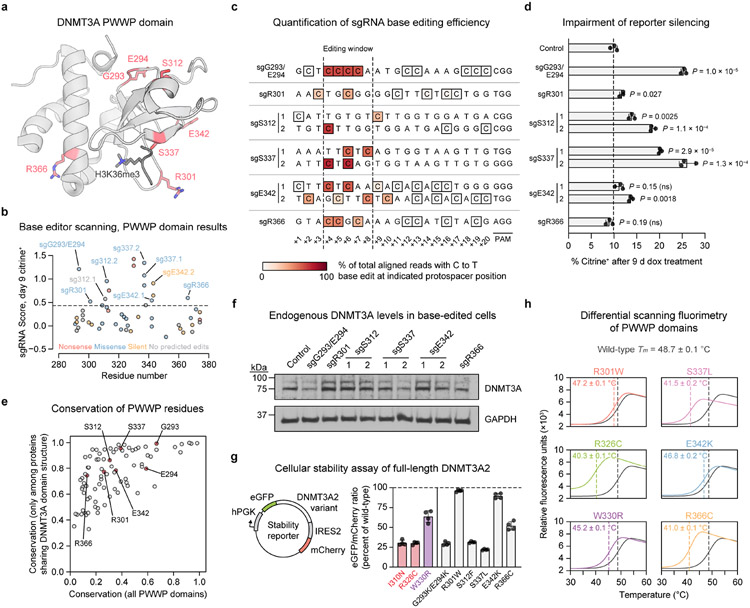
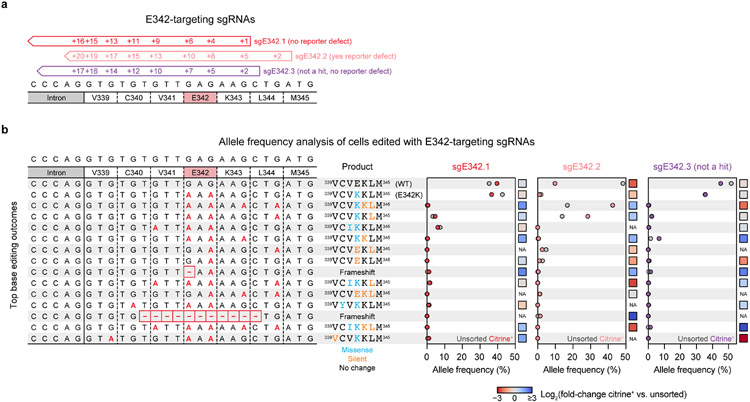
We next turned our attention to the sgRNA hits within the PWWP domain (Fig. 4a,b). These were particularly intriguing since our reporter, which involves artificial DNMT3A recruitment, was not expected to show sensitivity to loss of H3K36me2 targeting, the canonical role of the PWWP domain14,15. We validated a panel of these sgRNAs in reporter cells. Genotyping confirmed on-target C to T editing concomitant with dramatic defects in reporter silencing for sgG293/E294, sgS312.2, and sgS337.1/.2 (Figs. 4c,d and Extended Data Fig. 5a). A weaker effect was observed for sgR301, likely due to heterozygous editing (Extended Data Fig. 3a,b). Among these hits were also two sgRNAs annotated as causing no predicted edits (sgS312.1) or only a silent mutation (sgE342.2) (Fig. 4b). Genotyping revealed that these sgRNAs produce the corresponding mutations, S312F and E342K, by editing outside the predicted window at the +9 and +10 positions, respectively (Fig. 4c). sgE342.1 and sgR366 failed to validate.
Fig. 4 ∣. PWWP domain mutations have variable impacts on protein stability.
a. View of the DNMT3A PWWP domain (gray) with key residues targeted by hit sgRNAs shown as red sticks (PDB: 3LLR (PWWP domain), 5CIU (H3K36me3)).
b. Base editor scanning results within the PWWP domain. Dotted line indicates mean + 2 SD of intergenic control sgRNAs. Data are the same as in Fig. 2a,b and are the average of n = 3 replicates.
c. Base editing efficiency for selected PWWP sgRNAs. Each row depicts a protospacer sequence, with the heatmap intensity representing the C to T editing efficiency at each C measured by deep sequencing. Genotyping was performed once (n = 1). Allele tables are provided in Supplementary Data 5.
d. Flow cytometric quantification of sgRNA-treated cells remaining citrine+ after 9 d of dox treatment. Control, nontargeting sgLucA. Data are mean ± SD for n = 3 replicates. P values were calculated through two-tailed unpaired t tests comparing each sgRNA to control (ns, not significant). Citrine fluorescence histograms are shown in Extended Data Fig. 5a.
e. Conservation of each DNMT3A PWWP domain residue in two sets of related proteins. Raw values are provided in Supplementary Data 7.
f. Immunoblot for endogenous DNMT3A in reporter cells treated with the indicated sgRNAs (control, sgLucA). Uncropped images are provided as Source Data.
g. Stability of DNMT3A2 variants in K562 cells measured by a fluorescence reporter (schematic shown to left). Measurements are normalized to wild-type DNMT3A2 analyzed in parallel. Data are pooled from multiple experiments and are mean ± SD of n = 4 (two measurements of each of two independently transduced cell lines). Colored bars represent previously reported unstable (red) or stable (purple) disease-associated mutants5,19.
h. Thermal stability of purified PWWP domains measured by differential scanning fluorimetry. The wild-type curve (gray) is superimposed over that of each mutant. Curves represent mean of n = 3 replicates. Melting temperatures (Tm) are printed for each variant (mean ± SD) and indicated by dashed lines.
Results in d, f, and h are representative of two independent experiments. See also Extended Data Figs. 3-5.
We considered whether loss-of-function was due to simply disrupting structural integrity. Recent work has showed that a large fraction of PWWP clinical mutations destabilize DNMT3A5,19, suggesting this is a ubiquitous loss-of-function mechanism. Thus, we examined the evolutionary conservation of the residues targeted by these sgRNAs. We reasoned that residues universally conserved across PWWP-containing proteins likely serve important structural roles and therefore might be intolerant to mutation, while residues conserved only in DNMT3A-like proteins might play functional roles specific to DNMT3A. Both G293 and E294 were highly conserved across proteins with PWWP domains (Fig. 4e), suggesting that sgG293/E294 causes destabilization. However, the majority of validated sgRNAs targeted residues that were not conserved as broadly, namely R301, S312, S337, and E342.
Measuring the effects of sgRNA treatment on endogenous DNMT3A2 levels confirmed that sgG293/E294 destabilizes DNMT3A, as does sgS337.2 (Fig. 4f). However, many of the mutations tested did not obviously lower DNMT3A2 levels. To more quantitatively assess effects on stability, we measured DNMT3A2 expression in K562 cells using a dual-fluorescence stability reporter39 (Fig. 4g and Supplementary Fig. 1). Here, eGFP is fused to DNMT3A2 so its fluorescence reports on the amount of DNMT3A2 in the cell, while mCherry is expressed co-transcriptionally as a control. The ratio of eGFP to mCherry fluorescence thus provides a normalized measure of DNMT3A2 levels. We first tested several disease-associated mutations whose effects on stability have been characterized. Consistent with previous reports5,19, the I310N TBRS and R326C clonal hematopoiesis mutations were highly destabilizing, while the W330R HESJAS mutation preserved over 60% of wild-type expression (Fig. 4g). As expected from our prior results, the G293K/E294K and S337L mutants (products of sgG293/E294 and sgS337.1/.2) were highly unstable, as was the S312F mutant. In light of our conservation analysis, these results suggest that S337 and S312 play structural roles specific to the DNMT3A architecture. Notably, however, both the R301W (product of sgR301) and E342K mutations preserved stability.
To rigorously evaluate the impacts of these mutations on biochemical stability, we purified recombinant PWWP domains (Extended Data Fig. 4b) and performed differential scanning fluorimetry (Fig. 4h). These results were highly concordant with the cellular stability reporter data. In particular, the R326C, S337L, and R366C mutants all had a greatly lowered melting temperature, indicating that they are intrinsically less stable than wild-type. Interestingly, the R366C mutant caused a larger effect here than in the stability reporter assay (Fig. 4g), suggesting its biochemical instability may be attenuated in the context of full-length DNMT3A2. Importantly, the melting temperatures of the R301W and E342K mutants were similar to that of wild-type (Fig. 4h), confirming that these disease-associated mutations4,19 do not adversely impact protein stability. Thus, these results point to the existence of additional mechanisms by which the stable R301W and E342K mutations impact function.
DNA binding by the PWWP domain modulates DNMT3A activity
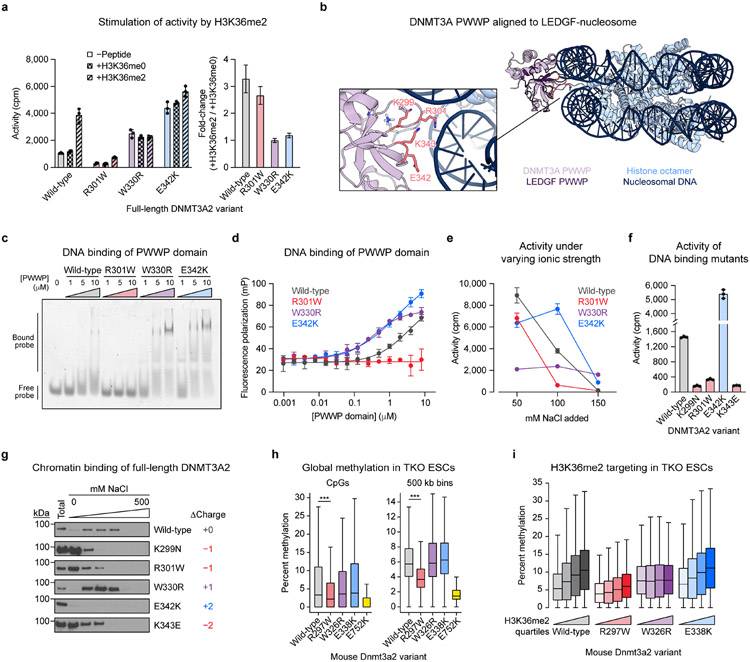
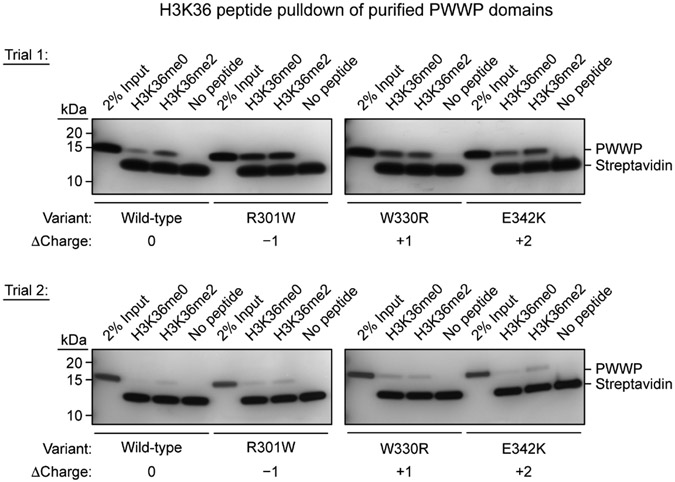
We next considered whether R301W and E342K directly impair catalysis. Activity assays showed opposing effects, with the R301W mutant losing catalytic activity and the E342K mutant surprisingly gaining activity (Fig. 5a). To assess whether these mutations impact the PWWP domain’s histone reader function, we tested for stimulation by H3K36me215,16. As expected, wild-type DNMT3A2 was stimulated selectively by H3K36me2 peptide (H3 residues 21–44), but not by H3K36me0 peptide. As a control, DNMT3A2 W330R, which lacks histone reader function due to a binding pocket mutation5, was refractory to histone stimulation and hyperactive at baseline like DNMT3A2 E342K. Notably, the R301W mutant was stimulated selectively by H3K36me2 like wild-type, suggesting it retains H3K36me2 recognition, while the E342K mutant was not stimulated. Pulldown assays confirmed that wild-type PWWP domain selectively bound H3K36me2 over H3K36me0, while PWWP W330R showed no preference (Extended Data Fig. 6). We note that unexpected signal was observed for PWWP W330R, possibly due to nonspecific interactions arising from the negatively charged truncation used here. PWWP R301W produced high signal with both histone modification states, suggesting loss of binding specificity; however, given that DNMT3A2 R301W was stimulated by H3K36me2 only (Fig. 5a), this may have been due to enhanced nonspecific binding resulting from increased net negative charge. Interestingly, PWWP E342K bound H3K36me2 more strongly than H3K36me0 (Extended Data Fig. 6), indicating that E342K preserves H3K36me2 recognition despite abrogating peptide stimulation. Since DNMT3A2 E342K’s basal activity was as high as stimulated wild-type enzyme (Fig. 5a), these results suggested that E342K may pre-activate DNMT3A.
Fig. 5 ∣. DNA binding by the PWWP domain modulates DNMT3A activity.
a. Activity of purified DNMT3A2 in the presence or absence of H3K36 peptide (residues 21–44). Right, calculated fold-change in activity observed with H3K36me2 versus H3K36me0.
b. View of the DNMT3A PWWP domain (PDB: 3LLR) aligned to the structure of the LEDGF-nucleosome complex (PDB: 6S01). Inset shows a close-up of the predicted DNA binding interface (LEDGF not shown in inset).
c–d. Binding of purified PWWP domains to a Cy3-labeled 30 bp oligonucleotide probe measured by (c) electrophoretic mobility shift assay and (d) fluorescence polarization assay.
e. Activity of purified DNMT3A2 under varying ionic strength (see Methods for details).
f. Activity of purified DNMT3A2, comparing effects of mutating residues at the PWWP-DNA interface shown in b.
g. Sequential salt extraction assay showing stepwise elution of FLAG-DNMT3A2 from HEK293T nuclear extracts using increasing concentrations of NaCl. Total refers to total lysate obtained in parallel with benzonase nuclease treatment. Immunoblots were processed in parallel.
h. Genome-wide CpG methylation in TKO ESCs ectopically expressing Dnmt3a2. Left, CpG-level methylation, excluding CpGs with zero methylation in all samples (n = 683,371). Right, methylation averaged across 500 kb bins (n = 5,204). P values (***, P < 2.3 × 10−308) were calculated through two-sided Wilcoxon signed-rank tests.
i. CpG methylation within 10 kb genomic bins ranked into quartiles based on normalized H3K36me2 ChIP-seq signal (n = 23,379 bins per quartile).
Data in a and d–f are mean ± SD for n = 3 replicates. Fold-change errors in a were propagated from the individual SDs. Unprocessed images for c and g are provided as Source Data. Results in a and c–g are representative of two independent experiments. For h and i, only CpGs with 5× coverage across all samples were considered, methylation values represent the average of two biological replicates, and boxplot components are as follows: center line, median; box, interquartile range; whiskers, up to 1.5 × interquartile range per the Tukey method; outliers not shown. See also Extended Data Figs. 4 and 6-9.
The hyperactivity of the E342K variant biochemically was unexpected, since sgE342.2 registered as loss-of-function in our reporter assay (Fig. 4d). Since sgE342.2 edited outside of the editing window (Fig. 4c), we considered whether bystander mutations might account for this discrepancy. To identify specific mutations associated with loss-of-function, we genotyped cells edited with three sgRNAs targeting the E342 codon and measured changes in allele frequency upon sorting for citrine+ dox-treated cells (Extended Data Fig. 7a,b). Counterintuitively, the E342K allele was depleted in citrine+ cells for both sgE342.1 and sgE342.2, revealing a lack of correlation with loss-of-function. Moreover, E342K was abundant in cells treated with sgE342.3, a third sgRNA not enriched in our screen. In the case of sgE342.2, sorting for citrine+ cells enriched for alleles containing E342K and additional silent mutations, suggesting that reporter loss-of-function may have arisen from synonymous mutations impairing expression. Nonetheless, we further investigated both the E342K and R301W mutants biochemically due to their stark differences.
The prior results suggest that R301W and E342K affect function through a mechanism distinct from H3K36me2 recognition. Since these mutations alter charged surface residues in opposing ways (Fig. 4a), their contrasting effects on enzyme activity might arise from changes in electrostatic interactions. Prior studies have demonstrated that the DNMT3A PWWP domain nonspecifically binds DNA40,41, though this phenomenon has not been shown to promote activity. Structural alignment to the PWWP-containing protein LEDGF complexed to the nucleosome42 revealed that R301 and E342 likely interact with or repel DNA, respectively (Fig. 5b). To test this, we conducted electrophoretic mobility shift and fluorescence polarization assays to measure binding of purified PWWP domains to a DNA probe (Fig. 5c,d). In both assays, R301W decreased DNA affinity while E342K increased it, as predicted by the net changes in charge. Similar to E342K, the W330R mutation also increased DNA affinity, consistent with increasing net positive charge. Interestingly, this suggested a potential link between DNA binding and the increased basal activity of the W330R and E342K mutants (Fig. 5a).
To test whether altered DNA binding indeed accounts for these differences in catalytic activity, we measured activity profiles under increasing ionic strength, which attenuates charged interactions with DNA43. Compared to wild-type, the R301W mutation sensitized DNMT3A2 to increasing concentrations of NaCl, while both the W330R and E342K mutations conferred insensitivity—consistent with weaker and stronger DNA binding, respectively (Fig. 5e). We next considered whether impairment of PWWP domain DNA affinity is a general loss-of-function mechanism beyond the R301W mutant. We identified two additional positively charged residues, K299 and K343, predicted to interact with DNA (Fig. 5b), and tested the impact of mutating them on activity. Both mutations tested, K299N and K343E, impaired activity (Fig. 5f). Notably, the K299N mutation has been identified in AML37, indicating this loss-of-function mechanism may be relevant to DNMT3A’s role in disease. Taken together, our in vitro data demonstrate that mutations impacting PWWP domain DNA binding can modulate catalytic activity.
DNA binding is a noncanonical PWWP domain role in cells
We next considered whether the PWWP mutations affect binding to chromatin in DNMT3A’s native cellular context. To investigate this, we performed sequential salt extraction assays to test how these mutations affect the strength of DNMT3A2 association to chromatin. Consistent with our prior results, the K299N, R301W, and K343E mutations all impaired chromatin association, as reflected by elution at a lower concentration of salt compared to wild-type (Fig. 5g). The W330R mutant was as strongly bound to chromatin as wild-type. Notably, we did not observe elution of the E342K mutant, possibly due to very tight binding to chromatin.
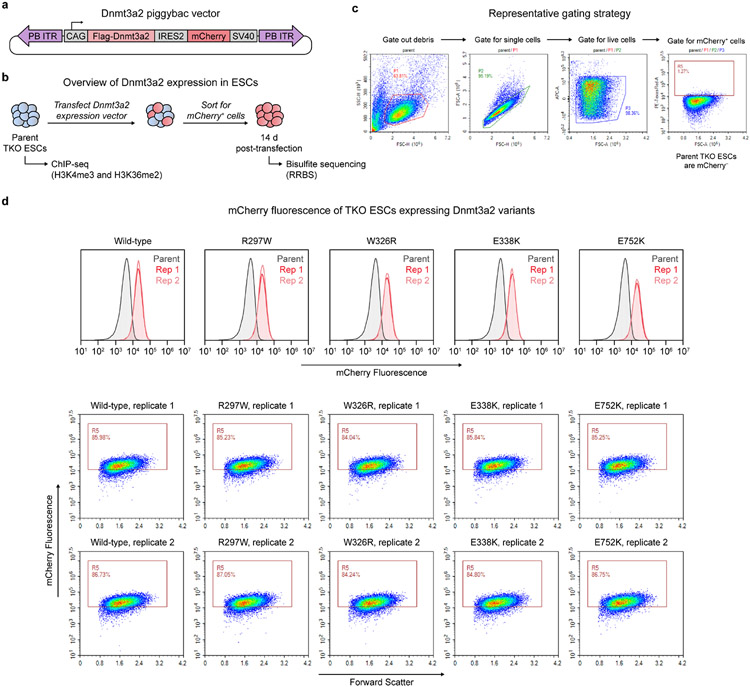
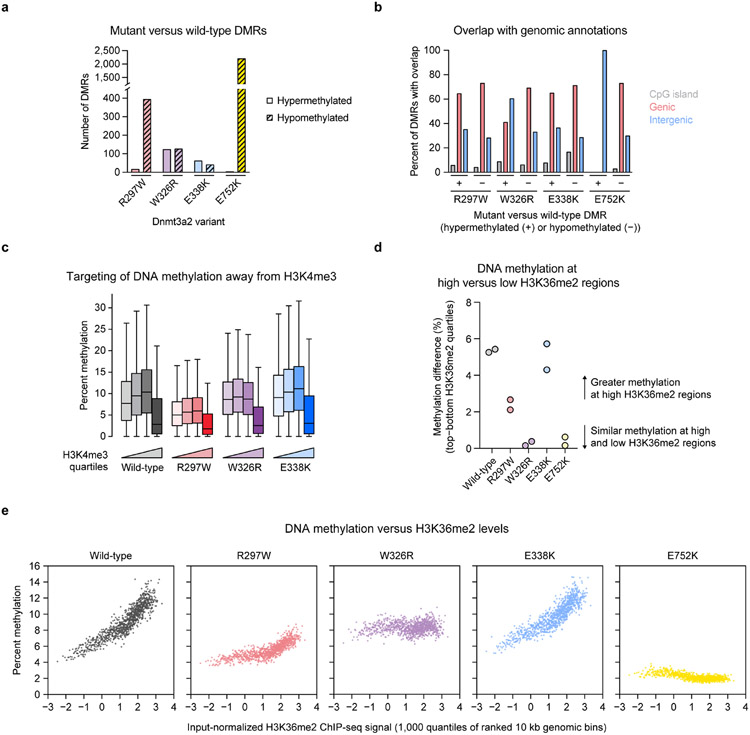
We next examined whether these mutations affect de novo DNA methylation in cells. We ectopically expressed mouse Dnmt3a2 variants in Dnmt1/Dnmt3a/Dnmt3b-triple knockout (TKO) mouse embryonic stem cells (ESCs)44 (Extended Data Fig. 8a-d) and conducted reduced representation bisulfite sequencing (RRBS)45 to measure DNA methylation. These cells are severely hypomethylated at baseline and rapidly lose methylation due to a lack of Dnmt1 maintenance, enabling us to evaluate methylation with high stringency. Consistent with in vitro results, Dnmt3a2 R297W (human R301W) produced less CpG methylation genome-wide than wild-type (Fig. 5h), while nevertheless retaining partial activity compared to catalytically dead E752K mutant (human E756K). As expected, Dnmt3a2 R297W produced predominately hypomethylated differentially methylated regions (DMRs) (Extended Data Fig. 9a). By contrast, both W326R and E338K mutants (human W330R and E342K, respectively) produced similar amounts of methylation compared to wild-type (Fig. 5h), though higher replicate variability was observed for the E338K mutant. Although similar numbers of hypo- and hypermethylated DMRs were called for the W326R mutant, they differed in genomic localization, consistent with mistargeting5,46 (Extended Data Fig. 9b).
To investigate whether these mutations affect histone targeting in cells, we mapped H3K4me3 and H3K36me2 in parental TKO ESCs by ChIP-seq. All active variants displayed a sharp reduction in methylation at regions with high H3K4me3 levels, as expected (Extended Data Fig. 9c). Dnmt3a2 W326R was equally able to methylate regions regardless of their H3K36me2 levels, and produced higher methylation at low H3K36me2 regions than the other variants (Fig. 5i and Extended Data Fig. 9d,e). This is consistent with mistargeting and aberrant spreading of methylation5,46, as well as the biochemical hyperactivity of DNMT3A2 W330R (Fig. 5a). By contrast, the wild-type, R297W, and E338K variants all displayed a positive correlation between DNA methylation and H3K36me2 levels (Fig. 5i), confirming that neither R297W nor E338K abrogate the PWWP domain’s canonical histone reader function. Therefore, our biochemical and genomic results together show that DNA binding is distinct from the PWWP domain’s canonical H3K36me2 reader function and plays an important role in cellular methylation (Fig. 6).
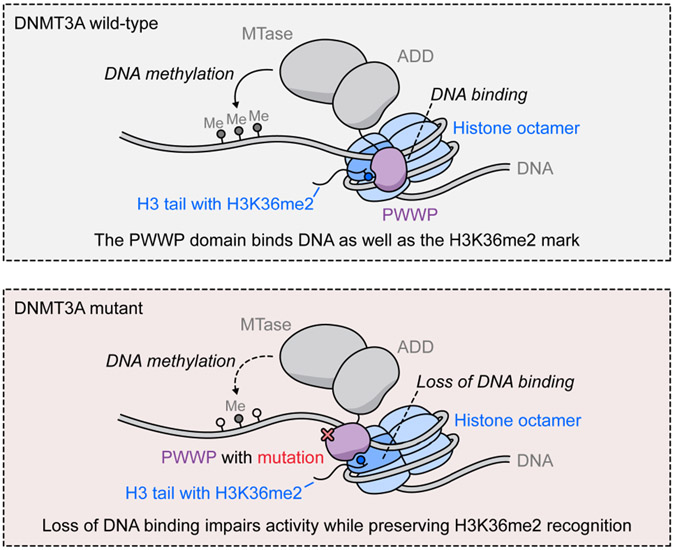
Fig. 6 ∣. PWWP domain DNA binding is required for full activity.
Model showing the role of PWWP domain DNA binding in DNMT3A methylation. Top, wild-type PWWP domain binds DNA in addition to its canonical role as a H3K36me2 reader. Bottom, mutations in the PWWP domain that disrupt DNA binding, such as R301W, lead to impaired methylation while preserving H3K36me2 targeting.
Discussion
The regulation of DNA methylation is of paramount importance to human health. Here, we innovate base editor screening to interrogate DNMT3A within its endogenous cellular ecosystem, identifying mutations impacting the diverse repertoire of functions mediated by its three domains. For instance, we show that the ADD-MTase interface is important for full allosteric stimulation. Additionally, like the recent large-scale DNMT3A mutational study19, we find an abundance of loss-of-function mutations within the MTase and PWWP domains. This prior study showed that destabilization was a common loss-of-function mechanism19, and indeed, we find that many of our PWWP domain hits also cause instability.
A key finding of our study is that PWWP domain DNA binding is required for optimal DNMT3A function. PWWP domains are known to simultaneously bind methylated H3K36 and nucleosomal DNA42. With DNMT3A, this DNA interaction causes substrate inhibition41 and promotes oligonucleosome binding and heterochromatic localization40. However, a direct role in promoting catalytic activity has not previously been shown. We demonstrate that the R301W mutation impairs DNA binding and thereby causes a loss of activity (Fig. 6). Although we cannot rule out the possibility of additional effects, this phenomenon is recapitulated by two additional mutations, including the K299N AML mutation, demonstrating its generality and potential clinical relevance. By contrast, increased DNA binding of the E342K mutant hyperactivates DNMT3A and phenocopies H3K36me2 stimulation in vitro. Given the physical proximity of H3K36 and DNA, this could suggest that H3K36me2 stimulation occurs by promoting DNA binding. These findings implicate the PWWP domain in direct allosteric regulation of MTase activity. Future insight into how the PWWP domain interacts with the rest of DNMT3A will be essential to address this, and may reveal novel therapeutic opportunities.
Recent work has established the importance of the PWWP domain in mediating crosstalk between DNA methylation and histone modifications. PWWP mutations disrupting recognition of methylated H3K36, such as D329A and W330R, cause DNA methylation to invade Polycomb-marked regions5,46-48 through a DNMT3A1 N-terminus-specific interaction with the Polycomb H2AK119ub mark46,49. Interestingly, the N-terminus has also been ascribed a DNA binding role43, highlighting how the N-terminus and PWWP domain’s roles overlap. Critically, we show that even in the absence of the N-terminal interaction, the W330R HESJAS mutation still promotes DNA binding and affects DNMT3A2’s catalytic behavior. Thus, a key question raised by our study is whether and how DNA binding—or other unknown interactions mediated by the PWWP domain—affect this complex crosstalk.
More broadly, our work highlights important considerations for future base editor scanning experiments. We show how bystander mutations and variable editing efficiency can complicate screen results, underscoring the need for rigorous validation. Notably, however, these concerns may be mitigated by recent advances integrating target sequences into sgRNA vectors to enable simultaneous readout of editing outcomes31,33. Additionally, recent work has applied adenine base editors and Cas9 variants with expanded targeting to base editor screening31-33. These tools will greatly increase coverage of the total mutational space, further improving the utility of this approach.
Importantly, the fluorescent reporter adapted here has been used to characterize writer enzymes of a variety of repressive chromatin modifications34, and more recently has been extended to a diverse set of repressive and activating transcriptional effector domains50, demonstrating its versatility. Although artificial recruitment assays cannot reveal certain aspects of protein function such as genomic targeting, our strategy nevertheless enables systematic interrogation of sequence-activity relationships and can yield biochemical insight for proteins that are challenging to study in vitro. At the same time, it is not limited by target essentiality in cells. We anticipate future studies employing base editor scanning will yield novel insights into chromatin regulators and the complex, intertwined mechanisms defining their activities in cells.
Methods
Cell culture
The following cell lines were used: K562 (ATCC), 293FT (Thermo Fisher Scientific), HEK293T (a gift from B.E. Bernstein, Massachusetts General Hospital), Dnmt1/Dnmt3a/Dnmt3b-triple knockout (TKO) mouse embryonic stem cells (ESCs) (a gift from A. Meissner, Max Planck Institute), and CD-1 mouse embryonic fibroblasts (MEFS) (Lonza, Cat#M-FB-481). All cell lines were cultured in a humidified 5% CO2 incubator at 37 °C and were tested for mycoplasma. All media were supplemented with 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco). Fetal bovine serum (FBS) was obtained from Peak Serum except for ESC medium. K562s were cultured in RPMI-1640 (Gibco) with 10% FBS. HEK293Ts were cultured in DMEM (Gibco) with 10% FBS. 293FTs were cultured in DMEM with 10% FBS, 2 mM GlutaMAX (Gibco), and 1× MEM non-essential amino acids (Gibco). ESCs were cultured in KnockOut DMEM (Gibco) with 15% FBS (Gibco, heat-inactivated), 2 mM GlutaMAX, 1× MEM non-essential amino acids, 103 U ml−1 ESGRO leukemia inhibitory factor (Millipore), and 55 μM 2-mercaptoethanol (Gibco). ESCs were cultured on a layer of MEF feeders (inactivated with 10 μg ml−1 mitomycin C (Sigma-Aldrich)) plated on 0.2% gelatin-coated vessels, and medium was changed daily.
Lentiviral transduction
To produce lentivirus, transfer plasmid was co-transfected with GAG/POL and VSVG plasmids into HEK293Ts or 293FTs using FuGENE HD (Promega, 3.33:1 FuGENE:DNA) and the medium was replaced 6–8 h after transfection. 48–60 h later, the medium was collected, passed through a 0.45 μm filter, snap-frozen, and stored at −80 °C. To generate reporter cell lines and introduce sgRNAs, K562s were transduced by spinfection (1,800g, 90 min, 37 °C) with 12 μg ml−1 polybrene (Santa Cruz Biotechnology) and the appropriate lentivirus. Transduced cells were selected using 2 μg ml−1 puromycin (Gibco) or FACS. A clonal methylation reporter cell line was first derived and used in all subsequent sgRNA transductions.
Plasmid construction
sgRNAs were ordered as synthetic oligonucleotides (Sigma-Aldrich), annealed, and ligated into the appropriate vector: lentiCRISPR v2 (Cas9 knockout), a gift from F. Zhang (Addgene #52961); or pRDA_254 (base editing), which expresses BE3.9max and is identical to pRDA_25629 (available at Addgene #158581) except lacking the guide capture sequence. Other plasmids were cloned by Gibson Assembly using NEBuilder HiFi (New England Biolabs). Cloning strains used were NEB Stable (lentiviral and PiggyBac vectors) and NEB 5-alpha (other plasmids) (New England Biolabs). For base editor cloning, bacterial cultures were grown at 30 °C. Final constructs were validated by Sanger sequencing (Quintara Biosciences). Plasmids and oligonucleotides are provided in Supplementary Tables 1-3. Key plasmids, including methylation reporter vectors, have been deposited to Addgene #186966–186970.
All DNMT3A expression plasmids encoded isoform 2 (human, residues 224–912; mouse, residues 220–908). Coding sequences of human DNMT3A and DNMT3L (mammalian expression) were amplified from pcDNA3/Myc-DNMT3A and pcDNA3/Myc-DNMT3L, respectively, gifts from A. Riggs (Addgene #35521, 35523). The methylation reporter was derived from PhiC31-Neo-ins-5xTetO-pEF-H2B-Citrine-ins and pEX1-pEF-H2B-mCherry-T2A-rTetR-KRAB, gifts from M. Elowitz (Addgene #78099, 78348). Reporter components (5xTetO-pEF-H2B-Citrine-SV40, pEF-H2B-mCherry-T2A-rTetR-DNMT3L-SV40) were each cloned into LT3REVIR, a gift from J. Zuber (Addgene #111176). For stability reporter constructs, DNMT3A2 was cloned into a modified Cilantro2, a gift from B. Ebert (Addgene #74450). For transfection constructs, FLAG-DNMT3A2 was cloned into pcDNA3. For PWWP domain bacterial expression, DNMT3A residues 278–427 were cloned into pET28b with a TEV protease-cleavable N-terminal His6-MBP tag. For PiggyBac constructs, the mouse Dnmt3a2 coding sequence (NM_007872.4) was synthesized in two pieces by Twist Bioscience and cloned as a CAG-Flag-Dnmt3a2-Ires2-mCherry-SV40 cassette into pSLQ2817, a gift from S. Qi (Addgene #84239).
Reporter silencing assays
Reporter cells were plated in triplicate at 1 × 105 cells ml−1 in medium with or without 100 ng ml−1 dox (Sigma-Aldrich). Every 3 d, samples were removed for analysis and cells were passaged by dilution into fresh medium. For washout, cells were washed once with phosphate-buffered saline (PBS) and resuspended in medium without dox. For flow cytometry, Helix NP NIR viability dye (BioLegend) was added and data were collected on a NovoCyte 3000RYB and analyzed using NovoExpress (ACEA). Gates were set based on reference reporter cells cultured in parallel without dox (gating scheme in Extended Data Fig. 1b). Assays were performed twice using independently transduced cells.
Genotyping
Genomic DNA was purified using the QIAamp DNA Blood Mini or UCP DNA Micro kits (Qiagen), unless otherwise specified. 100 ng DNA was subjected to a first round of PCR (25–27 cycles, Q5 hot start high-fidelity DNA polymerase, New England Biolabs) to amplify the locus of interest and attach common overhangs. PCR products were purified by 1.5× SPRI clean-up (Mag-Bind Total Pure NGS beads, Omega Bio-Tek), and 5 ng of each was amplified in a second round of PCR (8 cycles) to attach barcoded adapters. Primer sequences are provided in Supplementary Table 4. Final amplicons were purified by gel extraction (Zymo) and sequenced on an Illumina MiSeq. Data were processed using CRISPResso251 using the following parameters: --quantification_window_size 20 --quantification_window_center −10 --plot_window_size 20 -- exclude_bp_from_left 0 --exclude_bp_from_right 0 --min_average_read_quality 30 -- n_processes 12 --base_editor_output. Custom python scripts were used for downstream analysis.
Reporter bisulfite sequencing
Reporter cells were treated with 100 ng ml−1 dox starting at staggered timepoints for simultaneous harvest. Genomic DNA was extracted and subjected to bisulfite conversion using the EZ DNA Methylation kit (Zymo). Sequencing libraries were prepared as above, except EpiMark hot start Taq DNA polymerase (New England Biolabs) was used. Primer sequences are provided in Supplementary Table 4. Data were processed using CRISPResso251 (default settings). A custom python script was used to compute the percent methylation at each position containing a bisulfite-convertible base in the reference sequence (C or G depending on primer design, elsewhere set to zero), defined as the following read count ratios: C/(C+T)×100 or G/(G+A)×100.
Immunoblotting
Cells were washed three times with cold PBS and lysed on ice in RIPA buffer (Boston BioProducts) with 1× Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific), 1 mM phenylmethylsulfonylfluoride (PMSF), and 2 mM EDTA (Thermo Fisher Scientific) (50 μl per million cells). Then, an equivalent volume of wash buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 2 mM MgCl2, 1× Halt Protease Inhibitor Cocktail, 1 mM PMSF) with 1:500 benzonase (Sigma-Aldrich) was added, and samples were rotated at room temperature for 20 min. Lysate was clarified by centrifugation and total protein concentration was measured with the BCA Protein Assay (Thermo Fisher Scientific). Samples were electrophoresed and transferred to an Immobilon-P membrane (Millipore) for endogenous DNMT3A blots or a 0.45 μm nitrocellulose membrane (Bio-Rad) otherwise. Membranes were blocked with tris-buffered saline tween (TBST) with 5% bovine serum albumin (BSA) and incubated with primary antibody: DNMT3A (Cell Signaling Technology, Cat#32578, D2H4B, 1:5,000), DNMT3B (Cell Signaling Technology, Cat#67259, D7O7O, 1:1,000), FLAG (Sigma-Aldrich, Cat#F1804, M2, 1:2,000), GAPDH (Santa Cruz Biotechnology, Cat#sc-47724, 0411, 1:2,000). Membranes were washed 3× with TBST and incubated with secondary antibody: anti-rabbit IgG HRP conjugate (Promega, Cat#W4011, 1:100,000 for DNMT3A, 1:20,000 for DNMT3B), anti-mouse IgG HRP conjugate (Promega, Cat#W4021, 1:40,000 for FLAG, 1:100,000 for GAPDH). Following 3× washes with TBST, immunoblots were visualized using SuperSignal West Femto (DNMT3A) or SuperSignal West Pico PLUS (others) chemiluminescent substrates (Thermo Fisher Scientific).
Base editor scanning
The sgRNA library was designed as described previously29 to include all sgRNAs (NGG protospacer-adjacent motif) targeting exonic and flanking intronic regions of DNMT3A isoform 2 (NM_153759.3, ENST00000380746), excluding promiscuous sgRNAs and those with TTTT sequences. Negative (nontargeting, intergenic) and positive (essential splice site) controls were included. The library was synthesized as an oligonucleotide pool (Twist Biosciences) and cloned into pRDA_254 following published workflows24,52. Lentivirus was produced and titered by measuring cell counts after transduction and puromycin selection. 21 × 106 reporter cells were transduced with library lentivirus at a multiplicity of infection <0.3 and selected with puromycin for 7 d. Cells were then expanded and split into three replicate subcultures and treated with 100 ng ml−1 dox. 3 d, 6 d, and 9 d after starting dox treatment, cells were sorted on a FACSAria II (BD), collecting citrine+, citrine−, and unsorted (all mCherry+) cells. Genomic DNA was isolated using the QIAamp DNA Blood Mini kit and sgRNA sequences were amplified using barcoded primers, purified by gel extraction, and sequenced on an Illumina MiSeq as previously described22,24. At all steps, sufficient coverage of the library was maintained in accordance with published recommendations52.
Data analysis was performed using Python (v3.7.3) with Biopython (v1.76), Pandas (v1.0.1), NumPy (v1.18.1), and SciPy (v1.2.0). sgRNA scores were calculated as previously described22,24. Briefly, sequencing reads matching each sgRNA were quantified as reads per million, increased by a pseudocount of 1, log2-transformed, normalized to the plasmid library, and replicate-averaged. One sgRNA was excluded because zero reads were detected at day 0. Citrine+ and citrine− abundances were normalized to matched-timepoint unsorted abundances. The mean value for intergenic controls was subtracted to calculate the final sgRNA score. sgRNAs with scores >2 SD above or below the mean of intergenic negative controls were considered “enriched” or “depleted,” respectively. sgRNAs targeting DNMT3A were classified based on expected editing outcome, assuming any C within the editing window (protospacer +4 to +8) is converted to T, regardless of sequence context. sgRNAs were placed in one of 7 mutually exclusive classes: in order of assignment priority, (1) nonsense; (2) splice site; (3) missense; (4) silent; (5) exon, no predicted edits (no Cs); (6) intron/UTR; (7) intron/UTR, no predicted edits. Library sgRNA annotations and base editor scanning data are provided in Supplementary Data 1-3.
PWES analysis
The PWES for a given pair of sgRNAs and was calculated as previously described22 using the following formula:
where is the sum of the day 9 citrine+ sgRNA scores for and , is the Euclidean distance between their targeted residues, and , , . Only missense sgRNAs editing residues resolved in the structures of active (PDB: 4U7T) and autoinhibited (PDB: 4U7P) DNMT3A were considered. sgRNAs predicted to mutate two residues were assigned to the even-numbered residue. Hierarchical clustering of the active conformation scores (PWES4U7T) was performed as described previously22. ΔPWES was defined as ΔPWES = ∣PWES4U7T∣ − ∣PWES4U7P∣. For each sgRNA, this metric was summed over all other sgRNAs to calculate the summed ΔPWES (equivalent to a column sum of the ΔPWES matrix, excluding the diagonal). See Supplementary Data 4.
Conservation analysis
The protein sequence of full-length human DNMT3A (uniprot Q9Y6K1) was used as a query for five iterations of jackhmmer53 (v3.3) to search in the uniref database for sequences with homology. hmm searches were performed within these sequences using the hmm profiles from pfam for the PWWP domain (v17), ADD domain (v1) and MTase domain (v17), identifying subsets containing each domain. Subsets were intersected to generate a list of sequences containing homology to all three DNMT3A domains. Then, the same PWWP hmm profile was used to search in uniref to gather sequences from a wide diversity of proteins containing PWWP domains. Clustal omega54 (v1.2.0) was used to align both sets of sequences, and the relative level of conservation at each position in the DNMT3A PWWP domain was assessed from each alignment using Python with Biopython, Pandas, NumPy, and Alignment object from EVcouplings (v0.0.5). Raw conservation scores are provided in Supplementary Data 7.
Stability reporter assay
Wild-type K562 cells were transduced with the appropriate lentivirus, selected with puromycin, and analyzed by flow cytometry. After gating for eGFP+ mCherry+ cells (gating scheme in Supplementary Fig. 1), the ratio of eGFP to mCherry geometric mean fluorescences was calculated. Mutant ratios were normalized to that of wild-type DNMT3A2 measured in parallel. Each mutant was assessed by two measurements of each of two independently generated cell lines.
Protein expression and purification
Full-length DNMT3A2 was expressed recombinantly in Rosetta2(DE3)pLysS cells (Novagen). Freshly transformed cells were grown in LB with 50 μg ml−1 kanamycin and 50 μg ml−1 chloramphenicol at 37 °C to an optical density at 600 nm of 0.6–0.8. Cells were cooled on ice, induced with 1 mM isopropyl-β-D-thiogalactoside (IPTG) (Research Products International) at 16 °C overnight, and harvested. Purification was performed according to published protocol11 with some modifications. Cells were resuspended in lysis buffer (base buffer (50 mM Tris-HCl, pH 8.0 cold, 300 mM NaCl, 1 mM TCEP) with 10 mM imidazole and 0.1% Triton X-100) and sonicated. Lysate was clarified by centrifugation, further diluted with lysis buffer to a final volume of 175 ml per liter expression culture, and incubated with His60 Ni Superflow affinity resin (Takara). Resin was washed with base buffer containing a stepwise gradient of 20–100 mM imidazole, followed by elution using base buffer with 200 mM imidazole. Eluate was exchanged into storage buffer (base buffer with no imidazole) using an Econo-Pac 10DG desalting column (Bio-Rad) and then concentrated to 0.5–1 mg ml−1 using Amicon Ultra 30 kDa centrifugal filters (Millipore) with resuspension of the sample in between 2 min spins at 2,500g. Frequent resuspension was necessary to prevent over-concentration, which resulted in reduced activity and/or precipitation of the protein.
DNMT3A PWWP domain was expressed in BL21(DE3) cells (New England Biolabs) as above, except cells were grown in TB with 50 μg ml−1 kanamycin and induced with 0.2 mM IPTG. Cells were resuspended and sonicated in lysis buffer (PWWP base buffer (20 mM Tris-HCl, pH 7.5 cold, 300 mM NaCl, 5% glycerol) with 10 mM imidazole and cOmplete, EDTA-free protease inhibitor (Roche)). Clarified lysate was subjected to Ni affinity purification, eluting with PWWP base buffer with 250 mM imidazole. The eluate was exchanged into cleavage buffer (50 mM Tris-HCl, pH 7.5 cold, 75 mM NaCl, 5% glycerol, 1 mM dithiothreitol (DTT), 0.5 mM EDTA) and incubated with TEV protease overnight at 4 °C. The cleaved His6-MBP tag was removed by incubation with Ni resin. The cleaved PWWP domain was subsequently purified using a HiTrap Heparin HP (Cytiva) to remove nucleic acid contamination (20 mM sodium phosphate, pH 7.5, 5% glycerol, 0–1.5 M NaCl gradient). Finally, protein was polished on a Superdex 200 Increase 10/300 GL column (Cytiva), eluting in storage buffer (20 mM Tris-HCl, pH 7.5 cold, 150 mM NaCl, 5% glycerol). We note that the S337L mutant was not purified by Heparin column.
Purified proteins were quantified by absorbance at 280 nm using calculated extinction coefficients (Expasy ProtParam). Proteins were flash frozen in liquid N2 and stored at −80 °C. For subsequent assays, thawed proteins were normalized to a common concentration in storage buffer before dilution into assay buffer.
Activity assays
DNMT3A activity was measured using a previously described protocol11 with some modifications. Reactions were conducted in triplicate in 50 μl volume using 10 μM base pairs poly(dI-dC) (Sigma-Aldrich); 0.5 μM adenosyl-L-methionine, S-[methyl-3H] (3H-SAM) (PerkinElmer, 16.5–18.0 Ci mmol−1); and 0.1 μM purified DNMT3A2 in assay buffer (50 mM sodium phosphate, pH 7.5, 20 mM NaCl, 1 mM TCEP-HCl, 0.1 mg ml−1 BSA, 0.01% Triton X-100). Reactions were incubated at room temperature for 2 h and terminated with excess nonradioactive SAM (New England Biolabs) in 300 μl assay buffer. 40 μl DEAE sepharose fast flow resin (Cytiva) was added, and samples were rotated at room temperature. Resin was recovered using a Pierce spin column (Thermo Fisher Scientific), washed with 2 × 250 μl wash buffer (50 mM sodium phosphate, pH 7.5, 20 mM NaCl, 1 mM TCEP), resuspended in 200 μl H2O, and mixed with 4 ml Ultima Gold scintillation cocktail (PerkinElmer). Each sample was counted for 5 min on a Beckman LS 6000SC liquid scintillation counter. Raw measurements were corrected for background signal by subtracting the average of three mock (no enzyme) reactions processed in parallel.
For histone stimulation experiments, 1 μM peptide was added prior to enzyme. The following peptides were used: H3K4me0 (ARTKQTARKSTG-NH2, Biomatik), H3K36me0 and H3K36me2 (histone H3 (21–44)-GK(biotin), Anaspec, Cat#AS-64440 and AS-64442).
For the NaCl titration experiment, a modified buffer was used during the reaction incubation (20 mM HEPES, pH 7.5, 1 mM TCEP-HCl, 0.1 mg ml−1 BSA, 0.01% Triton X-100, supplemented with 50 mM, 100 mM, or 150 mM NaCl) to minimize background ionic strength. After terminating reactions, NaCl was added to equalize concentration across samples before adding DEAE resin. Termination and subsequent steps were conducted using unmodified buffer.
Differential scanning fluorimetry
5 μM purified PWWP domain was incubated in triplicate with 5× SYPRO Orange (Thermo Fisher Scientific) in assay buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5% glycerol). Samples were heated from 10 °C to 95 °C (10 s at each 0.5 °C step) using a CFX Connect qPCR with CFX Manager software (Bio-Rad). Melting temperatures were calculated with DSFWorld55 (by sigmoid fitting, model 1).
Electrophoretic mobility shift assay
Purified PWWP domain at varying concentrations was incubated with 50 nM Cy3-labeled 30 bp DNA probe (Supplementary Table 5) in assay buffer (20 mM HEPES, pH 7.5, 1 mM EDTA, 1 mg ml−1 BSA, 8% glycerol) for 20 min at room temperature. 5 μl of each reaction was subjected to electrophoresis (≤20 V cm−1, 4 °C) using a 6% acrylamide DNA retardation gel (Thermo Fisher Scientific). Gels were imaged using a Sapphire Biomolecular Imager with Sapphire Capture Software (Azure Biosystems).
Fluorescence polarization assay
Purified PWWP domain was diluted to 8 μM in assay buffer (20 mM HEPES, pH 7.5, 1 mM EDTA, 0.5 mg ml−1 BSA, 8% glycerol) containing 20 nM Cy3-labeled 30 bp DNA probe (Supplementary Table 5). This was aliquoted in triplicate into a black 384-well plate (Corning), followed by 2-fold serial dilution in assay buffer containing 20 nM probe (final volume, 20 μl). The plate was incubated at room temperature for 1 h and read (1,700 ms integration) using a SpectraMax i3x with a rhodamine fluorescence polarization cartridge and SoftMax Pro software (Molecular Devices). Wells containing only assay buffer were used for background subtraction. The G-factor was adjusted to set the polarization of assay buffer and 20 nM probe only to a reference value of 27 mP. Curves were fit to the sigmoidal, 4PL model in GraphPad Prism.
Peptide pulldown assay
1.5 nmol purified PWWP domain was incubated with 80 pmol biotinylated H3K36me0 or H3K36me2 peptide (same as in histone stimulation assays) in interaction buffer5 (50 mM Tris-HCl, pH 7.5 cold, 100 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, fresh 0.5 mM DTT, fresh 0.2 mM PMSF, fresh 1× cOmplete EDTA-free protease inhibitor) overnight at 4 °C with rotation. 10 μl Dynabeads MyOne T1 streptavidin beads were then added, followed by rotation at 4 °C for 4 h. Beads were washed 5× with interaction buffer and boiled in loading buffer (95 °C, 5 min, 1000 rpm shaking). Eluted proteins were resolved by SDS-PAGE using a 10% tricine gel (Novex) and visualized by silver staining (Pierce).
Sequential salt extraction assay
For each condition, 3.5 × 106 HEK293Ts were plated in a 10 cm dish and transfected the following day with 3 μg FLAG-DNMT3A2 expression vector using 20 μl FuGENE HD. Two days post-transfection, cells were trypsinized and washed twice with cold PBS. 2 × 106 cells were lysed with benzonase as described above to measure total exogenous expression, while 8 × 106 cells were subjected to sequential salt extraction according to published protocol56. Briefly, cells were resuspended in 1 ml modified buffer A (25 mM HEPES, pH 7.6, 25 mM KCl, 5 mM MgCl2, 0.05 mM EDTA, 0.1% NP-40 substitute, 10% glycerol), rotated for 10 min at 4 °C, and centrifuged (6,000g, 5 min, 4 °C) to isolate nuclei. Then, chromatin was extracted with increasing NaCl. For each extraction, nuclear material was resuspended in 200 μl mRIPA (50 mM Tris-HCl, pH 8.0, 1% NP-40 substitute, 0.25% sodium deoxycholate (NaDOC)) with the appropriate NaCl concentration, incubated on ice for 15 min, and centrifuged (6,500g, 3 min, 4 °C) to collect supernatant. Equal volumes of each extraction fraction were subjected to SDS-PAGE and immunoblotting.
ChIP-seq
MEF-depleted parent TKO ESCs were crosslinked with 1% formaldehyde (Sigma-Aldrich), quenching with 125 mM glycine. Cells were lysed, sonicated (Branson sonifier, 0.7 s on, 1.3 s off, 5 min total on, 50% amplitude), and clarified by centrifugation. Lysates were rotated overnight at 4 °C with the following antibodies: anti-H3K4me3 (Millipore, Cat#07-473, Lot#3394198, 1:240, 2.5 μg antibody for 4 × 106 cells), anti-H3K36me2 (Cell Signaling Technologies, Cat#2901, Lot#5, 1:150, 1 μg antibody for 10 × 106 cells). Dynabeads Protein G (Thermo Fisher Scientific) were added, and following incubation, beads were isolated and washed. Immunoprecipitated chromatin was eluted, treated with RNase (37 °C, 30 min) and Proteinase K (63 °C, overnight), and purified by 2× SPRI. 2.5 ng each sample was subjected to end-repair (End-It DNA End-Repair kit, Lucigen), A-tailing (Klenow fragment, 3’-5’ exo–, New England Biolabs), ligation to barcoded adapters (KAPA), and PCR library amplification (NEB Ultra 2× master mix), followed by SPRI size-selection and purification. Libraries were sequenced using a NovaSeq SP kit (Illumina).
ChIP-seq data analysis
Reads were aligned to the mm10 genome (UCSC) using bwa-backtrack (v0.7.17-r1188). Samtools (v1.10) was used to convert output to bam format, followed by deduplication using Picard (v2.26.9) (https://github.com/broadinstitute/picard). DeepTools bamCompare57 (v3.5.1) was used to create bigWig files for input-normalized ChIP signal (parameters: --extendReads --operation log2 --skipZeroOverZero -bs 200 --smoothLength 1000 --scaleFactorsMethod SES). See Supplementary Table 6 for coverage statistics.
Generation of Dnmt3a2-expressing TKO ESCs
3.6 × 104 TKO ESCs were plated per well in a 12-well plate and transfected the following day with a 1:2 molar ratio of PBase:vector plasmids (1.1 μg total DNA, medium without antibiotics) using 3.83 μl FuGENE HD. PBase was a gift from A. Meissner (Max Planck Institute). Two rounds of FACS were used to isolate successfully transposed mCherry+ cells. 14 d after transfection, cells were MEF-depleted and harvested.
RRBS
RRBS was performed according to a published enhanced protocol45 with modifications. Genomic DNA was isolated using the QIAamp DNA Blood Mini kit, and 80 ng was digested overnight at 37 °C with 150 units MspI (New England Biolabs). 0.5% unmethylated lambda phage DNA (Promega) was spiked in before digestion for assessment of bisulfite conversion efficiency. The digest was purified by phenol/chloroform extraction, subjected to end-repair and A-tailing, and ligated to barcoded adapters (xGen Methyl UDI-UMI Adapters, Integrated DNA Technologies) using concentrated T4 ligase (New England Biolabs). Adapter-ligated DNA was purified by 1× SPRI, size-selected by gel extraction, bisulfite-converted (EZ DNA Methylation kit, conversion with 55 cycles of 95 °C for 30 s, 50 °C for 15 min), and amplified using EpiMark hot start Taq DNA polymerase. Libraries were sequenced using a NovaSeq SP kit.
RRBS data analysis
Sequencing reads were trimmed using Trim Galore (v0.6.7) (https://github.com/FelixKrueger/TrimGalore) with parameters --illumina --rrbs --paired --length 21. Reads 1 and 2 were swapped for trimming because the adapters used flip insert strandedness. Trimmed reads were aligned to the mm10 genome using Bismark58 (v0.23.1) (bowtie2 default). MethylDackel (v0.6.1) (https://github.com/dpryan79/MethylDackel) was used to extract methylation, retaining only CpGs with at least 5× coverage (parameters: -d 5 --mergeContext -- keepDupes). These were further filtered for CpGs meeting coverage across all samples using BedTools2 intersect59. The resulting bedGraph files were converted to bigWig format using bedGraphToBigWig (UCSC) and then processed using DeepTools multiBigwigSummary57 (v3.5.1) along with ChIP-seq bigWigs. A custom python script was used to perform additional analysis. Differentially methylated regions were called using Defiant60 (v1.1.9) with parameters - c 5 -p 0.05 -s 5 -CpN 3 -d 2 -P 4 -S 2 -G 5000. Intersections with genomic annotations (UCSC) were performed using BedTools2 intersect59. See Supplementary Table 7 for coverage statistics.
Visualization
Data were visualized using Adobe Illustrator CS6, NovoExpress, GraphPad Prism and Microsoft Excel, Seaborn (v0.11.2), and Matplotlib (v3.1.3). Protein structures were visualized using PyMOL (Schrödinger). Color inversions and brightness adjustments of gels and blots were applied to the entire image using Adobe Photoshop CS6.
Statistics and replication
All statistical tests were two-sided, and are described in the figure legends. Tests were performed using SciPy or GraphPad Prism. Spearman correlations were calculated using Pandas. Unless otherwise noted, plots with error bars depict the mean ± SD of n = 3 replicates. Error bars are not shown in scatterplots where smaller than the data points themselves. Methylation reporter assays and biochemical experiments were performed in two independent trials with similar results. Experiments involving next-generation sequencing were conducted once.
Extended Data
Extended Data Fig. 1 ∣. Reporter silencing depends on DNMT3A and is concomitant with DNA methylation.
a, Schematic of lentiviral methylation reporter vectors. LTR, lentiviral long terminal repeat; TetO, tetracycline operator; SV40, simian virus 40 poly(A) sequence; rTetR, reverse Tet repressor. b, Representative gating scheme for flow cytometric analysis of reporter silencing assays. Helix NP NIR was used as a viability dye. Citrine fluorescence and mCherry fluorescence were monitored on the FITC and PE-Texas Red channels, respectively. c, Reporter methylation levels after varying duration of dox treatment measured by targeted bisulfite sequencing. In each plot, lines represent the percent cytosine methylation at each position (non-cytosine positions are set to 0). CpG sites are highlighted by dots. A schematic of the reporter is shown below, indicating the location of each sequenced region. This experiment was performed once (n = 1). d, Full timecourse for DNMT3-knockout silencing experiment shown in Fig. 1f (top), with representative histograms of citrine fluorescence from each day (bottom). e, Full timecourse for sgW698 silencing experiment shown in Fig. 1h (top), with representative histograms of citrine fluorescence from each day (bottom). f, Deep sequencing of cells edited with sgW698 after 9 d of dox treatment, either sorted for citrine+ cells (yellow) or unsorted (gray). Plot shows the percentage of aligned reads with C to T base edits at each indicated protospacer position, or the percentage of aligned reads with indels. Sequencing was performed once (n = 1). For d and e, data and error bars (where larger than data point) are mean ± SD of n = 3 replicates, and results are representative of two independent experiments.
Extended Data Fig. 2 ∣. Analysis and validation of DNMT3A base editor scanning results.
a–d, DNMT3A base editor scanning results: (a) heatmap depicting Spearman correlations between sgRNA scores at different timepoints and for citrine+ or citrine− cells, (b) correlation between day 9 citrine+ sgRNA scores and either day 3 citrine− sgRNA scores (left) or day 6 citrine+ sgRNA scores (right), (c) day 9 citrine+ sgRNA scores for select versus all missense sgRNAs (active site, residues within 5 angstroms of zebularine or SAH (PDB: 5YX2); near 3A-3L interface, residues called by the InterfaceResidues.py script (https://pymolwiki.org/index.php/InterfaceResidues) as at the DNMT3A-DNMT3L interface (PDB: 5YX2) and those adjacent to these residues in the linear sequence), (d) comparison of day 9 citrine+ sgRNA scores for missense sgRNAs targeting annotated domains versus any non-domain region of DNMT3A. Data are the average of n = 3 replicates. For c and d, dotted lines indicate ±2 SD of intergenic control sgRNAs, and boxplot components are as follows: center line, median; box, interquartile range; whiskers, up to 1.5 × interquartile range per the Tukey method. e–g, Structural views of the DNMT3A MTase domain (PDB: 5YX2) highlighting residues targeted by several top enriched missense sgRNAs. Day 9 citrine+ sgRNA scores for the corresponding sgRNAs are printed below (where multiple sgRNAs target the same residue(s), the top sgRNA is shown). h, Full timecourse for the silencing experiment shown in Fig. 2e. Data are mean ± SD of n = 3 replicates, and results are representative of two independent experiments. i, Deep sequencing of cells edited with sgE756 after 9 d of dox treatment (left) or with sgG532 after 3 d of dox treatment (right), either unsorted or sorted as indicated. Plots show the percentage of aligned reads with C to T base edits at each indicated protospacer position, or the percentage of aligned reads with indels. Sequencing was performed once (n = 1).
Extended Data Fig. 3 ∣. Clonal analysis of base editing outcomes.
a, Barplots showing the frequencies of wild-type (blue) and base-edited (other colors as defined in the legend of each plot) alleles in clones derived from sgRNA-treated reporter cells (n = 24 clones for each sgRNA). Single cells were plated using FACS and expanded to derive clonal populations, followed by isolation of genomic DNA using QuickExtract DNA extraction solution (Lucigen). Library preparation, sequencing, and analysis were performed as for other genotyping experiments. In plots, each bar represents a clone, and theoretical allele frequencies are indicated by dotted lines (note that K562 is triploid for DNMT3A and therefore three alleles are expected for each clone). All alleles with less than 5% allele frequency were pooled and designated as “Other.” Alleles containing both missense and silent mutations were classified based on the missense mutation. Allele tables for each clone are shown in Supplementary Data 6. b, Summary of results in a showing the fractions of clones for each sgRNA that contain only wild-type or silent alleles (blue), only nonsynonymous edited alleles (red), or a combination of wild-type/silent and nonsynonymous edited alleles (blue/red checkered).
Extended Data Fig. 4 ∣. SDS-PAGE of purified proteins.
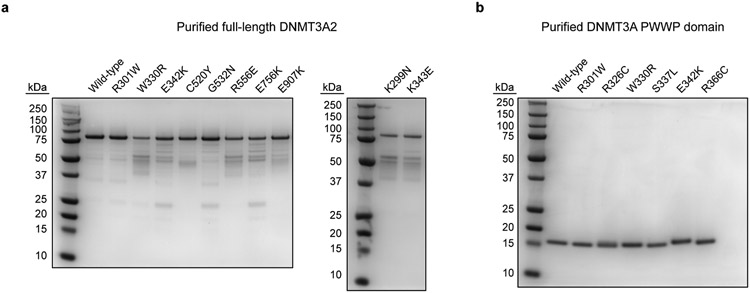
a–b, Purified (a) full-length DNMT3A2 (80 kDa, residues 224–912 with N-terminal 6×His tag) and (b) PWWP domains (17 kDa, residues 278–427, untagged). Proteins were electrophoresed on Novex 10% acrylamide tricine gels (Thermo Fisher Scientific) and visualized by Coomassie staining. 1 μg protein was loaded in each lane. Protein purifications were generally performed once, although wild-type DNMT3A2 was purified more than once and verified to have comparable activity across purifications. SDS-PAGE was performed twice independently for each purified protein with similar results, except for PWWP R326C, which was analyzed once.
Extended Data Fig. 5 ∣. Individual validation of sgRNA hits.
a, Citrine fluorescence of base-edited cells after 9 d of dox treatment for 17 sgRNAs targeting DNMT3A (red or light red). Histograms are representative of n = 3 replicates. Top and bottom plots show data from two independent experimental trials (independent transductions). The citrine fluorescence histogram of nontargeting sgLucA control cells treated with dox in parallel (gray) is overlaid in each plot. Control data shown are identical for samples analyzed in the same experiment. Data shown in Fig. 4d corresponds to trial 2 shown here. b, Next-generation sequencing analysis of base editing efficiency at each C within the target sites of the indicated sgRNAs. Allele tables are provided in Supplementary Data 5. c, Flow cytometric quantification of cells remaining citrine+ after 9 d of dox treatment. Data correspond to those shown in a (trial 2), and are mean ± SD of n = 3 replicates. P values were calculated through two-tailed unpaired t tests comparing each sgRNA to sgLucA control. d, Aggregate base editing outcomes for the 17 sgRNAs presented in a (includes PWWP, ADD, and MTase hit sgRNAs). The efficiency of base editing is plotted at each protospacer position for all sgRNAs containing a C at that position. Horizontal lines indicate the median at each position. The number of sgRNAs (n) with a C at each position is printed above the plot. Protospacer positions within the editing window (+4 to +8) are highlighted in red. The indel frequencies for all sgRNAs (n = 17) are shown to the right in dark gray. Genotyping was performed once for each sgRNA.
Extended Data Fig. 6 ∣. H3 peptide binding of purified PWWP domains.
Binding of purified PWWP domain variants to biotinylated H3K36me0 or H3K36me2 peptides (H3 residues 21–44). Bound proteins were captured by streptavidin pulldown, resolved by SDS-PAGE, and visualized by silver staining. This experiment was performed in two independent trials, and both are shown. Within each experimental trial, gels were electrophoresed and stained in parallel. A longer silver stain exposure was used for trial 1 than for trial 2. The PWWP truncation used here (residues 278–427, untagged, pI = 5.45 (Expasy ProtParam)) is negatively charged at the assay pH (pH = 7.5), which could promote nonspecific interactions with the positively charged histone peptide. Uncropped gels are provided as Source Data.
Extended Data Fig. 7 ∣. Analysis of base editing outcomes for three sgRNAs targeting the E342 codon of DNMT3A.
a, Schematic of sgRNAs targeting the E342 codon. sgE342.1 (red) and sgE342.2 (light red) correspond to the screening hits presented in Fig. 4 and Extended Data Fig. 5. sgE342.3 (purple) is an additional library sgRNA that did not score as a hit in the screen but also targets the E342 codon. b, Allele frequencies in base-edited reporter cells after 15 days of dox treatment, comparing citrine+ cells to unsorted cells. Genomic DNA was harvested using QuickExtract DNA extraction solution (Lucigen) and libraries were prepared and deep sequenced as for other genotyping experiments. Each row represents an allele (for the purposes of this analysis, alleles were merged if they were identical within the region depicted here). All alleles having at least 1% allele frequency in at least one sample are depicted. Left, nucleotide sequence of each allele, with C to T base edits shown in red (these appear as G to A because the protospacers are along the opposite strand) and deletions represented by dashes. Middle, amino acid sequence corresponding to the translation of the region shown, with missense and silent mutations colored blue and orange, respectively. Right, allele frequencies in unsorted or citrine+ cells for each sgRNA. Colored dots, citrine+ cells; gray dots, unsorted cells. Colored squares to the right of each plot indicate the log2(fold-change in allele frequency in citrine+ vs. unsorted cells). NA indicates undefined log2(fold-changes) where one or both of the allele frequencies is zero. This experiment was conducted once (n = 1).
Extended Data Fig. 8 ∣. Generation of Dnmt3a2-complemented TKO ESCs.
a, Schematic of PiggyBac (PB) vector used for ectopic Dnmt3a2 expression in TKO ESCs. ITR, inverted terminal repeat; CAG, CAG promoter; IRES2, internal ribosome entry site 2; SV40, simian virus 40 poly(A) sequence. b, Overview of Dnmt3a2 complementation experiment. c, Representative gating strategy for flow cytometric analysis and FACS of TKO ESCs. d, Flow cytometric analysis of mCherry fluorescence in ESCs transposed with the Dnmt3a2 expression vector. Top, histograms of mCherry fluorescence. Parent TKO ESCs are shown in gray (same data for all plots). Bottom, mCherry fluorescence versus forward scatter showing the percentage of cells that are gated as mCherry+. n = 2 biological replicates (separately transposed cells).
Extended Data Fig. 9 ∣. Additional analysis of de novo DNA methylation in Dnmt3a2-complemented TKO ESCs.
a, Counts of hypermethylated and hypomethylated differentially methylated regions (DMRs) called for each mutant Dnmt3a2 compared to wild-type Dnmt3a2. b, Overlap of called DMRs with three genomic annotations: CpG islands (gray), genic regions (red), and intergenic regions (blue). Hypermethylated and hypomethylated DMRs for each mutant were considered separately. The plot displays the percentage of DMRs in each group with any overlap with each genomic annotation. c, CpG methylation within 10 kb genomic bins ranked into quartiles based on normalized H3K4me3 ChIP-seq signal (n = 23,379 bins per quartile). Center line, median; box, interquartile range; whiskers, up to 1.5 × interquartile range per the Tukey method; outliers not shown. d, Difference in CpG methylation between top and bottom H3K36me2 quartiles for each sample. 10 kb genomic bins (n = 93,516 total) were grouped into quartiles, and the average methylation in the median bins from the top and bottom quartiles were compared. Biological replicates are shown separately (n = 2). e, CpG methylation within 10 kb genomic bins ranked into quantiles (n = 1,000 quantiles) based on normalized H3K36me2 ChIP-seq signal. The average bin methylation for each quantile is plotted against the H3K36me2 signal. For a–e, only CpGs with 5× coverage across all samples were considered. Methylation values in c and e represent the average of two biological replicates.
Supplementary Material
Acknowledgements
We thank members of the Liau Lab for helpful discussions and comments on the manuscript, in particular A. Siegenfeld, A. Waterbury, H.S. Kwok, S. Roseman, and P. Gosavi. We thank D. Bolduc for advice regarding biochemistry experiments, Z. Niziolek and J. Nelson for assistance with FACS, A. Meissner for providing TKO ESCs, A. Mattei and E. Jung for advice regarding stem cell culture, S. Berry for computational advice, V. Baidin for advice regarding radiometric assays, and T. Haining and K. Richards-Corke for additional assistance. N.Z.L., E.M.G., and K.C.N. were supported by National Science Foundation Graduate Research Fellowships (grant no. DGE1745303). E.M.G. was also supported by a Landry Cancer Biology Fellowship. C.L. was supported by a Herchel Smith Graduate Fellowship. B.B.L. is a Damon Runyon-Rachleff Innovator supported in part by the Damon Runyon Cancer Research Foundation (grant no. DDR 60S-20). This research was additionally supported by award no. 1DP2GM137494 from the National Institute of General Medical Sciences and startup funds from Harvard University.
Footnotes
Competing Interests
B.B.L. is on the scientific advisory board of H3 Biomedicine, holds a sponsored research project with H3 Biomedicine, is a scientific consultant for Imago BioSciences, and is a shareholder and member of the scientific advisory board of Light Horse Therapeutics. J.G.D. consults for Microsoft Research, Abata Therapeutics, Servier, Maze Therapeutics, BioNTech, Sangamo, and Pfizer. J.G.D. consults for and has equity in Tango Therapeutics. J.G.D. serves as a paid scientific advisor to the Laboratory for Genomics Research, funded in part by GlaxoSmithKline. J.G.D. receives funding support from the Functional Genomics Consortium: Abbvie, Bristol Myers Squibb, Janssen, Merck, and Vir Biotechnology. J.G.D.’s interests were reviewed and are managed by the Broad Institute in accordance with its conflict of interest policies. The remaining authors declare no competing interests.
Data Availability
RRBS and ChIP-seq data have been deposited to NCBI GEO (GSE199890). Base editor scanning data, genotyping and conservation analysis results, and oligonucleotide sequences are provided as Supplementary Information. Unprocessed gel and immunoblot images, as well as additional data generated by this study, are provided as Source Data. Key plasmids reported in this study have been deposited to Addgene #186966–186970. The following publicly available datasets were used: PDB accession codes 4U7T, 4U7P, 3LLR, 5YX2, 5CIU, and 6S01; uniref and pfam databases; and the mm10 genome and genomic annotations from UCSC.
Code Availability
Custom code used to analyze base editor scanning data, RRBS and ChIP-seq data, genotyping data, reporter bisulfite sequencing data, and PWWP evolutionary conservation is available at https://github.com/liaulab/DNMT3A_base_editor_scanning.
References
- 1.Mattei AL, Bailly N & Meissner A DNA methylation: a historical perspective. Trends Genet. 38, 676–707 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Schübeler D Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Okano M, Bell DW, Haber DA & Li E DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Tatton-Brown K. et al. The Tatton-Brown-Rahman Syndrome: A clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 3, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heyn P. et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet 51, 96–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong M. et al. Loss of Dnmt3a Immortalizes Hematopoietic Stem Cells In Vivo. Cell Rep. 23, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayle A. et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125, 629–638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunetti L, Gundry MC & Goodell MA DNMT3A in Leukemia. Cold Spring Harb. Perspect. Med 7, a030320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suetake I, Shinozaki F, Miyagawa J, Takeshima H & Tajima S DNMT3L Stimulates the DNA Methylation Activity of Dnmt3a and Dnmt3b through a Direct Interaction. J. Biol. Chem 279, 27816–27823 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Holz-Schietinger C, Matje DM & Reich NO Mutations in DNA Methyltransferase (DNMT3A) Observed in Acute Myeloid Leukemia Patients Disrupt Processive Methylation. J. Biol. Chem 287, 30941–30951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen T-V et al. The R882H DNMT3A hot spot mutation stabilizes the formation of large DNMT3A oligomers with low DNA methyltransferase activity. J. Biol. Chem 294, 16966–16977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu T-H et al. Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature 586, 151–155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X. et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Weinberg DN et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W. et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell 11, 150–154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bröhm A. et al. Methylation of recombinant mononucleosomes by DNMT3A demonstrates efficient linker DNA methylation and a role of H3K36me3. Commun. Biol 5, 192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H. et al. Structural and Histone Binding Ability Characterizations of Human PWWP Domains. PLoS One 6, e18919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z-M et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y-H et al. Systematic profiling of DNMT3A variants reveals protein instability mediated by the DCAF8 E3 ubiquitin ligase adaptor. Cancer Discov. 12, 220–235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C. et al. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol. Cell 60, 847–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinyard ME et al. CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nat. Chem. Biol 15, 529–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sher F. et al. Rational targeting of a NuRD subcomplex guided by comprehensive in situ mutagenesis. Nat. Genet 51, 1149–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosavi PM et al. Profiling the Landscape of Drug Resistance Mutations in Neosubstrates to Molecular Glue Degraders. ACS Cent. Sci 8, 417–429 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kweon J et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene 39, 30–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jun S, Lim H, Chun H, Lee JH & Bang D Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commun. Biol 3, 154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Després PC, Dubé AK, Seki M, Yachie N & Landry CR Perturbing proteomes at single residue resolution using base editing. Nat. Commun 11, 1871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanna RE et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Cuella-Martin R. et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez-Rivera FJ et al. Base editing sensor libraries for high-throughput engineering and functional analysis of cancer-associated single nucleotide variants. Nat. Biotechnol 40, 862–873 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangree AK et al. Benchmarking of SpCas9 variants enables deeper base editor screens of BRCA1 and BCL2. Nat. Commun 13, 1318 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y. et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nat. Biotechnol 40, 874–884 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bintu L. et al. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reither S, Li F, Gowher H & Jeltsch A Catalytic Mechanism of DNA-(cytosine-C5)-methyltransferases Revisited: Covalent Intermediate Formation is not Essential for Methyl Group Transfer by the Murine Dnmt3a Enzyme. J. Mol. Biol 329, 675–684 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Tate JG et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B-Z et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 21, 1172–1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers QL, Gasser JA, Cowley GS, Fischer ES & Ebert BL Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood 132, 1293–1303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dukatz M. et al. H3K36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J. Mol. Biol 431, 5063–5074 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Purdy MM, Holz-Schietinger C & Reich NO Identification of a second DNA binding site in human DNA methyltransferase 3A by substrate inhibition and domain deletion. Arch. Biochem. Biophys 498, 13–22 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Farnung L, Dienemann C & Cramer P Structure of H3K36-methylated nucleosome–PWWP complex reveals multivalent cross-gyre binding. Nat. Struct. Mol. Biol 27, 8–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suetake I. et al. Characterization of DNA-binding activity in the N-terminal domain of the DNA methyltransferase Dnmt3a. Biochem. J 437, 141–148 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Haggerty C. et al. Dnmt1 has de novo activity targeted to transposable elements. Nat. Struct. Mol. Biol 28, 594–603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrett-Bakelman FE et al. Enhanced Reduced Representation Bisulfite Sequencing for Assessment of DNA Methylation at Base Pair Resolution. J. Vis. Exp e52246 (2015) doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinberg DN et al. Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands. Nat. Genet 53, 794–800 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sendžikaitė G, Hanna CW, Stewart-Morgan KR, Ivanova E & Kelsey G A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun 10, 1884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kibe K. et al. The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes. PLoS Genet. 17, e1009570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu T. et al. The disordered N-terminal domain of DNMT3A recognizes H2AK119ub and is required for postnatal development. Nat. Genet 54, 625–636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tycko J. et al. High-Throughput Discovery and Characterization of Human Transcriptional Effectors. Cell 183, 2020–2035.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 51.Clement K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canver MC et al. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat. Protoc 13, 946–986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eddy SR Accelerated Profile HMM Searches. PLoS Comput. Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievers F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu T. et al. Three Essential Resources to Improve Differential Scanning Fluorimetry (DSF) Experiments. bioRxiv (2020) doi: 10.1101/2020.03.22.002543. [DOI] [Google Scholar]
- 56.Porter EG, Connelly KE & Dykhuizen EC Sequential Salt Extractions for the Analysis of Bulk Chromatin Binding Properties of Chromatin Modifying Complexes. J. Vis. Exp 55369 (2017) doi: 10.3791/55369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramírez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Condon DE et al. Defiant: (DMRs: easy, fast, identification and ANnoTation) identifies differentially Methylated regions from iron-deficient rat hippocampus. BMC Bioinform. 19, 31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RRBS and ChIP-seq data have been deposited to NCBI GEO (GSE199890). Base editor scanning data, genotyping and conservation analysis results, and oligonucleotide sequences are provided as Supplementary Information. Unprocessed gel and immunoblot images, as well as additional data generated by this study, are provided as Source Data. Key plasmids reported in this study have been deposited to Addgene #186966–186970. The following publicly available datasets were used: PDB accession codes 4U7T, 4U7P, 3LLR, 5YX2, 5CIU, and 6S01; uniref and pfam databases; and the mm10 genome and genomic annotations from UCSC.
Custom code used to analyze base editor scanning data, RRBS and ChIP-seq data, genotyping data, reporter bisulfite sequencing data, and PWWP evolutionary conservation is available at https://github.com/liaulab/DNMT3A_base_editor_scanning.