Abstract
Background
The gut microbiota is increasingly recognized as a crucial factor in human health and disease. Metformin, a commonly prescribed medication for type 2 diabetes, has been studied for its potential impact on the gut microbiota in preclinical models. However, the effects of metformin on the gut microbiota in humans remain uncertain.
Scope of review
We conducted a systematic review of clinical trials and observational studies to assess the existing knowledge on the impact of metformin on the gut microbiota in humans. The review focused on changes in bacterial composition and diversity following metformin treatment.
Major conclusions
Thirteen studies were included in the analysis. The results revealed alterations in the abundance of bacterial genera from various phyla, suggesting that metformin may selectively influence certain groups of bacteria in the gut microbiota. However, the effects on gut microbiota diversity were inconsistent across populations, with conflicting findings on changes in alpha and beta diversity measures.
Overall, the use of metformin was associated with changes in the abundance of specific bacterial genera within the gut microbiota of human populations. However, the effects on gut microbiota diversity were not consistent, highlighting the need for further research to understand the underlying mechanisms and clinical significance of these changes.
Keywords: Metformin, Gut microbiota, Diabetes, Dysbiosis, Microbial diversity
1. Introduction
The gut microbiota is a diverse and dynamic community of microorganisms that inhabit the gastrointestinal tract and have a significant impact on human health and disease [1]. These microorganisms play a vital role in the digestion and absorption of nutrients, the regulation of the immune system, and the production of essential metabolites [2]. The composition and function of the gut microbiota are influenced by a variety of factors, including diet, lifestyle, and medication use [3].
Metformin is a first-line medication widely used for the treatment of type 2 diabetes [[4], [5], [6], [7]]. In recent years, there has been growing interest in the potential effects of metformin on the gut microbiota [[8], [9], [10], [11]]. Preclinical studies have demonstrated that metformin alters the gut microbiota composition and function, and there is evidence to suggest that these changes may be beneficial for metabolic and immune health [12].
Despite the growing interest in this topic, the impact of metformin on the gut microbiota in humans remains unclear [13]. While some studies have reported significant changes in the gut microbiota following metformin treatment, others have found no significant effects [14,15]. Furthermore, there is conflicting evidence regarding the direction and magnitude of the effect. Therefore, a comprehensive and up-to-date systematic review is needed to synthesize the available evidence from human studies and provide a critical assessment of the effects of metformin on the gut microbiota.
This systematic review aims to summarize and evaluate the existing knowledge on the impact of metformin on the gut microbiota in humans. By synthesizing the available evidence, this review will identify knowledge gaps and inform future research directions. The results of this review may also have important implications for the use of metformin as a therapeutic agent, as the gut microbiota plays a critical role in metabolic and immune functions. Overall, the findings of this review will provide valuable insights into the potential effects of metformin on the gut microbiota and their impact on human health.
2. Materials and methods
2.1. Literature search
Our search strategies were designed to incorporate the PICOS framework, which focused on population, intervention, comparisons, outcomes, and study design. Specifically, we aimed to investigate the impact of metformin on the gut microbiota of humans, including both healthy individuals and those with conditions such as obesity, prediabetes, and diabetes. We conducted a systematic search of the PubMed, EMBASE, and SCOPUS databases from January 1, 2000, to January 1, 2023, using relevant keywords and search terms aligned with the PICOS criteria [16]. Additionally, we manually searched reference lists of relevant review papers to identify any additional publications of interest. Our search focused primarily on clinical trials and observational studies, as recommended for systematic reviews.
2.2. Study selection criteria
We adhered to the guidelines outlined in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) to ensure that our review was conducted in a transparent and rigorous manner [17]. Our inclusion criteria specified that we only included human studies that reported original data on the gut microbiota following treatment with metformin, and that analyzed gut microbiota from feces or colonic content specimens. Additionally, studies had to be written in English. We excluded studies that did not provide data on individual bacterial taxa or were only available in the form of conference abstracts or proceedings.
2.3. Selection of studies
At the outset, we screened the titles and abstracts of all potentially relevant articles, and subsequently evaluated the full-text articles to determine their eligibility. Two authors carried out this process independently, with any disagreements being resolved through consensus between the two authors, and, if needed, by consulting two additional authors.
2.4. Data extraction
Data extraction was conducted using a standardized form in a Microsoft Excel file, which included details such as study characteristics, participant information, treatment and comparison details, microbiome analysis methods, and outcome measures. One author extracted the data, while another reviewed it. Any discrepancies were resolved through consensus among the authors involved, and if necessary, a third or fourth author was consulted.
2.5. Quality assessment
To evaluate the risk of bias in the selected randomized trials, we utilized the Robvis tool [18]. Three authors extracted the risk of bias data, and in cases of disagreement, the remaining authors reached a consensus.
2.6. Outcomes of assessment
In addition to summarizing the features of the analyzed human studies, our main focus was to investigate the variation in the levels or alterations of specific intestinal bacterial taxa, classified into two widely used taxonomic groups [Phylum (P), Genus (G)], in relation to metformin intake, across the included human studies. The secondary outcomes of interest were disparities in microbial diversity following the administration of metformin.
2.7. Data synthesis
The primary and secondary outcomes were divided into three categories: significant increase, significant decrease, and no significant difference. At least two human studies were used to synthesize the changes of each taxon. Specifically, we compared the effects of metformin on specific taxa among the evaluated human studies. These results were further categorized based on the targeted research populations, such as obese, pre-diabetic, newly diagnosed type 2 diabetes (T2D), and prevalent T2D.
3. Results
3.1. Reviewed studies
A total of 2301 citations were initially identified, and after screening and evaluation, 13 human studies were included in the final analysis (see Figure 1). Most of the studies were published in or after 2017, and the duration of metformin treatment varied across the studies, ranging from a few days to several months. Out of the 13 studies, six (46.15%) were randomized controlled trials, while the remaining seven studies (53.8%) included either newly diagnosed or prevalent type 2 diabetes patients [13,[19], [20], [21], [22], [23], [24]]. The other participants in the studies were obese individuals [[25], [26], [27]], healthy participants [28,29], and gestational diabetes mellitus patients [30] (Table 1).
Figure 1.
PRISMA 2020 flow diagram.
Table 1.
Characteristics of included studies.
| Author, year | Study design | The country of origin of the population | Participants | Number of participants | Metformin dose | Duration | Analysis method | Comparison |
|---|---|---|---|---|---|---|---|---|
| Ejtahed et al., 2019 [25] | Randomized trials | Iran | Obese | 20/16 | 1000 mg | 2 months | 16S rRNA | Post vs. pre |
| Tong et al., 2018 [13] | Randomized trials | China | Newly T2D | 100/100 | 750 mg | 12 weeks | 16S rRNA V3–V4 region | Post vs. pre |
| Wu et al., 2017 [19] | Randomized trials | EU | Newly T2D | 22/18 | 425–1700 mg | 4 months | DNA shotgun metagenomics | Post vs. pre |
| Bryrup et al., 2019 [29] | Quasi-experimental studies | EU | Healthy | 25 | 500–2000 mg | 6 weeks | 16S rRNA V4 region | Post vs. pre |
| Sun et al., 2018 [20] | Quasi-experimental studies | China | Newly T2D | 22 | 2000 mg | 3 days | DNA metagenomics | Post vs. pre |
| Elbere et al., 2018 [28] | Quasi-experimental studies | EU | Healthy | 18 | 1700 mg | 7 days | 16S rRNA V3 region | Post vs. pre |
| Napolitano et al., 2014 [21] | Quasi-experimental studies | UK | Prevalent T2D | 14/14 | As usual | NA | 16S rRNA V1, V2, V3 regions | On vs. off |
| Barengolts et al., 2018 [22] | Cross-sectional studies | USA | Prevalent T2D | 25/16 | As usual | – | 16S rRNA V3–V4 region | With vs. without |
| De La Cuesta-Zuluaga et al., 2017 [23] | Cross-sectional studies | Columbia | Prevalent T2D | 14/14 | As usual | – | 16S rRNA V4 region | With vs. without |
| Forslund et al., 2015 [24] | Cross-sectional studies | EU | Prevalent T2D | 58/17 | As usual | – | 16S rDNA shotgun metagenomics | With vs. without |
| Noel T Mueller et al., 2021 [26] | Randomized trials | USA | Overweight or obese adults with a history of a malignant solid tumor diagnosis | 42/39/40 | 2000 mg | 1 year | DNA metagenomics | With vs. without |
| María Molina-Vega et al., 2021 [30] | Randomized trials | EU | Pregnant women with GDM | 30/28 | 850 mg | During pregnancy | 16S rRNA | With vs. without |
| Belén Pastor-Villaescusa et al., 2021 [27] | Randomized trials | EU | Children with obesity | 15/18 | 1000 mg | 6 months | Metagenomic analysis | With vs. without |
3.2. Methods of analysis for gut microbiota composition in human studies
In all the human studies included in the analysis, fecal specimens were collected and analyzed for gut microbiota composition. Among the various methods used for analysis, 16S rRNA gene sequencing was found to be the most commonly used method, as presented in Table 1.
3.3. The risk of bias
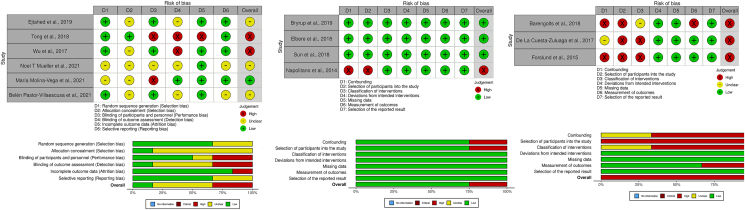
In the selected randomized trials, two studies had a high risk of bias in performance, detection, and attrition. Four studies had unclear risks in most domains. Among quasi-experimental studies, three had a low risk of bias in all domains, while one had a serious risk of bias in confounding, selection of participants, and classification of interventions. All three cross-sectional studies had a serious risk of bias in several domains, including confounding, selection of participants, and classification of intervention (as illustrated in Figure 2)
Figure 2.
Risk of bias.
3.4. Outcomes of assessment
3.4.1. Changes in bacterial genera induced by metformin treatment
Thirteen studies were included in this systematic review to investigate the effects of metformin on the gut microbiota. Overall, the use of metformin was associated with changes in the abundance of several bacterial genera (Figure 3) (Figure 4).
Figure 3.
Effects of metformin on specific taxa in human gut microbiota.
Figure 4.
Venn diagram illustrating changes in gut microbiota abundance with metformin treatment.
In the Bacteroidetes phylum, the abundance of Bacteroides was found to increase in one study [25] and decrease in another [13,20], while Parabacteroides showed an increase in one study [22]. Alistipes decreased in one study [13], while Prevotella decreased in one study [25] and increased in another [23].
In the Firmicutes phylum, Bacillus decreased in one study [27], while Blautia [13,25] and Butyrivibrio [23,25] increased in two studies each. Faecalibacterium decreased in one study [25], while Ruminococcus increased in one study [26]. Streptococcus increased in one study [25], while Oscillibacter [13] and Lactobacillus decreased in one study [25]. Catenibacterium increased in one study [22], while Clostridium decreased in two studies [28,29], and Intestinibacter in four studies [[24], [25], [26],29]. Roseburia decreased in one study [26] and increased in another [25].
In the Proteobacteria phylum, Escherichia increased in two studies [24,26]. In the Actinobacteria phylum, one study reported a decrease in this phylum [27], while Adlercreutzia increased in one study [21] and Bifidobacterium increased in one study [23].
Finally, in the Verrucomicrobia phylum, Akkermansia decreased in two studies [13,25] and increased in two studies [19,23], while in the Thermodesulfobacteriota phylum, Bilophila increased in two studies [19,29].
3.4.2. Effects of metformin on diversity of human gut microbiota
The effects of metformin on the diversity of human gut microbiota have been investigated through various studies, as presented in Table 2.
Table 2.
Effects of metformin on diversity of human gut microbiota.
| Author, year | Analysis method | Diversity indices or measures | Findings |
|---|---|---|---|
| Ejtahed et al., 2019 [25] | 16S rRNA | Richness, Simpson indices | Negative correlation of Simpson index with BMI, waist, insulin, and HOMA. No effect on global gut microbial diversity. |
| Tong et al., 2018 [13] | 16S rRNA V3–V4 region | Simpson's diversity index | Metformin increased diversity (Chao1 richness estimates). |
| Wu et al., 2017 [19] | DNA shotgun metagenomics | N/A | Not specified. |
| Bryrup et al., 2019 [29] | 16S rRNA V4 region | Richness, evenness, diversity | No effect of metformin on gut microbiota richness, evenness, or diversity. |
| Sun et al., 2018 [20] | DNA metagenomics | Shannon diversity index | Slight decrease in alpha diversity after metformin treatment. |
| Elbere et al., 2018 [28] | 16S rRNA V3 region | Shannon index, NMDS | Significant reduction in gut microbiota diversity 24 h after metformin. |
| Napolitano et al., 2014 [21] | 16S rRNA V1–V3 regions | Beta-diversity analysis | Gut communities are patient-specific. No specific metformin effect mentioned. |
| Barengolts et al., 2018 [22] | 16S rRNA V3–V4 region | Shannon index | Trend towards higher diversity. No beta-diversity differences. |
| De La Cuesta-Zuluaga et al., 2017 [23] | 16S rRNA V4 region | Beta-diversity estimates | No significant differences except for metformin users vs. nonusers. |
| Forslund et al., 2015 [24] | 16S rDNA shotgun metagenomics | N/A | Not specified. |
| Noel T Mueller et al., 2021 [26] | DNA metagenomics | Shannon diversity, metagenomic functional pathway richness | No change in Shannon diversity, but increased metagenomic pathway richness with metformin. |
| María Molina-Vega et al., 2021 [30] | 16S rRNA | Phylogenetic diversity | MET group had lower phylogenetic diversity compared to INS group. |
| Belén Pastor-Villaescusa et al., 2021 [27] | Metagenomic analysis | Alpha indexes and beta diversity in studied children at the phylum level. | No significant differences in diversity, Simpson index, inverse Simpson index, evenness, or beta diversity between placebo and metformin groups. A trend towards higher alpha index and species richness in the metformin group (not statistically significant). |
Among the studies conducted on healthy individuals, Elbere et al. [28] reported a significant reduction in gut microbiota diversity 24 h after metformin administration using 16S rRNA V3 region sequencing and Shannon index. Conversely, Bryrup et al. [29]found no significant changes in gut microbial richness, evenness, or diversity in response to metformin treatment.
In obese individuals, Ejtahed et al. [25] observed no significant changes in alpha or beta diversity but reported a negative correlation of Simpson index with BMI, waist, insulin, and HOMA, indicating potential metabolic associations.
Regarding patients with newly diagnosed T2D, Tong et al. [13] reported increased diversity, as indicated by Chao1 richness estimates based on 16S rRNA V3–V4 region sequencing, while Sun et al. [20] observed a slight decrease in alpha diversity using DNA metagenomics.
Barengolts et al. [22] showed a trend towards higher diversity with metformin treatment, and De La Cuesta-Zuluaga et al. [23] found no significant differences in diversity indices between metformin users and nonusers.
In other populations, Noel T Mueller et al. [26] reported no change in Shannon diversity but observed increased metagenomic pathway richness with metformin treatment. María Molina-Vega et al. [30] found lower phylogenetic diversity in the metformin group compared to the control group, and Belén Pastor-Villaescusa et al. [27] reported no significant differences in diversity indices or beta diversity.
4. Discussion
Metformin is a widely used medication for the treatment of T2D, and its effects on the gut microbiota have been a topic of interest in recent years [9,15]. Our systematic review of 13 studies found that metformin treatment was associated with changes in the abundance of several bacterial genera across different phyla.
In the Bacteroidetes phylum, Bacteroides showed inconsistent results, with one study reporting an increase and two studies reporting a decrease in its abundance. Parabacteroides showed an increase in one study, while Alistipes and Prevotella both showed decreases in one study and increases in another. In the Firmicutes phylum, Blautia and Butyrivibrio increased in two studies each, while Bacillus and Faecalibacterium decreased in one study each. Streptococcus increased in one study, while Lactobacillus and Oscillibacter decreased in another study. In the Proteobacteria phylum, Escherichia increased in two studies. In the Actinobacteria phylum, Bifidobacterium increased in one study, while Adlercreutzia decreased in one study. Finally, in the Verrucomicrobia phylum, Akkermansia showed inconsistent results, with two studies reporting an increase and two studies reporting a decrease in its abundance.
Our findings are consistent with previous studies that have reported changes in gut microbiota composition following metformin treatment. For example, Wu et al. [19] reported an increase in Akkermansia in individuals with T2D treated with metformin. However, Tong et al. [13] found a decrease in Akkermansia and no significant changes in Escherichia abundance in newly diagnosed T2D patients treated with metformin. These discrepancies may be due to differences in study design, sample size, metformin dosage, or duration of treatment.
Furthermore, it is important to highlight the necessity of future multicenter microbiome studies, standardization of protocols among multicenter studies, as well as in vitro microbiome-drug assays. These initiatives are crucial for advancing our understanding of metformin's impact on the gut microbiota and its implications for T2D.
The role of specific bacterial genera in the pathogenesis of T2D has been a subject of investigation, with some studies suggesting that alterations in gut microbiota composition may contribute to the development of insulin resistance and inflammation [[31], [32], [33]]. For instance, a decrease in butyrate-producing bacteria, such as Faecalibacterium, has been associated with T2D and metabolic syndrome [34]. Similarly, an increase in Escherichia coli has been linked to inflammation and insulin resistance in individuals with T2D [35]. On the other hand, Akkermansia muciniphila has been shown to have beneficial effects on glucose metabolism and gut barrier function [[36], [37], [38]]. Thus, the changes in gut microbiota composition associated with metformin treatment may have implications for the pathogenesis and management of T2D.
In our previous study, we investigated the gut microbiome composition of patients with T2D and COVID-19 who received metformin treatment, with or without antibiotic treatment. Our findings indicated that metformin-treated patients with T2D and COVID-19 who did not receive antibiotics had a significantly different gut microbiome composition compared to those who did receive antibiotics. Specifically, we observed a higher abundance of the genera Bacteroides, E. coli, and Lactobacillus, and a lower abundance of the genera Clostridium and Enterococcus in the group that did not receive antibiotics [39].
In conclusion, our systematic review provides further evidence of the effects of metformin on gut microbiota composition. The observed changes in bacterial genera may have implications for the pathogenesis and management of T2D, and further research is warranted to elucidate the underlying mechanisms and potential therapeutic targets.
5. Confounding factors and interpretation of bacterial taxa changes
The observed variability in the effects of metformin on gut microbiota composition may be attributed to several confounding factors inherent in the included studies. These factors, including study design, disease type, metformin treatment duration, and sequencing methods, played a crucial role in shaping the reported changes in bacterial taxa.
When comparing the results across studies, it becomes evident that different factors contributed to the specific outcomes observed in each study. For instance:
-
1.
Disease type: Tong et al. and Sun et al., both focusing on newly diagnosed T2D patients, reported contrasting changes in bacterial taxa. While Tong et al. found decreases in Alistipes and Prevotella, Sun et al. observed a decrease in Bacillus and an increase in Escherichia. These discrepancies highlight how disease-specific characteristics might influence the response of gut microbiota to metformin treatment.
-
2.
Study duration: The duration of metformin treatment varied widely across the included studies. Elbere et al. reported reduced gut microbiota diversity within 24 h of metformin administration. In contrast, other studies, such as those by Ejtahed et al. and Barengolts et al., did not find significant changes in diversity despite longer-term treatment. These findings suggest that the impact of metformin on gut microbiota might be more pronounced in the short term, while long-term administration may require extended periods to induce noticeable changes.
-
3.
Sequencing methods: Variation in sequencing methods might have contributed to the differences in observed changes. For example, Elbere et al. used 16S rRNA V3 region sequencing and found reduced diversity, while Bryrup et al. did not observe significant changes using 16S rRNA V4 region sequencing. This highlights the importance of standardized methodologies to accurately compare results across studies.
-
4.
Participant characteristics: Variation in participant characteristics, such as healthy individuals (Bryrup et al., 2019), obese individuals (Ejtahed et al., 2019; Bryrup et al., 2019), and specific disease populations (Noel T Mueller et al., 2021; María Molina-Vega et al., 2021; Belén Pastor-Villaescusa et al., 2021), further underscores the potential influence of baseline microbiota composition on the response to metformin.
6. Limitations
While this systematic review aims to provide a comprehensive assessment of the impact of metformin on the gut microbiota in humans, several limitations must be acknowledged.
-
1.
Heterogeneity of included studies: The studies included in this review exhibited substantial heterogeneity in terms of study design, participant characteristics, metformin dosage, treatment duration, and sequencing methods. This heterogeneity could introduce variability in the observed effects of metformin on the gut microbiota. The diversity of the included populations and methodologies limits the ability to draw definitive conclusions across all studies.
-
2.
Risk of bias: The assessment of risk of bias revealed varying levels of methodological quality among the included studies. While efforts were made to minimize bias through the use of robust systematic review methods, the presence of studies with high risk of bias may have influenced the overall findings.
-
3.
Taxonomic resolution: Our analysis focused on taxonomic changes at the genus level, which may not fully capture the diversity and complexity of the gut microbiota. Within a single genus, there can be substantial variation at the species or strain level, leading to different responses to metformin. Future studies employing higher taxonomic resolution or metagenomic/metatranscriptomic approaches could provide more detailed insights into the specific changes within microbial communities.
-
4.
Confounding factors: The included studies varied in terms of confounding factors such as disease type, metformin treatment duration, and co-administration of other medications. These factors could influence the observed effects of metformin on the gut microbiota. While subgroup analyses were conducted to account for some of these factors, residual confounding remains a potential limitation.
-
5.
Translation to clinical implications: While the observed changes in gut microbiota composition are of interest, the clinical implications of these changes remain to be fully elucidated. The complex interplay between gut microbiota, metformin treatment, and human health requires further investigation to understand the functional consequences of these alterations.
-
6.
Temporal considerations: Most of the included studies were published in recent years, and the duration of metformin treatment varied across the studies. Longer-term effects of metformin on the gut microbiota may not be fully captured in studies with shorter treatment durations.
Author contributions
P.P. and A.K. conceptualized and designed the study. I.K. conducted the literature search and data extraction. P.P. and A.K. analyzed and interpreted the data. P.P. drafted the manuscript, and all authors provided critical feedback and revisions. All authors have read and approved the final version of the manuscript.
Funding
Grant Agreement #2025-VSE “Virtual Scientist Engagement Fellowship Program for Vulnerable Ukrainian Scientists”
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Pavlo Petakh, Email: pavlo.petakh@uzhnu.edu.ua.
Aleksandr Kamyshnyi, Email: alexkamyshnyi@gmail.com.
Data availability
Data will be made available on request.
References
- 1.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thursby E., Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7 doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holman R. Journees annuelles de diabetologie de l'Hotel-Dieu; 2007. Metformin as first choice in oral diabetes treatment: the UKPDS experience; pp. 13–20. [PubMed] [Google Scholar]
- 5.Petakh P., Griga V., Mohammed I.B., Loshak K., Poliak I., Kamyshnyiy A. Effects of metformin, insulin on hematological parameters of COVID-19 patients with type 2 diabetes. Med Arch. 2022;76(5):329–332. doi: 10.5455/medarh.2022.76.329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petakh P., Isevych V., Mohammed I., Loshak K., Poliak I., Kamyshnyiy A. Association between use of metformin and insulin with hematological parameters in COVID-19 patients with type 2 diabetes: a single center, cross-sectional study. Clin Diabetol. 2022;11(6):432–433. [Google Scholar]
- 7.Petakh P., Loshak K., Kamyshnyi A. Hematological features of patients with type 2 diabetes depending on the variant of SARS-COV-2. Fiziolohichnyi Zh. 2023;69(1):35–42. [Google Scholar]
- 8.Ma W., Chen J., Meng Y., Yang J., Cui Q., Zhou Y. Metformin alters gut microbiota of healthy mice: implication for its potential role in gut microbiota homeostasis. Front Microbiol. 2018;9:1336. doi: 10.3389/fmicb.2018.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petakh P., Kamyshna I., Oksenych V., Kainov D., Kamyshnyi A. Metformin therapy changes gut microbiota alpha-diversity in COVID-19 patients with type 2 diabetes: the role of SARS-CoV-2 variants and antibiotic treatment. 2023;16(6):904. doi: 10.3390/ph16060904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petakh P., Oksenych V., Kamyshnyi A. The F/B ratio as a biomarker for inflammation in COVID-19 and T2D: impact of metformin. Biomed Pharmacother. 2023;163 doi: 10.1016/j.biopha.2023.114892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petakh P., Kamyshna I., Nykyforuk A., Yao R., Imbery J.F., Oksenych V., et al. Immunoregulatory intestinal microbiota and COVID-19 in patients with type two diabetes: a double-edged sword. J Viruses. 2022;14(3):477. doi: 10.3390/v14030477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollak M. The effects of metformin on gut microbiota and the immune system as research frontiers. Diabetologia. 2017;60(9):1662–1667. doi: 10.1007/s00125-017-4352-x. [DOI] [PubMed] [Google Scholar]
- 13.Tong X., Xu J., Lian F., Yu X., Zhao Y., Xu L., et al. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. mBio. 2018;9(3) doi: 10.1128/mBio.02392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C.B., Chae S.U., Jo S.J., Jerng U.M., Bae S.K. The relationship between the gut microbiome and metformin as a key for treating type 2 diabetes mellitus. Int J Mol Sci. 2021;22(7) doi: 10.3390/ijms22073566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Hu N. Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. Diabetes, Metab Syndrome Obes Targets Ther. 2020;13:5003–5014. doi: 10.2147/DMSO.S286430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scells H., Zuccon G., Koopman B., Deacon A., Azzopardi L., Geva S. Proceedings of the 2017 ACM on conference on information and knowledge management. Association for Computing Machinery; Singapore, Singapore: 2017. Integrating the framing of clinical questions via PICO into the retrieval of medical literature for systematic reviews; pp. 2291–2294. [Google Scholar]
- 17.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuinness L.A., Higgins J.P.T. Risk-of-bias VISualization (robvis): an R package and Shiny web app for visualizing risk-of-bias assessments. 2021;12(1):55–61. doi: 10.1002/jrsm.1411. [DOI] [PubMed] [Google Scholar]
- 19.Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Mannerås-Holm L., et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 20.Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Napolitano A., Miller S., Nicholls A.W., Baker D., Van Horn S., Thomas E., et al. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barengolts E., Green S.J., Eisenberg Y., Akbar A., Reddivari B., Layden B.T., et al. Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Cuesta-Zuluaga J., Mueller N.T., Corrales-Agudelo V., Velásquez-Mejía E.P., Carmona J.A., Abad J.M., et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 24.Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejtahed H.S., Tito R.Y., Siadat S.D., Hasani-Ranjbar S., Hoseini-Tavassol Z., Rymenans L., et al. Metformin induces weight loss associated with gut microbiota alteration in non-diabetic obese women: a randomized double-blind clinical trial. Eur J Endocrinol. 2019;180(3):165–176. doi: 10.1530/EJE-18-0826. [DOI] [PubMed] [Google Scholar]
- 26.Mueller N.T., Differding M.K., Zhang M., Maruthur N.M., Juraschek S.P., Miller E.R., 3rd, et al. Metformin affects gut microbiome composition and function and circulating short-chain fatty acids: a randomized trial. Diabetes Care. 2021;44(7):1462–1471. doi: 10.2337/dc20-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor-Villaescusa B., Plaza-Díaz J., Egea-Zorrilla A., Leis R., Bueno G., Hoyos R., et al. Evaluation of the gut microbiota after metformin intervention in children with obesity: a metagenomic study of a randomized controlled trial. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;134 doi: 10.1016/j.biopha.2020.111117. [DOI] [PubMed] [Google Scholar]
- 28.Elbere I., Kalnina I., Silamikelis I., Konrade I., Zaharenko L., Sekace K., et al. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0204317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryrup T., Thomsen C.W., Kern T., Allin K.H., Brandslund I., Jørgensen N.R., et al. Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia. 2019;62(6):1024–1035. doi: 10.1007/s00125-019-4848-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molina-Vega M., Picón-César M.J., Gutiérrez-Repiso C., Fernández-Valero A., Lima-Rubio F., González-Romero S., et al. Metformin action over gut microbiota is related to weight and glycemic control in gestational diabetes mellitus: a randomized trial. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2022;145 doi: 10.1016/j.biopha.2021.112465. [DOI] [PubMed] [Google Scholar]
- 31.Adeshirlarijaney A., Gewirtz A.T. Considering gut microbiota in treatment of type 2 diabetes mellitus. Gut Microb. 2020;11(3):253–264. doi: 10.1080/19490976.2020.1717719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham A.L., Stephens J.W., Harris D.A. Gut microbiota influence in type 2 diabetes mellitus (T2DM) Gut Pathog. 2021;13(1):50. doi: 10.1186/s13099-021-00446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allin K.H., Nielsen T., Pedersen O. Mechanisms in endocrinology: gut microbiota in patients with type 2 diabetes mellitus. Eur J Endocrinol. 2015;172(4):R167–R177. doi: 10.1530/EJE-14-0874. [DOI] [PubMed] [Google Scholar]
- 34.Maioli T.U., Borras-Nogues E., Torres L., Barbosa S.C., Martins V.D., Langella P., et al. Possible benefits of Faecalibacterium prausnitzii for obesity-associated gut disorders. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.740636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen C., Ijaz U.Z., Gallagher E., Horton F., Ellis R.J., Jaiyeola E., et al. Fecal Enterobacteriales enrichment is associated with increased in vivo intestinal permeability in humans. Physiological reports. 2018;6(7) doi: 10.14814/phy2.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues V.F., Elias-Oliveira J., Pereira Í S., Pereira J.A., Barbosa S.C., Machado M.S.G., et al. Akkermansia muciniphila and gut immune system: a good friendship that attenuates inflammatory bowel disease, obesity, and diabetes. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.934695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasani A., Ebrahimzadeh S., Hemmati F., Khabbaz A., Hasani A., Gholizadeh P. The role of Akkermansia muciniphila in obesity, diabetes and atherosclerosis. J Med Microbiol. 2021;70(10) doi: 10.1099/jmm.0.001435. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Ni Y., Qian L., Fang Q., Zheng T., Zhang M., et al. Decreased abundance of Akkermansia muciniphila leads to the impairment of insulin secretion and glucose homeostasis in lean type 2 diabetes. 2021;8(16) doi: 10.1002/advs.202100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petakh P., Kobyliak N., Kamyshnyi A. Vol. 13. 2023. (Gut microbiota in patients with COVID-19 and type 2 diabetes: a culture-based method). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.