Summary
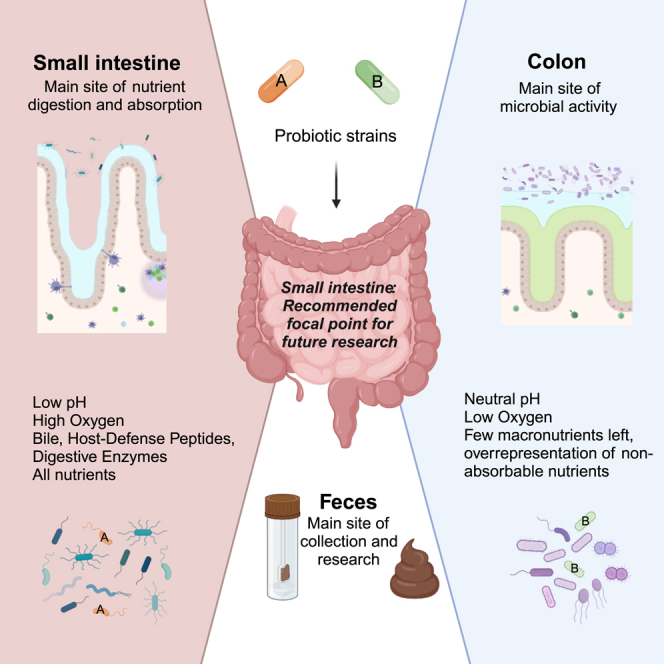
Research on gut microbiota has generally focused on fecal samples, representing luminal content of the large intestine. However, nutrient uptake is restricted to the small intestine. Abundant immune cell populations at this anatomical site combined with diminished mucus secretion and looser junctions (partly to allow for more efficient fluid and nutrient absorption) also results in intimate host-microbe interactions despite more rapid transit. It is thus crucial to dissect key differences in both ecology and physiology between small and large intestine to better leverage the immense potential of human gut microbiota imprinting, including probiotic engraftment at biological sensible niches. Here, we provide a detailed review unfolding how the physiological and anatomical differences between the small and large intestine affect gut microbiota composition, function, and plasticity. This information is key to understanding how gut microbiota manipulation, including probiotic administration, may strain-dependently transform host-microbe interactions at defined locations.
Graphical abstract
Jensen et al. provide a detailed review unfolding how the physiological and anatomical differences between the small and large intestine affect gut microbiota composition, function, and plasticity. This information is key to understanding how gut microbiota manipulation, including probiotic administration, may transform host-microbe interactions at defined locations.
Introduction
The gut microbiota—defined as the trillions of microbes living in the gastrointestinal (GI) tract—orchestrates human health.1 While the term “gut microbiota” typically refers to the multitude of bacterial species residing in the GI tract, non-bacterial microbes, including fungi and bacteriophages, have emerged as equally important modulators of host-microbe interactions.2,3 Unwarranted fluctuations in gut microbiota composition, as well as alterations in the crosstalk between the host and its microbiome—comprising the microbiota, its components and products, and the environment4—tend to influence disease development. The spectrum ranges from metabolic disorders5 and inflammatory bowel disease (IBD)6,7,8 to cancer9 and major depressive disorders.10,11
In addition, the gut microbiota can enzymatically transform drug structures, altering their bioavailability, bioactivity, and toxicity.12,13,14 Molecules secreted by human gut bacteria can further metabolize a range of host-mediated products, including bile acids, and ultimately coordinate GI immune homeostasis.15
To this end, the administration of natural or designer probiotics that carry relevant enzymes may help to prevent unwarranted immunity toward the gut microbiota and facilitate physiological processes such as lactose degradation in otherwise intolerant subjects. Indeed, emerging research points toward advanced microbial therapeutics16 (AMT) as a promising strategy to alleviate human disease traits.17,18 A key concept of AMT is microbial engineering.19 Although bacteria are attractive tools for genetic modulation, the eucaryotic nature of probiotic yeasts offers additional opportunities for complex expression systems, including post-translational modifications, resistance to antibiotics,20 and advanced containment systems.21 Despite the promising potential of (1) the gut mycobiome,22 (2) engineered yeasts as delivery vehicles for requested products, as well as (3) microbiomes beyond the GI tract, this review focusses its narrative on the intimate relationship between the host and its bacterial constituents of the small and large intestine.
Nutrient digestion and absorption: Exploring the anatomy of the small intestine and colon
The physiology and biochemical environment, including oxygen concentrations, pH, and redox potential, vary significantly throughout the intestine.23 pH varies not only along the GI tract (Figure 1) but also horizontally, where, e.g., the ventricle is protected by a diffusion barrier enabling luminal proton concentrations to exceed the concentrations at the epithelial surface by >1 million-fold. This, together with oxygen fluctuations shaping the epithelial barrier along intestine,24 is paramount for compartmentalized microbial colonization, nutrient digestion, and host metabolism. Considering the separate locations, the duodenum represents the most dynamic part of the gut, receiving acidic chyme from the stomach, neutralizing it with bicarbonate, and adding pancreatic secretions and bile. In addition to the acidity (defined by proton concentrations, cf. above), the bile secreted into the duodenum can reach concentrations between 2.6 mM (fasted) and 11.2 mM (fed),25 which is toxic to many microbial species. The pancreatic secretions further contain a cocktail of proteases, lipases, and glycosidases that may generate antimicrobial compounds from foods.26
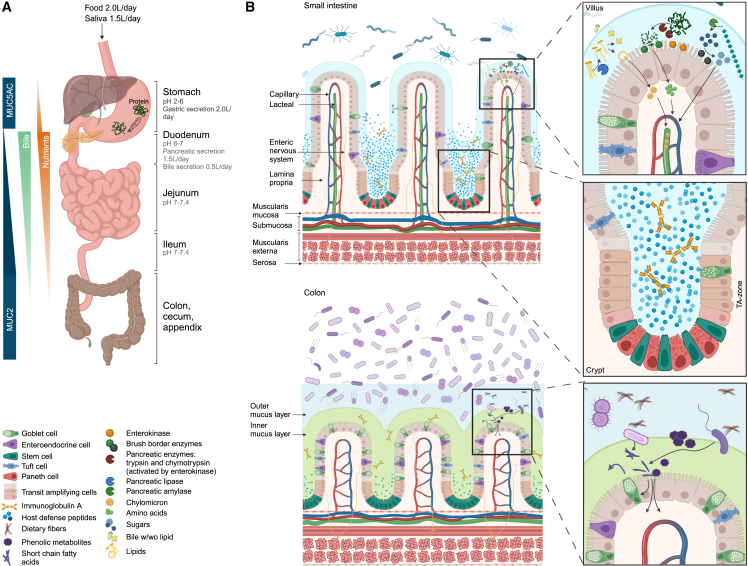
Figure 1.
Anatomy and physiology of the small intestine (SI) versus colon
(A) Overview of the gastrointestinal tract displaying spatial distribution of key processes influencing microbial colonization. In the healthy individual, liquid volume presented to the lumen of the SI is 8.5 L per day, for which pancreatic and bile secretion accounts for 1.5 and 0.5 L, respectively; ∼6.5 L are reabsorbed in the SI, while ∼2 L enters the colon. Here ∼1.9 L are reabsorbed and ∼0.1 L are lost in fecal secretions. Both secretion and absorption of liquids are affected by diet and diseases. Acidic ventricle secretion is neutralized by natrium hydrogen carbonate-buffered pancreatic juice in the upper duodenum. Proximal duodenum is also the site of bile acid (BA) secretion from the gall bladder. Nominal amounts of bile are passively absorbed through the entire length of the intestine, while the majority is recirculated by active transport in the distal ileum. Simple carbohydrates and protein are absorbed in the SI with a gradually decreasing absorption from proximal duodenum to distal ileum. Complex carbohydrates and proteins escaping digestion in the SI are converted into short-chain fatty acids (SCFAs) by microbial fermentation in the large intestine.
(B) Inner mucus layer (colon): green. Loosely attached mucus layer (outer layer in colon and only layer in SI): blue. Villus: in healthy individuals, most dietary lipids are absorbed in the first 60 cm of the SI, where BA concentrations are highest. As chylomicrons are too large to pass through the fenestrae of villus blood capillaries, they instead enter the lymph through larger inter-endothelial channels of the lacteals. Dietary proteins and carbohydrates are digested by pancreatic and brush border enzymes, respectively. Crypt: crypt-residing Paneth cells secreting abundant amounts of HDPs further protect and nurture the neighboring stem cells. In the transit-amplifying zone, lineage-committed progenitor cells swiftly divide to fuel the rapid intestinal cell turnover. Colon: the outer mucus layer provides a niche for mucolytic bacteria, many of which can ferment complex carbohydrates/dietary fibers, thereby generating SCFAs fortifying barrier function, including goblet cell-mediated mucus secretion. IgA, immunoglobulin A; TA-zone, transit-amplifying zone.
The mucosal environment in the proximal small intestine (SI) also diverges from the more distal parts. As it is the main site of digestion and absorption, the mucosal surface is covered in long villi (1–2 mm),27 maximizing the absorptive surface area (Figure 1). The cells in the intestinal epithelial layer develop from epithelial stem cells in the base of the crypts of Lieberkühn and gradually differentiate as they migrate up the villi, where they are shed and undergo apoptosis at the tip.28 In humans, this process takes 3–5 days and creates significant amounts of cellular debris, such as high-viscosity DNA, in the region around the villi tips.29
The mucus layer covering the intestinal epithelium is secreted by goblet cells. Its thickness gradually increases from ∼120 μm in the duodenum to 850 μm in the colon30 (Figure 1). In addition, the colonic mucus forms a bilayer, where the dense inner adherent layer of the healthy colon is largely devoid of bacteria, while the loose outer layer nourishes the resident microbiota.31 In the duodenum, the loosely adherent mucus is broken up by the movement of the intestinal content, allowing chyme to penetrate between the villi.32
The limited discontinuous mucus barrier in the SI gains immunological support from host defense peptides (HDPs), such as defensins, lysozyme, and regenerating islet-derived protein III-gamma (REG3g) released by crypt-resident Paneth cells. In this way, the crypts remain largely sterile,31,33 a process that is further substantiated by goblet cell mass-secretion essentially purging the crypts upon acetylcholine-mediated neural stimulation.34 Trypsin, a key protease in pancreatic secretions, processes human alpha defensins, such as HD5, from the pro-peptide to mature forms. The reducing environment near the epithelial barrier renders the peptides sensitive to proteolytic cleavage,35 which, instead of peptide degradation, may generate novel bioactive fragments36 exceeding the bactericidal repertoire of the full-length peptide.37 This, along with the high concentration of other toxic compounds such as bile acids, antimicrobial peptides and bioactive substrates of food origin, and secretory immunoglobulin A (IgA), limits microbial growth.
More than 90% of lipids, proteins, absorbable sugars, and lipid-soluble micronutrients have been removed by the time the digesta leaves the SI, while the concentration of non-absorbable components, including dietary fiber, has increased. This becomes important when considering region-specific engraftments of substrate-specific probiotics. A balance must be found between overgrowth and failure to establish due to the presence of either nutrients or antimicrobial components.
Microbiota distribution and function: Implications for probiotics
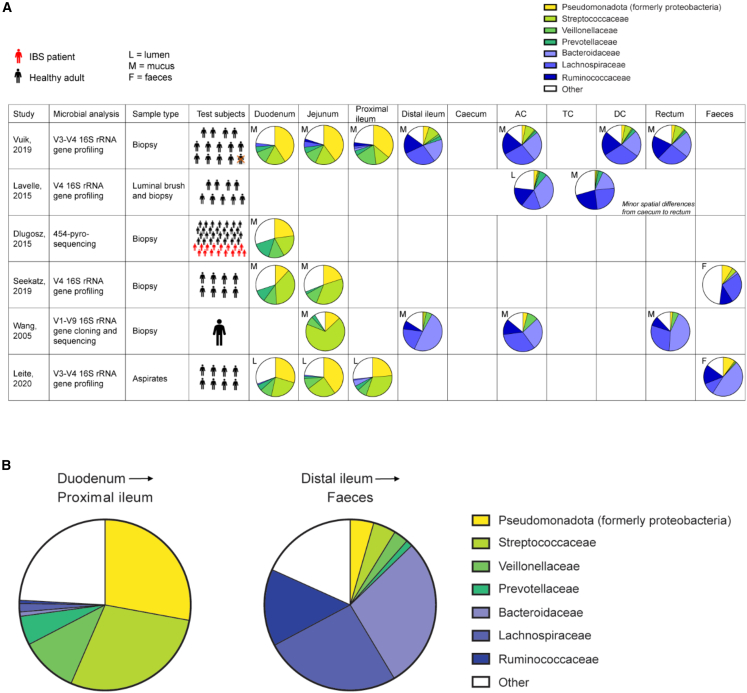
Along the various regions of the gut, microbes colonize the intestinal content (lumen) and the gut mucosa.38 Microbial colonization of these environments has rarely been studied in humans as sampling in the intestine has traditionally required invasive procedures. Among the limited number of studies that have analyzed the microbial composition of the human gut in vivo, samples were taken from widely different regions of the small and large intestine. Figure 2 provides a detailed overview of the characteristics of the studies considered for the current review along with the microbial taxa identified in specific regions. Overall, despite the variations in sampling sites, and sampling and analysis techniques, these studies consistently demonstrate the existence of different microbial communities along the human GI tract. That is, one community colonizes the duodenum down to the proximal ileum and one colonizes the distal ileum down to the rectum (Figure 2). These findings are beautifully mirrored by two recent studies that developed an ingestible capsule device to sample multiple regions of the GI tract in 15 healthy individuals during normal digestion.39,40 It is important to note, however, that mentioned studies primarily focus on healthy individuals and that differences may also depend on disease status. Probably this is where the most important implications for the use of probiotics lies. Not only are there substantial differences in the individual microbiota composition, hence posing different levels and types of competition with the probiotic administered, there is also compositional variation along and across the GI tract, which may determine where a probiotic exhibits its main activity.
Figure 2.
Longitudinal differences along human gastrointestinal (GI) tract
(A) The abundance of key taxa in a specific gut area, as averaged across all individuals tested in a given study. Six studies are included based on the requirement that they investigated different gut regions within specific individuals. Despite the differences in sampling and microbial analysis techniques, this revealed the consistent difference between microbial communities of the upper and lower GI tract. One community colonizes the duodenum down to the proximal ileum (generally dominated by Pseudomonadota, Streptococcaceae, and Veillonellaceae) and one colonizes the distal ileum down to the rectum (generally dominated by Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae).
(B) Weighted average of all the studies included in (A).
From a functional perspective, these studies indicate that the small-intestinal microbiota—ranging from 104–105 microbes/mL in the duodenum to 107–108 microbes/mL in the ileum—consists of fast-growing, primary fermenting bacteria, such as lactic acid bacteria and enterobacteria, and secondary fermenting bacteria, such as Veillonella, which grow on the lactate produced by primary fermenters.41
Metagenomic and metatranscriptomic analyses of ileostomy microbiota have indeed indicated that the rapid uptake and conversion of simple carbohydrates drives the microbial ecosystem in the SI.42 This contrasts with the colonic ecosystem, which is driven by the conversion of complex carbohydrates. A likely explanation this dichotomy is the enormous difference in transit time between the small and large intestine combined with the continuous removal of nutrients by the small-intestinal epithelium during transit. An in vitro modeling study has, in fact, shown that an ileum-like community can be obtained from a fecal inoculum when mimicking small-intestinal characteristics, including transit time.43 When developing novel probiotics that target the SI, it will thus be relevant to consider their ability to thrive in an environment fundamentally different from the colon.
In addition to easily fermentable carbohydrates, numerous other potential energy sources enter the SI that can be used not only by the host but also by the SI microbes. Human data on how, e.g., fats and proteins interact with the SI microbiota are limited. Fats are generally processed and absorbed in the duodenum and jejunum44 but, as energy-rich substrates, they can also be used by intestinal microbes. Ingestion of fats may exhibit a further indirect effect on the microbiota via the release of bile acids, which may inhibit microbes depending on their ability to hydrolyze bile salts45 and thus influence the survival of consumed probiotics. Proteins are another potential energy source for proteolytic microbes. While they are mainly metabolized in the SI, any remaining proteins may undergo colonic fermentation, resulting in the production of hazardous components, such as amines, sulfide, and phenolic compounds. The physiological impact of intestinal protein fermentation by Prevotella copri and other intestinal microbes can also increase the occurrence and cross-epithelial transport of by-products, such as branched-chained amino acids,5 exhibiting negative impact on insulin sensitivity in humans and mice.5
Microbial crosstalk and its impact on probiotic engraftment potential
One of the main implications of the intestinal microbiota is its resistance to incoming microbes. This effect, commonly known as competitive exclusion or colonization resistance, protects the host from infections with pathogens but also affects the ability of probiotic organisms to persist in the GI tract. Fast-growing organisms or organisms with a high affinity for available nutrients have an advantage to outcompete other, less well-adapted counterparts. Still, growth rate is not the only competitive edge observed in microbial communities. Speedy microbes (high motility), even when growing slow, may colonize new microhabitats faster than their immediate competitors,46 presenting speed, in either growth or motility, as a key evolutionary trait to facilitate efficient colonization.
In addition to competition, there is also collaboration between intestinal microbes. One of the most common examples of this is cross-feeding. As briefly alluded to above, metabolites from primary fermenters can be consumed by secondary fermenters as energy sources while simultaneously relieving primary fermenters from metabolite inhibition. For example, lactate is commonly formed by many primary fermenter species, among others lactobacilli and bifidobacteria, and is metabolized further by secondary fermenters to, e.g., propionate and butyrate. Besides carbon and nitrogen cross-feeding, there is also cross-feeding of micronutrients, including vitamins.47 To what extent this affects local probiotic persistence remains to be determined. A general observation is, however, that probiotics cannot be detected in feces 1–2 weeks after consumption has ceased,48 although exceptions exist.49
The chit-chatting behavior between the microbial community and host immunity
Apart from an apparent vertical variation in GI physiology and microbial colonization as described above, the anatomy of the intestinal immune system also exhibits profound variation from the SI to the colon. Specifically, the SI harbors visible Peyer’s patches (PPs), which mature in response to the microbiota. PPs are characterized by a follicle-associated epithelium interspersed with microfold cells (M cells), which are important for antigen uptake from the lumen into the underlying lymphoid tissue. As their presence reflects microbial loads, PPs are more abundant in the ileum than in the proximal SI.23
The large intestine contains colonic patches and the cecal patch. Isolated lymphoid follicles are also present, covering all stages of maturity from immature cryptopatches to mature isolated lymphoid follicles.23,50 These lymphoid structures are collectively known as gut-associated lymphoid tissue (GALT). The GALT comprises the immune-inductive site of adaptive immunity and moderates a tolerogenic or pathogen-protective immune response toward microorganisms and dietary antigens.51 The constant balancing act between tolerance and pathogen suppression may explain why the intestinal immune system is the largest and most diverse in the body.
The intestinal lamina propria, located between epithelium and underlying smooth muscle cells (muscularis mucosae), consists of loose connective tissue made of extracellular matrix protein produced by mesenchymal stromal cells. This nurtures the epithelial layer and provides a structure for the diverse immune cell populations to migrate along and become regionalized along the length of the intestine.52 Certain immune cell subsets then become dominant in different parts. For example, interleukin (IL)-17-producing CD4+ T helper (Th17) cells are more abundant in the SI, while the number of peripherally induced IL-17-secreting FoxP3+RORyt+ regulatory T cells (iTregs) is higher in the colon.23 Notably, this Treg subset is induced by the gut microbiota.53,54 Exhibiting increased immunosuppressive capabilities compared to thymus-derived counterparts,55 it is an eminent example of host-microbe interactions.
The bidirectional interplay between gut microbes and host immunity was further corroborated by a recent investigation of the immunomodulatory potential of microbial antigens used as a source of dietary protein.56 This dietary intervention facilitated iTreg blooms in the small and large intestine and corrected gut dysbiosis in obese mice, restoring it to a state resembling that of lean counterparts. The latter was observed as a substantial, >5-fold increase in Parabacteroides, known for their immune-regulating capabilities.57 The increase was dependent on a functional adaptive immune system,56 suggesting that, while mutualistic microbes can orchestrate mucosal immunity, adaptive immune cells can dictate microbial composition.
In addition to the regionalization of mucosal immune subsets, intraepithelial lymphocytes also exhibit spatially separated phenotypic characteristics. Similarly, the gut epithelium comprises distinct compositions of epithelial cell types from the proximal to the distal intestine. In general, the crypts harbor the intestinal epithelial stem cell (IESC) niche, which provides all types of mature epithelial cells. These cells develop as they migrate along the crypt and through the transit-amplifying zone, away from the stem cell-maintaining factors in the crypt area.28
The only mature epithelial cell interspersed with the IESCs in the crypt is the small-intestinal-specific Paneth cell.28 Paneth cells provide IESC niche factors and secrete HDPs into the crypts, protecting this crucial area and potentially making up for the thin and discontinuous mucus layer of the SI. During IBD, ectopic Paneth cells have been reported in the colon, probably a response to the dysbiotic nature of such diseases.58 This observation is of critical importance to probiotic usage as bacteria, such as Ruminococcus gnavus, otherwise thriving in the healthy colon and even exhibit tolerogenic capabilities,59 can promote inflammation when exposed to Paneth cell derived products. In other words, disease activity is aggravated when the inflammation-driven occurrence of Paneth cells in the large intestine release lysozyme into the R. gnavus habitat, disrupting the membrane of the bacterium and releasing intracellular proinflammatory components.60
How to collect microbiota samples from the SI?
The properties of the microbiota in the human SI are not well characterized, primarily because of the challenges involved in obtaining samples from this part of the digestive tract. To enhance resolution of engraftment potential and interrogate in situ dynamics in the SI, we are in desperate need of better sampling techniques, allowing us to, e.g., investigate how the microbiome is assembled in the SI from early life to aging in the healthy population. Such information would aid in characterizing disruptions caused by GI conditions and help elucidate how probiotic administration may counter or even reverse these effects.
A recent study demonstrated that probiotic supplementation altered the antibiotic-resistance gene reservoir along the human GI tract, including in the SI, in a person-specific and antibiotic-dependent manner.61 However, as this research is still in its infancy, standardized sampling techniques with spatial and temporal resolution are urgently needed. Specifically, rather than the biopsy-related techniques frequently used in the clinic, non-invasive access to the SI microbiota will ultimately improve our understanding of SI community dynamics and enable more precise studies on the efficacy of selected probiotics at this location. An appealing option is the use of swallowable smart capsule systems that allow samples to be taken directly on site without need of endoscopy.62 The newest development of such capsules enables multi-omics analyses of the intestinal environment under physiological conditions at spatially separated locations.40 Selected sampling devices for the SI are shown in Table S1.
Future research should address the development of affordable sampling tools to provide a detailed picture of probiotic activity in the SI under physiological conditions, both among different sex and age groups of the healthy population and among those with a disease. These tools could initially be used in probiotic clinical trials and ultimately made available to practitioners and consumers for monitoring probiotic survival, engraftment, and activity.
In vivo models for studying the small-intestinal and colonic microbiota
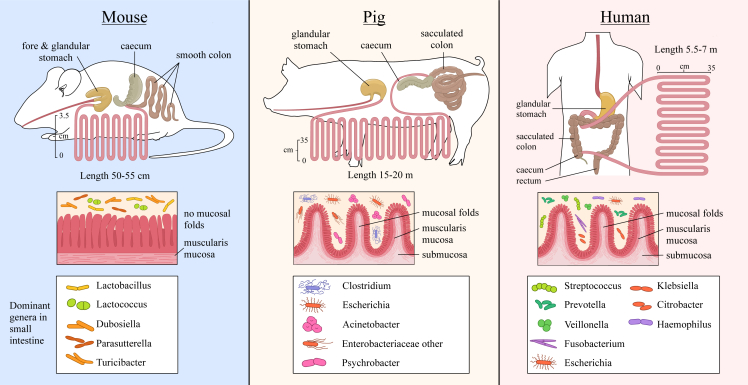
As an alternative to human samples, animals can be used to obtain valuable insights about the human microbiota. Figure 3 and Table S2 summarize specific characteristics of the human, mouse, and pig GI tract.
Figure 3.
Differences in anatomy and microbiota (on genus level) of the small intestine (SI) between two animal models (mouse and pig) vs. humans
Upper part: the length of the SI in comparison with the other gut compartments is shown. Mice have a forestomach and a glandular stomach, while pigs and humans have a glandular stomach, but the pig’s stomach is two to three times larger than the human stomach. Middle part: the anatomical structure of the luminal small-intestinal wall is shown, highlighting the absence of plicae circulares or mucosal folds in mice in contrast to pigs and humans. In mice, the finger-shaped villi are directly oriented on the small-intestinal muscle layer (muscularis mucosa), making the mucosal surface smooth. In pigs and humans, there are plicae circulares in distal duodenum, jejunum, and proximal ileum. Lower part: dominant bacterial genera in the SI.
While mice generally surpass most other preclinical models in experimental flexibility, due to their easy accessibility, modest housing requirements, short breeding cycles, and a multitude of available genetic models for delicate mode-of-action (MoA) studies, their GI tract does not resemble human physiology in adulthood. The developing intestine does, however, share marked similarities.63 The most notable differences in the adult mouse intestine relate to the lack of a transverse segment, a substantially enlarged caecum used as a fermentation sack, and a smooth outer mucosa in the SI, unlike the circular folds in the human mucosa.64 Therefore, it is reasonable to speculate whether host-microbe interactions in mouse models are relevant for studying the engraftment potential of probiotics selected for human use.
A catalog of the mouse gut metagenome was produced in 201665 and expanded in 2021.66 Although the mouse gut shares only 4% of its microbial genes (95% identity, 90% coverage) with those of the human gut microbiome, a 95% similarity was found between functional pathways. It is important to note that the mouse provider and, to some extent, housing conditions seem to have a pronounced effect on the composition of the gut microbiota,67,68 similar to household69 and/or enterotype70,71,72 differences in human studies. This potential drawback introduces heterogeneity to an otherwise homogeneous setup. On the upside, however, it can be used to (1) corroborate scientific relevance and rigor, if the data are reproduced in mice from various vendors and preferably also different strains or genotypes,56,73 and (2) tease out critical information about specific communities and, potentially, identify driver species of relevant phenotypes.74
Despite the limited overlap between the mouse and human microbiota, mice have repeatedly been used as translators in microbiome research where seminal discoveries of host-microbe interactions have been established.75,76,77,78,79 This has confirmed the relevance of mice as a notable tool for, at least, colonic microbiota research.
When evaluating the applicability of mouse models to probiotic engraftment, a key point to consider relates to the dietary composition. As gut microbiota community structures fluctuate depending on the available substrates, it is no wonder that the microbial composition in mice fed regular mouse feed differs from the microbial communities in human subjects with substantially different dietary habits. This point is evident from the few mouse studies where experimental animals were fed human diets. An experimental strategy to improve probiotic engraftment could, therefore, be to feed experimental mice a humanized diet.80 Such an approach was indeed recently employed to tease out how experimental diet affects the metabolic benefits of probiotics in obesity,81 a concept that was further corroborated in a randomized controlled human trial,82 thus advocating clinical translatability.83
Notable limitations to the above are the lack of small-intestinal samples and the fact that most samples included in the gene catalogs originate from C57BL/6J mice, which could affect their applicability in studies of other mouse strains. A recent study of the regional diversity of the gut microbiota in mice showed a high abundance of Lactobacillaceae and Muribaculaceae in the SI; this is quite different from humans.84 Another drawback of using mice is their coprophagic behavior. It has been shown that continuous self-exposure to the fecal microbiota has substantial quantitative and qualitative effects on the SI microbiota.85 By contrast, the SI of non-coprophagic mice had a lower microbial load, reduced abundance of anaerobic microbes, and bile acids predominantly in the conjugated form, resembling the human SI more closely.
As it is both difficult and ethically challenging to ensure non-coprophagic behavior in mice, other, more appropriate models may be used to study the intricate relationship between SI microbes, probiotic engraftment, and host phenomics. To this end, pigs appear as a suitable model.
Like humans, pigs are monogastric, omnivorous animals. They have a simple stomach and a relatively long SI (Figure 3).86 Although the large intestinal morphology and physiology of pigs resembles the human large intestine more closely than that of mice, conclusions about the effects of orally dosed probiotics should be made with caution. Pigs lack an appendix, but their caecum is more developed compared to humans, and the colon has a spiral arrangement.87 Moreover, their large intestinal fermentation may yield up to 30% of their maintenance energy requirement, compared to an estimated 7% in humans.88
A reference gene catalog of the pig gut microbiome was collated in 201689 and recently expanded to include samples from a wide range of sources.90 The pig metagenome exhibited a higher alpha diversity than both the human and mouse microbiomes. The relative abundances of the same bacterial species were seen to differ significantly in different gut locations. A modest 9.5% overlap between the human and pig microbiome was found, while 96% of the functional pathways identified in the human catalog were also present in the pig catalog and hence were close to the mouse-human overlap described above.65 However, compared to mice, the use of pigs has notable limitations, including the higher cost of animal maintenance and husbandry, longer reproductive cycles, and retentive time to maturity. Their genetic background is also more difficult to control.87 Still, the use of cannulated pigs makes it possible to study nutrient digestibility and the dynamics of intestinal microbiota composition through repeated sampling. In addition to the collection of chyme, this model also allows easy endoscopic examination, enabling the collection of ileal effluent and fecal samples from the same individual, which is not possible in human ileostomy patients. Besides animal welfare concerns, a disadvantage could be that the cannulation itself affects GI functioning.91
This said, accurate and representative models are crucial to the study of the intestine and its interaction with the host. For this reason, in vivo models remain indispensable. By comparison, in vitro models cannot adequately simulate the full spectrum of host-microbe interactions, both within and beyond the GI tract, as discussed below.
Simulating the gut in vitro
Various in vitro systems have been developed to simulate different parts of the intestine. The systems range from batch-fermentation models for parts of the SI or colon to continuous-flow models for the upper gut and/or colon.92,93,94,95 While the SI models typically mimic host physiology (digestive enzymes, bile, pH), allowing studies of digestion and absorption, the lower-gut models simulate the colonic microbiota, enabling studies of microbial fermentation processes. Although these models do not generally simulate the host response or the mucosal layer, they can simulate the in vivo situation reasonably well, thus providing an opportunity to investigate the influence of different environments on, for example, probiotic survival, fiber fermentation, and microbiota composition and activity.
An improvement to these models would be to include a simulated SI microbiota, which has only been attempted in a limited number of studies. Cieplak et al.96 developed “the smallest ntestine” (TSI) stand-alone model where seven strains from six genera (Escherichia, Streptococcus, Enterococcus, Bacteroides, Veillonella, Flavonifractor) were incubated for 8 h to simulate the duodenum, jejunum, and ileum. The growth and activity of the various species were not investigated, however, which is necessary to understand how closely the TSI model represents the colonization of SI microbiota. Stolaki et al.43 developed a simulation model of the ileal microbiota using TNO gastro-intestinal model (TIM)-1 and TIM-2 technology.97,98 Here, human ileostomy effluent and fecal inocula were incubated in environmental conditions representative of the ileum (3.5-h transit time, pH 7.2) over a period of 14 days, resulting in the establishment of an in vitro microbiota with a degree of similarity to in vivo ileal samples. However, the microbiota was dominated by phyla such as Bacteroidota that typically enrich colonic in vitro models, possibly due to the high content of fibers in the simulation medium.99 Finally, Roussel et al.100 developed an extension of the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME) model101 in which an ileum reactor was seeded with a suspension from the simulated ascending colon reactor. Following a 20-day incubation period, during which a fill-and-draw principle was applied to simulate short transit, the simulated ileal microbiota was found to be strongly enriched with Pseudomonadota. This was potentially due to the addition of simple sugars to the nutritional medium.
Compared to in vitro models for the human colon microbiota, models for the small-intestinal microbiota are at an early stage of development. While the studies described above have conducted essential pioneering work, a method to preserve the composition and functionality of an in vivo-derived small-intestinal microbial community has yet to be developed. Biorelevance for the in vivo situation must be demonstrated before in vitro models may be used to broaden our understanding of how probiotics thrive in the SI environment.
Intestinal epithelial in vitro and ex vivo models can be valuable to study host-microbe interactions, as summarized elsewhere.102,103 Although widely used, immortalized epithelial cell lines, such as Caco-2 and HT29, possess many non-physiological characteristics. This can be overcome in part using 3D stem cell-based organoids or organ-on-a-chip approaches. Comprising different cell types and embedment in an extracellular matrix, these can enhance the reflection of in vivo microarchitecture.102,104 Use of primary intestinal cells has the additional advantage that host genetic features, including disease specific traits, can be captured. The models mentioned facilitate studies of the direct interactions between probiotic strains or their metabolites and epithelial cells under normal and stressed conditions. They have been used to study the preventive effects of probiotics on bacterial adhesion or invasion and barrier dysfunction, including underlying mechanisms.103 Still, major drawbacks relate to their limited robustness for high-throughput analyses, poor accessibility to human specimens, and, especially, the oxygen requirements that hinder exposure to strict anaerobic bacteria.
Host-diet-microbe-disease interactions in the SI
Diet is the most obvious factor affecting the composition and function of the GI microbiota. In addition to dietary factors, a multitude of intestinal diseases influence microbial composition both directly (disease-microbe interactions) and indirectly (drug-microbe interactions). To this end, celiac disease (CeD) is an inflammatory disorder that primarily affects the SI, triggered by dietary gluten exposure in genetically predisposed individuals. While recognized that gluten is the trigger in CeD, evidence suggests that genetics only explain 55% of disease susceptibility, pointing toward a strong environmental influence, with intestinal microbiota involved in the pathogenesis, progression, and clinical symptoms.105 Consequently, probiotics has been proposed to relieve the condition. This includes the possibility to rebalance the altered microbial composition in the SI, thereby improving intestinal barrier function, or to detoxify gliadin peptides to reduce the load of peptides with immunogenic properties.106 Clinical trials that have studied the effect of various probiotics on the prevention or treatment of CeD, however, lack homogeneity. Despite the demonstration of good safety profiles, symptom management appears to be a greater opportunity than disease prevention.107,108
Another well-defined disease affecting the SI is food allergy (FA), starting in early childhood and representing a major public health concern.109 It is well established that gut leakiness enhances unwarranted immunity toward dietary antigens in susceptible individuals.110 However, while a perturbed barrier may increase the likelihood of an allergic response, it does not seem to be the primary cause of disease onset.110 A recent study has shown that a gradually evolving dysbiosis drives FA in infants.111 Gut microbial dysbiosis—characterized by diminished abundances of a consortium of species belonging to the Clostridiales order—inhibits the formation of iTregs in the infant GI tract. Fecal transplants from FA infants to predisposed recipient mice replicated the diminished iTreg phenotype observed in human donors. Conversely, bacteriotherapy with a consortium of species belonging to the Clostridiales order has been seen to suppress FA in experimental mice while promoting enhanced retinoid orphan receptor (ROR)-γt+ iTreg abundances in their mesenteric lymph nodes.111
ROR-γt is required to antagonize effector programs in GI Tregs,112 and Treg-specific deletion of ROR-γt removed Clostridiales-mediated protection against FA.111 This suggests that ROR-γt+ iTregs are imperative to protect against FA, a finding that lends further credence to the hypothesis of probiotic treatment as an effective strategy in FA treatment. Further support comes from conceptual findings that cell surface polysaccharides from Bifidobacterium bifidum engage with lamina propria-residing dendritic cells to promote a 2-fold upregulation of colonic iTregs.113 As one study has shown, a French-press-lysed non-intestinal bacterium, Methylococccus capsulatus Bath, facilitated a 3- to 5-fold increase in ROR-γt+ iTregs in both small and large intestine.56 The suggestion is that exogenously administered, relevant bacteria strains can not only nourish intestinal immunity but also stimulate the induction of the immune subset required to suppress FA at the preferred anatomical niche.77
Probiotic trials in IBD: Lessons learned and the potential for other GI tract diseases
IBD is the umbrella name for Crohn’s disease (CD), ulcerative colitis (UC), and IBD unclassified (IBDU), which are chronic disorders characterized by relapsing inflammation of temporal intensity in the GI tract. The multifactorial nature of IBD, including its complex underlying genetics, represents a challenge to diagnosis and therapy. A defective and leaky gut lining, contributing to chronic inflammation associated with defects in the innate and adaptive immune responses, is a hallmark of this disease. Apart from the varying symptoms, clinical diagnosis mainly distinguishes CD from UC by the fact that CD can precipitate transmural inflammation in the entire GI tract, whereas UC is restricted to mucosal inflammation in the colon.
The steep increase in the number of IBD cases over the last 50 years clearly indicates that the environment and microbiota are contributing factors alongside genetic predisposition. The microbial impact on IBD has been suspected for over a century,114 supported by animal models where disease severity is microbiota dependent,115 just as IBD-associated communities can transfer the donor phenotype to recipients of fecal microbiota transplants.116 The latter was further corroborated in recent work using multi-omics analyses of the UC fecal microbiota to identify bacterially shed proteases that promote disease severity. Bacteroides vulgatus proteases disturbed colon barrier function both in vitro and in mono-colonized, IL-10-deficient, colitis-prone mice. Disease activity was curbed by simultaneous administration of broad-spectrum protease inhibitors. The importance of host-microbe interactions in disease etiology is further rooted in the empiric observation that antibiotic use, especially at early life stages, is associated with IBD prevalence.117,118 Apart from changes in bacterial119 and viral120 populations, reduced diversity in the fungal121 community has also been observed in IBD patients.
The molecular mechanism of conventional probiotics administered in clinical trials to IBD patients has rarely been investigated, although some potential MoAs are occasionally supported by animal models. It is, however, worth noting that it is inherently difficult to translate murine findings to human IBD based on the vastly diverging disease etiology between species. Still, there is general consensus that both strain and dose are paramount for treatment efficacy. Perhaps more importantly, disease severity seems equally imperative.122,123 To gauge this disparity, it is important to consider the underlaying cause and multifactorial consequences of mentioned diseases. Both UC and CD are characterized by mucosal barrier defects that imbalance host-microbe interactions,124 and while altered community structures by probiotic administration may curb host immunity, it does not treat the underlying cause of disease, namely the barrier defect. Thus, if disease activity is too pronounced, probiotic administration may not only fail to dampen excessive immunity but on the contrary may succeed to penetrate the compromised mucosa, hence further fueling the inflammatory process, which—although installed to avoid bacterial translocation—may further compromise barrier function. Moreover, as both the molecular etiology and intestinal manifestation of the disease differs between UC and CD,125,126 both strain (predictive for its MoA) and location (small versus large intestine) are essential parameters to consider. Based on the evidence available from clinical trials to date, it seems that patients with active UC may benefit mainly from treatment with Escherichia coli Nissle 1917 and the probiotic mixture VSL#3 (now marketed as “De Simone Formulation”). The latter is a probiotic cocktail of eight live freeze-dried bacterial species comprising different lactobacilli and bifidobacteria mainly provided in high doses. The most recent Cochrane Review performed on this topic concluded that “low-certainty evidence suggests that probiotics may induce clinical remission in active UC when compared to placebo.”127 However, the same review also noted that “no evidence exists to assess whether probiotics are effective in people with severe and more extensive disease, or if specific preparations are superior to others.” Interestingly, and further corroborating the importance of location, the route of probiotic administration significantly alters its efficacy. Rectally administered Lacticaseibacillus casei DG-modified colonic microbiota, while significantly elevating mucosal IL-10 levels in UC patients.128 However, these effects were not replicated by oral administration of this strain at the same dosage, corroborating that the delivery of active strains at biologically sensible niches is crucial to maximize probiotic efficacy. The situation is, however, considerably more complicated in CD than in UC. As CD can involve any part of the GI tract, the microbial community will differ between the sites affected. For this reason, it is challenging—if not impossible—to establish generic mechanistic links to benefit patients. While animal models have supported the use of probiotics to manage CD,129 the effectiveness of probiotics, prebiotics, and synbiotics in inducing or maintaining remission in CD patients has not been confirmed.130 A 2020 Cochrane Review on the use of probiotics for the induction of CD remission concluded that “The available evidence is very uncertain about the efficacy or safety of probiotics, when compared with placebo, for induction of remission in Crohn’s disease.”131 There is a lack of well-designed randomized clinical trials in this area and further research is needed. An earlier Cochrane Review from 2006 suggested there was a lack of clear evidence to support the use of probiotics for the maintenance of remission in CD.132 However, the included studies were small. More adequately powered studies are necessary to determine if probiotics are of benefit for CD patients as a whole or subgroups thereof.
A potential confounder of probiotic trials in CD relates to genetic polymorphisms, which potentially interfere with probiotic MoA. For example, frameshift mutations in genes responsible for innate immune sensing of the intestinal microbiota, such as NOD2, IL-23R, and ATG16L1, increase the risk of IBD in humans.133,134,135 NOD2 polymorphism is a particularly interesting case that may provide conceptual learnings applicable to probiotic treatment of other GI diseases. NOD2, the first IBD susceptibility gene to be identified,134 and its polymorphism have been linked to poor effectiveness of some probiotics in CD.136 Specifically, the anti-inflammatory effect of Ligilactobacillus salivarius Ls-33 peptidoglycan correlated with the release of a specific muropeptide sensed by NOD2.137 In Paneth cells, NOD2 mediates the activation of nuclear factor κB (NF-κB), leading to the induction of defensins. Therefore, NOD2 polymorphisms in CD may reduce the production of defensins such as HD5, HD6, and hBD-2.138,139,140
Defensins are innate defense molecules produced at a variety of epithelial surfaces to balance protection against pathogens, while maintaining tolerance toward the intestinal microbiota. Attenuated expression of defensins will compromise host immunity, as seen in CD where reduced alpha defensin levels are observed in patients with ileal disease and reduced hBD-2 levels in those with a colonic form of the disease.141 In mechanistic studies, recombinant hBD-2 has been shown to curb inflammation in murine and human peripheral blood mononuclear cells via the Toll-like receptor 4 (TLR4)/NF-κB pathway. Consequently, it was possible to mitigate disease activity in three different colitis models.142 Confirming a niche for probiotic administration, flagellin shed by probiotic E. coli Nissle 1917 but not the apathogenic E. coli strain, ATCC 25922, promotes hBD-2 expression.143 As such induction is bound to be compromised in patients with frameshift mutation in the NOD2 gene, this patient group may benefit from orally administered hBD-2 but is unlikely to respond to probiotic treatments intended to stimulate endogenous defensin production. However, patients without this mutation may benefit substantially from intake of probiotic strains capable of eliciting an endogenous defensin (or other HDP) response. Such hypothesis is further supported by the above-described observation that rectally administered E. coli Nissle 1917 reduce inflammation in UC,128 which is not characterized by the mentioned frameshift mutation but rather decreased colonic mucus secretion and thus is fully capable of eliciting an appropriate hBD-2 response upon stimulation. Indeed, 3-week oral administration of Symbioflor 2, containing one strain of several viable genotypes of E. coli, to 23 healthy individuals stimulated hBD-2 synthesis and fecal excretions.144 Subsequent in vitro analyses revealed that this effect was mediated by a single genotype performing on par with E. coli Nissle 1917. Interestingly, the fecal hBD-2 peptide was still increased 9 weeks post treatment cessation, pointing toward sufficient engraftment potential of this probiotic.
Mitigating risks of potential adverse effects from probiotic use
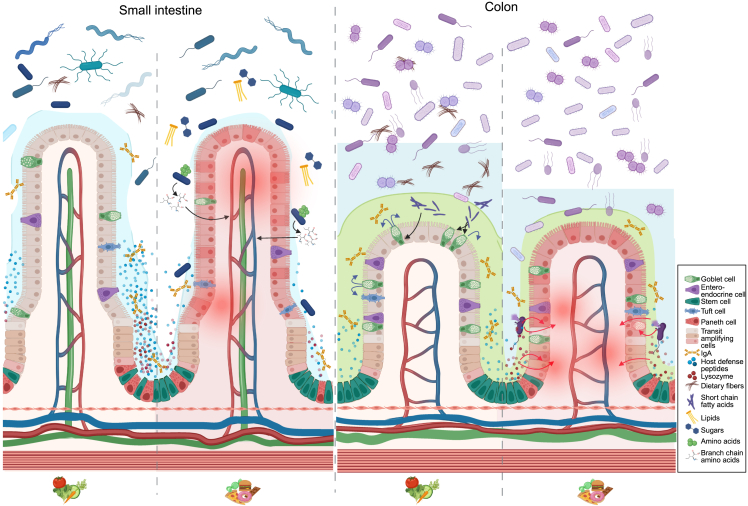
Safety is one of the prime characteristics of probiotics. However, being live microorganisms, they may cause harm under unfortunate conditions.145 For example, a recent report demonstrated that over-the-counter probiotic usage curbed the efficacy of checkpoint inhibitor immunotherapy.146,147 During clinical trials, it is important to assess such risks and act promptly in the event of adverse effects. As studies of small intestinal bacterial overgrowth (SIBO) show, the endogenous microbiota may also become detrimental to health. While the organisms associated with SIBO are similar to those commonly detected in the SI, their higher numbers cause symptoms to develop,148 driven by a breakdown in defense mechanisms, such as the gastric acid barrier.149 Similar breakdowns in microbial control mechanisms due to diet, lifestyle, or disease may increase sensitivity to otherwise benign endogenous microbes.150 For this reason, it is of crucial importance to monitor and discuss the potential side effects of probiotics. Context-dependent microbial traits observed for P. copri, Akkermansia muciniphila, and R. gnavus (Figure 4), to name a few, may cause intestinal microbes generally regarded as safe to turn into foes.77 This is especially relevant in susceptible hosts, including those with a leaky intestinal barrier or altered immune function.
Figure 4.
Regional differences in host-diet-microbe interactions
In the healthy SI, conditioned by high-fiber dietary intake, crypt-residing Paneth cells secrete an ample amount of host defense peptides (HDPs). Barrier integrity is further bolstered by the loosely adherent mucus layer ensured by well-functioning goblet cells. Changing dietary habits from a balanced high-fiber diet to a typical westernized diet (low in fiber; high in fat, animal protein, and simple sugars) disrupts immune balance, mucus production, Paneth cell function, and microbiota composition. On a westernized diet, but not a balanced fiber-rich diet, Prevotella copri was able to enhance cross-epithelial transport of branched-chain amino acids (BCAAs), potentiating metabolic syndrome. Next, while dietary fibers are passing through the SI, they are metabolized by colonic microbes. The microbial metabolites from this process, SCFAs, stimulate goblet cell function, hence enhancing barrier integrity by increased mucus production. Conversely, in the inflamed colon—often mediated by disease activity potentiated by westernized diets—decreased mucus production enables bacterial encroachment to the otherwise sterile inner mucus layer. As a host-mediated counterresponse, this process fuels Paneth cell hyperplasia, enabling this cell type that is otherwise restricted to the SI to steadily emerge in the inflamed colon. Here, they secrete HDPs and lysozyme to ward of potential introducers. Since the microbial composition in colon diverges from the SI, so will the physiological response to, e.g., host-mediated defense mechanisms. As an example, Paneth cell-secreted lysozyme is one of the most important protectors of bacterial invasion in the SI. However, when these cells emerge in the inflamed colon, their secreted lysozyme promotes bacterial lysis of microbes. This includes the otherwise beneficial Ruminococcus gnavus, which is overly abundant in colon but not SI. Lysis of, e.g., R. gnavus liberates inflammatory effector molecules potentiating intestinal inflammation.41
Conclusions
Most of our understanding of the human intestinal microbiota is based on analyses of fecal samples. For this reason, there is a need for tools such as capsule systems to study the intestinal microbiota in the upper GI tract. In vitro and in vivo models provide substantial opportunities to study the interaction between the intestinal microbiota and various organ systems. However, these models need to be further developed, particularly to increase translatability. Finally, to improve our understanding of the mechanisms of probiotic action in general and in the SI in particular, our knowledge of the intestinal microbiota and its activity needs to be expanded beyond the colon to also include the upper parts of the GI tract.
Acknowledgments
This work was conducted by an expert group (EG) of ILSI Europe. Composition of the EG is listed on the ILSI Europe website at https://ilsi.eu/scientific-activities/nutrition/probiotics/. ILSI Europe facilitated scientific meetings and coordinated the overall project management and administrative tasks relating to the completion of this work.
This manuscript was copy edited by Cath Mersh. Traveling and accommodation expenses were covered by ILSI Europe.
Author contributions
All authors contributed to design and writing of the first manuscript draft. B.A.H.J. and A.C.O. led the process of editing and contextualizing the body text while integrating the contributions of each individual author. B.A.H.J. was responsible for the final editing of the manuscript. First and last authors are positioned according to standard guidelines. Remaining authors are ordered alphabetically, emphasizing the joint effort.
Declaration of interests
A.C.O., M.N., B.P., D.S., L.G.W.S., and P.V.D.A. work full time for IFF, ADM, Yakult, Novozymes, Caelus Health, and Cryptobiotix respectively. These companies were not involved in carrying out this research.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101190.
Contributor Information
Benjamin Anderschou Holbech Jensen, Email: benjamin.jensen@sund.ku.dk.
Naomi Vita Venlet, Email: publications@ilsieurope.be.
Supplemental information
References
- 1.Lynch S.V., Pedersen O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Hallen-Adams H.E., Suhr M.J. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaswal K., Todd O.A., Behnsen J. Neglected gut microbiome: interactions of the non-bacterial gut microbiota with enteric pathogens. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2226916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg G., Rybakova D., Fischer D., Cernava T., Vergès M.C.C., Charles T., Chen X., Cocolin L., Eversole K., Corral G.H., et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A.H., Forslund K., Hildebrand F., Prifti E., Falony G., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 6.Ni J., Shen T.C.D., Chen E.Z., Bittinger K., Bailey A., Roggiani M., Sirota-Madi A., Friedman E.S., Chau L., Lin A., et al. A role for bacterial urease in gut dysbiosis and Crohn's disease. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., Zeng L., Chen J., Fan S., Du X., et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol. Psychiatr. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y., Wang W., Tang W., Tan Z., Shi J., et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Davar D., Dzutsev A.K., McCulloch J.A., Rodrigues R.R., Chauvin J.M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruch E.N., Youngster I., Ben-Betzalel G., Ortenberg R., Lahat A., Katz L., Adler K., Dick-Necula D., Raskin S., Bloch N., et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 14.Weersma R.K., Zhernakova A., Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–1519. doi: 10.1136/gutjnl-2019-320204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paik D., Yao L., Zhang Y., Bae S., D’Agostino G.D., Zhang M., Kim E., Franzosa E.A., Avila-Pacheco J., Bisanz J.E., et al. Human gut bacteria produce Η17-modulating bile acid metabolites. Nature. 2022;603:907–912. doi: 10.1038/s41586-022-04480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Toole P.W., Marchesi J.R., Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 17.Kurtz C.B., Millet Y.A., Puurunen M.K., Perreault M., Charbonneau M.R., Isabella V.M., Kotula J.W., Antipov E., Dagon Y., Denney W.S., et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 18.Isabella V.M., Ha B.N., Castillo M.J., Lubkowicz D.J., Rowe S.E., Millet Y.A., Anderson C.L., Li N., Fisher A.B., West K.A., et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 19.Pedrolli D.B., Ribeiro N.V., Squizato P.N., de Jesus V.N., Cozetto D.A., Team AQA Unesp at iGEM 2017. Gracindo A., Cesar M.B., Freire P.J.C., da Costa A.F.M., et al. Engineering Microbial Living Therapeutics: The Synthetic Biology Toolbox. Trends Biotechnol. 2019;37:100–115. doi: 10.1016/j.tibtech.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Hedin K.A., Rees V.E., Zhang H., Kruse V., Vazquez-Uribe R., Sommer M.O.A. Effects of broad-spectrum antibiotics on the colonisation of probiotic yeast Saccharomyces boulardii in the murine gastrointestinal tract. Sci. Rep. 2022;12:8862. doi: 10.1038/s41598-022-12806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedin K.A., Kruse V., Vazquez-Uribe R., Sommer M.O.A. Biocontainment strategies for in vivo applications of Saccharomyces boulardii. Front. Bioeng. Biotechnol. 2023;11:1136095. doi: 10.3389/fbioe.2023.1136095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hager C.L., Ghannoum M.A. The mycobiome: Role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig. Liver Dis. 2017;49:1171–1176. doi: 10.1016/j.dld.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Mowat A.M., Agace W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 24.Konjar Š., Pavšič M., Veldhoen M. Regulation of Oxygen Homeostasis at the Intestinal Epithelial Barrier Site. Int. J. Mol. Sci. 2021;22:9170. doi: 10.3390/ijms22179170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalantzi L., Goumas K., Kalioras V., Abrahamsson B., Dressman J.B., Reppas C. Characterization of the human upper gastrointestinal contents under conditions simulating bioavailability/bioequivalence studies. Pharm. Res. (N. Y.) 2006;23:165–176. doi: 10.1007/s11095-005-8476-1. [DOI] [PubMed] [Google Scholar]
- 26.Rutherfurd-Markwick K.J., Moughan P.J. Bioactive peptides derived from food. J. AOAC Int. 2005;88:955–966. [PubMed] [Google Scholar]
- 27.Dailey M.J. Nutrient-induced intestinal adaption and its effect in obesity. Physiol. Behav. 2014;136:74–78. doi: 10.1016/j.physbeh.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 29.Umar S. Intestinal stem cells. Curr. Gastroenterol. Rep. 2010;12:340–348. doi: 10.1007/s11894-010-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atuma C., Strugala V., Allen A., Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 31.Johansson M.E.V., Hansson G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macierzanka A., Mackie A.R., Krupa L. Permeability of the small intestinal mucus for physiologically relevant studies: Impact of mucus location and ex vivo treatment. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bevins C.L., Salzman N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 34.Dolan B., Ermund A., Martinez-Abad B., Johansson M.E.V., Hansson G.C. Clearance of small intestinal crypts involves goblet cell mucus secretion by intracellular granule rupture and enterocyte ion transport. Sci. Signal. 2022;15 doi: 10.1126/scisignal.abl5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder B.O., Stange E.F., Wehkamp J. Waking the wimp: Redox-modulation activates human beta-defensin 1. Gut Microb. 2011;2:262–266. doi: 10.4161/gmic.2.4.17692. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder B.O., Wu Z., Nuding S., Groscurth S., Marcinowski M., Beisner J., Buchner J., Schaller M., Stange E.F., Wehkamp J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 37.Ehmann D., Wendler J., Koeninger L., Larsen I.S., Klag T., Berger J., Marette A., Schaller M., Stange E.F., Malek N.P., et al. Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc. Natl. Acad. Sci. USA. 2019;116:3746–3751. doi: 10.1073/pnas.1817376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniel N., Lécuyer E., Chassaing B. Host/microbiota interactions in health and diseases-Time for mucosal microbiology. Mucosal Immunol. 2021;14:1006–1016. doi: 10.1038/s41385-021-00383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folz J., Culver R.N., Morales J.M., Grembi J., Triadafilopoulos G., Relman D.A., Huang K.C., Shalon D., Fiehn O. Human metabolome variation along the upper intestinal tract. Nat. Metab. 2023;5:777–788. doi: 10.1038/s42255-023-00777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shalon D., Culver R.N., Grembi J.A., Folz J., Treit P.V., Shi H., Rosenberger F.A., Dethlefsen L., Meng X., Yaffe E., et al. Profiling the human intestinal environment under physiological conditions. Nature. 2023;617:581–591. doi: 10.1038/s41586-023-05989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kastl A.J., Jr., Terry N.A., Wu G.D., Albenberg L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2020;9:33–45. doi: 10.1016/j.jcmgh.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoetendal E.G., Raes J., van den Bogert B., Arumugam M., Booijink C.C.G.M., Troost F.J., Bork P., Wels M., de Vos W.M., Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolaki M., Minekus M., Venema K., Lahti L., Smid E.J., Kleerebezem M., Zoetendal E.G. Microbial communities in a dynamic in vitro model for the human ileum resemble the human ileal microbiota. FEMS Microbiol. Ecol. 2019;95 doi: 10.1093/femsec/fiz096. [DOI] [PubMed] [Google Scholar]
- 44.Volk N., Lacy B. Anatomy and Physiology of the Small Bowel. Gastrointest. Endosc. Clin. N. Am. 2017;27:1–13. doi: 10.1016/j.giec.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Jones B.V., Begley M., Hill C., Gahan C.G.M., Marchesi J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gude S., Pinçe E., Taute K.M., Seinen A.B., Shimizu T.S., Tans S.J. Bacterial coexistence driven by motility and spatial competition. Nature. 2020;578:588–592. doi: 10.1038/s41586-020-2033-2. [DOI] [PubMed] [Google Scholar]
- 47.Culp E.J., Goodman A.L. Cross-feeding in the gut microbiome: Ecology and mechanisms. Cell Host Microbe. 2023;31:485–499. doi: 10.1016/j.chom.2023.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taverniti V., Koirala R., Dalla Via A., Gargari G., Leonardis E., Arioli S., Guglielmetti S. Effect of Cell Concentration on the Persistence in the Human Intestine of Four Probiotic Strains Administered through a Multispecies Formulation. Nutrients. 2019;11 doi: 10.3390/nu11020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merenstein D., Pot B., Leyer G., Ouwehand A.C., Preidis G.A., Elkins C.A., Hill C., Lewis Z.T., Shane A.L., Zmora N., et al. Emerging issues in probiotic safety: 2023 perspectives. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2185034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenton T.M., Jørgensen P.B., Niss K., Rubin S.J.S., Mörbe U.M., Riis L.B., Da Silva C., Plumb A., Vandamme J., Jakobsen H.L., et al. Immune Profiling of Human Gut-Associated Lymphoid Tissue Identifies a Role for Isolated Lymphoid Follicles in Priming of Region-Specific Immunity. Immunity. 2020;52:557–570.e6. doi: 10.1016/j.immuni.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pabst O., Mowat A.M. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roulis M., Flavell R.A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation. 2016;92:116–131. doi: 10.1016/j.diff.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Sefik E., Geva-Zatorsky N., Oh S., Konnikova L., Zemmour D., McGuire A.M., Burzyn D., Ortiz-Lopez A., Lobera M., Yang J., et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ⁺ regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt⁺ T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 55.Yang B.H., Hagemann S., Mamareli P., Lauer U., Hoffmann U., Beckstette M., Föhse L., Prinz I., Pezoldt J., Suerbaum S., et al. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 56.Jensen B.A.H., Holm J.B., Larsen I.S., von Burg N., Derer S., Sonne S.B., Pærregaard S.I., Damgaard M.V., Indrelid S.A., Rivollier A., et al. Lysates of Methylococcus capsulatus Bath induce a lean-like microbiota, intestinal FoxP3(+)RORγt(+)IL-17(+) Tregs and improve metabolism. Nat. Commun. 2021;12:1093. doi: 10.1038/s41467-021-21408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai H.C., Lin T.L., Chen T.W., Kuo Y.L., Chang C.J., Wu T.R., Shu C.C., Tsai Y.H., Swift S., Lu C.C. Gut microbiota modulates COPD pathogenesis: role of anti-inflammatory Parabacteroides goldsteinii lipopolysaccharide. Gut. 2022;71:309–321. doi: 10.1136/gutjnl-2020-322599. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka M., Saito H., Kusumi T., Fukuda S., Shimoyama T., Sasaki Y., Suto K., Munakata A., Kudo H. Spatial distribution and histogenesis of colorectal Paneth cell metaplasia in idiopathic inflammatory bowel disease. J. Gastroenterol. Hepatol. 2001;16:1353–1359. doi: 10.1046/j.1440-1746.2001.02629.x. [DOI] [PubMed] [Google Scholar]
- 59.Zigdon M., Bel S. Lysozyme: A Double-Edged Sword in the Intestine. Trends Immunol. 2020;41:1054–1056. doi: 10.1016/j.it.2020.10.010. [DOI] [PubMed] [Google Scholar]
- 60.Yu S., Balasubramanian I., Laubitz D., Tong K., Bandyopadhyay S., Lin X., Flores J., Singh R., Liu Y., Macazana C., et al. Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine. Immunity. 2020;53:398–416.e8. doi: 10.1016/j.immuni.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montassier E., Valdés-Mas R., Batard E., Zmora N., Dori-Bachash M., Suez J., Elinav E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021;6:1043–1054. doi: 10.1038/s41564-021-00920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rios-Morales M., van Trijp M.P.H., Rösch C., An R., Boer T., Gerding A., de Ruiter N., Koehorst M., Heiner-Fokkema M.R., Schols H.A., et al. A toolbox for the comprehensive analysis of small volume human intestinal samples that can be used with gastrointestinal sampling capsules. Sci. Rep. 2021;11:8133. doi: 10.1038/s41598-021-86980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanford A.H., Gong H., Noonan M., Lewis A.N., Gong Q., Lanik W.E., Hsieh J.J., Lueschow S.R., Frey M.R., Good M., McElroy S.J. A direct comparison of mouse and human intestinal development using epithelial gene expression patterns. Pediatr. Res. 2020;88:66–76. doi: 10.1038/s41390-019-0472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao L., Feng Q., Liang S., Sonne S.B., Xia Z., Qiu X., Li X., Long H., Zhang J., Zhang D., et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J., Ren H., Zhong H., Li X., Zou Y., Han M., Li M., Madsen L., Kristiansen K., Xiao L. An Expanded Gene Catalog of Mouse Gut Metagenomes. mSphere. 2021;6 doi: 10.1128/mSphere.01119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S., Deng L., Bry L., Gordon J.I., Kahn C.R. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metabol. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hildebrand F., Nguyen T.L.A., Brinkman B., Yunta R.G., Cauwe B., Vandenabeele P., Liston A., Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song S.J., Lauber C., Costello E.K., Lozupone C.A., Humphrey G., Berg-Lyons D., Caporaso J.G., Knights D., Clemente J.C., Nakielny S., et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2 doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knights D., Ward T.L., McKinlay C.E., Miller H., Gonzalez A., McDonald D., Knight R. Rethinking “enterotypes”. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsen I.S., Jensen B.A.H., Bonazzi E., Choi B.S.Y., Kristensen N.N., Schmidt E.G.W., Süenderhauf A., Morin L., Olsen P.B., Hansen L.B.S., et al. Fungal lysozyme leverages the gut microbiota to curb DSS-induced colitis. Gut Microb. 2021;13 doi: 10.1080/19490976.2021.1988836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanov I.I., Frutos R.d.L., Manel N., Yoshinaga K., Rifkin D.B., Sartor R.B., Finlay B.B., Littman D.R. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chassaing B., Compher C., Bonhomme B., Liu Q., Tian Y., Walters W., Nessel L., Delaroque C., Hao F., Gershuni V., et al. Randomized Controlled-Feeding Study of Dietary Emulsifier Carboxymethylcellulose Reveals Detrimental Impacts on the Gut Microbiota and Metabolome. Gastroenterology. 2022;162:743–756. doi: 10.1053/j.gastro.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., Gewirtz A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jensen S.K., Pærregaard S.I., Brandum E.P., Jørgensen A.S., Hjortø G.M., Jensen B.A.H. Rewiring host-microbe interactions and barrier function during gastrointestinal inflammation. Gastroenterol. Rep. 2022;10:goac008. doi: 10.1093/gastro/goac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruff W.E., Greiling T.M., Kriegel M.A. Host-microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 2020;18:521–538. doi: 10.1038/s41579-020-0367-2. [DOI] [PubMed] [Google Scholar]
- 79.Viennois E., Chassaing B. First victim, later aggressor: How the intestinal microbiota drives the pro-inflammatory effects of dietary emulsifiers? Gut Microb. 2018;9:1–4. doi: 10.1080/19490976.2017.1421885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lundsgaard A.M., Holm J.B., Sjøberg K.A., Bojsen-Møller K.N., Myrmel L.S., Fjære E., Jensen B.A.H., Nicolaisen T.S., Hingst J.R., Hansen S.L., et al. Mechanisms Preserving Insulin Action during High Dietary Fat Intake. Cell Metabol. 2019;29:50–63.e4. doi: 10.1016/j.cmet.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 81.Larsen I.S., Choi B.S.Y., Föh B., Kristensen N.N., Ouellette A., Haller R.F., Olsen P.B., Saulnier D., Sina C., Jensen B.A.H., Marette A. Experimental diets dictate the metabolic benefits of probiotics in obesity. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2192547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wastyk H.C., Perelman D., Topf M., Fragiadakis G.K., Robinson J.L., Sonnenburg J.L., Gardner C.D., Sonnenburg E.D. Randomized controlled trial demonstrates response to a probiotic intervention for metabolic syndrome that may correspond to diet. Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2178794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rytter H., Combet E., Chassaing B. Probiotic: is diet part of the efficacy equation? Gut Microb. 2023;15 doi: 10.1080/19490976.2023.2222438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lkhagva E., Chung H.-J., Hong J., Tang W.H.W., Lee S.-I., Hong S.-T., Lee S. The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol. 2021;21:44. doi: 10.1186/s12866-021-02099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bogatyrev S.R., Rolando J.C., Ismagilov R.F. Self-reinoculation with fecal flora changes microbiota density and composition leading to an altered bile-acid profile in the mouse small intestine. Microbiome. 2020;8:19. doi: 10.1186/s40168-020-0785-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kararli T.T. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm. Drug Dispos. 1995;16:351–380. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 87.Ziegler A., Gonzalez L., Blikslager A. Large Animal Models: The Key to Translational Discovery in Digestive Disease Research. Cell. Mol. Gastroenterol. Hepatol. 2016;2:716–724. doi: 10.1016/j.jcmgh.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heinritz S.N., Mosenthin R., Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr. Res. Rev. 2013;26:191–209. doi: 10.1017/s0954422413000152. [DOI] [PubMed] [Google Scholar]
- 89.Xiao L., Estellé J., Kiilerich P., Ramayo-Caldas Y., Xia Z., Feng Q., Liang S., Pedersen A.Ø., Kjeldsen N.J., Liu C., et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 90.Chen C., Zhou Y., Fu H., Xiong X., Fang S., Jiang H., Wu J., Yang H., Gao J., Huang L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021;12:1106. doi: 10.1038/s41467-021-21295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Millet S., Van Oeckel M.J., Aluwé M., Delezie E., De Brabander D.L. Prediction of in vivo short-chain fatty acid production in hindgut fermenting mammals: problems and pitfalls. Crit. Rev. Food Sci. Nutr. 2010;50:605–619. doi: 10.1080/10408390802565939. [DOI] [PubMed] [Google Scholar]
- 92.Dupont D., Alric M., Blanquet-Diot S., Bornhorst G., Cueva C., Deglaire A., Denis S., Ferrua M., Havenaar R., Lelieveld J., et al. Can dynamic in vitro digestion systems mimic the physiological reality? Crit. Rev. Food Sci. Nutr. 2019;59:1546–1562. doi: 10.1080/10408398.2017.1421900. [DOI] [PubMed] [Google Scholar]
- 93.Pérez-Burillo S., Molino S., Navajas-Porras B., Valverde-Moya Á.J., Hinojosa-Nogueira D., López-Maldonado A., Pastoriza S., Rufián-Henares J.Á. An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality. Nat. Protoc. 2021;16:3186–3209. doi: 10.1038/s41596-021-00537-x. [DOI] [PubMed] [Google Scholar]
- 94.Van de Wiele T., Van den Abbeele P., Ossieur W., Possemiers S., Marzorati M. In: The Impact of Food Bioactives on Health: in vitro and ex vivo models. Verhoeckx K., Cotter P., López-Expósito I., Kleiveland C., Lea T., Mackie A., Requena T., Swiatecka D., Wichers H., editors. Springer Copyright; 2015. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME(®)) pp. 305–317. The Author(s). [DOI] [PubMed] [Google Scholar]
- 95.van Trijp M.P.H., Rösch C., An R., Keshtkar S., Logtenberg M.J., Hermes G.D.A., Zoetendal E.G., Schols H.A., Hooiveld G.J.E.J. Fermentation Kinetics of Selected Dietary Fibers by Human Small Intestinal Microbiota Depend on the Type of Fiber and Subject. Mol. Nutr. Food Res. 2020;64 doi: 10.1002/mnfr.202000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cieplak T., Wiese M., Nielsen S., Van de Wiele T., van den Berg F., Nielsen D.S. The Smallest Intestine (TSI)—a low volume in vitro model of the small intestine with increased throughput. FEMS Microbiol. Lett. 2018;365 doi: 10.1093/femsle/fny231. [DOI] [PubMed] [Google Scholar]
- 97.Minekus M., Marteau P., Havenaar R., Veld J.H.H.i. A Multicompartmental Dynamic Computer-controlled Model Simulating the Stomach and Small Intestine. Altern. Lab. Anim. 1995;23:197–209. doi: 10.1177/026119299502300205. [DOI] [Google Scholar]
- 98.Minekus M., Smeets-Peeters M., Bernalier A., Marol-Bonnin S., Havenaar R., Marteau P., Alric M., Fonty G., Huis in't Veld J.H. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 1999;53:108–114. doi: 10.1007/s002530051622. [DOI] [PubMed] [Google Scholar]
- 99.Van den Abbeele P., Grootaert C., Marzorati M., Possemiers S., Verstraete W., Gérard P., Rabot S., Bruneau A., El Aidy S., Derrien M., et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 2010;76:5237–5246. doi: 10.1128/aem.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roussel C., De Paepe K., Galia W., De Bodt J., Chalancon S., Leriche F., Ballet N., Denis S., Alric M., Van de Wiele T., Blanquet-Diot S. Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 2020;18:141. doi: 10.1186/s12915-020-00860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van den Abbeele P., Belzer C., Goossens M., Kleerebezem M., De Vos W.M., Thas O., De Weirdt R., Kerckhof F.M., Van de Wiele T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elzinga J., van der Oost J., de Vos W.M., Smidt H. The Use of Defined Microbial Communities To Model Host-Microbe Interactions in the Human Gut. Microbiol. Mol. Biol. Rev. 2019;83 doi: 10.1128/mmbr.00054-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anjum M., Laitila A., Ouwehand A.C., Forssten S.D. Current Perspectives on Gastrointestinal Models to Assess Probiotic-Pathogen Interactions. Front. Microbiol. 2022;13:831455. doi: 10.3389/fmicb.2022.831455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shin W., Kim H.J. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat. Protoc. 2022;17:910–939. doi: 10.1038/s41596-021-00674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Sousa Moraes L.F., Grzeskowiak L.M., de Sales Teixeira T.F., Gouveia Peluzio M.d.C. Intestinal microbiota and probiotics in celiac disease. Clin. Microbiol. Rev. 2014;27:482–489. doi: 10.1128/cmr.00106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caminero A., Galipeau H.J., McCarville J.L., Johnston C.W., Bernier S.P., Russell A.K., Jury J., Herran A.R., Casqueiro J., Tye-Din J.A., et al. Duodenal Bacteria From Patients With Celiac Disease and Healthy Subjects Distinctly Affect Gluten Breakdown and Immunogenicity. Gastroenterology. 2016;151:670–683. doi: 10.1053/j.gastro.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 107.Marasco G., Cirota G.G., Rossini B., Lungaro L., Di Biase A.R., Colecchia A., Volta U., De Giorgio R., Festi D., Caio G. Probiotics, Prebiotics and Other Dietary Supplements for Gut Microbiota Modulation in Celiac Disease Patients. Nutrients. 2020;12 doi: 10.3390/nu12092674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pecora F., Persico F., Gismondi P., Fornaroli F., Iuliano S., de'Angelis G.L., Esposito S. Gut Microbiota in Celiac Disease: Is There Any Role for Probiotics? Front. Immunol. 2020;11:957. doi: 10.3389/fimmu.2020.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Branum A.M., Lukacs S.L. Food allergy among children in the United States. Pediatrics. 2009;124:1549–1555. doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 110.Heyman M. Gut barrier dysfunction in food allergy. Eur. J. Gastroenterol. Hepatol. 2005;17:1279–1285. doi: 10.1097/00042737-200512000-00003. [DOI] [PubMed] [Google Scholar]
- 111.Abdel-Gadir A., Stephen-Victor E., Gerber G.K., Noval Rivas M., Wang S., Harb H., Wang L., Li N., Crestani E., Spielman S., et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 2019;25:1164–1174. doi: 10.1038/s41591-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhaumik S., Mickael M.E., Moran M., Spell M., Basu R. RORγt Promotes Foxp3 Expression by Antagonizing the Effector Program in Colonic Regulatory T Cells. J. Immunol. 2021;207:2027–2038. doi: 10.4049/jimmunol.2100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verma R., Lee C., Jeun E.J., Yi J., Kim K.S., Ghosh A., Byun S., Lee C.G., Kang H.J., Kim G.C., et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aat6975. [DOI] [PubMed] [Google Scholar]
- 114.Hawkins H.P. An Address ON THE NATURAL HISTORY OF ULCERATIVE COLITIS AND ITS BEARING ON TREATMENT. Br. Med. J. 1909;1:765–770. doi: 10.1136/bmj.1.2517.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Low D., Tran H.T., Lee I.A., Dreux N., Kamba A., Reinecker H.C., Darfeuille-Michaud A., Barnich N., Mizoguchi E. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology. 2013;145:602–612.e9. doi: 10.1053/j.gastro.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weingarden A.R., Vaughn B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microb. 2017;8:238–252. doi: 10.1080/19490976.2017.1290757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manichanh C., Borruel N., Casellas F., Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 118.Theochari N.A., Stefanopoulos A., Mylonas K.S., Economopoulos K.P. Antibiotics exposure and risk of inflammatory bowel disease: a systematic review. Scand. J. Gastroenterol. 2018;53:1–7. doi: 10.1080/00365521.2017.1386711. [DOI] [PubMed] [Google Scholar]
- 119.Khan I., Ullah N., Zha L., Bai Y., Khan A., Zhao T., Che T., Zhang C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens. 2019;8:126. doi: 10.3390/pathogens8030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zuo T., Lu X.-J., Zhang Y., Cheung C.P., Lam S., Zhang F., Tang W., Ching J.Y.L., Zhao R., Chan P.K.S., et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sokol H., Leducq V., Aschard H., Pham H.-P., Jegou S., Landman C., Cohen D., Liguori G., Bourrier A., Nion-Larmurier I., et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yan F., Polk D.B. Probiotics and Probiotic-Derived Functional Factors—Mechanistic Insights Into Applications for Intestinal Homeostasis. Front. Immunol. 2020;11:1428. doi: 10.3389/fimmu.2020.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jakubczyk D., Leszczyńska K., Górska S. The Effectiveness of Probiotics in the Treatment of Inflammatory Bowel Disease (IBD)—A Critical Review. Nutrients. 2020;12:1973. doi: 10.3390/nu12071973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stange E.F., Schroeder B.O. Microbiota and mucosal defense in IBD: an update. Expet Rev. Gastroenterol. Hepatol. 2019;13:963–976. doi: 10.1080/17474124.2019.1671822. [DOI] [PubMed] [Google Scholar]
- 125.Jäger S., Stange E.F., Wehkamp J. Inflammatory bowel disease: an impaired barrier disease. Langenbeck's Arch. Surg. 2013;398:1–12. doi: 10.1007/s00423-012-1030-9. [DOI] [PubMed] [Google Scholar]
- 126.Wehkamp J., Koslowski M., Wang G., Stange E.F. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn's disease. Mucosal Immunol. 2008;1:S67–S74. doi: 10.1038/mi.2008.48. [DOI] [PubMed] [Google Scholar]
- 127.Kaur L., Gordon M., Baines P.A., Iheozor-Ejiofor Z., Sinopoulou V., Akobeng A.K. Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst. Rev. 2020;3:CD005573. doi: 10.1002/14651858.CD005573.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.D'Incà R., Barollo M., Scarpa M., Grillo A.R., Brun P., Vettorato M.G., Castagliuolo I., Sturniolo G.C. Rectal Administration of Lactobacillus casei DG Modifies Flora Composition and Toll-Like Receptor Expression in Colonic Mucosa of Patients with Mild Ulcerative Colitis. Dig. Dis. Sci. 2011;56:1178–1187. doi: 10.1007/s10620-010-1384-1. [DOI] [PubMed] [Google Scholar]
- 129.Liang Y., Liu M., Pu J., Zhu Z., Gao Z., Zhou Q., Gu Q., Li P. Probiotics and Their Metabolites Ameliorate Inflammatory Bowel Disease: A Critical Review. Infect. Microbes Dis. 2021;3:4–13. [Google Scholar]
- 130.Akutko K., Stawarski A. Probiotics, Prebiotics and Synbiotics in Inflammatory Bowel Diseases. J. Clin. Med. 2021;10:2466. doi: 10.3390/jcm10112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Limketkai B.N., Akobeng A.K., Gordon M., Adepoju A.A. Probiotics for induction of remission in Crohn's disease. Cochrane Database Syst. Rev. 2020;7:CD006634. doi: 10.1002/14651858.CD006634.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rolfe V.E., Fortun P.J., Hawkey C.J., Bath-Hextall F.J. Probiotics for maintenance of remission in Crohn's disease. Cochrane Database Syst. Rev. 2006:CD004826. doi: 10.1002/14651858.CD004826.pub2. [DOI] [PubMed] [Google Scholar]
- 133.Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 134.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O'Morain C.A., Gassull M., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 135.Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 136.Macho Fernandez E., Pot B., Grangette C. Beneficial effect of probiotics in IBD. Gut Microb. 2011;2:280–286. doi: 10.4161/gmic.2.5.18255. [DOI] [PubMed] [Google Scholar]
- 137.Macho Fernandez E., Valenti V., Rockel C., Hermann C., Pot B., Boneca I.G., Grangette C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–1059. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 138.Wehkamp J., Wang G., Kübler I., Nuding S., Gregorieff A., Schnabel A., Kays R.J., Fellermann K., Burk O., Schwab M., et al. The Paneth Cell α-Defensin Deficiency of Ileal Crohn’s Disease Is Linked to Wnt/Tcf-4. J. Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 139.Voss E., Wehkamp J., Wehkamp K., Stange E.F., Schröder J.M., Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- 140.Fellermann K., Wehkamp J., Herrlinger K.R., Stange E.F. Crohn's disease: a defensin deficiency syndrome? Eur. J. Gastroenterol. Hepatol. 2003;15:627–634. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 141.Ramasundara M., Leach S.T., Lemberg D.A., Day A.S. Defensins and inflammation: the role of defensins in inflammatory bowel disease. J. Gastroenterol. Hepatol. 2009;24:202–208. doi: 10.1111/j.1440-1746.2008.05772.x. [DOI] [PubMed] [Google Scholar]
- 142.Koeninger L., Armbruster N.S., Brinch K.S., Kjaerulf S., Andersen B., Langnau C., Autenrieth S.E., Schneidawind D., Stange E.F., Malek N.P., et al. Human β-Defensin 2 Mediated Immune Modulation as Treatment for Experimental Colitis. Front. Immunol. 2020;11:93. doi: 10.3389/fimmu.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schlee M., Wehkamp J., Altenhoefer A., Oelschlaeger T.A., Stange E.F., Fellermann K. Induction of Human β-Defensin 2 by the Probiotic Escherichia coli Nissle 1917 Is Mediated through Flagellin. Infect. Immun. 2007;75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Möndel M., Schroeder B.O., Zimmermann K., Huber H., Nuding S., Beisner J., Fellermann K., Stange E.F., Wehkamp J. Probiotic E. coli treatment mediates antimicrobial human β-defensin synthesis and fecal excretion in humans. Mucosal Immunol. 2009;2:166–172. doi: 10.1038/mi.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Besselink M.G., van Santvoort H.C., Buskens E., Boermeester M.A., van Goor H., Timmerman H.M., Nieuwenhuijs V.B., Bollen T.L., van Ramshorst B., Witteman B.J., et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008;371:651–659. doi: 10.1016/s0140-6736(08)60207-x. [DOI] [PubMed] [Google Scholar]
- 146.Binda S., Hill C., Johansen E., Obis D., Pot B., Sanders M.E., Tremblay A., Ouwehand A.C. Criteria to Qualify Microorganisms as “Probiotic” in Foods and Dietary Supplements. Front. Microbiol. 2020;11:1662. doi: 10.3389/fmicb.2020.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Spencer C.N., McQuade J.L., Gopalakrishnan V., McCulloch J.A., Vetizou M., Cogdill A.P., Khan M.A.W., Zhang X., White M.G., Peterson C.B., et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–1640. doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sorathia S.J., Chippa V., Rivas J.M. StatPearls. StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC.; 2022. Small Intestinal Bacterial Overgrowth. [PubMed] [Google Scholar]
- 149.Zafar H., Jain A.G., Khetpal N., Ali S., Rashid M.U., Ahmad S. Small Intestinal Bacterial Overgrowth: A Critical Review of an Underrecognized but Disrupting Entity. Crit. Rev. Oncog. 2020;25:365–379. doi: 10.1615/CritRevOncog.2020036017. [DOI] [PubMed] [Google Scholar]
- 150.Bekkers M., Stojkovic B., Kaiko G.E. Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.