Highlights
-
•
Regulons ETS1 and MEF2C were exclusively upregulated in CAFs.
-
•
Two ccRCC molecular subclusters based on the communication signature, manifested distinct prognoses, immune cell infiltration landscapes, and immune-related characteristics.
-
•
Signature-derived model demonstrated high accuracy and robustness in predicting prognosis and ICIs therapy responses.
-
•
SLC38A5 was related to decreased survival probabilities, inflamed TME, and upregulated inhibitory checkpoints.
-
•
SLC38A5 inhibition could attenuate the pro-tumoral abilities of CAFs in terms of proliferation, migration, and invasion. Mechanically, CCL5 could restore these properties induced by SLC38A5 inhibition.
Keywords: Immunotherapy, Clear cell renal cell carcinoma, Cancer-associated fibroblast, Tumor microenvironment, SlC38A5
Abstract
Cancer-associated fibroblasts paly critical roles in regulating cancer cell biological properties by intricate and dynamic communication networks. But the mechanism of CAFs in clear cell renal cell carcinoma (ccRCC) is not clear. In our study, we identified CAFs and malignant cells from the integrated scRNA-seq datasets and establish a CAF-derived communication signature based on the highly activated regulons ETS1 and MEF2C. We stratified the ccRCC TME into two molecular subtypes with distinct prognoses, immune cell infiltration landscapes, and immune-related characteristics. The model derived from signature demonstrated high accuracy and robustness in predicting prognosis and ICIs therapy responses. Subsequently, the SLC38A5 of the model was found upregulated in CAFs and was related to decreased survival probabilities, inflamed TME, and upregulated inhibitory checkpoints. SLC38A5 inhibition could attenuate the pro-tumoral abilities of CAFs in terms of proliferation, migration, and invasion. Mechanically, CCL5 could restore these properties induced by SLC38A5 inhibition. In conclusion, our communication signature and its derived model enabled a more precise selection of ccRCC patients who were potential beneficiaries of ICIs. Besides, the SLC38A5-CCL5 axis may serve as a promising target for ccRCC treatment.
Graphical abstract
Introduction
Renal cell carcinoma (RCC) ranks third among all urological malignancies, with an estimated 431,288 new incidences and 179,368 deaths globally in 2020 [1]. Clear cell RCC (ccRCC) is the most common type of RCC in adults, accounting for about 70 % of the newly diagnosed [2]. The primary management of localized ccRCC is partial or radical nephrectomy, contributing to 91 % and 94 % five-year disease-free (DFS) and overall survival (OS) probabilities, respectively [3]. However, the remaining about 30 % of ccRCC patients are at an advanced stage or have developed metastasis when diagnosed [4]. For these patients, the available treatments are limited and the five-year OS decreases to less than 20 % [5].
Recently, immune checkpoint inhibitors (ICIs) therapy has revolutionized cancer treatment. Motzer et al. in their CheckMate-214 trial investigated 1096 previously untreated ccRCC patients who were randomly assigned to receive ICIs or sunitinib and found that OS and objective response rates were significantly higher with immunotherapy than with target therapy [6]. However, due to tumoral heterogeneity and the “cold” tumor microenvironment (TME), not all candidates could benefit from this strategy [7]. It has been well-documented that cancer-associated fibroblasts (CAFs) play crucial roles in promoting tumor progression and metastasis. Moreover, CAFs could secret various chemokines like CXCL12 that recruit tumor-associated macrophage (TAM) infiltration [8]. Consequently, the intricate communication networks among CAFs, TAMs, and tumor cells could induce the formation of an immunosuppressive TME that hinders the delivery of drug molecules and promotes immune escape [9]. Therefore, decoding the intercellular talk between CAFs and tumor cells can shed light on molecular mechanisms of tumor development and immunotherapeutic resistance.
To address these concerns, we unraveled the intricate communication signature between CAFs and tumor cells. Based on the signature genes, we stratified the ccRCC TME into two molecular subclusters with distinct properties. We then applied 101 machine-learning combinations of 10 algorithms to establish a consensus model based on 10-fold cross-validation. Subsequently, the hub gene SLC38A5 was identified and was related to the proliferation and migration of RCC cells. Mechanically, CCL5 could restore these properties induced by SLC38A5 inhibition.
Methods
RAN-seq datasets, scRNA-seq datasets, and ICIs cohorts
Two public ccRCC scRNA-seq datasets were procured from the GEO database (www.ncbi.nlm.nih.gov/gds/?term=) with accession numbers GSE171306 and GSE111360. TCGA-KIRC and E-MTAB-1980 RNA-seq datasets were downloaded from the GDC portal of TCGA (portal.gdc.cancer.gov/) and ArrayExpress (www.ebi.ac.uk/arrayexpress/) databases, respectively. Six ICIs cohorts including Braun cohort [10], IMvigor210 cohort [11], Liu cohort [12], Gide cohort [13], Rose cohort [14], and Van cohort [15], were obtained from either Tumor Immune Dysfunction and Exclusion (TIDE) online framework (tide.dfci.harvard.edu/) [16] or supplementary materials of the original studies.
scRNA-seq analysis
The scRNA-seq analytic procedures were conducted utilizing Seurat 4.2.0 R package [17]. Default parameters were used if not specifically listed. In brief, we performed quality control to remove low-quality cells by calculating the percentage of erythrocyte and mitochondrial genes. After normalization by the “NormalizeData” function, the top 2000 highly variable features of each dataset were identified by the “FindVariableFeatures” function. Then, we performed integration and generated an integrated data assay based on anchors selected by the “FindIntegrationAnchors” function. Subsequently, dimensionality reduction and visualization were conducted by principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) algorithms. The “FindClusters” function and previously reported markers were used to identify and annotate malignant cells and CAFs.
Pseudotime analysis, cell-cell communication, and gene regulatory network (GRN)
After malignant cells and CAFs identification, we presented the developmental trajectories by pseudotime analysis implemented in Monocle R package [18]. The procedures have been reported in previous studies [19]. Default parameters were used if not specifically listed. In brief, the subclusters of CAFs and malignant cells were loaded into the R environment. The “newCellDataSet” function was used to develop a CellDataSet Object. Cells were sorted in pseudo-time order based on genes that satisfied the thresholds (mean_expression ≥ 0.1 and dispersion_empirical ≥ 1 * dispersion_fit). The “plot_cell_trajectory” function was applied to visualize the trajectory in the reduced dimensional space.
Based on the expressions of known ligands, receptors, and their cofactors, we can calculate the number of interactions and the interaction weights between cell subpopulations. The analytic procedures were performed by CellChat R package [20]. NicheNet was employed to infer the active ligands and their targets on CAFs and malignant cells, respectively [21].
GRN analysis was performed using SCENIC R package [22]. SCENIC was a framework for GRNs reconstruction and identification of stable cell states from the scRNA-seq dataset. GRN was developed based on DNA motif and coexpression analysis. The GRN activity of each cell was calculated to infer the recurrent cellular states.
Enrichment analysis
Over Representation Analysis (ORA) was conducted to evaluate whether known biological processes or functions are enriched in marker genes of each subpopulation of CAFs. Gene Set Enrichment Analysis (GSEA) was employed to determine whether a defined gene set shows remarkable differences between two biological states. The clusterProfiler R package was employed to perform the analyses and visualize the results [23].
Gene set variation analysis (GSVA) was used to determine the pathway enrichment for each sample based on the gene sets by the sample matrix. The GSVA R package was utilized to perform the analysis [24].
Immune cell infiltration abundances were evaluated by several published methodologies including CIBERSORT, TIMER, xCell, MCPcounter, ESTIMATE, EPIC, and quanTIseq. These methodologies have been curated in the IOBR R package [25].
Unsupervised consensus clustering
Based on communication signature genes, we performed unsupervised consensus clustering of the TCGA-KIRC dataset by ConsensusClusterPlus R package [26]. A consensus matrix was generated and samples with high similarities were divided into one cluster. The optimal number of clustering was determined by cumulative distribution function (CDF) curves and the proportion of ambiguous clustering (PAC).
Communication signature-based model establishment
A total of 101 combinations of ten machine-learning algorithms, including survival support vector machine (survival-SVM), random survival forest (RSF), elastic network (Enet), generalized boosted regression modeling (GBM), supervised principal components (SuperPC), partial least squares regression for Cox (plsRcox), CoxBoost, stepwise Cox, Ridge, and Lasso based on 10-fold cross-validation were used to train and validate the model in TCGA-KIRC and E-MTAB-1980 datasets, respectively. The optimal model was the one with the highest C-index.
Immunotherapeutic response prediction
The online TIDE framework was employed to predict immunotherapeutic response. Besides, the SubMap module of the GenePattern online tool [27] was used to compare the similarities between our identified molecular clusters and the anti-PD-1 or anti-CTLA-4 checkpoint-treated melanoma cohort by Chen et al. [28].
Cell lines and culture
The experiments were approved by the Ethics Committee of Peking Union Medical College Hospital. The RCC cell lines (786-O and ACHN) came from the Cancer Institute of the Chinese Academy of Medical Sciences (Beijing, China). Primary fibroblasts were isolated from ccRCC tissue and tumor-adjacent tissue as described in a previous study [29]. In brief, tissues were first excised and minced with scissors and forceps and then washed with PBS (Beyotime, Shanghai, China) containing 20 % antibiotics (Beyotime, Shanghai, China). Collagenase type I and IV (Sigma, Saint Louis, MO, USA) were subsequently used to digest the tissues to isolate CAFs and normal fibroblasts (NFs). All cells were cultured with Dulbecco's modified eagle medium (DMEM, Gibco, Grand Island, NY, USA) with 10 % fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) and 1 % penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C with a humidity of 5 % CO2.
RNA isolation, reverse transcription, and quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted from NFs and CAFs with Trizol (Life Technologies, Gaithersburg, MD, USA) following the manufacturer's instructions. The concentration and quality of extracted RNA of each sample were evaluated by a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at a ratio of A260/A280. One Step SYBR PrimeScript RT-PCR Kit II (Takara Biomedical Technology, Tokyo, Japan) was used to qualify the expression level of SLC38A5. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference for the analysis. All qRT-PCR reactions were performed using the 7500 Fast RT-PCR System (Applied Biosystems, Waltham, MA, USA). The relative quantification 2-ΔΔCt method was applied to calculate the gene expression values. The primers of SLC38A5 were 5′-GCTACAGGCAAGAACGTGAGG- 3′ (Forward) and 5′-ATTCCAAACGATGTCTTCCCC- 3′ (Reverse).
Western blotting
Cells were harvested and lysed in RIPA buffer (Beyotime, Shanghai, China) to extract total protein. After protein concentration measurement, equivalent amounts of lysates were separated by 10 % Sodium Dodecyl Sulfate – polyacrylamide gel electrophoresis (SDS-PAGE, NCM Biotech, Suzhou, China) and then transferred to a polyvinylidene difluoride (PVDF) membrane using a semi-dry Western Blot set-up. The membrane was incubated with primary antibody overnight at 4 °C following 20 min in blocking buffer (Beyotime, Shanghai, China). The membrane was washed three times with Tris-buffered saline with Triton X-100 (TBST) and then incubated with a secondary antibody (anti-Rabbit IgG) for 2 h at room temperature. The protein bands were detected by an Enhanced Chemiluminescent (ECL) kit (NCM Biotech, Suzhou, China) and normalized to the GAPDH protein expression level. Primary antibodies used: anti-SLC38A5, anti-αSMA, anti-CCL5, anti-GAPDH. All antibodies were purchased from Abcam (Cambridge, UK).
Cell transfection, RNA-seq, and analysis
CAFs were plated in triplicates in 6-well plates (Sigma, Saint Louis, MO, USA) at a seeding density of 2 × 106 cells/well. Cells were transfected with si-SLC38A5 or si-NC (GenePharma, Shanghai, China) using Lipofectamine 3000 reagent (Life Technologies, Paisley, UK) and cultured for 48 h. RNA was extracted and samples were stored at a − 80 °C refrigerator (Haier, Qingdao, China) until analysis.
The mRNA library preparation and RNA-seq were performed as described in previous studies [30]. In brief, after quality and integrity evaluation, the mRNA library was prepared via poly-A enrichment according to the Illumina TruSeq RNA Sample Prep Kit instructions. RNA-seq was employed to detect the expression levels between si-SLC38A5 and si-NC samples using Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA).
Following quality control, the clean reads were mapped to the human genome GRCh38/ hg38 by bowtie2. DESeq2 R package was utilized to perform the differential expression analysis.
Conditioned medium (CM) and enzyme-linked immunosorbent assay (ELISA)
CAFs at approximately 80 % confluence were washed with PBS (Beyotime, Shanghai, China) and then cultured in DMEM medium without FBS. The CM was collected and filtered for further analysis.
The concentrations of CCL5 of CM derived from CAFs with conditioned SLC38A5 were determined by ELISA kit (Thermo Fisher Scientific, Waltham, MA, USA).
Cell proliferation assay
The cell proliferation assay was performed by Cell Counting Kit 8 (CCK-8, Abcam, Cambridge, UK). RCC cells (1 × 104) were seeded in 96-well plates supplemented with CM derived from CAFs and cultured for 48 h. Cells were then cultured with CCK-8 solution for another 4 h. All samples were measured at 450 nm.
Wound-healing and transwell assays
For the wound-healing assay, the wound of confluent RCC cells was created by a 200 µL pipette tip. Cells were incubated in DMEM supplemented with 2 % FBS and 50 % CM. Cell migration into wound areas was photographed every 6 h. ImageJ software was used to analyze the cell migration rate.
For the Transwell assay, RCC cells (2 × 104) were suspended in 200 μL of the CM and were plated into the upper chamber that was coated with an extracellular matrix (BD Biosciences, San Jose, CA, USA). 500 μL DMEM medium with 10 % FBS was added into the lower well. After 12 h incubation, the cells of the upper surface of the filter were removed and cells under the filter were fixed and stained with a 0.5 % crystal violet solution and counted under the microscope (Nikon, Tokyo, Japan).
Statistical analysis
All statistical analyses were performed using R (Version 4.1.3). The Shapiro–Wilk test was used to evaluate the normality property of two continuous variables. Student's t-test or Wilcoxon test were used to compare the paired independent samples. The Spearman or Pearson correlation analysis were employed to assess the relationship between the two continuous variables. Chi-squared test was used to compare categorical variables. Survival differences between the two groups were determined by Kaplan–Meier curves with the log-rank test. Multivariate cox regression analysis was used to explore the independent risk or protective factors. The time-dependent receiver operating characteristic curve (ROC) was used to evaluate the performance of the variable in predicting survival. Statistical significance was set two-tail p-value < 0.05.
Results
scRNA-seq analysis unravels the heterogeneity of ccRCC
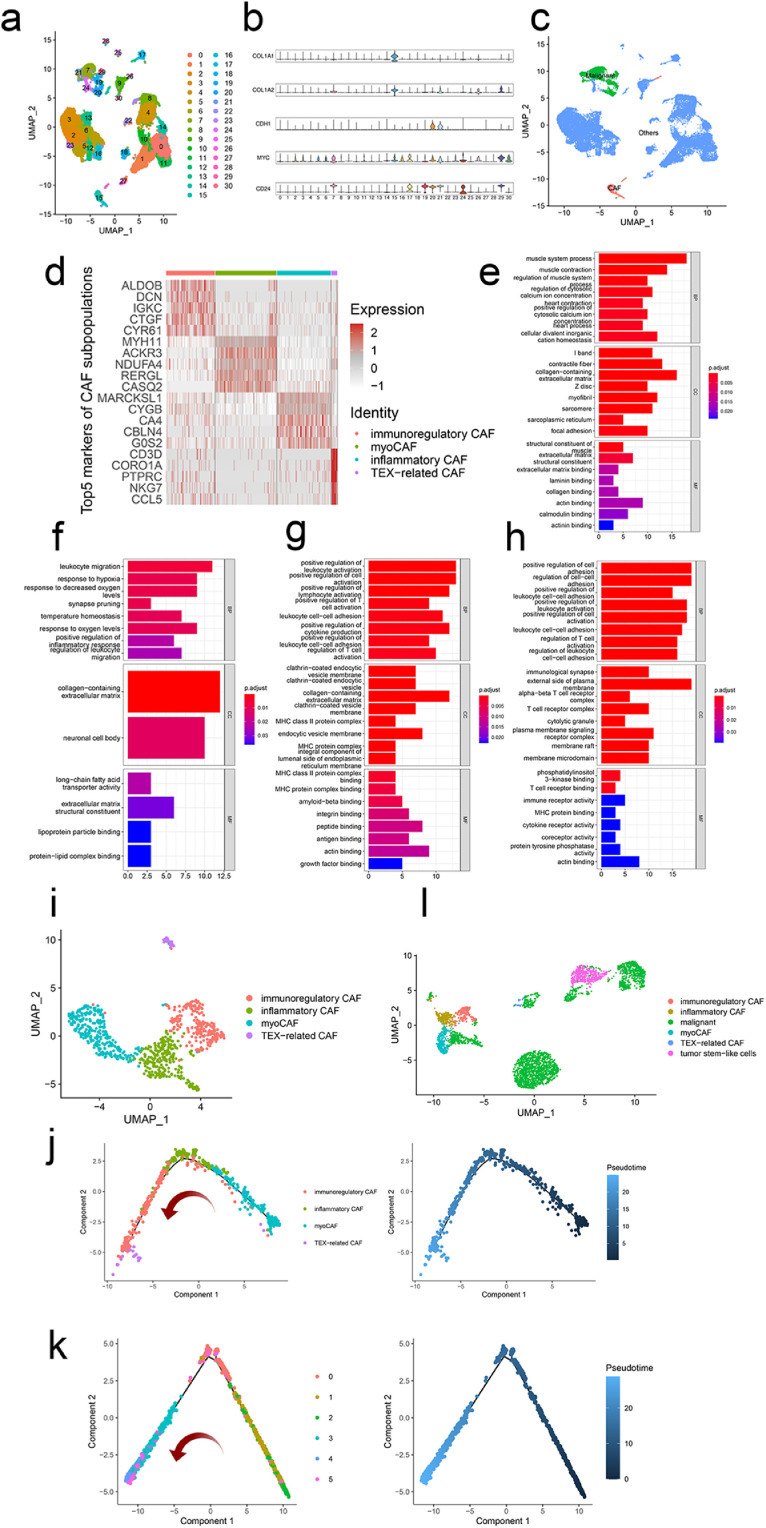
Two ccRCC scRNA-seq datasets were normalized and integrated following the Seurat pipeline (https://satijalab.org/seurat/articles/integration_introduction.html). After dimensional reduction and clustering, we obtained 31 clusters (Fig. 1a). Based on previously documented cell-type specific markers (CAFs: COL1A1 and COL1A2, malignant cells: CDH1, MYC, and CD24, Fig. 1b), cluster 15 was annotated as CAFs while clusters 7 and 19 to 21were regarded as ccRCC (Fig. 1c). Previous studies have reported the heterogeneity of CAFs and diverse properties [8]. Therefore, we re-clustered the CAFs and successfully identified four subpopulations. Fig. 1d presented the top five markers of each subpopulation. GO enrichment analysis showed that cluster 0 was related to biological processes of the muscle system and extracellular matrix (Fig. 1e). Thus, we named cluster 0 “myoCAF”. GO terms including leukocyte migration and regulation of inflammatory response were enriched in cluster 1 (Fig. 1f). Thus, we referred cluster 1 as “inflammatory CAF”. As for cluster 2, functional annotation results revealed several biological processes and pathways that are involved in immune regulation, including regulation of T cell activation and MHC family proteins (Fig. 1g). Thus, we named cluster 2 “immunoregulatory CAF”. Intriguingly, most GO-enriched terms of cluster 3 were consistent with those of cluster 2 (Fig. 1e), indicating the immunoregulatory property of cluster 3. However, KEGG analysis demonstrated the PD-L1 expression and PD-1 expression checkpoint pathway was related to cluster 3, suggesting the possible T cells exhaustion (TEX)-related characteristic of these cells (Fig. S1c). Therefore, cluster 3 was named “TEX-related CAF” (Fig. 1i).
Fig. 1.
scRNA-seq analysis unravels the heterogeneity of ccRCC. (a) 31 clusters were identified in the integrated scRNA-seq dataset. (b) Expression levels of cell type-specific markers (CAFs: COL1A1 and COL1A2, malignant cells: CDH1, MYC, and CD24). (c) Identification of CAFs and malignant cells. (d) The top five markers of each subcluster of CAFs. GO enrichment terms for subcluster 0 (e), 1 (f), 2 (g), and 3 (h) of CAFs. (i) Annotation of four subclusters of CAFs. The trajectory path of CAFs (j) and malignant cells (k). (l) Annotation of subclusters of CAFs and malignant cells.
Pseudotime analysis was employed to verify our hypothesis. Fig. 1j showed that myoCAF may be the origin of three other subtypes as these cells were located at the beginning of the trajectory path. Interestingly, immunoregulatory and inflammatory CAFs were intermixed on the trajectory of these lineages. The similar KEGG enrichment analysis results further support the findings (Fig. S1a, b), indicating the intimate relations between these two subtypes in the TME. TEX-related CAF was in the terminal end, consistent with the transformation of anti-tumor immunity that was highly activated at the early stage of the tumor whereas inhibited at the late stage.
For the trajectory of ccRCC, cluster 2 was located at the beginning of the trajectory path and was regarded as the tumor stem-like cells [31]. Clusters 3 to 5 were mixed in the terminal end, indicating the heterogeneity of tumor cells of ccRCC (Fig. 1k).
Considering the critical roles of CAFs in promoting tumor development and progression, it is of great necessity of deciphering the reciprocal communication networks. Therefore, CAFs and malignant cells were extracted for the following analysis (Fig. 1l).
Regulons ETS1 and MEF2C are exclusively activated in CAFs
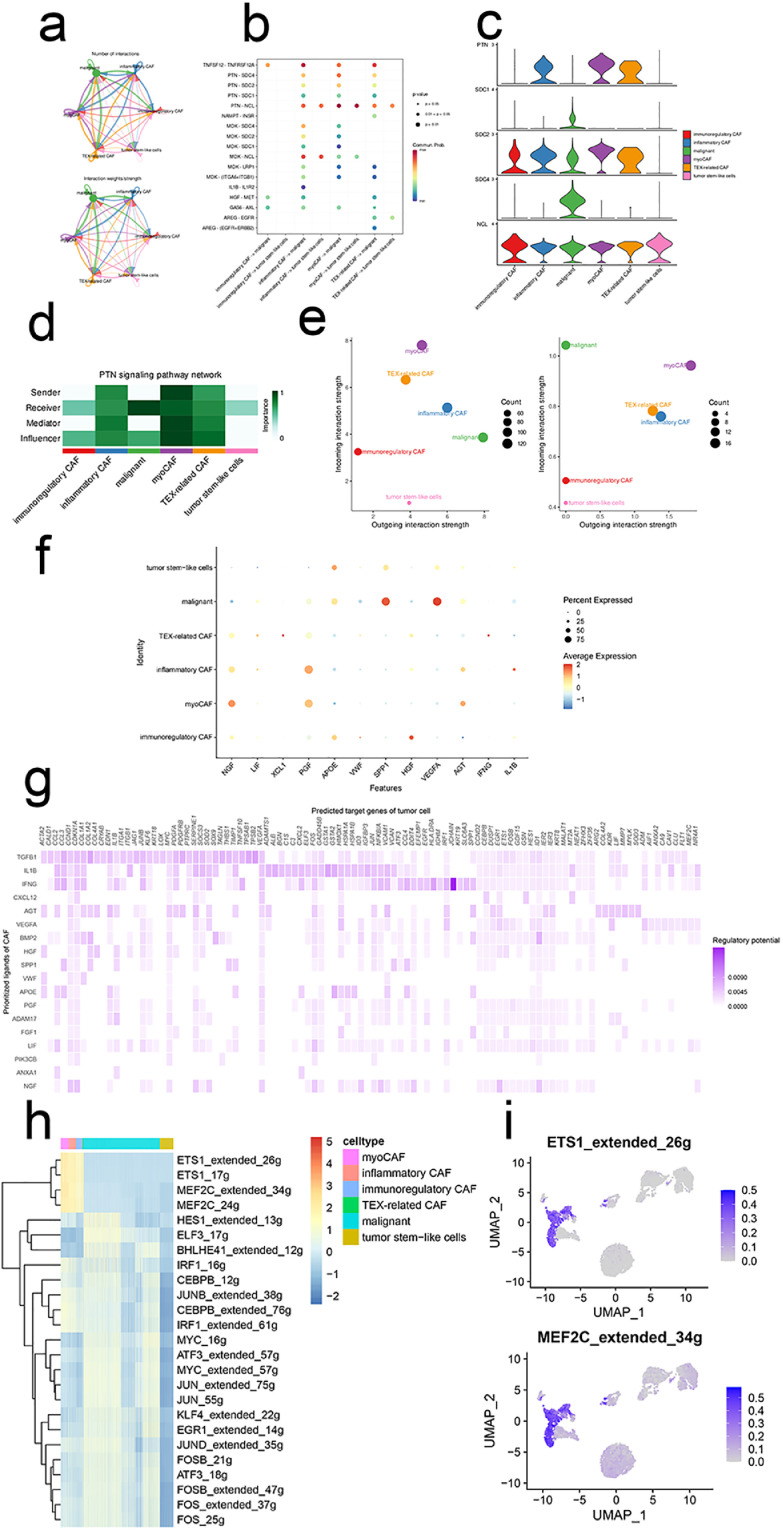
Intercellular signaling pathways among four subtypes of CAFs, tumor stem-like cells, and malignant cells were demonstrated in Fig. 2a. Tumor stem-like cells were in the dormancy-like state that had less reciprocal crosstalk with other cell types. Mounting evidence demonstrated the roles of CAFs-secreted molecules in facilitating tumor cell progression [32]. Our ligand-receptor (LR) networks demonstrated that the PTN—NCL pair was upregulated (Fig. 2b-e), indicating that targeting this signaling axis may have therapeutic potential. Shi and colleagues uncovered that upregulated PTN derived from M2 TAM promoted glioma stem cells-driven tumor growth while blocking the LR signaling potently abated tumor growth and prolonged animal survival [33].
Fig. 2.
Regulons ETS1 and MEF2C are exclusively activated in CAFs. (a) The intercellular in-teractions between subclusters of CAFs and malignant cells. (b)The ligand-receptor pairs between CAFs and malignant cells. (c)Expression profiles of PTN signaling pathway in CAFs and malig-nant cells. (d)The importance of each subcluster of CAFs and malignant cells in the PTN signaling pathway. (e)The incoming/outgoing strength of each subcluster of CAFs and malignant cells in the PTN signaling pathway (left) and the whole signaling pathways (right). (f)Top ligands in the communication network. (g)Ligand-target gene matrix denoting the potential regulatory rela-tionships between ligands and target genes among CAFs and malignant cells. The color intensity represented the regulatory potentials. (h)Heatmap demonstrated the activity of each regulon in CAFs and malignant cells. (i)UMAP illustrated the activities of regulons ETS1 and MEF2C.
To unravel the ligand-target genes network, we employed the NicheNet analysis. Growth factors (e.g., NGF, PGF, and HGF), chemokines (e.g., XCL1), and cytokines (e.g., INF-γ) were regarded as the top ligands in the network, suggesting the inflamed TME of ccRCC (Fig. 2f). A ligand-targets heatmap was illustrated in Fig. 2g. Then, GRN analysis was adopted to further identify the upregulated regulons within the targets by SCENIC framework. As demonstrated in Fig. 2h and 2i, regulons ETS1 and MEF2C were exclusively upregulated in CAFs whereas the tumor stem-like cells were less activated. Subsequently, target genes of the two regulons were obtained and regarded as the cell communication gene signature.
Signature stratifies ccRCC TME into two subclusters with distinct prognoses and biological features
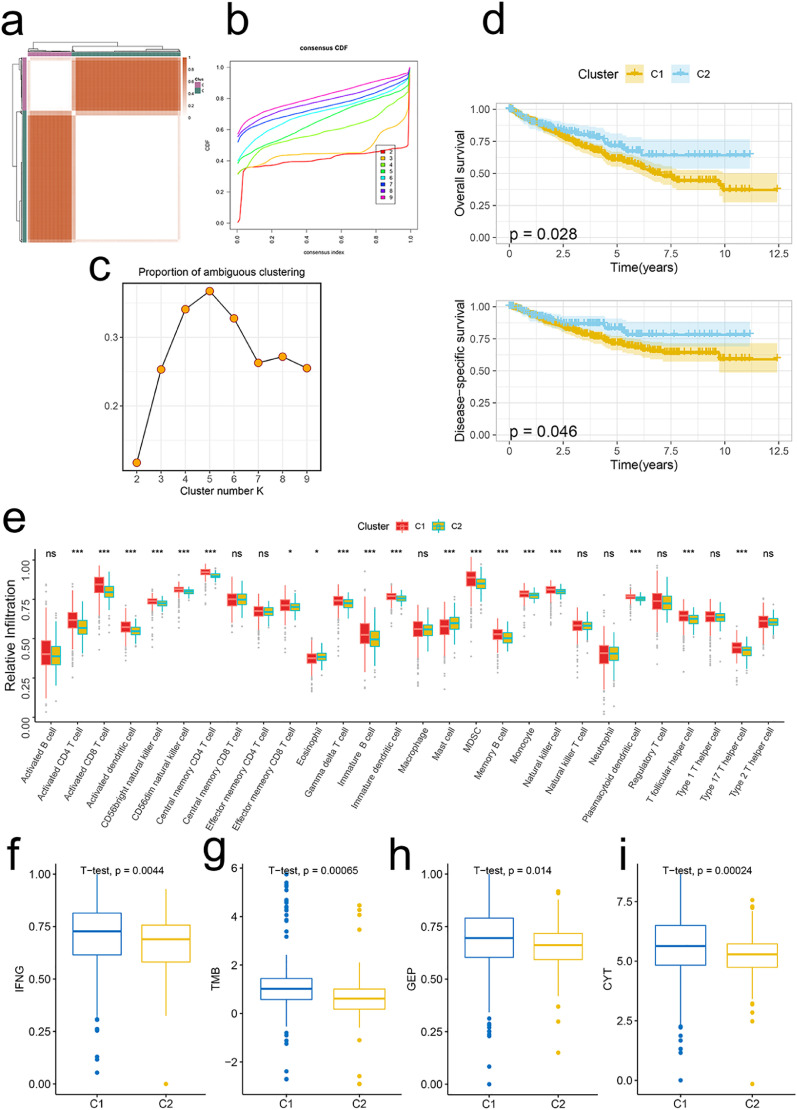
Based on the signature genes, we divided the TCGA-KIRC into two molecular subclusters (Fig. 3a). Both CDF curves and PAC scores determined the optimal clustering number of 2 (Fig. 3b, c). Survival analyses demonstrated that cluster 1 was related to decreased OS and DSS (Fig. 3d).
Fig. 3.
Signature stratifies ccRCC TME into two subclusters with distinct prog-noses and biological features. (a)The consensus score matrix of all samples when k = 2. A higher consensus score denotes higher similarity. (b)The CDF curves of the consensus matrix for each k (indicated by colors). (c)The PAC score for each k. (d)KM survival curves with log-rank test demonstrate survival discrepancies between two clusters. (e)Relative infiltration abundances of 28 immune cell subsets in two clusters. P values are determined by the Wilcoxon test. ns: non-significant; * p < 0.05; *** p < 0.001. The activities of IFNG (f), TMB (g), GEP (h), and CYT (i) between two clusters.
Considering the inflamed TME of ccRCC (Fig. 2f), we next explored the immunological landscapes of two clusters. 28 immune cells were highly infiltrated in cluster 1 compared to those in cluster 2 (Fig. 3e). And the similar results were addressed by other six TME deconvolution methodologies (Fig. S2a). Besides, cluster 1 was related to most immune modulators including chemokines, immune inhibitors, immunostimulators, antigen presentation, and receptors (Fig. S2b).
To further explore the immunotherapy responses between two clusters, we evaluated the activities of several immunotherapy predictors including IFN-γ, tumor mutation burden (TMB), T cell-inflamed gene expression profile (GEP), and cytotoxic activity (CYT). Results showed that these predictors were all significantly higher in cluster 1 (Fig. 3f-i). In addition, seven steps of the anti-cancer immunity cycle (Fig. S3a) and most immunotherapy-predicted pathways were highly activated in cluster 1 (Fig. S3b). Together, ccRCC patients in cluster 1 may have an inflamed TME and benefit from immunotherapy.
To assess the dysregulated pathways between two clusters, GSEA analysis was employed. As listed in Fig. S3c, several signaling pathways were highly activated in cluster 1. Additionally, cancer hallmarks including angiogenesis and epithelial-mesenchymal transition were related to cluster 1 (Fig. S3d).
The signature-based model demonstrates high accuracy and robust performance in predicting prognosis
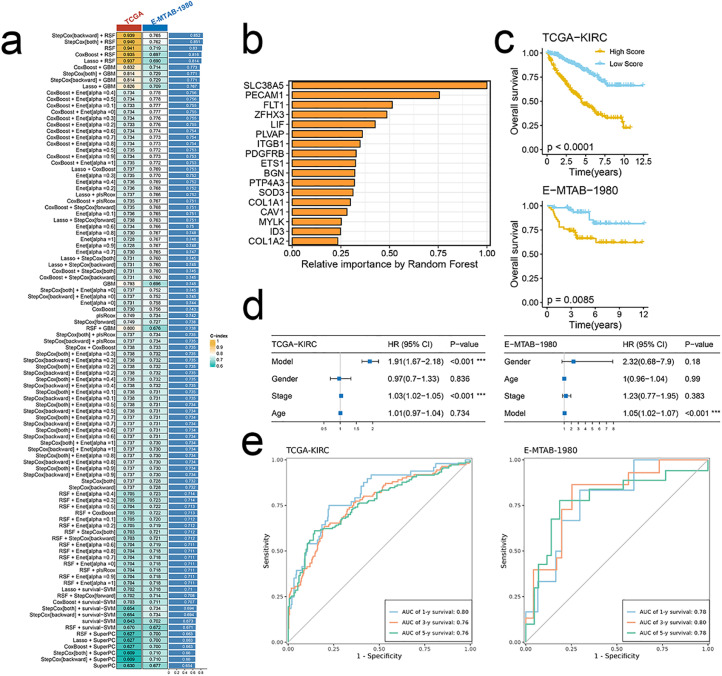
Benefiting from advances in high throughput sequencing and bioinformatic methodologies, various biological processes-associated mRNA/lncRNA/miRNA signatures have been established to predict the prognosis and response to therapy [34,35]. However, these multigene signatures are hardly applied to clinical practices due to inappropriate machine-learning methods and underutilized data information. Therefore, we developed an integrated machine-learning framework of 101 combinations based on 10 machine-learning algorithms. Models were fitted in TCGA-KIRC and then validated in E-MTAB-1980. Our optimal signature-based model of seventeen genes was established based on the combination of backward StepCox and RSF (Fig. 4a, b). To assess the performance of the established model in predicting prognosis, both KM survival analysis and multivariate cox regression analysis were conducted. Results showed that a high score predicted poorer prognosis in both datasets (Fig. 4c). Besides, the model was an independent risk factor for survival (Fig. 4d). Time-dependent ROC analysis showed the stability and high accuracy of the model (Fig. 4e).
Fig. 4.
Signature-based model demonstrates high accuracy and robust perfor-mance in predicting prognosis. (a)C-index of 101 kinds of prediction models in TCGA-KIRC and E-MTAB-1980 datasets. (b)The importance of the 17 most valuable signature genes based on the RSF algorithm. (c)KM survival curves with log-rank test demonstrate survival discrepancies between high- and low-score groups of TCGA-KIRC (up) and E-MTAB-1980 (bottom) datasets. (d) The model is an independent risk factor for OS in TCGA-KIRC (left) and E-MTAB-1980 (right) datasets by multivariate cox re-gression analysis. (e) Time-dependent ROC analysis demonstrates the performance of the model in TCGA-KIRC (left) and E-MTAB-1980 (right) datasets at 1y-, 3y-, and 5y- time points.
The signature-based model is a predictive biomarker for immunotherapy response
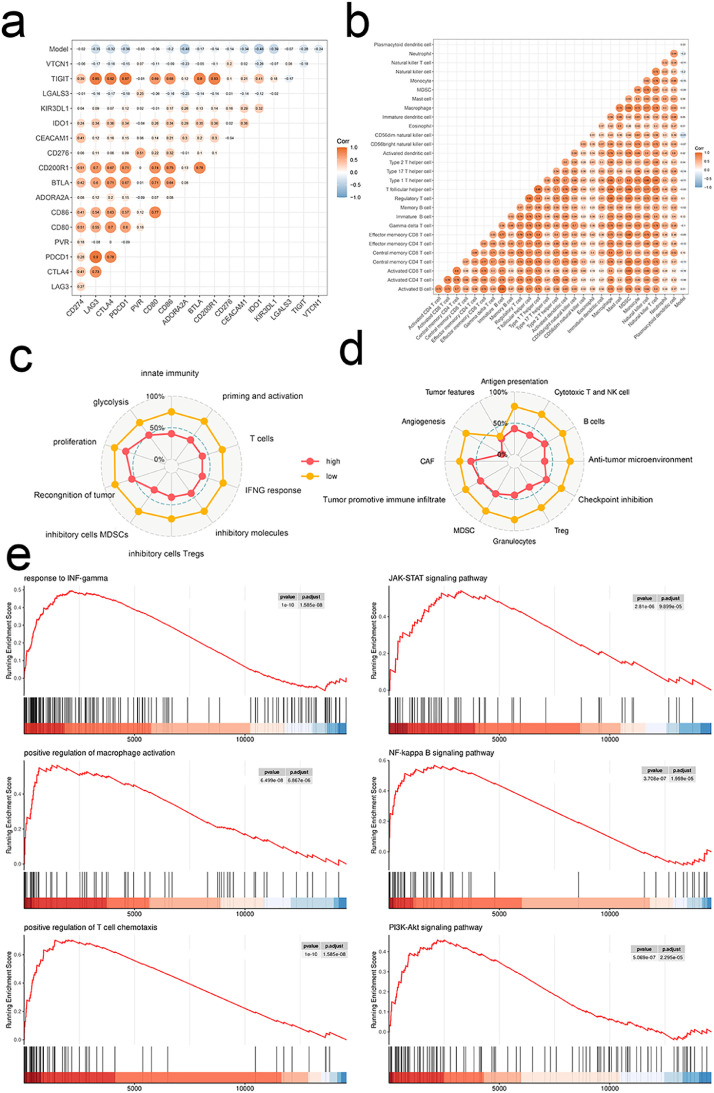
The high score of the model was negatively correlated with inhibitory immune checkpoints and immune cell infiltration abundances (Fig. 5a, b). And a high score was associated with lower levels of various TME signatures (Fig. 5c, d). Additionally, GO and KEGG terms of GESA were less enriched in the high score group (Fig. 5e). As predicted, a high score predicted a less favorable response to immunotherapy in TCGA-KIRC (Fig. 6a, b).
Fig. 5.
The signature-based model is linked to non-inflamed TME. Correlations between model and immune checkpoints (a), infiltration levels of 28 immune cell sub-sets (b), and TME signatures developed by Kobayashi (c) and Bagaev (d). e Dysregulated GO (left) and KEGG (right) terms in TME of ccRCC by GSEA.
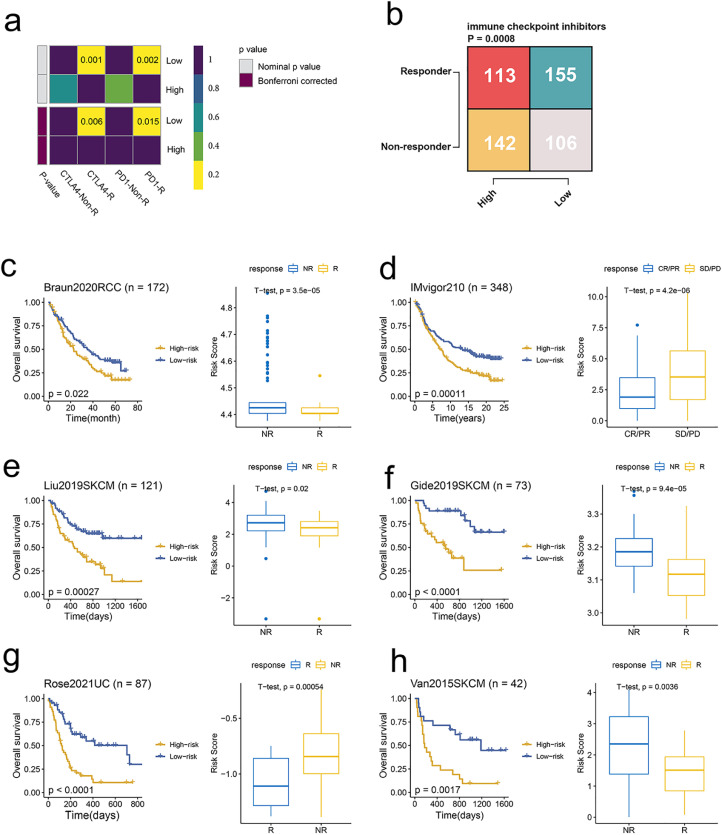
Fig. 6.
The signature-based model predicts response to immunotherapy. (a) Contingency table between immunotherapy responses and model score groups based on the TIDE algorithm. (b) Contingency table between immunotherapy responses (an-ti-PD-1 and anti-CTLA-4) and model score groups based on submap analysis. (c) KM curves of OS between patients with a high score and a low score in the Braun dataset (left). Box plot displaying the model score in patients with different immunotherapy responses in the Braun dataset (right). (d) KM curves of OS between patients with a high score and a low score in the IMvigor210 dataset (left). Box plot displaying the model score in patients with different immunotherapy responses in the IMvigor210 dataset (right). (e) KM curves of OS between patients with a high score and a low score in the Liu dataset (left). Box plot displaying the model score in patients with different im-munotherapy responses in the Liu dataset (right). (f) KM curves of OS between patients with a high score and a low score in the Gide dataset (left). Box plot displaying the model score in patients with different immunotherapy responses in the Gide dataset (right). (g) KM curves of OS between patients with a high score and a low score in the Rose dataset (left). Box plot displaying the model score in patients with different im-munotherapy responses in the Rose dataset (right). (h) KM curves of OS between pa-tients with a high score and a low score in the Van dataset (left). Box plot displaying the model score in patients with different immunotherapy responses in the Van dataset (right).
To further explore the performance of the model in ICIs therapy cohorts, we analyzed the survival and scores between responders and non-responders. Consistent with predictions, a high score was related to a more unfavorable prognosis compared to a low score. Moreover, scores were significantly higher in non-responder across three cancer types including RCC, urothelial carcinoma, and melanoma (Fig. 6c-h).
Collectively, the signature-based model is a predictive biomarker for immunotherapy response.
SLC38A5 dominates an inflamed and immunosuppressive TME
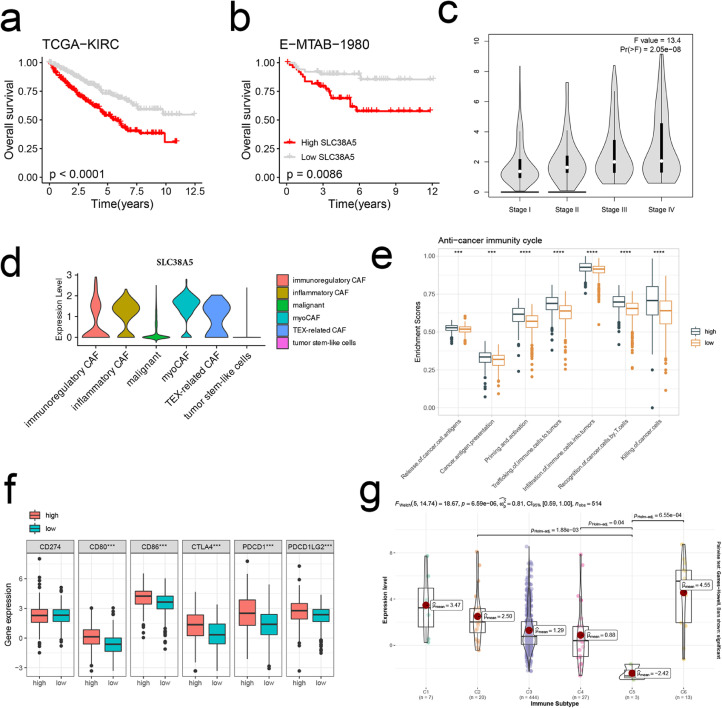
As the most important variable of the model, SLC38A5 was explored in subsequent analysis. SLC38A5 was related to decreased OS in the TCGA-KIRC and E-MTAB-1980 cohorts (Fig. 7a, b). In addition, the expression increased gradually from stage I to IV (Fig. 7c). SLC38A5 was exclusively upregulated in CAFs (Fig. 7d). Analysis of immune-related characteristics indicated that anti-cancer immunity cycles and inhibitory checkpoints were highly upregulated in high expression group (Fig. 7e, f). Thorsson et al. classified TME of pan-cancer into six immune subtypes [36]. In our study, SLC38A5 remarkably downregulated in the “immunologically quiet” subtype (C5) whereas upregulated in the “TGF-β dominant” (C6) and “IFN-γ dominant” (C2) subtypes (Fig. 7g). Both C2 and C6 subtypes indicated the inflamed and immunosuppressive TME [37,38]. Furthermore, SLC38A5 was positively correlated with immune scores across TCGA-KIRP (r = 0.44), TCGA-KIPAN (r = 0.55), TCGA-KIRC (r = 0.38), and TCGA-KICH (r = 0.56) (Fig. S4a).
Fig. 7.
SLC38A5 dominates an inflamed and immunosuppressive TME. (a, b) KM survival curves of OS between patients with a high expression of SLC38A5 and with a low expression of SLC38A5 in the TCGA-KIRC (a) and E-MTAB-1980 cohorts (b). (c) The distribution of the expression of SLC38A5 across stages I to IV. (d) The distribution of the expression of SLC38A5 across subclusters of CAFs and malignant cells. (e) The distribution of activi-ties of anti-cancer immunity cycles between patients with a high expression of SLC38A5 and with a low expression of SLC38A5. (f) The expression levels of immune checkpoints CD274, CD80, CD86, CTLA-4, PDCD1 (PD-1), and PDCD1LG2 (PD-L2) between patients with a high expression of SLC38A5 and with a low expression of SLC38A5. (g) The distribution of the expression of SLC38A5 across immune subtypes C1 to C6. C1: wound healing, C2: IFN-γ dominant, C3: inflammatory, C4: lymphocyte depleted, C5: immu-nologically quiet, and C6: TGF-βdominant.
We also evaluated the biological roles of SLC38A5 in pan-cancer. Correlation analysis showed that SLC38A5 was positively correlated with most immunoregulators and immune checkpoints (Fig. S4b, c). Besides, positive correlations with TMB were addressed in KIPAN, KIRC, and KICH (Fig. S4d).
CAFs promote tumor cell proliferation, invasion, and migration through the SLC38A5-CCL5 axis
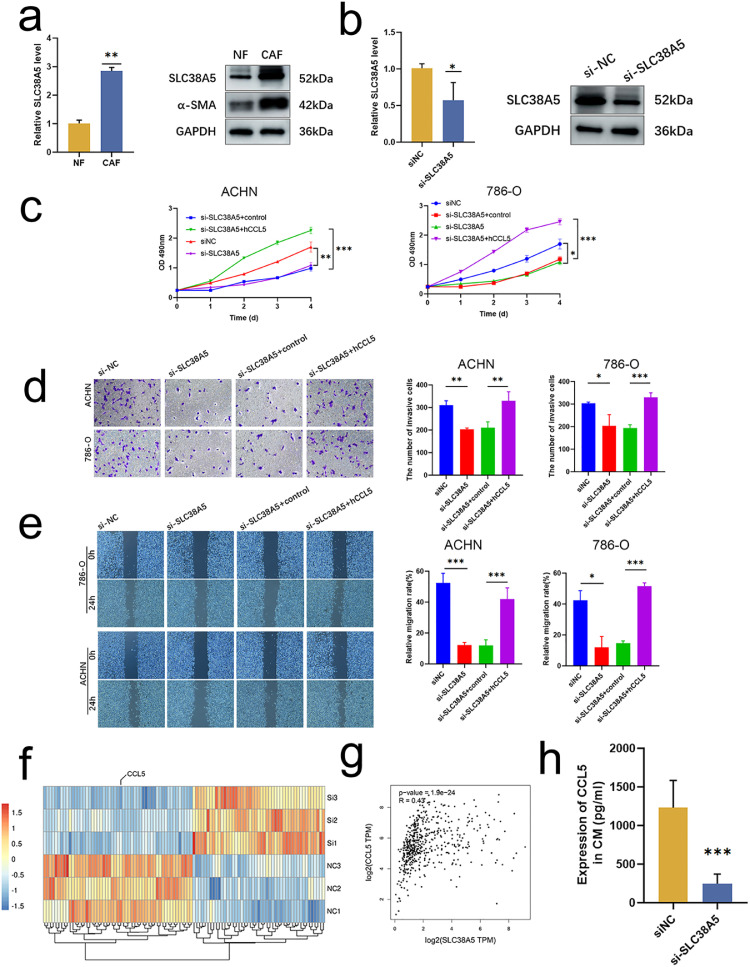
Considering the remarkable roles of SLC38A5 in ccRCC, we performed a series of phenotypic assays. SLC38A5 was upregulated in CAFs at both mRNA and protein levels (Fig. 8a). The efficacy of siRNA targeting SLC38A5 was verified by qRT-PCR and blotting (Fig. 8b). CCK-8 assay revealed that SLC38A5 deficiency inhibited the proliferative abilities of ACHN and 786-O cell lines (Fig. 8c). The similar trends were found in transwell invasion (Fig. 8d) and wound-healing assays (Fig. 8e). Collectively, these results indicated that SLC38A5 promoted the proliferation and metastasis of tumor cell.
Fig. 8.
CAFs promote tumor cell proliferation, invasion, and migration through the SLC38A5-CCL5 axis. (a) The expression of SLC38A5 between NFs and CAFs at both mRNA (left) and protein (right) levels. (b) The efficacy of siRNA targeting SLC38A5 is validated by qRT-PCR (left) and blotting (right). (c) RCC cell lines ACHN (left) and 786-O (right) viability detected by a CCK-8 assay. (d) RCC cells lines ACHN (up) and 786-O (bottom) invasion when co-cultured with CAFs; detected by a transwell assay. (e) RCC cells lines ACHN (bottom) and 786-O (up) migration when co-cultured with CAFs; detected by a wound-healing assay. (f) Heatmap shows the decreased CCL5 expression in si-SLC38A5 CAFs. (g) Correlation between SLC38A5 and CCL5 (GEPIA, http://gepia.cancer-pku.cn/index.html, accessed on 19 December 2022). (h) The de-creased protein level of CCL5 after SLC38A5 inhibition by ELISA. * p < 0.05, ** p < 0.01, *** p < 0.001.
To unveil the mechanisms of how SLC38A5 affects tumor cells, we performed RNA-seq on si- SLC38A5 CAFs and identified the soluble factor CCL5 that was decreased after SLC38A5 inhibition (Fig. 8f). Positive correlation between SLC38A5 and CCL5 was addressed (Fig. 8g). ELISA showed that the protein level of CCL5 was dramatically decreased after SLC38A5 inhibition (Fig. 8h). A series of subsequent phenotypic assays were conducted to uncover the functional relationship between SLC38A5 and CCL5. The CCK-8 assay showed that rIL32 could restore the proliferative (Fig. 8c), invasive (Fig. 8d), and migrative abilities (Fig. 8e) of ACHN and 786-O cell lines. In the co-culture system with CM derived from SLC38A5 intact CAFs, anti-IL32 could inhibit the tumor cell's proliferative, migration, and invasive properties (Fig. S5). These findings indicated that CAFs promoted tumor cell proliferation, invasion, and migration through SLC38A5-CCL5 axis.
Discussion
TME represents an intricate and dynamic ecosystem composed of cellular components including malignant and stromal cells [39]. CAFs account for a majority of the stromal cells across all most all solid tumors [40]. The pro-tumorigenic biological properties of CAFs have been reported in a body of researches including, but not limited to, tumoral migration and invasion, self-renewal of cancer stem cells, and resistance to chemotherapy and immunotherapy [41]. Recently, single-cell technology, well-designed databases, and sophisticated tools have promoted our understanding of molecular mechanisms at single-cell levels, especially in the respect of intercellular communications [42]. Therefore, an in-depth understanding the of communication networks of CAFs may pave the way for cancer management.
CAFs originate from various cell types including normal resident fibroblasts, and they may be characterized by anti-tumoral functions at an early stage. With tumor development, CAFs, however, are educated by tumor cells to promote tumor growth and progression [43]. Therefore, uncovering the dual role of CAFs can shed light on cancer behavior and therapeutic perspectives. In the study, four subtypes of CAFs were identified and the pseudotime analysis re-constructed the trajectory path where myoCAF was located at the origin whereas TEX-related CAF was projected onto the terminal. Functional enrichment analysis showed that myoCAF was related to biological processes of the muscle system and extracellular matrix that are similar to normal resident fibroblasts [44]. Along with the trajectory path, myoCAF differentiated into the intermixed inflammatory and immunoregulatory CAFs that may dominate an inflamed TME and hot tumors. Inflamed TME or hot tumors have been reported to be associated with sensitivity to ICIs therapy [45]. This evidence may suggest the tumor-suppressing role of the two subtypes of CAFs in ccRCC. Terminally, the phenotype of CAFs transformed into the TEX-related that may facilitate immune escape and benefit tumor development. Future studies are needed to address the cellular state transformation-dependent genes.
Deciphering the communication network showed the remarkably activated regulons ETS1 and MEF2C. ETS1 is involved in cancer progression and linked to decreased survival in most malignancies including breast cancer [46], gastric carcinoma [47], and ccRCC [48]. For MEF2C, the dual roles depend on cancer types. In hepatocellular carcinoma, highly expressed MEF2C promotes tumor development via VEGF whereas inhibits cancer proliferation by blockade of Wnt/β-catenin signaling [49]. However, the biological roles of MEF2C remain largely unknown in ccRCC.
Considering the heterogenous subtypes and dual-role regulons of CAFs, we performed consensus clustering to stratify the TME of ccRCC into two molecular clusters. It was impressive that the two clusters had dramatically different prognoses, infiltration levels of immune cells, expression patterns of immunoregulators, and dysregulated pathways and cancer hallmarks. Six TME-deconvolution methodologies including CIBERSORT, EPIC, MCP-counter, quanTIseq, TIMER, and xCell discovered that cluster 1 was linked to an inflamed TME. Consistent findings were addressed in the upregulated expression profiles of most immunoregulators in this cluster. As we mentioned before, patients with an inflamed TME indicated favorable responses to ICIs therapy. That is to say, ccRCC patients in cluster 1 may have a higher response rate compared to those in cluster 2. The highly activated anti-cancer immunity cycles and four immunotherapeutic predictors in cluster 1 verified these findings. Considering the limited ICIs efficacy in solid tumors, application ICIs to potential candidates with an inflamed TME may be promising in clinical settings.
To translate the findings from the bench to bedside, we developed an integrated machine-learning framework to establish an optimal model with high accuracy and stable performance. Each ccRCC patient was assigned a risk score and patients in the high-score group suffered from poorer survival compared to those in the low-score group. Moreover, the model was an independent risk factor for OS, suggesting the appealing application in the early selection of high-risk patients and providing personalized intervention to prolong survival. Immunological characteristics demonstrated that a high score was negatively correlated with immune cell infiltration and various TME signatures, suggesting ccRCC patients with an inflamed TME also had high-risk scores. To explore the predictive values in ICIs therapy, a possible response ratio was inferred. As expected, a high score was linked to resistance to ICIs therapy of anti-PD-1 and anti-CTLA-4. This may be further explained by the negative correlation with inhibitory checkpoints. In addition, the analytic results of six ICI-treated cohorts across three cancer types verified our predictions. Collectively, the model had promising clinical applications in the selection of high-risk ccRCC and ICIs candidates.
In the seventeen model genes, SLC38A5 was the most important gene as uncovered by RSF. It was overexpressed in CAFs and associated with poor OS, DFS, and high stages. The oncogenetic role of SLC38A5 was validated by a series of phenotypic assays. Experiments showed that SLC38A5 overexpression CAFs promoted RCC cell proliferation, migration, and invasion. Previous studies have addressed the CAFs-derived soluble factors in cancer development. Chen et al. found that IL32 regulated the biological properties of bladder cancer cells induced by CAFs [29]. CCL5 was found to restore the RCC cells proliferation, migration, and invasion that were mitigated by SLC38A5 inhibiting CAFs. Similar results were reported in hepatocellular carcinoma that CAFs-derived CCL5 promoted cancer cell metastasis by activating HIF1α/ZEB1 axis [50]. Besides, inspired by the appealing application of predicting therapeutic responses of the model, we found that highly expressed SLC38A5 also indicated an inflamed TME with simultaneously upregulated inhibitory checkpoints. To sum up, the SLC38A5-CCL5 axis can regulate ccRCC development, and targeting the axis may promote cancer management.
There were some limitations in our study. First, the performance of our model was not further validated in our cohorts in terms of survival and treatment response prediction. Second, in vivo study using the SLC38A5 genetic mouse model will be more encouraging and highlight its therapeutic significance.
Conclusions
Through integrated scRNA-seq and bulk RNA-seq analyses, we established a CAFs-derived communication signature based on which the TME of ccRCC can be divided into two clusters with distinct prognoses, immune cell infiltration landscapes, and immune-related characteristics. Based on the signature and a curated machine learning framework, a risk model was established with appealing applications in predicting prognosis and ICI-therapy response. Hug gene SLC38A5 demonstrated its oncogenetic roles by promoting RCC cell lines proliferation, migration, and invasion. Mechanically, rescue experiments showed that the biological functions of SLC38A5 were mediated by the CCL5 protein. Our findings provide the cornerstones for further elucidating the mechanism of the CAFs in promoting ccRCC progression, contributing to unveil potential therapeutic targets and novel clinical treatment of ccRCC.
Author contributions
Hualin Chen and Zhigang Ji conceived and designed the study; Hualin Chen contributed to date acquisition; Hualin Chen, Wenjie Yang, Lin Ma, and Yingjie Li analyzed data; Hualin Chen wrote the manuscript. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors report no conflict of interest.
Acknowledgments
Acknowledgments
We thank Dr.Jianming Zeng(University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
Funding
This work was supported by the National High Level Hospital Clinical Research Funding (2022-PUMCH-B-008).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101790.
Appendix. Supplementary materials
Data availability
All data used in this study are publicly available as described in the Method section. The unique identifiers for public cohorts are described in the paper. The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Pan Y., Jin X., Chen G. Identification of a four hypoxia-associated long non-coding RNA signature and establishment of a nomogram predicting prognosis of clear cell renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.713346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouracade P., Kara O., Maurice M.J., Dagenais J., Malkoc E., Nelson R.J., et al. Patterns and predictors of recurrence after partial nephrectomy for kidney tumors. J. Urol. 2017;197(6):1403–1409. doi: 10.1016/j.juro.2016.12.046. [DOI] [PubMed] [Google Scholar]
- 4.Zhong W., Zhang F., Huang C., Lin Y., Huang J. Identification of epithelial-mesenchymal transition-related lncrna with prognosis and molecular subtypes in clear. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.591254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serzan M.T., Atkins M.B. Current and emerging therapies for first line treatment of metastatic clear cell renal cell carcinoma. J Cancer Metastasis Treat. 2021;7 doi: 10.20517/2394-4722.2021.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer R.J., Tannir N.M., McDermott D.F., Arén Frontera O., Melichar B., Choueiri T.K., et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonaventura P., Shekarian T., Alcazer V., Valladeau-Guilemond J., Valsesia-Wittmann S., Amigorena S., et al. Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol. 2019;10:168. doi: 10.3389/fimmu.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z., Zhou L., Liu L., Hou Y., Xiong M., Yang Y., et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020;11(1):5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H., Chen G. Dissecting immunosuppressive cell communication patterns reveals junb proto-oncogene (JUNB) Shaping a non-inflamed tumor microenvironment. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.883583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun D.A., Hou Y., Bakouny Z., Ficial M., Sant' Angelo M., Forman J., et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 2020;26(6):909–918. doi: 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019;25(12):1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gide T.N., Quek C., Menzies A.M., Tasker A.T., Shang P., Holst J., et al. Distinct immune cell populations define response to anti-pd-1 monotherapy and anti-PD-1/Anti-CTLA-4 combined therapy. Cancer Cell. 2019;35(2):238–255. doi: 10.1016/j.ccell.2019.01.003. e6. [DOI] [PubMed] [Google Scholar]
- 14.Rose T.L., Weir W.H., Mayhew G.M., Shibata Y., Eulitt P., Uronis J.M., et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br. J. Cancer. 2021;125(9):1251–1260. doi: 10.1038/s41416-021-01488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J., Li K., Zhang W., Wan C., Zhang J., Jiang P., et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21. doi: 10.1186/s13073-020-0721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Q., Chen S., Hu Y., Huang W. Single-cell RNA transcriptome reveals the intra-tumoral heterogeneity and regulators underlying tumor progression in metastatic pancreatic ductal adenocarcinoma. Cell Death Discov. 2021;7(1):331. doi: 10.1038/s41420-021-00663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17(2):159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 22.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng D., Ye Z., Shen R., Yu G., Wu J., Xiong Y., et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkerson M.D., Hayes D.N. Consensusclusterplus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 28.Chen P.L., Roh W., Reuben A., Cooper Z.A., Spencer C.N., Prieto P.A., et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6(8):827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H., Yang W., Xue X., Li Y., Jin Z., Ji Z. Integrated analysis revealed an inflammatory cancer-associated fibroblast-based subtypes with promising implications in predicting the prognosis and immunotherapeutic response of bladder cancer patients. Int. J. Mol. Sci. 2022;23(24):15970. doi: 10.3390/ijms232415970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren I., Moeller M.M., Guiggey D., Chiang A., Maloy M., Ogoke O., et al. FOXA1/2 depletion drives global reprogramming of differentiation state and metabolism in a human liver cell line and inhibits differentiation of human stem cell-derived hepatic progenitor cells. FASEB J. 2023;37(1):e22652. doi: 10.1096/fj.202101506RRR. [DOI] [PubMed] [Google Scholar]
- 31.Wang L., He T., Liu J., Tai J., Wang B., Chen Z., et al. Pan-cancer analysis reveals tumor-associated macrophage communication in the tumor microenvironment. Exp Hematol Oncol. 2021;10(1):31. doi: 10.1186/s40164-021-00226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen S., Hou Y., Fu L., Xi L., Yang D., Zhao M., et al. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin β3–p38 MAPK signalling. Cancer Lett. 2019;442:320–332. doi: 10.1016/j.canlet.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y., Ping Y.F., Zhou W., He Z.C., Chen C., Bian B.S.J., et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 2017;8(1):15080. doi: 10.1038/ncomms15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H., Pan Y., Jin X., Chen G. An immune cell infiltration-related gene signature predicts prognosis for bladder cancer. Sci. Rep. 2021;11(1):16679. doi: 10.1038/s41598-021-96373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W., Ma L., Dong J., Wei M., Ji R., Chen H., et al. Machine learning to improve prognosis prediction of metastatic clear-cell renal cell carcinoma treated with cytoreductive nephrectomy and systemic therapy. Bosn. J. Basic Med. Sci. 2022 doi: 10.17305/bjbms.2022.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derynck R., Turley S.J., Akhurst R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021;18(1):9–34. doi: 10.1038/s41571-020-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kretzschmar K., Clevers H. IFN-γ: the T cell's license to kill stem cells in the inflamed intestine. Sci. Immunol. 2019;4(42) doi: 10.1126/sciimmunol.aaz6821. [DOI] [PubMed] [Google Scholar]
- 39.Galbo P.M., Jr., Zang X., Zheng D. Molecular Features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin. Cancer Res. 2021;27(9):2636–2647. doi: 10.1158/1078-0432.CCR-20-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T., Han C., Wang S., Fang P., Ma Z., Xu L., et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019;12(1):86. doi: 10.1186/s13045-019-0770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X.H., Jin X., Malladi S., Zou Y., Wen Y.H., Brogi E., et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154(5):1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grün D., van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163(4):799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 43.Toledo B., Picon-Ruiz M., Marchal J.A., Perán M. Dual role of fibroblasts educated by tumour in cancer behavior and therapeutic perspectives. Int. J. Mol. Sci. 2022;23(24) doi: 10.3390/ijms232415576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Csapo R., Gumpenberger M., Wessner B. Skeletal muscle extracellular matrix – what do we know about its composition, regulation, and physiological roles? a narrative review. Front Physiol. 2020:11. doi: 10.3389/fphys.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trujillo J.A., Sweis R.F., Bao R., Luke J.J. T Cell-inflamed versus non-t cell-inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol. Res. 2018;6(9):990–1000. doi: 10.1158/2326-6066.CIR-18-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim G.C., Kwon H.K., Lee C.G., Verma R., Rudra D., Kim T., et al. Upregulation of Ets1 expression by NFATc2 and NFKB1/RELA promotes breast cancer cell invasiveness. Oncogenesis. 2018;7(11):91. doi: 10.1038/s41389-018-0101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y., Zhang Y.C., Zhang W.Z., Shen L.S., Hertzog P., Wilson T.J., et al. Ets1 as a marker of malignant potential in gastric carcinoma. World J. Gastroenterol. 2003;9(10):2154–2159. doi: 10.3748/wjg.v9.i10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu Y., Meng M., Cao C., Zhang J., Cheng X., Huang Y., et al. RNA-binding protein MEX3A controls G1/S transition via regulating the RB/E2F pathway in clear cell renal cell carcinoma. Mol. Ther. Nucl. Acids. 2022;27:241–255. doi: 10.1016/j.omtn.2021.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai X.L., Zhang Q., Ye L.Y., Liang F., Sun X., Chen Y., et al. Myocyte enhancer factor 2C regulation of hepatocellular carcinoma via vascular endothelial growth factor and Wnt/β-catenin signaling. Oncogene. 2015;34(31):4089–4097. doi: 10.1038/onc.2014.337. [DOI] [PubMed] [Google Scholar]
- 50.Xu H., Zhao J., Li J., Zhu Z., Cui Z., Liu R., et al. Cancer associated fibroblast–derived CCL5 promotes hepatocellular carcinoma metastasis through activating HIF1α/ZEB1 axis. Cell Death. Dis. 2022;13(5):478. doi: 10.1038/s41419-022-04935-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are publicly available as described in the Method section. The unique identifiers for public cohorts are described in the paper. The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.