Abstract
In the past few years, application of the PCR to the detection of herpes simplex virus (HSV) DNA in the cerebrospinal fluid (CSF) from patients with encephalitis and meningitis has become standard laboratory practice. However, from an operational perspective, the true diagnostic value of PCR in this setting is yet to be realized because most laboratories subject the amplification products to lengthy probe hybridization procedures by Southern blotting. As alternatives to Southern blotting, we evaluated colorimetric microtiter plate (MTP) systems from ViroMed Laboratories, Inc. (PrimeCapture), CPG, Inc. (Quanti-PATH), and Incstar Corp. (GEN-ETI-K), in addition to a system developed at the Mayo Clinic with the PCR ELISA system (Boehringer Mannheim Corp.). We tested PCR products from 86 clinical CSF specimens submitted to our Molecular Microbiology Laboratory. The CSF specimens used had to have sufficient volume for comparative analysis. By conventional Southern blotting methods, 54 were positive and 32 were negative for HSV DNA. Compared with Southern blotting, the sensitivity and specificity were 63.0 and 100.0%, respectively, for the PrimeCapture system, 98.2 and 96.9%, respectively, for the Quanti-PATH system, 98.2 and 100.0%, respectively, for the GEN-ETI-K system, and 100.0 and 96.9%, respectively, for the Mayo system. All four MTP systems had turnaround times 12 to 24 h less than that for Southern blotting. There were no significant differences in costs or technologist time between the Mayo system and Southern blotting. Other features of the Mayo system include type-specific genotypic identification of HSV and the potential for determination of drug resistance by DNA sequencing. Overall, we found that colorimetric MTP systems were likely to improve test turnaround times and patient care at no additional cost.
Herpes simplex virus (HSV) is a ubiquitous agent responsible for a wide variety of human infections. In addition to epithelial infections such as gingivostomatitis, pharyngitis, genital herpes, whitlow, conjunctivitis, and keratitis, HSV is an important cause of central nervous system (CNS) infections and accounts for 2 to 19% of human encephalitis cases (9, 33, 38). The clinical spectrum of CNS diseases has recently been expanded; for example, most cases of benign recurrent aseptic meningitis (Mollaret’s meningitis) are caused by HSV (39), especially HSV type 2 (HSV-2) (36). Because specific antiviral therapy is available, the rapid, definitive laboratory diagnosis of HSV is important to support clinical findings. Moreover, in the setting of possible HSV encephalitis, patients are often managed as inpatients while awaiting test results.
Although cell culture is considered the standard method for laboratory diagnosis of ulcerative HSV infections, HSV is rarely recovered in cell cultures inoculated with cerebrospinal fluid (CSF). Brain biopsy specimens may yield culturable virus, but this invasive surgical procedure is controversial when performed solely to collect specimens for the laboratory diagnosis of infectious disease. The sensitivities of HSV antigen and antibody assays for CNS infections are very low (13). In addition, antibodies may appear in the CSF as a consequence of the breakdown in the blood-brain barrier, leading to false-positive results (23). Since 1990, several studies have shown that PCR detection of HSV DNA in CSF should be considered the new standard for the laboratory diagnosis of CNS disease caused by this virus (8, 11, 12, 15, 22, 24, 28–30).
Although the technology underlying PCR is relatively rapid, the PCR product (amplicon) must be identified definitively as the sequence of interest to provide adequate diagnostic specificity. The conventional technique for this is hybridization of a specific probe to a Southern blot, which increases both the sensitivity and the specificity of the test. This step, however, takes an additional 12 to 24 h to complete, delaying the use of test results for clinical intervention. The ideal postamplification detection system would combine the increased sensitivity and specificity of Southern blotting with a rapid turnaround time (2, 14, 19, 20, 32). For this purpose, enzyme-linked adsorbent microtiter plate (MTP) systems have been adapted for amplicon detection (40).
We report a comparison of four colorimetric MTP systems: the PrimeCapture system from ViroMed Laboratories, Inc., the Quanti-PATH system from CPG, Inc., the GEN-ETI-K system from Incstar Corp., and a system developed at the Mayo Clinic by PCR ELISA (Boehringer Mannheim Corp.). We tested all four systems against standard Southern blotting for the detection of HSV PCR products resulting from clinical CSF specimens.
(This study was presented in part at the 14th Annual Meeting of the Pan American Society of Clinical Virology, 26 to 29 April 1998, Clearwater Beach, Fla.)
MATERIALS AND METHODS
Specimens.
Eighty-six CSF specimens submitted to the Molecular Microbiology Laboratory at the Mayo Clinic for the diagnosis of HSV CNS disease by PCR were selected retrospectively for the study.
Colorimetric MTP systems.
We evaluated four colorimetric MTP systems. Three were commercial kits specifically designed for the detection of HSV DNA. In one type of format (PrimeCapture [lot 9705309116; ViroMed Laboratories, Inc., Minneapolis, Minn.]; Quanti-PATH [lot 116; CPG, Inc., Lincoln Park, N.J.]), amplification of the HSV target was performed with the biotinylated primers included in the kit. The amplified product was denatured and hybridized to the MTP wells, which were precoated with a sequence-specific capture probe. Unbound amplicons were removed by washing, and a streptavidin-enzyme conjugate was added followed by colorimetric detection (17, 20, 37). A second format (GEN-ETI-K [lot 98B04; Incstar Corp., Stillwater, Minn.]) also had HSV-specific probes bound to the MTP wells. In this system, however, the target (also amplified with primers included in the kit) was not biotinylated. Instead, an enzyme-linked antibody recognized target DNA which has hybridized to the specific probes on the MTP well surface (21). In another format, developed by Boehringer Mannheim Corp. (the PCR ELISA kit), digoxigenin-dUTP was incorporated with the PCR product. A sequence-specific, biotinylated capture probe was hybridized to the denatured amplicon, and the complexes were captured in avidin-coated MTP wells. Detection was completed with enzyme-linked anti-digoxigenin antibodies (10, 16, 27). We developed the Mayo system with the PCR ELISA kit. Type-specific 5′-biotinylated capture probes for HSV-1 (TK-G, 5′-ACAAACATCGTGTTGGGGGC-3′) and HSV-2 (TK-H, 5′-ACGAACCTGGTCCTGGGTGT-3′) were designed to differentiate HSV genotypes (7, 34).
Extraction of nucleic acid.
Nucleic acid from 200 μl of CSF was extracted with IsoQuick (Orca Research, Inc., Bothell, Wash.) according to the manufacturer’s instructions. Extracted DNAs were resuspended in 50 μl of water. Five microliters of each specimen extract was used for PCR amplification.
PCR amplification.
When MTP systems from ViroMed Laboratories, Inc., CPG, Inc., Incstar Corp., or Boehringer Mannheim Corp. were used, the manufacturers’ recommendations were followed. For the Mayo-developed system, the reaction mixtures were placed in a Perkin-Elmer 9600 thermal cycler programmed for a three-step PCR procedure by a previously described protocol (5, 22, 35). The primer set used at Mayo (TK-A, [5′-GACMAGCGCCCAGATAACAA-3′] and TK-B [5′-MCAGCATRGCCAGGTCAAGC-3′]) amplified a 335-bp portion of the thymidine kinase (TK) gene (22).
Amplicon identification.
All specimens were amplified by PCR followed by gel electrophoresis, conventional Southern blotting, and probe hybridization as described previously (5, 22, 35). Amplicon identification with MTP systems from ViroMed Laboratories, Inc., CPG, Inc., and Incstar Corp. was performed according to the manufacturers’ instructions.
The Mayo system was developed with Boehringer Mannheim’s PCR ELISA kit for the detection of digoxigenin-labeled PCR products. Aliquots of the denatured amplicon were mixed with each of two hybridization solutions, one of which contained a 5′ biotin-labeled DNA capture probe (TK-G) specific for HSV-1 and the other of which contained a probe (TK-H) specific for HSV-2. If the corresponding target DNA sequence was present, the probes hybridized and the resulting biotinylated DNA complexes were captured on streptavidin-coated microtiter plate wells. HSV-specific DNA complexes were detected with an anti-digoxigenin-peroxidase conjugate, which recognized digoxigenin-11-dUTP substitutions incorporated into the amplicon during PCR. The peroxidase substrate was then added for color development. A positive result was defined as an optical density (OD) at 405 nm (OD405) − OD490 value greater than or equal to 0.1. The HSV genotype was determined by which probe gave positive results. A uracil-N-glycosylase-based inactivation system was adapted to control for possible amplicon carryover contamination (5, 18).
Turnaround time and cost assessment.
The turnaround time for amplicon identification was calculated for each protocol. Direct cost was determined on an annualized basis and included the costs for test kits, materials, reagents, and equipment and laboratory personnel salaries. We added 8% to each direct cost to cover utilities. Variable allied health effort (hands-on time) was calculated per specimen. Fixed effort (specimen processing, buffer preparation, maintenance, bench cleaning, and data entry) was calculated on a per-day basis. Each procedure was outlined on a flowchart and timed by laboratory personnel in a manner consistent with other cost analyses performed routinely in the Department of Laboratory Medicine and Pathology at the Mayo Clinic.
Statistics.
Comparisons of sensitivities (based on rate ratios) were performed with Epiinfo software (version 6; Centers for Disease Control and Prevention, Atlanta, Ga.).
RESULTS
Performance evaluation.
Of 86 CSF specimens tested, 54 were HSV DNA positive and 32 were HSV DNA negative, as determined by detection of an amplification product in ethidium bromide-stained agarose gels or by film exposure after Southern blotting with probe hybridization. This technique is referred to as the “conventional method” in the present study. All four MTP systems had high specificities (≥96.9%) compared to the results of conventional methods (Table 1). The MTP systems from CPG, Inc., Incstar Corp., and Mayo also had excellent sensitivities (≥98.2%) compared with the sensitivity of the system from ViroMed Laboratories, Inc., which was significantly lower (63.0%; 95% confidence interval, 51 to 77%).
TABLE 1.
Sensitivities and specificities of colorimetric MTP systems for HSV amplicon identification
| Test source | No. of specimens with the following resultsa:
|
Sensitivity (%) | Specificity (%) | |||
|---|---|---|---|---|---|---|
| SB+, MTP+ | SB+, MTP− | SB−, MTP+ | SB−, MTP− | |||
| CPG, Inc. | 53 | 1 | 1b | 31 | 98.2 | 96.9 |
| Incstar Corp. | 53 | 1 | 0 | 32 | 98.2 | 100.0 |
| Mayo | 54 | 0 | 1b | 31 | 100.0 | 96.9 |
| ViroMed Laboratories, Inc. | 34 | 20 | 0 | 32 | 63.0 | 100.0 |
SB, Southern blotting followed by probe hybridization; MTP, colorimetric MTP systems; +, positive result; −, negative result.
These two results are for a single specimen from a patient clinically diagnosed as having aseptic meningitis.
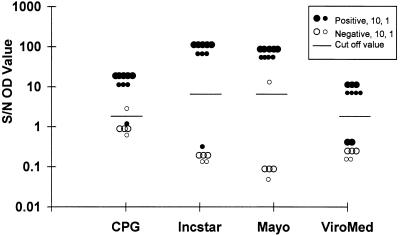
With MTP assays, the difference in OD between positive and negative controls is the objective basis for interpreting colorimetric data. All four MTP systems had mean ratios of the OD for a specimen versus the average OD for a negative control (S/N ratios) of >10 for specimens positive by conventional methods and mean S/N ratios of ≤1 for specimens negative by conventional methods (Fig. 1). Thus, the mean cutoff values which distinguished positive results from negative results were well separated for all four systems evaluated.
FIG. 1.
S/N ratios determined with four MTP systems. HSV DNA positivity or negativity was determined by Southern blotting followed by probe hybridization. The large dots represent 10 specimens, and the small dots represent 1 specimen. A short bar stands for the cutoff value for each test.
The Mayo system was able to differentiate HSV genotypes by using type-specific capture probes. Among 55 CSF specimens which yielded positive tests with the Mayo system, 20 (36.4%) were infected with HSV-1, 32 (58.2%) were infected with HSV-2, and 3 (5.5%) were infected with both genotypes of the virus. Of the 54 CSF specimens identified as positive both with the Mayo system and by the conventional method, the MTP system from ViroMed Laboratories, Inc., was positive for 13 (65.0%) of 20 HSV-1-positive specimens, 20 (64.5%) of 31 HSV-2-positive specimens, and 1 (33.3%) of 3 HSV-1- and HSV-2-positive specimens. Hence, the prevalence of false-negative results for the system from ViroMed Laboratories, Inc., was not associated with a specific genotype.
Turnaround time estimation and cost analysis.
Southern blotting followed by probe hybridization required a minimum of 15 h for amplicon identification in our laboratory, including sample loading and running of the gel (2 h), denaturation and neutralization (1 h), blotting (6 h), prehybridization (30 min), hybridization (4 h), two washings (40 and 10 min), and film exposure and development (30 min). All four MTP systems needed a test time of <4 h for completion of the identification of the HSV amplicon (Table 2). The cost and technologist time including specimen processing and amplification steps for the Mayo-developed MTP system were compared with those for conventional methods on the basis of an estimated laboratory volume of 3,040 procedures in 1996. Costs and technologist times were similar for the Mayo assay and the conventional methods (Table 3).
TABLE 2.
Contrast of test time among colorimetric MTP systems for amplicon identification
| Test source | Time (min) required for the following step:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Denaturation | Hybridization | Washing | Conjugation | Washing | Substrate incubation | Colorimetric detection | Total | |
| CPG, Inc. | 2 | 20 | 5 | 10 | 5 | 30 | 10 | 82 |
| Incstar Corp.a | 15 | 60 | 5 | 30 × 2 | 5 × 2 | 30 | 10 | 190 |
| Mayo | 10 | 60 | 5 | 30 | 5 | 40 | 5 | 155 |
| ViroMed Laboratories, Inc. | 2 | 20 | 5 | 10 | 5 | 30 | 10 | 82 |
Additional precoating step not included.
TABLE 3.
Cost analysis of HSV PCR: Southern blotting versus the Mayo colorimetric MTP systema
| Test | Direct cost ($/procedure)
|
Total annual costb | Technologist time (min/procedure)
|
Total technologist time (h) annuallyb | |||
|---|---|---|---|---|---|---|---|
| Cost | Total cost with cost of capital | Variable para- medical effect | Fixed effort | Total technol- ogist time | |||
| SBc | 53.75 | 58.05 | 176,472.00 | 13.30 | 2.14 | 15.44 | 782.29 |
| Mayo MTP | 52.75 | 56.95 | 173,128.00 | 7.03 | 8.22 | 15.25 | 772.67 |
Volume and expenses are based on information for 1996.
The estimated annual volume is 3,040 procedures.
SB, Southern blotting.
DISCUSSION
The generation of rapid laboratory test results has had a major impact on the clinical management of patients with CNS infections. This is especially true for HSV infections since early treatment with acyclovir is very effective, reducing the rate of mortality from HSV encephalitis from 70% to 19 to 30% (33, 38). In the present study, we used Southern blotting followed by probe hybridization as our evaluation standard, which has a demonstrated sensitivity of 97.7 to 98.1% compared to the detection of HSV antigens or recovery of the virus from brain biopsy specimens (1, 15). PCR performed with CSF has a reduced turnaround time and a reduced level of patient risk for the diagnosis of HSV infection of the CNS compared to those for cell culture technology; however, 2 days is usually required to complete conventional Southern blotting and probe hybridization in our laboratory.
In contrast, the turnaround time for colorimetric MTP systems was about 10 h, including <4 h for amplicon identification. Providing that CSF specimens were processed in the morning, test results could be available on the same day as specimen submission. If test results were positive, unnecessary antibacterial treatment could be eliminated and appropriate antiviral therapy could be provided. In addition, negative PCR results for HSV DNA in some cases provide sufficient evidence to mitigate against unnecessary intravenous acyclovir treatment ($180.00/day) and would expand the diagnostic consideration for other etiologic causes of CNS infection.
Colorimetric MTP systems and Southern blotting use a sequence-specific probe that provides two technical enhancements to PCR (10, 20, 21). First, the sensitivity for the detection of PCR products with MTP systems is 10- to 100-fold greater than that with ethidium bromide-stained agarose gels. Second, the use of a specific probe confirms the specificity of the PCR product. Unlike Southern blotting and probe hybridization, use of the MTP allows the simultaneous rapid analysis of multiple PCR mixtures. Importantly, the assay is potentially automatable when performed in a 96-well format. Finally, the physical containment of the amplification products in the MTP is superior to that by Southern blotting-based methods and thus may be less prone to contamination problems.
Additional advantages of the Mayo-developed system include identification of virus genotypes and the recognition of polymorphisms that may be responsible for drug resistance. The TK gene of HSV has been targeted by the Mayo MTP system. Because significant heterogeneity between HSV-1 and HSV-2 exists in the 335-bp amplicon, allele-specific probes can be used for genotype determination. Interestingly, our results have indicated that some HSV-amplified DNAs react with both type-specific probes, suggesting mixed infections with the two genotypes. Point mutations in the TK gene may also be responsible for acyclovir resistance (3, 4, 25), and polymorphisms in this locus may be associated with neurotropism (31). Determination of acyclovir resistance or neurotropism by detection of these point mutations may be important for patients undergoing long-term therapy and immunocompromised hosts (6, 26). A single test that includes amplification of the TK gene followed by determination of sequence polymorphisms may be able to predict the HSV genotype and possibly drug resistance and neurotropism without the need for a second amplification reaction. Therefore, by using separate MTP wells for HSV-1- and HSV-2-specific hybridizations, the viral type can be identified immediately. Acyclovir resistance could be determined by direct sequencing of the amplicon based on clinical management.
Our data indicate that one need not sacrifice sensitivity to obtain rapid results. Three of the four MTP systems tested had sensitivities of >98% compared with the results of Southern blotting followed by probe hybridization. The Mayo system had the best sensitivity (100%) of the four systems tested. We noted that among 36 CSF specimens determined to be HSV DNA negative by conventional methods, one was positive by MTP analysis with the systems from both CPG, Inc., and Mayo. This result was obtained for a patient who was clinically diagnosed with aseptic meningitis on the basis of a headache, increased leukocyte counts, and elevated total protein levels in her CSF, suggesting a likely false-negative result by the conventional method.
Colorimetric MTP analysis for HSV PCR amplicon identification can be performed in less than 4 h. These assays do not require toxic chemical agents or an electrophoretic apparatus. Substitution of MTP for Southern blot analysis is cost neutral, with no compromise in test sensitivity. MTP systems may be adapted for automation, which is a compelling requirement for PCR testing, which, in many laboratories, is expanding at a rapid rate. Collectively, these factors make the implementation of this technology in routine diagnostic testing a fundamental goal in our laboratory.
ACKNOWLEDGMENTS
We thank Jill Thorvilson, T. J. Berg, Carl Greiner, Heather Skarhus, Paul Heimgartner, Ann Wimmer, and David Majewski for technical assistance and Jonathan Hibbs for critically reviewing the manuscript.
ADDENDUM IN PROOF
The data generated with the PrimeCapture HSV DNA detection system were derived using a system lot number recalled by the manufacturer. The data may not accurately reflect the current performance of the assay system.
REFERENCES
- 1.Aurelius E, Johansson B, Skoldenberg B, Staland A, Forsgren M. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet. 1991;337:189–192. doi: 10.1016/0140-6736(91)92155-u. [DOI] [PubMed] [Google Scholar]
- 2.Buck G E. Detection of Bordetella pertussis by rapid-cycle PCR and colorimetric microwell hybridization. J Clin Microbiol. 1996;34:1355–1358. doi: 10.1128/jcm.34.6.1355-1358.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coen D M. Acyclovir-resistant, pathogenic herpesviruses. Trends Microbiol. 1994;2:481–485. doi: 10.1016/0966-842x(94)90652-1. [DOI] [PubMed] [Google Scholar]
- 4.Englund J A, Zimmerman M E, Swierkosz E M, Goodman J L, Scholl D R, Balfour H H., Jr Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann Intern Med. 1990;112:416–422. doi: 10.7326/0003-4819-76-3-112-6-416. [DOI] [PubMed] [Google Scholar]
- 5.Espy M J, Smith T F, Persing D H. Dependence of polymerase chain reaction product inactivation protocols on amplicon length and sequence composition. J Clin Microbiol. 1993;31:2361–2365. doi: 10.1128/jcm.31.9.2361-2365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field A K, Biron K K. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev. 1994;7:1–13. doi: 10.1128/cmr.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham D, Larder B A, Inglis M M. Evidence that the ‘active centre’ of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J Gen Virol. 1986;67:753–758. doi: 10.1099/0022-1317-67-4-753. [DOI] [PubMed] [Google Scholar]
- 8.Jeffery K J, Read S J, Peto T E, Mayon-White R T, Bangham C R. Diagnosis of viral infections of the central nervous system: clinical interpretation of PCR results. Lancet. 1997;349:313–317. doi: 10.1016/S0140-6736(96)08107-X. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R T. Acute encephalitis. Clin Infect Dis. 1996;23:219–226. doi: 10.1093/clinids/23.2.219. [DOI] [PubMed] [Google Scholar]
- 10.Jonas D, Rosenbaum A, Weyrich S, Bhakdi S. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J Clin Microbiol. 1995;33:1247–1252. doi: 10.1128/jcm.33.5.1247-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H, Futamura M, Kito H, Ando T, Goto M, Kuzushima K, Shibata M, Morishima T. Detection of viral DNA in neonatal herpes simplex virus infections: frequent and prolonged presence in serum and cerebrospinal fluid. J Infect Dis. 1991;164:289–293. doi: 10.1093/infdis/164.2.289. [DOI] [PubMed] [Google Scholar]
- 12.Klapper P E, Cleator G M, Dennett C, Lewis A G. Diagnosis of herpes encephalitis via Southern blotting of cerebrospinal fluid DNA amplified by polymerase chain reaction. J Med Virol. 1990;32:261–264. doi: 10.1002/jmv.1890320413. [DOI] [PubMed] [Google Scholar]
- 13.Kohl S. Herpes simplex virus encephalitis in children. Pediatr Clin N Am. 1988;35:465–483. doi: 10.1016/s0031-3955(16)36466-5. [DOI] [PubMed] [Google Scholar]
- 14.Lage A P, Fauconnier A, Burette A, Glupczynski Y, Bollen A, Godfroid E. Rapid colorimetric hybridization assay for detecting amplified Helicobacter pylori DNA in gastric biopsy specimens. J Clin Microbiol. 1996;34:530–533. doi: 10.1128/jcm.34.3.530-533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakeman F D, Whitley R J. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. J Infect Dis. 1995;171:857–863. doi: 10.1093/infdis/171.4.857. [DOI] [PubMed] [Google Scholar]
- 16.Lear W, McDonnel M, Kashyap S, Boer P H. Random primer p(dN)6-digoxigenin labeling for quantitation of mRNA by Q-RT-PCR and ELISA. BioTechniques. 1995;18:78–83. [PubMed] [Google Scholar]
- 17.Loeffelholz M J, Lewinski C A, Silver S R, Purohit A P, Herman S A, Buonagurio D A, Dragon E A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992;30:2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 19.Luneberg E, Jensen J S, Frosch M. Detection of Mycoplasma pneumoniae by polymerase chain reaction and nonradioactive hybridization in microtiter plates. J Clin Microbiol. 1993;31:1088–1094. doi: 10.1128/jcm.31.5.1088-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansy F, Hoyois B, De Vos M J, Van Elsen A, Bollen A, Godfroid E. Colorimetric solid-phase capture hybridization assay for detection of amplified Borrelia burgdorferi DNA. BioTechniques. 1996;21:122–125. doi: 10.2144/96211rr03. [DOI] [PubMed] [Google Scholar]
- 21.Mantero G, Zonaro A, Albertini A, Bertolo P, Primi D. DNA enzyme immunoassay: general method for detecting products of polymerase chain reaction. Clin Chem. 1991;37:422–429. [PubMed] [Google Scholar]
- 22.Mitchell P S, Espy M J, Smith T F, Toal D R, Rys P N, Berbari E F, Osmon D R, Persing D H. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J Clin Microbiol. 1997;35:2873–2877. doi: 10.1128/jcm.35.11.2873-2877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nahmias A J, Whitley R J, Visintine A N, Takei Y, Alford C A., Jr Herpes simplex virus encephalitis: laboratory evaluations and their diagnostic significance. J Infect Dis. 1982;145:829–836. doi: 10.1093/infdis/145.6.829. [DOI] [PubMed] [Google Scholar]
- 24.Nicoll J A, Maitland N J, Love S. Autopsy neuropathological findings in ‘burnt out’ herpes simplex encephalitis and use of the polymerase chain reaction to detect viral DNA. Neuropathol Appl Neurobiol. 1991;17:375–382. doi: 10.1111/j.1365-2990.1991.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 25.Palu G, Gerna G, Bevilacqua F, Marcello A. A point mutation in the thymidine kinase gene is responsible for acyclovir-resistance in herpes simplex virus type 2 sequential isolates. Virus Res. 1992;25:133–144. doi: 10.1016/0168-1702(92)90105-i. [DOI] [PubMed] [Google Scholar]
- 26.Pease A C, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poljak M, Seme K. Rapid detection and typing of human papillomaviruses by consensus polymerase chain reaction and enzyme-linked immunosorbent assay. J Virol Methods. 1996;56:231–238. doi: 10.1016/0166-0934(95)01969-3. [DOI] [PubMed] [Google Scholar]
- 28.Powell K F, Anderson N E, Frith R W, Croxson M C. Non-invasive diagnosis of herpes simplex encephalitis. Lancet. 1990;335:357–358. doi: 10.1016/0140-6736(90)90648-o. [DOI] [PubMed] [Google Scholar]
- 29.Puchhammer-Stockl E, Popow-Kraupp T, Heinz F X, Mandl C W, Kunz C. Establishment of PCR for the early diagnosis of herpes simplex encephalitis. J Med Virol. 1990;32:77–82. doi: 10.1002/jmv.1890320202. [DOI] [PubMed] [Google Scholar]
- 30.Rowley A H, Whitley R J, Lakeman F D, Wolinsky S M. Rapid detection of herpes-simplex-virus DNA in cerebrospinal fluid of patients with herpes simplex encephalitis. Lancet. 1990;335:440–441. doi: 10.1016/0140-6736(90)90667-t. [DOI] [PubMed] [Google Scholar]
- 31.Rozenberg F, Lebon P. Analysis of herpes simplex virus type 1 glycoprotein D nucleotide sequence in human herpes simplex encephalitis. J Neurovirol. 1996;2:289–295. doi: 10.3109/13550289609146892. [DOI] [PubMed] [Google Scholar]
- 32.Seesod N, Nopparat P, Hedrum A, Holder A, Thaithong S, Uhlen M, Lundeberg J. An integrated system using immunomagnetic separation, polymerase chain reaction, and colorimetric detection for diagnosis of Plasmodium falciparum. Am J Trop Med Hyg. 1997;56:322–328. doi: 10.4269/ajtmh.1997.56.322. [DOI] [PubMed] [Google Scholar]
- 33.Skoldenberg B, Forsgren M, Alestig K, Bergstrom T, Burman L, Dahlqvist E, Forkman A, Fryden A, Lovgren K, Norlin K, et al. Acyclovir versus vidarabine in herpes simplex encephalitis. Randomised multicentre study in consecutive Swedish patients. Lancet. 1984;ii:707–711. doi: 10.1016/s0140-6736(84)92623-0. [DOI] [PubMed] [Google Scholar]
- 34.Swain M A, Galloway D A. Nucleotide sequence of the herpes simplex virus type 2 thymidine kinase gene. J Virol. 1983;46:1045–1050. doi: 10.1128/jvi.46.3.1045-1050.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Y W, Espy M J, Persing D H, Smith T F. Molecular evidence and clinical significance of herpesvirus coinfection in central nervous system. J Clin Microbiol. 1997;35:2869–2872. doi: 10.1128/jcm.35.11.2869-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tedder D G, Ashley R, Tyler K L, Levin M J. Herpes simplex virus infection as a cause of benign recurrent lymphocytic meningitis. Ann Intern Med. 1994;121:334–338. doi: 10.7326/0003-4819-121-5-199409010-00004. [DOI] [PubMed] [Google Scholar]
- 37.Vesanen M, Piiparinen H, Kallio A, Vaheri A. Detection of herpes simplex virus DNA in cerebrospinal fluid samples using the polymerase chain reaction and microplate hybridization. J Virol Methods. 1996;59:1–11. doi: 10.1016/0166-0934(95)01991-x. [DOI] [PubMed] [Google Scholar]
- 38.Whitley R J, Alford C A, Hirsch M S, Schooley R T, Luby J P, Aoki F Y, Hanley D, Nahmias A J, Soong S J. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N Engl J Med. 1986;314:144–149. doi: 10.1056/NEJM198601163140303. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto L J, Tedder D G, Ashley R, Levin M J. Herpes simplex virus type 1 DNA in cerebrospinal fluid of a patient with Mollaret’s meningitis. N Engl J Med. 1991;325:1082–1085. doi: 10.1056/NEJM199110103251507. [DOI] [PubMed] [Google Scholar]
- 40.Yang B, Viscidi R, Yolken R. Quantitative measurement of nonisotopically labeled polymerase chain reaction product. Anal Biochem. 1993;213:422–425. doi: 10.1006/abio.1993.1441. [DOI] [PubMed] [Google Scholar]