Abstract
The Children’s Oncology Group (COG) Epidemiology Committee has a primary focus on better understanding the etiologies of childhood cancers. Over the past 10 years, the Committee has leveraged the Childhood Cancer Research Network, and now more recently Project:EveryChild (PEC), to conduct epidemiologic assessments of various childhood cancers including osteosarcoma, neuroblastoma, germ cell tumors, Ewing sarcoma, rhabdomyosarcoma, and Langerhans cell histiocytosis. More recent studies have utilized questionnaire data collected as part of PEC to focus on specific characteristics and/or features, including the presence of congenital disorders and the availability of stored cord blood. Members of the COG Epidemiology Committee have also been involved in other large-scale National Institutes of Health efforts, including the Childhood Cancer Data Initiative and the Gabriella Miller Kids First Pediatric Research Program, which are improving our understanding of the factors associated with childhood cancer risk. Future plans will focus on addressing questions surrounding health disparities, utilizing novel biospecimens in COG epidemiology studies, exploring the role of environmental factors on the etiologies and outcomes of childhood cancer, collaborating with other COG committees to expand the role of epidemiology in childhood cancer research, and building new epidemiologic studies from the Molecular Characterization Initiative - all with the ultimate goal of developing novel prevention and intervention strategies for childhood cancer.
I. Introduction and Rationale of the Committee
Epidemiology is broadly defined as the study of the distribution and determinants of health-related states or events in specified populations. Studies evaluating the distribution of childhood cancers are commonly referred to as “descriptive” and largely rely on cancer registry data. For example, by using data from the National Program of Cancer Registries,1 we can observe relative differences in the incidence of childhood cancer across the U.S. (Figure 1a). Furthermore, if we focus on the incidence of acute lymphoblastic leukemia (ALL; Figure 1b), we can not only see differences across states but also differences from childhood cancer overall. While these data are important in describing trends in childhood cancer, they may also inform hypotheses surrounding the determinants of childhood cancer (e.g., differences in population characteristics, environmental exposures). Evaluating such patterns, generating hypotheses, and developing studies related to the development of childhood cancer has been the primary focus of the Children’s Oncology Group (COG) Epidemiology Committee. Our goal is to understand the etiologies of childhood cancer so that we can inform prevention measures or novel screening and early detection protocols.
Figure 1.
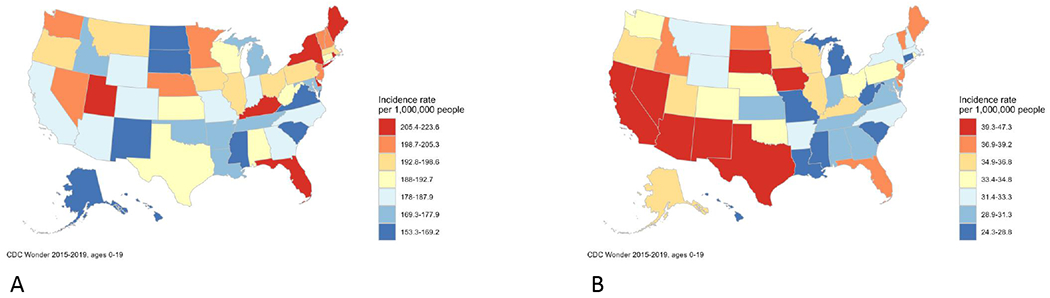
Incidence of childhood cancer overall (A) and childhood acute lymphoblastic leukemia (B) using data from the National Program of Cancer Registries, 2015-2019. The distributional characteristics of childhood cancer can inform epidemiologic assessments of potential risk factors.
The central challenge to conducting epidemiologic studies of childhood cancer is the rarity of these malignancies. This often precludes the use of prospective study designs. Because of this, our knowledge on the etiologies of childhood cancers comes mainly from case-control studies and must be interpreted within the limitations of that particular study design (e.g., recall bias). Furthermore, childhood cancers are heterogeneous, and it must be recognized that each tumor has its individual risk factor profile. However there are some commonalities.2 For instance, incidence is higher in males for nearly all cancers, and there is evidence that many childhood cancers originate in utero. Some risk factors, like birth weight, are associated with several tumors but differ in the direction and magnitude of association, while others (e.g., hernias and Ewing sarcoma) are unique to a particular cancer.2 It is also the case that the literature on risk factors is roughly proportional to incidence, so that we know far more about the etiology of ALL (36 cases per million) than hepatoblastoma (2 cases per million).
Members of the Epidemiology Committee have leveraged these and other observations to evaluate both established and novel risk factors for childhood cancer. Studies conducted in the past 15 years have largely been based on the case-parent trio design with a focus on understanding the role of germline genetic variants on the risk of childhood cancer. The trio for which this design is named is composed of the affected individual (i.e., case) and both biological parents. This design is useful for studies of childhood cancer and conditions with early disease onset, as parents are usually available and willing to participate.3 The case-parent trio design also has the advantage (as compared to case-control studies) of being immune to population stratification bias when assessing the effects of the inherited genotypes, allowing for the inclusion of genetically diverse populations. This design can be used to assess maternal genetic effects, which governs the in utero environment of the developing offspring and could be important in the etiologies of childhood cancer. Furthermore, the case-parent trio design allows for the assessment of other genetic effects, including gene-environment interactions and the role of de novo variants on disease susceptibility.
In this COG Blueprint for Research, we will highlight the achievements of the Epidemiology Committee, which has been guided by a collaborative approach with the COG Disease Committees. This includes a number of studies focused on specific childhood cancers that were developed using the COG groupwide consenting protocol ACCRN07 (the Childhood Cancer Research Network or CCRN), which allowed for the recruitment of families for non-therapeutic studies (see Table 1 for examples). Additionally, the Epidemiology Committee has been at the forefront of leveraging APEC14B1 (Project:EveryChild or PEC), which also includes consent for recontact, as well as some basic information which can be leveraged for epidemiologic assessments (e.g., family history of cancer) and the storage of biospecimens. We will also outline our strategic plan for the next decade. This includes building from existing resources including PEC and exploring new approaches for understanding the etiologies of childhood cancer and improving outcomes for these children.
Table 1.
Disease-focused COG Epidemiology Committee studies using the case-parent trio design
| Tumor type | Name of study | Years of diagnosis | Number of cases enrolled | Number of family members enrolled |
|---|---|---|---|---|
| Osteosarcoma | GO | 2007-2010 | 289 | 514 |
| Neuroblastoma | NENA | 2007-2013 | 611 | 1,151 |
| Germ cell tumor | GaMETES | 2008-2015 | 866 | 1,517 |
| Ewing sarcoma | GENESIS | 2008-2015 | 430 | 646 |
| Rhabdomyosarcoma | GEARS | 2008-2020 | 299 | 853 |
| LCH* | GECHO | 2008-2023 | 226 | 412 |
Still enrolling as of May 2023
II. Key Achievements
1. Childhood Cancer Research Network and Case-Parent Trio Studies (Table 1)
a. Osteosarcoma
The Genetics of Osteosarcoma study (GO; AEPI05N2; U01CA122371) was the first COG epidemiology study to use the case-parent trio design. The purpose was to study genetic variation in 1) the insulin-like growth factor/growth hormone axis and estrogen metabolism pathways, which relate to bone growth and 2) genomic integrity. DNA samples were successfully obtained for 289 probands, with both parents of 229 patients providing DNA samples and one parent of 56 patients doing so. Investigation of 798 variants in 42 genes in the aforementioned pathways uncovered several associations which survived correcting for multiple testing.4 These samples later were assayed by genome-wide SNP arrays, exome sequencing, and/or targeted sequencing of cancer susceptibility genes. The array data have contributed to a genome-wide association study (GWAS) for metastasis at diagnosis5 and survival for osteosarcoma.6
Sequencing data have contributed to a survey of pathogenic/likely pathogenic (P/LP) variants in cancer predisposition genes,7 which revealed that nearly 30% of osteosarcoma patients harbored these variants. Lastly, in one of the first assessments of de novo variants among childhood cancer case-parent trios, it was discovered that nearly half of P/LP germline TP53 variants among osteosarcoma patients were de novo,8 which has important implications for genetic counseling strategies.
b. Neuroblastoma
The Neuroblastoma Epidemiology in North America study (NENA; AEPI07N1; R01CA132887 and R01CA124709) was a case-parent trio study designed to evaluate the nutrient-related exposures and candidate genes. Cases were diagnosed before six years of age at a U.S. or Canadian COG institution from December 24, 2007 to July 31, 2013. Saliva samples were collected for 592 children, 626 biological mothers, and 525 biological fathers; blood samples were obtained for 19 deceased children. Variants related to folate and choline metabolism were investigated for main genetic effects and for interactions with maternal intake of these nutrients. There were no significant associations with folate metabolism genes but a handful of variants remained significant for gene-choline interactions.9 In another analysis there was little evidence to suggest that genetic variation related to the metabolism of vitamin A played a role in neuroblastoma susceptibility.10 A more recent study used case-parent trios to validate a 16p11.2 microdeletion among children with neuroblastoma11 and to examine de novo variants among these children.
c. Germ cell tumors
The Germ Cell Tumor Epidemiology Study (GaMETES; AEPI10N1; R01CA151284) was established to understand genetic susceptibility to germ cell tumors (GCT) in the pediatric and adolescent age group and to understand patterns of DNA methylation in tumor samples. Children were eligible for the study if they had a GCT diagnosed at age < 20 years between July 1, 2008 and December 31, 2015. Enrollment was open from 2011-2015, and probands from 865 families were enrolled in the study during this time period. Of these families, 682 (79%) were complete trios with DNA available for the proband and both parents, 160 (18%) included the proband and one parent with or without siblings, and 23 (2.6%) were case only families. Maternal and paternal questionnaires were also completed, and the majority of cases have questionnaire data from at least one parent (91%).
Given the paucity of molecular and epidemiologic data for GCT, the data from the GaMETES study have made a large contribution to understanding of disease etiology. Family history data collected from mothers and fathers of affected GCT cases was used to evaluate heritability of pediatric GCT and showed a higher than expected number of GCT cases in both male and female relatives of cases.12 The data were used to identify common variants that increase risk of developing GCT and provided evidence of overlapping genetic susceptibility to pediatric and adult GCT.13 We also quantified the association between Klinefelter syndrome and risk of GCT,14 evaluated the role of predicted leukocyte telomere length in GCT risk,15 and reported on the higher prevalence of birth defects in GCT cases. Finally, tumor methylation data were used to describe patterns of methylation by tumor histology16 and to describe aberrations in WNT signaling and their potential role in predicting outcomes.17 Of note, in collaboration with the COG Outcomes and Survivorship Committee, the GaMETES study has been used to establish a new survivorship cohort to study outcomes and late effects of children and adolescent survivors of GCT.18 It is hoped this will be a template for converting epidemiology studies focused on etiology to survivorship cohorts.
d. Ewing sarcoma
The goals of the Genetics of Ewing Sarcoma International Study (Project GENESIS; AEPI10N5; R01CA161780) were to examine GGAA microsatellite repeat length in patients, conduct admixture mapping among non-European patients, and to determine whether variants related to hernia development increase risk of Ewing sarcoma. DNA was obtained for 430 probands and 602 parents. Data from GENESIS was recently used to examine germline predisposition to Ewing sarcoma, and suggested enrichment of pathogenic or likely pathogenic variants in cancer susceptibility genes, particularly FANCC, in probands.19 Several studies related to Project GENESIS are ongoing and are likely to lead to a deeper understanding of the genetic epidemiology of this sarcoma.
e. Rhabdomyosarcoma
The primary goal of the Genetics of Embryonal and Alveolar Rhabdomyosarcoma Study (GEARS; AEPI15N1, CPRIT RP170071) was to evaluate the role of cancer predisposition genes among children diagnosed with rhabdomyosarcoma and determine differences in the frequency of P/LP variants in these genes by demographic and clinical factors. Recruitment for GEARS started in April 2018 and ended May 2022. Overall, 299 probands were enrolled, along with 853 family members. Additionally, as part of GEARS, 901 germline samples were obtained through the Soft Tissue Sarcoma Committee biology protocol. Findings from GEARS include a report among 615 rhabdomyosarcoma patients indicating that 7.3% harbored a P/LP variant in a cancer predisposition gene.20 Also genetic etiology differed with histology, as germline variants were more frequent in embryonal versus alveolar rhabdomyosarcoma patients (10% vs 3%, P = 0.02). Although patients with a cancer predisposition variant tended to be younger at diagnosis (P = 9.9 × 10-4), 40.0% of germline variants were identified in those older than 3 years of age, which is in contrast to current genetic testing recommendations based on early age at diagnosis. Another report published among those with anaplastic rhabdomyosarcoma indicated that 11% of cases (5/46) harbored a pathogenic germline variant in TP53,21 which while high is lower than previous estimates.
As with the GO and GaMETES study, efforts have expanded to evaluating outcomes among patients with rhabdomyosarcoma. A recent example includes a GWAS of survival, which implicated common germline variants are associated with EFS and OS among individuals with rhabdomyosarcoma.22 Additional studies are focusing on the role of rare variants on survival, as well as the influence of genetic variation on toxicities during therapy.
f. Langerhans cell histiocytosis
One of the most recent case-parent trio studies to build from CCRN is the Genetic Epidemiology of Childhood Histiocytosis (GECHO; AEPI17N1, R01CA233719). Compared to other tumors included in recent protocols, much less is known about the epidemiology of Langerhans cell histiocytosis (LCH),which is an inflammatory myeloid neoplasia characterized by lesions including pathogenic CD207+ dendritic cells among an inflammatory infiltrate. Sequencing studies have found recurrent, mutually exclusive somatic activating mutations in MAPK pathway genes in ~85% of LCH lesions, including BRAFV600E in 50-65%. Despite advances to elucidate the somatic mutational landscape underlying LCH pathogenesis, germline risk factors remain largely unknown. Because of this, members of the Epidemiology and Non-Hodgkin Lymphoma Committees started a collaboration to comprehensively characterize germline genetic susceptibility to LCH through COG. As a first step, a novel LCH susceptibility variant in SMAD6 was discovered through an institutional GWAS.23 Findings from this assessment served as the impetus for the GECHO study, with the primary objectives to characterize the role of SMAD6 on LCH susceptibility and identify germline genomic regions associated with LCH somatic mutations. While recruitment is ongoing, to date, 226 probands have been enrolled, along with 412 family members. Early findings suggest additional variants in SMAD6 are important for LCH risk.
2. PEC and Novel Epidemiologic Assessments
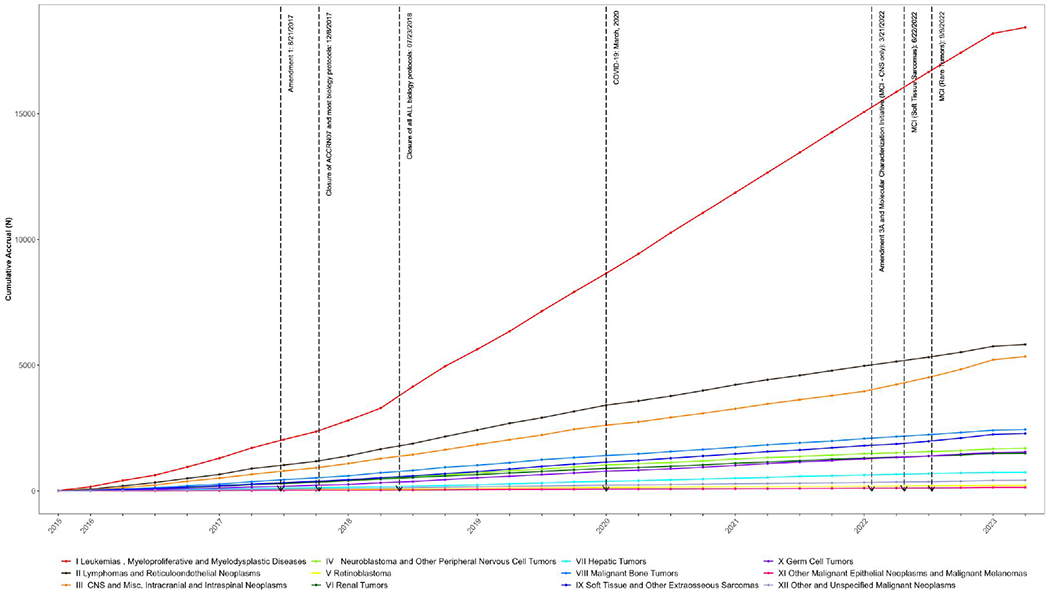
PEC is the combined biobanking and research registration protocol which succeeded the CCRN. Accrual to PEC since inception in December, 2015, is shown in Figure 2; panel A shows accrual by stratum while panel B shows accrual by disease class (using the International Classification of Childhood Cancer system) with several key dates in the history of PEC noted. One which visibly affected the pace of accrual of leukemias was closure of ALL biology protocols in July, 2018. Notably, and perhaps surprisingly, COVID-19 restrictions which began in March, 2020 seem not to have affected accrual to a large degree. Note that while neuroblastoma and renal tumor patients are eligible for PEC (as are all patients treated by COG institutions), accrual is lower than for other diseases as biology protocols specific to these tumors remain open currently but will close in favor of PEC enrollment by 2024.
Figure 2A.
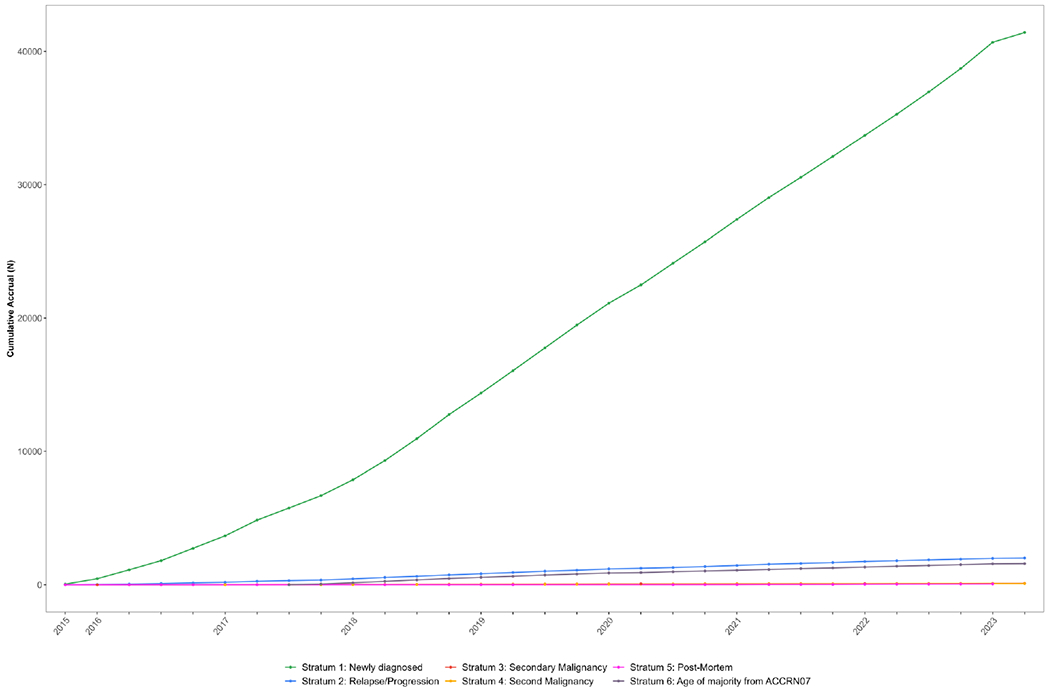
Accrual to Project:EveryChild (APEC14B1) by enrollment stratum.
Figure 2B. Accrual to Project:EveryChild (APEC14B1) by disease class.
In addition to obtaining consent to contact patients for future non-therapeutic research, all PEC enrollees are invited to complete an up-front questionnaire regarding some critical epidemiologic variables. Thus for thousands of PEC enrollees there is information on family history of cancer in first degree relatives, the presence of genetic syndromes, the presence of structural birth defects, history of autoimmune disease, storage of cord blood at birth, membership in a twin pregnancy, and conception by in vitro fertilization.
This information has been used to design and execute several studies which would have been nearly impossible to conduct before the advent of PEC. For example, ReCord (https://record.umn.edu/; AEPI20N1, R01CA262012) is collecting stored cord blood and matched tumor samples from children with leukemia in order to conduct a state-of-the-art “backtracking” of leukemia-typical translocations to birth. Additionally, there are currently two funded studies building on observations related to the risk of cancer in children with congenital disorders identified as part of the PEC questionnaire. This includes 1) the Genetic Overlap Between Anomalies and Cancer in Kids study (GOBACK; AEPI19N1, R03CA272955), which is focused on the role of structural birth defects on a spectrum of childhood cancers, and 2) Evaluating the association between co-occurring birth defects and acute lymphoblastic leukemia risk in children with Down syndrome: The DS phenotyping acute leukemia study (DS-PALS; AEPI20N2, R01CA249867), which is focused on the risk of ALL in children with DS. These examples demonstrate the power of PEC for COG epidemiology studies and the collaborative research conducted in COG.
3. Gabriella Miller Kids First Pediatric Research Program and COG Epidemiology Studies
The goal of the Gabriella Miller Kids First Pediatric Research Program (Kids First; https://commonfund.nih.gov/kidsfirst) is to help researchers uncover new insights into the biology of structural birth defects and childhood cancer, including the discovery of shared genetic pathways between these disorders. Over 2015-2022, the program selected 63 cohorts (19 focused on childhood cancer) for whole-genome sequencing through a peer-review process, representing 21,000 patients and 55,000 genomes. Notably, a focus of the Kids First was the inclusion of case-parent trios. Building from that, of the 19 childhood cancer cohorts selected for inclusion to date, 10 have been led by members of the COG Epidemiology Committee. These efforts include patients enrolled in NENA (neuroblastoma), GaMETES (germ cell tumors), GENESIS (Ewing sarcoma), GEARS (rhabdomyosarcoma), GECHO (LCH), GOBACK (cancer in children with structural birth defects), and DS-PALS (DS-associated ALL). Findings from Kids First are already leading to new insights into genetic predisposition to Ewing sarcoma19 and the genetic underpinnings of DS-ALL (Figure 3). Furthermore, these data will be leveraged for years to come to elucidate the molecular etiologies of childhood cancer.
Figure 3.
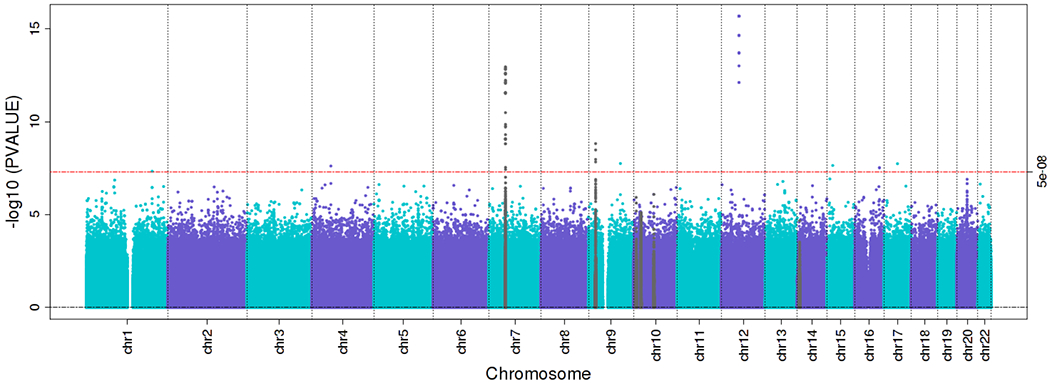
Common variant analysis of whole-genome sequencing data from Kids First on children with Down syndrome-associated acute lymphoblastic leukemia (ALL) compared to controls (children with Down syndrome who did not develop ALL) point to the role of these factors on susceptibility. These data point to the utility of Kids First data and the collaborative efforts in COG to understand the etiology of ALL in children with Down syndrome.
IV. Strategic Plan
1. Addressing Health Disparities
There are numerous disparities among racial and ethnic groups in the incidence, clinical presentation, and outcomes of childhood cancer. For example, Black and Asian Pacific Islander children have lower incidence rates of most major tumor types compared to non-Hispanic White children, while Hispanic children have notably higher incidence rates of ALL, retinoblastoma, and hepatoblastoma.24 Additionally, non-Hispanic Black, Asian, and Hispanic children are more likely to present with advanced stage cancer than non-Hispanic White children25 and may have higher rates of molecular subtypes that confer a poor prognosis.26 Compared with non-Hispanic White children, non-Hispanic Black and Hispanic children experience lower survival from many cancers, including leukemias,27 lymphomas,28 central nervous system (CNS) tumors,29 and non-CNS solid tumors.30 The underlying causes of these differences are poorly understood and may vary by cancer type, and both biological and socioeconomic pathways have been proposed. There is mounting evidence supporting an association between higher socioeconomic status (SES) and increased childhood cancer survival regardless of race/ethnicity.31
There are ongoing efforts to identify and address health disparities utilizing COG infrastructure and existing data. For example, investigators are currently leveraging CCRN data, which provides an opportunity to examine the influence of small-area based SES and distance to care on outcomes (AEPI21N1, R01CA266105). There are also groupwide initiatives to embed SES and other social determinants of health data into clinical trials to enable future research.32
2. Leveraging the Molecular Characterization Initiative
The Molecular Characterization Initiative (MCI) is part of National Cancer Institute’s Childhood Cancer Data Initiative (CCDI) and is a national collaboration involving the childhood cancer community building from the PEC. It provides state-of-the-art molecular characterization at the time of diagnosis that helps participants and doctors select the best and most appropriate treatment. For the initial phase of the MCI, the focus has been on less frequent childhood cancers and/or those where little is known in terms of molecular features. This includes CNS tumors, soft tissue sarcomas, and various rare tumors (e.g., several pediatric carcinomas). Members of the Epidemiology Committee have been leaders in the CCDI and a priority for the Committee in the coming years will be to build from the MCI for novel epidemiologic assessments. This will include 1) developing cohorts focused on unique populations (e.g., rare tumors, children with germline findings), 2) leveraging existing germline data to better characterize genetic susceptibility to childhood cancer - especially those where less is known (e.g., non-rhabdomyosarcoma soft tissue sarcomas), and 3) incorporating molecular features of tumors and somatic data in novel epidemiologic assessments to identify subtype-related risk factors.
3. Incorporating Blood Spots
Residual newborn dried blood spots (DBS) offer a unique and extremely valuable resource to advance epidemiologic research, as they allow for the assessment of genetic and epigenetic factors, as well as environmental exposures present at birth, with possible extrapolation to earlier in utero exposures. In fact, the utilization of DBS to elucidate causal agents and biological response associated with childhood cancer risk has the potential to dramatically increase knowledge of the etiology of these malignancies.
Newborn screening is a mandatory public health program in the U.S. and other counties that tests newborns for selected genetic, endocrine, and metabolic disorders. Some states retain residual DBS which can be released for research purposes after screening is complete, although the duration of retention and storage practice varies widely by state, from 30 days to indefinite, and policies regarding their use for research have evolved over the past decade.33
The potential uses of DBS in studies of childhood cancer are extensive (Table 2). For example, DBS can be used to measure amino acids, enzymes, viral DNA, antibodies, inflammation markers, steroids, metals, small molecules, protein adducts, and some drugs, as well as certain environmental exposures, including epigenetic signatures of exposures.34 Furthermore, DBS also serve as a source of germline DNA for use in studies of genetic risk factors of childhood cancers.
Table 2.
Publications utilizing blood spots for studies of childhood cancer
| First author | Year | Title | Journal | PMID |
|---|---|---|---|---|
| KB Gale | 1997 | Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots | Proceedings of the Academy of Sciences | 9391133 |
| JL Wiemels | 1999 | Prenatal origin of acute lymphoblastic leukemia in children | Lancet | 10551495 |
| JW Taub | 2004 | The prenatal origin of childhood acute lymphoblastic leukemia | Leukemia and Lymphoma | 15061193 |
| LG Spector | 2007 | Detection of cotinine in newborn dried blood spots | Cancer Epidemiology Biomarkers and Prevention | 17855712 |
| JS Chang | 2011 | Profound deficit of IL10 at birth in children who develop acute lymphoblastic leukemia | Cancer Epidemiology Biomarkers and Prevention | 21653647 |
| AP Chokkalingam | 2013 | Blood levels of folate at birth and risk of childhood leukemia | Cancer Epidemiology Biomarkers and Prevention | 23576692 |
| WL Ma | 2014 | Analysis of polychlorinated biphenyls and organochlorine pesticides in archived dried blood spots and its application to track temporal trends of environmental chemicals in newborns | Environmental Research | 24968082 |
| AM Dahlin | 2015 | CCND2, CTNNB1, DDX3X, GLI2, SMARCA4, MYC, MYCN, PTCH1, TP53, and MLL2 gene variants and risk of childhood medulloblastoma | Journal of Neurooncology | 26290144 |
| SS Francis | 2017 | In utero cytomegalovirus infection and development of childhood acute lymphoblastic leukemia | Blood | 27979823 |
| LM Petrick | 2017 | An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies | Metabolomics | 29706849 |
| LM Morimoto | 2018 | Neonatal hormone concentration and risk of testicular germ cell tumors (TGCT) | Cancer Epidemiology Biomarkers and Prevention | 29475970 |
| LM Petrick | 2019 | Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatrics acute lymphoblastic leukemia and potential effects of early-life nutrition | Cancer Letters | 30904619 |
Given the value of DBS in etiologic research and the need to prospectively collect and store DBS in states with short retention time, a possibility for expanding the collection of pre-diagnostic biospecimens for biomarker assessments would be linking PEC to state-based DBS repositories. Members of the Epidemiology Committee are exploring these options through the Childhood Cancer Data Initiative and the National Childhood Cancer Registry.
V. Conclusions
COG allows for unique and unparalleled opportunities for studying the epidemiology of childhood cancers. During the past decade, the Epidemiology Committee has been responsible for the development of studies focused on specific tumors, including those for which little is known in terms of risk factors. Additionally, there has been an effort to fully utilize the resources available as part of PEC and build from cross-committee collaborative efforts. Future plans will focus on addressing questions surrounding health disparities, utilizing novel biospecimens in COG epidemiology studies (e.g., DBS), exploring the role of environmental factors on the etiologies and outcomes of childhood cancer, collaborating with other COG committees to expand the role of epidemiology in childhood cancer research, and building new epidemiologic studies from the MCI - all with the ultimate goal of developing novel prevention and intervention strategies for childhood cancer.
Funding Information
St. Baldrick’s Foundation; Children’s Oncology Group Foundation; Isabella Santos Foundation, Grant/Award Numbers: U10CA098543, U10CA098413, U10CA180899, U10CA180886, U01CA122371, R01CA132887, R01CA124709, R01CA151284, R01CA161780, CPRIT RP170071, R01CA233719, R01CA262012, R03CA272955.
Table of Abbreviations
- COG
Children’s Oncology Group
- PEC
Project:EveryChild
- ALL
Acute lymphoblastic leukemia
- CCRN
Childhood Cancer Research Network
- GO study
Genetics of Osteosarcoma study
- GWAS
Genome-wide association study
- P/LP
Pathogenic/likely pathogenic
- NENA study
Neuroblastoma Epidemiology in North America study
- GaMETES
The Germ Cell Tumor Epidemiology Study
- GCT
Germ cell tumors
- GENESIS
Genetics of Ewing Sarcoma International Study
- GEARS
Genetics of Embryonal and Alveolar Rhabdomyosarcoma Study
- GECHO study
Genetic Epidemiology of Childhood Histiocytosis study
- LCH
Langerhans cell histiocytosis
- GOBACK study
Genetic Overlap Between Anomalies and Cancer in Kids study
- DS-PALS
DS Phenotyping Acute Leukemia Study
- CNS
Central nervous system
- MCI
Molecular Characterization Initiative
- CCDI
Childhood Cancer Data Initiative
- DBS
Dried blood spots
References
- 1.United States Department of Health and Human Services. United States Cancer Statistics - Incidence: 1999-2019. 2022. Accessed 5/27, 2023.
- 2.Lupo PJ, Spector LG. Cancer Progress and Priorities: Childhood Cancer. Cancer Epidemiol Biomarkers Prev. 2020;29(6):1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62(4):969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musselman JR, Bergemann TL, Ross JA, et al. Case-parent analysis of variation in pubertal hormone genes and pediatric osteosarcoma: a Children’s Oncology Group (COG) study. Int J Mol Epidemiol Genet. 2012;3(4):286–293. [PMC free article] [PubMed] [Google Scholar]
- 5.Mirabello L, Koster R, Moriarity BS, et al. A Genome-Wide Scan Identifies Variants in NFIB Associated with Metastasis in Patients with Osteosarcoma. Cancer Discov. 2015;5(9):920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koster R, Panagiotou OA, Wheeler WA, et al. Genome-wide association study identifies the GLDC/IL33 locus associated with survival of osteosarcoma patients. Int J Cancer. 2018;142(8):1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirabello L, Zhu B, Koster R, et al. Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma. JAMA Oncol. 2020;6(5):724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diessner BJ, Pankratz N, Hooten AJ, et al. Nearly Half of TP53 Germline Variants Predicted To Be Pathogenic in Patients With Osteosarcoma Are De Novo: A Report From the Children’s Oncology Group. JCO Precis Oncol. 2020;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazul AL, Siega-Riz AM, Weinberg CR, et al. A family-based study of gene variants and maternal folate and choline in neuroblastoma: a report from the Children’s Oncology Group. Cancer Causes Control. 2016;27(10):1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazul AL, Weinberg CR, Engel SM, et al. Neuroblastoma in relation to joint effects of vitamin A and maternal and offspring variants in vitamin A-related genes: A report of the Children’s Oncology Group. Cancer Epidemiol. 2019;61:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egolf LE, Vaksman Z, Lopez G, et al. Germline 16p11.2 Microdeletion Predisposes to Neuroblastoma. Am J Hum Genet. 2019;105(3):658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poynter JN, Richardson M, Roesler M, Krailo M, Amatruda JF, Frazier AL. Family history of cancer in children and adolescents with germ cell tumours: a report from the Children’s Oncology Group. Br J Cancer. 2018;118(1):121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcotte EL, Pankratz N, Amatruda JF, et al. Variants in BAK1, SPRY4, and GAB2 are associated with pediatric germ cell tumors: A report from the children’s oncology group. Genes Chromosomes Cancer. 2017;56(7):548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams LA, Pankratz N, Lane J, et al. Klinefelter syndrome in males with germ cell tumors: A report from the Children’s Oncology Group. Cancer. 2018;124(19):3900–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cigan SS, Meredith JJ, Kelley AC, et al. Predicted leukocyte telomere length and risk of germ cell tumours. Br J Cancer. 2022;127(2):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams LA, Mills L, Hooten AJ, et al. Differences in DNA methylation profiles by histologic subtype of paediatric germ cell tumours: a report from the Children’s Oncology Group. Br J Cancer. 2018;119(7):864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Pierce JL, Sanchez A, et al. Integrated genomic analysis reveals aberrations in WNT signaling in germ cell tumors of childhood and adolescence. Nat Commun. 2023;14(1):2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lone DW, Sadak KT, Miller BS, et al. Growth Hormone Deficiency in Childhood Intracranial Germ Cell Tumor Survivors. J Endocrinol Metab. 2022;12(3):79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillani R, Camp SY, Han S, et al. Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes. Am J Hum Genet. 2022;109(6):1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Sisoudiya SD, Martin-Giacalone BA, et al. Germline Cancer Predisposition Variants in Pediatric Rhabdomyosarcoma: A Report From the Children’s Oncology Group. J Natl Cancer Inst. 2021;113(7):875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fair D, Maese L, Chi YY, et al. TP53 germline pathogenic variant frequency in anaplastic rhabdomyosarcoma: A Children’s Oncology Group report. Pediatr Blood Cancer. 2023:e30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Giacalone BA, Richard MA, Scheurer ME, et al. Germline Genetic Variants and Pediatric Rhabdomyosarcoma Outcomes: A Report from the Children’s Oncology Group. J Natl Cancer Inst. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peckham-Gregory EC, Chakraborty R, Scheurer ME, et al. A genome-wide association study of LCH identifies a variant in SMAD6 associated with susceptibility. Blood. 2017;130(20):2229–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcotte EL, Domingues AM, Sample JM, Richardson MR, Spector LG. Racial and ethnic disparities in pediatric cancer incidence among children and young adults in the United States by single year of age. Cancer. 2021;127(19):3651–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Brown DS, Cao Y, Ekenga CC, Guo S, Johnson KJ. The impact of health insurance coverage on racial/ethnic disparities in US childhood and adolescent cancer stage at diagnosis. Cancer. 2022;128(17):3196–3203. [DOI] [PubMed] [Google Scholar]
- 26.Marcotte EL, Spector LG, Mendes-de-Almeida DP, Nelson HH. The Prenatal Origin of Childhood Leukemia: Potential Applications for Epidemiology and Newborn Screening. Front Pediatr. 2021;9:639479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hossain MJ, Xie L, Caywood EH. Prognostic factors of childhood and adolescent acute myeloid leukemia (AML) survival: evidence from four decades of US population data. Cancer Epidemiol. 2015;39(5):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grubb WR, Neboori HJ, Diaz AD, Li H, Kwon D, Panoff J. Racial and Ethnic Disparities in the Pediatric Hodgkin Lymphoma Population. Pediatr Blood Cancer. 2016;63(3):428–435. [DOI] [PubMed] [Google Scholar]
- 29.Austin MT, Hamilton E, Zebda D, et al. Health disparities and impact on outcomes in children with primary central nervous system solid tumors. J Neurosurg Pediatr. 2016;18(5):585–593. [DOI] [PubMed] [Google Scholar]
- 30.Henderson TO, Bhatia S, Pinto N, et al. Racial and ethnic disparities in risk and survival in children with neuroblastoma: a Children’s Oncology Group study. J Clin Oncol. 2011;29(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pui CH, Pei D, Pappo AS, et al. Treatment outcomes in black and white children with cancer: results from the SEER database and St Jude Children’s Research Hospital, 1992 through 2007. J Clin Oncol. 2012;30(16):2005–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz-Bose R, Zheng DJ, Umaretiya PJ, et al. Feasibility of oncology clinical trial-embedded evaluation of social determinants of health. Pediatr Blood Cancer. 2022;69(11):e29933. [DOI] [PubMed] [Google Scholar]
- 33.Linabery AM, Slater ME, Spector LG, et al. Feasibility of neonatal dried blood spot retrieval amid evolving state policies (2009–2010): a Children’s Oncology Group study. Paediatr Perinat Epidemiol. 2011;25(6):549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupo PJ, Petrick LM, Hoang TT, et al. Using primary teeth and archived dried spots for exposomic studies in children: Exploring new paths in the environmental epidemiology of pediatric cancer. Bioessays. 2021;43(9):e2100030. [DOI] [PMC free article] [PubMed] [Google Scholar]


