Abstract
Clarithromycin-susceptible and clarithromycin-resistant Helicobacter pylori isolates from the same patient were investigated for the mode of development and mechanism of clarithromycin resistance. The clarithromycin-resistant strain UA1182 harbors homozygous A-to-G mutations at position 2143 in both copies of the 23S rRNA gene and has a phenotype of resistance to clarithromycin and clindamycin but no significant resistance to streptogramin B. Pulsed-field gel electrophoresis patterns of NruI- and NotI-digested genomic DNA from the Clas and Clar isolates demonstrated that they are genetically distinct, suggesting that the development of clarithromycin resistance is not from the mutation of the existing Clas strain but from a completely new strain.
To eradicate Helicobacter pylori, a human gastric pathogen, antibacterial treatment including a proton pump inhibitor (e.g., omeprazole) in association with other antibiotics, such as clarithromycin, metronidazole, or amoxicillin, is commonly recommended (11, 18). In recent years, many cases in which clarithromycin resistance developed after treatment with this macrolide antibiotic (clarithromycin) were reported (2, 6). Genetic studies have revealed that clarithromycin resistance can most often be attributed to A-to-G transition mutations at either position 2142 or 2143 of 23S rRNA genes (5, 13, 14, 16, 19, 20).
The prevalence of clarithromycin-resistant H. pylori varies with geographic location (6). In Alberta, Canada, only a single clarithromycin-resistant H. pylori strain (UA1182) has so far been isolated in this lab (in 1993) from an adult patient (15). Several years earlier (in 1990), another H. pylori strain (UA799) that was shown to be susceptible to clarithromycin was cultured from a gastric biopsy specimen that was obtained from the same patient. At that time, the patient had symptoms of bloating, epigastric pain, and nausea and was diagnosed as having gastritis and a duodenal ulcer. After initial diagnosis, the patient was treated with ranitidine, bismuth, and metronidazole. As the clinical consequence of the drug treatment, the ulcer disappeared and other symptoms, including nonulcer dyspepsia, remained but were less severe. The patient then traveled in Saudi Arabia on a regular basis before the isolation of the second strain, UA1182.
To examine the genetic basis of clarithromycin resistance, the nucleotide sequence within the peptidyltransferase-encoding region of the 23S rRNA gene from both strains was determined. Briefly, a 300-bp-long PCR fragment was amplified from the chromosomal DNA by using primers DP1 and ZGE23 (16), and the fragment was then sequenced with primer DP1. The DNA sequence in this region of strain UA799 is identical to that reported for the clarithromycin-susceptible wild-type strain, UA802 (16). In strain UA1182, there is an A-to-G transition mutation at position 2143 of its 23S rRNA gene sequence (Table 1), and the mutation occurs in both copies of the gene. This type of mutation was reported in many cases to be associated with clarithromycin resistance (5, 13, 16, 19–21). In addition, a T-to-C mutation at position 2182 was revealed in the 23S rRNA gene sequence of strain UA1182. However, in vitro site-directed mutagenesis experiments suggested that this additional mutation is not associated with clarithromycin resistance (data not shown).
TABLE 1.
MLSa phenotypes and 23S rRNA genotypes of H. pylori UA799 and UA1182
| H. pylori strain | MICb (μg/ml) of:
|
23S rRNA genotype | ||
|---|---|---|---|---|
| Clarithromycin | Clindamycin | Quinupristin | ||
| UA799 | 0.01 | 32 | 4 | Wild type |
| UA1182 | 4–8 | 256 | 4 | A2143G, T2182C |
MLS, macrolide-lincosamide-streptogramin B.
MICs were determined by the agar dilution method.
Macrolide resistance due to mutation in the peptidyltransferase-encoding region of the 23S rRNA is often associated with cross-resistance to lincosamide and streptogramin B antibiotics (macrolide-lincosamide-streptogramin B phenotype) (4). We tested the MICs of three representative antibiotics, clarithromycin, clindamycin, and quinupristin, for both strains by the agar dilution method (Table 1). UA1182 was shown to be resistant to clarithromycin and clindamycin but not significantly resistant to quinupristin (the MIC was identical to that of reference strains UA802 and UA799).
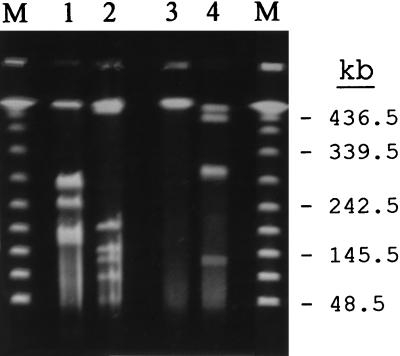
To investigate whether UA1182 developed as a result of a point mutation in strain UA799 after drug treatment, the overall genotypic characteristics of both strains were analyzed by pulsed-field gel electrophoresis (PFGE) as previously described (3, 9). Briefly, H. pylori was grown for 48 h, and the cells were then embedded in low-melting-point agarose blocks and lysed by treatment with N-lauroylsarcosine and proteinase K. Subsequently, chromosomal DNA was digested with restriction endonuclease NruI or NotI and then separated in a 1% agarose gel by using a contour-clamped homogenous electric field system (CHEF-DR-II; Bio-Rad). PFGE patterns of each strain (Fig. 1) showed that they were significantly different from one another, suggesting that the two strains were genetically unrelated. Therefore, clarithromycin-resistant UA1182 did not develop from the clarithromycin-susceptible strain UA799.
FIG. 1.
PFGE patterns of NruI (lanes 1 and 3)- and NotI (lanes 2 and 4)-digested genomic DNA from H. pylori UA799 (lanes 1 and 2) and UA1182 (lanes 3 and 4). The sizes of lambda DNA ladder standards (lanes M) are indicated on the right.
To explain the occurrence of UA1182, the following two possibilities could be considered. (i) UA1182 may have existed at the time of the first isolation in a mixed infection with UA799, but in a minor fraction or at a different site in the gastric mucosa so that it escaped identification in the first isolation. It has been documented that some patients can be concomitantly colonized by multiple H. pylori strains, even though this is rare (7, 17). (ii) Since the patient had traveled to Saudi Arabia, where a high proportion of the population is infected with H. pylori (1, 10), between the times of the two isolations, we could also speculate that the second strain may have been acquired as a new infection. Although the mode of transmission of H. pylori remains uncertain and the acquisition of infection in adults is rare, some studies have suggested a continuous risk of acquisition in adults, especially when the person has been exposed to an environment with a high incidence rate of H. pylori infection (8, 12). In conclusion, the data presented suggest that a clarithromycin-resistant H. pylori strain found in a patient may not necessarily be derived from a mutation of an existing strain identified in that patient; it may be a different strain, one which is involved in a multiple infection, or it may result from an entirely new infection.
Acknowledgments
This work was supported in part by funding from the Canadian Bacterial Diseases Network (Centers of Excellence Program) to D.E.T., who is a Medical Scientist with the Alberta Heritage Foundation for Medical Research (AHFMR), and by a Postdoctoral Fellowship from the Canadian Association of Gastroenterology and Astra Canada in association with an MRC-PMAC award to G.W., who also held a fellowship from AHFMR.
We thank S. Salama and R. N. Fedorak for helpful discussions.
REFERENCES
- 1.Al-Moagel M A, Evans D G, Abdulghani M E, Adam E, Evans D J, Jr, Malaty H M, Graham D Y. Prevalence of Helicobacter (formerly Campylobacter) pylori infection in Saudi Arabia, and comparison of those with and without upper gastrointestinal symptoms. Am J Gastroenterol. 1990;85:944–948. [PubMed] [Google Scholar]
- 2.Cayla, R., F. Zerbib, P. Talbi, F. Megraud, and H. Lamouliatte. 1995. Pre- and post-treatment clarithromycin resistance of Helicobacter pylori strains: a key factor of treatment failure. Gut 37(Suppl. 1):A55.
- 3.Chang N, Jiang Q, Taylor D E. Construction of a genetic map of H. pylori by pulsed-field gel electrophoresis (PFGE) In: Clayton C L, Mobley H L T, editors. Methods in molecular medicine, Helicobacter pylori protocols. Totowa, N.J: Humana Press Inc.; 1997. pp. 165–176. [DOI] [PubMed] [Google Scholar]
- 4.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dalberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 5.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M J E. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 6.Goddard A F, Logan R P H. Antimicrobial resistance and Helicobacter pylori. J Antimicrob Chemother. 1996;37:639–643. doi: 10.1093/jac/37.4.639. [DOI] [PubMed] [Google Scholar]
- 7.Hirschl A M, Richter M, Makristathis A, Pruckl P M, Willinger B, Schutze K, Rotter M L. Single and multiple strain colonization in patients with Helicobacter pylori-associated gastritis: detection by macrorestriction DNA analysis. J Infect Dis. 1994;170:473–475. doi: 10.1093/infdis/170.2.473. [DOI] [PubMed] [Google Scholar]
- 8.Hyams K C, Taylor D N, Gray G C, Knowles J B, Hawkins R, Malone J D. The risk of Helicobacter pylori infection among U.S. military personnel deployed outside the United States. Am J Trop Med Hyg. 1995;52:109–112. doi: 10.4269/ajtmh.1995.52.109. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 10.Parsonnet, J. 1995. The incidence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 9(Suppl. 2):45–51. [PubMed]
- 11.Rene W M, Hulst V D, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 12.Sander J O, Zanten V, Pollak P T, Best L M, Bezanson G S, Marrie T. Increasing prevalence of Helicobacter pylori infection with age: continuous risk of infection in adults rather than cohort effect. J Infect Dis. 1994;169:434–437. doi: 10.1093/infdis/169.2.434. [DOI] [PubMed] [Google Scholar]
- 13.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Ghoneim A T, Tanaka K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone G G, Shortridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 15.Taylor D E, Jiang Q, Fedorak R N. Antibiotic susceptibilities of Helicobacter pylori strains isolated in the province of Alberta, Canada. Can J Gastroenterol. 1998;12:295–298. doi: 10.1155/1998/672746. [DOI] [PubMed] [Google Scholar]
- 16.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of the two copies of 23S rRNA genes from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaira D, Holton J, Miglioli M, Menegatti M, Mule P, Barbara L. Peptic ulcer disease and Helicobacter pylori infection. Curr Opin Gastroenterol. 1994;10:98–104. [Google Scholar]
- 19.Versalovic J, Shortridge D, Kibler K, Griffy M V, Bryer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Versalovic J, Osato M S, Spakovsky K, Dore M P, Reddy R, Stone G G, Shortridge D, Flamm R K, Tanaka S K, Graham D Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 21.Wang, G., and D. E. Taylor. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob. Agents Chemother. 42:1952–1958. [DOI] [PMC free article] [PubMed]