Abstract
In this paper, we developed a highly sensitive approach to detect interchromosomal rearrangements in cattle by searching for abnormal linkage disequilibrium patterns between markers located on different chromosomes in large paternal half-sib families genotyped as part of routine genomic evaluations. We screened 5571 families of artificial insemination sires from 15 breeds and revealed 13 putative interchromosomal rearrangements, 12 of which were validated by cytogenetic analysis and long-read sequencing. These consisted of one Robertsonian fusion, 10 reciprocal translocations, and the first case of insertional translocation reported in cattle. Taking advantage of the wealth of data available in cattle, we performed a series of complementary analyses to define the exact nature of these rearrangements, investigate their origins, and search for factors that may have favored their occurrence. We also evaluated the risks to the livestock industry and showed significant negative effects on several traits in the sires and in their balanced or aneuploid progeny compared with wild-type controls. Thus, we present the most comprehensive and thorough screen for interchromosomal rearrangements compatible with normal spermatogenesis in livestock species. This approach is readily applicable to any population that benefits from large genotype data sets, and will have direct applications in animal breeding. Finally, it also offers interesting prospects for basic research by allowing the detection of smaller and rarer types of chromosomal rearrangements than GTG banding, which are interesting models for studying gene regulation and the organization of genome structure.
Interchromosomal rearrangements (IRs) are a variable group of cytogenetic abnormalities characterized by the transfer of genetic material between two nonhomologous chromosomes. These types of mutations can occur at different stages of cell life (Weckselblatt and Rudd 2015). Robertsonian fusions are known to occur mostly during mitosis (Matveevsky et al. 2020), whereas complex rearrangements would be the result of defective meiosis (Burssed et al. 2022). Simple centromeric fusions are generally better tolerated than more complex structural variations, the latter being a common cause of reproductive problems and other pathogenic effects owing to meiotic defects and abnormal gene dosage or regulation (Popescu 1989; Scherer et al. 2007; Poot and Haaf 2015). To illustrate this fact, in humans, cytogenetic abnormalities are detected in ∼0.5% of newborns (Forabosco et al. 2009; Vasilevska et al. 2013), but their frequency increases to 5%–15% in infertile couples (Gekas et al. 2001; Clementini et al. 2005; Poornima et al. 2020), ∼25% in miscarriages and stillbirths, and up to 50%–60% in first-trimester miscarriages (Shaffer and Lupski 2000; Yatsenko and Rajkovic 2019).
Despite their potential impact on productivity, animal welfare, and economics, chromosomal aberrations in livestock species have received much less attention than in humans. In cattle, for example, a recent literature review counted only 81 IRs (Iannuzzi et al. 2021), whereas hundreds of thousands would be expected given a global bovine population of 1.5 billion heads at the end of 2022 (https://www.fao.org/faostat/en/#data/QCL, last accessed October 27, 2022) and an estimated prevalence of at least 0.14% for reciprocal translocations alone (De Lorenzi et al. 2012). Several reasons may explain this situation. First of all, because cattle breeds are characterized by high inbreeding, the attention of clinical geneticists has been focused primarily on recessive defects over the past decades at the expense of other pathologies (Bourneuf et al. 2017). Moreover, there is no systematic cytogenetic analysis of reproducers, except in a few breeds segregating for an ancestral Robertsonian (Rob(1;29)) translocation, which is easily identifiable without the use of GTG banding (Ducos et al. 2000a). This means that other chromosomal aberrations, such as reciprocal translocations, may be missed. In addition, certain breeding practices have acted as a filter to limit the prevalence of IR. Indeed, in selected populations, most of the gene pool is transmitted to the next generation through artificial insemination (AI) with semen from elite sires that have been declared normozoospermic after drastic control of sperm concentration, morphology, and motility (Kastelic and Thundathil 2008). Furthermore, these sires were historically subject to progeny testing in order to estimate their genetic merit. They were made to produce about 100 offspring whose performance, including female fertility, was evaluated for various traits before deciding if it was worth to inseminate tens to hundreds of thousands of females with their semen. At this step, the reproductive careers of males with low conception rates or repeated instances of congenital malformations, presumably owing to genetic defects, ended prematurely and their progeny excluded from the breeding programs.
From 2010, the implementation of genomic evaluation in livestock production, which consists in predicting the genetic value of each animal based on its genotype at thousands of genetic markers, has opened new perspectives. This technological revolution has led to the wide dissemination of semen from elite sires without prior progeny testing and, above all, to the accumulation of large data sets of SNP array genotypes, whole-genome sequences, and phenotypes that can be used to address a wide variety of scientific questions (Bourneuf et al. 2017; Zhao et al. 2022; Besnard et al. 2023).
In this article, we propose a population-wide method to detect IR carriers among 5571 normozoospermic bulls by leveraging the routine genotyping of their offspring for genomic evaluation and exploiting the assumed absence of linkage disequilibrium (LD) between markers from different chromosomes. In addition, we perform cytogenetic validation, long-read sequencing, and a series of complementary analyses to define the exact nature of the rearrangements identified, investigate their origins, and describe their clinical consequences.
Altogether, we present the most comprehensive and thorough screen for IRs compatible with normal spermatogenesis in livestock species.
Results
Estimation of LD significance thresholds by simulation
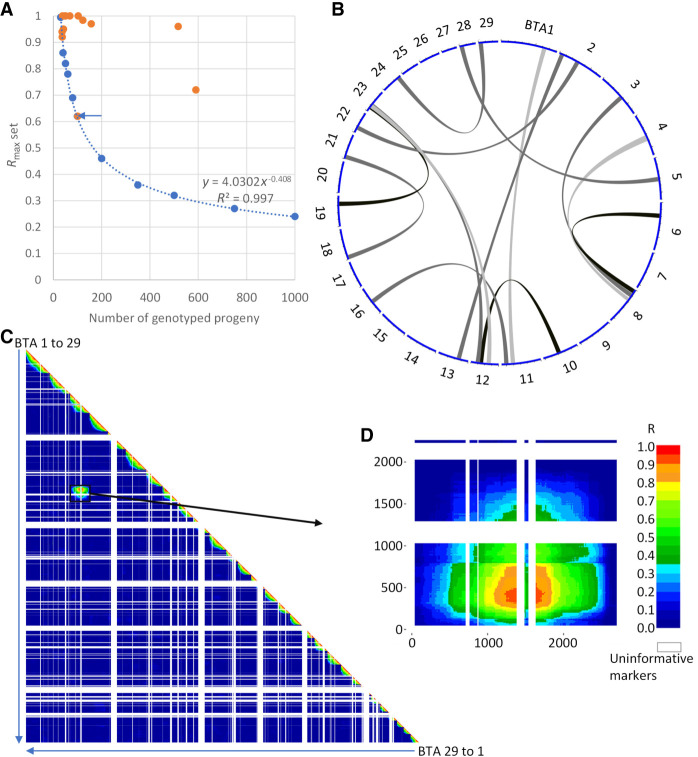
As a first step, we conducted a series of simulations using progeny genotypes of 100 bulls considered free of chromosomal abnormalities in order to determine the significance threshold (P ≤ 0.001) of LD between markers from different chromosomes according to the size of the half-sib families (see Methods) (Fig. 1A). Obviously, these thresholds were higher when the number of offspring was lower and the minimum size of the sire families was set to more than 30 half-siblings.
Figure 1.
A novel approach to detect interchromosomal rearrangements (IRs) using SNP array genotypes in large half-sib families. (A) Regression curve of linkage disequilibrium (LD) significance thresholds (P = 0.001) obtained from series of 10,000 simulations. Blue dots indicate the results of simulations with 10 progeny group sizes ranging from 30 to 1000. Orange dots point to the maximal significant LD observed for 13 paternal half-sib families. (B) Circos plot (Yu et al. 2018; yimingyu.shinyapps.io/shinycircos/) representing the 13 rearrangements found. (C) Whole-genome LD map for a bull assumed to have an IR between BTA3 and BTA8. (D) Detail of the LD map between markers from these two chromosomes.
Large-scale screening for IRs
In the next step, we analyzed 5571 paternal half-sib families from 15 different breeds and found significant LD patterns for 13 of them (2.33‰) (Fig. 1; Supplemental Figs. S1A–S13A). They consisted of nine Holstein, two Charolais, one Normande, and one Abondance pedigrees accounting for 1.56‰–7.57‰ of the bulls tested within their breeds. Overall, there was no significant difference in the proportions of putative IR carriers between the breeds studied.
All rearrangements detected were unique in terms of the chromosomes and regions involved. Several chromosomes were overrepresented, with three occurrences for Chromosomes BTA12 and BTA23 (including two putative t(12;23) rearrangements) and two for BTA2, BTA8, and BTA11 (Fig. 1B). However, observing two or three times the same chromosome and even two times the same chromosome association out of 13 IRs was not unexpected by chance based on 100,000 series of random sampling with replacement (P > 0.05). In 11 cases, the maximum LD (R2) was >0.9, whereas it only reached 0.72 and 0.61 for the bulls Nt and Ra, respectively (Fig. 1A).
Cytogenetic confirmation
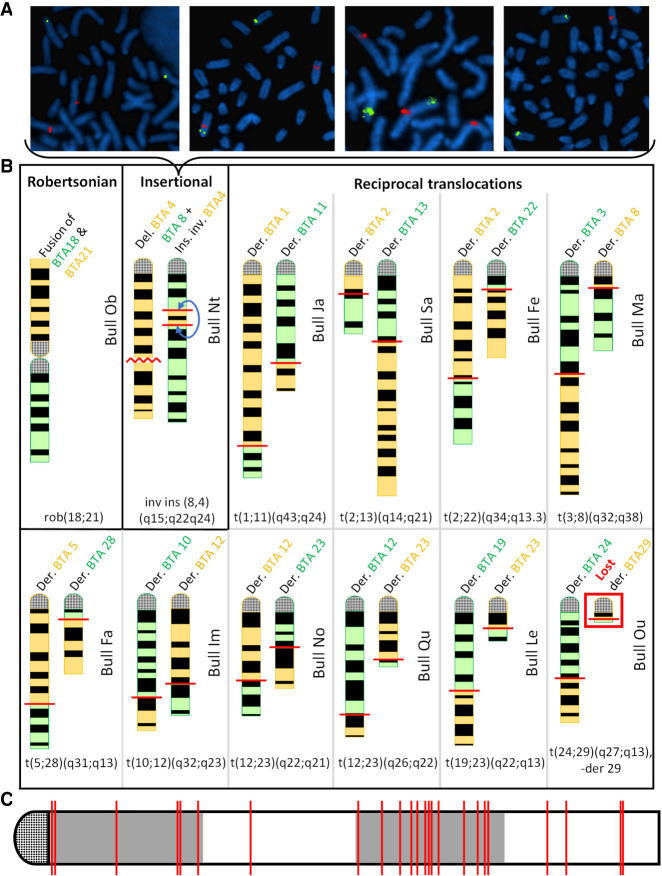
For verification, we conducted cytogenetic analyses either directly on the sires, when they were still alive (applicable for one bull), or on a selection of their progeny representing each of the genotype combinations observed around putative chromosomal fusion points (sons and daughters for 10 bulls, granddaughters for two bulls). Analysis of blood lymphocytes in metaphase confirmed 12 of the 13 suspected rearrangements (Fig. 2B; Supplemental Figs. S1B–S13B, S1C–S13C, S1D–S13D). Thus, the validated prevalence of IR in our cohort of 5571 normozoospermic AI bulls was 2.15‰.
Figure 2.
Validation of 12 IRs by cytogenetic analyses. (A) FISH mapping on cultured fibroblasts from four daughters of Nt with BAC clones located in the translocated segment from BTA4 (labeled in red) and in the centromeric region of BTA8 (labeled in green). From left to right, these animals show a normal karyotype, a balanced rearrangement, a partial BTA4 trisomy, and a partial BTA4 monosomy. (B) Schematic representation of the rearranged chromosomes based on GTG-banding karyotypes. Chromosomes with a loss or gain of material are shown in yellow and green, respectively. The schematics of the original chromosomes are available in Supplemental Figures S1D–S13D. (C) Location of the breakpoints on a theoretical chromosome. Coordinates are expressed as a percentage of chromosome length. Note the presence of 13 out of 24 breakpoints in the third quarter, whereas only six would be expected by chance.
No karyotypic abnormality was observed among four daughters of the 13th bull, Ra, whereas the size of the segments putatively involved in the rearrangement exceeded the resolution limit of GTG banding (5–10 Mb). Note that this bull had the lowest significant LD value of the 13 putative IRs carriers (point with blue arrow in Fig. 1A). For two validated rearrangements, complementary fluorescence in situ hybridization (FISH) analyses were performed to increase the resolution. In one bull (Nt), a small segment of BTA4 inserted in BTA8 was barely visible. Using two probes targeting this BTA4 segment and the centromere of BTA8, we further confirmed the existence of this rearrangement and documented the presence of live monosomic and trisomic animals among its progeny (Fig. 2A; Supplemental Fig. S7J). For another bull (Ou), we suspected a reciprocal translocation between BTA24 and BTA29 and a loss of the derivative Chromosome 29. Marking the centromeres of all chromosomes confirmed our assumption (Supplemental Fig. S10J).
At the end of the cytogenetic analysis, we confirmed 12 IRs, consisting of one Robertsonian translocation, one insertional translocation, and 10 reciprocal translocations, including one with derivative loss. By analyzing the location of the breakpoints in more detail, we found that they were more frequently located in the third-quarter of the chromosomes than expected by chance (P = 0.04; chi-square) (Fig. 2C).
Determining the origin and possible causes of rearrangement events
We then sought to determine in which animal and at what stage of development these mutations occurred. To achieve this goal, we set up haplotype tests to trace the segments involved in the rearrangements through generations, taking into account various sources of information such as the parental origin of these segments, the co-occurrence (or not) of the two segments in the same ancestor, and the results of our initial screen for these ancestors. Indeed, the sires and maternal grandsires of the 12 IR carriers and, in some cases, more distant male ancestors were included in this analysis and were predicted to have normal karyotypes (for further details of the available data and the investigations performed within each pedigree, see Supplemental Figs. S1F–S13F). We determined that the Robertsonian translocation was probably the result of a chromosomal fusion at the zygote stage in the maternal granddam of the sire Ob. Ou's reciprocal translocation occurred during female meiosis, whereas the nine other reciprocal translocations and the insertional translocation were the result of abnormal male meiosis (Table 1; Supplemental Figs. S1F–S13F). Nine of the carriers of reciprocal translocations belonged to the first generation of mutants, whereas the last one was a second-generation mutant that inherited the two rearranged chromosomes from its carrier dam. Finally, it is worth noting that two carriers of distinct reciprocal translocations resulting from abnormal paternal meiosis (t(5;28) in Fa and t(2;22) in Fe) shared the same sire, which was tested as wild type along with 46 other of its sons in our initial screen. With a prevalence of 41.67‰ (2/48) of IR in its progeny versus 2.15‰ in the total cohort, this elite AI sire was a clear outlier.
Table 1.
Origins of interchromosomal rearrangements (IRs)
Given the high proportion of IR caused by abnormal male meiosis (n = 10/12) and the general trend to collect semen of ever younger bulls, we compared the age of the sires at the conception of their mutant progeny with the weighted mean age at the conception of their 118 wild-type progeny included in our initial screen. We found no significant difference (3.33 ± 2.14 yr vs. 3.42 ± 2.24 yr, respectively; Student's t-test P-value = 0.93), indicating that the age of the sire at conception was not a factor influencing the risk of producing IR-affected progeny.
Characterization of the pathological consequences of IRs
To gain insight into the pathological consequences of these cytogenetic abnormalities, we then mined the French national database containing information on various aspects of the life of cows (such as their date of birth, death, insemination, and calving). The bulls used for this analysis had more than 30 progeny with available genotypes and were therefore all tested for the presence of IR.
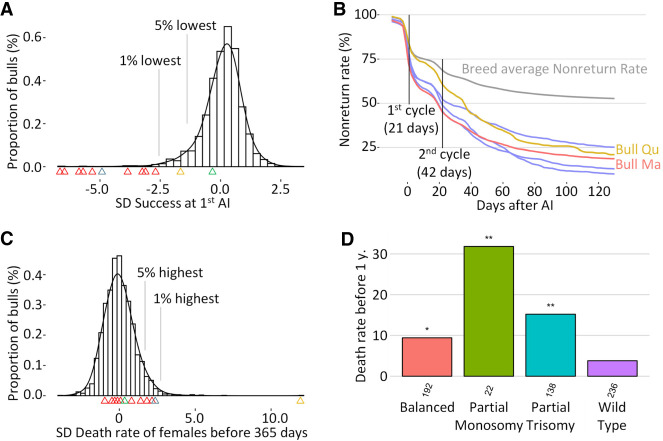
First, we calculated conception rates (i.e., the proportion of inseminations that resulted in the birth of a calf) for 2900 bulls of the Holstein, Normande, Charolaise, and Abondance breeds, including the 12 IR carriers (Fig. 3A). Carriers of reciprocal translocations were all in the lowest percentile of their breed for this trait, and together, they explained 35% of the normozoospermic bulls affected by idiopathic subfertility (i.e., in the lowest 1% of their breed for conception rate).
Figure 3.
Analysis of conception rate, nonreturn rate (NRR), and juvenile mortality. (A) Conception rate of first artificial inseminations (1st AI) of 2900 bulls with nulliparous females, conventional semen, expressed as SD from the mean to allow comparison between bulls from different breeds. Arrowheads indicate carriers of the following IRs: red for reciprocal translocations, yellow for insertional translocation, gray for reciprocal with derivative loss, and green for Robertsonian translocation. (B) NRR for five affected Holstein bulls (i.e., the proportion of females that were not reinseminated after an insemination with the semen of the bull in question) and mean NRR of 2152 Holstein bulls. Two extreme bulls, Qu and Ma, are highlighted in red and yellow. (C) Mortality rates over the first year of life among the daughters of 2854 bulls considered, expressed as the SD from the mean of their respective breeds (same color code as in vignette A). (D) Distribution of mortality rates at 1 yr of age for Nt's progeny (n below bars). (*) Fisher comparison with wild-type group P < 0.05, (**) P < 0.01.
A detailed analysis of the nonreturn rates (NRRs; i.e., the proportion of females probably pregnant at a given time after the insemination, given that they have no new AI) revealed different patterns of embryonic loss for these subfertile IR-affected bulls (Fig. 3B; Supplemental Figs. S1E–S11E). For example, the bull Ma showed an early drop in NRR compatible with a loss of aneuploid conceptuses at the beginning of embryogenesis, whereas the loss for Qu was more progressive and extended over a period covering organogenesis and beyond.
We also calculated mortality rates between birth and 1 yr of age in the female offspring of 2854 bulls, including the same 12 IR carriers (Fig. 3C), and found that four IR carriers were in the worst 5% of their breed. The bull Nt, carrying a large insertional translocation, was the worst sire of all breeds, with as much as 44% of daughters dying of natural causes before the age of 1 yr. Following the observation of live aneuploid progeny for this bull during cytogenetic analyses (see FISH in Fig. 2A), we estimated the proportion of normal and abnormal karyotypes based on haplotype test in 588 of its descendants that had been genotyped for genomic evaluation. We found that 40% of them were wild type, 33% had a balanced karyotype like their sire, and only 23% and 4% showed partial BTA4 trisomy and monosomy, respectively. We also observed significantly higher mortality rates <1 yr of age in carriers of abnormal karyotypes than in wild-type daughters (Fig. 3D).
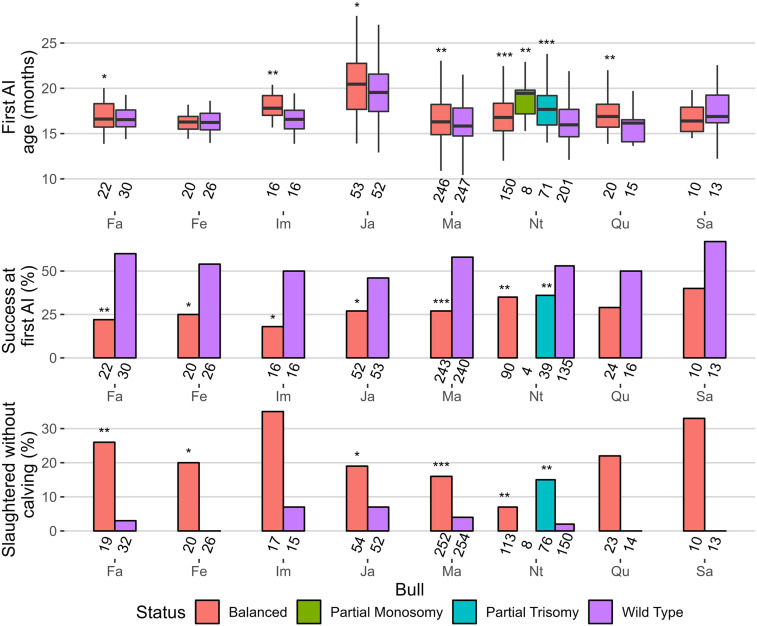
Finally, still based on haplotype tests, we analyzed the performance of the progeny of eight IR-affected dairy bulls that had sufficient data for three traits recorded in heifers (Fig. 4). For six of them, we observed a significant delay in age at first insemination, which is a proxy for growth, considering that dairy heifers are usually inseminated when they reach 60% of the expected adult weight. Field examination of the progeny of the bull Nt further confirmed this result (Supplemental Fig. S7K). Carriers of abnormal karyotypes also showed a significantly lower insemination success rate for six out of eight bulls studied. Consistent with the low performance observed for the previous two traits, we also observed a significant increase in the proportion of females slaughtered before starting a productive life among carriers of abnormal karyotypes versus half-sib controls for five bulls. Note that the progeny of bulls Ou and Ob were too young at the time of writing to reach significant numbers. We have not included Charolais phenotypes in this part because delaying AI is often a breeder's choice in this breed.
Figure 4.
Comparison of the performance of daughters with normal and abnormal karyotypes among the progeny of nine IR-affected bulls. Cohort sizes are given below each bar. Significance levels refer to comparisons with wild type groups. Student's t-test for first AI, Fisher's test for the other traits): (*) P < 0.1; (**) P < 0.05; (***) P < 0.01.
Long-read sequencing of seven IR carriers
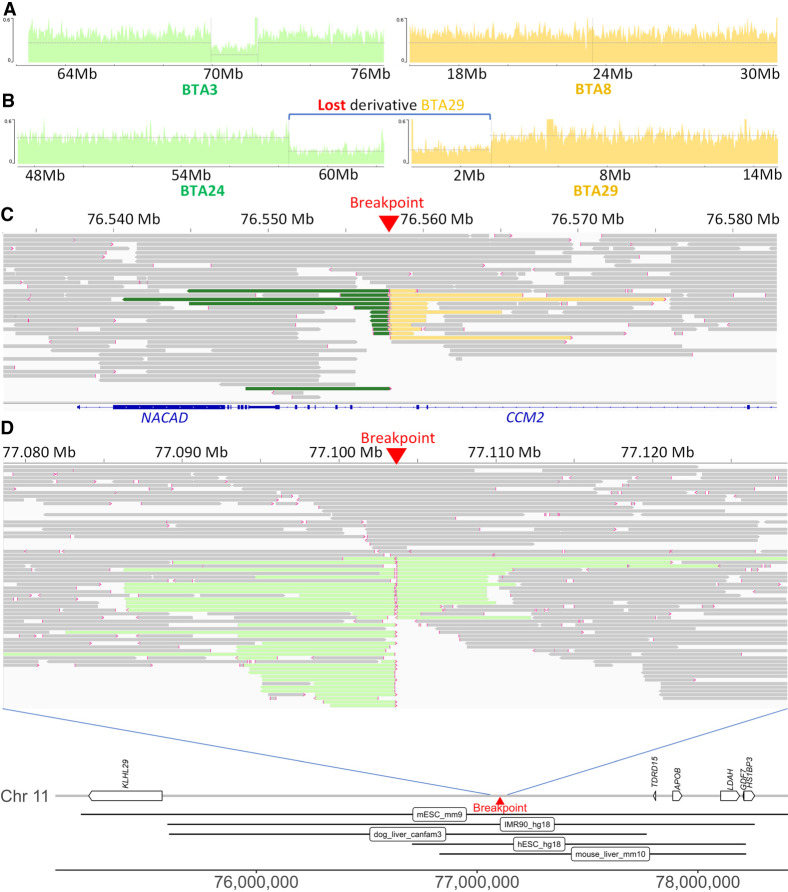
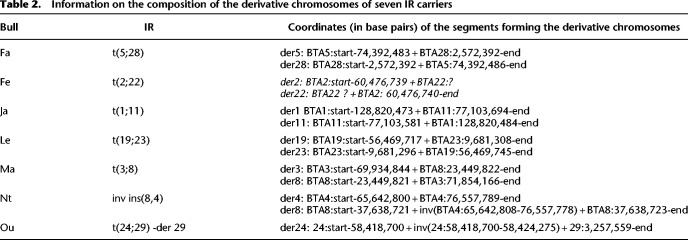
To characterize the fusion points of the rearrangements, we sequenced the genomes of seven affected bulls at 32× unique molecular yield (UMY) coverage using Pacific Biosciences’ (PacBio's) continuous long-read (CLR) technology (Fig. 5C,D; Supplemental Figs. S1G–S10G). These bulls were affected by an insertional translocation (Nt), a reciprocal translocation with derivative loss (Ou), and five different reciprocal translocations (Fe, Fa, Ja, Le, and Ma). In doing so, we were able to retrieve chimeric reads encompassing each of the fusion points, thus allowing determination of the coordinates of the segment forming the derivative chromosomes (Table 2) except for Fe (affected by a t(2;22)). For the latter, we observed a breakpoint on BTA2 around position 60,476,739 bp and chimeric reads containing up to 32 kb of BTSAT2 repeats on each side. We assumed that this material came from BTA22, but we were not able to confirm this hypothesis because of the highly repetitive nature of this component of the centromeres and because of the fact that centromeric regions are largely absent from the ARS-UCD1 assembly. We then investigated the exact nature of the fusion points and their possible consequences on the expression of neighboring genes. Information about breakpoint locations, neighboring candidate genes, and the possible consequences of their haploinsufficiency is presented in Tables 2 and 3. Annotated maps of chromosomal segments affected by the breakpoints are provided in panels H of Supplemental Figures S1–S10. For Ou, we confirmed the results of FISH experiments (Fig. 2B) supporting the absence of the derived Chromosome 29 and determined that it was partially monosomic for the last 3.9 Mb of BTA24 and the first 3.2 Mb of BTA29 (Fig. 5B; Supplemental Fig. S10I). The same bull presented an unexpected inversion of ∼6 kb at the BTA24 breakpoint level. We also observed a heterozygous deletion of 1.9 Mb at the breakpoint on BTA3 in Ma (t(3;8); Fig. 5A). The regions deleted in these two bulls contained 29 and five genes, respectively, including, for Ou, several whose haploinsufficiency has previously been implicated in various mouse and human pathologies (Table 3). Apart from these cases, the other breakpoints and fusion points consisted of deletions or insertions of only a few nucleotides. Three of them resulted in gene disruption, and all were predicted to affect a topologically associated domain (TAD) based on orthology comparison with human, mouse, and dog (Fig. 5E; Table 3; Supplemental Figs. S1H–S10H).
Figure 5.
Contribution of PacBio CLR sequencing to the characterization of IR break points and fusion points. (A) Sequence coverage plots around the breakpoints of t(3;8) supporting a heterozygous deletion of ∼2 Mb on BTA3 of bull Ma. (B) Sequence coverage plots around the breakpoints of t(24;29),-der29 supporting a loss of the derivative chromosome of bull Ou. For vignettes A and B, control alignments are provided in Supplemental Figures S6I and S10I. (C) Read alignment around the second breakpoint of inv ins(8,4) at 76.6 Mb on BTA4 (bull Nt). Split reads that also map to BTA8 and to BTA4 around position 65.9 Mb are colored in green and yellow, respectively. Note that the breakpoint resulted in the disruption of CCM2. (D, top) Read alignment around the breakpoint of t(1;11) on BTA11 with split read aligning to BTA1 colored in light green (bull Ja). (D, bottom) Details of the genes and putative TADs (according to Wang et al. 2018) located around the breakpoint of t(1;11) on BTA11.
Table 2.
Information on the composition of the derivative chromosomes of seven IR carriers
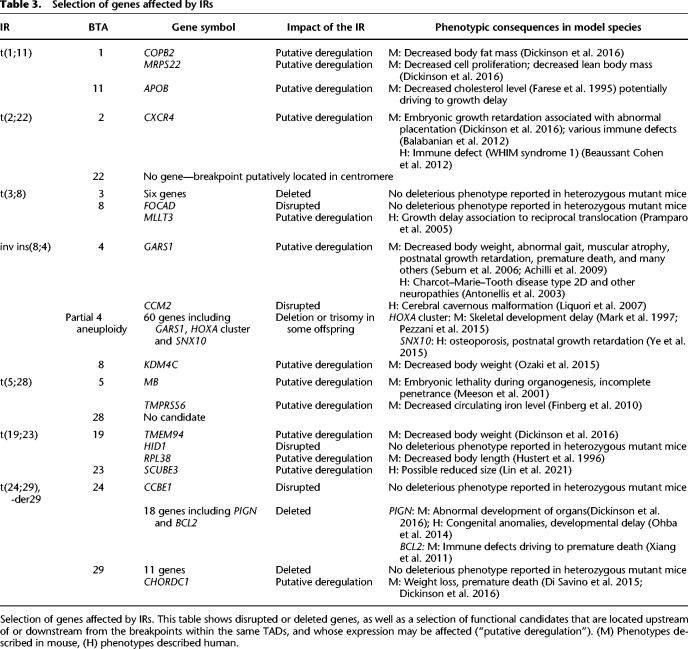
Table 3.
Selection of genes affected by IRs
In addition, for each of the seven bulls, we observed that one breakpoint was located in a repeated element, whereas the second was not. These repeats were of different types (Supplemental Table S4) and even differed between the two paternal half-brothers Fa and Fe.
As a final attempt to identify potential genetic factors promoting IR, we sought to determine whether the breakpoints were located within recombination hotspots. For five IRs resulting from abnormal male meiosis with precise breakpoint coordinates, we analyzed the LD between the closest informative SNPs on either side of the breakpoint among the genotyped descendants of the sires that produced the mutant spermatozoid. This LD was then ranked in terms of percentiles compared with the LD between all informative SNPs separated by a similar distance along the same chromosome and along all other chromosomes. We did not observe any ranking <5% and therefore concluded that none of the breakpoints were located in recombination hotspots (see Supplemental Table S3).
Discussion
On the power of our approach
Analyzing 5571 paternal half-sib families with 31 or more genotyped progeny, we highlighted 13 putative IR (P ≤ 0.001) using the approach developed in this study. Subsequent cytogenetic analyses revealed only one false positive, which is quite satisfactory, as five or six were expected given the sample size and the false-discovery rate chosen. Therefore, we estimated the prevalence of IR in our cohort of AI sires to be 2.15‰ (12/5571), which is similar to findings in normozoospermic males from other species. Indeed, 2.06‰ of 10,202 human sperm donors and 4.7‰ of 7700 AI boars were affected by IR according to Ravel et al. (2006) and Ducos et al. (2007), respectively.
Our strategy has led to significant advances in the detection of cytogenetic abnormalities in cattle, with the first report of an insertional translocation in this species (inv ins(8,4)(q15;q22q24)) and the identification of 10 new reciprocal translocations (rcp), whereas only 20 had been described previously (Iannuzzi et al. 2021). We also identified a new Robertsonian fusion (Rob(18;21)). These 12 events are unique. The proportions of the different types of IR highlighted in our study differ markedly from previous findings (0 inv ins, 20 rcp, and 61 Rob reviewed by Iannuzzi et al. 2021; P = 1.3 × 10−5 for Fisher's exact test for comparison of Rob vs. non-Rob translocations) but are consistent with the predictions of De Lorenzi et al. (2012), who estimated that reciprocal translocations would be at least five times more frequent than Robertsonian fusions in terms of de novo mutations events. This result reflects a major bias toward the detection of Robertsonian fusion in cattle, which is because of the lack of routine cytogenetic screening outside of a few breeds segregating for the Rob(1;29) translocation. Because Rob(1;29) and other submetacentric chromosomes formed by Robertsonian fusion are easily identifiable among the acrocentric bovine autosomes, they are tested by simple Giemsa staining, which hinders the detection of other types of translocations if they do not cause major changes in long arm length. To detect them, more accurate methods are needed, such as the GTG banding used for validation purpose in this study. However, even these are not infallible. In our experience, the insertion of an 11-Mb segment from BTA4 into BTA8 (inv ins(8,4)) would have been missed in a blind test because of its size, which is close to the resolution limit of banding (∼5–10 Mb). The approximate knowledge of the breakpoint provided by LD analysis and FISH was instrumental in showing the existence of this novel type of IR in cattle.
As designed, our strategy is sensitive enough to detect translocations involving segments of only a few kilobases (containing at least one marker), with at least 30 genotyped progeny under the best conditions. As explained in the Methods, we recorded for each family the highest LD value between markers from different chromosomes (absolute correlation between alleles, also called Rmax), and if the latter was significant (P < 0.001), we applied a filter to avoid spurious and punctual LD. We kept Rmax only if the two markers with the highest correlation on each chromosome were surrounded by at least 19 consecutive markers with R > Rmax * 0.75. This filter is very robust and flexible because the 19 neighboring informative markers on each chromosome can be located on one or both sides of the breakpoints and up to several megabases away, allowing detection of IRs even when they affect the telomeres or when they are in runs of homozygosity (RoHs) (Fig. 1C, white bands in the LD heatmap). In case of long RoHs, however, more than 30 progeny might be required to obtain a significant Rmax if the first informative marker is located >10 Mb from the breakpoint. These large RoHs classically represent >3% of genomes in inbred breeds (Marras et al. 2015).
Another factor that may also contribute to reduced power of detection is the genotyping of a small proportion of aneuploid conceptuses, although in a general manner most of them die during embryogenesis, and the survivors are less likely to be genotyped for genomic evaluation because of their poor appearance. In the case of the bull Nt, the genotyping of a small proportion of calves affected by partial BTA4 mono- or trisomy resulted in imputation and phasing errors, which explains why this sire had the lowest LD max value among confirmed IR carriers. The deletions of several megabases associated with t(24;29)-der 29 and t(3;8) also caused local phasing and imputation errors because of Mendelian transmission errors between hemizygous sires and hemizygous progeny. An example of this can be seen in the LD heat map between the BTA3 and BTA8 markers (Fig. 1D), where a region of low LD corresponding to the deletion site (green) is surrounded by two regions of high LD (red bars).
In any case, LD-based screening for IRs is more sensitive than GTG banding, and we anticipate that its routine use as a by-product of genomic evaluations will allow the discovery of even rarer and smaller chromosomal alterations in the future.
Effects on phenotypes and implications for the industry
Taking advantage of the wealth of information available in the French national bovine databases, we investigated the phenotypic effects of IRs on several traits in male carriers and their female offspring.
We observed strong negative effects of the insertional and reciprocal translocations on male fertility, in agreement with previous studies reporting that these types of IR form quadrivalents at meiosis, which after segregation produce relatively high proportions of unbalanced gametes and nonviable embryos. For example, Ogilvie and Scriven (2002) found only 47.7% (71/149) of human embryos with normal or balanced karyotypes in a preimplantation genetic diagnosis study of 18 couples carrying 15 different reciprocal translocations. The fact that the 10 reciprocal translocation carriers were in the worst percentile for the success at first insemination and that they represent 35% of this worst percentile in our study also agrees with the observations of Ducos et al. (2007) in pigs. The latter detected IR in about half of the boars referred to their cytogenetic platform by the breeding organization for hypoprolificacy during the period of 2002–2006. Therefore, our approach adds to the tools set up in the past decade to manage bovine fertility using genomic data (Capitan et al. 2015). In parallel, we recommend that breeding companies consider low NRR as a potential sign of IR and take the necessary steps to verify this hypothesis and stop marketing the semen of these bulls.
It should also be noted that the kinetics of embryo loss varied between bulls. For example, Ma showed an early decrease in NRR compatible with a loss of aneuploid conceptuses at the beginning of embryogenesis, whereas for Qu, the loss was more progressive and extended over a period covering organogenesis and beyond (see Fig. 3B). We assume that segment size and gene content influence the onset and the penetrance of embryonic lethality, as these two bulls carried one of the largest and smallest reciprocal translocations, respectively. Further supporting this hypothesis, the bull Nt, affected by a small insertional translocation of 11 Mb, was less infertile than carriers of reciprocal translocations, being only in the worst 5% for the success at first insemination. Conversely, it had the highest juvenile mortality rate in our data set, with 44% of females dying between birth and 365 d. We found low proportions of carriers of partial BTA4 trisomy (23%) and monosomy (4%) among its genotyped offspring, indicating that some of them were able to survive until sampling, which is usually performed between 2 and 6 mo of age for routine genomic evaluation. In addition, we showed that these animals had higher mortality rates compared with wild type and balanced controls (Fig. 3D). These results are in line with the recent report by Besnard et al. (2023) of high mortality rates and partial monosomy of the first 37 Mb of BTA29 in the offspring of a bull carrier of a t(26;29) reciprocal translocation, which had a slightly reduced conception rate. Taking all these elements into account, we assume that the higher mortality rates observed for three other IR carriers (in the worst 5%) (see Fig. 3C) are the result of a small proportion of aneuploid births that probably died at an early age or had a particularly poor appearance that prevented their genotyping.
Finally, the rob(18;21) carrier was within the normal range for both male fertility and juvenile mortality. Therefore, we assume that this bull produced only a low proportion of chromosomally unbalanced spermatozoa resulting from adjacent meiotic segregation, that is, possibly ∼≤5%, as previously reported, for example, for carriers of rob(1;29), rob(1;21), rob(7;21), and rob(3;16) (Tateno et al. 1994; Bonnet-Garnier et al. 2006; Barasc et al. 2019).
Regarding the consequences of IRs on female offspring, we found significant effects on at least one of the phenotypes studied (age at the first insemination, success at first insemination, and rate of slaughtered animals without calving), except for the bull Sa (t(2;11)), which had the lowest number of phenotyped progeny (n = 23). For the latter, we obtained suggestive but not significant effects owing to statistical power limitations. Conversely, the two bulls with the highest number of available phenotypes (Ma and Nt, 506 and 347 complete phenotypes, respectively) showed highly significant differences for all traits studied.
In six of the eight bulls studied, we found that balanced daughters had significantly delayed first inseminations compared with their wild-type sisters, thus supporting growth retardation. This effect, probably because of the disruption, deletion, or deregulation of genes located near the breakpoints (see section “Long-read sequencing and candidate genes”), was not expected as their balanced sires had gone through a selection process to become AI bulls. This calls into question the criteria used to select candidate bulls, which may be more focused on genomic indices and semen quality than on physical appearance. In contrast, the detrimental effects of IRs on the success at first insemination were expected. Indeed, female carriers are predicted to produce rates of unbalanced gametes as high as those of their own sires, resulting in the loss of aneuploid embryos. Our study also provides an overview of the technical consequences of infertility, with a significant increase in the culling of balanced and aneuploidy females before they calve and start their productive life. Of note, the decision to cull an unproductive animal is costly for farmers, as the sale of meat generally does not cover the rearing costs.
In this paper, we have not estimated the financial costs of IRs, but they seem to be colossal. Lewis et al. (2022) proposed an estimation of the cost to the industry. Their estimation reaches US $1.5 million per year per AI bull carrier, based on an average fertility reduction of 10 points and the use of 23,000 semen straws per bull per year (which only Ma and Nt reached in the 12 IRs). In this study, we found higher effects on fertility, with a reduction in conception rates of more than 20 points for most IR carriers (Fig. 3B), and highlighted other deleterious effects on subsequent generations, such as reduced fertility and growth and increased mortality of balanced and aneuploid offspring, which were not considered in the latter estimate. Therefore, the costs estimated by Lewis et al. (2022) are underestimated despite their high level. Finally, in addition to the economic aspects, we would also like to emphasize the negative effects of IRs on animal welfare, on the working conditions of the breeders, and the environmental footprint of livestock production.
In light of these elements, we strongly recommend the early and systematic karyotype screening of future insemination bulls using a GTG banding protocol. As a by-product of genomic selection, our approach provides an easy to implement and very sensitive tool to detect IRs that may be missed by cytogenetic analyses and, most importantly, to track carriers through the generations (e.g., down to great-granddaughters for Fe and Fa) (Supplemental Figs. S1F, S2F) for eradication purposes.
Long-read sequencing and candidate genes
To better characterize IRs, we sequenced the genomes of seven carrier bulls using long-read technology and were able to identify the exact breakpoint locations for six of them. In one case (t(2;22)), we identified the breakpoint on only one chromosome because of the presence of several kilobases of BTSAT2 satellite material, leading us to believe that the breakpoint was located in the centromere of the second chromosome. Thus, our success rate was similar to that of Bouwman et al. (2020), who found breakpoints in six out of seven boars affected by reciprocal translocations using 30× short-read paired-end sequencing. It is worth noting that the Bos taurus genome contains a higher proportion of repeated elements (49.4%) than that of Sus scrofa (44.5%; http://repeatmasker.org/genomicDatasets/RMGenomicDatasets.html). A posteriori analyses revealed that using short-read sequencing, we would have missed the breakpoint on BTA2 for t(2;22) and also another breakpoint located in repeated elements whose size exceeds that of paired-end inserts (typically 350–500 bp). This was the case for the second breakpoint of t(3;8) in BTA3, which was located in a 3-kb segment consisting of three long interspersed nuclear elements (LINEs), a short interspersed nuclear element (SINE), and a long terminal repeat sequence (LTR). It is also likely that we would have missed other features of the IRs studied, such as a 6-kb inversion at the fusion point of BTA24 for t(24;29),-der29 (Table 2), without the reconstruction of local alignments with nucleotide resolution around the breakpoints made possible by long-read sequencing technology.
We then sought to explain the phenotypic differences observed between wild-type and affected progeny by analyzing the function of the genes located on and around the breakpoints.
For only two of seven IRs, we found convincing disrupted or deleted functional candidates (Table 3). Regarding t(24;29),-der29, heterozygous mutations in PIGN have been associated with developmental delay and progressive cerebellar atrophy in human (Ohba et al. 2014) and mice (Dickinson et al. 2016). In addition, BCL2 haploinsufficiency was found to cause premature death in mouse. All affected animals died before 2 yr of age, compared with only 20% for wild-type animals (Xiang et al. 2011). For the inv ins(8;4), mutations in CCM2 are responsible for dominant defects of brain vascularization in humans (Liquori et al. 2007) and in mice (Plummer et al. 2006), leading to an increased risk of hemorrhagic stroke and seizures. This could explain the 2.5-fold higher juvenile mortality rate in the balanced compared with wild-type heifers.
In addition to the disrupted and deleted genes, we identified 12 genes located upstream of or downstream from the breakpoints whose expression may be affected by the alteration of the TAD boundaries and that have been associated with deleterious phenotypes in haploinsufficient mice and/or humans. Among them, GARS1, which is located in the segment of BTA4 inserted into BTA8 (inv ins(8;4) in bull Nt) at only 60 kb from the most proximal breakpoint, caught our attention. Numerous heterozygous mutations of this gene have been identified in human patients affected by Charcot–Marie-Tooth disease, type 2D, and other neuropathies, the most severe form of which is infantile spinal muscular atrophy, James type (Antonellis et al. 2003; James et al. 2006; Forrester et al. 2020). Similar observations have been made in the mouse with reports of muscle atrophy, decreased body weight, and premature death in heterozygotes for several different variants (Seburn et al. 2006; Achilli et al. 2009; Dickinson et al. 2016). Therefore, in addition to CCM2 disruption, GARS1 down-regulation may also contribute to the retarded growth and high mortality rate of balanced daughters (see Fig. 3D; Supplemental Fig. S7K). Together with untolerated aneuploidy, some of the putatively deregulated genes may also increase embryonic lethality and partially explain the very low level of fertility of IR carriers (see Fig. 3A,B; Supplemental Table S1). For example, the gene MB, located at 562 kb from the BTA5 breakpoint of t(5;28), has been identified in mice as causing embryonic lethality at the organogenesis stage in heterozygous mutant embryos (Meeson et al. 2001).
Regarding the putative causes of reduced survival in aneuploid progeny, we would like to mention that the 11-Mb segment of BTA4, for which we observed live monosomic and trisomic progeny, contains 60 genes, including 11 members of the HOXA cluster. Hox genes are key determinants of the identity of the morphological segments of each body part along the anteroposterior axis in most bilaterians (Hughes and Kaufman 2002; Mallo and Alonso 2013), and their deletion, duplication, or ectopic expression has been shown to cause a wide variety of body plan modifications in numerous species, including domestic animals (e.g. chicken, goat, and sheep, to name a few) (Wang et al. 2012; Allais-Bonnet et al. 2021). Haploinsufficiency for the HOXA gene cluster may be the cause of limb malformations reported in two progeny of the bull Nt (carrier of the inv ins(8;4)) that were euthanized at birth several months before the start of this study. In addition to GARS1 and CCM2 previously presented above, SNX10 haploinsufficiency could also contribute to the higher mortality rate of monosomic offspring of Nt between genotyping and the first year of life and to their significant underrepresentation, supporting prenatal or early juvenile mortality. Indeed, mice heterozygous for a mutation in SNX10 were found to display postnatal growth retardation (Ye et al. 2015). This matches with the high delay at first insemination age and with the developmental delay observed in a partially monosomic female (Supplemental Fig. S7K). Note that trisomic females have also been described as having delayed growth (M. Van Berghe, pers. comm.).
To conclude this section, the combination of analyses of large phenotype data sets with long-read sequencing and functional annotations suggests that most of the rearrangements have deleterious consequences in balanced animals even in the absence of direct deletion or disruption of candidate genes. Although further analyses are needed to describe the mechanisms involved, the IRs we identified are interesting models for studying gene regulation and the organization of genome structure.
Search for specific characteristics of IR
Throughout this study, we have attempted to highlight specific characteristics of IR in order to contribute to a better understanding of the processes responsible for their occurrence. Despite our limited data set, we obtained encouraging results. For example, we determined that 11 of the 12 detected IRs were caused by female (n = 1) and male (n = 10) meiotic defects. This result is consistent with observations in humans, where it has been found that 87% of unequal crossing-overs resulting in a viable conceptus occur in male meiosis (Shaffer and Lupski 2000).
We also observed a significant overrepresentation of the breakpoints in the third-quarter of the chromosomes (P < 0.04). At this stage, we do not have any explanations to propose to explain this phenomenon because this region did not show higher recombination rates in the general population (Supplemental Fig. S14), and the breakpoints did not colocalize with recombination hotspots based on LD analyses in the progeny of the bulls that produced the mutant gametes.
Furthermore, by analyzing long-read sequences, we did not identify any redundant repeat elements or specific motifs around the breakpoints that could have promoted the occurrence of IRs. In fact, we found repeat elements of various types in about half of the breakpoints, which is not unlikely considering that such DNA material makes up 49.4% of the bovine genome. The sire's age at conception had no effect, as also reported by Sloter et al. (2004) in a review of human and mouse sperm production. Finally, we found no significant increase in chromosome or chromosome pair representation in our reduced data set of 12 IRs. Taken together, these results suggest that IRs are a random process with an increased risk of occurring during male gametogenesis following intense meiotic activity. However, the question of the existence of genetic predisposition factors remains, which was raised by the observation of two carriers of different reciprocal translocations out of 48 sons of a single elite sire (t(5;28) and t(2;22) in the bulls Fa and Fe, respectively). We have no doubt that such a question will be answered as our easy-to-implement approach is applied to other populations and species and as an increasing number of well-characterized IRs will accumulate.
In conclusion, our results show the value of the large genotypic, pedigree, and phenotypic data sets generated by routine genomic evaluations in cattle to detect IRs and characterize their phenotypic consequences. LD-based screening for IRs is readily applicable to any population that benefits from similar data sets, and will have direct applications in plant and animal breeding. This approach also offers interesting prospects in terms of basic research, by allowing the detection of smaller and rarer types of chromosomal rearrangements than those identified by GTG banding, which are interesting models for studying gene regulation and the organization of genome structure.
Methods
Screening for IRs using SNP array genotypes
Genotypes
The initial data set consisted of 1,749,746 cattle from 15 breeds that were genotyped together with their sire using various Illumina SNP arrays over time (LD, ∼7 K SNPs; custom LD, ∼10 K to 20 K; BovineSNP50, ∼50 K; EuroGMD, ∼63 K; and HD, ∼777 K). Raw genotypes were imputed and phased for 44,307 autosomal SNPs by FImpute (Sargolzaei et al. 2014) as part of the French routine genomic evaluation of cattle, as described by Mesbah-Uddin et al. (2019).
Simulation
To estimate significance thresholds for LD between pairs of markers from different chromosomes, we developed a specific simulation procedure. One hundred AI bulls were selected based on their large number of genotyped progeny (n ≥ 1500) and phenotypic information suggesting that they were free of IR (percentiles 11 to 89 for conception rate and juvenile mortality). For each sire, 100 samples of n offspring were randomly drawn, resulting in 10,000 simulations for a given n. LD was calculated between pairs of markers from different chromosomes using the R (R Core Team 2021) package gaston 1.5.7 (https://cran.r-project.org/package=gaston) and map information from the ARS-UCD1.2 bovine genome assembly. The highest LD value (absolute correlation between alleles, also called Rmax) was scored for each simulation. To avoid spurious and punctual LD, we retained Rmax only if the two markers with the highest correlation were surrounded by at least 19 consecutive markers with R > Rmax * 0.75. The 10th highest Rmax found in the 10,000 simulations was taken as the threshold value for P < 0.001. We chose a false-discovery rate of 1‰, which is slightly lower than the prevalence of IR reported in two large studies focusing on phenotypically normal and fertile males of the human (2.06‰) (Ravel et al. 2006) and pig (4.7‰) (Ducos et al. 2007) species, in order to limit false positives.
The same process was repeated for 12 values of n (n = 25, 30, 40, 50, 60, 80, 100, 200, 350, 500, 750, and 1000), and a regression curve was fitted to obtain the appropriate threshold for each number of progeny per bull.
Screening of the population
To detect putative rearrangements in the population, we followed the same procedure as for simulations and calculated LD in 5571 paternal half-sib families with more than 30 progeny (mean = 292 ± 565 individuals; max = 13,347) (Supplemental Table S2). Thirty was found to be the minimum number of progeny to reach a significant LD (P < 0.001) based on simulation results. On average, 14,561 ± 607 markers per sire family were considered to be informative (i.e., heterozygous in the sire and with a minor allele frequency set at 20% in the paternal phases of the progeny; this rate allows avoidance of imputation errors and was fixed using data from the same bull's progeny as in the simulations). To be considered as significant, the LD values between pairs of markers from different chromosomes had to exceed the threshold determined by the equation obtained by simulation: y = 4.0302x−0.0408 (where x is the number of progeny). Analyses were performed on 1000 randomly selected offspring when the number of genotyped progeny exceeded this threshold. For more details on the available material, see Supplemental Table S2.
Finally, it should be noted that Chromosome X has not been considered in this study for two reasons. First, markers from the X-specific region are hemizygous in the sires and therefore not informative in paternal half-sib families, and, second, there is a sex ratio bias in the genotyped progeny with an average of 92% of females, which would affect calculations for markers located on the pseudoautosomal region.
Cytogenetic analyses
To verify our findings based on LD, we conducted a series of cytogenetic analyses on the bulls themselves (for bull Ma), their daughters (for bulls Im, Ja, Le, Nt, No, Ob, Ou, Qu, Ra, and Sa), or even their granddaughters (for bulls Fa and Fe) depending on the availability of material at the time of the study. Among the descendants of a bull, our goal was to sample at least two carrier and two noncarrier progeny of the haplotypic combination observed around the putative breakpoints. A total of 42 animals were included in these analyses. At first, we studied the karyotypes observed in metaphase II and stained by GTG banding. These were obtained from blood lymphocytes as described by Ducos et al. (1998). Breakpoints were estimated according to the standard GTG banded bovine karyotype (Cribiu et al. 2001).
In two cases, the GTG banding resolution (5–10 Mb) was not sufficient, and we performed FISH with chromosome-specific DNA painting probes to confirm the rearrangements and highlight cases of aneuploidy. Skin biopsies were obtained from ear punches. Fibroblast cultures and metaphases were obtained according to the method of Ducos et al. (2000b). For the inv ins(8;4) rearrangement, the nucleotide sequences of the segments adjacent to the breakpoints were aligned against bovine bacterial artificial chromosome (BAC) end sequences using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed January 24, 2022). Two INRA BAC clones were selected and obtained from the biological resources of the @BRIDGe facilities (http://abridge.inrae.fr): CH240-444F5, targeting the segment deleted on BTA4, and CH240-267G16, targeting the centromeric region of BTA8 that is duplicated and inserted in BTA4. FISH experiments were performed according to the method of Yerle et al. (1994). The two BACs were labeled with biotin and digoxygenin, respectively, using the BioPrime DNA labeling system kit (Invitrogen). Finally, they were revealed by Alexa Fluor 594 conjugated to streptavidin (Molecular Probes S32356A) and FITC conjugated mouse antidigoxygenin antibodies (Sigma-Aldrich F3523).
For the reciprocal translocation involving BTA24 and BTA29, an INRA BAC clone targeting the centromeric regions of the acrocentric chromosomes was selected: CH240-369L1. This BAC was also labeled with biotin using the BioPrime DNA labeling system kit (Invitrogen) and then revealed with Alexa Fluor 594 streptavidin (Molecular Probes).
Age of sire at conception
For 10 IRs resulting from abnormal male meiosis, we calculated the age of the sires at conception of their first mutant carriers. In parallel, we calculated their mean age at conception of their wild-type progeny (n = 118 in total) available in our initial screen of 5571 bulls. Finally, we compared the means of the two groups using a Student's t-test. For the sire of bulls Fa and Fe, the control group was randomly split in two.
Haplotype tests
To predict the status of the genotyped progeny of IR carrier bulls and to trace the affected segments in their pedigrees, we developed haplotype tests using 10 informative SNPs on each chromosome. These were chosen to be as close as possible to the breakpoints when sequence information was available or, by default, to retain the highest LD values between markers from nonhomologous chromosomes.
Phenotypic effects of IR
To characterize the phenotypic effects of IR, a variety of information was obtained from the French national bovine databases. In addition to individual ID, sex, pedigree, and herd, these data included information on insemination (date, female and male ID) and other life events such as dates of birth, death, and sale, as well as cause of death (natural or slaughter). Only records between January 1, 2010, and July 31, 2022, were considered. Conception and NRRs were calculated for IR carriers and for 2900 wild-type bulls (based on our initial screen for IR) with at least 100 registered AIs, and expressed as standard deviations from the mean of their breed to allow compilation of results from different breeds. Only first inseminations within parity (i.e., the first AI for heifers or the first AI after each calving for adult cows) performed with conventional semen were included, resulting in a total of 26,372,760 AI records. AIs were considered successful if calving occurred between 265 and 295 d later, unsuccessful if the calving occurred after 295 d, and filtered out if the female died before 295 d. NRRs after n days were scored as one if no subsequent insemination occurred within n days after and as zero otherwise. Mortality rates per bull were calculated as the number of females that died before 365 d of age divided by the number of females born and then expressed as standard deviations from the mean of their breed.
Additional indicators were calculated for eight IR carriers that had at least 20 genotyped and inseminated daughters. In the absence of routine growth recording in dairy heifers, we used age at insemination as a proxy, as it is generally recommended to inseminate Holstein heifers at 380–400 kg (i.e., ∼60% of adult liveweight) (Le Cozler et al. 2008). Finally, the proportion of females slaughtered without calving was calculated as the number of heifers alive at 20 mo that were later slaughtered without a calving record divided by the total number of daughters alive at 20 mo.
Long-read sequencing
DNA extraction
DNA from seven IR-affected bulls was extracted from 200-µL semen straws using the Gentra Puregene tissue kit (Qiagen). The straws were rinsed with 1 mL of PBS 1× buffer, and the resulting solutions were centrifuged for 5 min at 21,130g at room temperature. The sperm pellets were resuspended in 500 µL of Qiagen RLT buffer containing 50 mM of Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; pH = 7), before adding 500 µL of nuclei lysis solution and incubating for 3 hours at 55°C with slow shaking. The lysates were then cooled for 5 min on ice before 200 µL of protein precipitation solution was added, and they were incubated for a further 5 min on ice. After 10 sec of vigorous vortexing, the proteins were pelleted by centrifugation at 21,130g for 5 min. The supernatants were then transferred to clean tubes containing 600 µL of isopropanol. DNA pellets were obtained by gently inverting the tubes, centrifuging at 21,130g for 5 min, rinsing in 70% ethanol, and resuspending in TE buffer.
PacBio CLR sequencing
Library preparation and sequencing were performed at the INRAE GeT-PlaGe core facility in Toulouse (https://get.genotoul.fr/la-plateforme/get-plage/) according to the manufacturer's instructions (procedure & checklist–preparing HiFi SMRTbell libraries using the SMRTbell express template prep kit 2.0). At each step, DNA samples were purified using AMPure PB beads (PacBio), and DNA concentration, purity, size distribution, and degradation were assessed using a Qubit fluorometer (Thermo Fisher Scientific), a NanoDrop spectrophotometer (Thermo Fisher Scientific), and a Femto pulse system (Agilent), respectively. For each library, 10 µg of high-molecular-weight DNA was sheared to ∼ 30 kb using the Megaruptor3 system (Diagenode). Single-strand overhangs were removed, and DNA fragments were damage-repaired using the SMRTbell express template prep kit 2.0. Blunt hairpin adapters were ligated to the library. Fragments of ∼10–15 kb were filtered using the BluePippin size selection system (Sage Science). The SMRTbell libraries were annealed (1 h) and bound (4 h) with sequencing primer v5 and the Sequel II DNA Polymerase 2.0 from the Sequel II binding kit 2.0. The final sample-bound complexes were sequenced on a Sequel II instrument (PacBio) using one SMRT cell each and the sequencing kit 2.0 (loading concentration of 80 to 90 pM, movie time of 15 h). On average, a mean CLR coverage of 58× and a mean UMY coverage of 32× were obtained for the seven genomes.
Sequence analysis
Reads were aligned to the bovine ARS-UCD1.2 reference assembly (https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_002263795.1/) using the pbmm2 software (https://github.com/PacificBiosciences/pbmm2), and structural variations were detected using pbsv (https://github.com/PacificBiosciences/pbsv). The translocation breakpoints were identified as breakend (BND) records, which document the two breakends of a new adjacency (i.e., an adjacency not present in the reference genome; https://samtools.github.io/hts-specs/VCFv4.3.pdf). To confirm and characterize the different rearrangements at the nucleotide level, local (haplotype resolved) assemblies were constructed. For each new adjacency, the corresponding chimeric reads were extracted and assembled using the flye software (Kolmogorov et al. 2019). These assemblies were used to characterize the breakpoint region in more detail. Finally, the Integrative Genomics Viewer (IGV) v2.15.2 (Thorvaldsdóttir et al. 2013) was used to visualize the reads supporting the existence of IR.
Annotation
Various sources of information were used to annotate the genomic regions affected by the break points and fusion points and to predict the putative consequences of the rearrangements on neighboring genes. These included Ensembl (v108) annotated genes extracted using BioMart (https://www.ensembl.org/biomart/martview/), repeat elements available in the UCSC Genome Browser (https://genome-euro.ucsc.edu/; track “Repeat elements by RepeatMasker”), clinical signs reported in mutants from different species (human, https://www.omim.org; mouse, https://www.informatics.jax.org; and other animals, https://omia.org/), and, finally, a map of bovine TADs predicted by synteny with other mammals (Wang et al. 2018). For the latter, we used the UCSC liftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver) to convert positions from the UMD3.1 to the ARS-UCD1.2 bovine genome assembly. The list of genes obtained is available in Supplemental Table S5.
Recombination hotspots
We estimated the locations of recombination hotspots using the same LD as in the previous part of the Methods section. We considered the closest informative markers (i.e., heterozygous) surrounding breakpoints found in sequences in the sires of bulls affected by a meiotic-origin IR. The LD between these two SNPs was then compared with the LD along the affected chromosome in the same progeny cohort. The SNPs considered for the comparison were at similar distances as the two informative neighboring SNPs around the breakpoints (±10%).
To compare rearrangement positions with LD along a theoretical chromosome, we used LD computed in simulations and plotted it, expressing chromosome-wide positions as a percentage of their physical position (for more details, see Supplemental Fig. S14).
Data access
The long-read sequences data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) under accession number PRJEB59364. The genotype data used in this study have been submitted to the Recherche Data Gouv (https://entrepot.recherche.data.gouv.fr/) under the DOI https://doi.org/10.57745/ELW696. No accession number is required.
Supplementary Material
Acknowledgments
We thank the many breeders, veterinarians, agricultural technicians, and breeding companies involved in this study for providing blood, semen samples, and phenotypic information. We also thank @BRIDGe for providing BAC clones. Finally, we thank the members of the SeqOccIn Consortium for their valuable help and advice (i.e., Camille Eché, Christine Gaspin, Clément Birbes, Andreea Dreau, Quentin Boone, Christophe Klopp, Matthias Zytnicki, and Denis Milan from INRAE, the Genotoul GeT-PlaGe facility, or the Genotoul Bioinformatics facility). J.J. is supported by a CIFRE PhD grant from Eliance, with financial support from the Association Nationale de la Recherche et de la Technologie and APIS-GENE (Paris, France). This study was also supported by the Fertiligest and Effitness projects funded by APIS-GENE and by the SeqOccIn project cofunded by the European Union and the Occitania Region (FEDER-FSE MIDI-PYRENEES ET GARONNE 2014-2020).
Author contributions: A.Cap. and J.J. conceived the study with insights from H.B., A.P., and D.B. on specific points. H.B., A.Cal., N.B., and A.P. performed the cytogenetic analyses. J.J., S.T., F.B., A.B., C.H., and A.Cap. analyzed the genotypic and phenotypic data. C.M., A.S., C.K., C.I., C.D., and C.G. participated in DNA extraction, library preparation, and long-read sequencing. T.F., J.J., and A.Cap. analyzed long-read sequences. J.J. and A.Cap. wrote the manuscript with input from all the coauthors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277787.123.
Competing interest statement
The authors declare no competing interests.
References
- Achilli F, Bros-Facer V, Williams HP, Banks GT, AlQatari M, Chia R, Tucci V, Groves M, Nickols CD, Seburn KL, et al. 2009. An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Dis Model Mech 2: 359–373. 10.1242/dmm.002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais-Bonnet A, Hintermann A, Deloche M-C, Cornette R, Bardou P, Naval-Sanchez M, Pinton A, Haruda A, Grohs C, Zakany J, et al. 2021. Analysis of polycerate mutants reveals the evolutionary co-option of HOXD1 for horn patterning in Bovidae. Mol Biol Evol 38: 2260–2272. 10.1093/molbev/msab021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin S-Q, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. 2003. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 72: 1293–1299. 10.1086/375039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanian K, Brotin E, Biajoux V, Bouchet-Delbos L, Lainey E, Fenneteau O, Bonnet D, Fiette L, Emilie D, Bachelerie F. 2012. Proper desensitization of CXCR4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood 119: 5722–5730. 10.1182/blood-2012-01-403378 [DOI] [PubMed] [Google Scholar]
- Barasc H, Mouney-Bonnet N, Peigney C, Calgaro A, Revel C, Mary N, Ducos A, Pinton A. 2019. Analysis of meiotic segregation pattern and interchromosomal effects in a bull heterozygous for a 3/16 Robertsonian translocation. Cytogenet Genome Res 156: 197–203. 10.1159/000494289 [DOI] [PubMed] [Google Scholar]
- Beaussant Cohen S, Fenneteau O, Plouvier E, Rohrlich P-S, Daltroff G, Plantier I, Dupuy A, Kerob D, Beaupain B, Bordigoni P, et al. 2012. Description and outcome of a cohort of 8 patients with WHIM syndrome from the French Severe Chronic Neutropenia Registry. Orphanet J Rare Dis 7: 71. 10.1186/1750-1172-7-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Leclerc H, Boussaha M, Grohs C, Jewell N, Pinton A, Barasc H, Jourdain J, Femenia M, Dorso L, et al. 2023. Detailed analysis of mortality rates in the female progeny of 1,001 Holstein bulls allows the discovery of new dominant genetic defects. J Dairy Sci 106: 439–451. 10.3168/jds.2022-22365 [DOI] [PubMed] [Google Scholar]
- Bonnet-Garnier A, Pinton A, Berland HM, Khireddine B, Eggen A, Yerle M, Darré R, Ducos A. 2006. Sperm nuclei analysis of 1/29 Robertsonian translocation carrier bulls using fluorescence in situ hybridization. Cytogenet Genome Res 112: 241–247. 10.1159/000089877 [DOI] [PubMed] [Google Scholar]
- Bourneuf E, Otz P, Pausch H, Jagannathan V, Michot P, Grohs C, Piton G, Ammermüller S, Deloche M-C, Fritz S, et al. 2017. Rapid discovery of de novo deleterious mutations in cattle enhances the value of livestock as model species. Sci Rep 7: 11466. 10.1038/s41598-017-11523-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman AC, Derks MFL, Broekhuijse MLWJ, Harlizius B, Veerkamp RF. 2020. Using short read sequencing to characterise balanced reciprocal translocations in pigs. BMC Genom 21: 576. 10.1186/s12864-020-06989-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssed B, Zamariolli M, Bellucco FT, Melaragno MI. 2022. Mechanisms of structural chromosomal rearrangement formation. Mol Cytogenet 15: 23. 10.1186/s13039-022-00600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitan A, Michot P, Baur A, Saintilan R, Hozé C, Valour D, Guillaume F, Boichon D, Barbat A, Boichard D, et al. 2015. Genetic tools to improve reproduction traits in dairy cattle. Reprod Fertil Dev 27: 14–21. 10.1071/RD14379 [DOI] [PubMed] [Google Scholar]
- Clementini E, Palka C, Iezzi I, Stuppia L, Guanciali-Franchi P, Tiboni GM. 2005. Prevalence of chromosomal abnormalities in 2078 infertile couples referred for assisted reproductive techniques. Hum Reprod 20: 437–442. 10.1093/humrep/deh626 [DOI] [PubMed] [Google Scholar]
- Cribiu EP, Di Berardino D, Di Meo GP, Eggen A, Gallagher DS, Gustavsson I, Hayes H, Iannuzzi L, Popescu CP, Rubes J, et al. 2001. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet Cell Genet 92: 283–299. 10.1159/000056917 [DOI] [PubMed] [Google Scholar]
- De Lorenzi L, Morando P, Planas J, Zannotti M, Molteni L, Parma P. 2012. Reciprocal translocations in cattle: frequency estimation. J Anim Breed Genet 129: 409–416. 10.1111/j.1439-0388.2011.00983.x [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Savino A, Panuzzo C, Rocca S, Familiari U, Piazza R, Crivellaro S, Carrà G, Ferretti R, Fusella F, Giugliano E, et al. 2015. Morgana acts as an oncosuppressor in chronic myeloid leukemia. Blood 125: 2245–2253. 10.1182/blood-2014-05-575001 [DOI] [PubMed] [Google Scholar]
- Ducos A, Berland HM, Pinton A, Guillemot E, Seguela A, Blanc MF, Darre A, Darre R. 1998. Nine new cases of reciprocal translocation in the domestic pig (Sus scrofa domestica L.). J Hered 89: 136–142. 10.1093/jhered/89.2.136 [DOI] [PubMed] [Google Scholar]
- Ducos A, Berland HM, Pinton A, Seguela A, Brun-Baronnat C, Darre A, Darre R. 2000a. Contrôle chromosomique des populations animales d’élevage. INRAE Prod Anim 13: 25–35. 10.20870/productions-animales.2000.13.1.3765 [DOI] [Google Scholar]
- Ducos A, Dumont P, Séguéla A, Pinton A, Berland H, Brun-Baronnat C, Darré A, Marquant-Le Guienne B, Humblot P, Boichard D, et al. 2000b. A new reciprocal translocation in a subfertile bull. Genet Sel Evol 32: 589–598. 10.1186/1297-9686-32-6-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducos A, Berland H-M, Bonnet N, Calgaro A, Billoux S, Mary N, Garnier-Bonnet A, Darré R, Pinton A. 2007. Chromosomal control of pig populations in France: 2002–2006 survey. Genet Sel Evol 39: 583–597. 10.1186/1297-9686-39-5-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Ruland SL, Flynn LM, Stokowski RP, Young SG. 1995. Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes. Proc Natl Acad Sci 92: 1774–1778. 10.1073/pnas.92.5.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg KE, Whittlesey RL, Fleming MD, Andrews NC. 2010. Down-regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood 115: 3817–3826. 10.1182/blood-2009-05-224808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forabosco A, Percesepe A, Santucci S. 2009. Incidence of non-age-dependent chromosomal abnormalities: a population-based study on 88965 amniocenteses. Eur J Hum Genet 17: 897–903. 10.1038/ejhg.2008.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester N, Rattihalli R, Horvath R, Maggi L, Manzur A, Fuller G, Gutowski N, Rankin J, Dick D, Buxton C, et al. 2020. Clinical and genetic features in a series of eight unrelated patients with neuropathy due to Glycyl-tRNA synthetase (GARS) variants. J Neuromuscul Dis 7: 137–143. 10.3233/JND-200472 [DOI] [PubMed] [Google Scholar]
- Gekas J, Thepot F, Turleau C, Siffroi JP, Dadoune JP, Briault S, Rio M, Bourouillou G, Carré-Pigeon F, Wasels R, et al. 2001. Chromosomal factors of infertility in candidate couples for ICSI: an equal risk of constitutional aberrations in women and men. Hum Reprod 16: 82–90. 10.1093/humrep/16.1.82 [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. 2002. Hox genes and the evolution of the arthropod body plan. Evol Dev 4: 459–499. 10.1046/j.1525-142X.2002.02034.x [DOI] [PubMed] [Google Scholar]
- Hustert E, Scherer G, Olowson M, Guénet JL, Balling R. 1996. Rbt (rabo torcido), a new mouse skeletal mutation involved in anteroposterior patterning of the axial skeleton, maps close to the ts (tail-short) locus and distal to the sox9 locus on chromosome 11. Mamm Genome 7: 881–885. 10.1007/s003359900261 [DOI] [PubMed] [Google Scholar]
- Iannuzzi A, Parma P, Iannuzzi L. 2021. Chromosome abnormalities and fertility in domestic bovids: a review. Animals (Basel) 11: 802. 10.3390/ani11030802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PA, Cader MZ, Muntoni F, Childs A-M, Crow YJ, Talbot K. 2006. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology 67: 1710–1712. 10.1212/01.wnl.0000242619.52335.bc [DOI] [PubMed] [Google Scholar]
- Kastelic J, Thundathil J. 2008. Breeding soundness evaluation and semen analysis for predicting bull fertility. Reprod Domest Anim 43: 368–373. 10.1111/j.1439-0531.2008.01186.x [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Le Cozler Y, Lollivier V, Lacasse P, Disenhaus C. 2008. Rearing strategy and optimizing first-calving targets in dairy heifers: a review. Animal 2: 1393–1404. 10.1017/S1751731108002498 [DOI] [PubMed] [Google Scholar]
- Lewis NM, Canedo-Ribeiro C, Rathje CC, Jennings RL, Danihel M, Bosman LM, Silvestri G, Griffin DK. 2022. The economic burden of chromosome translocations and the benefits of enhanced screening for cattle breeding. Animals (Basel) 12: 1982. 10.3390/ani12151982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Niceta M, Muto V, Vona B, Pagnamenta AT, Maroofian R, Beetz C, van Duyvenvoorde H, Dentici ML, Lauffer P, et al. 2021. SCUBE3 loss-of-function causes a recognizable recessive developmental disorder due to defective bone morphogenetic protein signaling. Am J Hum Genet 108: 115–133. 10.1016/j.ajhg.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Berg MJ, Squitieri F, Leedom TP, Ptacek L, Johnson EW, Marchuk DA. 2007. Deletions in CCM2 are a common cause of cerebral cavernous malformations. Am J Hum Genet 80: 69–75. 10.1086/510439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Alonso CR. 2013. The regulation of Hox gene expression during animal development. Development 140: 3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Mark M, Rijli FM, Chambon P. 1997. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res 42: 421–429. 10.1203/00006450-199710000-00001 [DOI] [PubMed] [Google Scholar]
- Marras G, Gaspa G, Sorbolini S, Dimauro C, Ajmone-Marsan P, Valentini A, Williams JL, Macciotta NPP. 2015. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim Genet 46: 110–121. 10.1111/age.12259 [DOI] [PubMed] [Google Scholar]
- Matveevsky S, Kolomiets O, Bogdanov A, Alpeeva E, Bakloushinskaya I. 2020. Meiotic chromosome contacts as a plausible prelude for Robertsonian translocations. Genes (Basel) 11: 386. 10.3390/genes11040386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeson AP, Radford N, Shelton JM, Mammen PP, DiMaio JM, Hutcheson K, Kong Y, Elterman J, Williams RS, Garry DJ. 2001. Adaptive mechanisms that preserve cardiac function in mice without myoglobin. Circ Res 88: 713–720. 10.1161/hh0701.089753 [DOI] [PubMed] [Google Scholar]
- Mesbah-Uddin M, Hoze C, Michot P, Barbat A, Lefebvre R, Boussaha M, Sahana G, Fritz S, Boichard D, Capitan A. 2019. A missense mutation (p.Tyr452Cys) in the CAD gene compromises reproductive success in French normande cattle. J Dairy Sci 102: 6340–6356. 10.3168/jds.2018-16100 [DOI] [PubMed] [Google Scholar]
- Ogilvie CM, Scriven PN. 2002. Meiotic outcomes in reciprocal translocation carriers ascertained in three-day human embryos. Eur J Hum Genet 10: 801–806. 10.1038/sj.ejhg.5200895 [DOI] [PubMed] [Google Scholar]
- Ohba C, Okamoto N, Murakami Y, Suzuki Y, Tsurusaki Y, Nakashima M, Miyake N, Tanaka F, Kinoshita T, Matsumoto N, et al. 2014. PIGN mutations cause congenital anomalies, developmental delay, hypotonia, epilepsy, and progressive cerebellar atrophy. Neurogenetics 15: 85–92. 10.1007/s10048-013-0384-7 [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Fujiwara K, Ikeda M, Ozaki T, Terui T, Soma M, Inazawa J, Nagase H. 2015. The oncogenic role of GASC1 in chemically induced mouse skin cancer. Mamm Genome 26: 591–597. 10.1007/s00335-015-9592-9 [DOI] [PubMed] [Google Scholar]
- Pezzani L, Milani D, Manzoni F, Baccarin M, Silipigni R, Guerneri S, Esposito S. 2015. HOXA genes cluster: clinical implications of the smallest deletion. Ital J Pediatr 41: 31. 10.1186/s13052-015-0137-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer NW, Squire TL, Srinivasan S, Huang E, Zawistowski JS, Matsunami H, Hale LP, Marchuk DA. 2006. Neuronal expression of the Ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm Genome 17: 119–128. 10.1007/s00335-005-0098-8 [DOI] [PubMed] [Google Scholar]
- Poornima S, Daram S, Devaki RK, Qurratulain H. 2020. Chromosomal abnormalities in couples with primary and secondary infertility: genetic counseling for assisted reproductive techniques (ART). J Reprod Infertil 21: 269–274. 10.18502/jri.v21i4.4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Haaf T. 2015. Mechanisms of origin, phenotypic effects and diagnostic implications of complex chromosome rearrangements. Mol Syndromol 6: 110–134. 10.1159/000438812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu PC. 1989. Cytogénétique des mammifères d’élevage. INRAE, Paris. [Google Scholar]
- Pramparo T, Grosso S, Messa J, Zatterale A, Bonaglia MC, Chessa L, Balestri P, Rocchi M, Zuffardi O, Giorda R. 2005. Loss-of-function mutation of the AF9/MLLT3 gene in a girl with neuromotor development delay, cerebellar ataxia, and epilepsy. Hum Genet 118: 76–81. 10.1007/s00439-005-0004-1 [DOI] [PubMed] [Google Scholar]
- Ravel C, Berthaut I, Bresson JL, Siffroi JP, the Genetics Commission of the French Federation of 818 CECOS. 2006. Prevalence of chromosomal abnormalities in phenotypically normal and fertile adult males: large-scale survey of over 10,000 sperm donor karyotypes. Hum Reprod 21: 1484–1489. 10.1093/humrep/del024 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Sargolzaei M, Chesnais JP, Schenkel FS. 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genom 15: 478. 10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. 2007. Challenges and standards in integrating surveys of structural variation. Nat Genet 39: S7–S15. 10.1038/ng2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. 2006. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 51: 715–726. 10.1016/j.neuron.2006.08.027 [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. 2000. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34: 297–329. 10.1146/annurev.genet.34.1.297 [DOI] [PubMed] [Google Scholar]
- Sloter E, Nath J, Eskenazi B, Wyrobek AJ. 2004. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil Steril 81: 925–943. 10.1016/j.fertnstert.2003.07.043 [DOI] [PubMed] [Google Scholar]
- Tateno H, Miyake Y-I, Mori H, Kamiguchi Y, Mikamo K. 1994. Sperm chromosome study of two bulls heterozygous for different Robertsonian translocations. Hereditas 120: 7–11. 10.1111/j.1601-5223.1994.00007.x [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinf 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilevska M, Ivanovska E, Kubelka Sabit K, Sukarova-Angelovska E, Dimeska G. 2013. The incidence and type of chromosomal translocations from prenatal diagnosis of 3800 patients in the republic of Macedonia. Balkan J Med Genet 16: 23–28. 10.2478/bjmg-2013-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao Y, Imsland F, Gu X, Feng C, Liu R, Song C, Tixier-Boichard M, Gourichon D, Li Q, et al. 2012. The crest phenotype in chicken is associated with ectopic expression of HOXC8 in cranial skin. PLoS One 7: e34012. 10.1371/journal.pone.0034012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Hancock TP, Chamberlain AJ, Vander Jagt CJ, Pryce JE, Cocks BG, Goddard ME, Hayes BJ. 2018. Putative bovine topological association domains and CTCF binding motifs can reduce the search space for causative regulatory variants of complex traits. BMC Genom 19: 395. 10.1186/s12864-018-4800-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckselblatt B, Rudd MK. 2015. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet 31: 587–599. 10.1016/j.tig.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, Noonan EJ, Wang J, Duan H, Ma L, Michie S, Boxer LM. 2011. The immunoglobulin heavy chain gene 3′ enhancers induce Bcl2 deregulation and lymphomagenesis in murine B cells. Leukemia 25: 1484–1493. 10.1038/leu.2011.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Rajkovic A. 2019. Genetics of human female infertility. Biol Reprod 101: 549–566. 10.1093/biolre/ioz084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Morse LR, Zhang L, Sasaki H, Mills JC, Odgren PR, Sibbel G, Stanley JRL, Wong G, Zamarioli A, et al. 2015. Osteopetrorickets due to Snx10 deficiency in mice results from both failed osteoclast activity and loss of gastric acid-dependent calcium absorption. PLoS Genet 11: e1005057. 10.1371/journal.pgen.1005057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerle M, Goureau A, Gellin J, Le Tissier P, Moran C. 1994. Rapid mapping of cosmid clones on pig chromosomes by fluorescence in situ hybridization. Mamm Genome 5: 34–37. 10.1007/BF00360565 [DOI] [PubMed] [Google Scholar]
- Yu Y, Ouyang Y, Yao W. 2018. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34: 1229–1231. 10.1093/bioinformatics/btx763 [DOI] [PubMed] [Google Scholar]
- Zhao R, Talenti A, Fang L, Liu S, Liu G, Chue Hong NP, Tenesa A, Hassan M, Prendergast JGD. 2022. The conservation of human functional variants and their effects across livestock species. Commun Biol 5: 1003. 10.1038/s42003-022-03961-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.