Abstract
Large-scale high-throughput sequencing data sets have been transformative for informing clinical variant interpretation and for use as reference panels for statistical and population genetic efforts. Although such resources are often treated as ground truth, we find that in widely used reference data sets such as the Genome Aggregation Database (gnomAD), some variants pass gold-standard filters, yet are systematically different in their genotype calls across genotype discovery approaches. The inclusion of such discordant sites in study designs involving multiple genotype discovery strategies could bias results and lead to false-positive hits in association studies owing to technological artifacts rather than a true relationship to the phenotype. Here, we describe this phenomenon of discordant genotype calls across genotype discovery approaches, characterize the error mode of wrong calls, provide a list of discordant sites identified in gnomAD that should be treated with caution in analyses, and present a metric and machine learning classifier trained on gnomAD data to identify likely discordant variants in other data sets. We find that different genotype discovery approaches have different sets of variants at which this problem occurs, but there are characteristic variant features that can be used to predict discordant behavior. Discordant sites are largely shared across ancestry groups, although different populations are powered for the discovery of different variants. We find that the most common error mode is that of a variant being heterozygous for one approach and homozygous for the other, with heterozygous in the genomes and homozygous reference in the exomes making up the majority of miscalls.
Although massively parallel sequencing technologies have been transformative for genomics research, they have an appreciable error rate (Ma et al. 2019) as a cost of their high-throughput capacity. To account for this, sophisticated pipelines have been developed for the detection and removal of incorrect sequencing calls (Anderson et al. 2010; McKenna et al. 2010; Highnam et al. 2015; Adelson et al. 2019; Lam et al. 2019; Li et al. 2019). However, even with gold-standard filtering, spurious genotype calls can infiltrate data sets and potentially skew results. This is of particular importance with data sets that aggregate calls generated by multiple genotype discovery approaches, as different strategies have distinct error modes. Identifying variants that have technical artifacts affecting genotype calls is of major importance, as such loci give misleading information regarding population allele frequencies (AFs) and could be incorrectly identified as being phenotypically meaningful in gene discovery.
By leveraging the unprecedented size and depth of the Genome Aggregation Database (gnomAD) (Lek et al. 2016; Karczewski et al. 2020), we comprehensively characterize trends in genotype calling depending on the sequencing technology. Specifically, we note that a subset of variants, despite passing standard quality filters (Karczewski 2017), produce discordant AFs in data generated using different genotype discovery approaches, stemming from unreliable variant calling. This cannot be explained by population stratification, as this effect is observed even when looking at the same set of individuals. Such unreliably genotyped variants should therefore be screened out of analyses. Including these variants in gene discovery efforts, particularly in study designs in which case and control data are represented by different combinations of sequencing platforms or genotype discovery approaches, could result in their appearance as false-positive associations.
In this article, we comprehensively characterize the observation of discordant genotyping depending on a genotype discovery approach using a large set of diverse individuals from the gnomAD database, including a subset of participants who underwent both whole-exome and whole-genome sequencing (WES/WGS). We then validate our findings in two external data sets for which data from multiple genotype discovery approaches are available: the 1000 Genomes Project and the All of Us Research Program (Auton and Salcedo 2015; The All of Us Research Program Investigators 2019). Correcting for this technical error, whether by removing the gnomAD discordant variants provided here or by identifying user-identified spurious calls with our freely distributed machine learning predictor, should be incorporated as a step in QC pipelines to avoid spurious associations, particularly in large-scale studies aggregating data from multiple sources.
Results
Discordance in genotype calls across genotype discovery approaches replicates across ancestries
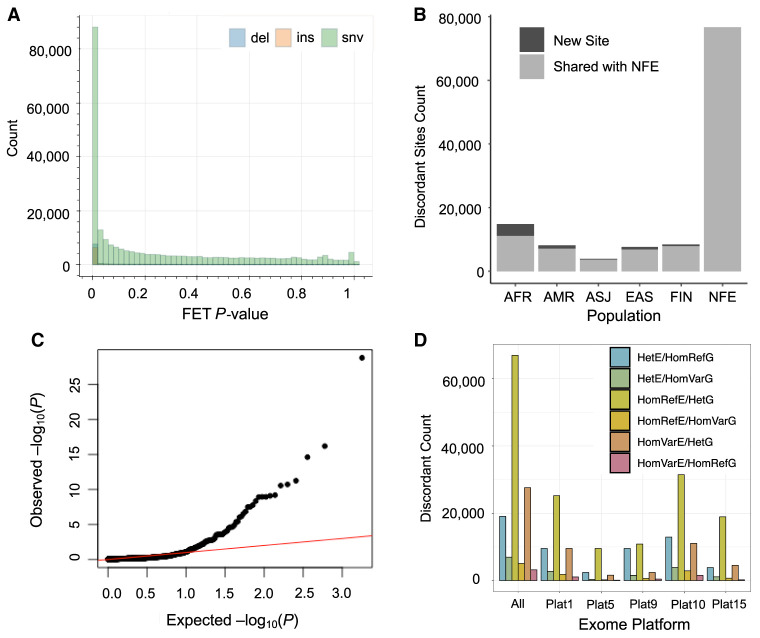
We sought to compare AFs across variants found within exome and genome sequencing data sets in gnomAD to test whether there are regions with significant bias associated with genotype discovery approach. We first focused on the largest population represented in gnomAD 0.2: the non-Finnish Europeans (NFE). Using the full release of gnomAD version 2.1.1, we filtered the data to include only sites that were present and had a quality determination of PASS in both the genomes and exomes (Karczewski et al. 2020). To ensure sufficient power, we filtered for sites with allele count (AC) greater than 10 and ran a Fisher's exact test on the difference in the number of alternate AC to total alleles (allele number [AN]) between these two data sets. A nonnegligible fraction of sites was significantly discordant in their calls (Fig. 1A). We also tested other less-stringent AC thresholds (AC > 1 and AC > 5) and observed that the trends of discordance between the genotype discovery approaches were consistent across AC cutoffs (Supplemental Fig. S1).
Figure 1.
Discordance in genotype calls across high-throughput genotype discovery approaches. (A) Fisher's exact test concordance test P-value for shared, PASS sites in the gnomAD NFE exomes and genomes. Bars are colored by variant type: insertion (ins), deletion (del), or single-nucleotide variant (snv). (B) “Bad” sites are replicated across ancestry groups in gnomAD. Sites flagged as discordant in both the NFE and another ancestry group are plotted in gray; those new sites not in the NFE are shown in black. (C) QQ plot for the Fisher's exact test P-value of shared variants in a set of 946 individuals for whom both WES and WGS data were available. (D) Different exome captures’ contribution to discordant sites. Bars are colored by the error mode that was observed for the discordant genotype call: heterozygous (Het), homozygous reference (HomRef), or homozygous variant (HomVar) in either the exomes (E) or genomes (G).
When comparing AFs between sequencing strategies, it is critical to control for ancestry, as populations will have differing frequencies at many loci simply owing to demography (Gravel et al. 2011; Auton and Salcedo 2015; Bergström et al. 2020). To assess if ancestry affected discordance rates, we ran Fisher's exact test concordance checks across all gnomAD continental groups (Fig. 1B). The total discordant variant counts were directly related to the sample size of the population in question and were not enriched for any given ancestry (Supplemental Fig. S2). Although novel discordant variants were discovered in each population, most variants observed in other populations were shared with the NFE (86.9%) (Fig. 1B; Supplemental Table S1). Replication of the same variants across multiple ancestries strengthens the argument of a shared technical artifact and suggests that ancestral bias between exome and genome data sets is unlikely to be a confounding factor. Future work may wish to investigate other biological and nonbiological factors for an impact on discordance.
Error mode of discordant calls
Next, we aimed to classify the typical error mode that results in discordant calls using individuals with both WES and WGS data. As 946 gnomAD individuals underwent both WES and WGS, we were able to examine the rates and error modes of discordant genotype calls without concern over population structure or differing sample composition; because these are the same individuals, any difference in AF and/or genotype calls can be conclusively determined to be owing to technical artifacts. We tallied the number of sites in each pairwise exome–genome genotype category (6333 sites) (Supplemental Fig. S3) to classify miscall error mode. Of the six possible error modes (homozygous reference/heterozygous, homozygous reference/homozygous variant, or homozygous variant/heterozygous for both data set directions), we find that the majority of calls, 57.7%, are a heterozygous genotype call in the genomes but a homozygous reference genotype call in the exomes (Fig. 1D; Supplemental Fig. S4). We also note that different sequencing platforms have different rates of discordant calls; although because of sharing restrictions, we cannot identify platform names with certainty. Overall, ∼16% of the variants that were present and PASS in both exomes and genomes in the overlapping individuals had at least one discordant call.
Next, we examined the discordance of AFs, again with a Fisher's exact test. Usually in cohort-based comparisons, the expected distribution of Fisher's exact test P-values is represented by a uniform distribution. Because we are looking at the same individuals, it is expected that AFs should be identical (i.e., P = 1). We observe the presence of many variants substantially deviating from expectations, representing loci with significantly different MAF in the exomes versus the genomes (Fig. 1C).
Identification of discordant sites
We next sought to identify problematic sites failing a Fisher's exact test of concordant WGS/WES frequency estimates in the largest subset of gnomAD, the NFE. Based on the distribution of P-values from this test, we decided upon a threshold of P < 1 × 10−5 to determine the classification of a variant as “bad” or “good” (Fig. 1; Supplemental Fig. S5). Of the 283,287 PASS/PASS variants tested with MAF > 0.01 and AC > 10, 51,255 (18.1%) failed the Fisher's exact test and were deemed “bad,” whereas 231,631 (81.8%) passed and were deemed “good.” Distributions of metadata features for the good versus bad sites do show trends in several features, although no feature alone perfectly explains the phenomenon (Supplemental Fig. S6). They also highlight a difference in discordance patterns of indels versus SNVs (Supplemental Fig. S7). Specifically, SNVs show a pattern of higher AF in genomes compared with exomes, whereas indels do not have this trend. Indels are also generally less stable in AF estimates than SNVs. It therefore appears that two distinct technical error modes might be affecting miscalls in indels versus SNVs, rather than one shared mechanism. As many of these indels fell in the low complexity regions of the genome, it is likely that a mapping issue is responsible for their miscalls. A comprehensive description of gnomAD structural variant calling and considerations is published and can be found in a gnomAD blog post (Collins et al. 2020; https://gnomad.broadinstitute.org/news/2019-03-structural-variants-in-gnomad/). To correct this, we therefore recommend excluding the low complexity regions from stringent analyses. In general, when there was discordance, the genomes were found to have a higher MAF than the exomes (Supplemental Fig. S1B). The trend in MAF difference aligns with the most commonly observed error mode in genotyping.
Having confirmed that there was a systematic and significant AF discordance between genotype discovery approach, we used our Fisher's exact tests to generate a list of sites harboring this technical artifact that may be excluded from analyses. Again, these discordant sites represent variants that were a PASS in gnomAD QC in both the exomes and genomes but are unreliably genotyped depending on the sequencing technology used. Given a situation in which, for example, a case cohort has been exome-sequenced and the control cohort has been genome-sequenced, such sites could give false-positive associations owing to the resulting AF differences. We, therefore, recommend they be treated with caution or broadly excluded (in addition to standard cohort QC) unless thorough confirmation of their validity in a particular data set has been performed.
Our analysis of variant AF discordance reflects technical differences between whole-exome and whole-genome high-throughput sequencing approaches in recovering coding DNA variation. Similarly, we performed this concordance analysis on the All of Us Research Program data set to compare AFs between WGS and microarray genotyping to quantify any similar effect arising between these genotype discovery approaches in primarily noncoding variation. We subsampled the All of Us primary release cohort down to the 95,596 samples who have both WGS and microarray genotyping data available. Call rate, HWE, and MAF > 0.05 filters were applied to ensure only good-quality common variants entered the analysis. Out of 102,631 variants (7944 coding), 2344 had Fisher's exact test P < 0.05 (Supplemental File S1). Note that because of identical samples being analyzed, the expected P-value distribution is centered at one (Supplemental Fig. S8). We evaluated the overlap between the variants flagged in All of Us and gnomAD, finding that only seven out of them were found in both samples, likely owing to the focus of gnomAD on coding variation (given comparisons included WES) versus on noncoding variation in All of Us. Out of these seven variants, rs4951250 was found to be significantly discordant in both data sets (P < 1 × 10−16 genome vs. exome; P = 4 × 10−5 genome vs. array).
Recovering filtered concordant sites
In addition to generating this discordant list of bad sites that should be excluded or treated with caution despite being a PASS in gnomAD QC, we investigated whether additional trustworthy sites could be rescued from the “non-PASS” list based on our AF concordance criteria. Non-PASS variants are those that did not meet all required passing criteria in the gnomAD QC pipeline (Karczewski et al. 2020). We tested this by conditioning on PASS in one data set, non-PASS in the other, and reran the concordance pipeline, requiring the following threshold in both data sets to add a higher level of stringency for recovering sites: AC > 1, DP > 10, and AF > 0.01%. In total, there were 41,584 sites that met these criteria, of which 30,683 were instances in which the genomes are a non-PASS and the exomes are a PASS. Approximately half of these sites had P-values greater than 1 × 10−5, which we consider to be reliable. The exomes represent the vast majority of sequences in gnomAD, which may make their results more stable. For analyses that require less stringent QC, we provide these sites that can be optionally retained, given that they pass cohort QC in the individual data set.
Predicting technical bias for variants
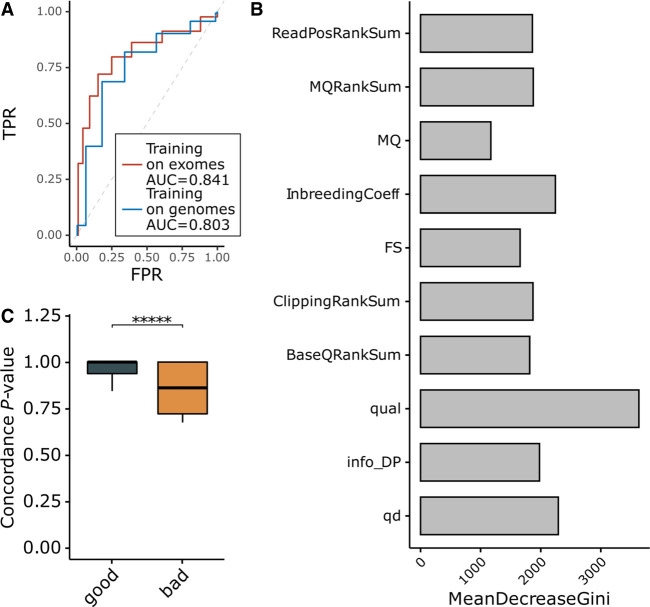
We used features based on variant annotations generated during variant calling (e.g., variant quality, mapping quality, etc.) to build a random forest predictor that detects the presence of technical bias for a particular variant (Supplemental Table S2). Model training and validation were performed using several approaches. First, we used a leave-one-out cross-validation procedure using the exome data set from gnomAD. In 22 trials (one for each of the 22 autosomes), we set aside one chromosome and used the other 21 for model training. Then, the model was tested on the variants from the chromosome that was not used for training. Using the Fisher's exact test P-value threshold for the discordance analysis, we classified the variants into “bad” (with P < threshold) and “good” (P ≥ threshold) groups. By varying the Fisher's exact test P-value threshold discriminating the groups, we performed ROC analysis. Because representation of the classes varies depending on the selected threshold, we used the class weights for balancing the classes (in this and the following tests, the weight for the “bad” class was set as the fraction of “bad” variants in the training data, and the weight for the “good” class was set to one). The area under the ROC curve (ROC AUC) for such a model was estimated as 0.841 (Fig. 2A).
Figure 2.
Reliable classification of discordant variants based on variant annotations. (A) ROC analysis for two random forest predictor validation approaches either using leave-one-out analysis on the exomes or using the genomes as a training set to classify exome variants. (B) Feature importance analysis for the random forest model. (C) Comparison of concordance analysis Fisher's exact test P-values for variants from the 1000 Genomes classified using the random forest predictor trained on the gnomAD genomes data set. MeanDecreaseGini is a measure of how each variable contributes to the homogeneity of the nodes and leaves in the resulting random forest. The higher the value of mean decrease accuracy or mean decrease Gini score, the higher the importance of the variable in the model.
Next, we used variant annotations from the gnomAD genomes data set for training and variant annotations from the gnomAD exomes as a test sample. These are two separate data sets that are well powered to detect biases in AFs and ensure full independence between test and training samples. In this setting, our model again reliably predicted discordant variants, with ROC AUC = 0.803 (Fig. 2A). Feature importance analysis of the model suggests that variant quality, inbreeding coefficient, and quality by depth are the key parameters discriminating variants with and without evidence for technical bias. Therefore, current protocols for alignment and variant calling are leaving a notable footprint that can be used to detect platform biases (Fig. 2B).
Finally, we used data from The 1000 Genomes Project (The 1000 Genomes Project Consortium 2012) as an independent public data set for testing the predictor. Exome sequences from 1393 samples (Supplemental Table S3) were used to create the variant call set following GATK best practices. Variant annotations were used to classify variants using the gnomAD genomes data as a training sample. Because of sample size, 1000 Genomes data are significantly less powered to detect technical biases compared with gnomAD. Therefore, it is harder to confidently identify the ground truth for discordant variants. Identical 1000 Genomes samples from exome and genome sequencing were used to detect variants with a signature of technical bias using the AF concordance Fisher's exact test described here. Because of the power limitations, instead of performing ROC analysis, we compared the Fisher's exact test P-values for the variants classified as having evidence of technical bias (“bad”) and those without such evidence (“good”) (Fig. 2C).
This random forest model trained on the gnomAD genome v2.1 data set was incorporated into a freely distributed R package called DNA DISCOrdant Variant IdentifiER (DNAdiscover) (R Core Team 2021; https://github.com/na89/DNAdiscover). The package uses variant annotations to predict whether a variant is likely to be “discordant” or “concordant” in user input data and performs well with both genome and exome sequencing data.
Discordant variants are reported in published studies and are predicted to be functionally important
We first observed the phenomenon of variant discordance through the investigation of GWAS variants in the COVID host genetics initiative and the UK Biobank (COVID-19 Host Genetics Initiative 2021). When inspecting top associated variants that did not have strong LD friends, we noticed that many had discordant frequencies between GWAS arrays and gnomAD, and this often corresponded to variants that had discrepant frequencies between the gnomAD exomes and genomes. We thus suggest that the discordant site list provided herein can be used for quality control of GWAS variants.
To investigate whether discordant variants may be spuriously attributed phenotypic relevance, we annotated all variants with their predicted functional consequence using the Ensembl Variant Effect Predictor (VEP) (McLaren et al. 2016). Discordant variants appear in all functional consequence categories, including 11,536 that are annotated as missense (Supplemental Fig. S9; Supplemental Table S4). Such variants are tempting to prioritize for functional follow-up given their apparent functional importance despite having GWAS signal likely driven by the observed genotype calling artifact. Technical artifacts are actually expected to be enriched in functionally important categories given that they are immune from the effects of natural selection, a phenomenon that has been previously observed for putatively loss-of-function somatic variation (Buckley et al. 2017).
To see if discordant sites have been reported in peer-reviewed publications, we intersected genome-wide significant variants (P < 5 × 10−8) from the GWAS catalog (Welter et al. 2014) with our discordant sites list. Seventeen bad variants were found in the GWAS catalog, underscoring the importance of controlling for this artifact, as it may impact downstream interpretation of association findings (Supplemental Table S5). Variants in this list have been associated with multiple health-related phenotypes, including schizophrenia, telomere length, and blood protein levels. Of these 17, half are multiallelic and approximately a third are indels, echoing our earlier results of a higher discordance rate for these variant types than biallelic SNVs. Additionally, more than half of the 17 are present in the first megabase of the chromosome, suggesting that areas flanking the telomeres should be treated with caution.
Discussion
The need for extremely large sample sizes to obtain sufficient statistical power in genetic studies requires the creation of data sets that may go beyond the financial capabilities of many individual research groups. This leads to the creation of metadata sets that have contributions from many individual studies, thus creating heterogeneity in the genotype discovery approaches that were used for genotyping. Therefore, identification of DNA variants that are susceptible to technical bias when genotypes originate from multiple discovery strategies is vital in order to avoid false-positive associations and analyses of the artificially inflated AFs. This is of particular concern in instances in which cases may originate from one data generation effort and controls from another.
Here, we identify and describe a technical artifact arising in various genotype discovery approaches that may affect cohort data variant quality despite the following of gold-standard QC procedures. We present our metric for the identification of discordant sites, provide a list of the discordant variants identified in gnomAD that should be treated with caution, and release an openly available software package containing our random forest predictor that reliably classifies untrustworthy variants in user cohort data. Excluding variants with signals of discordance across sequencing platforms results in higher-quality results and reduces the risk of spurious associations in gene discovery. This is particularly important as we observe that technical artifacts are enriched in functionally important annotations.
Additionally, we show that discordance in AFs is also present in the All of Us Research Program data set when comparing WGS to microarray genotyping for overlapping samples. This finding indicates that variants in both coding and noncoding DNA could have discordant genotype calls. Importantly, in our predictor, we use the variant annotations, which often are used in variant quality score recalibration and filtration pipelines. Our results indicate that stricter filtration thresholds might be helpful for the elimination of some discordant variants; however, more cautious consideration of discordance is warranted in heterogeneous data sets.
We note that although we provide a discordant list of variants failing our discordance test in the gnomAD v2 data set for ready exclusion, the specific sites that are discordant in a given cohort depends on the genotype discovery approach used and data set composition. Therefore, for optimal precision, we recommend identification of discordant sites within user cohorts with the provided classifier rather than a blanket restriction of variants identified in gnomAD. We freely provide an R package with a predictor trained on gnomAD WGS data, DNAdiscover, for such use in other cohort data to identify cohort-specific sites with features indicative of unreliable genotype calls.
Our work is primarily aimed to show that our methodology is effective in detecting technical biases in high-throughput sequencing approaches and to call attention to this important consideration for aggregated data sets. We also believe that our findings can pave the way for even more robust approaches to detect such artifacts in the future. Specifically, we propose that a meta-analysis could be performed across all ancestral groups simultaneously, which would provide increased statistical power in identifying discordant variants. This would allow for the detection of smaller biases and could potentially extend this approach to less common variants.
Based on the examinations presented in this paper, we recommend that researchers using aggregated cohort data implement the following conservative QC procedures to ensure the elimination of discordant sites:
Drop any variant that fails in both the gnomAD exomes and genomes;
Consider dropping any variant that fails in the gnomAD exomes, as these represent the bulk of gnomAD data;
Drop the discordant list variants presented here that are PASS in both the gnomAD exomes and genomes but that are discordant in frequency across the genotype discovery approach;
Drop variants that are flagged by our random forest predictor, DNAdiscover, in an independent data set, as each genotype discovery approach has a distinct genotyping error mode;
Remove the low complexity regions; and
Optionally, skeptically retain sites that are on the “recovered” list here.
Methods
Characterizing discordance in genotype calls across gnomAD exomes and genomes
All analyses were conducted using the Hail software program on the Google Cloud platform (GCP 2021). Plots were created using Bokeh and ggplot2 (Wickham 2011; Jolly 2018). Concordance metrics for genotype calls were generated from the overlapping individuals with the command hail.methods.concordance(). Using the full release of gnomAD version 2.1 (Karczewski et al. 2020), we filtered to include only sites that were both present and had a quality determination of PASS in the genomes and exomes. We split multiallelic variants and retained only sites that were present and PASS in both exomes and genomes, filtering to only biallelic sites with AC > 1 and AF > 0.01% in either data set for the NFE. Starting with all sites with at least one alternate allele, and subsequently for sites with AC > 5 and 10, we calculated the AF in the genomes and exomes separately and ran a Fisher's exact test on the difference in the number of alternate AC to total alleles (AN) between these two data sets. Specific filters for various steps are described in their relevant Results section. gnomAD summary data are freely available at gnomAD (https://gnomad.broadinstitute.org). Additional information and a discussion of the best practices for using gnomAD can be found at https://macarthurlab.org/blog/. The list we have curated of variants failing the discordance test in gnomAD is provided with this paper in the Supplemental Materials. Further details regarding data treatment are described throughout the paper for context.
All of Us data were subsampled to 95,596 with both WGS and microarray genotyping (WGA) available. MAF > 0.05, HWE > 0.0001, and MAC > 10 filters were applied to keep only common variants. Multiallelic variants were split. A variant call rate >0.8 was required in both the WGS and WGA data sets. Variants with a call rate difference between data sets greater than 0.05 were also eliminated from analysis. The R libraries dplyr, reshape2, pROC, ROCR, and RandomForest were used to process variant annotations, evaluate predictor quality, and build our classifier package, DNAdiscover, using R-4.0.3 (Liaw and Wiener 2002; Sing et al. 2005; Robin et al. 2011; https://github.com/hadley/reshape; https://CRAN.R-project.org/package=dplyr).
Software availability
Our DNAdiscover package for prediction of the presence of technical bias in variants coming from high-throughput sequencing, alongside a user manual, is available at GitHub (https://github.com/na89/DNAdiscover), and source code is presented as Supplemental File S2.
Supplementary Material
Acknowledgments
We thank the members of the gnomAD and Hail teams for their assistance with this project. This project was supported by the National Institute of Mental Health (K01 MH121659 and T32 MH017119 to E.G.A.) and the National Human Genome Research Institute (U24HG011450). E.G.A. was additionally supported by the Caroline Wiess Law Fund for Research in Molecular Medicine and the ARCO Foundation Young Teacher-Investigator Fund at Baylor College of Medicine. A.A.L. was supported by the Ministry of Science and Higher Education of the Russian Federation (agreement no. 075-15-2022-301, institutional grant to Almazov National Medical Research Center). M.A. was supported by the Aging Biology Foundation and Nationwide Foundation Pediatric Innovation Fund. The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549, 1 OT2 OD026554, 1 OT2 OD026557, 1 OT2 OD026556, 1 OT2 OD026550, 1 OT2 OD 026552, 1 OT2 OD026553, 1 OT2 OD026548, 1 OT2 OD026551, 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205, 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277, 3 OT2 OD025315, 1 OT2 OD025337, 1 OT2 OD025276. In addition, the All of Us Research Program would not be possible without the partnership of its participants.
Author contributions: E.G.A. and M.A. designed and implemented pipelines, ran analyses, and drafted the primary manuscript. K.J.K. and A.A.L. aided in code implementation and interpretation. H.L.R., D.G.M., B.M.N., and M.J.D. supervised and advised on the project. All authors reviewed and approved the final draft.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277908.123.
Competing interest statement
M.J.D. is a founder of Maze Therapeutics. B.M.N. is a member of the Deep Genomics Scientific Advisory Board and serves as a consultant for the Camp4 Therapeutics Corporation, Takeda Pharmaceutical, and Biogen. K.J.K. is a consultant for Vor Biopharma. The remaining authors declare no competing interests.
References
- The 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelson RP, Renton AE, Li W, Barzilai N, Atzmon G, Goate AM, Davies P, Freudenberg-Hua Y. 2019. Empirical design of a variant quality control pipeline for whole genome sequencing data using replicate discordance. Sci Rep 9: 16156. 10.1038/s41598-019-52614-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The All of Us Research Program Investigators. 2019. The “All of Us” Research Program. N Engl J Med 381: 668–676. 10.1056/NEJMsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. 2010. Data quality control in genetic case-control association studies. Nat Protoc 5: 1564–1573. 10.1038/nprot.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Salcedo T. 2015. The 1000 genomes project. In Assessing rare variation in complex traits: design and analysis of genetic studies (ed. Zeggini E, Morris A), pp. 71–85. Springer, New York. 10.1007/978-1-4939-2824-8_6 [DOI] [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, et al. 2020. Insights into human genetic variation and population history from 929 diverse genomes. Science 367: eaay5012. 10.1126/science.aay5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley AR, Standish KA, Bhutani K, Ideker T, Lasken RS, Carter H, Harismendy O, Schork NJ. 2017. Pan-cancer analysis reveals technical artifacts in TCGA germline variant calls. BMC Genomics 18: 458. 10.1186/s12864-017-3770-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, et al. 2020. A structural variation reference for medical and population genetics. Nature 581: 444–451. 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative. 2021. Mapping the human genetic architecture of COVID-19. Nature 600: 472–477. 10.1038/s41586-021-03767-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, 1000 Genomes Project, Bustamante CD. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci 108: 11983–11988. 10.1073/pnas.1019276108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highnam G, Wang JJ, Kusler D, Zook J, Vijayan V, Leibovich N, Mittelman D. 2015. An analytical framework for optimizing variant discovery from personal genomes. Nat Commun 6: 6275. 10.1038/ncomms7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly K. 2018. Hands-on data visualization with Bokeh: interactive web plotting for Python using Bokeh. Packt Publishing Ltd., Birmingham, UK. [Google Scholar]
- Karczewski K. 2017. The genome aggregation database (gnomAD). MacArthur Lab Blog. https://macarthurlab.org/2017/02/27/the-genome-aggregation-database-gnomad/ [accessed May 13, 2020]. [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V, Karlsson R, Frei O, Fan C-C, De Witte W, et al. 2019. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics 36: 930–933. 10.1093/bioinformatics/btz633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jew B, Zhan L, Hwang S, Coppola G, Freimer NB, Sul JH. 2019. ForestQC: quality control on genetic variants from next-generation sequencing data using random forest. PLoS Comput Biol 15: e1007556. 10.1371/journal.pcbi.1007556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2: 18–22. [Google Scholar]
- Ma X, Shao Y, Tian L, Flasch DA, Mulder HL, Edmonson MN, Liu Y, Chen X, Newman S, Nakitandwe J, et al. 2019. Analysis of error profiles in deep next-generation sequencing data. Genome Biol 20: 50. 10.1186/s13059-019-1659-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941. 10.1093/bioinformatics/bti623 [DOI] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001–D1006. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2011. ggplot2. Wiley Interdiscip Rev Comput Stat 3: 180–185. 10.1002/wics.147 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.