Abstract
The severe respiratory syndrome 2019 novel coronavirus disease (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread explosively, raising global health concerns. Luteolin shows antiviral properties, but its effect on SARS-CoV-2 and the associated mechanisms are not elucidated. We used network pharmacology, molecular docking and molecular dynamics to provide potential molecular support of luteolin (3,4,5,7-tetrahydroxyflavone) (LUT) against COVID-19. We employed network pharmacology, molecular docking, and molecular dynamics techniques to investigate how LUT affected COVID-19. Several databases were queried to determine potential target proteins related to LUT and COVID-19. Protein-protein interaction network was constructed, and core targets were filtered by degree value. Following that, functional enrichment was conducted. Molecular docking was utilized to ensure LUT was compatible with core target proteins. Finally, molecular dynamics was used to analyze the effects of the LUT on the optimal hub target. A total of 64 potential target genes for treating COVID-19 were identified, of which albumin, RAC-alpha serine/threonine-protein kinase, caspase-3, epidermal growth factor receptor, heat shock protein HSP 90-alpha, and mitogen-activated protein kinase 1 might be the most promising. In addition, molecular docking results showed that LUT could interact with SARS-CoV-2 major protease 3CL. LUT can bind to the active sites of 3CL protease and mitogen-activated protein kinase 1, showing an anti-SARS-CoV-2 potential.
Keywords: COVID-19, luteolin, molecular docking, network pharmacology, molecular dynamics
1. Introduction
Viral pneumonia caused by a highly contagious novel coronavirus (SARS-CoV-2) that first emerged at the end of 2019 is referred to by the World Health Organization as 2019 novel coronavirus disease (COVID-19),[1] and its rapid spread has posed severe challenges to healthcare systems in hundreds of countries worldwide. According to the World Health Organization Daily Report (https://covid19.who.int/), 492,189,439 COVID-19 cases have been confirmed, with 6159,474 deaths recorded as of April 6, 2022. The pandemic is ongoing and even intensifying, so it is critical to accelerate the development of novel preventive and therapeutic drugs.[2]
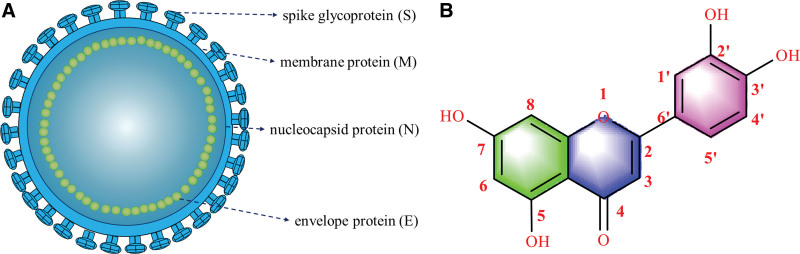
Individuals who contract COVID-19 typically present with fever, difficulty breathing, and fatigue, and a minority may have no symptoms.[3] COVID-19 confers severe conditions, including pneumonia, acute respiratory distress syndrome, multiorgan failure, and death.[4,5] Through these clinical consequences, combined with its globally rapid spread, the virus poses a serious challenge to public health.[6] So far, there exist no specific medicine available for this viral pneumonia and its complications. As a single-stranded RNA virus,[7] SARS-CoV-2 is constituted of 4 structural proteins, including nucleocapsid (N), envelope (E), membrane (M), and spike (S), suggested by the genome sequencing results (Fig. 1). In addition, RNA replication and reverse transcription are influenced by SARS-CoV-2-3CL hydrolase (Mpro) and 3CL proteinase.[8,9]
Figure 1.
. The structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (A) and luteolin (B).
Patients affected by COVID-19 have been reported to benefit from herbal treatments, highlighting the possibility of herbal compounds as potential agents for COVID-19 and its comorbidities.[10] 3′,4′,5,7-tetrahydroxyflavonoids, commonly known as luteolin (3,4,5,7-tetrahydroxyflavone) (LUT, PubChem CID: 5280445), is a natural flavonoid extracted from diverse vegetables and herbs with various biological benefits, including immunomodulatory, anti-inflammatory, and antioxidant properties.[11–15] Importantly, luteolin exerts significant antiviral effects as it can inhibit dengue virus, hepatitis B virus, and Epstein–Barr virus.[16–19] As shown in Figure 1, luteolin possesses hydroxyl (−OH) groups at positions 5, 7, 3′, and 4′ in the flavone backbone structure.
With network pharmacology, we can decode mechanisms of drugs with an overall viewpoint by focusing on “multiple components, multiple protein targets, multiple pathways” rather than single ones.[20–22] Network pharmacology is an effective and efficient tool extensively utilized to identify multiple targets for various diseases and explore other unknown mechanisms. More intriguingly, network pharmacology can be organically integrated with multidisciplinary technologies to construct complicated drug-target-diseases networks and clarify the mechanisms by which agents work.[23]
As a computational technology of drug design for traditional Chinese medicine (TCM), molecular docking applies stoichiometric calculation methods to simulate molecular geometry and the intermolecular interactions.[24] This technique can aid in the search for the optimal binding pattern between small-molecule agents with proteins of interest. Through the above 2 methods, published literatures have reported several Chinese medicines with potential favorable effects on COVID-19.[25] Molecular dynamics simulation allows capturing the behavior of biomolecules in atomic detail at excellent spatial and temporal resolutions. It determines particle velocity and position within the evolving system by generating successive system configurations over time.[26]
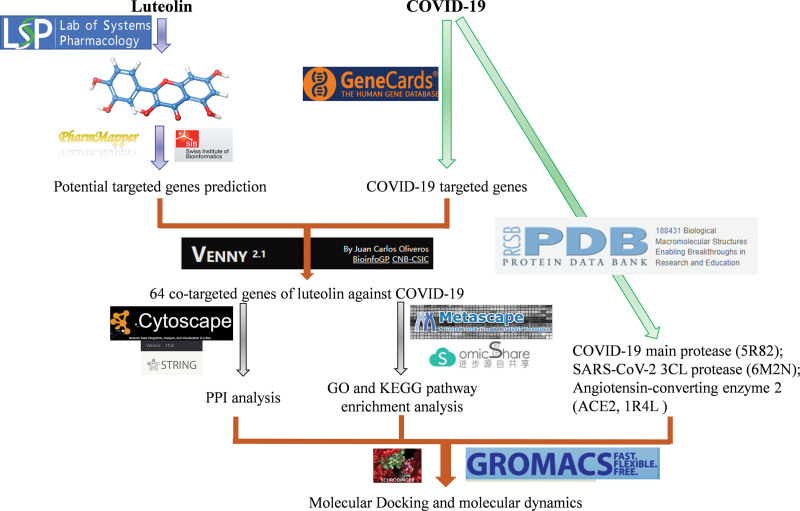
This study explored LUT and predicted its effective targets using network pharmacology and related technology. Consequently, we proposed potential molecular mechanisms underlying the effects of LUT in treating COVID-19, which might provide theoretical implications for the broader application of LUT in fighting COVID-19. The workflow is depicted in Figure 2.
Figure 2.
. Study workflow.
2. Materials and methods
2.1. Target proteins associated with LUT and their intersection with COVID-19
The interaction between active compounds and targets is the most critical stage of drug discovery.[27] Drug discovery pipelines begin to identify and validate drug-target interactions accurately.[28] Developing protein targets with traditional methods requires extensive workforce, material, and financial resources. Traditional Chinese medicine systems pharmacology database and analysis platform database (http://tcmspw.com/tcmsp.php) provided pharmacokinetics information like absorption, distribution, metabolism, and excretion, as well as oral bioavailability (OB) and drug-likeness (DL) related to LUT.[29] The potential targets for LUT were identified by searching 2 databases. We entered the simplified molecular input line entry system format into the SwissTargetPrediction database (http://swisstargetprediction.ch/) and the SDF format into the Pharmmapper Server database (http://www.lilab-ecust.cn/pharmmapper/) to access the target proteins of LUT. We profiled COVID-19-related targets through keyword searching including “coronavirus 2019, 2019-nCoV, COVID-19” in the GeneCards database (https://www.genecards.org/).[30] A literature search was then conducted to determine the authenticity of the related genes restricted to “homo sapiens.” The visualization of Venn plot was achieved by Venny 2.1.0 online tool (https://bioinfogp.cnb.csic.es/tools/venny/).
2.2. Protein-protein interaction construction and functional analysis
We established a protein-protein interaction (PPI) network by analyzing COVID-19 related target proteins on STRING 11.5 (https://string-db.org).[31] Based on degree values, the hub targets were screened. In the treatment of COVID-19, we judged the reliability of the target by the degree value, as a more significant value indicated that the target was more likely to be the core target of the LUT. Six core targets were identified with the highest degree in this study. Moreover, Cytoscape 3.8.2 was employed to construct, analyze, and visualize the PPI network.[32] We performed enrichment analysis regarding gene ontology (GO) and the pathway of proteins within the PPI network using the Metascape database.[33] P values derived from the Metascape database could represent the enrichment level, and smaller P values indicated more significant enrichment. Additionally, GO and Kyoto encyclopedia of genes and genomes (KEGG) enrichment and visualization were achieved using OmicShare (http://www.omicshare.com/tools, OmicShare).
2.3. Molecular docking
A molecular docking technique was utilized for verifying the affinity between LUT and the core target protein. We retrieved the crystal structures of selected targets from the RCSB PDB database (https://www.rcsb.org/).[34] To verify the network pharmacological results, we employed a professional software Maestro 11.1 for docking simulation and molecular pathway prediction. We further accessed the SDF format of LUTs via the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). In brief, we transformed the candidate target proteins into PDB ID and subsequently imported them into Maestro 11.1 for molecular docking. Low energy conformation of the ligand and receptor suggests a significant possibility of interaction. A docking score ≥4.25 is generally considered meaningful.[35] Six crucial targets and 3 joint receptors in clinical practice related to COVID-19 were selected to assess the binding activity between LUT and the target proteins. We set the ligand of related protein targets as positive controls.[36]
2.4. Molecular dynamics simulation
We validated the docking results using molecular dynamics simulations of the protein-ligand complex with the lowest binding energy. Target proteins with the most affinity for binding to LUT determined in dockings, such as the crucial target in the PPI network and 3CLpro enzyme, were selected to evaluate their inhibitory impact on LUT using molecular dynamics simulation. Binding free energies were calculated using the gmmpbsa program,[37] with the MM-PBSA method adapted for GROMACS. The best-docking scored models of the most promising LUT complexed with 3CLpro and mitogen-activated protein kinase 1 (MAPK1) were chosen as starting coordinates for 100 ns molecular dynamics simulations via the “GROMACS 2020.6 molecular dynamics” package (GNU, General Public License; http://www.gromacs.org) and CHARMM 36 force field.[38] In all processes, the electrostatic force was treated by the long-range electrostatic particle mesh (Particle Mesh Ewald PME) means, the intercept of van der Waals force was 1.2 nm, and the bonds of the hydrogen atoms were constrained by the Shake method. V-rescale and Berendsen controlled the temperature and pressure throughout the system. The step size is 2 fs during the molecular dynamic simulation, and the time interval of each dynamic trajectory saving is 1 ps. Finally, the root mean-square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bond, and free energy of the system were analyzed after 100 ns molecular dynamics simulation.
3. Results
3.1. Potential LUT-related COVID-19 target genes
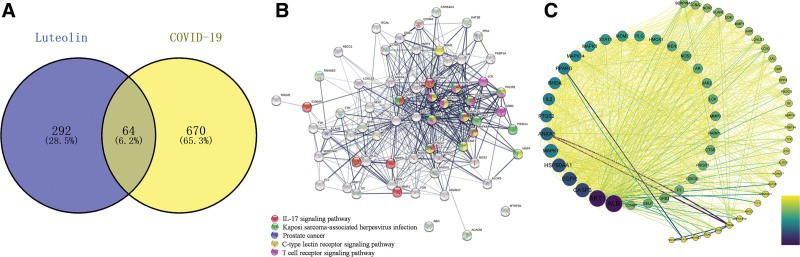
The form of luteolin is yellow needle-like crystal, and the pharmacokinetic properties of LUT can be found in Supplemental Digital Content (Table S1), http://links.lww.com/MD/J771. The OB and DL of LUT in our study were 36.16% and 0.25%, respectively. Finally, we collected 356 COVID-19-related target proteins (and corresponding genes) from the SwissTargetPrediction and PharmMapper databases and 734 target genes from the GeneCards databases. Furthermore, 64/1090 (5.9%) genes/proteins were identified as significant mediators for the effective therapy of LUT for COVID-19 (Fig. 3A), and Supplemental Digital Content (Table S2), http://links.lww.com/MD/J773 provides detailed information about these target genes. LUT may affect the combination of SARS-CoV-2 by acting on these 64 targets and eventually achieve its therapeutic effect on COVID-19.
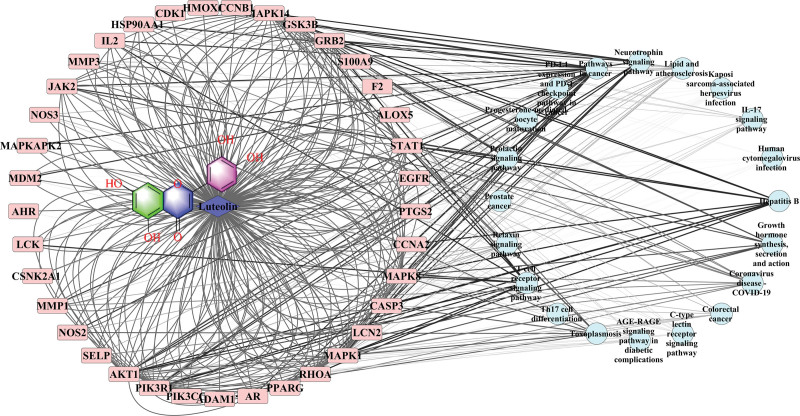
Figure 3.
. Protein-protein interaction (PPI) network (A) Venn diagram showing common targets of luteolin (3,4,5,7-tetrahydroxyflavone) (LUT) and COVID-19; (B) construction via String 11.5, the node color represented different signaling pathways as shown above; (C) constructed using Cytoscape 3.8.2; the node color had a positive association with its contribution to COVID-19).
3.2. PPI network construction
The interprotein interactions among those 64 targets were retrieved from a string database. The PPI network (Fig. 3B) was established to expound on the relationships among the 64 COVID-19-related targets; the PPI network incorporates 64 nodes, and 515 edges (Fig. 3C). The color of nodes is positively associated with their contribution degree in the network (Table 1). These targets might be significantly involved in COVID-19 development.
Table 1.
Degree values of crucial target proteins.
| Genes | Targets | Degree | UniProt ID | Betweenness centrality | Neighborhood connectivity |
|---|---|---|---|---|---|
| ALB | Albumin | 50 | P02768 | 4.258096 | 18.1 |
| AKT1 | RAC-alpha serine/threonine-protein kinase | 48 | P31749 | 4.295016 | 18.58333 |
| CASP3 | Caspase-3 | 38 | P42574 | 1.428342 | 21.5 |
| EGFR | Epidermal growth factor receptor | 37 | P00533 | 1.443815 | 21.05405 |
| HSP90AA1 | Heat shock protein HSP 90-alpha | 36 | P07900 | 1.437741 | 20.86111 |
| MAPK1 | Mitogen-activated protein kinase 1 | 29 | P28482 | 0.806769 | 22.96552 |
3.3. GO and KEGG enrichment analysis for targets
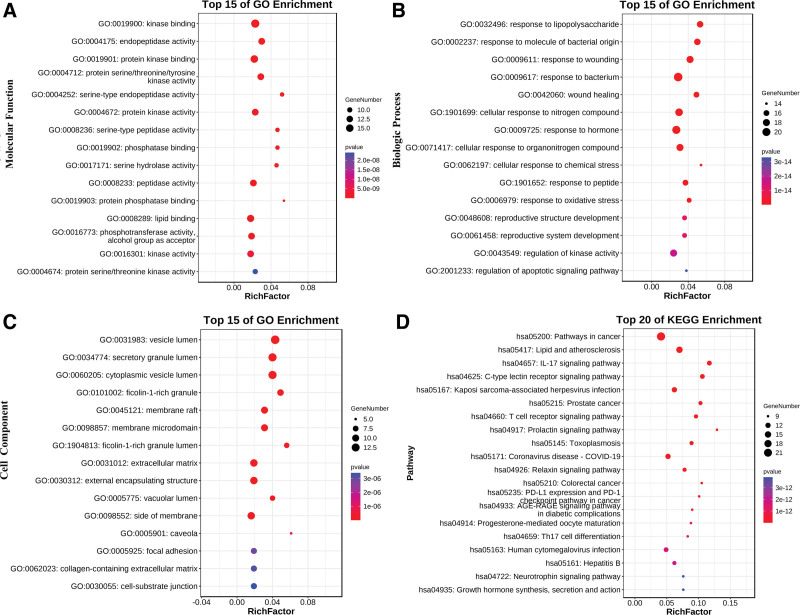
A total of 921 GO enrichment entries (top 15 were shown according to P values, Fig. 4A–C) were identified, of which 778 entries were associated with biological processes (Supplemental Digital Content, Table S3), http://links.lww.com/MD/J775, mainly including response to bacterium, response to molecule of bacterial origin, response to lipopolysaccharide, and response to tumor necrosis factor. Eighty-one terms fell in the context of molecular functions (Supplemental Digital Content, Table S4), http://links.lww.com/MD/J776, and 62 entries belonged to cell components entries (Supplemental Digital Content, Table S5), http://links.lww.com/MD/J777. A total of 137 pathways (Supplemental Digital Content, Table S6), http://links.lww.com/MD/J778 correlated to 64 target proteins were screened (top 20 were shown according to P value < .05, Fig. 4D) through KEGG analysis, of which 137 pathways mainly including lipid and atherosclerosis, IL-17 signaling pathway, coronavirus disease-COVID-19, and C-type lectin receptor signaling pathway.
Figure 4.
. Gene ontology (GO) function and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis results of molecular function (A), biological processes (B), cell composition (C), and KEGG pathway (D).
3.4. Compound-targets-pathways network analysis of LUT against COVID-19
We used KEGG pathway enrichment analysis to establish a component–targets–pathways network and further confirm the potential targets (Fig. 5). This network indicated that LUT interacted with 36 targets associated with COVID-19 through 20 pathways. These 36 target proteins might exert a crucial impact on the development of BC. However, further animal and cellular studies and clinical trials are necessary to confirm these speculations.
Figure 5.
. Network construction of component-targets-pathways.
3.5. Molecular docking
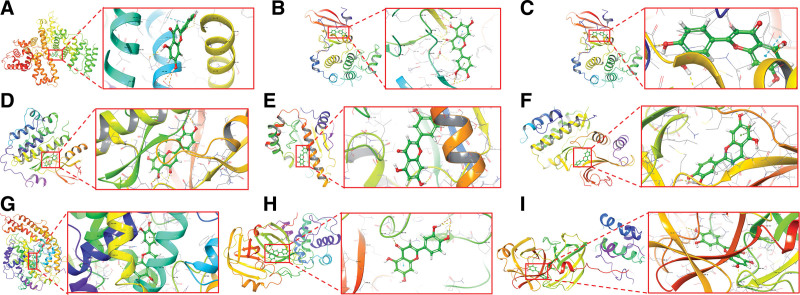
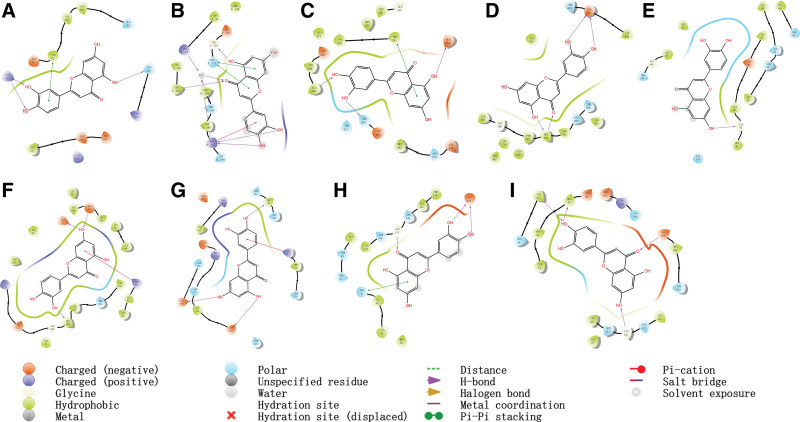
In this paper, molecular docking was utilized to simulate LUT with 6 corresponding crucial targets and 3 clinical targets, and we display the docking scores of LUT in Table 2. Finally, based on the docking score, we obtain the top 6 target proteins and 3 clinical targets, and the highest-scoring model is visualized in Figure 6 (3D) and Figure 7 (2D). Moreover, LUT forms hydrogen bonds with ASN 429 and LYS195 residues on albumin, with ALA230, GLU234, and THR291 residues on RAC-alpha serine/threonine-protein kinase, with GLY122 and ARG207 residues on caspase-3, with ASP855 and MET793 residues on epidermal growth factor receptor, with GLY97 residues on heat shock protein HSP 90-alpha, with GLU71, MET108, and ASP167 residues on MAPK1, with GLU406, PRO346, and ASP367 residues on angiotensin I converting enzyme 2, with GLU166 and GLY143 residues on the main protease, with GLU166, MET49, CYS44 and GLY143 residues on 3CL protease (Fig. 7). Molecular docking results revealed a negative binding free energy (DG in kcal/mol) of LUT to the core target, indicating a spontaneous ligand-receptor binding process. The crucial amino in these protein targets and the key sites of LUT is depicted in Figure 7.
Table 2.
Docking scores of luteolin (3,4,5,7-tetrahydroxyflavone) (LUT) bound to 6 crucial target proteins and 3 common clinical targets.
| Proteins | Compound | Glide gscore | Glide hbond | Glide evdw | Glide ecoul | Glide energy |
|---|---|---|---|---|---|---|
| ALB (1HK4) | Contrast | −6.123 | −0.442 | −36.688 | −9.173 | −45.862 |
| Luteolin | −7.285 | −0.289 | −31.373 | −9.605 | −40.978 | |
| AKT1 (3CQU) | Contrast | −9.465 | −0.608 | −46.455 | −7.047 | −53.503 |
| Luteolin | −8.122 | 0 | −34.412 | −11.862 | −46.274 | |
| CASP3 (1NMS) | Contrast | −5.558 | −0.674 | −32.781 | −6.181 | −38.962 |
| Luteolin | −5.85 | −0.152 | −23.962 | −13.848 | −37.81 | |
| EGFR (3POZ) | Contrast | −10.391 | −0.318 | −59.946 | −6.215 | −66.161 |
| Luteolin | −8.526 | −0.320 | −24.478 | −18.665 | −43.143 | |
| HSP90AA1 (2YI7) | Contrast | −7.609 | −0.172 | −41.046 | −10.810 | −51.856 |
| Luteolin | −7.161 | −0.320 | −35.367 | −7.523 | −42.890 | |
| MAPK1 (1PME) | Contrast | −7.914 | −0.370 | −37.564 | −9.106 | −46.669 |
| Luteolin | −9.015 | −0.320 | −46.420 | −5.544 | −51.964 | |
| ACE2 (1R4L) | Contrast | −8.602 | −0.874 | −39.865 | −17.627 | −57.492 |
| Luteolin | −6.328 | −0.144 | −32.543 | −14.439 | −46.982 | |
| Main protease (5R82) | Contrast | −5.559 | −0.32 | −20.16 | −5.642 | −25.802 |
| Luteolin | −6.045 | −0.289 | −18.973 | -−17.841 | −36.813 | |
| 3CL protease (6M2N) | Contrast | −7.732 | −0.391 | −34.23 | −17.627 | −43.793 |
| Luteolin | −8.020 | −0.596 | −38.311 | −14.439 | −45.522 |
Glide hbond refers to hydrogen bonding. Glide evdw refers to Van der Waals interactions. Glide ecoul refers to Coulomb energy. All items are components of the Glide gscore algorithm.
ACE2 = angiotensin I converting enzyme 2, AKT1 = RAC-alpha serine/threonine-protein kinase, ALB = albumin, CASP3 = caspase-3, EGFR = epidermal growth factor receptor, HSP90AA1 = heat shock protein HSP 90-alpha, MAPK1 = mitogen-activated protein kinase 1.
Figure 6.
. Molecular docking of luteolin and key targets (3D) (A) ALB_1H4K; (B) CASP3_1NMS; (C) AKT1_3CQU; (D) EGFR_3POZ; (E) HSP90AA1_2YI7; (F) MAPK1_1PME; (G) ACE2_1R4L; (H) Main protease_5R82; (I) 3CL protease_6M2N). ACE2 = angiotensin I converting enzyme 2, AKT1 = RAC-alpha serine/threonine-protein kinase, ALB = albumin, CASP3 = caspase-3, EGFR = epidermal growth factor receptor, HSP90AA1 = heat shock protein HSP 90-alpha, MAPK1 = mitogen-activated protein kinase 1.
Figure 7.
. Molecular docking of luteolin and key targets (2D) (A) ALB_1H4K; (B) CASP3_1NMS; (C) AKT1_3CQU; (D) EGFR_3POZ; (E) HSP90AA1_2YI7; (F) MAPK1_1PME; (G) ACE2_1R4L; (H) Main protease_5R82; (I) 3CL protease_6M2N). ACE2 = angiotensin I converting enzyme 2, ALB = albumin, CASP3 = caspase-3, EGFR = epidermal growth factor receptor, HSP90AA1 = heat shock protein HSP 90-alpha, MAPK1 = mitogen-activated protein kinase 1.
3.6. Molecular dynamics simulation
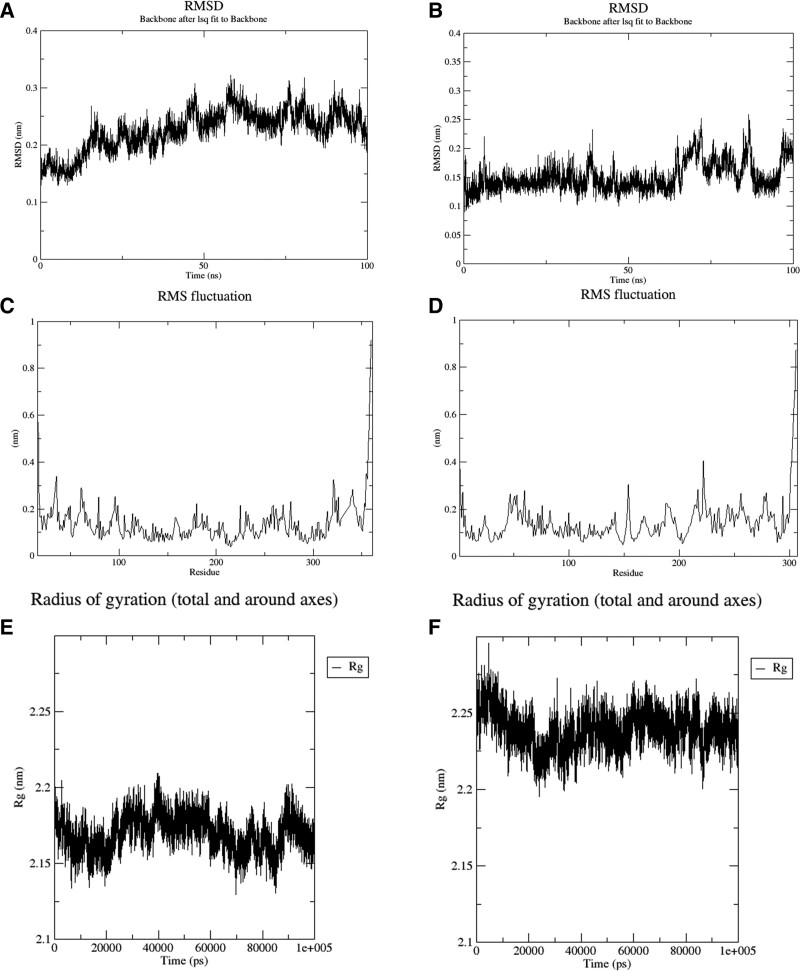
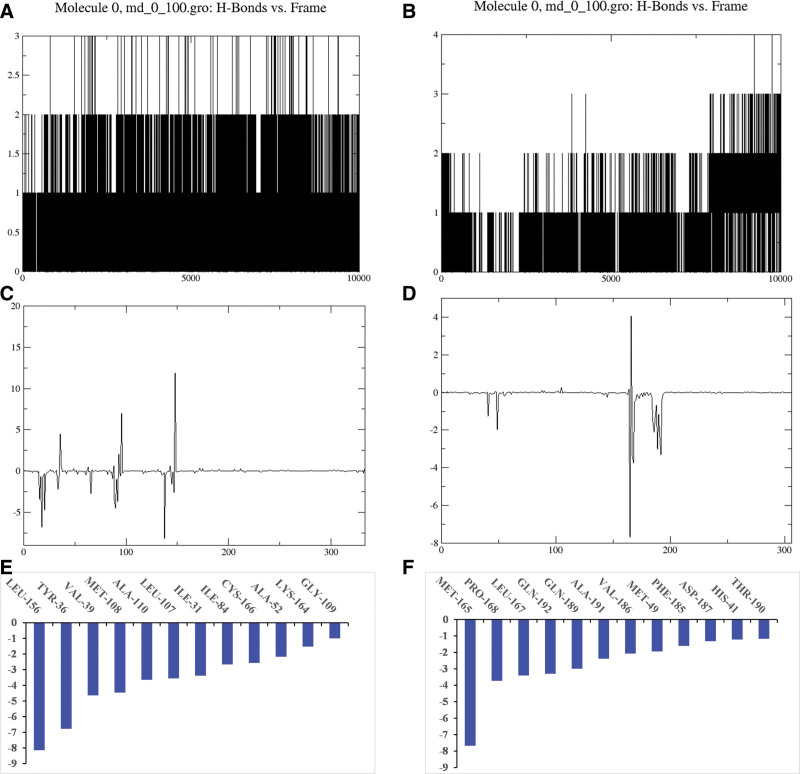
From the molecular docking results, MAPK1 and 3CL protease were selected for molecular dynamics simulation studies with LUT. While conducting all-atom MD simulation studies to validate the stability of the predicted protein complex, this study also provided interesting information on the dynamic behavior of ligands and target proteins and identified the ligand crucial interactions with recognized catalytic site residues. In a nutshell, the predicted ligand-protein complexes for LUT and the glycosylated MAPK1 protein and 3CL protease were simulated by MD simulation in 100 ns. Figure 8 depicts the changes in the RMSD, RMSF values and radius of gyration in the simulation of 3CL protease and MAPK1 at 100 ns simulation time. Although oscillation and instability were observed at the beginning of the simulation time, the system reached a steady-state after 10 ns, and the amount of RMSD fluctuations (<0.1 nm) decreased. Therefore, LUT is in the acceptable range of RMSD, which suggests that LUT is tightly connected within the cavity of MAPK1 and 3CL proteases. Similarly, LUT is in an acceptable range of RMSF and radius of gyration, which indicates that the binding pocket of MAPK1 and 3CL protease are stable during the MD simulation period. Moreover, Figure 9 revealed H-bond interactions during 100 ns simulation of LUT in complex with MAPK1 and 3CL protease protein. We also analyzed the positive effect on the free energy of the complex formed by LUT with MAPK1 and 3CL protease to determine the contribution of crucial amino acids to binding free energy. A lower binding free energy of the ligand and the protein receptor suggested a more significant promotion effect on the free energy of the complex formed between LUT, MAPK1, and 3CL protease.
Figure 8.
. Root mean-square deviation (RMSD), root mean square fluctuation (RMSF) values, and radius of gyration (Rg) during 100 ns simulation of MAPK1 (A, C, and E) and 3CL protease (B, D, and F) protein in complex with luteolin (3,4,5,7-tetrahydroxyflavone) (LUT). MAPK1 = mitogen-activated protein kinase 1.
Figure 9.
. H-bond interactions and binding free energy analysis during 100 ns MD simulation of luteolin (3,4,5,7-tetrahydroxyflavone) (LUT) in complex with MAPK1 (A, C, and E) and 3CL protease (B, D, and F) protein. MAPK1 = mitogen-activated protein kinase 1.
4. Discussion
The OB and DL of LUT in our study were 36.16% and 0.25% respectively, meeting the standard criteria in pharmacology: OB ≥ 30% and DL ≥ 0.18.[39] Nodes with darker shades could be readily noticed in the PPI network,[40] their degree values were 50, 48, 38, 37, 36, and 29, respectively. Then, the results of GO enrichment analysis suggested LUT with the potential in treating COVID-19 by regulating biological processes, molecular functions, cell components, and signaling pathways.[33] Moreover, molecular docking sheds new insight on the interaction between ligands and receptors and the structural characterization of ligands within the active site of receptors.[41] In December 2019, several hospitals successively received cases with a definite history of exposure to the seafood market in Wuhan, China, defined as an acute respiratory infectious disease caused by a novel coronavirus. Some modern agents have offered protection to vulnerable populations,[42] but the remarkable curative properties of TCM on the COVID-19 cannot be overlooked.
Currently, there is a limited arsenal available to help combat coronaviruses. Although several options, including vaccines, peptides, interferon therapies, and monoclonal antibodies, could be developed to control or prevent emerging infections of COVID-19, it requires months to years for ultimate approval. Generally, phytochemicals, notably polyphenols, possess antiviral activities against several viral components and actions.[43] Many recent studies have used molecular docking to screen for the ability of quercetin to interfere with the coronavirus S-protein, the ligand binding to angiotensin I converting enzyme 2.[44] Like quercetin, luteolin has also been shown to regulate cytokine storms in COVID-19 patients. Recently, a domestic efficacy analysis of TCM also suggested that luteolin contributed to the recovery of COVID-19.[45] Moreover, many researches could prove this point of view.
Network pharmacology, an intersectional discipline including classical pharmacology, molecular biology, and bioinformatics, is highly effective in identifying potential targets and pathways and drug-target interactions.[46] In addition, network pharmacology can be used to study complexities among drugs, target proteins, and diseases.[47] Currently, computer-aided drug design is considered a pivotal approach in modern drug discovery due to its beneficial properties, such as minimizing costs, accelerating the drug development process, and nonrequirement of specialist operators.[48] Therefore, molecular docking could accelerate the screening and design of drugs for future laboratory testing. Interestingly, in the recent pandemic, such technology has been able to determine the ability of various drugs and compounds and their potential mechanism to alleviate SARS-CoV-2 cytotoxicity. Molecular docking reliably predicts major binding trends between ligands and a specific protein, indicating how ligands inhibit it.
Potentially, the inactivation of p38 MAPK could serve as a therapeutic strategy for eliminating SARS-infected cells.[49] In addition, it could induce the production of cytokines, including interleukin-10, tumor necrosis factor-alpha, and interferon-gamma.[50] The MAPK inhibitor can alleviate COVID-19-induced inflammatory responses. Furthermore, inhibition of MAPK3 reduces the inflammatory response to lung injury, as MAPK3 exacerbates the production of pro-inflammatory cytokines.[51] This interaction between LUT and MAPK1 protein may be directly related to the antagonistic effect of LUT on COVID since we investigated and proved LUT interaction with MAPK1.
3CL protease is regarded as a potential target due to its unique function in the replication cycle of viruses.[52] Thus, we analyzed the interactions between LUT and 3CL protease. LUT fitted perfectly into the active pocket of 3CL protease, revealing its critical role in suppressing the virus. 3CL protease is an essential receptor for coronavirus, mediates endocytosis of SARS-CoV-2, and is necessary for drug profiling with the hope to impede COVID-19 progression. Structural theory and existing findings consistently suggest that targeting the 3CL protease, a critical factor in the viral replication cycle, might be a therapeutic target to address viral RNA translation. Therefore, we screened the interaction between 3CL protease and LUT and identified the active sites where LUT docked with the enzyme and 4 bonds involving amino acid residues CYS44, MET49, GLY143, and GLU166. Researches on the binding mode of the major protease of SARS-CoV-2 suggest that LUTs have a potential clinical application, and mutations in these interacting amino acids may inactivate the function of the major protease of the virus. The binding patterns found in this study demonstrate LUTs with potential anti-COVID-19 clinical applications and that mutations in the amino acids responsible for interaction could also hamper the functional activation of the major viral proteases.
Subsequent calculations of the binding free energy were conducted to elucidate the information on kinetic and thermodynamic properties and validate the docking results. Probably, the findings shaped an encouraging lead candidate for treatment agents against COVID-19. Moreover, molecular dynamics simulations with high temporal resolution and accuracy allow understanding of the behavior of biologically active molecules in atomic detail. In simple terms, by creating a continuous configuration of the evolving system, can generate trajectories that help determine the velocity and position of certain particles over time.[26]
We routinely selected ligand-protein complexes with the lowest binding energy for MD simulation to validate docking results and determine the impact of hydrogen-bonding interactions on complex stability. To ensure the complex stability, the system was simulated for 100 ns. And RMSD, RMSF, and RG were utilized to evaluate the potential of each trajectory. A duration of 100 ns allows the COVID-19 proteases to rearrange the C-a atoms in complexes with selected inhibitors. During 100 ns of simulation, we evaluated the thermodynamic conformational stability according to the RMSD and RMSF values. According to all results, LUT may have the most potential as an inhibitor of COVID-19.
There are some limitations to our study, even though we have described some interesting findings. First, the lack of a complete Chinese herbal medicine database confers uncertainty about the accuracy and integrity of our conclusions. Second, traditional Chinese medicine systems pharmacology database and analysis platform provides active compounds with OB ≥ 30% and DL ≥ 0.18, which may not be well absorbed. In addition, all the findings in this paper are drawn through in silico analysis that hampers ruling out false-positive and false-negative interactions between proteins. Our results require further experimental verification.
5. Conclusion
This study strives to offer a possible treatment option for COVID-19 and to predict the mechanism of LUT mitigation of COVID-19. Sixty-four target genes of LUT made significant contributions to COVID-19, and albumin, RAC-alpha serine/threonine-protein kinase, caspase-3, epidermal growth factor receptor, heat shock protein HSP 90-alpha, and MAPK1 were the crucial genes. The primary mechanism by which LUT inhibited COVID-19 was associated with the inhibition of 20 signaling pathways, specifically the inhibition of inflammation response through inactivating IL-17 signaling pathways and C-type lectin receptor signaling pathways. We proposed potential molecular mechanisms underlying the effects of LUT in treating COVID-19, which might provide theoretical support for the broader application of LUT in fighting COVID-19. At length, our results provided evidence for that LUT had potential anti-SARS-CoV-2 activity via integrating 3CL protease and MAPK1 protein compared with respective ligand, as shown by molecular dynamic simulation. These results may help elucidate the interactions between LUT and COVID-19-related proteins, as well as the fundamental applications of network pharmacology, molecular docking, and molecular dynamics in drug discovery.
Author contributions
Conceptualization: Wenxiang Wang, Wei Xiong.
Data curation: Wenxiang Wang, Ce Yang, Jing Xia.
Funding acquisition: Wenxiang Wang, Ning Li.
Writing – original draft: Wenxiang Wang, Ce Yang.
Writing – review & editing: Ning Li, Wei Xiong.
Supplementary Material
Abbreviations:
- ALB
- albumin
- CASP3
- caspase-3
- COVID-19
- 2019 novel coronavirus disease
- DL
- drug-likeness
- EGFR
- epidermal growth factor receptor
- GO
- gene ontology
- KEGG
- Kyoto encyclopedia of genes and genomes
- LUT
- luteolin (3,4,5,7-tetrahydroxyflavone)
- MAPK1
- mitogen-activated protein kinase 1
- OB
- oral bioavailability
- PPI
- protein-protein interaction
- RMSD
- root mean-square deviation
- RMSF
- root mean square fluctuation
- SARS-CoV-2
- severe acute respiratory syndrome coronavirus 2
- TCM
- traditional Chinese medicine
This work was supported by the Science and Technology Research Program of Chongqing Municipal Education Commission [grant number KJZD-K202202701, KJQN202002710, KJQN2022027], Natural Science Foundation of Chongqing (grant number cstc2020jcyj-msxmX1049), Doctor Direct Train Project of Wanzhou District (wzstc-20220125), and the Natural Science Major Project of Chongqing Three Gorges Medical College [grant number XJ2021000301].
Supplemental Digital Content is available for this article.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
How to cite this article: Wang W, Yang C, Xia J, Li N, Xiong W. Luteolin is a potential inhibitor of COVID-19: An in silico analysis. Medicine 2023;102:38(e35029).
Contributor Information
Wenxiang Wang, Email: wangwenxiang@cqtgmc.edu.cn.
Ce Yang, Email: yangce2024@163.com.
Jing Xia, Email: xiajing0510@163.com.
Ning Li, Email: lining20230508@163.com.
References
- [1].Zhu N, Zhang DY, Wang WL, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wu Y, Wang FR, Shen CG, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].WHO. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [access date December 23, 2020].
- [4].Chen T, Wu D, Chen HL, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ghaebi M, Tahmasebi S, Jozghorban M, et al. Risk factors for adverse outcomes of COVID-19 patients: possible basis for diverse responses to the novel coronavirus SARS-CoV-2. Life Sci. 2021;277:119503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu CR, Liu Y, Yang YY, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang LL, Lin DZ, Sun XYY, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang Y, Islam MS, Wang J, et al. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16:1708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hytti M, Piippo N, Korhonen E, et al. Fisetin and luteolin protect human retinal pigment epithelial cells from oxidative stress- induced cell death and regulate inflammation. Sci Rep. 2015;5:17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bai L, Nong YH, Shi Y, et al. Luteolin inhibits hepatitis B virus replication through extra cellular signal regulated kinase mediated down-regulation of hepatocyte nuclear factor 4α expression. Mol Pharm. 2016;13:568–77. [DOI] [PubMed] [Google Scholar]
- [13].Yan JQ, Wang Q, Zheng XL, et al. Luteolin enhances TNF-related apoptosis-inducing ligand’s anticancer activity in a lung cancer xenograft mouse model. Biochem Biophys Res Commun. 2012;417:842–6. [DOI] [PubMed] [Google Scholar]
- [14].Lin Y, Shi RX, Wang X, et al. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr Cancer Drug Targets. 2008;8:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Miean KH, Mohamed S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. J Agric Food Chem. 2001;49:3106–12. [DOI] [PubMed] [Google Scholar]
- [16].Wu CC, Fang CY, Hsu HY, et al. Luteolin inhibits Epstein-Barr virus lytic reactivation by repressing the promoter activities of immediate-early genes. Antiviral Res. 2016;132:99–110. [DOI] [PubMed] [Google Scholar]
- [17].Peng MH, Watanabe S, Chan KWK, et al. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antiviral Res. 2017;143:176–85. [DOI] [PubMed] [Google Scholar]
- [18].Fan WC, Qian SH, Qian P, et al. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–6. [DOI] [PubMed] [Google Scholar]
- [19].Manvar D, Mishra M, Kumar S, et al. Identification and evaluation of anti hepatitis C virus phytochemicals from Eclipta alba. J Ethnopharmacol. 2012;144:545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oh KK, Adnan M, Ju I, et al. A network pharmacology study on main chemical compounds from Hibiscus cannabinus L. leaves. RSC Adv. 2021;11:11062–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oh KK, Adnan M, Cho DH. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS One. 2020;15:e0240873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Oh KK, Adnan M, Cho DH. Active ingredients and mechanisms of Phellinus linteus (grown on Rosa multiflora) for alleviation of Type 2 diabetes mellitus through network pharmacology. Gene. 2021;768:145320. [DOI] [PubMed] [Google Scholar]
- [23].Luo TT, Lu Y, Yan SK, et al. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med. 2020;26:72–80. [DOI] [PubMed] [Google Scholar]
- [24].Wang ZF, Hu YQ, Wu QG, et al. Virtual screening of potential antifatigue mechanism of Polygonati Rhizoma based on network pharmacology. Comb Chem High Throughput Screen. 2019;22:612–24. [DOI] [PubMed] [Google Scholar]
- [25].Gu YY, Zhang M, Cen H, et al. Quercetin as a potential treatment for COVID-19-induced acute kidney injury: based on network pharmacology and molecular docking study. PLoS One. 2021;16:e0245209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron. 2018;99:1129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Núñez S, Venhorst J, Kruse CG. Target-drug interactions: first principles and their application to drug discovery. Drug Discov Today. 2012;17:10–22. [DOI] [PubMed] [Google Scholar]
- [28].Chen X, Clarence Yan CG, Zhang XT, et al. Drug-target interaction prediction: databases, web servers and computational models. Brief Bioinform. 2016;17:696–712. [DOI] [PubMed] [Google Scholar]
- [29].Ru JL, Li P, Wang JN, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. [DOI] [PubMed] [Google Scholar]
- [31].Kushwaha SK, Shakya M. Protein interaction network analysis-approach for potential drug target identification in Mycobacterium tuberculosis. J Theor Biol. 2010;262:284–94. [DOI] [PubMed] [Google Scholar]
- [32].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou YY, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Joosten RP, Te Beek TAH, Krieger E, et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8:e83922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Deng WN, Wang LN, Cui YL. Research progress on drug targets of anti-corona virus disease 2019. Drugs Clin. 2020;35:401–4. [Google Scholar]
- [37].Kumari R, Kumar R, Lynn A. g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–62. [DOI] [PubMed] [Google Scholar]
- [38].da Silva TU, de Carvalho Pougy K, Albuquerque MG, et al. Development of parameters compatible with the CHARMM36 force field for [Fe4S4]2+ clusters and molecular dynamics simulations of adenosine-5’-phosphosulfate reductase in GROMACS 2019. J Biomol Struct Dyn. 2022;40:3481–91. [DOI] [PubMed] [Google Scholar]
- [39].Ursu O, Rayan A, Goldblum A, et al. Understanding drug-likeness. WIREs Comput Mol Sci. 2011;1:760–81. [Google Scholar]
- [40].Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl4):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. 2019;20:4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li XQ, Zhang CB, Liu LY, et al. Existing bitter medicines for fighting 2019-nCoV-associated infectious diseases. FASEB J. 2020;34:6008–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Levy E, Delvin E, Marcil V, et al. May phytotherapy with polyphenols serve as a powerful approach for the prevention and therapy tool of novel coronavirus disease 2019 (COVID-19)? AJP Endocrinol Metab. 2020;319:E689–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sargiacomo C, Sotgia F, Lisanti MP. COVID-19 and chronological aging: senolytics and other anti-aging drugs for the treatment or prevention of corona virus infection? Aging (Milano). 2020;12:6511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Luo E, Zhang DY, Luo H, et al. Treatment efficacy analysis of traditional Chinese medicine for novel coronavirus pneumonia (COVID-19): an empirical study from Wuhan, Hubei Province, China. Chin Med. 2020;15:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–90. [DOI] [PubMed] [Google Scholar]
- [47].Zheng CL, Wang JN, Liu JL, et al. System-level multi-target drug discovery from natural products with applications to cardiovascular diseases. Mol Divers. 2014;18:621–35. [DOI] [PubMed] [Google Scholar]
- [48].Islam R, Parves MR, Paul AS, et al. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2021;39:3213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mizutani T, Fukushi S, Saijo M, et al. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun. 2004;319:1228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].De Souza AP, Costa Vale VL, Costa Silva MD, et al. MAPK involvement in cytokine production in response to Corynebacterium pseudotuberculosis infection. BMC Microbiol. 2014;14:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Di Paola R, Crisafulli C, Mazzon E, et al. Effect of PD98059, a selective MAPK3/MAPK1 inhibitor, on acute lung injury in mice. Int J Immunopathol Pharmacol. 2009;22:937–50. [DOI] [PubMed] [Google Scholar]
- [52].Zumla A, Chan JF, Azhar EI, et al. Coronaviruses-drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.