Abstract
Factors that render patients with cardiovascular disease at high risk for heart failure remain incompletely defined. Recent insights into molecular genetic causes of myocardial diseases have highlighted the importance of single-gene defects in the pathogenesis of heart failure. Through analyses of the mechanisms by which a mutation selectively perturbs one component of cardiac physiology and triggers cell and molecular responses, studies of human gene mutations provide a window into the complex processes of cardiac remodeling and heart failure. Knowledge gleaned from these studies shows promise for defining novel therapeutic targets for genetic and acquired causes of heart failure.
Introduction
Despite considerable public awareness and technological advances that foster early diagnosis and aggressive therapeutic interventions for cardiovascular disease, heart failure, a pathophysiologic state in which blood delivery is inadequate for tissue requirements, remains a critical unsolved problem. Reaching almost epidemic proportions, heart failure currently affects 4.8 million Americans, and each year over 500,000 new cases are diagnosed. These statistics are associated with considerable social and economic costs. In 2003 heart failure contributed to over 280,000 deaths and accounted for 17.8 billion health care dollars (1).
Heart failure is a complex pathophysiologic state in which delivery of blood and nutrients is inadequate for tissue requirements. Heart failure almost universally arises in the context of antecedent cardiovascular disease: atherosclerosis, cardiomyopathy, myocarditis, congenital malformations, or valvular disease. Knowledge of why some patients with these conditions are at high risk for heart failure is incomplete, and deciphering critical mediators from nonspecific factors is often difficult in the context of underlying heart disease. The study of single-gene mutations that trigger heart failure provides an opportunity for defining important molecules involved in these processes. Although these monogenic disorders account for only a small subset of overall heart failure cases, insights into the responses triggered by gene mutations are likely to also be relevant to more common etiologies of heart failure.
The earliest clinical manifestation of an underlying genetic cause of heart failure is ventricular remodeling. One of 2 distinct morphologies occurs: left ventricular hypertrophy (increased wall thickness without chamber expansion) or dilation (normal or thinned walls with enlarged chamber volumes). Each is associated with specific hemodynamic changes. Systolic function is normal, but diastolic relaxation is impaired in hypertrophic remodeling; diminished systolic function characterizes dilated remodeling. Clinical recognition of these cardiac findings usually prompts diagnosis of hypertrophic cardiomyopathy (HCM) or dilated cardiomyopathy (DCM). These were once considered as specific diagnoses, but there is now considerable evidence that many different gene mutations can cause these pathologies (Figure 1), and with these discoveries has come recognition of distinct histopathologic features that further delineate several subtypes of remodeling. While compilation of the full repertoire of genes that remodel the heart remains a work in progress, the current compendia already suggest a multiplicity of pathways by which the human heart can fail. To facilitate a discussion, we have grouped known cardiomyopathy genes according to the probable functional consequences of mutations on force generation and transmission, metabolism, calcium homeostasis, or transcriptional control. This approach, while convenient for organization, is undoubtedly somewhat arbitrary; gene mutations in one functional category inevitably have an impact on multiple myocyte processes. Indeed, the eventual delineation of signals between functional groups may be critical to understanding nodal points that result in, or protect against, cardiac decompensation and heart failure development.
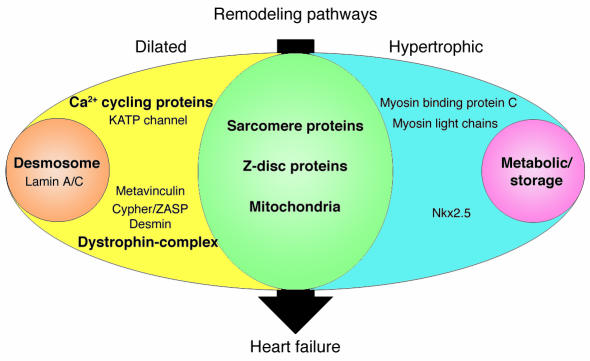
Figure 1.
Human gene mutations can cause cardiac hypertrophy (blue), dilation (yellow), or both (green). In addition to these two patterns of remodeling, particular gene defects produce hypertrophic remodeling with glycogen accumulation (pink) or dilated remodeling with fibrofatty degeneration of the myocardium (orange). Sarcomere proteins denote β-myosin heavy chain, cardiac troponin T, cardiac troponin I, α-tropomyosin, cardiac actin, and titin. Metabolic/storage proteins denote AMP-activated protein kinase γ subunit, LAMP2, lysosomal acid α 1,4–glucosidase, and lysosomal hydrolase α-galactosidase A. Z-disc proteins denote MLP and telethonin. Dystrophin-complex proteins denote δ-sarcoglycan, β-sarcoglycan, and dystrophin. Ca2+ cycling proteins denote PLN and RyR2. Desmosome proteins denote plakoglobin, desmoplakin, and plakophilin-2.
Force generation and propagation
Generation of contractile force by the sarcomere and its transmission to the extracellular matrix are the fundamental functions of heart cells. Inadequate performance in either component prompts cardiac remodeling (hypertrophy or dilation), produces symptoms, and leads to heart failure. Given the importance of these processes for normal heart function and overt clinical manifestations of deficits in either force generation or transmission, it is not surprising that more single-gene mutations have been identified in molecules involved in these critical processes than in those of other functional classes (Figure 2).
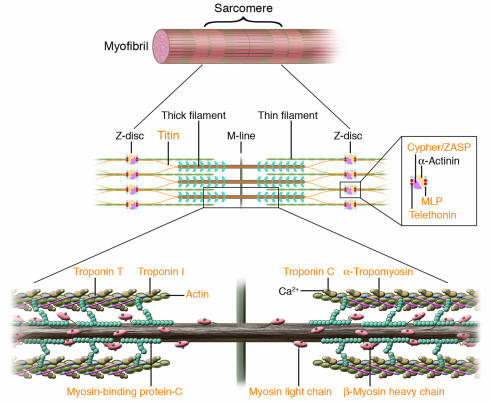
Figure 2.
Human mutations affecting contractile and Z-disc proteins. The schematic depicts one sarcomere, the fundamental unit of contraction encompassing the protein segment between flanking Z discs. Sarcomere thin filament proteins are composed of actin and troponins C, T, and I. Sarcomere thick filament proteins include myosin heavy chain, myosin essential and regulatory light chains, myosin-binding protein-C and titin. The sarcomere is anchored through titin and actin interactions with Z disc proteins α-actinin, calsarcin-1, MLP, telethonin (T-cap), and ZASP. Human mutations (orange text) in contractile proteins and Z-disc proteins can cause HCM or DCM.
Sarcomere protein mutations.
Human mutations in the genes encoding protein components of the sarcomere cause either HCM or DCM. While progression to heart failure occurs with both patterns of remodeling, the histopathology, hemodynamic profiles, and biophysical consequences of HCM or DCM mutations suggest that distinct molecular processes are involved.
Over 300 dominant mutations in genes encoding β-cardiac myosin heavy chain (MYH7), cardiac myosin-binding protein-C (MYBPC3), cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), essential myosin light chain (MYL3), regulatory myosin light chain (MYL2), α-tropomyosin (TPM1), cardiac actin (ACTC), and titin (TTN) have been reported to cause HCM (Figure 2) (2, 3). Recent reports of comprehensive sequencing of sarcomere protein genes in diverse patient populations indicate that MYBPC3 and MYH7 mutations are most frequent (4, 5). Sarcomere gene mutations that cause HCM produce a shared histopathology with enlarged myocytes that are disorganized and die prematurely, which results in increased cardiac fibrosis. The severity and pattern of ventricular hypertrophy, age at onset of clinical manifestations, and progression to heart failure are in part dependent on the precise sarcomere protein gene mutation. For example, TNNT2 mutations are generally associated with a high incidence of sudden death despite only mild left ventricular hypertrophy (6, 7). While only a small subset (10–15%) of HCM patients develop heart failure, this end-stage phenotype has a markedly poor prognosis and often necessitates cardiac transplantation. Accelerated clinical deterioration has been observed with MYH7 Arg719Trp, TNNT2 Lys273Glu, TNNI3 Lys183del, and TPM1 Glu180Val mutations (8–11).
Most HCM mutations encode defective polypeptides containing missense residues or small deletions; these are likely to be stably incorporated into cardiac myofilaments and to produce hypertrophy because normal sarcomere function is disturbed. Many HCM mutations in MYBPC3 fall within carboxyl domains that interact with titin and myosin; however, the exact biophysical properties altered by these defects remain unknown (Figure 2). HCM mutations in myosin are found in virtually every functional domain, which suggests that the biophysical consequences of these defects may vary. Genetic engineering of some human myosin mutations into mice has indicated more consistent sequelae. Isolated single-mutant myosin molecules containing different HCM mutations had increased actin-activated ATPase activity and showed greater force production and faster actin-filament sliding, biophysical properties that may account for hyperdynamic contractile performance observed in HCM hearts and that suggest a mechanism for premature myocyte death in HCM (12–14). Uncoordinated contraction due to heterogeneity of mutant and normal sarcomere proteins, increased energy consumption, and changes in Ca2+ homeostasis could diminish myocyte survival and trigger replacement fibrosis. With insidious myocyte loss and increased fibrosis, the HCM heart transitions from hypertrophy to failure.
Mice that are engineered to carry a sarcomere mutation replicate the genetics of human disease; heterozygous mutations cause HCM. One exception is a deletion of proximal myosin-binding protein-C sequences; heterozygous mutant mice exhibited normal heart structure while homozygous mutant mice developed hypertrophy (15). Remarkably, while most heterozygous mouse models with a mutation in myosin heavy chain, myosin-binding protein-C, or troponin T developed HCM (16–18), homozygous mutant mice (19, 20) developed DCM with fulminant heart failure and, in some cases, premature death. These mouse studies might indicate that HCM, DCM, and heart failure reflect gradations of a single molecular pathway. Alternatively, significant myocyte death caused by homozygous sarcomere mutations may result in heart failure. Human data suggest a more complicated scenario. The clinical phenotype of rare individuals who carry homozygous sarcomere mutations in either MYH7 (21) or in TNNT2 (22) is severe hypertrophy, not DCM. Furthermore, individuals with compound heterozygous sarcomere mutations exhibit HCM, not DCM. The absence of ventricular dilation in human hearts with 2 copies of mutant sarcomere proteins is consistent with distinct cellular signaling programs that remodel the heart into hypertrophic or dilated morphologies.
DCM sarcomere protein gene mutations affect distinct amino acids from HCM-causing mutations, although the proximity of altered residues is remarkable. The histopathology of sarcomere DCM mutations is quite different from those causing HCM, and is remarkably nonspecific. Degenerating myocytes with increased interstitial fibrosis are present, but myocyte disarray is notably absent. There are 2 mechanisms by which sarcomere mutations may cause DCM and heart failure: deficits of force production and deficits of force transmission. Diminished force may occur in myosin mutations (e.g., MYH7 Ser532Pro) that alter actin-binding residues involved in initiating the power stroke of contraction. Impaired contractile force may also occur in DCM troponin mutations (TNNT2 ΔLys210, ref. 23; and TNNI3 Ala2Val, ref. 24) that alter residues implicated in tight binary troponin interactions. Because troponin molecules modulate calcium-stimulated actomyosin ATPase activity, these defects may cause inefficient ATP hydrolysis and therein decrease contractile power.
Other DCM sarcomere mutations are more likely to impair force transmission (Figure 2). For example, a myosin mutation (at residue 764) located within the flexible fulcrum that transmits movement from the head of myosin to the thick filament is likely to render ineffectual the force generated by actomyosin interactions (23). DCM TPM1 mutations (25) are predicted to destabilize actin interactions and compromise force transmission to neighboring sarcomere. Likewise, ACTC mutations (26) that impair binding of actin to Z-disc may compromise force propagation. TTN mutations provide quintessential evidence that deficits in force transmission cause DCM and heart failure. By spanning the sarcomere from Z-disc to M-line, this giant muscle protein assembles contractile filaments and provides elasticity through serial spring elements. Titin interacts with α-actinin and telethonin (T-cap) at the Z-disc, with calpain3 and obscurin at the I-band (the extensible thin filament regions flanking Z-discs), and with myosin-binding protein-C, calmodulin, and calpain3 at the M-line region. Human mutations identified in the Z-disc–I-band transition zone (27), in the telethonin and α-actinin–binding domain, and in the cardiac-specific N2B domain (an I-band subregion; ref. 28) each cause DCM and heart failure.
Intermediate filaments and dystrophin-associated glycoprotein mutations.
Intermediate filaments function as cytoskeletal proteins linking the Z-disc to the sarcolemma. Desmin is a type III intermediate filament protein, which, when mutated, causes skeletal and cardiac muscle disease (Figure 3). The hearts of mice deficient in desmin (29) are more susceptible to mechanical stress, which is consistent with the function of intermediate proteins in force transmission.
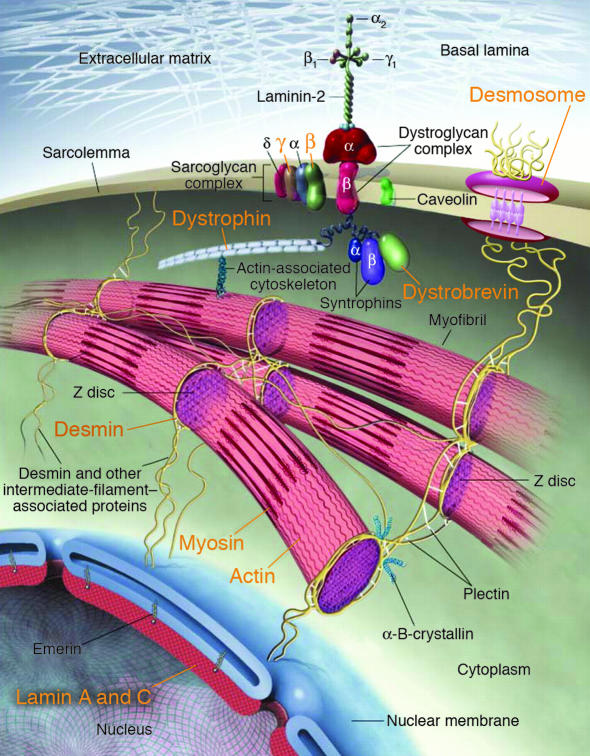
Figure 3.
Human mutations (orange text) in components of myocyte cytoarchitecture cause DCM and heart failure. Force produced by sarcomeric actin-myosin interactions is propagated through the actin cytoskeleton and dystrophin to the dystrophin-associated glycoprotein complex (composed of α- and β-dystroglycans, α-, β-, γ- and δ-sarcoglycans, caveolin-3, syntrophin, and dystrobrevin). Desmosome proteins plakoglobin, desmoplakin, and plakophilin-2, provide functional and structural contacts between adjacent cells and are linked through intermediate filament proteins, including desmin, to the nuclear membrane, where lamin A/C is localized. Adapted from ref. 96.
Through dystrophin and actin interactions, the dystrophin-associated glycoprotein complex (composed of α- and β-dystroglycans, α-, β-, γ- and δ-sarcoglycans, caveolin-3, syntrophin, and dystrobrevin) provides stability to the sarcomere and transmits force to the extracellular matrix. Human mutations in these proteins cause muscular dystrophy with associated DCM and heart failure (Figure 3). Skeletal muscle manifestations can be minimal in female carriers of X-linked dystrophin defects, and some individuals present primarily with heart failure (30). In the mouse experiment, coxsackievirus B3–encoded protease2A, which can cleave dystrophin, was shown to produce sarcolemmal disruption and cause DCM, which suggests that dystrophin is also involved in the pathologic mechanism of DCM and heart failure that follow viral myocarditis (31).
While deficiencies of proteins that link the sarcomere to the extracellular matrix are likely to impair force transmission, recent studies of mice engineered to carry mutations in these molecules indicate other mechanisms for heart failure. A model of desmin-related cardiomyopathies (32) uncovered striking intracellular aggresomes, electron dense accumulations of heat shock and chaperone protein, α-B-crystalline, desmin, and amyloid in association with sarcomeres. While particularly abundant in the amyloid heart, aggresomes were also found in some DCM and HCM specimens, which suggests that excessive degenerative processing induced by myocyte stress or gene mutation may be toxic to sarcomere function.
Analyses of δ-sarcoglycan null mice (33) also yielded unexpected disease mechanisms, primary coronary vasospasm and myocardial ischemia. Selective restoration of δ-sarcoglycan to the cardiac myocytes extinguished this pathology, thereby implicating chronic ischemia as a contributing factor to heart failure development in patients with sarcoglycan mutations.
Mutations in intercalated and Z-disc proteins.
To generate contraction, one end of each actin thin filament must be immobilized. The Z-disc defines the lateral boundary of the sarcomere, where actin filaments, titin, and nebulette filaments are anchored. Metavinculin provides attachment of thin filaments to the plasma membrane and plays a key role in productive force transmission. Two metavinculin gene mutations cause DCM by disruption of disc structure and actin-filament organization (34).
Other Z-disc protein constituents may also function as mechano-stretch receptors (35). Critical components include α-actinin, which aligns actin and titin from neighboring sarcomeres and interacts with muscle LIM protein (MLP encoded by CSRP3), telethonin (encoded by TCAP), which interacts with titin and MLP to subserve overall sarcomere function, and Cypher/Z-band alternatively spliced PDZ-motif protein (Cypher/ZASP), a striated muscle-restricted protein that interacts with α-actinin–2 through a PDZ domain and couples to PKC-mediated signaling via its LIM domains (Figure 2). Mutations in these molecules cause either DCM (35, 36) or HCM (37, 38) and predispose the affected individuals to heart failure. Genetically engineered mice with MLP deficiency (39) help to model the mechanism by which mutations in distinct proteins cause disease. Without MLP, telethonin is destabilized and gradually lost from the Z-disc; as a consequence, MLP-deficient cardiac papillary muscle shows an impairment in tension generation following the delivery of a 10% increase in passive stretch of the muscle and a loss of stretch-dependent induction of molecular markers (e.g., brain natriuretic peptide), which suggests that an MLP-telethonin–titin complex is an essential component of the cardiac muscle mechanical stretch sensor machinery. An important question is how signaling proteins (e.g., Cyper/ZASP) within the Z-disc translate mechanosensing into activation of survival or cell death pathways.
Lamin A/C mutations.
The inner nuclear-membrane protein complex contains emerin and lamin A/C. Defects in emerin cause X-linked Emery-Dreifuss muscular dystrophy, joint contractures, conduction system disease, and DCM. Dominant lamin A/C mutations exhibit a more cardiac-restricted phenotype with fibrofatty degeneration of the myocardium and conducting cells, although subclinical involvement of skeletal muscles and contractures are sometimes apparent. The remarkable electrophysiologic deficits (progressive atrioventricular block and atrial arrhythmias) observed in mutations of lamin A/C and emerin indicate the particular importance of these proteins in electrophysiologic cells. A recent study of lamin A/C mutant mice showed evidence of marked nuclear deformation, fragmentation of heterochromatin, and defects in mechanotransduction (40, 41), all of which likely contribute to reduced myocyte viability. The similarities of cardiac histopathology (fibrofatty degeneration) observed in mutations of the nuclear envelope and desmosomes raise the possibility that these structures may both function as important mechanosensors in myocytes (Figure 3).
Desmosome protein mutations.
Arrhythmogenic right ventricular cardiomyopathy (ARVD) identifies an unusual group of cardiomyopathies characterized by progressive fibrofatty degeneration of the myocardium, electrical instability, and sudden death (42). While right ventricular dysplasia predominates, involvement of the left ventricle also occurs. Progressive myocardial dysfunction is seen late in the course of disease, often with right-sided heart failure. ARVD occurs in isolation or in the context of Naxos syndrome, an inherited syndrome characterized by prominent skin (palmar-plantar keratosis), hair, and cardiac manifestations. Mutations in protein components of the desmosomes (Figure 3) (plakoglobin, ref. 43; desmoplakin, refs. 44, 45; and plakophilin-2, ref. 46) and in the cardiac ryanodine receptor (RyR2) (ref. 47; discussed below) cause syndromic and nonsydromic ARVD. Desmosomes are organized cell membrane structures that provide functional and structural contacts between adjacent cells and that may be involved in signaling processes. Whether mutations in the desmosomal proteins render cells of the heart (and skin) inappropriately sensitive to normal mechanical stress or cause dysplasia via another mechanism is unknown.
Energy production and regulation
Mitochondrial mutations.
Five critical multiprotein complexes, located within the mitochondria, synthesize ATP by oxidative phosphorylation. While many of the protein components of these complexes are encoded by the nuclear genome, 13 are encoded by the mitochondrial genome. Unlike nuclear gene mutations, mitochondrial gene mutations exhibit matrilineal inheritance. In addition, the mitochondrial genome is present in multiple copies, and mutations are often heteroplasmic, affecting some but not all copies. These complexities, coupled with the dependence of virtually all tissues on mitochondrial-derived energy supplies, account for the considerable clinical diversity of mitochondrial gene mutations (Figure 4). While most defects cause either dilated or hypertrophic cardiac remodeling in the context of mitochondrial syndromes such as Kearns-Sayre syndrome, ocular myopathy, mitochondrial encephalomyopathy with lactic-acidosis and stroke-like episodes (MELAS), and myoclonus epilepsy with ragged-red fibers (MERFF) (48), there is some evidence that particular mitochondrial mutations can produce predominant or exclusive cardiac disease (49, 50). An association between heteroplasmic mitochondrial mutations and DCM has been recognized (51).
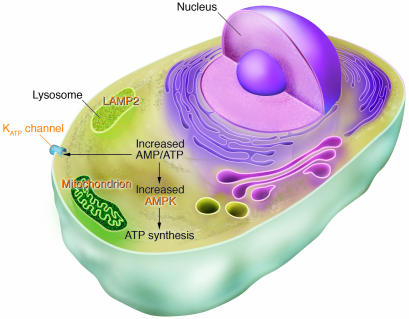
Figure 4.
Human gene mutations affecting cardiac energetics and metabolism. Energy substrate utilization is directed by critical metabolic sensors in myocytes, including AMP-activated protein kinase (AMPK), which, in response to increased AMP/ATP levels, phosphorylates target proteins and thereby regulates glycogen and fatty acid metabolism, critical energy sources for the heart. Glycogen metabolism involves a large number of proteins including α-galactosidase A (mutated in Fabry disease) and LAMP2 (mutated in Danon disease). Glycogen and fatty acids are substrates for multiprotein complexes located within the mitochondria for the synthesis of ATP. KATP channels composed of an enzyme complex and a potassium pore participate in decoding metabolic signals to maximize cellular functions during stress adaptation. Human mutations (orange text) that cause cardiomyopathies have been identified in the regulatory SUR2A subunit of KATP, the γ2 subunit of AMPK, mitochondrial proteins, α-galactosidase A, and LAMP2.
Nuclear-encoded metabolic mutations.
Nuclear gene mutations affecting key regulators of cardiac metabolism are emerging as recognized causes of hypertrophic cardiac remodeling and heart failure (Figure 4). Mutations in genes encoding the γ2 subunit of AMP-activated protein kinase (PRKAG2), α-galactosidase A (GLA), and lysosome-associated membrane protein-2 (LAMP2) can cause profound myocardial hypertrophy in association with electrophysiologic defects (52). AMP-activated protein kinase functions as a metabolic-stress sensor in all cells. This heterotrimeric enzyme complex becomes activated during energy-deficiency states (low ATP, high ADP) and modulates (by phosphorylation) a large number of proteins involved in cell metabolism and energy (53). Most GLA mutations can cause multisystem classic Fabry disease (angiokeratoma, corneal dystrophy, renal insufficiency, acroparesthesia, and cardiac hypertrophy), but some defects produce primarily cardiomyopathy. LAMP2 mutations can also produce either multisystem Danon disease (with skeletal muscle, neurologic, and hepatic manifestations) or a more restricted cardiac phenotype.
Cardiac histopathology reveals that, unlike sarcomere gene mutations, which cause hypertrophic remodeling, the mutations in PRKAG2, LAMP2, and GLA accumulate glycogen in complexes with protein and/or lipids, thereby defining these pathologies as storage cardiomyopathies. Progression from hypertrophy to heart failure is particularly common and occurs earlier with LAMP2 mutations than with other gene mutations that cause metabolic cardiomyopathies. Since both GLA and LAMP2 are encoded on chromosome X, disease expression is more severe in men, but heterozygous mutations in women are not entirely benign, perhaps due to X-inactivation that equally extinguishes a normal or mutant allele. The cellular and molecular pathways that produce either profound hypertrophy or progression to heart failure from PRKAG2, GLA, or LAMP2 mutations are incompletely understood. While accumulated byproducts are likely to produce toxicity, animal models indicate that mutant proteins cause far more profound consequences by changing cardiac metabolism and altering cell signaling. This is particularly evident in PRKAG2 mutations that increase glucose uptake by stimulating translocation of the glucose transporter GLUT-4 to the plasma membrane, increase hexokinase activity, and alter expression of signaling cascades (54).
The cooccurrence of electrophysiologic defects in metabolic mutations raises the possibility that pathologic cardiac conduction and arrhythmias contribute to cardiac remodeling and heart failure in these gene mutations. One mechanism for electrophysiologic defects appears to be the direct consequence of storage: transgenic mice that express a human PRKAG2 mutation (55) developed ventricular pre-excitation due to pathologic atrioventricular connections by glycogen-filled myocytes that ruptured the annulus fibrosis (the normal anatomic insulator which separates atrial and ventricular myocytes). A second and unknown mechanism may be that these gene defects are particularly deleterious to specialized cells of the conduction system. Little is known about the metabolism of these cells, although historical histopathologic data indicate glycogen to be particularly more abundant in the conduction system than in the working myocardium (56–58).
Ca2+ cycling
Considerable evidence indicates the presence of abnormalities in myocyte calcium homeostasis to be a prevalent and important mechanism for heart failure. Protein and RNA levels of key calcium modulators are altered in acquired and inherited forms of heart failure, and human mutations in molecules directly involved in calcium cycling have been found in several cardiomyopathies (Figure 5).
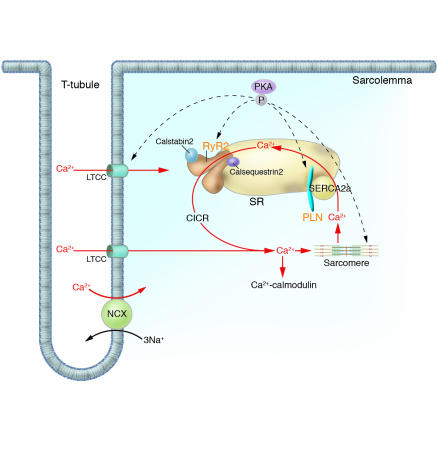
Figure 5.
Human mutations affecting Ca2+ cycling proteins. Intracellular Ca2+ handling is the central coordinator of cardiac contraction and relaxation. Ca2+ entering through L-type channels (LTCC) triggers Ca2+ release (CICR) from the SR via the RyR2, and sarcomere contraction is initiated. Relaxation occurs with SR Ca2+ reuptake through the SERCA2a. Calstabin2 coordinates excitation and contraction by modulating RyR2 release of Ca2+. PLN, an SR transmembrane inhibitor of SERCA2a modulates Ca2+ reuptake. Dynamic regulation of these molecules is effected by PKA-mediated phosphorylation. Ca2+ may further function as a universal signaling molecule, stimulating Ca2+-calmodulin and other molecular cascades. Human mutations (orange text) in molecules involved in calcium cycling cause cardiac remodeling and heart failure. NCX, sodium/calcium exchanger.
Calcium enters the myocyte through voltage-gated L-type Ca2+ channels; this triggers release of calcium from the sarcoplasmic reticulum (SR) via the RyR2. Emerging data define FK506-binding protein (FKBP12.6; calstabin2) as a critical stabilizer of RyR2 function (59), preventing aberrant calcium release during the relaxation phase of the cardiac cycle (Figure 5). Stimuli that phosphorylate RyR2 (such as exercise) by protein kinase A (PKA) dissociate calstabin2 from the receptor, thereby increasing calcium release and enhancing contractility. At low concentrations of intracellular calcium, troponin I and actin interactions block actomyosin ATPase activity; increasing levels foster calcium binding to troponin C, which releases troponin I inhibition and stimulates contraction. Cardiac relaxation occurs when calcium dissociates from troponin C, and intracellular concentrations decline as calcium reuptake into the SR occurs through the cardiac sarcoplasmic reticulum Ca2+-ATPase pump (SERCA2a). Calcium reuptake into SR is regulated by phospholamban (PLN), an inhibitor of SERCA2a activity that when phosphorylated dissociates from SERCA2a and accelerates ventricular relaxation.
RyR2 mutations.
While some mutations in the RyR2 are reported to cause ARVD (47) (see discussion of desmosome mutations), defects in this calcium channel are more often associated with catecholaminergic polymorphic ventricular tachycardia (60, 61), a rare inherited arrhythmic disorder characterized by normal heart structure and sudden cardiac death during physical or emotional stress. Mutations in calsequestrin2, an SR calcium-binding protein that interacts with RyR2, also cause catecholaminergic polymorphic ventricular tachycardia (62, 63). Whether the effect of calsequestrin2 mutations directly or indirectly alters RyR2 function is unknown (Figure 5).
While RyR2 mutations affect residues in multiple functional domains of the calcium channel, those affecting residues involved in calstabin2-binding provide mechanistic insights into the substantial arrhythmias found in affected individuals. Mutations that impair calstabin2-binding may foster calcium leak from the SR and trigger depolarization. Diastolic calcium leak can also affect excitation-contraction coupling and impair systolic contractility.
Studies of mice deficient in FKBP12.6 (64) confirmed the relevance of SR calcium leak from RyR2 to clinically important arrhythmias. RyR2 channel activity in FKBP12.6-null mice was significantly increased compared with that of wild-type mice, consistent with a diastolic Ca2+ leak. Mutant myocytes demonstrated delayed after-depolarizations, and exercise-induced syncope, ventricular arrhythmias, and sudden death were observed in FKBP12.6-null mice.
Calcium dysregulation is also a component of hypertrophic remodeling that occurs in sarcomere gene mutations. Calcium cycling is abnormal early in the pathogenesis of murine HCM (65, 66): SR calcium stores are decreased and calcium-binding proteins and RyR2 levels are diminished. Whether calcium changes contribute to ventricular arrhythmias in mouse and human HCM remains an intriguing question.
Related mechanisms may contribute to ventricular dysfunction and arrhythmias in acquired forms of heart failure, in which chronic phosphorylation of RyR2 reduces calstabin2 levels in the channel macromolecular complex and increases calcium loss from SR stores. These data indicate the potential benefit of therapeutics that improve calstabin2-mediated stabilization of RyR2 (67, 68); such agents may both improve ventricular contractility and suppress arrhythmias in heart failure.
PLN mutations.
Rare human PLN mutations cause familial DCM and heart failure (69, 70). The pathogenetic mechanism of one mutation (PLN Arg9Cys) was elucidated through biochemical studies, which indicated unusual PKA interactions that inhibited phosphorylation of mutant and wild-type PLN. The functional consequence of the mutation was predicted to be constitutive inhibition of SERCA2a, a result confirmed in transgenic mice expressing mutant, but not wild-type, PLN protein. In mutant transgenic mice, calcium transients were markedly prolonged, myocyte relaxation was delayed, and these abnormalities were unresponsive to β-adrenergic stimulation. Profound biventricular cardiac dilation and heart failure developed in mutant mice, providing clear evidence of the detrimental effects of protracted SERCA2a inhibition due to excess PLN activity.
The biophysical consequences accounting for DCM in humans who are homozygous for a PLN null mutation (Leu39stop; ref. 70) are less clear. PLN-deficient mice show increased calcium reuptake into the SR and enhanced basal contractility (71). Indeed, these effects on calcium cycling appear to account for the mechanism by which PLN ablation rescues DCM in MLP-null mice (72). However, normal responsiveness to β-adrenergic stimulation is blunted in PLN-deficient myocytes, and cells are less able to recover from acidosis that accompanies vigorous contraction or pathologic states, such as ischemia (73). The collective lesson from human PLN mutations appears to be that too little or too much PLN activity is bad for long-term heart function.
Acquired causes of heart failure are also characterized by a relative decrease in SERCA2a function due to excessive PLN inhibition. Downregulation of β-adrenergic responsiveness attenuates PLN phosphorylation, which compromises calcium reuptake and depletes SR calcium levels, which may impair contractile force and enhance arrhythmias. Heterozygote SERCA2 null mice are a good model of this phenotype and exhibit impaired restoration of SR calcium with deficits in systolic and diastolic function (74).
Cardiac ATP-sensitive potassium channel mutations.
In response to stress such as hypoxia and ischemia, myocardial cells undergo considerable changes in metabolism and membrane excitability. Cardiac ATP-sensitive potassium channels (KATP channels) contain a potassium pore and an enzyme complex that participate in decoding metabolic signals to maximize cellular functions during stress adaptation (Figure 4) (75). KATP channels are multimeric proteins containing the inwardly rectifying potassium channel pore (Kir6.2) and the regulatory SUR2A subunit, an ATPase-harboring, ATP-binding cassette protein. Recently, human mutations in the regulatory SUR2A subunit (encoded by ABCC9) were identified as a cause of DCM and heart failure (76). These mutations reduced ATP hydrolytic activities, rendered the channels insensitive to ADP-induced conformations, and affected channel opening and closure. Since KATP-null mouse hearts have impaired response to stress and are susceptible to calcium overload (75), some of the pathophysiology of human KATP mutations (DCM and arrhythmias) may reflect calcium increases triggered by myocyte stress.
Transcriptional regulators
Investigation of the molecular controls of cardiac gene transcription has led to the identification of many key molecules that orchestrate physiologic expression of proteins involved in force production and transmission, metabolism, and calcium cycling. Given that mutation in the structural proteins involved in these complex processes is sufficient to cause cardiac remodeling, it is surprising that defects in transcriptional regulation of these same proteins have not also been identified as primary causes of heart failure. Several possible explanations may account for this. Transcription factor gene mutations may be lethal or may at least substantially impair reproductive fitness so as to be rapidly lost. The consequences of transcription factor gene mutations may be so pleiotropic that these cause systemic rather than single-organ disease. Changes in protein function (produced by a structural protein mutation) may be more potent for remodeling than changes in levels of structural protein (produced by transcription factor mutation). While many other explanations may be relevant, the few human defects discovered in transcriptional regulators that cause heart failure provide an important opportunity to understand molecular mechanisms for heart failure.
Nkx2.5 mutations.
The homeodomain-containing transcription factor Nkx2.5, a vertebrate homolog of the Drosophila homeobox gene tinman, is one of the earliest markers of mesoderm. When Nkx2.5 is deleted in the fly, cardiac development is lost (77). Targeted disruption of Nkx2.5 in mice (Nkx2.5–/–) causes embryonic lethality due to the arrested looping morphogenesis of the heart tube and growth retardation (78, 79). Multiple human dominant Nkx2.5 mutations have been identified as causing primarily structural malformations (atrial and ventricular septation defects) accompanied by atrioventricular conduction delay, although cardiac hypertrophic remodeling has also been observed (80). Although the mechanism for ventricular hypertrophy in humans with Nkx2.5 mutations is not fully understood, the pathology is unlike that found in HCM, which perhaps indicates that cardiac hypertrophy is a compensatory event. Several human Nkx2.5 mutations have been shown to abrogate DNA binding (81), which suggests that the level of functional transcription factor is the principle determinant of structural phenotypes. Heterozygous Nkx2.5+/– mice exhibit only congenital malformations with atrioventricular conduction delay (82, 83). Remarkably, however, transgenic mice expressing Nkx2.5 mutations develop profound cardiac conduction disease and heart failure (84) and exhibit increased sensitivity to doxorubicin-induced apoptosis (85), which suggests that this transcription factor plays an important role in postnatal heart function and stress response.
Insights into transcriptional regulation from mouse genetics.
Dissection of the combinatorial mechanisms that activate or repress cardiac gene transcription has led to the identification of several key molecules that directly or indirectly lead to cardiac remodeling. While human mutations in these genes have not been identified, these molecules are excellent candidates for triggering cell responses to structural protein gene mutations.
Hypertrophic remodeling is associated with reexpression of cardiac fetal genes. Molecules that activate this program may also regulate genes that directly cause hypertrophy. Activation of calcineurin (Ca2+/calmodulin-dependent serine/threonine phosphatase) results in dephosphorylation and nuclear translocation of nuclear factor of activated T cells 3 (NFAT3), which, in association with the zinc finger transcription factor GATA4, induces cardiac fetal gene expression. Transgenic mice that express activated calcineurin or NFAT3 in the heart develop profound hypertrophy and progressive decompensation to heart failure (86), responses that were prevented by pharmacologic inhibition of calcineurin. Although these data implicated NFAT signaling in hypertrophic heart failure, pharmacologic inhibition of this pathway fails to prevent hypertrophy caused by sarcomere gene mutations in mice and even accelerates disease progression to heart failure (65). Mice lacking calsarcin-1, which is localized with calcineurin to the Z-disc, showed an increase in Z-disc width, marked activation of the fetal gene program, and exaggerated hypertrophy in response to calcineurin activation or mechanical stress, which suggests that calsarcin-1 plays a critical role in linking mechanical stretch sensor machinery to the calcineurin-dependent hypertrophic pathway (87).
Histone deacetylases (HDACs) are emerging as important regulators of cardiac gene transcription. Class II HDACs (4/5/7/9) bind to the cardiac gene transcription factor MEF2 and inhibit MEF2-target gene expression. Stress-responsive HDAC kinases continue to be identified but may include an important calcium-responsive cardiac protein, calmodulin kinase. Kinase-induced phosphorylation of class II HDACs causes nuclear exit, thereby releasing MEF2 for association with histone acetyltransferase proteins (p300/CBP) and activation of hypertrophic genes. Mice deficient in HDAC9 are sensitized to hypertrophic signals and exhibit stress-dependent cardiac hypertrophy. The discovery that HDAC kinase is stimulated by calcineurin (88) implicates crosstalk between these hypertrophic signaling pathways.
Recent attention has also been focused on Hop, an atypical homeodomain-only protein that lacks DNA-binding activity. Hop is expressed in the developing heart, downstream of Nkx2-5. While its functions are not fully elucidated, Hop can repress serum response factor–mediated (SRF-mediated) transcription. Mice with Hop gene ablation have complex phenotypes. Approximately half of Hop-null embryos succumb during mid-gestation with poorly developed myocardium; some have myocardial rupture and pericardial effusion. Other Hop-null embryos survive to adulthood with apparently normal heart structure and function. Cardiac transgenic overexpression of epitope-tagged Hop causes hypertrophy, possibly by recruitment of class I HDACs that may inhibit anti-hypertrophic gene expression (89–92).
PPARα plays important roles in transcriptional control of metabolic genes, particularly those involved in cardiac fatty acid uptake and oxidation. Mice with cardiac-restricted overexpression of PPARα replicate the phenotype of diabetic cardiomyopathy: hypertrophy, fetal gene activation, and systolic ventricular dysfunction (93). Heterozygous PPARγ-deficient mice, when subjected to pressure overload, developed greater hypertrophic remodeling than wild-type controls, implicating the PPARγ-pathway as a protective mechanism for hypertrophy and heart failure (94).
Retinoid X receptor α (RXRα) is a retinoid-dependent transcriptional regulator that binds DNA as an RXR/retinoic acid receptor (RXR/RAR) heterodimer. RXRα-null mice die during embryogenesis with hypoplasia of the ventricular myocardium. In contrast, overexpression of RXRα in the heart does not rescue myocardial hypoplasia but causes DCM (95).
Integrating functional and molecular signals
Study of human gene mutations that cause HCM and DCM provides information about functional triggers of cardiac remodeling. In parallel with evolving information about molecular-signaling cascades that influence cardiac gene expression, there is considerable opportunity to define precise pathways that cause the heart to fail. To understand the integration of functional triggers with molecular responses, a comprehensive data set of the transcriptional and proteomic profiles associated with precise gene mutations is needed. Despite the plethora of information associated with such studies, bioinformatic assembly of data and deduction of pathways should be feasible and productive for defining shared or distinct responses to signals that cause cardiac remodeling and heart failure. Accrual of this data set in humans is a desirable goal, although confounding clinical variables and tissue acquisition pose considerable difficulties that can be more readily addressed by study of animal models with heart disease. With more knowledge about the pathways involved in HCM and DCM, strategies may emerge to attenuate hypertrophy, reduce myocyte death, and diminish myocardial fibrosis, processes that ultimately cause the heart to fail.
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institute and the NIH.
Footnotes
Nonstandard abbreviations used: ARVD, arrhythmogenic right ventricular cardiomyopathy; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyo-pathy; HDAC, histone deacetylase; KATP channel, cardiac ATP-sensitive potassium channel; LAMP2, lysosome-associated membrane protein-2; MLP, muscle LIM protein; NFAT, nuclear factor of activated T cells; PLN, phospholamban; RXRα, retinoid X receptor α; RyR2, cardiac ryanodine receptor; SERCA2a, cardiac sarcoplasmic reticulum Ca2+-ATPase pump; SR, sarcoplasmic reticulum; ZASP, Z-band alternatively spliced PDZ-motif protein.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.American Heart Association. 2004. Heart disease and stroke statistics – 2004 update. American Heart Association. http://www.americanheart.org.
- 2.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 3.CardioGenomics. Genomics of cardiovascular development, adaptation, and remodeling. NHLBI program for genomic applications. Harvard Medical School. http://cardiogenomics.med.harvard.edu.
- 4.Morita H, et al. Molecular epidemiology of hypertrophic cardiomyopathy. Cold Spring Harb. Symp. Quant. Biol. 2002;67:383–388. doi: 10.1101/sqb.2002.67.383. [DOI] [PubMed] [Google Scholar]
- 5.Richard P, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 6.Watkins H, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N. Engl. J. Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 7.Moolman JC, et al. Sudden death due to troponin T mutations. J. Am. Coll. Cardiol. 1997;29:549–555. doi: 10.1016/s0735-1097(96)00530-x. [DOI] [PubMed] [Google Scholar]
- 8.Anan R, et al. Prognostic implications of novel β cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J. Clin. Invest. 1994;93:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujino N, et al. A novel mutation Lys273Glu in the cardiac troponin T gene shows high degree of penetrance and transition from hypertrophic to dilated cardiomyopathy. Am. J. Cardiol. 2002;89:29–33. doi: 10.1016/s0002-9149(01)02158-0. [DOI] [PubMed] [Google Scholar]
- 10.Kokado H, et al. Clinical features of hypertrophic cardiomyopathy caused by a Lys183 deletion mutation in the cardiac troponin I gene. Circulation. 2000;102:663–669. doi: 10.1161/01.cir.102.6.663. [DOI] [PubMed] [Google Scholar]
- 11.Regitz-Zagrosek V, Erdmann J, Wellnhofer E, Raible J, Fleck E. Novel mutation in the alpha-tropomyosin gene and transition from hypertrophic to hypocontractile dilated cardiomyopathy. Circulation. 2000;102:E112–E116. doi: 10.1161/01.cir.102.17.e112. [DOI] [PubMed] [Google Scholar]
- 12.Tyska MJ, et al. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ. Res. 2000;86:737–744. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 13.Palmer BM, et al. Differential cross-bridge kinetics of FHC myosin mutations R403Q and R453C in heterozygous mouse myocardium. Am. J. Physiol. Heart Circ. Physiol. 2004;287:H91–H99. doi: 10.1152/ajpheart.01015.2003. [DOI] [PubMed] [Google Scholar]
- 14.Palmer BM, et al. Effect of cardiac myosin binding protein-C on mechanoenergetics in mouse myocardium. Circ. Res. 2004;94:1615–1622. doi: 10.1161/01.RES.0000132744.08754.f2. [DOI] [PubMed] [Google Scholar]
- 15.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 16.Geisterfer-Lowrance AA, et al. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 17.Carrier L, et al. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc. Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Oberst L, et al. Dominant-negative effect of a mutant cardiac troponin T on cardiac structure and function in transgenic mice. J. Clin. Invest. 1998;102:1498–1505. doi: 10.1172/JCI4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatkin D, et al. Neonatal cardiomyopathy in mice homozygous for the Arg403Gln mutation in the α cardiac myosin heavy chain gene. J. Clin. Invest. 1999;103:147–153. doi: 10.1172/JCI4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConnell BK, et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishi H, et al. Possible gene dose effect of a mutant cardiac beta-myosin heavy chain gene on the clinical expression of familial hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 1994;200:549–556. doi: 10.1006/bbrc.1994.1483. [DOI] [PubMed] [Google Scholar]
- 22.Ho CY, et al. Homozygous mutation in cardiac troponin T: implications for hypertrophic cardiomyopathy. Circulation. 2000;102:1950–1955. doi: 10.1161/01.cir.102.16.1950. [DOI] [PubMed] [Google Scholar]
- 23.Kamisago M, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 24.Murphy RT, et al. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 25.Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2001;33:723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 26.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 27.Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 28.Itoh-Satoh M, et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;291:385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- 29.Milner DJ, Weitzer G, Tran D, Bradley A, Capetanaki Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J. Cell Biol. 1996;134:1255–1270. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franz WM, et al. Association of nonsense mutation of dystrophin gene with disruption of sarcoglycan complex in X-linked dilated cardiomyopathy. Lancet. 2000;355:1781–1785. doi: 10.1016/S0140-6736(00)02266-2. [DOI] [PubMed] [Google Scholar]
- 31.Badorff C, et al. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat. Med. 1999;5:320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 32.Sanbe A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler MT, et al. Smooth muscle cell–extrinsic vascular spasm arises from cardiomyocyte degeneration in sarcoglycan-deficient cardiomyopathy. J. Clin. Invest. 2004;113:668–675. doi:10.1172/JCI200420410. doi: 10.1172/JCI20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson TM, et al. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 35.Knoll R, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 36.Vatta M, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Geier C, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi T, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 39.Arber S, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 40.Nikolova V, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C–deficient mice. J. Clin. Invest. 2004;113:357–369. doi:10.1172/JCI200419448. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi:10.1172/JCI200419670. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna WJ, et al. Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Task Force of the Working Group Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology. Br. Heart J. 1994;71:215–218. doi: 10.1136/hrt.71.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKoy G, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 44.Norgett EE, et al. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 2000;9:2761–2766. doi: 10.1093/hmg/9.18.2761. [DOI] [PubMed] [Google Scholar]
- 45.Rampazzo A, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am. J. Hum. Genet. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerull B, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 47.Tiso N, et al. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum. Mol. Genet. 2001;10:189–194. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 48.Anan R, et al. Cardiac involvement in mitochondrial diseases. A study on 17 patients with documented mitochondrial DNA defects. Circulation. 1995;91:955–961. doi: 10.1161/01.cir.91.4.955. [DOI] [PubMed] [Google Scholar]
- 49.Santorelli FM, et al. Maternally inherited cardiomyopathy and hearing loss associated with a novel mutation in the mitochondrial tRNA(Lys) gene (G8363A) Am. J. Hum. Genet. 1996;58:933–939. [PMC free article] [PubMed] [Google Scholar]
- 50.Santorelli FM, et al. Maternally inherited cardiomyopathy: an atypical presentation of the mtDNA 12S rRNA gene A1555G mutation. Am. J. Hum. Genet. 1999;64:295–300. doi: 10.1086/302188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arbustini E, et al. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am. J. Pathol. 1998;153:1501–1510. doi: 10.1016/S0002-9440(10)65738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 2002;11:2499–2506. doi: 10.1093/hmg/11.20.2499. [DOI] [PubMed] [Google Scholar]
- 53.Kemp BE, et al. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 2003;31:162–168. doi: 10.1042/bst0310162. [DOI] [PubMed] [Google Scholar]
- 54.Tian R, Musi N, D’Agostino J, Hirshman MF, Goodyear LJ. Increased adenosine monophosphate-activated protein kinase activity in rat hearts with pressure-overload hypertrophy. Circulation. 2001;104:1664–1669. doi: 10.1161/hc4001.097183. [DOI] [PubMed] [Google Scholar]
- 55.Arad M, et al. Transgenic mice overexpressing mutant PRKAG2 define the cause of Wolff-Parkinson-White syndrome in glycogen storage cardiomyopathy. Circulation. 2003;107:2850–2856. doi: 10.1161/01.CIR.0000075270.13497.2B. [DOI] [PubMed] [Google Scholar]
- 56.Henry CG, Lowry OH. Enzymes and metabolites of glycogen metabolism in canine cardiac Purkinje fibers. Am. J. Physiol. 1985;248:H599–H605. doi: 10.1152/ajpheart.1985.248.5.H599. [DOI] [PubMed] [Google Scholar]
- 57.Kubler W, Schomig A, Senges J. The conduction and cardiac sympathetic systems: metabolic aspects. J. Am. Coll. Cardiol. 1985;5:157B–161B. doi: 10.1016/s0735-1097(85)80548-9. [DOI] [PubMed] [Google Scholar]
- 58.Gabrielli F, Aita M, Arturi E, Alcini E. A comparative enzyme histochemical study of glucose metabolism in the conduction system of mammalian hearts. Cell. Mol. Biol. 1992;38:449–455. [PubMed] [Google Scholar]
- 59.Wehrens XH, et al. Protection from cardiac arrhythmia through ryanodine receptor-stabilizing protein calstabin2. Science. 2004;304:292–296. doi: 10.1126/science.1094301. [DOI] [PubMed] [Google Scholar]
- 60.Laitinen PJ, et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103:485–490. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- 61.Priori SG, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 62.Lahat H, et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am. J. Hum. Genet. 2001;69:1378–1384. doi: 10.1086/324565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Postma AV, et al. Absence of calsequestrin 2 causes severe forms of catecholaminergic polymorphic ventricular tachycardia. Circ. Res. 2002;91:e21–e26. doi: 10.1161/01.res.0000038886.18992.6b. [DOI] [PubMed] [Google Scholar]
- 64.Wehrens XH, et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 65.Fatkin D, et al. An abnormal Ca2+response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J. Clin. Invest. 2000;106:1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semsarian C, et al. The L-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J. Clin. Invest. 2002;109:1013–1020. doi:10.1172/JCI200214677. doi: 10.1172/JCI14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doi M, et al. Propranolol prevents the development of heart failure by restoring FKBP12.6-mediated stabilization of ryanodine receptor. Circulation. 2002;105:1374–1379. doi: 10.1161/hc1102.105270. [DOI] [PubMed] [Google Scholar]
- 68.Yano M, et al. FKBP12.6-mediated stabilization of calcium-release channel (ryanodine receptor) as a novel therapeutic strategy against heart failure. Circulation. 2003;107:477–484. doi: 10.1161/01.cir.0000044917.74408.be. [DOI] [PubMed] [Google Scholar]
- 69.Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 70.Haghighi K, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J. Clin. Invest. 2003;111:869–876. doi:10.1172/JCI200317892. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo W, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ. Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 72.Minamisawa S, et al. Chronic phospholamban-sarcoplasmic reticulum calcium ATPase interaction is the critical calcium cycling defect in dilated cardiomyopathy. Cell. 1999;99:313–322. doi: 10.1016/s0092-8674(00)81662-1. [DOI] [PubMed] [Google Scholar]
- 73.DeSantiago J, Maier LS, Bers DM. Phospholamban is required for CaMKII-dependent recovery of Ca transients and SR Ca reuptake during acidosis in cardiac myocytes. J. Mol. Cell. Cardiol. 2004;36:67–74. doi: 10.1016/j.yjmcc.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 74.Periasamy M, et al. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J. Biol. Chem. 1999;274:2556–2562. doi: 10.1074/jbc.274.4.2556. [DOI] [PubMed] [Google Scholar]
- 75.Zingman LV, et al. Kir6.2 is required for adaptation to stress. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13278–13283. doi: 10.1073/pnas.212315199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bienengraeber M, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat. Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 79.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 80.Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 81.Kasahara H, Benson DW. Biochemical analyses of eight NKX2.5 homeodomain missense mutations causing atrioventricular block and cardiac anomalies. Cardiovasc. Res. 2004;64:40–51. doi: 10.1016/j.cardiores.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 82.Biben C, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ. Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- 83.Tanaka M, et al. A mouse model of congenital heart disease: cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor Csx/Nkx2.5. Cold Spring Harb. Symp. Quant. Biol. 2002;67:317–325. doi: 10.1101/sqb.2002.67.317. [DOI] [PubMed] [Google Scholar]
- 84.Kasahara H, et al. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J. Clin. Invest. 2001;108:189–201. doi:10.1172/JCI200112694. doi: 10.1172/JCI12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toko H, et al. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J. Biol. Chem. 2002;277:24735–24743. doi: 10.1074/jbc.M107669200. [DOI] [PubMed] [Google Scholar]
- 86.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frey N, et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 88.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 90.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 91.Kook H, et al. Cardiac hypertrophy and histone deacetylase–dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 2003;112:863–871. doi:10.1172/JCI200319137. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKinsey TA, Olson EN. Cardiac histone acetylation--therapeutic opportunities abound. Trends Genet. 2004;20:206–213. doi: 10.1016/j.tig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 93.Finck BN, et al. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 2002;109:121–130. doi:10.1172/JCI200214080. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asakawa M, et al. Peroxisome proliferator-activated receptor gamma plays a critical role in inhibition of cardiac hypertrophy in vitro and in vivo. Circulation. 2002;105:1240–1246. doi: 10.1161/hc1002.105225. [DOI] [PubMed] [Google Scholar]
- 95.Subbarayan V, et al. RXRα overexpression in cardiomyocytes causes dilated cardiomyopathy but fails to rescue myocardial hypoplasia in RXRα-null fetuses. J. Clin. Invest. 2000;105:387–394. doi: 10.1172/JCI8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dalakas MC, et al. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N. Engl. J. Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]