Abstract
Chondrocyte senescence is an important mechanism underlying osteoarthritis in the senile population and is characterized by reduced expressions of the extracellular matrix proteins. The involvement of glycolysis and the tricarboxylic acid cycle in the development of osteoarthritis is inclusive. The present study aims to investigate the role of the glycolytic enzyme M2 isoform of pyruvate kinase (PKM2) in chondrocytes in senescence and inflammation. Primary chondrocytes are isolated from the knee joints of neonatal mice. Small interfering RNAs (siRNAs) against PKM2 are transfected using lipofectamine. RNA sequencing is conducted in primary chondrocytes with the PKM2 gene deleted. Cell apoptosis, autophagy, reactive oxygen species measurement, and senescent conditions are examined. The glycolytic rate in cells is measured by Seahorse examination. Interleukin 1-β (IL-1β) increases the protein expressions of matrix metallopeptidases (MMP)13 and PKM2 and reduces the protein expression of collagen type II (COL2A1) in primary chondrocytes. Silencing of PKM2 alters the protein expressions of MMP13, PKM2, and COL2A1 in the same pattern in quiescent and stimulated chondrocytes. RNA sequencing analysis reveals that PKM2 silencing reduces senescent biomarker p16 INK4a expression. Compared with low-passage chondrocytes, high-passage chondrocytes exhibit increased expression of p16 INK4a and reduced expression of COL2A1. Silencing of PKM2 reduces SA-β-Gal signals and increases COL2A1 expression in high-passage chondrocytes. Seahorse assay reveals that PKM2 deletion favors the tricarboxylic acid cycle in mitochondria in low- but not in high-passage chondrocytes. In summary, the glycolytic enzyme PMK2 modulates chondrocyte senescence but does not participate in the regulation of inflammation.
Keywords: chondrocyte, PKM2, p16 INK4a , senescence, metabolic reprogramming
Introduction
Osteoarthritis is a common degenerative joint disease, especially in the senile population, showing joint pain, stiffness, and even disability. Pathological examination of osteoarthritis joints reveals cartilage destruction, subchondral sclerosis, osteophyte formation, and synovial inflammation [ 1, 2]. Cartilage is composed of chondrocytes and chondrocyte-produced extracellular matrix, including type II collagen (COL2A1), aggrecan, and proteoglycan 4 (Prg4, also known as lubricin) [3]. Chondrocyte senescence is an important mechanism underlying the development of osteoarthritis, resulting in reduced production of extracellular matrix proteins and an increase in the release of proinflammatory factors [ 3– 5]. Removal of senescent chondrocytes decelerates the process of osteoarthritis in surgery-induced osteoarthritis mice [6] .
Cartilage is located inside articular capsules and nourished mainly by synovial fluid [7], indicating that chondrocytes are exposed to long-term hypoxia. It is assumed that adenosine triphosphate (ATP) in chondrocytes is mainly produced by glycolysis and compensated by the tricarboxylic acid cycle in mitochondria under pathological conditions [8]. Pharmacological inhibition of glycolysis by monosodium iodoacetate accelerates the progression of osteoarthritis [9], whereas inhibition of cyclooxygenase-2 attenuates the progression of osteoarthritis [10]. It has also been reported that deleting the isoform of pyruvate kinase (PKM2), the last rate-limiting enzyme in the process of glycolysis [11], induces cell loss and reduces extracellular matrix in cultured chondrocytes under basal conditions [12] but inhibits proinflammatory cytokine interleukin-1 beta (IL-1β)-induced apoptosis [13]. Thus, the participation of glycolysis in osteoarthritis is inconclusive.
The present study was designed to investigate metabolic changes in cultured chondrocytes challenged with IL-1β. Moreover, cell apoptosis, autophagy, and reactive oxygen species were examined in cultured chondrocytes, since these are important mechanisms in the development of cartilage degeneration [ 14, 15]. It is difficult to separate the role of aging in the development of osteoarthritis since aging is a crucial player. Repeated passage of chondrocytes was used in the present study to mimic the pathological condition of osteoarthritis in the senile population. The glycolytic enzyme gene Pkm2 was deleted in both low- and high-passage chondrocytes under basal and proinflammatory conditions.
Materials and Methods
Reagents
Murine IL-1β, prepared in phosphate-buffered saline (PBS; HyClone, Logan, USA) containing 0.1% bovine serum albumin (BSA; Beyotime, Shanghai, China), was purchased from R&D Systems (Minneapolis, USA). Lipopolysaccharide (LPS) and tumor necrosis factor-alpha (TNF-α) were purchased from Sigma (St Louis, USA).
Isolation, culture, and identification of primary murine chondrocytes
Neonatal C57BL/6 mice (5 days old) were purchased from Shanghai Model Organisms Center (Shanghai, China). Immature murine articular chondrocytes (iMACs) were isolated from the knee joints of neonatal mice [16]. Briefly, after removal of the surrounding tissues, the mouse femoral condyle and tibial plateau were collected and prepared in small pieces. The cartilage fragments were digested in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12; Gibco, Carlsbad, USA) containing 0.2% collagenase II (Gibco) and 10% fetal bovine serum (FBS; Gibco) overnight at 37°C. After being filtered through a 70-μm strainer, chondrocytes were cultured in DMEM/F12 medium containing 10% FBS. All animal studies were approved by the Ethics Committee for Animal Research of Zhongshan Hospital (Shanghai, China).
The isolated primary chondrocytes were validated by examining the presence of sulfate proteoglycan and collagen II [16]. Cultured cells were fixed with 4% paraformaldehyde at room temperature for 15 min. To detect the presence of sulfate proteoglycan, cells were incubated in 1% Alcian blue (Servicebio, Wuhan, China) in 1 N HCl for 30 min ( Supplementary Figure S1). To examine the presence of COL2A1, the cells were permeabilized with 0.1% Triton-X-100 solution (Sigma) and blocked with 3% goat serum (Beyotime) for 1 h, followed by incubation with primary antibody against COL2A1 overnight at 4°C, and then with fluorescence-conjugated secondary antibodies for 1 h ( Supplementary Figure S1). Images were taken using an optical microscope (Olympus, Tokyo, Japan).
Small interfering RNA transfection
When cultured cells reached 60%‒80% confluency, cells were transfected with small interfering RNAs (siRNAs) against mouse Pkm2 and their scrambled sequences (GenePharma, Shanghai, China) which are shown in Supplementary Table S1). According to the manufacturer’s instructions, siRNA (50 nM) was delivered into cultured murine chondrocytes using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, USA). The presence of PKM2 was examined 48 h after transfection. Based on the efficiency, the third siPkm2 sequence was used in the present study ( Supplementary Figure S2D–F).
Cell culture experiments
Passage 1 chondrocytes were used in the present study, and passage 3 and 6 cells were used for the senescence study. In the present study, mRNA samples were collected 6 h after IL-1β stimulation (1 ng/mL), and protein levels, oxidative stress, and cell apoptosis were detected after 24 h of incubation.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol (Sigma) and mRNAs were prepared with the First Strand cDNA synthesis kit (TaKaRa, Dalian, China). Real-time qPCR was performed by using Hieff qPCR SYBR Green Master Mix (Yeasen, Shanghai, China). The detailed information of primers is listed in Supplementary Table S2.
Western blot analysis
Cell lysates were prepared using RIPA buffer (Beyotime). Protein samples were separated by 7.5%‒15% SDS-polyacrylamide electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, USA). After being blocked in 5% nonfat milk for 1 h at room temperature, the membranes were incubated with primary antibodies ( Supplementary Table S3) overnight at 4°C, followed by incubation with corresponding secondary antibodies for 1 h at room temperature. Protein blots were visualized using a Tanon Imager 4600 system (Tanon, Shanghai, China).
Reactive oxygen species detection
Reactive oxygen species levels were measured by cell fluorescence and fluorescence-activated cell sorting (FACS). Cultured cells were incubated with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; Sigma) at 37°C for 30 min. The fluorescent signals were detected with a fluorescence microscope (Olympus). Alternatively, 1–2×10 5 cells were collected and incubated with CellROX Detection Reagent (1 μM; Thermo Fisher Scientific, Waltham, USA) for 45 min. SYTOX Dead Cell Stain solution was added to the cell suspension and incubated for 15 min. The FACS signals were examined by flow cytometry on a BD-FACSAria-III flow cytometer (BD Biosciences, Franklin Lakes, USA) and analyzed using FlowJo software (Ashland, USA).
Cell apoptosis
Cell apoptosis was quantified using an Annexin V-FITC/PI Apoptosis Detection kit (Vazyme, Nanjing, China). A total of 1–5×10 5 cells were collected using non-EDTA trypsin (Yeasen). Then, 400 μL of cell suspension was stained with 5 μL of Annexin V and 5 μL of PI for 10 min at room temperature. Apoptotic cells were identified by flow cytometry on a BD-FACSAria-III flow cytometer (BD Biosciences).
Senescence-associated β-galactosidase (SA-β-Gal) staining assay
Cell senescence was examined using SA-β-Gal staining kit (Beyotime). Briefly, cells were fixed with 4% paraformaldehyde and incubated with SA-β-Gal staining solution at 37°C. Images were obtained using an optical microscope (Olympus).
RNA-seq analysis
Cultured chondrocytes were transfected with siNC or siPkm2 and RNA was collected. RNA was first extracted from the cells using Trizol(Sigma). After quantification and quality control with ND-2000 (NanoDrop Technologies, Waltham, USA), 1 μg of high-quality total RNA sample was used for the transcriptome library construction by Shanghai Majorbio Biopharm Technology (Shanghai, China). Briefly, mRNA was enriched and prepared using fragmentation buffer. Reversed transcription was carried out to synthesize cDNA using random hexamer primers (Illumina, San Diego, USA) with a SuperScript double-stranded cDNA synthesis kit (Invitrogen). Then, end repair, phosphorylation and ′A′ base addition were performed to generate cDNA according to the library construction protocol from Illumina. An Illumina NovaSeq 6000 sequencer (2×150 bp read length) was used to sequence the RNA-seq sequencing library after quantification with Qubit 4.0 (Thermo Fisher Scientific). The trim and quality control were performed by fastq with default parameters. The clean reads were then separately aligned to the mouse genome using HISAT2 software. Differentially expressed genes (DEGs) were defined as |log2(fold change)|≥1 and adjusted Pvalue≤0.05. The online platform of Majorbio Cloud was used for analyses. The R/Bioconductor packages pheatmap and ggplot2 were used.
Measurement of cellular glycolytic rates by Seahorse assay
Primary chondrocyte cells (1×10 5 cells/well) were seeded in the Seahorse XF cell culture microplate with DMEM/F12 medium (XF96 extracellular analyzer; Seahorse Bioscience, Wilmington, USA). Cell glycolytic rates were measured in the presence of rotenone and antimycin A (Rot/AA; 0.5 μM), as well as 2-Deoxy-D-glucose (2-DG; 50 nM), according to the manufacturer’s protocol. Wave software was used to analyze the data.
Measurement of intracellular ATP
Intracellular ATP was measured using an ATP assay kit (Beyotime). After incubation with IL-1β (10 ng/mL) for 24 h, cell lysates were collected for ATP detection. Luminance was measured with a luminescent plate reader (Thermo Fisher Scientific). The final readout of the samples was normalized to the protein concentration and expressed as nmol/mg protein.
Statistical analysis
All results were obtained from at least three independent experiments. Data were analyzed by GraphPad Prism 8 (San Diego, USA) and presented as the mean±SEM. Unpaired two-tailed Student’s t test and one-way analysis of variance (ANOVA) multiple comparisons test followed by Tukey’s test, were used for comparisons. P values less than 0.05 were considered statistically significant.
Results
PKM2 participates in regulating chondrocyte function.
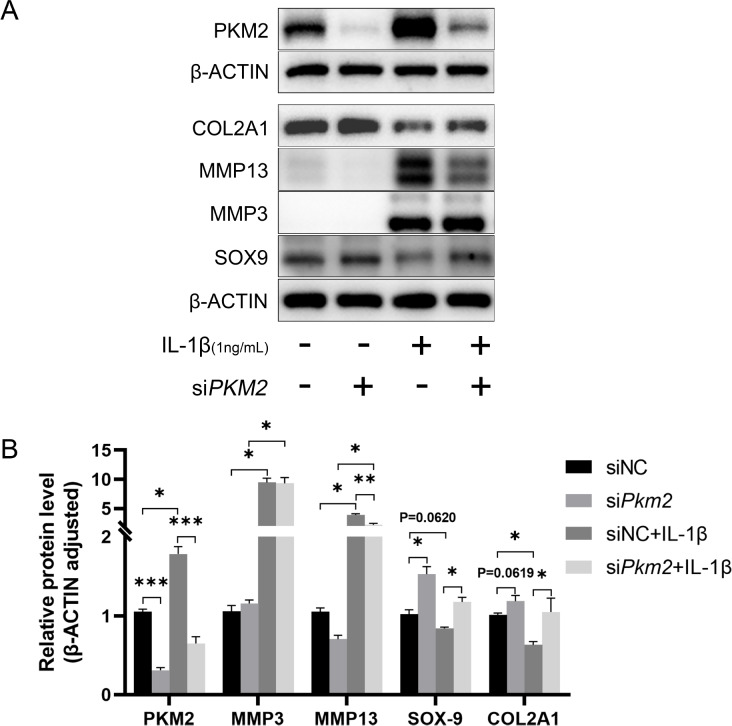
In the present study, the functions of cultured chondrocytes were examined by protein expressions of MMP3, MMP13, COL2A1, and SRY-Box transcription factor (SOX)-9. MMPs are responsible for the degradation of the extracellular matrix protein COL2A1, in which MMP13 is a dominant player in the progression of osteoarthritis [17]. COL2A1 is one of the main components of the extracellular matrix [18]. SOX-9 functions during chondrocyte differentiation and is a phenotype marker for chondrocyte proliferation and differentiation [19].
IL-1β stimulation significantly increased the protein expressions of MMP3 and MMP13 and downregulated COL2A1 and SOX-9 protein expressions ( Figure 1A,B). Silencing of Pkm2 reduced the upregulation of MMP13 but not MMP3 in cells challenged with IL-1β. Silencing of Pkm2 significantly increased COL2A1 and SOX-9 protein expressions under basal and stimulated conditions ( Figure 1A,B).
Figure 1 .
Pkm2 silencing alters chondrocyte functions in cells stimulated with IL-1β
Representative blots (A) and densitometric quantification (B) of PKM2, SOX-9, MMP3, MMP13, and COL2A1 proteins. n=6. *P<0.05, **P<0.01, ***P<0.001.
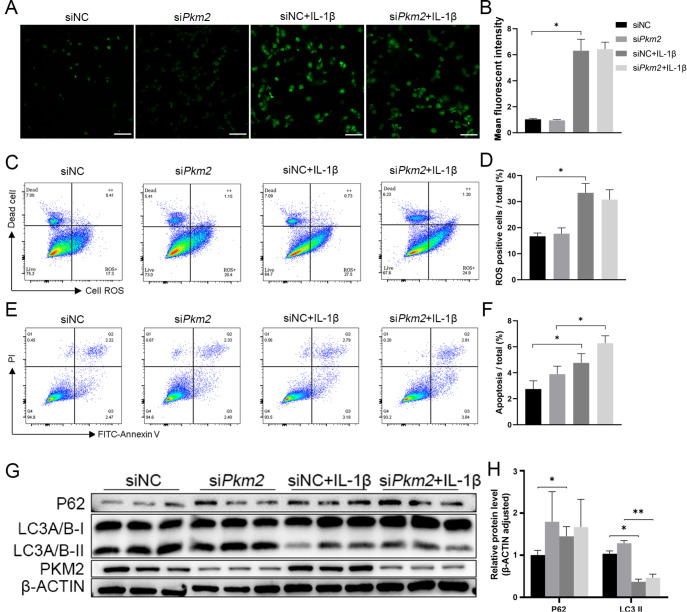
PKM2 does not participate in IL-1β-induced oxidative stress, autophagy, or apoptosis
IL-1β stimulation significantly increased oxidative stress in chondrocytes, as measured by cell fluorescence and FACS. Silencing of Pkm2 did not reduce IL-1β-induced oxidative stress ( Figure 2A–D). Stimulation with IL-1β significantly induced cell apoptosis, as shown by increased Annexin V signals. Silencing of Pkm2 further increased apoptotic signals in cells stimulated with IL-1β ( Figure 2E,F).
Figure 2 .
Pkm2 silencing does not alter ROS levels, cell apoptosis, or autophagy
(A) Measurement of reactive oxidative stress, a reduced form of H2DCFDA, in cultured cells stimulated with IL-1β and (B) quantification of mean fluorescence intensity, n=6. (C) Measurement of CellROX/SYTOX signals by FACS in cultured cells stimulated with IL-1β and (D) quantification of ROS-positive cells, n=3. (E) Annexin V/PI signals examined by FACS and (F) quantification of apoptotic cells, n=6. Representative blots (G) and densitometric quantification (H) of LC3A/B and p62 proteins, n=3. *P<0.05, **P<0.01. Scale bar=500 μm.
Stimulation with IL-1β significantly increased sequestosome-1 (p62) protein expression and decreased microtubule-associated protein 1A/1B light chain 3 (LC3A/B) protein expression, two key players in the process of autophagy. Silencing of Pkm2 did not affect the protein expressions of p62 and LC3A/B in cells stimulated with IL-1β ( Figure 2G,H).
Since silencing of Pkm2 did not play a role in inflammation, it is assumed that the protective role of Pkm2 silencing in chondrocytes occurs under quiescent conditions.
Silencing of Pkm2 affects senescent p16 INK4a protein in chondrocytes
To further understand the role of PKM2 in chondrocytes, RNA sequencing was conducted in primary chondrocytes with Pkm2 silencing. There were 95 differentially expressed genes (DEGs), 40 upregulated and 55 downregulated ( Figure 3A,B). Modules of extracellular matrix organization, extracellular structure organization, collagen-containing extracellular matrix, and extracellular matrix structural constituent were enriched ( Figure 3C). The ECM-receptor interaction pathway was markedly enriched in the KEGG pathway analysis ( Figure 3D).
Figure 3 .
Bioinformatics analysis of DEGs in primary chondrocytes with Pkm2 silencin
(A) Volcano plot of DEGs. (B) Heatmap showing the top 20 upregulated or downregulated DEGs. (C) GO terms enriched for DEGs. (D) KEGG pathway analysis for DEGs.
RNA sequencing analysis revealed that p16 INK4a, known as Cdkn2a, was downregulated in cells transfected with siPkm2 compared with that in siNC-transfected chondrocytes ( Figure 3 A,B).
RT-qPCR confirmed the increased mRNA expressions of Prg4 and extracellular heparin sulfate 6-O ( Sulf1) and reduced mRNA expression of a disintegrin and metalloproteinase 10 ( Adam10) and follistatin-like protein 1 ( Fstl1) in quiescent and stimulated chondrocytes with Pkm2 silencing ( Supplementary Figure S3A‒D).
PKM2 regulates chondrocyte senescence
Protein p16 INK4a is a biomarker of cellular aging [ 3, 4, 20]. High-passage chondrocytes had higher SA-β-Gal activity than low-passage chondrocytes ( Figure 4A,B), which was reduced by Pkm2 silencing ( Figure 4A,B).
Figure 4 .
Pkm2 silencing decelerates chondrocyte senescence
(A) SA-β-Gal activity and (B) quantification of SA-β-Gal-positive chondrocytes in cultured cells, n=6. (C) mRNA expressions of p16INK4a and PKM2 in low- and high-passage chondrocytes stimulated with IL-1β, n=3. Representative blots (D) and densitometric quantification (E) of PKM2, p16INK4a, and COL2A1 proteins in low- and high-passage chondrocytes stimulated with IL-1β, n=3. *P<0.05, **P<0.01. Scale bar=50 μm.
Both mRNA and protein expressions of p16 INK4a were upregulated in chondrocytes in a passage-dependent manner ( Figure 4C–E). In high-passage cells, IL-1β stimulated PKM2 and p16 INK4a upregulation, which were in the same pattern as those in low-passage chondrocytes. Silencing of PKM2 reduced p16 INK4a expression under quiescent and stimulated conditions ( Figure 4C–E).
Under quiescent conditions, high-passage chondrocytes had reduced protein expression of COL2A1. In line with the results of low-passage cells, silencing of Pkm2 increased COL2A1 protein expression under quiescent but not stimulated conditions in high-passage chondrocytes ( Figure 4D–E).
PKM2 is important in cell senescence but not in IL-1β-induced inflammation in chondrocytes
PKM2, a rate-limiting enzyme in glycolysis, is important for ATP generation [ 21, 22]. In the present study, silencing of Pkm2 reduced the mRNA levels of fructose-bisphosphate aldolase c ( Aldoc), glucose-6-phosphate isomerase 1 ( Gpi1), and phosphoglycerate kinase 1 ( Pgk1), but not lactate dehydrogenase A ( Ldha), phosphofructokinase muscle isoform ( Pfkm), or hexokinase 2 ( Hk2), in quiescent cells, both in low- and high-passage cells ( Supplementary Figure S4A–F). Stimulation with IL-1β significantly increased the mRNA expressions of these glycolytic enzymes, including Aldoc, Ldha, Gpi1, Hk2, and Pgk1, which were not affected by Pkm2 silencing ( Supplementary Figure S4A–F).
The mRNA expression levels of genes in the tricarboxylic acid cycle were examined. Silencing of Pkm2 increased the mRNA levels of citrate synthase ( Cs), dihydrolipoamide s-succinyltransferase ( Dlst), isocitrate dehydrogenase 2 ( Idh2), Idh3a, Idh3b, oxoglutarate dehydrogenase ( Ogdh), and mitochondrial transcription factor A ( Tfam) in low-passage cells but not in high-passage cells ( Supplementary Figure S4F‒P).
Stimulation with IL-1β significantly increased the mRNA expression of Dlst but not other players in the tricarboxylic acid cycle in low-passage cells ( Supplementary Figure S4F‒P). Stimulation with IL-1β did not change the mRNA expression of the above mentioned players in the tricarboxylic acid cycle in high-passage cells ( Supplementary Figure S4F‒P).
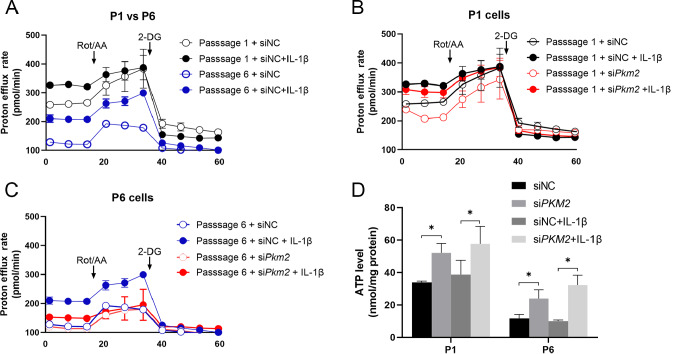
To further confirm the protective role of Pkm2 in chondrocytes, the glycolytic rates of cells under both basal and stimulated conditions were measured by Seahorse assay. Compared with low-passage cells, high-passage cells had reduced proton efflux rates, an indicator of the extracellular level of lactate ( Figure 5A). IL-1β stimulation significantly increased proton efflux rates in low- and high-passage chondrocytes ( Figure 5A).
Figure 5 .
Glycolytic rate measured by Seahorse assay in low- and high-passage chondrocytes stimulated with IL-1β
(A) Glycolytic rate measured by Seahorse assay in low- and high-passage chondrocytes stimulated with IL-1β. (B,C) Glycolytic rates in low-passage and high-passage cells stimulated with IL-1β, n=4. (D) ATP levels in low- and high-passage cells stimulated with IL-1β, n=6. *P<0.05.
The proton efflux rates in low-passage cells were comparable when the mitochondrial respiratory chain was blocked, comparing quiescent cells to the stimulated cells ( Figure 5B). In high-passage chondrocytes, IL-1β stimulated a higher proton efflux rate when the mitochondrial respiratory chain was blocked ( Figure 5B).
Silencing of Pkm2 reduced proton efflux rates in low-passage cells, which were not significantly affected when the mitochondrial respiratory chain was blocked ( Figure 5C). Silencing of Pkm2 reduced proton efflux rates in high-passage cells under both quiescent and stimulated conditions when the mitochondrial respiratory chain was blocked ( Figure 5A,B).
The basal levels of ATP were increased in both low- and high-passage chondrocytes with PKM2 silencing. IL-1β stimulation did not significantly increase ATP production in either low- or high-passage cells ( Figure 5D).
Discussion
In the present study, we revealed that glycolysis is the primary energy-producing setting of chondrocytes under quiescent conditions and shifts to the tricarboxylic acid cycle when cells are silenced with Pkm2. In the process of cell senescence, the glycolytic enzyme PKM2 is a key player, but not in the process of inflammation. The protective role of PKM2 in senescence is attributed to the downregulation of p16 INK4a protein.
MMPs and ADAMs degrade extracellular matrix proteins, including fibrillar collagen and aggrecan, respectively. In addition, the expressions of Prg4, Sulf1, Adam10, and Fstl1 were also examined in quiescent and stimulated chondrocytes. Prg4, an important component of extracellular matrix proteins, is synthesized and secreted by chondrocytes [23]. Prg4 protein modulates cartilage homeostasis by reducing friction between articular cartilage surfaces [24], since deleting Prg4 induces chondrocyte apoptosis [ 24, 25]. Sulf1 hydrolyses sulfates from heparan sulfate proteoglycans and regulates the contents of the extracellular matrix [ 26, 27]. Mice lacking the Sulf1 protein exhibit spontaneous cartilage degeneration and osteoarthritis [28]. The Adam10 protein, which degrades aggrecan, is increased in degenerated cartilage and osteoarthritis cartilage [ 29, 30]. Enhanced expression of Fstl1 suppresses chondrogenesis, resulting in cartilage destruction [ 31, 32]. In the present study, chondrocytes stimulated with IL-1β exhibited downregulation of COL2A1 and upregulation of MMP13, Prg4, Sulf1, Adam10, and Fstl1, supporting that enhanced inflammation is a critical mechanism underlying the progression of osteoarthritis [33].
Stimulation with IL-1β increased oxidative stress, enhanced autophagy, and induced cell apoptosis, confirming that these biological processes are involved in the development of osteoarthritis [ 14, 15, 34]. However, silencing of Pkm2 did not change IL-1β-induced oxidative stress, autophagy, or cell apoptosis. Silencing of Pkm2 exerted similar patterns of chondrocyte function proteins under both basal and stimulated conditions. Thus, these data imply that glycolysis is the dominant player under basal conditions.
The tricarboxylic acid cycle occurs in mitochondria and is the primary energy source for cell migration, proliferation, and function [35]. Nevertheless, when cells are exposed to hypoxia, glycolysis is more responsible for energy production [36]. Since the cartilage inside the articular capsules is exposed to long-term hypoxia, glycolysis is assumed to be the energy-producing setting of chondrocytes [37]. In the present study, increased level of ATP, together with the upregulation of genes in the tricarboxylic acid cycle in cells silenced with Pkm2, suggested that the tricarboxylic acid cycle in mitochondria compensates for the impairment of glycolysis.
It is acknowledged that laboratory-induced aging does not fully reproduce the pathophysiological changes in the course of nature. However, it is difficult to separate the role of aging in the process of osteoarthritis, since aging is an important underlying mechanism of the disease. In the laboratory, cell aging is induced by chronic oxidative stress and repeated passage, two common approaches. Chondrocytes are terminally differentiated cells, and their differentiated phenotypes are lost during senescence [16]. Indeed, primary chondrocytes rapidly lose their differentiated phenotype upon repeated passages, showing reduced expressions of COL2A1 and degrading proteins. Importantly, increased SA-β-Gal activity and enhanced p16 INK4a in high-passage chondrocytes were observed in the present study, supporting that chondrocyte senescence was successfully induced.
In addition to increased SA-β-Gal activity and enhanced p16 INK4a expression, in high-passage chondrocytes, deletion of PKM2 reduced p16 INK4a protein expression, decelerated chondrocyte senescence, and partially restored chondrocyte functions, as shown by decreased SA-β-Gal activity and increased expression of COL2A1 in cultured cells, indicating that PKM2 protein is beneficial for chondrocyte senescence.
To mimic the pathological conditions of osteoarthritis in the aged population, stimulation with IL-1β was also applied in high-passage chondrocytes. Consistent with the observation in low-passage cells, silencing of Pkm2 had similar patterns on protein changes of p16 INK4a under both basal and stimulated conditions, supporting the note that PKM2 does not participate in inflammation.
Notably, increased expression of p16 INK4a is also observed in quiescent cells stimulated with IL-1β, suggesting that the upregulation of p16 INK4a by proinflammatory cytokines or repeated passages occurs through different mechanisms, including the NF-κB signaling pathway [ 36, 38– 40] .
In summary, metabolic changes respond quickly to physiological and pathological stimulations. Glycolysis is vital in quiescent chondrocytes, and the tricarboxylic acid cycle steps forward when the protective effects of glycolysis recede under pathological conditions. Pkm2 silencing decelerates chondrocyte senescence by regulating p16 INK4a. Therefore, targeting the Pkm2 gene is a promising clinical therapy and sheds light on the treatment of age-related osteoarthritis.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grant from the National Natural Science Foundation of China (No. 81971308 to C.Z.).
References
- 1.Jansen MP, Mastbergen SC. Joint distraction for osteoarthritis: clinical evidence and molecular mechanisms. Nat Rev Rheumatol. . 2022;18:35–46. doi: 10.1038/s41584-021-00695-y. [DOI] [PubMed] [Google Scholar]
- 2.Latourte A, Kloppenburg M, Richette P. Emerging pharmaceutical therapies for osteoarthritis. Nat Rev Rheumatol. . 2020;16:673–688. doi: 10.1038/s41584-020-00518-6. [DOI] [PubMed] [Google Scholar]
- 3.Rahmati M, Nalesso G, Mobasheri A, Mozafari M. Aging and osteoarthritis: Central role of the extracellular matrix. Ageing Res Rev. . 2017;40:20–30. doi: 10.1016/j.arr.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. . 2016;12:412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coryell PR, Diekman BO, Loeser RF. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat Rev Rheumatol. . 2021;17:47–57. doi: 10.1038/s41584-020-00533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, Chung JW, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. . 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gastel N, Carmeliet G. Metabolic regulation of skeletal cell fate and function in physiology and disease. Nat Metab. . 2021;3:11–20. doi: 10.1038/s42255-020-00321-3. [DOI] [PubMed] [Google Scholar]
- 8.Stegen S, Laperre K, Eelen G, Rinaldi G, Fraisl P, Torrekens S, Van Looveren R, et al. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature. . 2019;565:511–515. doi: 10.1038/s41586-019-0874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang LW, Huang TC, Hu YC, Hsieh BS, Chiu PR, Cheng HL, Chang KL. Zinc protects chondrocytes from monosodium iodoacetate-induced damage by enhancing ATP and mitophagy. Biochem Biophys Res Commun. . 2020;521:50–56. doi: 10.1016/j.bbrc.2019.10.066. [DOI] [PubMed] [Google Scholar]
- 10.Yu SM, Kim SJ. Endoplasmic reticulum stress (ER-stress) by 2-deoxy-D-glucose (2DG) reduces cyclooxygenase-2 (COX-2) expression and N-glycosylation and induces a loss of COX-2 activity via a Src kinase-dependent pathway in rabbit articular chondrocytes. Exp Mol Med. . 2010;42:777–786. doi: 10.3858/emm.2010.42.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu MC, Hung WC. Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Mol Cancer. . 2018;17:35. doi: 10.1186/s12943-018-0791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Chen W, Zhao X, Chen L, Li W, Ran J, Wu L. Pyruvate kinase M2 modulates the glycolysis of chondrocyte and extracellular matrix in osteoarthritis. DNA Cell Biol. . 2018;37:271–277. doi: 10.1089/dna.2017.4048. [DOI] [PubMed] [Google Scholar]
- 13.Li ZZ, Wang F, Liu S, Li H, Wang Y. Ablation of PKM2 ameliorated ER stress-induced apoptosis and associated inflammation response in IL-1β-treated chondrocytes via blocking Rspo2-mediated Wnt/β-catenin signaling. J Cell Biochem. . 2020;121:4204–4213. doi: 10.1002/jcb.29611. [DOI] [PubMed] [Google Scholar]
- 14.Son YO, Kim HE, Choi WS, Chun CH, Chun JS. RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat Commun. . 2019;10:77. doi: 10.1038/s41467-018-08035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Vasheghani F, Li Y, Blati M, Simeone K, Fahmi H, Lussier B, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. . 2015;74:1432–1440. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 16.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. . 2008;3:1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 17.Neuhold LA, Killar L, Zhao W, Sung MLA, Warner L, Kulik J, Turner J, et al. Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J Clin Invest. . 2001;107:35–44. doi: 10.1172/JCI10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowlton RG, Katzenstein PL, Moskowitz RW, Weaver EJ, Malemud CJ, Pathria MN, Jimenez SA, et al. Genetic linkage of a polymorphism in the type II procollagen gene (COL2A1) to primary osteoarthritis associated with mild chondrodysplasia. N Engl J Med. . 1990;322:526–530. doi: 10.1056/NEJM199002223220807. [DOI] [PubMed] [Google Scholar]
- 19.Haseeb A, Kc R, Angelozzi M, de Charleroy C, Rux D, Tower RJ, Yao L, et al. SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc Natl Acad Sci USA. . 2021;118 doi: 10.1073/pnas.2019152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diekman BO, Sessions GA, Collins JA, Knecht AK, Strum SL, Mitin NK, Carlson CS, et al. Expression of p16 INK4a is a biomarker of chondrocyte aging but does not cause osteoarthritis . Aging Cell. . 2018;17:e12771. doi: 10.1111/acel.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F, Simmet T, et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Mol Cancer. . 2016;15:3. doi: 10.1186/s12943-015-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, Wang Y, et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. . 2017;19:1358–1370. doi: 10.1038/ncb3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruan MZC, Erez A, Guse K, Dawson B, Bertin T, Chen Y, Jiang MM, et al. Proteoglycan 4 expression protects against the development of osteoarthritis. Sci Transl Med. . 2013;5:176ra134. doi: 10.1126/scitranslmed.3005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki T, Alvarez-Garcia O, Mokuda S, Nagira K, Olmer M, Gamini R, Miyata K, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. . 2018;10 doi: 10.1126/scitranslmed.aan0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waller KA, Zhang LX, Elsaid KA, Fleming BC, Warman ML, Jay GD. Role of lubricin and boundary lubrication in the prevention of chondrocyte apoptosis. Proc Natl Acad Sci USA. . 2013;110:5852–5857. doi: 10.1073/pnas.1219289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao W, Sala-Newby GB, Dhoot GK. Sulf1 expression pattern and its role in cartilage and joint development . Dev Dyn. . 2006;235:3327–3335. doi: 10.1002/dvdy.20987. [DOI] [PubMed] [Google Scholar]
- 27.Otsuki S, Taniguchi N, Grogan SP, D′Lima D, Kinoshita M, Lotz M. Expression of novel extracellular sulfatases Sulf-1 and Sulf-2 in normal and osteoarthritic articular cartilage. Arthritis Res Ther. . 2008;10:R61. doi: 10.1186/ar2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, Wong CH, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci USA. . 2010;107:10202–10207. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou L, Huang W, Zhang T, Xu D, Kong D, Meng Y. Circular RNA circ_0114876 regulates osteoarthritis through upregulating ADAM10 via targeting miR-1227-3p. Transplant Immunol. . 2022;77:101747. doi: 10.1016/j.trim.2022.101747. [DOI] [PubMed] [Google Scholar]
- 30.Yang CY, Chanalaris A, Troeberg L. ADAMTS and ADAM metalloproteinases in osteoarthritis-looking beyond the ‘usual suspects’. Osteoarthritis Cartilage. . 2017;25:1000–1009. doi: 10.1016/j.joca.2017.02.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaly Y, Blair HC, Smith SM, Bushnell DS, Marinov AD, Campfield BT, Hirsch R. Follistatin-like protein 1 regulates chondrocyte proliferation and chondrogenic differentiation of mesenchymal stem cells. Ann Rheum Dis. . 2015;74:1467–1473. doi: 10.1136/annrheumdis-2013-204822. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Alahdal M, Deng Z, Liu J, Zhao Z, Cheng X, Chen X, et al. Molecular functions of FSTL1 in the osteoarthritis. Int Immunopharmacol. . 2020;83:106465. doi: 10.1016/j.intimp.2020.106465. [DOI] [PubMed] [Google Scholar]
- 33.Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, Sokolove J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol. . 2016;12:580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosseinzadeh A, Kamrava SK, Joghataei MT, Darabi R, Shakeri-Zadeh A, Shahriari M, Reiter RJ, et al. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J Pineal Res. . 2016;61:411–425. doi: 10.1111/jpi.12362. [DOI] [PubMed] [Google Scholar]
- 35.Vuoristo KS, Mars AE, Sanders JPM, Eggink G, Weusthuis RA. Metabolic engineering of TCA cycle for production of chemicals. Trends Biotechnol. . 2016;34:191–197. doi: 10.1016/j.tibtech.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Arra M, Swarnkar G, Ke K, Otero JE, Ying J, Duan X, Maruyama T, et al. LDHA-mediated ROS generation in chondrocytes is a potential therapeutic target for osteoarthritis. Nat Commun. . 2020;11:3427. doi: 10.1038/s41467-020-17242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng L, Zhang Z, Sheng P, Mobasheri A. The role of metabolism in chondrocyte dysfunction and the progression of osteoarthritis. Ageing Res Rev. . 2021;66:101249. doi: 10.1016/j.arr.2020.101249. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Yu Y, Wen Y, Chen J, Lin J, Sheng Z, Zhou W, et al. A high-resolution route map reveals distinct stages of chondrocyte dedifferentiation for cartilage regeneration. Bone Res. . 2022;10:38. doi: 10.1038/s41413-022-00209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Yin H, Yan Z, Li H, Wu J, Wang Y, Wei F, et al. The immune microenvironment in cartilage injury and repair. Acta Biomater. . 2022;140:23–42. doi: 10.1016/j.actbio.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Claus S, Aubert-Foucher E, Demoor M, Camuzeaux B, Paumier A, Piperno M, Damour O, et al. Chronic exposure of bone morphogenetic protein-2 favors chondrogenic expression in human articular chondrocytes amplified in monolayer cultures. J Cell Biochem. . 2010;111:1642–1651. doi: 10.1002/jcb.22897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.