Summary
Glioma, an aggressively growing and highly malignant brain tumor, poses substantial therapeutic challenges due to its resistance to radiotherapy and chemotherapy. Recent research has identified circRNAs as pivotal players in glioma formation and development. However, the roles of circRNA in the metabolic and immune regulation of glioma are unclear. In this study, circSOBP expression was significantly downregulated in glioma cells and specimens. Functionally, enhanced circSOBP expression mitigated cell proliferation, invasion, migration, and glycolysis in gliomas. Mechanistically, circSOBP inhibited glycolysis and activated the MDA5-mediated IKKε/TBK1/IRF3 signaling pathway by binding TKFC proteins. Furthermore, the elevated levels of IFN-I induced by the MDA5 pathway increased the number and activity of CD8+ T and NK cells in the immune response of the animal models. In summary, our findings have emphasized the critical role of circSOBP in binding and modulating TKFC protein, offering potential therapeutic avenue for targeting glioma metabolism and immunological reprogramming.
Subject areas: Biochemistry, Biological sciences, Cancer, Immunology, Molecular biology, Natural sciences
Graphical abstract
Highlights
-
•
CircSOBP expression was significantly decreased in glioma
-
•
CircSOBP regulated glioma glycolysis by binding TKFC protein
-
•
CircSOBP promoted antitumor immunity by activating TKFC/MDA5/IRF3 signaling
Biochemistry; Biological sciences; Cancer; Immunology ; Molecular biology; Natural sciences
Introduction
Glioma is the most common malignant tumor in the central nervous system, originating from the intrinsic component cells of the central nervous system, with an annual incidence rate of from 3 to 6.4 per one hundred thousand.1 Among them, high-grade glioblastoma (GBM) is the most malignant, with an annual incidence of 4.03/100,000, accounting for 48.6% of primary malignant brain tumors, and has biological characteristics such as invasive growth and frequent recurrence.1 In recent years, molecular typing based on molecular pathology has gradually gained attention and has become the basis for the precise diagnosis and treatment of gliomas.2 The primary treatment is high limit surgical resection along with chemotherapy and radiation therapy.3 However, since the detailed mechanisms involved in the development of GBM are still uncertain, patients cannot be eradicated with the median overall survival being less than 20 months and the 5-year survival rate being less than 3%.4
Circular RNAs are a type of covalent closed-loop single-stranded RNA molecules that are produced by various cis and/or trans factors that regulate back-splicing or other RNA circularization5,6; additionally, circularized exons are typically flanked by longer introns containing reverse complementary sequences, and some RNA-binding proteins, such as QKI, are involved in the circularization process.7 Because it lacks the 5' cap and the 3' poly-A tail, unlike linear RNA, it is not sensitive to exonuclease degradation, resulting in a more stable structure, giving it a natural advantage as a therapeutic target and biomarker for malignancies.5 Initially believed to be a byproduct of mis-splicing, growing researches suggest that circRNAs are participating in tumorigenesis and evolution through a variety of mechanisms. For example, circRNA CDR1as contained several specialized miRNA binding sites that could function through sponge miRNAs8; EIciRNAs (Exon and intron circular RNAs) might bind U1 snRNP through RNA-RNA interactions and cis-promote the transcription of RNA polymerase II.9 Furthermore, circRNAs could directly bind proteins10 or acted through cap-independent translation.11 A growing number of studies have shown that circRNAs play an essential regulatory role in glioma. For example, circNDC80 promoted glioblastoma development via the miR-139-5p/ECE1 pathway12; circNCAPG promoted the malignant phenotype of glioma stem cells via activation of the TGF-β pathway13; Hsa_circ_0072309 promoted autophagy by p53 signaling pathway and enhanced sensitivity of glioblastoma to temozolomide (TMZ) in p53 wild-type GBM.14 According to recent findings, circSOBP was a critical regulator of prostate cancer and was predicted to be used as a biomarker in the diagnosis and prognosis of gastric cancer.15,16 However, the role of circSOBP in glioma remains uncertain.
It is generally recognized that metabolic reprogramming is a significant characteristic and hallmark of malignancies.17 Even under aerobic environments, tumor cells mainly produce their adenosine triphosphate (ATP) by glycolysis, which is referred as the Warburg effect. Although aerobic glycolysis generates less ATP than oxidative phosphorylation, it supplies the biosynthetic molecules necessary for tumorigenesis.17 Lactic acid, for instance, is considered to be the final metabolic waste product of glycolysis, and its generation and accumulation in tumors can accelerate tumor growth and metastasis, as well as be associated with a poor prognosis.18 More and more research has shown that metabolic enzymes play an important role in tumor development, not only by establishing a pro-tumor metabolic environment, but also by influencing the involvement of multiple signaling pathways.19,20 As instance, IDH mutation was a crucial metabolic biological marker for gliomas, which was independent of tumor grade, head much higher progression-free survival rates than individuals with IDH wild type, and that suggested a different approach to therapy.21,22 Mutations in the FH gene caused fumaric acid accumulation in kidney cancer, and raised fumaric acid levels were implicated in tumor formation mechanisms such as antitumor factor inhibition and activation of oncogenic HIF signaling pathways.23
Immunosuppressive factors and immune cells in the immunological microenvironment provide the basis for glioma development.24 As a result, therapies targeting the immune microenvironment of cancers were important research directions.24 For example, the immune checkpoint PD-1 (programmed cell death protein 1)/PD-L1 (programmed cell death 1 ligand 1) was being used successfully in the therapeutic treatment of malignant cancers such as melanoma and breast cancer.25 CAR-T cell therapies caused by genetic engineering technologies were capable of effectively killing tumor cells precisely.26 According to studies, early IFN-I generation was crucial for the efficient activation of CD8+ T cells that were specific for tumor antigens as well as a necessary regulator for maintaining NK cell stability, activation, and antitumor activity.27 However, prolonged interferon activation could also cause immune depletion in the tumor microenvironment.27 Therefore, it is important to understand how to maintain the immunological stimulating impact of interferon while limiting the immune suppressive effect.
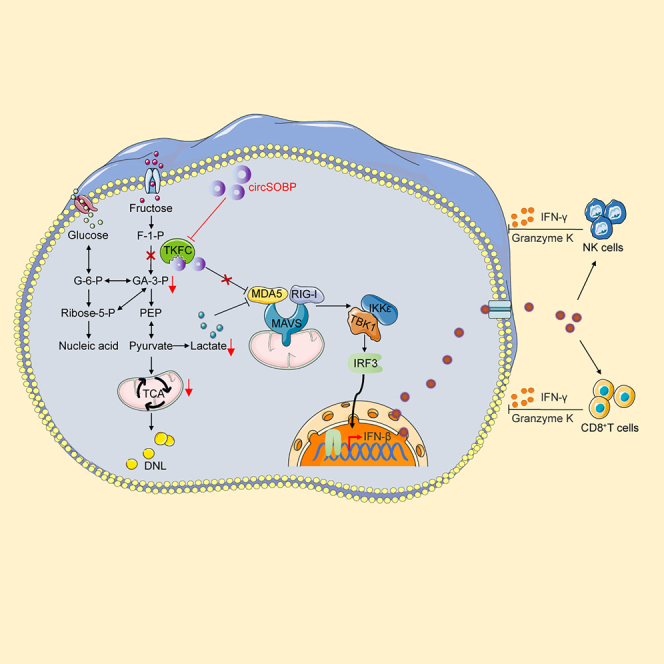
In this study, we identified a circRNA generated by SOBP gene circularization, named circSOBP, which was significantly downregulated in glioma. CircSOBP was able to suppress glycolysis, mitochondrial respiration, and the malignant phenotype of glioblastoma both in vitro and in vivo. Additionally, circSOBP binding regulation of TKFC not only decreased energy consumption in glioma but also activated the IKKε/TBK1/IRF3 signaling pathway, that promoted IFN-I transcription, and stimulated CD8+ T cell and NK cell activation. In conclusion, we have suggested a positive feedback mechanism between the regulation of glucose metabolic reprogramming and immune heterogeneity in glioma by circSOBP binding to TKFC protein, might provide a potential target for glioma treatment.
Results
CircRNA profiling in glioma and circSOBP characterization
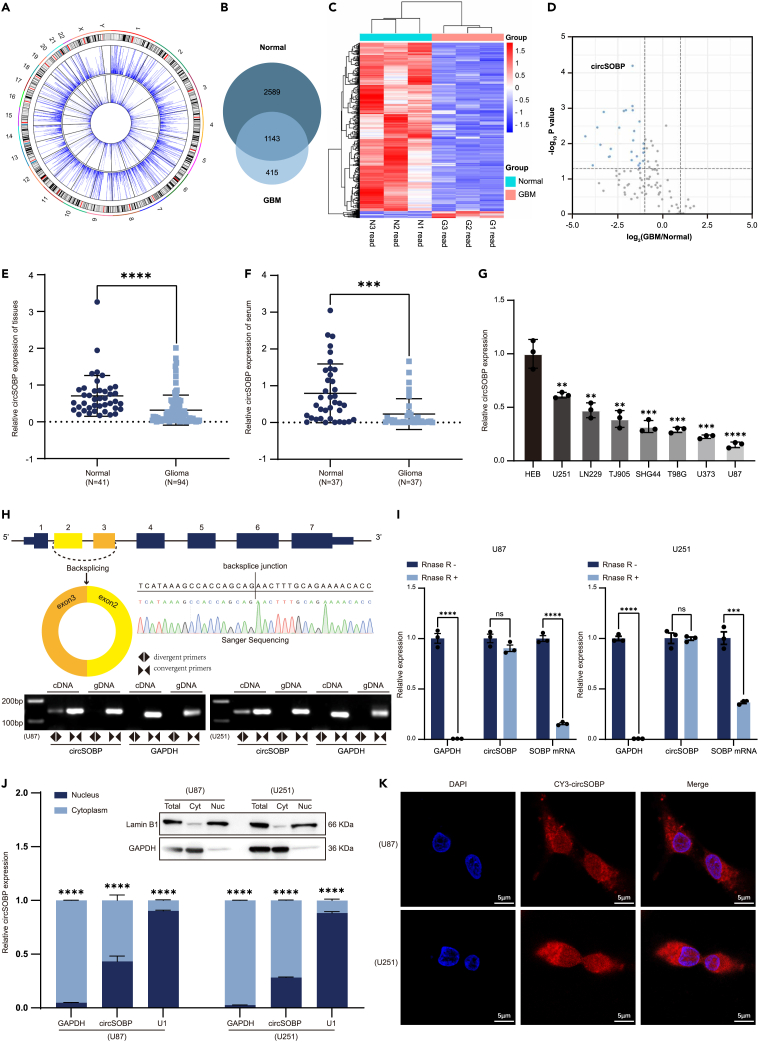
To thoroughly characterize the genome-wide distribution of circRNAs in glioma, we performed RNA sequencing (RNA-seq) of total RNA depleted of ribosomal RNA, and discovered that the expression levels of circRNA transcripts varied between three glioma tissues and three normal brain tissues (GSE153692). Circos plots globally displayed the 4147 circRNAs discovered in glioma tissue and controls (Figure 1A). Furthermore, we observed an overlap of circRNAs in the glioma and control groups, with 28% of all 4147 candidate circRNAs being detected in both groups (Figure 1B). Hierarchical cluster analysis was also conducted to reavel the differential expression of circRNAs in glioma tissues and controls. Among them 466 were significantly altered, with 448 decreased and 18 increased (Figure 1C). Notably, volcano plots showed a general decrease for circRNAs in glioma, of which circSOBP was the most evident (Figure 1D). To verify the findings, the expression of circSOBP was next investigated in 41 normal tissues and 94 glioma tissues, and defected considerably decrease in glioma tissues (Figure 1E; Table S1). Importantly, circSOBP expression was also significantly downregulated in the serum of patients with glioma, indicating that it might be used as a glioma biomarker (Figure 1F). Seven glioma cell lines were randomly selected for RT-qPCR analysis to test the endogenous expression level of circSOBP, and found that circSOBP expression was significantly lower in glioma cells than in normal glial cells (HEB) (Figure 1G). Based on this result, U87 and U251 were selected for the loss-of-function and gain-of-function assays in vitro.
Figure 1.
Characterization of circSOBP in glioma
(A) Circos plots of circRNAs in the human genome (HG19). The outer tracks represent the cytoband ideogram of chromosome. For the two blue tracks, the outer side represents the level of circRNAs in normal tissues and the inner one represents the level of circRNAs in glioma tissues.
(B) The Venn diagram revealed the overlap of candidate circRNAs between normal brain tissues (Normal group) and glioma tissues (GBM group). Backsplicing reads ≥2.
(C) Hierarchical clustering analysis of differentially expressed circRNAs in three normal and glioma tissues, with rows representing circRNAs and columns representing tissues. The red color indicates a high expression level, and the blue color indicates a low expression level.
(D) Volcano plots describing the differential changes of circRNAs for the top 100 in GBM and Normal samples. The blue dots represent circRNAs that are significantly downregulated and the gray dots represent circRNAs with no significant change.
(E) The expression levels of circSOBP in 41 normal brain tissues and 94 glioma tissues were detected by RT-qPCR.
(F) The expression levels of circSOBP in 37 normal serum cases and 37 glioma serum cases were detected by RT-qPCR.
(G) RT-qPCR analysis of the relative expression of circSOBP in HEB cell line (human brain normal glial cell) and seven glioma cell lines.
(H) CircSOBP gene locus and its circularization validation. The back-spliced junction fragment of circSOBP was detected in U87 and U251 cells by RT-PCR and validated by Sanger sequencing. GAPDH was used as a negative control.
(I) The abundance of circSOBP and SOBP mRNA by RT-qPCR in U87 and U251 cells. RNA samples were treated with RNase R+ or RNase R-treated without the enzyme. The contents of circSOBP and SOBP mRNA were normalized to the values of GAPDH.
(J) The expression of circSOBP in the nucleus and cytoplasm of U87 and U251 cells. Western blot showed that the nucleus and plasma of the cells were successfully separated. GAPDH and Lamin B1 are makers of cytoplasm and nucleus respectively in protein level; GAPDH and U1 are makers of cytoplasm and nucleus respectively in the RNA level.
(K) The localization of circSOBP was detected by RNA FISH in U87 and U251 cells. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). CircSOBP was labeled with Cyanine 3 (CY3) dye. Scale bar, 5 μm. All statistics of error bars, S.E.M. from three independent experiments. NS, not significant; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
The genome structure suggests that circSOBP (CircBase ID hsa-circ 0001633) is a 325nt splice sequence length consisting of the second and third exons of the human SOBP transcript (GenBank ID NM 018013). The putative back-splicing junction of circSOBP was amplified by PCR with divergent primers from complementary DNA (cDNA), but not from genomic DNA (gDNA), confirmed with sanger sequencing (Figure 1H). Then, the expression of linear SOBP mRNA and circSOBP were detected by RT-qPCR in U87 and U251 cell lines, respectively. Results confirmed that circSOBP was unaffected by Rnase R which showed the digestion-resistant characteristics of a circular structure (Figure 1I). Western blotting assays for expression of the nuclear marker protein Lamin B1 and cytoplasmic marker protein GAPDH demonstrate successful separation of nucleus and cytoplasm. Then, RT-qPCR was performed to analyze circSOBP expression in the nucleus and cytoplasm. Furthermore, RNA-FISH detected the localization of circSOBP. Results showed that circSOBP was mainly expressed in cytoplasm in glioma cells rather than in nuclear. (Figures 1J and 1K). All these results indicated that circSOBP had the potential to act as a predictive biomarker or therapeutic target for glioma.
CircSOBP inhibits the malignant phenotype and the mitochondrial respiratory glycolysis in glioma
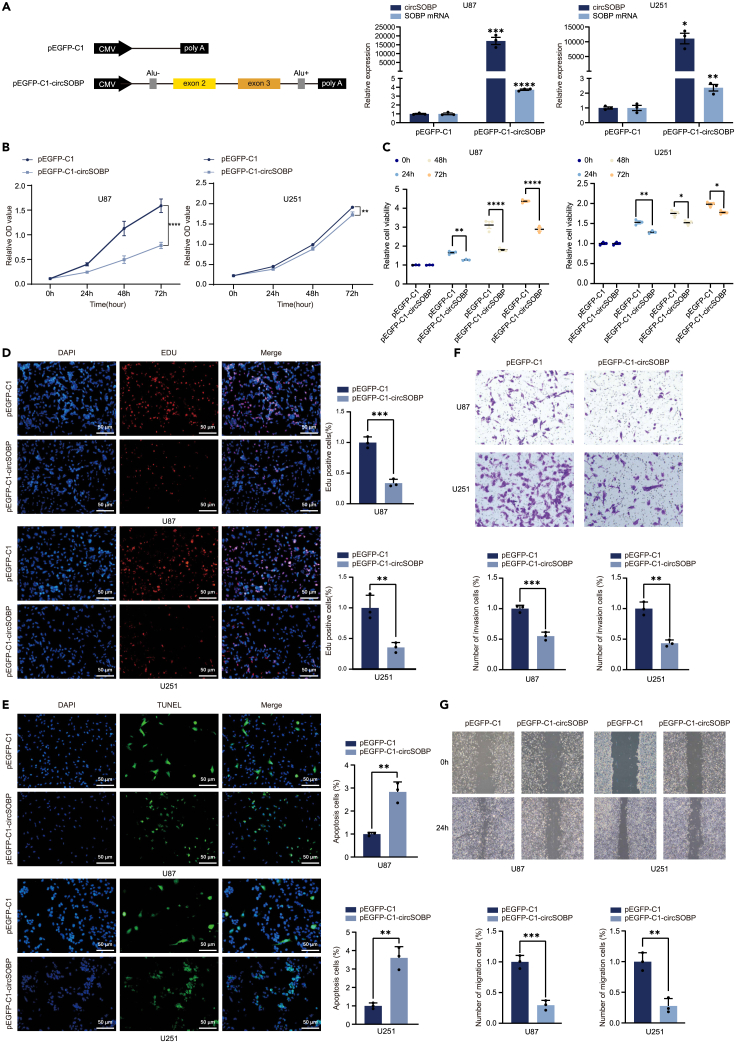
To explore the function roles of circSOBP in glioma, we transfected circSOBP overexpression vector with endogenous flanking sequences containing complementary Alu element pairs into U87 and U251 cells, and confirmed its overexpression efficiency (Figure 2A). In vitro overexpression of circSOBP significantly inhibited the cell proliferation rate (Figures 2B and 2C) and DNA replication activity (Figure 2D) of U87 and U251 cells. Furthermore, TUNEL staining indicated that increasing circSOBP expression greatly promoted the apoptosis of glioma cells (Figure 2E). Transwell and wound healing assays suggested that circSOBP overexpression inhibited cell invasion and migration abilities. (Figures 2F and 2G). In contrast, two small interfering RNAs (siRNAs) directed against the back-spliced junction site of circSOBP were performed, which effectively knockdown circSOBP but not its linear format in U87 and u251 cells (Figure S1A). CircSOBP knockdown enhanced glioma cell proliferation, invasion, and migration. It also suppressed apoptosis. (Figures S1B–S1E, S2A, and S2B).
Figure 2.
Increased expression of circSOBP inhibits the proliferation, invasion, and migration of glioma cells
(A) The spliced sequence of circSOBP was cloned into the pEGFP-C1 vector, with the upstream and downstream containing the identified active ALU elements. RT-qPCR analysis of circSOBP overexpression in U87 and U251 cells.
(B) CCK-8 analysis of the effect of circSOBP overexpression on the proliferation of U87 and U251 cells.
(C) Cell viability was detected at 0 h, 24 h, 48 h, and 72 h after transfection with pEGFP-C1 or pEGFP-C1-circSOBP plasmids in U87 and U251 cells.
(D) Edu assay to analyze the effect on cell replication after overexpression of circSOBP in U87 and U251 cells. Scale bars, 50 μm.
(E) TUNEL analysis of the effect of circSOBP overexpression on apoptosis of U87 and U251 cells. Scale bars, 50 μm.
(F) Transwell assay to assess the invasive ability of glioma cells after overexpression of circSOBP.
(G) Wound healing assay to detect the migration level of glioma cells after overexpression of circSOBP. All statistics of error bars, S.E.M. from three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test. Normalized by the value of the control group.
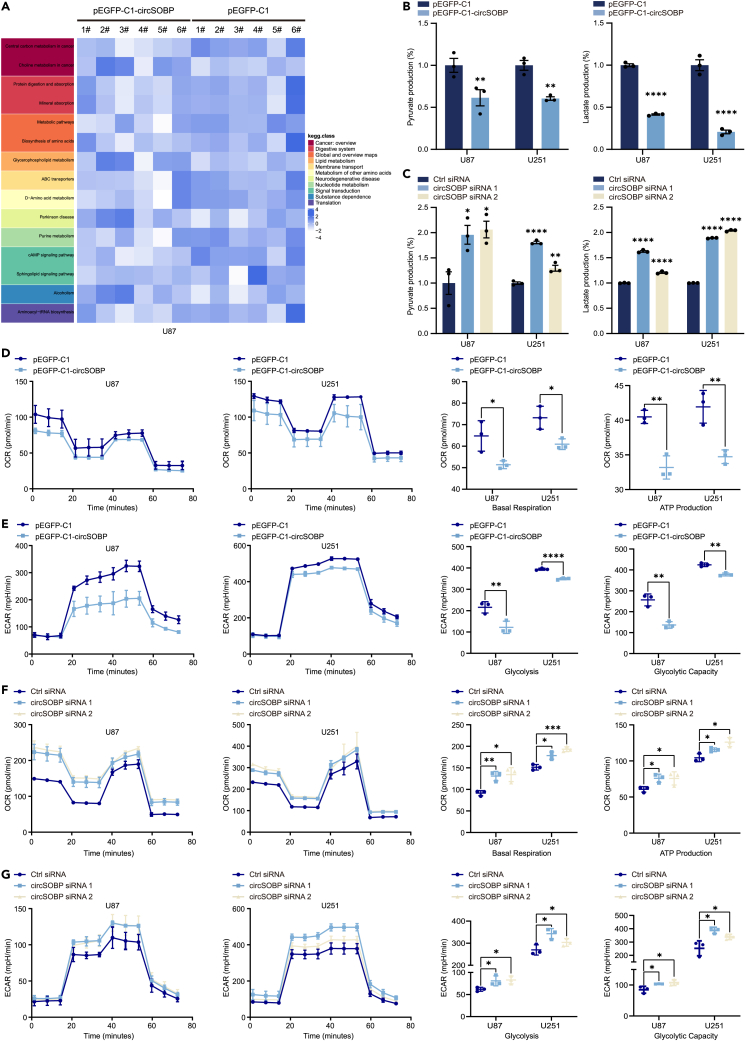
Metabolic reprogramming is a crucial characteristic in tumor formation and is essential for tumors to maintain their malignant phenotype.17 We performed metabolomic analysis after overexpression of circSOBP in glioma cells. Differential analysis revealed that the levels of metabolites were significantly altered after circSOBP overexpression (Table S2). For example, glycolysis and de novo lipogenesis (DNL) metabolites were downregulated (Figure S3A). Metabolomic data integration revealed metabolic pathways that were possibly changed in circSOBP overexpressing cells relative to controls. This included the most enriched metabolic pathways associated with glycolysis downstream of pyruvate and the major metabolites amino acids and purines produced by the TCA cycle (Figure S3B). In addition, we observed that the central carbon metabolic pathway was significantly downregulated in circSOBP overexpressing U87 cells (Figure 3A). Considering that tumor metabolic reprogramming is a series of metabolic alterations characterized by the Warburg effect as the main biochemical metabolic feature, we investigated the effect of circSOBP on glycolysis and mitochondrial respiration in glioma cells. CircSOBP overexpression inhibited pyruvate, lactate generation (Figure 3B) as well as mitochondrial respiration, glycolytic in U87 and U251 cells (Figures 3D and 3E). In contrast, circSOBP knockdown increased pyruvate and lactate production (Figure 3C) as well as increased cellular mitochondrial respiratory capacity and glycolysis by OCR and ECAR assays (Figures 3F and 3G). These results suggested that circSOBP had the potential to suppress the malignant phenotype of gliomas by regulating glycolysis or related genes.
Figure 3.
CircSOBP inhibits glycolysis and mitochondrial respiration in glioma cells
(A) Heatmap of pathway enrichment analysis of differential metabolites in circSOBP overexpression group (pEGFP-C1-circSOBP) and control group (pEGFP-C1) of U87 cells.
(B) Concentrations of pyruvate and lactate between circSOBP overexpression group and control cells.
(C) Concentrations of pyruvate and lactate between circSOBP-silenced and control cells.
(D) Oxygen consumption rate (OCR) indicators, including basal respiration and ATP production levels, were measured in U87 and U251 cells after overexpression of circSOBP.
(E) Overexpression of circSOBP in U87 and U251 cells and detection of extracellular acidification rate (ECAR) as an indicator of inferred glycolytic flux and glycolytic capacity.
(F) Oxygen consumption rate (OCR) was used as an indicator of oxidative phosphorylation (OXPHOS) to detect basal respiration and ATP production levels after silencing of circSOBP in U87 and U251 cells.
(G) Extracellular acidification rate (ECAR) was used as an index to infer glycolytic flux and glycolytic capacity to detect knockdown of circSOBP in U87 and U251 cells. All statistics of error bars, S.E.M. from three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
Identification of TKFC as a circSOBP binding protein
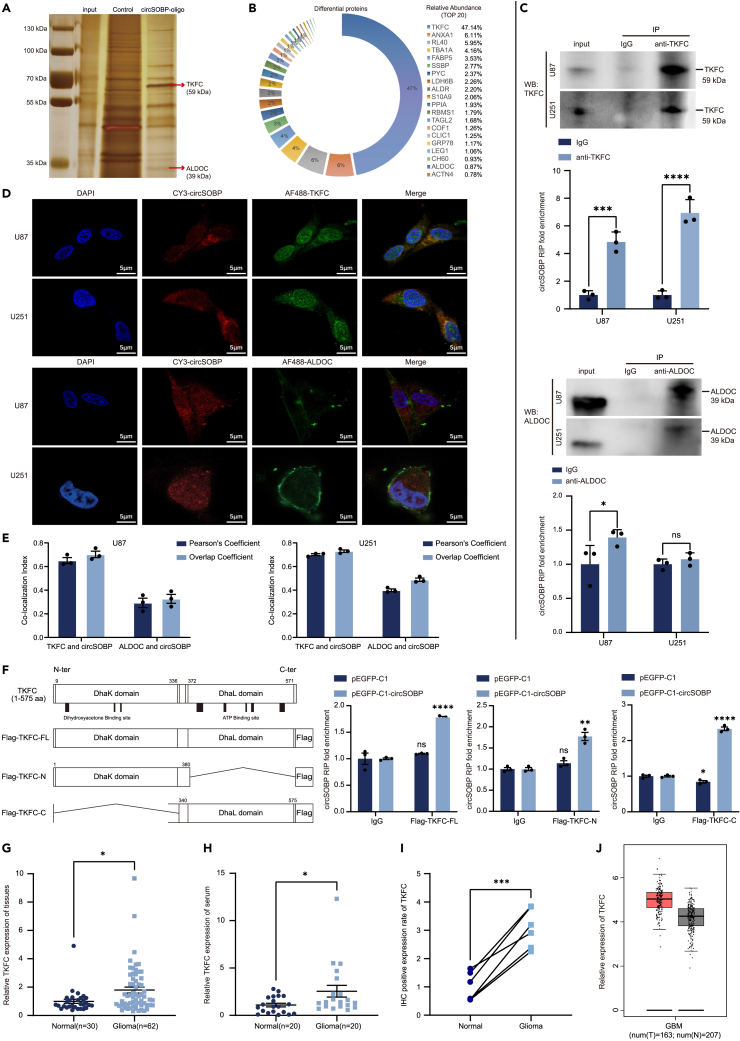
To explore the downstream mechanisms of circSOBP regulating glioma phenotype and metabolism, we performed RNA pulldown assays using biotin-labeled circSOBP oligo which against the junction site (Figure 4A). Mass spectrometry (MS) analysis showed 48 differentially expressed proteins, among which TKFC and ALDOC (Aldolase C) protein were consistent with silver staining results, and significantly, TKFC had a relative abundance of 47.14% (Figures 4B; Table S3). To further confirm the interaction, we performed RNA immunoprecipitation (RIP), the results showed that circSOBP was significantly enriched in the IPs of TKFC antibody, but not ALDOC antibody (Figure 4C). FISH results showed that circSOBP co-localized with TKFC in the cytoplasm of U87 and U251, but not ALDOC (Figures 4D and 4E). The above results confirmed that circSOBP binds to TKFC rather than ALDOC. According to the results of GO enrichment of differentially expressed binding proteins, circSOBP is primarily associated in the monosaccharide metabolic pathway (Figure S4A). Fructose was phosphorylated to fructose-1-phosphate (F1P), which was subsequently cleaved by ALDOC to dihydroxyacetone phosphate (DHAP) and glyceraldehyde. Glyceraldehyde was further phosphorylated to glyceraldehyde 3-phosphate (G3P) via TKFC. DHAP and G3P next enter the glycolytic/glycomimetic carbon pool (Figure S4B).28 In addition, circSOBP did not binding to TKFC and ALDOC transcripts (Figure S4C).
Figure 4.
Identification of TKFC as a binding protein for circSOBP
(A) Potential circSOBP-associated proteins were identified by SDS-PAGE and silver staining. Red arrows indicate bands identified by mass spectrometry as TKFC and ALDOC.
(B) Mass spectrometry (MS) to identify the relative abundance of proteins in specific bands (top 20 shown). The full list of proteins is shown in Table S3.
(C) Anti-TKFC and Anti-ALDOC antibody was used to pull down circSOBP in RIP. Western blotting showed effective pull down of TKFC and ALDOC in U87 and U251 cells. RT-qPCR determined the enrichment efficiency of circSOBP in TKFC and ALDOC pulldown samples, using IgG as a negative control.
(D) RNA in situ hybridization (FISH) and immunofluorescence (IF) double staining were performed to detect the co-localization of circSOBP with TKFC or ALDOC in U87 and U251 cells. Scale bar, 5 μm.
(E) ImageJ analysis of the intracellular distribution of circSOBP with TKFC or ALDOC. Pearson’s Coefficient >0.5 and Overlap Coefficient >0.6 indicate co-localization.
(F) Schematic diagram showing full-length TKFC and multiple truncated forms of TKFC. RT-qPCR analysis of enrichment levels of circSOBP in pulldown experiments of exogenously expressed full-length TKFC and multiple truncated forms. Using IgG as a negative control.
(G) The expression levels of TKFC in 30 normal brain tissues and 62 glioma tissues were detected by RT-qPCR.
(H) The expression levels of TKFC in 20 normal serum cases and 20 glioma serum cases were detected by RT-qPCR.
(I) Positive rate statistics of TKFC in normal brain tissue (n = 6) and glioma tissue (n = 6) by IHC staining.
(J) Expression levels of TKFC in gliomas were sourced from the GEPIA database. Red represents the glioma group (T) and gray represents the normal group (N). All statistics of error bars, S.E.M. from three independent experiments. NS, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
TKFC acts as a bifunctional RNA-binding protein that functions as a kinase through the C-terminus and inhibits the transcription of IFN-I through the N-terminus.29 To further characterize the interaction between cirSOBP and TKFC, we performed RNA immunoprecipitation assays following transfecting 293T cells with FLAG-tagged TKFC truncators expression plasmids. According to the results, circSOBP interacted with both the N-terminal and C-terminal of TKFC, with the C-terminal enrichment efficiency being the highest (Figures 4F and S4D). Our data showed increased levels of TKFC expression in glioma tissue and serum compared to normal samples (Figures 4G–4I and S4E), and consistent with the results of the GEPIA database analysis (Figure 4J). TKFC protein levels could be modified by circSOBP overexpression or knockdown in U87 and U251 cells but not transcript expression. Furthermore, circSOBP did not influence the protein or transcript levels of ALDOC (Figures S4F–S4H). According to the above results, TKFC acted as an RBP of circSOBP and its expression was regulated by circSOBP.
The effects of circSOBP and TKFC on the malignant phenotype and mitochondrial respiratory glycolysis of glioma
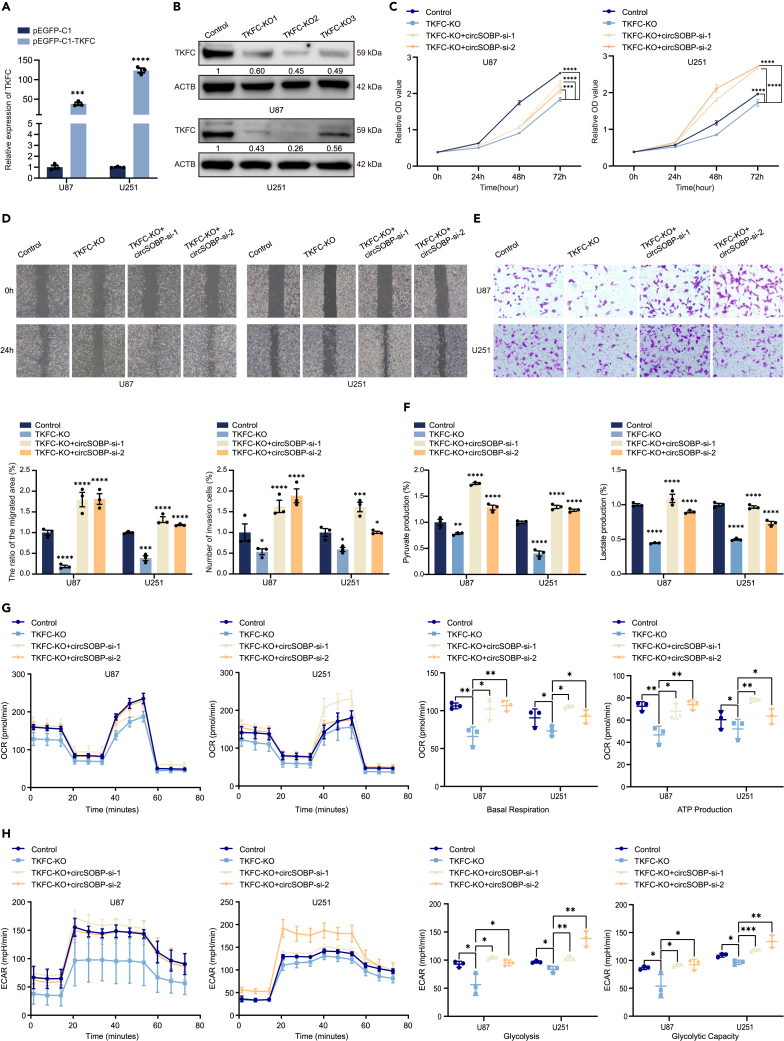
TKFC is a fructose metabolic pathway protease that has been linked to the development of multisystemic disorders, although it has not been studied in cancers.28 To investigate the role of TKFC in glioma, TKFC overexpression plasmids were constructed and their efficiency was verified (Figure 5A). Similarly, three pairs of TKFC sgRNAs were designed and tested for knockdown efficiency in U87 and U251 cells (Figure 5B). The highest knockdown efficiency of TKFC-KO2 was achieved and used for subsequent cell function experiments. Overexpression of TKFC increased the proliferation, migration, and invasion in U87 and U251 cells. More notably, circSOBP overexpression partly inhibited the promotion of TKFC on the malignant phenotype of glioma cells (Figures S5A and S6A–S6C). TKFC ablation, on the other hand, inhibited cell growth, migration, and invasion. We also discovered that silencing circSOBP simultaneously restored the negative effect of TKFC knockdown on U87 and U251 cell proliferation, migration, and invasion (Figures 5C–5E and S5B). These findings suggested that the function of circSOBP in cell proliferation, migration, and invasion was dependent on its interaction with TKFC protein.
Figure 5.
The effect of circSOBP binding TKFC on malignant phenotype as well as glycolysis and mitochondrial respiration in glioma
(A) RT-qPCR assay of TKFC overexpression efficiency in U87 and U251 cells.
(B) Western blotting analysis of TKFC knockdown efficiency by 3 independent pairs of gRNAs. ACTB was used as a negative control.
(C) TKFC knockdown alone or co-transfection of circSOBP siRNAs in U87 and U251 cells were analyzed by CCK-8.
(D) Wound-healing assays in U87 and U251 cells were independently knocked down with TKFC or co-transfected with circSOBP siRNAs.
(E) TKFC knockdown alone or co-transfection of circSOBP siRNAs in U87 and U251 cells were analyzed by transwell assays.
(F) Concentrations of pyruvate and lactate in cells knocked down with TKFC or simultaneously transfected with circSOBP siRNAs.
(G) OCR index assay in TKFC knockdown or co-transfected with circSOBP siRNAs in U87 and U251 cells.
(H) ECAR analysis of glycolytic flux and glycolytic capacity levels with TKFC knockdown or co-transfection of circSOBP siRNAs in U87 and U251 cells. All statistics of error bars, S.E.M. from three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
Furthermore, we performed rescue assays to identify the effects of circSOBP interaction with TKFC on glioma glycolysis and mitochondrial respiration. Overexpression of circSOBP reversed the promotion of pyruvate, lactate production (Figure S6D), mitochondrial respiration, and glycolysis (Figures S6E–S6F) by increasing TKFC expression. Meanwhile, we also observed that the effects of TKFC knockdown on pyruvate, lactate generation (Figure 5F), cellular OCR, and ECAR (Figures 5G and 5H) were reversed by knocking down circSOBP. These findings highlighted the significance of TKFC in glucose metabolism regulating by circSOBP.
CircSOBP promotes IFN-I signaling triggered by cytoplasmic poly (I:C)
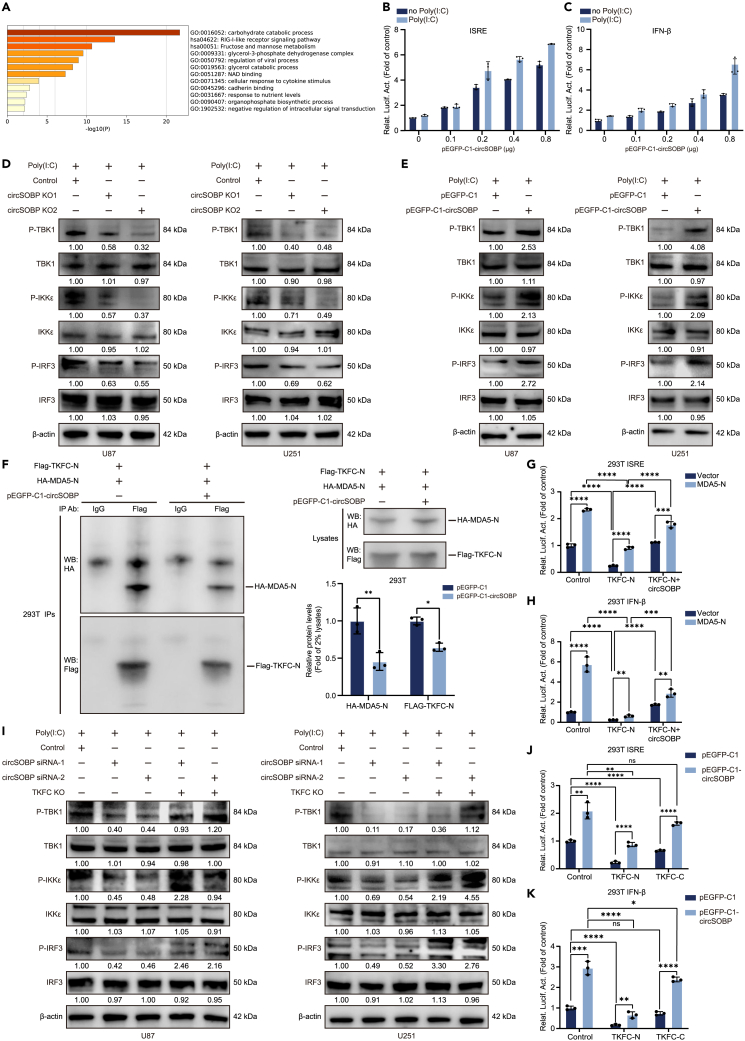
TKFC is not only a protein kinase for glucose metabolism, but is also associated with involvement in the RIG-I-like receptor signaling pathway (Figure 6A). We constructed ISRE and IFN β promoter reporter gene plasmids to determine whether circSOBP also regulated the transcriptional activity of IFN-I. In the dual luciferase reporter assay, circSOBP promoted poly(I:C)-induced activation of the ISRE and interferon promoters in a dose-dependent manner (Figures 6B and 6C). Furthermore, we created two sets of gRNAs to knock down the Alu repeat sequence adjacent to circSOBP to investigate the impact of circSOBP ablation on the RIG-I-like receptor signaling pathway (Figure S7A). We confirmed that abolishing Alu considerably reduced the formation of circSOBP (Figures S7B–S7F). Western blot analysis revealed that decreased circSOBP expression inhibited the phosphorylation of IRF3, IKKε and TKB1 in the RIG-I-like receptor signaling pathway (Figure 6D). Meanwhile, circSOBP overexpression boosted poly(I:C)-induced activation of these proteins (Figure 6E). We also observed that circSOBP promoted IFN-I transcription via the IRF-3 axis, yet not the NF-κB pathway (Figure S8A). These results demonstrated that the interaction of circSOBP with TKFC could regulate IFN-I expression.
Figure 6.
CircSOBP promotes IFN-I transcription by regulating TKFC
(A) KEGG enrichment analysis of the downstream pathways of TKFC and its interacting proteins.
(B and C) CircSOBP promoted Poly(I:C) mediated activation of ISRE and IFN promote in a dose-dependent manner. 293T cells were transfected with ISRE or IFN promoter luciferase reporter plasmid, pEGFP-C1-circSOBP expression plasmid or control plasmid and renilla luciferase reporter plasmid, respectively. The RLU values obtained from the renilla luciferase were used for normalization.
(D and E) Western blot showed the effect of circSOBP knockdown or overexpression on Poly(I:C)-mediated activation of RIG-I-like receptor signaling pathway-associated proteins. Using β-actin as a normalization.
(F) CircSOBP disrupted the binding between TKFC-N and MDA5-N. The indicated plasmids were co-transfected in 293T cells and cell lysates were immunoprecipitated with anti-FLAG antibody or control rabbit IgG. Immunoprecipitations were analyzed by western blotting with anti-HA (top) or anti-FLAG (bottom) antibodies. Western blot results of anti-HA and anti-FLAG antibodies in Lysates were used to do normalized to analyze the expression of transfected proteins.
(G and H) Dual luciferase reporter assay showed circSOBP rescued TKFC-N to inhibit MDA5-N-mediated activation of ISRE and IFN promoters in 293T cells.
(I) Expression levels of RIG-I-like receptor signaling pathway-related proteins in U87 and U251 cells treated with TKFC knockdown alone or co-transfected with circSOBP siRNAs.
(J and K) Effect of circSOBP on TKFC-N and TKFC-C to suppress ISRE and IFN promoter activation in 293T cells. The RLU values obtained from the renilla luciferase were used for normalization. All statistics of error bars, S.E.M. from three independent experiments. NS, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
Previous research revealed that TKFC functioned as a physiological MDA5 repressor, specifically inhibiting MDA5-mediated transcriptional activation of IFN-I.29 We transfected 293T cells with HA-tagged MDA5 or its truncators and FLAG-tagged TKFC or its truncators expression plasmids and performed co-immunoprecipitation assays. The results showed that MDA5 N-terminal card modules (MDA5-N) but not MDA5 C-terminal card modules (MDA5-C) interacted with TKFC (Figure S8B). Furthermore, rather than the ATP-rich C-terminal card modules (TKFC-C), the N-terminal card modules of TKFC (TKFC-N) bind to MDA5-N (Figure S8C). Dual luciferase reporter studies further revealed that the TKFC-N-terminal interacts with the MDA5-N-terminal to repress the transcription of the ISRE and IFN promoters, but not the C-terminal card module in 293T and glioma cells. (Figures S8D–S8E). It has been proposed that the interaction of TKFC to MDA5 has no impact on its enzymatic activity during glycolysis. However, it is unclear whether circSOBP mediates the interaction between TKFC and MDA5. In circSOBP overexpressing 293T and glioma cells, we co-transfected TKFC-N and MDA5-N expression plasmids. Immunoprecipitation assays revealed that circSOBP significantly blocked the interaction between TKFC-N and MDA5-N (Figures 6F, S9A, and S9B). Furthermore, reporter analysis revealed that circSOBP restored the function of TKFC-N, which inhibited MDA5-N-mediated transcriptional activation of the ISRE and IFN β promoter but not TKFC-C (Figures 6G–6H and S9C–S9D).
We performed rescue assays in U87 and U251 cells to determine how circSOBP interaction with TKFC affected RIG-I-like receptor signaling pathway proteins. The Western blot analysis showed that TKFC knockdown boosted phosphorylation of IRF3, IKKε, and TKB1, which was reversed by a decrease in circSOBP expression (Figure 6I). Meanwhile, enhanced circSOBP expression reversed TKFC’s inhibitory effect on RIG-I-like receptor signaling pathway proteins (Figure S9E). Dual luciferase analysis revealed that circSOBP triggered the ISRE and IFN β promoter via interaction with TKFC-N but not TKFC-C in 293T and glioma cells. (Figures 6J–6K and S9F). In summary, the above findings indicated that the circSOBP activation of IFN-I was regulated by interaction with the TKFC N-terminal card modules.
CircSOBP inhibits tumorigenesis, Warburg effect, and immunity of glioma in vivo
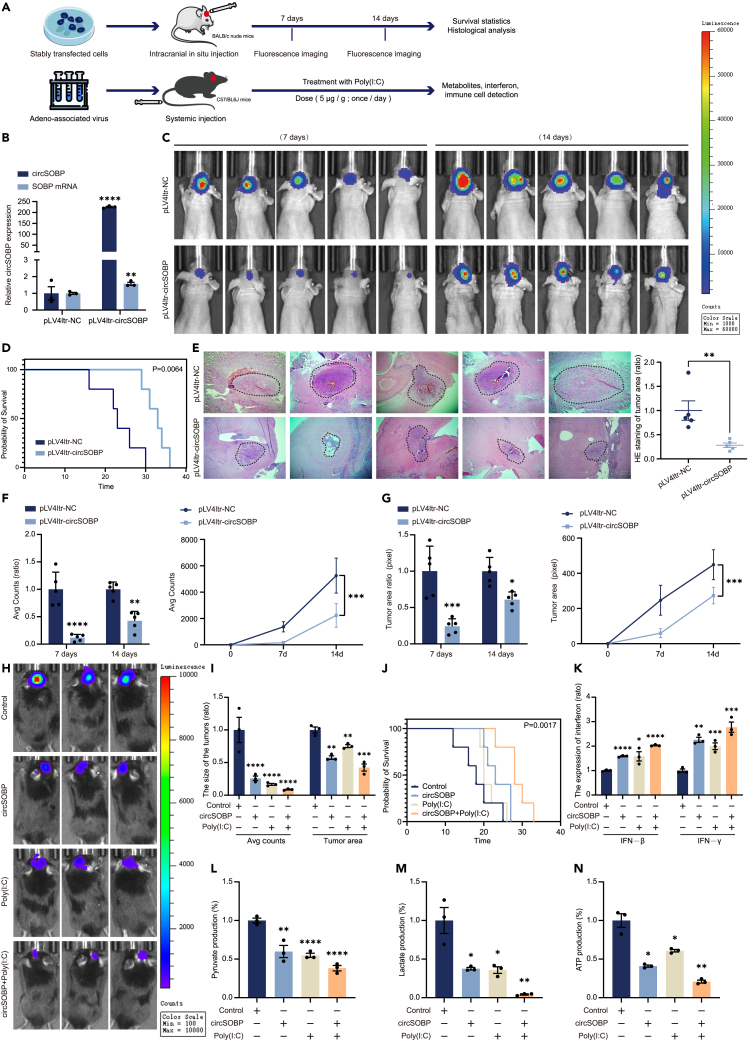
To investigate the effect of circSOBP binding to TKFC in glioma, we performed animal model studies using BALB/c nude mice and C57/BL6J mice (Figure 7A). The in vivo tumorigenic ability of glioma was investigated by cerebral in situ injection of circSOBP stable overexpression cell lines. The stability of circSOBP overexpression was verified by RT-qPCR analysis (Figure 7B). Increased expression of circSOBP significantly inhibited the tumorigenic ability of GBM in vivo, according to luciferase labeling of tumor cells as well as in vivo imaging of mice (photographed once every 7 days) (Figure 7C). The survival curve statistics showed that the circSOBP overexpression group had a longer life time compared to the control group (Figure 7D). HE staining showed that mice in the circSOBP overexpression group had less relative area of tumors in brain cross-sections compared to the negative control group (Figure 7E). Furthermore, fluorescence numerical statistics indicated that circSOBP overexpression reduced the growth and volume of mouse brain tumor in situ compared to the negative controls (Figures 7F and 7G). The IHC results showed that increased circSOBP expression significantly inhibited glioma proliferative ability in vivo (Figure S10A), and the protein level of TKFC was significantly decreased in the circSOBP overexpression group (Figure S10B). These findings suggested that circSOBP inhibited GBM tumorigenesis in vivo by downregulating TKFC expression.
Figure 7.
CircSOBP inhibits tumorigenesis and glycolysis as well as promotes the production of IFNs in vivo
(A) Illustration of the tumor transplantation timelines when treated with circSOBP.
(B) Expression level of circSOBP in glioma cells after transduction with overexpression lentivirus.
(C) Bioluminescence imaging of intracranial xenograft tumors in BALB/c nude mice is shown. Images were obtained after 5 min by intraperitoneal injection of 100 μL (25 mg/mL) of D-Luciferin. Seven days later, the same experimental procedure was performed to obtain images again.
(D) Kaplan-Meier plots of overall survival of mice with high circSOBP levels and controls.
(E) HE staining was used to detect the relative area of tumors in cross-sections of mouse brains.
(F) Quantitative analysis of the mean fluorescence intensity (left) and growth rate (right) of brain tumor areas in circSOBP overexpression and control groups of BALB/c nude mice.
(G) The size (left) and growth rate (right) of brain tumor areas (ROI Pixels) in circSOBP overexpression and control BALB/c nude mice were measured.
(H) Bioluminescence imaging of the C57/BL6J glioma model mouse with adeno-associated viral fluid alone or co-injected with Poly(I:C).
(I) Fluorescence intensity and area were used as indicators for the quantitative analysis of tumor size.
(J) Kaplan-Meier plots show overall survival of mice treated with circSOBP/Poly(I:C) together or alone.
(K) IFNs concentrations in the tumor tissues of the grouped mice indicated.
(L–N) The levels of glycolysis and energy metabolism in the tumor tissues of the grouped mice indicated. Data (L) suggest pyruvate contents. Data (M) imply lactate production. Data (N) are indicative of ATP concentrations. All statistics of error bars, S.E.M. from three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by two-tailed Student’s t test.
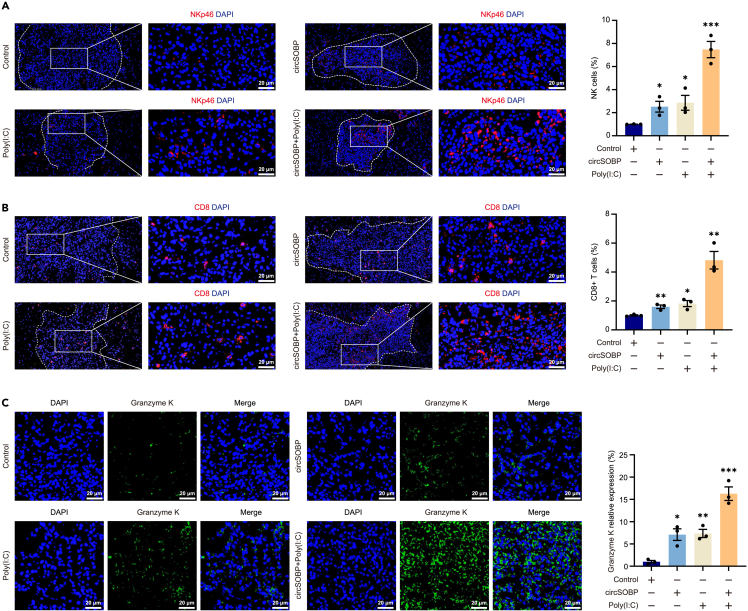
Sequence comparison showed that circSOBP and TKFC were highly conserved between human and mice (Figure S10C). Thus, we explored the activation of circSOBP on Warburg effect and immunity by adeno-associated virus (AAV) system in C57 glioma model. The GFAP is a protein specifically expressed brain astrocytes, and we constructed a shuttle plasmid containing the GFAP promoter sequence for packaging adeno-associated viruses (Figure S10D). IF staining of fusion-expressed EGFP confirmed the specific expression of circSOBP in the central system (Figure S10E). In vivo imaging showed that either circSOBP or Poly(I:C) treatment inhibited tumor growth. Notably, circSOBP enhanced the inhibitory effect of Poly(I:C) on glioma (Figures 7H and 7I). Survival curve statistics showed that the circSOBP+poly(I:C) group survived longer than mice treated with circSOBP or Poly(I:C) alone (Figure 7J). ELISA results showed that either circSOBP or Poly(I:C) could activate interferon transcription and synergistic effect of circSOBP with Poly(I:C) in vivo (Figure 7K). In vivo tumor metabolites analysis revealed that circSOBP or circSOBP-co-Poly(I:C) significantly suppressed the Warburg effect and ATP production (Figures 7L–7N). Furthermore, Poly(I:C) also suppressed glycolysis and ATP production in gliomas, which might result from the competitive binding of activated RIG-I to MAVS to inhibit the role of HK2 in glycolysis.30 To further investigate the effect of IFN-I activation on CD8+ T and NK cells, IF staining for CD8 and NKp46 markers was performed on tumor tissues. The results showed that synergistic treatment with circSOBP and Poly(I:C) significantly increased the number of CD8+ T cells, NK cells, and Granzyme K which is synthesized by immune cells and ability to act as a tumor-killing enzyme compared with circSOBP or Poly(I:C) treatment alone in glioma (Figures 8A–8C). These results implied that circSOBP could suppress tumorigenicity and the Warburg effect in glioma while also activating tumor immunity.
Figure 8.
CircSOBP treatment produces effective tumor-reactive CTL and NK cell responses
(A and B) IF staining analysis of circSOBP and Poly(I:C) treatment alone or in combination increased infiltration of NK cells and CD8+ T cells into the tumor. Values of control were used to normalize and quantify the rate of cell positivity. Scale bar, 20 μm.
(C) Increased expression of granzyme K in tumor-infiltrating CD8+ T cells or NK cells using immunofluorescence (IF) staining. Scale bar, 20 μm. All statistics of error bars, S.E.M. from three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by two-tailed Student’s t test. Normalized by the value of the control group.
Discussion
Most circRNAs were identified in low abundance in human normal brain tissue and glioma tissue in our RNA-seq data which was consistent with previous reports.31 However, certain circRNAs have characteristics as stability, high conservatism, tissue specificity, and particular functions.32 For example, circ-MAPK4 expression was elevated in gliomas and promoted the proliferation of glioma cells.33 circDIDO1 expression was downregulated in gastric cancer and inhibited tumor progression.34 In this work, circSOBP significantly decreased in glioma tissue compared to normal brain tissue and inhibited the abilities of glioma to proliferation, invasion, and migration. circSOBP is a highly conserved exonic circRNA molecule generated by back splicing of the SOBP exons 2 and 3. Previous research had shown that it significantly reduced and inhibited migration and invasion in prostate cancer.15 In addition, similar lower circSOBP expression in the serum of patients with gastric cancer,16 we also discovered it downregulated in the sera of patients with glioma. Considering that circRNA is a stable covalently closed loop structure, circSOBP has a natural advantage as a diagnostic and therapeutic marker for glioma. However, further information about the role of circSOBP is lacking, and we characterize the function and mechanism of circSOBP in glioma for the first time in a thorough and comprehensive manner herein. The type of circRNAs determines different subcellular distribution and biological mechanisms. Exonic circRNAs are mainly distributed in the cytoplasm and play biological roles as miRNA sponges or through RNA-binding proteins.32 Such as, circRNA cRAPGEF5 inhibited growth and metastasis of renal cell carcinoma by sponging miR-27a-3p.35 For instance, circURI1 inhibited gastric cancer metastasis via modulating invasion-related gene alternative splicing.36 circFOXP1 interacted with PTBP1 to promote tumor progression and Warburg effect in gallbladder cancer.37 In contrast, a few circRNAs retain introns during splicing, localize in the nucleus, and function through pre-transcriptional regulation.9 Our work confirmed that circSOBP bind to TKFC specifically and tightly in the cytoplasm, inhibiting its protein expression but not the transcript level. TKFC is upregulated in gliomas and can significantly promote the malignant phenotype of glioma, as well as its effect in glioma was rescued by circSOBP. However, we cannot deny the possibility that circSOBP functions through the ceRNA mechanism. Similar to CDR1as, which can bind to both P53 protein and sponge mir-7 to suppress tumor development.38,39
Malignancies consistently produce ATP preferentially by glycolysis under conditions of sufficient oxygen, which is known as the Warburg effect and is one of the most essential characteristics of tumors.40 It is characterized by increased glycolysis, increased glucose uptake and consumption, increased lipid and protein synthesis, and increased amino acid catabolism, such as glutamine. These metabolic modifications are intended to fulfill the tumor energy, biomolecule synthesis, and redox homeostasis demands.41 In addition, mutations or abnormal expression of some metabolism-related genes such as IDH, EGFR, and PTEN are considered to be key molecules in the development of gliomas and are crucial for the pathological diagnosis and molecular typing of gliomas.2 A growing number of studies have recently shown that circRNAs play an important role in regulating tumor cell metabolism. Li J. et al. found that circRPN2 bind to the ENO1 protein and inhibited glycolysis in hepatocellular carcinoma.42 Jiang X et al. discovered that EIF4A3-induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer.43 Song J et al. identified circHEATR5B encoding a novel protein, HEATR5B-881aa, that directly interacted with JMJD5 and inhibited glycolysis of GBM.44 The kinase TKFC plays a crucial role in the metabolism of monosaccharides via phosphorylating glyceraldehyde to glyceraldehyde 3-phosphate (GA3P), which contributes to the production of pyruvate, a major substrate of glycolysis and the tricarboxylic acid cycle (TCA).45 Considering that TKFC is a circSOBP binding protein, we investigated the effect of circSOBP on glioma glycolysis and mitochondrial respiration. On the one hand, circSOBP significantly reduced OCR, ECAR, pyruvate, and lactate levels in glioma cells. TKFC, on the other hand, was a specific target of circSOBP for regulating metabolic reprogramming in glioma, and its metabolic phenotype was rescued by circSOBP. Importantly, metabolomic analysis revealed that increasing circSOBP expression not only suppressed the Warburg effect in glioma, but also inhibited adipose heavy head production (DNL) via TCA. However, because of the interconversion of DHAP and GA3P, tumor cells were likely adjusting for the lack of 3-phosphate carbon pool caused by increased circSOBP expression through DHAP,and the later postulation should be further investigated in GBM in the future. We believe the above results suggested that circSOBP inhibited glycolysis and ATP production in gliomas by regulating TKFC, providing a candidate target for glioma treatment by blocking the Warburg effect.
Emerging evidence suggests that MDA5 activation induces IFN-I signaling, which in turn activates innate and adaptive immune responses, as the key to the antitumor efficacy of MDA5-targeted therapies.46 Our results indicated that TKFC, a natural MDA5 inhibitor,29 had a dual role in regulating the natural immune response and glycolysis. Upregulation of TKFC expression increased aerobic glycolysis in gliomas while inhibiting interferon-mediated immune system activation through binding to MDA5. Interestingly, the TKFC C-terminus, which included the ATP site necessary for glycolysis, was clipped without affecting the ability to inhibit IFN-I activation, indicated that TKFC was a critical modulator of metabolic immune reprogramming during glioma development. The effects of circSOBP intraction TKFC on gliomas in humans and mice were comparable, most probably due to sequence homology and evolutionary conservation. In this study, IFN-I signaling via two pathways could be considerably activated by the interaction of circSOBP with TKFC. Firstly, circSOBP directly enhanced IFN-I transcriptional activation of the IRF3 pathway by blocking TKFC binding to MDA5. Secondly, lactate was able to bind to MAVS to block its signaling, while the inhibition of the Warburg effect by circSOBP indirectly enhanced the transcription of IFN-I. Notably, MAVS-RIG-I recognition disrupted hexokinase binding to MAVS, resulting in impaired hexokinase mitochondrial localization and glycolysis. Thus, our study identified that circSOBP promoted IFN-I-mediated antitumor effects by disrupting the metabolic reprogramming of gliomas with a positive circular mechanism linking energy metabolism and innate immunity. Synergistic adeno-associated virus-mediated circSOBP overexpression appeared to stimulate IFN-I production and antitumor immunity more effectively than the double-stranded RNA analogue (poly(I:C) alone. Furthermore, this adeno-associated virus-mediated circSOBP overexpression had little toxicity to normal cells, which might be caused by the brain astrocyte-specific promoter and serotype.47 Furthermore, circSOBP adeno-associated viral therapy improved antitumor immune responses as evidenced by increased IFN-β, IFN-γ, and granzyme K levels, which were associated to an increase in the frequency of functionally activated, tumor-infiltrating cytotoxic CD8+ CTLs and NK cells. It seems that by modulating the crosstalk between glioma metabolism and immunity is more able to circumvent the protumorigenic effects caused by prolonged direct activation of interferon and promote its anti-tumor immunity.48 However, the complex mechanisms involved require more molecular studies.
In conclusion, we discovered that circSOBP was significantly downregulated in gliomas and had a cancer-suppressive function. By binding and regulating TKFC expression, circSOBP blocked the Warburg effect of glioma and stimulated interferon-initiated antitumor immunity, suggesting a positive feedback mechanism to reverse metabolic immunological reprogramming in malignancies. Our research not only gives important molecular insights into how energy metabolism and IFN-I interact to regulate diverse biological processes, but also offers novel therapeutic strategies for glioma.
Limitations of the study
Although our study shows that circSOBP regulates the RIG-I-like receptor signaling pathway and glycolysis in glioma cells by interacting with TKFC. However, the exact binding site of circSOBP interaction with TKFC and how it is regulated remains unknown. In addition, the regulatory role between lactate and IFN-I is a complex process involving multiple immune cells and signaling pathways in the tumor microenvironment. Therefore, more immunological studies are needed to reveal the specific mechanisms by which circSOBP regulates the crosstalk between metabolic alterations and immune responses in gliomas. Finally, the application of circSOBP as a biomarker or therapeutic target in clinical practice needs to be further explored.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TKFC | Proteintech | 12224-1-AP |
| Anti-Flag | Proteintech | 20543-1-AP |
| Anti-HA | Proteintech | 51064-2-AP |
| Anti-GFP | Proteintech | 50430-2-AP |
| Anti-TBK1 | Santa Cruz | A-6 |
| Anti-pTBK1 | CST | D52C2 |
| Anti-IKKε | CST | #2690 |
| Anti-pIKKε | CST | #8766 |
| Anti-IRF3 | Abclonal | A11373 |
| Anti-pIRF3 | Abclonal | AP0623 |
| Anti-NF-κB | CST | #8242 |
| Anti-pNF-κB | CST | #3031 |
| Anti-ALDOC | Proteintech | 14884-1-AP |
| Anti-Ki67 | CST | #9449 |
| Anti-CD8α | CST | #98941 |
| Anti-Granzyme K | Proteintech | 27894-1-AP |
| Anti-NCR1 | CST | #23732 |
| Anti-Lamin B1 | Santa Cruz | sc-377000 |
| Anti-GAPDH | Sangon | D110016 |
| Anti-β-actin | Sangon | D110001 |
| Bacterial and virus strains | ||
| DH5α,Escherichia coli | Sangon | B528413 |
| Adeno-associated virus | This paper | N/A |
| Lentivirus | This paper | N/A |
| Biological samples | ||
| Human glioma tissues and serums | The First Affiliated Hospital of USTC | N/A |
| Fetal bovine serum | Clark Bioscience | FB25015 |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | Invitrogen | 15596026 |
| DMEM medium | Gibco | C11995500BT |
| DAPI Stain Solution | Sangon | E607303 |
| Lipo8000 transfection reagent | Beyotime | C0533 |
| CCK-8 reagent | Dojindo | CK04 |
| Matrigel | BD | 356234 |
| RIPA buffer | Beyotime | P0013B |
| BSA | Beyotime | ST025 |
| yeast RNA | Beyotime | R0038 |
| Protein G Agarose | Thermo | 88848 |
| ECL chemiluminescent reagents | Tanon | 180-501 |
| M-280 Streptavidin | Thermo | 11206D |
| 1.25% tribromoethanol | Lab Anim Tech | N/A |
| Ploy(i:c) | InvivoGen | tlrl-pic |
| D-Luciferin potassium salt | Beyotime | ST196 |
| Critical commercial assays | ||
| TruSeq Ribo Profile Library Prep Kit | Illumina | N/A |
| Illumina Nextseq 500 system | Novogene | N/A |
| Deposited data | ||
| Raw and analyzed data (RNA-seq) | This paper | GEO:GSE153692 |
| Experimental models: cell lines | ||
| HEB | Immocell | IM-H209 |
| U251 | ECACC | 09063001 |
| LN229 | ATCC | CRL-2611 |
| TJ905 | Cellcook | CC1722 |
| SHG44 | Pricella | CL-0207 |
| T98G | ATCC | CRL-1690 |
| U373 | ATCC | HTB-17 |
| U87 | ATCC | HTB-14 |
| GL261 | DSMZ | ACC 802 |
| GBM-Y2 | This paper | N/A |
| Experimental models: organisms/strains | ||
| C57BL/6 | Gempharmatech | N000295 |
| BALB/c-Nude | Gempharmatech | D000521 |
| Recombinant DNA | ||
| pAAV-GFAabc1d-NC | This paper | N/A |
| pAAV-GFAabc1d-circSOBP | This paper | N/A |
| pLV4ltr-Puro-circSOBP | This paper | N/A |
| pEGFP-C1-circSOBP | This paper | N/A |
| pcDNA3-HA-MDA5 | This paper | N/A |
| pcDNA3-HA-MDA5-ΔN | This paper | N/A |
| pcDNA3-HA-MDA5-ΔC | This paper | N/A |
| pcDNA3-TKFC-Flag | This paper | N/A |
| pcDNA3-TKFC-ΔN-Flag | This paper | N/A |
| pcDNA3-TKFC-ΔC-Flag | This paper | N/A |
| pCMV-VSV-G | Beyotime | D8215 |
| pCAG-dR8.9 | Beyotime | D8216 |
| phRL-TK | Beyotime | D2760 |
| pISRE-TA-Luc | Beyotime | D2179 |
| IFN-Beta_pGL3 | Addgene | #102597 |
| pHelper | Fenghui Biotechnology | BR417 |
| pAAV-RC9 | Fenghui Biotechnology | BR227 |
| Software and algorithms | ||
| ImageJ | National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9.0 | GraphPad Prism Software, Inc | https://www.graphpad.com/ |
| SPSS Statistics | IBM | https://www.ibm.com/cn-zh/products/spss-statistics |
| Other | ||
| TransStart Green qPCR SuperMix kit | TransGen | AQ101-01 |
| Cytoplasmic and Nuclear Fractionation kit | Invent | SC-003 |
| RNAmax-T7 biotin-labeled transcription kit | Ribobio | C11002-1 |
| DIG-RNA labeling kit | Roche | 11175033910 |
| DIG Northern Starter Kit | Roche | 12039672910 |
| Primerstar Max DNA polymerase Mix | Takara | R045B |
| EdU Cell Proliferation Kit | Beyotime | C0078S |
| CellTiter-Lumi™ Luminescent Cell Viability Assay Kit | Beyotime | C0056S |
| One-Step TUNEL Apoptosis Kit | RiboBio | C11012-2 |
| Fast Silver Stain Kit | Beyotime | P0017S |
| Dual Luciferase Reporter Gene Assay Kit | Beyotime | RG088S |
| Seahorse XF Cell Mito Stress Test Kit | Agilent | 103015-100 |
| Seahorse XF Glycolysis Stress Test Kit | Agilent | 103020-100 |
| Pyruvate (PA) Content Assay Kit | Sangon | D799450 |
| Lactic Acid (LA) Content Assay Kit | Sangon | D799099 |
| ATP Assay Kit | Beyotime | S0026 |
| Mouse IFN-β(Interferon Beta) ELISA Kit | ElabScience | E-EL-M0033c |
| Mouse IFN-γ(Interferon Gamma) ELISA Kit | ElabScience | E-EL-M0048c |
| AAVpro® Purification Kit | Takara | 6675 |
| AAVpro® Titration Kit (for Real Time PCR) | Takara | 6233 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chaoshi Niu (niuchaoshi@ustc.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Human normal and glioma samples
This study was approved by the ethics committee of The First Affiliated Hospital of University of Science and Technology of China in accordance with the Declaration of Helsinki (approval No. 2019-N (H)-064). 94 glioma cases and 41 normal brain tissue case specimens were collected in the Department of Neurosurgery of the First Affiliated Hospital of USTC from 2020 to 2022 and were available from our institutional tissue bank. These samples diagnosed as gliomas by clinical and histopathological methods. Clinical information on all recruited patients was collected from the electronic medical records. Informed consents were obtained from all participants. All samples were washed with PBS after resection and stored in a -80°C refrigerator. The distribution of age, sex, and pathologic characteristics of the sample is shown in Table S1.
Cell culture
In this study, human-derived glioma cell lines including U251, LN229, TJ905, SHG44, T98G, U373, U87 and glioma primary cell line GBM-Y2, human brain astrocyte normal cells (HEB), and mouse-derived glioma cell line GL261 all had available information for cell line authentication. All cells were cultured in DMEM medium (Gibco, USA) containing 10% FBS (Clark Bioscience, USA) and maintained in a humidified incubator at 37°C with 5% CO2.
Animal experiments
All animal experiments were approved by the Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China (approval No. 2019-N (A)-100) and were conducted in accordance with the Guide for the Care and Use of Laboratory Animal by International Committees. For in vivo tumor xenograft assay, Primary glioma cells (GBM-Y2) with stable overexpression of circSOBP were constructed by adding lentiviral solution (MOI=3) and polybrene (a pro-infection reagent, 8μg/ml) to cells in logarithmic growth phase. Cells were infected for 48 hours then incubated for two weeks using medium containing puromycin (2 μg/ml). BALB/c nude mice (female, 4 weeks old) were purchased from Gempharmatech (China) and housed under specific pathogen-free conditions. After a week, nude mices were anesthetized with 1.25% tribromoethanol (20 μl/g) by intraperitoneal injection, and their heads were fixed in prone position on the brain stereotactic apparatus. The skin of the head was disinfected with 75% ethanol and then clipped to expose the bregma. The injection site was located 1 mm behind bregma, 2 mm from the midline, and the needle was inserted 3 mm vertically, using a micropump to push 10 μl of cells with or without circSOBP overexpression (3×105 per mice) at a rate of 2.5 μl/min. After the injection was completed, the incision was sutured and disinfected. The survival of the mice was observed and recorded, and tumor size was detected using a small animal live imaging system IVIS spectrum (Perkin Elmer, USA). Finally, the mice were executed and the brains were removed and fixed in 10% formaldehyde. For the detection of metabolites and immune cell experiments in mice, we used C57BL/6 mice (female, 4 weeks old) to construct a glioma model with GL261 cells and method was the same as above. Mice were divided into 4 groups: (a) poly(i:c) + pAAV-GFAabc1d-circSOBP; (b) no-poly(i:c) + pAAV-GFAabc1d-circSOBP; (c) poly(i:c) + pAAV-GFAabc1d-NC; (d) no-poly(i:c) + pAAV-GFAabc1d-NC according to whether they were systematically injected with AAV virus solution and poly(i:c) (5 μg/g/d). Tumor size was detected using a small animal live imaging system IVIS spectrum (Perkin Elmer, USA). Mice were killed by the anesthesia method and subjected to subsequent experiments.
Method details
Total RNA extraction and RNA-seq analysis
The total RNA was isolated from the tissue samples using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Measure the concentration of total RNA with Nanodrop and assess RNA integrity and DNA contamination by denaturing gel electrophoresis. The Ribo-minus transcriptome libraries were constructed with TruSeq Ribo Profile Library Prep Kit (Illumina, USA), according to the manufacturer’s instructions. The libraries were then subjected to 151-nt paired-end sequencing generating a depth of ∼100 million read pairs with an Illumina Nextseq 500 system (Novogene, China). Annotation of the identified circRNAs using the CircBase database and circ2Trait disease database. The differentially expressed circRNAs were determined by DEseq2 with the corresponding cutoff (P value <0.05, RPKM ≥1, |log2(fold change) | ≥1 ). Differentially expressed circRNAs between the glioma (Glioma tissue) and control groups (Normal brain tissue) were tested by t-test.
Real-time quantitative PCR (RT–qPCR)
The cDNA was prepared using the GoScript Reverse Transcription System (Promega, USA) according to the manufacturer’s protocol. RT-qPCR was performed using the TransStart Green qPCR SuperMix kit (TransGen, China) with a LC96 real-time PCR system (Roche, Switzerland). GAPDH was used as an internal control. The primers used are listed in Table S4, Supporting Information.
Nuclear and cytoplasmic extraction
Subcellular fractionation was performed to obtain cytoplasmic and nuclear fractions using Cytoplasmic and Nuclear Fractionation kit (Invent, USA) according to the manufacturer’s instructions. GAPDH was used as a cytoplasmic endogenous control and U1 as a nuclear endogenous control.
Fluorescence in situ hybridization (FISH)
The Cy3-labelled RNA probes spanning the splice junction specific to circSOBP were designed and synthesized by in vitro transcription. The assay was performed using RNAmax-T7 biotin-labeled transcription kit (Ribobio, China). Before the experiment, cells were inoculated at 60%-70% density per well, washed with 1X PBS for 5 min and fixed with 4% paraformaldehyde at room temperature for 10 min, then 1 ml of pre-chilled permeabilization solution (PBS with 0.5% Triton X-100) was added to each well and incubated for 5 min at 4°C. After three washes for 5 min, cells were equilibrated with pre-hybridization buffer for 30 min at 37°C. Then, cells were incubated overnight (avoid light) at 37°C in FISH probe hybridization buffer. After three times with Wash Buffer I (4 × SSC with 0.1% Tween-20), once with Wash Buffer II (2 × SSC), Wash Buffer III (1 × SSC) at 42°C inthe dark for 5 min and one wash with 1 × PBS at room temperature, the cell nucleus was stained with 1 × DAPI. Images were captured using an ZEISS LSM980 confocal microscope.
Northern blotting
The digoxin-labeled RNA probe was prepared using DIG-RNA labeling kit (Roche, Switzerland), which spanning the splice junction of circSOBP. Total RNA (20 ug/well) was electrophoresed (5 V/cm) by 2% formaldehyde denaturing agarose gel and checked using a UV imaging system. After the completion of electrophoresis, the RNA was transferred to the nylon membrane with the capillary transfer method. Washing, pre-hybridization and hybridization were performed using DIG Northern Starter Kit (Roche, Switzerland). After washing, the blot was detected with CDP-Star luminescent solution (Roche, Switzerland) and imaged with the AI680 chemiluminescent imaging system (GE, USA).
Vector construction
To construct the circSOBP expression vector, the exon 2 and 3 sequences of the SOBP (NM_018013) genome and its upstream (chr6:107823976-107824263) and downstream (chr6:107827998-107828284) repeats were amplified from U87 genomic DNA using Primerstar Max DNA polymerase Mix (Takara, Japan) and cloned into the transient expression plasmid pEGFP-C1, the lentiviral expression plasmid pLV4ltr-Puro or the adeno-associated virus expression plasmid pAAV-GFAabc1d. The gRNAs targeting the repeats of circSOBP were designed and cloned into the lentiCRISPRv2 vector for knocking down the expression of circSOBP. As above, the CDS sequence of TKFC was amplified and cloned in the pEGFP-C1 plasmid to construct a protein expression plasmid for TKFC. Truncates of TKFC and MDA5 were constructed by amplifying the C- or N-terminal sequences of the proteins that was cloned in the pcDNA3 plasmid and insertion of Flag and HA tags. phRL-TK and pISRE-TA-Luc plasmids purchased from Beyotime Biologicals. IFN-Beta_pGL3 plasmid provided by Addgene Plasmid Repository.
Cell transfection
The small interfering RNA (siRNA) oligonucleotides and the negative control siRNA were synthesized by Ribobio (RiboBio Biotechnology, China). Cells were inoculated in culture dishes for 24 hours and continued to be cultured for 48 hours after transfection of siRNA or vector using lipo8000 transfection reagent (Beyotime, China). Following the incubation period, they were collected for other assays.
Cell proliferation analysis
For the cell proliferation ability, cells were digested with 0.25% trypsin and seeded in 96-well plates, transfected and then remained under culture for 24, 48, and 72 hours at 37 degrees. 10 μl of cck-8 reagent (Dojindo Laboratories, Japan) was added to each well and the absorbance value at 450 nm was measured after incubation at 37 degrees for 4 hours. For the EdU assay, cells were trypsinized and seeded in 6-well plates at a density of 5×104 cells per well. After transfection for 48 hours, cell proliferation was measured using EdU Cell Proliferation Kit with Alexa Fluor 594 (Beyotime, China). Images were captured by Olympus' BX53 upright microscope and positive rates were counted by imageJ software. For Cell Titer Glo experiments, 2-3×104 cells were seeded in 96-well plates. Samples were prepared using the CellTiter-Lumi™ Luminescent Cell Viability Assay Kit (Beyotime, China) after 0 h, 24 h, 48 h and 72 h of transfection, respectively, and detected by chemiluminescence using a multi function measuring instrument (Switzerland, Tecan M200).
TUNEL assay
For the cell apoptotic rate, after transfection for 48 hours, cells were digested with 0.25% trypsin and seeded on poly-L-lysine coated slides. Next, TUNEL assay was performed using the One-Step TUNEL Apoptosis Kit (RiboBio, China) according to the manufacturer’s instructions. Images were captured by Olympus' BX53 upright microscope and positive rates were counted by imageJ software.
Invasion and migration assay
The cell migration and invasion abilities were determined using transwell chamber (8 μm, 6-well insert; Costar). In the invasion assay, matrigel (BD Biosciences, USA) was diluted (1:40) with serum-free medium, mixed and used to coat the insertion chamber membrane. In the migration assay, no matrix was used to coat the inserted chamber membrane. Then, cells (1 × 104) with 48 h transfection in serum-free medium were added to the upper chamber, and medium containing 10% FBS was added to the lower. Cells were continued to be incubated for 48 hours then fixed with 4% paraformaldehyde and the number of cells that migrated or invaded into the lower chambers was counted under an inverted microscope (Olympus, Japan).
RNA pulldown assays
The specific 5’biotin-labeled probe complementary to the sequence at the the splice junction of circSOBP was synthesized by Sangon Biotech (China). In brief, cells (2×107) were washed with 1×PBS to remove remaining medium and UV (1200 mJ) cross-linked on ice for 2 minutes. After lysis of the cells with RIPA buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM DTT, Complete protease inhibitor, and 0.1 U/μl RNase inhibitor) on ice for 10 min, the cells were harvested by cell scraper in RNase-free EP tubes and further fragmented by Ultrasonic Homogenizer (Scientz Biotechnology, Japan). Then, the samples were centrifuged (4°C) at 12,000g for 20 minutes and collect the supernatant in a new RNase-free EP tube. The probe (100 pmol/tube) was added to the supernatant and incubated for 2 h at 4°C with rotary shaker. Meanwhile, the M-280 streptavidin beads (Invitrogen, USA) were washed three times with RIPA buffer and blocked by mixing BSA (1 mg/ml) and yeast RNA (0.5 mg/ml) for 1 h at 4°C. The blocked beads were mixed with the cell supernatant which contained the probe and incubated overnight using rotary shaker at 4°C. Finally, we harvested the beads and assayed the probe enrichment efficiency by RT-qPCR. The binding proteins of circSOBP were identified by Fast Silver Stain Kit (Beyotime, China) and mass spectrometry (MS).
Immunoprecipitation
Cells were digested by 0.25% trypsin, washed twice with 1×PBS, and were sufficiently lysed on ice with RIP lysis buffer (20mM Tris (pH7.5),150mM NaCl,1% Triton X-100, Complete protease inhibitor, and 0.1 U/μl RNase inhibitor). Then, the samples were centrifuged (4°C) at 12,000g for 20 minutes and collect the supernatant in a new RNase-free EP tube. Protein G Agarose (Thermo, USA) were added to the cell supernatant (50 ul/tube) and incubated overnight by rotary shaker at 4°C. After washing the beads three times, the following antibodies: anti-TKFC (Proteintech, 12224-1-AP), anti-Flag (Proteintech, 20543-1-AP) or anti-HA (Proteintech, 51064-2-AP) was mixed with them and incubated 6h by rotary shaker at room temperature. Finally, we harvested the beads and assayed the enrichment efficiency by Western blot and RT-qPCR.
Luciferase reporter assay
The 293T cells of 1×104 per well density were seeded in 96-well white opaque plates 24 hours before transfection. In this study, reporter gene plasmids IFN-Beta_pGL3, phRL-TK, pISRE-TA-Luc, were transfected with 50 ng per well and expression gene plasmids pEGFP-C1, pEGFP-C1-circSOBP, pcDNA3, pcDNA3-TKFC-Flag, pcDNA3-TKFC-ΔC-Flag, pcDNA3-TKFC-ΔN-Flag, pcDNA3-HA-MDA5, pcDNA3-HA-MDA5-ΔN, pcDNA3-HA-MDA5-ΔC were transfected with 0.4 ug per well. After transfection for 24 hours, replace the medium that contains poly(i:c) (4 ug/ml) and continue incubation for 16 hours. Next, luciferase reporter assay was performed using the Dual Luciferase Reporter Gene Assay Kit (Beyotime, China) according to the manufacturer’s instructions. The luminescence (RLU) of each well was measured with the Infinite M200 (Tecan, Switzerland). The activation level of target reporter genes was calculated among different samples using renilla luciferase as an internal reference.
Seahorse analysis
The mitochondrial bioenergetics of U87 and U251 cells, in response to altered circSOBP or TKFC expression, was assessed using a Seahorse XFe96 Analyzer (Agilent Technologies, USA). The U87 or U251 cells of 2×104 per well density were seeded in 96-well transparent plates 24 hours before transfection. After transfection for 48 hours, change to Seahorse XF base medium (Agilent Technologies, USA) and incubate for 1 hour at 37°C in a CO2-free environment. Thereafter, the following chemicals were injected into the medium and OCR was measured: (a) oligomycin (OM; 10 mM), an ATP synthase inhibitor; (b) p-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP;10 mM), an accelerator that leads to maximum respiratory rate; (c) a mixture of rotenone (ROT) and antimycin A (AA) (50 mM) and mitochondrial complex I and III inhibitors, respectively. For the ECAR assay, the following chemicals were sequentially injected in the medium: (a) glucose (10 mM) glucose was added and the elevated value represents glycolysis; (b) oligomycin (OM; 10 mM), an ATP synthase inhibitor; (c) 2-deoxyglucose (2-DG, 50 mM)), a competitive inhibitor of hexokinase. Seahorse Wave softwarewas used to analyze the results. OCR and ECAR were estimated as pmol/min and mPH/min, respectively.
Determination of ATP, Pyruvic Acid and Lactic Acid content
Metabolic assays for cells or tissues were performed according to the operating instructions of the kits. In brief, we seeded U87 and U251 cells in 6-well plates and transfected them with plasmids or siRNAs. The level of PA, LA and ATP in cells or tissues was measured using Pyruvate (PA) Content Assay Kit, Micromethod (Sangon Biotech China), Lactic Acid (LA) Content Assay Kit, Micromethod (Sangon Biotech China) and ATP Assay Kit (Beyotime, China). Each study was conducted at least three times.
Western blotting
For western blots, protein samples were separated on SDS–PAGE gels and then transferred to PVDF membranes (Millipore, USA). The blots were imaged using ECL chemiluminescent reagents (Tanon, China) and the AI680 imaging system (GE, USA). The following antibodies were used in western blots: anti-Lamin B1 (Santa Cruz, sc-377000), anti-GAPDH (Sangon, D110016), anti-TKFC (Proteintech, 12224-1-AP), anti-Flag (Proteintech, 20543-1-AP), anti-TBK1 (Santa Cruz, A-6), anti-pTBK1 (CST, D52C2), anti-HA (Proteintech, 51064-2-AP), anti-β-actin (Sangon, D110001), anti-IKKε (CST, #2690), anti-pIKKε (CST, #8766), anti-IRF3 (Abclonal, A11373), anti-pIRF3 (Abclonal, AP0623), anti-NF-κB (CST, #8242), anti-pNF-κB (CST, #3031), anti-ALDOC (Proteintech, 14884-1-AP). Antibody validation is provided on the manufacturers’ websites. In this study, we repeated three independent experiments and loaded the same number of protein samples in each experiment. To quantitatively analyze the expression levels of RIG-Ⅰ pathway proteins, we measured the gray values of the bands separately using ImageJ software and corrected for differences in the amount of samples loaded and transmembrane using β-actin as an internal standard. A t-test was performed on the data from three independent experiments to determine whether the differences in protein expression in the subgroups were statistically significant (p < 0.05). Finally, we provided the raw data for Western Blot see Supplementary Material: Original Data File.
Enzyme-linked immunosorbent assay (ELISA)
Interferon was assayed in mouse brain tissue using an ELISA kit (E-EL-M0033c; E-EL-M0048c, ElabScience) according to the manufacturer’s instructions. Briefly, the tissues were washed with pre-cooled PBS to remove residual blood, weighed and then shredded. The shredded tissue was added to a glass homogenizer with a corresponding volume of PBS (generally at a weight-to-volume ratio of 1:9) and ground thoroughly on ice. To further lyse the tissue cells, the homogenate was repeatedly freeze-thawed or ultrasonically broken. Finally, the homogenate was centrifuged at 2-8°C for 5-10 min at 5000 × g, and the supernatant was retained. Protein samples are diluted 50-fold and incubated with the antibody for 2 hours at 37°C. And then, The absorbance of each well was measured at 450 nm with the Infinite M200 (Tecan, Switzerland). The standard curve was plotted with Origin software and the interferon content of each well was calculated.
Histological analysis
For Immunohistochemistry (IHC) analysis, paraffin sections (5 μm) were deparaffinized and rehydrated through a xylene and graded alcohol series. Sections were heat-induced antigen repair using citrate buffer (10 mM pH 6.0) and then endogenous peroxidase was removed with 3% hydrogen peroxide. Next, the non-specific protein binding sites were blocked with 5% goat serum and sections were incubated overnight at 4°C with the following antibodies: anti-Ki67 (CST, #9449), anti-TKFC (Proteintech, 12224-1-AP), anti-CD8α (CST, #98941), anti-Granzyme K (Proteintech, 27894-1-AP), anti-NCR1 (CST, #23732). Finally, DAB or fluorescence photography was performed using HRP secondary antibody or fluorescent secondary antibody.
Adeno-associated virus and lentivirus production
For lentivirus production, 293T cells were seeded in 6-well plates and cotransfected with pLV4ltr-Puro-circSOBP, pCMV-VSV-G, pCAG-dR8.9 plasmids using Lipo8000 transfection reagent (Beyotime, China). Cells were continued to be cultured for 72 hours, the medium was gathered and filtered (0.45 μm) for storage at -80°C. To obtain adeno-associated viruses, 293T cells (80% cell density per 10 cm dish) were co-transfected with pAAV-GFAabc1d-circSOBP, pHelper, and pAAV-RC9 plasmids using the lipo8000 transfection reagent. After 72 hours of incubation, the cells were collected by centrifugation for 5 minutes, 1000g. The precipitated cells were gathered into new centrifuge tubes to manufacture aav virus solution by the AAVpro® Purification Kit (All Serotypes) (Takara, Japan). AAV viral solution was quantified with the AAVpro® Titration Kit (for Real Time PCR) (Takara, Japan) and stored at -80°C.
Quantification and statistical analysis
All statistical analyses were performed with the SPSS 20.0 statistical software package. Data are expressed as mean ± S.E.M from at least three independent experiments. Differences were evaluated by student’s t test for two groups, one-way analysis of variance for multiple groups, and parametric generalized linear model with random effects for tumor growth. The chi-square test was applied to the examination of relationship between circSOBP expression levels and clinicopathologic characteristics. P values <0.05 were considered statistically significant and all statistical tests asterisks indicate statistical significance. ImageJ analysis of fluorescence co-localization with Pearson coefficient >0.5 and overlap coefficient >0.6 proved to be statistically significant. NS, not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No: 81902525, No: 82273281), the Fundamental Research Funds for the Central Universities (No: WK9110000145), Special Fund Project for Guiding Local Science and Technology Development by the Central Government (No: 2019b07030001) and the Excellent Youth Research Project of Natural Science in Anhui Province Universities (No: 2022AH030127). We would like to express our deepest appreciation to all those who have contributed to this project. A special thanks goes to our colleagues: Zhang Ning, Bao Zixu, Wang Zixuan, and Qiao Xiaolong, who have provided us with valuable insights and expertise. We would also like to thank our families and mentors for their unwavering support and encouragement throughout this journey. Finally, we are grateful to the funding agencies for their generous support.
Author contributions
C.S.N and S.S.H designed this project. M.L.M and W.X.N performed the experiments. F.C and Q.S.D collected specimen. M.L.M analyzed the data and wrote the article. All authors have discussed the results and made comments on the experiment.
Declaration of interests
The authors declare no conflict of interest.
Published: September 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107897.
Contributor Information
Shanshan Hu, Email: hss923@ustc.edu.cn.
Chaoshi Niu, Email: niuchaoshi@ustc.edu.cn.
Supplemental information
Data and code availability
-
•
The circRNA RNA-seq data in this study have been deposited in the GEO database with the accession codes GSE153692, which is publicly available.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molinaro A.M., Hervey-Jumper S., Morshed R.A., Young J., Han S.J., Chunduru P., Zhang Y., Phillips J.J., Shai A., Lafontaine M., et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol. 2020;6:495–503. doi: 10.1001/jamaoncol.2019.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J.B., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 6.Misir S., Wu N., Yang B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Wang S., Sun Z., Lei Z., Zhang H.-T. RNA-binding proteins and cancer metastasis. Semin. Cancer Biol. 2022;86:748–768. doi: 10.1016/j.semcancer.2022.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Wang B., Zhou F., Lv K., Xu X., Cao W. CircNDC80 promotes glioblastoma multiforme tumorigenesis via the miR-139-5p/ECE1 pathway. J. Transl. Med. 2023;21:22. doi: 10.1186/s12967-022-03852-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Wang H., Cui Y., Jiang W., Zhan H., Feng L., Gao M., Zhao K., Zhang L., Xie X., et al. The U2AF65/circNCAPG/RREB1 feedback loop promotes malignant phenotypes of glioma stem cells through activating the TGF-β pathway. Hereditas. 2023;160:23. doi: 10.1038/s41419-023-05556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan F., Zhang S., Sun Q., Ye L., Xu Y., Xu Z., Deng G., Zhang S., Liu B., Chen Q. Hsa_circ_0072309 enhances autophagy and TMZ sensitivity in glioblastoma. CNS Neurosci. Ther. 2022;28:897–912. doi: 10.1111/cns.13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao F., Song Z., Wang S., Ma Z., Zhuo Z., Meng T., Xu G., Chen G. Novel circular RNA circSOBP governs amoeboid migration through the regulation of the miR-141-3p/MYPT1/p-MLC2 axis in prostate cancer. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan X., Wang J., Bai Y. Potentials of circSOBP in the diagnosis and prognosis of gastric cancer. Scand. J. Gastroenterol. 2022;57:1344–1348. doi: 10.1080/00365521.2022.2088246. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi A., Wu S.-Y., Watabe K. Metabolism in the progression and metastasis of brain tumors. Cancer Lett. 2022;539 doi: 10.1016/j.canlet.2022.215713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.San-Millan I., Sparagna G.C., Chapman H.L., Warkins V.L., Chatfield K.C., Shuff S.R., Martinez J.L., Brooks G.A. Chronic Lactate Exposure Decreases Mitochondrial Function by Inhibition of Fatty Acid Uptake and Cardiolipin Alterations in Neonatal Rat Cardiomyocytes. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.809485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer I.-M., Hornick J.L., Bovée J.V.M.G. The role of metabolic enzymes in mesenchymal tumors and tumor syndromes: genetics, pathology, and molecular mechanisms. Lab. Invest. 2018;98:414–426. doi: 10.1038/s41374-017-0003-6. [DOI] [PubMed] [Google Scholar]
- 20.Sun L., Zhang H., Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877–919. doi: 10.1007/s13238-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koriyama S., Nitta M., Kobayashi T., Muragaki Y., Suzuki A., Maruyama T., Komori T., Masui K., Saito T., Yasuda T., et al. A surgical strategy for lower grade gliomas using intraoperative molecular diagnosis. Brain Tumor Pathol. 2018;35:159–167. doi: 10.1007/s10014-018-0324-1. [DOI] [PubMed] [Google Scholar]
- 22.Wick W., Hartmann C., Engel C., Stoffels M., Felsberg J., Stockhammer F., Sabel M.C., Koeppen S., Ketter R., Meyermann R., et al. NOA-04 Randomized Phase III Trial of Sequential Radiochemotherapy of Anaplastic Glioma With Procarbazine, Lomustine, and Vincristine or Temozolomide. J. Clin. Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 23.Ooi A. Advances in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) research. Semin. Cancer Biol. 2020;61:158–166. doi: 10.1016/j.semcancer.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. 328rv324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Depil S., Duchateau P., Grupp S.A., Mufti G., Poirot L. 'Off-the-shelf' allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020;19:185–199. doi: 10.1038/s41573-019-0051-2. [DOI] [PubMed] [Google Scholar]
- 27.von Locquenghien M., Rozalén C., Celià-Terrassa T. Interferons in cancer immunoediting: sculpting metastasis and immunotherapy response. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinman R.D., Fine E.J. Fructose in perspective. Nutr. Metab. 2013;10:45. doi: 10.1186/1743-7075-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diao F., Li S., Tian Y., Zhang M., Xu L.G., Zhang Y., Wang R.P., Chen D., Zhai Z., Zhong B., et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Wang G., Xu Z.-G., Tu H., Hu F., Dai J., Chang Y., Chen Y., Lu Y., Zeng H., et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell. 2019;178:176–189.e15. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 32.Chen L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 33.He J., Huang Z., He M., Liao J., Zhang Q., Wang S., Xie L., Ouyang L., Koeffler H.P., Yin D., Liu A. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Jiang J., Zhang J., Shen H., Wang M., Guo Z., Zang X., Shi H., Gao J., Cai H., et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol. Cancer. 2021;20:101. doi: 10.1186/s12943-021-01390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q., Liu T., Bao Y., Zhao T., Wang J., Wang H., Wang A., Gan X., Wu Z., Wang L. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Li J., Bian X., Wu C., Hua J., Chang S., Yu T., Li H., Li Y., Hu S., et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2012881118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Zhang Y., Cai Q., Ma M., Jin L.Y., Weng M., Zhou D., Tang Z., Wang J.D., Quan Z. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol. Cancer. 2019;18:145. doi: 10.1186/s12943-019-1078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., Yang J., Fei X., Wang X., Wang K. CircRNA ciRS-7: a Novel Oncogene in Multiple Cancers. Int. J. Biol. Sci. 2021;17:379–389. doi: 10.7150/ijbs.54292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lou J., Hao Y., Lin K., Lyu Y., Chen M., Wang H., Zou D., Jiang X., Wang R., Jin D., et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol. Cancer. 2020;19:138. doi: 10.1186/s12943-020-01253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 41.Martínez-Reyes I., Chandel N.S. Cancer metabolism: looking forward. Nat. Rev. Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 42.Li J., Hu Z.-Q., Yu S.-Y., Mao L., Zhou Z.-J., Wang P.-C., Gong Y., Su S., Zhou J., Fan J., et al. CircRPN2 Inhibits Aerobic Glycolysis and Metastasis in Hepatocellular Carcinoma. Cancer Res. 2022;82:1055–1069. doi: 10.1158/0008-5472.CAN-21-1259. [DOI] [PubMed] [Google Scholar]
- 43.Jiang X., Guo S., Wang S., Zhang Y., Chen H., Wang Y., Liu R., Niu Y., Xu Y. EIF4A3-Induced circARHGAP29 Promotes Aerobic Glycolysis in Docetaxel-Resistant Prostate Cancer through IGF2BP2/c-Myc/LDHA Signaling. Cancer Res. 2022;82:831–845. doi: 10.1158/0008-5472.CAN-21-2988. [DOI] [PubMed] [Google Scholar]
- 44.Song J., Zheng J., Liu X., Dong W., Yang C., Wang D., Ruan X., Zhao Y., Liu L., Wang P., et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J. Exp. Clin. Cancer Res. 2022;41:171. doi: 10.1186/s13046-022-02374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues J.R., Couto A., Cabezas A., Pinto R.M., Ribeiro J.M., Canales J., Costas M.J., Cameselle J.C. Bifunctional homodimeric triokinase/FMN cyclase: contribution of protein domains to the activities of the human enzyme and molecular dynamics simulation of domain movements. J. Biol. Chem. 2014;289:10620–10636. doi: 10.1074/jbc.M113.525626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X., Wang H., Li X., Guo C., Yuan F., Fisher P.B., Wang X.Y. Activation of the MDA-5-IPS-1 Viral Sensing Pathway Induces Cancer Cell Death and Type I IFN-Dependent Antitumor Immunity. Cancer Res. 2016;76:2166–2176. doi: 10.1158/0008-5472.CAN-15-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M., Liu Z., Wu Y., Zheng N., Liu X., Cai A., Zheng D., Zhu J., Wu J., Xu L., et al. In vivo imaging of astrocytes in the whole brain with engineered AAVs and diffusion-weighted magnetic resonance imaging. Mol. Psychiatry. 2022 doi: 10.1038/s41380-022-01580-0. [DOI] [PubMed] [Google Scholar]
- 48.Boukhaled G.M., Harding S., Brooks D.G. Opposing Roles of Type I Interferons in Cancer Immunity. Annu. Rev. Pathol. 2021;16:167–198. doi: 10.1146/annurev-pathol-031920-093932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The circRNA RNA-seq data in this study have been deposited in the GEO database with the accession codes GSE153692, which is publicly available.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.