Abstract
Oncogenic KRAS mutations occur in approximately 30% of lung adenocarcinoma. Despite several decades of effort, oncogenic KRAS-driven lung cancer remains difficult to treat, and our understanding of the regulators of RAS signaling is incomplete. To uncover the impact of diverse KRAS-interacting proteins on lung cancer growth, we combined multiplexed somatic CRISPR/Cas9-based genome editing in genetically engineered mouse models with tumor barcoding and high-throughput barcode sequencing. Through a series of CRISPR/Cas9 screens in autochthonous lung cancer models, we show that HRAS and NRAS are suppressors of KRASG12D-driven tumor growth in vivo and confirm these effects in oncogenic KRAS-driven human lung cancer cell lines. Mechanistically, RAS paralogs interact with oncogenic KRAS, suppress KRAS-KRAS interactions, and reduce downstream ERK signaling. Furthermore, HRAS and NRAS mutations identified in KRAS-driven human tumors partially abolished this effect. By comparing the tumor-suppressive effects of HRAS and NRAS in oncogenic KRAS- and oncogenic BRAF-driven lung cancer models, we confirm that RAS paralogs are specific suppressors of KRAS-driven lung cancer in vivo. Our study outlines a technological avenue to uncover positive and negative regulators of oncogenic KRAS-driven cancer in a multiplexed manner in vivo and highlights the role of the phenomenon we term RAS paralog imbalance in oncogenic KRAS-driven lung cancer.
INTRODUCTION
The RAS family genes KRAS, HRAS and NRAS are frequently mutated across cancers, and KRAS mutations occur in approximately 30% of lung adenocarcinomas1–3. RAS proteins are small GTPases that switch between a GTP-bound active state and a GDP-bound inactive state in response to upstream growth factor signaling4. RAS proteins regulate multiple downstream signaling pathways that control proliferation. Oncogenic mutations in codons 12, 13, and 61 reduce GTP hydrolysis and increase the fraction of RAS proteins in the GTP-bound state, which results in widespread changes in RAS protein-protein interactions5, 6 and hyper-activation of RAS effector pathways, culminating in cellular transformation and tumorigenesis7,8. Oncogenic KRAS therefore represents a key node in growth factor-induced signaling and a critical target for therapeutic intervention in lung adenocarcinoma9.
Genetic and proteomic mapping has revealed that KRAS interacts with a large network of proteins10, 11. These KRAS-interacting proteins include canonical regulators and effectors, as well as many proteins that remain poorly understood in the context of oncogenic KRAS-driven lung cancer. Much of our understanding of RAS signaling has stemmed from diverse cellular and cell-free systems12–14. Thus, while recent studies have mapped KRAS protein-protein interaction networks10, 11, 15, 16, it remains difficult to assess the relevance of these interactions to cancer growth in vivo. Genetically engineered mouse models of oncogenic KRAS-driven cancer uniquely recapitulate autochthonous tumor growth and have contributed to our understanding of KRAS signaling17. However, the development and use of such models has traditionally been insufficiently scalable to broadly assess modifiers of KRAS-driven tumor growth. The ability to uncover functional components of RAS signaling that affect lung cancer growth in vivo in a multiplexed manner would accelerate our understanding of RAS biology and could aid in the development of pharmacological strategies to counteract hyperactivated KRAS.
To enable the analysis of genetic modifiers of lung tumor growth in vivo, we recently integrated somatic CRISPR/Cas9-based genome editing with tumor barcoding and high-throughput barcode sequencing (Tuba-seq)18–20. This approach allows precise quantification of the effects of inactivating panels of genes of interest on lung tumor initiation and growth in a multiplexed manner. By employing Tuba-seq to assess the functions of KRAS-interacting proteins nominated by unbiased affinity purification/mass spectrometry (AP/MS), we show that wild-type HRAS and NRAS suppress the growth of oncogenic KRAS-driven lung adenocarcinoma. Competition between oncogenic KRAS and wild-type HRAS and NRAS diminishes KRAS-KRAS interaction and suppresses downstream signaling. In vivo screening across multiple oncogenic contexts revealed that HRAS and NRAS specifically suppress the growth of tumors driven by oncogenic KRAS. Our study reveals that changes in the ratio of RAS paralogs (which we term “RAS paralog imbalance”) is a driver of oncogenic KRAS-driven lung cancer.
RESULTS
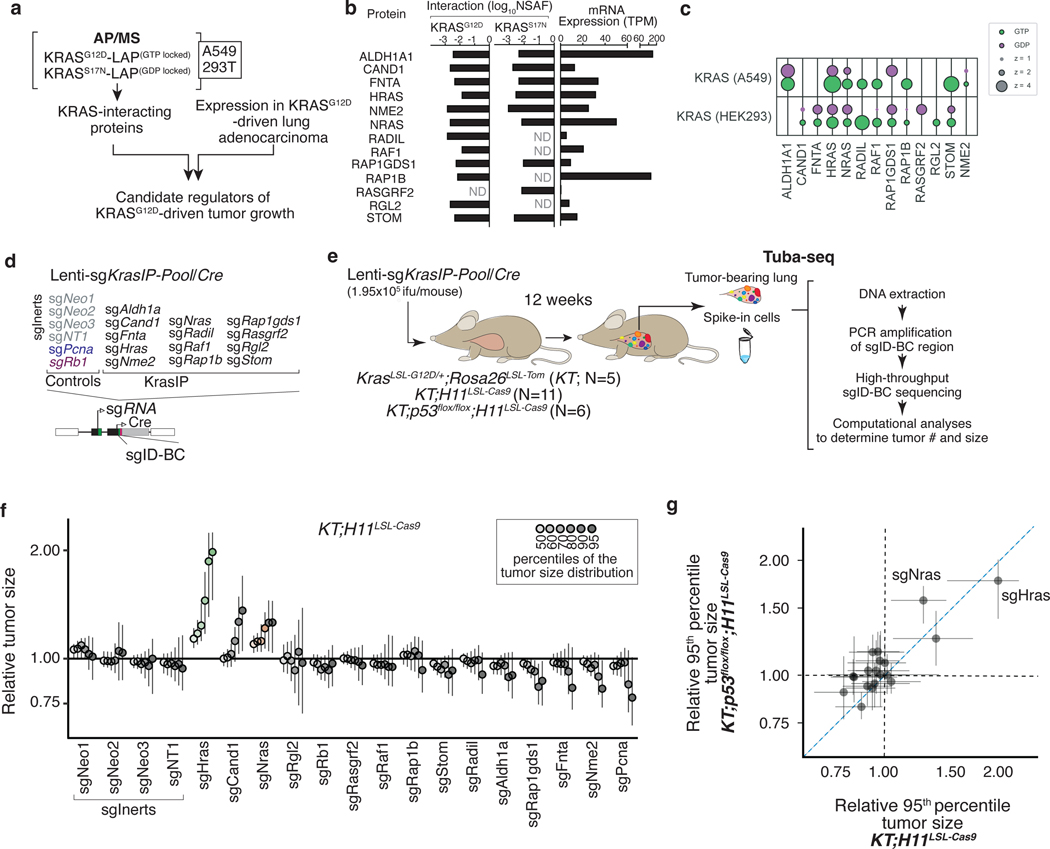
Selection of candidate KRAS-interacting proteins
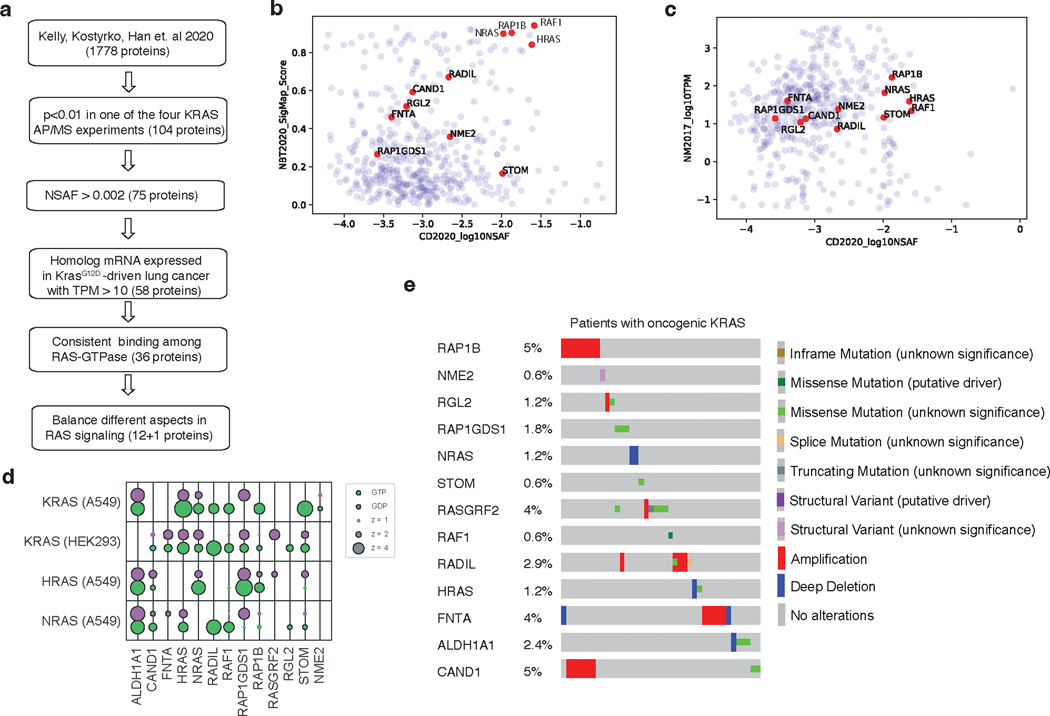
To identify KRAS-interacting proteins that could affect oncogenic KRAS-driven lung tumor growth in vivo, we integrated pre-existing proteomic data from AP/MS studies with gene expression data from cancer cells from autochthonous mouse models (Figure 1a)10, 21. We prioritized a list of candidate genes according to the probability of their protein products interacting with KRAS and other RAS GTPases, and their mRNA expression in mouse models of oncogenic KRASG12D-driven lung cancer (Figure 1b–c, Extended Data Figure 1a–d)10, 21. We selected 13 proteins that represent diverse aspects of RAS biology, including RAS paralogs (HRAS, NRAS – which were supported by the identification of paralog-specific peptides), RAS regulators (RASGRF2, RAP1GDS1)22, 23, a RAS farnesyltransferase (FNTA)24, 25, and RAS effectors (RAF1, RGL2)26, 27, as well as several proteins whose functions in RAS signaling are understudied. While the majority of these candidate genes trend towards amplification in oncogenic KRAS-driven lung adenocarcinoma, NRAS, HRAS, and ALDH1A1 have deep genomic deletions (Extended Data Figure 1e)28. Interestingly, some of these proteins bound preferentially to either GTP- or GDP-bound KRAS, while others interact with KRAS independently of its nucleotide state (Figure 1c).
Figure 1. Multiplexed identification of KRAS-interacting proteins that impact KRASG12D-driven lung cancer growth in vivo.
a. Candidate mediators of KRAS-driven lung tumor growth were identified on the basis of their interactions with GTP- and GDP-locked KRAS in multiple AP/MS-based protein-protein interaction screens and their expression in a mouse model of KRAS-driven lung adenocarcinoma.
b. Selected proteins interact with either GTP- or GDP-locked KRAS and are expressed in mouse KRASG12D-driven lung cancer. NSAF: normalized spectral abundance factor; TPM: transcripts per million; ND: undetected.
c. Plot of two AP/MS experiments with GTP- and GDP-locked mutant GTPases as baits (rows), showing the enrichment of selected candidate KRAS-interacting proteins (columns). Dark borders indicate FDR < 0.05.
d. Schematic of tumor initiation with a pool of barcoded Lenti-sgRNA/Cre vectors (Lenti-sgKrasIP-Pool/Cre). Each vector contains an sgRNA, Cre, and a two-component barcode composed of an sgRNA identifier (sgID) and a random barcode (BC). This design allows inactivation of multiple target genes in parallel followed by quantification of the resulting tumor size distributions through high-throughput sgID-BC sequencing.
e. Tumors were initiated in cohorts of KT, KT;H11LSL-Cas9 and KT;p53flox/flox;H11LSL-Cas9 mice through intratracheal delivery of Lenti-sgKrasIP-Pool/Cre. Tuba-seq was performed on each tumor-bearing lung 12 weeks after initiation to characterize the effects of inactivating each gene.
f. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles in KT;H11LSL-Cas9 mice. Genes are ordered by 95th percentile tumor size, with sgInerts on the left. sgInerts are in gray, and the line at y=1 indicates no effect relative to sgInert. Percentiles that are significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color.
g. Comparison of 95th percentile tumor size for each sgRNA relative to the 95th percentile tumor size of sgInert-containing tumors in KT;H11LSL-Cas9 mice versus KT;p53flox/flox;H11LSL-Cas9 mice. For panels f and g: Error bars indicate 95% confidence intervals around the point estimate of the test statistic. Confidence intervals and P-values in panels f and g were calculated using the nested bootstrap resampling approach across 11 KT;H11LSL-Cas9 mice and 6 KT;p53flox/flox;H11LSL-Cas9 mice.
KRAS-interacting proteins impact lung tumor growth in vivo
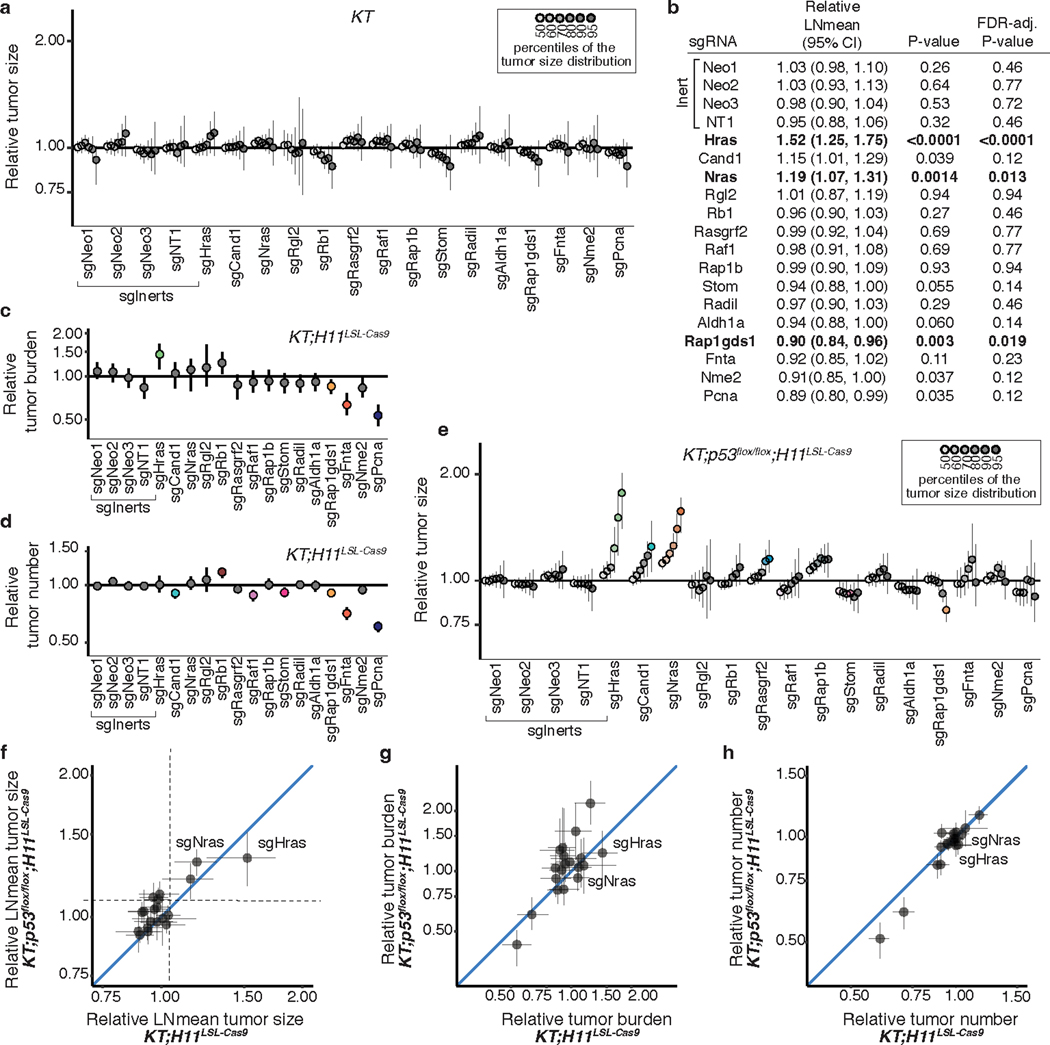
Given that KRAS-interacting proteins could have either positive or negative effects on tumor growth, we first assessed whether we could detect gene-targeting events that have deleterious effects on tumor fitness using Tuba-seq. We initiated tumors in KrasLSL-G12D/+;Rosa26LSL-tdTomato;H11LSL-Cas9 (KT;H11LSL-Cas9) and control KT mice with a pool of barcoded Lenti-sgRNA/Cre vectors targeting two essential genes (Pcna and Rps19), a known tumor suppressor (Apc)20, 29, and several inert sgRNAs (Lenti-sgEssential/Cre; Extended Data Figure 2a). After 12 weeks of tumor growth, we performed Tuba-seq on bulk tumor-bearing lungs (Extended Data Figure 2b). By incorporating measures of tumor number and size, we could confidently identify genetic deficiencies that reduced tumor fitness (Extended Data Figure 2c–g and Methods).
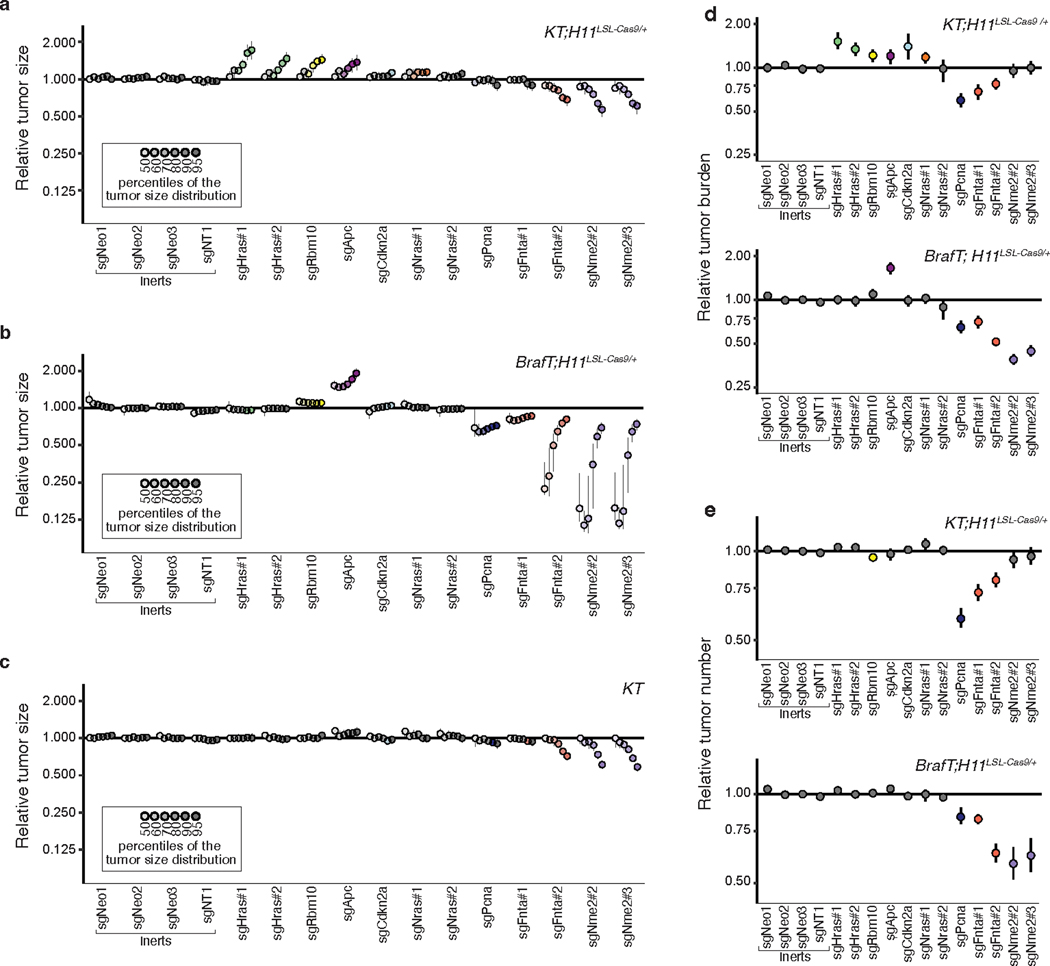
To quantify the impact of inactivating our panel of KRAS-interacting proteins on oncogenic KRASG12D-driven lung tumor growth in vivo, we generated a pool of barcoded Lenti-sgRNA/Cre vectors targeting the genes that encode these proteins, as well as sgInert controls and vectors targeting an essential gene (Pcna) and a known tumor suppressor (Rb1, Lenti-sgKrasIP/Cre; Figure 1d). Given the importance of farnesylation in KRAS localization and signaling, sgRNA targeting of Fnta served as a control for KRAS dependency30, 31. We initiated tumors with the Lenti-sgKrasIP/Cre pool in KT;H11LSL-Cas9 and KT mice and calculated metrics of tumor size and number after 12 weeks of tumor growth (Figure 1e). To our surprise, inactivation of the Kras paralogs Hras and Nras had the most dramatic effect on increasing tumor growth. Inactivation of Cand1 also increased tumor size, while inactivation of several genes including Fnta, Nme2, Rap1gds1, and Aldh1a decreased tumor size and/or number, suggesting reduced cancer cell fitness (Figure 1f and S3a–d).
Given the fundamental importance of the p53 tumor suppressor in oncogenic KRAS-driven lung cancer, as well as previous data suggesting crosstalk between RAS and p53 signaling19, 32, 33, we determined whether p53 deficiency changed the impact of inactivating KRAS-interacting proteins on tumor growth. We initiated tumors with the Lenti-sgKrasIP/Cre pool in KrasLSL-G12D/+;Rosa26LSL-tdTom;p53flox/flox;H11LSL-tdTom (KT;p53flox/flox;H11LSL-Cas9) mice and performed Tuba-seq after 12 weeks of tumor growth (Figure 1e). The effects of inactivating each gene encoding a KRAS-interacting protein were generally consistent between the p53-proficient and -deficient settings (Figure 1g, Extended Data Figure 3e–h). Notably, inactivation of either Hras or Nras significantly increased the growth of p53-deficient tumors (Figure 1g, Extended Data Figure 3e). Collectively, these results suggest that HRAS and NRAS are tumor suppressors within in vivo models of oncogenic KRAS-driven lung cancer, while several other KRAS-interacting proteins have less consistent effects on tumor growth between p53-proficient and -deficient backgrounds (Extended Data Figure 3e–h).
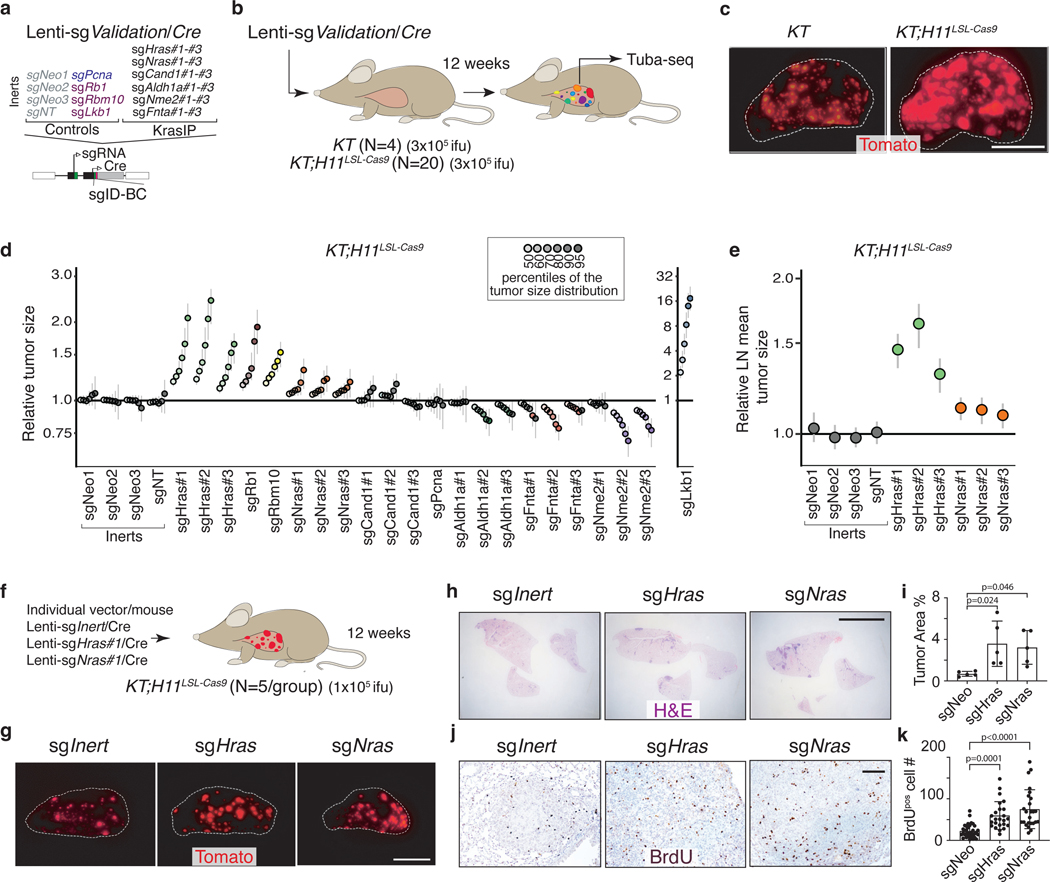
Validation of HRAS and NRAS as tumor suppressors
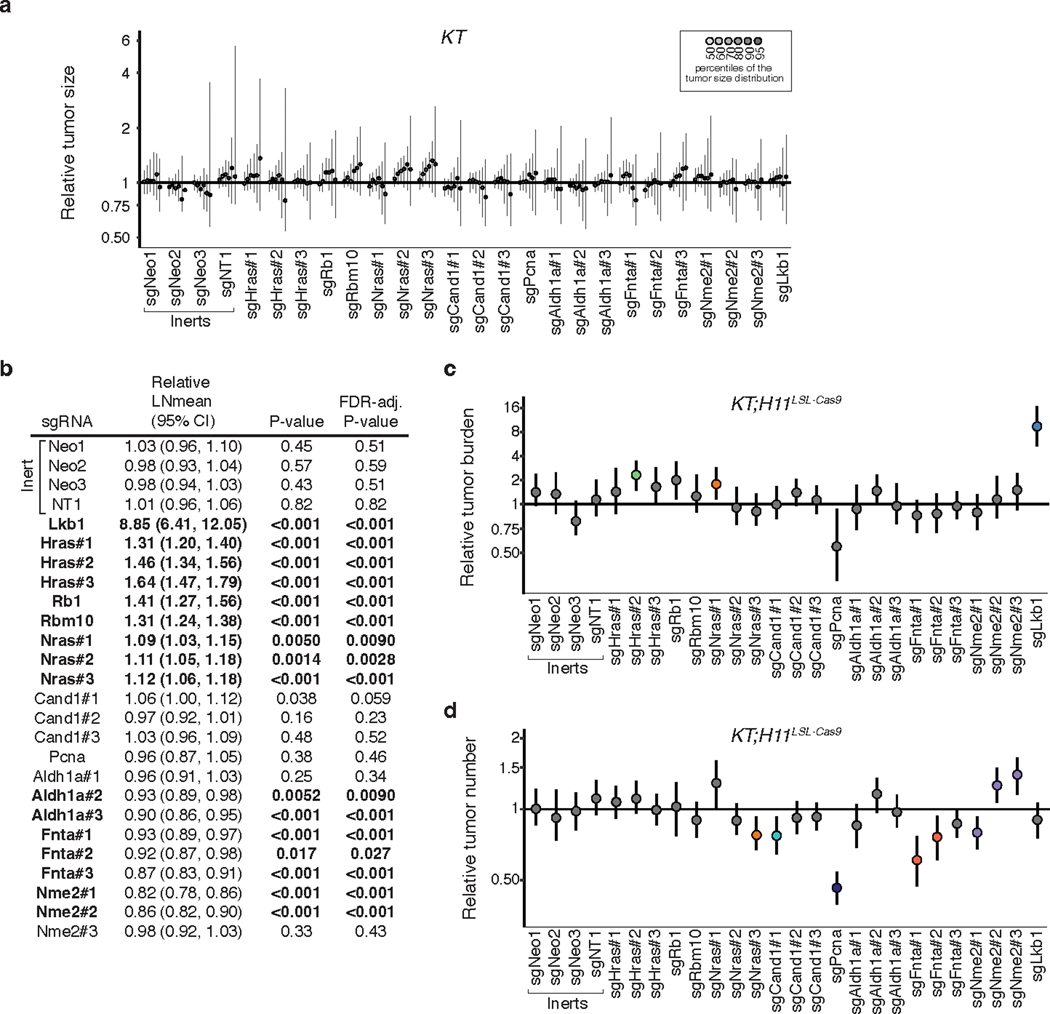
To further validate the effect of inactivating six top candidate genes (Hras, Nras, Cand1, Aldh1a, Fnta, and Nme2) on oncogenic KRAS-driven tumor growth in vivo and confirm that these results are driven by on-target effects, we generated three barcoded Lenti-sgRNA/Cre vectors targeting each gene. To contextualize the effects of inactivating these genes we also included vectors targeting three established tumor suppressors (Lkb1, Rbm10, and Rb1) in this pool (Lenti-sgValidation/Cre; Figure 2a)18, 20, 34. We initiated tumors with the Lenti-sgValidation/Cre pool in KT;H11LSL-Cas9 and KT mice and assessed metrics of tumor initiation and growth 12 weeks after tumor initiation (Figure 2b–c). Targeting Fnta consistently reduced growth across all three sgRNAs, while the impact of inactivating Aldh1a and Nme2 was more variable (Figure 2d, Extended Data Figure 4). Most importantly, all sgRNAs targeting Hras and Nras significantly increased tumor growth (Figure 2d–e, Extended Data Figure 4b). Notably, Hras inactivation increased tumor growth to a similar extent as inactivation of the Rb1 and Rbm10 tumor suppressors (Figure 2d, Extended Data Figure 4b). These results suggest a potentially pivotal role for wild-type HRAS and NRAS in suppressing oncogenic KRAS-driven lung tumor growth in vivo.
Figure 2. HRAS and NRAS are potent suppressors of KRASG12D-driven lung cancer growth in vivo.
a. Lenti-sgValidation/Cre targets candidate mediators of KRAS-driven lung tumor growth (3 sgRNAs/gene).
b. Tumors were initiated in KT and KT;H11LSL-Cas9 mice through intratracheal delivery of Lenti-sgValidation/Cre, and Tuba-seq was performed on each tumor-bearing lung. .
c. Fluorescence images of lung lobes 12 weeks after tumor initiation. Representative of 5 KT and 15 KT;H11LSL-Cas9 mice. Scale bars: 5 mm. Lung lobes are outlined with a white dashed line.
d. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles in KT;H11LSL-Cas9 mice. Genes are ordered by 95th percentile tumor size, with sgInerts on the left. Note that sgLkb1 is plotted on a separate scale to facilitate visualization. The line at y=1 indicates no effect relative to sgInert. Percentiles that are significantly different from sgInert (2-sided FDR-adjusted p<0.05) are in color.
e. Targeting Hras and Nras significantly increases mean tumor size relative to sgInerts, assuming a log-normal distribution of tumor sizes (LNmean). For panels d and e: Error bars indicate 95% confidence intervals around the point estimate of the test statistic. Confidence intervals and P-values were calculated using nested bootstrap resampling across 20 KT;H11LSL-Cas9 mice.
f. Schematic of tumor initiation with individual Lenti-sgRNA/Cre vectors.
g. Fluorescence images of lungs from KT;H11LSL-Cas9 mice 12 weeks after tumor initiation with Lenti-sgRNA/Cre vectors. Representative of 5 mice/group. Scale bar: 5 mm.
h. Representative H&E images of lungs from KT;H11LSL-Cas9 mice after tumor initiation with Lenti-sgRNA/Cre vectors. Scale bar: 5 mm.
i. Tumor burden in KT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors. Error bars are Mean±SD. Each dot represents relative tumor area (percentage of total lung area) from one mouse. N=5 animals (one-way ANOVA).
j. Representative BrdU staining images of lungs from KT;H11LSL-Cas9 mice after tumor initiation with Lenti-sgRNA/Cre vectors. Scale bar: 100 μm.
k. Quantification of proliferating cells in KT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors. Error bars are Mean±SD. Each dot is a tumor (sgNeo: N=34, sgHras: N=24, sgNras: N=25). N=5 animals (one-way ANOVA).
We also validated the tumor-suppressive functions of HRAS and NRAS by initiating tumors in KT;H11LSL-Cas9 mice with individual sgInert-, sgHras- or sgNras-containing Lenti-sgRNA/Cre vectors (Figure 2f). Inactivation of either Hras or Nras increased tumor growth as assessed by direct fluorescence and histological analyses (Figure 2g–k). Collectively, these results suggest that RAS paralogs constrain the growth of oncogenic KRASG12D-driven lung cancer.
HRAS and NRAS suppress growth of human lung cancer cells
To assess the relevance of HRAS and NRAS as tumor suppressors in human lung cancer, we tested the function of these proteins in oncogenic KRAS-driven human lung adenocarcinoma cell lines. Previous genome-scale CRISPR/Cas9 screens revealed that inactivating these genes is either detrimental or of no consequence to cancer cell line growth under standard culture conditions (Extended Data Figure 5a)10, 35. Interestingly, HRAS and NRAS suppressed the growth of oncogenic KRASG12S-driven A549 cells grown in 2D culture conditions, and were growth-suppressive in several oncogenic KRAS-driven lung cancer cell lines grown in 3D culture conditions, suggesting that these genes can function as tumor suppressors in certain contexts (Extended Data Figure 5b–c)10, 15. To further assess the functions of HRAS and NRAS in oncogenic KRAS-driven human adenocarcinoma cell lines, we performed gain and loss of function studies on H23 (KRASG12C) and H727 (KRASG12V) cells under growth factor-restricted growth conditions. We inactivated HRAS and NRAS using CRISPR/Cas9 and generated variants with doxycycline-inducible wild-type HRAS re-expression (Extended Data Figure 5d). Inactivation of HRAS or NRAS in oncogenic KRAS-driven cells increased proliferation when cells were grown with limited serum and increased clonal growth potential in anchorage-independent conditions (Figure 3a, c, d). Re-expression of HRAS in these HRAS-null cells impaired proliferation and clonal growth (Figure 3b, e, f; Extended Data Figure 5e). H23 cells with inactivated HRAS or NRAS also formed larger and more proliferative tumors after intravenous and subcutaneous transplantation (Figure 3g–k, Extended Data Figure 5f–i). These results demonstrate that wild-type HRAS and NRAS can suppress the growth of oncogenic KRAS-driven human lung cancer cells in vitro and in vivo, further suggesting that HRAS and NRAS are tumor suppressors in oncogenic KRAS-driven lung adenocarcinoma.
Figure 3. Wild-type HRAS and NRAS constrain the growth of human KRAS-driven cancer cell lines.
a. Wildtype (sgSAFE) or HRAS- or NRAS-knockout cells were seeded in 96 well plates and cultured under limited serum (1%). Cell numbers were measured via CCK8 assay. Points are Mean±SD of 12 wells normalized to Day 0 (one-way ANOVA).
b. Re-expression of wild-type HRAS suppresses proliferation of HRAS-null H23 and H727 cells. TRE-HRAS cells were seeded in 96-well plates and cultured under limited serum (1%) with or without 50 ng/ml Doxycycline (Dox) and cell numbers were measured via CCK8 assay. Points are Mean±SD of 8 wells normalized to Day 0 (one-tailed t-test).
c-d. Inactivation of HRAS or NRAS increases H23 colony formation. Wild-type (sgSAFE), HRAS-knockout (sgHRAS), or NRAS-knockout (sgNRAS) H23 cells were seeded at 1000 cells/well in 6-well plates and grown for two weeks. Cells were stained with crystal violet. C.
Representative images. Scale bar: 5mm. d. Mean±SD of colony number from 12 fields (one-way ANOVA).
e-f. Re-expression of wild type HRAS suppresses HRAS-null H23 cell colony formation. Cells were seeded at 1000 cells/well in 6-well plates and grown +/-50 ng/ml Dox for two weeks. Cells were stained with crystal violet. e. Representative images. Scale bar: 5mm. f. Mean±SD of colony number from 12 fields (one-way ANOVA).
g-k. Inactivation of wild-type HRAS or NRAS increases H23 cell growth after transplantation. g. Schematic of tumor initiation by transplantation of H23 cells with inactivation of HRAS or NRAS in NSG mice. h. Tumor weight from SubQ transplantation of indicated cells. Each dot represents a mouse. Mean value is shown. i. Ki67pos cell number in tumor sections from SubQ transplantation of indicated cells, shown as Mean±SD value of 20 view fields. j. Tumor area (percentage of human mitochondriapos area) from IV transplantation of indicated cells. Each dot represents a tumor. Mean value is shown. k. Ki67pos cell number in tumor sections from IV transplantation of indicated cells, shown as Mean±SD value of 15 view fields (200x). ns: not significant (one-way ANOVA).
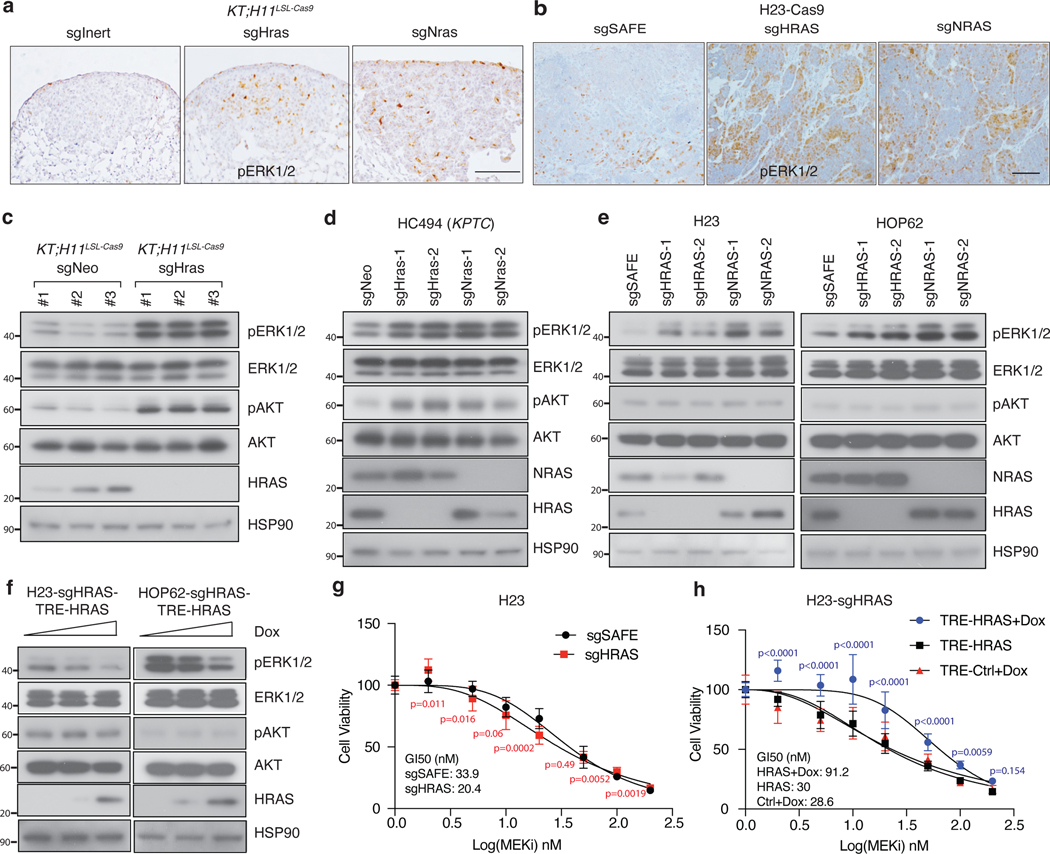
RAS paralog inactivation increases signaling downstream of oncogenic KRAS
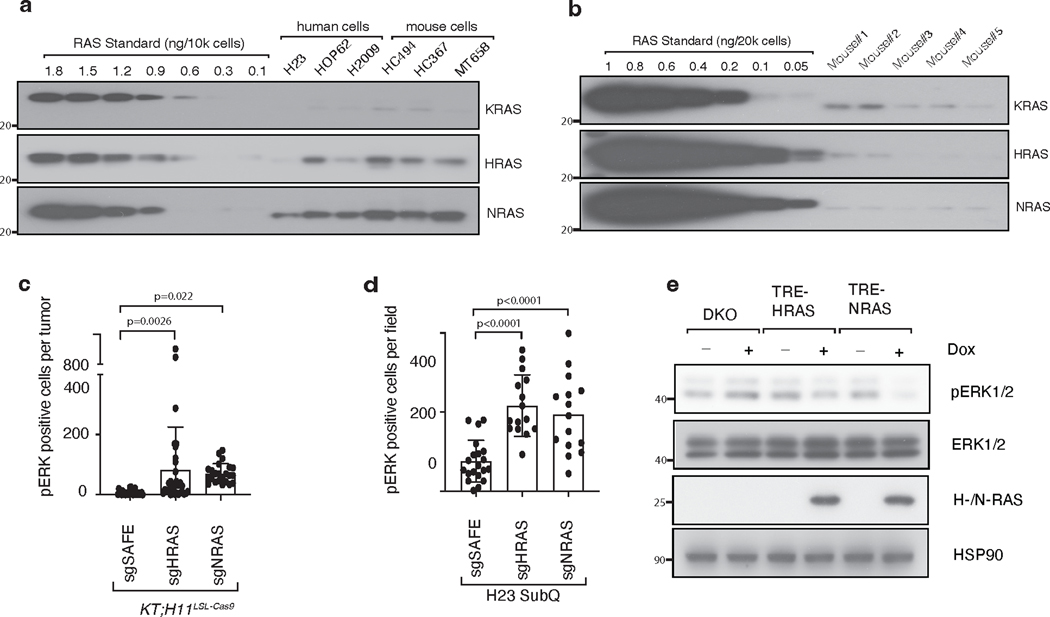
Wild-type KRAS has been shown to be tumor-suppressive in multiple experimental models of oncogenic KRAS-driven cancer, likely due to its ability to interact and compete with oncogenic KRAS36–38. We have demonstrated that wild-type HRAS and NRAS suppress oncogenic KRASG12D-driven lung cancer growth in vivo. We assessed the expression of K-, H-, and N-RAS in human and mouse lung cancer cells. HRAS and NRAS are more highly expressed than KRAS in KRAS-driven lung cancer cells, supporting their roles in regulating KRAS signaling (Extended Data Figure 6a–b). Thus, to further explore the molecular mechanism driving this effect, we assessed whether HRAS and NRAS alter signaling downstream of oncogenic KRAS. We performed pERK immunohistochemistry on lung tumors initiated with Lenti-sgRNA/Cre vectors containing sgInert, sgHras or sgNras in KT;H11LSL-Cas9 mice. Inactivation of HRAS or NRAS increased the number of pERK-positive cells in KRASG12D-driven lung cancer (Figure 4a, Extended Data Figure 6c). Subcutaneous tumors from H23 cells with inactivated HRAS or NRAS also contained more pERK-positive cells when compared to tumors from wildtype (sgSAFE) H23 cells (Figure 4b, Extended Data Figure 6d). Finally, sorted cancer cells from KT;H11LSL-Cas9 mice with lung tumors initiated with Lenti-sgHras/Cre also had greater pERK and pAKT compared to those from tumors initiated with Lenti-sgInert/Cre (Figure 4c).
Figure 4. Wildtype RAS paralogs suppress RAS signaling.
a. Images of pERK staining in KT;H11LSL-Cas9 mice with tumors initiated with indicated Lenti-sgRNA/Cre vectors. Representative of >20 tumors across 5 mice/group. Scale bar: 100 μm.
b. Images of pERK staining in H23 cell subcutaneous tumors. Representative of >15 tumors across 5 mice/group. Scale bar: 100 μm.
c. Western blot of sorted cancer cells from KT;H11LSL-Cas9 mice transduced with indicated Lenti-sgRNA/Cre vectors.
d. Western blot of a murine lung adenocarcinoma cell line transduced with indicated Lenti-sgRNA vectors and selected with puromycin to generate stable knockout cell lines. Cells were cultured under limited serum (1%) for 2 days before protein extraction. HSP90 is a loading control.
e. Western blot of cultured human lung adenocarcinoma cell lines transduced with indicated Lenti-sgRNA vectors and selected with puromycin to generate stable knockout cell lines. Cells were cultured under limited serum (1%) for 2 days before protein extraction.
f. Western blot of human lung adenocarcinoma cell lines re-expressing HRAS (TRE-HRAS) under Doxycycline (Dox) treatment. HRAS-null cells were re-transduced with lentiviral vector expressing TRE-HRAS to generate stable HRAS re-expressing cells (sgHRAS-TRE-HRAS). To re-express HRAS, cells were treated with 0, 1, or 2ng/ml Dox and cultured under limited serum (1%) for 2 days before protein extraction. c-f: HSP90 is a loading control.
g. GI50 values to the MEK inhibitor trametinib among wild-type and HRAS-null H23 cells treated with the indicated doses of trametinib for 4 days. Cell numbers were measured via CCK8 assay and normalized to cells treated with vehicle. Each data point is shown as Mean±SD of 12 wells (one-tailed t-test).
h. GI50 values to the MEK inhibitor trametinib among HRAS-null H23 cells (H23-sgHRAS) re-expressing HRAS in the presence (HRAS+Dox) or absence (HRAS) of Doxycycline plus the indicated dose of trametinib for 4 days. Cell numbers were measured via CCK8 assay and normalized to cells treated with vehicle. Each data point is shown as Mean±SD of 12 wells (one-way ANOVA).
Inactivation of either Hras or Nras in mouse (HC494) and human (H23 and HOP62) oncogenic KRAS-driven cell lines increased ERK phosphorylation, while their effects on AKT phosphorylation were more cell context dependent (Figure 4d–e). Re-expression of wild-type HRAS in HRAS-null H23 and HOP62 human lung cancer cells reduced ERK phosphorylation while again having a cell context dependent effect on AKT phosphorylation (Figure 4f, Extended Data Figure 6e). Furthermore, re-expression of either HRAS or NRAS in HRAS/NRAS double knock out HOP62 cells reduced pERK. Previous publications have shown that inactivating wild-type KRAS increases sensitivity to MEK inhibitors37, 39. Consistent with these studies, inactivation of HRAS in H23 cells modestly increased sensitivity to the MEK inhibitor trametinib while re-expression of HRAS made cells more resistant (Figure 4g, h). These data suggest that inactivation of HRAS or NRAS hyper-activates MAPK-ERK signaling in KRAS mutant cancer cells40–42.
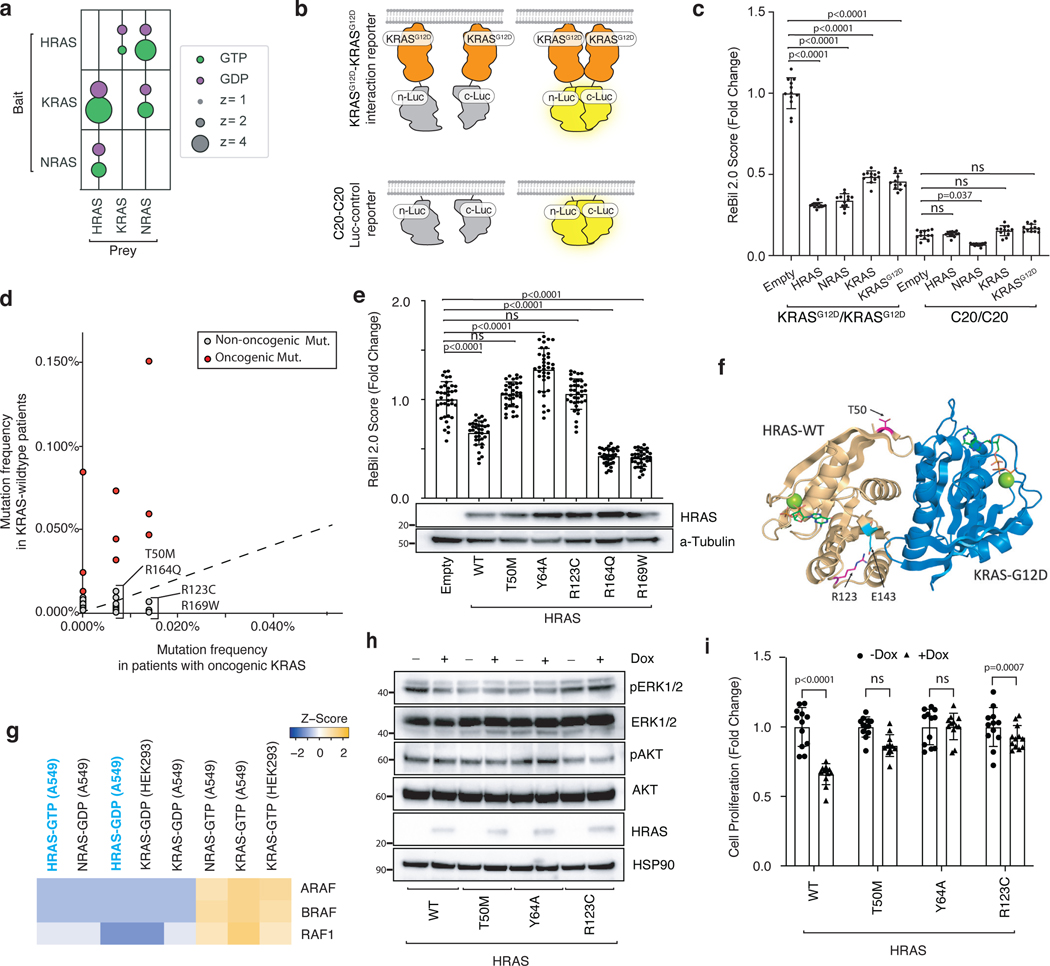
RAS paralogs suppress oncogenic KRAS-KRAS interaction
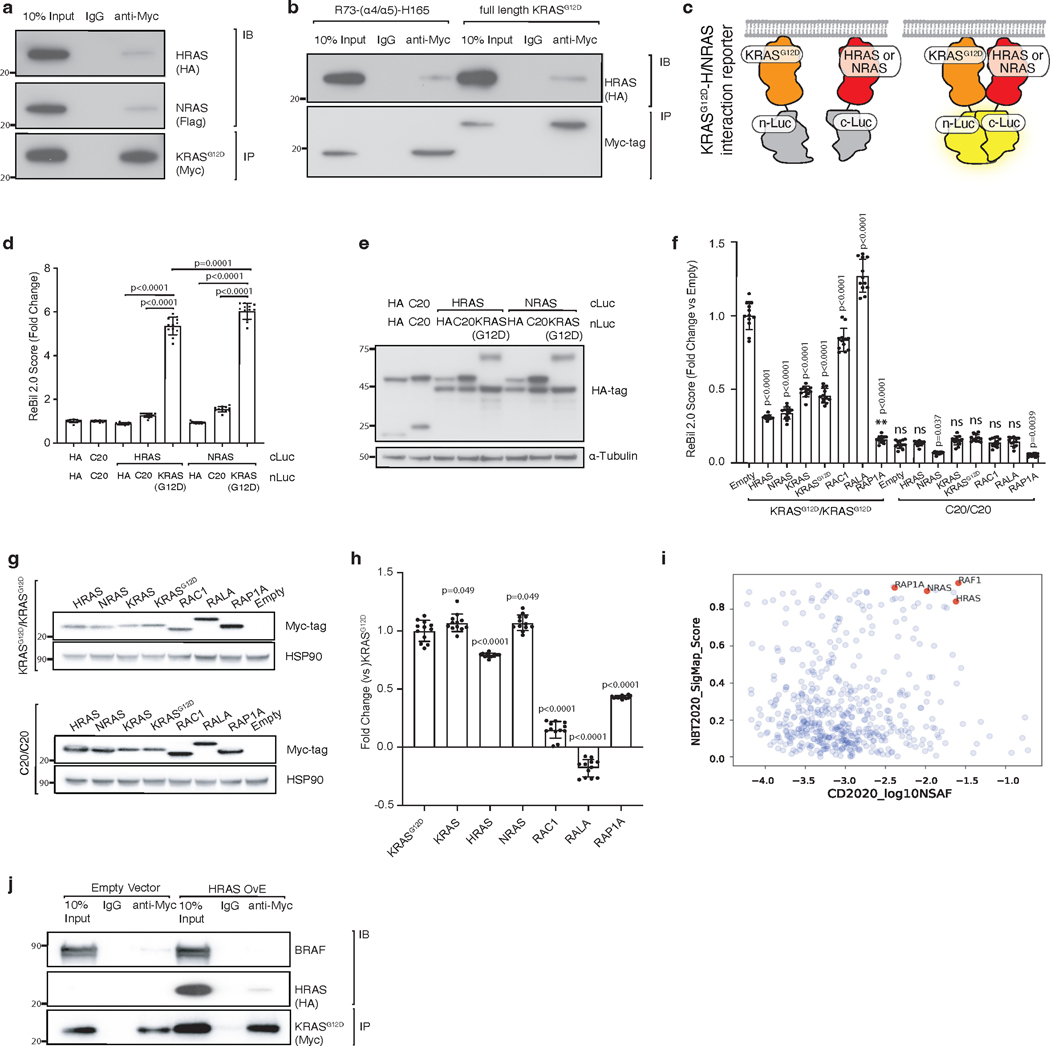
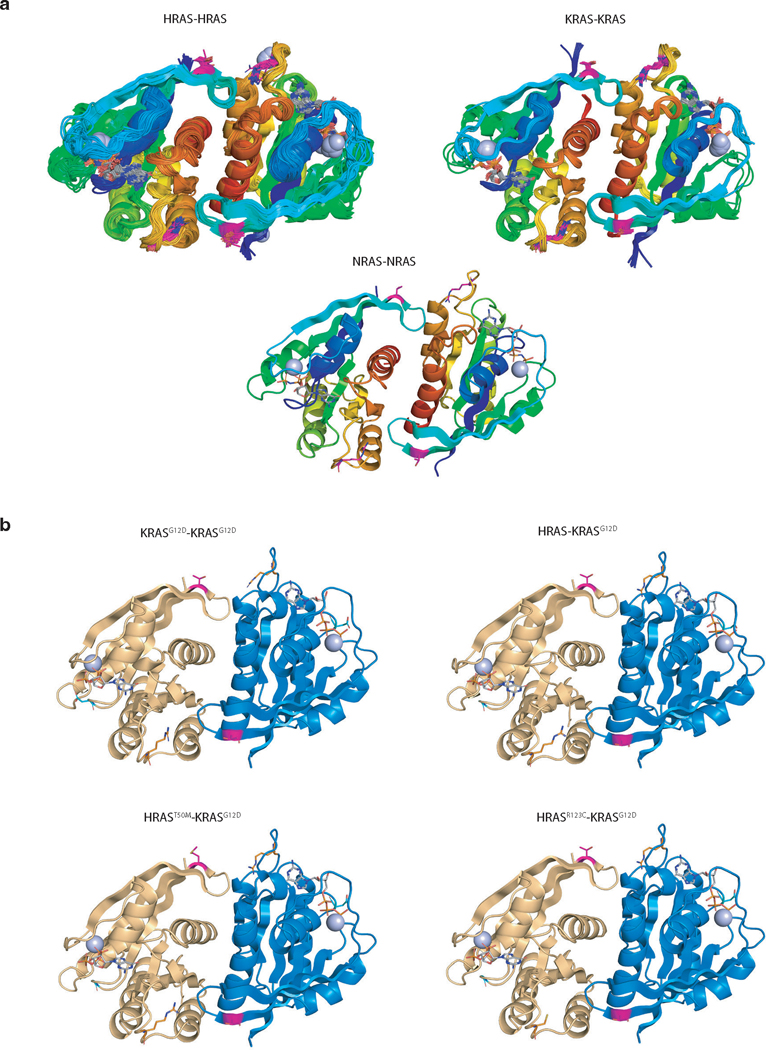
RAS proteins interact and form functional clusters on membranes to efficiently recruit downstream effectors43–45. Whether RAS proteins form dimers or oligomers through direct interactions or through close physical proximity is debated within the field16, 46–47. We next assessed whether HRAS and NRAS interact with KRAS. Both AP/MS data and co-immunoprecipitation experiments suggest that HRAS and NRAS interact with KRASG12D, supporting the existence of heterotypic RAS-RAS interactions, possibly through a domain containing the α4/α5 interface (Figure 5a and Extended Data Figure 7a–b). To assess the ability of RAS paralogs to interact with oncogenic KRASG12D, we adapted a split-luciferase reporter system, which relies on luciferase complementation to quantify RAS-RAS interactions in living cells (Figure 5b)16. We first used this split-luciferase reporter system to confirm the interaction between HRAS and NRAS with KRASG12D (Extended Data Figure 7c–e) 16. Through expression of wild-type KRAS, HRAS, or NRAS in KRASG12D-KRASG12D interaction reporter cells and control reporter cells, we found that all wild-type RAS paralogs can disrupt KRASG12D-KRASG12D interactions. While the other RAS family members RAC1 or RALA did not impact KRASG12D-KRASG12D interactions, we validated the RAP1A-KRASG12D interaction that was predicted from the initial AP/MS data (Figure 5c and Extended Data Figure 7f–i). Lastly, we overexpressed HRAS in KRASG12D-expressing 293T cells and found HRAS-KRASG12D interaction reduced BRAF-KRASG12D interactions (Extended Data Figure 7j).
Figure 5. Wildtype RAS paralogs fine-tune RAS signaling through interaction with oncogenic KRAS.
a. Enrichment of RAS paralogs in AP/MS experiments with HRAS, KRAS, and NRAS as baits.
b. Diagram of the ReBiL2.0 split luciferase system. KRASG12D-KRASG12D interactions were quantified by normalized luminescent signal generated by membrane-association-facilitated interaction of split-luciferase fused to the N-terminus of KRASG12D (upper). Split-luciferase fused to the last 20 amino acids of KRAS (C20) is used as a control for background split-luciferase interaction (lower).
c. 293T cells expressing nLuc-KRASG12D/cluc-KRASG12D or nLuc-C20/cluc-C20 with or without exogenous KRAS, HRAS, or NRAS were cultured in limited serum (1%) for 24 hours. Points are Mean±SD ReBiL2.0 score of 12 wells normalized to cells transduced with empty vector. ns: not significant (one-way ANOVA).
d. Pan-cancer frequency of HRAS mutations in wild-type and oncogenic KRAS-driven tumors. The dashed line indicates equal mutation frequency. Four mutations chosen for further validation in this study are highlighted.
e. U2OS-764 (nLuc-KRASG12D/cluc-KRASG12D) cells expressing wildtype or mutant HRAS were cultured in limited serum (1%) with100 ng/ml Dox for 24 hours. Points are Mean±SD ReBiL2.0 score of 36 wells normalized to cells transduced with empty lentiviral vector. HRAS protein expression level in corresponding cells is shown by western blot. ns: not significant (one-way ANOVA).
f. HRAST50M and HRASR123C are located close to the predicted HRAS-KRAS interaction interface. Residue R123 (magenta) makes an intrachain salt bridge with E143 (cyan).
g. Prey RAF proteins enriched in each experiment with the indicated baits in A549 cells or HEK293 cells.
h. Western blot of cultured HRAS-null HOP62 cells (HOP62-Cas9-sgHRAS) re-expressing wild-type or mutant HRAS under Dox treatment. Cells were cultured under limited serum (1%) for 2 days before protein extraction.
i. Proliferation of cultured HRAS-null HOP62 cells (HOP62-Cas9-sgHRAS) re-expressing wildtype or mutant HRAS under Dox treatment. Cells were cultured in limited serum (1%) with or without Dox for 4 days. Cell viability was measured via CCK8 assay and normalized to cells treated with vehicle. Re-expression of HRAS mutants had no effects on cell proliferation. Points are Mean±SD of 12 wells. ns: not significant (one-tailed t-test).
Patient-derived HRAS and NRAS mutations impair interaction with oncogenic KRAS
Our findings suggest that the tumor-suppressive function of wild-type HRAS and NRAS are mediated, at least in part by competitive interactions with oncogenic KRAS. We therefore hypothesized that there could be HRAS and NRAS mutations in human tumors with oncogenic KRAS that impair this interaction. To evaluate this possibility, we analyzed data from AACR Project GENIE48. Mutations in HRAS and NRAS were rare (pan-cancer frequency of non-synonymous mutations was 0.83% and 2.87%, respectively). The majority of these were oncogenic mutations in codons 12, 13 or 61 that occurred in samples lacking oncogenic KRAS (Extended Data Figure 8a–b). We did, however, identify multiple rare non-oncogenic HRAS and NRAS mutations (Figure 5d, Extended Data Figure 8c–d). We next assessed the ability of these mutants to interact with oncogenic KRAS. We measured the ability of four HRAS mutants and five NRAS mutants, as well as a control Y64A HRAS mutant that has been suggested to reduce HRAS-HRAS dimerization47, to inhibit KRASG12D-KRASG12D interactions. This identified two HRAS mutants (T50M and R123C) and one NRAS mutant (R102Q) that are unable to reduce KRASG12D-KRASG12D interactions (Figure 5e and S8e-f). Interestingly, both HRAST50 and HRASR123 are located close to the predicted HRAS-KRASG12D interface involving the α4 and α5 helices (Figure 5f, Extended Data Figure 9). These findings are consistent with a model in which interaction of wild-type RAS paralogs with oncogenic KRAS suppresses tumor growth, such that mutations that impair this interaction are beneficial to tumor growth.
Previous publications have shown that RAS proteins differentially bind to RAS effectors and thus could function differently in their downstream signaling10, 50. Re-analysis of HRAS and NRAS AP/MS datasets shows that the binding affinity of GTP-bound HRAS to RAF is more similar to GDP-bound KRAS than to its activated, GTP-bound form, suggesting that RAS heterodimers containing HRAS may be less able to activate downstream oncogenic signaling (Figure 5g)10. To test this hypothesis we re-expressed wild-type HRAS, HRASY64A, or the two patient-derived HRAST50M and HRASR123C mutants in HRAS-null lung cancer cells. Re-expression of wild-type HRAS, but not any of the three mutants, reduced ERK phosphorylation and cell proliferation (Figure 5h–i). Similarly, re-expression of wild-type NRAS, but not NRASR102Q, suppressed ERK phosphorylation and proliferation in NRAS-null lung cancer cells (Extended Data Figure 8g–h). These results further suggest that RAS paralog imbalance alters oncogenic KRAS signaling via oncogenic KRAS-wildtype RAS paralog interaction and is thus a driver of lung cancer growth.
HRAS and NRAS are specific suppressors of oncogenic KRAS-driven lung cancer growth
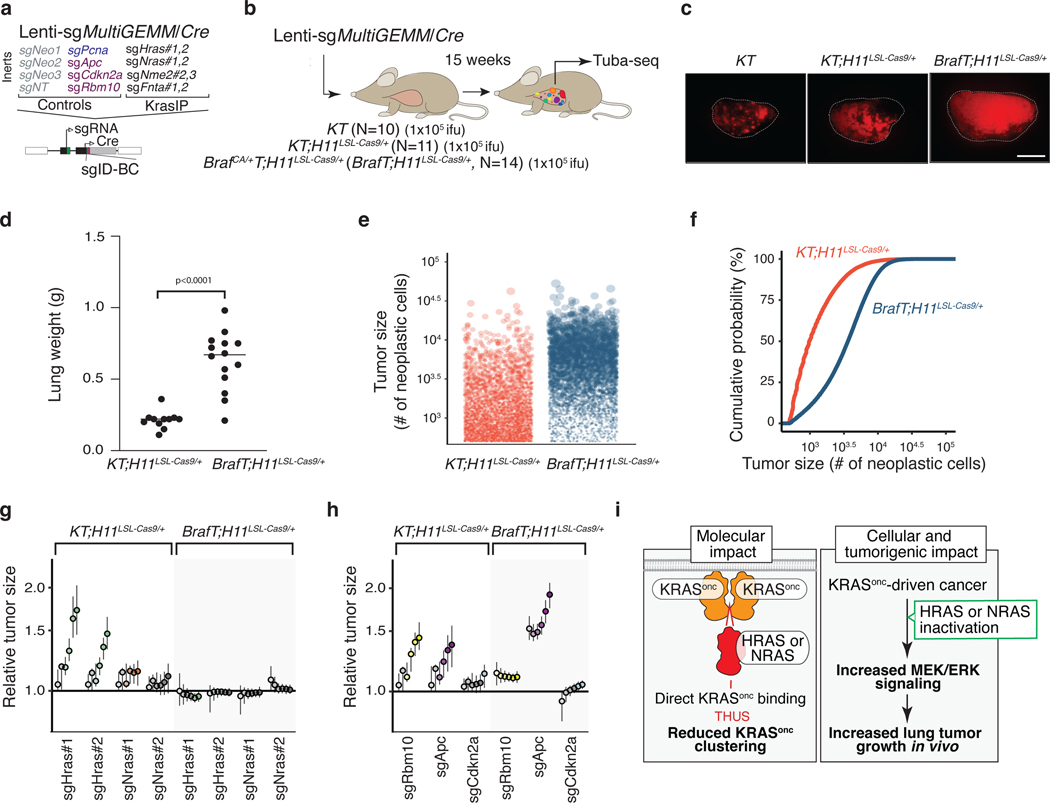
Our in vivo data demonstrate that HRAS and NRAS function as tumor suppressors, and our in vitro results suggest that these suppressive effects are mediated through the interaction of these RAS paralogs with oncogenic KRAS. If the mechanism by which HRAS and NRAS suppress tumor growth is mediated by interactions with oncogenic KRAS, then these proteins should not be tumor suppressors in lung adenocarcinomas in which activation of the RAS/RAF/MEK signaling pathway occurs downstream of KRAS. To test this directly in autochthonous tumors, we initiated tumors in mouse models of oncogenic KRAS-driven and oncogenic BRAF-driven lung cancer using a sub-pool of barcoded Lenti-sgRNA/Cre vectors (Lenti-sgMultiGEMM/Cre; Figure 6a). In addition to vectors targeting Hras and Nras, this pool contained vectors targeting several known tumor suppressors (Apc, Rbm10, and Cdkn2a) and other KRAS-interacting proteins (Aldh1a, Nme2), as well as control vectors (Figure 6a). We initiated tumors with the Lenti-sgMultiGEMM/Cre pool in KT and KT;H11LSL-Cas9 mice as well as in BrafT;H11LSL-Cas9 mice which contain a Cre-regulated allele of oncogenic BRAFV618E (the mouse equivalent of human BRAFV600E)(Figure 6b)51. 15 weeks after tumor initiation the two models had similar maximum tumor sizes, but BrafT;H11LSL-Cas9 mice had larger tumors of relatively uniform size which is consistent with previous results (Figure 6c–f)51.
Figure 6. Paired screens in KRAS-driven and BRAF-driven lung cancer models validate HRAS and NRAS as KRAS-specific tumor suppressors.
a-b. Schematic of pairwise screens to assess tumor-suppressive function in KRAS- and BRAF-driven lung cancer. Tumors were initiated in KT;H11LSL-Cas9/+ and BrafCA/+T;H11LSL-Cas9 /+ (BrafT;H11LSL-Cas9/+) mice through intratracheal delivery of Lenti-sgMultiGEMM/Cre and Tuba-seq was performed on each tumor-bearing lung 15 weeks after initiation.
c. Fluorescence images of representative lung lobes 15 weeks after tumor initiation. Scale bars: 5 mm. Lung lobes are outlined.
d. Lung weights in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice 15 weeks after tumor initiation. Each dot is a mouse, and mean values are indicated (one-tailed t-test).
e-f. Size distribution of sgInert tumors in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice. In e., each dot represents a tumor and has an area proportional to its size. A random sample of 1,000 tumors from each of five representative KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice is plotted. In f., the empirical cumulative distribution function of tumor sizes (>500 cells) across all KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice are plotted.
g. Inactivation of either Hras or Nras increases tumor size in KT;H11LSL-Cas9/+ but not BrafT;H11LSL-Cas9/+ models.
h. Comparison of the effects of inactivation of known tumor suppressors on tumor size in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ models. For panels g, h: Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice. Line at y=1 indicates no effect relative to sgInert. Error bars indicate 95% confidence intervals around the point estimate of the test statistic. Percentiles that are significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. Confidence intervals and P-values were calculated using a nested bootstrap resampling approach across 11 KT;H11LSL-Cas9/+ mice and 14 BrafT;H11LSL-Cas9/+ mice.
i. Schematic of the function of wild-type RAS paralogs as tumor suppressors in oncogenic KRAS-driven lung cancer. Left panel, wildtype RAS paralogs competitively interact with oncogenic KRAS and suppress oncogenic KRAS clustering. Right panel, inactivation of wild-type RAS alleles, or “RAS paralog imbalance”, hyper-activates oncogenic KRAS signaling and promotes lung cancer growth.
Our Tuba-seq data also allowed us to compare the impact of the CRISRP/Cas9-inactivated genes across oncogenic contexts. Importantly, while inactivation of Hras or Nras increased the growth of oncogenic KRAS-driven lung tumors, inactivation of Hras or Nras had no effect on the growth of oncogenic BRAF-driven lung cancer (Figure 6g, Extended Data Figure 10). These results were consistent for both Lenti-sgRNA/Cre vectors targeting each gene. The known tumor suppressor genes assayed (Apc, Cdkn2a, and Rbm10) generally retained their growth-suppressive effects in the BRAF-driven model, suggesting that the abrogation of effect observed for Hras and Nras is not due to a generic inability of additional alterations to increase BRAF-driven lung tumor growth (Figure 6h, Extended Data Figure 10). Thus, HRAS and NRAS specifically suppress oncogenic KRAS-driven tumor growth in vivo.
We also identified several other instances of oncogene-tumor suppressor epistasis. For instance, Apc inactivation has a greater effect on BRAF-driven lung cancer whereas Rbm10 inactivation has a greater effect on KRAS-driven lung cancer (Figure 6h, Extended Data Figure 10). In contrast, inactivation of Nme2, Fnta, and Aldh1a reduced initiation and growth of oncogenic KRAS-driven and oncogenic BRAF-driven lung cancer, suggesting that they are generally required for optimal lung cancer growth in vivo (Extended Data Figure 10). Thus, our paired screens not only localized the effect of Hras and Nras inactivation, but also highlighted the value of this approach in uncovering alterations that have effects within or across oncogenic contexts.
DISCUSSION
Oncogenic KRAS-driven lung cancer is a leading cause of cancer-related deaths. Despite the identification of oncogenic RAS almost half a century ago, the function of many RAS-interacting proteins remains largely unknown. KRAS and other RAS family proteins have been reported to form dimers and nanoclusters16, 46–47. Importantly, both in vitro and in vivo studies suggest that KRAS-KRAS interactions influence effector protein activation, cellular transformation, and tumor growth45. Multiple lines of evidence, including observation of loss of the wild-type KRAS allele and copy number gain of oncogenic KRAS in human tumors, as well as functional studies in mouse models, suggest that wild-type KRAS is tumor-suppressive (also called “RAS allelic imbalance”), although the exact role of wild-type KRAS in lung cancer is still debated3, 38, 41, 52, 53. Recent data suggest that interactions also occur among HRAS, NRAS and KRAS, thus raising the question of the roles of wild-type HRAS and NRAS in oncogenic KRAS-driven cancer10, 11, 16.
By integrating AP/MS data with multiplexed somatic cell CRISPR/Cas9-editing in autochthonous mouse models, we assess the functional impact of inactivating a panel of KRAS-interacting proteins including the RAS paralogs HRAS and NRAS on lung cancer in vivo. We show that wild-type HRAS and NRAS are potent KRAS-specific tumor suppressors that interact with oncogenic KRAS, disrupt KRAS-KRAS interactions, and suppress downstream RAS/MAPK signaling. These results support a model in which heterotypic interactions between RAS paralogs suppress oncogenic KRAS-driven lung cancer growth such that changes in the ratio of wild-type RAS paralogs to oncogenic KRAS can drive tumor growth, a phenomenon we term “RAS paralog imbalance”.
Interestingly, our data suggest that HRAS is a more potent suppressor of tumor growth than NRAS in mouse models of oncogenic KRAS-driven lung adenocarcinoma, while NRAS appears to have stronger effects on KRAS-KRAS dimerization and downstream signaling in human cells. Our data suggest that NRAS may be slightly more potent than HRAS at inhibiting oncogenic KRAS-KRAS interactions (Extended Data Figure 8c–g), but that the protein expression of the RAS paralogs can vary among cell types and cancer types (Extended Data Figure 7a–b). Thus, per-molecule ability to disrupt KRAS-KRAS interaction, preferential interactions with downstream effectors, and the stoichiometry of KRAS, HRAS and NRAS proteins likely integrate to drive the cellular and in vivo phenotypes.
The impact of RAS paralog imbalance may extend beyond lung cancer and KRAS codon 12 mutations. Germline HRAS deletion increases the development of KRAS-driven pancreatic cancer, skin papillomas, and carcinogen-induced KRASQ61 lung cancer53–55. However, the impact of inactivating RAS paralogs in cancers with different driver oncogenes (e.g. oncogenic EGFR-driven lung cancer) is likely to be different since RAS proteins also serve as important components in growth factor signaling pathways. Whether RAS paralog heterodimerization also impacts signaling during normal development, homeostasis, or other diseases states remains unknown. Collectively, these findings suggest that modulating RAS protein interactions, such as by skewing the stoichiometry of oncogenic to wild-type RAS or by forcing inter-paralog competition, could lead to therapeutic strategies.
Given the complexity of RAS signaling, other non-mutually exclusive mechanisms by which RAS paralogs could reduce oncogenic KRAS-driven cancer growth should be considered. For example, it has been reported that upstream regulators, such as SOS1, could bridge the interaction between oncogenic and wild-type RAS56. GDP-bound wild-type HRAS and NRAS could also compete with oncogenic KRAS for upstream guanine nucleotide exchange factors and thus reduce RAS signaling57. Additionally, it is possible that HRAS and NRAS compete with oncogenic KRAS for downstream effectors. Whether HRAS and NRAS also function through these alternative routes, and how different mechanisms are integrated to execute their tumor-suppressive functions, will require additional investigation.
The National Cancer Institute "RAS Pathway V2.0", contains more than 200 proteins known or suspected to be involved in RAS signaling. Characterizing the role of these proteins in tractable in vivo models of RAS-driven cancer remains a challenge. Our study outlines a technological avenue to study KRAS-specific signaling components in a multiplexed manner. By harnessing the power of Tuba-seq, we quantified the effects of more than a dozen putative RAS pathway genes on tumor growth simultaneously.. Furthermore, by performing paired screens in oncogenic KRAS-driven and BRAF-driven cancer models, we localized the growth-suppressive effects of these RAS paralogs. Our study demonstrates the feasibility of performing in vivo genetic interaction screens, and the power of such approaches to provide insight into the mechanisms of tumor suppression. Future studies of this type should enable a more quantitative understanding of the role of RAS pathway components in oncogenesis.
METHODS
Mouse research
The use of mice for the current study has been approved by the Institutional Animal Care and Use Committee at Stanford University, protocol number 26696. KrasLSL-G12D/+ (RRID:IMSR_JAX:008179), R26LSL-tdTomato (RRID:IMSR_JAX:007909), and H11LSL-Cas9 (RRID:IMSR_JAX:027632) mice have been previously described. They were on a C57BL/6:129 mixed background for the experiments in Figures 1, 2, 4; and were on a C57BL/6 background for the experiments in Figure 6. The B6.129P2(Cg)-Braftm1Mmcm/J (BRAF CA/+) mice were initially generated by Dankort et al. and obtained from the Jackson Laboratory (RRID:IMSR_JAX: 017837). We used balanced sex of animals with age ranging 8 to 15 weeks at the time of tumor initiation. Mice were housed at Stanford SIM1 barrier facility under a 12hr-12hr light-dark cycle with dark hours between 18:30–6:30. Housing temperature at 68–73F under 40–60% humidity.
Cells and Reagents:
NCI-H23 (CRL-5800), NCI-H727 (CRL-5815), and HEK293T (CRL-3216) were originally purchased from ATCC; HC494 (KPT), HC367 (KPT), and MT658 (KPT) lung adenocarcinoma cells were generated in the Winslow Lab; U2OS-134–764np (nLuc-KRASG12D cluc-KRASG12D; KRASG12D was fused to the N-termini of split luciferase proteins) cell were generated in the Wahl lab by Dr. Yao-Cheng Li (Salk Institute for Biological Studies). HC494, HC367, and MT658 cells were cultured in DMEM containing 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin. H23 and H727 cells were cultured in RPMI1640 medium containing 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin. U2OS cells were cultured in DMEM/F12 (Thermo Fisher; phenol-red free), 10% (vol/vol) FBS, and 10 μg/mL ciprofloxacin. All cell lines were confirmed to be mycoplasma negative (MycoAlert Detection Kit, Lonza).
Trametinib was purchased from MedChemExpress (HY-10999); 5-Bromo-2′-deoxyuridine (10280879001) and D-Luciferin (L9504–5MG) were purchased from Sigma-Aldrich. All plasmids used in this study were listed in Supplementary Table 1 and are available from our laboratory (key plasmids will be donated to Addgene).
Design, generation, barcoding, and production of lentiviral vectors
sgRNA sequences targeting the putative tumor suppressor genes were designed using CRISPick (https://portals.broadinstitute.org/gppx/crispick/public ). All sgRNA sequences are shown in Supplementary Table 2. Each desired sgRNA vector was modified from our previously published pll3-U6-sgRNA-Pgk-Cre vector via site-directed mutagenesis (New England Biolabs, E0554S). The generation of the barcode fragment containing the 8-nucleotide sgID sequence and 20-nucleotide degenerate barcode, and subsequent ligation into the vectors, were performed as previously described18–19.
Lentiviral vectors were produced using polyethylenimine (PEI)-based transfection of 293T cells with delta8.2 and VSV-G packaging plasmids in 150 mm cell culture plates. Sodium butyrate (Sigma Aldrich, B5887) was added 8 hours after transfection to achieve a final concentration of 20 mM. Media was refreshed 24 hours after transfection. 20 mL of virus-containing supernatant was collected 36, 48, and 60 hours after transfection. The three collections were then pooled and concentrated by ultracentrifugation (112,000 g for 1.5 hours), resuspended overnight in 100 μL PBS, then frozen at −80°C and were thawed and pooled at equal ratios immediately prior to delivery to mice.
Tumor initiation
Tumors were initiated by intratracheal delivery of 60 μl of lentiviral vectors dissolved in PBS. For the initial experiments in Figures 1 and 2, tumors were allowed to develop for 12 weeks after delivery of a lentiviral pool that contained 19 barcoded Lenti-sgRNA/Cre vectors (Lenti-sgKrasIP/Cre). Tumors were initiated in KrasLSL-G12D; R26LSL-tdTomato (KT), KT;H11LSL-Cas9, or KT;p53fl/fl;H11LSL-Cas9 mice with 1.95x105 infectious units (ifu)/mouse.
For the validation experiments in Figure 3, tumors were allowed to develop for 15 weeks after delivery of a lentiviral pool that contained 26 barcoded Lenti-sgRNA/Cre vectors (Lenti-sgValidation/Cre). Tumors were initiated in KrasLSL-G12D; R26LSL-tdTomato (KT) or KT;H11LSL-Cas9; mice with 3x105 ifu/mouse.
For the individual sgRNA tumor initiation experiments in Figure 3, tumors were allowed to develop for 12 weeks after delivery of individual sgRNA-expressing lentiviral vectors targeting Neo2, Hras, or Nras. Tumors were initiated in KT;H11LSL-Cas9; mice with 1x105 ifu/mouse.
For the paired screen experiments in Figure 6, tumors were allowed to develop for 15 weeks after delivery of a lentiviral pool that contained 15 barcoded Lenti-sgRNA/Cre vectors (Lenti-sgMultiGEMM/Cre). Tumors were initiated in KT;H11LSL-Cas9/+ or BrafV600E;R26LSL-tdTomato;H11LSL-Cas9/+ mice with 3x105 ifu/mouse. Note that KT;H11LSL-Cas9/+ rather than KT;H11LSL-Cas9/LSL-Cas9 mice were used in this experiment to match the Cas9 dosage of the BrafT;H11LSL-Cas9/+ mice, whereas KT;H11LSL-Cas9/LSL-Cas9 mice were used in all other experiments. To evaluate the effects of Cas9 dosage on the tumor-suppressive effects of the Lenti-sgMultiGEMM/Cre pool, we also initiated tumors in a small cohort of KT;H11LSL-Cas9/LSL-Cas9 mice. Reductions in the magnitude of the effects of various sgRNAs were observed in the KT;H11LSL-Cas9/+ cohort relative to the KT;H11LSL-Cas9/LSL-Cas9 cohort, underscoring the importance of matching Cas9 dosage and suggesting that Cas9 can be limiting in H11LSL-Cas9/+ mice.
Tuba-seq library generation
Genomic DNA was isolated from bulk tumor-bearing lung tissue from each mouse as previously described. Briefly, benchmark control cell lines were generated from LSL-YFP MEFs transduced by a barcoded Lenti-sgNT3/Cre vector (NT3: an inert sgRNA with a distinct sgID) and purified by sorting YFPpos cells. Three benchmark control cell lines (500,000 cells each) were added to each mouse lung sample prior to lysis to enable the calculation of the absolute number of neoplastic cells in each tumor from the number of sgID-BC reads. Following homogenization and overnight protease K digestion, genomic DNA was extracted from the lung lysates using standard phenol-chloroform and ethanol precipitation methods. Subsequently, Q5 High-Fidelity 2x Master Mix (New England Biolabs, M0494X) was used to amplify the sgID-BC region from 32 μg of genomic DNA in a total reaction volume of 800 μl per sample. The unique dual-indexed primers used were Forward: AAT GAT ACG GCG ACC ACC GAG ATC TAC AC-8 nucleotides for i5 index-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT-6 to 9 random nucleotides for increased diversity-GCG CAC GTC TGC CGC GCT G and Reverse: CAA GCA GAA GAC GGC ATA CGA GAT-6 nucleotides for i7 index- GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T-9 to 6 random nucleotides for increased diversity-CAG GTT CTT GCG AAC CTC AT. The PCR products were purified with Agencourt AMPure XP beads (Beckman Coulter, A63881) using a double size selection protocol. The concentration and quality of the purified libraries were determined using the Agilent High Sensitivity DNA kit (Agilent Technologies, 5067–4626) on the Agilent 2100 Bioanalyzer (Agilent Technologies, G2939BA). The libraries were pooled based on lung weight to ensure even sequencing depth, cleaned up again using AMPure XP beads, and sequenced (read length 2x150bp) on the Illumina HiSeq 2500 or NextSeq 500 platform (Admera Health Biopharma Services).
Generation of Stable Cell Lines:
Parental cells were seeded at 50% confluency in a 6-well plate the day before transduction (day 0). The cell culture medium was replaced with 2 mL fresh medium containing 8 μg/mL hexadimethrine bromide (Sigma Aldrich, H9268–5G), 20 μL ViralPlus Transduction Enhancer (Applied Biological Materials Inc., G698) and 40 μL concentrated lentivirus, then cultured overnight (Day 1). The medium was then replaced with complete medium and cultured for another 24 hours (Day 2). Cells were transferred into a 100 mm cell culture dish with appropriate amounts of antibiotic (Blasticidin doses: U2OS: 10 μg/mL; HOP62: 50 μg/mL; H727: 10 μg/mL; H23: 15 μg/mL; Puromycin doses: HC494: 5 μg/mL; U2OS: 1 μg/mL; HOP62: 5 μg/mL; H727: 5 μg/mL; H23: 5 μg/mL; 293T: 3ug/ml) and selected for 48 hours (Day 3).
Western Blot
Cells were lysed in RIPA buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, and 0.1% SDS) and incubated at 4 °C with continuous rotation for 30 minutes, followed by centrifugation at 12,000 × rcf for 10 minutes. The supernatant was collected, and the protein concentration was determined by BCA assay (Thermo Fisher Scientific, 23250). Protein extracts (10–50 μg) were separated on 4–12% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% non-fat milk in TBS with 0.1% Tween 20 (TBST) at room temperature for one hour, cut according to the molecular weight of the target protein (with at least two flanking protein markers), followed by incubation with primary antibodies diluted in TBST (1:1000) at 4 °C overnight. After three 10-minutes washes with TBST, the membranes were incubated with the appropriate secondary antibody conjugated to HRP diluted in TBST (1:10000) at room temperature for 1 hour. After three 10-minutes washes with TBST, protein expression was quantified with enhanced chemiluminescence reagents (Fisher Scientific, PI80196).
Antibodies used in this study: HSP90 (BD Biosciences, 610418), pAKT (Cell Signaling, 4060S), pERK (Cell Signaling, 4370L), ERK (Cell Signaling, 9102S), AKT (Cell Signaling, 4691S), HRAS (Thermo Fisher Scientific, 18295–1-AP), NRAS (Santa Cruz Biotechnology, sc-31), KRAS (EMD Millipore, OP-24), HA-tag (Cell Signaling, 2999S), Myc-tag (Cell Signaling, 2040S), Flag-tag (Cell Signaling, 86861S), BRAF (Cell Signaling, 14814S), α-Tubulin (Cell Signaling, 2144S), GAPDH (Cell Signaling, 5174S), Goat-anti-Rabbit IgG Antibody, HRP-conjugate (Sigma-Aldrich, 12–348), Goat-anti-Mouse IgG Antibody, HRP-conjugate (Thermo Fisher Scientific, 62–6520). Recombinant RAS proteins used in this study: Recombinant KRAS (Abcam, ab156968), Recombinant HRAS (Abcam, ab93949), Recombinant NRAS (Abcam, ab268821).
Coimmunoprecipitation assay.
293T cells with stable expression of myc-KRASG12D were transfected with plasmids expressing HA-tagged HRAS and Flag-tagged NRAS for 24 hrs before lysed with ice-cold IP lysis buffer (Thermo Scientific, 87788) containing protease inhibitor cocktail (Thermo Fisher Scientific 78442). The lysates were pre-cleared with magnetic beads (Thermo Fisher Scientific, 88802) at 4°C for 2hrs. Then protein concentration was determined by BCA assay (Thermo Fisher Scientific, 23250) and equal amount of protein lysis were incubated with anti-Myc- (Thermo Fisher Scientific, 88842) or IgG- (Cell Signaling, 5873S) magnetic beads at 4°C for 12 hrs. The immunoprecipitates were collected using a MACSiMAG Separator (Miltenyi Biotec, 130–092-168), washed for three times with IP lysis buffer and three times with TBST. The immunoprecipitates were eluted via incubating in 1x Non-Reducing Sample Buffer (Thermo Fisher Scientific, 39001) at 95°C for 10 mins before subjected to immunoblotting.
Histology and immunohistochemistry (IHC)
Lung lobes were fixed in 4% formalin and paraffin embedded. Hematoxylin and eosin (H&E) staining was performed using standard methods. IHC was performed on 4-μm sections using the Avidin/Biotin Blocking Kit (Vector Laboratories, SP-2001), Avidin-Biotin Complex kit (Vector Laboratories, PK-4001), and DAB Peroxidase Substrate Kit (Vector Laboratories, SK-4100) following standard protocols.
The following primary antibodies were used: Ki-67 (BD Pharmingen, 550609, 1:100), BrdU (BD Pharmingen, 555627, 1:100), human mitochondria (Abcam, ab92824, 1:100), pERK (Cell Signaling, 4370L, 1:50).
Total tumor burden (tumor area/total area × 100%), mitochondriapos tumor burden (mitochondriapos area/total area × 100%), BrdUpos cell number, Ki67pos cell number, and pERKpos cell number were calculated using ImageJ 1.52p.
Cell proliferation assay (CCK8)
For cell proliferation assays, cells were seeded in 96-well plates at a density of 5000 cells per well and allowed to adhere overnight in regular growth medium (Day 0). Cells were then cultured in medium as indicated on each figure panel for 7 days. Relative cell number were measured every other day using Cell Counting Kit-8 (Bimake, B34304) according to the manufacturer’s instructions.
Colony formation assay
For clonogenic assays, cells were seeded in 6-well plates at a density of 500 cells per well and allowed to adhere overnight in regular growth medium. Cells were then cultured in medium as indicated on each figure panel for 14 days. Growth medium with or without drugs was replaced every 2 days. At the end point, cells were stained with 0.5% crystal violet in 20% methanol. Colony numbers were calculated using ImageJ
Allograft studies in immunocompromised mice
For intravenous transplants into immunocompromised NSG mice, 5x105 H23 cells were injected into one of the lateral tail veins. Mice were sacrificed 28 days post-injection and lung lobes were fixed in 4% formalin and paraffin-embedded. For subcutaneous transplants into immunocompromised NSG mice, 2× 106 of each H23 cells (sgSAFE, sgHRAS, and sgNRAS) were re-suspended in 200uL Matrigel® Basement Membrane Matrix (Corning, 354234) and injected into three parallel sites per mouse. Mice were sacrificed 28 days post-injection. Tumors were dissected and the weight, height, width, and length, of each tumor was measured. Tumor volume was roughly calculated via the formula: V = (4/3) × π × (L/2) × (L/2) × (D/2).
Maximal tumor size/burden permitted by Institute of Medicine Animal Care and Use Committee is 1.75 cm3, the maximal tumor size/burden was not exceeded in our study. Institute of Medicine Animal Care and Use Committee approved all animal studies and procedures.
ReBiL2.0 assay
The ReBiL2.0 assay was performed as previously descried16. 293T cells were co-transfected with plasmids expressing i) nLuc- tethering with KRASG12D or C20 (last 20 amino acids of KRAS); ii) cLuc tethering with KRASG12D or C20 or wildtype H-/N-RAS; iii) myc-tagged RAS-GTPase; and iv) renilla luciferase for 24 hrs. Transfected cells were seeded in i) 96-well plates at a density of 2x104 cells per well, and ii) 6-well plates at a density of 1x106 cells per well, and allowed to adhere overnight in regular growth medium (DMEM, 10% FBS). The next day, cells were then cultured in serum limited medium (DMEM, 1% FBS) for 24 hours. For experiment in Figure 5e, stable ReBiL cells (U2OS-134–764np) were seeded in i) 96-well plates at a density of 2x104 cells per well, and ii) 6-well plates at a density of 1x106 cells per well and allowed to adhere overnight in regular growth medium (DMEM/F12, 10% FBS, and 10 μg/mL ciprofloxacin). The next day, cells were then cultured in serum limited medium (DMEM/F12, 1% FBS, and 10 μg/mL ciprofloxacin) containing 100 ng/mL doxycycline for 24 hours.
Upon termination of the ReBiL assay, i) to measure raw luciferase activity, 300 μM D-luciferin was added to 96-well plate cultures and incubated at 37°C for 30 minutes, and raw luminescent data for both renilla and firefly luciferase were collected by a Tecan microplate reader; ii) to quantify the expression of 1/2luc fusion proteins, ReBiL cells from 6-well plate cultures were harvested with RIPA lysis buffer for protein extraction, and western blots were performed for HA-tag, Myc-tag and HSP90 expression. Then the ReBiL2.0 score was calculated via the formula:
The strength of different RAS-GTPase in disrupting KRASG12D- KRASG12D interactions was calculated via the formula:
Analysis of human lung adenocarcinoma cancer genome sequencing data (for rare HRAS and NRAS mutations)
To assess evidence that HRAS and NRAS function as KRAS-specific tumor suppressors in human cancer, we queried publicly available cancer genomic datasets. GENIE Release 9.1-public was accessed through the Synapse platform and data on somatic mutations (data_mutations_extended.txt), sample- and patient-level clinical data (data_clinical_sample.txt and data_clinical_patient.txt), and genotyping panel information (genomic_information.txt) were downloaded. While it is unclear how our findings may extrapolate to cancer types beyond lung adenocarcinoma, HRAS and NRAS mutations are rare (occurring at frequencies of just 0.83% and 2.87% in GENIE samples, respectively) so we performed a pan-cancer analysis. Each sample was assigned to its patient of origin and annotated for the presence of both oncogenic KRAS mutations (defined as missense mutations in KRAS exons 12, 13 or 61) and for the presence of potentially functional HRAS or NRAS mutations (variants that were silent, intergenic, intronic, or fell in the 3’ or 5’ UTRs were excluded from this analysis). When multiple samples were derived from the same patient, the patient in question was annotated as having a mutation if it occurred in at least one of their associated samples. From this information we produced a list of the frequency of all HRAS and NRAS variants in patients with and without oncogenic KRAS in both datasets. The genotyping panel information was used to identify GENIE patients that were not genotyped at HRAS and/or NRAS and exclude these from the frequency calculation.
Analysis of DepMap data
Cancer cell line dependency data (DepMap Public 19Q4) and mutation data (Cancer Cell Line Encyclopedia) were acquired from the Broad Institute DepMap Portal (https://depmap.org/portal/ ). Lung adenocarcinoma cell lines were identified by their Project Achilles identification code and partitioned into KRAS mutant, if they contained a hotspot mutation, or wildtype groups. Subsequently, dependency scores for NRAS or KRAS were calculated for each cell line within the two groups. Finally, the distribution of dependency scores was plotted using GraphPad Prism 9.
Processing of paired-end reads to identify the sgID and barcode
Sequencing of Tuba-seq libraries produces reads that are expected to contain an 8-nucleotide sgID followed by a 30-nucleotide barcode (BC) of the form GCNNNNNTANNNNNGCNNNNNTANNNNNGC, where each of the 20 Ns represents a random nucleotide. Each sgID has a one-to-one correspondence with an sgRNA in the viral pool; thus, the sgID sequence identifies the gene targeted in a given tumor. Note that all sgID sequences in the viral pool differ from each other by at least three nucleotides such that incorrect sgID assignment (and thus, inference of tumor genotype) due to PCR or sequencing error is extremely unlikely. The random 20-nucleotide portion of the BC is expected to be unique to each lentiviral integration event, and thus tags all cells in a single clonal expansion. Note that the length of the barcode ensures a high theoretical potential diversity (~420 > 1012 barcodes per vector), so while the actual diversity of each Lenti-sgRNA/Cre vector is dictated by the number of colonies generated during the plasmid barcoding step, it is very unlikely that we will observe the same BC in multiple clonal expansions.
FASTQ files were parsed using regular expressions to identify the sgID and BC for each read. To minimize the effects of sequencing error on BC identification, we required the forward and reverse reads to agree completely within the 30-nucleotide sequence to be further processed. We also screened for barcodes that were likely to have arisen due to errors in sequencing the barcodes of genuine tumors. Given the low rate of sequencing error, we expect these spurious “tumors” to have read counts that are far lower than the read counts of the genuine tumors from which they arise. While it is impossible to eliminate these spurious tumors, we sought to minimize their effect by identifying small “tumors” with barcodes that are highly similar to the barcodes of larger tumors. Specifically, if a pair of “tumors” had barcodes that were within a Hamming distance of two, and if one of the tumors had fewer than 5% as many reads as the other, then the reads associated with the smaller tumor were attributed to the larger tumor. After these filtering steps, the read counts associated with each barcode were converted to absolute neoplastic cell numbers by normalizing to the number of reads in the “spike-in” cell lines added to each sample prior to lung lysis and DNA extraction. The median sequencing depth across experiments was ~1 read per 6.4 cells.
For statistical comparisons of tumor genotypes, we applied a minimum tumor size cutoff of 100 cells. In selecting a cutoff, we sought to include tumors that are large enough to be consistently detected despite differences in sequencing depth among mice, while using as many tumors as possible to maximize the statistical power. Importantly, we analyzed each Tuba-seq dataset with multiple minimum tumor size cut-offs (50, 100, 200, 500 cells) and found that our findings were robust.
Summary statistics for overall growth rate
To assess the extent to which a given gene (X) affects tumor growth, we compared the distribution of tumor sizes produced by vectors targeting that gene (sgX tumors) to the distribution produced by our negative control vectors (sgInert tumors). We relied on two statistics to characterize these distributions: the size of tumors at defined percentiles of the distribution (specifically the 50th, 60th, 70th, 80th, 90th, and 95th percentile tumor sizes), and the log-normal mean size (LN mean). The percentile sizes are nonparametric summary statistics of the tumor size distribution. In considering percentiles corresponding to the right tail of the distribution, we focus on the growth of larger tumors, thereby avoiding issues stemming from potential variation in cutting efficiency among guides. The LN mean is the maximum-likelihood estimate of mean tumor size assuming a log-normal distribution. Previous work found that this statistic represents the best parametric summary of tumor growth based on the maximum likelihood quality of fit of various common parametric distributions.
To quantify the extent to which each gene suppressed or promoted tumor growth, we normalized statistics calculated on tumors of each genotype to the corresponding inert statistic. The resulting ratios reflect the growth advantage (or disadvantage) associated with each tumor genotype relative to the growth of sgInert tumors.
For example, the relative ith percentile size for tumors of genotype X was calculated as:
Likewise, the relative LN mean size for tumors of genotype X was calculated as:
Summary statistics for relative tumor number and relative tumor burden
In addition to the tumor size metrics described above, we characterized the effects of gene inactivation on tumorigenesis in terms of the number of tumors and total neoplastic cell number (“tumor burden”) associated with each genotype. Unlike the aforementioned metrics of tumor size, tumor number and burden are linearly affected by lentiviral titer and are thus sensitive to underlying differences in the representation of each Lenti-sgRNA/Cre vector in the viral pool. Critically, each Tuba-seq experiment included a cohort of KT control mice. KT mice lack expression of Cas9, thus all Lenti-sgRNA/Cre vectors are functionally equivalent in these mice, and the observed tumor number and burden associated with each sgRNA reflects the make-up of the viral pool.
To assess the extent to which a given gene (X) affects tumor number, we therefore first normalized the number of sgX tumors in KT;H11LSL-Cas9 mice (also KT;p53flox/flox;H11LSL-Cas9 and BrafLSL-V600E/+T; H11LSL-Cas9 mice in the initial Kras-interacting protein screen and the paired screen, respectively) to the number of sgX tumors in the KT mice:
As with the tumor size metrics, we then calculated a relative tumor number by normalizing this statistic to the corresponding statistic calculated using sgInert tumors:
Genes that influence relative tumor number modify the probability of tumor initiation and/or the very early stages of oncogene-driven epithelial expansion, which prior work suggests are imperfectly correlated with tumor growth at later stages. Relative tumor number thus captures an additional and potentially important aspect of tumor suppressor gene function.
Analogous to the calculation of relative tumor number, we characterized the effect of each gene on tumor burden by first normalizing the sgX tumor burden in Cas9-expressing mice to the burden in KT mice:
We then calculated a relative tumor burden by normalizing this number to the corresponding statistic calculated using sgInert tumors:
Tumor burden is an integration over tumor size and number, and thus reflects the total neoplastic load in each mouse. Tumor burden is thus more strongly related to morbidity than are our metrics of tumor size and is closely related to traditional measurements of tumor progression such as duration of survival and tumor area. While intuitively appealing, tumor burden is notably nosier than our metrics of tumor size as it is strongly determined by the size of the largest tumors.
Calculation of confidence intervals and P-values for tumor growth and number metrics
Confidence intervals and P-values were calculated using bootstrap resampling to estimate the sampling distribution of each statistic. To account for both mouse-to-mouse variability and variability in tumor size and number within mice, we adopted a two-step, nested bootstrap approach where we first resampled mice, and then resampled tumors within each mouse in the pseudo-dataset. 10,000 bootstrap samples were drawn for all reported P-values. 95% confidence intervals were calculated using the 2.5th and 97.5th percentile of the bootstrapped statistics. Because we calculate metrics of tumor growth that are normalized to the same metrics in sgInert tumors, under the null model where genotype does not affect tumor growth, the test statistic is equal to 1. Two-sided p-values were thus calculated as followed:
Where T is the test statistic and Pr(T>1) and Pr(T<1) were calculated empirically as the proportion of bootstrapped statistics that were more extreme than the baseline of 1. To account for multiple hypothesis testing, p-values were FDR-adjusted using the Benjamini-Hochberg procedure as implemented in the Python package stats models. Summarized statistics of all Tuba-seq experiments in this study can be found in Supplementary Table 3–6.
AP-MS data visualization
AP-MS data were analyzed as described (Ding et al 2016). Briefly, protein spectral matches (PSMs; Kelly et al. 2020) were normalized by protein length and total spectral matches per experiment. These normalized spectral abundance factors (NSAFs) were then normalized to NSAFs of matched prey proteins from a large cohort of unrelated AP/MS experiments to produce a Z-score. Z-scores are proportional to the areas of circles in bubble plots. In cluster diagrams, NSAFs are binarized by statistical significance (FDR > 0.5), similarities between interactome profiles were determined by cosine distance, and dendrogram topology was determined by UPGMA.
Modeling RAS-RAS dimers
Potential templates for modeling RAS heterodimers were obtained from the ProtCID database. ProtCID is built from clustering interfaces of homologous proteins obtained from domain-domain contacts within protein crystals in the Protein Data Bank. Hierarchical clustering of interfaces is performed with a Jaccard-index similarity metric based on the contacts shared between different interfaces. Models for the structure of the HRAS/KRAS heterodimer were built by superposing a structure of KRAS-G12D (PDB: 5USJ) onto a monomer of the HRAS homodimer in PDB entry 3K8Y. All structural data files generated in this study can be accessed via zenodo: https://zenodo.org/record/7104280
Statistical analysis for non-Tuba-seq experiments
Sample or experiment sizes were estimated based on similar experiments previously performed in our laboratory, as well as in the literature. Biological replications (> 5 mice for each cohort, >10 wells per culture condition) of the experiments were as detailed in the manuscript. All values are presented as mean ± SD, with individual data points shown in the figure when possible. Comparisons of parameters between two groups were made by two-tailed Student’s t-tests. The differences among several groups were evaluated by one-way ANOVA with Tukey-Kramer post hoc evaluation. p-values less than 0.05 and 0.01 were considered significant (*) or very significant (**), respectively.
Statistics & Reproducibility
The statistical tests used for each analysis are described in detail in the sections above. All analyses of barcode sequencing data were performed in Python (3.4) and visualizations of data were performed in R (3.6.0). Sample sizes were determined based on our previous experience conducting similar experiments and, in the case of Tuba-seq experiments, based on previously published power analyses18. For experiments using western blot as a readout, at least three independent experiments were repeated with similar results. In all the experiments reported in this study, no data points were excluded. No randomization was used in this study. Data collection and analysis were not performed blind to the conditions of the experiments. Analyses of barcode sequencing data used non-parametric statistics; therefore no assumptions about the distribution of data were made. For all other analyses data distributions were assumed to be normal but this was not formally tested and individual data points are plotted to show distribution.
Extended Data
Extended Data Fig. 1. Prioritization of candidate KRAS-interacting proteins for this study.
a. Flow chart for prioritization of candidate KRAS-interacting proteins for this study. Candidate KRAS-interacting proteins were chosen based on multiple criteria including their interaction with KRAS, their homolog mRNA expression in a mouse model of KrasG12D-driven lung cancer, and the consistency with which they bind different RAS-GTPases. RADIL was added at the last step due to its validated importance in KRAS-mutant human cell lines. b. Candidate proteins interact with KRAS in two protein-protein interaction analyses (Kelly, Kostyrko, Han et al. 2020; Broyde, Simpson, Murray et al. 2020). KRAS-interacting proteins are shown as their log10NSAF and SigMap Scores. c. Homolog mRNA expression (TPM) of candidate KRAS-interacting proteins in a mouse model of KrasG12D-driven lung cancer (Chuang et al. 2017). d. Bubble plot of eight AP/MS experiments with GTP- and GDP-locked mutant GTPases as baits (rows), showing the enrichment of selected candidate KRAS-interacting proteins (columns). Dark borders indicate FDR < 0.05. e. Mutation and copy number alteration frequencies of the 13 candidate genes in lung adenocarcinomas with oncogenic KRAS (N = 152; data from TCGA PanCancer Atlas, Cell 2018).
Extended Data Fig. 2. Tumor barcoding coupled with barcode sequencing (Tuba-seq) can uncover engineered alterations that reduce tumor number and growth.
a-b. Schematic of the Tuba-seq approach to measure the effects of essential gene inactivation on tumor growth. Tumors were initiated with pool of barcoded lentiviral-sgRNA/Cre vectors targeting known essential genes and tumor suppressor Apc (Lenti-sgEssential/Cre) in KT and KT;H11LSL-Cas9 mice (a). Tuba-seq was performed on each tumor-bearing lung 12 weeks after initiation (b). c. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles. Percentiles that are significantly different from sgInert (two-sided FDR-corrected p < 0.05) are in color. d. The impact of each sgRNA on mean tumor size relative to sgInerts, assuming a log-normal distribution of tumor sizes (LNmean). sgRNAs with two-sided FDR-corrected P < 0.05 are in bold. e. Points denote the impact of each sgRNA on tumor burden relative to sgInerts and normalized to the same statistic in KT mice. Relative burdens significantly different from sgInert (two-sided FDR-corrected p < 0.05) are in color. f. Points denote the impact of each sgRNA on tumor number relative to sgInerts and normalized to the same statistic in KT mice. Relative tumor numbers significantly different from sgInert (two-sided FDR-corrected p < 0.05) are in color. g. Points denote the impact of each sgRNA on tumor number plotted against its impact on LNmean tumor size. The lines at y = 1 and x = 1 indicate no effect relative to sgInert on tumor number and size, respectively. For panels c and e-g: Error bars indicate 95% confidence intervals around point estimates of the test statistic. Confidence intervals and P-values were calculated using a nested bootstrap resampling approach across 9 KT;H11LSL-Cas9 mice and 2 KT mice. sgInerts are in gray and the line at y = 1 indicates no effect.
Extended Data Fig. 3. Inactivation of KRAS-interacting proteins has similar impacts on tumor growth in p53-proficient and p53-deficient contexts.
a. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponading percentiles in KT mice. Genes are ordered as in Fig. 1f. Line at y = 1 indicates no effect relative to sgInert. As expected, no percentiles were significantly different from sgInert (two-sided FDR-adjusted p < 0.05). b. The impact of each sgRNA on mean tumor size relative to sgInerts in KT;H11LSL-Cas9 mice, assuming a log-normal distribution of tumor sizes (LNmean). sgRNAs with two-sided P < 0.05 after FDR-adjustment are in bold. c-d. Points denote the impact of each sgRNA on tumor burden (c) and number (d) relative to sgInerts in KT;H11LSL-Cas9 mice, normalized to the corresponding statistic in KT mice to account for representation of each sgRNA in the viral pool. sgInerts are in gray and the line at y = 1 indicates no effect. Relative tumor burdens and numbers significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. e. Points denote tumor sizes at the indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors in KT;p53flox/flox;H11LSL-Cas9 mice. Genes are ordered as in Fig. 1f. The line at y = 1 indicates no effect relative to sgInert. Percentiles that are significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. f-h. Comparison of the impact of each sgRNA on relative LNmean tumor size (f), tumor burden (g) and tumor number (h) in KT;H11LSL-Cas9 and KT;p53flox/flox;H11LSL-Cas9 mice. For all panels: Error bars indicate 95% confidence intervals around point estimates of the test statistics. Confidence intervals and P-values were calculated using a nested bootstrap resampling approach described across 11 KT;H11LSL-Cas9 mice, 6 KT;p53flox/flox;H11LSL-Cas9 mice and 5 KT mice.
Extended Data Fig. 4. Top candidate KRAS-interacting proteins from initial Tuba-seq screen impact multiple metrics of tumor growth in validation cohort.
a. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles in KT mice. KT mice lack Cas9, thus all sgRNAs are functionally equivalent to sgInerts. Genes are ordered as in Fig. 2d, but note the change in axis scaling. Line at y = 1 indicates no effect relative to sgInerts. As expected, no percentiles were significantly different from sgInert (FDR-adjusted p < 0.05). b. The impact of each sgRNA on mean tumor size relative to sgInerts, assuming a log-normal distribution of tumor sizes (LNmean). sgRNAs with two-sided P < 0.05 after FDR-adjustment are in bold. Note that these data for the sgInerts, sgHras#1–3 and sgNras#1–3 are also plotted in Fig. 2e. c. Points denote the impact of each sgRNA on tumor burden relative to sgInerts in KT;H11LSL-Cas9 mice, normalized to the corresponding statistic in KT mice to account for the representation of each sgRNA in the viral pool. sgInerts are in gray and the line at y = 1 indicates no effect. Relative tumor burdens significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. d. Points denote the impact of each sgRNA on tumor number relative to sgInerts in KT;H11LSL-Cas9 mice, normalized to the corresponding statistic in KT mice to account for representation of each sgRNA in the viral pool. sgInerts are in gray and the line at y = 1 indicates no effect. Relative tumor numbers significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. For all panels: Error bars indicate 95% confidence intervals around point estimates of the test statistic. Confidence intervals and P-values were calculated using a nested bootstrap resampling approach across 20 KT;H11LSL-Cas9 mice and 4 KT mice.
Extended Data Fig. 5. Wild type RAS paralogs constrain the growth of human KRAS-driven cancer cell lines.
a. RAS family member dependency scores in human lung adenocarcinoma (LUAD) cell lines. b. Effects of RAS gene knockouts in A549 cells. The T-score represents the normalized effect of multiple sgRNAs targeting a gene. A positive T-score indicates a tumor-suppressive effect. The effects of each gene relative to SAFE sgRNAs were tested via Mann–Whitney U test, corrected via Benjamini-Hochberg procedure. (Data source: Kelly, Kostyrko, Han et al. 2020). c. Effects of RAS gene knockouts in KRAS-mutant human LUAD cells in 3D culture. The effects of each gene relative to SAFE sgRNAs were tested via two-sided Benjamini-Hochberg-corrected t-test. (Data source: Han et al. 2020). d. Indel rates in cell lines with the indicated sgRNAs. * denotes sgRNAs used for cell culture and transplantation experiments. e. Re-expression of wild-type HRAS or NRAS suppresses proliferation of HRAS and NRAS double knockout (DKO) HOP62 cells. Cells were seeded in 96-well plates and cultured under limited serum (1%) with or without Doxycycline (Dox). Cell numbers were measured via CCK8 assay. Points are Mean±SD of 16 wells normalized to Day 0. (one-tailed t-test). f. Representative images of subcutaneous tumors 4 weeks after transplantation with H23 cells. Quantification is in Fig. 3h. Scale bar: 2 mm. g. Representative images of Ki67 staining from subcutaneous tumors four weeks after transplantation with H23 cells. Quantification is shown in Fig. 3i. Scale bar: 100 μm. h. Representative images of H&E and human mitochondria staining on lung tumors 4 weeks after intravenous transplantation with H23 cells. Quantification is in Fig. 3j. Scale bar: 500 μm. i. Representative images of Ki67 staining from lung tumors 4 weeks after intravenous transplantation with H23 cells. Quantification is in Fig. 3k. Scale bar: 200 μm.
Extended Data Fig. 6. Wild-type RAS paralogs finetune RAS signaling.
a. Western blot analysis of three RAS paralogs’ expression per 10,000 human and mouse KRAS-driven lung cancer cell lines. Recombinant RAS proteins were used as a standard. b. Western blot analysis of three RAS paralogs’ expression per 20,000 sorted BrafV600E-driven mouse lung cancer cells. Recombinant RAS proteins were used as a standard. c. Quantification of pERKpos cells in KT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors as indicated in Fig. 4a. Each dot represents a tumor. (one-way ANOVA). d. Quantification of pERKpos cells per field of indicated cells from Fig. 4b. Each dot represents a view field. (one-way ANOVA). SubQ, subcutaneous. e. Western blot analysis of HRAS and NRAS double knockout (DKO) HOP62 cells re-expressing HRAS (TRE-HRAS) or NRAS (TRE-NRAS) under Doxycycline (Dox) treatment. DKO cells were generated as described in Fig. 3a. DKO cells were re-transduced with lentiviral vector expressing TRE-HRAS or TRE-NRAS at high MOI ( > 5) to generate stable re-expressing cells. To re-express HRAS, cells were treated with 10 ng/ml Dox. To re-express NRAS, cells were treated with 50 ng/ml Dox. All cells were cultured under limited serum (1%) for 2 days before protein extraction. HSP90 is blotted as loading control.
Extended Data Fig. 7. HRAS and NRAS directly interact with KRASG12D.
a. Co-immunoprecipitation of HRAS (HA-tagged) and NRAS (Flag-tagged) with KRASG12D (Myc-tagged), imaged by western blotting. 293T cells were co-transfected with Myc-KRASG12D, HA-HRAS, and Flag-NRAS for 24 hours before co-immunoprecipitation. b. Co-immunoprecipitation of HRAS (HA-tagged) with truncated (aa73-aa165) or full length KRASG12D (Myc-tagged), imaged by western blotting. 293T cells were co-transfected with Myc-KRASG12D and HA-HRAS for 24 hrs before co-immunoprecipitation. c. Diagram of the modified ReBiL2.0 system to assess direct KRASG12D-HRAS/NRAS interaction. d. HRAS and NRAS can directly interact with KRASG12D. 293 T cells expressing indicated cLuc- and nLuc- luciferase were cultured in limited serum (1%) for 24 hours and ReBiL2.0 assay was performed. Points are Mean±SD ReBiL2.0 score of 12 wells normalized to cells expressing free luciferase (cLuc-HA/nLuc-HA). (one-way ANOVA). e. Luciferase protein expression in c, imaged by western blotting for the HA-tag. α-Tubulin is loading control. f. Full data from experiment shown in Fig. 5c. 293T cells expressing nLuc-KRASG12D/cluc-KRASG12D or nLuc-C20/cluc-C20 with indicated Myc-tagged RAS-GTPases were cultured in limited serum (1%) for 24 hours and ReBiL2.0 assays were performed. Points are Mean±SD ReBiL2.0 score of 12 wells normalized to cells transduced with empty vector. ns: not significant (one-way ANOVA). g. RAS-GTPases protein expression in e, imaged by western blotting for the Myc-tag. HSP90 is loading control. h. Relative strength of RAS-GTPases in disrupting KRASG12D-KRASG12D interactions in f. Differences in ReBiL2.0 score between empty vector and indicated RAS-GTPases were normalized by their own protein expression via western blotting for the Myc-tag. (one-way ANOVA). i. RAP1A interacts with KRASG12D in two protein-protein interaction analyses (Kelly, Kostyrko, Han et al. 2020; Broyde, Simpson, Murray et al. 2020). j. Co-immunoprecipitation of BRAF with KRASG12D (Myc-tagged), imaged by western blotting. 293T cells were co-transfected with Myc-KRASG12D, BRAF, and with or without HA-HRAS for 24 hours before co-immunoprecipitation. Overexpression (OvE) of HRAS suppressed BRAF co-immunoprecipitation with KRASG12D.
Extended Data Fig. 8. Identification and analysis of rare RAS mutations in oncogenic KRAS-mutant tumors.
a-b. Pan-cancer frequency of HRAS (a) or NRAS (b) mutations in patients from Project GENIE. Mutations that are intergenic, intronic, silent, or in the 3’ or 5’ UTR were excluded. Oncogenic KRAS mutants were defined as tumors having missense mutations in codons 12, 13 or 61. Known oncogenic HRAS (a) or NRAS (b) mutations are highlighted. The dashed line indicates equal mutation frequency in samples with wild-type and mutant KRAS. Non-oncogenic mutations occurring at least once in patients with oncogenic KRAS mutations are annotated. Mutants selected for analysis of ability to disrupt KRASG12D-KRASG12D interactions are in bold. c-d. Characteristics of samples with rare HRAS (c) or NRAS (d) mutations selected for analysis of ability to disrupt KRASG12D-KRASG12D interactions using the ReBiL2.0 system. e. Identification of RAS-RAS interaction-deficient NRAS mutation. 293 T (nLuc-KRASG12D/cluc-KRASG12D) cells expressing wild-type or NRAS mutants were cultured in limited serum (1%) for 24 hours. Points are Mean±SD ReBiL2.0 score of 12 wells normalized to cells transfected with empty vector. ns: not significant. (one-way ANOVA). f. NRAS (wild-type and mutant) protein expression levels in a shown by anti-Myc tag western blot. HSP90 is loading control. g. Western blot of cultured NRAS-null HOP62 cells (HOP62-Cas9-sgNRAS) re-expressing sgRNA-resistant wild-type NRAS or NRASR102Q under Dox treatment. Cells were cultured under limited serum (1%) with or without Dox for 2 days before protein extraction. Re-expression of NRASR102Q had no effect on ERK phosphorylation. GAPDH is loading control. h. Proliferation of cultured NRAS-null HOP62 cells (HOP62-Cas9-sgNRAS) expressing sgRNA-resistant wild-type NRAS or NRASR102Q under Dox treatment. Cells were cultured in limited serum (1%) with or without Dox for 4 days. Cell viability was measured via CCK8 assay and normalized to cells treated with vehicle. Re-expression of NRASR102Q had no effect on cell proliferation. Points are Mean±SD of 10 wells. ns: not significant (one-tailed t-test).
Extended Data Fig. 9. Prediction of RAS-RAS dimer interfaces.
a. Homodimers of RAS present in crystals of HRAS, KRAS, and NRAS in the Protein Data Bank. Dimers were downloaded from the Protein Common Interface Database (ProtCID)58, which clusters interfaces present in different crystals of homologous proteins. The α4-α5 dimer shown is present in 84 entries of HRAS, 13 entries of KRAS, and one entry of NRAS (PDB 5UHV). b. Models of a homodimer of KRASG12D and heterodimers of KRASG12D with HRAS, HRAST50M, and HRASR123C. The α4-α5 HRAS dimer from PDB entry 3K8Y was used as a template. KRASG12D from PDB entry 5USJ was superposed with the program PyMol on one or both monomers of 3K8Y to form the heterodimers and the homodimer respectively. Residues T50 and R123 were mutated with PyMol. R123 is involved in an intrachain salt bridge with residue E143, which also participates in the RAS-RAS interface. Mutation to cysteine results in an uncompensated charge on E143, which may destabilize the RAS-RAS interaction. All four structures were relaxed with the program Rosetta using the FastRelax protocol with the Ref2015 scoring function59. Rosetta uses the backbone-dependent rotamer library of Shapovalov and Dunbrack to repack side chains around the mutated sites60. The resulting energies were: KRASG12D-KRASG12D, -1122.8 kcal/mol; HRAS-KRASG12D, -1144.8 kcal/mol; HRAST50M-KRASG12D, -1135.5 kcal/mol; HRASR123C-KRASG12D, -1130.9 kcal/mol. Residues T50 (magenta) and R123 (orange) are indicated in sticks.
Extended Data Fig. 10. Paired screen in KRAS-driven and BRAF-driven lung cancer models validates HRAS and NRAS as KRAS-specific tumor suppressors.
a-c. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponding percentiles in KT;H11LSL-Cas9/+ (a), BrafT;H11LSL-Cas9/+ (b) and KT mice (c). Genes are ordered by 95th percentile tumor size in KT;H11LSL-Cas9/+ mice, with sgInerts on the left. Percentiles that are significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. The negative effects of sgRNAs targeting Fnta and Nme2 in the KT mice (c) are unexpected and indicate a potential bias in the size distributions of tumors with these genotypes. We note that the same bias may be present in the KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ data; however, previous experiments showed consistent negative effects on tumor size for these sgRNAs, suggesting that the observed effects in this KT;H11LSL-Cas9/+ cohort are not solely the product of this bias. d. Points denote the impact of each sgRNA on tumor burden relative to sgInerts in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice, normalized to the corresponding statistic in KT mice to account for representation of each sgRNA in the viral pool. Relative tumor burdens significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. e. Points denote the impact of each sgRNA on tumor number relative to sgInerts in KT;H11LSL-Cas9/+ and BrafT;H11LSL-Cas9/+ mice, normalized to the corresponding statistic in KT mice to account for representation of each sgRNA in the viral pool. Relative tumor numbers significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. For all panels: Error bars indicate 95% confidence intervals around point estimates of the test statistic. sgInerts are in gray and the line at y = 1 indicates no effect relative to sgInerts. Confidence intervals and P-values were calculated using the nested bootstrap resampling approach described in the Methods across 11 KT;H11LSL-Cas9/+ mice, 14 BrafT;H11LSL-Cas9/+ mice and 10 KT mice.
Supplementary Material
ACKNOWLEDGEMNTS:
We thank the Stanford Veterinary Animal Care Staff for expert animal care, Human Pathology/Histology Service Center, Stanford Protein and Nucleic Acid Facility for experimental support; A. Orantes for administrative support; Members of the Winslow laboratory and Ian Prior for helpful comments. R.T. was supported by a Tobacco-Related Disease Research Program (TRDRP) Postdoctoral fellowship (27FT-0044), a Stanford Cancer Institute fellowship, a Stanford-Rambam Health Care collaboration grant (259348), and an NCI K99/R00 pathway way to independence grant (K99CA256039). C.W.M. was supported by the NSF Graduate Research Fellowship Program and an Anne T. and Robert M. Bass Stanford Graduate Fellowship. J.D.H was supported by a Stanford University School of Medicine Dean's Postdoctoral Fellowship and a TRDRP Postdoctoral fellowship (T31FT1619). H.C. was supported by a TRDRP Postdoctoral Fellowship (28FT-0019). N.W.H. was supported by the NSF Graduate Research Fellowship Program. M.I.P was supported by an NIH F30 fellowship (GM142263). Work in the laboratory of R.L.D was supported by the Fox Chase Cancer Center Support Grant (NIH P30 CA006927). Work in the laboratory of G.M.W. was supported, in part, by Cancer Center Core Grant CA014195, the Breast Cancer Research Foundation, the Freeberg Foundation and the NIH/National Cancer Institute (Grant R35 CA197687). This work was supported by NIH R01-CA230025 (to M.M.W), NIH R01-CA231253 (to M.M.W and D.A.P), NIH R01-CA234349 (to M.M.W and D.A.P.), TRDRP 27IP-0052 (to M.M.W), R01-CA250534 (to P.K.J), R35-GM122517 (to R.L.D.) and in part by the Stanford Cancer institute support grant (NIH P30-CA124435). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
COMPETING INTERESTS STATEMENT
M.M.W. and D.A.P are co-founders of, and hold equity in, D2G Oncology, Inc.
The remaining authors declare no competing interests.
CODE AVAILABILITY
The code used for data analysis in this study is available on GitHub (https://github.com/eshuldiner/KrasIP)
DATA AVAILABILITY
The human cancer genomic data analyzed for the presence of rare HRAS and NRAS variants in this manuscript were derived from the AACR’s Project GENIE (https://www.aacr.org/professionals/research/aacr-project-genie/) Release 9.1-public dataset. All data files that support the findings of this study are available through the Synapse platform (https://www.synapse.org/#!Synapse:syn24179657). Human cancer genomic data analyzed for alterations in KRAS-interacting proteins were derived from the TCGA PanCancer Atlas dataset, which is publicly available through cBioPortal at https://www.cbioportal.org/study/summary?id=luad_tcga_pan_can_atlas_2018 The protein templates used to model RAS heterodimers in this study are available through the ProtCID database (KRAS-G12D: PDB entry 5USJ, HRAS homodimer: PDB entry 3K8Y), and resulting structural data files can be accessed through Zenodo (https://zenodo.org/record/7104280 ). AP/MS data were derived from Kelly, Kostryko, Han et al. 2020 (PMID: 32727735, Supplemental Table 2) and Broyde, Simpson, Murray et al. 2020 (PMID: 32929263 Supplementary File 3). Gene expression data were derived from Chuang et al. 2017 (PMID: 28191885; Supplemental Table 1). Pre-existing CRISPR/Cas9 screen data were derived from Han et al. 2020 (PMID: 32238925, Supplementary Table 4). All barcode sequencing datasets are available through the NCBI’s Sequence Read Archive database under the BioProject accession number PRJNA763983 (https://www.ncbi.nlm.nih.gov/sra/PRJNA763983 ). Processed data plotted in figures has been available in the Supplementary Information. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
REFERENCES :
- 1.Karnoub AE & Weinberg RA Ras oncogenes: split personalities. Nature reviews Molecular cell biology 9, 517–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox AD, Fesik SW, Kimmelman AC, Luo J. & Der CJ Drugging the undruggable RAS: mission possible? Nature reviews Drug discovery 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou B, Der CJ & Cox AD in Seminars in cell & developmental biology, Vol. 58 60–69 (Elsevier, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wennerberg K, Rossman KL & Der CJ The Ras superfamily at a glance. Journal of cell science 118, 843–846 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hobbs GA, Der CJ & Rossman KL RAS isoforms and mutations in cancer at a glance. Journal of cell science 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephen AG, Esposito D, Bagni RK & McCormick F. Dragging ras back in the ring. Cancer cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Brose MS et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer research 62, 6997–7000 (2002). [PubMed] [Google Scholar]
- 8.Prior IA, Lewis PD & Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer research 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papke B. & Der CJ Drugging RAS: Know the enemy. Science 355, 1158–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Kelly MR et al. Combined proteomic and genetic interaction mapping reveals new RAS effector pathways and susceptibilities. Cancer discovery 10, 1950–1967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broyde J. et al. Oncoprotein-specific molecular interaction maps (SigMaps) for cancer network analyses. Nature biotechnology 39, 215–224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y. & Hancock JF Deciphering lipid codes: K‐Ras as a paradigm. Traffic 19, 157–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittinghofer A. & Pal EF The structure of Ras protein: a model for a universal molecular switch. Trends in biochemical sciences 16, 382–387 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Omerovic J, Hammond DE, Clague MJ & Prior IA Ras isoform abundance and signalling in human cancer cell lines. Oncogene 27, 2754–2762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han K. et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 580, 136–141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y-C et al. Analysis of RAS protein interactions in living cells reveals a mechanism for pan-RAS depletion by membrane-targeted RAS binders. Proceedings of the National Academy of Sciences 117, 12121–12130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hingorani SR et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Cai H. et al. A functional taxonomy of tumor suppression in oncogenic KRAS-driven lung cancer. Cancer Discovery (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers ZN et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nature genetics 50, 483–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers ZN et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nature methods 14, 737–742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang C-H et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109–Janus kinase–Stat axis. Nature medicine 23, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz S, Santos E. & Bustelo XR RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Molecular and cellular biology 27, 8127–8142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandt AC, Koehn OJ & Williams CL SmgGDS: An Emerging Master Regulator of Prenylation and Trafficking by Small GTPases in the Ras and Rho Families. Frontiers in Molecular Biosciences 8, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowell CA, Kowalczyk JJ, Lewis MD & Garcia AM Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. Journal of Biological Chemistry 272, 14093–14097 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Zhang FL et al. Characterization of Ha-ras, N-ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. Journal of Biological Chemistry 272, 10232–10239 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Takaya A. et al. R-Ras regulates exocytosis by Rgl2/Rlf-mediated activation of RalA on endosomes. Molecular biology of the cell 18, 1850–1860 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marais R, Light Y, Paterson H. & Marshall C. Ras recruits Raf‐1 to the plasma membrane for activation by tyrosine phosphorylation. The EMBO journal 14, 3136–3145 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell JD et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nature genetics 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Rivera FJ et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohl NE et al. Selective inhibition of ras-dependent transformation by a farnesyltransferase inhibitor. Science 260, 1934–1937 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Rowinsky EK, Windle JJ & Von Hoff DD Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. Journal of Clinical Oncology 17, 3631–3652 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Collisson E. et al. Comprehensive molecular profiling of lung adenocarcinoma: The cancer genome atlas research network. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldser DM et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray CW et al. An LKB1–SIK axis suppresses lung tumor growth and controls differentiation. Cancer discovery 9, 1590–1605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsherniak A. et al. Defining a cancer dependency map. Cell 170, 564–576. e516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staffas A, Karlsson C, Persson M, Palmqvist L. & Bergo M. Wild-type KRAS inhibits oncogenic KRAS-induced T-ALL in mice. Leukemia 29, 1032–1040 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Ambrogio C. et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868. e815 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Kong G. et al. Loss of wild-type Kras promotes activation of all Ras isoforms in oncogenic Kras-induced leukemogenesis. Leukemia 30, 1542–1551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgess MR et al. KRAS allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell 168, 817–829. e815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young A, Lou D. & McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer discovery 3, 112–123 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Grabocka E. et al. Wild-type H-and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer cell 25, 243–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y. et al. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Molecular and cellular biology 34, 862–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y. & Hancock JF Ras nanoclusters: Versatile lipid-based signaling platforms. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Henis YI, Hancock JF & Prior IA Ras acylation, compartmentalization and signaling nanoclusters. Molecular membrane biology 26, 80–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inouye K, Mizutani S, Koide H. & Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. Journal of Biological Chemistry 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Muratcioglu S. et al. GTP-dependent K-Ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin W-C et al. H-Ras forms dimers on membrane surfaces via a protein–protein interface. Proceedings of the National Academy of Sciences 111, 2996–3001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.AACR Project Genie Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer discovery 7(8), 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nan X. et al. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proceedings of the National Academy of Sciences 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terrell EM et al. Distinct binding preferences between ras and raf family members and the impact on oncogenic ras signaling. Molecular cell 76, 872–884. e875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dankort D. et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes & development 21, 379–384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dietrich P. et al. Neuroblastoma RAS viral oncogene homolog (NRAS) is a novel prognostic marker and contributes to sorafenib resistance in hepatocellular carcinoma. Neoplasia 21, 257–268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weyandt JD et al. Wild-type Hras suppresses the earliest stages of tumorigenesis in a genetically engineered mouse model of pancreatic cancer. PloS one 10, e0140253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.To MD, Rosario R, Westcott PM, Banta KL & Balmain A. Interactions between wild-type and mutant Ras genes in lung and skin carcinogenesis. Oncogene 32, 4028–4033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weyandt JD, Carney JM, Pavlisko EN, Xu M. & Counter CM Isoform-Specific effects of wild-type ras genes on carcinogen-Induced lung tumorigenesis in mice. Plos one 11, e0167205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeng H-H, Taylor LJ & Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nature communications 3, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller MS & Miller LD RAS mutations and oncogenesis: not all RAS mutations are created equally. Frontiers in genetics 2, 100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Q. & Dunbrack RL ProtCID: a data resource for structural information on protein interactions. Nature communications 11, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alford RF et al. The Rosetta all-atom energy function for macromolecular modeling and design. Journal of chemical theory and computation 13, 3031–3048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shapovalov MV & Dunbrack RL Jr A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19, 844–858 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human cancer genomic data analyzed for the presence of rare HRAS and NRAS variants in this manuscript were derived from the AACR’s Project GENIE (https://www.aacr.org/professionals/research/aacr-project-genie/) Release 9.1-public dataset. All data files that support the findings of this study are available through the Synapse platform (https://www.synapse.org/#!Synapse:syn24179657). Human cancer genomic data analyzed for alterations in KRAS-interacting proteins were derived from the TCGA PanCancer Atlas dataset, which is publicly available through cBioPortal at https://www.cbioportal.org/study/summary?id=luad_tcga_pan_can_atlas_2018 The protein templates used to model RAS heterodimers in this study are available through the ProtCID database (KRAS-G12D: PDB entry 5USJ, HRAS homodimer: PDB entry 3K8Y), and resulting structural data files can be accessed through Zenodo (https://zenodo.org/record/7104280 ). AP/MS data were derived from Kelly, Kostryko, Han et al. 2020 (PMID: 32727735, Supplemental Table 2) and Broyde, Simpson, Murray et al. 2020 (PMID: 32929263 Supplementary File 3). Gene expression data were derived from Chuang et al. 2017 (PMID: 28191885; Supplemental Table 1). Pre-existing CRISPR/Cas9 screen data were derived from Han et al. 2020 (PMID: 32238925, Supplementary Table 4). All barcode sequencing datasets are available through the NCBI’s Sequence Read Archive database under the BioProject accession number PRJNA763983 (https://www.ncbi.nlm.nih.gov/sra/PRJNA763983 ). Processed data plotted in figures has been available in the Supplementary Information. All other data supporting the findings of this study are available from the corresponding author on reasonable request.