Abstract
Introduction
In a consanguineous family, seven siblings born in three sibships showed a syndromic disorder characterized by obesity, seizures, and language impairment phenotypes, which appeared at early age or developed during early childhood.
Methods
By whole-exome sequencing and subsequent Sanger sequencing, a novel homozygous missense variant (c.3371 T>A [p.Ile1124Asn]) in exon 20 of the CNTNAP2 gene was identified.
Results
The pathogenic variant in this family is located within one of the laminin G-like 4 domains of CASPR2 and may cause loss of hydrophobic interactions of CASPR2 with its partner proteins. Single nucleotide and copy number variants in this gene have previously been related to Gilles de la Tourette syndrome, cortical dysplasia-focal epilepsy syndrome, schizophrenia, Pitt-Hopkins syndrome, and autism spectrum, attention deficit hyperactivity, and obsessive compulsive disorders. Yet, few studies described patients with CNTNAP2 variants showing diet-induced obesity.
Conclusion
This report expands the phenotypic spectrum of this rare syndrome and provides deeper insights by documenting the clinical features and genetic findings of the patients.
Keywords: CNTNAP2, Neurological disorders, Obesity, Syndrome, Autosomal recessive form
Introduction
Contactin-associated protein-like 2 or CNTNAP2 gene (OMIM # 604569) is located on chromosome 7q35, comprises 24 exons, and spans 2.3 Mb of genomic DNA [Nakabayashi and Scherer, 2021] with complete length of 3,996 nucleotides ORF. This gene encodes a full-length transcript protein (CASPR2) consisting of 1,331 amino acids [Rubio-Marrero et al., 2016]. This transmembrane (single-pass) glycoprotein contactin-associated protein-like 2 (CASPR2), is a member of the protein superfamily neurexin (a cell-adhesion protein), helps in interaction between cells [Velez Edwards et al., 2013; Rodenas-Cuadrado et al., 2014; Canali and Goutebroze, 2018; Freri et al., 2021]. CASPR2 protein comprises a large extracellular (i.e., eight), 1 transmembrane, and a small C-terminal intracellular component. Protein domains consist of four G domains (laminin) and two EGF-like domains (epidermal growth factor like), involved in cell adhesion, interactions, binding (receptor ligand), arborization of dendrites, differentiation, and migration [Rubio-Marrero et al., 2016; Traka et al., 2003; Canali and Goutebroze, 2018; Falsaperla et al., 2020].
CNTNAP2 is expressed in different parts of spinal cord and brain, all over the development up to adulthood, and plays an essential part in brain function and development [Alarcon et al., 2008; Rodenas-Cuadrado et al., 2016]. Both in myelinated axons of the CNS and the spinal cord, CASPR2 protein's primary function is the clustering of voltage-gated K+ channels (Shaker type) and transmission of axon potentials in the regions of juxtaparanodes [Varea et al., 2015; Flaherty et al., 2017; Freri et al., 2021].
CNTNAP2 is target for a wide range of structural rearrangements or genetic variants, involving CNVs (copy number variants), binding of altered transcription factor, inversions and translocations, epigenetic alterations, and exonic variants [Rodenas-Cuadrado et al., 2014; Poot, 2015; Falsaperla et al., 2020]. Mostly, CNTNAP2 gene variants are associated with neurodevelopmental disorders and other rare phenotypes such as language impairment, epilepsy, intellectual disabilities, and autism [Strauss et al., 2006; Velez Edwards et al., 2013; Saint-Martin et al., 2018; Canali and Goutebroze, 2018]. In some reports, it also exhibits additional phenotypes such as Pitt-Hopkins syndrome [Zweier et al., 2009], Tourette syndrome, language dysfunction, schizophrenia, and epilepsy syndromes [State, 2010; Poot, 2015; Rodenas-Cuadrado et al., 2016]. Moreover, a single copy of CNTNAP2 has also been reported to have coding variations in unaffected individuals, indicating that not all heterozygous variants are always fully penetrant [Rodenas-Cuadrado et al., 2016].
During the initial report, CNTNAP2-linked disorder was reported by Verkerk et al. [2003], in a family with Gilles de la Tourette syndrome showing an affected daughter and son of affected father by heterozygous chromosomal insertion (translocation) at the region 7q35-q36, disturbing the intron 8 of CNTNAP2 gene. Gilles de la Tourette syndrome is an inherited neuropsychiatric complex disorder showing intellectual disability and Tourette syndrome mostly accompanied by obsessive disorders.
Here, we studied a large consanguineous family from Balochistan, Pakistan, segregating a syndromic disorder characterized by obesity, seizures, and language impairment. Whole-exome sequencing (WES) and subsequent targeted sequencing identified a novel homozygous missense variant (c.3371T>A [p.Ile1124Asn]) in exon 20 of CNTNAP2. The unaffected parents and siblings were heterozygous while affected individuals were homozygous for this newly identified variant. We studied the structural effect through online bioinformatics software HOPE (https://www3.cmbi.umcn.nl/hope/) showing this change alters the CASPR2 protein function. Current pathogenic variant is located within a domain (laminin G-like 4) of CASPR2 protein; due to the large size and less hydrophobic properties of the mutant, residue can cause loss of hydrophobic interactions of laminin G-like 4 domain and might abolish the normal function of CASPR2. This variant is not reported in any of the publicly available databases, thus enhances the mutational spectrum of CNTNAP2 gene.
Materials and Methods
A large consanguineous family was recruited from Balochistan, Pakistan, and the study was approved by Institutional Review Board at Department of Biotechnology, BUITEMS, Quetta, Pakistan. All the participants (or their guardians) gave written informed consents for current study.
Family History and Clinical Phenotype
A consanguineous family was recruited from Balochistan, Pakistan, with syndromic disorder characterized by obesity, seizures, and language impairment phenotypes, inherited in an autosomal recessive form. Seven affected children were born in three sibships (loops); parents of affected children have cousin marriages and are healthy with no disease symptoms. Family history and clinical evaluation were attained by detailed interview from all members of the family. The affected individuals showed similar clinical phenotypes appearing at an early age or developed during early childhood.
All affected individuals were born at term with normal pregnancy and delivery (not observed any perinatal problems), with normal weight, height, and head circumference. Apgar scores were ≥7 in all patients. At birth, no clinical or facial deformities were observed. After few months, parents noticed developmental delay in their affected children as compared to their age fellows: delay in starting to crawl (at 12–16 months), walking (at 18–24 months). Parents also noticed delay in starting to talk (at the age of 2.5–4 years), talking slowly, trouble in interpreting, and in using language to express themselves, difficulties with learning and reading, hardly pronouncing the words; also academic progress was not good. Additional clinical phenotypes like obesity at the age of 2.9–4.5 years, and seizures at the age of 1.2–3.5 years, mild facial deformities were also observed. Electroencephalogram analysis was performed, showing ictal and interictal abnormalities. For diagnosis of language impairment, parents were interviewed using “Alberta Language and Development Questionnaire” (ALDeQ in Urdu translation); the questionnaire is divided into four parts, i.e., “early milestones, current first-language abilities, behaviour patterns and activity preferences, and family history” [Paradis et al., 2010]. Fourth edition “Peabody Picture Vocabulary Test” (PPVT-4), in Urdu translation, was performed to evaluate receptive vocabulary. “Clinical Evaluation of Language Fundamentals” (CELF-5), in Urdu translation, was performed to evaluate the receptive and expressive language. Obesity was confirmed by measuring BMI (≥30 kg/m2); lifestyle, diet, and family history were also recorded (Table 1).
Table 1.
Clinical description of the affected individuals of the family
| Clinical phenotype | Clinical observation | Patient |
||||||
|---|---|---|---|---|---|---|---|---|
| Vl:1 | VI:2 | VI:5 | VI:6 | VI:7 | VI:10 | VI:11 | ||
| Age | Present age | 20 years | 24 years | 22 years | 18 years | 14 years | 16 years | 12 years |
|
| ||||||||
| Epilepsy | Age of onset | 2.5 years | 3 years | 1.2 years | 2.5 years | 2 years | 1.8 years | 3.5 years |
|
|
||||||||
| Types of seizures | Focal onset | Focal onset | Generalized tonic clonic | Focal onset | Focal onset | Focal onset aware | Generalized tonic clonic | |
|
|
||||||||
| Treatment | Carbamazepine and lamotrigine | Carbamazepine and lamotrigine | Valproic acid and clonazepam | Carbamazepine and lamotrigine | Valproic acid, levetiracetam, clonazepam | Levetiracetam and carbamazepine | Carbamazepine and valproic acid | |
|
|
||||||||
| Outcome | No | No | No | No | Under minor control | No | No | |
|
| ||||||||
| Language impairment | Difficulty in receptive and expressive language | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
|
|
||||||||
| Type of language impairment | SLI | SLI | SLI | SLI | SLI | SLI | SLI | |
|
| ||||||||
| Obesity | Age of onset | 3.5 years | 4 years | 2.9 years | 3 years | 3.5 years | 4 years | 4.5 years |
|
|
||||||||
| BMI (kg/m2/percentile) | 37 kg/m2 | 34.4 kg/m2 | 42 kg/m2 | 98th centile | 96th centile | 99th centile | 95th centile | |
|
| ||||||||
| Observations at Birth | Delivery | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
|
|
||||||||
| Apgar scores | >7 | >7 | >7 | >7 | >7 | >7 | >7 | |
|
| ||||||||
| Developmental milestones | Crawling | 12–15 months | 14–16 months | 12–15 months | 13–16 months | 12–15 months | 14–16 months | 14–16 months |
|
|
||||||||
| Walking | 18–22 months | 20–24 months | 18–24 months | 18–24 months | 20–24 months | 20–24 months | 20–24 months | |
|
|
||||||||
| Talking | 3—4 years | 3–4 years | 3–4 years | 3–4 years | 2.5–3 years | 3–4 years | 3–4 years | |
Various AED treatments were given to patients to control the epilepsy; one of the patients (VI:7) showed to some degree effective response leading to minor control of seizures, while rest of the patients showed severe seizures. The anticonvulsant drug treatment (carbamazepine and lamotrigine) was provided to patients VI:1, VI:2, and VI:6, but the patients showed no response to treatment. Patient VI:5 was provided valproic acid treatment, he showed good response, but after few months, multiple occurrences of seizures per day were reported, subsequently clonazepam was added but no response to anticonvulsant drug treatment. Patient VI:7 was provided clonazepam, but no response was shown as well as frequency worsened day by day; subsequently, valproic acid and levetiracetam were added to anticonvulsant drug treatment that led to under minor control of seizures (frequency of seizures decreased). Patient VI:10 was provided levetiracetam and carbamazepine anticonvulsant drug treatment but showed no response to treatment. Initially, patient VI:11 was treated with carbamazepine. Later, valproic acid was added to the treatment but still unresponsive to treatment.
Peripheral blood samples of patients, their parents, and normal siblings were collected in EDTA containing vacutainer tubes. Genomic DNA was extracted by standard established protocol.
Whole-Exome Sequencing
WES was performed, using DNA of an affected and a healthy member of the family. Agilent SureSelect Human all-exon kit was used; preparation of libraries and sequencing were performed on the Illumina HiSeq 2000 system. The obtained reads were aligned by using Burrows-Wheeler Alignment tool, to the human genome hg19. Variant calling was performed by analysis tools SAMtools and PINDEL. Variant annotation was done on ANNOVAR. By considering consanguineous family and autosomal recessive inheritance, variants were filtered by applying appropriate filters, i.e., excluded MAF >0.01, synonymous, heterozygous variants. However, more precisely selected homozygous variants were considered for Sanger validation.
Sanger Sequencing
We confirmed the identified variant by Sanger sequencing. Primers for exon 20 of CNTNAP2 (OMIM # 604569) gene, including a minimum of 40–50 bp flanking exon-intron boundaries, were designed by using online software Primer3 (https://primer3.ut.ee/). Exon amplifications were performed by using standard PCR reactions. PCR products were purified by using a commercially available kit and bidirectional sequencing was performed by sequencing kit v3.1 “BigDye Terminator Ready Reaction Cycle” from Applied Biosystems. Results were analysed on an ABI3100 Avant (Applied Biosystems).
Bioinformatic Analysis
The resulting variant was confirmed by using online databases (online suppl. Fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000529427). This variant was not reported previously in any of the online databases, i.e., Human Gene Mutation Database, gnomAD, ClinVar, and Exome Variant Server.
Results
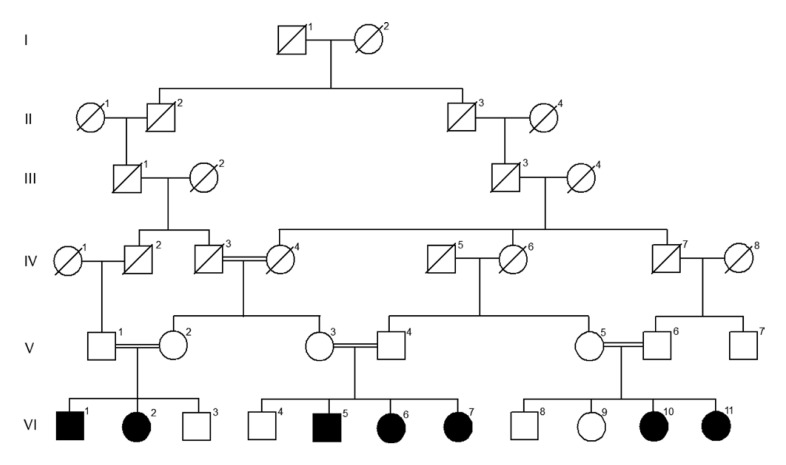
A large consanguineous family ascertained from Balochistan, Pakistan, segregating a syndromic disorder characterized by obesity, seizures, and language impairment phenotype, inherited in an autosomal recessive manner was investigated in the present study (Fig. 1). Parents of affected children have cousin marriages; seven children are affected in three sibships born to normal and healthy parents.
Fig. 1.
Pedigree of family with syndromic disorder (phenotypes, i.e., obesity, seizures, and language impairment), showing multiple affected individuals. Black symbols depict affected individuals while clear symbols represent unaffected members.
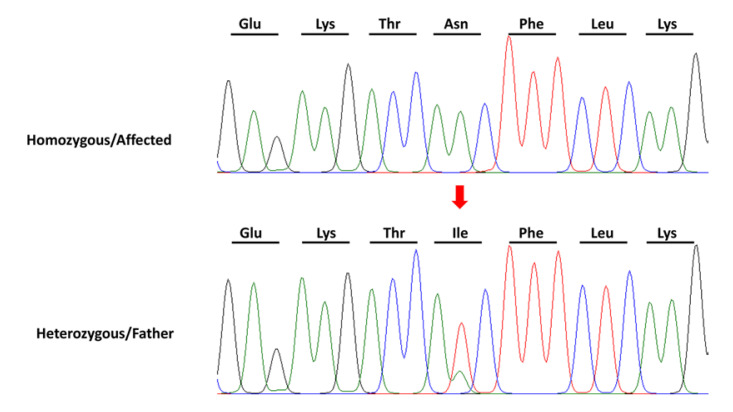
Clinically, all affected individuals have phenotypic symptoms, i.e., obesity, seizures, language impairment, and development delay. We performed WES to identify the causative gene. A novel variant was detected in exon 20 of CNTNAP2 gene (OMIM # 604569) at chromosome 7q35. We confirmed the identified variant (c.3371T>A [p.Ile1124Asn]) by targeted Sanger sequencing that showed “T” changes into “A” depicting unaffected parents and siblings were heterozygous while affected were homozygous (Fig. 2).
Fig. 2.
Sequencing chromatogram of exon 20 of gene CNTNAP2 showing homozygous missense substitution (c.3371T>A [p.Ile1124Asn]). Arrow indicates the variant.
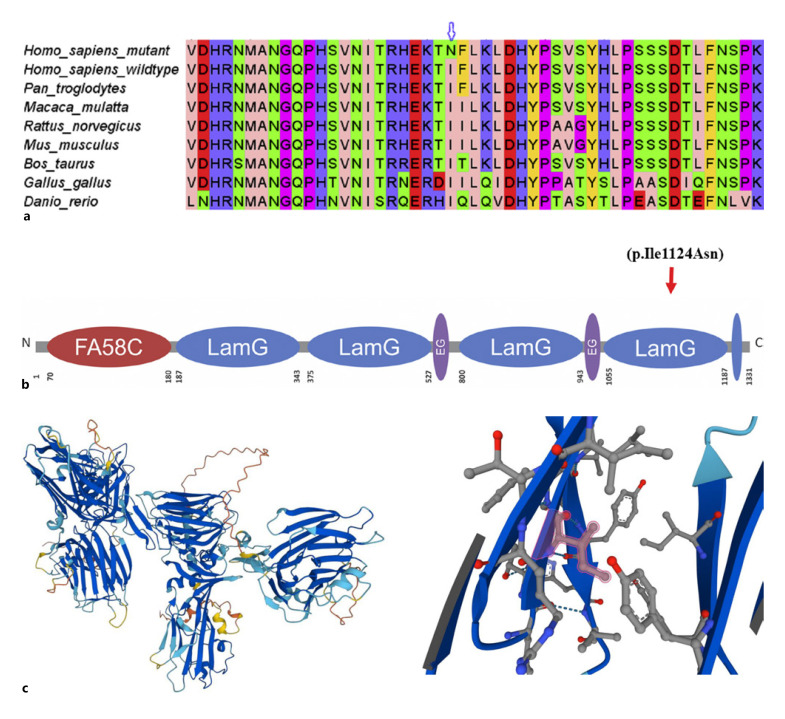
Confirmation of the identified variant was performed by online bioinformatics tools including MutationTaster, PolyPhen-2, and SIFT which predicted that the current substitution produces a pathogenic altered protein. The variant was not found in Human Gene Mutation Database, gnomAD, ClinVar, and Exome Variant Server. Alignment of amino acid sequences of CASPR2 from different species shows that current amino acid is present in a conserved region of the protein (Fig. 3a-c).
Fig. 3.
a Multiple alignment of a segment of the human CNTNAP2 protein with the corresponding segments in some of its homologues. The top sequence shows the mutated human sequence and the arrow indicates the I1124N mutated position. b Schematic protein structure of CASPR2/CNTNAP2 showing novel p.Ile1124Asn variant. c (Left) CASPR2 structure of human gene CNTNAP2; (right) purple highlighted is the residue that indicates the mutated position (I1124N). Current variant is located within a domain, annotated in laminin G-like 4. Due to the large size and less hydrophobic properties of the mutant, residue can cause the loss of hydrophobic interactions of this domain and would abolish the normal function of CASPR2.
Discussion
In the present study, we reported a novel variant c.3371T>A in exon 20 of CNTNAP2 that produces an altered protein (p.Ile1124Asn). The current variant segregates in family in an autosomal recessive mode of inheritance causing a syndromic disorder with clinical phenotypes, i.e., obesity, seizures, and language impairment. Sequence analysis revealed the affected individuals were homozygous while their unaffected parents and siblings were heterozygous. Current homozygous missense variant indicates loss of function of CASPR2 protein.
Mostly, loss of function due to CNTNAP2 gene variants is related with neurological diseases like intellectual disability, autism, and epilepsy disorders [Strauss et al., 2006; Alarcón et al., 2008; Freri et al., 2021], although previously a few studies described association of CNTNAP2 gene with metabolic diseases. CASPR2 protein's primary function is the clustering of voltage-gated K+ channels (Shaker type) and transmission of axon potentials in the regions of juxtaparanodes [Varea et al., 2015; Flaherty et al., 2017; Freri et al., 2021]. In humans, juxtaparanodal CASPR2 protein also controls diet-induced obesity [Buchner et al., 2012].
Buchner et al. [2012] reported two variants (non-synonymous) in CNTNAP2 gene associated with obesity by mapping of chromosome (QTLs in CSS) at higher resolution level, between 2 mouse strains (A/J and B6) and identified it to regulate diet-induced obesity, although no prior associations with metabolic diseases had been reported. Variant rs48516420 is conserved and falls within the protein-binding domain but not highly conserved as both valine and isoleucine were identified evolutionarily in different species at this position while rs13478735 is highly conserved residue as there is consistent presence of histidine in distantly related species. But they have not observed any obvious neurological disorder in any mice used in their study concluding that absence of neurological symptoms in mutant mice indicates that phenotype obesity resistance reported in their studies was not secondary to neurological defects. According to their findings, a mild variant in CNTNAP2 effects on body weight was highly dependent on genetics (CNTNAP2 affects both obesity resistant and obesity promoting in different genetic backgrounds). Therefore, CNTNAP2 is a single identified gene that codes protein in the Obrq3b (QTL) interval that controls the body weight because of CNTNAP2 gene variants. The Obrq3b-derived allele (A/J) shows 15% reduction in body weight of mice when high fat diet was given on background C57BL/6J, while it induces a 15% increase in body weight when second QTL (Obrq2)-derived allele (A/J) was also present within same genetic background [Buchner and Nadeau, 2015].
Velez Edwards et al. [2013] identified SNP rs4549702 in CNTNAP2 gene by GWAS (human obesity-related traits) and showed a correlation of body weight increase in population of African Americans by taking energy intake diet and decrease in Hispanics by interacting with physical activity. According to their results, CNTNAP2 gene is only strongest gene among African Americans that shows environmental interaction with body weight.
Canali and Goutebroze [2018] described in their study both effects; obesity boosting or obesity resistance increases the interesting possibility that CNTNAP2 mutants are vulnerable factors for obesity that intriguingly shows a significant correlation with autism spectrum disorder. Previous studies showed patients with autism have high risk of obesity at early ages [Curtin et al., 2010; Tyler et al., 2011]. Various variants were identified that confer the risk of several developmental impairments and obesity [Bochukova et al., 2010; Shinawi et al., 2011; Dykens et al., 2011]. A clear molecular association between obesity, neurological deregulation, and developmental disability is coherent with a function for CNTNAP2 gene in the vulnerability of obesity, though this relationship appears to be changed by environmental factors correlated to energy balance [Velez Edwards et al., 2013].
Next-generation sequencing performed by Freri et al. [2021] reported two novel CNTNAP2 genetic variants, i.e., insertion in exon 22 c.3697dupC (p.Leu1233ProfsTer14) and complete deletion of exon 2, causing Pitt-Hopkins-like syndrome with severe epilepsy. Patient was the daughter of healthy non-consanguineous parents showing clinical features like epilepsy, facial dysmorphisms, and psychomotor delay at the age of 3 years. In subsequent years, cognitive disability, behaviour disorder, severe myopia, uncertain gait, and obesity were also observed.
Vuillaume et al. [2014] performed array-CGH on cohort of 100 children from different families to identify new candidate loci or genes responsible for syndromic obesity with unknown aetiology; 60 CNVs were reported in 42% of children. They found CNV in CNTNAP2 gene in patient P2016 (in group 2, potentially clinically related), with incomplete inheritance.
Rodenas-Cuadrado et al. [2016] reported a deletion of exon 2 to 3 spanning 203 kb, i.e., hg38; Chr7:146,711,006–146,914,175 in CNTNAP2 gene, resulting in a premature stop codon (at 39 amino acid [L39X]), which produces frameshift in transcript and loss of protein function, causing a syndrome with phenotype of autism, epilepsy, and language impairment. Both affected females were homozygous for identified variant and unaffected parents (cousin marriage) were heterozygous. Both were born with normal height and weight, but at the age of 2 years, patient 2 was diagnosed with severe obesity resulting in limited movements and awkward gait.
The majority of CNTNAP2 disruptions are heterozygous, indicating that loss of single allele might be enough to cause disease condition [Rodenas-Cuadrado et al., 2014; Poot, 2015]. In patients with ASD, de novo heterozygous intragenic CNTNAP2 deletions were reported: deletion of intron 1 [Alarcón et al., 2008], Pitt-Hopkins syndrome [Zweier et al., 2009], stuttering (deletion of 1–3 exons) [Petrin et al., 2010], ADHD (deletion of intron 1) [Elia et al., 2010], and ID (deletion of exon 2–3) by Mikhail et al. [2011]. A patient with infantile epilepsy was described by Mefford et al. [2010]; both alleles of the exons 2–4 (153 amino acid) of DNA segment were deleted (in-frame) covering intron 2 and 3 of gene CNTNAP2. A heterozygous deletion of a comparable size was also observed in another affected individual with epilepsy and ID [Smogavec et al., 2016]. Inherited heterozygous deletion of CNTNAP2 has been revealed in two affected individuals with schizophrenia from two different families with numerous psychiatric disorders [Friedman et al., 2008]. There were 2 patients with ASD, minor proximal deletions in CNTNAP2 detected in ASD patients, inherited from their normal mothers. They also reported patient with ASD with 677 kb of DNA loss spanning 21–24 exons of CNTNAP2 and other genes (EZH2 and CUL1) [Gregor et al., 2011].
Single-nucleotide polymorphisms (SNPs) in the region of CNTNAP2 were convincingly associated to the most prevalent type of language impairment in children (specific language impairment). SLI is characterized as abnormality in development of language, when other medical conditions like autism or hearing loss are not present [Bishop et al., 2003]. An important connection was identified for non-word repetition scores and quantitative measures of receptive and expressive language abilities with a group of SNPs within 13-14 introns [Newbury et al., 2011; Vernes et al., 2008]. A cohort of dyslexia probands showed a similar association between non-word repetition with intron 13 variant (rs2710102) [Peter et al., 2011]. Different studies showed significant connection between ASD language endophenotypes and SNPs, including (rs2710102) age at talking first word [Alarcón et al., 2008] and (rs1718101) age at first sentence [Anney et al., 2012]. Intriguingly, primary communication skills in a large sample of phenotypically healthy individuals have been linked with a group of SNPs in 13-14 introns, i.e., rs2710102, rs2538976, rs17236239, rs759178, indicating that the genetic variants at this locus might contribute to individual disparities in the general population. Various evidence related with the SNP rs2710102 strongly supports the role of this part to language endophenotypes [Rodenas-Cuadrado et al., 2014]. Newbury et al. [2011] and Vernes et al. [2008] identified various SNPs associated with SLI with quantitative measures non-word repetition, receptive language, and expressive language.
Biallelic variants in CNTNAP2 gene, reported by Strauss et al. [2006] in an old-order Amish family, carry homozygous variation (3709delG) in exon 22 of CNTNAP2 identified in 3 patients. As a result of this frameshift variant, an early stop codon (I1253X) was introduced and is estimated to encode a non-functional protein (without cytoplasmic and transmembrane CASPR2 domains); this truncating variant was discovered in the C-terminal region of CNTNAP2. The clinical phenotypes in affected children presented with CDFE, frequent seizures, and mild delay in gross motor skills, followed by deterioration of learning skills, language, and social behaviours involving symptoms of autism and ADHD [Rodenas-Cuadrado et al., 2014; Poot, 2015]. Subsequently, a biallelic variant was reported by Zweier et al. [2009] in an autosomal recessive family, homozygous deletion of 2–9 exons (in-frame deletion) in gene CNTNAP2. This truncating variant produces loss of 33–500 amino acids residues. All affected individuals displayed severe ID, severe MR, infantile epilepsy (drug-fast) with associated language deterioration, autistic features, and poor communication skills, overlapping phenotypically with Pitt-Hopkins syndrome. Further, two more homozygous deletions in CNTNAP2 gene were reported, a homozygous deletion of 2-3 exon in a consanguineous family; affected family members display phenotype autism, epilepsy, severe intellectual disability, and language impairment [Rodenas-Cuadrado et al., 2016]. Watson et al. [2014] reported another case, a homozygous deletion of exon 3 in a Pakistani-origin UK family presenting with an autosomal recessive syndrome; three members of family were affected with phenotypes epilepsy, severe intellectual disability, obsessive compulsive behaviour, language impairments, autistic features, and wide ataxic gait. Deletion of exon 3 in CNTNAP2 results in frameshift variation and introduces a premature termination codon, i.e., A156X, producing out-of-frame mRNA deletion, resulting in a specific syndrome from biallelic losses of CNTNAP2 that has been reviewed by Poot [2017].
More recently, Scala et al. [2021] presented a case report of biallelic CNTNAP2 variants in a 7-year-old boy. Affected child was born (at term) to healthy non-consanguineous parents without a history of neurodevelopmental disorders in the family. Affected child displayed early infantile hyperkinetic stereotyped movements that remained throughout childhood. Irregular movements involved shaking of four limbs in a rhythmic, repeating manner with clear stereotypical characteristics. Other clinical phenotypes involved ADHD, ID, speech impairment, and ASD. Genetic variant identified by whole-genome array-CGH, a maternal origin duplication of 0.402 Mb (c.97 +? _209-? dup) includes intron 1, intron 2, and exon 2 of CNTNAP2; the disturbed intronic 1 region contains FOXP2 (transcription factor) binding site, predicted to abnormal expression regulation of CNTNAP2. Another variant in proband was detected by screening of exons of CNTNAP2 by Sanger sequencing, i.e., c.2752C>T (p.Leu918Phe), a missense variant that was inherited paternally.
In conclusion, we have identified a novel variant (c. 3371T>A [p.Ile1124Asn]) in CNTNAP2 gene in a Pakistani family having obesity, seizures, and language impairment. Present study expands the mutational spectrum of CNTNAP2 to be helpful in future research and diagnoses of patients with similar phenotypes.
Statement of Ethics
The study was ethically conducted in accordance with the World Medical Association Declaration of Helsinki. It was approved by Institutional Review Board (IRB) at Department of Biotechnology, BUITEMS, Quetta, Pakistan. All the participants (or their guardians) gave written informed consents to perform current study and publish research data.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Funding Sources
No funding was received for this study.
Author Contributions
Sara Naudhani: methodology and writing original draft; Adeel Ahmad, Fariya Khan, Azqa Zafar, and Nafeesa Raheem: clinical investigation and diagnosis of patients; Muhammad Tariq Pervez: bioinformatic analysis; Sajjad Ali Shah: sample and data collection; Abdul Hameed Baloch: informed consents from participants and data analyses; Muhammad Mushtaq: supervision and facilitated laboratory work; and Shakeela Daud: conceptualization, supervision, review, and editing of the manuscript. All authors read and approved the final manuscript.
Data Availability Statement
All data generated or analysed during this study are included in this article and its online supplementary material. Further enquiries can be directed to the corresponding author.
Supplementary Material
Supplemental Video
Supplementary data
Acknowledgments
We would like to express our gratitude to all of the patients and their family members who took part in this study. SN was supported through an “indigenous PhD fellowship programme” from Higher Education Commission (HEC) of Pakistan.
Funding Statement
No funding was received for this study.
References
- 1.Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82((1)):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop DV. Genetic and environmental risks for specific language impairment in children. Int Congr Ser. 2003;1254:225–245. doi: 10.1098/rstb.2000.0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463((7281)):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchner DA, Geisinger JM, Glazebrook PA, Morgan MG, Spiezio SH, Kaiyala KJ, et al. The juxtaparanodal proteins CNTNAP2 and TAG1 regulate diet-induced obesity. Mamm Genome. 2012;23((7-8)):431–442. doi: 10.1007/s00335-012-9400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchner DA, Nadeau JH. Contrasting genetic architectures in different mouse reference populations used for studying complex traits. Genome Res. 2015;25((6)):775–791. doi: 10.1101/gr.187450.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canali G, Goutebroze L. CNTNAP2 heterozygous missense variants: risk factors for autism spectrum disorder and/or other pathologies? J Exp Neurosci. 2018;12:117906951880966. doi: 10.1177/1179069518809666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtin C, Anderson SE, Must A, Bandini L. The prevalence of obesity in children with autism: a secondary data analysis using nationally representative data from the national survey of children's health. BMC Pediatr. 2010;10((1)):11. doi: 10.1186/1471-2431-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykens EM, Lee E, Roof E. Prader-Willi syndrome and autism spectrum disorders: an evolving story. J Neurodev Disord. 2011;3((3)):225–237. doi: 10.1007/s11689-011-9092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15((6)):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falsaperla R, Pappalardo XG, Romano C, Marino SD, Corsello G, Ruggieri M, et al. Intronic variant in CNTNAP2 gene in a boy with remarkable conduct disorder, minor facial features, mild intellectual disability, and seizures. Front Pediatr. 2020;8:550. doi: 10.3389/fped.2020.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty E, Deranieh RM, Artimovich E, Lee IS, Siegel AJ, Levy DL, et al. Patient-derived hiPSC neurons with heterozygous CNTNAP2 deletions display altered neuronal gene expression and network activity. NPJ Schizophr. 2017;3((1)):35–34. doi: 10.1038/s41537-017-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13((3)):261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 14.Freri E, Castellotti B, Canafoglia L, Ragona F, Solazzi R, Vannicola C, et al. Severe epilepsy in CNTNAP2-related Pitt-Hopkins-like syndrome successfully treated with stiripentol. Seizure. 2021;88:143–145. doi: 10.1016/j.seizure.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Gregor A, Albrecht B, Bader I, Bijlsma EK, Ekici AB, Engels H, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Genet. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 201020;6((5)):e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A((10)):2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 18.Nakabayashi K, Scherer SW. The human contactin-associated protein-like 2 gene (CNTNAP2) spans over 2 Mb of DNA at chromosome 7q35. Genomics. 2001;73((1)):108–112. doi: 10.1006/geno.2001.6517. [DOI] [PubMed] [Google Scholar]
- 19.Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41((1)):90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paradis J, Emmerzael K, Duncan TS. Assessment of English language learners: using parent report on first language development. J Commun Disord. 2010;43((6)):474–497. doi: 10.1016/j.jcomdis.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Peter B, Raskind WH, Matsushita M, Lisowski M, Vu T, Berninger VW, et al. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord. 2011;3((1)):39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrin AL, Giacheti CM, Maximino LP, Abramides DV, Zanchetta S, Rossi NF, et al. Identification of a microdeletion at the 7q33–q35 disrupting the CNTNAP2 gene in a Brazilian stuttering case. Am J Med Genet A. 2010;152A((12)):3164–3172. doi: 10.1002/ajmg.a.33749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poot M. Connecting the CNTNAP2 networks with neurodevelopmental disorders. Mol Syndromol. 2015;6((1)):7–22. doi: 10.1159/000371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poot M. Intragenic CNTNAP2 deletions: a bridge too far? Mol Syndromol. 2017;8((3)):118–130. doi: 10.1159/000456021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodenas-Cuadrado P, Ho J, Vernes SC. Shining a light on CNTNAP2: complex functions to complex disorders. Eur J Hum Genet. 2014;22((2)):171–178. doi: 10.1038/ejhg.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodenas-Cuadrado P, Pietrafusa N, Francavilla T, La Neve A, Striano P, Vernes SC. Characterisation of CASPR2 deficiency disorder-a syndrome involving autism, epilepsy and language impairment. BMC Med Genet. 2016;17:8. doi: 10.1186/s12881-016-0272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubio-Marrero EN, Vincelli G, Jeffries CM, Shaikh TR, Pakos IS, Ranaivoson FM, et al. Structural characterization of the extracellular domain of CASPR2 and insights into its association with the novel ligand contactin 1. J Biol Chem. 2016;291((11)):5788–5802. doi: 10.1074/jbc.M115.705681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saint-Martin M, Joubert B, Pellier-Monnin V, Pascual O, Noraz N, Honnorat J. Contactin-associated protein-like 2, a protein of the neurexin family involved in several human diseases. Eur J Neurosci. 2018;48((3)):1906–1923. doi: 10.1111/ejn.14081. [DOI] [PubMed] [Google Scholar]
- 29.Scala M, Anijs M, Battini R, Madia F, Capra V, Scudieri P, et al. Hyperkinetic stereotyped movements in a boy with biallelic CNTNAP2 variants. Ital J Pediatr. 2021;47((1)):208. doi: 10.1186/s13052-021-01162-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinawi M, Sahoo T, Maranda B, Skinner SA, Skinner C, Chinault C, et al. 11p14. 1 microdeletions associated with ADHD, autism, developmental delay, and obesity. Am J Med Genet A. 2011;155A((6)):1272–1280. doi: 10.1002/ajmg.a.33878. [DOI] [PubMed] [Google Scholar]
- 31.Smogavec M, Cleall A, Hoyer J, Lederer D, Nassogne MC, Palmer EE, et al. Eight further individuals with intellectual disability and epilepsy carrying bi-allelic CNTNAP2 aberrations allow delineation of the mutational and phenotypic spectrum. J Med Genet. 2016;53((12)):820–827. doi: 10.1136/jmedgenet-2016-103880. [DOI] [PubMed] [Google Scholar]
- 32.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68((2)):254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354((13)):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 34.Traka M, Goutebroze L, Denisenko N, Bessa M, Nifli A, Havaki S, et al. Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J Cell Biol. 2003;162((6)):1161–1172. doi: 10.1083/jcb.200305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyler CV, Schramm SC, Karafa M, Tang AS, Jain AK. Chronic disease risks in young adults with autism spectrum disorder: forewarned is forearmed. Am J Intellect Dev Disabil. 2011;116((5)):371–380. doi: 10.1352/1944-7558-116.5.371. [DOI] [PubMed] [Google Scholar]
- 36.Varea O, Martin-de-Saavedra MD, Kopeikina KJ, Schürmann B, Fleming HJ, Fawcett-Patel JM, et al. Synaptic abnormalities and cytoplasmic glutamate receptor aggregates in contactin associated protein-like 2/Caspr2 knockout neurons. Proc Natl Acad Sci U S A. 2015;112((19)):6176–6181. doi: 10.1073/pnas.1423205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velez Edwards DR, Naj AC, Monda K, North KE, Neuhouser M, Magvanjav O, et al. Gene-environment interactions and obesity traits among postmenopausal African-American and Hispanic women in the Women's Health Initiative SHARe Study. Hum Genet. 2013;132((3)):323–336. doi: 10.1007/s00439-012-1246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA, et al. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82((1)):1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 39.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359((22)):2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuillaume ML, Naudion S, Banneau G, Diene G, Cartault A, Cailley D, et al. New candidate loci identified by array-CGH in a cohort of 100 children presenting with syndromic obesity. Am J Med Genet A. 2014;164A((8)):1965–1975. doi: 10.1002/ajmg.a.36587. [DOI] [PubMed] [Google Scholar]
- 41.Watson CM, Crinnion LA, Tzika A, Mills A, Coates A, Pendlebury M, et al. Diagnostic whole genome sequencing and split-read mapping for nucleotide resolution breakpoint identification in CNTNAP2 deficiency syndrome. Am J Med Genet A. 2014;164A((10)):2649–2655. doi: 10.1002/ajmg.a.36679. [DOI] [PubMed] [Google Scholar]
- 42.Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85((5)):655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video
Supplementary data
Data Availability Statement
All data generated or analysed during this study are included in this article and its online supplementary material. Further enquiries can be directed to the corresponding author.