ABSTRACT
Staphylococcus aureus is an important human pathogen responsible for a variety of infections including skin and soft tissue infections, endocarditis, and sepsis. The combination of increasing antibiotic resistance in this pathogen and the lack of an efficacious vaccine underscores the importance of understanding how S. aureus maintains metabolic homeostasis in a variety of environments, particularly during infection. Within the host, S. aureus must regulate cellular levels of the cofactor heme to support enzymatic activities without encountering heme toxicity. Glutamyl tRNA reductase (GtrR), the enzyme catalyzing the first committed step in heme synthesis, is an important regulatory node of heme synthesis in Bacteria, Archaea, and Plantae. In many organisms, heme status negatively regulates the abundance of GtrR, controlling flux through the heme synthesis pathway. We identified two residues within GtrR, H32 and R214, that are important for GtrR-heme binding. However, in strains expressing either GtrRH32A or GtrRR214A, heme homeostasis was not perturbed, suggesting an alternative mechanism of heme synthesis regulation occurs in S. aureus. In this regard, we report that heme synthesis is regulated through phosphorylation and dephosphorylation of GtrR by the serine/threonine kinase Stk1 and the phosphatase Stp1, respectively. Taken together, these results suggest that the mechanisms governing staphylococcal heme synthesis integrate both the availability of heme and the growth status of the cell.
IMPORTANCE
Staphylococcus aureus represents a significant threat to human health. Heme is an iron-containing enzymatic cofactor that can be toxic at elevated levels. During infection, S. aureus must control heme levels to replicate and survive within the hostile host environment. We identified residues within a heme biosynthetic enzyme that are critical for heme binding in vitro; however, abrogation of heme binding is not sufficient to perturb heme homeostasis within S. aureus. This marks a divergence from previously reported mechanisms of heme-dependent regulation of the highly conserved enzyme glutamyl tRNA reductase (GtrR). Additionally, we link cell growth arrest to the modulation of heme levels through the post-translational regulation of GtrR by the kinase Stk1 and the phosphatase Stp1.
KEYWORDS: Staphylococcus aureus, heme synthesis
INTRODUCTION
Heme, a tetrapyrrole ring coordinating an iron atom, is a nearly ubiquitous molecule used as a cofactor in a variety of essential processes, including electron transport, redox sensing, cell signaling, and enzyme catalysis (1 - 4). Despite the utility of heme as a cofactor, the redox-active heme-iron can lead to the production of macromolecule-damaging radicals necessitating tight control over cellular heme levels (1). Thus, organisms that encounter and use heme encode regulatory systems to sense and alleviate heme toxicity (5).
Staphylococcus aureus is a gram-positive pathogen responsible for a myriad of diseases, ranging from skin and soft tissue infections to infectious endocarditis (6). In 2019, over one million deaths were attributable to S. aureus infection making S. aureus the leading bacterial cause of death worldwide (7). During infection, S. aureus likely experiences varying intracellular heme concentrations as well as requirements for hemoproteins as evidenced by multiple strategies employed by the bacterium to maintain heme homeostasis (5). In conditions of high environmental heme, the heme sensor two-component system (HssRS, TCS) activates the expression of the heme responsive transporter (HrtAB), a heme efflux pump, to alleviate heme toxicity (1, 8, 9). HssRS can also sense endogenously produced heme; thus, HssRS activation can be used as a readout for heme production (1, 8). The level of HssRS activation can be measured using a reporter of Phrt promoter activity or by assessing the ability of S. aureus to adapt to heme toxicity using a heme adaptation assay. In this assay, activation of the HssRS TCS by subinhibitory levels of heme in overnight cultures or by excess heme production enables robust growth in toxic concentrations of heme (5, 9).
Heme is an important contributor to the growth and survival of S. aureus within the hostile host environment (10 - 14). In conditions in which heme is required, S. aureus can acquire host heme using the Iron-regulated surface determinant (Isd) system or it can synthesize heme de novo (15). Strategies for maintaining appropriate heme levels are conserved among gram-positive bacteria (5, 15); however, the mechanisms by which the systems involved in heme uptake, synthesis, and detoxification are integrated to maintain heme homeostasis remain unknown. The work described here aimed to define the regulation of heme synthesis in S. aureus and to determine how the bacterium modulates heme levels.
In all organisms that synthesize heme, production of δ-aminolevulinic acid (ALA) represents the first committed step in heme synthesis and thus control of ALA production is critical to the regulation of heme synthesis (1, 4, 16 - 21). ALA synthesis occurs through one of two pathways. Members of the domains Bacteria, and Archaea, and the kingdom Plantae form ALA in a two-step process starting with the conversion of glutamate-charged tRNA (tRNAglu) to glutamate-1-semialdehyde (GSA) by glutamyl tRNA reductase [GtrR, formerly HemA (4)] and GSA to ALA by glutamate-1-semialdehyde amino-mutase [GsaM, formerly HemL (4)] through the C5 pathway (3, 22, 23). Metazoans, however, synthesize the precursor ALA through the one-step condensation of succinyl-CoA and glycine in the C4 or Shemin pathway (4, 23). Recently, an alternative branch of terminal heme synthesis was identified in gram-positive organisms in which coproporphyrinogen is oxidized to coproporphyrin, which is then loaded with iron and decarboxylated to heme. This process is named the coproporphyrin-dependent branch of heme synthesis (24). By contrast, other organisms synthesize protoporphyrin IX and then load the molecule with iron to form heme in the protoporphyrin-dependent pathway (24). These differences in heme synthesis strategies between gram-positive bacterial pathogens and the human host offer the potential for bacteria-specific heme synthesis-disrupting therapeutics. For example, inducing endogenous overproduction of ALA could drive the production of porphyrins, known photosensitizing molecules, in a bacterial-specific manner sparing host cells from phototoxicity (25, 26). Additionally, overproduction of heme depletes available iron within the cell hampering the ability of the bacterium to populate iron-requiring enzymes important for pathogenesis (1).
Previous studies in several organisms, including S. aureus, have identified GtrR as a key point of regulation in heme synthesis (1, 19, 27 - 29). Numerous studies in other organisms report that heme binding to GtrR impacts heme synthesis by altering the abundance or catalytic activity of GtrR (1, 16, 19, 20, 28, 30 - 34). GtrR protein abundance is negatively regulated by heme status and the conserved membrane protein HemX in S. aureus and Bacillus subtilis (1, 18). Since GtrR catalyzes one of the rate-limiting reactions of heme synthesis, accumulation of GtrR protein in a hemX mutant drives heme production. Elevated heme levels within a ΔhemX mutant are sufficient to activate the HssRS TCS, further illustrating how HssRS signaling can be used as a readout for cellular heme levels (1). Despite an abundance of literature describing heme-dependent regulation of GtrR and heme synthesis, the mechanism by which this occurs in S. aureus is unknown.
Another mechanism of heme synthesis regulation could occur through the post-translational modification of heme biosynthetic enzymes. Eukaryote-like serine/threonine (Ser/Thr) protein kinases (eSTK) and phosphatases (eSTP) represent a conserved strategy to tune cellular signaling and activities by altering the phosphorylation state of proteins (35). Most strains of S. aureus encode only one eSTK/eSTP pair (Stk1/Stp1) (36, 37). Upon sensing of lipid II, Stk1 dimerizes and autophosphorylation of activation loops induces kinase activity. Active Stk1 catalyzes the phosphorylation of serine and threonine residues of target proteins to induce a cellular response to stalled growth (38, 39). Phosphatases such as Stp1 antagonize Stk1 activity by dephosphorylating Stk1 and substrate proteins (40). Stk1 and Stp1 in S. aureus work together to modulate cell wall biosynthesis, virulence, resistance to antimicrobials, central metabolism, and tuning of TCS signaling in response to cell growth status (36). While several examples of Stk1-dependent phosphoregulation of enzymes have been reported (41 - 43), it is unknown whether GtrR is regulated by phosphorylation.
We hypothesized that heme-bound GtrR is unstable and that this interaction enables heme-dependent regulation of GtrR abundance in S. aureus. In this work, we identified two residues within GtrR that are important for heme binding to GtrR in vitro. Surprisingly, we found that the heme-GtrR interaction does not impact heme production in vivo. We also identified GtrR as a substrate of Stk1 and found that inactivation of the genes encoding either Stk1 or Stp1 impacts baseline levels of heme stress signaling, implicating the pair in the regulation of heme synthesis. Together, the data presented here describe a divergence from known mechanisms of heme synthesis regulation and report the phosphorylation of a heme biosynthetic enzyme by a master cellular regulator, connecting heme biosynthesis to cell growth status in S. aureus.
RESULTS
Recombinant S. aureus GtrR binds heme
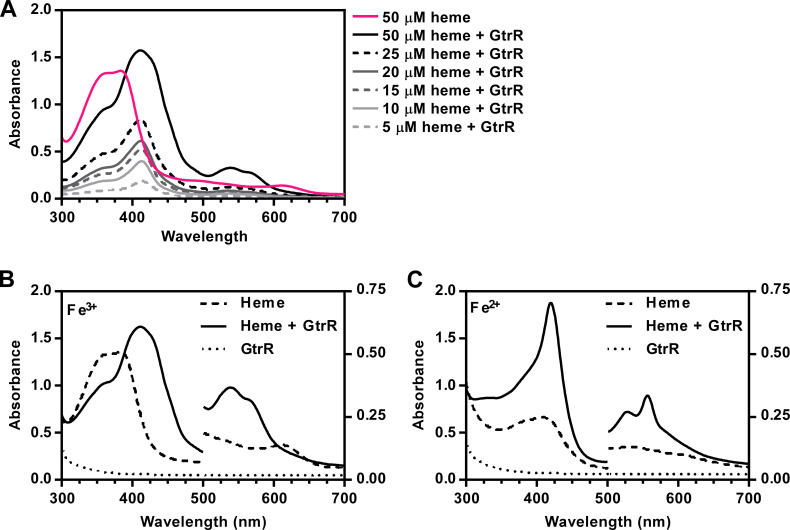
We hypothesized that the heme-dependent regulation of GtrR abundance occurs through direct interactions between heme and GtrR. To test this hypothesis, we purified recombinantly expressed S. aureus GtrR from Escherichia coli and assessed whether GtrR binds heme in vitro. GtrR was not bound to heme upon purification, evidenced by the lack of a Soret peak. Addition of GtrR to a hemin chloride (heme) solution induced shift in the maximal absorbance of heme from 380 nm to approximately 414 nm. The presence of this Soret peak upon the addition of a protein is indicative of heme binding (Fig. 1). The absence of a charge transfer maximum and the presence of α/β-bands at 540 and 567 nm, respectively, indicates that GtrR is bound to a low-spin (LS) heme species (Fig. 1A) (44, 45). Since the electronic spin state of heme often correlates with the degree of heme coordination (46), excess dithiothreitol was used to reduce the heme-GtrR solution. Upon reduction, the Soret band red-shifted to 421 nm and the α/β-bands were sharpened at 526 and 557 nm also supporting the presence of an LS hexacoordinate heme in both oxidation states (Fig. 1B) (45 - 50).
Fig 1.
S. aureus GtrR binds heme in vitro. (A) Absorption spectra of heme in solution and in the presence of purified WT GtrR in dialysis buffer. The spectrum corresponding to 50 µM free heme is shown as a solid pink line. Increasing concentrations of heme (5–50 µM, in grayscale from light to dark) were incubated in the presence of 5 µM purified protein. (B and C) Absorption spectra of heme in dialysis buffer (B) and after reduction with the addition of excess dithionite (C). Absorbance values at wavelengths below 500 nm are plotted on the left Y axis and absorbance values above 500 nm are plotted on the right Y axis for visualization of α/β bands.
Conservation, domain organization, and homology model of S. aureus GtrR
We next sought to identify the residues important for GtrR binding to heme. While canonical heme-binding motifs have been described, heme can also be bound by proteins through several general mechanisms. Heme ligation involves amino acid residues that coordinate heme-iron such as cysteine, methionine, histidine, lysine, and tyrosine (51 - 54). Heme-protein interactions are also mediated and/or strengthened by π-π stacking, hydrogen bonding, and Van der Waals interactions (51, 55). Thus, it is difficult to predict specific residues that mediate heme binding by sequence gazing alone (53).
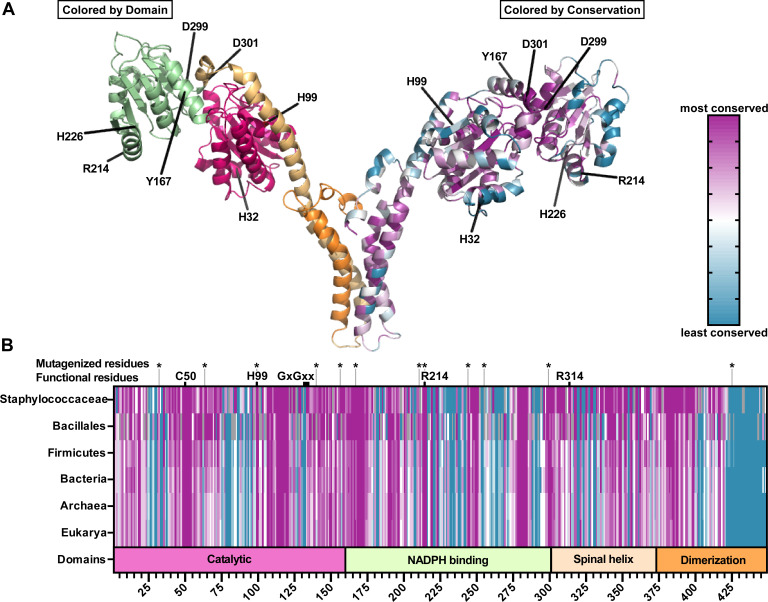
To expand our search for heme-binding motifs in GtrR, we employed a bioinformatics approach. A multiple sequence alignment (MSA) and phylogenetic tree were generated using GtrR protein sequences from 275 gtrR-encoding species including S. aureus strain Newman. The ConSurf Server was then used to generate conservation scores for each residue within S. aureus GtrR at increasing taxonomic levels (Staphylococcaceae, Bacillales, Firmicutes, Bacteria, prokaryotes, and all other organisms encoding GtrR) (56 - 59), (Fig. 2; Fig. S1). To identify residues within S. aureus GtrR that mediate heme-GtrR interactions, a directed alanine screening strategy was employed. We previously found that heme status impacts the protein levels of GtrR (1) and predicted that heme binding to GtrR impacts abundance rather than activity. To this end, most residues that were highly conserved or with predicted roles in catalysis were excluded from mutagenesis while residues known to mediate heme-protein interactions were prioritized (51, 56 - 58). The amino acid H99 was selected despite its conservation and predicted catalytic function because heme inhibits GtrR activity in Hordeum vulgare and structure prediction by Huang and Wang identified H99 as a likely heme-binding residue (Fig. 2) (30, 60). Using these strategies, the S. aureus GtrR residues H32, H63, H99, H140, T156, Y167, R214, H226, Y255, D299, D301, and H425 were selected for alanine mutagenesis (Fig. 2).
Fig 2.
Conservation, domain organization, and homology model of S. aureus GtrR. (A) Homology model of a S. aureus GtrR homodimer generated using SWISS-MODEL with GtrR from Arabidopsis thaliana as a template (61, 62). Residues selected for mutagenesis are highlighted. On the left monomer, domains are colored to match the domain organization modeled in B. On the right, amino acids are colored according to the degree of conservation. A scale of variable (teal) to conserved (purple) residues is indicated in the figure. (B) Domain organization (bottom bar: Catalytic, pink; NADPH binding, green; Spinal Helix, light orange; Dimerization, dark orange) and conservation of S. aureus GtrR primary sequence compared to increasing taxonomic levels (top bars, colored as in A).
Histidine 32 and arginine 214 of S. aureus GtrR are individually important for heme binding in vitro
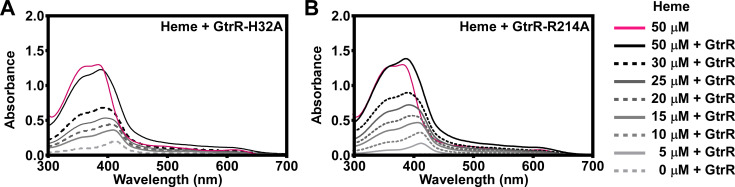
To test whether the predicted heme-binding amino acid residues were important for GtrR-heme interactions, 12 alanine substitution variant GtrRs were expressed, purified, and their ability to bind heme was assessed. Only 7 out of the 12 variant GtrRs were purified successfully (Table 1), likely due to the amino acid substitution resulting in an unstable protein. Out of the six substitutions evaluated for the ability to bind heme (Fig. 3; Fig. S2A through E), GtrRH32A and GtrRR214A were the only variants that did not induce a spectral shift in absorbance upon exposure to heme (Fig. 3A and B). The absence of the Soret peak and the α/β-bands seen in the GtrRH32A and GtrRR214A variants indicates a lack of heme binding. To ensure equal amounts of GtrR protein were used in each binding assay, aliquots were separated using SDS-PAGE, and protein was visualized using Coomassie staining (Fig. S2F). From these experiments, we hypothesized that heme binding to residues H32 and R214 enables negative regulation of heme synthesis through GtrR-heme interactions in S. aureus. Therefore, we created ΔgtrR ΔhemX mutant strains expressing HemX and either GtrRH32A or GtrRR214A in cis to test this hypothesis.
TABLE 1.
Expression outcome for mutant GtrR proteins
| Mutant GtrR | Expression and purification outcome |
|---|---|
| WT GtrR | Expressed and purified |
| H32A | Expressed and purified |
| H63A | Did not express |
| H99A | Expressed and purified |
| H140A | Did not express |
| T156A | Did not express |
| Y167A | Expressed and purified |
| R214A | Expressed and purified |
| H226A | Expressed and purified |
| D244A | Did not express |
| Y255A | Did not express |
| D299A | Expressed and purified |
| D301A | Expressed and purified |
| H425A | Did not express |
Fig 3.
Histidine 32 and arginine 214 are individually important for GtrR binding to heme in vitro. Absorption spectra of increasing concentrations of heme (5–50 µM) in the presence of 5 µM purified GtrRH32A (A) or GtrRR214A (B). The spectrum corresponding to 50 µM free heme is shown as a solid pink line. Heme concentrations ranging from 5 to 50 µM of heme are graphed in grayscale from light to dark.
GtrR variants expressed in S. aureus ΔgtrR ΔhemX are catalytically active and heme produced is utilized in cellular hemoproteins
To test whether abrogation of heme binding to GtrR alters heme homeostasis, we constructed ΔgtrR ΔhemX strains that express HemX and myc-tagged GtrR variants in cis from the SAPI locus, a neutral site in the genome (Table 4) (63). Table 4 contains detailed descriptions of each strain, but we abbreviated the genotypic annotations to simplify legends and references in the text. For example, we refer to the ΔgtrR ΔhemX strain that expresses HemX and WT N-terminally myc-tagged GtrR as “ΔΔPgtrRmyc-gtrR.hemX.”
TABLE 4.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant feature or genotype | Ref. |
|---|---|---|
| Staphylococcus aureus Newman WT | Wild-type, methicillin-sensitive clinical isolate | (94) |
| Escherichia coli DH5α | Cloning strain | (95) |
| E. coli XL1-Blue | Cloning strain | (96) |
| E. coli BL21(DE3) | Strain used for expression of recombinant protein | (97) |
| S. aureus RN4220 | Restriction-deficient cloning intermediate strain | (98) |
|
S. aureus Newman ΔgtrR ΔhemX |
In-frame unmarked deletion of gtrR (NWMN_1566) and hemX (NWMN_1565). Referred to as ΔΔ | (1) |
| S. aureus JE2 hemX::erm | hemX::Tn, NE1501 | (99) |
| S. aureus Newman katA::erm | katA::Tn. Catalase negative | (1) |
| S. aureus Newman Δstk1 | In-frame unmarked deletion of stk1 (NWMN_1130) | This study |
| S. aureus Newman Δstk1 hemX::erm | hemX::Tn transduced into Newman Δstk1 | This study |
| S. aureus Newman Δstp1 | In-frame unmarked deletion of stp1 (NWMN_1129) | This study |
| S. aureus Newman Δstp1 hemX::erm | hemX::Tn transduced into Newman Δstp1 | This study |
| pJC1306 | Plasmid used for genomic integration into the S. aureus SaPI1 attachment site | (63) |
| S. aureus RN9011 | RN4220 harboring the pRN7023 integrase plasmid. Intermediate cloning strain | (63) |
|
S. aureus Newman ΔgtrR ΔhemX attC::PgtrRmyc-gtrR.hemX |
S. aureus Newman ΔgtrR ΔhemX with PgtrRmyc-gtrR.hemX integrated into the chromosome using the pJC1306 vector. Referred to as ΔΔPgtrRmyc-gtrR.hemX |
This study |
|
S. aureus Newman ΔgtrR ΔhemX attC::PgtrRgtrR-myc.hemX |
S. aureus Newman ΔgtrR ΔhemX with PgtrRgtrR-myc.hemX integrated into the chromosome using the pJC1306 vector. Referred to as ΔΔPgtrRgtrR-myc.hemX |
This study |
|
S. aureus Newman ΔgtrR ΔhemX attC::PgtrRmyc-gtrR |
S. aureus Newman ΔgtrR ΔhemX with pJC1306.PgtrRmyc-gtrR.hemX integrated into the chromosome using the pJC1306 vector. Referred to as ΔΔPgtrRmyc-gtrR |
This study |
|
S. aureus Newman ΔgtrR ΔhemX attC::PgtrRmyc-gtrR-H32A.hemX |
S. aureus Newman ΔgtrR ΔhemX with pJC1306.PgtrRmyc-gtrR-H32A.hemX integrated into the chromosome using the pJC1306 vector. Referred to as ΔΔPgtrRmyc-gtrR(H32A).hemX |
This study |
|
S. aureus Newman ΔgtrR ΔhemX attC::PgtrRmyc-gtrR-R214A.hemX |
S. aureus Newman ΔgtrR ΔhemX with pJC1306.PgtrRmyc-gtrR-R214A.hemX integrated into the chromosome using the pJC1306 vector. Referred to as ΔΔPgtrRmyc-gtrR(R214A).hemX |
This study |
| pXen1 PhrtluxABCDE | Luminescence-producing operon luxABCDE cloned from Photorhabdus luminescens controlled by the HssRS-regulated promoter Phrt | (1) |
| pET15b.gtrR | GtrR expression plasmid | This study |
| pET15b.gtrR-H63A | GtrR-H63A expression plasmid | This study |
| pET15b.gtrR-H99A | GtrR-H99A expression plasmid | This study |
| pET15b.gtrR-H140A | GtrR-H140A expression plasmid | This study |
| pET15b.gtrR-T156A | GtrR-H156A expression plasmid | This study |
| pET15b.gtrR-Y167A | GtrR-Y167A expression plasmid | This study |
| pET15b.gtrR-R214A | GtrR-R214A expression plasmid | This study |
| pET15b.gtrR-Y230A | GtrR-Y230A expression plasmid | This study |
| pET15b.gtrR-H226A | GtrR-H226A expression plasmid | This study |
| pET15b.gtrR-D244A | GtrR-D244A expression plasmid | This study |
| pET15b.gtrR-Y255A | GtrR-Y255A expression plasmid | This study |
| pET15b.gtrR-D299A | GtrR-D299A expression plasmid | This study |
| pET15b.gtrR-D301A | GtrR-D299A expression plasmid | This study |
| pET15b.gtrR-H425A | GtrR-H425A expression plasmid | This study |
| pGEX-2T.stk1 | Stk1 kinase domain (1-348) expression plasmid | (100) |
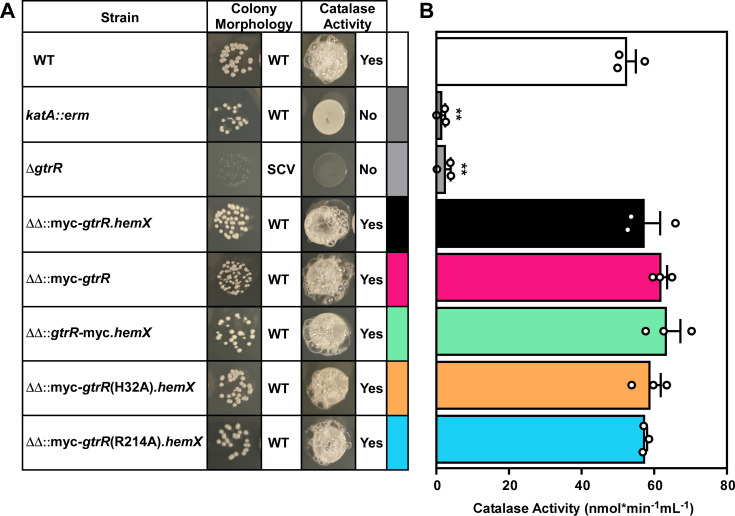
To validate the strains expressing GtrR variants, we initially assessed GtrR expression in these strains using immunoblots probing for the myc tag. Unfortunately, we were unable to detect myc-tagged GtrR in any of the constructed strains. Therefore, we evaluated whether the expression of the GtrR variants in S. aureus complements the heme-deficient phenotype of the ΔgtrR ΔhemX mutant. Substitution of either R214 or H32 in GtrR for alanine could result in the destabilization of the protein in vivo or impact catalytic efficiency. These phenotypes could impact heme synthesis and heme trafficking through mechanisms other than loss of heme-dependent regulation of GtrR. To ensure that the H32A and R214A substitutions within GtrR did not result in defects in heme synthesis or heme insertion into hemoproteins, strains were spotted on tryptic soy agar to assess colony morphology and catalase status. All strains adopted a normal colony morphology except for ΔgtrR, a strain incapable of heme synthesis that forms punctate colonies (14, 64, 65). The absence of heme synthesis produces respiration-deficient “small colony variants.” Both quantitative and qualitative assays of catalase activity were used to assess the production of functional catalase, an abundant hemoprotein whose activity requires heme (1). All tested strains possessed comparable catalase activity except for the heme-deficient ΔgtrR mutant and the katA::erm catalase mutant (Fig. 4B). Addition of ALA chemically complements the heme deficiency of the ΔgtrR mutant and rescues catalase activity. Combined, these data demonstrate that the GtrRH32A and GtrRR214A variants are functional and that expression of these variants enables the production and incorporation of heme into representative hemoproteins at levels similar to wild type (WT).
Fig 4.
GtrR variants expressed in a S. aureus ΔgtrR ΔhemX mutant are catalytically active and heme produced is utilized in cellular hemoproteins. (A) S. aureus strains were serially diluted and spot plated on TSA. After growth, hydrogen peroxide was pipetted onto the resulting colonies. Colony morphology and qualitative catalase activity are summarized on the left (A). Strains with wild-type colony size are annotated as “WT” and small colony variants “SCV.” (B) Catalase activity of strains used in (A) was quantified using a Catalase Assay Kit. Bars are representative of the averages from three independent experiments performed in biological triplicate with error bars representing the SEM. Significance was calculated in Graphpad Prism 8 using the Brown-Forsythe and Welch ANOVA tests and Dunnett’s T3 multiple comparisons tests with individual variances computed for each comparison with WT; **P < 0.01; non-significant comparisons are not shown.
Disruption of heme binding to GtrR does not correlate with activation of the HssRS two-component system
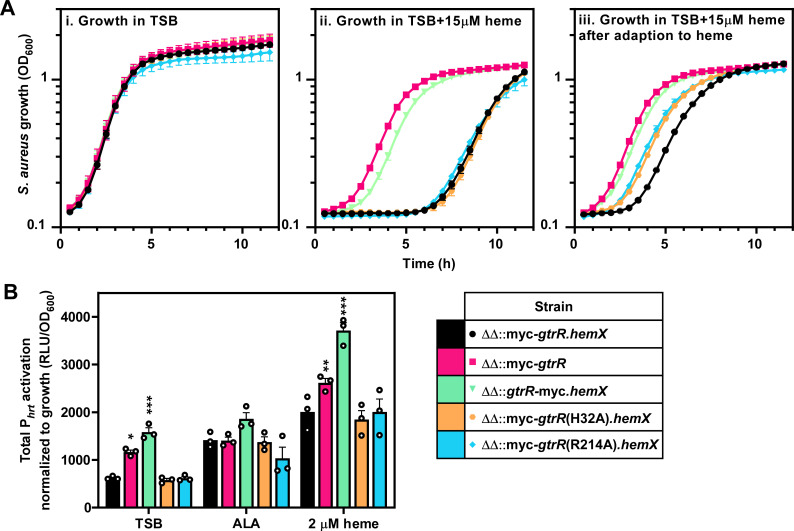
Dysregulation of GtrR abundance in S. aureus results in activation of the heme stress response due to overproduction of heme (1, 9). To test whether abrogation of heme binding to GtrR results in a similar stress response, we measured the ability of S. aureus expressing either GtrRH32A or GtrRR214A to adapt to heme toxicity using a heme adaptation assay. Activation of the HssRS TCS by subinhibitory levels of heme in overnight cultures or by high endogenous heme levels enables robust growth under conditions of toxic heme levels compared to non-adapted cultures (5, 9). Thus, HssRS activation serves as a readout for heme concentrations in the cell (1, 5, 8, 66 - 71). A strain expressing N-terminally myc-tagged WT GtrR in cis (ΔΔPgtrRmyc-gtrR.hemX) from the SAPI locus experiences a delay in entrance into exponential phase when not adapted to heme while expression of only myc-tagged GtrR (ΔΔPgtrRmyc-gtrR) recapitulates the pre-adaptation phenotype of a ΔhemX mutant (Fig. 5A) (1). Interestingly, a strain expressing C-terminally myc-tagged WT GtrR (ΔΔPgtrRgtrR-myc.hemX) enters logarithmic growth phase when grown in toxic levels of heme faster than the strain expressing N-terminally tagged WT GtrR ΔΔPgtrRmyc-gtrR.hemX (Fig. 5A). This increase in resistance to heme toxicity suggests that the addition of a tag on the C-terminus affects GtrR abundance or activity. Strains expressing GtrR variants with either the non-heme-binding substitutions H32A or R214A both grew similar to the ΔΔPgtrRmyc-gtrR.hemX strain in high levels of heme, indicating that disrupting heme binding to GtrR does not result in overproduction of heme (Fig. 5A). Activation of the hrtAB promoter (Phrt) by HssRS, measured using a luminescent transcriptional reporter, mirrors the phenotypes seen in the heme adaptation growth curves with significantly higher HssRS signaling in the ΔΔPgtrRgtrR-myc.hemX and the ΔhemX-like ΔΔPgtrRmyc-gtrR compared to ΔΔPgtrRmyc-gtrR.hemX grown in tryptic soy broth (Fig. 5B; Fig. S3). Together, these data establish H32 and R214 as residues involved in heme binding and preclude the role of heme binding through these residues in regulating GtrR activity.
Fig 5.
Disruption of heme binding to GtrR does not result in activation of the HssRS two-component system. (A) Growth as measured by OD600 was monitored over time for S. aureus strains grown in TSB containing either vehicle (i) or 15 µM heme (ii, iii). Prior to the measured growth, the strains were pre-grown to stationary phase in TSB containing vehicle (i, ii) or 2 µM heme (iii). (B) Total activation of the hrtAB promoter (Phrt) over 20 h of strains grown in TSB containing 2 mM δ-aminolevulinic acid, 2 µM heme, or vehicle. Time course data used to determine total promoter activation are graphed in Fig. S3. (A and B) Graphs represent the averages of means from three independent experiments, performed in biological triplicate ± SEM. Significance was calculated using two-way ANOVA with Dunnett’s test for multiple comparisons between ΔΔ::myc-gtrR.hemX and each strain, with *P < 0.05; **P < 0.005; ***P = 0.001; non-significant comparisons are not shown.
Stk1 and Stp1 impact heme stress signaling within S. aureus
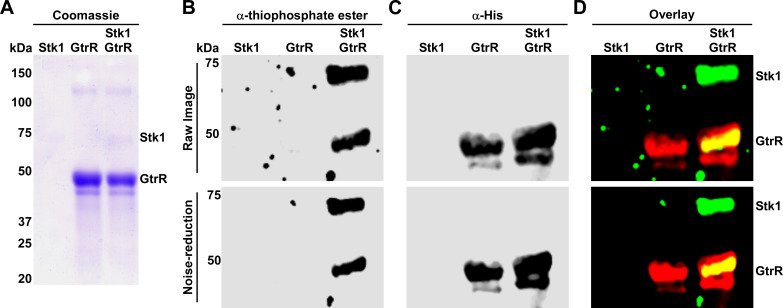
GtrR was previously identified as a phosphoprotein in Synechococcus sp. and Shigella flexneri (72, 73), suggesting that post-translational modification of GtrR serine or tyrosine residues could be an additional mechanism to regulate heme biosynthesis in bacteria. Recent S. aureus phosphoproteomics data sets do not identify GtrR as a phosphoprotein (41, 74, 75) although in silico analysis of GtrR identified 15 predicted serine/threonine phosphosites, several of which are highly conserved (Table 2; Fig. 2) (76). This led us to investigate whether phosphorylation of GtrR by the S/T kinase and global regulator Stk1 impacts heme homeostasis in S. aureus. To assess Stk1-dependent phosphorylation of GtrR in vitro, purified Stk1 was loaded with ATPγS, an ATP analog that, upon phosphorylation of a target protein, can be modified by treatment with the alkylating agent p-nitrobenzyl mesylate to produce a thiophosphate ester that can be detected by immunoblotting. ATPγS-loaded Stk1 was incubated with purified GtrR for 2 h, and then, the reactions were separated by SDS-PAGE. Total protein was visualized by Coomassie staining, phosphorylated proteins were detected using an anti-thiophosphate ester antibody, and the His tag on GtrR was detected using an anti-His antibody (Fig. 6). Control reactions containing only a single protein were included to demonstrate Stk1-specific phosphorylation of GtrR. GtrR alone was not phosphorylated as illustrated by the lack of a ~50 kDa band in the anti-thiophosphate immunoblot. Upon addition of Stk1, however, a band which overlaps with His-tagged GtrR appears on the anti-thiophosphate immunoblot (Fig. 6). These data indicate that Stk1 modulates the phosphorylation status of GtrR.
TABLE 2.
Predicted serine/threonine phosphosites within S. aureus GtrR
| Position | Amino acid | NetPhosBac score |
Predicted domain | Consurf score across taxonomic level | |||||
|---|---|---|---|---|---|---|---|---|---|
| Staphylococcaceae | Bacillales | Firmicutes | Bacteria | Archaea | Eukarya | ||||
| 40 | S | 0.511 | Catalytic | 7 | 8 | 8 | 8 | 7 | 7 |
| 64 | T | 0.512 | Catalytic | 9 | 9 | 8 | 8 | 7 | 7 |
| 109 | S | 0.515 | Catalytic | 9 | 9 | 9 | 9 | 9 | 9 |
| 182 | S | 0.506 | NADPH binding | 1 | 1 | 1 | 1 | 1 | 1 |
| 210 | T | 0.502 | NADPH binding | 8 | 9 | 7 | 6 | 6 | 6 |
| 235 | S | 0.593 | NADPH binding | 1 | 3 | 3 | 3 | 2 | 2 |
| 249 | S | 0.59 | NADPH binding | 9 | 9 | 9 | 9 | 9 | 9 |
| 251 | S | 0.546 | NADPH binding | 9 | 8 | 8 | 8 | 7 | 7 |
| 273 | S | 0.808 | NADPH binding | 6 | 6 | 6 | 6 | 5 | 5 |
| 290 | S | 0.611 | NADPH binding | 1 | 1 | 1 | 1 | 1 | 1 |
| 371 | S | 0.775 | Spine | 7 | 7 | 6 | 6 | 6 | 6 |
| 379 | S | 0.75 | Dimerization | 9 | 7 | 6 | 6 | 6 | 6 |
| 434 | S | 0.604 | Dimerization | 1 | 1 | 1 | 1 | 1 | 1 |
| 440 | S | 0.732 | Dimerization | 1 | 1 | 1 | 1 | 1 | 1 |
| 446 | S | 0.664 | Dimerization | 4 | 7 | 1 | 4 | 1 | 1 |
Fig 6.
GtrR is a substrate of Stk1 kinase. The cytoplasmic kinase domain of S. aureus Stk1 was expressed and purified as a GST-tagged fusion and incubated with purified His-tagged GtrR in the presence of the ATP analog ATPγS. Following PNBM alkylation, the labeled substrates were separated by SDS-PAGE, (A) coomassie stained, and immunoblotting used to detect (B) phosphorylated protein and (C) His-tagged GtrR. Phosphorylated protein and His-tagged GtrR were detected using an anti-thiophosphate ester and anti-His antibodies, respectively. An overlay of the His and thiophosphate ester signals is shown in (D). Bands corresponding to phosphorylated Stk1 and GtrR are annotated. (B–D) Raw images with noise were processed using noise reduction software to improve background clarity. Both the raw (top) and processed images (bottom) are shown for each immunoblot.
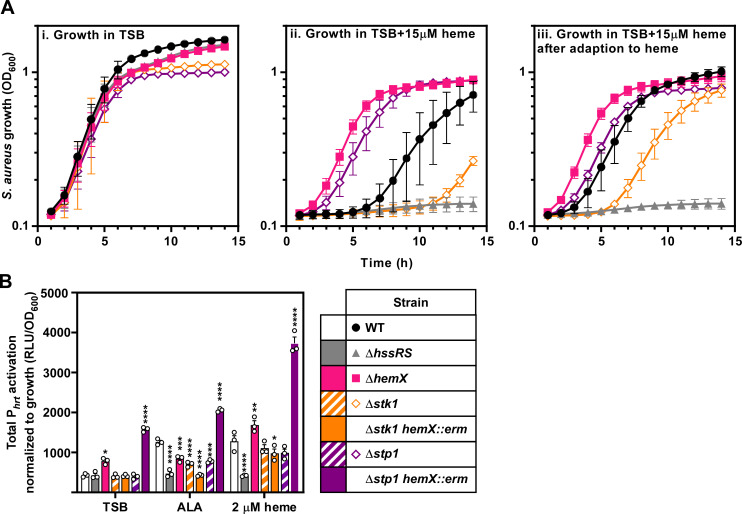
Deletion of stk1 sensitizes S. aureus to heme toxicity, but a Δstk1 mutant is still capable of being adapted to heme (Fig. 7A). We hypothesize that in the absence of Stk1, GtrR is no longer phosphorylated which results in lower levels of heme production. This could increase the amount of heme required to adapt a Δstk1 mutant to heme toxicity and account for the mutant’s growth defect in toxic concentrations of heme, even when exposed to heme overnight. To test this, we deleted stp1 and assessed the growth phenotype of the mutant in media containing toxic levels of heme. Interestingly, the Δstp1 mutant grew equally well in the presence of high concentrations of heme regardless of whether the strain was adapted to heme toxicity similar to a ΔhemX mutant (1). Since deletion of stk1 and stp1 alters the permeability of the cell envelope, we also assessed heme stress levels using a luminescent reporter of hrtAB promoter (Phrt) activity (77, 78). We found that the Δstk1 and Δstp1 mutant strains have similar levels of Phrt activation in both TSB and 2 µM heme, suggesting that the growth differences in the presence of high heme levels may be due to increased flux of heme across the cell envelope rather than differences in baseline Phrt promoter activation (Fig. 7B; Fig. S4; Table 3). To genetically raise endogenous heme levels, hemX was inactivated in both the Δstk1 and Δstp1 backgrounds and HssRS-dependent Phrt activation was assessed. A stk1 hemX double mutant has similar Phrt activation to the Δstk1 mutant, implying that deletion of stk1 is epistatic to the overproduction of heme caused by disruption of hemX (Fig. 7B; Fig. S4; Table 3). Again, disruption of hemX in a Δstp1 mutant background has the opposite phenotype, with high levels of Phrt activation; higher even, than a ΔhemX mutant (Fig. 7B; Fig. S4; Table 3). These results suggest that Stk1 and Stp1 work antagonistically to regulate GtrR activity and modulate heme levels in the cell.
Fig 7.
Stk1 and Stp1 impact heme stress signaling within S. aureus. (A) Growth as measured by OD600 was monitored over time for S. aureus strains grown in TSB containing either vehicle (i) or 15 µM heme (ii, iii). Prior to the measured growth, the strains were pre-grown to stationary phase in TSB containing vehicle (i, ii) or 2 µM heme (iii). (B) Total activation of luciferase expression from the hrtAB promoter (Phrt) from strains grown in TSB containing 2 mM δ-aminolevulinic acid (ALA), 2 µM heme, or vehicle. Time course data used to determine total promoter activation are graphed in Fig. S4. (A and B) Graphed are the averages of means from three independent experiments performed in biological triplicate ± SEM. Significance was calculated using two-way ANOVA with Tukey’s test for multiple comparisons using the comparisons listed in Table 3.
TABLE 3.
| Condition | Strains compared | Significance | |
|---|---|---|---|
| TSB | WT | ΔhssRS | ns |
| TSB | WT | ΔhemX | * |
| TSB | WT | Δstk1 | ns |
| TSB | WT | Δstk1 hemX::erm | ns |
| TSB | WT | Δstp1 | ns |
| TSB | WT | Δstp1 hemX::erm | **** |
| TSB | ΔhemX | Δstk1 hemX::erm | ** |
| TSB | ΔhemX | Δstp1 hemX::erm | **** |
| TSB | Δstk1 | Δstk1 hemX::erm | ns |
| TSB | Δstp1 | Δstp1 hemX::erm | **** |
| ALA | WT | ΔhssRS | **** |
| ALA | WT | ΔhemX | ** |
| ALA | WT | Δstk1 | **** |
| ALA | WT | Δstk1 hemX::erm | **** |
| ALA | WT | Δstp1 | *** |
| ALA | WT | Δstp1 hemX::erm | **** |
| ALA | ΔhemX | Δstk1 hemX::erm | ** |
| ALA | ΔhemX | Δstp1 hemX::erm | **** |
| ALA | Δstk1 | Δstk1 hemX::erm | ns |
| ALA | Δstp1 | Δstp1 hemX::erm | **** |
| Heme | WT | ΔhssRS | **** |
| Heme | WT | ΔhemX | ** |
| Heme | WT | Δstk1 | ns |
| Heme | WT | Δstk1 hemX::erm | ns |
| Heme | WT | Δstp1 | ns |
| Heme | WT | Δstp1 hemX::erm | **** |
| Heme | ΔhemX | Δstk1 hemX::erm | **** |
| Heme | ΔhemX | Δstp1 hemX::erm | **** |
| Heme | Δstk1 | Δstk1 hemX::erm | ns |
| Heme | Δstp1 | Δstp1 hemX::erm | **** |
Significance was calculated in Graphpad Prism using a two-way ANOVA with Tukey’s test for multiple comparisons test for multiple comparisons between the strain pairs listed in the table on the right with *P = 0.05; **P =0.01; ***P = 0.001; ****P < 0.0001; ns, non-significant.
We also used ALA supplementation to drive heme synthesis to determine whether Stk1 and Stp1 may regulate other steps in the heme synthesis pathway. The ΔhemX, Δstk1, Δstk1 hemX::erm, and Δstp1 mutants all have lower overall Phrt activation when treated with ALA compared to WT and the Δstp1 hemX::erm mutant (Fig. 7B; Fig. S4: Table 3). However, the comparison of time course data for these strains shows a similar peak in luminescent signal at 5 h, indicating that the magnitude of Phrt activation upon ALA supplementation is comparable between WT and the ΔhemX, Δstk1, Δstk1 hemX::erm, and Δstp1 mutants (Fig. S4). ALA treatment of the Δstk1 hemX::erm double mutant does not significantly increase the levels of Phrt activation compared to the Δstk1 mutant. The Δstp1 hemX::erm double mutant, however, has significantly higher Phrt activation upon ALA supplementation.
Stk1 and Stp1 impact heme levels in S. aureus
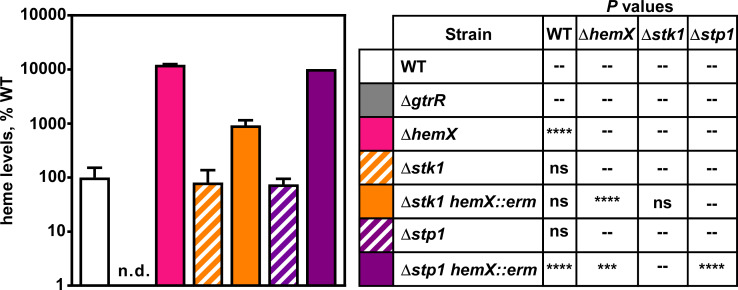
Since the heme-related phenotypes of Δstk1 and Δstp1 mutant strains differed between the heme adaptation experiments and luminescent Phrt reporter experiments, we measured heme levels using high-performance liquid chromatography (HPLC). Like what was observed in Phrt reporter experiments, deletion of either Δstk1 or Δstp1 does not significantly impact heme levels compared to WT (Fig. 8). The Δstp1 hemX::erm strain produces significantly more heme than the Δstp1 single mutant (Fig. 8). However, while the Δstk1 hemX::erm mutant produces slightly more heme than a Δstk1 mutant, both the Δstk1 hemX::erm and Δstp1 hemX::erm mutants produce significantly less heme compared to a ΔhemX mutant (Fig. 8). These data illustrate that deletion of stk1 and stp1 impact the regulation of heme synthesis by HemX.
Fig 8.
Stk1 is important for full heme synthesis in a hemX mutant. Heme was quantified by HPLC in S. aureus WT and mutant strains. Data are representative of the averages of means normalized to the amount of heme measured in WT from two independent experiments performed in biological triplicate ± SEM. “n.d.” indicates that no signal was detected. Significance was calculated in Graphpad Prism 8 using a two-way ANOVA with Welch’s test for multiple comparisons between the strain pairs listed in the table on the right with ***P = 0.001; ****P < 0.0001; ns, non-significant; --, comparison not made. No comparison was made between WT and ΔgtrR since Welch’s test cannot be performed on samples with identical values.
DISCUSSION
For optimal growth and survival, heme-utilizing organisms must maintain homeostatic levels of heme. This is accomplished by modulating heme acquisition, synthesis, degradation, and export (1). For organisms that initiate heme synthesis with the C5 pathway of ALA production, the enzyme GtrR represents a key point in the regulation of heme production (1, 28, 79). In many of these organisms, heme binding to GtrR impacts the regulation of GtrR, resulting in the post-translational regulation of GtrR protein abundance or GtrR activity (20, 30, 34, 80, 81).
Heme binding to GtrR often results in the modulation of GtrR protein abundance (1, 19, 33). In E. coli, the residue C170 of GtrR mediates heme binding and GtrRC170A accumulates in the cell, suggesting that heme binding targets GtrR for degradation (28). In H. vulgare, H99 is predicted to bind heme to enable heme-dependent regulation of GtrR activity (60). In this work, we identified two residues within S. aureus GtrR, H32 and R214, that mediate heme binding (Fig. 3), but disruption of heme binding has no significant impact on heme homeostasis (Fig. 4 and 5, S3).
Spectroscopic studies of heme bound to WT GtrR are suggestive of a hexacoordinated heme, meaning that two residues coordinate the iron contained within heme (Fig. S2). Out of the two residues found to be important for heme-GtrR binding (H32 and R214), H32 is likely one of the axial residues mediating the coordination of heme-iron, but arginine is not known to function in this way. While the amino acid residues flanking H32 (LRIAHEDLY) share properties with heme regulatory motifs identified by Kuhl et al. in a peptide-based screen for MRM, including lysine/alanine at the N-terminus and glutamic acid/aspartic acid at the C-terminus, this peptide was not identified as a strong heme-binding motif (82). Perhaps, upon heme binding, the NADPH binding and catalytic domains are assembled such that a non-adjacent amino acid serves as a second axial heme ligand. The formation of a clamp-like structure induced by heme binding could impact GtrR catalytic activity by bringing the two domains in closer proximity (83). This could also explain why substitution of alanine for H32 does not seem to have a heme binding-dependent phenotype in vivo as nearby residues such as Y36 could substitute for H32 in the heme-binding interaction. Because arginine is not likely an axial heme ligand and substitution of R214 with alanine in S. aureus does not have an appreciable impact on heme levels, we hypothesize that R214 forms a salt bridge with a propionate group within heme to increase the affinity of GtrR for heme (51, 84).
The expression of several GtrR protein variants in E. coli yielded little or no GtrR protein (Table 1). Like others (85), we found overexpression of GtrR to be difficult, with low yields after purification and a propensity of purified GtrR to precipitate out of solution. It is possible low yields could be due to conserved mechanisms of negative regulation of GtrR abundance between E. coli and S. aureus (85, 86). It is also possible that GtrR is unstable in solution alone since co-expression of GtrR from several species with the downstream enzyme GsaM improves protein yield and solubility of GtrR (17, 86). Finally, one of the mutants with poor yields could be more important for mediating negative regulation of GtrR by heme. This could drive heme synthesis to toxic levels or cause sequestration of aggregated GtrR to inclusion bodies. This would have led to the omission of the residue responsible from subsequent studies.
Although we demonstrate that GtrR binds heme in vitro, it is unknown if GtrR binds heme in vivo. Whether mutations sufficient to abrogate heme binding in vitro also disrupt in vivo heme binding remains an open question, particularly since strains expressing the GtrRH32A and GtrRR214A variants lack an appreciable phenotype. Since recombinant GtrR was not bound to heme upon purification from E. coli, we hypothesize that GtrR would not be bound to heme when immunoprecipitated from S. aureus. It is likely that the heme-GtrR interaction is weak since heme binding is not thought to play a structural or catalytic role for GtrR. We hypothesize that the primary function of heme binding is to impact GtrR stability since GtrR protein levels are elevated in a heme-deficient mutant (1). This does not exclude the possibility of heme regulation of GtrR activity, though, so future work could investigate the activity of GtrR in vitro.
In Salmonella enterica Serovar Typhimurium, heme binding to GtrR results in Lon- and ClpAP-mediated proteolytic degradation under heme-replete conditions (19, 33). This regulation relies on a stretch of positively charged amino acids on the C-terminus of GtrR, which are thought to target GtrR for proteolysis (33). Interestingly, we found that the addition of a C-terminal tag to GtrR in S. aureus impacts heme production, suggesting that a similar mechanism of proteolytic regulation may be occurring. S. aureus lacks a Lon protease but does encode Clp protease and adapter proteins. Trapping experiments identified GtrR as a substrate of the Clp protease in S. aureus, implying a similar mechanism of proteolytic regulation (87). Moreover, a hemX mutant accumulates GtrR, indicating that HemX may participate in the proteolytic regulation of GtrR, perhaps through the recruitment of the Clp protease (1, 18).
Eukaryote-type Ser/Thr protein kinases and phosphatases represent a conserved strategy used by bacteria to modulate enzyme and signaling activities in response to cell growth status (88). While there have been many reports of Stk1 and Stp1 modulating cellular signaling, Stk1 can also phosphorylate enzymes to modulate their activity (35, 89). For example, phosphorylation of PurA in S. aureus results in decreased enzymatic activity in vitro (41). We identified GtrR as a substrate of Stk1 (Fig. 6) and found that deletion of stk1 or stp1 impacts the sensitivity of S. aureus to heme toxicity (Fig. 7A). Additionally, in silico analysis of S. aureus GtrR identified multiple residues which could be phosphorylated by Stk1 (Table 2). However, activation of the Phrt promoter was comparable among the Δstk1 and Δstp1 mutants when measured using a reporter construct suggesting that differences in cell envelope integrity may be responsible for the growth difference in the presence of toxic levels of heme (Fig. 7B; Fig. S4) (77, 78).
To circumvent issues with envelope permeability, we tried to increase cellular heme levels by inactivating hemX, the gene encoding a negative regulator of GtrR abundance. In the Δstk1 hemX::erm mutant, we hypothesize that the protein levels of GtrR are high. However, without phosphorylation by Stk1, GtrR does not function optimally, so the deletion of stk1 is epistatic to the absence of HemX. Conversely, disruption of hemX in a Δstp1 background results in high levels of GtrR that is maximally phosphorylated resulting in extremely high heme levels and elevated HssRS signaling (Fig. 7B and 8, Fig. S4). Since supplementation of ALA does not circumvent deletion of stk1 or stp1 individually but does increase Phrt activation in the Δstp1 hemX::erm double mutant, it is likely that Stk1 and Stp1 also regulate other steps in heme synthesis (Fig. 7B). To our knowledge, neither Stk1 nor Stp1 impacts the expression of HssRS or HrtAB suggesting that the pair do not modulate HssRS signaling (41, 75, 90 - 93). This, combined with the differences in heme production in Δstk1 hemX::erm and Δstp1 hemX::erm double mutants, implies that Stk1 and Stp1 regulate heme synthesis rather than heme sensing or export.
Here, we report an instance of a heme-GtrR interaction that plays an unknown role in regulation of heme synthesis within the pathogen S. aureus. We have also identified Stk1 and Stp1 as regulators of heme synthesis, linking heme production with a system that responds to cell growth arrest. Further work is needed to define the function of heme binding to GtrR, the conditions in which Stk1 and Stp1 alter the phosphostatus of GtrR to modulate heme synthesis and to uncover the mechanistic link between cell envelope damage and heme synthesis.
MATERIALS AND METHODS
Bacterial strains, culturing methods, and reagents
Bacterial strains and plasmids (Table 4) and oligonucleotides (Table 5) are listed in the specified tables. S. aureus strains were grown on tryptic soy agar (TSA) or in broth (TSB) supplemented with antibiotics when appropriate. Prior to inoculation of liquid cultures, strains were streaked to agar plates and incubated for 24 h. Heme (hemin chloride, Cayman Chemical) was prepared fresh at a concentration of 10 mM in 0.1 N NaOH; for experiments in which heme was used, an equal volume of 0.1 N NaOH was used as a vehicle control. E. coli strains were grown on lysogeny broth (LB) or LB agar (LBA), supplemented with antibiotics when necessary. For standard S. aureus cultures, individual colonies were inoculated into 4 mL of TSB media in aerated 15-mL round-bottomed polypropylene tubes and incubated at a 45° angle in an Innova44 incubator shaking at 180 rpm for 15–16 h. Since heme can inhibit chloramphenicol acetyltransferase activity (83), strains harboring the pXen1 plasmid were grown in the presence of 10 µg/mL chloramphenicol during routine culture and diluted into media without antibiotics for growth and reporter experiments. For cloning and mutagenesis, all constructs were confirmed by Sanger sequencing (GeneWiz) and strains were confirmed using whole-genome sequencing (WGS, SeqCenter). Unless otherwise noted, all chemicals were from Sigma and molecular biology reagents were from New England Biolabs. Phusion 2X Hi-fidelity master mix was used for all cloning PCRs, and GoTaq Green Master Mix (Promega) was used for all screening PCRs.
TABLE 5.
Oligonucleotides used in this study
| Primer name | Primer sequence | Ref. |
|---|---|---|
| CSL032 | agcagtagaagctttattgcgtgtcacttc | This study |
| CSL033 | tcgtcccccacttttact | This study |
| CSL059 | attcgagctcggtacccggggatcctcaattcacaaaatgtgttg | This study |
| CSL059 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL059 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL063 | attcgagctcggtacccggggatccttattcaaaactaaagatacgtcg | This study |
| CSL209 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL209 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL209 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL209 | ggccgctgcatgcctgcaggtcgacttgaaacattttgatattcctactttag | This study |
| CSL210 | tagttttgaataagcatattaatatggtgataatatg | This study |
| CSL211 | aagatcttcttcactaataagtttttgttccatttgaaacgcccccatatatac | This study |
| CSL212 | gaacaaaaacttattagtgaagaagatcttcattttattgcaattagtataaatcatc | This study |
| CSL213 | aatatgcttattcaaaactaaagatacgtcg | This study |
| CSL214 | gtttcaaatgcattttattgcaattagtataaatcatc | This study |
| CSL215 | aagatcttcttcactaataagtttttgttcttcaaaactaaagatacgtcg | This study |
| CSL216 | gaacaaaaacttattagtgaagaagatctttaagcatattaatatggtgataatatg | This study |
| CSL217 | acgaattgccgcagaagatttatatgaaactaaatc | This study |
| CSL218 | aaggcatcatctctaaaag | This study |
| CSL219 | tgatcaaattgcaacaggtcgttactatattcaac | This study |
| CSL220 | acaacagcatatacttcag | This study |
| CSL223 | agtagtaaatgcaacaattgaaaatgctatg | This study |
| CSL224 | gtaatatcagtaattccagaac | This study |
| CSL225 | tcaagtgaaagcagatgaactatcatcattaccaaatttac | This study |
| CSL226 | tgctttgctgctaatttc | This study |
| CSL227 | ctttaattatgcagttgatgacttaaaaggtttag | This study |
| CSL228 | atgtttgtgatggcactaatac | This study |
| JC101 | gcggcatatggccatgcattttattgcaattagtat | This study |
| JC102 | gcggggatccgccttattcaaaactaaagatacgtc | This study |
| JC365 | aatttttaatgcactatttaaacaggcaattacttttgcaaaaag | This study |
| JC366 | gttcctgtcgtacctgtg | This study |
| JC369 | acataatgaagcagatatagctg | This study |
| JC370 | gctctttttgcaaaagtaattg | This study |
| JC387 | tgaaagtgcagcaattgtgattagttc | This study |
| JC388 | agtaaatttggtaatgatgatag | This study |
| JC389 | tgcacaatctgcaatcattacaaatgaaatgatag | This study |
| JC390 | ctcgttgaactaatcacaatatc | This study |
| JC408 | atatcaaatatattttgaaatagctc | This study |
| JC409 | tgaatgtcctgcagagcaagcgaagc | This study |
| JCO717 | gtgcttcaccagcaccacatgctg | (63) |
| JCO719 | ggtattagtttgagctgtcttggttcattgattgc | (63) |
| JC291 | gggcccgagcttaagact | (1) |
| JC292 | gatatcccctatagtgagtcgtattac | (1) |
| CSL285 | gactcactataggggatatcaatatggtctgtttggtggtg | This study |
| CSL286 | attatatttacatactttatcaccttcaatagcc | This study |
| CSL287 | tgataaagtatgtaaatataattgaagtaaatgtaccgag | This study |
| CSL288 | ccagtcttaagctcgggcccttaatatctaacctttctatttgaaatttc | This study |
| CSL293 | gttgagaagttaatcgaagc | This study |
| CSL294 | accatccataacatc | This study |
| CSL295 | cactataggggatatctcgagtagattcgattgatgaaatgac | This study |
| CSL296 | ccttcaatagcatttgtctttacctcgtttctac | This study |
| CSL297 | aagacaaatgctattgaaggtgataaagtatgatagg | This study |
| CSL298 | cttaagctcgggcccgatccagcacaatctttcgtgttg | This study |
| CSL301 | aaacactacccccttgtttggatccttatacatcatcatagctgacttc | This study |
| CSL302 | gagacataagccggagattaag | This study |
| JC155 | gggcggatccggctcaattcacaaaatgtgttgc | This study |
| JC157 | ggggggcatatgcaagaaaacctgtttattcgattcaatg | This study |
| CSL229 | ttatgatgttgcagacttaaaaggtttag | This study |
| CSL230 | ttaaagatgtttgtgatggc | This study |
PgtrR.gtrR.hemX chromosomal integration
Chromosomal complementation of ΔgtrR ΔhemX (ΔΔ) was performed by cloning PgtrR gtrR.hemX with N- or C-terminal myc tags into pJC1306, a vector that integrates at the S. aureus SaPI1 attachment site (63). For all constructs, pJC1306 was linearized with the restriction endonucleases BamHI and SalI. PCR amplicons were generated from S. aureus Newman genomic DNA using the primers listed in Table 5 and then inserted into digested pJC1306 using NEBuilder HiFi DNA Assembly Master Mix: pJC1306.PgtrR.gtrR.hemX (CSL209/059), pJC1306.PgtrR.myc-gtrR.hemX (CSL209/211, CSL212/213, CSL210/059), pJC1306.PgtrR.gtrR-myc.hemX (CSL209/213, CSL214/215, CSL216/059), pJC1306.PgtrR.myc-gtrR (CSL209/211, CSL212/063). Site-directed mutagenesis of pJC1306.PgtrR.myc-gtrR.hemX was performed using NEB’s Q5 SDM Kit according to the manufacturer’s instructions, using the mutagenic primers CSL217/218 (H32A) and CSL223/224 (R214A) (Table 5). pJC1306 plasmids were integrated into the chromosome of strain RN9011 as described previously (63) and then transduced into the S. aureus Newman ΔgtrR ΔhemX strain using the bacteriophage φ85, plating to TSA + 10 mg/mL tetracycline + 40 mM sodium citrate (1). Transductants were patched to TSA + tetracycline + sodium citrate and TSA + 10 µg/mL chloramphenicol + sodium citrate to confirm tetracycline resistance and chloramphenicol sensitivity to ensure loss of the helper plasmid harbored by RN9011 (19). Strains were screened for chromosomal integration using primers JCO719/JCO717 (63), and those with integrated pJC1306 were passaged 2× on TSA + sodium citrate to eliminate remaining phage particles. The integrity of final strains was assessed using WGS. The pXen1Phrt.lux reporter plasmid was moved into S. aureus strains by phage transduction as described above (1).
Genetic manipulation of S. aureus by allelic exchange and transduction
Deletion of stk1 and stp1 was performed as described previously with a few modifications (1, 101). pKOR1 plasmids containing approximately 1 kb regions flanking the gene to be deleted were constructed using NEB Hi-Fi assembly. The pKOR1 backbone was amplified by PCR using JC291/292, and the ~1 kb flanking regions including the start and stop codons ATG and TAA were amplified from S. aureus Newman genomic DNA using the following primer pairs: CSL285 + 286 (stk1 upstream), CSL287 + 288 (stk1 downstream), CSL295 + 296 (stp1 upstream), CSL297 + 298 (stp1 downstream). Upon confirmation of construct sequence, plasmids were electroporated into the shuttle strain RN4220 (63) and then moved by phage transduction into Newman WT. Resultant strains confirmed by PCR (stk1, 293 + 294; stp1, 301 + 302, Table 5) and WGS. The NARSA hemX::erm transposon allele was moved by phage transduction from JE2 into Newman Δstk1 and Δstp1 and strain integrity was confirmed using primers 155 + 157 (Table 5) (1, 99).
Bioinformatic analysis of S. aureus GtrR
Homologs of S. aureus GtrR across bacterial, archaeal, and eukaryotic species were identified, and protein sequences downloaded using NCBI’s Standard Protein BLAST and multiple sequence alignments of protein sequences were generated using Qiagen’s CLC Genomics Workbench (102). The resulting MSAs were input into the Consurf Server to score the conservation of each residue within S. aureus GtrR among the taxonomic levels (56 - 58). SWISS-MODEL was used to construct a homology model of a S. aureus GtrR homodimer using glutamyl-tRNA reductase from Arabidopsis thaliana as a template (PDBe: 5yjl.1.A) (61, 103 - 106). Using the MSAs generated, a phylogenetic tree was constructed using CLC Genomics Workbench (Qiagen) and domains and important catalytic residues were mapped onto the primary structure of S. aureus GtrR. To identify predicted phosphorylation sites within S. aureus GtrR, the primary sequence of GtrR was input into the NetPhosBac 1.0 server found at http://www.cbs.dtu.dk/services/NetPhosBac (76). For this analysis, a score above 0.5 is indicative of a predicted phosphosite (76).
Expression and characterization of recombinant GtrR
Protein expression plasmids
Protein expression plasmids for GtrR were generated by amplifying the S. aureus GtrR coding sequence flanked by BamHI and NdeI using primers JC101/102 and cloning into the multiple cloning site of pET15b after digestion (Table 5). Point mutant generation in pET15b.gtrR was performed using the Q5 SDM Kit using the mutagenic primers CSL217/218 (H32A), CSL219/220 (H63A), CSL032/033 (H99A), JC365/366(H140A), JC369/370 (T156A), CSL221/222 (Y167A), CSL223/224 (R214A), CSL225/226 (Y230A), JC387/388 (D244A), JC389/390 (Y255A), CSL227/228 (D299A), CSL229/230 (D301A), and JC409/409 (H425A) (Table 5). Constructs were confirmed by Sanger sequencing using T7 sequencing primers (GeneWiz).
GtrR protein expression and purification
A single colony of E. coli BL21 (DE3) pREL harboring pET15b.gtrR was inoculated into 5 mL of LB + 50 µg/mL carbenicillin + 30 µg/mL chloramphenicol and grown for 15 h at 37°C. Cells were diluted 1:100 into 1 L Terrific broth (ThermoFisher Scientific) + 50 µg/mL carbenicillin + 30 μg/mL chloramphenicol and grown at 37°C with shaking at 200 rpm to an OD600 = 0.8 prior the addition of isopropyl-1-thiol-d-galactopyranoside to 1 mM. Growth was continued at 30°C for 16 h after which cells were harvested by centrifugation (5,300 × g for 30 min) and then resuspended in lysis buffer (25 mL 50 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 10 µg/mL DNaseI, 0.2 mg/mL lysozyme). Cell suspensions were stirred for 30 min on ice and then lysed using a 550 Sonic Dismembrator (Fischer Scientific). Samples were sonicated 6× on ice at an amplitude of 4 for 30 s with 2 min in between each cycle. All steps following sonication were performed at 4°C. Intact cells were removed by centrifugation (11,000 × g for 30 min) and the supernatant was clarified by centrifugation (30,000 × g for 1 h). Twenty-five millilitersof clarified lysate was loaded onto 3 mL column volume (CV) equilibrated Nuvia IMAC Resin (BioRad) and rotated end over end for 30 min prior to transfer to a gravity column. The column was washed 2× with 5 CV of wash buffer (25 mL 50 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol) followed by elution with 2 CV of elution buffer (25 mL 50 mM Tris, pH 8.0, 300 mM NaCl, X mM imidazole, 10% glycerol) in a gradient from 40 to 150 mM imidazole. Purity was assessed using SDS-PAGE, and buffer exchange and protein concentration were performed on selected fractions using 10 kDa MWCO Amicon ultra centrifugal filters (Millipore). The dialysis buffer comprised 1 mM EDTA, 5% glycerol, 50 mM Tris pH 8. Purified protein was quantified by BCA, purity assessed using SDS-PAGE, and stored at −80°C in dialysis buffer.
Heme-binding experiments
Heme binding to GtrR was determined by measuring the absorption spectrum of increasing amounts of hemin (0–50 µM) in the presence or absence of 5 µM purified GtrR protein in 150 µL dialysis buffer in a 96-well flat-bottomed plate. Samples were incubated at room temperature (RT) in the dark for 5 min prior to reading the absorbance spectrum between 300 and 700 nm at 1 nm increments on an Epoch II microplate reader (BioTek) using the “scanning” function. For measurement of reduced spectra, dithiothreitol was added to a final concentration of 20 mM and the spectrum read again.
Complementation analysis and qualitative catalase assays
Overnight cultures of S. aureus strains Newman WT, katA::erm, ΔgtrR, ΔΔPgtrRmyc-gtrR.hemX, ΔΔPgtrRmyc-gtrR, ΔΔPgtrRgtrR-myc.hemX, ΔΔPgtrRmyc-gtrR(H32A).hemX, and ΔΔPgtrRmyc-gtrR(R214A).hemX were serially diluted and spotted on TSA (Table 4). After incubation at 37°C for 24 h, the more dilute spots were imaged to illustrate colony morphology and 2 µL of 30% hydrogen peroxide was added to the less dilute spots to qualitatively assess catalase activity.
Quantitative catalase assay
Overnight cultures of S. aureus strains were grown in TSB for 15 h at 37°C with shaking. The Cayman Chemical Catalase Assay Kit was used to quantitate catalase activity according to the kit’s instructions. For each measurement, 10 µL of overnight culture was used, and catalase activity was normalized to the optical density of the culture at 600 nm (OD600).
In vitro phosphorylation experiments
GST-tagged protein expression and purification
The stk1 kinase domain (1–348) was cloned into pGEX-2T expression vector in E. coli XL1-Blue and expressed in E. coli BL21 cells as previously described (100). Bacterial cells were pelleted by centrifugation, resuspended in PBS, and stored at −80°C for 15 h. Cells were thawed; supplemented with 25 µg/mL PMSF, 1 mg/mL lysozyme, and 60 µM E-64 protease inhibitor; and lysed by sonication. All steps after sonication were performed at 4°C. Cell debris was removed by centrifugation (3,220 × g for 45 min) and the cell lysate was filtered through a 0.2-µm PES membrane. The filtered lysate was loaded onto a prepacked Glutathione Sepharose 4 Fast Flow column (Sigma Millipore). The column was washed twice with 10 column volumes of cleavage buffer (25 mM Tris pH8, 100 mM NaCl, 1 mM DTT) before elution. Purified Stk1 was eluted with 1 mL of elution buffer (25 mM Tris pH 8, 100 mM NaCl, 1 mM DTT, 20 mM reduced glutathione) and dialyzed overnight in kinase buffer (50 mM Tris pH 7.4, 5 mM MgCl2). Stk1 purity was assessed by SDS-PAGE, yield quantified by BCA assay (Pierce), and stored at −80°C.
ATPγS phosphorylation assay and immunoblotting
In vitro phosphorylation assays were adapted by Allen et al. (107). Thirty microliters phosphorylation reactions were prepared with 0.1 µM GST-tagged Stk1 kinase and 5 µM purified GtrR, 1 mM ATPγS (Abcam), and kinase buffer (50 mM Tris pH 7.4, 5 mM MgCl2, 0.5 mM DTT). Reactions were incubated for 2 h at room temperature after which the reactions were quenched using 10 mM EDTA. After quenching, 1.5 µL of 50 mM p-nitrobenzyl mesylate (PNBM, Abcam) was added and the reactions were gently vortexed and incubated for an additional 2 h at room temperature. Negative control experiments were conducted under the same conditions without the addition of Stk1 kinase. Labeling reactions were terminated by the addition of 6× SDS loading buffer and the samples boiled for 10 min.
Two 15 µL aliquots of each sample were separated by SDS-PAGE in parallel, with one set stained with Imperial Protein Stain (ThermoFisher) and the second set transferred to a nitrocellulose membrane and subjected to immunoblotting. To detect PNBM labeling, proteins were transferred to a nitrocellulose membrane, washed in TBS-T (20 mM Tris pH 7.4, 150 mM NaCl, 0.05% Tween 20) 3× for 5 min each, and blocked with TBS-T + 5% skim milk for 20 min. The membrane was again washed with TBS-T and then incubated with a rabbit monoclonal anti-thiophosphate ester antibody (ab92570, Abcam) diluted 1:5,000 and a mouse anti-6× His tag monoclonal antibody (Invitrogen) diluted 1:5,000 in TBS-T + 5% skim milk for 15 h at 4°C. The membrane was washed with TBS-T and then incubated with DyLight 800 goat anti-rabbit IgG (ThermoFisher) and DyLight 680 rabbit anti-mouse IgG (Fisher Scientific) both diluted 1:10,000 in TBS-T + 5% skim milk for 1 h at room temperature. After a final wash with TBS-T, the membrane was imaged using a Li-Cor Odyssey XF and analyzed with Empiria Studio 2.3 software (Li-Cor).
Probing heme levels
Heme toxicity growth experiments
Freshly streaked bacterial colonies were used to inoculate TSB containing 2 µM heme or vehicle and grown for 15–16 h at 37°C with shaking. Cultures were diluted 1:100 into 200 µL TSB containing either vehicle or 15 µM heme in a 96-well round-bottomed plate. S. aureus growth was measured every 30 min in an Epoch II microplate reader (BioTek) using the OD600.
Luminescent HssRS reporter experiments
Overnight cultures of strains harboring luminescent Phrt reporter constructs were grown in 4 mL TSB + chloramphenicol were diluted 1:200 into 200 μl TSB + 2 mM ALA, 2 µM heme, or vehicle and growth followed by measuring OD600 and Phrt activation followed using relative luminescence every 30 min over 20 h. Total activation of Phrt over 20 h was quantified using Graphpad Prism 8 using area under the curve analysis of the luminescence normalized to OD600.
Heme quantitation sample prep
Overnight cultures of WT and mutant strains of S. aureus were grown in 10 mL TSB in 50 mL conical tubes at 37°C with shaking. After 16 h of growth, the OD600 of the cultures was recorded and the entire culture was pelleted by centrifugation at 11,500 × g for 10 min. Cell pellets were transferred to 15 mL conical tubes and incubated for 1 h in 500 µL digestion buffer (PBS + 10 mM MgCl2 + 40 µg/mL lysostaphin). Prior to incubation, a 50 µL aliquot was transferred to a 2 mL, screw-cap tube containing ~50 µL lysing matrix B (MP Biologicals) for protein quantitation. After digestion, aliquots were diluted in 200 µL PBS + 2% IGEPAL and the protoplasts were disrupted by bead beating for 45 s at 4.5 m/s in a MP FastPrep bead beater. Cellular debris and lysing matrix were pelleted by centrifugation at 15,000 × g for 5 min and protein content of the supernatant was measured using the Pierce BCA protein assay kit. Digested samples for heme extraction were sonicated 10 s on power level 2 using a Fisher Scientific 550 Sonic Dismembrator resting on ice after sonication. Samples were stored at −80°C until processing.
Heme quantitation by HPLC
Heme was extracted from samples with 5 mL ethyl acetate + 0.1% trifluoroacetic acid (TFA). Samples were turned end over end at RT for 30 min in the dark. Phase separation was accelerated by centrifugation at 11,500 × g for 15 min. Four milliliters of the organic (top) layer was transferred to a glass tube and dried under inert gas at RT. Samples were dissolved in 200 µL methanol and immediately analyzed on an Agilent 1260 Infinity HPLC. For quantitation, 5–10 µL sample or hemin standard (0, 1, 3, 10, 30 mM in MeOH) was separated using a Supelco Ascentis Express C18 (50 × 2.1 mm, 5 mm particles) column with a Phenomenex SecurityGuard (C18 cartridge 3.2 × 8 mm) guard column in place. Solvent A was 0.1% TFA in water, while solvent B was 0.1% TFA in acetonitrile. The flow rate was set at 0.4 mL/min at room temperature for a total run time of 20 min using the following successive linear gradient settings for run time in minutes versus B: 0.0, 25%; 0.5, 25%; 10.0, 100%; 16.0, 100%; 16.01, 25%; 20.0, 25%. Absorbance was detected at 210, 380, and 398 nm. Under these conditions, heme elutes around 4 min, and we use 398 nm absorbance for quantification. Peak area at ~4 min was calculated for each sample, scaled to account for the volume analyzed and heme loss during extraction, and normalized to the mass of protein in the sample.
ACKNOWLEDGMENTS
The authors are grateful to the Skaar lab for their thoughtful review of this manuscript.
The work presented was supported by the following sources of funding: R01AI069233 (E.P.S.), T32AI112541 (C.S.L.), F31AI126662/T32GM065086 (J.E.C.), AH830263 (C.M.G.), and R01AI137070/R21AI144060 (J.D.S.). The pJC1306 vector and S. aureus RN9011 intermediate cloning strain were a gift from Dr. Victor Torres. Strain NE1501 was obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH [Nebraska Transposon Mutant Library (NTML) Screening Array, NR-48501].
Contributor Information
John-Demian Sauer, Email: sauer3@wisc.edu.
Eric P. Skaar, Email: eric.skaar@vumc.org.
Michael J. Federle, University of Illinois Chicago, Chicago, Illinois, USA
DATA AVAILABILITY
All genomic data are available upon request.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00171-23.
Supplemental figures and figure legends.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Choby JE, Grunenwald CM, Celis AI, Gerdes SY, DuBois JL, Skaar EP, Kline KA, Richardson A, Heinrichs D. 2018. Staphylococcus aureus hemX modulates glutamyl-tRNA reductase abundance to regulate heme biosynthesis. mBio 9:e02287-17. doi: 10.1128/mBio.02287-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponka P. 1999. Cell biology of Heme. Am J Med Sci 318:241–256. doi: 10.1097/00000441-199910000-00004 [DOI] [PubMed] [Google Scholar]
- 3. Celis AI, DuBois JL. 2019. Making and breaking heme. Curr Opin Struct Biol 59:19–28. doi: 10.1016/j.sbi.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dailey HA, Dailey TA, Gerdes S, Jahn D, Jahn M, O’Brian MR, Warren MJ. 2017. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol Mol Biol Rev 81:e00048-16. doi: 10.1128/MMBR.00048-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stauff DL, Skaar EP. 2009. The heme sensor system of staphylococcus aureus. Contrib Microbiol 16:120–135. doi: 10.1159/000219376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 7. Ikuta KS, Swetschinski LR, Robles Aguilar G, Sharara F, Mestrovic T, Gray AP, Davis Weaver N, Wool EE, Han C, Gershberg Hayoon A, Aali A, Abate SM, Abbasi-Kangevari M, Abbasi-Kangevari Z, Abd-Elsalam S, Abebe G, Abedi A, Abhari AP, Abidi H, Aboagye RG, Absalan A, Abubaker Ali H, Acuna JM, Adane TD, Addo IY, Adegboye OA, Adnan M, Adnani QES, Afzal MS, Afzal S, Aghdam ZB, Ahinkorah BO, Ahmad A, Ahmad AR, Ahmad R, Ahmad S, Ahmad S, Ahmadi S, Ahmed A, Ahmed H, Ahmed JQ, Ahmed Rashid T, Ajami M, Aji B, Akbarzadeh-Khiavi M, Akunna CJ, Al Hamad H, Alahdab F, Al-Aly Z, Aldeyab MA, et al. 2022. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. 2007. A staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119. doi: 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakamura H, Hisano T, Rahman M, Tosha T, Shirouzu M, Shiro Y. 2022. Structural basis for heme detoxification by an ATP-binding cassette–type efflux pump in gram-positive pathogenic bacteria. Proc Natl Acad Sci U S A 119:e2123385119. doi: 10.1073/pnas.2123385119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP, Gilmore MS. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific Colonization of the vertebrate host. mBio 4:e00241-13. doi: 10.1128/mBio.00241-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cosgrove K, Coutts G, Jonsson I-M, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189:1025–1035. doi: 10.1128/JB.01524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nobre LS, Todorovic S, Tavares AFN, Oldfield E, Hildebrandt P, Teixeira M, Saraiva LM. 2010. Binding of azole antibiotics to Staphylococcus aureus flavohemoglobin increases intracellular oxidative stress. J Bacteriol 192:1527–1533. doi: 10.1128/JB.01378-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sapp AM, Mogen AB, Almand EA, Rivera FE, Shaw LN, Richardson AR, Rice KC. 2014. Contribution of the NOS-PDT operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLOS ONE 9:e108868. doi: 10.1371/journal.pone.0108868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X, Li W, Wang W, Wang S, Xu T, Chen J, Zhang W. 2021. Involvement of small colony variant-related heme biosynthesis genes in Staphylococcus aureus Persister formation in vitro. Front Microbiol 12:756809. doi: 10.3389/fmicb.2021.756809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skaar EP, Schneewind O. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect 6:390–397. doi: 10.1016/j.micinf.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 16. de Armas-Ricard M, Levicán G, Katz A, Moser J, Jahn D, Orellana O. 2011. Cellular levels of Heme affect the activity of dimeric glutamyl-tRNA reductase. Biochem Biophys Res Commun 405:134–139. doi: 10.1016/j.bbrc.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 17. Nardella C, Boi D, di Salvo ML, Barile A, Stetefeld J, Tramonti A, Contestabile R. 2019. Isolation of a complex formed between Acinetobacter baumannii hemA and HemL, key enzymes of tetrapyrroles biosynthesis. Front Mol Biosci 6:6. doi: 10.3389/fmolb.2019.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schröder I, Johansson P, Rutberg L, Hederstedt L. 1994. The hemX gene of the Bacillus subtilis hemAXCDBL Operon Encodes a membrane protein, negatively affecting the steady-state cellular concentration of Hema (glutamyl-tRNA reductase). Microbiology (Reading) 140 ( Pt 4):731–740. doi: 10.1099/00221287-140-4-731 [DOI] [PubMed] [Google Scholar]
- 19. Wang LY, Brown L, Elliott M, Elliott T. 1997. Regulation of heme biosynthesis in Salmonella typhimurium: activity of glutamyl-tRNA reductase (HemA) is greatly elevated during Heme limitation by a mechanism which increases abundance of the protein. J Bacteriol 179:2907–2914. doi: 10.1128/jb.179.9.2907-2914.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srivastava A, Beale SI. 2005. Glutamyl-tRNA reductase of chlorobium vibrioforme is a dissociable homodimer that contains one tightly bound heme per subunit. J Bacteriol 187:4444–4450. doi: 10.1128/JB.187.13.4444-4450.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. May BK, Dogra SC, Sadlon TJ, Bhasker CR, Cox TC, Bottomley SS. 1995. Molecular regulation of heme biosynthesis in higher vertebrates. Prog Nucleic Acid Res Mol Biol 51:1–51. doi: 10.1016/s0079-6603(08)60875-2 [DOI] [PubMed] [Google Scholar]
- 22. Layer G, Reichelt J, Jahn D, Heinz DW. 2010. Structure and function of enzymes in heme biosynthesis. Protein Sci 19:1137–1161. doi: 10.1002/pro.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamza I, Dailey HA. 2012. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta 1823:1617–1632. doi: 10.1016/j.bbamcr.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. 2015. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci U S A 112:2210–2215. doi: 10.1073/pnas.1416285112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nitzan Y, Salmon-Divon M, Shporen E, Malik Z. 2004. ALA induced photodynamic effects on gram positive and negative bacteria. Photochem Photobiol Sci 3:430–435. doi: 10.1039/b315633h [DOI] [PubMed] [Google Scholar]
- 26. Agrawal T, Avci P, Gupta GK, Rineh A, Lakshmanan S, Batwala V, Tegos GP, Hamblin MR. 2015. Harnessing the power of light to treat staphylococcal infections focusing on MRSA. Curr Pharm Des 21:2109–2121. doi: 10.2174/1381612821666150310102318 [DOI] [PubMed] [Google Scholar]
- 27. Bibb LA, Kunkle CA, Schmitt MP. 2007. The ChrA-ChrS and HrrA-HrrS signal Transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect Immun 75:2421–2431. doi: 10.1128/IAI.01821-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones AM, Elliott T. 2010. A purified mutant HemA protein from Salmonella enterica Serovar Typhimurium lacks bound Heme and is defective for heme-mediated regulation in vivo. FEMS Microbiol Lett 307:41–47. doi: 10.1111/j.1574-6968.2010.01967.x [DOI] [PubMed] [Google Scholar]
- 29. Ilag LL, Kumar AM, Söll D. 1994. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 6:265–275. doi: 10.1105/tpc.6.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pontoppidan B, Kannangara CG. 1994. Purification and partial characterisation of barley glutamyl-tRNAGlu reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur J Biochem 225:529–537. doi: 10.1111/j.1432-1033.1994.00529.x [DOI] [PubMed] [Google Scholar]
- 31. Schauer S, Lüer C, Moser J. 2003. Large scale production of biologically active Escherichia coli glutamyl-tRNA reductase from inclusion bodies. Protein Expr Purif 31:271–275. doi: 10.1016/s1046-5928(03)00184-0 [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Wilson S, Elliott T. 1999. A mutant hema protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J Bacteriol 181:6033–6041. doi: 10.1128/JB.181.19.6033-6041.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L, Elliott M, Elliott T. 1999. Conditional stability of the hemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol 181:1211–1219. doi: 10.1128/JB.181.4.1211-1219.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang WY, Huang DD, Stachon D, Gough SP, Kannangara CG. 1984. Purification, characterization, and fractionation of the Δ-Aminolevulinic acid synthesizing enzymes from light-grown Chlamydomonas reinhardtii cells. Plant Physiol 74:569–575. doi: 10.1104/pp.74.3.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pereira SFF, Goss L, Dworkin J. 2011. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev 75:192–212. doi: 10.1128/MMBR.00042-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pensinger DA, Schaenzer AJ, Sauer J-D. 2018. Do shoot the messenger: PASTA kinases as virulence determinants and antibiotic targets. Trends Microbiol 26:56–69. doi: 10.1016/j.tim.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galperin MY, Higdon R, Kolker E. 2010. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol Biosyst 6:721–728. doi: 10.1039/b908047c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Djorić D, Minton NE, Kristich CJ. 2021. The enterococcal PASTA kinase: a sentinel for cell envelope stress. Mol Oral Microbiol 36:132–144. doi: 10.1111/omi.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hardt P, Engels I, Rausch M, Gajdiss M, Ulm H, Sass P, Ohlsen K, Sahl H-G, Bierbaum G, Schneider T, Grein F. 2017. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int J Med Microbiol 307:1–10. doi: 10.1016/j.ijmm.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 40. Dworkin J. 2015. Ser/Thr phosphorylation as a regulatory mechanism in bacteria. Curr Opin Microbiol 24:47–52. doi: 10.1016/j.mib.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donat S, Streker K, Schirmeister T, Rakette S, Stehle T, Liebeke M, Lalk M, Ohlsen K. 2009. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol 191:4056–4069. doi: 10.1128/JB.00117-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen F, Di H, Wang Y, Peng C, Chen R, Pan H, Yang C-G, Liang H, Lan L. 2023. The enzyme activity of sortase a is regulated by phosphorylation in Staphylococcus aureus. Virulence 14:2171641. doi: 10.1080/21505594.2023.2171641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cluzel M-E, Zanella-Cléon I, Cozzone AJ, Fütterer K, Duclos B, Molle V. 2010. The Staphylococcus aureus autoinducer-2 synthase luxs is regulated by Ser/Thr phosphorylation. J Bacteriol 192:6295–6301. doi: 10.1128/JB.00853-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghiladi RA, Medzihradszky KF, Rusnak FM, Ortiz de Montellano PR. 2005. Correlation between isoniazid resistance and superoxide reactivity in mycobacterium tuberculosis Katg. J Am Chem Soc 127:13428–13442. doi: 10.1021/ja054366t [DOI] [PubMed] [Google Scholar]
- 45. Ishimori K, Watanabe Y. 2014. Unique heme environmental structures in heme-regulated proteins using heme as the signaling molecule. Chem Lett 43:1680–1689. doi: 10.1246/cl.140787 [DOI] [Google Scholar]
- 46. Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. 2010. Structure and reactivity of hexacoordinate hemoglobins. Biophys Chem 152:1–14. doi: 10.1016/j.bpc.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, Johnston AWB, Le Brun NE. 2010. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem 285:16023–16031. doi: 10.1074/jbc.M109.067215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trent JT, Hargrove MS. 2002. A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem 277:19538–19545. doi: 10.1074/jbc.M201934200 [DOI] [PubMed] [Google Scholar]
- 49. Arredondo-Peter R, Hargrove MS, Sarath G, Moran JF, Lohrman J, Olson JS, Klucas RV. 1997. Rice hemoglobins. gene cloning, analysis, and O2-binding kinetics of a recombinant protein synthesized in Escherichia coli. Plant Physiol 115:1259–1266. doi: 10.1104/pp.115.3.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L. 2001. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J Biol Chem 276:38949–38955. doi: 10.1074/jbc.M106438200 [DOI] [PubMed] [Google Scholar]
- 51. Brewitz HH, Hagelueken G, Imhof D. 2017. Structural and functional diversity of transient heme binding to bacterial proteins. Biochim Biophys Acta Gen Subj 1861:683–697. doi: 10.1016/j.bbagen.2016.12.021 [DOI] [PubMed] [Google Scholar]
- 52. Kühl T, Sahoo N, Nikolajski M, Schlott B, Heinemann SH, Imhof D. 2011. Determination of hemin-binding characteristics of proteins by a combinatorial peptide library approach. Chembiochem 12:2846–2855. doi: 10.1002/cbic.201100556 [DOI] [PubMed] [Google Scholar]
- 53. Shimizu T, Lengalova A, Martínek V, Martínková M. 2019. Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem Soc Rev 48:5624–5657. doi: 10.1039/c9cs00268e [DOI] [PubMed] [Google Scholar]
- 54. Smith LJ, Kahraman A, Thornton JM. 2010. Heme proteins-diversity in structural characteristics, function, and folding. Proteins 78:2349–2368. doi: 10.1002/prot.22747 [DOI] [PubMed] [Google Scholar]
- 55. Guallar V, Olsen B. 2006. The role of the heme propionates in heme biochemistry. J Inorg Biochem 100:755–760. doi: 10.1016/j.jinorgbio.2006.01.019 [DOI] [PubMed] [Google Scholar]
- 56. Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. Consurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–50. doi: 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. Consurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–33. doi: 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. 2013. Consurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem 53:199–206. doi: 10.1002/ijch.201200096 [DOI] [Google Scholar]
- 59. Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brody SS, Gough SP, Kannangara CG. 1999. Predicted structure and fold recognition for the glutamyl tRNA reductase family of proteins. Proteins 37:485–493. doi: [DOI] [PubMed] [Google Scholar]
- 61. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao A, Fang Y, Chen X, Zhao S, Dong W, Lin Y, Gong W, Liu L. 2014. Crystal structure of arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein. Proc Natl Acad Sci U S A 111:6630–6635. doi: 10.1073/pnas.1400166111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen J, Yoong P, Ram G, Torres VJ, Novick RP. 2014. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76:1–7. doi: 10.1016/j.plasmid.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Götz F. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol 179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Balwit JM, van Langevelde P, Vann JM, Proctor RA. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J Infect Dis 170:1033–1037. doi: 10.1093/infdis/170.4.1033 [DOI] [PubMed] [Google Scholar]
- 66. Juárez-Verdayes MA, González-Uribe PM, Peralta H, Rodríguez-Martínez S, Jan-Roblero J, Escamilla-Hernández R, Cancino-Diaz ME, Cancino-Diaz JC. 2012. Detection of hssS, hssR, hrtA, and hrtB genes and their expression by hemin in Staphylococcus epidermidis. Can J Microbiol 58:1063–1072. doi: 10.1139/w2012-086 [DOI] [PubMed] [Google Scholar]
- 67. Mike LA, Dutter BF, Stauff DL, Moore JL, Vitko NP, Aranmolate O, Kehl-Fie TE, Sullivan S, Reid PR, DuBois JL, Richardson AR, Caprioli RM, Sulikowski GA, Skaar EP. 2013. Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc Natl Acad Sci U S A 110:8206–8211. doi: 10.1073/pnas.1303674110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Keppel M, Piepenbreier H, Gätgens C, Fritz G, Frunzke J. 2019. Toxic but tasty - temporal dynamics and network architecture of heme-responsive two-component signaling in corynebacterium glutamicum. Mol Microbiol 111:1367–1381. doi: 10.1111/mmi.14226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stauff DL, Torres VJ, Skaar EP. 2007. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J Biol Chem 282:26111–26121. doi: 10.1074/jbc.M703797200 [DOI] [PubMed] [Google Scholar]
- 70. Surdel MC, Dutter BF, Sulikowski GA, Skaar EP. 2016. Bacterial nitric oxide synthase is required for the Staphylococcus aureus response to heme stress. ACS Infect Dis 2:572–578. doi: 10.1021/acsinfecdis.6b00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wakeman CA, Stauff DL, Zhang Y, Skaar EP. 2014. Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J Bacteriol 196:1335–1342. doi: 10.1128/JB.01067-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Standish AJ, Teh MY, Tran ENH, Doyle MT, Baker PJ, Morona R. 2016. Unprecedented abundance of protein tyrosine phosphorylation modulates Shigella flexneri virulence. J Mol Biol 428:4197–4208. doi: 10.1016/j.jmb.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 73. Yang M, Qiao Z, Zhang W, Xiong Q, Zhang J, Li T, Ge F, Zhao J. 2013. Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium synechococcus sp. strain PCC 7002. J Proteome Res 12:1909–1923. doi: 10.1021/pr4000043 [DOI] [PubMed] [Google Scholar]
- 74. Bäsell K, Otto A, Junker S, Zühlke D, Rappen G-M, Schmidt S, Hentschker C, Macek B, Ohlsen K, Hecker M, Becher D. 2014. The phosphoproteome and its physiological dynamics in Staphylococcus aureus. Int J Med Microbiol 304:121–132. doi: 10.1016/j.ijmm.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 75. Prust N, van der Laarse S, van den Toorn HWP, van Sorge NM, Lemeer S. 2021. In-depth characterization of the Staphylococcus aureus phosphoproteome reveals new targets of Stk1. Mol Cell Proteomics 20:100034. doi: 10.1074/mcp.RA120.002232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Miller ML, Soufi B, Jers C, Blom N, Macek B, Mijakovic I. 2009. Netphosbac - a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics 9:116–125. doi: 10.1002/pmic.200800285 [DOI] [PubMed] [Google Scholar]
- 77. Beltramini AM, Mukhopadhyay CD, Pancholi V. 2009. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun 77:1406–1416. doi: 10.1128/IAI.01499-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liebeke M, Meyer H, Donat S, Ohlsen K, Lalk M. 2010. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: involvement in cell wall biosynthesis. Chem Biol 17:820–830. doi: 10.1016/j.chembiol.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 79. Schröder I, Hederstedt L, Kannangara CG, Gough P. 1992. Glutamyl-tRNA reductase activity in Bacillus subtilis is dependent on the hemA gene product. Biochem J 281 ( Pt 3):843–850. doi: 10.1042/bj2810843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Moser J, Lorenz S, Hubschwerlen C, Rompf A, Jahn D. 1999. Methanopyrus kandleri glutamyl-tRNA reductase. J Biol Chem 274:30679–30685. doi: 10.1074/jbc.274.43.30679 [DOI] [PubMed] [Google Scholar]
- 81. Rieble S, Beale SI. 1991. Purification of glutamyl-tRNA reductase from synechocystis sp. PCC 6803. J Biol Chem 266:9740–9745. [PubMed] [Google Scholar]
- 82. Kühl T, Imhof D. 2014. Regulatory FeII/III heme: the reconstruction of a molecule's biography. Chembiochem 15:2024–2035. doi: 10.1002/cbic.201402218 [DOI] [PubMed] [Google Scholar]
- 83. Brewitz HH, Goradia N, Schubert E, Galler K, Kühl T, Syllwasschy B, Popp J, Neugebauer U, Hagelueken G, Schiemann O, Ohlenschläger O, Imhof D. 2016. Heme interacts with histidine- and tyrosine-based protein motifs and inhibits enzymatic activity of chloramphenicol acetyltransferase from Escherichia coli. Biochim Biophys Acta 1860:1343–1353. doi: 10.1016/j.bbagen.2016.03.027 [DOI] [PubMed] [Google Scholar]
- 84. Moriwaki Y, Terada T, Caaveiro JMM, Takaoka Y, Hamachi I, Tsumoto K, Shimizu K. 2013. Heme binding mechanism of structurally similar iron-regulated surface determinant near transporter domains of Staphylococcus aureus exhibiting different affinities for heme. Biochemistry 52:8866–8877. doi: 10.1021/bi4008325 [DOI] [PubMed] [Google Scholar]
- 85. Verkamp E, Jahn M, Jahn D, Kumar AM, Söll D. 1992. Glutamyl-tRNA reductase from Escherichia coli and synechocystis 6803. gene structure and expression. J Biol Chem 267:8275–8280. [PubMed] [Google Scholar]
- 86. Lüer C, Schauer S, Möbius K, Schulze J, Schubert W-D, Heinz DW, Jahn D, Moser J. 2005. Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J Biol Chem 280:18568–18572. doi: 10.1074/jbc.M500440200 [DOI] [PubMed] [Google Scholar]
- 87. Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, Ingmer H, Frees D. 2013. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res 12:547–558. doi: 10.1021/pr300394r [DOI] [PubMed] [Google Scholar]
- 88. Ohlsen K, Donat S. 2010. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol 300:137–141. doi: 10.1016/j.ijmm.2009.08.016 [DOI] [PubMed] [Google Scholar]
- 89. Jarick M, Bertsche U, Stahl M, Schultz D, Methling K, Lalk M, Stigloher C, Steger M, Schlosser A, Ohlsen K. 2018. The serine/threonine kinase Stk and the phosphatase Stp regulate cell wall synthesis in Staphylococcus aureus. Sci Rep 8:13693. doi: 10.1038/s41598-018-32109-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. 2008. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial ser/thr/tyr phosphorylation. Mol Cell Proteomics 7:299–307. doi: 10.1074/mcp.M700311-MCP200 [DOI] [PubMed] [Google Scholar]
- 91. Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium bacillus subtilis. Mol Cell Proteomics 6:697–707. doi: 10.1074/mcp.M600464-MCP200 [DOI] [PubMed] [Google Scholar]
- 92. Soufi B, Gnad F, Jensen PR, Petranovic D, Mann M, Mijakovic I, Macek B. 2008. The ser/thr/tyr phosphoproteome of Lactococcus lactis Il1403 reveals multiply phosphorylated proteins. Proteomics 8:3486–3493. doi: 10.1002/pmic.200800069 [DOI] [PubMed] [Google Scholar]
- 93. Liang C, Rios-Miguel AB, Jarick M, Neurgaonkar P, Girard M, François P, Schrenzel J, Ibrahim ES, Ohlsen K, Dandekar T. 2021. Staphylococcus aureus transcriptome data and metabolic modelling investigate the interplay of ser/thr kinase PknB, its phosphatase Stp, the glmR/yvcK Regulon and the cdaA Operon for metabolic adaptation. Microorganisms 9:2148. doi: 10.3390/microorganisms9102148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. DUTHIE ES, LORENZ LL. 1952. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol 6:95–107. doi: 10.1099/00221287-6-1-2-95 [DOI] [PubMed] [Google Scholar]
- 95. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 96. Yang JE, Choi YJ, Lee SJ, Kang K-H, Lee H, Oh YH, Lee SH, Park SJ, Lee SY. 2014. Metabolic engineering of Escherichia coli for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from glucose. Appl Microbiol Biotechnol 98:95–104. doi: 10.1007/s00253-013-5285-z [DOI] [PubMed] [Google Scholar]
- 97. Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi S-H, Couloux A, Lee S-W, Yoon SH, Cattolico L, Hur C-G, Park H-S, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21 (DE3). J Mol Biol 394:644–652. doi: 10.1016/j.jmb.2009.09.052 [DOI] [PubMed] [Google Scholar]
- 98. Kreiswirth BN, Löfdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 99. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW, Bush K. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schaenzer AJ, Wlodarchak N, Drewry DH, Zuercher WJ, Rose WE, Striker R, Sauer J-D. 2017. A screen for kinase inhibitors identifies antimicrobial imidazopyridine aminofurazans as specific inhibitors of the Listeria monocytogenes PASTA kinase PrkA. J Biol Chem 292:17037–17045. doi: 10.1074/jbc.M117.808600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 102. McEntyre J, Ostell J. 2002. The NCBI Handbook. National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 103. Zhao A, Han F. 2018. Crystal structure of Arabidopsis thaliana glutamyl-tRNAGlu reductase in complex with NADPH and glutamyl-tRNAGlu reductase binding protein. Photosynth Res 137:443–452. doi: 10.1007/s11120-018-0518-8 [DOI] [PubMed] [Google Scholar]
- 104. Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. 2017. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep 7:10480. doi: 10.1038/s41598-017-09654-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T. 2017. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res 45:D313–D319. doi: 10.1093/nar/gkw1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-Pdb Viewer: a historical perspective. Electrophoresis 30 Suppl 1:S162–73. doi: 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- 107. Lo H-C, Hollingsworth NM. 2011. Using the semi-synthetic epitope system to identify direct substrates of the meiosis-specific budding yeast kinase, Mek1. Methods Mol Biol 745:135–149. doi: 10.1007/978-1-61779-129-1_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and figure legends.
Data Availability Statement
All genomic data are available upon request.