SUMMARY
Microbiomes have highly important roles for ecosystem functioning and carry out key functions that support planetary health, including nutrient cycling, climate regulation, and water filtration. Microbiomes are also intimately associated with complex multicellular organisms such as humans, other animals, plants, and insects and perform crucial roles for the health of their hosts. Although we are starting to understand that microbiomes in different systems are interconnected, there is still a poor understanding of microbiome transfer and connectivity. In this review we show how microbiomes are connected within and transferred between different habitats and discuss the functional consequences of these connections. Microbiome transfer occurs between and within abiotic (e.g., air, soil, and water) and biotic environments, and can either be mediated through different vectors (e.g., insects or food) or direct interactions. Such transfer processes may also include the transmission of pathogens or antibiotic resistance genes. However, here, we highlight the fact that microbiome transmission can have positive effects on planetary and human health, where transmitted microorganisms potentially providing novel functions may be important for the adaptation of ecosystems.
KEYWORDS: microbiome interconnectedness, microbiome transfer
INTRODUCTION
Microorganisms are (almost) everywhere on our planet and provide critical contributions to the establishment and functioning of terrestrial, marine, and freshwater environments. Furthermore, eukaryotes, including plants and animals, are typically associated with complex microbial communities that are pivotal for health and functioning of their host. These microbial communities are also referred to as microbiomes, defined as assemblages of bacteria, archaea, fungi, viruses, protozoans, and other microeukaryotes, as well as their activities in the context of a given (a)biotic habitat (1). Microorganisms drive local and global elemental cycles on our planet; for example, they determine soil fertility at a local scale, but also drive and react to changes acting at a global scale, such as greenhouse gas emissions, climate change, and climate change mitigation. Microbiomes play an essential role in many elements of our society, such as the microbiomes associated particularly with the digestive tract of humans and other animals, in the treatment of domestic, agricultural, and industrial waste streams, in fermentative food production, and in the biotechnological production of bulk and fine chemicals (2). Hence, microbiomes occupy and shape the vast array of ecological niches available in natural and engineered environments. The microbial composition and functional capacity in many of these environments is a major theme of current research, often with the goal of understanding the contributions of microbiomes to the functioning and health of these environments. Although it is widely accepted that microorganisms are transmitted between ecosystems, microbial connections between ecosystems have not yet been explored at large scale, at least in part due to fragmentation of resources (3, 4). Such interconnectivity is now recognized in what has been coined the One Health approach (5, 6). Nevertheless, to date this approach has often focused on negative aspects of microbial transmission, such as the spread of (zoonotic) pathogens and antibiotic resistance genes or organisms related to the production of molecules with adverse activities, such as mycotoxins (7). In contrast, the potential and extent of more positive aspects of microbial transmission have not been addressed with equal attention, even though their impact and importance were recently emphasized (5). Such knowledge, however, and particularly quantitative aspects of microbial transmission routes as well as the conditions that determine these, would be essential for the optimization and/or de novo design of microbiome-inspired intervention strategies that can allow safer, more sustainable, and healthier food and feed production (3). To this end, this review will provide an up-to-date summary of our current understanding of microbial transmission within and across different environments (Fig. 1), including both the supportive and negative aspects of microbiome transmission and circularity. This analysis is illustrated with examples from the different domains within food production systems and beyond. We conclude with predictions of the future directions needed to exploit microbiomes to their full potential.
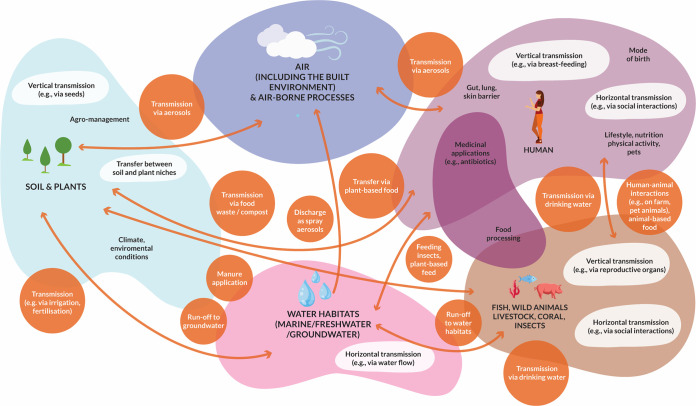
FIG 1.
Microbiome transfer between environments and modes of transfer.
MICROBIOME TRANSFER AND INTERACTIONS IN ENVIRONMENTAL MICROBIOMES
The Soil-Plant Continuum
The soil-plant system represents a continuum of microorganisms, which are able to survive both in the plant and in the soil environment and may be exchanged between the two. Because of this close relationship, the type of vegetation, soil management practices or environmental conditions greatly influence microbiome diversity and composition of soil as well as of plant-associated microbiomes (8, 9). This connection has been extensively investigated, although connectivity over large distances or to environments beyond the soil-plant system is less well understood.
Dispersal of microorganisms or microbiomes may occur locally, within a field or site, as well as more widely between different environments. Typical dispersal routes in the soil environment include dispersal from the air above the vegetation, from nearby vegetation and leaf litter near the soil surface or from litter below the top layer (10). Microbial dispersal may also occur via pollen, seeds, or soil-associated animals or mobile fungi (11, 12). Different dispersal routes transport distinct microbial communities that differentially influence microbiota composition in the recipient environment (6). Dispersed microorganisms may establish over the long term or may only exist transiently, depending on the microbiome already present (5, 13, 14). However, even transient invaders may drive microbial community shifts (15).
Soils serve as major reservoirs of plant-associated microbiota comprising plant beneficial, neutral, or pathogenic microorganisms. Particularly prominent is the symbiosis of plants with mycorrhizal fungi or of legumes with nitrogen-fixing rhizobia. Other beneficial microorganisms may have direct effects, e.g., by mobilizing and providing important plant nutrients, alleviating plant stress (such as drought), or by protecting plants from pests and pathogens through competition, antibiosis, or the production of enzymes or metabolites (16). Indirect benefits include, for example, the induction of plant responses leading to improved resistance to pathogens. The soil microbiome, the environmental parameters, as well as the physiology of plants all determine which microorganisms are transferred to and establish within and upon plants.
Within the plant, there are multiple niches enabling the growth of diverse microbial communities. In roots, for example, a microbial continuum extends from the rhizosphere soil to the rhizoplane and different niches within the endosphere. Microorganisms colonizing the plant endosphere can comprise obligate or facultative endophytes (17). The latter are often environmentally derived, utilizing the presence of a compatible plant tissue as an interim habitat and resource rather than being dependent on it. Thus, depending on the plant species and genotype, microbial properties, and environmental conditions, different subsets of rhizosphere microbial communities enter and colonize roots as endophytes (18).
A range of formal interactions and opportunistic events enable rhizosphere microorganisms to reach inner root tissues (19). These include intricate “chemical dialogues” between the plant and compatible microorganisms (e.g., legumes and rhizobia) that lead to modification of the host and microorganisms, colonization of root hairs, and formation of new organs. Less formally, compatible or opportunistic microbes can enter root systems through cracks (e.g., when lateral roots form) or by cell wall degradation (18). Once inside the plant, microorganisms can disseminate to below- and above-ground tissues by colonizing the apoplast or the vascular system. Overall, a plethora of opportunities exist for members of the soil microbiome to enter and colonize plant root systems, spread within the plant, and even be disseminated to new environments and generations of plants by movement of pollen, seed, or other tissues.
Plant Seeds as Vehicles of Microbiota Transmission
Some endophytes colonize reproductive organs such as flowers, fruits, and seeds (20), the latter in particular, being increasingly recognized as habitats for functionally important microorganisms. Microorganisms colonizing seeds and the spermosphere, i.e., the area around the germinating seed, can improve germination and increase seedling vigor, but also protect seeds against rotting or the emerging seedling against disease (21–23). Seed microorganisms are, to a great extent, horizontally acquired, as many of them derive from the soil environment (23, 24), where soil microorganisms colonize and then enter roots and then systemically colonize plant tissues and seeds. In addition, microorganisms from alternative sources (e.g., insects, air, and rain [25, 26] may colonize reproductive and disseminative plant organs by using stems, flowers, or fruits horizontally [27–29]). The colonization of pollen grains by microorganisms may result in the subsequent colonization of the ovule and the seed after pollination (30).
The vertical transmission of seed microbiota has been increasingly identified as an important route for delivering microorganisms to the next generation plants, especially at early vegetation stages. Well-known examples of vertical transmission of plant endophytes are members of the fungal genus Epichloë (Neotyphodium for anamorphs) (31). Similarly, the vertical transmission or microbial inheritance of bacteria has received considerable attention. Abdelfattah et al. (32) identified two consecutive stages of vertical transmission – from parents to seeds and from seeds to seedlings. The authors also defined a third stage, i.e., the phase of seed dormancy, which requires that microorganisms are able to survive the harsh conditions of limited nutrient and water availability. Vertical transmission of seed microbiomes has been demonstrated in many different plant species, including maize, rice, wheat, barley, sugarcane, soybean, tomato, and oak, as well as in model plants (11, 24, 25, 33–35). A survey of seed microbiomes and their transmission routes in several monocot and dicot plant species showed that the bacterial family Enterobacteriaceae, particularly members of the genera Pantoea, Enterobacter, Klebsiella, and Massilia, are vertically transmitted (13, 24). However, it was shown that pathogenic Escherichia coli was not able to penetrate seed embryonic tissue, neither via the parental vascular tissue, nor via the flower receptacle (36). Among fungal endophytes, nonpathogenic Fusarium and Alternaria were commonly vertically transmitted. Seeds may be widely dispersed (e.g., by wind, water, or animals) in nature and are much more widely disseminated in the frame of agricultural systems going hand in hand with a wide dissemination of seed-borne microbiomes.
The Plant Phyllosphere and Exchange of Airborne Microbiota
The phyllosphere refers to the plant leaf as a microbiome habitat. The phyllosphere is an open system which is exposed to, and thereby connected with, the surrounding environment (37). As such, microbial immigration to the phyllosphere can originate from multiple sources, both local and remote. Importantly, arrival of microorganisms from the surrounding environment potentially represents a constant flow of new microorganisms (38), where areas such as agricultural and horticultural land, forests, grasslands, and even urban environments are sources of microbial inocula (39, 40). Overall, the phyllosphere community composition is therefore the outcome of multiple factors, such as host-based selection, priority effects, natural successional processes, and stochastic influences (41). Given its exposure to the environment, the composition of the phyllosphere may be in a constant state of flux.
While environmental sources of phyllosphere microbiomes can be diverse, two are of particular importance: (i) the local vegetation, including both living plants and decomposing plant material such as leaf-litter or fallen wood, and (ii) the soil. Movement of the microbiome from these sources to the phyllosphere can occur via direct physical interactions, for example, movement and contact of leaves across the surfaces of adjacent plants (42), or via transmission vectors such as invertebrates or other animals. However, air transport is often the primary mechanism for movement of material from neighboring environments to the phyllosphere, particularly when wind combined with other disturbances, such as rain droplets, drive microorganisms into the atmosphere (43–46).
The efficiency and randomness of airborne transport provide an effective way for dispersal and exchange of plant-associated microbiomes. This has been well characterized for foliar plant pathogens (38, 44). Indeed, movement of microbiome members from plant surfaces into the air, and then transport within the air column, is an effective means to overcoming geographic barriers. Bacteria, because of their small size, may have extended atmospheric residence times and, thereby, have potential for long distance transport (see below). However, bacterial cells are often clumped, and/or attached to plant fragments such as leaf material. While this is anticipated to protect their viability during transport, it also limits potential dispersive capability (47).
Water droplets are important for microbial transport into and subsequent survival within the atmosphere. Using population genomics, Monteil et al. (48) demonstrated that Pseudomonas sp. strains pathogenic to cantaloupe plants could be identified within the atmospheric water cycle (e.g., rain and snow). Similarly, rain has also been shown to be a key reservoir of phyllosphere microbiota for other plant species (e.g., tomato) (49).
It is increasingly apparent there is a reciprocal connection between the phyllobiome and the atmospheric microbiome, driven by atmospheric processes occurring at global scale. The phyllosphere is theoretically immense: globally, plant leaf surface area (adaxial and abaxial) is estimated between 2×108 and 1×109 km2 (50). For perspective, this is up to twice the Earth’s entire surface area. On these leaves, and directly exposed to the atmosphere, some 1024 to 1026 microbial cells are thought to be present (38) and therefore potentially accessible to atmospheric transport.
Understanding the microbiomes of these environments and the processes that affect reciprocal exchange, assembly and function of these microbiomes, is critical to understanding plant health and agriculture. Indeed, it is time to consider these compartments – the microbiome of the plant (and other terrestrial or aquatic organisms) and the atmosphere – as a singular holobiome, where especially the plants, microorganisms, and the atmosphere have coevolved and are therefore to some extent interdependent.
Airborne Transport of Microorganisms
Microorganisms are transported long distances by aeolian (wind) processes (51, 52). Transport can be as autonomous bodies such as fungal spores, as cells adsorbed to the external and internal surfaces of mineral particles (dust), or in liquid microbodies (aerosols) (46, 53). Both the extent of transport and the distance transported are likely to be affected by both the particle size of the dust, and cell body size of the microorganism (54). Dust plumes generated from farmlands, drylands and deserts can transport microorganisms hundreds or thousands of kilometers from their emissive source (51). Dust can rise to very high altitudes within the Earth’s troposphere (as high as 38 km above sea-level) and persist in the atmosphere for long periods (55).
The air masses of the Earth impose some constraints on long distance aeolian transport. The major tropospheric air masses circulate in the northern and southern hemispheres, with limited mixing at the equatorial boundary (56). This barrier might limit transfer of particulate material, aerosols, and microbial cells between the two hemispheres (57). Another barrier is the limited vertical mixing above the troposphere due to thermal inversion (58). Similarly, the westerly airflows over the Southern Ocean probably limit transfer of aerial particulates from the lower latitudes of the Southern Hemisphere to the Antarctic continent.
While the process of aeolian dissemination and deposition of microorganisms is recognized (59), the ecological consequences of these processes are still being determined. Microbial activities in the atmosphere impact cloud formation, hydrologic cycles (60), atmospheric chemistry, and processes integral to climate regulation (61). Cloud condensation and ice nucleation, for example, are common traits across a range of bacteria, fungi, and other microorganisms, particularly those associated with plants (62). Furthermore, there is evidence that plant, animal, and human pathogens are present in long-distance aeolian transported microbiomes (63, 64). These also might be associated with disease outbreaks (65). For instance, a recent study by Björnham et al. (64) showed that the foot-and-mouth disease virus could be transmitted over distances of up to 50 km. Dust-associated microbiomes harboring diverse antibiotic resistance genes have also been detected downwind (c.f. upwind) of cattle feed yards (albeit from near-surface sampling) (66). Human pathogen signals have also been detected in aerial microbiomes (63). These include the presence of, e.g., Neisseria meningitides in Saharan dust (67), which was associated with an outbreak of meningococcal meningitis in Barcelona (68). Microorganisms are not only dispersed via bioaerosols, but have also shown to be metabolically active (69, 70).
Fungal taxa are prevalent in the atmosphere due to their resilience and ease of airborne (spore and conidia) transmission, and several fungal pathogens have been detected in atmosphere microbiomes. This includes plant pathogens such as Blumeria graminis (corn mildew disease) and Sclerotinia sclerotiorum (white mold) (71, 72). Fungal allergens such as those found in species of Aspergillus and Alternaria have also been detected at high concentrations in dust (73, 74), and the presence of these has been linked to increased risk of respiratory diseases (75). Despite these reports on the transfer of pathogens, there is little understanding on the transfer of nonpathogenic microorganisms via aeolian transport but this is very likely to occur. Phyllosphere inhabitants, including plant beneficial microorganisms may be lifted into the atmosphere and transported over long distances and return to phyllosphere habitats. Overall, aeolian transport may mediate long-distance transfer of microbiomes with potential impact on weather, as well as plant, animal, and human health.
Insects and Microorganism Transmission
Hundreds of microbial symbioses are known to exist with insects. Many of those are ancient, extending back 30 to 250 million years (76). Given the time for coevolution, it is not surprising that the symbionts span a range of different relationships, from transient pathogens to obligate, highly specialized mutualists (77). Microorganisms play diverse and unexpected roles in the functioning and life of insects such as allowing host diet specialization (78), provision of specific nutrients or detoxification of chemicals (79, 80), as well as enhancing resistance to pathogens and parasitoids (81 and references therein). Generally, given the diversity of insects and the importance of microbial endosymbionts in conferring many traits and impacting their fitness (81), we have not yet even begun to realize the full extent of microbiome symbiosis among insects.
Many microbial symbionts, especially those colonizing reproductive organs, are inherited via vertical transmission. These are also termed primary endosymbionts. Vertical transmission via eggs is common in endosymbionts such as Wolbachia spp., Ricksettia spp., Spiroplasma spp., Buchnera spp., certain yeast-like symbionts, protists, and viruses (77). There is increasing interest in understanding the role of microbial-microbial interactions in vertical transmission. For instance, the type of hereditary microorganisms positioning themselves first in the next generation of offspring may influence community assembly and composition (priority effects) of other microorganisms, thereby influencing offspring health and evolutionary fitness (77, 82).
Secondary endosymbionts are often facultative symbionts. These may be transmitted vertically as well as horizontally (83). They colonize different tissues and organs of their hosts, such as the gut system, muscles or the hemocoel, and usually show lower abundances than primary endosymbionts. Primary endosymbionts are highly adapted to their hosts, whereas secondary endosymbionts may be transferred intra- and interspecifically (84). Horizontal transmission of endosymbionts has been postulated to provide direct fitness effects to the insect host (81). An additional form is social transmission, i.e., via transmission between colony mates (85) and sexual partners (86).
Insects and other invertebrates represent important vectors of plant pathogens, including bacteria, fungi, protists, and viruses. The importance of insect vectors in transmission of numerous phytopathogens is well understood (87). However, insects may also transmit mutualists or entire microbial communities. Lòpez-Fernàndez et al. (27), for example, demonstrated that the American sap-feeding leafhopper Scaphoideus titanus mediated the transfer of entire plant endophytic bacterial communities between grapevine plants. At the same time, the endophyte communities influenced the leafhopper’s microbiome (27). Such transfer events can have implications for plant fitness and performance. Furthermore, plants shape the soil microbiome and insects feeding on plants which are grown on these differently conditioned soils respond to these changes (88). Hannula et al. (89) performed a study with herbivorous caterpillars fed on dandelion leaves and showed that the soil microbiome was partly transferred to the feeding insects.
Herbivore-associated bacteria have been reported to suppress plant defenses. For instance, the Colorado potato beetle secretes symbiotic bacteria capable of manipulating plant defense responses. These microorganisms elicit salicylic acid-regulated defense, which counteracts jasmonate signaling. This disruption makes plants unable to fully activate their jasmonate-mediated resistance against the herbivore (90). Furthermore, different volatile organic compounds emitted by microorganisms may affect insect behavior. For instance, Fusarium proliferatum, F. poae, and F. culmorum can attract Tenebrio molitor larvae, whereas F. avenaceum can repel the same insect (91). Another study showed, that variations in chlorosis caused by Russian wheat aphid (Diuraphis noxia) feeding are determined, in part, by aphid-associated bacteria (92).
Many angiosperm plant species are visited by honeybees (Apis mellifera L.) which collect nectar from flowers. While feeding, the external surface of bees contacts the nectar, allowing microbial exchange between the bee and nectar. Similarly, microorganisms in the nectar may be ingested by bees (93, 94). Altogether, these findings suggest that flowers may act as key hot spots for microbial exchange, including horizontal gene transfer (HGT) events. Different insects visiting a flower may all receive similar microbiomes. For example, Manirajan et al. (95) found a Lactobacillus species in flowers as well as in adults and larvae of seven megachilid bee species. Furthermore, pollen microbiomes of insect-pollinated plant species were found to be more similar than those of wind-pollinated plants (95). This indicates that insects and the transmission of microorganisms play an important role for pollen-associated microbiomes and a single flower may be involved in a series of transmission events.
Despite high sugar content and osmotic pressure, a range of microorganisms such as yeasts, yeast-like fungi, filamentous fungi, and bacteria are found in nectar (93, 96). This “nectar microbiome” has been shown to be functional, modifying sugar and amino acids content (97), and impacting volatile release (98). This is important, as different nectar properties can impact attractiveness of a given flower to pollinators, thus affecting the plants’ success. Indeed, alteration of the nectar microbiome may impact visitation frequency of insects (99) and reproductive success of the plant.
Fungal growing termites (containing the fungus Termitomyces) and leaf-cutting ants (containing the fungus Leucoagaricus) collect plant materials, respectively, dry straw, and green leaves, and bring it to their subterranean nests (100) where it is used as the substrate for a mutualistic basidiomycetous fungal colony. In return, the termites and the leaf-cutting ants harvest specialized fungal structures, rich in protein and sugars, and use these as feed for their larvae. This unique biomass converting system has been intensely studied, a strong interconnectedness between the microbiomes of the plant materials, the subterranean fungal colony, and the insect has been observed.
We have provided only a few of the many known examples of insect-microbiome transmissions. Nevertheless, they demonstrate the fundamental importance of microbiomes to insects, and how coevolutionary processes between insects and their microbiomes are not only important for the animal host, but also other components of the ecosystem such as plants. Indeed, it is clear, that the connection of the microbiomes across these systems can influence critical outcomes that affect pollinator and plant success, and thereby ecosystem functioning.
Microbiome Interconnectedness in Aquatic Environments
Microorganisms are discharged as spray aerosols over water bodies (e.g., sea, lakes, and rivers). These are produced at the surface of water bodies by wind or transported into the atmosphere over long-range distances. Certain taxa such as Actinobacteria, certain Gammaproteobacteria, and lipid-enveloped viruses show high transfer rates to sea sprays, whereas Flavobacteriia and some Alphaproteobacteria are transferred less frequently (101). Understanding the role of marine and other water bodies as a source and sink of microorganisms and the transfer of airborne bacteria could deliver important understanding of microbial diversity, spatial distribution, and the interaction between aquatic and terrestrial microbiomes. A survey on the genetic diversity of airborne and ocean-surface bacterial communities across the northwest Pacific and subtropical north Atlantic showed that 3% of all taxa identified were shared between both oceans (102). This study also showed that the atmospheric microbial community composition over the Atlantic Ocean was dominated by terrestrial, typically dust-associated microorganisms.
The ocean environment harbors microbiomes that have evolved and adapted through convergent evolution. Coral reef ecosystems are highly productive and diversified marine habitats that have photosynthetic and primary production features in common with terrestrial ecosystems. The coral itself is a holobiont and represents a well-recognized model system for symbiosis. In particular, recruiting or shuffling stress-tolerant microbial symbionts in corals are important for the recovery from stress events (e.g., coral-reef bleaching) (103). These “symbiosis shuffles” can also alter the metabolic repertoire of the coral at large (104). Similarly, sponges are filter-feeding animals hosting extensive microbial assemblages, where the microbial component may represent up to 35% of the sponge biomass (105). Most sponge-associated fungi are likely to be sourced from the surrounding environment and belong to the genera of terrestrial fungi adapted to the marine ecosystem (106). Comparative genome analysis of Actinobacteria associated with a marine sponge showed genomic signatures of environmental niche adaptation, indicating both terrestrial affiliation and sponge niche adaptation (107).
The coastal marsh soil microbiome sits at the interface of the terrestrial and marine ecosystems. Both ecosystems host a wide range of microorganisms involved in critical biogeochemical cycles. At this interface, sea level rise is a threat potentially leading to the loss of marshes and their associated microbiomes. For instance, increased salinity due to a rising sea level could negatively impact the microbial metabolism of organic matter by suppressing carbon cycling genes and their metabolites (108). Salt marshes, which are located at intertidal wetlands in temperate zones, are one of the marine-terrestrial transition zones for microorganisms.
Transmission of Human Pathogens and Antimicrobial Resistance Genes in Agricultural Production Systems
The agricultural ecosystem is a congruence, where microbiomes from soil, plants, and livestock (including manure) come together. Often these systems also include microbiomes originating from irrigation water, wildlife, wastewater, food chains (e.g., waste and residues fed to livestock) and humans. How the microbial communities from these different sources amalgamate in agricultural production systems has consequences for food and feed production, as well as the health of humans, livestock, and wild animals. The focus here is on the transmission of foodborne pathogens and antimicrobial resistance.
Human pathogens can be present in animal manure and other products of animal origin used for soil fertilization in some countries. They can be transmitted from irrigation water or airborne propagules, including open water bodies (e.g., surface water, collected rainwater) that stay in contact with wildlife such as migratory birds, or which are mixed with agricultural runoff water or sewage effluent after episodes of severe rainfall (109). Crops that are consumed fresh, especially those that are harvested after short production cycles, are of greatest concern for transmission of pathogens to humans. The most commonly observed pathogens in products of plant origin (e.g., vegetables, fruits, herbs, spices, and nuts) are zoonotic pathogens; e.g., pathogenic E. Coli strains, Salmonella Typhimurium, Campylobacter jejuni, and Listeria monocytogenes. These species prevail in the plant environment (110, 111), and it is now generally accepted that plants serve as secondary habitats for these zoonotic species (112). The persistence of human pathogens within plant systems raises concerns for the emergence of new and possibly more virulent or resistant lines. The rhizosphere is considered a hot spot for HGT and when microorganisms from different sources accumulate in this environment, new traits making human pathogens better adapted to selective circumstances that prevail in primary food production systems may develop (113).
A serious foodborne disease outbreak related to consumption of a plant-derived product with a huge impact from human and economic perspectives, occurred in Hamburg in 2011. This outbreak was caused by an unusual enterohemorrhagic E. coli O104:H4 (EHEC) type that was presumably present on, or inside fenugreek seeds used for sprout production. The origin of the outbreak strain was human and not zoonotic, indicating that contact must have taken place between the fenugreek seeds and sewage either at the production site or at seed storage or transport locations. From genomic studies it became clear that this strain must have acquired new traits via HGT, making it more aggressive but also more resistant to particular antibiotic classes (114).
Large foodborne disease outbreaks related to consumption of plant-derived products are rare in comparison to outbreaks associated with consumption of animal products, but the impact of such outbreaks can be significant. Microbiome interconnectivity within primary food production pipelines is therefore critical in understanding the consequences of mixing microorganisms from different ecosystems in relation to food safety. Similarly, it is important for human and animal health to understand how antimicrobial resistant microorganisms are transmitted via food or feed to our microbiome. Food safety and health aspects related to microbiome interconnectivity should therefore be taken into account in existing (intensive) agro-production systems, and particularly in more extensive systems that receive inputs from side-stream materials obtained from other production systems.
Contamination of soils with antibiotics and antibiotic resistance (AMR) genes is a global health concern. Soil contamination is mostly due to the utilization of animal manure (115) or contaminated water used for irrigation (116). Recent surveys documented the role of HGT in movement of AMR genes from and among microbiomes in soil to plant tissues (117, 118). Agricultural management practices such as fertilizer application favored HGT. Plant microbiomes may also host microorganisms resistant to antibiotics and may serve as a gateway for the transfer of AMR to human or animal microbiomes (119–122).
Livestock and livestock production are also important sources and zones of AMR genes and HGT, with rumen and gut microbiomes being hot spots for HGT. Comparisons of microbiomes and antimicrobial resistance patterns in animals have revealed a higher abundance and diversity of AMR genes in intensive farming compared to extensive farming (123). In particular, antibiotic administration to animals during intensive farming exerts a strong selection pressure leading to the enrichment of AMR in agricultural systems (124). Enrichment of AMR genes within the food chain, especially when these genes are located on mobile genetic elements (MGEs), is a significant risk for a downstream transfer into the food chain. Even within foods such as fermented meat and dairy products, horizontal transfer of genomic elements (e.g., via bacteriophages [125]) can further induce exchange of AMR genes within dietary microbiomes. Given the importance of food microbiomes in human health, a more holistic understanding of the exchange of pathogens and AMR genes from the environment to plants, animals, food, livestock, and human populations is needed.
FOOD DERIVED MICROBIOTA AND THE HUMAN (GUT) MICROBIOME
The Edible Microbiome
Plant microorganisms, particularly endophytes, are a fundamental component of human diets and animal feed. In human diets, fresh vegetables are often eaten raw and contain different microbiomes: reflecting the plant species and its origin (126). Fresh vegetables and fruits are therefore an important route for the introduction of microorganisms in the gut (127–129). For instance, Wassermann et al. (128) calculated that approximately 100 million bacterial cells are consumed with each apple. However, postharvest of fruit can dramatically change the number, types and type of microorganisms ingested (128, 130). Even after processing (e.g., air-drying, boiling, or preparing a puree), about one third of the original microbial load was maintained, but with a substantial compositional shift (e.g., higher abundances of Pseudomonas spp. and Ralstonia spp., and lower abundances of Bacillus spp.) (131).
There is a growing body of evidence indicating that both the soil and plant microbiomes may influence the flavor of food products (132). Winemakers have long known that the soil is central to the physiology of the grapevine and the production of flavor compounds in the wine itself. Grapes of the same variety grown in different regions (132) have different metabolic and flavor profiles. The famous terroir of the wine has a microbiome element that extends from the soil microbiome, through direct and indirect impacts on vine physiology and health, to microbiomes in and on grapes that impact their metabolome during growth and into fermentation (133, 134).
Microorganisms ingested from plants can at least transiently colonize the human gut but this largely depend on their ability to survive stress conditions of the gastrointestinal tract (135), although it is not yet clear how food microorganisms interact with and influence the human gut microbiome (136). The processes of transfer and persistence of microorganisms in the food system have generally not been explored in depth, and current studies are mostly limited to pathogens (reviewed in 137) or probiotics (138). Food-associated fermentative bacteria, including probiotics, may temporarily complement resident microbial communities, thus forming part of our transient microbiome (138). The application of longitudinal multiomics approaches, including high-throughput cultivation, confirmed the hypothesis that bacteria (in particular bifidobacteria) of parmesan cheese possess the ability to colonize and persist in the human gut (139). More studies of this nature are required if our fundamental understanding of the links between food ingested and the gut microbiome is to progress.
Along with bacteria, fungi are ingested from food and are transferred to, and interact with, the gut microbiome. As a part of the gut mycobiota, the genera Penicillium, Aspergillus, and Saccharomyces are typically ingested with plant-based foods (140), Yarrowia with fermented meats (141), and Kluyveromyces with dairy products (142). Complex relationships between gastrointestinal bacteria and fungi from food origins have been reported in humans. For example, the cooccurrence of pathogenic fungi and inflammatory bacteria and of potentially antiinflammatory fungi and bacteria clearly showed how the different components of the mycobiota interacted and suggested that these organisms my impact the inflammatory process in the human gut (143). These findings demonstrate the importance of a wider view of the microbiome rather than focusing on bacteria only. More studies bringing together prokaryotes, fungi, protozoa and viral components of the entire microbiome, and addressing how these interact and impact the host gut system, are required.
Microorganism Transfer at the Interface between Environments and Foods of Animal Origin
The transfer of microorganisms from the environment to food of animal origin (including fish) is an important factor for the understanding and prevention of food spoilage. While fresh meat and fish products harbor bacterial communities from the gut and skin of animals as well as from food processing, they also host a core microbiome often derived from the environment (144). For example, cod and salmon meat samples were shown to contain different core microbiota, with cod containing more bacteria from seawater than salmon. In cod, an uncharacterized taxon of Fusobacteria was identified, which was also found as a dominant taxon in the spoiled cod fillet (144). Overall, the transfer of microbiota from the environment at the initial stages of production of foodstuffs of animal or plant origin is not the result of a simple “contamination” but of microbiome exchange in the environment.
The connection between microbiomes within the dairy production/processing chain has been widely investigated. This has, for example, included tracing origins of microorganisms present in raw milk. One such study highlighted differences in the raw milk microbiomes in connection to production systems, comparing those with stock predominantly located indoors (winter) or outdoors. Regardless of these systems, the teat surface and, to a lesser extent, feces were identified as the primary sources of raw milk microorganisms (145). Consumption of raw milk has the potential to expose the consumers to many food pathogens and is generally not recommended. Much of the world’s milk production is processed before consumption or production of other dairy products, but its microbiome can, in some circumstances, have a major influence on the final dairy product. In some cases, the microbiomes found within the processing facility can also have a considerable influence (146). Overall, these studies indicate the important link between environmental and animal microbiomes influencing food safety and food production processes, ultimately all determining food quality and nutritional value.
Microbiome Exchange in the Food/Feed (Production) Environment
The food microbiome derives from the interaction of microorganisms from primary production, raw materials, operators, environment, and production systems (147). While these microorganisms may be present at a low relative abundance in the environment, their levels and contribution to food and feed safety and quality can be considerable. These relationships between different types of microorganisms can be illustrated by the fermentation process, which is one of the oldest forms of food processing, where fermented foods are a natural reservoir of complex microbiomes. Fermentation processes involve interactions between different types of microorganisms as well as multiple metabolic reactions, including food biomass conversion. The specific role of microorganisms present in fermented foods in human health is not always clearly evidenced. The fact that many of these microorganisms are lactic acid bacteria (LAB), and are related to probiotic strains, suggests that at least some confer health benefits (148). Indeed, a study of the overlap between LAB strains found in fermented food and human gut (via fecal sampling) microbiomes has highlighted that closely related strains occur in both food and gut environments, providing evidence that fermented foods can be indeed regarded as a possible source of LAB for the gut microbiome (136). LAB in fermented foods are not exempt from the risk of transfer of AMR genes, as evidenced by microbial transfer events and pointed out as concern by the European Food Safety Agency (149).
Many types of microorganisms can be exchanged in the food-producing environment. Fungi, protozoa, bacteria, and viruses can all be transferred in food systems; e.g., from humans (150), materials (151), animals and plants (152) as well as soil and water. In some cases, their transfer can change the microbial diversity of food ingredients, potentially contributing to fermentation characteristics and/or modifying the sensorial characteristics of a food product (153). However, most research has focused on the risk of transfer of pathogens. For example, transfer of foodborne pathogens from contaminated hands to food represents a potential risk to human health (154). Similarly, human pathogens can be transferred from animal sources to humans via poor hygiene of food handlers or contaminated equipment (155). As such, food service establishments are frequent places of microorganism transfer (156).
Microbiomes of built environments, from stable walls, floors, and instrument surfaces are a key source of inoculum to food/feed production. Particularly in industrial meat production facilities, the built environment provides both a contact source for exchange of microbiomes to foods and also a route for inocula. For instance, in a production facility housing pigs with unhealthy gut systems and attendant diarrhea, treatment by changing feed alone is inefficient. However, when the animals are initially treated with pro- and prebiotics, accompanied by a change in the feeding regime (containing more gut health-promoting feed), improvements in livestock health can be realized (157). Such practices can result in a stronger and more resilient piglet health and less reliance on antibiotics, hereby also lowering risk of antimicrobial resistance (157).
Exchange of microorganisms and ARM genes can also involve sources such as silage, which is often used to enhance the storage stability of animal fodder. However, this feed source may also facilitate the transfer of microorganisms from the plant microbiome to the animal gut. Most silage is produced by a conversion of the animal feed carried out by the microbiome already present in and on the harvested plant materials (viz a mixed culture, via anaerobic fermentation). Silage conditions favor specific types of bacteria, e.g., different types of LAB, potentially contributing to a more diverse animal gut microbiome (158). Finally, it is well known that food microbiomes can also be a hot spot of MGEs, including ARM genes. These microbiomes can be readily exchanged between environments, operators, among foods, and finally to consumers (159). The processing systems of meat and in particular fermented meat are considered one on the main source of ARM genes (160).
Microbial exchange in the feed/food production environment has been mostly investigated in light of food safety and potential contamination with pathogens and/or AMR. Nevertheless, there is also exchange of nonpathogenic and potentially beneficial microorganisms, such as, e.g., in fermentation processes, playing a role for the production process itself, but also for providing unique features like taste or nutritional value.
Vertical Transmission and Breastfeeding as Driver for Microbiome Development at Early Stages of Life
The human gut hosts diverse microbial communities which are subject to microorganism exchange between humans. Already at birth, about 50% of the infant’s gut microorganisms originate from the mother's gut, vagina, or skin. Within just 2 to 5 days after birth, mother and infant microbiomes can have up to 72% of shared species (161).
During and after birth, an infant is exposed to maternal vaginal, fecal, and skin microbes, and exposure depends on the mode of birth. However, vaginal and skin microorganisms are usually only transiently found in infant fecal samples, whereas the infant gut is permanently colonized by gut bacteria that are partly of maternal origin (162). Predominantly, Bifidobacterium spp. and Bacteroides spp. are transferred from mother to child (163): both taxa have the ability to utilize human milk oligosaccharides (164). Due to their oxygen sensitivity and lack of spore formation, it seems that these taxa rely mostly on vertical transmission at birth, after which they persist indefinitely (162), unless antibiotics or toxins are ingested (165). Conversely, caesarean-born babies, harbor distinct microbial communities, that are more similar to skin surface and allow colonization of nosocomial opportunistic bacterial pathogens causing short- and long-term adverse health effects (166, 167). Post birth, a significant fraction of the infant gut microbiota is derived from breast milk (first transport route of microorganisms by “food”) during the first year of life. Breast milk may provide over 800,000 bacterial cells per day, serving as pioneer colonizers of the infant's gut (168). Microbial signatures shared between breast milk and infant stools were 88% 1 week after birth, declining to 70% at week 12 (169). The percentage microbiome shared between mother and infant increased with frequency of breast milk sconsumption (170). Key shared microorganisms include Escherichia/Shigella, Bifidobacterium longum, Bacteroides fragilis, Bacteroides thetaiotaomicron, Bilophila wadsworthia, and Enterococcus faecalis (171).
Many other animals exchange microorganisms in a similar way: for example, some of the calf fecal microbiota seem to derive from inoculation from the birth canal of the dam (172). Other routes of transmission can include the calf licking the dam, from the environment during and immediately after birth, and from the dam licking the calf clean immediately post birth. Taxa which showed the highest abundance in calf mouth samples taken within first 30 min of life included Acinetobacter spp. and Solibacillus spp., also detected in fecal calf and cow samples. However, their abundance in fecal samples decreased with time (172).
Postbirth vertical transmission routes provide the first exposure of newborns to rich and diverse microbiomes and thereby provide the initial inoculum for the development of their own gut microbiome. Due the overall importance of a healthy gut microbiome for human/animal health and well-being, breast-feeding and similar exchanges represent highly important transmission routes of microbiomes.
OTHER LIFESTYLE FACTORS WHICH INFLUENCE THE HUMAN MICROBIOME
The Relevance of the Environment and Social Interactions for Microorganism Transfer
Environmental microorganisms are thought to play an important role in triggering the immune system at early stages of life, making the human immune system more resilient toward challenges as adults (173). Children growing up on farms, for example, are exposed to a high microbiome diversity from the environment, and subsequently develop a more diverse gut and body microbiome. Evidence suggests that such children are less prone to allergy development in later life than children from urban areas (174). The exposure of children to diverse food and environmental microbiomes is thus important (175) and is the basis of the “hygiene theory” (176), i.e., an intimate connection between microbial diversity in the environment, microbial community structure and function at barrier organs of the human body and subsequent health and wellbeing, potentially via the interaction of microorganisms and the immune system (173).
Social interactions represent a route for microorganism exchange between individuals with systems-level implications. Studies in humans have shown that proximity and frequent social physical contact result in microorganism exchange. Individuals living together showed increased gut microbial diversity and abundance of potentially beneficial microorganisms (177, 178). Kort et al. (179) reported that the salivary microbiome may be influenced by intimate kissing. Also, there is evidence that oral and gut microbiota are shared in close social networks (mothers and infants and marital partners), as well between females but less between males (178). Furthermore, there is increasing evidence that microbiota are transmitted along the gastrointestinal tract, and that the large intestine can be colonized by oral microorganisms (180), When comparing the gut microbiomes of spouses to those of sibling pairs, spouses had more microbial species in common than siblings, even after accounting for dietary factors (177). Humans sharing the same household, including unrelated individuals, harbored more similar gut microbiota than individuals living in different houses (181). A recent study by Valles-Colomer et al. (182) analyzed more than 9,700 human metagenomes and computational strain-level profiling revealed extensive bacterial strain sharing across individuals. Different transmission patterns were identified for mother-to-infant, intrahousehold, and intrapopulation transmission patterns. Overall, these findings indicate that social interactions are important in shaping the human microbiome, and that this factor may exert an even stronger influence than shared genetic factors and early life environments supporting previous findings (177). The patterns observed within households extends to other socially shared spaces such as schools, workplaces, and public transportation (183).
From the current evidence for microbial interconnections between hosts and ecosystems, the concept of the social microbiome (collective metacommunity) has emerged. It is defined as the microbiome of a given group that can be transmitted horizontally across members of a group or acquired from the environment where it socializes (178).
Poor social integration relates to an increased risk of developing diseases, ranging from metabolic disorders to mental conditions. The links between alterations in the human microbiome and mental health (the gut-brain-axis) are well described (184). On this basis, it has been hypothesized that social life may bring health benefits (and sometimes disbenefits; SARS-CoV-2, for example) through microorganism transmission among members of a social group. Furthermore, microbiomes may influence the ability of their hosts to cooperate and interact, for example in displaying paternal care behavior (185). The connection between the gut and brain means that microbiomes can influence social behavior and decision-making through emotions and cognitive processes (186).
Theories about the implications of socialization in microbial transmission are still grounded on preliminary evidence. To date, only a few studies have controlled for relevant variables (dietary, environmental, and genetic), and even fewer have investigated microbial transmission via strain tracking and linked transmission with health outcomes. Yet emerging work on primate populations highlights the intimate connection between microbiota composition, functional links to immune status (e.g., antiinflammatory taxa), and social behavior (187). Inevitably, some key research questions have emerged from animal and human studies: how social-microbial communities of (mammal) hosts participate in their selection by modifying the host’s or the group’s behavior; does this phenomenon transcend individual and closely living groups; and is there a role of coevolution of humans and microbiomes of social behavior, demographic changes, and global health?
Exchange of Microbiota between Pets and Humans
Humans have been sharing living spaces and food resources with companion animals for millennia. Dogs were domesticated 30 thousand and cats 10 thousand years ago. Humans and horses have been in close proximity for over six thousand years. Such long periods of mutual exposure have most likely enabled coevolution of the microbiomes of both humans and animals: it is not just the pets that were domesticated, but their microbiomes, too.
Short-term studies have shown that cohabitation with pets results in an alteration of gut microbiota diversity and composition in both humans and animals (188). These alterations have functional implications. For instance, Du et al. (188) showed that cat ownership was associated significantly changed metabolic pathways, e.g., increased metabolism of amino acids, nucleotides, biological oxidation carbohydrates, vitamins, and lipids. Also, intriguing interactions were observed for microbiome exchange between cats and their owners with respect to the gender and physiology of owners. To this end, differences in the exchange of different bacterial families from cats to human females and males have been observed, and between feline pets as well as between healthy and overweight owners (188).
Wetzels et al. (189) analyzed skin bacterial communities of wolves and dogs living in outdoor packs and compared these with human care-takers and their pet dogs. Even though humans had more distinct and less diverse bacterial communities than other studied groups, bacterial communities of individuals in close contact with outdoor pack animals showed more similarities to the bacterial communities of these animals. In particular, both the ratio of Gram-negative to Gram-positive microorganisms on the skin and the phylum-level diversity were increased.
The intimate relationship between pets and owners potentially represents a public health concern in terms of AMR gene development and transmission (190). Indicative evidence has been provided in several studies where AMR genes present in fecal samples of humans and their companion animals were characterized (191–193). In a more comprehensive metagenomic study, Zhao et al. (194) compared the gut AMR genes, the MGEs and the microbiota among dogs and their owners as well as kennel dogs. Owned dogs shared 70% of AMR genes with their owners, whereas only 52% of observed AMR genes were shared between kennel dogs and owners. More detailed analysis focusing on dog-owner pairs has indicated that AMR genes, MGEs, and microbiota composition correlated significantly with each other. The shared microbiome (sensu bacterial community) between the owner and pet was considered to be the main basis of the cooccurrence in AMR genes. Despite the increasing knowledge of the microbiome exchange between pets and their owners involving also the exchange of AMR genes, little understanding exists on the functional consequences of these transfer events.
Microorganism Transfer in the Built Environment
Buildings are typically complex ecosystems that not only provide shelter for their inhabitants, but also harbor trillions of microorganisms that can interact with each other (195). The two primary mechanisms of microbiome transfer in the built environment are (i) bioaerosols, and (ii) via physical contact/exchange from surfaces.
As outlined earlier in this review, bioaerosols are airborne particles of biological origin. They can include bacteria, fungi, archaea, viruses, pollen, and their cell wall components and/or metabolic products. Overall, bioaerosols can be considered an imprint of the environment (58) where they derive from. They are important in the transmission of pathogenic organisms and/or their metabolic products to plants, animals, and humans, resulting in the spread of diseases within populations (196, 197).
In built environments, the quantity of air circulation and the type of environment will affect the mechanism and magnitude of the transfer of microorganisms via bioaerosols (198). For instance, Triadó-Margarit et al. (199) showed a 22% overlap of bacterial taxa in microbiomes found in different locations of the Barcelona subway, such as inside trains, the platform, or the lobby. This is indicative of the flow of microorganisms between different locations.
Fungal spores are efficiently spread in many environments. While airborne spores of pathogens such as Aspergillus fumigatus are typically inhaled without harmful consequences, for immunocompromised people, airborne fungal spores may result in invasive aspergillosis or skin infections (200). Similarly, immunocompromised patients can more readily acquire fungal skin infections caused by dermatophytic fungi. Dermatophytic fungi were shown to have a set of keratin-degrading enzymes, enabling them to invade through the skin of humans or animals (201). In contrast, healthy skin microbiota (particularly bacteria and yeasts) do not have such set of keratin-degrading proteases (201).
Residents leave their microbial fingerprint mainly from their skin (202), but less is known about the transfer of microbiota from plants to the built environment. First indications that plants substantially contribute to the microbial abundance and diversity in the built environment were found in a study on the surface microbiome of intensive care units of a university hospital (203). Transfer of microorganisms from plants to surfaces in a building may be mediated by window ventilation. Kembel et al. (195) showed that the phylogenetic diversity of airborne bacterial communities was lower indoors than outdoors, and mechanically ventilated rooms contained less diverse microbial communities than window-ventilated rooms. The initial observations were later experimentally confirmed by analyzing the microbiome of the spider plant Chlorophytum comosum and its surrounding environment (204). The abundance of archaea, bacteria, and fungi increased on the floor and wall surfaces near the plant within 6 months, whereas the microbial abundance on plant leaves and in the indoor air remained stable. Moreover, a clear shift in the composition of the microbiota was observed; bacterial diversity on surfaces increased significantly, while fungal diversity decreased. This study demonstrated for the first time that indoor plants can alter the microbiome of a built environment, which supports the significance of plants and provides insights into the complex interplay of plants, microbiomes, and human beings (204).
Kozdrój et al. (198) evaluated the exposure of visitors and workers to airborne bacteria and fungi at different botanical garden sites, including within the garden glasshouses. Not surprisingly, the concentrations of bioaerosol microbiota and their diversity were higher in the glasshouses compared to those found in the outdoor air of the garden area. The bacterial taxa present in adjacent streets were also found in the glasshouse and garden, suggesting substantial microorganism exchange.
Exposure to diverse environmental microbiota has been suggested to confer protection against immune-mediated disorders (205). For example, vegetation around homes was shown to be associated with health-related changes in gut microbiota composition suggesting a transmission route via built environments (206). However, additional studies will be required to understand how to specifically utilize indoor plants to modulate the indoor microbiota for health benefits. Interestingly, such potential links equally inspire scientists and artists, in the attempt to define human identity in the broader perspective of the surrounding environment and biosphere (207).
Transfer of microorganisms from the built environment to humans is particularly impactful in the hospital environment. According to the World Health Organization, 7 to 15% of patients in acute-care hospitals acquire at least one health care-associated infection during their hospital stay, often with severe or fatal outcomes that are augmented when resistant microorganisms are involved (208). Recently, the persistence of pathogens on inanimate surfaces was reviewed (209) and direct transfer to patients was demonstrated in several studies. For example, a human-pathogenic plant pathogen (Exserohilum rostratum) originated from the laboratory where corticosteroid injection fluids were produced, and caused the deaths of many patients treated with this corticosteroid in various hospitals (210). Cason et al. (211) used whole-genome sequencing-based typing of vancomycin-resistant enterococci to analyze the genetic relationships between bacterial isolates originating from patients and the hospital environment. Five out of eight identified clusters of closely related strains (≤3 alleles differing between the genotypes) contained both environmental and patient isolates, providing strong evidence of the exchange of microorganisms between hospital environments and humans.
Microbiomes in the built environment and their dispersal in bioaerosols have only been recently recognized as an important issue for human health. Apart from the transmission of pathogens or allergenic molds, airborne microorganisms are also likely to beneficially influence human health and well-being, e.g., by out-competition or antagonism of pathogens. Microbiome dynamics in the built environment and its impact on air quality requires further understanding, and bioaerosol microbiomes may be subject of new approaches to improve the health and well-being of humans in their home and working environments.
Microbial Transmission from Humans to the Environment
Vast numbers of microorganisms (primarily prokaryotes) are discharged from humans to wastewater treatment plants. Typically, 0.5 kg feces are discharged per person per day, with each gram containing 109 bacterial cells: i.e., ~5 × 1011 bacterial cells per person per day. With a world population of 7.8 billion people, 3.9 × 1024 bacterial cells are released from humans into the environment daily, from fecal waste alone.
Given the sheer numbers of bacteria discharge by humans, perhaps it is no surprise there are overlaps between the microbiomes of the human gastrointestinal tract, municipal sewer systems (212), and municipal wastewater treatment plants (213). For example, phylogenetically related members of the family Lachnospiraceae, which currently comprises 80 genera and 176 species (https://lpsn.dsmz.de/family/lachnospiraceae), have been recovered from both human feces and environmental samples (214).
A unique example of horizontal interkingdom transfer of a human opportunistic pathogen (Propionibacterium acnes) to the domesticated grapevine (Vitis vinifera L.) was shown by Campisano et al. (215). The authors showed that this bacterium colonizes different plant tissues, such as bark and inside pith tissues, both inter- and intracellularly. Phylogenetic and comparative genomics analyses indicated that the establishment of the grapevine-associated P. acnes as an obligate endophyte was due to a recent transfer event, likely during the Neolithic period when the grapevine was domesticated.
THE EXCHANGE OF VIROMES BETWEEN ENVIRONMENTS
The COVID-19 pandemic has focused attention on the role and impact of the environmental virome on human health, socio-political and economic systems, and on planetary health. During the COVID-19 pandemic, the testing of municipal wastewater by qPCR and genome sequencing were invaluable in the surveillance and informed responses to the disease. These tools allowed science to inform decision-making which directly impacted human health and wellbeing, while seeking to maintain essential services and business where possible. The application of these tools enabled estimating the extent of SARS-CoV-2 infection within populations, and determining the relative abundance of genetic variants that arose over time (216).
To better understand viral transfer processes, virome surveillance can potentially be applied in a more generalized manner, for example to educational institutions, long-term-care facilities and hospitals, cruise ships, farms, airports, and aircraft (217). The technology and approaches are widely applicable to human and animal viruses, whether they be RNA viruses, such as SARS-CoV-2, or DNA viruses, such as monkey pox (218).
Viruses that impact human health directly are of high priority. However, human health is dependent on a safe and abundant food supply. As such, environmental virome surveillance is an emerging tool for detection of virus outbreaks more broadly. Shotgun metagenomics analysis has revealed that the most abundant RNA viruses in municipal wastewater, by far, are plant viruses such as the widespread tobamoviruses (219). These infect a wide range of common crop plants, and new variants commonly arise (e.g., the tomato brown rugose fruit virus [220]). The presence of these plant viruses is almost certainly of dietary origin, rather than agricultural sources such as runoff from vegetable greenhouse operations. This suggests that the incidence and distribution of viruses infecting crop plants is far greater than previously thought. Routine surveillance through testing of plant tissue samples would be laborious and would suffer from inadequate coverage. Alternatively, sampling of the proximate environment such as irrigation runoff has the potential to detect viruses and other pathogens far more effectively.
Viromes in the environment are still poorly understood, however, the COVID-19 pandemic and other viral diseases have increased the awareness of their importance. Virome transfer may not only indicate the presence of a disease but may also induce microbiome changes in the receiving environment with yet unknown effects on ecosystem functioning.
MICROBIOME ESTABLISHMENT AND ADAPTATION IN NEW ENVIRONMENTS
Establishment of microorganisms in new ecosystems occurs via a phased process (221). First, microorganisms have to be transferred to a new habitat via microbial dispersal (i.e., by horizontal or vertical transmission). Then, the introduced microorganisms need to establish and adapt to their new environments which may, or may not, lead to a viable and self-sustaining population. As the system stabilises and processes such as initial environmental filtering have been completed, the importance of longer-term environmental selection and species competition become stronger (222).
Establishment of new microorganisms and reassembly of microbiomes in a new ecosystem mainly depend on the selectivity of the environmental conditions, the original structure of the microbiome, the dilution rate upon transfer, the availability of free ecological niches in the place of transfer, and the evolutionary capacity and fitness of the transferred strains. Selectivity of the new environmental conditions spans a continuum of neutral to highly selective; these have differing impacts depending on microbial diversity (222). For example, in the case of the soil microbiome, it has been shown that soil pH is a key predictor of microbial community structure before or after transfer (7). In human milk, oligosaccharides may act as a selective factor for the outgrowth of Bacteroides spp. and Bifidobacterium spp. upon maternal fecal microbiota transplantation to caesarean-born infants (223). Most fermented foods go through ecological succession with early growth of prevailing autochthonous bacteria. As the pH of the food decreases, overgrowth of LAB occurs (224). Refeeding of sourdough (back-slopping propagation) shapes the microbiome and selects specific LAB species depending on the age and other parameters of the starter and receiving materials. For example, Fructilactobacillus sanfranciscensis is a dominant species in mature wheat and rye sourdoughs with short fermentation times, whereas more acid-tolerant Limosilactobacillus reuteri is found in sourdough with long fermentation cycles at higher temperatures (225).
The resident microbiome may prevent the colonization of newcomers in the ecosystem by providing resistance and competition against the intruders, which is well known for the establishment of pathogens. For example, pathogens do not readily establish in soils due to soil suppression (226). Likewise, in the human gut, the microbiome prevents the establishment of pathogens via colonization resistance (227). Both processes are mediated by the resident microbiome.
The availability of unoccupied niches in the place of transfer may increase the likelihood of invasion by transferable microorganisms. Availability of niches to occupy can enable direct colonization or adaptive radiation processes. This is observed in the efficacy of fecal transplantation treatments for patients suffering from recurrent infection with Clostridioides difficile. In these patients, the very low diversity of colonic microbiota with limited microbial interaction networks enables higher transplantation success and better health outcomes than for patients with nonintestinal infectious diseases (228). Success of the transplantation also depends on the characteristics of the donor’s microbiota when comparing subjects with the same condition (229). The outcome of microbiome transfer also depends on the size of the transferable aliquot. A dilution effect could significantly reduce the taxonomic and functional diversity of reassembled bacterial communities (230).
Upon transfer to the new environment individual microbiome members can become maladapted. At the strain level, adaptation to new conditions could include alteration of the physiological state, HGT, or the selection of new mutations. The process of strain adaptation to the new environment depends not only on intrinsic abilities of the strain (e.g., genome size and encoded life strategies), but also on the members of the surrounding community. It has been shown that there is stronger evolutionary response in low-diversity communities (231).
Microbiome establishment is key to a long-lasting microbiome transfer. It is therefore not only important to understand how individual strains can establish but also how a complex microbiome can establish and how this is influenced by microbial interactions or environmental effects.
CONCLUDING REMARKS
The ecology and functioning of microbial communities are typically studied in one specific environment at a time, and microbiome transmission between different environments has been generally overlooked. Most studies on microbiome transmission have addressed horizontal or vertical transmission routes of microbiomes associated with higher organisms, such as humans (181, 182), plants (24, 32) or insects (77, 85). Also, transmission routes of pathogens and of AMR genes have been investigated in some detail, particularly the transfer from primary habitats to those affecting human health. However, microbiome connectivity between different environments is vast (Fig. 1) and has a magnitude that has been little considered or understood.
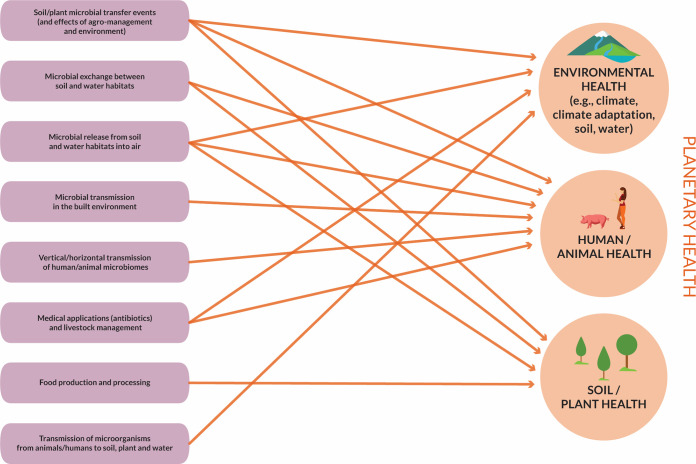
Some transmission events have detrimental effects, such as the transmission of pathogens, whereas many microbiome connections have positive effects on ecosystem functioning or human health (Fig. 2). Examples of the positive effects of microbiome transfer include the methods used to, e.g., promote plant health and productivity (232), improve environmental health (233), and establish diverse and healthy human gut microbiomes (probiotic treatments, fecal transplants) (228, 234).
FIG 2.
Microbiome connectivity between environments and impact on environmental health, human/animal health, and plant health.
It is evident that microbiome transmission between environments occurs continuously and between most environments on Earth (Fig. 1). Considering the ubiquitous nature of microbiome transfer, we can reasonably assume that the consequences of microbiome transfer on global ecosystem functioning and the health of our planet are very large, even if we do not yet fully understand the magnitude of the process.
As microbiomes play key roles in most, if not all, global environments, the fluidity of microbiome diversity and composition may be associated with a gain or loss of functions, with potentially positive or negative consequences for the environment. Transmission of microbiomes can provide a mechanism for ecosystem or holobiont adaptation. For instance, plants receiving new microbiome members from other plants, soils, insects, or bioaerosols may acquire positive attributes such as increased pathogen or stress resistance. Similarly, the human acquisition of microorganisms from other humans, animals or pets, plant-derived food, or even bioaerosols may affect their immune status or even social behavior.
The development of high-throughput nucleic acid sequencing technologies has facilitated the rapid analysis of complex microbiomes and their transfer between environments. While the majority of such studies are based on amplicon sequencing of phylogenetic markers, there is an increasing trend of whole-metagenome analysis (e.g., 183). Metagenome-based studies make it possible to investigate the functional potential of microbiomes, but also facilitate strain-level profiling of microbiomes. The latter is important for investigations of the transmission of individual strains, e.g., either pathogens (such as specific outbreak strains) or beneficial microbiota. Metagenome information may be also used to study the effect of MGEs on the transmission of particular traits, for example, those enhancing ecological competence in the new environment.
There is a critical need to link microbiome and metagenome information with phenotypic or functional data to better understand the functional consequences of microbiome transmission events for the microbial community as well as the ecosystem/host. Metatranscriptomics of source and sink communities can address the issues of gain or loss of functionality during or after transfer events. Other “omics” approaches, including metaproteomics, metabolomics, and high-throughput cultivation, can all generate critical information on microbiome functionality.
It is widely accepted that a more holistic approach to understanding microbiome transmission processes would be advantageous, even if technically demanding. Given that most microbiomes contain taxa from all three kingdoms (e.g., bacteria, fungi, protozoa, archaea, and viruses, in addition to MGEs), and given that many of these may interact in multiple, complex and poorly understood ways, a multitaxon approach to studying microbiome transfer processes is likely to reveal novel and potentially exciting results. There is also yet limited knowledge of the adaptation processes of individual microbiome members upon transfer to a new and different environment.
The fact that microbiomes of different environments are connected and exchanged needs to be considered in global practices such as agricultural management. Microbiome interconnectedness also has potential to be used as an approach to modulate microbiomes in the selection or provision of desirable traits. For instance, the human gut microbiome could be modulated by “designing” plant microbiota of plant-derived food (e.g., fermented food or salads). Similarly, plants enriched in probiotic strains could help to enrich bioaerosols to support the human immune system, supporting the respiratory system or skin functions. Such microbiota-enriched plants could potentially be used in the built environment, on green walls or other type of plant “installations.” As there is a connection between microbiomes, atmosphere and dispersal, local weather, and global climate, it is important to increase our understanding of the role microorganisms have in hydrological cycles, and how phyllosphere microorganisms impact and are impacted by weather. Such advanced understanding will not only help to quantitate the role of microorganisms in the global climate but may assist in the design of novel strategies employing transferrable microbiomes to define new solutions for improving human health and the health of our planet.
ACKNOWLEDGMENT
All authors have received funding from the European Union’s H2020 Research and Innovation Program under grant no. 818116 (MicrobiomeSupport). The grant of the Spanish Ministry of Science and Innovation (MCIN/AEI) to IATA-CSIC as Accredited Research Center of Excellence (CEX2021-001189-S/MCIN/AEI/10.13039/501100011033) is acknowledged.
REFERENCES
- 1.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, Souza R, van Overbeek L, Singh BK, Wagner M, Walsh A, Sessitsch A, Schloter M. 2020. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleerebezem R, Stouten G, Koehorst J, Langenhoff A, Schaap P, Smidt H. 2021. Experimental infrastructure requirements for quantitative research on microbial communities. Curr Opin Biotechnol 67:158–165. doi: 10.1016/j.copbio.2021.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Meisner A, Wepner B, Kostic T, van Overbeek LS, Bunthof CJ, de Souza RSC, Olivares M, Sanz Y, Lange L, Fischer D, Sessitsch A, Smidt H, MicrobiomeSupport C . 2022. Calling for a systems approach in microbiome research and innovation. Curr Opin Biotechnol 73:171–178. doi: 10.1016/j.copbio.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Singh BK, Liu H, Trivedi P. 2020. Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ Microbiol 22:564–567. doi: 10.1111/1462-2920.14900. [DOI] [PubMed] [Google Scholar]
- 5.van Bruggen AHC, Goss EM, Havelaar A, van Diepeningen AD, Finckh MR, Morris JG. Jr., 2019. One Health - Cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health. Sci Total Environ 664:927–937. doi: 10.1016/j.scitotenv.2019.02.091. [DOI] [PubMed] [Google Scholar]
- 6.Yan Z, Xiong C, Liu H, Singh BK. 2022. Sustainable agricultural practices contribute significantly to One Health. J Sust Agri & Env 1:165–176. doi: 10.1002/sae2.12019. [DOI] [Google Scholar]
- 7.Banerjee S, van der Heijden MGA. 2023. Soil microbiomes and one health. Nat Rev Microbiol 21:6–20. doi: 10.1038/s41579-022-00779-w. [DOI] [PubMed] [Google Scholar]
- 8.Trivedi P, Batista BD, Bazany KE, Singh BK. 2022. Plant–microbiome interactions under a changing world: responses, consequences and perspectives. New Phytol 234:1951–1959. doi: 10.1111/nph.18016. [DOI] [PubMed] [Google Scholar]
- 9.Custódio V, Gonin M, Stabl G, Bakhoum N, Oliveira MM, Gutjahr C, Castrillo G. 2022. Sculpting the soil microbiota. Plant J 109:508–522. doi: 10.1111/tpj.15568. [DOI] [PubMed] [Google Scholar]
- 10.Walters KE, Capocchi JK, Albright MBN, Hao Z, Brodie EL, Martiny JBH. 2022. Routes and rates of bacterial dispersal impact surface soil microbiome composition and functioning. ISME J 16:2295–2304. doi: 10.1038/s41396-022-01269-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelfattah A, Wisniewski M, Schena L, Tack AJM. 2021. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ Microbiol 23:2199–2214. doi: 10.1111/1462-2920.15392. [DOI] [PubMed] [Google Scholar]
- 12.Choudoir MJ, DeAngelis KM. 2022. A framework for integrating microbial dispersal modes into soil ecosystem ecology. iScience 25:103887. doi: 10.1016/j.isci.2022.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu G, Cevallos-Cevallos JM, Vallad GE, van Bruggen AHC. 2013. Organically managed soils reduce internal colonization of tomato plants by Salmonella enterica serovar Typhimurium. Phytopathology 103:381–388. doi: 10.1094/PHYTO-04-12-0072-FI. [DOI] [PubMed] [Google Scholar]
- 14.Semenov MV, Krasnov GS, Semenov VM, Ksenofontova N, Zinyakova NB, van Bruggen AHC. 2021. Does fresh farmyard manure introduce surviving microbes into soil or activate soilborne microbiota? J. J Environ Manage 294:113018. doi: 10.1016/j.jenvman.2021.113018. [DOI] [PubMed] [Google Scholar]
- 15.Amor DR, Ratzke C, Gore J. 2020. Transient invaders can induce shifts between alternative stable states of microbial communities. Sci Adv 6:eaay8676. doi: 10.1126/sciadv.aay8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. 2020. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 17.Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compant S, Cambon MC, Vacher C, Mitter B, Samad A, Sessitsch A. 2021. The plant endosphere world – bacterial life within plants. Environ Microbiol 23:1812–1829. doi: 10.1111/1462-2920.15240. [DOI] [PubMed] [Google Scholar]
- 19.Kandel S, Joubert P, Doty S. 2017. Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. doi: 10.3390/microorganisms5040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glassner H, Zchori-Fein E, Compant S, Sessitsch A, Katzir N, Portnoy V, Yaron S. 2015. Characterization of endophytic bacteria from cucurbit fruits with potential benefits to agriculture in melons (Cucumis melo L). FEMS Microbiol Ecol 91:fiv074. doi: 10.1093/femsec/fiv074. [DOI] [PubMed] [Google Scholar]
- 21.Nelson EB. 2018. The seed microbiome: origins, interactions, and impacts. Plant Soil 422:7–34. doi: 10.1007/s11104-017-3289-7. [DOI] [Google Scholar]
- 22.Escobar Rodríguez C, Antonielli L, Mitter B, Trognitz F, Sessitsch A. 2020. Heritability and functional importance of the Setaria viridis bacterial seed microbiome. Phytobiomes J 4:40–52. doi: 10.1094/PBIOMES-04-19-0023-R. [DOI] [Google Scholar]
- 23.Matsumoto H, Fan X, Wang Y, Kusstatscher P, Duan J, Wu S, Chen S, Qiao K, Wang Y, Ma B, Zhu G, Hashidoko Y, Berg G, Cernava T, Wang M. 2021. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants 7:60–72. doi: 10.1038/s41477-020-00826-5. [DOI] [PubMed] [Google Scholar]
- 24.Johnston-Monje D, Gutiérrez JP, Lopez-Lavalle LAB. 2021. Seed-transmitted bacteria and fungi dominate juvenile plant microbiomes. Front Microbiol 12:737616. doi: 10.3389/fmicb.2021.737616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escobar Rodríguez C, Mitter B, Antonielli L, Trognitz F, Compant S, Sessitsch A. 2018. Roots and panicles of the C4 model grasses Setaria viridis (L). and S. pumila host distinct bacterial assemblages with core taxa conserved across host genotypes and sampling sites. Front Microbiol 9:2708. doi: 10.3389/fmicb.2018.02708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frago E, Dicke M, Godfray HCJ. 2012. Insect symbionts as hidden players in insect–plant interactions. Trends Ecol Evol 27:705–711. doi: 10.1016/j.tree.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Lòpez-Fernàndez S, Mazzoni V, Pedrazzoli F, Pertot I, Campisano A. 2017. A phloem-feeding insect transfers bacterial endophytic communities between grapevine plants. Front Microbiol 8:834. doi: 10.3389/fmicb.2017.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compant S, Clément C, Sessitsch A. 2010. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 29.Mitter B, Pfaffenbichler N, Flavell R, Compant S, Antonielli L, Petric A, Berninger T, Naveed M, Sheibani-Tezerji R, von Maltzahn G, Sessitsch A. 2017. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol 8:11. doi: 10.3389/fmicb.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal VK, Sinclair JB. 1997. Principles of seed pathology, 2nd ed. CRC Press, Boca Raton, FL. doi: 10.1201/9781482275650. [DOI] [Google Scholar]
- 31.Saikkonen K, Ion D, Gyllenberg M. 2002. The persistence of vertically transmitted fungi in grass metapopulations. Proc Biol Sci 269:1397–1403. doi: 10.1098/rspb.2002.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdelfattah A, Tack AJM, Lobato C, Wassermann B, Berg G. 2023. From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends Microbiol 31:346–355. doi: 10.1016/j.tim.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Wassermann B, Abdelfattah A, Wicaksono WA, Kusstatscher P, Müller H, Cernava T, Goertz S, Rietz S, Abbadi A, Berg G. 2022. The Brassica napus seed microbiota is cultivar-specific and transmitted via paternal breeding lines. Microb Biotechnol 15:2379–2390. doi: 10.1111/1751-7915.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Ma Y-N, Wang X, Liao K, He S, Zhao X, Guo H, Zhao D, Wei H-L. 2022. Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome 10:216. doi: 10.1186/s40168-022-01422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh CM, Becker-Uncapher I, Carlson M, Fierer N. 2021. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. ISME J 15:2748–2762. doi: 10.1038/s41396-021-00967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Overbeek LS, Lombaers-van der Plas C, van der Zouwen P. 2020. The role of pea (pisum sativum) seeds in transmission of entero-aggregative Escherichia coli to growing plants. Microorganisms 8:1271. doi: 10.3390/microorganisms8091271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lymperopoulou DS, Adams RI, Lindow SE. 2016. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl Environ Microbiol 82:3822–3833. doi: 10.1128/AEM.00610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindow SE, Brandl MT. 2003. Microbiology of the Phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujiyoshi S, Tanaka D, Maruyama F. 2017. Transmission of airborne bacteria across built environments and its measurement standards: a review. Front Microbiol 8:2336. doi: 10.3389/fmicb.2017.02336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan Z-Z, Chen Q-L, Zhang Y-J, He J-Z, Hu H-W. 2020. Industrial development as a key factor explaining variances in soil and grass phyllosphere microbiomes in urban green spaces. Environ Pollut 261:114201. doi: 10.1016/j.envpol.2020.114201. [DOI] [PubMed] [Google Scholar]
- 41.Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE. 2016. The phyllosphere: microbial jungle at the plant–climate interface. Annu Rev Ecol Evol Syst 47:1–24. doi: 10.1146/annurev-ecolsys-121415-032238. [DOI] [Google Scholar]
- 42.Koskella B. 2020. The phyllosphere. Curr Biol 30:R1143–R1146. doi: 10.1016/j.cub.2020.07.037. [DOI] [PubMed] [Google Scholar]
- 43.Burrows SM, Butler T, Jöckel P, Tost H, Kerkweg A, Pöschl U, Lawrence MG. 2009. Bacteria in the global atmosphere – part 2: modeling of emissions and transport between different ecosystems. Atmos Chem Phys 9:9281–9297. doi: 10.5194/acp-9-9281-2009. [DOI] [Google Scholar]
- 44.Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2:321–334. doi: 10.1038/ismej.2007.113. [DOI] [PubMed] [Google Scholar]
- 45.Cevallos-Cevallos JM, Danyluk MD, Gu G, Vallad GE, van Bruggen AHC. 2012. Dispersal of Salmonella by rain splash onto tomato plants. J Food Prot 75:472–479. doi: 10.4315/0362-028X.JFP-11-399. [DOI] [PubMed] [Google Scholar]
- 46.Cevallos-Cevallos JM, Gu G, Danyluk MD, Dufault NS, van Bruggen AHC. 2012. Salmonella enterica Typhimurium can reach tomato fruits on plants exposed to aerosol formed by rain. Int J Food Microbiol 158:140–146. doi: 10.1016/j.ijfoodmicro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Tong Y, Lighthart B. 2000. The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Sci Technol 32:393–403. doi: 10.1080/027868200303533. [DOI] [Google Scholar]
- 48.Monteil CL, Yahara K, Studholme DJ, Mageiros L, Méric G, Swingle B, Morris CE, Vinatzer BA, Sheppard SK. 2016. Population-genomic insights into emergence, crop adaptation and dissemination of Pseudomonas syringae pathogens. Microb Genom 2. doi: 10.1099/mgen.0.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mechan Llontop ME, Tian L, Sharma P, Heflin L, Bernal-Galeano V, Haak DC, Clarke CR, Vinatzer BA. 2021. Experimental evidence pointing to rain as a reservoir of tomato phyllosphere microbiota. Phytobiomes J 5:382–399. doi: 10.1094/PBIOMES-04-21-0025-R. [DOI] [Google Scholar]
- 50.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 51.Acosta-Martínez V, Van Pelt S, Moore-Kucera J, Baddock MC, Zobeck TM. 2015. Microbiology of wind-eroded sediments: current knowledge and future research directions. Aeolian Res 18:99–113. doi: 10.1016/j.aeolia.2015.06.001. [DOI] [Google Scholar]
- 52.Maki LR, Galyan EL, Chang-Chien M-M, Caldwell DR. 1974. Ice nucleation induced by Pseudomonas syringae. Appl Microbiol 28:456–459. doi: 10.1128/am.28.3.456-459.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tignat-Perrier R, Dommergue A, Vogel TM, Larose C. 2020. microbial ecology of the planetary boundary layer. Atmosphere 11:1296. doi: 10.3390/atmos11121296. [DOI] [Google Scholar]
- 54.Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I. 2012. Modelling the effect of size on the aerial dispersal of microorganisms. J Biogeogr 39:89–97. doi: 10.1111/j.1365-2699.2011.02569.x. [DOI] [Google Scholar]
- 55.Bryan NC, Christner BC, Guzik TG, Granger DJ, Stewart MF. 2019. Abundance and survival of microbial aerosols in the troposphere and stratosphere. ISME J 13:2789–2799. doi: 10.1038/s41396-019-0474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton JF, Allen G, Watson NM, Lee JD, Saxton JE, Lewis AC, Vaughan G, Bower KN, Flynn MJ, Crosier J, Carver GD, Harris NRP, Parker RJ, Remedios JJ, Richards NAD. 2008. Observations of an atmospheric chemical equator and its implications for the tropical warm pool region. J Geophys Res 113:D20313. doi: 10.1029/2008JD009940. [DOI] [Google Scholar]
- 57.Archer S, Lee K, Caruso T, Leung M, Tong X, Salter SJ, Hinchliffe G, Maki T, Santl-Temkiv T, Warren-Rhodes K, Gomez-Silva B, Hyde K, Liu C, Alcami A, Al-Mailem D, Araya J, Cary S, Cowan D, Dempsey J, Etchebehere C, Gantsetseg B, Hartery S, Harvey M, Hayakawa K, Hogg I, Inoue M, Kansour M, Lawrence T, Lee C, Leopold M, McKay C, Nagao S, Poh YH, Ramond J-B, Rastrojo A, Sekiguchi T, Sim JH, Stahm W, Sun H, Tang N, Vandenbrink B, Walther C, Lee P, Pointing SB. 2022. Global biogeography of atmospheric microorganisms reflects diverse recruitment and environmental filtering. Reseach Square. doi: 10.21203/rs.3.rs-244923/v4. [DOI] [Google Scholar]
- 58.Šantl-Temkiv T, Amato P, Casamayor EO, Lee PKH, Pointing SB. 2022. Microbial ecology of the atmosphere. FEMS Microbiol Rev 46:fuac009. doi: 10.1093/femsre/fuac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearce DA, Alekhina IA, Terauds A, Wilmotte A, Quesada A, Edwards A, Dommergue A, Sattler B, Adams BJ, Magalhães C, Chu W-L, Lau MCY, Cary C, Smith DJ, Wall DH, Eguren G, Matcher G, Bradley JA, de Vera J-P, Elster J, Hughes KA, Cuthbertson L, Benning LG, Gunde-Cimerman N, Convey P, Hong SG, Pointing SB, Pellizari VH, Vincent WF. 2016. Aerobiology over antarctica – a new initiative for atmospheric ecology. Front Microbiol 7:16. doi: 10.3389/fmicb.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM, Barazesh JM, Anderson BE, Beyersdorf AJ, Ziemba LD, Bergin M, Nenes A, Konstantinidis KT. 2013. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci USA 110:2575–2580. doi: 10.1073/pnas.1212089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bringel F, Couée I. 2015. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front Microbiol 6:486. doi: 10.3389/fmicb.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christner BC. 2012. Cloudy with a chance of microbes. Microbe 7:70–75. doi: 10.1128/microbe.7.70.1. [DOI] [Google Scholar]
- 63.Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev 20:459–477. doi: 10.1128/CMR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Björnham O, Sigg R, Burman J. 2020. Multilevel model for airborne transmission of foot-and-mouth disease applied to Swedish livestock. PLoS One 15:e0232489. doi: 10.1371/journal.pone.0232489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vergadi E, Rouva G, Angeli M, Galanakis E. 2022. Infectious diseases associated with desert dust outbreaks: a systematic review. IJERPH 19:6907. doi: 10.3390/ijerph19116907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McEachran AD, Blackwell BR, Hanson JD, Wooten KJ, Mayer GD, Cox SB, Smith PN. 2015. Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ Health Perspect 123:337–343. doi: 10.1289/ehp.1408555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meola M, Lazzaro A, Zeyer J. 2015. Bacterial composition and survival on sahara dust particles transported to the european alps. Front Microbiol 6:1454. doi: 10.3389/fmicb.2015.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tobías A, Caylà JA, Pey J, Alastuey A, Querol X. 2011. Are Saharan dust intrusions increasing the risk of meningococcal meningitis? Int J Infect Dis 15:e503. doi: 10.1016/j.ijid.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Krumins V, Mainelis G, Kerkhof LJ, Fennell DE. 2014. Substrate-dependent rRNA production in an airborne bacterium. Environ Sci Technol Lett 1:376–381. doi: 10.1021/ez500245y. [DOI] [Google Scholar]
- 70.Dillon KP, Krumins V, Deshpande A, Kerkhof LJ, Mainelis G, Fennell DE. 2022. Characterization and DNA Stable-Isotope Probing of Methanotrophic Bioaerosols. Microbiol Spectr 10:e03421-22. doi: 10.1128/spectrum.03421-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicolaisen M, West JS, Sapkota R, Canning GGM, Schoen C, Justesen AF. 2017. Fungal communities including plant pathogens in near surface air are similar across northwestern Europe. Front Microbiol 8:1729. doi: 10.3389/fmicb.2017.01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rogers SL, Atkins SD, West JS. 2009. Detection and quantification of airborne inoculum of Sclerotinia sclerotiorum using quantitative PCR. Plant Pathol 58:324–331. doi: 10.1111/j.1365-3059.2008.01945.x. [DOI] [Google Scholar]
- 73.Khalaf NF, Al-Obaidi MJ, Mohammed SW, Al-Malkey MK, Nayyef HJ, Al-Hur FJA, Sameer FO, Mesheal KI, Taqi IA, Ad’hiah AH. 2022. Indoor house dust-borne fungi and risk of allergic respiratory diseases in Baghdad city. Rev Franç D’Allergologie 62:401–406. doi: 10.1016/j.reval.2021.05.002. [DOI] [Google Scholar]
- 74.Salawu-Rotimi A, Lebre PH, Vos HC, Fister W, Kuhn N, Eckardt FD, Cowan DA. 2021. Gone with the wind: microbial communities associated with dust from emissive farmlands. Microb Ecol 82:859–869. doi: 10.1007/s00248-021-01717-8. [DOI] [PubMed] [Google Scholar]
- 75.Wardlaw AJ, Rick E-M, Pur Ozyigit L, Scadding A, Gaillard EA, Pashley CH. 2021. New perspectives in the diagnosis and management of allergic fungal airway disease. J Asthma Allergy 14:557–573. doi: 10.2147/JAA.S251709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 77.Perlmutter JI, Bordenstein SR. 2020. Microorganisms in the reproductive tissues of arthropods. Nat Rev Microbiol 18:97–111. doi: 10.1038/s41579-019-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 79.Brockhurst MA, Harrison E, Hall JPJ, Richards T, McNally A, MacLean C. 2019. The Ecology and Evolution of Pangenomes. Curr Biol 29:R1094–R1103. doi: 10.1016/j.cub.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 80.Gong L, He H, Li D, Cao L, Khan TA, Li Y, Pan L, Yan L, Ding X, Sun Y, Zhang Y, Yi G, Hu S, Xia L. 2019. A new isolate of Pediococcus pentosaceus (SL001) with antibacterial activity against fish pathogens and potency in facilitating the immunity and growth performance of grass carps. Front Microbiol 10:1384. doi: 10.3389/fmicb.2019.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543. doi: 10.1111/j.1365-2311.2011.01318.x. [DOI] [Google Scholar]
- 82.Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y, Patt AA, Cui L, Nossa CW, Barry RM, Sakamoto JM, Hornett EA, Rasgon JL. 2014. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA 111:12498–12503. doi: 10.1073/pnas.1408888111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 84.Vorburger C, Gehrer L, Rodriguez P. 2010. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol Lett 6:109–111. doi: 10.1098/rsbl.2009.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Engel P, Moran NA. 2013. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 86.Smith CC, Mueller UG. 2015. Sexual transmission of beneficial microbes. Trends Ecol Evol 30:438–440. doi: 10.1016/j.tree.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Weintraub PG, Beanland L. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol 51:91–111. doi: 10.1146/annurev.ento.51.110104.151039. [DOI] [PubMed] [Google Scholar]
- 88.Heinen R, van der Sluijs M, Biere A, Harvey JA, Bezemer TM. 2018. Plant community composition but not plant traits determine the outcome of soil legacy effects on plants and insects. J Ecol 106:1217–1229. doi: 10.1111/1365-2745.12907. [DOI] [Google Scholar]
- 89.Hannula SE, Zhu F, Heinen R, Bezemer TM. 2019. Foliar-feeding insects acquire microbiomes from the soil rather than the host plant. Nat Commun 10:1254. doi: 10.1038/s41467-019-09284-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW. 2013. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc Natl Acad Sci USA 110:15728–15733. doi: 10.1073/pnas.1308867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo Z, Döll K, Dastjerdi R, Karlovsky P, Dehne H-W, Altincicek B. 2014. Effect of fungal colonization of wheat grains with fusarium spp. on food choice, weight gain and mortality of meal beetle larvae (Tenebrio molitor). PLoS One 9:e100112. doi: 10.1371/journal.pone.0100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luna E, van Eck L, Campillo T, Weinroth M, Metcalf J, Perez-Quintero AL, Botha A-M, Thannhauser TW, Pappin D, Tisserat NA, Lapitan NLV, Argueso CT, Ode PJ, Heck ML, Leach JE. 2018. Bacteria associated with russian wheat aphid (Diuraphis noxia) enhance aphid virulence to wheat. Phytobiomes J 2:151–164. doi: 10.1094/PBIOMES-06-18-0027-R. [DOI] [Google Scholar]
- 93.Aizenberg-Gershtein Y, Izhaki I, Halpern M. 2013. Do honeybees shape the bacterial community composition in floral nectar? PLoS One 8:e67556. doi: 10.1371/journal.pone.0067556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McFrederick QS, Thomas JM, Neff JL, Vuong HQ, Russell KA, Hale AR, Mueller UG. 2017. Flowers and wild megachilid bees share microbes. Microb Ecol 73:188–200. doi: 10.1007/s00248-016-0838-1. [DOI] [PubMed] [Google Scholar]
- 95.Manirajan AB, Ratering S, Rusch V, Schwiertz A, Geissler-Plaum R, Cardinale M, Schnell S. 2016. Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ Microbiol 18:5161–5174. doi: 10.1111/1462-2920.13524. [DOI] [PubMed] [Google Scholar]
- 96.Vannette RL. 2020. The floral microbiome: plant, pollinator, and microbial perspectives. Annu Rev Ecol Evol Syst 51:363–386. doi: 10.1146/annurev-ecolsys-011720-013401. [DOI] [Google Scholar]
- 97.Canto A, Herrera CM. 2012. Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Ann Bot 110:1173–1183. doi: 10.1093/aob/mcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rering CC, Beck JJ, Hall GW, McCartney MM, Vannette RL. 2018. Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. New Phytol 220:750–759. doi: 10.1111/nph.14809. [DOI] [PubMed] [Google Scholar]
- 99.Junker RR, Romeike T, Keller A, Langen D. 2014. Density-dependent negative responses by bumblebees to bacteria isolated from flowers. Apidologie 45:467–477. doi: 10.1007/s13592-013-0262-1. [DOI] [Google Scholar]
- 100.Grell MN, Linde T, Nygaard S, Nielsen KL, Boomsma JJ, Lange L. 2013. The fungal symbiont of Acromyrmex leaf-cutting ants expresses the full spectrum of genes to degrade cellulose and other plant cell wall polysaccharides. BMC Genomics 14:928. doi: 10.1186/1471-2164-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Michaud JM, Thompson LR, Kaul D, Espinoza JL, Richter RA, Xu ZZ, Lee C, Pham KM, Beall CM, Malfatti F, Azam F, Knight R, Burkart MD, Dupont CL, Prather KA. 2018. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat Commun 9:2017. doi: 10.1038/s41467-018-04409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lang-Yona N, Flores JM, Haviv R, Alberti A, Poulain J, Belser C, Trainic M, Gat D, Ruscheweyh H-J, Wincker P, Sunagawa S, Rudich Y, Koren I, Vardi A. 2022. Terrestrial and marine influence on atmospheric bacterial diversity over the north Atlantic and Pacific Oceans. Commun Earth Environ 3:121. doi: 10.1038/s43247-022-00441-6. [DOI] [Google Scholar]
- 103.Thinesh T, Meenatchi R, Jose PA, Kiran GS, Selvin J. 2019. Differential bleaching and recovery pattern of southeast Indian coral reef to 2016 global mass bleaching event: occurrence of stress-tolerant symbiont Durusdinium (Clade D) in corals of Palk Bay. Mar Pollut Bull 145:287–294. doi: 10.1016/j.marpolbul.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 104.Avila-Magaña V, Kamel B, DeSalvo M, Gómez-Campo K, Enríquez S, Kitano H, Rohlfs RV, Iglesias-Prieto R, Medina M. 2021. Elucidating gene expression adaptation of phylogenetically divergent coral holobionts under heat stress. Nat Commun 12:5731. doi: 10.1038/s41467-021-25950-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hentschel U, Piel J, Degnan SM, Taylor MW. 2012. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10:641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 106.Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Almeida EL, Carrillo Rincón AF, Jackson SA, Dobson ADW. 2019. Comparative genomics of marine sponge-derived Streptomyces spp. Isolates SM17 and SM18 with their closest terrestrial relatives provides novel insights into environmental niche adaptations and secondary metabolite biosynthesis potential. Front Microbiol 10:1713. doi: 10.3389/fmicb.2019.01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J, Fu Q, Huang Y, Fan Y, Liang M, Chen H, Yu S. 2022. Negative impacts of sea-level rise on soil microbial involvement in carbon metabolism. Sci Total Environ 838:156087. doi: 10.1016/j.scitotenv.2022.156087. [DOI] [PubMed] [Google Scholar]
- 109.Hamilton AJ, Stagnitti F, Kumarage SC, Premier RR. 2007. RIRA: a tool for conducting health risk assessments for irrigation of edible crops with recycled water. Comp Electron Agricult 57:80–87. doi: 10.1016/j.compag.2007.02.004. [DOI] [Google Scholar]
- 110.Sahlström L, Aspan A, Bagge E, Danielsson- Tham M-L, Albihn A. 2004. Bacterial pathogen incidences in sludge from Swedish sewage treatment plants. Water Res 38:1989–1994. doi: 10.1016/j.watres.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 111.Franz E, van Bruggen AHC. 2008. Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit Rev Microbiol 34:143–161. doi: 10.1080/10408410802357432. [DOI] [PubMed] [Google Scholar]
- 112.Gibb R, Redding DW, Chin KQ, Donnelly CA, Blackburn TM, Newbold T, Jones KE. 2020. Zoonotic host diversity increases in human-dominated ecosystems. Nature 584:398–402. doi: 10.1038/s41586-020-2562-8. [DOI] [PubMed] [Google Scholar]
- 113.van Overbeek LS, van Doorn J, Wichers JH, van Amerongen A, van Roermund HJW, Willemsen PTJ. 2014. The arable ecosystem as battleground for emergence of new human pathogens. Front Microbiol 5:104. doi: 10.3389/fmicb.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the german enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu P, Jia S, He X, Zhang X, Ye L. 2017. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 188:455–464. doi: 10.1016/j.chemosphere.2017.08.162. [DOI] [PubMed] [Google Scholar]
- 116.Han X-M, Hu H-W, Shi X-Z, Wang J-T, Han L-L, Chen D, He J-Z. 2016. Impacts of reclaimed water irrigation on soil antibiotic resistome in urban parks of Victoria, Australia. Environ Pollut 211:48–57. doi: 10.1016/j.envpol.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 117.Cerqueira F, Matamoros V, Bayona JM, Berendonk TU, Elsinga G, Hornstra LM, Piña B. 2019. Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ Res 177:108608. doi: 10.1016/j.envres.2019.108608. [DOI] [PubMed] [Google Scholar]
- 118.Margenat A, Matamoros V, Díez S, Cañameras N, Comas J, Bayona JM. 2017. Occurrence of chemical contaminants in peri-urban agricultural irrigation waters and assessment of their phytotoxicity and crop productivity. Sci Total Environ 599-600:1140–1148. doi: 10.1016/j.scitotenv.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 119.Cernava T, Erlacher A, Soh J, Sensen CW, Grube M, Berg G. 2019. Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa Mill.). Microbiome 7:13. doi: 10.1186/s40168-019-0624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haahtela T. 2019. A biodiversity hypothesis. Allergy 74:1445–1456. doi: 10.1111/all.13763. [DOI] [PubMed] [Google Scholar]
- 121.Tadić Đ, Bleda Hernandez MJ, Cerqueira F, Matamoros V, Piña B, Bayona JM. 2021. Occurrence and human health risk assessment of antibiotics and their metabolites in vegetables grown in field-scale agricultural systems. J Hazard Mater 401:123424. doi: 10.1016/j.jhazmat.2020.123424. [DOI] [PubMed] [Google Scholar]
- 122.Wassermann B, Abdelfattah A, Müller H, Korsten L, Berg G. 2022. The microbiome and resistome of apple fruits alter in the post-harvest period. Environ Microbiome 17:10. doi: 10.1186/s40793-022-00402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mencía-Ares O, Cabrera-Rubio R, Cobo-Díaz JF, Álvarez-Ordóñez A, Gómez-García M, Puente H, Cotter PD, Crispie F, Carvajal A, Rubio P, Argüello H. 2020. Antimicrobial use and production system shape the fecal, environmental, and slurry resistomes of pig farms. Microbiome 8:164. doi: 10.1186/s40168-020-00941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun J, Liao XP, D’Souza AW, Boolchandani M, Li SH, Cheng K, Luis MJ, Li L, Feng YJ, Fang LX, Huang T, Xia J, Yu Y, Zhou YF, Sun YX, Deng XB, Zeng ZL, Jiang HX, Fang BH, Yan QL, Dantas G, Liu YH. 2020. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nature Commun 11:1–11. doi: 10.1038/s41467-020-15222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Somerville V, Lutz S, Schmid M, Frei D, Moser A, Irmler S, Frey JE, Ahrens CH. 2019. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol 19:143. doi: 10.1186/s12866-019-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leff JW, Fierer N. 2013. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marco ML, Hutkins R, Hill C, Fulgoni VL, Cifelli CJ, Gahche J, Slavin JL, Merenstein D, Tancredi DJ, Sanders ME. 2022. A classification system for defining and estimating dietary intake of live microbes in us adults and children. J Nutr 152:1729–1736. doi: 10.1093/jn/nxac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wassermann B, Müller H, Berg G. 2019. An apple a day: which bacteria do we eat with organic and conventional apples? Front Microbiol 10:1629. doi: 10.3389/fmicb.2019.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang H, Zhang Q, Chen S, Zhang Z, Song J, Long Z, Yu Y, Fang H. 2020. Enterobacteriaceae predominate in the endophytic microbiome and contribute to the resistome of strawberry. Sci Total Environ 727:138708. doi: 10.1016/j.scitotenv.2020.138708. [DOI] [PubMed] [Google Scholar]
- 130.Abdelfattah A, Whitehead SR, Macarisin D, Liu J, Burchard E, Freilich S, Dardick C, Droby S, Wisniewski M. 2020. Effect of washing, waxing and low-temperature storage on the postharvest microbiome of apple. Microorganisms 8:944. doi: 10.3390/microorganisms8060944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wicaksono WA, Buko A, Kusstatscher P, Sinkkonen A, Laitinen OH, Virtanen SM, Hyöty H, Cernava T, Berg G. 2022. Modulation of the food microbiome by apple fruit processing. Food Microbiol 108:104103. doi: 10.1016/j.fm.2022.104103. [DOI] [PubMed] [Google Scholar]
- 132.Knight S, Klaere S, Fedrizzi B, Goddard MR. 2015. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep 5:14233–14210. doi: 10.1038/srep14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gilbert JA, van der Lelie D, Zarraonaindia I. 2014. Microbial terroir for wine grapes. Proc Natl Acad Sci USA 111:5–6. doi: 10.1073/pnas.1320471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gobbi A, Acedo A, Imam N, Santini RG, Ortiz-Álvarez R, Ellegaard-Jensen L, Belda I, Hansen LH. 2022. A global microbiome survey of vineyard soils highlights the microbial dimension of viticultural terroirs. Commun Biol 5:241. doi: 10.1038/s42003-022-03202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pasolli E, De Filippis F, Mauriello IE, Cumbo F, Walsh AM, Leech J, Cotter PD, Segata N, Ercolini D. 2020. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat Commun 11:1–12. doi: 10.1038/s41467-020-16438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Larsen MH, Dalmasso M, Ingmer H, Langsrud S, Malakauskas M, Mader A, Møretrø T, Smole Možina S, Rychli K, Wagner M, Wallace JR, Zentek J, Jordan K. 2014. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Contr 44:92–109. doi: 10.1016/j.foodcont.2014.03.039. [DOI] [Google Scholar]
- 138.Derrien M, van Hylckama Vlieg JET. 2015. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 139.Milani C, Duranti S, Napoli S, Alessandri G, Mancabelli L, Anzalone R, Longhi G, Viappiani A, Mangifesta M, Lugli GA, Bernasconi S, Ossiprandi MC, Van Sinderen D, Ventura M, Turroni F. 2019. Colonization of the human gut by bovine bacteria present in Parmesan cheese. Nat Commun 10:12. doi: 10.1038/s41467-019-09303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hallen-Adams HE, Suhr MJ. 2017. Fungi in the healthy human gastrointestinal tract. Virulence 8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mamaev D, Zvyagilskaya R. 2021. Yarrowia lipolytica: a multitalented yeast species of ecological significance. FEMS Yeast Res 21:1–18. doi: 10.1093/femsyr/foab008. [DOI] [PubMed] [Google Scholar]
- 142.Fadda ME, Mossa V, Deplano M, Pisano MB, Cosentino S. 2017. In vitro screening of Kluyveromyces strains isolated from Fiore Sardo cheese for potential use as probiotics. LWT - Food Sci Technol 75:100–106. doi: 10.1016/j.lwt.2016.08.020. [DOI] [Google Scholar]
- 143.Ferrocino I, Ponzo V, Pellegrini M, Goitre I, Papurello M, Franciosa I, D’Eusebio C, Ghigo E, Cocolin L, Bo S. 2022. Mycobiota composition and changes across pregnancy in patients with gestational diabetes mellitus (GDM). Sci Rep 12:1–13. doi: 10.1038/s41598-022-13438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chaillou S, Chaulot-Talmon A, Caekebeke H, Cardinal M, Christieans S, Denis C, Desmonts MH, Dousset X, Feurer C, Hamon E, Joffraud J-J, La Carbona S, Leroi F, Leroy S, Lorre S, Macé S, Pilet M-F, Prévost H, Rivollier M, Roux D, Talon R, Zagorec M, Champomier-Vergès M-C. 2015. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J 9:1105–1118. doi: 10.1038/ismej.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Doyle CJ, Gleeson D, O'Toole PW, Cotter PD. 2017. Impacts of seasonal housing and teat preparation on raw milk microbiota: a high-throughput sequencing study. Appl Environ Microbiol 83:e02694-16. doi: 10.1128/AEM.02694-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McHugh AJ, Feehily C, Fenelon MA, Gleeson D, Hill C, Cotter PD. 2020. Tracking the dairy microbiota from farm bulk tank to skimmed milk powder. mSystems 5:e02694-16. doi: 10.1128/mSystems.00226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ferrocino I, Rantsiou K, Cocolin L. 2022. Investigating dairy microbiome: an opportunity to ensure quality, safety and typicity. Curr Opin Biotechnol 73:164–170. doi: 10.1016/j.copbio.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 148.Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligné B, Gänzle M, Kort R, Pasin G, Pihlanto A, Smid EJ, Hutkins R. 2017. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol 44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 149.EFSA. 2022. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J 20:7209. doi: 10.2903/j.efsa.2022.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Montville R, Chen Y, Schaffner DW. 2001. Glove barriers to bacterial cross-contamination between hands to food. J Food Prot 64:845–849. doi: 10.4315/0362-028x-64.6.845. [DOI] [PubMed] [Google Scholar]
- 151.Midelet G, Carpentier B. 2002. Transfer of microorganisms, including Listeria monocytogenes, from various materials to beef. Appl Environ Microbiol 68:4015–4024. doi: 10.1128/AEM.68.8.4015-4024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arora A, Dogra A, Dogra A, Goyal B, Sharma MA. 2018. Nipah virus: an outbreak of deadly paramyxvirus. Biomed Pharmacol J 11:1177–1185. doi: 10.13005/bpj/1479. [DOI] [Google Scholar]
- 153.Reese AT, Madden AA, Joossens M, Lacaze G, Dunn RR. 2020. Influences of ingredients and bakers on the bacteria and fungi in sourdough starters and bread. MSphere 5:e00950-19. doi: 10.1128/mSphere.00950-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Luber P, Brynestad S, Topsch D, Scherer K, Bartelt E. 2006. Quantification of Campylobacter species cross-contamination during handling of contaminated fresh chicken parts in kitchens. Appl Environ Microbiol 72:66–70. doi: 10.1128/AEM.72.1.66-70.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zwirzitz B, Wetzels SU, Dixon ED, Fleischmann S, Selberherr E, Thalguter S, Quijada NM, Dzieciol M, Wagner M, Stessl B. 2021. Co-occurrence of Listeria spp. and spoilage associated microbiota during meat processing due to cross-contamination events. Front Microbiol 12:632935. doi: 10.3389/fmicb.2021.632935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schaffner DW, Jensen D, Gerba CP, Shumaker D, Arbogast J. 2018. Influence of soap characteristics and food service facility type on the degree of bacterial contamination of open, refillable bulk soaps. J Food Prot 81:218–225. doi: 10.4315/0362-028X.JFP-17-251. [DOI] [PubMed] [Google Scholar]
- 157.Olmo R, Wetzels SU, Armanhi JSL, Arruda P, Berg G, Cernava T, Cotter PD, Araujo SC, de Souza RSC, Ferrocino I, Frisvad JC, Georgalaki M, Hansen HH, Kazou M, Kiran GS, Kostic T, Krauss-Etschmann S, Kriaa A, Lange L, Maguin E, Mitter B, Nielsen MO, Olivares M, Quijada NM, Romaní-Pérez M, Sanz Y, Schloter M, Schmitt-Kopplin P, Craven Seaton S, Selvin J, Sessitsch A, Wang M, Zwirzitz B, Selberherr E, Wagner M. 2022. Microbiome research as an effective driver of success stories in agrifood systems – a selection of case studies. Front Microbiol 13:834622. doi: 10.3389/fmicb.2022.834622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bach A, Joulie I, Chevaux E, Elcoso G, Ragués J. 2021. Milk performance and rumen microbiome of dairy cows as affected by the inclusion of corn silage or corn shredlage in a total mixed ration. Animal 15:100014. doi: 10.1016/j.animal.2020.100014. [DOI] [PubMed] [Google Scholar]
- 159.Belloso Daza MV, Milani G, Cortimiglia C, Pietta E, Bassi D, Cocconcelli PS. 2022. Genomic Insights of Enterococcus faecium UC7251, a Multi-Drug Resistant Strain From Ready-to-Eat Food, Highlight the Risk of Antimicrobial Resistance in the Food Chain. Front Microbiol 13:894241. doi: 10.3389/fmicb.2022.894241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pérez-Rodríguez F, Mercanoglu Taban B. 2019. A state-of-art review on multi-drug resistant pathogens in foods of animal origin: risk factors and mitigation strategies. Front Microbiol 10:2091. doi: 10.3389/fmicb.2019.02091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Ramnik X, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Korpela K, de Vos WM. 2022. Infant gut microbiota restoration: state of the art. Gut Microbes 14:2118811. doi: 10.1080/19490976.2022.2118811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mitchell CM, Mazzoni C, Hogstrom L, Bryant A, Bergerat A, Cher A, Pochan S, Herman P, Carrigan M, Sharp K, Huttenhower C, Lander ES, Vlamakis H, Xavier RJ, Yassour M. 2020. Delivery mode affects stability of early infant gut microbiota. Cell Rep Med 1:100156. doi: 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marcobal A, Sonnenburg JL. 2012. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect 18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Bruggen AHC, Finckh MR, He M, Ritsema CJ, Harkes P, Knuth D, Geissen V. 2021. Indirect effects of the herbicide glyphosate on plant, animal and human health through its effects on microbial communities. Front Environ Sci 9:763917. doi: 10.3389/fenvs.2021.763917. [DOI] [Google Scholar]
- 166.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ríos-Covian D, Langella P, Martín R. 2021. From short- to long-term effects of c=C-section delivery on microbiome establishment and host health. Microorganisms 9:2122. doi: 10.3390/microorganisms9102122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Doare KL, Holder B, Bassett A, Pannaraj PS. 2018. Mother’s milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol 9:e00361. doi: 10.3389/fimmu.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Murphy K, Curley D, O'Callaghan TF, O'Shea C-A, Dempsey EM, O'Toole PW, Ross RP, Ryan CA, Stanton C. 2017. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep 7:40597. doi: 10.1038/srep40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, Bailey A, Bushman FD, Sleasman JW, Aldrovandi GM. 2017. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Jun W. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 172.Klein-Jöbstl D, Quijada NM, Dzieciol M, Feldbacher B, Wagner M, Drillich M, Schmitz-Esser S, Mann E. 2019. Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves’ gastrointestinal microbiota. PLoS One 14:e0220554. doi: 10.1371/journal.pone.0220554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Renz H, Holt PG, Inouye M, Logan AC, Prescott SL, Sly PD. 2017. An exposome perspective: early-life events and immune development in a changing world. J Allergy Clin Immunol 140:24–40. doi: 10.1016/j.jaci.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 174.Pechlivanis S, Von Mutius E. 2020. Effect of Farming on Asthma Introduction Asthma is a chronic airway inflammation that causes coughing, wheezing, chest tightness or Prevention (CDC) from the United States. ama 49:144–155. doi: 10.5644/ama2006-124.293. [DOI] [PubMed] [Google Scholar]
- 175.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, Ledford JG, Marques dos Santos M, Anderson RL, Metwali N, Neilson JW, Maier RM, Gilbert JA, Holbreich M, Thorne PS, Martinez FD, von Mutius E, Vercelli D, Ober C, Sperling AI. 2016. Innate immunity and asthma risk in amish and hutterite farm children. N Engl J Med 375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pongsiri MJ, Bickersteth S, Colón C, DeFries R, Dhaliwal M, Georgeson L, Haines A, Linou N, Murray V, Naeem S, Small R, Ungvari J. 2019. Planetary health: from concept to decisive action. Lancet Planet Health 3:e402–e404. doi: 10.1016/S2542-5196(19)30190-1. [DOI] [PubMed] [Google Scholar]
- 177.Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, Palloni A, Sorenson T, Rey FE, Herd P. 2019. Close social relationships correlate with human gut microbiota composition. Sci Rep 9:703. doi: 10.1038/s41598-018-37298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Brito IL, Gurry T, Zhao S, Huang K, Young SK, Shea TP, Naisilisili W, Jenkins AP, Jupiter SD, Gevers D, Alm EJ. 2019. Transmission of human-associated microbiota along family and social networks. Nat Microbiol 4:964–971. doi: 10.1038/s41564-019-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kort R, Caspers M, van de Graaf A, van Egmond W, Keijser B, Roeselers G. 2014. Shaping the oral microbiota through intimate kissing. Microbiome 2:41. doi: 10.1186/2049-2618-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, de Beaufort C, Sobhani I, Heintz-Buschart A, Sunagawa S, Zeller G, Wilmes P, Bork P. 2019. Extensive transmission of microbes along the gastrointestinal tract. Elife 8:e42693. doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sarkar A, Harty S, Johnson KVA, Moeller AH, Archie EA, Schell LD, Carmody RN, Clutton-Brock TH, Dunbar RIM, Burnet PWJ. 2020. Microbial transmission in animal social networks and the social microbiome. Nat Ecol Evol 4:1020–1035. doi: 10.1038/s41559-020-1220-8. [DOI] [PubMed] [Google Scholar]
- 182.Valles-Colomer M, Blanco-Míguez A, Manghi P, Asnicar F, Dubois L, Golzato D, Armanini F, Cumbo F, Huang KD, Manara S, Masetti G, Pinto F, Piperni E, Punčochář M, Ricci L, Zolfo M, Farrant O, Goncalves A, Selma-Royo M, Binetti AG, Becerra JE, Han B, Lusingu J, Amuasi J, Amoroso L, Visconti A, Steves CM, Falchi M, Filosi M, Tett A, Last A, Xu Q, Qin N, Qin H, May J, Eibach D, Corrias MV, Ponzoni M, Pasolli E, Spector TD, Domenici E, Collado MC, Segata N. 2023. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614:125–135. doi: 10.1038/s41586-022-05620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Settanni CR, Ianiro G, Bibbò S, Cammarota G, Gasbarrini A. 2021. Gut microbiota alteration and modulation in psychiatric disorders: current evidence on fecal microbiota transplantation. Prog Neuropsychopharmacol Biol Psychiatry 109:110258. doi: 10.1016/j.pnpbp.2021.110258. [DOI] [PubMed] [Google Scholar]
- 185.Lewin-Epstein O, Hadany L. 2020. Host–microbiome coevolution can promote cooperation in a rock–paper–scissors dynamics. Proc Biol Sci 287:20192754. doi: 10.1098/rspb.2019.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee S-H, Yoon S-H, Jung Y, Kim N, Min U, Chun J, Choi I. 2020. Emotional well-being and gut microbiome profiles by enterotype. Sci Rep 10:20736. doi: 10.1038/s41598-020-77673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Johnson KV-A, Watson KK, Dunbar RIM, Burnet PWJ. 2022. Sociability in a non-captive macaque population is associated with beneficial gut bacteria. Front Microbiol 13:1032495. doi: 10.3389/fmicb.2022.1032495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Du G, Huang H, Zhu Q, Ying L. 2021. Effects of cat ownership on the gut microbiota of owners. PLoS One 16:e0253133. doi: 10.1371/journal.pone.0253133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wetzels SU, Strachan CR, Conrady B, Wagner M, Burgener IA, Virányi Z, Selberherr E. 2021. Wolves, dogs and humans in regular contact can mutually impact each other’s skin microbiota. Sci Rep 11:17106. doi: 10.1038/s41598-021-96160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.EMA. 2020. Reflection paper on the risk of antimicrobial resistance transfer from companion animals. https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-risk-antimicrobial-resistance-transfer-companion-animals_en.pdf.
- 191.Belas A, Menezes J, Gama LT, Pomba C. 2020. Sharing of clinically important antimicrobial resistance genes by companion animals and their human household members. Microb Drug Resist 26:1174–1185. doi: 10.1089/mdr.2019.0380. [DOI] [PubMed] [Google Scholar]
- 192.Menezes J, Moreira da Silva J, Frosini S-M, Loeffler A, Weese S, Perreten V, Schwarz S, Telo da Gama L, Amaral AJ, Pomba C. 2022. mcr-1 colistin resistance gene sharing between Escherichia coli from cohabiting dogs and humans, Lisbon, Portugal, 2018 to 2020. Eurosurveillance 27:2101144. doi: 10.2807/1560-7917.ES.2022.27.44.2101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Naziri Z, Poormaleknia M, Ghaedi Oliyaei A. 2022. Risk of sharing resistant bacteria and/or resistance elements between dogs and their owners. BMC Vet Res 18:203. doi: 10.1186/s12917-022-03298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao R, Hao J, Yang J, Tong C, Xie L, Xiao D, Zeng Z, Xiong W. 2022. The co-occurrence of antibiotic resistance genes between dogs and their owners in families. IMeta 1:21. doi: 10.1002/imt2.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJM, Brown GZ, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J 6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Joung YS, Ge Z, Buie CR. 2017. Bioaerosol generation by raindrops on soil. Nat Commun 8:14668. doi: 10.1038/ncomms14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Peltola J, Andersson MA, Haahtela T, Mussalo-Rauhamaa H, Rainey FA, Kroppenstedt RM, Samson RA, Salkinoja-Salonen MS. 2001. Toxic-metabolite-producing bacteria and fungus in an indoor environment. Appl Environ Microbiol 67:3269–3274. doi: 10.1128/AEM.67.7.3269-3274.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kozdrój J, Frączek K, Ropek D. 2019. Assessment of bioaerosols in indoor air of glasshouses located in a botanical garden. Build Environ 166:106436. doi: 10.1016/j.buildenv.2019.106436. [DOI] [Google Scholar]
- 199.Triadó-Margarit X, Veillette M, Duchaine C, Talbot M, Amato F, Minguillón MC, Martins V, de Miguel E, Casamayor EO, Moreno T. 2017. Bioaerosols in the Barcelona subway system. Indoor Air 27:564–575. doi: 10.1111/ina.12343. [DOI] [PubMed] [Google Scholar]
- 200.O’Gorman CM. 2011. Airborne Aspergillus fumigatus conidia: a risk factor for aspergillosis. Fung Biol Rev 25:151–157. doi: 10.1016/j.fbr.2011.07.002. [DOI] [Google Scholar]
- 201.Lange L, Huang Y, Busk PK. 2016. Microbial decomposition of keratin in nature—a new hypothesis of industrial relevance. Appl Microbiol Biotechnol 100:2083–2096. doi: 10.1007/s00253-015-7262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Grice EA, Segre JA. 2011. The skin microbiome. Nat Rev Microbiol 9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Oberauner L, Zachow C, Lackner S, Högenauer C, Smolle K-H, Berg G. 2013. The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Sci Rep 3:1413. doi: 10.1038/srep01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mahnert A, Moissl-Eichinger C, Berg G. 2015. Microbiome interplay: plants alter microbial abundance and diversity within the built environment. Front Microbiol 6:e00887. doi: 10.3389/fmicb.2015.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hui N, Parajuli A, Puhakka R, Grönroos M, Roslund MI, Vari HK, Selonen VAO, Yan G, Siter N, Nurminen N, Oikarinen S, Laitinen OH, Rajaniemi J, Hyöty H, Sinkkonen A. 2019. Temporal variation in indoor transfer of dirt-associated environmental bacteria in agricultural and urban areas. Environ Int 132:105069. doi: 10.1016/j.envint.2019.105069. [DOI] [PubMed] [Google Scholar]
- 206.Parajuli A, Hui N, Puhakka R, Oikarinen S, Grönroos M, Selonen VAO, Siter N, Kramna L, Roslund MI, Vari HK, Nurminen N, Honkanen H, Hintikka J, Sarkkinen H, Romantschuk M, Kauppi M, Valve R, Cinek O, Laitinen OH, Rajaniemi J, Hyöty H, Sinkkonen A, ADELE study group (all additional members of the ADELE study group in Lahti and Tampere) . 2020. Yard vegetation is associated with gut microbiota composition. Sci Total Environ 713:136707. doi: 10.1016/j.scitotenv.2020.136707. [DOI] [PubMed] [Google Scholar]
- 207.Bäumel S, Tytgat HLP, Nemec B, Schmidt R, Chia LW, Smidt H. 2018. Fifty Percent Human – how art brings us in touch with our microbial cohabitants. Microb Biotechnol 11:571–574. doi: 10.1111/1751-7915.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.WHO. 2022. Global report on infection prevention and control. ISBN: 978–92-4–005116-4. https://www.who.int/publications/i/item/9789240051164.
- 209.Jabłońska-Trypuć A, Makuła M, Włodarczyk-Makuła M, Wołejko E, Wydro U, Serra-Majem L, Wiater J. 2022. Inanimate surfaces as a source of hospital infections caused by fungi, bacteria and viruses with particular emphasis on SARS-CoV-2. IJERPH 19:8121. doi: 10.3390/ijerph19138121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sharma K, Goss EM, Dickstein ER, Smith ME, Johnson JA, Southwick FS, van Bruggen AHC. 2014. Exserohilum rostratum: characterization of a cross-kingdom pathogen. PLoS One 9:e108691. doi: 10.1371/journal.pone.0108691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cason C, D’Accolti M, Campisciano G, Soffritti I, Ponis G, Mazzacane S, Maggiore A, Risso FM, Comar M, Caselli E. 2021. Microbial contamination in hospital environment has the potential to colonize preterm newborns’ nasal cavities. Pathogens 10:615. doi: 10.3390/pathogens10050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Roguet A, Newton RJ, Eren AM, McLellan SL. 2022. Guts of the urban ecosystem: microbial ecology of sewer infrastructure. mSystems 7:e00118-22. doi: 10.1128/msystems.00118-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wu L, Ning D, Zhang B, Li Y, Zhang P, Shan X, Zhang Q, Brown MR, Li Z, Van Nostrand JD, Ling F, Xiao N, Zhang Y, Vierheilig J, Wells GF, Yang Y, Deng Y, Tu Q, Wang A, Zhang T, He Z, Keller J, Nielsen PH, Alvarez PJJ, Criddle CS, Wagner M, Tiedje JM, He Q, Curtis TP, Stahl DA, Alvarez-Cohen L, Rittmann BE, Wen X, Zhou J, Global Water Microbiome Consortium . 2019. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat Microbiol 4:1183–1195. doi: 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- 214.Abdugheni R, Wang W, Wang Y, Du M, Liu F, Zhou N, Jiang C, Wang C, Wu L, Ma J, Liu C, Liu S. 2022. Metabolite profiling of human-originated Lachnospiraceae at the strain level. IMeta 1:e58. doi: 10.1002/imt2.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Campisano A, Ometto L, Compant S, Pancher M, Antonielli L, Yousaf S, Varotto C, Anfora G, Pertot I, Sessitsch A, Rota-Stabelli O. 2014. Interkingdom transfer of the acne-causing agent, propionibacterium acnes, from human to grapevine. Mol Biol Evol 31:1059–1065. doi: 10.1093/molbev/msu075. [DOI] [PubMed] [Google Scholar]
- 216.Crits-Christoph A, Kantor RS, Olm MR, Whitney ON, Al-Shayeb B, Lou YC, Flamholz A, Kennedy LC, Greenwald H, Hinkle A, Hetzel J, Spitzer S, Koble J, Tan A, Hyde F, Schroth G, Kuersten S, Banfield JF, Nelson KL. 2021. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio 12:e02703-20. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Karthikeyan S, Nguyen A, McDonald D, Zong Y, Ronquillo N, Ren J, Zou J, Farmer S, Humphrey G, Henderson D, Javidi T, Messer K, Anderson C, Schooley R, Martin NK, Knight R. 2021. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems 6:e00793-21. doi: 10.1128/mSystems.00793-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tiwari A, Adhikari S, Kaya D, Islam MA, Malla B, Sherchan SP, Al-Mustapha AI, Kumar M, Aggarwal S, Bhattacharya P, Bibby K, Halden RU, Bivins A, Haramoto E, Oikarinen S, Heikinheimo A, Pitkänen T. 2023. Monkeypox outbreak: wastewater and environmental surveillance perspective. Sci Total Environ 856:159166. doi: 10.1016/j.scitotenv.2022.159166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rothman JA, Whiteson KL. 2022. Sequencing and variant detection of eight abundant plant-infecting tobamoviruses across southern California wastewater. Microbiol Spectr 10:e03050-22. doi: 10.1128/spectrum.03050-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Salem N, Mansour A, Ciuffo M, Falk BW, Turina M. 2016. A new tobamovirus infecting tomato crops in Jordan. Arch Virol 161:503–506. doi: 10.1007/s00705-015-2677-7. [DOI] [PubMed] [Google Scholar]
- 221.Mallon CA, van Elsas JD, Salles JF. 2015. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol 23:719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 222.Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. 2015. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA 112:E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Korpela K, Helve O, Kolho K-L, Saisto T, Skogberg K, Dikareva E, Stefanovic V, Salonen A, Andersson S, de Vos WM. 2020. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: a proof-of-concept study. Cell 183:324–334.e5. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 224.Wolfe BE, Dutton RJ. 2015. Fermented foods as experimentally tractable microbial ecosystems. Cell 161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 225.Gänzle MG, Zheng J. 2019. Lifestyles of sourdough lactobacilli – do they matter for microbial ecology and bread quality? Int J Food Microbiol 302:15–23. doi: 10.1016/j.ijfoodmicro.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 226.Raaijmakers JM, Mazzola M. 2016. Soil immune responses. Science 352:1392–1393. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 227.Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, Sim CK, Lim AI, Link VM, Enamorado M, Trinchieri G, Segre JA, Rehermann B, Belkaid Y. 2021. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 184:615–627.e617. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hanssen NMJ, de Vos WM, Nieuwdorp M. 2021. Fecal microbiota transplantation in human metabolic diseases: from a murky past to a bright future? Cell Metab 33:1098–1110. doi: 10.1016/j.cmet.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 229.Benítez-Páez A, Hartstra AV, Nieuwdorp M, Sanz Y. 2022. Species- and strain-level assessment using rrn long-amplicons suggests donor's influence on gut microbial transference via fecal transplants in metabolic syndrome subjects. Gut Microbes 14:2078621. doi: 10.1080/19490976.2022.2078621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, Emani R, Cani PD, De Vos WM. 2018. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut 67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 231.Scheuerl T, Hopkins M, Nowell RW, Rivett DW, Barraclough TG, Bell T. 2020. Bacterial adaptation is constrained in complex communities. Nat Commun 11:754. doi: 10.1038/s41467-020-14570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li J, Wang J, Liu H, Macdonald CA, Singh BK. 2022. Application of microbial inoculants significantly enhances crop productivity: a meta-analysis of studies from 2010 to 2020. J Sust Agri & Env 1:216–225. doi: 10.1002/sae2.12028. [DOI] [Google Scholar]
- 233.Tarfeen N, Nisa KU, Hamid B, Bashir Z, Yatoo AM, Dar MA, Mohiddin FA, Amin Z, Ahmad RA, Sayyed RZ. 2022. Microbial remediation: a promising tool for reclamation of contaminated sites with special emphasis on heavy metal and pesticide pollution: a review. Processes 10:1358. doi: 10.3390/pr10071358. [DOI] [Google Scholar]
- 234.Cunningham M, Azcarate-Peril MA, Barnard A, Benoit V, Grimaldi R, Guyonnet D, Holscher HD, Hunter K, Manurung S, Obis D, Petrova MI, Steinert RE, Swanson KS, van Sinderen D, Vulevic J, Gibson GR. 2021. Shaping the future of probiotics and prebiotics. Trends Microbiol 29:667–685. doi: 10.1016/j.tim.2021.01.003. [DOI] [PubMed] [Google Scholar]