SUMMARY
Type III secretion systems (T3SSs) are utilized by Gram-negative pathogens to enhance their pathogenesis. This secretion system is associated with the delivery of effectors through a needle-like structure from the bacterial cytosol directly into a target eukaryotic cell. These effector proteins then manipulate specific eukaryotic cell functions to benefit pathogen survival within the host. The obligate intracellular pathogens of the family Chlamydiaceae have a highly evolutionarily conserved nonflagellar T3SS that is an absolute requirement for their survival and propagation within the host with about one-seventh of the genome dedicated to genes associated with the T3SS apparatus, chaperones, and effectors. Chlamydiae also have a unique biphasic developmental cycle where the organism alternates between an infectious elementary body (EB) and replicative reticulate body (RB). T3SS structures have been visualized on both EBs and RBs. And there are effector proteins that function at each stage of the chlamydial developmental cycle, including entry and egress. This review will discuss the history of the discovery of chlamydial T3SS and the biochemical characterization of components of the T3SS apparatus and associated chaperones in the absence of chlamydial genetic tools. These data will be contextualized into how the T3SS apparatus functions throughout the chlamydial developmental cycle and the utility of heterologous/surrogate models to study chlamydial T3SS. Finally, there will be a targeted discussion on the history of chlamydial effectors and recent advances in the field.
KEYWORDS: chlamydia, type III secretion
INTRODUCTION
Chlamydiae have an evolutionarily conserved nonflagellar type III secretion system (T3SS), which is dedicated to virulence and is an absolute requirement for intracellular survival. Only Gram-negative bacteria, both pathogens and symbionts, express T3SSs to deliver effector proteins from the bacterial cytosol to the eukaryotic cell cytosol (reviewed in reference 1). A T3SS is comprised of the main secretion apparatus, chaperones, and effector proteins. The T3SS apparatus is a needle-like structure that originates in the bacterial cytosol and spans the inner membrane, periplasmic space, and outer membrane. The internal diameter of the needle is too narrow to accommodate fully folded effector proteins, and hence, the T3SS chaperone proteins function to usher effectors in a partially folded or unfolded state to the apparatus. To enhance pathogen survival, the effector proteins target specific eukaryotic cell functions to subvert host defense mechanisms or commandeer normal host functions (i.e., inhibition of Toll-like receptor signaling pathways or the promotion of endocytosis of the bacteria, respectively). For some Gram-negative pathogens, the T3SS is one of many virulence factors. But for Chlamydia, the genes encoding components of the T3SS, including effectors, comprise 10 to 14% of a highly reduced genome and is regarded as the main, and if not the most important, virulence factor (2).
Chlamydiae exhibit a biphasic developmental cycle (reviewed in reference 3), which is initiated by an infectious elementary body (EB) entering the host cell within a vacuole that ultimately forms the chlamydial inclusion. The EB then undergoes primary differentiation into a no-infectious reticulate body (RB) that grows and divides by a polarized budding mechanism within the inclusion (4). At later time points postentry, some RBs cease to divide and undergo an asynchronous secondary differentiation forming new infectious EBs. Chlamydial genes are expressed temporally at distinct stages of the chlamydial developmental cycle (5). A functional T3SS apparatus and effectors have been associated with chlamydial entry, establishment of the inclusion, interception of host trafficking pathways to acquire nutrients, and egress—essentially at all stages of chlamydial development and pathogenesis (reviewed in reference 6 and 7).
The chlamydial T3SS is thought to have been acquired about 700 million years ago, when the last common ancestor of phylum Chlamydiae had already adapted to intracellular survival (8, 9). Recent studies have indicated that Chlamydia diverged from a superphylum and ancient endosymbionts (Planctomycetes-Verrucomicrobia-Chlamydiae [PVC]) 1 to 2 billion years ago. Genetic analysis of the last common ancestor of Chlamydiae revealed that genes encoding components of T3SS were acquired within the same time frame as genes involved in energy parasitism and biphasic development (9). Hence, this information highlights the rationale for why those types of genes were maintained after Chlamydia underwent reductive evolution (10–12). These characteristics contrast with those of other pathogenic Gram-negative bacteria, as their T3SSs were acquired primarily to enhance pathogenesis. Within pathogenic or symbiotic Gram-negative bacteria, T3SSs are often encoded by genes organized in genomic islands (commonly referred to as pathogenicity islands in pathogenic bacteria) or found on plasmids. Furthermore, these genomic islands are flanked with transposon or insertion sequence elements, and often these stretches of genes differ in G+C content compared with the overall G+C content of the genome of the bacteria (13). Consistent with when Chlamydiales acquired T3SS, there are no discernible differences in G+C content of chlamydial T3SS genes compared with the rest of the genome and no evidence of horizontal gene transfer elements (2, 14, 15). Genes encoding proteins associated with the main T3SS apparatus and some chaperones are organized in 4 to 6 gene clusters, depending on the chlamydial species. Genes encoding chaperones associated with effector proteins and effectors proteins are found within various operons or as single open reading frames distributed throughout the genome (2, 15). This type of genetic organization has been dubbed a “pathogenicity archipelago” to distinguish it from the tight and distinct genomic organization of a genomic or pathogenicity island (16).
Of note, the nonflagellar T3SS arose from an exaptation of bacterial flagella, which were found the free-living members of the ancient PVC superphylum (9). The intermediate ancestral form of a nonflagellar T3SS was found in Myxococcales, where this system lacked essential elements associated with motility but retained features required for protein translocation, as in the ability to secrete flagellin subunits to form a structure. Again, this information is consistent with the nonmotile nature of pathogenic chlamydial species. The nonflagellar T3SS became fully functional after a second adaptation that involved the acquisition of secretins (17). Given the evolutionary history of nonflagellar T3SSs generally, it is easy to understand how nonmotile chlamydial species have annotated genes (e.g., fliI and flhA) that are more commonly associated with functional flagella (16). It is possible that the chlamydial T3SS represents an original prototype of T3SSs from which all others evolved; however, given the genomic organization of the chlamydial T3SS, it is more likely an example of how indispensable genes become permanently organized within a genome.
DISCOVERY OF THE CHLAMYDIAL T3SS
Electron microscopy images from the late 1970s and early 1980s revealed spike-like projections, often in a rosette formation, on both chlamydial developmental forms (EB and RB) (18–21). These projections or rosettes were hypothesized initially to be adhesins or porins. However, several of these transmission electron micrographs captured clear projections that emanated from RBs through the inclusion membrane and into the host cytosol (19). These images inspired the “soup through straw” hypothesis first presented by Richard Stephens in 1992 in which he proposed that these projections helped the pathogens siphon metabolites from the host cell cytosol without triggering detection from the endogenous major histocompatibility complex (MHC) class I pathways (22).
Also, in the early 1990s, characterization of novel Yersinia Yop proteins led to the discovery of a new secretion system for Gram-negative bacteria (23). A large gene cluster within the virC virulence plasmid-associated locus that encoded 13 genes was absolutely required for Yop secretion (24). This study was the first description of the Ysc (Yop secretion) secretion machinery (24) and the foundational data responsible for the eventual naming of the novel secretion system, now commonly known as the type III secretion system (T3SS) (25). Furthermore, T3SS gene expression was induced upon a temperature shift to 37°C and inhibited by Ca2+ (26). A pinnacle study demonstrated how T3SS worked with contact between Yersinia and a eukaryotic host cell stimulating the expression and polarized transfer of YopE into the host cell cytosol (27). Ultrastructural analysis of the T3SS apparatus by electron microscopy revealed a needle-like structure that spanned the inner and outer bacterial membranes and extended beyond the bacteria, thus allowing the needle to pierce opposing eukaryotic membranes, such as the plasma membrane (28). Hence, the chlamydial projections potentially had another purpose that did not involve siphoning nutrients from the host cytosol.
By the mid-1990s, T3SSs were identified in many Gram-negative pathogens and appreciated as key virulence mechanisms (13). Some nonpathogenic symbiotic Gram-negative bacteria also carry T3SSs that are integral toward establishing a positive relationship with their hosts (29–33). An altered G+C content within genes associated with the T3SS, the common organization of these T3SS on virulence plasmids or within chromosomally organized genomic/pathogenicity islands, and the ability to genetically modify these regions all facilitated T3SS discovery in various Gram-negative bacteria. As discussed above, the early challenges with understanding if Chlamydia also carried and expressed T3SSs was that the genetic composition and organization were entirely different. Examination of a fragment of the Chlamydia psittaci strain guinea pig inclusion conjunctivitis (GPIC) revealed 4 genes that were associated with virulence and had high homology to the contact-dependent (cds) or T3SS apparatus of Yersiniae (14). Within this gene cluster, the genes were compared to genes expressed by Yersinae and encoded the following proteins: Cds1, which is an ortholog to known T3SS apparatus component YcsU; Cds2, which is orthologous to LcrD, a known regulator of Yop secretion; CopN, which is orthologous to YopN, the T3SS Ca2+ sensor; and Scc1, which is an ortholog to a T3SS chaperone. Furthermore, these genes were conserved in Chlamydia trachomatis serovars L2 and D (2), Chlamydia pecorum strain IB1, and Chlamydia pnuemoniae strain TWAR (14). Western blot analysis demonstrated the presence of Cds2 in lysates harvested at 44 h postinfection, which suggested the possibility of a functional T3SS in Chlamydia (14). Even though this study identified only 4 genes out of a possible ~40+ genes associated with T3SS, these data were significant and consistent with the conservation of T3SS genes across other intracellular pathogens (14). Importantly, data from this initial study were the first to suggest that if Chlamydiae had an intact T3SS, its genomic organization and characteristics would be different than those observed with other Gram-negative bacteria.
As confirmation of the original observation by Hsia, Bavoil, and colleagues, genomic sequencing of C. trachomatis serovar D strain UW-3/CX (D/UW-3/CX) revealed T3SS components, including orthologs to chaperones and apparatus components, dispersed throughout the genome (2). A comparative genomic analysis of the C. trachomatis and C. pneumoniae genomes revealed that indeed genes encoding the T3SS were found in 4 or more gene clusters. And while this scattered organization was consistent between chlamydial species, the specific organization of these individual clusters was not necessarily conserved (15). Importantly, though, the genes encoded proteins that were structurally similar to proteins of other Gram-negative bacteria, and the core chlamydial T3SS components shared 48 to 92% amino acid identity to other T3SS orthologs (15). T3SS structures could be produced when genes from the 6 C. trachomatis gene clusters were incorporated into 3 individual plasmids and transformed into nonpathogenic Escherichia coli (a strain that would naturally be devoid of a T3SS). Expression of the genes was induced with the addition of EGTA (to chelate Ca2+) or fetal bovine serum (FBS) and then needle-like projections were visualized by electron microscopy (34). This structure was unable to secrete known chlamydial T3SS effectors; however, not all chlamydial chaperones were included in these E. coli transformants (34).
AN ASIDE ON NOMENCLATURE
As discussed, the initial observation of a chlamydial T3SS occurred by examining genetic sequences of C. psittaci (14), which was later classified as Chlamydia caviae strain GPIC. Other labs initially studying chlamydial T3SS were using the C. trachomatis serovar L2 strain 434/Bu (L2/434/Bu), given its ease of culture compared with C. pneumoniae and the biosafety restrictions required to work with C. psittaci. The first C. trachomatis strain to be sequenced and annotated was serovar D strain UW-3/CX (2). About a decade later, when strain L2/434/Bu was sequenced and annotated, a different methodology was used, creating new gene numbers, despite the similarity in genetic organization between the strains (35). Because many chlamydial researchers were working with strain L2/434/Bu prior to its annotation, many studies had been (and are being) published using strain D/UW-3/CX nomenclature. This practice remains a common one in the field even though the original genetic tools were developed for C. trachomatis strain L2/434/Bu (6). For clarity, defined common names as well as C. trachomatis serovars D/UW-3/CX (CT) and L2/434/Bu (CTL) genomic annotations will be used, where appropriate. Table 1 (T3SS apparatus genes) and Table 2 (chlamydial chaperone genes) can also be referenced for further clarification. A comprehensive table of chlamydial effector proteins is not provided in this review because one has been published recently (6).
TABLE 1.
Chlamydial orthologs to proteins of bacterial T3SS
| Annotation by strain or species |
Common gene nameb | Proposed function | Expression during the developmental cyclec | |||
|---|---|---|---|---|---|---|
| D/UW-3CX | L2/434/Bu | C. pneumoniae | GPICa | |||
| CT669 | CTL0038 | CPn0707 | CCa00035 | cdsN /yscN | Inner membrane ATP synthase, part of Hub complex | 16 hpi; peak, 24–40 hpi |
| CT717 | CTL0086 | CPn0858 | CCa00909 | fliI | Inner membrane ATP synthase, part of Hub complex | 8 hpi; peak, 40 hpi |
| CT670 | CTL0039 | CPn0706 | CCa00036 | cdsO /yscO | Stalk | 8 hpi; peak, 24 hpi; slightly decreased at 40 hpi |
| CT561 | CTL0824 | CPn0826 | CCa00937 | cdsL /yscL | Support protein “spokes,” inner membrane basal body | 16 hpi; peak, 24–40 hpi |
| CT672 | CTL0041 | CPn0704 | CCa00038 | cdsQ /yscQ | C-ring of inner membrane complex/cytoplasmic sorting platform | 16 hpi; peak, 40 hpi |
| CT719 | CTL0088 | CPn0860 | CCa00907 | fliF | Flagellar M-ring protein | 8 hpi; peak, 40 hpi |
| CT060 | CTL0316 | CPn0363 | CCa00428 | cdsV /flhA | Export gate | 8 hpi; steadily increases to 40 hpi |
| CT091 | CTL0346 | CPn0322 | CCa00460 | cdsU /yscU | Inner membrane basal body component | 8 hpi; peak, 24–40 hpi |
| CT562 | CTL0825 | CPn0825 | CCa00938 | cdsR /yscR | Export apparatus | 16 hpi; peak, 24–40 hpi |
| CT563 | CTL0826 | CPn0824 | CCa00939 | cdsS /yscS | Export apparatus | 16 hpi; peak, 24–40 hpi |
| CT564 | CTL0827 | CPn0823 | CCa00940 | cdsT /yscT | Export apparatus | 24 hpi; peak, 40 hpi |
| CT718 | CTL0087 | CPn0859 | CCa00908 | fliH | Flagellar assembly protein; inner ring component | 8 hpi; peak, 24–40 hpi |
| CT664 | CTL0033 | CPn0712 | CCa00030 | cdsD /yscD | Inner membrane ring of basal body; orthologs: PrgH, MxiG | 16 hpi; peak, 24 hpi and remains at this level through 40 hpi |
| CT559 | CTL0822 | CPn0828 | CCa00935 | cdsJ /yscJ | Basal body component that spans periplasm | 16 hpi; peak, 24–40 hpi |
| CT674 | CTL0043 | CPn0702 | CCa00040 | cdsC /yscC | Outer membrane ring | 16 hpi; peak, 40 hpi |
| CT666 | CTL0035 | CPn0710 | CCa00032 | cdsF | Needle protein, SctF | 8 hpi; peak, 24–40 hpi |
| CT671 | CTL0040 | CPn0705 | CCa00837 | cdsP /yscP | Molecular ruler | 8 hpi; peak, 24 hpi; slightly decreased at 40 hpi |
| CT584 | CTL0847 | CPn0803 | CCa00960 | none | Needle tip; IpaD LcrV | 16 hpi; peak, 24–40 hpi |
| CT578 | CTL0841 | CPn0809 | CCa00954 | copB | Needle tip; translocator | 24 hpi; peak, 40 hpi |
| CT579 | CTL0842 | CPn0808 | CCa00955 | copD | Needle tip; translocator | 24 hpi; peak, 40 hpi |
| CT861 | CTL0236 | CPn1020 | CCa00741 | copB2 | Needle tip; translocator | 16 hpi; peak, 24–40 hpi |
| CT860 | CTL0235 | CPn1019 | CCa00742d | copD2 | Needle tip: translocator | 8 hpi; steadily increases to 40 hpi |
| CT398 | CTL0655 | CPn0525 | CCa00220 | cdsZ | FlgZ protein; flagellar-associated zinc-ribbon domain protein | 8 hpi; steadily increases to 40 hpi |
| CT301 | CTL0553 | CPn0095 | CCa00677 | pknD | Serine/threonine kinase targets CdsD | 3 hpi; remains at steady high levels 16 hpi–40 hpi |
| CT090 | CTL0345 | CPn0323 | CCa00459 | IcrD | Low calcium sensor | 16 hpi; peak, 40 hpi |
| CT089 | CTL0344 | CPn0324 | CCa00458 | copN /lcrE | Low calcium response protein | 16 hpi; peak, 24-40 hpi |
GPIC C. caviae strain ATCC VR-813/DSM 19441.
If there are multiple common names given for an individual gene, the bolded name is used in the text.
Times are relevant for C. trachomatis serovars D and L2. Information is from reference 5.
CCa00742 was annotated as the CarD transcriptional regulator.
TABLE 2.
Candidate chlamydial T3SS chaperone proteins
| Annotation by strain or species |
Common gene namee | Class | Associated operon/gene clustera | Expression during the developmental cycleb | Orthologs with homology to chlamydial chaperone proteinsc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D/UW-3CX | L2/434/Bu | C. pneumoniae | GPICd | |||||||||
| Y. pestis | Y. pseudotuberculosis | Y. enterocolitica | Shigella flexneri | Salmonella Typhimurium | ||||||||
| CT088 | CTL0343 | CPn0325 | CCa00457 | scc1; sycE | IA | CT091/CTL0346 gene cluster encoding CdsU, LcrD, CopN | Begins 16 hpi; peaks 24 hpi, slight drop 40 hpi | None | None | None | None | SicP |
| CT663 | CTL0032 | CPn0713 | CCa00029 | scc4; sycE; slc\ill\ | IB | CT663/CTL0032; largest T3SS gene cluster encoding 3 chaperones, 7 T3SS proteins, serine/threonine kinase pkn5 | Begins 16 hpi; peaks 24 hpi, slight drop 40 hpi | None | None | None | None | None |
| CT043 | CTL0299 | CPn0387 | CCa00409 | slc1 | IB | None | Begins 8 hpi; peak 24 hpi, slight drop 40 hpi | None | None | None | None | SicP |
| CT260 | CTL0512 | CPn0409 | CCa00385 | mcsc | IB | 5′ of 4 Gene cluster encoding DnaQ, MqnD, and an uncharacterized protein | Begins 8 hpi; peak 24 hpi, slight drop 40 hpi | None | None | None | None | None |
| CT584 | CTL0847 | CPn0803 | CCa00960 | unassigned | IB | 3′ End of 3 gene cluster encoding MinD and gp6D | Begins 16 hpi; peak 40 hpi | None | None | None | None | None |
| CT576 | CTL0839 | CPn0811 | CCa00952 | lcrH_1; scc2 | II | CT576/CTL0839 gene cluster encoding CopB and CopD | Begins 16 hpi; peak 40 hpi | SycD/LcrH | SycD/LcrH | SycD/LcrH | IpgC/CesD/Syc D/LcrH family | SicA/SscA/CesD/S ycD/LcrH family |
| CT274 | CTL0526 | CPn0423 | CCa00370 | unassigned | II | 3′ End of 2 gene cluster; first gene encodes an uncharacterized protein | Begins 8 hpi; peak 24 hpi, slight drop 40 hpi | None | None | None | None | SscB/CesD/SycD/L crH family |
| CT862 | CTL0237 | CPn1021 | CCa00740 | lcrH_2; scc3 | II | copB2/copD2 Gene cluster | Begins 16 hpi; peak 40 hpi | SycD/LcrH family | SycD/LcrH family | SycD/LcrH family | CesD | SycD/LcrH family |
| CT665 | CTL0034 | CPn0711 | CCa00031 | cdsE | III/V? | CT663/CTL0032; largest T3SS gene cluster encoding 3 chaperones, 7 T3SS proteins, serine/threonine kinase pkn5 | Begins 8 hpi; peak 24 hpi, slight drop 40 hpi | None | None | None | None | None |
| CT667 | CTL0036 | CPn0709 | CCa00033 | cdsG | III | CT663/CTL0032; largest T3SS gene cluster encoding 3 chaperones, 7 T3SS proteins, serine/threonine kinase pkn5 | Begins 8 hpi; peak 40 hpi | None | None | None | None | None |
| CT114 | CTL0369 | CPn0145 | CCa00623 | unassigned | unkn, likely II; TPR domain aa261-462 | None, but precedes incD operon | Begins 3 hpi; peaks 24 hpi, slight drop 40 hpi | None | None | None | None | None |
Determined by C. trachomatis serovar D genome organization.
Times are relevant for C. trachomatis serovars D and L2. Information is from reference 5.
BLAST searches using CTL protein sequences, including PSI-BLAST.
GPIC C. caviae strain ATCC VR-813/DSM 19441.
Bolded names are the most commonly used in the field.
THE ELUSIVE SEARCH FOR AN INHIBITOR OF THE CHLAMYDIAL T3SS
As briefly mentioned above, the genetic tractability of pathogenic E. coli (including Shigella), Salmonella spp., and Yersinia eased the ability to characterize the spatial organization and function of T3SS gene products (13). Genetic manipulation of Chlamydia was not achieved successfully until 2011 (36); therefore, those interested in studying chlamydial T3SS had to look at alternative means to disrupt its function. Given the importance of chlamydial T3SS to every stage of the developmental cycle, this endeavor was complicated. Hence, there was a concerted effort in the field to identify a pharmacological inhibitor that targeted chlamydial T3SS (37–52). Initial studies focused on inhibitors that inhibited T3SS in other Gram-negative pathogens, such as Yersinia (39). However, in cell culture, these same inhibitors limit/chelate iron, which leads to chlamydial persistence that, in turn, halts many chlamydial biological processes, including T3SS (42). Furthermore, chlamydial strains that have mutations in hemG, a protoporphyrinogen oxidase that requires iron, are resistant to these T3SS inhibitors (41). To date, there have been no chemical inhibitors that are capable of specifically targeting chlamydial T3SS versus triggering a general persistence phenotype. The recently developed CRISPR interference (CRISPRi) or inducible knockdown of chlamydial genes (53–59) may provide a much-needed tool to temporally inhibit specific components of the chlamydial T3SS. Thus, inducible knockdown of gene expression provides a more direct means to characterize the spatial temporal organization and function of the chlamydial T3SS. The stability of proteins encoded by chlamydial T3SS genes is unknown, which may be a complicating factor to this approach.
THE CHLAMYDIAL T3SS APPARATUS: INTRODUCTION TO THE EYE OF THE NEEDLE
Core Components of a T3SS Apparatus
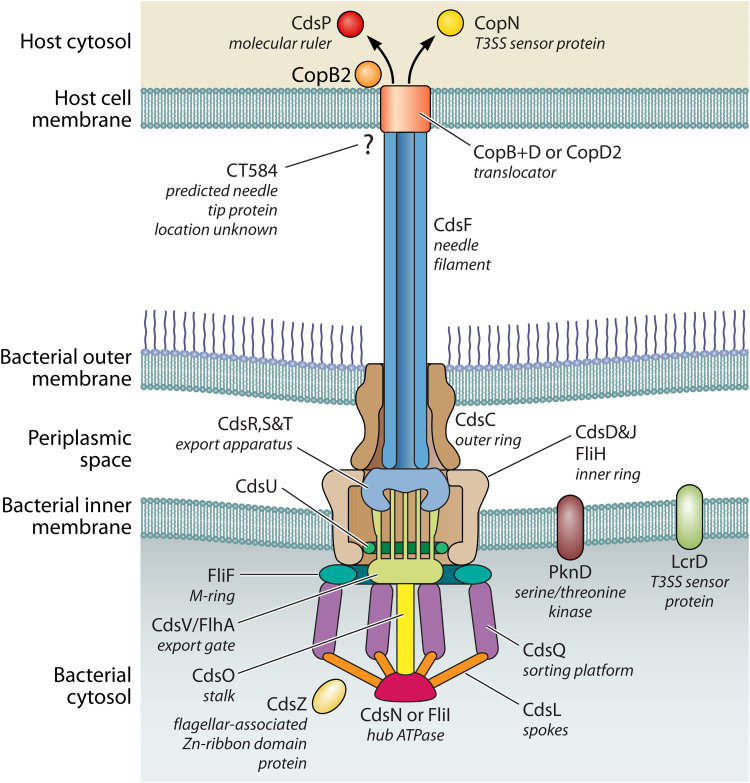
The T3SS apparatus has the following two core components: the basal body and the needle. Chlamydial specific T3SS genes and their proposed functions are provided in Table 1. A proposed chlamydial T3SS structure is depicted in Fig. 1, and the localization of the individual proteins is based on the similarity of these proteins to orthologs in other T3SSs and limited biochemical data. In general, the basal body of the apparatus is comprised of 14 to 15 different proteins and spans the inner membrane, periplasmic space, and the outer membrane. It is composed of the Hub or main ATPase that originates in the bacterial cytosol and attaches, via junction proteins (e.g., CdsL and CdsQ) and a stalk protein (e.g., CdsO), to the inner ring that forms in the inner membrane and extends into the periplasmic space (Fig. 1). The inner ring is comprised of an oligomerized structure that supports and houses the export gate and export apparatus. Within the periplasmic space, the inner ring connects to the outer ring, which then inserts and spans the outer membrane, which is the final component of the T3SS basal body (Fig. 1). The base of the needle is housed within the outer ring and is sometimes connected to the export apparatus by inner rod proteins. Chlamydia bacteria have orthologs to all these components except for an inner rod protein as PSI-BLAST or other standard bioinformatic methods did not reveal any obvious candidates (E. A. Rucks unpublished observation).
FIG 1.
Graphical representation of components that comprise the chlamydial T3SS based on similarity to other bacterial T3SSs and limited biochemical data.
Basal Body Assembly
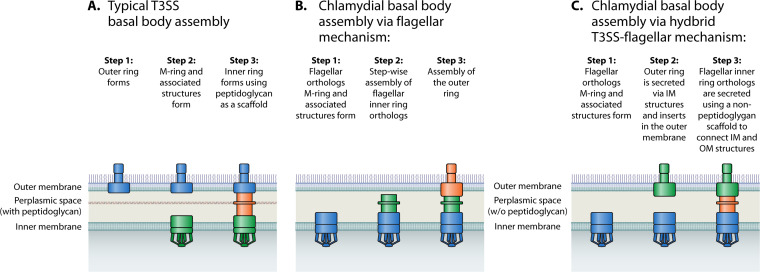
The formation of the T3SS apparatus basal body is different from that of a flagellum, despite the structural similarities of the core components of these structures. Assembly of the basal body of the T3SS apparatus starts at the outer membrane with the outer ring and is followed by the formation of structures within the inner membrane. Components of the inner membrane and outer membrane are then connected by a protein secreted through the inner membrane structure that forms a channel, binding the periplasmic side of the outer membrane ring (Fig. 2A). The portion of the T3SS apparatus that spans the periplasm is supported by peptidoglycan within the periplasmic space. The sequential nature of the T3SS apparatus assembly is proposed to be in of itself a quality control-related process, as components are added only after the previous components have been assembled successfully (60).
FIG 2.
Graphical representation depicting models of chlamydial basal body assembly. (A) Diagrams of how typical T3SS basal bodies are formed. (B) Diagrams of how the chlamydial basal body may form via the mechanism associated with flagellar basal body assembly. (C) Proposes a novel hybrid mechanism that shares characteristics between both flagellar and T3SS basal body assembly.
Hypothetical mechanisms of chlamydial T3SS basal body assembly.
An interesting consideration in T3SS assembly is that in Chlamydia, the peptidoglycan is detected at the division septum but is not detected throughout the periplasm (61). With no detectable peptidoglycan that would act as a scaffold for the assembly of the periplasmic portion of the chlamydial T3SS apparatus, it may occur similarly to the basal bodies of flagella. Assembly of the flagellar basal body begins in the inner cytoplasmic membrane, and each new component is ushered through the central channel and attached at the outermost protein of the previously assembled apparatus. In support of this idea, key components of the chlamydial T3SS basal body are orthologs to flagellar proteins (62) (Fig. 2B). Specifically, these orthologs are FliI, an ATP synthase or the Hub ATPase; FliH, a flagellar assembly protein that is part of the inner ring; FliF, the M-ring; and FlhA, the export gate (62) (Fig. 1). Chlamydia bacteria also encode a highly conserved flagellar-associated zinc-ribbon domain protein, CdsZ (CT398), that has similarity to FlgZ (63), which is required for flagellar assembly and motility in Helicobacter pylori but is not required for T3SS apparatus assembly (64). Given that the chlamydial T3SS basal body has both flagellar and T3SS orthologs, there is the interesting possibility that the formation of the chlamydial T3SS basal body is formed by a novel, hybrid mechanism (Fig. 2C). In this scenario, the flagellar orthologs form the inner membrane structures first. Then, the outer membrane component, which has greater similarity to T3SS orthologs than flagellar proteins, is secreted through the inner membrane structure. Finally, an unknown periplasmic molecule forms a scaffold to allow inner ring assembly (Fig. 2C).
(i) Clarification of Previously Annotated M-ring and ATP Synthase orthologs. With respect to this novel hypothesis of chlamydial T3SS basal body assembly, there has been some confusion in the field relative to the annotation of predicted M-ring and ATP synthase homologs. In several different databases, the fliF inner membrane ring orthologs (referred to as M-ring) in C. trachomatis serovars D and L2 have been referred to as ct719 and ctl0086, respectively; however, in the literature, ct719 and ctl0086 have been annotated as FliA ATP synthase homologs (62). While Chlamydia bacteria have been known to combine two functionally distinct proteins into a single protein, it is unlikely in this scenario due to structural limitations of a functional T3SS. To clarify their possible functions and correct mistakes in previous annotations, the proteins encoded by these genes were examined via InterPro analysis (https://www.ebi.ac.uk/interpro/). The protein encoded by ct719 is 334 amino acids (aa) in length and 37,306 Da in size, which is a size consistent with flagellar motor proteins (65). An InterPro analysis of CT719 revealed that it has homology to the FliG flagellum motor family of proteins over the entire length of the protein and homology to the flagellar M-ring protein N terminus within amino acids 69 to 114. These characteristics are very similar to the proteins encoded by the C. pneumoniae and C. caviae fliF flagellar M-ring homologs cpn0860 (341 aa; 38,293 Da) and cca00907 (337aa; 37,503 Da), respectively. In contrast, the protein encoded by ctl0086 is larger at 434 aa and 47,584 Da and, by InterPro analysis, has a predicted N-terminal ATPase domain (aa 24 to 86), ATPase nucleotide binding domain (aa 144 to 351), and C-terminal T3SS EscN ATPase domain (aa 360 to 429). Combined, these characteristics are consistent with family members belonging to the FliI inner membrane ATPases (66). Furthermore, the proteins encoded by the annotated homologs in C. pneumoniae (cpn0858) and C. caviae (cca00909) demonstrate similar sizes and ATPase domains as CTL0086. To summarize, CT719 is likely a candidate flagellar M-ring protein, not an inner membrane ATP synthase, while CTL0086 demonstrates similarity to an ATP synthase of the FliI protein family. Furthermore, examination of adjacent genes within the same cluster revealed that in C. trachomatis serovar D, the gene encoding the fliI ATP synthase ortholog is ct717 and the C. trachomatis serovar L2 gene encoding the fliF M-ring ortholog is ctl0088. These findings contributed to entries in Table 1.
Needle Assembly
In general, the proteins that oligomerize to form the needle filament, the second major component of the T3SS apparatus, are added only after the basal body has been completed. This aspect of T3SS needle assembly mirrors the assembly process of a flagellum, with each additional needle filament subunit being secreted through the central core of the apparatus and polymerizing to the outermost tip of the needle. Needle length is tightly controlled by an accessory protein or “molecular ruler,” CdsP, the Yersinia YscP ortholog. Chlamydiae also have genes that are orthologs to those known to encode needle tip proteins, such as ipaD and lcrV (Table 1) (67). In general, needle tip proteins are thought to sense contact with the host cell to prevent constitutive or wasteful secretion of T3S effector proteins. C. trachomatis carries one possible needle tip protein, CT584, as it structurally resembles other T3SS needle tip proteins (67); although, there is no direct experimental evidence that supports CT584 functioning as a needle tip protein. Additional candidate needle accessory proteins are CopB, CopB2 and CopD, CopD2, which are orthologs to Yop B and D, respectively, and comprise the translocon, which is a needle component required for translocation of effector proteins across eukaryotic membranes. The significance of the duplication of the Cop proteins is unknown and discussed in depth later in this review. In Yersinia, the Yop proteins affix to the tip of the needle, help to prevent premature translocation of effector proteins, and are thought to form pores within the target mammalian membrane (68). Direct contact between the T3SS apparatus and the host membrane is one of the signals associated with the permissiveness of effector protein secretion (23).
BIOCHEMICAL CHARACTERIZATION OF CHLAMYDIAL T3SS APPARATUS COMPONENTS
After the initial discovery of T3SS homologs in Chlamydia (14), and the subsequent sequencing of C. trachomatis serovar D (2), many initial studies were designed to understand if homology dictated function. These early studies did not have the benefit of genetic tools, as transformation of Chlamydia was not developed until 2011 (36). Therefore, these studies focused on bioinformatic analysis and elegant biochemical characterization of purified proteins. As such, there are biochemical characterizations of the Hub ATPases (CdsN and FliI), spoke protein (CdsL), stalk protein (CdsO), sorting platform (CdsQ), export gate (FlhA), inner membrane ring component (CdsD), needle filament protein (CdsF), predicted needle tip protein (CT584), and translocon proteins (CopB and CopD). The transformation of some of these genes for expression in orthologous or surrogate T3SS systems will be discussed in a later section. These proteins were cloned and purified from either C. trachomatis serovars D or L2 or from C. pneumoniae. Given the conserved nature of these genes across chlamydial species, it is likely that a purified protein from one chlamydial species will likely translate to how that protein is functioning in the other species.
Hub ATPases and Adjacent Structures
Both Hub ATPases associated with the chlamydial T3SS were biochemically characterized with purified C. pneumoniae proteins CPn0707 (CdsN) and CPn0858 (FliI). Specifically, the C-terminal fragment of CdsN demonstrated ATPase activity (69). Furthermore, by bacterial two-hybrid and glutathione S-transferase (GST)-pulldowns, CdsN was found to interact with CdsL (spoke protein), CdsQ (sorting platform protein), and CopN (effector). Interestingly, it did not interact with the proposed molecular ruler CdsP (CPn0705) but did interact with CdsO (stalk protein), which was initially, but incorrectly, identified as a putative chaperone (69). The ATPase activity of FliI is slightly better characterized as it has confirmed Walker A and B motifs and can interact with some of the same proteins as CdsN, namely, CdsL, FlhA, and CopN. The N terminus of FliI cannot interact directly with M-ring protein FliF (62), but these data suggest that there may be a series of intermediate proteins, such as CdsL and CdsQ, connecting the main ATPase with the M-ring (Fig. 1). Within Chlamydia, the expression of cdsN and fliI is slightly different, with fliI transcription beginning before cdsN transcription (5). These expression data fail to illuminate whether these proteins ultimately function as Hub ATPases in distinct T3SS apparatus, if they collaborate, or if are interchangeable. These possibilities are not mutually exclusive.
There are also data to suggest that the activity of these ATPases may be regulated posttranslationally. In in vitro assays with purified proteins, increasing concentrations of the CdsL spoke protein inhibited CdsN ATPase activity, and CdsL was found to bind to CdsN within its catalytic domain (70). These data suggest that CdsL has a structural function that links the Hub ATPase to the sorting platform and/or a regulatory function. Furthermore, a peptide mimetic of CdsN inhibited invasion of C. pneumoniae presumably by inhibiting the function of the T3SS and highlighting the importance of this protein in chlamydial pathogenesis (70). Consistent with these proteins being present at the time of invasion/entry, both CdsN and CdsL were identified in a proteome analysis of C. pneumoniae EBs (71). The assembly or function of the CdsL spoke proteins may be regulated by another protein, CdsZ (CT398), which is a flagellar-associated zinc-ribbon domain protein whose orthologs facilitate flagellar synthesis. CdsZ was found to interact with CdsL and inner ring component FliH (export gate) by bacterial adenylate cyclase two-hybrid assays (BACTH), although the biological function of these interactions remain uncharacterized (63).
The Spoke and Stalk Structure
If the chlamydial basal body is structured similarly to characterized T3SS basal body structures of other Gram-negative bacteria (60), then the Hub ATPase is linked to the cytoplasmic ring (C-ring) by two separate proteins, namely, CdsL and CdsO (Fig. 1). As a candidate stalk protein, CdsO presumably binds the central core of ATPase and links it directly to the M-ring FliF. Biochemical evidence demonstrating the likely function of CdsO as a stalk protein is mixed. Studies of the C. pneumoniae CdsO stalk protein CPn0706 indicated that CdsO is not secreted (72) and that it forms a dimer (69), which is consistent with characterized stalk proteins of other Gram-negative bacteria. Chlamydial CdsO homologs are unique in amino acid content from other organisms; however, Clustal IX 1.83 alignment of C. trachomatis serovar D CdsO protein CT670 indicated that it is likely a YscO ortholog, which is the Yersinia stalk protein. Furthermore, CT670 and the Yersinia YscO have similar molecular weights and isoelectric points (pI), with CT670 being 20.1 kDa with 8.22 pI and Yersinia YscO being 18.8 kDa with a 7.89 pI (73). Analytical centrifugation of purified CT670 indicated that it can form monomers or dimers, but size exclusion chromatography was less straightforward as one of the forms was “cigar shaped,” which is inconsistent with forms of orthologous stalk proteins. An analysis of crystal structures of CT670 revealed symmetric units of monomers with two coil-coiled helical domains, with charged residues facing out of the structure and hydrophobic residues residing between the helices, which are consistent with the orthologous YscO structures (73). Bacterial two-hybrid studies demonstrated that CT670 did not interact with expected binding partners, including the needle filament CdsF or an inner membrane basal body component, CdsU. The interaction between CT670 and the FliF M-ring protein CT719 was not tested, but CT670 did have a positive interaction with the molecular ruler CdsP (73). These interaction data complicate the understanding of how CdsO may be functioning or positioned within the chlamydial T3SS. At the same time, negative data are difficult to interpret.
The Sorting Platform or C-ring, M-ring, Export Gate, and Export Apparatus
Similar to the flagellar basal body, the portion of the T3SS basal body that originates in the bacterial cytosol and extends into the inner membrane is composed of a series of rings, which includes the C-ring, commonly referred to as the sorting platform, and the M-ring. Yeast two-hybrid analysis supports that CdsQ is a likely T3SS C-ring, as the C. trachomatis serovar D homolog was identified as interacting with multiple chlamydial T3SS proteins, which is consistent with its function as a sorting platform (74). The M-ring is distal to the sorting platform, and these proteins are thought to bind directly (Fig. 1), although to this author’s knowledge, no biochemical studies have directly assessed an interaction between CdsQ and FliF. A bacterial two-hybrid assay and pulldowns with purified proteins revealed that the C. pneumoniae M-ring protein FliF (CPn0860) interacts with the export gate FlhA (CPn0363) (62). The same protein-protein interaction studies demonstrated that FlhA (CPn0363) can interact with CdsU (CPn0322), the next adjacent inner ring component (62) (Fig. 1).
Inner Ring Structure
Based strictly on sequence similarity to other T3SS homologs, three proteins, namely, CdsD, CdsJ, and FliH, are proposed to form the inner ring, which is the structure distal to the M-ring and spans the inner membrane and periplasmic space. Also based on sequence similarity, the inner ring likely houses the export gate (FlhA), CdsU, and the export apparatus (CdsR, CdsS, and CdsT). Within Chlamydia, it is unclear if CdsD, CdsJ, and FliH function together or individually, as fliH is expressed at 8 hours postinfection (hpi) and cdsD and cdsJ are not expressed until 16 hpi. All 3 genes reach peak expression between 24 and 40 hpi (5).
CdsD and possible posttranslation modification of the inner ring.
In C. pneumoniae, the CdsD (CPn0712) homolog was found to be a target of serine/threonine kinase, PknD (CPn0095) (75). Although, the molecular consequences of this phosphorylation are unclear. PknD was characterized as an integral membrane protein after expression in E. coli followed by differential centrifugation (75). Furthermore, an inhibitor of PknD phosphorylation activity inhibited phosphorylation of CdsD and suppressed replication of C. pneumoniae (76). CdsD is a large protein with 829 aa and is 89 kDa in size. The last 400 aa are orthologous to the YscD/HrpQ protein family. When amino acid residues 558 to 771 were crystalized, the resulting crystals formed oligomeric complexes that were mediated by disulfide bridges (77). This structural observation may have importance for how this protein functions during the chlamydial developmental cycle, as it has been noted that changes in disulfide bonds occur in other T3SS proteins during chlamydial development (78). Further analysis of the CdsD crystal structure revealed three trefoil domains of αββαβ topology which was similar in structure to PrgH of Salmonella enterica serovar Typhimurium and YscD of Yersinia enterocolitica. Although the structure of CdsD is more extended than PrgH and less extended than YscD, the functional implications of these differences are unclear (79). A Western blot analysis revealed that CdsD is present in C. pneumoniae EBs and is also localized within RBs by indirect immunofluorescence (80). These data are consistent with the identification of CdsD in the proteome of C. pneumoniae EBs (71). CdsD does have a transmembrane domain between aa 530 to 552 (77), but TX-144 fractionation found CdsD in the aqueous fractions of EBs and RBs (80). TX-114 is used to determine the hydrophobicity of proteins via phase partitioning/separation; for example, integral membrane proteins will segregate to the TX-114 phase not the aqueous or soluble phase (81, 82). These data suggest that CdsD is not an integral membrane protein that spans the inner membrane and that most of the protein may reside within the periplasmic space (77, 80). Sarkosyl solubilization of purified C. trachomatis serovar L2 EBs demonstrated that CdsJ was found in the soluble fraction (83), indicating that it is not found in the chlamydial outer membrane (81, 82). However, CdsJ is found in TX-114 insoluble fractions of EB lysate (84), which is consistent with it being an integral membrane protein found in the inner membrane (81, 82). CdsJ is present in both EBs and RBs, suggesting that it may have functions throughout the chlamydial developmental cycle (84). Furthermore, cryo-electron tomography has revealed that inner membrane T3SS components create an invagination of the inner membrane (85). These data suggest that proteins associated with the inner ring are associated with chlamydial inner membrane but also protrude into the periplasmic space (Fig. 1).
Outer Ring Components
Sarkosyl purification of chlamydial outer membrane complexes (COMCs), found CdsC, a candidate outer ring component, in the insoluble fraction (83). Consistent with CdsC being an integral membrane protein, it was found in the TX-114 phase of EB lysates (84). These data are consistent with the hypothesis that this protein resides in the outer membrane and forms the outer ring of the T3SS basal body complex. Of note, the chlamydial CdsC protein is larger than orthologous T3SS outer ring proteins and is phylogenetically distinct from other secretins. Specifically, the first 250 amino acids in the N-terminal region comprise a hydrophilic domain that is unique to Chlamydia, with the remainder of the protein demonstrating similarity to other outer membrane secretins (86).
Needle Filament
The needle filament protein CdsF is unique among other orthologous T3SS needle proteins as it is not identified as a T3SS needle protein by standard database searches and has cysteine residues (83). These inconsistencies led Fields and colleagues to examine CdsF thoroughly. Thus, it is likely the best-characterized component of the chlamydial T3SS apparatus. Sarkosyl purification of COMCs from purified EBs demonstrated CdsF in the insoluble fraction (83). Furthermore, by indirect immunofluorescence, CdsF can be localized to the inclusion membrane in distinct puncta adjacent to RBs (83, 87). This localization was also confirmed by immunoelectron microscopy (83). In a separate study, cryo-electron tomography captured needle structures that were polarized on one side of EBs, namely, the side facing HeLa cell filopodium. A CdsF antibody confirmed that these structures were T3SS needles (85).
Similar to other T3SS needle proteins, CdsF can polymerize. CdsF is a 9-kDa monomer, and with the addition of cross-linker bismaleimido-hexane, it forms a laddered pattern of 13 different CdsF-containing complexes, ranging in size from 30 kDa to 100 kDa (83). These data are consistent with a previous study that imaged isolated needles (referred to as “rods” in this publication) by electron microscopy and determined that they were helical in nature and arranged in subunits with an estimated size of 50 kDa (88). These subunit estimates fall within the size associated with CdsF-containing complexes (83). Furthermore, CdsF polymerization occurs only at the bacterial surface. However, polymerization patterns may be different between EB and RBs as the predominant cross-linked form in the EB was a dimer and in the RB it was a trimer. These data suggest that there is a confirmational change in the needle relative to its environment that is likely due to the cysteines within the protein (83).
Possible significance of a lack of an annotated inner rod protein.
Related to the discussion of how CdsF was characterized, it is important to point out that Chlamydia bacteria do not have annotated inner rod protein, which is the internal structure found in many T3SS systems (89). This information could reflect another similarity between the chlamydial T3SS apparatus and the flagellum in that the inner rod structure of a flagellum is composed of the M-ring protein (90), which in Chlamydia would be the FliF protein. Another possibility is that this gene was lost or consolidated in Chlamydia during reductive evolution. Yet another possibility is that the gene encoding this protein has not been annotated due to the lack of similarity to other genes or proteins in standard databases. A possible candidate for an inner rod protein is encoded by ct716/cyl0085/cpn0857/caa00910, which is a gene that encodes an uncharacterized protein and is found within a gene cluster encoding other T3SS components. But a PSI-BLAST analysis of these proteins failed to demonstrate similarity to YscI/HrpB inner rod protein family members, and InterPro analysis failed to reveal key structural features consistent with inner rod proteins (E. A. Rucks, unpublished observation).
Molecular ruler.
In Chlamydia, there are orthologs to YscP, the Yersinia molecular ruler (Table 1), but the functional or biochemical characterization of CdsP has not been elucidated. In Y. pestis, yscP expression is associated with controlling Yop (effector protein) secretion (91), ostensibly to prevent premature Yop secretion. Further studies in Y. enterocolitica indicated that YscP formed a blockade within the T3S apparatus that was relieved once the needle length or secretion conditions were optimal (92). A more detailed analysis of YscP helical content suggested that YscP could determine needle length via two proposed models (93). In the first model, YscP remains within the needle apparatus, attached to both the tip of the growing needle and the export apparatus. As the needle grows, YscP would stretch, and at a certain tension (determined by structural helices), a confirmational change would dislodge YscP from the export apparatus and promote its secretion and “unblock” the apparatus to allow secretion of effector proteins. In the second model, YscP acts as a “timer” in that the amount of time that YscP is being progressively threaded through the apparatus would determine when the C-terminal switch domain of YscP would encounter its binding partner. The confirmational change to YscP upon binding to its partner would then promote secretion and unblock the apparatus (93).
Proteins that cap the T3SS needle.
T3SS needles are capped by several proteins, as follows: the needle tip and the translocon.
(i) The needle tip protein. Based on current data, it is entirely unclear if the IpaD/LcrV chlamydial orthologs function as needle tip proteins. The characterization of ct584, ctl0847, cpn0803, and cca00960 as genes predicted to encode chlamydial needle tip proteins was based on sequence similarity and biophysical properties of C. trachomatis serovar D protein CT584. Data from size exclusion chromatography (ability of the purified CT584 to form higher order complexes of ~150 kDa) and circular dichroism (evidence of alpha helical structures with at least two folding domains) were consistent with CT584 potentially acting as a needle tip protein. But Fourier transform infrared spectroscopy found no evidence of standard coil-coiled domains associated with characterized needle tip proteins. Furthermore, an empirical phase diagram analysis, which uses multiple parameters to compare proteins of similar function regardless of sequence similarity, was indeterminant of CT584 belonging to the IpaD or LcrV subfamily (67). Furthermore, crystal structures of the CT584 homolog CPn0803 from C. pnuemoniae did not align with LcrV but did form hexamers composed of 3 dimers, consistent with structures associated with needle tip proteins (94, 95). These studies could not conclude if CPn0803 was a member of the T3SS apparatus, a chaperone, or an effector (94). However, CPn0803 can interact with components of the T3SS apparatus, including CdsN, CdsQ, and CdsF via an in vitro GST-plate assay and pulldowns from C. pneumoniae EB lysates (95). Purified CPn0803 was also incubated with lipid strips and found to bind to phosphatidylinositol and phosphatic acid (95), which are both components of eukaryotic cell membranes. Of note, CT584 has also been studied as a possible chlamydial chaperone (96) and will be discussed as such in a later section. Currently, there are no data supporting that the chlamydial T3SS apparatus contains a needle tip protein.
(ii) Translocon proteins. In other Gram-negative bacteria, when T3SS is activated, the needle tip protein undergoes a confirmational change and the translocon proteins are T3SS secreted and bind the needle tip and pierce the target mammalian membrane. Chlamydia bacteria have 4 possible translocon proteins, namely, CopB/CopB2 and CopD/CopD2, with biochemical studies focusing on CopB and CopD. Although CopB and CopD have not been explicitly shown to interact with one another during chlamydial infection, it is hypothesized that these proteins bind to one another and function together to form the translocon, similarly to orthologs of YopB and YopD (97). Known interactions of CopB and CopD were determined using the C. pneumoniae homologs CPn0809 and CPn0808, respectively. They were cloned with a GST tag, immobilized on beads, and incubated with an E. coli lysate overexpressing specific His-tagged chlamydial proteins. These studies demonstrated that CopB interacts with CdsF (needle filament protein) and Scc2/LcrH_1 (putative chlamydial T3SS chaperone) (98). CopD (CPn0808) of C. pneumoniae was found to interact with CopN (a secreted T3SS sensor protein), CdsN (Hub ATPase), CdsF (needle filament protein), and Scc2/LcrH_1 (putative chlamydial T3SS chaperone) (99). Consistent with their function as translocons, antibodies against CopB or CopD decreased C. pneumoniae infectivity, presumably by inhibiting the translocation of effectors required for entry (98, 99). Furthermore, peptides that inhibited the interaction of CopB with its cognate T3SS chaperone LcrH_1 also inhibited C. pneumoniae infectivity (98). These studies are consistent with CopB or CopD being critical functionality for T3SS (97, 100–102).
The Use of Electron Microscopy to Characterize the Chlamydial T3SS Apparatus
As biochemical assays and ultrastructural analysis were used to interrogate specific components of the needle apparatus, electron microscopy has also been used to contextualize the biochemical findings. As noted above, electron microscopy originally found spike-like projections on both EBs and RBs (18–21, 88, 103). One of the original ultrastructural studies via transmission electron microscopy of chlamydial EBs revealed projections that were 60 to 80 Å in diameter and ~500 Å in length that were inserted into a ring structure in the outer membrane (88). The authors of these studies conclude that the diameter of these structures was too small for DNA or protein to pass through and likened them to ion transporters (88), thus supporting the early soup through straw hypothesis that was discussed in an earlier section (22). Technologically advanced cryo-electron tomography studies performed by Hayward and colleagues revisited these projections and demonstrated that these structures are consistent with T3SS needles. They revealed detailed images of needle-like structures projecting from the RBs, contacting the inclusion membrane, and connecting to membrane segments of the rough endoplasmic reticulum (87). These data were consistent with a previous study, which indicated that the chlamydial inclusion membrane creates membrane contact sites with the endoplasmic reticulum (104). Nans et al. (85) quantified that there are typically 14 to 20 T3SS needles, spaced on average 56.5 nm apart on one side of the EB. Cryo-soft X-ray tomography has also captured needle structures of RBs inside the chlamydial inclusion (105), which clarified earlier electron micrograph images demonstrating direct connections via projections between RBs and the inclusion membrane that then extended into the cytosol (19). Building upon their original study, Hayward and colleagues used cryo-electron tomography with subtomogram averaging to derive intact structures by measuring 515 chlamydial T3SS needle complexes (86). In comparison to other T3SS, the chlamydial basal body is distinct in that it is elongated with a convex curvature. This basal body measures 34 nm in length and 14 to 20 nm in diameter. The measurement of these structures when Chlamydia was in contact with host cells revealed that the basal body contracts by at least 4 to 5 nm, which suggests that this conformational change may be associated with a pumping action to help propel effector proteins through the structure. As noted above, the C-ring protein CdsC is larger than other T3SS orthologs, and the EM images revealed that the outer ring surrounds the needle at the face of the outer membrane, which is also unique from other described T3SSs. The T3SS needle length is also shorter than other T3SS needles. When not in contact with host cells, the chlamydial needle averages 28 nm in length (86). For comparison, the Salmonella T3SS needle measures 60 nm in length (106). Upon contact with a eukaryotic membrane, the needle lengthens to about 30 to 33 nm (86). Furthermore, pores consistent with the insertion of the translocon formed by CopB and CopD are observed after contact between the chlamydial T3SS needle and the target mammalian membrane (86).
T3SS FOR ALL STAGES OF CHLAMYDIAL DEVELOPMENT
Basic Considerations in the Context of the Chlamydial Developmental Cycle
Chlamydia bacteria use their T3SS at every stage of their developmental cycle. It is unclear if the T3SS apparatus associated with the EB is the same as that associated with the RB, but both engage two different types of membranes, namely, the plasma membrane and the inclusion membrane, respectively. Furthermore, the cell walls of these two developmental forms are radically different, as the outer membrane of the EB is highly cross-linked and likely impermeable to many extracellular ions. In contrast, the outer membrane of the RB is not cross-linked and is likely permeable to extracellular ions that gain access to the lumen of the chlamydial inclusion. These differences will be highlighted in the following section where possible chlamydial T3SS (calcium) sensors are discussed.
There are several unanswered questions regarding T3SS in Chlamydia. After entry and the EB differentiates into an RB, does the T3SS apparatus of the EB get degraded and a new apparatus form in the newly differentiated RB? What happens to the RB T3SS apparatus during chlamydial division? Are new daughter cells equipped with an existing T3SS apparatus that is donated by the mother cell or are they made anew after the final steps of division? What happens to the T3SS apparatus of RBs that are undergoing secondary differentiation and forming infectious EBs? Some organisms like Salmonella have solved similar dilemmas by have having two distinct T3SSs. The T3SS encoded by the Salmonella pathogenicity island I (SPI1) is associated with entry (107), while the T3SS encoded by the Salmonella pathogenicity island II (SPI2) functions within the host cell (108, 109). But as obligate intracellular pathogens, Chlamydia bacteria have a highly reduced genome, and genome sequencing data do not support the presence of 2 entirely unique T3SSs. However, clues to this quandary may reside within the existence of a few redundant genes (cdsN and fliI; copB/copD and copB2/copD2) (2), the localization of chlamydial T3SS-secreted inclusion membrane proteins (Incs) in the inclusion membrane several hours after entry (110), and a temporal transcriptional program that is linked with progression through the chlamydial developmental cycle (5).
The EB is not transcriptionally competent due to the highly condensed chromosome. However, within a newly differentiated RB, the chromosome is relaxed, and transcription commences, typically at ~2 hpi, which is the start of early gene transcription. Early genes are typically transcribed until 8 to 10 hpi, after which the RBs undergo multiple rounds of exponential division and the midcycle genes are transcribed. Starting around 16 hpi, some RBs undergo a secondary differentiation creating new EBs as other RBs continue to divide. Even though, for C. trachomatis, egress will not occur for another 32 h, from a transcriptional viewpoint, 16 hpi marks the onset of late gene transcription (3, 5). Because 16 hpi has also been discussed as being midcycle (111, 112), this review will consider early-, mid-, and late-cycle transcription by the time points identified above. For C. pneumoniae, the developmental cycle is longer, as egress does not occur until 72 hpi. But gene transcription occurs in a similar time frame as C. trachomatis, with early gene transcription occurring at ~1.5 to 8 hpi, midcycle gene transcription occurring at ~12 to 18 hpi, and late-cycle gene transcription beginning at ~24 hpi (113).
By the above definitions, the majority (~60%) chlamydial T3SS apparatus genes are considered late-midcycle or late-cycle genes, as their transcription does not commence until 16 hpi (Table 1). This timing suggests that these genes may be transcribed to preload EBs with apparatus proteins. Notably, once transcription begins for these genes, it continues for the remainder of chlamydial developmental cycle. In C. trachomatis, the earliest T3SS apparatus-associated gene that is transcribed at 3 hpi and is pknD, which encodes the serine/threonine kinase that targets inner ring protein CdsD (75). Of the other T3SS genes that are transcribed early, most transcription commences around 8 hpi and are associated with the expression of core T3SS apparatus components FliI (Hub ATP synthase), CdsO (stalk), FliF (M-ring), CdsU (inner membrane basal body component), FliH (inner ring component), CdsF (needle filament), YscP (molecular ruler), and CopD2 (Table 1) (5).
Redundant Structural Elements in the Chlamydial T3SS
Understanding the timing of chlamydial T3SS apparatus assembly from current literature is complicated by inherent lab-to-lab differences. Data emerging from the same lab using the same chlamydial strain and culture conditions indicate that for C. trachomatis strain L2/434/Bu, secretion of effector proteins Incs occur within the first few hours postinfection (84). Additionally, the chlamydial inclusion is restricted from fusion with the endocytic pathway that occurs within the first hours postinfection (114, 115), and a subset of Incs are transcribed during the early stages of infection and appear on the inclusion membranes within the first 2 to 8 h of infection (110, 112). Combined, these data indicate that Chlamydia bacteria have a functional T3SS during the first few hours of infection, suggesting that some of the T3SS apparatus associated with the infectious EB may remain after primary differentiation to an RB or that premade proteins are available to quickly assemble a new apparatus. Furthermore, the 8-hpi time point is also associated with the first division events in Chlamydia (4), which suggests that a new apparatus is made during these events. It is entirely unclear if transcription events are limited to the mother or daughter cell or if they are occurring in both.
One of the two structural elements of the chlamydial T3SS apparatus that is redundant is the core Hub ATP synthases FliI and CdsN. FliI transcription begins during the mid-developmental cycle and peaks at 40 hpi. In contrast, cdsN is synthesized only during the late stages of C. trachomatis development and also peaks between 24 and 40 hpi (5). The expression pattern of the C. pneumoniae CdsN ortholog is similar to that in C. trachomatis (113), indicating that these expression patterns are conserved and therefore likely important toward when the proteins function. Genes that are expressed during the late stages of the chlamydial developmental cycle are thought to synthesize proteins required for either the secondary differentiation of an RB to an EB or for the protein to be prepackaged into EBs. Supporting the latter, CdsN is found in chlamydial EBs of both C. trachomatis (116) and C. pneumoniae (71). A proteomic analysis of C. trachomatis EBs and RBs revealed that most T3SS apparatus proteins were found in EBs and not in RBs (116); although, we know that the secretion of Inc proteins occurs temporally throughout the chlamydial developmental cycle (110, 112, 117). A separate study that performed a deep sequencing analysis of the C. trachomatis transcriptome revealed that transcripts of cdsJ, which encodes the candidate inner membrane ring protein, and cdsC, which encodes the candidate outer membrane ring, were enriched in RBs (118). Confounding these types of analyses is the fact that some chlamydial T3SS proteins have been identified in uninfected HeLa lysates either because they share homology to a mitochondrial protein (e.g., CdsC) or they were identified due to unknown reasons (e.g., LcrD and CopB) (119). Thus, it is possible that chlamydial T3SS proteins associated with RBs may have been identified in some of these other proteomic studies but excluded as possible background contaminants. In a previous review, Ferrell and Fields (120) hypothesized that the redundant chlamydial T3SS apparatus components may represent two functionally distinct and separate basal core units, which may provide Chlamydia with a potential mechanism to control effector secretion. Given that there are chlamydial effectors expressed and secreted at each stage of development, the timing of secretion and effector hierarchy are critical toward supporting chlamydial growth and development. Previous studies performed in the Schneewind laboratory demonstrated that Hub ATPase can recognize effector proteins as a mechanism to control effector secretion hierarchy (121). It is also well-characterized that the Hub ATPases recognize chaperone and effector complexes which results in effector secretion and represents another mechanism controlling effector secretion hierarchy (120, 122).
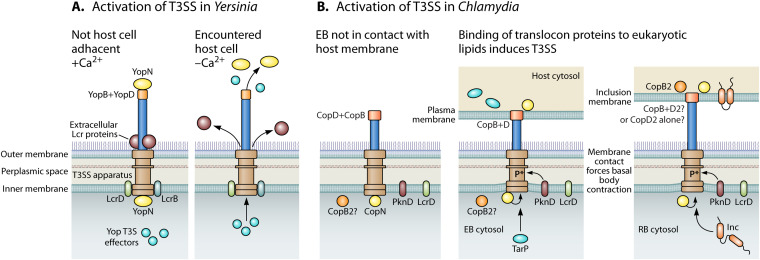
An interesting duplication, which is conserved across chlamydial species, is that of copB and copB2 and that of copD and copD2. Transcription of copB and copD occurs during the late stages of the developmental cycle, whereas the transcription of copB2 and copD2 occurs during the later stages of the early- or mid-developmental cycle (5). These data suggest that these proteins may be functioning at different stages of chlamydial development. Using antibodies and Western blot analysis, CopB was detected in C. trachomatis within the first few hours of infection and then there was no detectable signal again until 20 hpi, a time when some RBs are undergoing secondary differentiation to form new EBs (123). These data are consistent with a proteomic analysis that found CopB associated with the proteomes of EBs only (116) and also in lysates of purified EBs (84). CopB is also associated with the membranes of mature inclusions, indicating that it has functions other than early T3SS events (124). Antibodies against CopB or CopD reduced the infectivity of C. pneumoniae (98, 99) Although, it should be noted that antibodies against CopB2 or CopD2 have not been tested in similar assays. In contrast, a Western blot analysis revealed that CopB2 was constitutively expressed, at least at the protein level (123). Further indirect immunofluorescence revealed that CopB2 was found associated with the inclusion membrane, during time points when CopB was absent (123). Selective permeabilization of the plasma membrane followed by the addition of a nonmembrane permeable cross-linking biotin reagent demonstrated that CopB2 was exposed to the cytosolic side of the chlamydial inclusion (124). CopB and CopB2 separate differently after TX-114 extraction. CopB partitions in the TX-114 insoluble fraction, consistent with an integral membrane protein, such as a protein component of a translocon. In contrast, CopB2 partitioned in the TX-114 soluble fraction along with the soluble protein control GroEL. Furthermore, ectopic expression wild-type or deletion mutant CopB2 revealed that deletion of several coil-coil domains rendered CopB2 unable to localize to chlamydial inclusions. These data indicated that a CopB2 association with the inclusion membrane was driven by protein-protein interactions and not by membrane (lipid) binding, suggesting that CopB2 functions in a different capacity from a typical translocon protein (123). Collectively, these studies suggest that the most likely scenario is that CopB functions as a translocon component, while CopB2 may function as an additional accessory or, even, as a sensor protein. The hypothesis that CopD or CopD2 associate with CopB within the translocon of the T3SS apparatus of the EB (CopD) or RB (CopD2) is supported by transcription data, with similarity to the Yersinia protein YopD, and in vitro pulldown data. In this scenario, the presence of CopD or CopD2 may be one of the defining or determinant features of the T3SS translocon associated with specific developmental forms (Fig. 3). This model has yet to be tested experimentally.
FIG 3.
Activation of T3SS. (A) Depicts how the Yersinia T3SS is activated by host cell contact, which was originally characterized by chelating Ca2+ from bacterial growth medium (127, 256–258). (B) Depicts a likely mechanism of chlamydial T3SS activation, which is contact with lipids of the plasma membrane or the inclusion membrane (145). This model further depicts possible differences in the translocon proteins of an EB (CopB and CopD) versus an RB (CopD2 only or CopB and CopD2). CopB2 is modeled as a peripheral inclusion membrane protein as is consistent with current data (123).
POSSIBLE TRIGGERS THAT MAY ACTIVATE THE CHLAMYDIAL T3SS
Description of the Classical Low Calcium Response that Induces T3SS in Yersinia
Significant questions regarding how secretion through the chlamydial T3SS apparatus is controlled are what are the environmental cues that trigger chlamydial T3SS and how is effector secretion regulated? The presumption is that the ultimate control mechanism for Chlamydia is similar to the originally described contact-dependent secretion (68). We do know that the needle protein CdsF does not polymerize until it is on the surface of the organisms (83). It has been reported for other T3SSs that upon contact with the host membrane, the needle itself can transmit an activation signal (125, 126). Environmental cues, such as the ability to recognize the temperature of a human host (e.g., 37°C) and extracellular calcium levels consistent with being in contact with a host cell, activate the T3SSs of other Gram-negative bacteria. This mechanism associated with environmental cues was first described in Yersinia cultured in broth. The responsive genes were characterized as the low calcium response genes or lcr genes that encode proteins to block secretion at the tip of needle [LcrQ, LcrE(YopN), and LcrG], sense extracellular calcium levels from inside the organism (LcrD), sequester effectors and chaperones in the bacterial cytosol (LcrQ, LcrE, and LcrG), or suppress the expression of T3SS-related genes (LcrH) (127). It was later recognized that chelation of Ca2+ in bacterial medium artificially mimics the microenvironment of a bacterium coming in close contact with a host cell (27). Of note, Chlamydia bacteria have orthologs to two lcr genes, specifically lcrD and lcrE (commonly referred to as copN in C. trachomatis) (Table 1) (Fig. 3A). What was important about the early studies characterizing the lcr genes in Yersinia was not necessarily that a low Ca2+ environment and human body temperatures induced the T3SS (although these were helpful mimics for the identification of T3SSs in other bacteria [128]); it was the notion that the expression and implementation of the T3SS was responsive to environmental queues. Based on the number of host defense mechanisms, it is not conducive to pathogen survival if effector proteins are constitutively secreted.
Chlamydial Homologs to Low Calcium Response Proteins
As Chlamydia bacteria are obligate intracellular pathogens, it is challenging to modify the intracellular environment to artificially trigger chlamydial T3SS. Temperatures compatible with tissue culture remain relatively stable, and chelating ions like Ca2+ is not practical. In general, Ca2+ concentrations within the eukaryotic cytosol are low compared with those in the extracellular space, but Ca2+ concentrations also exist in a gradient that is controlled by ATPases within the endoplasmic reticulum and plasma membrane. Furthermore, the ability of Ca2+ ions to readily cross into the chlamydial periplasmic space is unknown. In EBs, their outer membrane is highly cross-linked rendering it is unlikely that Ca2+ ions would readily translocate to the EB periplasmic space (82, 129–132). In RBs, Ca2+ would have to cross both the inclusion membrane and then the more permeable outer membrane. Therefore, by understanding how the Yersinia LcrD and YopN proteins function may provide clues to how the chlamydial orthologs may function. Chlamydial LcrD is found in the proteomes of both EBs and RBs but is enriched in EBs (116). The Yersinia ortholog is a protein found in the inner membrane and responds to Ca2+ levels as ions enter the periplasmic space (127, 133). In Chlamydia, LcrD localization has not been resolved; although, Fig. 1 depicts chlamydial LcrD as residing within the inner membrane. However, given the retention and the conservation of the lcrD gene, LcrD likely functions in some capacity to help regulate chlamydial T3SS.
More experimental data are available about CopN, which is a candidate protein that may help regulate the function of chlamydial T3S. CopN is an ortholog to SctW, which represents the family of T3SS gate keeper proteins (134). This protein family includes the Yersinia YopN (also known as LcrE) protein (127, 135), which has great similarity to CopN (14, 136). In Yersinia, yopN was characterized as part of a locus that became transcriptionally active in response to low Ca2+ growth conditions in broth medium (127, 135). A Western blot analysis revealed that YopN was surface localized and T3SS secreted (135) and possibly associated with the outer bacterial membrane (136). Strains that carried mutations or deletions in lcrE/yopN were impervious to Ca2+ levels and constitutively secreted Yop effectors (136, 137). These data indicated that in Ca2+-rich environments, YopN played a pivotal role in suppressing T3SS by plugging the apparatus. Upon encountering low Ca2+ environments, which mimics host cell contact, T3SS was activated and YopN was secreted (135). The removal of the plug YopN resulted in the secretion of other Yop proteins (127) (Fig. 3A). Supporting the notion that Ca2+ functions as an artificial or proxy signal to activate Yersinia T3SS, YopN proteins do not have a binding affinity for Ca2+ that is typical of calcium-binding proteins (135). But results obtained from characterizing YopN function directly informed experiments that examined CopN function in chlamydial T3SS.
Using the Y. enterocolitica heterologous T3SS, CopN was the first chlamydial protein demonstrated to be T3SS secreted (138). Additional studies using S. enterica serovar Typhimurium demonstrated that CopN localized to the host cytosol via T3SS (139). Immunofluorescence studies localized CopN to the cytosol in C. trachomatis- or C. pneumoniae-infected cells (138, 140). Similar to the function of YopN, CopN is thought to be held in the plug position within the chlamydial cytosol by chlamydial chaperone proteins Scc1 and Scc4 (141) or Scc3 (141, 142). CopN is detected in TX-114 soluble fractions of C. trachomatis EB lysates, consistent with it not being an integral membrane protein (84). Proteome analysis also revealed an enrichment of CopN in C. trachomatis EBs with no peptides of CopN being identified in RBs (116). These protein data are consistent with copN expression occurring during the late stages of the chlamydial developmental cycle to produce CopN to be prepackaged into EBs (5). In C. pneumoniae-infected cells, CopN is associated with the sequestration of fructose bisphosphate aldose A to benefit bacterial growth (140) and may interact with α/β-tubulin of microtubules (142, 143). Of note, the ability of CopN to bind to microtubules is recognized only with the C. pneumoniae homolog and not with the C. trachomatis homolog (143). Furthermore, exogenous expression of C. pneumoniae CopN in yeast or mammalian cells caused cell cycle arrest which was linked to the noticeable alterations of the microtubule cytoskeleton (144). Based on these combined observations, it is likely that CopN has T3SS-related functions upon chlamydial entry and early inclusion establishment but also in potentially modulating chlamydial-host interactions.
Possible Role of Lipids as Activators of Chlamydial T3SS
In thinking about likely molecular signals that would activate chlamydial T3SS, lipids likely play a role (Fig. 3B). Small molecules are unlikely to passively cross the highly cross-linked outer membrane of chlamydial EBs, but contact with a lipid bilayer would create confirmation or biophysical changes that could be key to activating chlamydial T3S. This hypothesis is supported by previously discussed electron tomography data (86). In a different study, liposomes enriched in sphingolipids and cholesterol induced secretion of chlamydial effector TarP from purified C. trachomatis EBs (145). It is also established that cholesterol and sphingolipids are required for pore formation to T3SS translocon proteins (e.g., CopB and CopD) (146). Lastly, these are lipids that are components of both the plasma membrane and the chlamydial inclusion membrane.
Linking Induction of T3SS and Transcription
Any environmental trigger, including contact between the needle and the target membrane, will impact the expression of the T3SS at the transcriptional level. In Chlamydia, there are σ66 promoter regions upstream of many T3SS genes (147, 148), but the transcriptional control of other chlamydial T3SS genes may be promoted by an alternative σ factor, σ54 (RpoN) (149). Transcriptional regulation of chlamydial T3SS is poorly understood, but there are several candidate proteins that may function in this capacity. For example, CdsZ is a contact-dependent secretion-zinc ribbon binding domain, and bacterial two-hybrid assays revealed that CdsZ binds to RpoN, spoke protein CdsL, and inner ring protein FliH. CdsZ interactions with CdsL and FliH were confirmed via GST-pulldowns from E. coli lysates. While RpoN was exogenously expressed in C. trachomatis and coprecipitated with CdsZ (63). These data, in addition to cdsZ being transcribed during late stages of the chlamydial development cycle (5), contributed to Barta et al. (63) hypothesizing that CdsZ is involved in regulating early T3SS events. Another chlamydial T3SS protein that interacts with RpoN is chaperone Scc4 (141, 150), which will be discussed fully within the next section. Recent studies have also identified a possible transcriptional regulator, ChxR (CT630), that can regulate the expression of genes associated with virulence, which ostensibly includes T3SS genes (151). For this study, Caldwell and colleagues used chemical mutagenesis to create a ChxR null mutant chlamydial strain via chemical mutagenesis. A comparative proteomic analysis of lysates from cells infected with either a wild-type or ChxR null strain harvested at 36 hpi revealed a significant decrease in 5 known chlamydial T3SS effector proteins. Reverse transcription-quantitative PCR (qRT-PCR) analysis confirmed that the decrease in these proteins was occurring at the level of transcription (151).
CHLAMYDIAL T3SS CHAPERONES: USHERS TO THE INJECTISOME
Introduction to the 5 Structural and Functional Classes of T3SS Chaperone Proteins
Generally, chaperone proteins are accessory cytosolic proteins that are critical for T3SS and are categorized into 5 structural and functional classes (89). The functions of chaperones are varied, as they are required for proper localization and secretion of T3SS machinery, establishment of a hierarchy of effector secretion, promotion of T3SS-associated protein or effector stability, maintenance of certain effectors in an unfolded or partially folded state, and/or modulation of transcription in response to T3SS events (152–154). Class I chaperones are involved in the secretion of effector proteins, have a distinct identifiable structural motif (α-β-β-β-α-β-β), and are typically homologous to the larger CesT family of bacterial chaperone proteins (155–157). Class I chaperones are further classified as binding to either single effector proteins (class IA chaperones) or multiple effector proteins (class IB chaperones) (157). Class II chaperones bind to translocon proteins (e.g., CopB and CopD) and are also typified by alpha-helical structures and the presence of tetratricopeptide repeat (TPR) domains (158, 159). TPR domains are defined as 3 to 16 tandem repeats of up to 34 amino acids (158) Class III chaperones bind to proteins that form the T3SS apparatus (160). Very few chaperones are classified as types IV or V because they are both structurally distinct from other chaperone proteins and are involved in binding to needle filament proteins (89). Class IV chaperones are typified by enteropathogenic E. coli protein CesA, which binds to the EspA needle filament protein (161). Class V chaperones are typified by yersiniae protein YscE, which contains a unique structural fold (162). YscE and Pseudomonas aeruginosa homolog PscE require cochaperones YscG and PscG, respectively, to bind to and to prevent premature polymerization of respective needle filament proteins YscF or PscF (163). Furthermore, YscE binds to the C-terminal region of YscF (163), not within the first 10 to 15 amino acids that is typical of chaperone-substrate interactions (164, 165). Because T3SS chaperone proteins have very little amino acid similarities across bacterial species, secondary structural features (e.g., α-β-β-β-α-β-β or TPR domains) are more commonly used to identify candidate chaperone proteins (89).
Candidate Chlamydial T3SS Chaperones
Chlamydiae have 11 genes that encode candidate T3SS chaperones (Table 2). These genes are conserved across chlamydial species and are often organized within or adjacent to gene clusters that encode other T3SS proteins (Table 2) (2). In the initial studies that characterized these chaperones, the proteins were often given different common names depending on individual preference or which chlamydial species was being studied. For example, Scc3 (CT862/CTL0237/CPn1021/CCa00740) as studied in C. trachomatis has also been referred to as LcrH_2 when studied in C. pneumoniae (148); herein, this protein will be referred to as Scc3. The chaperone Mcsc (CT260/CTL0512/CPn0409/CCa00385) is not an acronym commonly associated with T3SS chaperones; however, when Mcsc was initially characterized, it was found capable of binding multiple effector proteins, and hence it was aptly named, multiple cargo secretion chaperone (166). In instances where there is divergence in the literature regarding the common name of a chaperone, the one used most frequently in the literature will be used here; these names are also bolded in Table 2. Lastly, there are examples of chlamydial proteins being labeled as novel putative chaperones only because they were demonstrated to bind to known chlamydial T3SS effectors (e.g., CT635 binding to effector CT622 [167]). These proteins are not considered in the following discussion because further bioinformatic analysis could not identify common structural motifs or other characteristics consistent with known chaperones (E. A. Rucks, personal observation).
Chlamydiae have at least five class I chaperones, three (and a probable fourth) class II chaperones, and two class III chaperones (although one of these may be a class V chaperone; see below) (Table 2). Of note, chaperones are expressed at lower levels than effectors, and chaperone availability is typically a limiting factor associated with T3SS effector secretion (168, 169). Mass spectrometry analysis of the protein content of C. trachomatis serovar L2 EBs found that 2% of the total protein mass consisted of chlamydial chaperones (166), while T3S effectors exist in a 10-fold molar mass excess over chaperone proteins (116, 166). The most abundant chaperone proteins are Slc1, Scc2, and Mcsc, and the least abundant ones are Scc1, CT274/CTL0526, Scc3, and Scc4 (166). Given the importance of chaperone function to T3SS, generally, a conventional hypothesis is that the chaperones in Chlamydia are essential genes. Two recent transposon mutagenesis studies challenge this thought. A study by LaBrie et al. (170) in 2019 was able to generate a transposon mutant in C. trachomatis L2/434/Bu that disrupted the gene encoding Scc1. A separate study performed by Ian Clarke’s group generated transposon mutants in C. trachomatis serovar L2 strain SWFP for Scc1 and Slc1 (171). In both studies, the functional impact of these mutations was not experimentally tested to determine if Scc1 or Slc1 activity was abolished. The fact that Scc1 could be targeted by transposon mutagenesis by two separate groups and experimental methodologies does suggest that it may not be an essential gene as originally hypothesized or that there is redundancy in chaperone function. The transposon mutation in Slc1 is also curious, as Slc1 has been implicated in assisting the secretion of entry-associated effectors (discussed below), and yet, viable mutants were recovered indicating that the entry and induction of the developmental cycle were not completely abolished. These data are consistent with chlamydial entry being essential for survival and the existence of redundant mechanisms.
Class I chaperones.
Three of the five class I chaperones (slc1, scc1, and scc4) were identified as members of the CesT family of chaperones via PSI-BLAST (Table 2). CesT family members are based on sequence similarity to the Tir chaperone in EPEC (172). Scc1 and Scc4 are likely class IA chaperones, as they have been demonstrated to assist in the secretion of a single substrate, CopN (138, 141, 150). Chlamydial proteins that are likely class IB chaperones are Slc1, CT584, and Mcsc, as they can bind to multiple substrates (74, 96, 166). To promote the secretion of their substrates, T3SS chaperones often homodimerize, but there is some evidence that there are often heterodimer interactions from two different chaperones that promote substrate or effector secretion (89). Chlamydial chaperones that can form homodimers are Slc1, Mcsc, CT584, and Scc4 (96). Homodimers formed by Scc1 have not been demonstrated, but Scc1 forms a heterodimer complex with Scc4 (141, 150). When expressed by itself in E. coli, Scc1 is not soluble; however, when it is coexpressed with Scc4, soluble Scc1 can be purified from cultures (141). Of note, the chaperones that are responsible for the TT3S of most chlamydial effectors during chlamydial infection are unknown.
(i) Scc4 can form heterodimer complexes with Scc1 or RNA polymerase. The best-characterized chaperone-effector pairing is between CopN and its chaperones Scc1 and Scc4. To shed light on possible chaperone-substrate/effector interactions, the field has used yeast two-hybrid or bacterial two-hybrid systems, in conjunction with pulldowns from purified chlamydial organisms or from coexpression in heterologous systems. Scc1 and Scc4 function together to promote the secretion of CopN (141, 150, 173). In a seminal study, both scc1 and copN were transformed into Y. enterocolitica and under T3SS-inducing conditions, CopN, was secreted when coexpressed with Scc1 (138). Later studies revealed that C. trachomatis and C. pnuemoniae homologs of CopN can bind to respective homologs of Scc1, Scc3 (class II; discussed below), and Scc4 in C. trachomatis or C. pnuemoniae, respectively (141, 173, 174). Scc4 is required for the secretion of CopN from Y. pestis, and coexpression of Scc1 and Scc4 enhances CopN secretion (175). The Scc1 and Scc4 complex was the first example of heterodimeric chaperone interactions promoting secretion of an effector. It is also one of the few chaperone effector complexes that has been demonstrated to occur during chlamydial infection. Treatment of infected cells with membrane-permeable chemical cross-linker dithiobis(succinimidyl propionate) (DSP), followed by coimmunoprecipitation, revealed that Scc4, Scc1, and CopN are complexed within chlamydial organisms. Furthermore, Scc1 and Scc4 colocalize within chlamydial organisms starting at 24 hpi (141).
This Scc1-Scc4 heterodimer complex also has regulatory implications relative to chlamydial transcription. Scc4 has been demonstrated to interact with the β- and α-subunits of RNA polymerase (RNAP) (176). In vitro transcription assays revealed that the interaction between Scc4 and RNAP inhibited E. coli σ70- and chlamydial σ66-dependent transcription but not chlamydial σ28 transcription (176); σ66 belongs to the σ70 family of bacterial transcription factors (177, 178), while σ28 and σ54 are alternative σ factors in Chlamydia (2). Consistent with the above findings, expression of Scc4 alone in E. coli inhibited transcription of recA, idnT, and cysG and halted E. coli growth. Coexpression of Scc4, Scc1, and CopN alleviated both the inhibition of transcription and growth (150). These studies highlight that Scc4 has two distinct functions, as follows: binding to RNAP and promoting the secretion of CopN. Nuclear magnetic resonance studies revealed that Scc4 must undergo a structural rearrangement to bind Scc1. In turn, Scc1 has a dynamic region that allows for this binding but must also be partially unfolded; this last step likely requires additional accessory proteins in Chlamydia (179).
Data from Shen and colleagues support a model in which Scc1 and Scc4 bind to CopN during the early and late stages of chlamydial development. After the secretion of CopN, Scc4 is free to bind to RNAP during mid-developmental cycle time points (179). To examine the impact of overexpression of Scc4, scc4 was cloned into an anhydrotetracycline (aTc)-inducible expression plasmid (180) which was then transformed into C. trachomatis serovar L2 (175). Chlamydial-infected cells were treated with 10 ng/mL aTc at the time of infection, which resulted in larger inclusions, which were noticeable at 16 hpi, and overall, more abundant RBs and fewer EBs produced between 16 and 24 hpi. However, there were no noticeable differences in the total number of EBs at 32 hpi between induced and uninduced samples. Furthermore, overexpression of Scc4 resulted in an increase of transcription of late genes (e.g., scc1, copN, and slc1), while the transcription of early- and midcycle genes (e.g., incD or euo) were not impacted. Combined, these data suggested that overexpression of Scc4 accelerated chlamydial growth but not necessarily the duration or pace of the chlamydial developmental cycle (175). It is unclear if excess Scc4 binding to RNAP directly resulted in changes of the transcription of genes examined, of which many are related to chlamydial T3SS. There are σ66 promoter regions upstream of many T3SS genes (147, 148); however, the activities of alternative σ factor σ54 may also play a role in the expression of other chlamydial T3SS genes (149).
(ii) Class IB chaperones. Pulldowns from C. trachomatis serovar L2 EB lysates were performed to elucidate the function of class 1B chaperone Slc1. These studies demonstrated that Slc1 can bind to chlamydial entry effectors TarP (166, 181), TmeA, TmeB, and TepP (166). These data are consistent with slc1 transcription occurring late in the chlamydial developmental cycle to be prepackaged into EBs (5). These studies were further confirmed by coimmunoprecipitation of TarP, TmeA, and TmeB with Slc1 that were coexpressed by Y. enterocolitica (96). Additionally, coexpression of only Slc1 with various chlamydial effector proteins during T3SS-inducing culture conditions, enhanced the secretion of TarP from Y. pseudotuberculosis (181) and that of TarP, TmeA, TmeB, TepP from Y. pestis (166). Mass spectrometry analysis from Slc1 pulldowns from purified EB lysate revealed that Slc1 can also weakly bind to another chaperone protein, Mcsc (166). The biological significance and function of this interaction are unknown because previous studies demonstrated that Slc1 forms homodimers to promote the secretion of chlamydial effectors (96), and the effector TarP is the preferred binding partner of Slc1 (166).
There is less understood about the last two class IB chaperones, namely, Mcsc and CT584. Both proteins are hypothesized to function, in part, to stabilize effectors and protect them from premature proteolysis (74, 96). In vitro biochemical studies revealed that Mcsc can form a single homodimer and binds to at least two chlamydial T3SS effectors, namely, CT618 and Cap1. These Mcsc-effector complexes can then bind the CdsQ sorting platform (74). Bacterial two-hybrid analysis revealed that CT584 is capable of binding to 6 different chlamydial T3SS effectors but could assist only in the translocation of one (CT082) via the Y. enterocolitica T3SS (96). It is unknown whether coexpression of another chlamydial chaperone would allow the secretion of more effectors from Y. enterocolitica. Unpublished data from the Rucks lab demonstrate by a bacterial two-hybrid assay that CT584 can form heterodimers with Slc1 and Mcsc (L. Knight and E. A. Rucks unpublished data). The further use of the Y. enterocolitica surrogate T3SS revealed that when CT584 binds to a central region within CT082, it stabilizes CT082 protein levels in Yersinia (96). These data support that CT584 acts as a chaperone and not as a needle tip protein, as inferred previously (67, 94, 95). The ability of the protein to be able to form homodimers (96) is consistent with both functions.
Class II chaperones.
Class II T3SS chaperones are likely responsible for establishing hierarchy in the secretion of effector proteins in part by promoting the secretion of translocon proteins. Class II chaperones are typified by the presence of distinctive structural motifs called tetratricopeptide repeat (TPR) domains. Examples of chlamydial class II chaperones are Scc2, Scc3, CT274, and possibly CT114. There are no functional data associated with CT274, other than it contains TPR domains and is orthologous to class II chaperones of other bacteria (Table 2). CT114 was discovered in a Phyre prediction search as having TPR domains between amino acids 261 and 462. The TPR domain was confirmed with InterPro analysis of the CT114 (CTL0369) amino acid sequence (182) (S. P. Ouellette and E. A. Rucks, unpublished observations) (Fig. 4). In the chlamydial chromosome, ct114 is organized immediately upstream but separate from the incD operon, which encodes four chlamydial T3SS effectors (2, 35). Furthermore, the transcription of ct114 follows a similar pattern to the incD operon (5).
FIG 4.
Model of CT114 as a class II chaperone. (A) Shows a high-resolution three-dimensional (3D) model of a CTL0369 amino acid sequence generated by the Phyre2 server (259). CTL0369 is the C. trachomatis L2/434/Bu homolog to CT114 in C. trachomatis serovar D. The N-terminus is in red and the structure follows the coloring of the rainbow with the C terminus appearing in dark blue. The alpha helical structures are consistent with structures associated with chaperone proteins. (B) Shows a comparison between CT114 and known Class II chaperone, Scc2 (CTL0839). CT114 is twice as large as Scc2 but contains a characteristic TPR domain (aa 261 to 462) within the C-terminal half of the protein similar to Scc2 (aa 56 to 204). Proximal to the TPR domain in CT114 is a transglutamase-like domain (aa 157 to 268), which is not consistent with the function or structure of a T3SS chaperone protein. These domains were defined by analyzing the sequences of these proteins (source, C. trachomatis serovar L2 strain 424/Bu) by InterPro (182).
Consistent with a function associated with establishing a hierarchy of substrate secretion, Scc2 and Scc3 have both been implicated in promoting the secretion of translocon proteins CopB/CopD. Translocon proteins form pores in the target membrane, and therefore, their secretion must precede any other effectors. In lysates obtained from C. trachomatis-infected cells, CopB coimmunoprecipitates with Scc2 and Scc3 (124). Also, purified proteins from C. pneumoniae subjected to GST-pulldowns revealed interactions between CopB and CopD with Scc2 (98, 99). Combined, these data suggest that these translocon-chaperone interactions are conserved across chlamydial species.
As Scc2 and Scc3 are orthologs to Yersinia SycD, it was originally hypothesized that these proteins would complement the function of SycD in the secretion of YopD. But when either scc2 or scc3 were cloned into a ΔsycD mutant, only Scc3 could partially restore wild-type levels of YopD secretion. Both Scc2 and Scc3 are capable of binding YopD in vitro, even though Scc2 could not complement the ΔsycD mutant (124). Of note, both Scc2 and Scc3 are larger in size than SycD and also have unique amphipathic helices in their amino-terminal regions which may indicate Scc2 and Scc3 have unique functions not typically associated with SycD proteins (124).
In addition to being in involved in the T3SS of translocon proteins, Scc3 also can play an inhibitory role in chlamydial T3SS. Specifically, Scc3 binds to the C-terminal region of CopN and reduces CopN secretion (173). Of note, CopN is capable of binding to Scc3 or the Scc4-Scc1 chaperone complex (141). The inhibitory function may play a regulatory role in establishing the hierarchy of effector secretion, as CopN may be the first effector that is secreted after the translocons have formed the pore in the target membrane. Scc3 was the first example of a chlamydial class II chaperone demonstrating a function consistent with establishing hierarchy or titrating the amount of effectors that is ultimately secreted. Hence, CopN and Scc3 serve the gatekeeper function that does not allow premature effector secretion before that apparatus is fully assembled, specifically with the translocon in place at the tip of the needle (142). As such, chlamydial class II chaperones may function in multiple individual or synergistic capacities to support chlamydial T3SS.
Class III and a candidate class V chaperone.
Yeast two-hybrid and yeast three-hybrid assays have revealed the possible function of class III chaperones CdsE and CdsG. Specifically, CdsE and CdsG can bind to one another and form a three-protein complex with CdsF, the needle filament protein. This chaperone-substrate pairing is consistent with CdsE and CdsG functioning to promote the secretion of T3SS apparatus proteins. Further coexpression of CdsE and CdsG with CdsF is required for CdsF to be stably expressed and not degraded in E. coli. These data are consistent with CdsE and CdsG collaborating to promote the secretion of CdsF (83). This heterodimer arrangement is similar to the YscE/YscG and PscE/PscG chaperone heterodimers of Yersinia and Pseudomonas, respectively (163), which may indicate that CdsE may be more appropriately defined as a class V T3SS chaperone (89).
WILL YOU SECRETE MY PROTEIN? THE USE OF SURROGATE T3SS MODELS TO STUDY CHLAMYDIAL T3SS EFFECTOR PROTEINS
Overview of Model Organisms
The use of heterologous or surrogate models to study chlamydial T3SS by specifically cloning chlamydial T3SS genes into a different Gram-negative pathogen was introduced briefly in the previous section. The first heterologous system used to study chlamydial T3SS was Y. enterocolitica, and it was also the first study to provide definitive evidence that C. trachomatis contained genes that encoded functional components of a T3SS (138). For the first time, Fields et al. (138) demonstrated that under conditions that induced Y. enterocolitica T3SS, CopN when coexpressed with Scc1 was secreted. This study also highlighted the following primary utility of these heterologous systems: to screen possible uncharacterized chlamydial proteins to determine if they were chlamydial T3SS effectors. Since these initial studies, other groups have used Y. enterocolitica to probe chaperone-effector interactions (96, 124) and to identify novel chlamydial effectors (183, 184). Other heterologous systems used to study chlamydial T3SS include the following: Y. pseudotuberculosis (84, 124, 181, 185, 186), Y. pestis (166), S. enterica serovar Typhimurium (139), and Shigella flexneri (167, 187).
Comparison of pathogen lifestyles to that of Chlamydia spp.
While the main components of T3SS are homologous between species (13), there are key differences in pathogen lifestyle, which in turn impact the types of effectors and cognate chaperones. Typically, chaperone proteins are specific for the effector or group of effectors of which they are promoting the secretion, and while structural aspects of chaperones are conserved, specific amino acid content is not (89). Further T3SS effectors are typically bacterium specific, with few orthologs across different bacterial species (13). The amino acid content of proteins does directly impact protein-protein interactions; therefore, differences in effector amino acid content may alter the interaction with an orthologous chaperone. In consideration of pathogen lifestyles relative to those of Chlamydia, the Yersiniae species used are all considered extracellular pathogens. S. flexneri is intracellular but localizes to the cytosol of infected cells and is not a membrane-bound vacuole. Only, S. enterica Typhimurium is both intracellular and resides within a membrane-bound vacuole.
(i) The similarities and differences in chaperone proteins. These differences are further highlighted by the fact that a PSI-BLAST search between chlamydial chaperone proteins and the species that have been used in orthologous systems revealed few similarities between chlamydial chaperones and those of other bacterial species (Table 2). This lack of similarity is important in contextualizing data derived from these systems. For example, highlighting the fact that existing Y. pestis chaperones could not entirely compensate for the chlamydial chaperones, the addition of slc1 to the Y. pestis surrogate model enhanced the T3SS of several effectors (166). Furthermore, cloning of full-length effectors into these systems was recognized as problematic for their T3SS as the candidate chlamydial effectors got stuck or degraded without the appropriate chlamydial chaperone (96, 186). None of these pathogens have developmental cycles, and hence, nuances to temporal chaperone-effector or chaperone-chaperone interactions would not be realized in these systems.
Bioinformatic Tools Used to Identify Chlamydial T3SS Effector Proteins
It is estimated that Chlamydia bacteria have dedicated greater than 1/10 of their genome to T3SS effectors (2, 6). The largest family of effectors is the Inc family. Incs are structurally distinct (188), which facilitates the use of bioinformatics to identify putative Inc proteins across chlamydial species and then test for their secretion via surrogate T3SSs (189). There have been several strategies employed to identify additional chlamydial T3SS effectors. Several of these strategies have employed bioinformatic tools. In one study, proteins of unknown function that were conserved across chlamydial species were tested to see if they contained T3SS signal sequences. The first 20 to 65 codons were cloned and fused to cyaA and then tested for secretion by S. flexneri. This methodology revealed 24 possible non-Inc chlamydial T3SS effectors (187). In a separate study, a computational algorithm-based support vector machine (SVM) or SVM-based Identification and Evaluation of Virulence Effectors (known as SIEVE) (190) was trained against the chlamydial genome. SIEVE successfully identified many Inc proteins, which are known chlamydial T3SS effectors, but also some Pmp proteins, which are not exported via T3SS (190). In the second iteration of SIEVE, the algorithm was tweaked and “retrained.” Candidate genes were then cloned for the expression and T3SS testing in Y. pseudotuberculosis. This robust study yielded a list of 100 possible T3SS effectors (186).
There have also been studies that tried to identify secreted chlamydial proteins localized to the host cell cytosol (184, 191). In one such study, cells were infected with C. trachomatis and permeabilized with perfringolysin O (PFO), which will essentially leak the contents of the cytosol into the extracellular milieu. Shotgun protein mass spectrometry analysis revealed 13 possible chlamydial effectors that were then tested for T3SS compatibility using Y. enterocolitica, which further narrowed the list to 4 possible novel effectors (184). Initial approaches to identify or screen for possible chlamydial type III effectors utilized surrogate or heterologous T3SS to test for compatibility. A recent study highlights the complication of the use of such systems to evaluate whether a chlamydial protein is secreted by T3SS (192). Specifically, bioinformatic predictions identified 382 possible C. pneumoniae T3SS effectors, and validation of these proteins using a Yersinia or Shigella surrogate model gave many false positives. Ultimately, candidate genes were cloned with a GSK epitope tag, which becomes phosphorylated in the host cytosol, and transformed into C. trachomatis, which identified 49 possible effector proteins (192).
Advances in the field of chlamydial genetics will eliminate the dependence on surrogate models to study the chlamydial T3SS. Currently, there is the ability to express a T3SS effector with a C-terminal epitope tag from a plasmid (180, 193, 194) or directly from the chromosome in C. trachomatis serovar L2 (434/Bu) (195). In addition to the GSK-tag described above (192), a separate study utilized a β-lactamase reporter fusion (BlaM) to demonstrate the secretion of specific effectors into the host cytosol (196). Similarly, creating an Inc-APEX2 fusion that can be inducibly expressed from C. trachomatis allowed for visualization by transmission electron microscopy of the orientation of the fusion construct within the inclusion membrane (197). While the FLAG epitope tag has no intrinsic enzymatic activity to determine whether a candidate effector is localized to the host cytosol, it remains a common epitope tag used to study the secretion of chlamydial T3SS effectors (7, 195, 197–207). Recent studies in the Rucks lab have demonstrate that additional epitope tags can also be successfully secreted by C. trachomatis L2/434/Bu, such as 6×His, HA, StrepTagII, and Spot tags (L. Knight and E. A. Rucks, unpublished observation). The use of these epitope tags will remain an important tool in studying chlamydial effector secretion and function as antibodies against these proteins are rare and finite resources.
CHLAMYDIA’S GUIDE TO OCCUPYING A HOST CELL: A T3SS EFFECTOR FOR EVERY OCCASION
Overview of Chlamydial Effector Proteins
As thoroughly discussed, the components of the T3SS apparatus are highly conserved between different species of Gram-negative bacteria (13). Effector proteins are highly specific to the pathogen or symbiont in question, as they are related to their specific survival needs. The number and types of effectors also vary greatly among and within bacterial species. It is widely accepted that within a bacterial species that differ in effector proteins between individual serovars or isolates there are “core effectors” or effectors that are common to all bacteria within a given species (208). In examining the total number of effectors for some of the pathogens used in surrogate models for chlamydial T3SS, the average number of effectors is 16. Specifically, the S. enterica SPI-1 T3SS encodes 2 effectors (209) and the SPI-2 T3SS translocates an estimated 28 effectors (208). S. flexneri is estimated to have about 19 effectors, while 11 effectors have been identified in Yersiniae (210). Based on the total protein-coding genes found in these bacterial species, these numbers represent 0.67% (S. enterica [211]), 0.4% (S. flexneri [212]), and 0.2% (Yersinae [213]) of the genome encoding T3SS effectors. In contrast to these pathogens, C. trachomatis is estimated to produce about 100 T3SS effector proteins (6); therefore, Chlamydia devotes about 11% of its limited coding capacity to producing effector proteins (2). While this large percentage might reflect some redundancy, it does highlight the necessity of a veritable menagerie of effectors required to coax a host cell into being an effective growth incubator. A fundamental distinction between the pathogens used in surrogate models of T3SS and Chlamydia is the fact that it is helpful to the other pathogens to subvert normal host cell function, but for Chlamydia, this is a necessity. There have been two very recent and comprehensive reviews written about C. trachomatis T3SS effectors (6, 7). Furthermore, Andersen et al. (6) provide a comprehensive up-to-date table outlining all known chlamydial T3SS effectors. This section will be used to highlight broader function of these effectors and the recent data regarding effector function.
Function of Effectors in Chlamydial Entry
Chlamydial entry occurs in two distinct sequential steps (214, 215). The first step is reversible and is guided by electrostatic interactions and heparan sulfate glycosaminoglycans (216–218). Within the female reproductive tract, estrogen is thought to enhance attachment (219). Additionally, coatamer complex I (COPI) enhances the presentation of heparan sulfate on the cell surface and is required for chlamydial entry into host cells (220). The second step is irreversible and thought to involve a receptor (221), with one possible receptor being the estrogen receptor (222). It is during this second step that chlamydial T3SS is deployed and results in the translocation of TarP, TmeA (CT694), and TmeB (CT695) (185, 196, 223). Small interfering RNA (siRNA) knockdown of COPI decreases the amount of TarP that is T3SS secreted/translocated into the host cell, supporting that a tight association between Chlamydia and the host cells is required for T3SS (220). The function of TarP locally remodels the actin cytoskeleton to promote chlamydial entry (224, 225). Interestingly, TarP is considered a core effector, with the basic gene being conserved across chlamydial serovars and species; but the structures of TarP (e.g., number of tyrosine-rich repeat regions or actin binding domains) are specific to the species or serovar of Chlamydia (226). TmeA has been demonstrated to activate N-WASP to promote actin polymerization and likely functions synergistically with TarP during chlamydial entry (227). Of interest, TmeA has a membrane localization domain (MLD), which is similar to the MLDs found in Yersinia effector YopE and Pseudomonas effector ExoS (228). The role of TmeB in chlamydial entry is less clear, but the deletion of this gene and the gene downstream resulted in a 2-log decrease in progeny (229).
The Largest Family of Chlamydial T3SS Effectors: Incs
Within the first few hours of infection, the EB differentiates into the RB, and protein synthesis commences. During this same time period, Chlamydia bacteria begin to remodel their inclusion membrane with T3SS chlamydial proteins (110, 230), which prevents the inclusion from progressing through the endosomal-lysosomal pathway (230). The chlamydial proteins that are commonly associated with the chlamydial inclusion are the Incs. These proteins were some of the first bacterial proteins localized to the chlamydial inclusion and have a unique topology, as follows: a bilobed hydrophobic transmembrane domain (188), with the N termini and C termini being exposed to the host cell cytosol (231). There are about 50 estimated candidate Inc proteins in C. trachomatis, and all chlamydial species have a various number of genes that encode Inc proteins, with 23 Incs being conserved across chlamydial species, as in a subset of “core effectors” (232). In 2001, the use of a S. flexneri surrogate T3SS model demonstrated that Inc proteins are T3SS secreted (233). Because members of the Inc family are easily identified due to key secondary structure features, they are the best studied ones of the known chlamydial T3SS effectors.
Temporal expression.
Like all chlamydial genes, the expression of Incs is temporal. Some incs are clustered within operons (e.g., incD-incG) or gene clusters (e.g., ct229-ct223), while others are scattered throughout the chromosome (e.g., incA and ct147) (2). The genes encoded on the incD operon, which include incD, incE, incF, and incG, are some of the first Incs to be synthesized after chlamydial entry (110). It is widely accepted that these early Incs help to establish the chlamydial inclusion; while midcycle Incs, such as CT813/InaC, help to maintain inclusion integrity and chlamydial acquisition of nutrients (207); and Incs found on the inclusion later in infection, such as CT228, are involved in chlamydial egress (234, 235).
Incs directing host-chlamydial interactions.
Given the positioning of the Incs, with their N termini and C termini being exposed to the host cell cytosol, and the fact that some of the C-terminal regions of Incs are quite large (greater than 60 amino acids), these proteins are also the focus of many studies examining host-chlamydial interactions. There have been several affinity purification-based proteomic studies that have increased our understanding of the web of host-chlamydial interactions that surround the chlamydial inclusion. One such study mapped possible interactions between exogenously expressed strep-tagged Incs in uninfected HeLa cells, which gave an impressive interaction map between Incs and possible eukaryotic protein targets (236). Other groups have used proximity labeling to understand protein networks that are proximal to the inclusion (197, 237, 238). Both of these studies constructed genetic fusions between Inc proteins and APEX2, an ascorbate peroxidase that creates a biotin-phenoxy radical in the presence of H2O2, which then binds to target amino acids of proteins that are within a 20- to 40-nm spatial radius of the APEX2-tagged protein of interest (239). In both studies, inc-APEX2 constructs were cloned into an anhydrotetracylcine (aTc)-inducible chlamydial expression plasmid (180). And upon the addition of aTc, these fusion proteins are expressed and secreted from Chlamydia and localize to the inclusion membrane (197, 237, 238). To understand if Inc-APEX2 proteins were oriented properly in the inclusion membrane, HeLa cells were infected with the Inc-APEX2-expressing strains, treated with diaminobenzidine (DAB) and horseradish peroxidase (HRP) prior to H2O2 treatment, and then processed for imaging by transmission electron microscopy (TEM). The TEM images revealed electron dense deposits on the cytosolic side of the inclusion, which indicated that Inc-APEX2 proteins were labeling the cytosolic side of the inclusion and that they were correctly oriented in the inclusion membrane (197).
To gain an understanding of possible protein networks, one of these APEX2 studies used several different Inc-APEX2 proteins and discovered common core proteins (e.g., LRRF1 and BASP1) and several overlapping protein networks (e.g., 14-3-3) that are proximal to the chlamydial inclusion (119, 197). These data highlight the fact that protein-protein interactions that occur at the chlamydial inclusion likely have ripple effects into these various signaling pathways that help rewire the host cell generally. The first Inc-host protein interaction described was between an early Inc, IncG, which becomes phosphorylated in the host cytosol and interacts with 14-3-3β (240). One of the more interesting interactions has been noted between another early Inc, IncD, and an ER protein known as CERT (104, 241), which also occurs in membrane contact sites between the ER and the inclusion membrane (104). These studies highlight how the inclusion likely integrates itself into host cell pathways and may modulate or engage with cross-talk between organelles to help maintain host cell health to maximize chlamydial growth and development. This idea is buttressed by soluble (or non-Inc) chlamydial effectors that localize to either the Golgi (242), mitochondria (243), or nucleus (244) to specifically alter the function of these organelles. Furthermore, Inc-host protein interactions help to integrate the chlamydial inclusion into the overall function of the host cell, which contributes toward maintaining host cell health until the point of egress (245).
Despite advances toward understanding the networks of protein interactions, the majority Inc-host protein interactions have been studied between a single Inc and a single eukaryotic protein target, as highlighted by two such examples above. However, a recent study demonstrated for the first time a single eukaryotic protein, VAMP3, interacting with up to five different Inc proteins in a temporal manner (199). VAMP3 is a eukaryotic soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein that helps to fuse two opposing membranes and functions within trans-Golgi, recycling endosome, plasma membrane trafficking pathways (246, 247). VAMP3 localizes to the chlamydial inclusion during the mid-developmental cycle and remains localized to the inclusion throughout the remainder of chlamydial development (199). The first Inc interaction detected was between VAMP3 and IncF, which becomes less robust, at which point VAMP3 interacts strongly with IncG. The longest sustained interaction between VAMP3 and any Inc is with Inc CT449. This interaction occurs during a period when VAMP3-IncF or -IncG interactions are no longer detected and before the time point that VAMP3 binds to yet another Inc partner, CT813/InaC. VAMP3 also interacts with a late Inc, CT442, during the late chlamydial developmental cycle (199). These studies were the first to demonstrate the dynamic nature of some protein-protein interactions that occur at the chlamydial inclusion. The function or consequences of these interactions are currently unclear, but if the inclusion is integrating into the host cell, these types of interactions are to be expected, as they are common in subcellular organelles.
Understanding how Incs may be organized in the inclusion membrane.
One of the original goals of the proximity labeling studies was to better understand how Incs are organized in the inclusion membrane, which is difficult to purify (116, 248). Indirect immunofluorescent studies have found that some Incs localize in small discrete microdomains (i.e., CT223/IpaM, IncB, and CT228) (188, 206, 249), while other Incs localize uniformly around the periphery of the inclusion membrane (i.e., IncA, IncG, and IncF) (110, 231, 250). In the first published proximity labeling study, Dickinson et al. (237) created a chlamydial strain that expressed an IncB-APEX2 construct, which localized around the periphery of the inclusion, and identified only four Inc proteins. In a separate study using three different Inc-APEX2 (IncA and IncF-based) constructs, four Inc proteins were also identified (197). The Inc that was commonly identified between the two studies was CT223/IpaM (197, 237), which may indicate that even though this Inc localizes primarily in membrane microdomains (188, 249), it may also be an abundant Inc in the inclusion membrane or its identification is related to the fact that it contains 14 APEX2-modifiable amino acids (which are cysteine, tyrosine, tryptophan, or histidine) (119, 239). There are several limiting factors in using APEX2 to determine Inc organization in the inclusion membrane. In addition to the relatively low number of APEX2 modifiable amino acid targets in Inc proteins, there is also the general problem of the low abundance of chlamydial proteins versus host proteins, which are the bulk of the protein identifications received in these types of studies (119).
(i) How genetic dysregulation of Inc expression alters inclusion membrane composition. Incs likely have multiple functions, of which one ostensibly is to mediate host-pathogen interactions and another function is likely to organize the inclusion membrane. For example, overexpression of IncF results in smaller inclusions and excess IncF tends to form aggregates that are then excluded from the inclusion membrane (205). In contrast, overexpression of IncA was not associated with these same defects (205). Deletion of IncA, which is responsible for homotypic inclusion fusion (251), also results in CT223/IpaM no longer being organized in membrane microdomains (199), an indication that other Inc proteins are also likely organized differently in the absence of IncA. Combined, these studies raised questions about how Inc proteins function in inclusion membrane organization and also if altered expression of certain Incs impacted the expression of other Incs.
A recent study examined the overexpression of several different Inc proteins. The overexpression of certain Incs (e.g., IncF or CT813/InaC) resulted in smaller inclusions, decreased progeny, and decreased expression of some endogenous Incs at the transcriptional level (204). Specifically, overexpression of ct813 resulted in reduced IncE and IncG proteins on the inclusion membrane, which were then linked to decreased expression of incE and incG. Of note, the IncA protein and incA transcript levels remained unchanged (204). Furthermore, previous studies found that IncE recruits sorting nexin-6 to the chlamydial inclusion (236). Overexpression of ct813, which results in more CT813/InaC and less IncE in the chlamydial inclusion membrane, reduced sorting nexin-6 recruitment to the chlamydial inclusion (204). These data highlight that while we often study chlamydial effectors as single entities, there is likely an interconnectedness at the level of effector expression and secretion. There are several possible explanations for these findings. For example, a disorganized inclusion membrane might lead to altered host cell interactions, which negatively impacts chlamydial development and possibly gene expression generally. As mentioned previously, inc expression is temporal. Endogenous ct813 is expressed after incE (5, 252). Therefore, an overproduction of CT813/InaC might trigger a premature reduction in the expression of genes encoding early Incs, such as incE. The chaperones for specific Inc proteins is unknown, but Mcsc is a candidate (74). Therefore, too much of an Inc can saturate its cognate chaperone, potentially altering heterodimer chaperone or chaperone-effector interactions, of which some have implications (e.g., Scc4) toward regulating gene expression. However, these data also suggest that too much of a single effector can globally disrupt T3SS function, so these phenotypes may also be indicative of systemic imbalances (e.g., lack of secretion of other T3SS effectors) versus a phenomenon that is strictly limited to a disorganized inclusion membrane. The menagerie of possibilities also highlights the complexity of studying T3SS in Chlamydia.
CONCLUDING REMARKS
There are many interesting questions regarding chlamydial T3SS that remain to be answered. While many of us are focused on the chlamydial effectors that localize to the inclusion membrane, the field has only begun to study chlamydial T3SS effectors that are secreted into the cytosol or localize to specific subcellular compartments or organelles. These proteins likely have important functions that promote a collaborative co-option of host cell function that enhances chlamydial growth and development because the chlamydial T3SS exists primarily to support the progression of the developmental cycle. Directly linking T3SS to developmental cycle progression was an early hypothesis in the field that posited that the T3SS projections from the RB mediated intimate contact with the inclusion membrane. As the number of projections decreased, the RB would be released from the inclusion. CopN would no longer be secreted into the host cytosol but into the inclusion lumen, an event that would trigger secondary differentiation (253). This hypothesis may be applicable to C. trachomatis and C. psittaci as RBs often line the edges of the inclusion and abut the inclusion membrane (254). But it is inconsistent with the manner in which C. pneumoniae develop, with RBs localized slightly away from the inclusion membrane within the lumen of the inclusion (255). Experiments designed to understand whether a T3SS-associated signal is the elusive molecular trigger that results in secondary differentiation are inherently complicated as the state of the host and the organism must be considered. For example, overexpression of a mid-cycle Inc, CT813/InaC, results in downregulation of other Incs at the transcriptional level and altered chlamydial development (204). Are these data a reflection of altered chlamydial-host interactions and/or a reflection of haywire signaling within the T3SS, including effector expression? With the exciting advances in chlamydial genetics, it may become possible to better understand the function of the proteins that are assumed to make up the chlamydial T3SS apparatus and the effector proteins. Do these proteins function as their orthologs do? Or do they have some unique functions to account for the reduced genome of Chlamydia? There are several candidate chlamydial T3SSs that may have unique transcription factor (Scc4) or environmental sensor (CopB2) functions that need to be experimentally elucidated. A better understanding of the regulatory mechanisms that control chlamydial T3SS at the transcriptional and posttranslational level, including the factors that govern effector hierarchy, are necessary to devise strategies to inhibit these processes. These scientific questions are interesting from the standpoint of understanding basic host-pathogen interactions and their consequences. But, also, answering these questions may lead to developing therapeutics that will efficiently treat Chlamydia while not disturbing the healthy microbial flora.
ACKNOWLEDGMENTS
I express much gratitude to Joan C. Olson for mentorship over the years and for the critical review and comments in the earlier drafts of the manuscript. A special thanks to Scot Ouellette for constructive and insightful conversations. Lindsey Knight is appreciated for assisting with the generation of unpublished preliminary data involving chlamydial chaperones and epitope tagged effectors. Patrick Lane from ScEYEnce Studios is acknowledged for his excellent help with the figures. Thanks also to Nathan Hatch and Jeonghoon Lee for critical review of the manuscript and for the thoughtful conversations and support of other members in the Rucks/Ouellette lab group.
This work is supported by 1R01 AI132406 to E.A.R.
REFERENCES
- 1.Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 3.AbdelRahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 4.AbdelRahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Gemomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen SE, Bulman LM, Steiert B, Faris R, Weber MM. 2021. Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins. Pathog Dis 79:ftaa078. doi: 10.1093/femspd/ftaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugalhão JN, Mota LJ. 2019. The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microb Cell 6:414–449. doi: 10.15698/mic2019.09.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 9.Dharamshi JE, Köstlbacher S, Schön ME, Collingro A, Ettema TJG, Horn M. 2023. Gene gain facilitated endosymbiotic evolution of Chlamydiae. Nat Microbiol 8:40–54. doi: 10.1038/s41564-022-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson JO, Andersson SG. 1999. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev 9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 11.Binet R, Fernandez RE, Fisher DJ, Maurelli AT. 2011. Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. mBio 2:e00051-11. doi: 10.1128/mBio.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT. 2006. l,l-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci USA 103:17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433. doi: 10.1128/MMBR.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsia RC, Pannekoek Y, Ingerowski E, Bavoil PM. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol 25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 15.Subtil A, Blocker A, Dautry-Varsat A. 2000. Type III secretion system in Chlamydia species: identified members and candidates. Microbes Infect 2:367–369. doi: 10.1016/s1286-4579(00)00335-x. [DOI] [PubMed] [Google Scholar]
- 16.Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. 2007. Type III secretion à la Chlamydia. Trends Microbiol 15:241–251. doi: 10.1016/j.tim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Abby SS, Rocha EP. 2012. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet 8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto A. 1981. Isolation and electron microscopic observations of intracytoplasmic inclusions containing Chlamydia psittaci. J Bacteriol 145:605–612. doi: 10.1128/jb.145.1.605-612.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto A. 1982. Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J Bacteriol 150:358–364. doi: 10.1128/jb.150.1.358-364.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto A, Fujiwara E, Higashi N. 1976. Observations of the surface projections of infectious small cell of Chlamydia psittaci in thin sections. J Electron Microsc 5:169–170. [PubMed] [Google Scholar]
- 21.Stokes GV. 1978. Surface projections and internal structure of Chlamydia psittaci. J Bacteriol 133:1514–1516. doi: 10.1128/jb.133.3.1514-1516.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens RS. 1992. Challenge of Chlamydia research. Infect Agents Dis 1:279–293. [PubMed] [Google Scholar]
- 23.Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G. 1990. Secretion of Yop proteins by Yersiniae. Infect Immun 58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michiels T, Vanooteghem JC, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis GR. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol 173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmond GP, Reeves PJ. 1993. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci 18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 26.Price SB, Straley SC. 1989. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun 57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosqvist R, Magnusson KE, Wolf-Watz H. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J 13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella Typhimurium type III protein secretion system. Science 280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 29.Dale C, Plague GR, Wang B, Ochman H, Moran NA. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA 99:12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale C, Young SA, Haydon DT, Welburn SC. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci USA 98:1883–1888. doi: 10.1073/pnas.98.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding JY, Shiu JH, Chen WM, Chiang YR, Tang SL. 2016. Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33(T) with its coral host. Front Microbiol 7:251. doi: 10.3389/fmicb.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadal-Jimenez P, Siozios S, Halliday N, Cámara M, Hurst GDD. 2022. Symbiopectobacterium purcellii, gen. nov., sp. nov., isolated from the leafhopper Empoasca decipiens. Int J Syst Evol Microbiol 72. doi: 10.1099/ijsem.0.005440. [DOI] [PubMed] [Google Scholar]
- 33.Zboralski A, Biessy A, Filion M. 2022. Bridging the gap: type III secretion systems in plant-beneficial bacteria. Microorganisms 10:187. doi: 10.3390/microorganisms10010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao X, Beatty WL, Fan H. 2012. Exploration of chlamydial type III secretion system reconstitution in Escherichia coli. PLoS One 7:e50833. doi: 10.1371/journal.pone.0050833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O'Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong S, Lei L, Chang X, Belland R, Zhong G. 2011. Chlamydia trachomatis secretion of hypothetical protein CT622 into host cell cytoplasm via a secretion pathway that can be inhibited by the type III secretion system inhibitor compound 1. Microbiology (Reading) 157:1134–1144. doi: 10.1099/mic.0.047746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 103:14566–14571. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf K, Betts HJ, Chellas-Géry B, Hower S, Linton CN, Fields KA. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol Microbiol 61:1543–1555. doi: 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zetterström CE, Hasselgren J, Salin O, Davis RA, Quinn RJ, Sundin C, Elofsson M. 2013. The resveratrol tetramer (-)-hopeaphenol inhibits type III secretion in the gram-negative pathogens Yersinia pseudotuberculosis and Pseudomonas aeruginosa. PLoS One 8:e81969. doi: 10.1371/journal.pone.0081969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engström P, Nguyen BD, Normark J, Nilsson I, Bastidas RJ, Gylfe A, Elofsson M, Fields KA, Valdivia RH, Wolf-Watz H, Bergström S. 2013. Mutations in hemG mediate resistance to salicylidene acylhydrazides, demonstrating a novel link between protoporphyrinogen oxidase (HemG) and Chlamydia trachomatis infectivity. J Bacteriol 195:4221–4230. doi: 10.1128/JB.00506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slepenkin A, Enquist PA, Hägglund U, de la Maza LM, Elofsson M, Peterson EM. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect Immun 75:3478–3489. doi: 10.1128/IAI.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ur-Rehman T, Slepenkin A, Chu H, Blomgren A, Dahlgren MK, Zetterström CE, Peterson EM, Elofsson M, Gylfe Å. 2012. Pre-clinical pharmacokinetics and anti-chlamydial activity of salicylidene acylhydrazide inhibitors of bacterial type III secretion. J Antibiot (Tokyo) 65:397–404. doi: 10.1038/ja.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zigangirova NA, Kost EA, Didenko LV, Kapotina LN, Zayakin ES, Luyksaar SI, Morgunova EY, Fedina ED, Artyukhova OA, Samorodov AV, Kobets NV. 2016. A small-molecule compound belonging to a class of 2,4-disubstituted 1,3,4-thiadiazine-5-ones inhibits intracellular growth and persistence of Chlamydia trachomatis. J Med Microbiol 65:91–98. doi: 10.1099/jmm.0.000189. [DOI] [PubMed] [Google Scholar]
- 45.Bailey L, Gylfe A, Sundin C, Muschiol S, Elofsson M, Nordström P, Henriques-Normark B, Lugert R, Waldenström A, Wolf-Watz H, Bergström S. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett 581:587–595. doi: 10.1016/j.febslet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Dahlgren MK, Zetterström CE, Gylfe S, Linusson A, Elofsson M. 2010. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg Med Chem 18:2686–2703. doi: 10.1016/j.bmc.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 47.Enquist PA, Gylfe A, Hägglund U, Lindström P, Norberg-Scherman H, Sundin C, Elofsson M. 2012. Derivatives of 8-hydroxyquinoline–antibacterial agents that target intra- and extracellular Gram-negative pathogens. Bioorg Med Chem Lett 22:3550–3553. doi: 10.1016/j.bmcl.2012.03.096. [DOI] [PubMed] [Google Scholar]
- 48.Fedina ED, Kolkova NI, Koroleva EA, Shabalina LA, Grabko VI, Zigangirova NA. 2012. Influence of Chlamydia trachomatis type III secretion system on regulation of cytokine response. Zh Mikrobiol Epidemiol Immunobiol 26–32. [PubMed] [Google Scholar]
- 49.Grishin AV, Luyksaar SI, Kapotina LN, Kirsanov DD, Zayakin ES, Karyagina AS, Zigangirova NA. 2018. Identification of chlamydial T3SS inhibitors through virtual screening against T3SS ATPase. Chem Biol Drug Des 91:717–727. doi: 10.1111/cbdd.13130. [DOI] [PubMed] [Google Scholar]
- 50.Lam HN, Lau T, Lentz A, Sherry J, Cabrera-Cortez A, Hug K, Lalljie A, Engel J, Lokey RS, Auerbuch V. 2021. Developing cyclic peptomers as broad-spectrum type III secretion system inhibitors in Gram-negative bacteria. Antimicrob Agents Chemother 65:e0169020. doi: 10.1128/AAC.01690-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muschiol S, Normark S, Henriques-Normark B, Subtil A. 2009. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol 9:75. doi: 10.1186/1471-2180-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sunduru N, Salin O, Gylfe Å, Elofsson M. 2015. Design, synthesis and evaluation of novel polypharmacological antichlamydial agents. Eur J Med Chem 101:595–603. doi: 10.1016/j.ejmech.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 53.Ouellette SP. 2018. Feasibility of a conditional knockout system for chlamydia based on CRISPR interference. Front Cell Infect Microbiol 8:59. doi: 10.3389/fcimb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouellette SP, Blay EA, Hatch ND, Fisher-Marvin LA. 2021. CRISPR interference to inducibly repress gene expression in Chlamydia trachomatis. Infect Immun 89:e0010821. doi: 10.1128/IAI.00108-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood NA, Blocker AM, Seleem MA, Conda-Sheridan M, Fisher DJ, Ouellette SP. 2020. The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. mBio 11. doi: 10.1128/mBio.02016-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood NA, Swoboda AR, Blocker AM, Fisher DJ, Ouellette SP. 2022. Tag-dependent substrate selection of ClpX underlies secondary differentiation of Chlamydia trachomatis. mBio 13:e0185822. doi: 10.1128/mbio.01858-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swoboda AR, Wood NA, Saery EA, Fisher DJ, Ouellette SP. 2023. The periplasmic tail-specific protease, Tsp, is essential for secondary differentiation in Chlamydia trachomatis. J Bacteriol 205:e00099-23. doi: 10.1128/jb.00099-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan S, Jensen AA, Wood NA, Henrichfreise B, Brötz-Oesterhelt H, Fisher DJ, Sass P, Ouellette SP. 2023. Molecular characterization of the ClpC AAA+ ATPase in the biology of Chlamydia trachomatis. mBio 14:e00075-23. doi: 10.1128/mbio.00075-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouellette SP, Fisher-Marvin LA, Harpring M, Lee J, Rucks EA, Cox JV. 2022. Localized cardiolipin synthesis is required for the assembly of MreB during the polarized cell division of Chlamydia trachomatis. PLoS Pathog 18:e1010836. doi: 10.1371/journal.ppat.1010836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diepold A, Wagner S. 2014. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev 38:802–822. doi: 10.1111/1574-6976.12061. [DOI] [PubMed] [Google Scholar]
- 61.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB. 2010. Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae. BMC Microbiol 10:18. doi: 10.1186/1471-2180-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barta ML, Battaile KP, Lovell S, Hefty PS. 2015. Hypothetical protein CT398 (CdsZ) interacts with σ(54) (RpoN)-holoenzyme and the type III secretion export apparatus in Chlamydia trachomatis. Protein Sci 24:1617–1632. doi: 10.1002/pro.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan KA, Karim N, Worku M, Moore SA, Penn CW, O'Toole PW. 2005. HP0958 is an essential motility gene in Helicobacter pylori. FEMS Microbiol Lett 248:47–55. doi: 10.1016/j.femsle.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 65.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 66.Minamino T, Tame JR, Namba K, Macnab RM. 2001. Proteolytic analysis of the FliH/FliI complex, the ATPase component of the type III flagellar export apparatus of Salmonella. J Mol Biol 312:1027–1036. doi: 10.1006/jmbi.2001.5000. [DOI] [PubMed] [Google Scholar]
- 67.Markham AP, Jaafar ZA, Kemege KE, Middaugh CR, Hefty PS. 2009. Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry 48:10353–10361. doi: 10.1021/bi901200y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. 2008. What’s the point of the type III secretion system needle? Proc Natl Acad Sci USA 105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stone CB, Johnson DL, Bulir DC, Gilchrist JD, Mahony JB. 2008. Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J Bacteriol 190:6580–6588. doi: 10.1128/JB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stone CB, Bulir DC, Emdin CA, Pirie RM, Porfilio EA, Slootstra JW, Mahony JB. 2011. Chlamydia pneumoniae CdsL regulates CdsN ATPase activity, and disruption with a peptide mimetic prevents bacterial invasion. Front Microbiol 2:21. doi: 10.3389/fmicb.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vandahl BB, Birkelund S, Demol H, Hoorelbeke B, Christiansen G, Vandekerckhove J, Gevaert K. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22:1204–1223. doi:. [DOI] [PubMed] [Google Scholar]
- 72.Herrmann M, Schuhmacher A, Mühldorfer I, Melchers K, Prothmann C, Dammeier S. 2006. Identification and characterization of secreted effector proteins of Chlamydophila pneumoniae TW183. Res Microbiol 157:513–524. doi: 10.1016/j.resmic.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Lorenzini E, Singer A, Singh B, Lam R, Skarina T, Chirgadze NY, Savchenko A, Gupta RS. 2010. Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO homolog). J Bacteriol 192:2746–2756. doi: 10.1128/JB.01479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spaeth KE, Chen YS, Valdivia RH. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson DL, Mahony JB. 2007. Chlamydophila pneumoniae PknD exhibits dual amino acid specificity and phosphorylates Cpn0712, a putative type III secretion YscD homolog. J Bacteriol 189:7549–7555. doi: 10.1128/JB.00893-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson DL, Stone CB, Bulir DC, Coombes BK, Mahony JB. 2009. A novel inhibitor of Chlamydophila pneumoniae protein kinase D (PknD) inhibits phosphorylation of CdsD and suppresses bacterial replication. BMC Microbiol 9:218. doi: 10.1186/1471-2180-9-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meriläinen G, Wierenga RK. 2014. Crystallization and preliminary X-ray diffraction studies of the C-terminal domain of Chlamydia trachomatis CdsD. Acta Crystallogr F Struct Biol Commun 70:1431–1433. doi: 10.1107/S2053230X14019712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Betts-Hampikian HJ, Fields KA. 2011. Disulfide bonding within components of the Chlamydia type III secretion apparatus correlates with development. J Bacteriol 193:6950–6959. doi: 10.1128/JB.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meriläinen G, Koski MK, Wierenga RK. 2016. The extended structure of the periplasmic region of CdsD, a structural protein of the type III secretion system of Chlamydia trachomatis. Protein Sci 25:987–998. doi: 10.1002/pro.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson DL, Stone CB, Mahony JB. 2008. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol 190:2972–2980. doi: 10.1128/JB.01997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bavoil P, Ohlin A, Schachter J. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun 44:479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Everett KD, Hatch TP. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol 177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Betts HJ, Twiggs LE, Sal MS, Wyrick PB, Fields KA. 2008. Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J Bacteriol 190:1680–1690. doi: 10.1128/JB.01671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fields KA, Mead DJ, Dooley CA, Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol 48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- 85.Nans A, Saibil HR, Hayward RD. 2014. Pathogen-host reorganization during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol 16:1457–1472. doi: 10.1111/cmi.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nans A, Kudryashev M, Saibil HR, Hayward RD. 2015. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat Commun 6:10114. doi: 10.1038/ncomms10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dumoux M, Clare DK, Saibil HR, Hayward RD. 2012. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic 13:1612–1627. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang JJ, Leonard KR, Zhang YX. 1997. Structural studies of the surface projections of Chlamydia trachomatis by electron microscopy. J Med Microbiol 46:1013–1018. doi: 10.1099/00222615-46-12-1013. [DOI] [PubMed] [Google Scholar]
- 89.Wilharm G, Dittmann S, Schmid A, Heesemann J. 2007. On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int J Med Microbiol 297:27–36. doi: 10.1016/j.ijmm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 90.Ueno T, Oosawa K, Aizawa S. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella Typhimurium are composed of subunits of a single protein, FliF. J Mol Biol 227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 91.Payne PL, Straley SC. 1999. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J Bacteriol 181:2852–2862. doi: 10.1128/JB.181.9.2852-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Riordan KE, Sorg JA, Berube BJ, Schneewind O. 2008. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J Bacteriol 190:6204–6216. doi: 10.1128/JB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner S, Sorg I, Degiacomi M, Journet L, Dal Peraro M, Cornelis GR. 2009. The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Mol Microbiol 71:692–701. doi: 10.1111/j.1365-2958.2008.06556.x. [DOI] [PubMed] [Google Scholar]
- 94.Barta ML, Hickey J, Kemege KE, Lovell S, Battaile KP, Hefty PS. 2013. Structure of CT584 from Chlamydia trachomatis refined to 3.05 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:1196–1201. doi: 10.1107/S1744309113027371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stone CB, Sugiman-Marangos S, Bulir DC, Clayden RC, Leighton TL, Slootstra JW, Junop MS, Mahony JB. 2012. Structural characterization of a novel Chlamydia pneumoniae type III secretion-associated protein, Cpn0803. PLoS One 7:e30220. doi: 10.1371/journal.pone.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pais SV, Milho C, Almeida F, Mota LJ. 2013. Identification of novel type III secretion chaperone-substrate complexes of Chlamydia trachomatis. PLoS One 8:e56292. doi: 10.1371/journal.pone.0056292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosqvist R, Persson C, Håkansson S, Nordfeldt R, Wolf-Watz H. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib Microbiol Immunol 13:230–234. [PubMed] [Google Scholar]
- 98.Bulir DC, Waltho DA, Stone CB, Liang S, Chiang CK, Mwawasi KA, Nelson JC, Zhang SW, Mihalco SP, Scinocca ZC, Mahony JB. 2015. Chlamydia Outer Protein (Cop) B from Chlamydia pneumoniae possesses characteristic features of a type III secretion (T3S) translocator protein. BMC Microbiol 15:163. doi: 10.1186/s12866-015-0498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bulir DC, Waltho DA, Stone CB, Mwawasi KA, Nelson JC, Mahony JB. 2014. Chlamydia pneumoniae CopD translocator protein plays a critical role in type III secretion (T3S) and infection. PLoS One 9:e99315. doi: 10.1371/journal.pone.0099315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frithz-Lindsten E, Holmström A, Jacobsson L, Soltani M, Olsson J, Rosqvist R, Forsberg A. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol Microbiol 29:1155–1165. doi: 10.1046/j.1365-2958.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- 101.Håkansson S, Schesser K, Persson C, Galyov EE, Rosqvist R, Homblé F, Wolf-Watz H. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J 15:5812–5823. doi: 10.1002/j.1460-2075.1996.tb00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67:914–920. doi: 10.1128/IAI.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bavoil PM, Hsia RC. 1998. Type III secretion in Chlamydia: a case of déjà vu? Mol Microbiol 28:860–862. doi: 10.1046/j.1365-2958.1998.00861.x. [DOI] [PubMed] [Google Scholar]
- 104.Derre I, Swiss R, Agaisse H. 2011. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog 7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phillips P, Parkhurst JM, Kounatidis I, Okolo C, Fish TM, Naismith JH, Walsh MA, Harkiolaki M, Dumoux M. 2021. Single cell cryo-soft X-ray tomography shows that each Chlamydia trachomatis inclusion is a unique community of bacteria. Life 11:842. doi: 10.3390/life11080842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kubori T, Sukhan A, Aizawa SI, Galán JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella Typhimurium type III protein secretion system. Proc Natl Acad Sci USA 97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mills DM, Bajaj V, Lee CA. 1995. A 40 kb chromosomal fragment encoding Salmonella Typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol 15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 108.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 109.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella Typhimurium. Proc Natl Acad Sci USA 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. 1999. Identification and characterization of Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol 33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 111.Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol 185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 113.Slepenkin A, Motin V, de la Maza LM, Peterson EM. 2003. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect Immun 71:2555–2562. doi: 10.1128/IAI.71.5.2555-2562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scidmore MA, Fischer ER, Hackstadt T. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol 134:363–374. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saka HA, Thompson JW, Chen Y-S, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol 82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect 4:333–340. doi: 10.1016/s1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- 118.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Olson MG, Ouellette SP, Rucks EA. 2020. A meta-analysis of affinity purification-mass spectrometry experimental systems used to identify eukaryotic and chlamydial proteins at the Chlamydia trachomatis inclusion membrane. J Proteomics 212:103595. doi: 10.1016/j.jprot.2019.103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ferrell JC, Fields KA. 2016. A working model for the type III secretion mechanism in Chlamydia. Microbes Infect 18:84–92. doi: 10.1016/j.micinf.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sorg JA, Blaylock B, Schneewind O. 2006. Secretion signal recognition by YscN, the Yersinia type III secretion ATPase. Proc Natl Acad Sci USA 103:16490–16495. doi: 10.1073/pnas.0605974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allison SE, Tuinema BR, Everson ES, Sugiman-Marangos S, Zhang K, Junop MS, Coombes BK. 2014. Identification of the docking site between a type III secretion system ATPase and a chaperone for effector cargo. J Biol Chem 289:23734–23744. doi: 10.1074/jbc.M114.578476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chellas-Géry B, Wolf K, Tisoncik J, Hackstadt T, Fields KA. 2011. Biochemical and localization analyses of putative type III secretion translocator proteins CopB and CopB2 of Chlamydia trachomatis reveal significant distinctions. Infect Immun 79:3036–3045. doi: 10.1128/IAI.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fields KA, Fischer ER, Mead DJ, Hackstadt T. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J Bacteriol 187:6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem 280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 126.Torruellas J, Jackson MW, Pennock JW, Plano GV. 2005. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol 57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 127.Straley SC, Plano GV, Skrzypek E, Haddix PL, Fields KA. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol 8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 128.Yahr TL, Mende-Mueller LM, Friese MB, Frank DW. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol 179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Y, Watkins N, Stewart S, Caldwell H. 1987. The low-molecular-mass, cysteine-rich outer membrane protein of Chlamydia trachomatis possesses both biovar- and species-specific epitopes. Infect Immun 55:2570–2573. doi: 10.1128/iai.55.11.2570-2573.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Everett KD, Hatch TP. 1991. Sequence analysis and lipid modification of the cysteine-rich envelope proteins of Chlamydia psittaci 6BC. J Bacteriol 173:3821–3830. doi: 10.1128/jb.173.12.3821-3830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hatch TP, Miceli M, Sublett JE. 1986. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol 165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Plano GV, Barve SS, Straley SC. 1991. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol 173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cherradi Y, Schiavolin L, Moussa S, Meghraoui A, Meksem A, Biskri L, Azarkan M, Allaoui A, Botteaux A. 2013. Interplay between predicted inner-rod and gatekeeper in controlling substrate specificity of the type III secretion system. Mol Microbiol 87:1183–1199. doi: 10.1111/mmi.12158. [DOI] [PubMed] [Google Scholar]
- 135.Forsberg A, Viitanen AM, Skurnik M, Wolf-Watz H. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol 5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 136.Viitanen AM, Toivanen P, Skurnik M. 1990. The lcrE gene is part of an operon in the lcr region of Yersinia enterocolitica O:3. J Bacteriol 172:3152–3162. doi: 10.1128/jb.172.6.3152-3162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rosqvist R, Forsberg A, Rimpiläinen M, Bergman T, Wolf-Watz H. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol 4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 138.Fields KA, Hackstadt T. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol 38:1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- 139.Ho TD, Starnbach MN. 2005. The Salmonella enterica serovar Typhimurium-encoded type III secretion systems can translocate Chlamydia trachomatis proteins into the cytosol of host cells. Infect Immun 73:905–911. doi: 10.1128/IAI.73.2.905-911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ishida K, Matsuo J, Yamamoto Y, Yamaguchi H. 2014. Chlamydia pneumoniae effector chlamydial outer protein N sequesters fructose bisphosphate aldolase A, providing a benefit to bacterial growth. BMC Microbiol 14:330. doi: 10.1186/s12866-014-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shen L, Macnaughtan MA, Frohlich KM, Cong Y, Goodwin OY, Chou CW, LeCour L, Jr., Krup K, Luo M, Worthylake DK. 2015. Multipart chaperone-effector recognition in the type III secretion system of Chlamydia trachomatis. J Biol Chem 290:28141–28155. doi: 10.1074/jbc.M115.670232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Archuleta TL, Spiller BW. 2014. A gatekeeper chaperone complex directs translocator secretion during type three secretion. PLoS Pathog 10:e1004498. doi: 10.1371/journal.ppat.1004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nawrotek A, Guimarães BG, Velours C, Subtil A, Knossow M, Gigant B. 2014. Biochemical and structural insights into microtubule perturbation by CopN from Chlamydia pneumoniae. J Biol Chem 289:25199–25210. doi: 10.1074/jbc.M114.568436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang J, Lesser CF, Lory S. 2008. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456:112–115. doi: 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jamison WP, Hackstadt T. 2008. Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb Pathog 45:435–440. doi: 10.1016/j.micpath.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, Rout S, Stark M, von Mering C, Pelkmans L, Hardt WD. 2011. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol 7:474. doi: 10.1038/msb.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hefty PS, Stephens RS. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol 189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ouellette SP, AbdelRahman YM, Belland RJ, Byrne GI. 2005. The Chlamydia pneumoniae type III secretion-related lcrH gene clusters are developmentally expressed operons. J Bacteriol 187:7853–7856. doi: 10.1128/JB.187.22.7853-7856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Soules KR, LaBrie SD, May BH, Hefty PS. 2020. Sigma 54-regulated transcription is associated with membrane reorganization and type III secretion effectors during conversion to infectious forms of Chlamydia trachomatis. mBio 11. doi: 10.1128/mBio.01725-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hanson BR, Slepenkin A, Peterson EM, Tan M. 2015. Chlamydia trachomatis type III secretion proteins regulate transcription. J Bacteriol 197:3238–3244. doi: 10.1128/JB.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang C, Kari L, Sturdevant GL, Song L, Patton MJ, Couch CE, Ilgenfritz JM, Southern TR, Whitmire WM, Briones M, Bonner C, Grant C, Hu P, McClarty G, Caldwell HD. 2017. Chlamydia trachomatis ChxR is a transcriptional regulator of virulence factors that function in in vivo host-pathogen interactions. Pathog Dis 75:ftx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fattori J, Prando A, Martins AM, Rodrigues FH, Tasic L. 2011. Bacterial secretion chaperones. Protein Pept Lett 18:158–166. doi: 10.2174/092986611794475048. [DOI] [PubMed] [Google Scholar]
- 153.Feldman MF, Cornelis GR. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol Lett 219:151–158. doi: 10.1016/S0378-1097(03)00042-9. [DOI] [PubMed] [Google Scholar]
- 154.Parsot C, Hamiaux C, Page AL. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol 6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 155.Birtalan S, Ghosh P. 2001. Structure of the Yersinia type III secretory system chaperone SycE. Nat Struct Biol 8:974–978. doi: 10.1038/nsb1101-974. [DOI] [PubMed] [Google Scholar]
- 156.Luo Y, Bertero MG, Frey EA, Pfuetzner RA, Wenk MR, Creagh L, Marcus SL, Lim D, Sicheri F, Kay C, Haynes C, Finlay BB, Strynadka NC. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat Struct Biol 8:1031–1036. doi: 10.1038/nsb717. [DOI] [PubMed] [Google Scholar]
- 157.Page AL, Parsot C. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol Microbiol 46:1–11. doi: 10.1046/j.1365-2958.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- 158.Büttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. 2008. Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J Mol Biol 375:997–1012. doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 159.Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. 2009. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci USA 106:9661–9666. doi: 10.1073/pnas.0812900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. 2002. Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis. Acta Crystallogr D Biol Crystallogr 58:398–406. doi: 10.1107/s090744490200015x. [DOI] [PubMed] [Google Scholar]
- 161.Yip CK, Finlay BB, Strynadka NC. 2005. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol 12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 162.Phan J, Austin BP, Waugh DS. 2005. Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci 14:2759–2763. doi: 10.1110/ps.051706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, Pastor A, Elsen S, Dessen A, Attree I. 2005. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J Biol Chem 280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 164.Lloyd SA, Sjöström M, Andersson S, Wolf-Watz H. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol Microbiol 43:51–59. doi: 10.1046/j.1365-2958.2002.02738.x. [DOI] [PubMed] [Google Scholar]
- 165.Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA 92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. 2014. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cossé MM, Barta ML, Fisher DJ, Oesterlin LK, Niragire B, Perrinet S, Millot GA, Hefty PS, Subtil A. 2018. The loss of expression of a single type 3 effector (CT622) strongly reduces Chlamydia trachomatis infectivity and growth. Front Cell Infect Microbiol 8:145. doi: 10.3389/fcimb.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Forde CE, Rocco JM, Fitch JP, McCutchen-Maloney SL. 2004. Real-time characterization of virulence factor expression in Yersinia pestis using a GFP reporter system. Biochem Biophys Res Commun 324:795–800. doi: 10.1016/j.bbrc.2004.08.236. [DOI] [PubMed] [Google Scholar]
- 169.Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, Garnham JB, Sokhansanj BA, Ott LL, Coleman MA, Elliott JM, Kegelmeyer LM, Wyrobek AJ, Slezak TR, Brubaker RR, Garcia E. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J Bacteriol 186:6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ, Hefty PS. 2019. Transposon mutagenesis in Chlamydia trachomatis identifies CT339 as a ComEC homolog important for DNA uptake and lateral gene transfer. mBio 10. doi: 10.1128/mBio.01343-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.O'Neill CE, Skilton RJ, Forster J, Cleary DW, Pearson SA, Lampe DJ, Thomson NR, Clarke IN. 2021. An inducible transposon mutagenesis approach for the intracellular human pathogen Chlamydia trachomatis. Wellcome Open Res 6:312. doi: 10.12688/wellcomeopenres.16068.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Creasey EA, Delahay RM, Bishop AA, Shaw RK, Kenny B, Knutton S, Frankel G. 2003. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol Microbiol 47:209–221. doi: 10.1046/j.1365-2958.2003.03290.x. [DOI] [PubMed] [Google Scholar]
- 173.Silva-Herzog E, Joseph SS, Avery AK, Coba JA, Wolf K, Fields KA, Plano GV. 2011. Scc1 (CP0432) and Scc4 (CP0033) function as a type III secretion chaperone for CopN of Chlamydia pneumoniae. J Bacteriol 193:3490–3496. doi: 10.1128/JB.00203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Slepenkin A, de la Maza LM, Peterson EM. 2005. Interaction between components of the type III secretion system of Chlamydiaceae. J Bacteriol 187:473–479. doi: 10.1128/JB.187.2.473-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gao L, Cong Y, Plano GV, Rao X, Gisclair LN, Schesser Bartra S, Macnaughtan MA, Shen L. 2020. Context-dependent action of Scc4 reinforces control of the type III secretion system. J Bacteriol 202. doi: 10.1128/JB.00132-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rao X, Deighan P, Hua Z, Hu X, Wang J, Luo M, Wang J, Liang Y, Zhong G, Hochschild A, Shen L. 2009. A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev 23:1818–1829. doi: 10.1101/gad.1784009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lonetto M, Gribskov M, Gross CA. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biol 4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ukwaththage TO, Keane SM, Shen L, Macnaughtan MA. 2020. Chain-selective isotopic labeling of the heterodimeric type III secretion chaperone, Scc4:Scc1, reveals the total structural rearrangement of the Chlamydia trachomatis bi-functional protein, Scc4. Biomolecules 10:1480. doi: 10.3390/biom10111480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brinkworth AJ, Malcolm DS, Pedrosa AT, Roguska K, Shahbazian S, Graham JE, Hayward RD, Carabeo RA. 2011. Chlamydia trachomatis Slc1 is a type III secretion chaperone that enhances the translocation of its invasion effector substrate TARP. Mol Microbiol 82:131–144. doi: 10.1111/j.1365-2958.2011.07802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, Gough J, Haft DH, Letunić I, Marchler-Bauer A, Mi H, Natale DA, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A. 2023. InterPro in 2022. Nucleic Acids Res 51:D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.da Cunha M, Milho C, Almeida F, Pais SV, Borges V, Maurício R, Borrego MJ, Gomes JP, Mota LJ. 2014. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 14:40. doi: 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kebbi-Beghdadi C, Pilloux L, Martin V, Greub G. 2020. Eukaryotic cell permeabilisation to identify new putative chlamydial type III secretion system effectors secreted within host cell cytoplasm. Microorganisms 8:361. doi: 10.3390/microorganisms8030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hovis KM, Mojica S, McDermott JE, Pedersen L, Simhi C, Rank RG, Myers GS, Ravel J, Hsia RC, Bavoil PM. 2013. Genus-optimized strategy for the identification of chlamydial type III secretion substrates. Pathog Dis 69:213–222. doi: 10.1111/2049-632X.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Subtil A, Delevoye C, Balana ME, Tastevin L, Perrinet S, Dautry-Varsat A. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 56:1636–1647. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- 188.Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol 2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 189.Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. 2011. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics 12:109. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog 5:e1000375. doi: 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kleba B, Stephens RS. 2008. Chlamydial effector proteins localized to the host cell cytoplasmic compartment. Infect Immun 76:4842–4850. doi: 10.1128/IAI.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yanatori I, Miura K, Chen YS, Valdivia RH, Kishi F. 2021. Application of a C. trachomatis expression system to identify C. pneumoniae proteins translocated into host cells. J Bacteriol 203. doi: 10.1128/JB.00511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Agaisse H, Derre I. 2014. The expression of the effector protein IncD in C. trachomatis mediates the recruitment of the lipid transfer protein CERT and the ER-resident protein VAPB to the inclusion membrane. Infect Immun 82:2037–2047. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. 2013. Conditional gene expression in using the Tet system. PLoS One 8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cortina ME, Derré I. 2023. Homologues of the Chlamydia trachomatis and Chlamydia muridarum inclusion membrane protein IncS are interchangeable for early development but not for inclusion stability in the late developmental cycle. mSphere 8:e0000323. doi: 10.1128/msphere.00003-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mueller KE, Fields KA. 2015. Application of beta-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Olson MG, Widner RE, Jorgenson LM, Lawrence A, Lagundzin D, Woods NT, Ouellette SP, Rucks EA. 2019. Proximity labeling to map host-pathogen interactions at the membrane of a bacterium-containing vacuole in Chlamydia trachomatis-infected human cells. Infect Immun 87. doi: 10.1128/IAI.00537-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bishop RC, Derré I. 2022. The Chlamydia trachomatis inclusion membrane protein CTL0390 mediates host cell exit via lysis through STING activation. Infect Immun 90:e0019022. doi: 10.1128/iai.00190-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bui DC, Jorgenson LM, Ouellette SP, Rucks EA. 2021. Eukaryotic SNARE VAMP3 dynamically interacts with multiple chlamydial inclusion membrane proteins. Infect Immun 89:e00409-20. doi: 10.1128/IAI.00409-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Carpenter V, Chen YS, Dolat L, Valdivia RH. 2017. The effector TepP mediates recruitment and activation of phosphoinositide 3-kinase on early Chlamydia trachomatis vacuoles. mSphere 2. doi: 10.1128/mSphere.00207-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cortina ME, Bishop RC, DeVasure BA, Coppens I, Derré I. 2022. The inclusion membrane protein IncS is critical for initiation of the Chlamydia intracellular developmental cycle. PLoS Pathog 18:e1010818. doi: 10.1371/journal.ppat.1010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ende R, Derré I. 2019. A coinfection model to evaluate chlamydia Inc protein interactions. Methods Mol Biol 2042:205–218. doi: 10.1007/978-1-4939-9694-0_14. [DOI] [PubMed] [Google Scholar]
- 203.Han Y, Derre I. 2017. A co-infection model system and the use of chimeric proteins to study chlamydia inclusion proteins interaction. Front Cell Infect Microbiol 7:79. doi: 10.3389/fcimb.2017.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Olson-Wood MG, Jorgenson LM, Ouellette SP, Rucks EA. 2021. Inclusion membrane growth and composition are altered by overexpression of specific inclusion membrane proteins in Chlamydia trachomatis L2. Infect Immun 89:e0009421. doi: 10.1128/IAI.00094-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rucks EA, Olson MG, Jorgenson LM, Srinivasan RR, Ouellette SP. 2017. Development of a proximity labeling system to map the Chlamydia trachomatis inclusion membrane. Front Cell Infect Microbiol 7:40. doi: 10.3389/fcimb.2017.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Weber MM, Bauler LD, Lam J, Hackstadt T. 2015. Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83:4710–4718. doi: 10.1128/IAI.01075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wesolowski J, Weber MM, Nawrotek A, Dooley CA, Calderon M, St Croix CM, Hackstadt T, Cherfils J, Paumet F. 2017. Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and Golgi complex positioning. mBio 8. doi: 10.1128/mBio.02280-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jennings E, Thurston TLM, Holden DW. 2017. Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22:217–231. doi: 10.1016/j.chom.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 209.Hensel M, Nikolaus T, Egelseer C. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol 31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 210.Pinaud L, Sansonetti PJ, Phalipon A. 2018. Host cell targeting by enteropathogenic bacteria T3SS effectors. Trends Microbiol 26:266–283. doi: 10.1016/j.tim.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 211.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, Liu X. 2015. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun 83:2897–2906. doi: 10.1128/IAI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang L, Zhu Z, Qian H, Li Y, Chen Y, Ma P, Gu B. 2019. Comparative genome analysis of 15 clinical Shigella flexneri strains regarding virulence and antibiotic resistance. AIMS Microbiol 5:205–222. doi: 10.3934/microbiol.2019.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang X, Li Y, Jing H, Ren Y, Zhou Z, Wang S, Kan B, Xu J, Wang L. 2011. Complete genome sequence of a Yersinia enterocolitica “Old World” (3/O:9) strain and comparison with the “New World” (1B/O:8) strain. J Clin Microbiol 49:1251–1259. doi: 10.1128/JCM.01921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Moulder JW. 1991. Interactions of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wyrick PB, Choong J, Davis CH, Knight ST, Royal MO, Maslow AS, Bagnell CR. 1989. Entry of genital Chlamydia trachomatis into polarized human epithelial cells. Infect Immun 57:2378–2389. doi: 10.1128/iai.57.8.2378-2389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Su H, Caldwell HD. 1998. Sulfated polysaccharides and a synthetic sulfated polymer are potent inhibitors of Chlamydia trachomatis infectivity in vitro but lack protective efficacy in an in vivo murine model of chlamydial genital tract infection. Infect Immun 66:1258–1260. doi: 10.1128/IAI.66.3.1258-1260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Su H, Raymond L, Rockey DD, Fischer E, Hackstadt T, Caldwell HD. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc Natl Acad Sci USA 93:11143–11148. doi: 10.1073/pnas.93.20.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Taraktchoglou M, Pacey AA, Turnbull JE, Eley A. 2001. Infectivity of Chlamydia trachomatis serovar LGV but not E is dependent on host cell heparan sulfate. Infect Immun 69:968–976. doi: 10.1128/IAI.69.2.968-976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Maslow AS, Davis CH, Choong J, Wyrick PB. 1988. Estrogen enhances attachment of Chlamydia trachomatis to human endometrial epithelial cells in vitro. Am J Obstet Gynecol 159:1006–1014. doi: 10.1016/s0002-9378(88)80189-3. [DOI] [PubMed] [Google Scholar]
- 220.Park JS, Helble JD, Lazarus JE, Yang G, Blondel CJ, Doench JG, Starnbach MN, Waldor MK. 2019. A FACS-based genome-wide CRISPR screen reveals a requirement for COPI in Chlamydia trachomatis invasion. iScience 11:71–84. doi: 10.1016/j.isci.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Carabeo RA, Hackstadt T. 2001. Isolation and characterization of a mutant chinese hamster ovary cell line that is resistant to Chlamydia trachomatis infection at a novel step in the attachment process. Infect Immun 69:5899–5904. doi: 10.1128/IAI.69.9.5899-5904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Davis CH, Raulston JE, Wyrick PB. 2002. Protein disulfide isomerase, a component of the estrogen receptor complex, is associated with Chlamydia trachomatis serovar E attached to human endometrial epithelial cells. Infect Immun 70:3413–3418. doi: 10.1128/IAI.70.7.3413-3418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hower S, Wolf K, Fields KA. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. 2002. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun 70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Caven L, Carabeo RA. 2019. Pathogenic puppetry: manipulation of the host actin cytoskeleton by Chlamydia trachomatis. Int J Mol Sci 21:90. doi: 10.3390/ijms21010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lutter EI, Bonner C, Holland MJ, Suchland RJ, Stamm WE, Jewett TJ, McClarty G, Hackstadt T. 2010. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect Immun 78:3678–3688. doi: 10.1128/IAI.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Keb G, Ferrell J, Scanlon KR, Jewett TJ, Fields KA. 2021. Chlamydia trachomatis TmeA directly activates N-WASP to promote actin polymerization and functions synergistically with TarP during invasion. mBio 12. doi: 10.1128/mBio.02861-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bullock HD, Hower S, Fields KA. 2012. Domain analyses reveal that Chlamydia trachomatis CT694 protein belongs to the membrane-localized family of type III effector proteins. J Biol Chem 287:28078–28086. doi: 10.1074/jbc.M112.386904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.McKuen MJ, Mueller KE, Bae YS, Fields KA. 2017. Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect Immun 85. doi: 10.1128/IAI.00640-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun 64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rockey DD, Grosenbach D, Hruby DE, Peacock MG, Heinzen RA, Hackstadt T. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol Microbiol 24:217–228. doi: 10.1046/j.1365-2958.1997.3371700.x. [DOI] [PubMed] [Google Scholar]
- 232.Lutter EI, Martens C, Hackstadt T. 2012. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp Funct Genomics 2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Subtil A, Parsot C, Dautry-Varsat A. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol Microbiol 39:792–800. doi: 10.1046/j.1365-2958.2001.02272.x. [DOI] [PubMed] [Google Scholar]
- 234.Lutter EI, Barger AC, Nair V, Hackstadt T. 2013. Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms. Cell Rep 3:1921–1931. doi: 10.1016/j.celrep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shaw JH, Key CE, Snider TA, Sah P, Shaw EI, Fisher DJ, Lutter EI. 2018. Genetic inactivation of Chlamydia trachomatis inclusion membrane protein CT228 alters MYPT1 recruitment, extrusion production, and longevity of infection. Front Cell Infect Microbiol 8:415. doi: 10.3389/fcimb.2018.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, Jager S, Gopalakrishnan AM, Sherry J, Dunn JD, Olive A, Penn B, Shales M, Cox JS, Starnbach MN, Derre I, Valdivia R, Krogan NJ, Engel J. 2015. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dickinson MS, Anderson LN, Webb-Robertson BM, Hansen JR, Smith RD, Wright AT, Hybiske K. 2019. Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites. PLoS Pathog 15:e1007698. doi: 10.1371/journal.ppat.1007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Olson MG, Jorgenson LM, Widner RE, Rucks EA. 2019. Proximity labeling of the Chlamydia trachomatis inclusion membrane. Methods Mol Biol 2042:245–278. doi: 10.1007/978-1-4939-9694-0_17. [DOI] [PubMed] [Google Scholar]
- 239.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Scidmore MA, Hackstadt T. 2001. Mammalian 14–3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol Microbiol 39:1638–1650. doi: 10.1046/j.1365-2958.2001.02355.x. [DOI] [PubMed] [Google Scholar]
- 241.Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. 2011. Chlamydia trachomatis co-opts GBF-1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog 7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pais SV, Key CE, Borges V, Pereira IS, Gomes JP, Fisher DJ, Mota LJ. 2019. CteG is a Chlamydia trachomatis effector protein that associates with the Golgi complex of infected host cells. Sci Rep 9:6133. doi: 10.1038/s41598-019-42647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dimond Z, Bauler LD, Zhang Y, Carmody A, Hackstadt T. 2022. Chlamydia trachomatis alters mitochondrial protein composition and secretes effector proteins that target mitochondria. mSphere 7:e0042322. doi: 10.1128/msphere.00423-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hu C, Hardee D, Minnear F. 2007. Membrane fusion by VAMP3 and plasma membrane t-SNAREs. Exp Cell Res 313:3198–3209. doi: 10.1016/j.yexcr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol 156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Aeberhard L, Banhart S, Fischer M, Jehmlich N, Rose L, Koch S, Laue M, Renard BY, Schmidt F, Heuer D. 2015. The proteome of the isolated Chlamydia trachomatis containing vacuole reveals a complex trafficking platform enriched for retromer components. PLoS Pathog 11:e1004883. doi: 10.1371/journal.ppat.1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol 12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bannantine JP, Stamm WE, Suchland RJ, Rockey DD. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect Immun 66:6017–6021. doi: 10.1128/IAI.66.12.6017-6021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol 1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 252.Gauliard E, Ouellette SP, Rueden KJ, Ladant D. 2015. Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Front Cell Infect Microbiol 5:13. doi: 10.3389/fcimb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wilson DP, Timms P, McElwain DL, Bavoil PM. 2006. Type III secretion, contact-dependent model for the intracellular development of chlamydia. Bull Math Biol 68:161–178. doi: 10.1007/s11538-005-9024-1. [DOI] [PubMed] [Google Scholar]
- 254.Ward ME. 1988. The chlamydial developmental cycle, p 71–95. In Barron AL (ed), Microbiology of Chlamydia. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 255.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol Microbiol 62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 256.Agrain C, Sorg I, Paroz C, Cornelis GR. 2005. Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol Microbiol 57:1415–1427. doi: 10.1111/j.1365-2958.2005.04758.x. [DOI] [PubMed] [Google Scholar]
- 257.Coleman MA, Cappuccio JA, Blanchette CD, Gao T, Arroyo ES, Hinz AK, Bourguet FA, Segelke B, Hoeprich PD, Huser T, Laurence TA, Motin VL, Chromy BA. 2016. Expression and association of the Yersinia pestis translocon proteins, YopB and YopD, are facilitated by nanolipoprotein particles. PLoS One 11:e0150166. doi: 10.1371/journal.pone.0150166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dewoody RS, Merritt PM, Marketon MM. 2013. Regulation of the Yersinia type III secretion system: traffic control. Front Cell Infect Microbiol 3:4. doi: 10.3389/fcimb.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]