Abstract
We performed phylogenetic analysis on dengue virus serotype 2 Cosmopolitan genotype in Ho Chi Minh City, Vietnam. We document virus emergence, probable routes of introduction, and timeline of events. Our findings highlight the need for continuous, systematic genomic surveillance to manage outbreaks and forecast future epidemics.
Keywords: dengue virus, viruses, arboviruses, mosquito-borne infections, DENV serotype 2, climatic factors, Vietnam, vector-borne infections
Dengue virus (DENV) represents a major public health concern globally and in Vietnam, where an estimated 1.6 million cases occur each year (1). Clinical manifestations range from fever to severe organ dysfunction (2). DENV includes 4 distinct serotypes (DENV 1–4), which have evolved into distinguishable genotypes; all are transmitted primarily by Aedes aegypti mosquitoes (3). DENV is endemic in both urban and peri-urban areas of Vietnam with substantial seasonal temporal and spatial variation. Although all 4 DENV serotypes have circulated in Vietnam, DENV-1 and DENV-2 have been most prevalent over the past 20 years.
In 2022, Ho Chi Minh City, Vietnam, experienced a 3-fold increase in reported DENV cases compared with 2020 and 5-fold compared with 2021 (4,5). Possible drivers of transmission include climate factors because optimal temperatures and humidity increase vector abundance (6), reduced population immunity because of lower rates of transmission in previous years (6), and possible introduction of new serotypes or genotypes or diversification of circulating lineages (7). Scarcity of available data on DENV circulation by lineage in Vietnam limits testing those hypotheses. Our study explored the DENV lineages circulating in Ho Chi Minh City over the past 5 years, aiming to provide a detailed assessment of DENV dynamics in Vietnam.
We randomly selected 362 samples from dengue patients enrolled in research studies (reviewed by the ethics committee and approved by the internal review board of the hospital) at the Hospital for Tropical Diseases in Ho Chi Minh City during 2017–2022. Of those patients, 303 (83.7%) tested positive for dengue using quantitative reverse transcription PCR. DENV-2 was predominant (72.3%); DENV-1 accounted for 23.1% and DENV-4 for 4.6% of positive samples. From those samples, we sequenced 45 DENV-2 viral envelope (E) genes. We amplified E genes using PCR, and after PCR product purification, we conducted Sanger sequencing on samples with nucleic acid concentrations >10 ng/μL. We supplemented the new sequences with a background dataset of DENV-2 genome and E-gene sequences from southern and southeast Asia (Appendix 1).
We constructed maximum-likelihood trees for all DENV-2 genotypes, as well as a time-scaled tree in which we estimated ancestral node locations and conducted root-to-tip regression analyses. Our results showed DENV-2 replaced DENV-1 as the predominant serotype in 2019, and multiple DENV-2 genotypes were cocirculating (Appendix 1 Figure 1). Seventeen sequences belonged to Asia I genotype, which has been established in the region since 2006; another 28 sequences were identified as sporadically detected Cosmopolitan genotype. Cosmopolitan genotype virus strain has displayed signs of reemergence, with indicators of >3 distinct recent introductions of clades A, B, and C into southern Vietnam (Appendix 1 Figure 2). Cosmopolitan phylogeny (Figure), constructed similarly to the broader DENV-2 tree, shows most (27/28) sequences from this region clustered within these 3 clades; clades A and B share ancestry with sequences from Indonesia and clade C shares ancestry with sequences from Cambodia.
Figure.
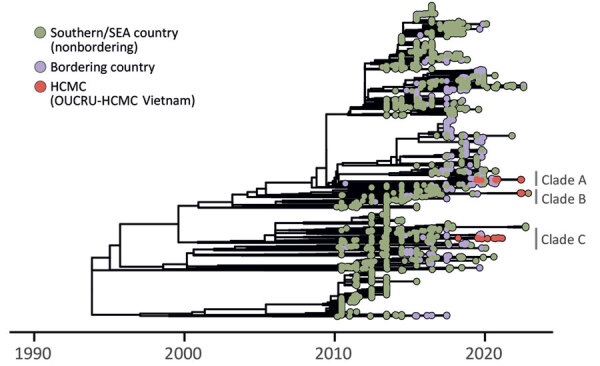
Time-scaled maximum-likelihood phylogenic tree showing emergence of the Cosmopolitan genotype of dengue virus serotype 2 in Vietnam. Clades A–C refer to transmission lineages. HCMC, Ho Chi Minh City; OUCRU, Oxford University Clinical Research Unit; SEA, southeast Asia
The clades were detected across several provinces in southern Vietnam, including all 3 clades in Ho Chi Minh City. Clade A circulated in provinces north and east of Ho Chi Minh City; clades B and C were found in provinces north and west of the city (Appendix 1 Figure 3). Estimated time to the most recent common ancestor suggests that clade A has been in circulation the longest, followed by clades B and C (Appendix 1 Figure 4). These findings indicate continuous introductions of the Cosmopolitan genotype into southern Vietnam over multiple years. Clades A and C have persisted locally for at least half a decade, with approximately 2 years between the earliest most recent common ancestor and the earliest sampling date, suggesting undetected Cosmopolitan genotype might have been cryptically transmitted during multiple dengue seasons.
Dynamics of DENV prevalence and spread within Vietnam might have been influenced by successive seedings of the Cosmopolitan genotype, thereby increasing likelihood of establishment and sustained transmission (8,9). Although the Cosmopolitan genotype has circulated in neighboring countries, likely since the late 1990s (Appendix 1 Figures 5, 6), it had not been reported in Vietnam until recently. Mechanisms underlying a specific genotype reemerging after new introductions and subsequent establishment warrant further investigation. Large-scale dengue surveillance in hyperendemic settings like Vietnam is a formidable challenge because it relies on syndromic surveillance and awareness of heterogeneous epidemiologic trends, and limited resources are available to support viral genomic sequencing to identify circulating strains. To refine our understanding of transmission pathways for specific lineages of DENV, improving surveillance using strategies that reduce sampling bias is critical (10). We emphasize the importance of continuous, systematic virus sequencing in urban centers in Vietnam and across southeast Asia to swiftly identify novel viral lineages. These strategies, paired with clinical and socioeconomic data, will support preventive measures and outbreak forecasting.
Additional information on reemergence of Cosmopolitan genotype of dengue virus serotype 2, southern Vietnam.
Additional list of acknowledgments.
Acknowledgments
We thank the researchers who provided sequences to GISAID (https://www.gisaid.org) that we used for this research. A list of sequences is provided in Appendix 2.
R.P.D.I. and M.U.G.K acknowledge funding from the European Union Horizon 2020 project MOOD (874850). M.U.G.K. and B.G. acknowledge funding from the Oxford Martin School Pandemic Genomics program. M.U.G.K. acknowledges funding from the Wellcome Trust, a Branco Weiss Fellowship, The Rockefeller Foundation, and Google.org. V.T.T., K.T.H.D., N.M.N. and S.Y. acknowledge funding from the Wellcome Trust (106680).
Contributions: R.P.D.I., V.T.T., S.Y., B.G.G., and M.U.G.K. developed the idea and planned the research. V.T.T. collected samples and performed sequencing. R.P.D.I., B.G., and I.R. developed the bioinformatic pipeline; R.P.D.I. conducting the analyses. R.P.D.I., V.T.T., S.Y., B.G., and M.U.G.K. wrote the first draft of the manuscript. K.T.H.D. contributed to the sequencing procedure for samples and initial data cleaning. N.M.N. contributed to areas related to clinical knowledge and patient recruitment. P.N.T. and T.C.T. contributed to management and oversight of conducting the study in the hospital. All authors interpreted the data, contributed to writing, and approved the manuscript.
Biography
Vi Thuy Tran is a senior research assistant working in the dengue research group at Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam. Her primary research interest focuses on the investigating evolution of dengue virus and its transmission dynamic.
Footnotes
Suggested citation for this article: Tran VT, Inward RPD, Gutierrez B, Nguyen NM, Nguyen PT, Rajendiran I, et al. Reemergence of Cosmopolitan genotype of dengue virus serotype 2, southern Vietnam. Emerg Infect Dis. 2023 Oct [date cited]. https://doi.org/10.3201/eid2910.230529
These authors contributed equally to the article.
References
- 1.Cattarino L, Rodriguez-Barraquer I, Imai N, Cummings DAT, Ferguson NM. Mapping global variation in dengue transmission intensity. Sci Transl Med. 2020;12:eaax4144. 10.1126/scitranslmed.aax4144 [DOI] [PubMed] [Google Scholar]
- 2.Yacoub S, Wills B. Dengue: an update for clinicians working in non-endemic areas. Clin Med (Lond). 2015;15:82–5. 10.7861/clinmedicine.15-1-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen-Tien T, Lundkvist Å, Lindahl J. Urban transmission of mosquito-borne flaviviruses - a review of the risk for humans in Vietnam. Infect Ecol Epidemiol. 2019;9:1660129. 10.1080/20008686.2019.1660129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho Chi Minh City Center for Disease Control. Ho Chi Minh City: warning about the risk of epidemic dengue fever is complicated [in Vietnamese]. [cited 2022 Apr 22] https://hcdc.vn/tphcm-canh-bao-nguy-co-dich-benh-sot-xuat-huyet-dien-bien-phuc-tap-4ac4d90b7e3c5ef9feaf468a4073b101.html
- 5.Ho Chi Minh City Center for Disease Control. Epidemic situation dengue, hand, foot and mouth updated to week 50 (as of 11/12/2022) [in Vietnamese]. [cited 2022 Dec 14]. https://hcdc.vn/tinh-hinh-dich-benh-sot-xuat-huyet-tay-chan-mieng-cap-nhat-den-tuan-50-tinh-den-ngay-11122022-8dfd9a4597e0425a7f0fc2d3ed204188.html
- 6.Rabaa MA, Simmons CP, Fox A, Le MQ, Nguyen TTT, Le HY, et al. Dengue virus in sub-tropical northern and central Viet Nam: population immunity and climate shape patterns of viral invasion and maintenance. PLoS Negl Trop Dis. 2013;7:e2581. 10.1371/journal.pntd.0002581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Mammen MP Jr, Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, et al. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol. 2005;79:15123–30. 10.1128/JVI.79.24.15123-15130.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasmono RT, Kalalo LP, Trismiasih S, Denis D, Yohan B, Hayati RF, et al. Multiple introductions of dengue virus strains contribute to dengue outbreaks in East Kalimantan, Indonesia, in 2015-2016. Virol J. 2019;16:93. 10.1186/s12985-019-1202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian H, Sun Z, Faria NR, Yang J, Cazelles B, Huang S, et al. Increasing airline travel may facilitate co-circulation of multiple dengue virus serotypes in Asia. PLoS Negl Trop Dis. 2017;11:e0005694. 10.1371/journal.pntd.0005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott P, Whitaker M, Tang D, Eales O, Steyn N, Bodinier B, et al. Design and implementation of a national SARS-CoV-2 monitoring program in England: REACT-1 study. Am J Public Health. 2023;113:545–54. 10.2105/AJPH.2023.307230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information on reemergence of Cosmopolitan genotype of dengue virus serotype 2, southern Vietnam.
Additional list of acknowledgments.


