Abstract
From 2015–2018 to 2019‒2021, hypertoxigenic M1UK lineage among invasive group A Streptococcus increased in the United States (1.7%, 21/1,230 to 11%, 65/603; p<0.001). M1UK was observed in 9 of 10 states, concentrated in Georgia (n = 41), Tennessee (n = 13), and New York (n = 13). Genomic cluster analysis indicated recent expansions.
Keywords: invasive group A Streptococcus, streptococci, bacteria, invasive disease, iGAS, M1UK lineage, clonal expansion, respiratory infections, zoonoses
The M1UK lineage of group A Streptococcus (GAS) is a hypertoxigenic clone within the serotype M1 GAS strain and has been associated with increased scarlet fever and invasive GAS (iGAS) disease incidence in the United Kingdom since 2014 (1–3). M1UK carries 27 characteristic lineage-defining single-nucleotide variants (SNVs) that distinguish it from other globally circulating M1 GAS clones (1). By 2020, M1UK had also became the dominant clone among M1 GAS in England (3), the Netherlands (4), and Australia (5) and showed substantial presence in Canada (6).
In the United States, M1UK was identified as a minor clone of M1 iGAS isolates in the Active Bacterial Core surveillance (ABCs) system, a laboratory- and population-based surveillance system for invasive bacterial infections that is currently implemented in 10 US states (7), in 2015–2018 (8). Using genomic surveillance data in ABCs, we investigated the trend of M1UK in 2019–2021 and documented the characteristics of iGAS infections caused by M1UK.
The Study
We identified iGAS cases through ABCs and mapped whole-genome sequencing reads of M1 isolates against the M1 reference genome MGAS5005 to identify M1UK based on previously reported characteristic M1UK SNVs (1). We constructed phylogenetic trees by using kSNP3.0 software (9). We identified genomic clusters by using a hierarchical cluster analysis with a cutoff value of 10 SNVs as described (10). We evaluated change of M1UK proportion among M1 iGAS over time by using the χ2 test for trend in proportions (trend test). We used the Fisher exact test to assess equality of proportions. All p values were 2 sided, and we considered p<0.05 statistically significant. We performed all analyses by using R software version 3.4.3 (The R Foundation for Statistical Computing, https://www.r-project.org).
We submitted all whole-genome sequencing data files of the study isolates to the National Center for Biotechnology Information Sequence Read Archive (BioProject no. PRJNA395240). Accession numbers of the 86 M1UK isolates are provided (Appendix Table 4).
During 2019‒2021, a total of 603 cases of M1 iGAS infections were documented through ABCs. Among those cases, 65 (11%) were caused by the M1UK clone (Figure 1, panel A), and the percentage was significantly higher than that observed during 2015–2018 (1.7%, 21/1,230; p<0.001). The trend test indicated a significant increasing trend in the M1UK proportion among M1 iGAS isolates during 2015‒2021 (p<0.001). During 2015–2021, most M1UK cases (67/86) were concentrated in 3 states: Georgia (41 cases,) Tennessee (13 cases), and New York (13 cases), although the M1UK clone was found in 9 of the 10 ABCs sites (Figure 1, panel B). Nearly one third of all M1UK infections (28/86) occurred in the first quarter of 2020 (Figure 1, panel B). During 2015–2021, a total of 12 iGAS isolates were identified as the intermediate lineages, containing 13 (n = 4) or 23 (n = 8) of the 27 characteristic M1UK SNVs (Appendix Figure 1), and they did not show significant expansion from 2015–2018 through 2019–2021 (p = 0.07).
Figure 1.
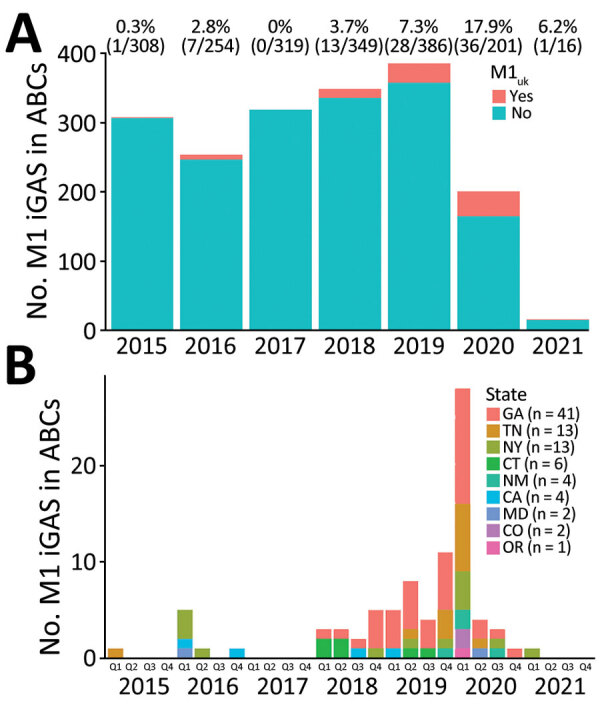
Expansion of M1UK lineage in serotype M1 iGAS in the United States, 2015–2021. A) Counts and percentages of M1UK isolates among M1 iGAS isolates in ABCs during 2015‒2021. B) Number of M1UK infections over time in 9 states that are part of the ABCs system. Key shows total number of M1UK infections during 2015‒2021 for each state. ABCs, Active Bacterial Core Surveillance System; iGAS, invasive group A Streptococcus disease; Q, quarter.
Phylogenetic analysis of the 86 M1UK isolates showed 9 distinctive genomic clusters (Figure 2). Each genomic cluster contained 2‒21 genomically closely related isolates, and collectively those clusters accounted for 74 (86%) of all M1UK isolates (Figure 2). For 2 M1UK isolates within a same cluster, the median pairwise genomic distance was 3 SNVs (interquartile range [IQR] 1.5‒6), consistent with continued transmission from a recent introduction event. However, for 2 M1UK isolates not in the same cluster, the median pairwise genomic distance was 33 SNVs (IQR 28‒38), indicating some degree of genomic diversity within M1UK, although not as much as the diversity observed among 100 randomly selected globally circulating M1 GAS clone isolates in ABCs, 2015–2021 (median 63 SNVs, IQR 49‒115). (Appendix Figure 1).
Figure 2.
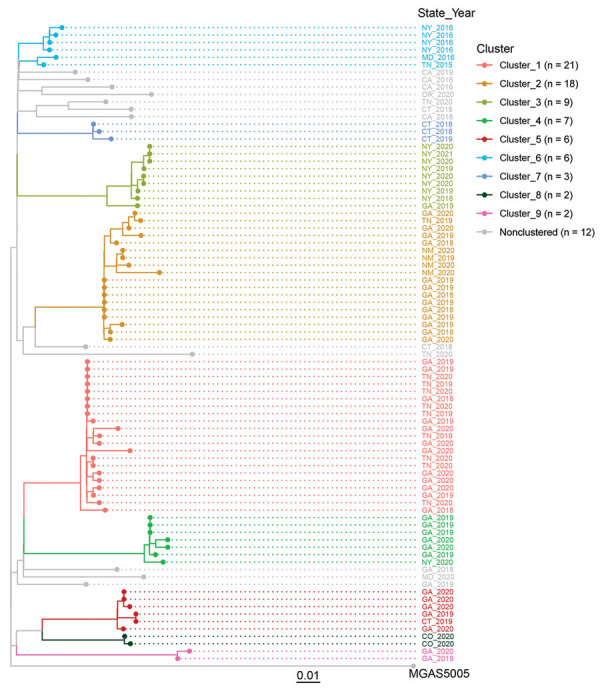
Genomic clusters of M1UK invasive group A Streptococcus disease, United States, 2015–2021. Core-genome phylogenetic tree of 86 M1UK invasive group A Streptococcus disease isolates and the reference M1 genome MGAS5005 was based on 462 core single-nucleotide variant sites generated by kSNP3.0 software (9). Tip colors indicate 9 groups of genomically closely related isolates (genomic clusters). Key shows total number of M1UK isolates in each cluster. Scale bar indicates expected nucleotide substitutions per site.
The clusters displayed clear signatures of temporal and geographic relatedness (Appendix Figure 2). For example, the largest cluster, cluster_1, showed a sharp increase of cases at the beginning of 2020, followed by a rapid decrease. The second largest cluster, cluster 2, showed relatively stable case numbers spanning from the third quarter of 2018 to the third quarter of 2020. Within a genomic cluster, most M1UK iGAS were identified in 1 or 2 states, suggesting a localized spread of the infection.
Overall, M1UK and non-M1UK M1 isolates had many common genetic features of the contemporary M1 S. pyogenes strain (Table). The M1UK clone had a higher proportion of isolates that had the streptococcal pyrogenic exotoxin gene speC (4.7% [4/86] vs. 1.4% [24/1,747]; p = 0.039), the super antigen A gene ssa (2.3% [2/86] vs. 0.1% (2/1,747]; p = 0.012), and the extracellular streptodornase D gene sda1 (also known as sdaD2; 100% [86/86] vs. 92% [1,613/1,747]; p = 0.002). The speC and ssa genes were found in 4 nonclustered M1UK isolates, of which 2 isolates had both genes, suggesting acquisition of prophage ΦHKU488.vir (5).
Table. Strain and patient features of M1UK iGAS compared with other M1 iGAS in ABCs, United States, 2015–2021.
| Characteristic | M1 iGAS, no. (%) cases |
p value† | |
|---|---|---|---|
| M1UK, n = 86 | Non-M1UK, n = 1,747 | ||
| Strain feature‡ | |||
| Antimicrobial susceptibility | |||
| Penicillin nonsusceptible | 0 | 0 | 1.000 |
| Erythromycin nonsusceptible | 0 | 21 (1.2) | 0.621 |
| Clindamycin nonsusceptible | 0 | 20 (1.1) | 1.000 |
| Tetracycline nonsusceptible | 0 | 18 (1) | 1.000 |
| Levofloxacin nonsusceptible |
0 |
9 (0.5) |
1.000 |
| Pyrogenic exotoxin genes | |||
| speA | 86 (100) | 1,720 (98.5) | 0.635 |
| speC | 4 (4.7) | 24 (1.4) | 0.039 |
| speG | 86 (100) | 1747 (100) | 1.000 |
| speH | 0 | 1 (0.1) | 1.000 |
| speI | 0 | 1 (0.1) | 1.000 |
| speJ | 86 (100) | 1,747 (100) | 1.000 |
| speK | 0 | 2 (0.1) | 1.000 |
| speL | 0 | 0 | 1.000 |
| speM | 0 | 0 | 1.000 |
| Ssa | 2 (2.3) | 2 (0.1) | 0.012 |
|
smeZ
|
85 (98.8) |
1,737 (99.4) |
0.411 |
| Other virulence factors | |||
| hasA hyaluronic acid synthetase, capsule | 86 (100) | 1,03 (97.5) | 0.266 |
| Virulence-associated DNase, SDA1§ |
86 (100) |
1,613 (92.3) |
0.002 |
| Patient characteristic | |||
| Age, y | |||
| <18 | 12 (14) | 233 (13.3) | 0.871 |
| 18–34 | 5 (5.8) | 178 (10.2) | 0.266 |
| 35–49 | 19 (22.1) | 333 (19.1) | 0.484 |
| 50–64 | 21 (24.4) | 445 (25.5) | 0.899 |
| 65–74 | 15 (17.4) | 289 (16.5) | 0.768 |
| >75 | 14 (16.3) | 269 (15.4) | 0.762 |
| Sex | |||
| M | 41 (47.7) | 955 (54.7) | 0.223 |
| F |
45 (52.3) |
792 (45.3) |
0.223 |
| Clinical syndrome | |||
| Cellulitis | 25 (29.1) | 633 (36.2) | 0.205 |
| Bacteremia without focus | 11 (12.8) | 276 (15.8) | 0.544 |
| Pneumonia | 28 (32.6) | 384 (22.0) | 0.033 |
| Necrotizing fasciitis | 5 (5.8) | 147 (8.4) | 0.546 |
| Streptococcal toxic shock syndrome | 7 (8.1) | 95 (5.4) | 0.328 |
| Death | 19 (22.1) | 260 (14.9) | 0.089 |
*ABCs, Active Bacterial Core Surveillance System; iGAS, invasive group A Streptococcus disease. †By Fisher exact test. ‡All strain features, including antimicrobial susceptibility, pyrogenic exotoxin genes, and other virulence factors, were inferred from whole-genome sequencing data. §Detecting genetic marker for the sda1 gene (GenBank accession no. AY452036.1), which is identical to the sdaD2 gene (M5005_Spy1415; GenBank accession no. AAZ52033.1).
Patients infected by the M1UK strain showed similar age, sex, and syndrome distribution compared with patients infected by non-M1UK M1 GAS (Table), except that M1UK isolates were more likely to be found in patients with pneumonia (33% vs. 22%; p = 0.033). The case-fatality rate was high for M1UK iGAS infection (22%) although it was not significantly different from that of non-M1UK M1 iGAS (15%; p = 0.089). In subgroup analysis stratified by time (2015–2018, 2019–2021) and location (GA, TN, and NY only), M1UK isolates were associated with higher proportions of speC, ssa, sda1, and pneumonia compared with non-M1UK isolates in all 3 subgroups (Appendix Tables 1‒3), except for speC in 2019–2021. The difference in subgroup analysis was generally not statistically significant, potentially caused by smaller sample size and reduced power in a subgroup.
Conclusions
This study demonstrates a substantial increase of M1UK lineage during 2019‒ 2021 in the ABCs sites in the United States. Additional data are needed to determine variance in M1UK iGAS incidence across states outside the 10 states in ABCs. The proportion of M1UK iGAS in ABCs remains much lower than that reported in England (3), Australia (5), and the Netherlands (4). We documented the mode of expansion for the M1UK lineage in the United States by determining whether the 86 M1UK iGAS cases could be explained by 1 recent introduction or multiple ones. We tracked the shape and characteristics of epidemiologic curves for each cluster, which could help understand different patterns of disease transmission. The increase was associated with the formation and expansion of multiple genomic clusters in which each cluster was mostly found in only 1 or 2 states. The results suggested that the M1UK clone might have been introduced and circulated in different geographic locations in the United States, rather than spreading from a recent single introduction event.
In 2020, emm1 was the leading cause of iGAS only in Georgia and New York in ABCs. In that year, the proportion of M1UK isolates among emm1 iGAS was 38% (16/42) in Georgia and 25% (5/20) in New York. It appeared that M1UK lineage followed the same state preferences as M1 in general. In recent years, there were rapidly expanding clusters of emm types that were not historically so prevalent within several different states, mostly pattern E lineages (emm11,49,82,92,60) and pattern D lineages (emm83,59,81), which was associated with increasing proportions of disadvantaged persons and led to drastic changes in emm type distributions in those states (11,12).
Although the speC and ssa genes were associated with M1UK isolates, they were present in <5% of these isolates, and the biologic role of this association is unclear. It is crucial to monitor the spread of this variant and the associated virulence determinants to inform development of effective prevention and treatment strategies.
Additional information on expansion of invasive group A Streptococcus M1UK lineage in active bacterial core surveillance, United States, 2019‒2021.
Acknowledgments
This study used the Streptococcus pyogenes multilocus sequence typing website (https://pubmlst.org/organisms/streptococcus-pyogenes) hosted at the University of Oxford.
This study was supported by the Centers for Disease Control and Prevention. Major support was provided by the Centers for Disease Control and Prevention Emerging Infections Program and the Advanced Molecular Detection Initiative.
Y.L., L.M., and B.B. contributed to study design; all authors contributed to data collection; Y.L. performed data analysis, produced the table and figures, and wrote the manuscript; and J.R., S.M., Z.L., S.C., B.J.M., B.B., J.O., C.J.G., and L.M. contributed to data analysis and interpretation. All authors reviewed and edited the manuscript.
Biography
Dr. Yuan Li is a microbiologist and bioinformatician in the National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA. His primary research interests are integration of laboratory and epidemiologic data to inform disease surveillance, outbreak responses, and vaccine strategies.
Footnotes
Suggested citation for this article: Li Y, Rivers J, Mathis S, Li Z, Chochua S, Metcalf BJ, et al. Expansion of invasive group A Streptococcus M1UK lineage in active bacterial core surveillance, United States, 2019‒2021. Emerg Infect Dis. 2023 Oct [date cited]. https://doi.org/10.3201/eid2910.230675
References
- 1.Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19:1209–18. 10.1016/S1473-3099(19)30446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcolea-Medina A, Snell LB, Alder C, Charalampous T, Williams TGS, Tan MKI, et al. ; Synnovis Microbiology Laboratory Group. The ongoing Streptococcus pyogenes (Group A Streptococcus) outbreak in London, United Kingdom, in December 2022: a molecular epidemiology study. Clin Microbiol Infect. 2023;29:887–90. 10.1016/j.cmi.2023.03.001 [DOI] [PubMed] [Google Scholar]
- 3.Zhi X, Li HK, Li H, Loboda Z, Charles S, Vieira A, et al. Emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020. Emerg Infect Dis. 2023;29:1007–10. 10.3201/eid2905.221887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rümke LW, de Gier B, Vestjens SMT, van der Ende A, van Sorge NM, Vlaminckx BJM, et al. Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes. Lancet Infect Dis. 2020;20:539–40. 10.1016/S1473-3099(20)30278-4 [DOI] [PubMed] [Google Scholar]
- 5.Davies MR, Keller N, Brouwer S, Jespersen MG, Cork AJ, Hayes AJ, et al. Detection of Streptococcus pyogenes M1UK in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14:1051. 10.1038/s41467-023-36717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect Dis. 2019;19:1284–5. 10.1016/S1473-3099(19)30622-X [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Active Bacterial Core Surveillance, Emerging Infections Program Network. 2020. [cited 2023 Aug 18]. https://www.cdc.gov/abcs
- 8.Li Y, Nanduri SA, Van Beneden CA, Beall BW. M1UK lineage in invasive group A streptococcus isolates from the USA. Lancet Infect Dis. 2020;20:538–9. 10.1016/S1473-3099(20)30279-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–8. 10.1093/bioinformatics/btv271 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Dominguez S, Nanduri SA, Rivers J, Mathis S, Li Z, et al. Genomic characterization of group A streptococci causing pharyngitis and invasive disease in Colorado, USA, June 2016‒April 2017. J Infect Dis. 2022;225:1841–51. 10.1093/infdis/jiab565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenciano SJ, Onukwube J, Spiller MW, Thomas A, Como-Sabetti K, Schaffner W, et al. Invasive group A streptococcal infections among people who inject drugs and people experiencing homelessness in the United States, 2010–2017. Clin Infect Dis. 2021;73:e3718–26. 10.1093/cid/ciaa787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalf B, Nanduri S, Chochua S, Li Y, Fleming-Dutra K, McGee L, et al. Cluster transmission drives invasive group A Streptococcus disease within the United States and is focused on communities experiencing disadvantage. J Infect Dis. 2022;226:546–53. 10.1093/infdis/jiac162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information on expansion of invasive group A Streptococcus M1UK lineage in active bacterial core surveillance, United States, 2019‒2021.


