Abstract
It is vital to ramp up crop production dramatically by 2050 due to the increasing global population and demand for food. However, with the climate change projections showing that droughts and heatwaves becoming common in much of the globe, there is a severe threat of a sharp decline in crop yields. Thus, developing crop varieties with inbuilt genetic tolerance to environmental stresses is urgently needed. Selective breeding based on genetic diversity is not keeping up with the growing demand for food and feed. However, the emergence of contemporary plant genetic engineering, genome-editing, and synthetic biology offer precise tools for developing crops that can sustain productivity under stress conditions. Here, we summarize the systems biology-level understanding of regulatory pathways involved in perception, signalling, and protective processes activated in response to unfavourable environmental conditions. The potential role of noncoding RNAs in the regulation of abiotic stress responses has also been highlighted. Further, examples of imparting abiotic stress tolerance by genetic engineering are discussed. Additionally, we provide perspectives on the rational design of abiotic stress tolerance through synthetic biology and list various bioparts that can be used to design synthetic gene circuits whose stress-protective functions can be switched on/off in response to environmental cues.
1. Introduction
Climate change is constantly altering the environment in which agricultural practices and crops evolved over the years [1]. Plant distribution and production are influenced by abiotic variables, which are natural components of the environment. Environmental conditions, drought, heat, cold, and high soil salinity, are considered abiotic stresses, and they confront crops in field conditions. These abiotic stressors restrict the global use of arable lands and negatively impact agricultural productivity [2]. Global food production must increase by 70% by 2050 to fulfil the demand imposed by the rising global population [3, 4]. Thus, the knowledge of mechanisms involved in plant abiotic stress sensing, signalling, and regulatory processes associated with adapting to stressful circumstances is crucial for global food security.
The cascades of regulatory pathways are activated in plants during an abiotic stress response, enabling them to react and adapt to their environment efficiently [5]. Understanding the stress-responsive molecular processes requires a better knowledge of the associated bioparts. Detailed molecular, genetic, and biochemical investigations have highlighted that complex and interconnected molecular networks are involved in stress perception/sensing, signalling, transcription, translation, RNA processing, protein processing, and epigenetic modifications [6] (Figure 1). These molecular responses elicited by different abiotic stresses can be shared or stress-specific [7]. Additionally, cross talk between diverse signalling and regulatory pathways lead to synergetic or antagonistic interactions critical for plant abiotic stress response. A comprehensive understanding of plants’ response to environmental stressors will aid in developing methods for imparting abiotic stress resistance in crops, thereby assuring plant survival and increased productivity.
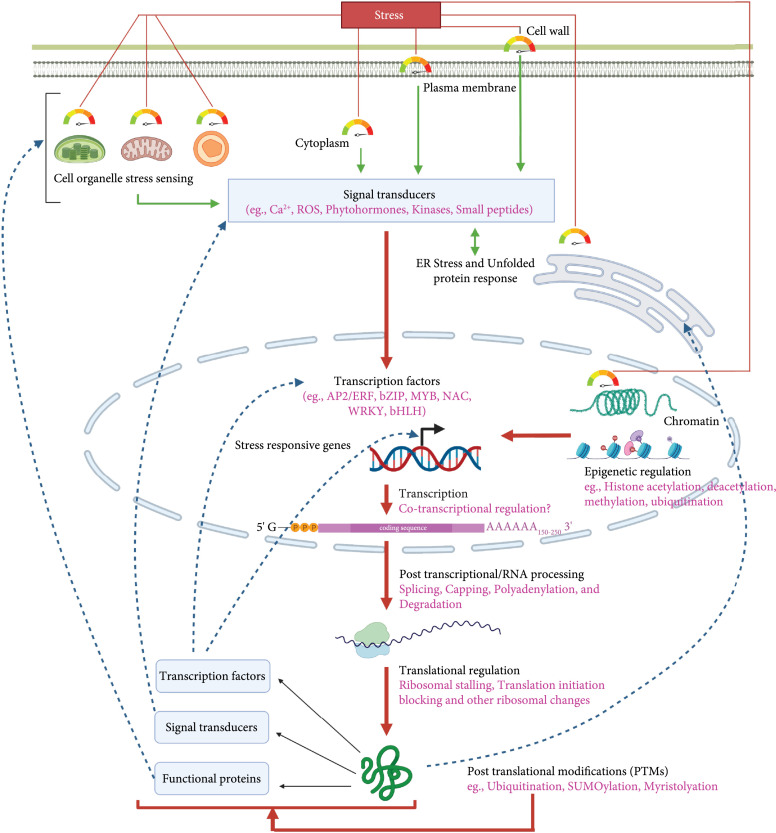
Figure 1.
An overview of abiotic stress response sensing, signal transduction, and regulation in plant cells. Plant cells can perceive/sense abiotic stress in several cellular compartments, and the signal transducers (e.g., secondary messengers such as Ca2+, ROS, phytohormones, kinases, and signalling (small) peptides) trigger the regulatory pathways involving transcription, posttranscription modifications, translation, posttranslational modifications, and epigenetic regulation. Multiple stress signals activate the stress-responsive transcription factors, which then regulate the stress-inducible gene expression cascade. Some stress-inducible genes code for functional proteins that directly impact role in stress tolerance; others encode regulatory proteins such as signal transducers.
Traditional breeding strategies are constrained by limited genetic diversity with higher productivity under stress and the finite efficient selection methodology. Using traditional breeding, few varieties have been introduced with enhanced abiotic stress tolerance in field conditions [8]. Genetic modification and engineering techniques are considered more precise and reliable for imparting stress tolerance in crops than conventional approaches [9, 10]. These techniques are centred on endogenous system enhancement by intervening at various phases of the abiotic stress response, including signal transducers, regulatory elements, transcription factors, sensors, effectors, and genes involved in metabolism. However, an abiotic stress response is a multigenic trait, and genetic modification approaches instead regulate individual components [11]. Therefore, there is a requirement for rational and efficient approaches for improving abiotic stress tolerance in crops.
The upstart field of synthetic biology (SynBio) can play a major role in overcoming these complex challenges [12, 13]. Plant synthetic biology is now trailing behind bacterial, yeast, and mammalian systems, where these methods are already altering basic research and the biotechnology industry [14–16]. The standardisation of genetic components and the development of modular cloning techniques in the plant sector were the initial steps towards broader synthetic biology technologies [17, 18]. Synthetic techniques for regulating gene expression and cellular processes, particularly chemically inducible systems, CRISPR/Cas9-based technologies, and other advancements in genome engineering, are critical for advancing plant synthetic biology in the future [12, 19–21].
The effective design of genetic circuits is a prerequisite for producing sentinel plants with desirable characteristics [18]. Plant genetic functions are complicated and influenced by various environmental signals, affecting synthetic gene circuit regulation. The genetic components should be able to act independently of the plant’s endogenous regulatory system. Furthermore, genetic circuits can be activated by external regulation, which potentially assists in switching the desired trait ON/OFF as and when required [22]. Control over synthetic genetic circuits can be further improved by introducing additional regulatory components (e.g., terminators and insulators). Thus, SynBio is a promising tool that can be widely utilised to develop plants with the ability to detect specific, combined, or multiple abiotic stressors displaying enhanced stress tolerance and overall increased crop productivity in the field.
Thus, this chapter will discuss the potential applications of synthetic biology approaches for improving abiotic stress tolerance in crops. In particular, we will focus on the current understanding of the molecular mechanisms involved in the regulation of the major abiotic stresses, namely, heat, cold, drought, and salinity in plants, followed by summarizing the validated and predicted bioparts which can be further explored for improving abiotic stress tolerance in crops by adopting synthetic biology.
2. Abiotic Stress Sensing/Perception
A primary stress sensing/perceiving mechanism translates the abiotic stress stimuli into a biological signal. In plants, the identification of abiotic stress sensors has been a challenging task due to the redundant nature of multiple sensors and the criteria used to define primary sensors. Addressing these limitations, four principal characteristics for defining a stress sensor have been proposed [23, 24]: (1) the true stress sensor must sense the abiotic stress by only perceiving the alterations in the environmental conditions, (2) the structure and activity of the cellular component must be directly altered in response to an abiotic stress stimulus, (3) the alterations in the cellular component must trigger a signal transduction pathway, and (4) the alterations lead to adaptive changes upon abiotic stress exposure. The identification of the abiotic stress sensing mechanisms has been a challenging task. Based on the outcomes of several studies adopting indirect approaches to identify abiotic stress sensors, putative stress sensors can be defined for the major abiotic stresses, namely: temperature, drought, and salt stress (Figure 2).
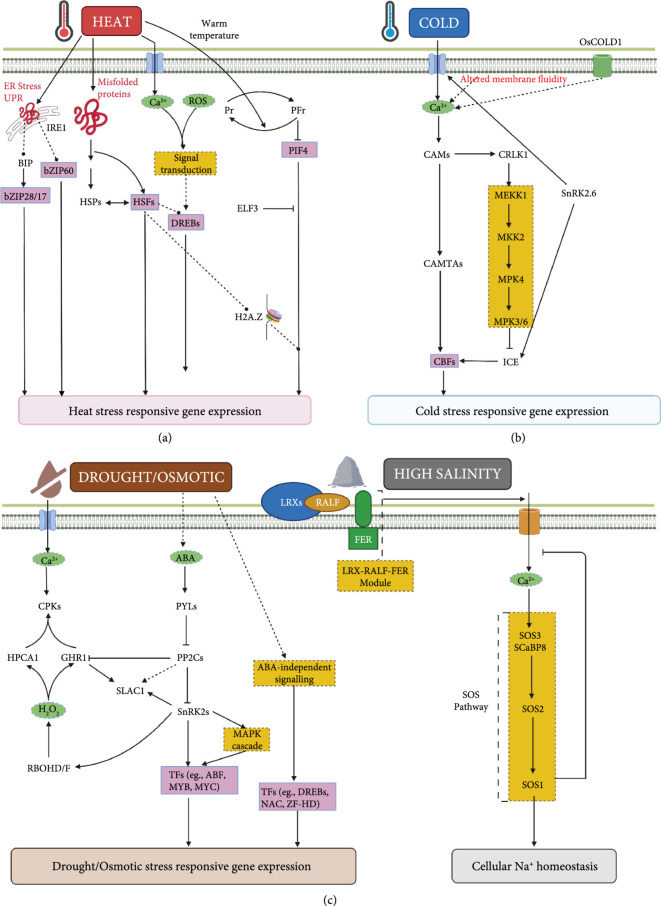
Figure 2.
A schematic representation of sensing and signalling cascades associated with various abiotic stresses. (a) Heat: heat induces misfolding of proteins that bind with HEAT SHOCK PROTEINS (HSPs), releasing HEAT SHOCK FACTORS (HSFs), which are then free to mediate heat-responsive gene expression. Misfolded proteins caused by heat stress can also activate the unfolded protein response (UPR) signalling pathway in the endoplasmic reticulum (ER). The ER-associated UPR signalling pathway has two arms, one involving two ER membrane–associated TFs, bZIP17, and bZIP28, and the other involving the RNA-splicing factor IRE1 and its target bZIP60 mRNA. When unfolded proteins attach to the luminal domain of IRE1, they dimerize (or oligomerize) and activate RNase activity, which cleaves bZIP60(u) mRNA, resulting in a spliced form of bZIP60. The spliced variant’s translation creates of active bZIP60 TF protein, which transport to the nucleus activates the stress-responsive genes. When BiP is separated from the ER-anchored transcription factors bZIP28/17, they are mobilised to the Golgi and delivered to the nucleus. bZIP28/17 binds to ER stress response elements in the nucleus to increase the transcription of UPR genes. Phytochrome-mediated signalling may detect warm temperatures. Heat-induced conversion of PhyB from the active Pfr form to the inactive Pr form frees PIF4 from Pfr inhibition, resulting in the activation of heat-responsive genes. When exposed to heat, ELF3 undergoes a phase change and aggregates, losing its capacity to suppress transcription of heat-responsive genes. Heat-induced replacement of H2A.Z by H2A in nucleosomes at specific genes enhances chromatin accessibility for transcription. Heat-induced Ca2+ spikes and ROS accumulation detect changes in membrane lipid fluidity. (b) Cold: calcium ion channels may contribute to cold-induced Ca2+ spikes by detecting altered membrane fluidity. In rice, OsCOLD1 is required for cold-induced Ca2+ increases. Cold stress activates the MEKK1–MKK2–MPK4 module (yellow box), linked to Ca2+ signalling via protein–protein interactions between CRLK1 and MEKK1. Additionally, cold induces the release of SnRK2.6, which results in the production of CBFs, which control the transcription of cold-responsive genes. Cold-induced Ca2+ signalling can directly activate the CBF regulon via the CAMTAs. (c) Drought and salinity both induce hyperosmotic stress, which is thought to alter the tension of the bilipid membrane, which may be recognised by Ca2+ channels leading to the induction of Ca2+ spikes. Both ABA-dependent and ABA-independent signalling are initiated in response to hyperosmotic stress. Additionally, stress-induced H2O2 is likely recognised by the Leucine-rich repeat receptor-like kinase (LRR-RLK) gene HPCA1 and, more particularly, by GUARD CELL HYDROGEN PEROXIDE RESISTANT1 (GHR1) in guard cells to produce Ca2+ signals via Ca2+ channel activation. This Ca2+ signal is sent to guard cells by protein kinases CPKs, which phosphorylate ABA-response effectors such as SLOW ANION CHANNEL-ASSOCIATED 1. (SLAC1), potentially enhancing stomatal closure in response to osmotic stress sensing. The signalling network demonstrates the critical functions of protein phosphorylation, calcium signalling, and ABA signalling in response to hyperosmotic stress. Salinity stress degrades the integrity of the cell wall, which may be detected by the LRX–RALF–FER module. Ca2+ stimulates the SOS3–SOS2–SOS1 pathway, which is responsible for maintaining cellular Na+ homeostasis.
2.1. Perception of Temperature Stress
Plants get exposed to temperature changes that vary in range, intensity, and duration daily and seasonal. Temperature changes affect enzyme kinetics, membrane fluidity, and protein folding makes it challenging to distinguish temperature-induced physiological changes from the actual sensing mechanism [23]. Plants respond in a variety of ways when temperatures rise over optimum levels. In Arabidopsis, exposure to warm ambient temperatures of up to 30°C induces changes in morphology and development known as thermo-morphogenesis, which may help avoid future heat stress [25]. Upon warm heat stress, temperature-dependent switching of phytochrome B (PhyB) from active to inactive state results in inhibition of phyB-mediated repression of the transcription factor PHYTOCHROME INTERACTING FACTOR-4 (PIF4) [26, 27]. This leads to the accumulation of PIF4, which promotes thermo-morphogenesis, such as promoting hypocotyl elongation [28, 29]. The activity of phyB as a thermo-sensor needs light activation [27]; thus, it is hypothesised that a separate and unknown thermo-sensing mechanism occurs in the root system. Recently, it was also proposed that warm ambient temperature sensing involves condensation of EARLY FLOWERING 3 (ELF3), which inhibits the transcriptional binding of ELF3 with its target genes [30]. Since ELF3 acts as a transcriptional repressor, its failure to bind to its target genes promotes their expression. The temperature responsiveness of the ELF3 was attributed to a polyglutamine (poly Q) repeat, entrenched within a prion domain (PrD). Moderate (20-38°C) to severe (>40°C) heat stress results in the accumulation of misfolded proteins, which induces the expression of HEAT SHOCK PROTEINS (HSPs) in an attempt to achieve cellular protein homeostasis [31, 32]. The binding of HSPs to misfolded proteins releases HEAT SHOCK FACTORS (HSFs), which then bind to the heat shock elements (HSEs) of their downstream targets, thereby regulating heat stress-responsive gene expression [33, 34].
Several potential sensors in the cold stress sensing pathway have been postulated, but their role as true cold sensors still requires verification [35]. The decrease in cell membrane fluidity after cold stress is commonly regarded as a key cold sensing mechanism [36]. DIACYLGLYCEROL KINASE (DAGK) activity, which occurs within seconds of cooling exposure, is linked to membrane fluidity [37]. Furthermore, the amount of desaturated fatty acids in the plasma membrane is related to its fluidity and is associated with FATTY ACID DESATURATION2 (FAD2) gene encoding the oleate desaturase. Mutations in FAD2 mutation reduce several physiological responses to cold stress [38]. In mammals, temperatures below optimum levels are sensed by TRANSIENT RECEPTOR POTENTIAL (TRP) ion channels [39, 40]. Ion channels orthologous to TRP are not known in plants. Cold-induced gene expression in plants is Ca2+ dependent [41, 42]. As a result, it is conceivable that ion channels (such as Ca2+ channels) and electrophysiological responses also play a role in low-temperature sensing in plants.
2.2. Perception of Drought Stress
Drought causes osmotic stress in plants; thus, reduction in osmotic potential is likely the earliest sign of water limiting conditions. Even though several basic drought sensors have been postulated, the intricacy of plant responses to water-limiting situations and the presence of potential multiple redundant osmo-sensors make it difficult to identify true osmo-sensors. Turgor loss caused by hyperosmotic stress modifies lateral tension on the bilipid membrane. Research indicates that increasing membrane tension in response to drought stress activates OSCA1, which encodes for a membrane hyperosmolality-gated calcium channel, resulting in the influx of Ca2+ ions [43]. In Arabidopsis, osca1 mutants, seedlings were grown under osmotic stress decreased primary root length and leaf area, indicating an enhanced susceptibility to osmotic stress. OSCA1 has a transmembrane domain similar to the Domain of Unknown Function221 (DUF221) domain present in the drought-responsive protein EARLY RESPONSIVE TO DEHYDRATION4 (ERD4; Ganie, Pani [44]). Additionally, CALCIUM PERMEABLE STRESS-GATED CATION CHANNEL1 (CSC1), an OSCA1 homolog (OSCA1.2), is depicted to be involved in osmotic stress sensing [45]. However, the precise function and subcellular localisation of CSC1A in plants are unclear.
2.3. Perception of Salt Stress
Upon exposure to salt stress, along with hyperosmotic stress, the plant also experiences ionic stress. While osmotic changes caused by salt stress may be detected using sensing mechanisms similar to those described above for drought stress, a different mechanism would be essential to detect the ionic stress. Ionic stress induces salt stress-specific Ca2+ signatures, which were recently investigated to understand the salt sensing mechanisms in plants [46]. It was proposed that Na+ might be detected by membrane lipid microdomains containing the sphingolipid Glycosyl Inositol Phosphoryl Ceramide (GIPC), which MOCA1 generates and binds Na+, resulting in salt-induced Ca2+ spikes. The channels that mediate the Ca2+ spikes are unknown; however, ANNEXINS1 (ANN1) and ANN4 are plausible candidates [47, 48].
The salt stress-triggered spike in intracellular Ca2+ is perceived by the classical Salt Overlay Sensitive (SOS) pathway [49]. The plant SOS pathway components: SOS3 and SOS3-LIKE CALCIUM BINDING PROTEIN8 (SCaBP8) acting as a Ca2+ sensor, SOS2 encoding a serine/threonine kinase and SOS2-LIKE PROTEIN KINASE (PKB5), and SOS1 encoding a plasma membrane Na+/H+ antiporter [50, 51]. Within a few seconds of salt stress exposure, the Ca2+ sensors of the SOS pathway are activated, which in turn activates SOS2. Through direct phosphorylation, the SOS3-SOS2 complex regulates SOS1 expression and function [52]. Salt stress-specific Ca2+ signatures regulate the SOS1 activation. The SOS3-SOS2-SOS1 cascade thereby initiates Na+ export to maintain cellular Na+ homeostasis.
3. Signalling Pathways
Stress perception or sensing triggers intricate response machinery involving a well-adjusted orchestration of signalling molecules, transcription factors, metabolic compounds/molecules, and other regulatory molecules. The sessile nature of plants has directed the evolution of highly robust, flexible, and sophisticated signalling networks which either utilise functionally redundant genes or multiple pathways existing and functioning parallelly. Thus, in this section, based on the available research findings, the molecular mechanisms involved in the regulation of abiotic stress signalling will be discussed in detail.
3.1. Calcium Signalling
Abiotic stress increases calcium ions into the cytosol beyond the threshold concentration inducing damage to the cell membrane and organelles [53]. Calcium homeostasis in the cell is then regulated by several ion channels, transporters, and intracellular organelles. The fluctuations in calcium concentration are stress/stimuli specific and spatially and temporally discrete signatures [54]. These calcium signatures are decrypted by calcium-binding proteins, namely, CALMODULIN (CaM), CAM-LIKE (CML), CALCINEURIN B-LIKE (CBL), CALCIUM-DEPENDENT PROTEIN KINASE (CDPK/CPK), and CALCIUM- AND CALMODULIN-DEPENDENT PROTEIN KINASE (CCaMK), which then bind to downstream effector molecules [55, 56]. CPKs, CBLs, and CMLs have been identified in protozoans and plants, but CaMs are extensively conserved across all eukaryotes [55]. The proteins associated with calcium signalling have characteristic EF-hands motif with distinct patterns. Most of the abovementioned calcium-binding proteins have four EF-hands, except CBL, which has three [57].
CPKs play a major regulatory function in the Ca2+-sensing protein families by binding directly to Ca2+ [58]. CDPK phosphorylate downstream protein targets in response to dynamic variations in cytoplasmic Ca2+ concentrations induced by hormones and abiotic stressors to control growth and stress responses [59, 60]. The significant role of CPKs in abiotic stress tolerance was validated via loss-of-function and gain-of-function experiments. CPK activity is verified by global expression studies, which reveal that many CPK members demonstrate stress-specific expression. Studies targeting abiotic stress tolerance in crops have identified CPKs as potential candidates [61]. For instance, in rice, drought tolerance was imparted by overexpression of OsCPK9 [62].
The activation of the SOS pathway exemplifies how Ca2+ signatures trigger particular intracellular Ca2+ sensing proteins, thereby regulating downstream transcription, translation, and further interactions in response to abiotic stress. Similarly, Ca2+ signals are transduced to the calmodulin-binding transcriptional activators Calmodulin-binding transcription activator (CAMTA)—CAMTA1, CAMTA2, and CAMTA3 stimulating CBF genes expression by binding to their promoters and thus mediate cold stress responses [63]. The stress response generated by specific calcium signatures is also governed by the colocalization and timely expression of calcium sensors and their putative partner and downstream proteins.
3.2. ROS-Mediated Signalling
Reactive oxygen species—ROS (O2-, H2O2, OH radical, and O2) —formerly considered as entirely harmful to plant life are produced in nearly all cell components during various enzymatic processes and upon exposure to abiotic stress [64]. Respiratory burst oxidase homologs (RBOHs), peroxidases, and oxalate oxidase are the proteins responsible for most ROS generation [65–67]. Elevated ROS levels are reduced to maintain cellular homoeostasis by the scavenging activity of enzymes, superoxide dismutase (SOD), catalase (CAT), glutathione reductase (GR), glutathione peroxidase (GPX), ascorbate peroxidase (APX), and peroxiredoxin (PRX) [68]. Plants also generate antioxidant compounds such as thiols, ascorbic acid, carotenes, and flavonoids to neutralise excess ROS [69]. ROS act as an effective signalling molecule both at the single-cell and cell-to-cell levels because of the mechanism involved in maintaining a fine balance of ROS in plant cells.
Under osmotic stress, ROS can increase unaided of stress-induced ABA accumulation; however, H2O2 generation is controlled by abscisic acid (ABA) signalling pathway via SNF1-related protein kinase 2 (SnRK2) and protein kinase-mediated activation of the NADPH oxidases (RbohD and RbohF) [70, 71]. Furthermore, extracellular H2O2 is likely sensed across the plant by the Leucine-rich repeat receptor-like kinase (LRR-RLK) gene HPCA1 and specifically in guard cells by GUARD CELL HYDROGEN PEROXIDE RESISTANT1 (GHR1) to induce Ca2+ signals via Ca2+ channel activation. This Ca2+ signal is transduced to guard cells by protein kinases CPKs which can phosphorylate ABA-response effectors, including SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1) [72, 73]. Thus, upon osmotic stress sensing, in addition to the ABA-dependent signalling module (discussed later in the section), stomatal closure is facilitated by an H2O2 HPCA1/GHR1–Ca2+–CPK module.
According to recent research, ROS build-up and Ca2+ generation both increase the induction of the other in response to abiotic stress exposure [74, 75]. Superoxide anions generated by RBOHD stimulates Ca2+ channels, activating the TWO PORE CHANNEL1 (TPC1, a vacuolar Ca2+ channel). TPC1 transfers Ca2+ accumulated in the vacuoles, which then activates RBOHD [76]. This feedback loop is potentially crucial for rapidly transmitting of stress-responsive ROS and Ca2+ waves (especially during salt stress) [77]. Abiotic stress such as drought and heat produces similar Ca2+ and ROS signatures across the plasma membrane [74, 75]; however, the elaborative mechanism is unclear.
3.3. Protein Kinase-Mediated Signalling
In eukaryotes and prokaryotes, protein phosphorylation acts as a ubiquitous signalling pathway. Protein kinases are divided into many groups based on their ability to phosphorylate specific amino acid residues. Experimentally validated two-component system (TCS) comprising of a histidine kinase (HK; signal sensor) and a nuclear effector response regulator (RR; transcription factor); play key roles in abiotic stress-induced signalling via a phosphorylation process [78]. The phosphoryl group is transferred from a conserved histidine (His) residue on the HK to a conserved aspartate (Asp) residue on the RR in the sensor-regulator coupling process between these two components [79]. There are also sophisticated TCSs with a multistep His-Asp phosphorelay in plants potentially providing an additional regulatory checkpoint.
The mitogen-activated protein kinase (MAPK) module is also part of the protein kinase family and is triggered by various stimuli including mitogens, phytohormones, and environmental stressors [80–82]. A typical MAPK module comprises three protein kinases that activate each other via relay phosphorylation. These protein kinases are a MAP kinase kinase kinase (MKKK or MEKK), a MAP kinase kinase (MKK or MEK), and a MAP kinase (MAPK or MPK). An active MEKK activates downstream MKK via phosphorylating two serine and/or threonine residues in its activation loop (S/T-X3 5-S/T) [83]. MKK activation leads to dual phosphorylation of a conserved motif, T-X-Y, in the activation loop of MAPK, thereby activating it. The activated MAPK then phosphorylates and changes the activity of the downstream target, allowing for downstream reactions [84].
3.4. Phytohormone-Mediated Signalling
Phytohormones are generated in extremely low quantities yet can control a wide range of cellular activities in plants [85]. They function as chemical messengers in higher plants, communicating cellular processes, and therefore, they perform critical functions in the abiotic-stress response, coordinating different signal transduction pathways [86]. Their essential functions of facilitating plant acclimation to the environments through plant growth, development, and nutrient allocation are thoroughly appreciated [87].
ABA, a key phytohormone, plays a vital role in regulating the abiotic stress response. It also functions in developmental processes like seed germination, seed dormancy, stomatal closure, and flowering [88–91]. In plants, the ABA signal transduction involves ABA receptors (PYR/PYL/RCAR), SnRK2 kinases (positive regulators), and type 2C protein phosphatases (PP2C) [92–95]. Under the lack of ABA conditions, PP2Cs bind to SnRK2s and block them from activating. Because inactive SnRK2s cannot phosphorylate downstream substrates, signal transduction does not proceed. In the presence of ABA, PYR/PYL/RCAR receptors bind to ABA and, through the interaction with PP2Cs, release SnRK2s. Autophosphorylation of the activation loop then activates the SnRK2s. The activated SnRK2s can phosphorylate substrate proteins such as ion channels, transcription factors, and enzymes (NADPH oxidases), triggering ABA responses. Other protein kinases control the activity of SnRK2s. SnRK2 may be activated by a Raf-like kinase (B3-MAPKKK) by activation loop phosphorylation; however, casein kinase 2 (CK2) can phosphorylate SnRK2’s carboxyl-terminal serine residues, increasing SnRK2-PP2C binding and resulting in inactivating SnRK2 [96]. ABA acts as a promoter of abiotic stress tolerance [97]. Exogenous administration of ABA or synthetic ABA mimics (i.e. ABA receptor agonists) is reported to elicit a stress response in plants, which improves their adaptability, showing the relevance of its activity under stress circumstances [98, 99].
Plants are also reported to produce ethylene in response to a variety of environmental stressors. Ethylene biosynthesis involves two steps. The first step is the transition of S-adenosyl-L-methionine (SAM) into 1-aminocyclopropane-1-carboxylic acid (ACC) via ACC-Synthase. In contrast, the second step involves the conversion of AAC to ethylene catalysed by ACC oxidase (ACO) [100]. Ethylene activates ER-located membrane protein ETHYLENE INSENSITIVE 2 (EIN2), which targets EIN3-BINDING F-BOX 1 (EBF1) mRNA to the cytoplasmic processing body (P-body) [101]. EIN2-mediated ethylene signalling also leads to translational inhibition of F-box binding proteins, EBF1, and EBF2 [101]. The function of ethylene as a signalling molecule is influenced by reactive oxygen species (ROS) quantity. Previous findings that ein2 and etr1 mutants had poor basal thermotolerance [102] and freezing tolerance [103], and ectopic overexpression of ERF74 improved heat tolerance and other abiotic stress tolerance [104], offer evidence that ethylene plays a significant role in abiotic stress response. Recently, EIN3-ERF95/ERF97-HSFA2 transcriptional cascade was shown to play an essential role in regulating basal thermotolerance and heat stress-responsive gene expression in plants [105].
Another phytohormone class, brassinosteroids (BRs) plays many plant growth and development roles. Plant-specific BR ligands bind directly to the membrane-bound LRR-RLK, BRASSINOSTEROID INSENSITIVE 1 (BRI1), and BRI1 ASSOCIATED RECEPTOR KINASE (BAK1), triggering signalling through cytoplasmic phosphorylation cascades including phosphorylation of serine /threonine phosphatase protein (BSU1) protein and proteasomal destruction of BIN2 (BRASSINOSTEROID INSENSITIVE 2) protein kinases [106–108]. Inactivation of BIN2 allows BRI1 EMS SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT 1 (BZR1) to gain entry in the nucleus and activate the expression of target genes [109]. BR interacts with other phytohormones in all of these signalling pathways. Plant growth and survival in drought stress are regulated by BR signalling via BIN2, which interacts with the autophagy system [110]. BR-pretreatment triggers the synthesis of ethylene under salinity [111], and therefore, signalling is increased by increasing the production of 1-ACS [112]. Upon exposure to high salinity, BR exogenous application also enhances the expression of ethylene signalling genes in cucumber, canola, and wheat [113–115]. Furthermore, the BR signal promotes ROS generation by NADPH oxidase, which activates MAPKs, causes protein phosphorylation, and targets genes involved in cellular defence [116].
Cytokinins are reported to perform a critical and multifaceted role in abiotic stress response. At the plasma membrane and ER, HISTIDINE KINASES (AHK2, AHK3, and AHK4/CRE1/WOL) detect cytokinins [117]. Recently, a small proportion of plasma membrane located AHKs can mediate the extracellular cytokinin signal has been reported [118]. Cytokinins bind to the CHASE domain of the receptor and stimulate intracellular histidine kinase (HK) activity, which leads to sensor autophosphorylation [119]. Cytokinin is often thought to regulate plant stress response negatively; however, this is not always firmly substantiated. Transgenic tobacco plants expressing the isopentenyl transferase (IPT, sourced from Agrobacterium tumfaciens), preceded by a stress-inducible promoter, showed improved tolerance to water-deficit conditions due to boosted cytokinin levels [120]. These findings were reproduced in transgenic rice [121] and peanut [122] plants utilising the same stress-induced cytokinin circuit. However, in contrast to the above findings, Arabidopsis ipt mutants with lower cytokinin levels are drought tolerant than the wild type [123]. Similarly, reduced cytokinin levels, obtained by constitutive or root-specific overproduction of cytokinin oxidase (CKX), the cytokinin-degrading enzyme, have a beneficial effect on drought tolerance [124, 125]. Furthermore, heat stress regulates the expression of several CK responsive genes [126], and exogenous cytokinins enhance plant heat tolerance [127]. The reduction of photosynthesis and chloroplast growth caused by heat stress is alleviated by exogenous administration of cytokinins and increased endogenous cytokinin levels.
Other phytohormones also perform regulatory roles in plants’ abiotic stress response [128, 129]. Salicylic acid (SA) is linked to the control of a variety of physiological activities, including photosynthesis, the formation of the antioxidant glycine betaine, proline metabolism, the plant-water relationship during stressful situations, and stress tolerance against abiotic stressors. The accumulation of SA causes reduced plant development, which reduces plant fitness. In response to stress, SA signalling is also reported to be linked with the accumulation of ROS. Similarly, gibberellins (GA) are phytohormones that control cell division and elongation, making them necessary for plant growth and development [130]. They also govern cellular redox equilibrium, which is essential in stress signalling via ROS signalling pathways. One of the most significant components involved in stress signalling is the DELLA protein which negatively regulates GA signalling [131]. The DELLA protein controls the production of ROS-scavenging proteins in plants, preventing oxidative damage and extending plant life and fitness [132, 133]. Another class of phytohormones—jasmonic acid (JAs) and methyl jasmonates (MeJAs)—have also been linked to a variety of physiological functions, including abiotic stress response [134]. Exogenous administration of JAs has been shown to improve plant stress resistance when tested on several plants under abiotic stressors such as salt, drought, and temperature (low/high) conditions.
3.5. G-Protein Coupled Receptors Mediated Signalling
The G protein (guanine nucleotide-binding protein) coupled receptors signalling module includes the Gα, Gβ, and Gγ subunits and is an evolutionarily conserved extracellular signal route [135]. In humans, the G-protein complex comprises 23 Gα, five Gβ, and 14 Gγ subunits [136]. Plant G proteins, on the other, include only one Gα subunit, three different Gα-like subunits (XLGs), one Gβ, and varying numbers of Gγ subunits depending on species [137]. Plant heterotrimeric G protein signal transduction pathway differs from animals. In contrast to animal G proteins, plant G proteins can self-activate without the help of GPCRs (G-protein-coupled receptors). For instance, Gα protein AtGPA1 can exchange GDP with GTP without the need for a GPCR, thereby activating it [138]. However, GTPase activity-accelerating proteins (GAPs) are involved in hydrolysing GTP and deactivating the Gα protein, AtGPA1 [139]. Additionally, activation of Gα or atypical Gα -like subunits in plants is ineffective in dissociating the G protein heterotrimer.
Studies have identified K+ and Ca2+ channels as key downstream effectors of heterotrimeric G protein. For instance, when plants are exposed to low temperatures, COLD1 interacts with a subunit of G protein to activate Ca2+ channels and boost G protein’s GTPase activity; in turn regulating the transcriptional expression of several stress-related genes, including OsAP2, OsDREB1A, OsDREB1B, and OsDREB1C [140]. Under drought stress, Gβ subunits are reported to upregulate NCED gene expression, favourably regulates ABA production. ABA-responsive genes (e.g., AtMPK6, AtVIP1, and AtMYB44) in agb1-2 Arabidopsis mutants are significantly upregulated after ABA or drought treatment [141]. Another subunit of the G-Protein module; Gα controls plant responses to salt stress potential by either attenuating cell cycle regulation in response to hyperosmotic stress or regulating cellular senescence in response to ionic stress [142].
Plant G protein activation/deactivation mechanisms are unclear, as are their direct effectors and connections with different transcriptional or protein networks. When comparing various plant lineages, there is also variation in the components and mechanisms of action. Thus, further studies targeting crop species are required to understand the G Proteins mediated abiotic stress signal transduction fully.
3.6. Signalling Peptides
Signalling peptides are short 5-10 or 40-100 amino acid long peptides, recently identified as abiotic stress-responsive signalling molecules [143, 144]. A major class of plant signalling peptides, CLAVATA3(CLV)/EMBRYO-SURROUNDING REGION RELATED (CLE) peptides, are ~12–14 amino acids long [145]. In Arabidopsis, CLE25 and CLE9 are involved in drought stress response [146, 147]. CLE25 is a transportable peptide that connects dehydration stress tolerance to abscisic acid- (ABA-) mediated tolerance by plausibly transmitting dehydration signals via CLE25–BAM modules from the roots to the leaves. This module acts via long-distance signalling to increase ABA accumulation by upregulating NCED3 expression [146]. By controlling stomatal closure, CLE9 helps to improve drought resistance by potentially interacting with the OST1 and anion channel protein SLAC1 protein [147]. In Arabidopsis, another member of this class, CLE-45, associated with the CLE45-STERILITY-REGULATING KINASE MEMBER1 (SKM1)/SMK2 receptor module promotes pollen tube development and results in effective seed setting in response to heat stress response in plants [148].
Another class of signalling peptides, RALF peptides, are 5 kDa cysteine-rich peptides involved in salt stress signalling [149]. The module involving LRX, FERONIA (FER), and RALF in Arabidopsis is suggested to detect high salinity-induced cell wall defects. LRX3/4/5 proteins have been found to bind with the peptide ligands RALF22 and RALF23, blocking their interaction with FER, a plasma membrane-localized receptor-like kinase (RLK) that potentially interacts with cell wall pectins. Salt stress disrupts these connections, leading to FER-dependent Ca2+ surge in the early elongation zone of roots [150]. The mechanism of how salt stress influences the interaction of LRXs with cell wall pectin and RALFs requires to be validated through biochemical experiments. Although few other signalling peptides have been discovered to coordinate plant abiotic stress, the molecular processes of this peptide signalling still need to be elucidated in detail.
4. Metabolic Pathways
Plants respond to diverse abiotic stimuli in different ways, and one of the most prevalent reactions is alterations in primary metabolism. ROS accumulation occurs under abiotic stress due to a disruption in PSII's electron transport chain [151]. Accumulation of ROS harms cells by causing membrane lipid peroxidation, and thus, plants have developed various methods to regulate lipid peroxidation, including the production/accumulation of numerous metabolites [152]. Similarly, the levels of secondary metabolites are also regulated in response to abiotic stress [153], but these changes are species- and stress-dependant.
4.1. Carbohydrate Metabolism
Plants are both producers and consumers of carbohydrate molecules. Photosynthesis produces a variety of sugars to maintain plant growth and development. They are essential regulators of abiotic stress responses in the cell, and their well-known function in numerous physiological processes. Tolerance to different environmental stresses is conferred by accumulating of soluble sugar molecules and sugar polyols and different levels of starch-sugar interconversion [154, 155]. These molecules stabilise cellular integrity (structure and osmotic potential) by serving as an osmolyte/osmoprotectant. These molecules also get interlinked into stress signalling pathways and assist in maintaining redox equilibrium [156, 157].
4.1.1. Sugar Metabolism
Sugars are the main products of photosynthesis, and they help plants grow and develop by providing energy or synthesising storage and structural components. Adverse environmental circumstances cause differential expression of genes involved in several processes such as photosynthesis, respiration, starch-sucrose metabolism, and cell cycle control, resulting in optimum carbon and energy use. The primary glucose sensor, HEXOKINASE 1 (HXK1), reacts to glucose concentrations under stress and regulates gene expression appropriately [158]. Because invertases are intimately linked to abiotic stress tolerance, glucose derived from invertase activity keeps HXK active, therefore, maintaining mitochondrial ROS equilibrium [156]. In plants, the HXK-independent glucose-sensing pathway has been documented; however, it is not well understood. Furthermore, some plants have fructokinases, which may play stress-induced sugar sensing [159]. Another sugar molecule, trehalose, which is present in low quantities in plants, show elevated levels upon abiotic stress exposure [160]. Endogenous trehalose levels are critical for maintaining development under stressful conditions. When given exogenously in small doses, trehalose reduces physiological and biochemical abnormalities caused by different abiotic stressors in plants by plausibly mediating ROS homeostasis and upregulating the stress-responsive genes in plants.
SnRK1 is another key mediator of stress signalling in abiotic stress reactions leading to the build-up of protective metabolites and defensive chemicals [161]. Stress can cause sugar imbalances, leading to ABA build-up and the activation of a special sugar signalling system. ABI4 is a key ABA sugar signalling downstream effector that regulates sugar sensitive gene expression. ABI4 also promotes the production of ANAC060, which inhibits the ABA signalling pathway in sucrose [162]. Carbohydrates like glucose and sucrose also influence auxin signalling and biosynthesis. The disaccharide sucrose interacts with the GA signalling system by stabilising the DELLA proteins, a negative regulator of GA signalling [131, 162].
4.1.2. Starch Metabolism
In response to abiotic stress, starch metabolism regulation can increase cellular carbohydrates or increase starch storage. Starch breakdown releases a range of sugars upon stress exposure, thereby boosting carbon flow into the hexose phosphate pool in a species-, tissue-, and stress-dependent manner [155]. In spinach, barley, and rice leaves, drought stress has been shown to suppress starch production and increase sugars [163–165]. Drought can cause starch-degrading enzymes to become active, increasing in sugars. Similarly, starch degradation is known to be triggered by cold stress [166]. Cold activation of certain β-amylase (BMY) isoforms has been frequently demonstrated based on expression and functional investigations [167]. In cereals, a mild drought postanthesis can activate important sucrose to starch conversion pathway associated enzymes, including Sucrose synthase (SuS), Starch branching enzymes (SBE), and AGPase [168]. Upon salinity stress exposure, a salt-tolerant rice cultivar, “Pokkali,” stored more starch in leaves than the sensitive cultivars examined, allowing the tolerant genotype to maintain photosynthesis [169]. In tomatoes, a similar effect was reported [170]. Furthermore, heat-tolerant tomato cultivars retained pollen starch content upon heat stress exposure, resulting in increased fertility in contrast to sensitive genotypes [171]. However, heat negatively regulates the activity of starch enzymes as the stress proceeds, resulting in a decrease in starch content. Thus, the starch-sugar interconversion in source and sink tissues plays a critical regulatory role in abiotic stress response. However, the current understanding of stress-induced carbohydrate alterations and the process behind these changes remains inadequate.
4.2. Amino Acid Metabolism
In plants subjected to abiotic stress, a general build-up of free amino acids has been documented [172]. Autophagy and ABA-triggered protein turnover may potentially lead to this rise in free amino acids levels. Plants can utilise amino acids as an alternate substrate for mitochondrial respiration in instances where there is a lack of glucose supply owing to a drop in photosynthesis rates in response to stress exposure. Plant fitness and, as a result, crop output is potentially affected by not just metabolic adaptations to stress but also by the proficiency of continuing growth processes.
Proline is the most prevalent water-soluble amino acid, and its metabolism in plants has been researched extensively in abiotic stress response. Proline accumulation can rise several folds under abiotic stress compared to nonstressed plants, indicating its involvement in abiotic stress regulation [173]. However, it is still unknown why proline accumulates during stressful situations. Proline has been found to accumulate in the cytosol in response to hyperosmotic stressors, suggesting that it can act as a suitable osmolyte, aiding plants in maintaining an optimal water balance [174]. Proline is also important for maintaining redox equilibrium in plants and preserving cellular integrity [175].
Another essential and effective solute is glycine betaine (GB). By maintaining an appropriate osmotic equilibrium, GB protects cells against the consequences of different stressors [176]. GB also helps to keep the quaternary structure of proteins stable. GB biosynthesis for stress tolerance induction is species/cultivar specific. Under diverse stressors, GB has a variety of protective benefits that are mediated by distinct metabolic processes. A considerable increase in GB accumulation was linked to the preservation of photosynthetic pigments and other biochemical characteristics that were beneficial in maintaining improved development in maize plants grown under osmotic stress [177].
4.3. Phenylpropanoid Metabolism
One of the most well-studied secondary metabolic pathways is the phenylpropanoid pathway [178]. The phenylpropanoid pathway involves enzymatic reactions: phenylalanine ammonia-lyase (PAL) catalyse phenylalanine deamination to trans-cinnamic acid, trans-cinnamic acid hydroxylation to 4-coumarate by cinnamic acid 4-hydroxylase (C4H) activity, and 4-coumarate conversion to 4-coumaroyl-CoA by 4-coumarate-CoA ligase (4CL). Various offshoots exist downstream of the main phenylpropanoid route, with the lignin and flavonoid pathways being two of the most important. Lignin deposition aids cell wall thickening during drought stress, allowing plants to retain cell turgor even under drought conditions. Upregulation of genes involved in lignin production (CAD, C4H, C3H, HCT, F5H, 4CL, CCR, COMT, and CCoAOMT) lead to the build-up of lignin, the secondary cell wall thickening, and thereby improving salt, cold, and drought stress tolerance in several plant species [179, 180]. Flavonoids operate as antioxidants, reducing the oxidative damage produced by ROS, which is triggered by abiotic stressors [181, 182]. In rice [183] and tobacco [184], treatment with flavonoids reduces oxidative damage and improves tolerance to salt and drought stress. Additionally, in rice [185], canola [186], and tobacco [184], flavonoid structural gene (CHS and DFR) overexpression enhances anthocyanins, and intermediate flavanol species production decreases ROS generation, thereby conferring salt stress tolerance. Furthermore, overexpression of F3H and DFR resulted in increased drought tolerance in alfalfa [187] and Arabidopsis [188]. Flavanols are also crucial for maintaining redox homeostasis and also enhancing pollen tube development and integrity during high-temperature exposure [189].
5. Regulatory Pathways
5.1. Transcriptional Regulation
The perception of abiotic stress and the signalling cascade that follows leads to the reprogramming of genome-wide transcription. Additional defensive strategies, such as osmotic adjustment, detoxification, repair of stress-induced damage, and attenuation of stress signalling, are triggered by the regulation of stress-responsive genes. Transcription factors belonging to the bZIP, bHLH, MYB, NAC, AP2/ERF, and WRKY families link stress-specific gene expression to upstream signalling [190]. A common strategy for imparting or improving abiotic stress tolerance in crops is to manipulate the expression of TFs genetically.
5.1.1. bZIP TFs
The bZIP TFs, one of the largest and evolutionary conserved TF family, can efficiently activate downstream gene expression upon abiotic stress exposure. These TFs are characterised by the bZIP domain comprising a basic domain and a leucine zipper domain [191]. The highly conserved DNA binding-basic region contains an invariant N-X7-R/K-X9 motif that usually binds to particular ACGT core nucleotide sequences such as A-box, C-box, G-box, and ABRE-elements. The basic region, site of a nuclear localization signal is composed of ∼16 amino acid residues. On the other hand, the less conserved leucine zipper domain comprises heptad repetitions of Leu or other hydrophobic amino acids that play a key role in dimerization and specific DNA sequence recognition. The role of bZIP TFs in stress-specific transcriptional regulation has been established through genetic screening studies in Arabidopsis. AtbZIP17 acts as a positive regulator of the salinity stress response by activating the expression of the salt stress-responsive genes ATHB-7 and SES1 [192], while AtbZIP24 was a negative regulator [193]. Furthermore, Arabidopsis salt tolerance is negatively controlled by AtbZIP62, which inhibits the transcription of SOS pathway genes [194].
bZIPs have been extensively studied in several crops, and they have been targeted using transgenic methods for imparting abiotic stress tolerance in crops. Overexpression of GmbZIP2 improved soybean tolerance to drought and salt stress by increasing stress-responsive genes (GmMYB48, GmWD40, GmDHN15, GmGST1, and GmLEA) expression [195]. In rice, OsbZIP05/OSBZ8 showed a higher transcription level in salt-tolerant cultivars than sensitive cultivars, suggesting a beneficial role of OsbZIP05/OSBZ8 in response to abiotic stress conditions [196]. Similarly, in response to drought stress, OsbZIP71 activates transcription of OsNHX1 and COR413-TM1 through binding to their promoters. The increased expression of these genes enhances drought tolerance in transgenic rice [197]. bZIP TFs can also regulate stress response by the regulation of plant metabolites. For instance, in soybean, GmbZIP44, GmbZIP62, and GmbZIP78 TFs, activate downstream genes ERF5, KIN1, CORl5A, and COR78 expression to control and stimulate the synthesis of proline which potentially enhances cold stress tolerance [198].
A small number of bZIP TF family members are also considered vital genes in UPR and the ER upon stress exposure. Plant cells have two subdivisions of the UPR signalling pathway: one comprises two ER membrane-associated TFs -bZIP17 and bZIP28, and the other involves the RNA-splicing factor IRE1 and its target bZIP60 mRNA [199, 200]. In one ER stress responsive UPR pathway, BiP (chaperone) is recruited to aid folding and protection of unfolded proteins, resulting in its separation from bZIP28. Two proteases cleave bZIP28 once it is transported to Golgi bodies. The cytosolic component of the protein is released as a result of this processing, and it subsequently translocates to the nucleus to activate downstream genes. Thus, bZIP28 acts both as a sensor and a signal transducer. Salinity stress activates bZIP17, which enhances the transcription of genes involved in salt stress tolerance and response [192]. In the other ER stress-responsive, UPR pathway IRE spliced the transmembrane domain of bZIP60. The spliced bZIP60 mRNA encodes a nucleus localized protein and induces UPR-related genes transcription. Recently, in maize, bZIP60 was reported to activate the production of an array of HSPs, thereby acting as a key connection between the UPR in the ER in addition to the nuclear/cytoplasmic heat shock system [201].
5.1.2. WRKY TFs
WRKY TFs, one of the largest plant-specific TF families [202], have a characteristic N-terminus located DNA-binding Domain (DBD) with an invariant heptad WRKYGQK motif and a C-terminus located zinc-binding motif. In the abiotic stress response, the various members of the WRKY TF family either interact with the ABA signalling pathway or ROS signalling pathway or act autonomously [203]. In tomatoes, SlWRKY81 improves drought tolerance by reducing H2O2 build-up and thus acting as a negative regulator of stomatal closure [204]. WRKY TFs usually regulate the expression of the target genes through their binding to the W-box cis-regulatory element [(T)TGAC(C/T)] to establish cellular homoeostasis. For instance, SbWRKY30 in sorghum, for example, controls the drought-responsive gene SbRD19 by binding to the W-box cis-elements and thereby protects plant cells from ROS-induced damage [205].
Functional characterisation of WRKY TF family members in different crop species highlights their potential role in regulating tolerance to single, combined, or multiple abiotic stress. GmWRKY49 expression was found to be different in salt-tolerant v/s salt-susceptible soybean genotypes [206]. Overexpressing GmWRKY49 in soybean and Arabidopsis conferred improved resistance to salt stress, with enhanced germination rate, survival rate, root length, and proline content. Further, in cucumbers, cold tolerance was enhanced by overexpressing WRKY46, which modulated the cold signalling system in an ABA-dependent manner [207]. Furthermore, transgenic rice expressing OsWRKY11 driven by the HSP101 promoter showed heat and drought tolerance [208].
5.1.3. MYB TFs
The largest TF family in plants is the MYB TFs, which are characterised by a conserved N-terminal MYB DNA-binding domain (DBD) repeat [209]. Each repetition (Rs) is made up of 52 amino acid residues folded into three -helices (R1, R2, R3), resulting a helix-turn-helix (HTH) structure. MYB transcription factors have one to four DNA-binding repeats in plants. The MYB TF family is classified into- R1-, R2R3-, R1R2R3-, and 4R-MYB TFs based on the position and number of repeats. The bulk of MYB proteins is members of the R2R3–MYB subfamily [210]. MYB transcription factors have been researched extensively and have been shown to regulate the production of secondary metabolites in plants. MYB proteins also perform various functions in the transcriptional regulation of abiotic stress response [211]. However, the regulation mechanism of MYB proteins upon abiotic stress exposure is yet unclear.
Functional characterisation studies have elucidated MYB TF to be potential candidates for imparting abiotic stress tolerance in crops. In Arabidopsis, AtMYB44 overexpression improves drought tolerance by increasing ABA sensitivity and ABA-induced stomatal closure, whereas atmyb44 knockout plants showed higher sensitivity to drought stress [212]. Furthermore, overexpression of AtMYB96 led to improved drought resistance by activating cuticular wax production, which prevented leaf surface water loss [213, 214]. Similar cuticular wax accumulation-based enhancement of drought tolerance observed in Camelina sativa plants are showing heterologous overexpression of AtMYB96 [215]. MYBs also have a role in salt stress response. Salt stress increases the expression of AtMYB20, and transgenic plants overexpressing AtMYB20 exhibited better salt tolerance. Suppression of AtMYB20, on the other hand, led to hypersensitivity to salt stress [216]. Furthermore, in response to heat stress, MYB30 inhibits the expression of ANN1 and ANN4 through binding directly to their promoters [217]. ANNs encode membrane Ca2+ transporter proteins that modulate cytosolic calcium signatures, and therefore, the regulation of ANN by MYB30 controls calcium signalling.
5.1.4. AP2/ERF TF
APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) TFs have emerged as key regulators of abiotic stress responses [218]. The distinguishing feature of these TFs is the presence of the APETALA2 (AP2)/Ethylene Responsive Element Binding Factor (EREB) DNA-binding domain comprising a conserved domain of 40–70 amino acids. APETALA2 (AP2), RELATED TO ABSCISIC ACID INSENSITIVE 3/VIVIPAROUS 1 (RAV), DEHYDRATION-RESPONSIVE ELEMENT BINDING proteins (DREBs) (subgroup A1–A6), and ETHYLENE RESPONSIVE FACTORS (ERFs) are the four main subfamilies of AP2/ERFs (subgroup V-X).
DREBs detect Dehydration-Responsive or C-Repeat Element (DRE/CRT) on stress-responsive genes with the A/GCCGAC core sequence to impart resistance to drought, cold, and heat abiotic stressors [219, 220]. Overexpression of DREB1s improves Arabidopsis plant tolerance to freezing stress. Drought and heat induce DREB2s, which upregulate the expression of DRE-containing drought-responsive genes, LEAs and heat-responsive genes, and heat chaperones [221]. Furthermore, members of the DREB-A4 family, e.g., HARDY (HRD), and the DREB-A6 family, e.g., ERF53, TG/RAP2.4A, and RAP2.4, favourably regulate salt and drought tolerance [222]. HRD overexpression in Arabidopsis or rice enhanced plant drought and salt tolerance dramatically [223]. DREBs are thought to control response to abiotic stress in plants through an ABA-independent mechanism. However, mounting data indicates that ABA-dependent stress responses are mediated via a number of stress-responsive AP2/ERFs. Furthermore, the AP2/ERF transcription factor RAV1 controls ABA sensitivity by directly interacting with SnRK2s, the essential kinases governing the ABF activity [96]. AP2/ERFs potentially regulate hormone sensitivity and gene expression by collaborating or antagonistically interacting with different hormone signalling components.
5.1.5. bHLH TFs
The bHLH family, extensively found in plants, is the second-largest TF family after the MYBs [224] are characterised by the occurrence of the bHLH domain comprising a DNA-binding N-terminal stretch of amino acids and HLH (helix loop helix) domain required for dimerization. More than half of the plant bHLHs identified contain a conserved HER motif (His5-Glu9-Arg13) which regulates DNA binding and transcriptional control of downstream genes. Although binding selectivity varies, bHLH TFs usually bind with E-box sequences (CANNTG), such as the G-box (CACGTG) cis-elements.
Abiotic stress response and tolerance regulation by bHLH TFs are highly conserved in plants [225]. The bHLH TFs control plant drought tolerance primarily via modulation of ABA sensitivity or regulation of stomata, leaf trichomes, and root hair production. ZmPTF1 promotes root growth and ABA synthesis in maize, which controls drought tolerance [226]. Controlling ROS balance through direct regulation of the expression of a few peroxidase genes is the significant way bHLH TFs contribute to salt tolerance. To improve Arabidopsis’ tolerance to salt stress, AtbHLH112 enhanced the expression of the POD and SOD genes while simultaneously decreasing the P5CDH and ProDH gene expression [227]. Another path for bHLH based enhancement of plant salt tolerance is through controlling the accumulation of secondary metabolites. A MAPK cascade regulates AtMYC2 in response to salt stress which binds the P5CS1 gene promoter (P5CS1 enzyme is the rate-limiting in proline biosynthesis). The promoter binding activates P5CS1 leading to enhanced proline biosynthesis and improved salt tolerance [228]. bHLH genes are also involved in plant cold tolerance, linked to increased proline accumulation, lower malondialdehyde levels, and less electrolyte leakage. AtICE1/AtbHLH116 interacts with the CBF promoter in Arabidopsis at low temperatures, affecting transcription, and the transgenic plants overexpressing AtICE1/AtbHLH116 exhibited increased cold tolerance [229]. Rice OrbHLH001, a homolog of ICE1, may improve transgenic Arabidopsis freezing stress resistance [230]. However, OrbHLH001, on the other hand, has a distinct function from ICE1 and is not reliant on the CBF/DREB1 cold-response pathway.
5.1.6. NAC TFs
NAC TF family name, NAC, comes from three genes (No Apical Meristem: NAM, Arabidopsis Transcription Activation Factor: ATAF, and Cup-Shaped Cotyledon: CUC), where the NAC domain was discovered for the first time [231]. The N-terminal DNA binding region of NAC transcription factors has a conserved NAC domain, while the C-terminal DNA binding region contains a regulatory domain. The C terminal region directs the interaction of NACs with diverse targets, including but not limited to lipoxygenase, DEAD/DEAH box helicase, PME or PMEIs, and Homeobox-related genes.
Across plant species, stress-responsive NACs function in a conserved manner. Abiotic stress activates the production of OsNAC5, OsNAC9, and OsNAC10, and overexpression of these TFs enhanced drought tolerance substantially [232–234]. Additionally, under stress circumstances, transgenic rice plants overexpressing OsNACs showed higher grain yields than wild-type control plants. In tomato, increased abiotic stress tolerance was observed plants with heterologous overexpression of Arabidopsis ANAC042/AtJUB1 [235, 236]. Furthermore, NACs potentially work in tandem with JA and ABA to regulate responses and tolerance to abiotic stress in plants. For instance, in Arabidopsis, ANAC096 regulates osmotic stress and dehydration responses by directly interacting with ABF2 and ABF4, key TFs of ABA signalling [237].
5.2. RNA Processing (Co- and Posttranscriptional Regulation)
RNA processing pathways such as splicing, capping, polyadenylation, and degradation are central to plant stress responses. Protein components, such as core spliceosomal proteins, proteins involved in spliceosome assembly, and splicing regulators, are largely conserved in plants. The failure of various elements of the RNA processing pathways is reported to significantly impair resistance to abiotic stresses while having no substantial impacts on plant function under stress-free conditions [238].
The spliceosome, a massive macromolecular complex of five ribonucleoprotein subcomplexes, removes introns during splicing (U snRNPs). U1snRNP-associated proteins, including U1-A and LUC7 zinc finger proteins, are required for abiotic stress tolerance. Arabidopsis mutants for spliceosomal protein U1A showed a salt stress hypersensitive phenotype in vitro and soil and increased in salt stress-induced reactive oxygen species (ROS) accumulation compared with wild-type. This mutant presented splicing defects associated with 5 SS recognition and transcripts encoding ROS detoxification enzymes, such as CSD1 and ACO1 [239].
Highly elevated expression of certain stress-responsive genes under stress conditions makes their transcripts particularly susceptible to RNA processing defects and, therefore, effective processing mechanisms are necessary to produce functional mature transcripts. Gene encoding HSFA2 has been shown to give rise to different splicing isoforms depending on the environmental temperature. HsfA2 contains two exons and a single intron. Under moderate heat, an additional exon within the intron is transcribed, introducing a pretermination stop codon (PTC). This HsfA2-II variant presents an incomplete DNA binding domain and is degraded through nonsense-mediated decay (NMD) [240]. Severe heat induces the formation of a different splice variant, HsfA2-III, that encodes a small, truncated protein due to a cryptic 5 SS in the intron. Interestingly, this isoform can bind the HsfA2 promoter to activate positive self-regulation.
Sm core protein SmEb, another spliceosome component, is involved in ABA signalling [241]. The expression of SmEb is upregulated after ABA treatment. SmEb enhances the HAB1.1 splicing variant while suppressing HAB1.2 through regulating the alternative splicing of the ABA signalling component HAB1. Contrary to HAB1.2, HAB1.1 overexpression can restore the ABA-hypersensitive phenotype of smeb mutants. ABA hypersensitivity of smeb mutants is reduced during seed germination when mutations in the transcription factors ABI3, ABI4, or ABI5. SmEb is therefore important for ABA-dependent regulation of seed germination and early seedling growth.
While RNA splicing has been regarded as a posttranscriptional process, recent evidence revealed that the intron could be cotranscriptionally spliced (cotranscriptional splicing, CTS). Cotranscriptional splicing has been reported to be a widespread phenomenon occurring at a high frequency in human cells [242]. It was recently reported that splicing is initiated during transcription for nearly all the introns in Arabidopsis [243, 244]. In addition, the processing of alternatively spliced introns was less efficient than constitutively spliced introns. Also, the cotranscriptional splicing was more efficient for protein-coding genes than for those in ncRNAs [243]. In Arabidopsis, native elongating transcript sequencing (NET-seq) revealed that phosphorylation of Polymerase II facilitates interaction with the spliceosome, influencing both constitutive and alternative splicing [245, 246]. Additional proteins involved in CTS include the RNA binding protein, HIGH OSMOTIC STRESS GENE EXPRESSION 5 (HOS5) and RS40 and RS41 (two arginine-rich splicing factors), which appear to promote efficient splicing of stress-related genes [247]. CTS efficiency is influenced by the expression level and the number of introns and exons within genes and chromatin modifications [248]. In accordance, a mutant in maize chromatin remodelling complex component ZmCHB101 showed defects in alternative splicing profiles under control and abiotic stress conditions [249]. Altogether, CTS is emerging as an important layer of regulation of alternative splicing, and its impact on abiotic stress responses is under investigation.
Additionally, abiotic stress also triggers alternative polyadenylation. In response to abiotic stress in sorghum, changes in polyadenylation result in the accumulation of nonfunctional transcripts and translational products [250]. Salt stress causes Arabidopsis to utilise alternate poly(A) sites in the coding and 5 untranslated regions of transcripts enriched for ABA signalling activities [251]. Plant heat tolerance is likewise adversely regulated by alternative polyadenylation in two rice landraces, Azucena and Tadukan98 [252].
5.3. Translational Regulation
Modulation of mRNA translation rates seems to be a conserved feature of cellular responses to diverse stress conditions [253]. The translation is one of the most energy-intensive processes making it the key cellular process to be downregulated under stress conditions. The immediate cellular stress responses occur at the translational apparatus, including ribosomal stalling, translation initiation blocking, and other ribosomal changes. Few reports have elucidated somewhat discordant protein and mRNA expression dependent on the duration, intensity, and type of abiotic stress [254–256]. Translational levels of downstream mORFs are affected by their sequence characteristics such as length, GC content, and minimum free energy that determines the structural stability of RNA secondary structures [254].
In Arabidopsis, exposure to heat stress shows similarity with an identified pattern in mammalian cells; induction of 5 ribosome pausing (ribosomal stalling) leads to degradation of mRNA preferentially targeting mRNA encoding HSP70/HSC [257]. This mRNA degradation likely contributes to plant acclimation and survival under chronic heat stress conditions due to XRN4 dysfunction, an exoribonuclease that degrades the mRNA downregulates tolerance of Arabidopsis plants to prolonged moderate-high temperature (35°C) exposure [258]. Conversely, the same exoribonuclease degrades mRNA encoding the key heat stress transcription factor, HSFA2, and without functional AtXRN4 gene, plants displayed enhanced survivability following short-term extreme heat stress (43.5°C) exposure [259], pointing to negative impact in plant response to acute heat stress caused by the heat-triggered mRNA. Furthermore, heat stress induces a block in translation initiation leading to preferential storage of mRNA encoding ribosomal protein (RPs) stress granules. These stored mRNAs are released during stress recovery, and their translation is restored by a process dependent on HSP101/CLB1 [260].
5.4. Posttranslational Regulation
Protein posttranslational modifications (PTMs), the covalent postsynthetic modifications influence the protein activities, cellular localization, and/or accumulation, thereby playing important functions in stress response regulation [261]. Various abiotic stress conditions are known to induce posttranslational modifications [262]. However, the functional significance of these modifications has not been addressed.
Rapid changes in plant growth behaviour in response to stress conditions are underpinned by the degradation of preexisting regulatory proteins and the synthesis of new ones. The Ubiquitin-Proteasome Pathway (UPP) plays a significant role in this function—allowing rapid response and adaptation of plants to ever-changing environmental cues. The proteolytic function of the UPP involves two discrete stages: ubiquitylation of the substrates and degradation of the tagged protein [263]. E3 Ubiquitin ligases catalyse the attachment of small protein modifier Ubiquitin to target selected proteins for degradation [264]. In consistency with the role of the UPP in plants stress response, a large group of E3 ligases are encoded in plant genomes. The specificity of the ubiquitin-proteasome degradation pathway can be attributed to at least the following proteins—E3 ubiquitin ligase and the matching substrate [265–267]. The stress-related proteins that are potential substrates of ubiquitylation include important TFs, epigenetic regulators, and enzymes involved in ABA signalling and metabolism.
Plant response to environmental stresses can be expedited by conjugating of Small Ubiquitin-like Modifiers (SUMO) to intracellular proteins. SUMO targets are the second most common kind of protein subjected to posttranslational changes. The SUMOylation of protein substrates is significantly enhanced by plant exposure to heat, cold, drought, and oxidative stresses. Short periods of exposure to abiotic stress conditions such as cold, heat, or oxidative stress (H2O2) trigger the sumoylation of a wide range of substrates [268–272]. Plant recovery from stress conditions is accompanied by rapid desumoylation of this massive pool of sumoylated proteins. SUMOylation is identified as the most significant posttranslational modification during abiotic stresses exposure in crops such as rice [273, 274], tomato [275], maize [276], and soybean [277]. For instance, in cotton, the Rice SUMO E3 LIGASE, OsSIZ1 overexpression enhanced water-deficit tolerance, improved net photosynthetic rate, as well as improved cotton growth and fibre yield [273].
Myristolyation is a protein-lipid modification that plays an essential role in membrane targeting [278]. The ubiquitous eukaryotic enzyme, N-myristoyltransferase, catalyses the myristoylation process. The N-myristoylation is the normal state of Arabidopsis phosphatase EGR2 that enables efficient interaction with and inhibition of SnRK2.6 protein kinase [279]. However, cold stress conditions lead to enhancement of EGR2 (Plasma membrane-localized clade-E growth-regulating 2) expression and weakening its interaction with the N-myristoyltransferase NMT1, resulting in the suppression of N-myristoylation of EGR2 [279]. Consequently, EGR2-mediated inhibition of SnRK2.6 activity is released, resulting its regulatory role in freezing tolerance. PTMs also influence the activity of several other proteins that are critical for stress tolerance but are not part of stress signalling. Osmotic stress conditions and ABA-dependent signalling activates SnRK2s protein kinases. The activated SnRK2s then phosphorylate TFs, transporters, and many enzymes, including enzymes associated with maintaining ROS homeostasis and biosynthesis of osmoprotectants/osmolytes [280]. For example, the phosphorylation of SLAC1 triggered by ABA results in stomatal closure due to reduced turgor pressure in guard cells [281]. Stress-induced accumulation of ROS, NOx (nitrogen oxides), and SO2 (sulphur dioxide) can trigger PTMs involving redox-based modifications such as oxidation, S-nitrosylation, nitration, glycations, S-glutathionylation, persulfidation, and carbonylation [282–284]. Nitric oxide-based modification is an important PTM involving cysteine residue modification of target proteins called S-nitrosylation [285]. In Arabidopsis, S-nitrosylation of PROTEIN ARGININE METHYLTRANSFERASE5 (PRMT5) enhances its methyltransferase activity essential for accurate splicing of pre-mRNAs upon stress exposure [286].
5.5. Epigenetic Processes in Abiotic Stress Tolerance
Epigenetic modifications lead to changes in specific chromatin domains to permit or repress transcription of a certain set of genes. Recently, it has been reported that a reversible epigenetic regulation of chromatin architecture can underpin genomic, transcriptional, and metabolic changes for different cellular processes [287–289]. Investigations on epigenetic control of abiotic stress response in plants have uncovered an additional layer of control exerted by epigenetic elements [290, 291]. The main epigenetic control elements include histone variants, histone modifications, chromatin remodelling, regulatory RNAs (e.g., noncoding RNA), and DNA methylation [292].
Histone acetylation is modulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). These two counteracting enzyme families regulate the acetylation state of lysine residues, particularly those within the N-terminal extensions of core histone proteins [293]. In Arabidopsis, salinity stress induces expression of histone acetyltransferase GCN5, and plants with mutations in this gene show enhanced salt stress sensitivity due to a deformation of cell wall integrity. GCN5 exerts its control via activation of a CTL1, a gene encoding a chitinase-like (CTL) protein through H3K9/K14 acetylation [294]. CTL1 plays a crucial role in cell walls biosynthesis and salt stress tolerance. In addition, gcn5 mutants exhibit severe heat stress sensitivity [295]. Hu et al. [295] propose that GCN5 mediates H3K9/K14ac enrichment in HsfA3 promoter and ULTRAVIOLET HYPERSENSITIVE6 genes. Transcriptome studies point towards the important role of HATs in the abiotic stress response of crop plants [296].
Histone deacetylases (HDACs) also play a significant role in drought and salt stress responses. The Arabidopsis genome contains 18 HDACs, and out of these, HDA9 and HDA19 enhance salt sensitivity [297–299], while HDA6, HD2C, and HD2D enhance salt tolerance [298, 300]. HDA19 modulates ABA signalling by regulating the expression level of ABA receptor genes [297].
Histone methyltransferases mediate the transfer of the methyl group to lysine residues of histones, whereas the removal is mediated by demethylases (HDM) [301, 302]. HDMs are classified into two groups, Lys-specific demethylases (LSD), and JumonjiC (JmjC) domain-containing protein family. JMJ15 demethylases have been reported to enhance salinity tolerance, while JMJ17 demethylases are reported to participate in water-deficit conditions [303, 304].
Histone ubiquitination is a reversible epigenetic modification that adds or removes the ubiquitin moiety from histones [305]. It has been shown that monoubiquitination of H2B is associated with abiotic stress response in rice and Arabidopsis. Enhanced drought tolerance has been observed in cotton plants overexpressing an Arabidopsis E3 ligase AtHUB2 [306]. In rice, the OsHUB2 overexpression unravelled that H2Bub1 (Histone H2B monoubiquitination) plays a role in positively modulating of ABA sensitivity and resistance to drought stress [307].
In plants, abiotic stress can induce the synthesis of histone variants that can modify the chromatin architecture by replacing their canonical forms [308]. Histone variant H2A.Z can exert positive or negative control on transcription depending upon its accumulation in gene bodies on the transcriptional start site [309]. The variant H2A.Z plays a significant role in regulating plant responses to cold and heat stress conditions [310].
Investigations on how histone modifiers are targeted to specific gene loci have revealed that some histone modifiers are targeted to specific chromatin sites via transcription factors. At the same time, in other cases, the targeting is achieved through lncRNAs [311, 312]. In the case of rice, INDETERMINATE SPIKELET1 (IDS1) and in Arabidopsis MYB96 are reported to recruit HDAC in response to high-salt and drought conditions, respectively [313, 314]. In the case of poplar (Populus trichocarpa), AREB1 acts as a recruiter of HAT in drought stress response [315]. Furthermore, in rice, OsbZIP46 acts as a recruiter of both an H2B ubiquitinase and deubiquitinase in response to water-deficit conditions [307].
DNA methylation, a conserved epigenetic mechanism, has also been reported to regulate abiotic stress response in plants. DNA methylation in plants mainly occurs by adding of a methyl group to the 5th position of the Cytosine’s pyrimidine ring (5mC: 5-methylcytosine) or the 6th position of the Adenine’s purine ring (6mA: N6-methyladenine). In plants, the RNA-directed DNA methylation (RdDM) pathway establishes de novo 5mC DNA methylation, and various DNA methyltransferases such as DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) maintain DNA methylation on the sequence contexts CG, CHG (H can be A, C, or T), and CHH [316, 317]. Diverse alteration of 5mC DNA methylation in response to different abiotic stress has been reported in crop species [318]. In response to heat stress, higher DNA methylation levels are reported in the anthers of a heat-tolerant cotton line compared to a heat-sensitive line [319, 320]. Contrary to this, drought-sensitive genotypes exhibit an increase in the DNA methylation levels in rice, whereas drought-tolerant genotypes exhibit hypomethylation [321].
Furthermore, DNA methylation of key abiotic stress regulatory genes is potentially associated with the stress response. For example, salt stress significantly decreases the 5mC levels at the promoter of TF GmMYB84 in soybean, which potentially upregulates its expression. GmMYB84 interacts with the cis-regulatory regions of K+ TRANSPORTER 1 (GmAKT1), thereby enhancing salt stress tolerance [322]. Similarly, in Arabidopsis, variation in ICE1 5mC methylation most likely contributes to phenotypic variability in freezing tolerance [323]. Compared to the 5mC DNA methylation, the regulation of abiotic stress by 6mA DNA methylation is reported by very few studies. In rice, heat and salt stress response is associated with increased 6mA levels, and the fold change is more significant in the tolerant cultivars [324]. It is, however, unknown whether heat or salt stress-induced 6mA upregulation is preserved across species.
Recent studies have addressed the role of selected DNA methylation-related genes in regulating the abiotic stress response. Arabidopsis plants lacking NRPD2, the shared second-largest component of PoI IV and Pol V, are highly susceptible to acute heat stress (42°C for 24–34 h). Additionally, the loss of function of RdDM components RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), DICER-LIKE 3 (DCL3), and ARGONAUTE 4 (AGO4) resulted in a significant reduction in basal thermotolerance [325]. Arabidopsis plants with a mutation in RDM16, which encodes a pre-mRNA splicing factor 3 involved in the RdDM pathway, are hypersensitive to salt stress [326]. Additionally, suppressing SlAGO4A, a critical component of the RdDM pathway, significantly increased salt and drought tolerance in tomatoes compared to nontransgenic and SlAGO4A overexpressing plants [327]. Further research utilising forward and reverse genetic techniques and genome-wide profiling is required to elucidate the functions of DNA methylation-related genes in abiotic stress response regulation.
6. Role of ncRNAs in Abiotic Stress Response
Based on their origin, biogenesis, and mode of action, noncoding RNAs (ncRNAs) have been divided into various groups [328]. The two most common types of ncRNA transcripts are housekeeping and regulatory ncRNAs. MicroRNAs (miRNAs), short interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs) are all examples of regulatory ncRNAs. These regulatory ncRNAs are transcribed from DNA but cannot be translated into proteins [329]. Differential expression of ncRNAs in response to unfavourable environmental conditions has been documented in several studies. ncRNAs can regulate gene expression in interconnected cellular networks, or they can respond to abiotic stress directly.
6.1. miRNAs
Research during the past decade has progressively emphasised the importance of miRNAs in plants’ responses to abiotic stress as a rapid, effective, and tissue-specific method for restoring normal plant function [330, 331]. While certain miRNAs are reported to be stress-specific, others are differentially expressed under different abiotic stresses. Additionally, certain abiotic stress-responsive miRNAs are evolutionarily conserved across plant species. For instance, in response to drought stress, upregulation of miR160, miR162, miR395, and miR827, whereas downregulation of miR166, miR172, miR397, miR827, and miR1432 in maize, rice, wheat, and Arabidopsis has been reported [332–335]. miRNAs have emerged as promising targets for improving plants’ ability to respond to and endure abiotic stress.
miRNAs are suggested to play a key role at the crossroads of complex stress-responsive gene regulatory networks. miRNAs target gene expression via mRNA cleavage, translational repression, and DNA methylation [336]. Thus, if a miRNA is upregulated in response to abiotic stress, it will downregulate the expression of its target genes. In contrast, if a miRNA is downregulated, it will accumulate the target mRNAs. Furthermore, the presence of complex regulatory networks involving stress-responsive TFs and miRNA has been suggested in plants [337]. In rice, the miR164 and NAC TF network are essential for regulating drought tolerance, as highlighted in drought-sensitive transgenic lines where miR164 overexpression displays associated suppression of target NAC TFs [338]. Several other TFs, including ARF, AP2, HD-ZIP III, HSF, TCP/PCF, NF-YA5, WRKY/GRF, MYB, NAC, and SPL, have been associated with miRNA:TF module regulating abiotic stress response [339]. These networks orchestrate abiotic stress signalling by altering various metabolic, signalling, molecular, and regulatory pathways. For example, miR159, miR160, miR164, and miR167 are potentially linked to ABA, GA, JA, SA, auxin, and other key phytohormone signalling pathways [340, 341].
6.2. lncRNAs
Plant lncRNAs have recently been identified for their plausible role in regulating of abiotic stress [342]. lncRNAs are not highly conserved, and their expression pattern is species-dependent, as a consequence, identifying conserved lncRNAs among different plant species is less likely. Furthermore, in response to abiotic stress, in contrast to protein-coding genes, lncRNAs display expression patterns highly specific to tissue and stage of development [343, 344]. This disparity associated with the differential number of identified lncRNAs across plants species may potentially be explained by variations in the techniques used to screen and identify lncRNAs. For instance, a report in Arabidopsis identified 1832 lncRNAs to be sensitive to drought, cold, salt, and ABA, but the technique only identified intergenic lncRNAs [345]. On the other hand, in Medicago truncatula 5634 lncRNAs responsive to drought were identified based on an approach to identify all classes of lncRNAs [346].
Mechanism of abiotic stress response regulation mediated by lncRNAs can be varied [342, 347]. Certain plant lncRNAs engage in the abiotic stress response by mimicking their targets by functioning as miRNA-targeted competitive endogenous RNAs (ceRNAs), preventing miRNA interactions with their targets. For instance, in B. rapa, two lncRNAs were identified as endogenous target mimics for miR164a in response to high temperature [348]. Certain lncRNAs use the RdDM (RNA-directed DNA methylation) silencing pathway to react to environmental stress [349]. An Arabidopsis long intergenic noncoding RNA induced by auxin—AUXIN REGULATED PROMOTER LOOP (APOLO)—was transcribed by RNA polymerases II and V [350]. APOLO’s dual transcription controls the formation of a chromatin loop that includes the promoter of its nearby gene PINOID (PID), a major regulator of polar auxin transport, causing its transcripts to be downregulated. APOLO may also target distant nonassociated loci by generating R-loops (DNA-RNA duplexes), or APOLO-mediated LIKE HETEROCHROMATIC PROTEIN 1 (LHP1) decoy may induce target locus transcription initiation, thus, altering local 3D chromatin conformation and coregulation of auxin-responsive genes [351]. Further research is warranted to unravel the apparent complexity of RdDM and its role activating stress-responsive genes.
Furthermore, in cold weather, COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs (COOLAIR) and COLD ASSISTED INTRONIC NON-CODING RNA (COLDAIR) lncRNAs are reported to assist blooming in plants [352, 353]. COOLAIR is an alternatively spliced natural antisense transcript lncRNA transcribed from the FLC (a regulator of flowering time) gene, whereas COLDAIR is transcribed from the first intron of the FLC gene. FLC encodes a MADS-box TF that represses floral induction [354]. COOLAIR and COLDAIR expression are reported to inhibit FLC expression in cold-stressed Arabidopsis through lncRNA-mediated chromatin changes (lncR2Epi). COOLAIR represses the FLC locus during the early stages of cold stress via modifying the FLC locus’ chromatin by decreasing the active histone mark H3K36me3 and increasing the repressive histone mark H3K27me3 during vernalization. COLDAIR represses FLC by engaging the Polycomb Repressive Complex 2 (PRC2), which assists in FLC locus chromatin modification by increasing H3K27me3 methylation. An additional polycomb-binding lncRNA, COLDWRAP, is also suggested to contribute to the stable suppression of FLC during Arabidopsisvernalization [355].
DROUGHT INDUCED lncRNA (DRIR) is a lncRNA that responds to high salt and water-deficit stress in Arabidopsis [356]. DRIR acts as a positive stress regulator and transcriptionally regulates several drought stress-responsive genes, including but not limited to signalling genes (ABI5, P5CS1, RD29A, and RD29B), aquaporin genes (NIP1, TIP4), annexin gene (ANNAT7), and TFs (NAC3, WARKY8). Further, drought and salinity tolerance was enhanced in DRIR overexpressed plants. Although lncRNAs have been shown to be abiotic stress-responsive, their functional characterisation is mostly missing.
6.3. circRNAs
Abiotic stress control by circRNAs in plants has received less attention so far. Stress-responsive circRNA expression in agricultural plants has only been documented in a few studies. circRNAs that respond to drought and heat stress have been detected in Arabidopsis and a few economically important crops [357, 358]. Available research indicates that circRNA potentially modulates gene expression by acting as miRNA sponges or regulating translation [359–361]. The exact mechanism of abiotic stress response regulation by circRNA in plants is unknown, and recent research suggests that circRNAs either function as miRNA sponges or limit the synthesis of sRNAs, thus, preserving stress-sensitive transcripts from gene silencing [362, 363]. In Arabidopsis, overexpression of circGORK (Guard cell outward-rectifying K+-channel) led to the activation of many ABA-sensitive genes in transgenic lines indicating a positive modulation of drought tolerance [358]. Furthermore, the overexpression of Vv-circATS1 sourced from grape resulted in increased cold tolerance in Arabidopsis, while its linear equivalent had no effect [364]. These studies offer practical methods and a framework for elucidating the function of circRNAs in stress response control.
7. Engineering Abiotic Stress Tolerance in Crops
As discussed in previous sections, a sensing/perception of abiotic stress by plants activating complex interconnected regulatory networks that govern stress-responsive gene expression to counteract the negative consequences of abiotic stress exposure, thereby maintaining cellular equilibrium. There is a crosstalk between various regulatory, metabolic, and developmental processes. As a result, while acting upstream in the signalling network may enhance tolerance to certain types of stress, there is an enhanced risk of causing undesirable pleiotropic consequences such as growth defects and developmental alterations. Targeting the expression of direct-action genes usually only improves performance in response to specific types of stress [365]. These considerations are especially important since plants in natural habitats are frequently exposed to multiple stressors, such as heat and drought, that can have synergistic, neutral, or even antagonistic effects.
Successful transfer of characteristics proved to be effective in the lab; the performance of the improved abiotic stress tolerance trait in the field has proven difficult. With several studies reporting the development of abiotic stress-tolerant plants, most research has focused on vegetative development stages such as leaf or root physiology instead of reproductive stages leading to seed formation, development, and maturation. A stress scenario during the flowering phase leads to hefty yield penalties [366, 367]; therefore, the plant development stage is also a key element. Additionally, genetically engineering plants to impart abiotic stress tolerance entails intervening at many levels of the abiotic stress response (Table 1).
Table 1.
List of representative stress-tolerant crops developed by genetic modification (GM). OE: overexpression; TSE: tissue-specific expression; COE: co-overexpression.
| Crop | GM | Promoter | Gene | Details of the gene | Source | Tolerance trait | Physiological benefit in response to stress | Reference |
|---|---|---|---|---|---|---|---|---|
| Barley | OE | 35S | AtAVP1 | H+-pyrophosphatase | Arabidopsis | Salt | In field, higher grain yield and higher shoot biomass | [431] |
| Rice | OE | Maize Ubi1 | AtHsp101 | Chaperone | Arabidopsis | Heat | Better growth performance in the recovery phase following the stress | [384] |
| Rice | OE | Maize Ubi1 | TaMBF1c | MBF TF | Wheat | Heat | Tolerance during both vegetative and reproductive stages | [369] |
| Rice | OE | Maize Ubi1 | OsRab7 | Small GTP-binding protein | Rice | Heat | Higher yield | [432] |
| Rice | OE | SNAC3 | NAC TF | Rice | Heat, drought | Better survival rate | [433] | |
| Rice | COE | Tissue-specific or stress-dependent | otsA and otsB (Fusion) | Trehalose biosynthetic genes | E. coli | Salt, cold, drought | Sustained plant growth, less photo-oxidative damage, and more favourable mineral balance | [386] |
| Rice | OE | ABA-inducible promoter | SAMDC | S-adenosylmethionine decarboxylase | Tritordeum | Salt | Increased seedling growth | [434] |
| Rice | OE | 35S | P5CS | Proline synthesis gene | Vigna aconitifolia | Salt | Wild type died after 10 days of salt stress whereas, even after 4 weeks of stress transgenics were capable of flowering and seed-setting | [435] |
| Rice | OE | 35S | PpENA1 | Na+ pumping ATPase | Physcomitrella patens | Salt | Higher biomass production | [436] |
| Rice | CE | 35S (pyramiding) | SaSRP3-1, SaVHAc1 | Salt Responsive Protein 3-1, Vacuolar H+-ATPase subunit c1 | Spartina alterniflora | Salt | Higher grain yield weight, and improved shoot and root growth | [437] |
| Rice | OE | 35S | Gly I, Gly II | Glyoxalase pathway genes | Brassica, Rice | Multiple stress | High shoot and root biomass | [438] |
| Rice | OE | Native promoter | SNAC1 | NAC TF | Rice | Drought | In field 22–34% higher seed setting (stress during reproductive stage) | [368] |
| Rice | OE | Stress-inducible rd29A promoter | AtDREB1A | AP2/ERF TF | Arabidopsis | Drought | Tolerant during both the vegetative and reproductive stages | [391] |
| Rice | TSE | Root specific RCc3 promoter | OsERF71 | AP2/ERF TF | Rice | Drought | 23% to 42% higher grain yield over WT or whole-body OE transgenic lines | [370] |
| Rice | OE | PGD1 | OsNAC14 | NAC TF | Rice | Drought | In field higher number of panicle and filling rate | [439] |
| Rice | OE | Rice Actin1P/LEA3-1P (stress inducible) | NPK1 | MAPKKK | Tobacco | Drought | In field higher seed setting rate | [374] |
| Rice | OE | Rice Actin1P/LEA3-1P (stress inducible) | SOS2 | Serine/threonine kinase | Arabidopsis | Drought | In field higher seed setting rate | [374] |
| Rice | OE | rice Actin1P/LEA3-1P (stress inducible) | AtDREB1A | AP2/ERF TF | Arabidopsis | Drought | In field higher seed setting rate | [374] |
| Rice | OE | Rice Actin1P/LEA3-1P (stress inducible) | Zat10 | C2H2-EAR zinc finger | Rice | Drought | In field higher seed setting rate | [374] |
| Rice | OE | Rice Actin1P/LEA3-1P (stress inducible) | LOS5 | Molybdenum cofactor sulfurase | Arabidopsis | Drought | In field higher seed setting rate | [374] |
| Rice | OE | Rice Actin1P | AtNHX1 | Na+/H+ antiporter | Arabidopsis | Drought | In field higher seed setting rate | [374] |
| Rice | OE | Maize Ubi1 | OsbZIP23 | bZIP TF | Rice | Salt, drought | Better growth | [440] |
| Rice | RNAi | Maize Ubi1 | SQS | Squalene synthase | Maize | Drought | 14–39% higher yield | [441] |
| Rice | OE | OsSKIPa | Rice homolog of human Ski-interacting protein | Rice | Drought | Resistant at both vegetative and reproductive stages | [442] | |
| Rice | OE | 2X35S | OsLEA3-2 | Late embryogenesis abundant (LEA) proteins | Rice | Salt, drought | Stronger growth performance, recovery after 20 days of drought stress | [443] |
| Rice | OE | 35S, drought inducible promoter | OsLEA3-1 | Late embryogenesis abundant (LEA) proteins | Rice | Drought | Higher relative yield (yield under drought stress treatment/yield under normal growth conditions) | [383] |
| Rice | OE | 35S | OsIF | Intermediate filament | Rice | Heat, salt | Maintenance of yield | [444] |
| Rice | OE | Maize Ubi1 | OsMYB55 | MYB TF | Rice | Heat | Lesser yield reduction | [445] |
| Rice | COE | 35S | OsbZIP46CA1, SAPK6 | bZIP TF, ABA-Activated Protein Kinase | Rice | Drought, cold, heat | Higher yield, biomass, spikelet number, and grain number | [446] |
| Rice | OE | 35S | RGB1 | Beta subunit of G protein | Rice | Heat, salt | Higher germination rate, root length, shoot length and plant height | [447] |
| Rice | OE | Promoter of Rca-a from Oryza meridionalis | Rubisco activase | Rubisco activase | Oryza australiensis | Heat | Higher yield (stress during vegetative stage) | [448] |
| Rice | COE | Rice Cab promoter | Rubisco, Rubisco activase | Rubisco, Rubisco activase | Rice, maize | Heat | Higher biomass and improved photosynthesis | [449] |
| Rice | OE | Actin | miRNA319 | miRNA | Rice | Cold | Increased leaf width and vein number, and better acclimation | [378] |
| Rice | OE | 35S | TERF2 | AP2/ERF TF | Tomato | Cold | Better survival rate | [450] |
| Rice | OE | COLD1 | Regulator of G-protein signalling | Rice | Cold | Enhanced chilling tolerance | [451] | |
| Rice | OE | Ubiquitin | MYBS3 | MYB TF | Rice | Cold | In field no yield penalty | [452] |
| Rice | OE | Ubiquitin | OsMYB3R-2 | MYB TF | Rice | Cold | Higher cold resistance, higher levels of proline | [453] |
| Rice & Tomato (small fruit species) | OE | 35S | ERECTA | LRR-RLK | Arabidopsis | Heat | Increased biomass and yield | [454] |
| Rice | OE | OsMADS57 | MADS-box TF | Rice | Cold | Maintenance rice tiller growth | [455] | |
| Wheat | OE | Ubiquitin | betA | Choline dehydrogenase | E. coli | Salt | In field higher grain yields, more tillers, and higher germination rates | [456] |
| Wheat | OE | pIND | HaHB4 (mutated) | Homeodomain-leucine zipper I family gene | Helianthus annus | Drought | In field higher spikelet numbers per spike, tillers per plant, and fertile florets per plant | [457] |
| Wheat | OE | Barley Dhn8s | TaHsfC2a | HSF TF | Wheat | Heat | Enhanced thermotolerance | [458] |
| Wheat | OE | Maize Ubi1 | TaFER-5B | Ferritin gene | Wheat | Heat | Enhanced stress tolerance | [459] |
| Wheat | OE | Hordein B1 promoter | HvSUT1 | Sucrose Transporter | Barley | Heat | Better performance for many yield-related traits and enhanced sucrose transport | [460] |
| Wheat | OE | Maize Ubi1 | EF-Tu | Elongation factor | Maize | Heat | Reduced heat injury to photosynthetic membranes (thylakoids), and enhanced rate of CO2 fixation | [461] |
| Wheat | OE | Drought inducible Barley HVA1s | Hsf6a | HSF TF | Wheat | Heat | Enhanced thermotolerance | [462] |
| Wheat | OE | 35S | AtWRKY30 | WRKY TF | Arabidopsis | Heat, drought | Improved biomass and photosynthesis | [463] |
| Maize | OE | Rd29A | ZmbZIP4 | bZIP TF | Maize | Salt, drought | Enhanced root development and ABA synthesis | [464] |
| Maize | CRISPR | Maize GOS2 | ARGOS8 | Negative regulator of ethylene signalling | Maize | Drought | In field increased grain yield by five bushels per acre (stress exposure during flowering) | [465] |
| Maize | OE | 35S | NPK1 | MAPKKK | Tobacco | Drought | No yield loss under stress | [375] |
| Maize | OE | Novel promoter | LOS5 | Molybdenum cofactor sulfurase | Arabidopsis | Drought | Increased root system development and biomass yield after re-watering | [466] |
| Maize | OE | Maize Ubi1 | OsMYB55 | MYB TF | Rice | Heat, drought | Higher plant biomass and reduced leaf damage | [467] |
| Cotton | OE | AhCMO | Choline monooxygenase | Atriplex hortensis | Salt | Higher seed yield | [385] | |
| Cotton | COE | AtNHX1, AtAVP1 | Na+/H+ antiporter, H+-pyrophosphatase | Arabidopsis | Salt, drought | 24% and 35% higher fibre yield under low-irrigation and dryland conditions, respectively | [468] | |
| Cotton | OE | 35S | AtAVP1 | H+-pyrophosphatase | Arabidopsis | Salt, drought | In field 20% higher yield under dry-land conditions | [469] |
| Cotton | OE | 35S | GhDof1 | DOF TF | Cotton | Salt, cold | Higher oil content and reduced protein in seeds | [470] |
| Tomato | CE | 35S | PgNHX1, AVP1 | Na+/H+ antiporter, H+-pyrophosphatase | Pennisetum glaucum, Arabidopsis | Salt | Better survival rate | [471] |
| Tomato | OE | 35S | SbNHXLP | Na+/H+ antiporter-like protein | Sorghum bicolor | Salt | Higher fruit yield | [381] |
| Tomato | OE | APX (cytosolic) | Ascorbate peroxidase | Pea | Heat | Enhanced tolerance to heat stress in lab and field | [382] | |
| Tomato | OE | 35S | SAMDC | S-adenosyl-l-methionine decarboxylase | Saccharomyces cerevisiae | Heat | Improved the efficiency of CO2 assimilation | [472] |
| Tomato | OE | 35S | ShDHN | Dehydrin | Wild tomato | Cold, drought | Better survival rate | [473] |
| Tomato | OE | Stress inducible AtRd29A | BoCRP1 | Novel cold-responsive protein1 | Brassica oleracea | Cold | Higher seed germination, increased root length, reduced membrane damage and increased accumulation of osmo-protectants | [474] |
| Tomato | OE | 35S | LeCOR413PM2 | Cold-regulated gene | Tomato | Cold | Reduced damage to cell membrane, accumulation of ROS, and photoinhibition of PSII, but also maintain high activity of antioxidant enzymes and content of osmotic regulators | [475] |
7.1. Regulatory Genes as Potential Bioparts for Imparting Abiotic Stress Tolerance
Modifying the expressions of regulatory genes, such as protein kinases, phosphatase, and transcription factors (TFs), is an efficient strategy to improve stress resistance in plants due to activating stress signals and coordinately regulating many downstream genes. Several TF families, bHLH, MYB, AP2/ERF, bZIP, DREB, NAC, and WRKY, operate as downstream integrators of regulatory networks, influencing the expression of stress-responsive genes in a combinatorial and amplificatory approach. Overexpression of SNAC1 (a NAC TF) in rice enhanced salt and drought resistance during the vegetative stage and significantly increased yield by 22-34% upon exposure to water-deficit conditions in the field during the reproductive stage [368]. Transgenic rice overexpressing MBF1c TF (isolated from wheat) imparted thermotolerance during vegetative and reproductive stages [369]. Furthermore, overexpression of an AP2/ERF TF—OsERF71—in rice under the control of a root-specific promoter conferred drought tolerance and enhanced yield by 23-42% upon drought exposure during the reproductive stage [370]. Selected studies utilising TFs for improving abiotic stress tolerance in crops are summarized in Table 1.
Protein kinases and phosphatases play critical roles in the adaptability and growth of plants [371]. Members of the protein kinase families associated with Ca2+ mediated signalling are potential candidates for imparting abiotic stress tolerance; for example, overexpression of OsCIPK03, OsCIPK12, and OsCIPK15 in rice demonstrated significantly increased cold, drought, and salt stress tolerance, respectively [372]. Protein kinases associated with MAPK cascades can be potentially targeted to enhance tolerance to abiotic stress in plants. OsMAPK5 overexpressing rice lines exhibited improved drought, salt, and cold stress tolerance [373]. Similarly, Xiao et al. [374] reported higher yield in transgenic rice overexpressing OsMAPK5 (NPK1) upon drought exposure in field conditions. In maize, overexpression of a tobacco MAPKKK (NPK1) enhanced drought resistance by potentially improving photosynthesis rates [375]. Among protein phosphatases gene candidates, overexpression of OsPP1a in rice increased resistance to high salinity by potentially upregulating the expression of SnRK1A as well as OsNAC5 and OsNAC6 in transgenic lines [376].
Another significant target class in this area is miRNAs, which regulate abiotic stress response by specifically targeting the expression of stress-responsive genes. In rice, salt and alkali responsive osa-MIR393 potentially target expression of stress-responsive genes, namely, phytosulfokine receptor precursor (LOC Os02g06260), putative transport inhibitor response protein (LOC Os05g41010), and oxidoreductase (LOC Os05g05800). Overexpression of osa-MIR393 in rice and Arabidopsis increased the salt and alkali sensitivity, thus, targeting the expression of osa-MIR393 might enhance stress tolerance [377]. Overexpression of osa-MIR319 in rice enhanced cold tolerance after acclimation of rice seedlings to suboptimal temperature [378]. Similarly, overexpression of osa-MIR319 in creeping bentgrass improved drought stress tolerance [379]. Furthermore, miR166 knockdown lines in rice (generated by using the Short Tandem Target Mimic system) exhibited a higher survival rate in response to drought stress. Additionally, these lines showed significantly higher spikelet fertility under drought exposure in field conditions [380]. Thus, by identifying and targeting specific miRNAs implicated in abiotic stress regulation using precise genome editing methods, abiotic stress tolerance in crops might be improved. In addition to the abovementioned bioparts, manipulation of bioparts associated with processes such as PTMs of signalling and regulatory elements, epigenetic modification, among others, provide promising ways to achieve generalised stress tolerance while maintaining a higher control over stress response.
7.2. Structural or Functional Genes as Potential Bioparts for Imparting Abiotic Stress Tolerance
Numerous approaches for enhancing abiotic stress resistance in crops have been explored, including overproduction of ion transporters, antioxidant enzymes, chaperones, protective proteins, and enzymes involved in metabolite synthesis. Abiotic stress tolerance can be enhanced or imparted in crops by targeting bioparts that restore cellular ionic and redox homeostasis. Under salt stress, ion transporters have been shown to help preserve cellular ion homoeostasis. For example, transgenic tomatoes overexpressing a Na+/H+ antiporter-like protein (NHXLP) isolated from Sorghum bicolor L. (SbNHXLP) displayed enhanced salt tolerance, decreased Na+, and increased K+ accumulation in root and floral tissues, indicating its involvement in maintaining ion homoeostasis [381]. Maintaining ROS homeostasis in plant cells by targeting the expression of enzymes in the antioxidant machinery is a prevalent approach for boosting plants’ tolerance to direct and indirect oxidative stress and thus improving plant performance under stress conditions. Increased thermotolerance to heat (40°C) was observed in transgenic tomato overexpressing cytosolic APX, most likely owing to the elimination of excessive damaging ROS (especially H2O2) [382].
LATE EMBRYO ABUNDANT (LEA) proteins and HSPs (also other chaperones) are produced in response to diverse abiotic stresses and are involved in protecting functional proteins. Overexpression of the OsLEA3-1 gene in rice significantly increased rice grain yields under water-deficit stress in field conditions [383]. Similarly, AtHSP101 overexpression in the rice cultivar Pusa basmati 1 increased thermotolerance, and transgenic lines had considerably improved growth performance during the recovery phase following stress [384].
Metabolic engineering of compatible solute accumulation is a widely adopted means of improving crop abiotic stress tolerance. Transgenic cotton lines overexpressing a choline monooxygenase from Atriplex hortensis (AhCMO) demonstrated significantly higher seed yield under salt stress and were more salt-resistant than wild-type cotton due to increased accumulation of glycine betaine, which protected the cell membrane and photosynthetic ability [385]. Accumulation of another compatible solute—Trehalose—in transgenic rice overexpressing a fusion gene made up of trehalose biosynthetic genes (otsA and otsB; sourced from E.coli) enhanced tolerance to multiple abiotic stress, and transgenic lines displayed sustained plant growth, less photo-oxidative damage, and more favourable mineral balance upon abiotic stress exposure [386].
Studies utilising similar approaches for generating transgenic lines displaying higher survival rates, higher yield, and improved abiotic stress tolerance during both vegetative and reproductive stages in economically important crops are summarized in Table 1.
7.3. cis-Regulatory Elements (Promoters) as Potential Bioparts for Imparting Abiotic Stress Tolerance
cis-Regulatory sequences are critical for gene regulation because they promote TF recruitment. These sequences may be a potential target for generating nucleotide-level alterations that can potentially increase crop tolerance to abiotic stress. For instance, in Arabidopsis, ANAC069 is reported to suppress the expression of various stress-responsive genes such as ROS-scavenging genes, thereby adversely regulating stress response mainly by interacting with C[A/G]CG[T/G] cis-elements [387]. A mutation in this core region might result in the inability of ANAC069 to regulate genes, thus, improving stress tolerance.
Furthermore, cis-regulatory sequences are predominantly found in the promoter region of genes. Their presence/absence/variation in position/sequence can affect the expression of the gene, resulting in induction, decrease, or even lack of expression. The most often utilised constitutively overexpressed promoters include the 35S promoter of the cauliflower mosaic virus (CaMV), and promoters are derived from plant actin and ubiquitin genes. The constitutive expression can have unforeseen effects on plant growth and development. It might result in overexpression of a specific transgene at the incorrect developmental stage or in tissues that are not ordinarily expressed. For example, constitutive overexpression of rice DREB1 (OsDREB1) in Arabidopsis exhibited resistance to salt, cold, and drought [388, 389]; however, these transgenic plants exhibited growth retardations. To address the issues related to constitutive overexpression, stress-inducible or tissue-specific promoters with low background expression or tissue-specific expression under normal growth conditions have been utilised. Stress-induced overexpression of the transcription factor AtDREB1A under the control of an Arabidopsis stress-inducible promoter, AtRD29A, resulted in increased abiotic stress tolerance in transgenic Arabidopsis and rice, addressing the problem of growth retardation [390, 391].
8. Synthetic Biology for Improving or Redesigning Abiotic Stress Tolerance in Plants
Synthetic biology uses fundamental engineering principles by employing naturally existing components for the rational design of new biological modules [392, 393]. Such an approach enables the de novo fabrication of new gene circuits and switches and restructuring signalling pathways [394]. This new discipline has already been successful in biotechnological manipulations of bacterial, yeast, and mammalian cell systems to develop new materials, production of chemicals via metabolic engineering, and the design of advanced molecular biology and medicinal applications [395–397]. Synthetic engineering of prokaryotic systems has dominated the field, which may be due to their simple cell structures and well-defined components, rendering them attractive systems for the operation of synthetic circuits. Designing synthetic circuits for eukaryotic systems has proven problematic because of their overwhelming complexity compared to prokaryotes. This issue is particularly relevant to the plant systems where despite rapid DNA assembly throughput, the poor transformation efficiency remains a bottleneck for further advances in synthetic biology [398, 399].
Recent advances in understanding systems biology of plant and application of bioinformatics tools have revealed a thorough understanding of regulatory components and processes operating at the cellular level. Such knowledge empowers the construction of synthetic modules using an infinite source of biological parts. Synthetic biology uses a top-down strategy to change what is already in nature. Synthetic biology diverges from the past biological approaches as it aims to build and develop something novel but still inspired by nature [400]. This is a reductionist approach in which biological systems are broken into building components from which new organisms may be constructed and produced. These biological parts (bioparts) may be put together to produce fundamental biological “devices,” which are the simplest assemblies capable of performing a specific function, such as a simple biological circuit (e.g., an on/off switch) or controlling the translation of a certain protein-coding sequence. These replaceable modules may then be integrated into “systems” within a cell or organism to execute a controlled (programmable) higher-level function, such as producing a metabolite in response to particular environmental inputs [401, 402].
8.1. Examples of Plant Biodesign Using Multiple Bioparts
The first application of the synthetic biology approach in plant systems includes the inventing of synthetic regulatory elements (synthetic promoters and cis-elements, synthetic short RNAs) and switches to modify of spatiotemporal gene expression and the engineering of signalling networks [403, 404]. The most common switch used in synthetic plant systems is “based on a mammalian steroid signalling pathway” from rats. The chemical-inducible promoter comprises a dexamethasone-inducible pOp/LhGR switch [405]. In the absence of the inducer, there is a low-level expression of the gene of interest, but in the presence of a steroid called dexamethasone (DEX), a high level of gene expression is induced. The DEX-inducible construct was designed by fusing the steroid-binding domain of the DEX-binding region of the glucocorticoid receptor (GR) of rat with the DNA binding domain of E. coli lac repressor and the activation domain of Gal4. A tissue-specific promoter is used to drive the expression of the GR-LhG4 transcription factor that remains in the inactive state in the cytoplasm. The presence of DEX allows fusion protein to move into the nucleus, leading to transcription activation of the reporter gene controlled by a synthetic promoter pOp6 that includes 35S core promoter and six copies of the LhG4 binding site. This approach led to the generation of a suite of transgenic driver lines. Following crossing, these lines can induce tissue-specific expression of the reporter gene in the resulting progeny.
Synthetic biologists have also been working to enhance the photosynthetic rate in plants for maximizing plant productivity [406]. Reengineering the primary photosynthetic enzyme, RuBisCO, is one of the approaches being used for this aim. Because RuBisCO is a key enzyme in all carbon-assimilating activities, an increase in RuBisCO activity will directly bear on plant productivity. There has been a successful report of replacing native RuBisCO in the chloroplasts of tobacco plants chloroplast with an alternative form derived from a cyanobacterial source [407]. Compared to the natural tobacco plant, the transformed one containing cyanobacterial RuBisCO had a higher carbon-assimilating efficiency. It has been shown that increased CO2 concentrations in the proximity of the RuBisCO enzyme enhances the efficiency of CO2 fixation and thus have a positive implication for plant productivity. The cyanobacteria and algae possess innovative carbon-concentrating mechanisms (CCMs) to enhance RuBisCO efficiency of CO2 fixation. Therefore, introducing these microautotrophic CCMs to enhance the photosynthetic ability of plants has shown to be an effective strategy [408, 409]. Plant species with C4 type of photosynthesis have evolved innovative CCM that depends on unique tissue anatomy and metabolic pathways. Engineering C4 type of CCMs into less efficient C3 plants via synthetic biology has been the key goal [410]. However, changing C4 photosynthesis into C3 plants remains challenging as much remains to be discovered regarding genes and gene functions underlying the C4 pathway.
Photorespiration is another natural process where RuBisCO binds oxygen resulting in the release of CO2 from plants. To address this issue, enzymes that can convert glycolate to glycol-aldehyde have been engineered using synthetic biology approach. Under in vitro conditions, these engineered enzymes and other endogenous enzymes could recycle glycolate straight to RuBP (Ribulose 1,5-bisphosphate) without releasing CO2 [411]. In vivo, several studies have shown that tailored carbon-conserving Calvin cycles may synthesise acetyl-CoA directly from C3 sugars without releasing CO2 [412]. Another successful example has been the reconstruction of the CETCH cycle, a synthetic pathway for carbon-fixation through a highly efficient reductive carboxylation process. A synthetic carbon-fixing pathway was engineered optimizing 17 enzymes from nine different organisms [413]. Hence, the above approaches have shown to be highly valuable in enhancing plant yield by reengineering the critical process of carbon fixation in plants.
The creation of plant sentinel biosensors is also a key growing application of synthetic biology in the plant area [12]. The design of the plant biosensors is based on cellular physiological mechanisms that occur naturally in plants or have been artificially developed. They offer various advantages, including better stability and enzyme activity, and being less expensive and time-consuming, making them ideal for application. The development of these whole-plant biosensors relies on a genetic circuit comprising genetically encoded components, which include promoters that respond to external inputs. The invention of such a biosensor for detecting 2,4,6-trinitrotoluene (TNT) explosive is one such successful example of a plant biosensor [414]. This approach was developed a TNT receptor from bacteria into the plant’s degreening gene circuit, triggering the activation of rapid chlorophyll breakdown whenever TNT is detected, resulting in a visible colour shift for simple identification.
Similarly, crops with efficient water utilisation capacity are being developed by controlling the production of ABA via an engineered PRY1 receptor with sensitivity to a fungicide Mandipropomid. The spray of this agrochemical on the plants under drought conditions activates ABA [415]. These modified plants require less water and are more resistant to stress conditions. Synthetic biologists have built a number of very sensitive and durable plant sensors that can track the system's transcriptional output. The stimulation of this mechanism by cytokinin signalling is one example [416]. Cytokinins are a class of plant hormones, playing an essential role in plant physiology and growth; thus, such monitoring sensors might improve understanding of the complex plant developmental mechanisms. Another essential plant hormone, auxin, has also been effectively monitored using synthetic circuits [417]. It has become simpler for plant physiologists to comprehend complicated physiological processes by offering help to construct and regulate responses by critical elements of a plant system, thanks to these sophisticated genetic circuits. Thus, the development of biosensors for the examination of molecular and physical cue perception and signalling relays will aid in the knowledge of stress regulation networks and, as a result, will make it easier to identify effective intervention areas for genetic engineering procedures.
In addition to providing proof of concept, the development of these synthetic techniques offers increasing prospects to reprogram plant development and metabolism and improve agricultural traits. Because these synthetic devices and platforms are plug-and-play, they have the potential to modulate gene expression patterns at numerous levels while not interfering with plant growth, development, or fitness [18, 403]. Even though most plant synthetic biology tools have been designed in well-studied model plants, Arabidopsis, and tobacco, and such engineered devices are being progressively adapted for crops.
8.2. Perspectives on Improving or Redesigning Abiotic Stress Tolerance through Biodesign
Combining synthetic biology approaches with existing genetic engineering practices offers enticing opportunities for the rational development of abiotic stress-resistant crops. Orthogonality, or the capacity of genetic components and circuits to operating independently of one another and the host’s regulatory activities, is crucial in synthetic biology [418]. Orthogonal components, most commonly bacterial, yeast, or plant viral sequences, can be borrowed in whole or in part from systems other than the intended host species. In this direction, many useful plant bioparts have been sourced from bacterial and viral plant pathogens [18]. Algal, fungal, or photosynthetic bacteria can offer regulatory components that confer sensitivity to stimuli that plants regularly encounter, such as light, drought, and temperature, resulting in plant-like responses in an orthogonal manner.
Synthetic promoters and TFs must be well designed and employed to control of gene expression using the endogenous plant cell transcriptional machinery [419, 420]. Synthetic promoter design entails inserting noncoding cis-regulatory areas (promoter motifs) into existing promoters, either alone or in combination [421, 422]. Promoter motifs are sourced from their native promoters. They are placed upstream of a core promoter that often comprises a TATA box to constitute a transcription preinitiation complex with RNA polymerase II and generic TFs. The minimal 35S promoter has been employed widely; however, several other minimal plant promoters have also been functionally validated. Synthetic promoters are designed and have been tested in a variety of plant species; the design includes synthetic constitutive, tissue-specific, bidirectional, biotic-, abiotic-, or chemical-inducible/responsive promoters [419, 423]. Such synthetic promoters can be used as critical components of genetic circuits for abiotic stress tolerance machinery regulation.
Transcription factors (TFs) play an essential role as regulators of stress-related gene expression (Section 5.1). Each TF has a DNA binding domain and an activation domain that engages the cell’s transcription machinery, reflecting a unique modular architecture. These domains can be constructed in a plug-and-play manner to generate synthetic TFs. By integrating distinct activation, repression, and DNA-binding domains from yeast, a library of synthetic transcription regulators was recently produced, which was subsequently employed for transient gene expression in tobacco leaves and the generation of stably transformed Arabidopsis plants [424]. A systematic technique of screening synthetic regulators increased the number of DNA parts tested significantly, and it was easily adaptable to evaluate diverse transcription regulators in different plant species. Synthetic TFs can also be utilised to regulate the expression of numerous genes in tissue-specific and environmentally sensitive ways [420]. Thus, synthetic TFs can be designed to target broad-spectrum stress-responsive genes and therefore can act as potential regulators of genetic circuits whose function can be switched on/off in response to environmental cues.
Synthetic transcription regulators can also be constructed to use epigenetics to control gene expression at the transcriptional level. In transgenic Arabidopsis overexpressing these synthetic regulators, Lee et al.[425] reported novel CRISPR-based toolbox for targeted controlling gene expression at transcriptional and epigenetic levels. The authors coupled dCas9, a variant of Cas9 protein lacking nuclease activity, with several regulatory domains for epigenetic regulation of endogenous FLOWERING LOCUS T (FT, master regulator of flowering) expression. Variations in FT expression and/or epigenetic state given by synthetic regulators were linked to changes in flowering time in transgenic Arabidopsis lines. Similarly, after further refinement, this strategy could be utilised to maximize the epigenetic control of stress-responsive genes or master stress regulatory genes that are either less epigenetically regulated or regulated by several epigenetic regulators.
Another recently pursued technique is applying riboswitch, a stem-loop RNA structure with regulatory and ligand-binding domains, to modulate mRNA stability and translation in plants [426]. A synthetic riboswitch library with theophylline acting as a ligand has been created for Arabidopsis to regulate endogenous and transgenes via posttranscriptional regulation [427]. These riboswitches regulate mRNA stability reliably and efficiently. There is potential to develop novel riboswitches to control gene expression provided their ligands have no influence on plant growth and development and/or are benign for the environment, including humans. This method might potentially be used to create bioparts that can control abiotic stress responses in a spatiotemporal manner.
Engineering the crassulacean acid metabolism (CAM) pathway in plants to improve water-use efficiency and thrive in water-scarce conditions like semiarid deserts is another potential approach for imparting stress tolerance [428]. Drought-tolerance methods observed in resurrection plants, known to tolerate severe drought, or evolutionarily distant creatures with a capacity to anhydrobiosis and survive extreme desiccation, might be engineered using more advanced synthetic biology approaches [429]. Similarly, a sophisticated method to develop a dynamic multilayer protective response regulated, maybe through the circadian clock [430], might permit optimal energy usage by synchronising the abiotic stress-protective response with the diurnal cycle.
9. Conclusions
A deep understanding of plant abiotic stress perception, signalling, and response processes is a prerequisite for crops that can maintain yield stability under stress conditions. In recent years, intensive “omics” based investigations have revealed activation of complex stress-responsive regulatory networks upon the perception of external stressors by plants. These interconnected networks involve various biological parts such as sensor proteins, enzymes, transcription factors, epigenetic, and posttranslational modifiers. While genetic engineering approaches involving the modulation of individual bio parts have yielded promising outcomes, the rational design of stress-responsive genetic circuits based on synthetic biology principles is urgently required.
Various genes that can be selected as bioparts for the assembly of stress-protective genetic circuits have been identified. Genes that play significant roles in plant abiotic stress tolerance as functionally tested in transgenic plants are summarized in table, providing a valuable database of bioparts for the rational design of synthetic circuits. These include genes encoding transcription factors, chaperones, stress sensor protein kinases, enzymes that can scavenge reactive oxygen species, and enzymes that promote the accumulation of protective osmolytes.
Besides protein-coding genes, several noncoding RNAs have emerged as potential bioparts to be deployed as tools for enhancing plant abiotic stress tolerance. Among noncoding RNAs, various miRNAs have been functionally validated for imparting abiotic stress tolerance. One of the main issues of concern is that while enhancing the ability of plants to tolerate stress conditions, the constitutive expression of protective genes can have detrimental effects on plant growth phenotype and yield. An on-demand protective functional module that gets activated in a spatially and temporally regulated manner can be fabricated using conditional or tissue-specific promoters is desirable to avoid undesirable consequences. Also, as mentioned, the use of transcription factors or other regulatory components upstream of the protective network enhances the risk of unwanted pleiotropic effects. Additional research is thus warranted to characterise the function of additional direct action candidate genes based on omics and comparative genomics approaches.
Another consideration for designing abiotic stress-tolerant crop plants is the combinatorial action of various stressors in field situations. As discussed, these combined stresses such as heat and drought can have synergistic negative consequences for plant growth and yield. Further, there is a gap in our knowledge regarding stress-responsive regulatory circuits required to protect reproductive development in plants. It has become clear that exposure to environmental stressors such as heat and drought during the flowering phase of plant growth can lead to male sterility, failure of fertilization, and seed set. Thus, a priority is to design protective gene regulatory circuits that are temporally activated in the target reproductive tissues in responding to abiotic stresses. Thus, one of the primary foci for future research in abiotic stress tolerance is to achieve a comprehensive picture of the stress vulnerability of plant reproductive cells. The use of single-cell transcriptomics will enable uncovering of potentially significant cell-specific stress-responsive genes that may be yet undiscovered.
Translation of synthetic biology approaches in plant systems depends upon efficient protocols for genetic transformation and control over the expression of inserted genes. Fortunately, for most of the major food crops such as wheat, corn, canola, soybean, and relatively facile methods of transgene addition are now available. The CRISPR technology for genome editing has already been implemented in major crop genera. The availability of diverse methodologies for genome manipulation and the engineering of synthetic circuits hold the potential for ushering in a new era for developing crop genotypes that can sustain yield stability in the face of multiple abiotic stress linked with climate challenges to agricultural productivity.
Acknowledgments
The research support from the ARC Discovery Grant DP0988972 and the University of Melbourne Postgraduate Research Scholarship is gratefully acknowledged.
Data Availability
Not applicable.
Authors’ Contributions
PB and MS conceived the concept. NL collected the literature and compiled the information. NL and MS wrote the article. MS and PB extensively edited the article.
References
- 1.Peng B., Guan K., Tang J., Ainsworth E. A., Asseng S., Bernacchi C. J., Cooper M., Delucia E. H., Elliott J. W., Ewert F., Grant R. F., Gustafson D. I., Hammer G. L., Jin Z., Jones J. W., Kimm H., Lawrence D. M., Li Y., Lombardozzi D. L., Marshall-Colon A., Messina C. D., Ort D. R., Schnable J. C., Vallejos C. E., Wu A., Yin X., and Zhou W., “Towards a multiscale crop modelling framework for climate change adaptation assessment,” Nature Plants, vol. 6, no. 4, pp. 338–348, 2020 [DOI] [PubMed] [Google Scholar]
- 2.He M., He C.-Q., and Ding N.-Z., “Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance,” Frontiers in Plant Science, vol. 9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mc Carthy U., Uysal I., Badia-Melis R., Mercier S., O'Donnell C., and Ktenioudaki A., “Global food security - Issues, challenges and technological solutions,” Trends in Food Science & Technology, vol. 77, pp. 11–20, 2018 [Google Scholar]
- 4.Yerlikaya B. A., Ömezli S., and Aydoğan N.. Climate Change Forecasting and Modeling for the Year of 2050, Springer, Environment, Climate, Plant and Vegetation Growth, 2020 [Google Scholar]
- 5.Zhu J.-K., “Abiotic stress signaling and responses in plants,” Cell, vol. 167, no. 2, pp. 313–324, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Zhu J., Gong Z., and Zhu J.-K., “Abiotic stress responses in plants,” Nature Reviews Genetics, vol. 23, pp. 104–119, 2021 [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., and Sonnewald U., “Differences and commonalities of plant responses to single and combined stresses,” The Plant Journal, vol. 90, no. 5, pp. 839–855, 2017 [DOI] [PubMed] [Google Scholar]
- 8.Gilliham M., Able J. A., and Roy S. J., “Translating knowledge about abiotic stress tolerance to breeding programmes,” The Plant Journal, vol. 90, no. 5, pp. 898–917, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Bhalla P. L., and Singh M. B., “Agrobacterium -mediated transformation of Brassica napus and Brassica oleracea,” Nature Protocols, vol. 3, no. 2, pp. 181–189, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Bhalla P. L., “Genetic engineering of wheat - current challenges and opportunities,” TRENDS in Biotechnology, vol. 24, no. 7, pp. 305–311, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Vij S., and Tyagi A. K., “Emerging trends in the functional genomics of the abiotic stress response in crop plants,” Plant Biotechnology Journal, vol. 5, no. 3, pp. 361–380, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Wurtzel E. T., Vickers C. E., Hanson A. D., Millar A. H., Cooper M., Voss-Fels K. P., Nikel P. I., and Erb T. J., “Revolutionizing agriculture with synthetic biology,” Nature Plants, vol. 5, no. 12, pp. 1207–1210, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Kassaw T. K., Donayre-Torres A. J., Antunes M. S., Morey K. J., and Medford J. I., “Engineering synthetic regulatory circuits in plants,” Plant Science, vol. 273, pp. 13–22, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Teixeira A. P., and Fussenegger M., “Engineering mammalian cells for disease diagnosis and treatment,” Current Opinion in Biotechnology, vol. 55, pp. 87–94, 2019 [DOI] [PubMed] [Google Scholar]
- 15.French K., “Harnessing synthetic biology for sustainable development,” Nature Sustainability, vol. 2, no. 4, pp. 250–252, 2019 [Google Scholar]
- 16.Boehm C. R., and Bock R., “Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism,” Plant Physiology, vol. 179, no. 3, pp. 794–802, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Cushman J. C., Borland A. M., and Liu Q., “Editorial: Systems biology and synthetic biology in relation to drought tolerance or avoidance in plants,” Frontiers in Plant Science, vol. 11, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy D. M., and Medford J. I., “Quantitative and predictive genetic parts for plant synthetic biology,” Frontiers in Plant Science, vol. 11, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jusiak B., Cleto S., Perez-Piñera P., and Lu T. K., “Engineering synthetic gene circuits in living cells with CRISPR technology,” Trends in Biotechnology, vol. 34, no. 7, pp. 535–547, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Bradley R. W., Buck M., and Wang B., “Recognizing and engineering digital-like logic gates and switches in gene regulatory networks,” Current Opinion in Microbiology, vol. 33, pp. 74–82, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Wright R. C., and Nemhauser J., “Plant synthetic biology: quantifying the “known unknowns” and discovering the “unknown unknowns”,” Plant Physiology, vol. 179, no. 3, pp. 885–893, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brophy J. A., and Voigt C. A., “Principles of genetic circuit design,” Nature Methods, vol. 11, no. 5, pp. 508–520, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai V. L., Gevaert K., and De Smet I., “Feeling the heat: searching for plant thermosensors,” Trends in Plant Science, vol. 24, no. 3, pp. 210–219, 2019 [DOI] [PubMed] [Google Scholar]
- 24.Lamers J., Van Der Meer T., and Testerink C., “How plants sense and respond to stressful environments,” Plant Physiology, vol. 182, no. 4, pp. 1624–1635, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casal J. J., and Balasubramanian S., “Thermomorphogenesis,” Annual Review of Plant Biology, vol. 70, no. 1, pp. 321–346, 2019 [DOI] [PubMed] [Google Scholar]
- 26.Jung J.-H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A. K., Box M. S., Charoensawan V., Cortijo S., Kumar M., Grant A., Locke J. C. W., Schäfer E., Jaeger K. E., and Wigge P. A., “Phytochromes function as thermosensors in Arabidopsis,” Science, vol. 354, no. 6314, pp. 886–889, 2016 [DOI] [PubMed] [Google Scholar]
- 27.Legris M., Klose C., Burgie E. S., Rojas C. C. R., Neme M., Hiltbrunner A., Wigge P. A., Schäfer E., Vierstra R. D., and Casal J. J., “Phytochrome B integrates light and temperature signals in Arabidopsis,” Science, vol. 354, no. 6314, pp. 897–900, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Arya H., Singh M. B., and Bhalla P. L., “Genomic and molecular analysis of conserved and unique features of soybean PIF4,” Scientific Reports, vol. 8, no. 1, pp. 1–11, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arya H., Singh M. B., and Bhalla P. L., “Overexpression ofPIF4affects plant morphology and accelerates reproductive phase transitions in soybean,” Food and Energy Security, vol. 10, no. 3, 2021 [Google Scholar]
- 30.Jung J.-H., Barbosa A. D., Hutin S., Kumita J. R., Gao M., Derwort D., Silva C. S., Lai X., Pierre E., Geng F., Kim S.-B., Baek S., Zubieta C., Jaeger K. E., and Wigge P. A., “A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis,” Nature, vol. 585, no. 7824, pp. 256–260, 2020 [DOI] [PubMed] [Google Scholar]
- 31.Angelos E., Ruberti C., Kim S. J., and Brandizzi F., “Maintaining the factory: the roles of the unfolded protein response in cellular homeostasis in plants,” The Plant Journal, vol. 90, no. 4, pp. 671–682, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohani N., Singh M. B., and Bhalla P., “RNA-seq highlights molecular events associated with impaired pollen-pistil interactions following short-term heat stress in Brassica napus,” Frontiers in Plant Science, vol. 11, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohama N., Sato H., Shinozaki K., and Yamaguchi-Shinozaki K., “Transcriptional regulatory network of plant heat stress response,” Trends in Plant Science, vol. 22, no. 1, pp. 53–65, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Lohani N., Golicz A. A., Singh M. B., and Bhalla P. L., “Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus,” Functional & Integrative Genomics, vol. 19, no. 3, pp. 515–531, 2019 [DOI] [PubMed] [Google Scholar]
- 35.Ding Y., Shi Y., and Yang S., “Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants,” The New Phytologist, vol. 222, no. 4, pp. 1690–1704, 2019 [DOI] [PubMed] [Google Scholar]
- 36.Örvar B. L., Sangwan V., Omann F., and Dhindsa R. S., “Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity,” The Plant Journal, vol. 23, no. 6, pp. 785–794, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Vaultier M.-N., Cantrel C., Vergnolle C., Justin A.-M., Demandre C., and Benhassaine-Kesri G., “Desaturase mutants reveal that membrane rigidification acts as a cold perception mechanism upstream of the diacylglycerol kinase pathway in Arabidopsis cells,” FEBS Letters, vol. 580, no. 17, pp. 4218–4223, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Martinière A., Shvedunova M., Thomson A. J. W., Evans N. H., Penfield S., Runions J., and McWatters H. G., “Homeostasis of plasma membrane viscosity in fluctuating temperatures,” The New Phytologist, vol. 192, no. 2, pp. 328–337, 2011 [DOI] [PubMed] [Google Scholar]
- 39.Bautista D. M., Siemens J., Glazer J. M., Tsuruda P. R., Basbaum A. I., Stucky C. L., Jordt S.-E., and Julius D., “The menthol receptor TRPM8 is the principal detector of environmental cold,” Nature, vol. 448, no. 7150, pp. 204–208, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Dhaka A., Murray A. N., Mathur J., Earley T. J., Petrus M. J., and Patapoutian A., “TRPM8 is required for cold sensation in mice,” Neuron, vol. 54, no. 3, pp. 371–378, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Knight H., Trewavas A. J., and Knight M. R., “Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation,” The Plant Cell, vol. 8, no. 3, pp. 489–503, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Wang C., Xue Y., Liu X., Chen S., and Song C., “Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance,” Nature Communications, vol. 10, no. 1, pp. 1–12, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., Zhang J., Theprungsirikul L., Shrift T., Krichilsky B., Johnson D. M., Swift G. B., He Y., Siedow J. N., and Pei Z.-M., “OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis,” Nature, vol. 514, no. 7522, pp. 367–371, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Ganie S. A., Pani D. R., and Mondal T. K., “Genome-wide analysis of DUF221 domain-containing gene family in Oryza species and identification of its salinity stress-responsive members in rice,” PLoS One, vol. 12, no. 8, article e0182469, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou C., Tian W., Kleist T., He K., Garcia V., and Bai F., “DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes,” Cell Research, vol. 24, no. 5, pp. 632–635, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Z., Zhou X., Tao M., Yuan F., Liu L., Wu F., Wu X., Xiang Y., Niu Y., Liu F., Li C., Ye R., Byeon B., Xue Y., Zhao H., Wang H.-N., Crawford B. M., Johnson D. M., Hu C., Pei C., Zhou W., Swift G. B., Zhang H., Vo-Dinh T., Hu Z., Siedow J. N., and Pei Z.-M., “Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx,” Nature, vol. 572, no. 7769, pp. 341–346, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Laohavisit A., Richards S. L., Shabala L., Chen C., Colaço R. D. D. R., Swarbreck S. M., Shaw E., Dark A., Shabala S., Shang Z., and Davies J. M., “Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1,” Plant Physiology, vol. 163, no. 1, pp. 253–262, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma L., Ye J., Yang Y., Lin H., Yue L., Luo J., Long Y., Fu H., Liu X., Zhang Y., Wang Y., Chen L., Kudla J., Wang Y., Han S., Song C.-P., and Guo Y., “The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress,” Developmental Cell, vol. 48, no. 5, pp. 697–709.e5, 2019 [DOI] [PubMed] [Google Scholar]
- 49.Zhu J.-K., “Genetic analysis of plant salt tolerance using Arabidopsis,” Plant Physiology, vol. 124, no. 3, pp. 941–948, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H., and Zhu J.-K., “Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid,” Plant Molecular Biology, vol. 50, no. 3, pp. 543–550, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Chinnusamy V., Schumaker K., and Zhu J. K., “Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants,” Journal of Experimental Botany, vol. 55, no. 395, pp. 225–236, 2004 [DOI] [PubMed] [Google Scholar]
- 52.Gong D., Guo Y., Schumaker K. S., and Zhu J.-K., “The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis,” Plant Physiology, vol. 134, no. 3, pp. 919–926, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy A. S., Ali G. S., Celesnik H., and Day I. S., “Coping with stresses: roles of calcium-and calcium/calmodulin-regulated gene expression,” The Plant Cell., vol. 23, no. 6, pp. 2010–2032, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martí M. C., Stancombe M. A., and Webb A. A., “Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis,” Plant Physiology, vol. 163, no. 2, pp. 625–634, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edel K. H., Marchadier E., Brownlee C., Kudla J., and Hetherington A. M., “The evolution of calcium-based signalling in plants,” Current Biology, vol. 27, no. 13, pp. R667–R679, 2017 [DOI] [PubMed] [Google Scholar]
- 56.McAinsh M. R., and Pittman J. K., “Shaping the calcium signature,” The New Phytologist, vol. 181, no. 2, pp. 275–294, 2009 [DOI] [PubMed] [Google Scholar]
- 57.Day I. S., Reddy V. S., Ali G. S., and Reddy A., “Analysis of EF-hand-containing proteins in Arabidopsis,” Genome Biology, vol. 3, no. 10, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yip Delormel T., and Boudsocq M., “Properties and functions of calcium-dependent protein kinases and their relatives inArabidopsis thaliana,” The New Phytologist, vol. 224, no. 2, pp. 585–604, 2019 [DOI] [PubMed] [Google Scholar]
- 59.Mohanta T. K., Mohanta N., Mohanta Y. K., and Bae H., “Genome-wide identification of calcium dependent protein kinase gene family in plant lineage shows presence of novel D-x-D and D-E-L motifs in EF-hand domain,” Frontiers in Plant Science, vol. 6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luan S., “The CBL-CIPK network in plant calcium signaling,” Trends in Plant Science, vol. 14, no. 1, pp. 37–42, 2009 [DOI] [PubMed] [Google Scholar]
- 61.Atif R. M., Shahid L., Waqas M., Ali B., Rashid M. A. R., and Azeem F., “Insights on calcium-dependent protein kinases (CPKs) signaling for abiotic stress tolerance in plants,” International Journal of Molecular Sciences, vol. 20, no. 21, p. 5298, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei S., Hu W., Deng X., Zhang Y., Liu X., and Zhao X., “A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility,” BMC Plant Biology, vol. 14, no. 1, pp. 1–13, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doherty C. J., Van Buskirk H. A., Myers S. J., and Thomashow M. F., “Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance,” The Plant Cell, vol. 21, no. 3, pp. 972–984, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mittler R., “ROS are good,” Trends in Plant Science, vol. 22, no. 1, pp. 11–19, 2017 [DOI] [PubMed] [Google Scholar]
- 65.Miller G., Schlauch K., Tam R., Cortes D., Torres M. A., Shulaev V., Dangl J. L., and Mittler R., “The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli,” Science Signaling, vol. 2, no. 84, p. ra45, 2009 [DOI] [PubMed] [Google Scholar]
- 66.Voothuluru P., and Sharp R. E., “Apoplastic hydrogen peroxide in the growth zone of the maize primary root under water stress. I. Increased levels are specific to the apical region of growth maintenance,” Journal of Experimental Botany, vol. 64, no. 5, pp. 1223–1233, 2013 [DOI] [PubMed] [Google Scholar]
- 67.Chapman J. M., Muhlemann J. K., Gayomba S. R., and Muday G. K., “RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses,” Chemical Research in Toxicology, vol. 32, no. 3, pp. 370–396, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mittler R., Vanderauwera S., Gollery M., and Van Breusegem F., “Reactive oxygen gene network of plants,” Trends in Plant Science, vol. 9, no. 10, pp. 490–498, 2004 [DOI] [PubMed] [Google Scholar]
- 69.Dumanović J., Nepovimova E., Natić M., Kuča K., and Jaćević V., “The significance of reactive oxygen species and antioxidant defense system in plants: a concise overview,” Frontiers in Plant Science, vol. 11, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meinhard M., and Grill E., “Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis,” Febs Letters, vol. 508, no. 3, pp. 443–446, 2001 [DOI] [PubMed] [Google Scholar]
- 71.Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., and Yamaguchi-Shinozaki K., “Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis,” Proceedings of the National Academy of Sciences of the United States of America, vol. 106, no. 41, pp. 17588–17593, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hua D., Wang C., He J., Liao H., Duan Y., and Zhu Z., “A plasma membrane receptor kinase, GHR1, mediates abscisic Acid- and hydrogen peroxide-regulated stomatal movement inArabidopsis,” The Plant Cell, vol. 24, no. 6, pp. 2546–2561, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu F., Chi Y., Jiang Z., Xu Y., Xie L., and Huang F., “Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in _Arabidopsis_,” Nature, vol. 578, no. 7796, pp. 577–581, 2020 [DOI] [PubMed] [Google Scholar]
- 74.Gilroy S., Suzuki N., Miller G., Choi W.-G., Toyota M., Devireddy A. R., and Mittler R., “A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling,” Trends in Plant Science, vol. 19, no. 10, pp. 623–630, 2014 [DOI] [PubMed] [Google Scholar]
- 75.Steinhorst L., and Kudla J., “Calcium and reactive oxygen species rule the waves of signaling,” Plant Physiology, vol. 163, no. 2, pp. 471–485, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans M. J., Choi W.-G., Gilroy S., and Morris R. J., “A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress,” Plant Physiology, vol. 171, no. 3, pp. 1771–1784, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi W.-G., Toyota M., Kim S.-H., Hilleary R., and Gilroy S., “Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants,” Proceedings of the National Academy of Sciences of the United States of America, vol. 111, no. 17, pp. 6497–6502, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urao T., Yamaguchi-Shinozaki K., and Shinozaki K., “Two-component systems in plant signal transduction,” Trends in Plant Science, vol. 5, no. 2, pp. 67–74, 2000 [DOI] [PubMed] [Google Scholar]
- 79.Suzuki T., Ishikawa K., Yamashino T., and Mizuno T., “An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2 in plants results in hypersensitiveness to cytokinin,” Plant & Cell Physiology, vol. 43, no. 1, pp. 123–129, 2002 [DOI] [PubMed] [Google Scholar]
- 80.Sinha A. K., Jaggi M., Raghuram B., and Tuteja N., “Mitogen-activated protein kinase signaling in plants under abiotic stress,” Plant Signaling & Behavior, vol. 6, no. 2, pp. 196–203, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Zelicourt A., Colcombet J., and Hirt H., “The role of MAPK modules and ABA during abiotic stress signaling,” Trends in Plant Science, vol. 21, no. 8, pp. 677–685, 2016 [DOI] [PubMed] [Google Scholar]
- 82.Xu J., and Zhang S., “Mitogen-activated protein kinase cascades in signaling plant growth and development,” Trends in Plant Science, vol. 20, no. 1, pp. 56–64, 2015 [DOI] [PubMed] [Google Scholar]
- 83.MAPK Group (Kazuya Ichimura et al.), Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., Kreis M., Zhang S., Hirt H., Wilson C., Heberle-Bors E., Ellis B. E., Morris P. C., Innes R. W., Ecker J. R., Scheel D., Klessig D. F., Machida Y., Mundy J., Ohashi Y., and Walker J. C., “Mitogen-activated protein kinase cascades in plants: a new nomenclature,” Trends in Plant Science, vol. 7, no. 7, pp. 301–308, 2002 [DOI] [PubMed] [Google Scholar]
- 84.Cristina M. S., Petersen M., and Mundy J., “Mitogen-activated protein kinase signaling in plants,” Annual Review of Plant Biology, vol. 61, no. 1, pp. 621–649, 2010 [DOI] [PubMed] [Google Scholar]
- 85.Voß U., Bishopp A., Farcot E., and Bennett M. J., “Modelling hormonal response and development,” Trends in Plant Science, vol. 19, no. 5, pp. 311–319, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bari R., and Jones J. D., “Role of plant hormones in plant defence responses,” Plant Molecular Biology, vol. 69, no. 4, pp. 473–488, 2009 [DOI] [PubMed] [Google Scholar]
- 87.Lymperopoulos P., Msanne J., and Rabara R., “Phytochrome and phytohormones: working in tandem for plant growth and development,” Frontiers in Plant Science, vol. 9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta A., Rico-Medina A., and Caño-Delgado A. I., “The physiology of plant responses to drought,” Science, vol. 368, no. 6488, pp. 266–269, 2020 [DOI] [PubMed] [Google Scholar]
- 89.Nakashima K., and Yamaguchi-Shinozaki K., “ABA signaling in stress-response and seed development,” Plant Cell Reports, vol. 32, no. 7, pp. 959–970, 2013 [DOI] [PubMed] [Google Scholar]
- 90.Wong C. E., Singh M. B., and Bhalla P. L., “Molecular processes underlying the floral transition in the soybean shoot apical meristem,” The Plant Journal, vol. 57, no. 5, pp. 832–845, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong C. E., Singh M. B., and Bhalla P. L., “The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process,” PLoS One, vol. 8, no. 6, article e65319, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., and Grill E., “Regulators of PP2C phosphatase activity function as abscisic acid sensors,” Science, vol. 324, no. 5930, pp. 1064–1068, 2009 [DOI] [PubMed] [Google Scholar]
- 93.Park S.-Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.-f. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J.-K., Schroeder J. I., Volkman B. F., and Cutler S. R., “Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins,” Science, vol. 324, no. 5930, pp. 1068–1071, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belin C., de Franco P.-O., Bourbousse C., Chaignepain S., Schmitter J.-M., and Vavasseur A., “Identification of features regulating OST1 kinase activity and OST1 function in guard cells,” Plant Physiology, vol. 141, no. 4, pp. 1316–1327, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonzalez-Guzman M., Pizzio G. A., Antoni R., Vera-Sirera F., Merilo E., and Bassel G. W., “Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid,” The Plant Cell, vol. 24, no. 6, pp. 2483–2496, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maszkowska J., Szymańska K. P., Kasztelan A., Krzywińska E., Sztatelman O., and Dobrowolska G., “The multifaceted regulation of SnRK2 kinases,” Cell, vol. 10, no. 9, p. 2180, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tuteja N., “Abscisic acid and abiotic stress signaling,” Plant Signaling & Behavior, vol. 2, no. 3, pp. 135–138, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Du Y.-L., Wang Z.-Y., Fan J.-W., Turner N. C., He J., and Wang T., “Exogenous abscisic acid reduces water loss and improves antioxidant defence, desiccation tolerance and transpiration efficiency in two spring wheat cultivars subjected to a soil water deficit,” Functional Plant Biology, vol. 40, no. 5, pp. 494–506, 2013 [DOI] [PubMed] [Google Scholar]
- 99.Vaidya A. S., Helander J. D. M., Peterson F. C., Elzinga D., Dejonghe W., Kaundal A., Park S.-Y., Xing Z., Mega R., Takeuchi J., Khanderahoo B., Bishay S., Volkman B. F., Todoroki Y., Okamoto M., and Cutler S. R., “Dynamic control of plant water use using designed ABA receptor agonists,” Science, vol. 366, no. 6464, 2019 [DOI] [PubMed] [Google Scholar]
- 100.Wang K. L.-C., Li H., and Ecker J. R., “Ethylene biosynthesis and signaling networks,” The Plant Cell, vol. 14, Supplement 1, pp. S131–S151, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., and Guo H., “EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis,” Cell, vol. 163, no. 3, pp. 670–683, 2015 [DOI] [PubMed] [Google Scholar]
- 102.Larkindale J., Hall J. D., Knight M. R., and Vierling E., “Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance,” Plant Physiology, vol. 138, no. 2, pp. 882–897, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Popov V., Deryabin A., Astakhova N., Antipina O., Suvorova T., and Alieva G., “Ethylene-insensitive Arabidopsis mutants etr1-1 and ein2-1 have a decreased freezing tolerance,” Doklady Biochemistry and Biophysics, vol. 487, pp. 269–271, 2019 [DOI] [PubMed] [Google Scholar]
- 104.Yao Y., He R. J., Xie Q. L., Zhao X. H., Deng X. M., He J. B., Song L., He J., Marchant A., Chen X.-Y., and Wu A.-M., “Ethylene response factor 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis,” The New Phytologist, vol. 213, no. 4, pp. 1667–1681, 2017 [DOI] [PubMed] [Google Scholar]
- 105.Huang J., Zhao X., Bürger M., Wang Y., and Chory J., “Two interacting ethylene response factors regulate heat stress response,” The Plant Cell, vol. 33, no. 2, pp. 338–357, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., and Walker J. C., “BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling,” Cell, vol. 110, no. 2, pp. 213–222, 2002 [DOI] [PubMed] [Google Scholar]
- 107.Friedrichsen D. M., Joazeiro C. A., Li J., Hunter T., and Chory J., “Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase,” Plant Physiology, vol. 123, no. 4, pp. 1247–1256, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ryu H., Kim K., Cho H., and Hwang I., “Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling,” Molecules and Cells, vol. 29, no. 3, pp. 291–296, 2010 [DOI] [PubMed] [Google Scholar]
- 109.He J.-X., Gendron J. M., Yang Y., Li J., and Wang Z.-Y., “The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no. 15, pp. 10185–10190, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nolan T. M., Brennan B., Yang M., Chen J., Zhang M., and Li Z., “Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival,” Developmental Cell, vol. 41, no. 1, pp. 33–46.e7, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu T., Deng X., Zhou X., Zhu L., Zou L., Li P., Zhang D., and Lin H., “Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato,” Scientific Reports, vol. 6, no. 1, pp. 1–15, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yi H. C., Joo S., Nam K. H., Lee J. S., Kang B. G., and Kim W. T., “Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.),” Plant Molecular Biology, vol. 41, no. 4, pp. 443–454, 1999 [DOI] [PubMed] [Google Scholar]
- 113.Li P., Chen L., Zhou Y., Xia X., Shi K., Chen Z., and Yu J., “Brassinosteroids-induced systemic stress tolerance was associated with increased transcripts of several defence-related genes in the phloem in Cucumis sativus,” PLoS One, vol. 8, no. 6, article e66582, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eleiwa M. E., Bafeel S. O., and Ibrahim S., “Influence of brassinosteroids on wheat plant (Triticum aestivum L.) production under salinity stress conditions. I-Growth parameters and photosynthetic pigments,” Australian Journal of Basic and Applied Sciences, vol. 5, no. 5, pp. 58–65, 2011 [Google Scholar]
- 115.Liu J., Gao H., Wang X., Zheng Q., Wang C., and Wang X., “Effects of 24-epibrassinolide on plant growth, osmotic regulation and ion homeostasis of salt-stressed canola,” Plant Biology, vol. 16, no. 2, pp. 440–450, 2014 [DOI] [PubMed] [Google Scholar]
- 116.Xia X.-J., Wang Y.-J., Zhou Y.-H., Tao Y., Mao W.-H., Shi K., Asami T., Chen Z., and Yu J.-Q., “Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber,” Plant Physiology, vol. 150, no. 2, pp. 801–814, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., and Ueguchi C., “Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis,” The Plant Cell, vol. 16, no. 6, pp. 1365–1377, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Antoniadi I., Novák O., Gelová Z., Johnson A., Plíhal O., Simerský R., Mik V., Vain T., Mateo-Bonmatí E., Karady M., Pernisová M., Plačková L., Opassathian K., Hejátko J., Robert S., Friml J., Doležal K., Ljung K., and Turnbull C., “Cell-surface receptors enable perception of extracellular cytokinins,” Nature Communications, vol. 11, no. 1, p. 4284, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hothorn M., Dabi T., and Chory J., “Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4,” Nature Chemical Biology, vol. 7, no. 11, pp. 766–768, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rivero R. M., Kojima M., Gepstein A., Sakakibara H., Mittler R., Gepstein S., and Blumwald E., “Delayed leaf senescence induces extreme drought tolerance in a flowering plant,” Proceedings of the National Academy of Sciences of the United States of America, vol. 104, no. 49, pp. 19631–19636, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peleg Z., Reguera M., Tumimbang E., Walia H., and Blumwald E., “Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress,” Plant Biotechnology Journal, vol. 9, no. 7, pp. 747–758, 2011 [DOI] [PubMed] [Google Scholar]
- 122.Qin H., Gu Q., Zhang J., Sun L., Kuppu S., Zhang Y., Burow M., Payton P., Blumwald E., and Zhang H., “Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions,” Plant & Cell Physiology, vol. 52, no. 11, pp. 1904–1914, 2011 [DOI] [PubMed] [Google Scholar]
- 123.Nishiyama R., Watanabe Y., Fujita Y., Le D. T., Kojima M., Werner T., Vankova R., Yamaguchi-Shinozaki K., Shinozaki K., Kakimoto T., Sakakibara H., Schmülling T., and Tran L.-S. P., “Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis,” The Plant Cell, vol. 23, no. 6, pp. 2169–2183, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ts W., Motyka V., Laucou V., Smets R., Van Onckelen H., and Schmülling T., “Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity,” The Plant Cell, vol. 15, no. 11, pp. 2532–2550, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Werner T., Nehnevajova E., Köllmer I., Novák O., Strnad M., Krämer U., and Schmülling T., “Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco,” The Plant Cell, vol. 22, no. 12, pp. 3905–3920, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.CERNY M., Jedelský P. L., Novák J., Schlosser A., and Brzobohatý B., “Cytokinin modulates proteomic, transcriptomic and growth responses to temperature shocks inArabidopsis,” Plant, Cell & Environment, vol. 37, no. 7, pp. 1641–1655, 2014 [DOI] [PubMed] [Google Scholar]
- 127.Yang D., Li Y., Shi Y., Cui Z., Luo Y., Zheng M., Chen J., Li Y., Yin Y., and Wang Z., “Exogenous cytokinins increase grain yield of winter wheat cultivars by improving stay-green characteristics under heat stress,” PLoS One, vol. 11, no. 5, article e0155437, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peleg Z., and Blumwald E., “Hormone balance and abiotic stress tolerance in crop plants,” Current Opinion in Plant Biology, vol. 14, no. 3, pp. 290–295, 2011 [DOI] [PubMed] [Google Scholar]
- 129.Devireddy A. R., Zandalinas S. I., Fichman Y., and Mittler R., “Integration of reactive oxygen species and hormone signaling during abiotic stress,” The Plant Journal, vol. 105, no. 2, pp. 459–476, 2021 [DOI] [PubMed] [Google Scholar]
- 130.Liew L. C., Singh M. B., and Bhalla P. L., “A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development,” Scientific Reports, vol. 7, no. 1, pp. 1–16, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., and Moritz T., “Integration of plant responses to environmentally activated phytohormonal signals,” Science, vol. 311, no. 5757, pp. 91–94, 2006 [DOI] [PubMed] [Google Scholar]
- 132.Achard P., Renou J.-P., Berthomé R., Harberd N. P., and Genschik P., “Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species,” Current Biology, vol. 18, no. 9, pp. 656–660, 2008 [DOI] [PubMed] [Google Scholar]
- 133.Achard P., and Genschik P., “Releasing the brakes of plant growth: how GAs shutdown DELLA proteins,” Journal of Experimental Botany, vol. 60, no. 4, pp. 1085–1092, 2009 [DOI] [PubMed] [Google Scholar]
- 134.Wang Y., Mostafa S., Zeng W., and Jin B., “Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses,” International Journal of Molecular Sciences, vol. 22, no. 16, p. 8568, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trusov Y., Chakravorty D., and Botella J. R., “Diversity of heterotrimeric G-protein γ subunits in plants,” BMC Research Notes, vol. 5, no. 1, pp. 1–9, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Milligan G., and Kostenis E., “Heterotrimeric G-proteins: a short history,” British Journal of Pharmacology, vol. 147, Supplement 1, pp. S46–S55, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Urano D., Miura K., Wu Q., Iwasaki Y., Jackson D., and Jones A. M., “Plant morphology of heterotrimeric G protein mutants,” Plant & Cell Physiology, vol. 57, no. 3, pp. 437–445, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Urano D., and Jones A. M., “Heterotrimeric G protein–coupled signaling in plants,” Annual Review of Plant Biology, vol. 65, no. 1, pp. 365–384, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pandey S., “Heterotrimeric G-protein signaling in plants: conserved and novel mechanisms,” Annual Review of Plant Biology, vol. 70, no. 1, pp. 213–238, 2019 [DOI] [PubMed] [Google Scholar]
- 140.Luo W., Huan Q., Xu Y., Qian W., Chong K., and Zhang J., “Integrated global analysis reveals a vitamin E-vitamin K1 sub-network, downstream of COLD1, underlying rice chilling tolerance divergence,” Cell Reports, vol. 36, no. 3, article 109397, 2021 [DOI] [PubMed] [Google Scholar]
- 141.Xu D.-b., Chen M., Ma Y.-n., Xu Z.-s., Li L.-c., Chen Y.-f., and Ma Y.-z., “A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis,” PLoS One, vol. 10, no. 1, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Urano D., Colaneri A., and Jones A. M., “Gα modulates salt-induced cellular senescence and cell division in rice and maize,” Journal of Experimental Botany, vol. 65, no. 22, pp. 6553–6561, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim J. S., Jeon B. W., and Kim J., “Signaling peptides regulating abiotic stress responses in plants,” Frontiers in Plant Science, vol. 12, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsubayashi Y., “Posttranslationally modified small-peptide signals in plants,” Annual Review of Plant Biology, vol. 65, no. 1, pp. 385–413, 2014 [DOI] [PubMed] [Google Scholar]
- 145.Goad D. M., Zhu C., and Kellogg E. A., “Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function,” The New Phytologist, vol. 216, no. 2, pp. 605–616, 2017 [DOI] [PubMed] [Google Scholar]
- 146.Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N., Fukuda H., Yamaguchi-Shinozaki K., and Shinozaki K., “A small peptide modulates stomatal control via abscisic acid in long- distance signalling,” Nature, vol. 556, no. 7700, pp. 235–238, 2018 [DOI] [PubMed] [Google Scholar]
- 147.Zhang L., Shi X., Zhang Y., Wang J., Yang J., Ishida T., Jiang W., Han X., Kang J., Wang X., Pan L., Lv S., Cao B., Zhang Y., Wu J., Han H., Hu Z., Cui L., Sawa S., He J., and Wang G., “CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide inArabidopsis thaliana,” Plant, Cell & Environment, vol. 42, no. 3, pp. 1033–1044, 2019 [DOI] [PubMed] [Google Scholar]
- 148.Endo S., Shinohara H., Matsubayashi Y., and Fukuda H., “A novel pollen-pistil interaction conferring high-temperature tolerance during reproduction via CLE45 signaling,” Current Biology, vol. 23, no. 17, pp. 1670–1676, 2013 [DOI] [PubMed] [Google Scholar]
- 149.Zhao C., Zayed O., Yu Z., Jiang W., Zhu P., Hsu C.-C., Zhang L., Tao W. A., Lozano-Durán R., and Zhu J.-K., “Leucine-rich repeat extensin proteins regulate plant salt tolerance inArabidopsis,” Proceedings of the National Academy of Sciences of the United States of America, vol. 115, no. 51, pp. 13123–13128, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feng W., Kita D., Peaucelle A., Cartwright H. N., Doan V., Duan Q., Liu M.-C., Maman J., Steinhorst L., Schmitz-Thom I., Yvon R., Kudla J., Wu H.-M., Cheung A. Y., and Dinneny J. R., “The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling,” Current Biology, vol. 28, no. 5, pp. 666–675.e5, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gururani M. A., Venkatesh J., and Tran L. S. P., “Regulation of photosynthesis during abiotic stress-induced photoinhibition,” Molecular Plant, vol. 8, no. 9, pp. 1304–1320, 2015 [DOI] [PubMed] [Google Scholar]
- 152.Farmer E. E., and Mueller M. J., “ROS-mediated lipid peroxidation and RES-activated signaling,” Annual Review of Plant Biology, vol. 64, no. 1, pp. 429–450, 2013 [DOI] [PubMed] [Google Scholar]
- 153.Isah T., “Stress and defense responses in plant secondary metabolites production,” Biological Research, vol. 52, no. 1, p. 39, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sami F., Yusuf M., Faizan M., Faraz A., and Hayat S., “Role of sugars under abiotic stress,” Plant Physiology and Biochemistry, vol. 109, pp. 54–61, 2016 [DOI] [PubMed] [Google Scholar]
- 155.Dong S., and Beckles D. M., “Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response,” Journal of Plant Physiology, vol. 234-235, pp. 80–93, 2019 [DOI] [PubMed] [Google Scholar]
- 156.Bolouri-Moghaddam M. R., Le Roy K., Xiang L., Rolland F., and Van den Ende W., “Sugar signalling and antioxidant network connections in plant cells,” The FEBS Journal, vol. 277, no. 9, pp. 2022–2037, 2010 [DOI] [PubMed] [Google Scholar]
- 157.Depaepe T., Hendrix S., van Rensburg H. C. J., Van den Ende W., Cuypers A., and Van Der Straeten D., “At the Crossroads of Survival and Death: The Reactive Oxygen Species-Ethylene- Sugar Triad and the Unfolded Protein Response,” Trends in Plant Science, vol. 26, no. 4, pp. 338–351, 2021 [DOI] [PubMed] [Google Scholar]
- 158.Harrington G. N., and Bush D. R., “The bifunctional role of hexokinase in metabolism and glucose signaling,” The Plant Cell, vol. 15, no. 11, pp. 2493–2496, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Granot D., Kelly G., Stein O., and David-Schwartz R., “Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development,” Journal of Experimental Botany, vol. 65, no. 3, pp. 809–819, 2014 [DOI] [PubMed] [Google Scholar]
- 160.Kosar F., Akram N. A., Sadiq M., Al-Qurainy F., and Ashraf M., “Trehalose: a key organic osmolyte effectively involved in plant abiotic stress tolerance,” Journal of Plant Growth Regulation, vol. 38, no. 2, pp. 606–618, 2019 [Google Scholar]
- 161.Emanuelle S., Doblin M. S., Stapleton D. I., Bacic A., and Gooley P. R., “Molecular insights into the enigmatic metabolic regulator, SnRK1,” Trends in Plant Science, vol. 21, no. 4, pp. 341–353, 2016 [DOI] [PubMed] [Google Scholar]
- 162.Ljung K., Nemhauser J. L., and Perata P., “New mechanistic links between sugar and hormone signalling networks,” Current Opinion in Plant Biology, vol. 25, pp. 130–137, 2015 [DOI] [PubMed] [Google Scholar]
- 163.Zrenner R., and Stitt M., “Comparison of the effect of rapidly and gradually developing water-stress on carbohydrate metabolism in spinach leaves,” Plant, Cell & Environment, vol. 14, no. 9, pp. 939–946, 1991 [Google Scholar]
- 164.Savin R., and Nicolas M. E., “Effects of short periods of drought and high temperature on grain growth and starch accumulation of two malting barley cultivars,” Functional Plant Biology, vol. 23, no. 2, pp. 201–210, 1996 [Google Scholar]
- 165.Rudack K., Seddig S., Sprenger H., Köhl K., Uptmoor R., and Ordon F., “Drought stress-induced changes in starch yield and physiological traits in potato,” Journal of Agronomy and Crop Science, vol. 203, no. 6, pp. 494–505, 2017 [Google Scholar]
- 166.Thalmann M., and Santelia D., “Starch as a determinant of plant fitness under abiotic stress,” The New Phytologist, vol. 214, no. 3, pp. 943–951, 2017 [DOI] [PubMed] [Google Scholar]
- 167.Kaplan F., and Guy C. L., “β-Amylase induction and the protective role of maltose during temperature shock,” Plant Physiology, vol. 135, no. 3, pp. 1674–1684, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thitisaksakul M., Jiménez R. C., Arias M. C., and Beckles D. M., “Effects of environmental factors on cereal starch biosynthesis and composition,” Journal of Cereal Science, vol. 56, no. 1, pp. 67–80, 2012 [Google Scholar]
- 169.Pattanagul W., and Thitisaksakul M., “Effect of salinity stress on growth and carbohydrate metabolism in three rice (Oryza sativa L.) cultivars differing in salinity tolerance,” Indian Journal of Experintal Biology, vol. 46, no. 10, 2008 [PubMed] [Google Scholar]
- 170.Balibrea M. E., Dell'Amico J., Bolarín M. C., and Pérez-Alfocea F., “Carbon partitioning and sucrose metabolism in tomato plants growing under salinity,” Physiologia Plantarum, vol. 110, no. 4, pp. 503–511, 2000 [Google Scholar]
- 171.Pressman E., Peet M. M., and Pharr D. M., “The effect of heat stress on tomato pollen characteristics is associated with changes in carbohydrate concentration in the developing anthers,” Annals of Botany, vol. 90, no. 5, pp. 631–636, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Batista‐Silva W., Heinemann B., Rugen N., Nunes‐Nesi A., Araújo W. L., Braun H.‐. P., and Hildebrandt T. M., “The role of amino acid metabolism during abiotic stress release,” Plant, Cell & Environment, vol. 42, no. 5, pp. 1630–1644, 2019 [DOI] [PubMed] [Google Scholar]
- 173.Verslues P. E., and Sharma S., “Proline metabolism and its implications for plant-environment interaction,” The Arabidopsis Book/American Society of Plant Biologists, vol. 8, article e0140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Verbruggen N., and Hermans C., “Proline accumulation in plants: a review,” Amino Acids, vol. 35, no. 4, pp. 753–759, 2008 [DOI] [PubMed] [Google Scholar]
- 175.Skopelitis D. S., Paranychianakis N. V., Paschalidis K. A., Pliakonis E. D., Delis I. D., and Yakoumakis D. I., “Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine,” The Plant Cell, vol. 18, no. 10, pp. 2767–2781, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen T. H., and Murata N., “Glycinebetaine: an effective protectant against abiotic stress in plants,” Trends in Plant Science, vol. 13, no. 9, pp. 499–505, 2008 [DOI] [PubMed] [Google Scholar]
- 177.Kurepin L. V., Ivanov A. G., Zaman M., Pharis R. P., Hurry V., and Hüner N. P., “Interaction of glycine betaine and plant hormones: protection of the photosynthetic apparatus during abiotic stress,” Photosynthesis: Structures, Mechanisms, and Applications, Hou H., Najafpour M., Moore G., and Allakhverdiev S., Eds., Springer, Cham, pp. 185–202, 2017 [Google Scholar]
- 178.Vogt T., “Phenylpropanoid biosynthesis,” Molecular Plant, vol. 3, no. 1, pp. 2–20, 2010 [DOI] [PubMed] [Google Scholar]
- 179.Chun H. J., Baek D., Cho H. M., Lee S. H., Jin B. J., Yun D.-J., Hong Y.-S., and Kim M. C., “Lignin biosynthesis genes play critical roles in the adaptation ofArabidopsisplants to high-salt stress,” Plant Signaling & Behavior, vol. 14, no. 8, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chen X., Wang H., Li X., Ma K., Zhan Y., and Zeng F., “Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis,” BMC Plant Biology, vol. 19, no. 1, pp. 1–16, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Davies K. M., Albert N. W., Zhou Y., and Schwinn K. E., “Functions of flavonoid and betalain pigments in abiotic stress tolerance in plants,” Annual Plant Reviews Online, vol. 1, no. 1, pp. 21–62, 2018 [Google Scholar]
- 182.Dong N. Q., and Lin H. X., “Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions,” Journal of Integrative Plant Biology, vol. 63, no. 1, pp. 180–209, 2021 [DOI] [PubMed] [Google Scholar]
- 183.Zhan X., Shen Q., Chen J., Yang P., Wang X., and Hong Y., “Rice sulfoquinovosyltransferase SQD2. 1 mediates flavonoid glycosylation and enhances tolerance to osmotic stress,” Plant, Cell & Environment, vol. 42, no. 7, pp. 2215–2230, 2019 [DOI] [PubMed] [Google Scholar]
- 184.Chen S., Wu F., Li Y., Qian Y., Pan X., Li F., Wang Y., Wu Z., Fu C., Lin H., and Yang A., “NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness,” Frontiers in Plant Science, vol. 10, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cui L. G., Shan J. X., Shi M., Gao J. P., and Lin H. X., “ThemiR156-SPL9-DFRpathway coordinates the relationship between development and abiotic stress tolerance in plants,” The Plant Journal, vol. 80, no. 6, pp. 1108–1117, 2014 [DOI] [PubMed] [Google Scholar]
- 186.Kim J., Lee W. J., Vu T. T., Jeong C. Y., Hong S.-W., and Lee H., “High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L,” Plant Cell Reports, vol. 36, no. 8, pp. 1215–1224, 2017 [DOI] [PubMed] [Google Scholar]
- 187.Feyissa B. A., Arshad M., Gruber M. Y., Kohalmi S. E., and Hannoufa A., “The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa,” BMC Plant Biology, vol. 19, no. 1, pp. 1–19, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nakabayashi R., Yonekura‐Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., Michael A. J., Tohge T., Yamazaki M., and Saito K., “Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids,” The Plant Journal, vol. 77, no. 3, pp. 367–379, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Muhlemann J. K., Younts T. L., and Muday G. K., “Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress,” Proceedings of the National Academy of Sciences of the United States of America, vol. 115, no. 47, pp. E11188–E11197, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yoon Y., Seo D. H., Shin H., Kim H. J., Kim C. M., and Jang G., “The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants,” Agronomy, vol. 10, no. 6, p. 788, 2020 [Google Scholar]
- 191.Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., and Parcy F., “bZIP transcription factors in Arabidopsis,” Trends in Plant Science, vol. 7, no. 3, pp. 106–111, 2002 [DOI] [PubMed] [Google Scholar]
- 192.Liu J. X., Srivastava R., and Howell S. H., “Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress inArabidopsis,” Plant, Cell & Environment, vol. 31, no. 12, pp. 1735–1743, 2008 [DOI] [PubMed] [Google Scholar]
- 193.Yang O., Popova O. V., Süthoff U., Lüking I., Dietz K.-J., and Golldack D., “The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance,” Gene, vol. 436, no. 1-2, pp. 45–55, 2009 [DOI] [PubMed] [Google Scholar]
- 194.Rolly N. K., Imran Q. M., Lee I.-J., and Yun B.-W., “Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana,” International Journal of Molecular Sciences, vol. 21, no. 5, p. 1726, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yang Y., Yu T.-F., Ma J., Chen J., Zhou Y.-B., and Chen M., “The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants,” International Journal of Molecular Sciences, vol. 21, no. 2, p. 670, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mukherjee K., Choudhury A. R., Gupta B., Gupta S., and Sengupta D. N., “An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice,” BMC Plant Biology, vol. 6, no. 1, pp. 1–14, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liu C., Mao B., Ou S., Wang W., Liu L., and Wu Y., “OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice,” Plant Molecular Biology, vol. 84, no. 1-2, pp. 19–36, 2014 [DOI] [PubMed] [Google Scholar]
- 198.Liao Y., Zou H.-F., Wei W., Hao Y.-J., Tian A.-G., and Huang J., “Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis,” Planta, vol. 228, no. 2, pp. 225–240, 2008 [DOI] [PubMed] [Google Scholar]
- 199.Li Z., and Howell S. H., “Review: The two faces of IRE1 and their role in protecting plants from stress,” Plant Science, vol. 303, 2021 [DOI] [PubMed] [Google Scholar]
- 200.Singh M. B., Lohani N., and Bhalla P. L., “The role of endoplasmic reticulum stress response in pollen development and heat stress tolerance,” Frontiers in Plant Science, vol. 12, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li Z., Tang J., Srivastava R., Bassham D. C., and Howell S. H., “The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize,” The Plant Cell, vol. 32, no. 11, pp. 3559–3575, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Eulgem T., Rushton P. J., Robatzek S., and Somssich I. E., “The WRKY superfamily of plant transcription factors,” Trends in Plant Science, vol. 5, no. 5, pp. 199–206, 2000 [DOI] [PubMed] [Google Scholar]
- 203.Li W., Pang S., Lu Z., and Jin B., “Function and mechanism of WRKY transcription factors in abiotic stress responses of plants,” Plants, vol. 9, no. 11, p. 1515, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ahammed G. J., Li X., Yang Y., Liu C., Zhou G., Wan H., and Cheng Y., “Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2 -mediated stomatal closure,” Environmental and Experimental Botany, vol. 171, article 103960, 2020 [Google Scholar]
- 205.Yang Z., Chi X., Guo F., Jin X., Luo H., Hawar A., Chen Y., Feng K., Wang B., Qi J., Yang Y., and Sun B., “SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum,” Journal of Plant Physiology, vol. 246-247, article 153142, 2020 [DOI] [PubMed] [Google Scholar]
- 206.Xu Z., Raza Q., Xu L., He X., Huang Y., Yi J., Zhang D., Shao H. B., Ma H., and Ali Z., “GmWRKY49, a salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis,” Frontiers in Plant Science, vol. 9, p. 809, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang Y., Yu H., Yang X., Li Q., Ling J., Wang H., Gu X., Huang S., and Jiang W., “CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner,” Plant Physiology and Biochemistry, vol. 108, pp. 478–487, 2016 [DOI] [PubMed] [Google Scholar]
- 208.Wu X., Shiroto Y., Kishitani S., Ito Y., and Toriyama K., “Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter,” Plant Cell Reports, vol. 28, no. 1, pp. 21–30, 2009 [DOI] [PubMed] [Google Scholar]
- 209.Jin H., and Martin C., “Multifunctionality and diversity within the plant MYB-gene family,” Plant Molecular Biology, vol. 41, no. 5, pp. 577–585, 1999 [DOI] [PubMed] [Google Scholar]
- 210.Stracke R., Werber M., and Weisshaar B., “The R2R3-MYB gene family in Arabidopsis thaliana,” Current Opinion in Plant Biology, vol. 4, no. 5, pp. 447–456, 2001 [DOI] [PubMed] [Google Scholar]
- 211.Li C., Ng C. K.-Y., and Fan L.-M., “MYB transcription factors, active players in abiotic stress signaling,” Environmental and Experimental Botany, vol. 114, pp. 80–91, 2015 [Google Scholar]
- 212.Jung C., Seo J. S., Han S. W., Koo Y. J., Kim C. H., Song S. I., Nahm B. H., Choi Y. D., and Cheong J. J., “Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis,” Plant Physiology, vol. 146, no. 2, pp. 623–635, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Seo P. J., Xiang F., Qiao M., Park J.-Y., Lee Y. N., Kim S.-G., Lee Y. H., Park W. J., and Park C. M., “The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis,” Plant Physiology, vol. 151, no. 1, pp. 275–289, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Seo P. J., Lee S. B., Suh M. C., Park M.-J., Go Y. S., and Park C.-M., “The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis,” The Plant Cell, vol. 23, no. 3, pp. 1138–1152, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lee S. B., Kim H., Kim R. J., and Suh M. C., “Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation,” Plant Cell Reports, vol. 33, no. 9, pp. 1535–1546, 2014 [DOI] [PubMed] [Google Scholar]
- 216.Cui M. H., Yoo K. S., Hyoung S., Nguyen H. T. K., Kim Y. Y., Kim H. J., Ok S. H., Yoo S. D., and Shin J. S., “An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance,” FEBS Letters, vol. 587, no. 12, pp. 1773–1778, 2013 [DOI] [PubMed] [Google Scholar]
- 217.Liao C., Zheng Y., and Guo Y., “MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling inArabidopsis,” The New Phytologist, vol. 216, no. 1, pp. 163–177, 2017 [DOI] [PubMed] [Google Scholar]
- 218.Licausi F., Ohme-Takagi M., and Perata P., “APETALA 2/ethylene responsive factor (AP 2/ERF) transcription factors: mediators of stress responses and developmental programs,” The New Phytologist, vol. 199, no. 3, pp. 639–649, 2013 [DOI] [PubMed] [Google Scholar]
- 219.Agarwal P. K., Gupta K., Lopato S., and Agarwal P., “Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance,” Journal of Experimental Botany, vol. 68, no. 9, pp. 2135–2148, 2017 [DOI] [PubMed] [Google Scholar]
- 220.Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y., and Thomashow M. F., “Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network,” The Plant Journal, vol. 82, no. 2, pp. 193–207, 2015 [DOI] [PubMed] [Google Scholar]
- 221.Qin F., Kakimoto M., Sakuma Y., Maruyama K., Osakabe Y., Tran L. S. P., Shinozaki K., and Yamaguchi-Shinozaki K., “Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L,” The Plant Journal., vol. 50, no. 1, pp. 54–69, 2007 [DOI] [PubMed] [Google Scholar]
- 222.Xie Z., Nolan T. M., Jiang H., and Yin Y., “AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis,” Frontiers in Plant Science, vol. 10, p. 228, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Karaba A., Dixit S., Greco R., Aharoni A., Trijatmiko K. R., Marsch-Martinez N., Krishnan A., Nataraja K. N., Udayakumar M., and Pereira A., “Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene,” Proceedings of the National Academy of Sciences of the United States of America, vol. 104, no. 39, pp. 15270–15275, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Heim M. A., Jakoby M., Werber M., Martin C., Weisshaar B., and Bailey P. C., “The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity,” Molecular Biology and Evolution, vol. 20, no. 5, pp. 735–747, 2003 [DOI] [PubMed] [Google Scholar]
- 225.Qian Y., Zhang T., Yu Y., Gou L., Yang J., Xu J., and Pi E., “Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses,” Frontiers in Plant Science, vol. 12, p. 1143, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li Z., Liu C., Zhang Y., Wang B., Ran Q., and Zhang J., “The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis,” Journal of Experimental Botany, vol. 70, no. 19, pp. 5471–5486, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu Y., Ji X., Nie X., Qu M., Zheng L., Tan Z., Zhao H., Huo L., Liu S., Zhang B., and Wang Y., “ArabidopsisAtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box andGCG‐box motifs,” The New Phytologist, vol. 207, no. 3, pp. 692–709, 2015 [DOI] [PubMed] [Google Scholar]
- 228.Verma D., Jalmi S. K., Bhagat P. K., Verma N., and Sinha A. K., “A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis,” The FEBS Journal, vol. 287, no. 12, pp. 2560–2576, 2020 [DOI] [PubMed] [Google Scholar]
- 229.Chinnusamy V., Ohta M., Kanrar S., Lee B.-h., Hong X., Agarwal M., and Zhu J. K., “ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis,” Genes & Development, vol. 17, no. 8, pp. 1043–1054, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li F., Guo S., Zhao Y., Chen D., Chong K., and Xu Y., “Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis,” Plant Cell Reports, vol. 29, no. 9, pp. 977–986, 2010 [DOI] [PubMed] [Google Scholar]
- 231.Olsen A. N., Ernst H. A., Leggio L. L., and Skriver K., “NAC transcription factors: structurally distinct, functionally diverse,” Trends in Plant Science, vol. 10, no. 2, pp. 79–87, 2005 [DOI] [PubMed] [Google Scholar]
- 232.Jeong J. S., Kim Y. S., Redillas M. C., Jang G., Jung H., Bang S. W., Choi Y. D., Ha S. H., Reuzeau C., and Kim J. K., “OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field,” Plant Biotechnology Journal, vol. 11, no. 1, pp. 101–114, 2013 [DOI] [PubMed] [Google Scholar]
- 233.Redillas M. C., Jeong J. S., Kim Y. S., Jung H., Bang S. W., Choi Y. D., Ha S. H., Reuzeau C., and Kim J. K., “The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions,” Plant Biotechnology Journal, vol. 10, no. 7, pp. 792–805, 2012 [DOI] [PubMed] [Google Scholar]
- 234.Jeong J. S., Kim Y. S., Baek K. H., Jung H., Ha S.-H., Do Choi Y., Kim M., Reuzeau C., and Kim J. K., “Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions,” Plant Physiology, vol. 153, no. 1, pp. 185–197, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Thirumalaikumar V. P., Devkar V., Mehterov N., Ali S., Ozgur R., Turkan I., Mueller-Roeber B., and Balazadeh S., “NAC transcription factor JUNGBRUNNEN 1 enhances drought tolerance in tomato,” Plant Biotechnology Journal, vol. 16, no. 2, pp. 354–366, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Alshareef N. O., Wang J. Y., Ali S., Al-Babili S., Tester M., and Schmöckel S. M., “Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato,” Plant Physiology and Biochemistry, vol. 140, pp. 113–121, 2019 [DOI] [PubMed] [Google Scholar]
- 237.Xu Z.-Y., Kim S. Y., Hyeon D. Y., Kim D. H., Dong T., Park Y., Jin J. B., Joo S. H., Kim S. K., Hong J. C., Hwang D., and Hwang I., “The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses,” The Plant Cell, vol. 25, no. 11, pp. 4708–4724, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Punzo P., Grillo S., and Batelli G., “Alternative splicing in plant abiotic stress responses,” Biochemical Society Transactions, vol. 48, no. 5, pp. 2117–2126, 2020 [DOI] [PubMed] [Google Scholar]
- 239.de Francisco Amorim M., Willing E.-M., Szabo E. X., Francisco-Mangilet A. G., Droste-Borel I., Maček B., Schneeberger K., and Laubinger S., “The U1 snRNP subunit LUC7 modulates plant development and stress responses via regulation of alternative splicing,” The Plant Cell, vol. 30, no. 11, pp. 2838–2854, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sugio A., Dreos R., Aparicio F., and Maule A. J., “The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis,” The Plant Cell, vol. 21, no. 2, pp. 642–654, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hong Y., Yao J., Shi H., Chen Y., Zhu J.-K., and Wang Z., “The Arabidopsis spliceosomal protein SmEb modulates ABA responses by maintaining proper alternative splicing of HAB1,” Stress Biology, vol. 1, no. 1, pp. 1–8, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gehring N. H., and Roignant J.-Y., “Anything but Ordinary - Emerging Splicing Mechanisms in Eukaryotic Gene Regulation,” Trends in Genetics, vol. 37, no. 4, pp. 355–372, 2021 [DOI] [PubMed] [Google Scholar]
- 243.Li S., Wang Y., Zhao Y., Zhao X., Chen X., and Gong Z., “Global Co-transcriptional Splicing in Arabidopsis and the Correlation with Splicing Regulation in Mature RNAs,” Molecular Plant, vol. 13, no. 2, pp. 266–277, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Golicz A. A., Allu A. D., Li W., Lohani N., Singh M. B., and Bhalla P. L., “A dynamic intron retention program regulates the expression of several hundred genes during pollen meiosis,” Plant Reproduction, vol. 34, no. 3, pp. 225–242, 2021 [DOI] [PubMed] [Google Scholar]
- 245.Zhu J., Liu M., Liu X., and Dong Z., “RNA polymerase II activity revealed by GRO-seq and pNET-seq in Arabidopsis,” Nature Plants, vol. 4, no. 12, pp. 1112–1123, 2018 [DOI] [PubMed] [Google Scholar]
- 246.Jabre I., Reddy A. S., Kalyna M., Chaudhary S., Khokhar W., Byrne L. J., Wilson C. M., and Syed N. H., “Does co-transcriptional regulation of alternative splicing mediate plant stress responses?,” Nucleic Acids Research, vol. 47, no. 6, pp. 2716–2726, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen T., Cui P., Chen H., Ali S., Zhang S., and Xiong L., “A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis,” PLoS Genetics, vol. 9, no. 10, article e1003875, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhu D., Mao F., Tian Y., Lin X., Gu L., Gu H., Qu L. J., Wu Y., and Wu Z., “The Features and Regulation of Co-transcriptional Splicing in Arabidopsis,” Molecular Plant, vol. 13, no. 2, pp. 278–294, 2020 [DOI] [PubMed] [Google Scholar]
- 249.Yu X., Meng X., Liu Y., Wang X., Wang T.-J., Zhang A., Li N., Qi X., Liu B., and Xu Z. Y., “The chromatin remodeler ZmCHB101 impacts alternative splicing contexts in response to osmotic stress,” Plant Cell Reports, vol. 38, no. 2, pp. 131–145, 2019 [DOI] [PubMed] [Google Scholar]
- 250.Chakrabarti M., de Lorenzo L., Abdel-Ghany S. E., Reddy A. S., and Hunt A. G., “Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum,” The Plant Journal, vol. 102, no. 5, pp. 916–930, 2020 [DOI] [PubMed] [Google Scholar]
- 251.Téllez-Robledo B., Manzano C., Saez A., Navarro-Neila S., Silva-Navas J., de Lorenzo L., González-García M. P., Toribio R., Hunt A. G., Baigorri R., Casimiro I., Brady S. M., Castellano M. M., and Pozo J. C., “The polyadenylation factor FIP1 is important for plant development and root responses to abiotic stresses,” The Plant Journal, vol. 99, no. 6, pp. 1203–1219, 2019 [DOI] [PubMed] [Google Scholar]
- 252.Ye C., Zhou Q., Wu X., Ji G., and Li Q. Q., “Genome-wide alternative polyadenylation dynamics in response to biotic and abiotic stresses in rice,” Ecotoxicology and Environmental Safety, vol. 183, article 109485, 2019 [DOI] [PubMed] [Google Scholar]
- 253.Jobava R., Mao Y., Guan B.-J., Hu D., Krokowski D., Chen C.-W., Shu X. E., Chukwurah E., Wu J., Gao Z., Zagore L. L., Merrick W. C., Trifunovic A., Hsieh A. C., Valadkhan S., Zhang Y., Qi X., Jankowsky E., Topisirovic I., Licatalosi D. D., Qian S. B., and Hatzoglou M., “Adaptive translational pausing is a hallmark of the cellular response to severe environmental stress,” Molecular Cell, vol. 81, no. 20, pp. 4191–4208.e8, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lei L., Shi J., Chen J., Zhang M., Sun S., Xie S., Li X., Zeng B., Peng L., Hauck A., Zhao H., Song W., Fan Z., and Lai J., “Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress,” The Plant Journal, vol. 84, no. 6, pp. 1206–1218, 2015 [DOI] [PubMed] [Google Scholar]
- 255.Keller M., and Simm S., “The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen,” BMC Genomics, vol. 19, no. 1, pp. 1–20, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Poidevin L., Forment J., Unal D., and Ferrando A., “Transcriptome and translatome changes in germinated pollen under heat stress uncover roles of transporter genes involved in pollen tube growth,” Plant, Cell & Environment, vol. 44, no. 7, pp. 2167–2184, 2021 [DOI] [PubMed] [Google Scholar]
- 257.Merret R., Nagarajan V. K., Carpentier M.-C., Park S., Favory J.-J., Descombin J., Picart C., Charng Y. Y., Green P. J., Deragon J. M., and Bousquet-Antonelli C., “Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana,” Nucleic Acids Research, vol. 43, no. 8, pp. 4121–4132, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Merret R., Descombin J., Juan Y.-t., Favory J.-J., Carpentier M.-C., Chaparro C., Charng Y. Y., Deragon J. M., and Bousquet-Antonelli C., “XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress,” Cell Reports, vol. 5, no. 5, pp. 1279–1293, 2013 [DOI] [PubMed] [Google Scholar]
- 259.Nguyen A. H., Matsui A., Tanaka M., Mizunashi K., Nakaminami K., Hayashi M., Iida K., Toyoda T., Nguyen D. V., and Seki M., “Loss of Arabidopsis 5–3 exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress,” Plant & Cell Physiology, vol. 56, no. 9, pp. 1762–1772, 2015 [DOI] [PubMed] [Google Scholar]
- 260.Merret R., Carpentier M.-C., Favory J.-J., Picart C., Descombin J., Bousquet-Antonelli C., Tillard P., Lejay L., Deragon J. M., and Charng Y. Y., “Heat shock protein HSP101 affects the release of ribosomal protein mRNAs for recovery after heat shock,” Plant Physiology, vol. 174, no. 2, pp. 1216–1225, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Friso G., and van Wijk K. J., “Posttranslational protein modifications in plant metabolism,” Plant Physiology, vol. 169, no. 3, pp. 1469–1487, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hashiguchi A., and Komatsu S., “Impact of post-translational modifications of crop proteins under abiotic stress,” Proteome, vol. 4, no. 4, p. 42, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Moon J., Parry G., and Estelle M., “The ubiquitin-proteasome pathway and plant development,” The Plant Cell, vol. 16, no. 12, pp. 3181–3195, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hua Z., and Vierstra R. D., “The cullin-RING ubiquitin-protein ligases,” Annual Review of Plant Biology, vol. 62, no. 1, pp. 299–334, 2011 [DOI] [PubMed] [Google Scholar]
- 265.Melo F. V., Oliveira M. M., Saibo N. J. M., and Lourenço T. F., “Modulation of abiotic stress responses in rice by E3-ubiquitin ligases: a promising way to develop stress-tolerant crops,” Frontiers in Plant Science, vol. 12, p. 368, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lee J.-H., and Kim W. T., “Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis,” Molecules and Cells, vol. 31, no. 3, pp. 201–208, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Shu K., and Yang W., “E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses,” Plant & Cell Physiology, vol. 58, no. 9, pp. 1461–1476, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Miura K., Jin J. B., Lee J., Yoo C. Y., Stirm V., Miura T., Ashworth E. N., Bressan R. A., Yun D. J., and Hasegawa P. M., “SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis,” The Plant Cell., vol. 19, no. 4, pp. 1403–1414, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Saracco S. A., Miller M. J., Kurepa J., and Vierstra R. D., “Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential,” Plant Physiology, vol. 145, no. 1, pp. 119–134, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., and Hay R. T., “System-wide changes to SUMO modifications in response to heat shock,” Science Signaling, vol. 2, no. 72, p. 24, 2009 [DOI] [PubMed] [Google Scholar]
- 271.Miller M. J., Scalf M., Rytz T. C., Hubler S. L., Smith L. M., and Vierstra R. D., “Quantitative Proteomics Reveals Factors Regulating RNA Biology as Dynamic Targets of Stress-induced SUMOylation in Arabidopsis,” Molecular & Cellular Proteomics, vol. 12, no. 2, pp. 449–463, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Augustine R. C., York S. L., Rytz T. C., and Vierstra R. D., “Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress,” Plant Physiology, vol. 171, no. 3, pp. 2191–2210, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mishra N., Sun L., Zhu X., Smith J., Prakash Srivastava A., Yang X., Pehlivan N., Esmaeili N., Luo H., Shen G., Jones D., Auld D., Burke J., Payton P., and Zhang H., “Overexpression of the rice SUMO E3 ligase gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions,” Plant & Cell Physiology, vol. 58, no. 4, pp. 735–746, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Srivastava A. K., Zhang C., Caine R. S., Gray J., and Sadanandom A., “Rice SUMO protease Overly tolerant to salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice,” The Plant Journal, vol. 92, no. 6, pp. 1031–1043, 2017 [DOI] [PubMed] [Google Scholar]
- 275.Zhang S., Zhuang K., Wang S., Lv J., Ma N., and Meng Q., “A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco,” Journal of Integrative Plant Biology, vol. 59, no. 2, pp. 102–117, 2017 [DOI] [PubMed] [Google Scholar]
- 276.Wang H., Wang M., and Xia Z., “The maize class-I SUMO conjugating enzyme ZmSCE1d is involved in drought stress response,” International Journal of Molecular Sciences, vol. 21, no. 1, p. 29, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Li S., Lin M., Wang J., Zhang L., Lin M., Hu Z., Qi Z., Jiang H., Fu Y., Xin D., Liu C., and Chen Q., “Regulation of soybean SUMOylation system in response to Phytophthora sojae infection and heat shock,” Plant Growth Regulation, vol. 87, no. 1, pp. 69–82, 2019 [Google Scholar]
- 278.Udenwobele D. I., Su R.-C., Good S. V., Ball T. B., Varma Shrivastav S., and Shrivastav A., “Myristoylation: an important protein modification in the immune response,” Frontiers in Immunology, vol. 8, p. 751, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ding Y., Lv J., Shi Y., Gao J., Hua J., Song C., Gong Z., and Yang S., “EGR2 phosphatase regulatesOST1 kinase activity and freezing tolerance inArabidopsis,” The EMBO Journal, vol. 38, no. 1, article e99819, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang P., Hsu C.-C., du Y., Zhu P., Zhao C., Fu X., Zhang C., Paez J. S., Macho A. P., Tao W. A., and Zhu J. K., “Mapping proteome-wide targets of protein kinases in plant stress responses,” Proceedings of the National Academy of Sciences of the United States of America, vol. 117, no. 6, pp. 3270–3280, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., al-Rasheid K. A. S., Romeis T., and Hedrich R., “Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair,” Proceedings of the National Academy of Sciences of the United States of America, vol. 106, no. 50, pp. 21425–21430, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Willems P., Horne A., van Parys T., Goormachtig S., de Smet I., Botzki A., van Breusegem F., and Gevaert K., “The plant PTM viewer, a central resource for exploring plant protein modifications,” The Plant Journal, vol. 99, no. 4, pp. 752–762, 2019 [DOI] [PubMed] [Google Scholar]
- 283.Feng J., Chen L., and Zuo J., “Protein S-nitrosylation in plants: current progresses and challenges,” Journal of Integrative Plant Biology, vol. 61, no. 12, pp. 1206–1223, 2019 [DOI] [PubMed] [Google Scholar]
- 284.Matamoros M. A., and Becana M., “Molecular responses of legumes to abiotic stress: post-translational modifications of proteins and redox signaling,” Journal of Experimental Botany, vol. 72, no. 16, pp. 5876–5892, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kovacs I., and Lindermayr C., “Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation,” Frontiers in Plant Science, vol. 4, p. 137, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hu J., Yang H., Mu J., Lu T., Peng J., Deng X., Kong Z., Bao S., Cao X., and Zuo J., “Nitric oxide regulates protein methylation during stress responses in plants,” Molecular Cell, vol. 67, no. 4, pp. 702–710.e4, 2017 [DOI] [PubMed] [Google Scholar]
- 287.Ueda M., and Seki M., “Histone modifications form epigenetic regulatory networks to regulate abiotic stress response,” Plant Physiology, vol. 182, no. 1, pp. 15–26, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Liew L. C., Singh M. B., and Bhalla P. L., “An RNA-seq transcriptome analysis of histone modifiers and RNA silencing genes in soybean during floral initiation process,” PLoS One, vol. 8, no. 10, article e77502, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Jung C. H., O'brien M., Singh M. B., and Bhalla P. L., “Epigenetic landscape of germline specific genes in the sporophyte cells of Arabidopsis thaliana,” Frontiers in Plant Science, vol. 6, p. 328, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lämke J., and Bäurle I., “Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants,” Genome Biology, vol. 18, no. 1, pp. 1–11, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Asensi-Fabado M.-A., Amtmann A., and Perrella G., “Plant responses to abiotic stress: the chromatin context of transcriptional regulation,” Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms., vol. 1860, no. 1, pp. 106–122, 2017 [DOI] [PubMed] [Google Scholar]
- 292.Goldberg A. D., Allis C. D., and Bernstein E., “Epigenetics: a landscape takes shape,” Cell, vol. 128, no. 4, pp. 635–638, 2007 [DOI] [PubMed] [Google Scholar]
- 293.Lusser A., Kölle D., and Loidl P., “Histone acetylation: lessons from the plant kingdom,” Trends in Plant Science, vol. 6, no. 2, pp. 59–65, 2001 [DOI] [PubMed] [Google Scholar]
- 294.Zheng M., Liu X., Lin J., Liu X., Wang Z., Xin M., Yao Y., Peng H., Zhou D. X., Ni Z., Sun Q., and Hu Z., “Histone acetyltransferase GCN 5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes,” The Plant Journal, vol. 97, no. 3, pp. 587–602, 2019 [DOI] [PubMed] [Google Scholar]
- 295.Hu Z., Song N., Zheng M., Liu X., Liu Z., Xing J., Ma J., Guo W., Yao Y., Peng H., Xin M., Zhou D. X., Ni Z., and Sun Q., “Histone acetyltransferase GCN 5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis,” The Plant Journal, vol. 84, no. 6, pp. 1178–1191, 2015 [DOI] [PubMed] [Google Scholar]
- 296.Choudhary A., Singh S., and Verma P. C., “Role of Histone Acetyltransferases in Plant Abiotic Stress,” Molecular Approaches in Plant Biology and Environmental Challenges. Energy, Environment, and Sustainability, Singh S., Upadhyay S., Pandey A., and Kumar S., Eds., Springer, Singapore, pp. 103–112, 2019 [Google Scholar]
- 297.Mehdi S., Derkacheva M., Ramström M., Kralemann L., Bergquist J., and Hennig L., “The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling,” The Plant Cell, vol. 28, no. 1, pp. 42–54, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Zheng Y., Ding Y., Sun X., Xie S., Wang D., Liu X., Su L., Wei W., Pan L., and Zhou D. X., “Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis,” Journal of Experimental Botany, vol. 67, no. 6, pp. 1703–1713, 2016 [DOI] [PubMed] [Google Scholar]
- 299.Ueda M., Matsui A., Tanaka M., Nakamura T., Abe T., Sako K., Sasaki T., Kim J. M., Ito A., Nishino N., Shimada H., Yoshida M., and Seki M., “The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response,” Plant Physiology, vol. 175, no. 4, pp. 1760–1773, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Han Z., Yu H., Zhao Z., Hunter D., Luo X., Duan J., and Tian L., “AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana,” Frontiers in Plant Science, vol. 7, p. 310, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Liu C., Lu F., Cui X., and Cao X., “Histone methylation in higher plants,” Annual Review of Plant Biology, vol. 61, no. 1, pp. 395–420, 2010 [DOI] [PubMed] [Google Scholar]
- 302.Lu F., Cui X., Zhang S., Jenuwein T., and Cao X., “Arabidopsis REF6 is a histone H3 lysine 27 demethylase,” Nature Genetics, vol. 43, no. 7, pp. 715–719, 2011 [DOI] [PubMed] [Google Scholar]
- 303.Shen Y., Conde e Silva N., Audonnet L., Servet C., Wei W., and Zhou D.-X., “Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis,” Frontiers in Plant Science, vol. 5, p. 290, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Huang S., Zhang A., Jin J. B., Zhao B., Wang T. J., Wu Y., Wang S., Liu Y., Wang J., Guo P., Ahmad R., Liu B., and Xu Z. Y., “Arabidopsis histone H3K4 demethylase JMJ 17 functions in dehydration stress response,” The New Phytologist, vol. 223, no. 3, pp. 1372–1387, 2019 [DOI] [PubMed] [Google Scholar]
- 305.Sridhar V. V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P. M., Bressan R. A., and Zhu J. K., “Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination,” Nature, vol. 447, no. 7145, pp. 735–738, 2007 [DOI] [PubMed] [Google Scholar]
- 306.Chen H., Feng H., Zhang X., Zhang C., Wang T., and Dong J., “An Arabidopsis E3 ligase HUB 2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton,” Plant Biotechnology Journal, vol. 17, no. 3, pp. 556–568, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ma S., Tang N., Li X., Xie Y., Xiang D., Fu J., Shen J., Yang J., Tu H., Li X., Hu H., and Xiong L., “Reversible histone H2B monoubiquitination fine-tunes abscisic acid signaling and drought response in rice,” Molecular Plant, vol. 12, no. 2, pp. 263–277, 2019 [DOI] [PubMed] [Google Scholar]
- 308.Zhou B.-R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., and Bai Y., “Structural insights into the histone H1-nucleosome complex,” Proceedings of the National Academy of Sciences of the United States of America, vol. 110, no. 48, pp. 19390–19395, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Sura W., Kabza M., Karlowski W. M., Bieluszewski T., Kus-Slowinska M., Pawełoszek Ł., Sadowski J., and Ziolkowski P. A., “Dual role of the histone variant H2A. Z in transcriptional regulation of stress-response genes,” The Plant Cell, vol. 29, no. 4, pp. 791–807, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kumar S. V., and Wigge P. A., “H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis,” Cell, vol. 140, no. 1, pp. 136–147, 2010 [DOI] [PubMed] [Google Scholar]
- 311.Deng X., Qiu Q., He K., and Cao X., “The seekers: how epigenetic modifying enzymes find their hidden genomic targets in Arabidopsis,” Current Opinion in Plant Biology, vol. 45, Part A, pp. 75–81, 2018 [DOI] [PubMed] [Google Scholar]
- 312.Golicz A. A., Bhalla P. L., and Singh M. B., “lncRNAs in plant and animal sexual reproduction,” Trends in Plant Science, vol. 23, no. 3, pp. 195–205, 2018 [DOI] [PubMed] [Google Scholar]
- 313.Cheng X., Zhang S., Tao W., Zhang X., Liu J., Sun J., Zhang H., Pu L., Huang R., and Chen T., “INDETERMINATE SPIKELET1 recruits histone deacetylase and a transcriptional repression complex to regulate rice salt tolerance,” Plant Physiology, vol. 178, no. 2, pp. 824–837, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lee H. G., and Seo P. J., “MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis,” Nature Communications, vol. 10, no. 1, pp. 1–14, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Li S., Lin Y.-C. J., Wang P., Zhang B., Li M., Chen S., Shi R., Tunlaya-Anukit S., Liu X., Wang Z., Dai X., Yu J., Zhou C., Liu B., Wang J. P., Chiang V. L., and Li W., “The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance inPopulus trichocarpa,” The Plant Cell, vol. 31, no. 3, pp. 663–686, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Law J. A., and Jacobsen S. E., “Establishing, maintaining and modifying DNA methylation patterns in plants and animals,” Nature Reviews Genetics, vol. 11, no. 3, pp. 204–220, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zhang H., Lang Z., and Zhu J.-K., “Dynamics and function of DNA methylation in plants,” Nature Reviews Molecular Cell Biology, vol. 19, no. 8, pp. 489–506, 2018 [DOI] [PubMed] [Google Scholar]
- 318.Liu J., and He Z., “Small DNA methylation, big player in plant abiotic stress responses and memory,” Frontiers in Plant Science, vol. 11, p. 1977, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Min L., Li Y., Hu Q., Zhu L., Gao W., Wu Y., Ding Y., Liu S., Yang X., and Zhang X., “Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton,” Plant Physiology, vol. 164, no. 3, pp. 1293–1308, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ma Y., Min L., Wang M., Wang C., Zhao Y., Li Y., Fang Q., Wu Y., Xie S., Ding Y., Su X., Hu Q., Zhang Q., Li X., and Zhang X., “Disrupted genome methylation in response to high temperature has distinct affects on microspore abortion and anther indehiscence,” The Plant Cell, vol. 30, no. 7, pp. 1387–1403, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Gayacharan, and Joel A. J., “Epigenetic responses to drought stress in rice (Oryza sativa L.),” Physiology and Molecular Biology of Plants, vol. 19, no. 3, pp. 379–387, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Zhang W., Wang N., Yang J., Guo H., Liu Z., Zheng X., Li S., and Xiang F., “The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean,” Plant Science, vol. 291, article 110326, 2020 [DOI] [PubMed] [Google Scholar]
- 323.Xie H., Sun Y., Cheng B., Xue S., Cheng D., Liu L., Meng L., and Qiang S., “Variation in ICE1 methylation primarily determines phenotypic variation in freezing tolerance in Arabidopsis thaliana,” Plant & Cell Physiology, vol. 60, no. 1, pp. 152–165, 2019 [DOI] [PubMed] [Google Scholar]
- 324.Zhang Q., Liang Z., Cui X., Ji C., Li Y., Zhang P., Liu J., Riaz A., Yao P., Liu M., Wang Y., Lu T., Yu H., Yang D., Zheng H., and Gu X., “N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses,” Molecular Plant, vol. 11, no. 12, pp. 1492–1508, 2018 [DOI] [PubMed] [Google Scholar]
- 325.Popova O. V., Dinh H. Q., Aufsatz W., and Jonak C., “The RdDM Pathway Is Required for Basal Heat Tolerance in Arabidopsis,” Molecular Plant, vol. 6, no. 2, pp. 396–410, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Huang C.-F., Miki D., Tang K., Zhou H.-R., Zheng Z., Chen W., Ma Z. Y., Yang L., Zhang H., Liu R., He X. J., and Zhu J. K., “A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis,” PLoS Genetics, vol. 9, no. 9, article e1003779, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Huang W., Xian Z., Hu G., and Li Z., “SlAGO4A, a core factor of RNA-directed DNA methylation (RdDM) pathway, plays an important role under salt and drought stress in tomato,” Molecular Breeding, vol. 36, no. 3, p. 28, 2016 [Google Scholar]
- 328.Eddy S. R., “Non-coding RNA genes and the modern RNA world,” Nature Reviews Genetics, vol. 2, no. 12, pp. 919–929, 2001 [DOI] [PubMed] [Google Scholar]
- 329.Palazzo A. F., and Lee E. S., “Non-coding RNA: what is functional and what is junk?,” Frontiers in Genetics, vol. 6, p. 2, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Song X., Li Y., Cao X., and Qi Y., “MicroRNAs and their regulatory roles in plant–environment interactions,” Annual Review of Plant Biology, vol. 70, no. 1, pp. 489–525, 2019 [DOI] [PubMed] [Google Scholar]
- 331.Wong C. E., Zhao Y.-T., Wang X.-J., Croft L., Wang Z.-H., Haerizadeh F., Mattick J. S., Singh M. B., Carroll B. J., and Bhalla P. L., “MicroRNAs in the shoot apical meristem of soybean,” Journal of Experimental Botany, vol. 62, no. 8, pp. 2495–2506, 2011 [DOI] [PubMed] [Google Scholar]
- 332.Liu X., Zhang X., Sun B., Hao L., Liu C., Zhang D., Tang H., Li C., Li Y., Shi Y., Xie X., Song Y., Wang T., and Li Y., “Genome-wide identification and comparative analysis of drought-related microRNAs in two maize inbred lines with contrasting drought tolerance by deep sequencing,” PLoS One, vol. 14, no. 7, article e0219176, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhou L., Liu Y., Liu Z., Kong D., Duan M., and Luo L., “Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa,” Journal of Experimental Botany, vol. 61, no. 15, pp. 4157–4168, 2010 [DOI] [PubMed] [Google Scholar]
- 334.Akdogan G., Tufekci E. D., Uranbey S., and Unver T., “miRNA-based drought regulation in wheat,” Functional & Integrative Genomics, vol. 16, no. 3, pp. 221–233, 2016 [DOI] [PubMed] [Google Scholar]
- 335.Liu H.-H., Tian X., Li Y.-J., Wu C.-A., and Zheng C.-C., “Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana,” RNA, vol. 14, no. 5, pp. 836–843, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wang J., Mei J., and Ren G., “Plant microRNAs: biogenesis, homeostasis, and degradation,” Frontiers in Plant Science, vol. 10, p. 360, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Samad A. F., Sajad M., Nazaruddin N., Fauzi I. A., Murad A., Zainal Z., and Ismail I., “MicroRNA and transcription factor: key players in plant regulatory network,” Frontiers in Plant Science, vol. 8, p. 565, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Fang Y., Xie K., and Xiong L., “Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice,” Journal of Experimental Botany, vol. 65, no. 8, pp. 2119–2135, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rao S., Balyan S., Jha S., Bansal C., Das J. R., Gupta A., and Mathur S., “Orchestration of MicroRNAs and Transcription Factors in the Regulation of Plant Abiotic Stress Response,” Plant Stress Biology, Giri B., and Sharma M. P., Eds., Springer, Singapore, pp. 251–277, 2020 [Google Scholar]
- 340.Curaba J., Singh M. B., and Bhalla P. L., “miRNAs in the crosstalk between phytohormone signalling pathways,” Journal of Experimental Botany, vol. 65, no. 6, pp. 1425–1438, 2014 [DOI] [PubMed] [Google Scholar]
- 341.Singh P., Dutta P., and Chakrabarty D., “miRNAs play critical roles in response to abiotic stress by modulating cross-talk of phytohormone signaling,” Plant Cell Reports, vol. 40, no. 9, pp. 1617–1630, 2021 [DOI] [PubMed] [Google Scholar]
- 342.Jha U. C., Nayyar H., Jha R., Khurshid M., Zhou M., Mantri N., and Siddique K. H. M., “Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation,” BMC Plant Biology, vol. 20, no. 1, pp. 1–20, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Golicz A. A., Singh M. B., and Bhalla P. L., “The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome,” Plant Physiology, vol. 176, no. 3, pp. 2133–2147, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Golicz A. A., Bhalla P. L., and Singh M. B., “MCRiceRepGP: a framework for the identification of genes associated with sexual reproduction in rice,” The Plant Journal, vol. 96, no. 1, pp. 188–202, 2018 [DOI] [PubMed] [Google Scholar]
- 345.Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., and Chua N. H., “Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis,” The Plant Cell, vol. 24, no. 11, pp. 4333–4345, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wang T.-Z., Liu M., Zhao M.-G., Chen R., and Zhang W.-H., “Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing,” BMC Plant Biology, vol. 15, no. 1, pp. 1–13, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang K. C., and Chang H. Y., “Molecular mechanisms of long noncoding RNAs,” Molecular Cell, vol. 43, no. 6, pp. 904–914, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Wang A., Hu J., Gao C., Chen G., Wang B., Lin C., Song L., Ding Y., and Zhou G., “Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp.chinensis),” Scientific Reports, vol. 9, no. 1, pp. 1–14, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wierzbicki A. T., “The role of long non-coding RNA in transcriptional gene silencing,” Current Opinion in Plant Biology, vol. 15, no. 5, pp. 517–522, 2012 [DOI] [PubMed] [Google Scholar]
- 350.Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., and Crespi M., “Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop,” Molecular Cell, vol. 55, no. 3, pp. 383–396, 2014 [DOI] [PubMed] [Google Scholar]
- 351.Ariel F., Lucero L., Christ A., Mammarella M. F., Jegu T., Veluchamy A., Mariappan K., Latrasse D., Blein T., Liu C., Benhamed M., and Crespi M., “R-Loop Mediated trans Action of the APOLO Long Noncoding RNA,” Molecular Cell, vol. 77, no. 5, pp. 1055–1065.e4, 2020 [DOI] [PubMed] [Google Scholar]
- 352.Kim D.-H., Xi Y., and Sung S., “Modular function of long noncoding RNA, COLDAIR, in the vernalization response,” PLoS Genetics, vol. 13, no. 7, article e1006939, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Csorba T., Questa J. I., Sun Q., and Dean C., “Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization,” Proceedings of the National Academy of Sciences of the United States of America, vol. 111, no. 45, pp. 16160–16165, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Gu X., le C., Wang Y., Li Z., Jiang D., Wang Y., and He Y., “Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues,” Nature Communications, vol. 4, no. 1, pp. 1–10, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kim D.-H., and Sung S., “Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs,” Developmental Cell, vol. 40, no. 3, pp. 302–312.e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Qin T., Zhao H., Cui P., Albesher N., and Xiong L., “A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance,” Plant Physiology, vol. 175, no. 3, pp. 1321–1336, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Wang K., Wang C., Guo B., Song K., Shi C., Jiang X., Wang K., Tan Y., Wang L., Wang L., Li J., Li Y., Cai Y., Zhao H., and Sun X., “CropCircDB: a comprehensive circular RNA resource for crops in response to abiotic stress,” Database, vol. 2019, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Zhang P., Fan Y., Sun X., Chen L., Terzaghi W., Bucher E., Li L., and Dai M., “A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis,” The Plant Journal, vol. 98, no. 4, pp. 697–713, 2019 [DOI] [PubMed] [Google Scholar]
- 359.Babaei S., Singh M. B., and Bhalla P. L., “Circular RNAs repertoire and expression profile during brassica rapa pollen development,” International Journal of Molecular Sciences, vol. 22, no. 19, p. 10297, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Conn V. M., Hugouvieux V., Nayak A., Conos S. A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., and Conn S. J., “A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation,” Nature Plants, vol. 3, no. 5, pp. 1–5, 2017 [DOI] [PubMed] [Google Scholar]
- 361.Holdt L. M., Stahringer A., Sass K., Pichler G., Kulak N. A., Wilfert W., Kohlmaier A., Herbst A., Northoff B. H., Nicolaou A., Gäbel G., Beutner F., Scholz M., Thiery J., Musunuru K., Krohn K., Mann M., and Teupser D., “Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans,” Nature Communications, vol. 7, no. 1, pp. 1–14, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S. D., Gregersen L. H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., and Rajewsky N., “Circular RNAs are a large class of animal RNAs with regulatory potency,” Nature, vol. 495, no. 7441, pp. 333–338, 2013 [DOI] [PubMed] [Google Scholar]
- 363.Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., and Kjems J., “Natural RNA circles function as efficient microRNA sponges,” Nature, vol. 495, no. 7441, pp. 384–388, 2013 [DOI] [PubMed] [Google Scholar]
- 364.Gao Z., Li J., Luo M., Li H., Chen Q., Wang L., Song S., Zhao L., Xu W., Zhang C., Wang S., and Ma C., “Characterization and cloning of grape circular RNAs identified the cold Resistance-Related Vv-circATS1,” Plant Physiology, vol. 180, no. 2, pp. 966–985, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Lohani N., Jain D., Singh M. B., and Bhalla P. L., “Engineering multiple abiotic stress tolerance in canola, Brassica napus,” Frontiers in Plant Science, vol. 11, p. 3, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lohani N., Singh M. B., and Bhalla P. L., “High temperature susceptibility of sexual reproduction in crop plants,” Journal of Experimental Botany, vol. 71, no. 2, pp. 555–568, 2020 [DOI] [PubMed] [Google Scholar]
- 367.Lohani N., Singh M. B., and Bhalla P. L., “Short-term heat stress during flowering results in a decline in canola seed productivity,” Journal of Agronomy and Crop Science, 2021
- 368.Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., and Xiong L., “Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice,” Proceedings of the National Academy of Sciences of the United States of America, vol. 103, no. 35, pp. 12987–12992, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Qin D., Wang F., Geng X., Zhang L., Yao Y., Ni Z., Peng H., and Sun Q., “Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice,” Plant Molecular Biology, vol. 87, no. 1-2, pp. 31–45, 2015 [DOI] [PubMed] [Google Scholar]
- 370.Lee D.-K., Jung H., Jang G., Jeong J. S., Kim Y. S., Ha S.-H., Do Choi Y., and Kim J. K., “Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance,” Plant Physiology, vol. 172, no. 1, pp. 575–588, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Knight H., and Knight M. R., “Abiotic stress signalling pathways: specificity and cross-talk,” Trends in Plant Science, vol. 6, no. 6, pp. 262–267, 2001 [DOI] [PubMed] [Google Scholar]
- 372.Xiang Y., Huang Y., and Xiong L., “Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement,” Plant Physiology, vol. 144, no. 3, pp. 1416–1428, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Xiong L., and Yang Y., “Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase,” The Plant Cell, vol. 15, no. 3, pp. 745–759, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Xiao B.-Z., Chen X., Xiang C.-B., Tang N., Zhang Q.-F., and Xiong L.-Z., “Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions,” Molecular Plant, vol. 2, no. 1, pp. 73–83, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Shou H., Bordallo P., and Wang K., “Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize,” Journal of Experimental Botany, vol. 55, no. 399, pp. 1013–1019, 2004 [DOI] [PubMed] [Google Scholar]
- 376.Liao Y.-D., Lin K.-H., Chen C.-C., and Chiang C.-M., “Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice,” Molecular Breeding, vol. 36, no. 3, p. 22, 2016 [Google Scholar]
- 377.Gao P., Bai X., Yang L., Lv D., Pan X., Li Y., Cai H., Ji W., Chen Q., and Zhu Y., “osa-MIR393: a salinity-and alkaline stress-related microRNA gene,” Molecular Biology Reports, vol. 38, no. 1, pp. 237–242, 2011 [DOI] [PubMed] [Google Scholar]
- 378.Yang C., Li D., Mao D., Liu X., Ji C., Li X., Zhao X., Cheng Z., Chen C., and Zhu L., “Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.),” Plant, Cell & Environment, vol. 36, no. 12, pp. 2207–2218, 2013 [DOI] [PubMed] [Google Scholar]
- 379.Zhao J., Yuan S., Zhou M., Yuan N., Li Z., Hu Q., Bethea F. G. Jr., Liu H., Li S., and Luo H., “Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance,” Plant Biotechnology Journal, vol. 17, no. 1, pp. 233–251, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Zhang J., Zhang H., Srivastava A. K., Pan Y., Bai J., Fang J., Shi H., and Zhu J. K., “Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development,” Plant Physiology, vol. 176, no. 3, pp. 2082–2094, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Kumari P. H., Kumar S. A., Sivan P., Katam R., Suravajhala P., Rao K., Varshney R. K., and Kishor P. B. K., “Overexpression of a plasma membrane bound Na+/H+ antiporter-like protein (SbNHXLP) confers salt tolerance and improves fruit yield in tomato by maintaining ion homeostasis,” Frontiers in Plant Science, vol. 7, p. 2027, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Wang Y., Wisniewski M., Meilan R., Cui M., and Fuchigami L., “Transgenic tomato (Lycopersicon esculentum) overexpressing cAPX exhibits enhanced tolerance to UV-B and heat stress,” Journal of Applied Horticulture, vol. 8, no. 2, pp. 87–90, 2006 [Google Scholar]
- 383.Xiao B., Huang Y., Tang N., and Xiong L., “Over-expression of a LEA gene in rice improves drought resistance under the field conditions,” Theoretical and Applied Genetics, vol. 115, no. 1, pp. 35–46, 2007 [DOI] [PubMed] [Google Scholar]
- 384.Katiyar-Agarwal S., Agarwal M., and Grover A., “Heat-tolerant basmati rice engineered by over-expression of hsp101,” Plant Molecular Biology, vol. 51, no. 5, pp. 677–686, 2003 [DOI] [PubMed] [Google Scholar]
- 385.Zhang H., Dong H., Li W., Sun Y., Chen S., and Kong X., “Increased glycine betaine synthesis and salinity tolerance in AhCMO transgenic cotton lines,” Molecular Breeding, vol. 23, no. 2, pp. 289–298, 2009 [Google Scholar]
- 386.Garg A. K., Kim J.-K., Owens T. G., Ranwala A. P., Do Choi Y., Kochian L. V., and Wu R. J., “Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no. 25, pp. 15898–15903, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.He L., Shi X., Wang Y., Guo Y., Yang K., and Wang Y., “Arabidopsis ANAC069 binds to C [A/G] CG [T/G] sequences to negatively regulate salt and osmotic stress tolerance,” Plant Molecular Biology, vol. 93, no. 4-5, pp. 369–387, 2017 [DOI] [PubMed] [Google Scholar]
- 388.Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., Seki M., Shinozaki K., and Yamaguchi-Shinozaki K., “OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression,” The Plant Journal, vol. 33, no. 4, pp. 751–763, 2003 [DOI] [PubMed] [Google Scholar]
- 389.Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., and Yamaguchi-Shinozaki K., “Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice,” Plant & Cell Physiology, vol. 47, no. 1, pp. 141–153, 2006 [DOI] [PubMed] [Google Scholar]
- 390.Kasuga M., Miura S., Shinozaki K., and Yamaguchi-Shinozaki K., “A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought-and low-temperature stress tolerance in tobacco by gene transfer,” Plant & Cell Physiology, vol. 45, no. 3, pp. 346–350, 2004 [DOI] [PubMed] [Google Scholar]
- 391.Ravikumar G., Manimaran P., Voleti S., Subrahmanyam D., Sundaram R., Bansal K., Viraktamath B. C., and Balachandran S. M., “Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice,” Transgenic Research, vol. 23, no. 3, pp. 421–439, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Andrianantoandro E., Basu S., Karig D. K., and Weiss R., “Synthetic biology: new engineering rules for an emerging discipline,” Molecular Systems Biology, vol. 2, no. 1, article 2006.0028, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Benner S. A., and Sismour A. M., “Synthetic biology,” Nature Reviews Genetics, vol. 6, no. 7, pp. 533–543, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Weber W., and Fussenegger M., “Molecular diversity--the toolbox for synthetic gene switches and networks,” Current Opinion in Chemical Biology, vol. 15, no. 3, pp. 414–420, 2011 [DOI] [PubMed] [Google Scholar]
- 395.Weber W., and Fussenegger M., “Emerging biomedical applications of synthetic biology,” Nature Reviews Genetics, vol. 13, no. 1, pp. 21–35, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Keasling J. D., “Synthetic biology for synthetic chemistry,” ACS Chemical Biology, vol. 3, no. 1, pp. 64–76, 2008 [DOI] [PubMed] [Google Scholar]
- 397.Clarke L., and Kitney R., “Developing synthetic biology for industrial biotechnology applications,” Biochemical Society Transactions, vol. 48, no. 1, pp. 113–122, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Pérez-González A., and Caro E., “Hindrances to the efficient and stable expression of transgenes in plant synthetic biology approaches,” Systems Biology Application in Synthetic Biology, Singh S., Ed., Springer, New Delhi, pp. 79–89, 2016 [Google Scholar]
- 399.Bhalla P. L., and Singh M. B.. Wheat Biotechnology: Methods and Protocols, Springer, 2017 [Google Scholar]
- 400.Roberts M., Cranenburgh R., Stevens M., and Oyston P., “Synthetic biology: biology by design,” Microbiology, vol. 159, Part 7, pp. 1219–1220, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Bradley R. W., and Wang B., “Designer cell signal processing circuits for biotechnology,” New Biotechnology, vol. 32, no. 6, pp. 635–643, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Baldwin G.Synthetic Biology: A Primer, Imperial College Press, 2012 [Google Scholar]
- 403.Liu W., and Stewart C. N. Jr., “Plant synthetic biology,” Trends in Plant Science, vol. 20, no. 5, pp. 309–317, 2015 [DOI] [PubMed] [Google Scholar]
- 404.Patron N. J., Orzaez D., Marillonnet S., Warzecha H., Matthewman C., Youles M., Raitskin O., Leveau A., Farré G., Rogers C., Smith A., Hibberd J., Webb A. A. R., Locke J., Schornack S., Ajioka J., Baulcombe D. C., Zipfel C., Kamoun S., Jones J. D. G., Kuhn H., Robatzek S., van Esse H. P., Sanders D., Oldroyd G., Martin C., Field R., O'Connor S., Fox S., Wulff B., Miller B., Breakspear A., Radhakrishnan G., Delaux P. M., Loqué D., Granell A., Tissier A., Shih P., Brutnell T. P., Quick W. P., Rischer H., Fraser P. D., Aharoni A., Raines C., South P. F., Ané J. M., Hamberger B. R., Langdale J., Stougaard J., Bouwmeester H., Udvardi M., Murray J. A. H., Ntoukakis V., Schäfer P., Denby K., Edwards K. J., Osbourn A., and Haseloff J., “Standards for plant synthetic biology: a common syntax for exchange of DNA parts,” The New Phytologist, vol. 208, no. 1, pp. 13–19, 2015 [DOI] [PubMed] [Google Scholar]
- 405.Schürholz A.-K., López-Salmerón V., Li Z., Forner J., Wenzl C., Gaillochet C., Augustin S., Barro A. V., Fuchs M., Gebert M., Lohmann J. U., Greb T., and Wolf S., “A comprehensive toolkit for inducible, cell type-specific gene expression in Arabidopsis,” Plant Physiology, vol. 178, no. 1, pp. 40–53, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Batista-Silva W., da Fonseca-Pereira P., Martins A. O., Zsögön A., Nunes-Nesi A., and Araújo W. L., “Engineering improved photosynthesis in the era of synthetic biology,” Plant Communications, vol. 1, no. 2, article 100032, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Long B. M., Hee W. Y., Sharwood R. E., Rae B. D., Kaines S., Lim Y.-L., Nguyen N. D., Massey B., Bala S., von Caemmerer S., Badger M. R., and Price G. D., “Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts,” Nature Communications, vol. 9, no. 1, pp. 1–14, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Rae B. D., Long B. M., Förster B., Nguyen N. D., Velanis C. N., Atkinson N., Hee W. Y., Mukherjee B., Price G. D., and McCormick A. J., “Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants,” Journal of Experimental Botany, vol. 68, no. 14, pp. 3717–3737, 2017 [DOI] [PubMed] [Google Scholar]
- 409.Mackinder L. C., “TheChlamydomonasCO2-concentrating mechanism and its potential for engineering photosynthesis in plants,” The New Phytologist, vol. 217, no. 1, pp. 54–61, 2018 [DOI] [PubMed] [Google Scholar]
- 410.Schuler M. L., Mantegazza O., and Weber A. P. M., “Engineering C4photosynthesis into C3chassis in the synthetic biology age,” The Plant Journal, vol. 87, no. 1, pp. 51–65, 2016 [DOI] [PubMed] [Google Scholar]
- 411.Trudeau D. L., Edlich-Muth C., Zarzycki J., Scheffen M., Goldsmith M., Khersonsky O., Avizemer Z., Fleishman S. J., Cotton C. A. R., Erb T. J., Tawfik D. S., and Bar-Even A., “Design and in vitro realization of carbon-conserving photorespiration,” Proceedings of the National Academy of Sciences of the United States of America, vol. 115, no. 49, pp. E11455–E11464, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Yu H., Li X., Duchoud F., Chuang D. S., and Liao J. C., “Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA- glycerate carbon fixation pathway,” Nature Communications, vol. 9, no. 1, pp. 1–10, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Schwander T., Schada von Borzyskowski L., Burgener S., Cortina N. S., and Erb T. J., “A synthetic pathway for the fixation of carbon dioxide in vitro,” Science, vol. 354, no. 6314, pp. 900–904, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Antunes M. S., Morey K. J., Smith J. J., Albrecht K. D., Bowen T. A., Zdunek J. K., Troupe J. F., Cuneo M. J., Webb C. T., Hellinga H. W., and Medford J. I., “Programmable ligand detection system in plants through a synthetic signal transduction pathway,” PLoS One, vol. 6, no. 1, article e16292, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Park S.-Y., Peterson F. C., Mosquna A., Yao J., Volkman B. F., and Cutler S. R., “Agrochemical control of plant water use using engineered abscisic acid receptors,” Nature, vol. 520, no. 7548, pp. 545–548, 2015 [DOI] [PubMed] [Google Scholar]
- 416.Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P. T., and Müller B., “A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta,” Plant Physiology, vol. 161, no. 3, pp. 1066–1075, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Khakhar A., Leydon A. R., Lemmex A. C., Klavins E., and Nemhauser J. L., “Synthetic hormone-responsive transcription factors can monitor and re-program plant development,” eLife, vol. 7, article e34702, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Medford J. I., and Prasad A., “Towards programmable plant genetic circuits,” The Plant Journal, vol. 87, no. 1, pp. 139–148, 2016 [DOI] [PubMed] [Google Scholar]
- 419.Huang D., Kosentka P. Z., and Liu W., “Synthetic biology approaches in regulation of targeted gene expression,” Current Opinion in Plant Biology, vol. 63, article 102036, 2021 [DOI] [PubMed] [Google Scholar]
- 420.Liu W., and Stewart C. N. Jr., “Plant synthetic promoters and transcription factors,” Current Opinion in Biotechnology, vol. 37, pp. 36–44, 2016 [DOI] [PubMed] [Google Scholar]
- 421.Ali S., and Kim W.-C., “A fruitful decade using synthetic promoters in the improvement of transgenic plants,” Frontiers in Plant Science, vol. 10, p. 1433, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Aysha J., Noman M., Wang F., Liu W., Zhou Y., Li H., and Li X., “Synthetic promoters: designing the cis regulatory modules for controlled gene expression,” Molecular Biotechnology, vol. 60, no. 8, pp. 608–620, 2018 [DOI] [PubMed] [Google Scholar]
- 423.Yang Y., Lee J. H., Poindexter M. R., Shao Y., Liu W., Lenaghan S. C., Ahkami A. H., Blumwald E., and Stewart C. N. Jr., “Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements,” Plant Biotechnology Journal, vol. 19, no. 7, pp. 1354–1369, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Belcher M. S., Vuu K. M., Zhou A., Mansoori N., Agosto Ramos A., Thompson M. G., Scheller H. V., Loqué D., and Shih P. M., “Design of orthogonal regulatory systems for modulating gene expression in plants,” Nature Chemical Biology, vol. 16, no. 8, pp. 857–865, 2020 [DOI] [PubMed] [Google Scholar]
- 425.Lee J. E., Neumann M., Duro D. I., and Schmid M., “CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants,” PLoS One, vol. 14, no. 9, article e0222778, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Ramakrishnan C., Kesharwani M., and Velmurugan D., “11 recent advances, challenges, and opportunities in riboswitches,” Advances in Synthetic Biology., vol. 187, 2020 [Google Scholar]
- 427.Shanidze N., Lenkeit F., Hartig J. S., and Funck D., “A theophylline-responsive riboswitch regulates expression of nuclear-encoded genes,” Plant Physiology, vol. 182, no. 1, pp. 123–135, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Głowacka K., Kromdijk J., Kucera K., Xie J., Cavanagh A. P., Leonelli L., Leakey A. D. B., Ort D. R., Niyogi K. K., and Long S. P., “Photosystem II Subunit S overexpression increases the efficiency of water use in a field-grown crop,” Nature Communications, vol. 9, no. 1, pp. 1–9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Cabello J. V., Lodeyro A. F., and Zurbriggen M. D., “Novel perspectives for the engineering of abiotic stress tolerance in plants,” Current Opinion in Biotechnology, vol. 26, pp. 62–70, 2014 [DOI] [PubMed] [Google Scholar]
- 430.Markham K. K., and Greenham K., “Abiotic stress through time,” The New Phytologist, vol. 231, no. 1, pp. 40–46, 2021 [DOI] [PubMed] [Google Scholar]
- 431.Schilling R. K., Marschner P., Shavrukov Y., Berger B., Tester M., Roy S. J., and Plett D. C., “Expression of theArabidopsisvacuolar H+-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field,” Plant Biotechnology Journal, vol. 12, no. 3, pp. 378–386, 2014 [DOI] [PubMed] [Google Scholar]
- 432.El-Esawi M. A., and Alayafi A. A., “Overexpression of rice Rab7 gene improves drought and heat tolerance and increases grain yield in rice (Oryza sativa L.),” Genes, vol. 10, no. 1, p. 56, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Fang Y., Liao K., du H., Xu Y., Song H., Li X., and Xiong L., “A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice,” Journal of Experimental Botany, vol. 66, no. 21, pp. 6803–6817, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Roy M., and Wu R., “Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance,” Plant Science, vol. 163, no. 5, pp. 987–992, 2002 [Google Scholar]
- 435.Karthikeyan A., Pandian S. K., and Ramesh M., “Transgenic indica rice cv. ADT 43 expressing a Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene from Vigna aconitifolia demonstrates salt tolerance,” Plant Cell, Tissue and Organ Culture, vol. 107, no. 3, pp. 383–395, 2011 [Google Scholar]
- 436.Jacobs A., Ford K., Kretschmer J., and Tester M., “Rice plants expressing the moss sodium pumping ATPase PpENA1 maintain greater biomass production under salt stress,” Plant Biotechnology Journal, vol. 9, no. 8, pp. 838–847, 2011 [DOI] [PubMed] [Google Scholar]
- 437.Biradar H., Karan R., and Subudhi P. K., “Transgene pyramiding of salt responsive protein 3-1 (SaSRP3-1) and SaVHAc1 from Spartina alterniflora L. enhances salt tolerance in rice,” Frontiers in Plant Science, vol. 9, p. 1304, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Gupta B. K., Sahoo K. K., Ghosh A., Tripathi A. K., Anwar K., Das P., Singh A. K., Pareek A., Sopory S. K., and Singla-Pareek S. L., “Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice,” Plant, Cell & Environment, vol. 41, no. 5, pp. 1186–1200, 2018 [DOI] [PubMed] [Google Scholar]
- 439.Shim J. S., Oh N., Chung P. J., Kim Y. S., Choi Y. D., and Kim J.-K., “Overexpression of OsNAC14 improves drought tolerance in rice,” Frontiers in Plant Science, vol. 9, p. 310, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Xiang Y., Tang N., Du H., Ye H., and Xiong L., “Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice,” Plant Physiology, vol. 148, no. 4, pp. 1938–1952, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Manavalan L. P., Chen X., Clarke J., Salmeron J., and Nguyen H. T., “RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice,” Journal of Experimental Botany, vol. 63, no. 1, pp. 163–175, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Hou X., Xie K., Yao J., Qi Z., and Xiong L., “A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance,” Proceedings of the National Academy of Sciences of the United States of America, vol. 106, no. 15, pp. 6410–6415, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Duan J., and Cai W., “OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance,” PLoS One, vol. 7, no. 9, article e45117, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Soda N., Gupta B. K., Anwar K., Sharan A., Singla-Pareek S. L., and Pareek A., “Rice intermediate filament, OsIF, stabilizes photosynthetic machinery and yield under salinity and heat stress,” Scientific Reports, vol. 8, no. 1, pp. 1–13, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.El-Kereamy A., Bi Y.-M., Ranathunge K., Beatty P. H., Good A. G., and Rothstein S. J., “The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism,” PLoS One, vol. 7, no. 12, article e52030, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Chang Y., Nguyen B. H., Xie Y., Xiao B., Tang N., Zhu W., Mou T., and Xiong L., “Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice,” Frontiers in Plant Science, vol. 8, p. 1102, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Biswas S., Islam M. N., Sarker S., Tuteja N., and Seraj Z. I., “Overexpression of heterotrimeric G protein beta subunit gene (OsRGB1) confers both heat and salinity stress tolerance in rice,” Plant Physiology and Biochemistry, vol. 144, pp. 334–344, 2019 [DOI] [PubMed] [Google Scholar]
- 448.Scafaro A. P., Atwell B. J., Muylaert S., Reusel B. V., Ruiz G. A., Rie J. V., and Gallé A., “A thermotolerant variant of Rubisco activase from a wild relative improves growth and seed yield in rice under heat stress,” Frontiers in Plant Science, vol. 9, p. 1663, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Qu Y., Sakoda K., Fukayama H., Kondo E., Suzuki Y., Makino A., Terashima I., and Yamori W., “Overexpression of both Rubisco and Rubisco activase rescues rice photosynthesis and biomass under heat stress,” Plant, Cell & Environment, vol. 44, no. 7, pp. 2308–2320, 2021 [DOI] [PubMed] [Google Scholar]
- 450.Tian Y., Zhang H., Pan X., Chen X., Zhang Z., Lu X., and Huang R., “Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings,” Transgenic Research, vol. 20, no. 4, pp. 857–866, 2011 [DOI] [PubMed] [Google Scholar]
- 451.Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., Pan Y., Lin X., Liu H., Zhang D., Xiao J., Guo X., Xu S., Niu Y., Jin J., Zhang H., Xu X., Li L., Wang W., Qian Q., Ge S., and Chong K., “COLD1 Confers Chilling Tolerance in Rice,” Cell, vol. 160, no. 6, pp. 1209–1221, 2015 [DOI] [PubMed] [Google Scholar]
- 452.Su C.-F., Wang Y.-C., Hsieh T.-H., Lu C.-A., Tseng T.-H., and Yu S.-M., “A novel MYBS3-dependent pathway confers cold tolerance in rice,” Plant Physiology, vol. 153, no. 1, pp. 145–158, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., Xiao J., Zhang D., Xu Z., Zhang X., and Chong K., “Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes,” Plant Physiology, vol. 150, no. 1, pp. 244–256, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Shen H., Zhong X., Zhao F., Wang Y., Yan B., Li Q., Chen G., Mao B., Wang J., Li Y., Xiao G., He Y., Xiao H., Li J., and He Z., “Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato,” Nature Biotechnology, vol. 33, no. 9, pp. 996–1003, 2015 [DOI] [PubMed] [Google Scholar]
- 455.Chen L., Zhao Y., Xu S., Zhang Z., Xu Y., Zhang J., and Chong K., “OsMADS57 together with OsTB1 coordinates transcription of its targetOsWRKY94andD14to switch its organogenesis to defense for cold adaptation in rice,” The New Phytologist, vol. 218, no. 1, pp. 219–231, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.He C., Yang A., Zhang W., Gao Q., and Zhang J., “Improved salt tolerance of transgenic wheat by introducing betA gene for glycine betaine synthesis,” Plant Cell, Tissue and Organ Culture, vol. 101, no. 1, pp. 65–78, 2010 [Google Scholar]
- 457.González F. G., Capella M., Ribichich K. F., Curín F., Giacomelli J. I., Ayala F., Watson G., Otegui M. E., and Chan R. L., “Field-grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type,” Journal of Experimental Botany, vol. 70, no. 5, pp. 1669–1681, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Hu X. J., Chen D., Lynne Mclntyre C., Fernanda Dreccer M., Zhang Z. B., Drenth J., Kalaipandian S., Chang H., and Xue G. P., “Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway,” Plant, Cell & Environment, vol. 41, no. 1, pp. 79–98, 2018 [DOI] [PubMed] [Google Scholar]
- 459.Zang X., Geng X., Wang F., Liu Z., Zhang L., Zhao Y., Tian X., Ni Z., Yao Y., Xin M., Hu Z., Sun Q., and Peng H., “Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging,” BMC Plant Biology, vol. 17, no. 1, pp. 1–13, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Weichert H., Högy P., Mora-Ramirez I., Fuchs J., Eggert K., Koehler P., Weschke W., Fangmeier A., and Weber H., “Grain yield and quality responses of wheat expressing a barley sucrose transporter to combined climate change factors,” Journal of Experimental Botany, vol. 68, no. 20, pp. 5511–5525, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Fu J., Momčilović I., Clemente T. E., Nersesian N., Trick H. N., and Ristic Z., “Heterologous expression of a plastid EF-Tu reduces protein thermal aggregation and enhances CO2 fixation in wheat (Triticum aestivum) following heat stress,” Plant Molecular Biology, vol. 68, no. 3, pp. 277–288, 2008 [DOI] [PubMed] [Google Scholar]
- 462.Xue G.-P., Drenth J., and McIntyre C. L., “TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets,” Journal of Experimental Botany, vol. 66, no. 3, pp. 1025–1039, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.El-Esawi M. A., Al-Ghamdi A. A., Ali H. M., and Ahmad M., “Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.),” Genes, vol. 10, no. 2, p. 163, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Ma H., Liu C., Li Z., Ran Q., Xie G., Wang B., Fang S., Chu J., and Zhang J., “ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development,” Plant Physiology, vol. 178, no. 2, pp. 753–770, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Shi J., Gao H., Wang H., Lafitte H. R., Archibald R. L., Yang M., Hakimi S. M., Mo H., and Habben J. E., “ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions,” Plant Biotechnology Journal, vol. 15, no. 2, pp. 207–216, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Lu Y., Li Y., Zhang J., Xiao Y., Yue Y., Duan L., Zhang M., and Li Z., “Overexpression of Arabidopsis molybdenum cofactor sulfurase gene confers drought tolerance in maize (Zea mays L.),” PLoS One, vol. 8, no. 1, article e52126, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Casaretto J. A., el-kereamy A., Zeng B., Stiegelmeyer S. M., Chen X., Bi Y.-M., and Rothstein S. J., “Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance,” BMC Genomics, vol. 17, no. 1, pp. 1–15, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Shen G., Wei J., Qiu X., Hu R., Kuppu S., Auld D., Blumwald E., Gaxiola R., Payton P., and Zhang H., “Co-overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants,” Plant Molecular Biology Reporter, vol. 33, no. 2, pp. 167–177, 2015 [Google Scholar]
- 469.Pasapula V., Shen G., Kuppu S., Paez-Valencia J., Mendoza M., Hou P., Chen J., Qiu X., Zhu L., Zhang X., Auld D., Blumwald E., Zhang H., Gaxiola R., and Payton P., “Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought-and salt tolerance and increases fibre yield in the field conditions,” Plant Biotechnology Journal, vol. 9, no. 1, pp. 88–99, 2011 [DOI] [PubMed] [Google Scholar]
- 470.Su Y., Liang W., Liu Z., Wang Y., Zhao Y., Ijaz B., and Hua J., “Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum,” Journal of Plant Physiology, vol. 218, pp. 222–234, 2017 [DOI] [PubMed] [Google Scholar]
- 471.Bhaskaran S., and Savithramma D. L., “Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato,” Journal of Experimental Botany, vol. 62, no. 15, pp. 5561–5570, 2011 [DOI] [PubMed] [Google Scholar]
- 472.Cheng L., Zou Y., Ding S., Zhang J., Yu X., Cao J., and Lu G., “Polyamine accumulation in transgenic tomato enhances the tolerance to high temperature stress,” Journal of Integrative Plant Biology, vol. 51, no. 5, pp. 489–499, 2009 [DOI] [PubMed] [Google Scholar]
- 473.Liu H., Yu C., Li H., Ouyang B., Wang T., Zhang J., Wang X., and Ye Z., “Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato,” Plant Science, vol. 231, pp. 198–211, 2015 [DOI] [PubMed] [Google Scholar]
- 474.Wani U. M., Majeed S. T., Raja V., Wani Z. A., Jan N., Andrabi K. I., and John R., “Ectopic expression of a novel cold-resistance protein 1 from Brassica oleracea promotes tolerance to chilling stress in transgenic tomato,” Scientific Reports, vol. 11, no. 1, article 16574, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Zhang L., Guo X., Zhang Z., Wang A., and Zhu J., “Cold-regulated gene LeCOR413PM2 confers cold stress tolerance in tomato plants,” Gene, vol. 764, article 145097, 2021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.