Abstract
Global demand for food and bioenergy production has increased rapidly, while the area of arable land has been declining for decades due to damage caused by erosion, pollution, sea level rise, urban development, soil salinization, and water scarcity driven by global climate change. In order to overcome this conflict, there is an urgent need to adapt conventional agriculture to water-limited and hotter conditions with plant crop systems that display higher water-use efficiency (WUE). Crassulacean acid metabolism (CAM) species have substantially higher WUE than species performing C3 or C4 photosynthesis. CAM plants are derived from C3 photosynthesis ancestors. However, it is extremely unlikely that the C3 or C4 crop plants would evolve rapidly into CAM photosynthesis without human intervention. Currently, there is growing interest in improving WUE through transferring CAM into C3 crops. However, engineering a major metabolic plant pathway, like CAM, is challenging and requires a comprehensive deep understanding of the enzymatic reactions and regulatory networks in both C3 and CAM photosynthesis, as well as overcoming physiometabolic limitations such as diurnal stomatal regulation. Recent advances in CAM evolutionary genomics research, genome editing, and synthetic biology have increased the likelihood of successful acceleration of C3-to-CAM progression. Here, we first summarize the systems biology-level understanding of the molecular processes in the CAM pathway. Then, we review the principles of CAM engineering in an evolutionary context. Lastly, we discuss the technical approaches to accelerate the C3-to-CAM transition in plants using synthetic biology toolboxes.
1. Introduction
The global population has quadrupled over the past 100 years and will continue to increase in the 21st century [1]. To feed the growing population, crop production must increase, either by expanding the amount of agricultural land for growing crops or by increasing crop yields on existing agricultural lands. Simultaneously, ongoing and projected climate changes are (1) affecting many sectors important to society, including human health, agricultural sustainability, water supply, energy security, and food supply and (2) becoming increasingly disruptive in the coming decades [2–4]. These opposing trends are threatening our global food and energy security [5]. To meet this challenge, various approaches have been explored to increase the productivity of agricultural crops [6–9]. Among them, one of the most direct approaches is engineering crassulacean acid metabolism (CAM) into C3 crops to enhance water-use efficiency (WUE) in plants [9] thereby allowing such crops to be grown on marginal lands with reduced fresh water inputs.
To adapt to various environments on Earth, plant species have evolved several photosynthetic chemistries—C3, C4, and CAM photosynthesis [10]. The way plants fix atmospheric CO2 is the key to distinguish different photosynthesis. C3 photosynthesis is a one-stage process that produces a three-carbon compound (3-phosphoglyceric acid) via the Calvin-Benson-Bassham (CBB) cycle, while C4 photosynthesis and CAM photosynthesis are two-stage processes, with the fisrt stage fixing CO2 into a series of four-carbon compounds from oxaloacetate to malate, followed by the secondary stage, where four-carbon compounds are decarboxylated, releasing CO2 to be refixed via the CBB cycle. In C4 plants, photosynthesis is separated spatially (mesophyll and bundle sheath cells), whereas in CAM photosynthesis CO2 fixation is separated temporally (day and night). In CAM plants, stomata close during part or all of the day to reduce water loss, and the CO2 is released from the malate generated during the first CO2-fixing stage, resulting in enhanced plant WUE in comparison with C3 or C4 plants. WUE is the crop’s ability to assimilate a unit of carbon per unit of water consumed [11]. However, gas exchange in the leaf to obtain CO2 inevitably results in water loss. The CAM solution to this problem is to open the stomata at night and fix carbon into malic acid, then close the stomata during the heat of the day, and release the stored CO2 to the CBB cycle, maximizing WUE. Typically, CAM species have very high WUE, at least six- and three-fold greater than that of C3 and C4 plants, respectively [12].
Fresh water is the most critical resource of sustainable agriculture, and approximately 42% of the land area on Earth is classified as dryland [13, 14], where precipitation is inadequate for major conventionally grown C3 or C4 crops. Bioengineering CAM into C3 plants is a potential solution to these challenges. However, engineering a major metabolic pathway like CAM is not a trivial task. Not only does it require a deep understanding of the metabolic and regulatory pathways during CAM photosynthesis, but also it requires precise regulation of the enzymatic activities, intracellular transporters, and stomatal conductance [9, 15, 16].
CAM species have been increasingly considered important climate-resilient species in the world and are a crucial driving force of ecosystem function in arid areas [17]. Recently, important achievements were made in CAM plant genomics research, significantly increasing our knowledge on the molecular mechanisms underlying CAM photosynthesis [17–20]. However, the application of this basic knowledge to CAM engineering is still limited due to technical challenges, including the lack of robust biosystems design capabilities for reconfiguring signaling and metabolic pathways in plants. Recently, biosystems design, integration of systems biology, and synthetic biology based on genome editing have emerged as innovative approaches for genetic improvement of complex biological systems in plants, microbes, and animals [21]. And as such, opportunities for revolutionizing agriculture with synthetic biology are emerging [22].
This review is intended to inspire the utilization of a biosystems design approach to accelerate C3-to-CAM progression. First, we provide a summary of the molecular mechanisms underpinning CAM photosynthesis based on systems biology research. Second, we discuss the principles of CAM engineering in an evolutionary context. Lastly, we integrate the capabilities of gene editing and synthetic biology for CAM engineering, with a focus on building a CAM-on-demand system to increase plant resistance to episodic or seasonal drought stress.
2. A Systems Biology-Level Understanding of CAM Photosynthesis
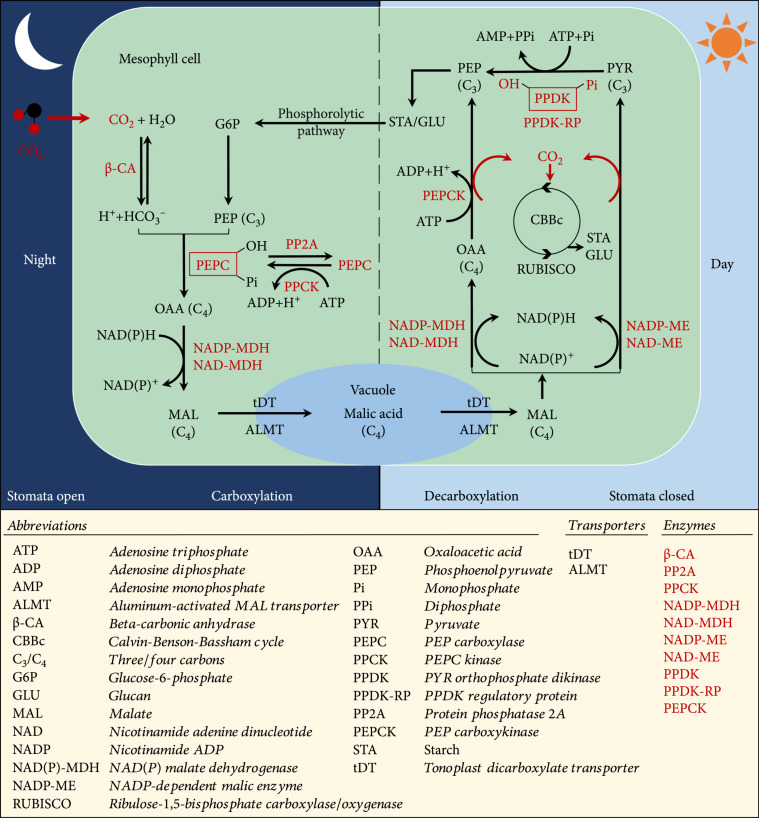
The exploration of the molecular mechanisms of CAM is critical for CAM engineering in C3 plant species. CAM features four core functional modules: (1) a carboxylation module to fix CO2 and accumulate malic acid in the vacuole during the nighttime, (2) a decarboxylation module to release CO2 from malic acid during the daytime for refixation mediated by ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) [9, 23] (Figure 1), (3) a stomatal control module to open stomata during the night and close them during the day, and (4) an anatomical module to increase the succulence of the leaf tissue [9]. A distinctive feature of CAM plants is that the stomata in the leaves remain closed during most or all of the daytime but open during the nighttime to take up CO2, reducing water loss and correspondingly increasing WUE due to the lower evapotranspiration rates at night. Over the past ten years, genes in these functional modules (Table 1) have been identified using systems biology approaches, which involved multiomics (e.g., genomics, transcriptomics, metabolomics, and proteomics), metabolic modeling, and molecular genetic technologies such as RNA interference (RNAi) and gene editing mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems.
Figure 1.
A simplified view of the crassulacean acid metabolism (CAM) photosynthetic pathway including key enzymes, regulatory proteins, and transporters.
Table 1.
List of known genes within the functional CAM modules.
| Protein name | Gene locus | Definition (subcellular location) | Species | Reference |
|---|---|---|---|---|
| Carboxylation module | ||||
| β-CA | Kaladp0018s0289 | β-Type carbonic anhydrase | K. fedtschenkoi | [20] |
| β-CA2 | Mcr010929t1 | β-Type carbonic anhydrase 2 (cytosol) | M. crystallinum | [30] |
| PEPC1 | Kaladp0095s0055 | Phosphoenolpyruvate carboxylase 1 | K. fedtschenkoi | [20] |
| PEPC1 | Mcr000915t1 | Phosphoenolpyruvate carboxylase 1 (cytosol) | M. crystallinum | [30] |
| PEPC1 | Kalax.0018s0056.1 | Phosphoenolpyruvate carboxylase 1 | K. laxiflora | [35] |
| Kalax.0021s0061.1 | ||||
| PEPC2 | Kaladp0048s0578 | Phosphoenolpyruvate carboxylase 2 | K. fedtschenkoi | [20] |
| PPCK | Kaladp0037s0517 | PEPC kinase | K. fedtschenkoi | [20] |
| PPCK1 | Mcr011042t1 | PEPC kinase 1 (cytosol) | M. crystallinum | [30] |
| NAD-MDH | Kaladp0022s0111 | NAD-malate dehydrogenase | K. fedtschenkoi | [20] |
| NAD-MDH1 | Mcr009416t1 | NAD-malate dehydrogenase 1 (cytosol) | M. crystallinum | [30] |
| NAD-MDH2 | Mcr008974t1 | NAD-malate dehydrogenase 2 (mitochondria) | M. crystallinum | [30] |
| NADP-MDH1 | Mcr006398t1 | NADP-malate dehydrogenase 1 (chloroplast) | M. crystallinum | [30] |
| ALMT6 | Kaladp0062s0038 | Tonoplast aluminum-activated malate transporter 6 | K. fedtschenkoi | [20] |
| ALMT4 | Tonoplast aluminum-activated malate transporter 4 (tonoplast membrane) | M. crystallinum | Lim et al., unpublished data | |
| tDT | Tonoplast dicarboxylate transporter | Agave | [37] | |
| tDT | Tonoplast dicarboxylate transporter (tonoplast membrane) | M. crystallinum | Lim et al., unpublished data | |
|
| ||||
| Decarboxylation module | ||||
| NAD-ME | NAD-dependent malic enzyme | K. fedtschenkoi | [20] | |
| NAD-ME1 | Mcr021367t1 | NAD-dependent malic enzyme 1, alpha subunit (mitochondria) | M. crystallinum | [30] |
| NAD-ME2 | Mcr003267t1 | NAD-dependent malic enzyme 2, beta subunit (mitochondria) | M. crystallinum | [30] |
| NADP-ME | Kaladp0092s0166 | NADP-dependent malic enzyme | K. fedtschenkoi | [20] |
| NADP-ME1 | Mcr003238t1 | NADP-dependent malic enzyme 1 (cytosol) | M. crystallinum | [30] |
| NADP-ME2 | Mcr002920t1 | NADP-dependent malic enzyme 2 (chloroplast) | M. crystallinum | [30] |
| PPDK | Mcr000976t1 | Pyruvate, orthophosphate dikinase (chloroplast) | M. crystallinum | [30] |
| PPDK-RP | Kaladp0010s0106 | Pyruvate, orthophosphate dikinase-regulatory protein | K. fedtschenkoi | [20] |
| PPDK-RP | Mcr007074t1 | Pyruvate, orthophosphate dikinase (chloroplast) | M. crystallinum | [30] |
| PEPCK | Phosphoenolpyruvate carboxykinase | K. fedtschenkoi | [20] | |
| PPCK1 | AF162661 | Phosphoenolpyruvate carboxykinase | K. fedtschenkoi | [25] |
| PEPCK | Phosphoenolpyruvate carboxykinase (cytosol) | M. crystallinum | [30] | |
|
| ||||
| Stomatal regulation module | ||||
| PHOT2 | Kaladp0033s0113 | Blue light receptor phototropin 2 | K. fedtschenkoi | [34] |
| AKT2 | Arabidopsis shaker family K+ channels 2/3 | Agave | [37] | |
| PP1 | Protein phosphatase 1 | K. pinnata, K. daigremontiana | [33] | |
| PM H+-ATPases | Plasma membrane H+-ATPase | K. pinnata, K. daigremontiana | [33] | |
|
| ||||
| Anatomical module | ||||
| VvCEB1 | Basic helix-loop-helix transcription factor | Vitis vinifera | [59, 60] | |
| PeXTH | The xyloglucan endotransglucosylase/hydrolase | P. euphratica | [57] | |
| Circadian clock module | ||||
| CCA1 | Circadian clock associated 1 | Agave | [37] | |
| TOC1 | Timing of cab expression1 | Agave | [37] | |
| RVE1 | Reveille 1 | Agave | [37] | |
2.1. Genes in the CAM Carboxylation Module
After atmospheric CO2 enters the mesophyll cells, it is converted to HCO3- by beta-carbonic anhydrase (β-CA), which is further, in combination with phosphoenolpyruvate (PEP), converted to oxaloacetate (OAA) by PEP carboxylase (PEPC) in the cytosol [24]. In most CAM plants, the reversible phosphorylation-dephosphorylation of PEPC mediated by PEPC kinase (PPCK) and possibly protein phosphatase 2A (PP2A) is understood to be under the control of the circadian clock (Figure 1) [25]. PEPC1 and PEPC2 are two most abundant PEPC transcripts in Kalanchoe fedtschenkoi. Two different patterns of convergent evolution are understood to be relevant to the carboxylation module. In the first pattern, the shift of PPCK expression from the light period to the dark period promoted the activation of PEPC1, as revealed in K. fedtschenkoi and Ananas comosus [20, 25]. In the second pattern, a single amino acid change from an arginine (R)/lysine (K)/histidine (H) to an aspartic acid (D) residue at the 509th position counting from N-terminal occurred to keep PEPC2 active without being phosphorylated, as observed in CAM species Phalaenopsis equestris and K. fedtschenkoi [20]. Then, NAD(P)-malate dehydrogenase (NAD(P)-MDH) converts OAA to malate, which is transported into the vacuole by an aluminum-activated malate transporter (ALMT) or a tonoplast dicarboxylate transporter (tDT) (Figure 1) [26–29]. Recently, ectopic overexpression of each of the five individual carboxylation proteins (β-CA2, NAD-MDH1, NAD-MDH2, PEPC1, and PPCK1) from Mesembryanthemum crystallinum, which is a facultative CAM species, enhanced leaf growth, along with an increase in organic acid accumulation and stomatal conductance in Arabidopsis thaliana [30]. The increased plant size and biomass yield in the transgenic Arabidopsis plants might arise from the release of intracellular CO2, which reduced photorespiration and consequently promoted plant growth [30].
2.2. Genes in the CAM Decarboxylation Module
During the daytime, the malic acid is moved out of the vacuole and subsequently decarboxylated to release CO2 for Rubisco-mediated refixation in the chloroplast, generating carbohydrates through the CBB cycle (Figure 1). Two different likely species-dependent processes for malate decarboxylation occur according to whether the plants contain high levels of PEP carboxykinase (PEPCK) or NAD(P)-malic enzyme (NAD(P)-ME) (Figure 1). In the NAD(P)-ME-mediated decarboxylation process, malate is converted by NAD(P)-ME to pyruvate, along with the release of CO2 in the cytosol (or mitochondria/chloroplast), followed by subsequent conversion of pyruvate to PEP mediated by pyruvate orthophosphate dikinase (PPDK). In this process, the reversible phosphorylation-dephosphorylation of PPDK, catalyzed by the PPDK regulatory protein (PPDK-RP), results in activation-inactivation of PPDK in the light-dark cycle [31]. In the PEPCK-mediated decarboxylation process, NAD(P)-MDH converts malate to OAA, which is subsequently decarboxylated to PEP and CO2 by PEPCK (Figure 1). PEP is then metabolized into starch or other storage glucans and stored in plants during the day. The starch or other stored carbohydrates can be converted back to PEP via glycolysis to fuel subsequent carboxylation at night (Figure 1). In the same study mentioned above [30], ectopic overexpression of five decarboxylation proteins (NADP-ME1, NADP-ME2, NAD-ME1, NAD-ME2, and PPDK) from M. crystallinum also increased plant size, along with a decrease in stomatal conductance and accumulation of organic acid caused by NADP-ME1 and NADP-ME2 in A. thaliana.
2.3. Genes Affecting Stomatal Movement
The typical gas exchange pattern in CAM plants shows extensive interspecific, intraspecific, and intraindividual variation, which complicates the study of stomatal movement. Multiple factors, including blue light, leaf-air vapor pressure deficit (VPD), leaf water status, and intercellular CO2 concentration (C), affect the regulation of a stomatal aperture [32]. Recently, protein phosphatase 1 (PP1) and plasma membrane (PM) H+-ATPase were shown to play crucial roles in the blue light-dependent stomatal opening in K. daigremontiana and K. pinnata, which are two obligate CAM species [33]. Furthermore, knocking out of blue phototropin 2 (KfePHOT2), a light receptor, reduced stomatal conductance and Rubisco-mediated CO2 fixation in the late afternoon when stomata are reopened and enhanced stomatal conductance and the nighttime CO2 fixation in the CAM species K. fedtschenkoi [34]. RNAi-mediated knockdown of the CAM PEPC isozyme (PEPC1) in K. laxiflora disrupts the dark period CO2 fixation and stomatal conductance and alters the temporal phasing of expression of genes controlling the movement of stomata, suggesting that inverse stomatal behavior is also likely to be dependent upon the activity of the primary carboxylation reaction [35]. Leaf water status usually acts on an ABA-dependent stomatal aperture in CAM plants [36–38]. C is a key driving force for CAM stomatal rhythm, which indicates the importance of metabolic control of stomatal movement in CAM plants [39, 40]. However, the key genes involved in leaf water status and C remain to be determined in CAM plants. Although the circadian oscillator can shape the rhythms of stomatal movement in CAM plants, it might not be as important as that in C3 plants [32]. Recently, numerous candidate genes were predicted to be involved in stomatal opening and closing in CAM plants [41]. More recently, over 200 K. fedtschenkoi genes were predicted to be relevant to stomatal movement [42]. Although it would be very challenging to engineer stomatal movement, there is precedence using small molecules to control stomata [43–45]. This could be used to provide proof-of-concept studies for CAM engineering.
2.4. Genes in the Anatomical Module
Besides the critical role of temporal gene expression in CAM plants, specific functional anatomical traits are thought to be associated with optimal CAM function [46–48]. Enlarged cells allow for a larger amount of organic acids to be stored in the vacuole during the nighttime [49] and also potentially enhance water uptake and remobilization in the chlorenchyma [50]. Densely packed mesophyll cells can reduce CO2 conductance () within the leaf and CO2 efflux from the leaf, increasing the capacity for performing CAM [46, 51, 52]. In typical CAM species, leaf thickness and cell size are increased whereas intracellular air space (IAS) and the length of mesophyll surface exposed to IAS per unit area () are reduced in comparison with non-CAM plant species [47]. For example, leaf thickness as a measure of tissue succulence has been associated with the performance of CAM in the Crassulaceae [53], the Orchidaceae [54], and other CAM families [47]. A comparative analysis of phylogenetically unrelated C3+CAM and strong CAM species revealed that cell size was not related to CAM, reduced IAS and were associated with CAM, and there was no difference in the proportion of IAS and between strong and weak CAM species [46]. Also, a comparative analysis of multiple Clusia species ranging from C3 to CAM with intermediates showed that the proportion of CO2 uptake during the nighttime was significantly correlated with the size of palisade mesophyll cells [55]. However, in Yucca gloriosa, which is a C3+CAM hybrid species, leaf anatomy and CAM function were not well correlated, suggesting that CAM evolution can proceed initially through different combinations of multiple traits, and then, more favorable trait combinations are selected to form strong CAM species [56].
Several strategies have proven successful in increasing leaf and tissue succulence in C3 species with beneficial traits. For example, overexpression of the Populus euphratica xyloglucan endotransglucosylase/hydrolase gene (PeXTH) in tobacco decreased IAS within the palisade parenchyma, along with an increase in both leaf water content and cell packing, leading to improved salinity tolerance, presumably due to a reduction in the content of intercellular NaCl within leaf tissues [57]. Also, ectopic expression of a codon-optimized form (VvCEB1opt) of the grape gene VvCEB1, which encodes a transcription factor in the basic helix-loop-helix (bHLH) family [58], increased organ and cell size, vegetative and reproductive biomass, and seed yield in A. thaliana [59]. Furthermore, overexpressing VvCEB1 was shown to increase tissue succulence and decrease intercellular air space (IAS), leading to a leaf anatomy that could potentially optimize the performance of CAM [60]. In the VvCEB1opt-overexpressing lines, the integrated and instantaneous WUE were increased, resulting in dramatically improved drought tolerance, along with enhanced salt tolerance due to a decrease in salinity uptake as well as a dilution of internal Na+ and Cl- within the succulent leaves [60].
Besides the gene products involved in the above CAM modules, many other gene products are also implicated to function in CAM, such as starch phosphorylase, which is involved in the formation of PEP by glycolysis [61], and gene products involved in the regeneration of storage carbohydrates. Recently, at least 60 genes that are potentially involved in CAM evolution were identified in a comparative analysis of three obligatory CAM species (K. fedtschenkoi, P. equestris, and A. comosus) and some non-CAM plant species [20] or by comparison of nonphotosynthetic and photosynthetic tissues in A. comosus [41]. Among these genes predicted to be involved in CAM evolution, 54 genes displayed rewired diel patterns of gene expression and 6 genes showed protein sequence mutations [20]. The functional analysis of individual CAM-related genes by either overexpression [30], knockdown [25, 35, 39], or knockout [34] has laid a solid foundation for CAM biodesign. A functional CAM pathway is unlikely to result from single-gene engineering in C3 plants [30]. Clearly, the engineering of multiple genes related to CAM in a modular manner is necessary to recapitulate partially or fully functional CAM modules or pathways. To move forward, the future effort for engineering CAM in C3 plants should focus on the coordinated expression of the genes involved in carboxylation and decarboxylation in a manner as displayed by CAM species. Also, CAM engineering requires precise dynamic control of carbohydrate transportation, degradation, and storage to supply PEP, which is required for the PEPC-mediated carboxylation process during the nighttime.
3. The Progress in the Understanding of CAM Evolution
C3 photosynthesis is the predominant route that plants take in CO2 and produce carbohydrates, representing approximately 95% of the Earth’s plant biomass [62]. In contrast, C4 and CAM species, derived from C3 ancestors, account for about 3% and 6% of flowering plant species, respectively [63, 64]. Among the angiosperms (flowering plants), C4 photosynthesis has evolved independently at least 61 times in 19 families, and CAM has evolved independently in more than 400 genera across more than 38 families [15, 17, 65, 66]. Therefore, CAM photosynthesis and C4 photosynthesis are thought to be the result of convergent evolution from independent C3 plant lineages [15]. Among the 60 candidate genes underpinning the convergent evolution of CAM from diverse lineages of C3 plants, 90% showed rewiring of diel gene expression [20]. Interestingly, all of the enzymes in CAM seem to have homologs in C3 species [67, 68]. Shared biochemical properties suggest that the repeated, independent CAM and C4 evolution is due to the reorganization of coopted and modified ancient metabolic pathways [69]. The involved modifications can be initiated by mutation(s) and then accommodated under selection by genomic change as the adaptive phenotype evolves [70]. Indeed, C4 evolution is thought to require an enabling mutation to form an initial C4 cycle, followed by selection for loss of high expression of photorespiratory genes in a certain cell type [71]. However, an enabling mutation is hypothesized not to be required for the evolution of CAM [67].
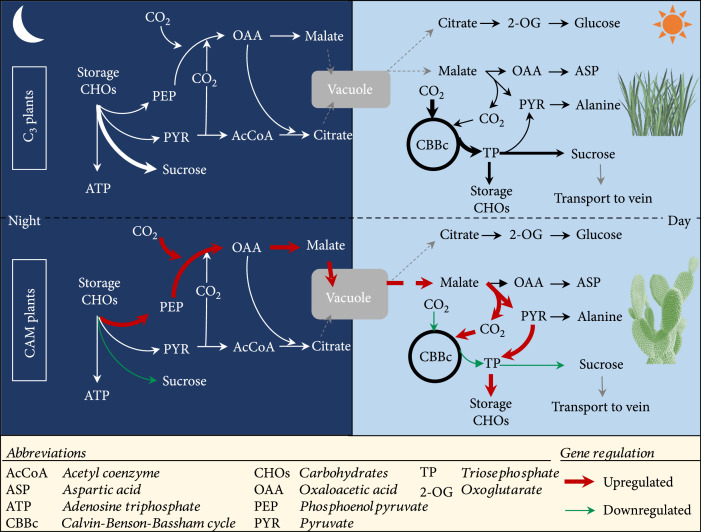
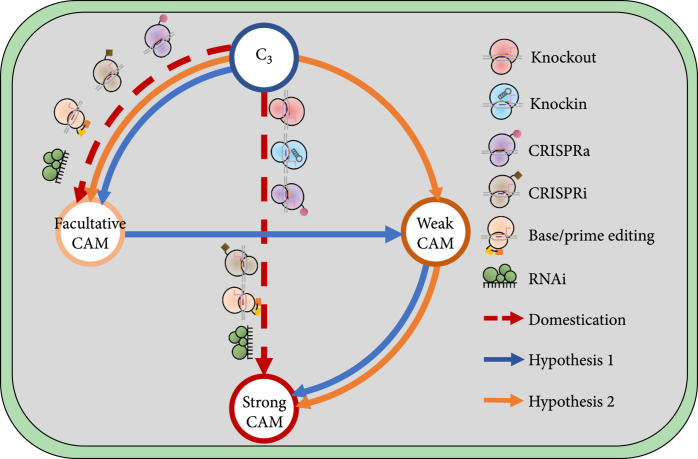
In different environments, ontogenies, and species, CAM-mediated CO2 fixation accounts for <1% to 100% of total carbon gain [72–74]. CAM plants may be facultative (i.e., reversible induction or upregulation of the CAM pathway by environmental stress) or obligatory (i.e., mature photosynthetic tissues always perform CAM photosynthesis as a result of a preprogrammed, irreversible developmental process) [72, 74]. In addition, strong CAM and weak CAM are also widely used to define CAM species, with strong CAM meaning that ~95% of carbon intake is through the CAM pathway [72]. A recent comparative analysis of key carbon fluxes between C3 and CAM pathways showed that C3 plants had metabolite fluxes similar to CAM fluxes [67] (Figure 2). More recently, two alternative models have been proposed to explain the evolution of the CAM pathway [18] (Figure 3). In hypothesis 1, C3 plants evolved forward to facultative CAM, weak CAM, and strong CAM in a linear manner. Under hypothesis 2, C3 plants evolved into facultative CAM and weak CAM independently, and then, weak CAM further evolved into strong CAM. The C3-to-CAM continuum might explain the reversible induction of CAM by environmental stress in facultative CAM plants [67]. These hypotheses are consistent with the view that the distribution of facultative CAM is wider among vascular plants than that reported previously [72]. However, the idea of a continuum must be tempered by the evident anatomical constraints placed on the evolutionary trajectories of CAM species reflected in the bimodal distributions of C3+CAM and CAM plants revealed by large-scale δ13C isotopic and leaf thickness surveys [75].
Figure 2.
Daytime and nighttime metabolism of organic acids in C3 and CAM plants. Arrow thickness denotes flux. Adapted from [67].
Figure 3.
An evolution-based conceptual framework for crassulacean acid metabolism (CAM) engineering guidance. Hypothesis 1: CAM evolution followed a linear course leading from facultative CAM to strong constitutive CAM. Hypothesis 2: facultative and constitutive CAM evolved independently. The hypotheses were adapted from [18].
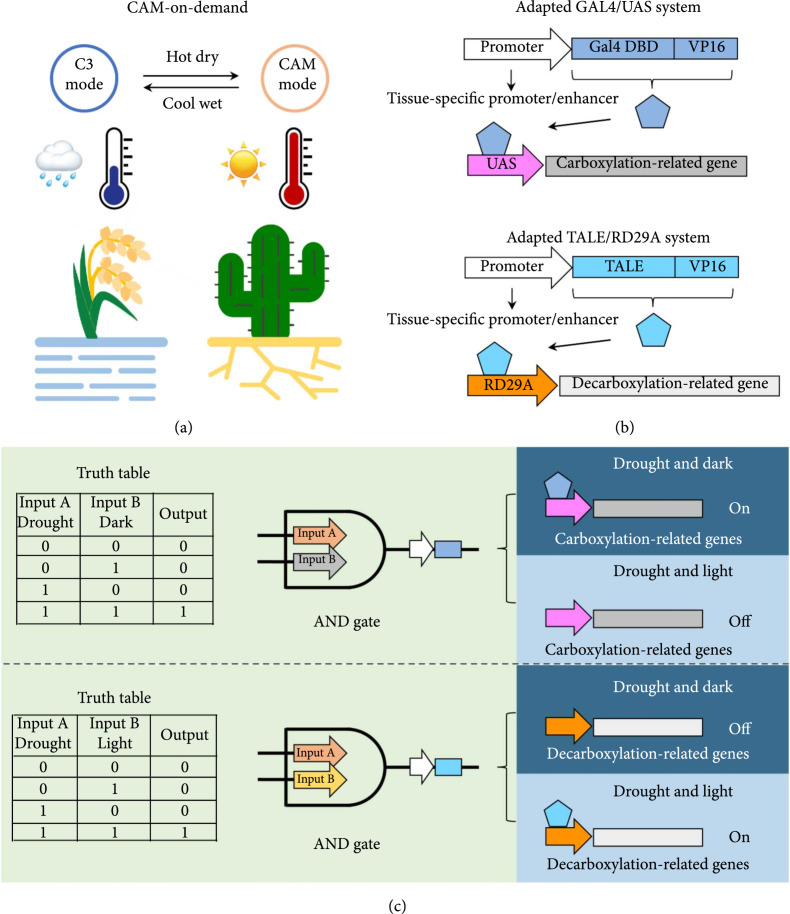
Seasonal drought stress is a widely existing challenge for crop production, and this challenge could be potentially addressed through engineering a drought-inducible CAM or CAM-on-demand system [9, 30, 76]. In facultative CAM plants, CAM metabolism can be induced and reverted to the C3 mode multiple times by water deficit, salinity, and high light [18, 72, 74], implying that C3 photosynthesis can be engineered to be metabolically compatible with the water-use efficient adaptation. A typical CAM-on-demand system represents an engineered photosynthesis system that enables reversible CAM induction in response to drought stress (Figure 4(a)). In particular, CAM-on-demand plants would operate in the C3 mode under moisture and cool conditions and temporarily switch to the CAM mode if the environment turns hot and dry. Such a system could not only possess a feature of drought tolerance under the CAM mode, but also maintain a relatively high growth rate of biomass accumulation under the C3 mode, resulting in a promising strategy in response to climate change. Therefore, from a CAM evolution-informed point of view, we can infer the following principles for CAM engineering: (1) there is no need to transfer a large number of genes from CAM species into C3 species (it is possible that the C3-to-CAM transition can be achieved through rewiring of temporal gene expression and rechanneling of existing metabolic flux) and (2) CAM-on-demand systems can be engineered through reversible drought-induced gene expression.
Figure 4.
An inducible system for C3 crop in response to drought. (a) A CAM-on-demand system. (b) Sequence-specific transcriptional activation systems. (c) Boolean logic gates mediated CAM signaling systems. A value of 1 represents a true answer, and 0 represents a false answer.
4. Installation of CAM-on-Demand Systems Using Gene Editing and Synthetic Biology Approaches
4.1. Genome Editing and Gene Regulation Approaches Required for CAM Engineering
There is a major difference in gene expression between facultative CAM and C3 plants, with facultative CAM plants featuring drought-inducible expression of genes related to CAM [72]. Recently, rapid development of the CRISPR technology has provided a very powerful toolbox for basic and applied biological research. For example, the CRISPR/Cas systems can be used to generate single- or multinucleotide replacements, insertions, and deletions in the genome using CRISPR/Cas9, base editors, and prime editors and to manipulate gene expression using CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) [77–79]. These tools have been used across different species, including E. coli, yeast, human cells, and plants. CRISPRa and CRISPRi can be used for the transcriptional gene regulation to facilitate C3-to-CAM transition, where compatible CRISPRa and CRISPRi systems promise to minimize the scale and complexity of biosystems design and engineering (Figure 3). Recently, CRISPRai was developed to simultaneously activate and inhibit gene expression in mammalian cells [80]. Developing such a CRISPRai tool in plants will clearly facilitate CAM bioengineering.
Alternately, small RNA-based RNA interference (RNAi), which regulates gene expression at the transcriptional and posttranscriptional levels, can be used together with CRISPRa for simultaneous gene activation and inhibition in plants [81, 82]. Additionally, a new gene editing method termed prime editing can perform targeted small insertions, deletions, and base swapping in a precise manner in yeast and human cells [83]. Recently, prime editing was successfully applied in plant species such as rice and wheat, although the editing efficiency is much lower than that of the mature CRISPR editing tools [84, 85]. In short, prime editing, together with base editing, will be useful for creating genomic mutations, such as single-nucleotide change and multinucleotide mutations (replacement, insertion, or deletion), required for C3-to-CAM progression [78]. Lastly, CRISPR knockout and knockin can be used for CAM engineering. Considering that the number of CAM-related genes is high, multiplex genome editing and regulation will be needed to accelerate the discovery and functional characterization of these genes, as well as to facilitate the engineering of functional CAM-related genes in C3 species. CRISPR/Cas9-enabled multiplex knockout and CRISPRa are available in plant systems [86, 87]. Engineering CAM in C3 plants will require biosystems design approaches such as CRISPR-based multiplex gene editing and gene regulation, engineering of drought-responsive gene circuits, and rewiring of metabolism.
4.2. Establishing a Drought Stress Signaling Pathway
A distinguishing feature of the CAM-on-demand system is that it requires drought-inducible transcription of CAM-related genes. Specifically, CAM-on-demand will require the regulatory expression of carboxylation-related genes under drought and dark conditions, while the expression of decarboxylation-related genes will be needed under drought and light conditions. Identifying a sensor that is capable of reading multiple inputs and transmitting them to a downstream network will be indispensable. In a general context, Boolean logic gates mediate synthetic genetic circuits that can convert multiple input signals. Such circuits have been successfully implemented in various biological systems, such as yeast and mammalian cells [88, 89]. Boolean logic gates convert multiple input signals into “truth” values, where a value of 1 represents a true answer and 0 represents a false answer [90]. Typically, following a set of algorithms, these synthetic genetic circuits can generate a defined response through an integration of multiple molecular input signals [91]. A synthetic gene circuit based on an AND gate, which generates an output only when two input signals are present, can be used for drought-inducible expression of CAM-related genes. As illustrated in Figure 4(c), if inputs A and B are defined as drought and dark signals, respectively, then the downstream carboxylation-related genes cannot be activated unless both drought and dark conditions are met. In this case, drought-induced positive transcriptional regulators and dark-inducible switches for the regulation of gene expression can be integrated to define the inputs A and B. In the context of CAM, the family of abiotic stress-responsive transcription factors (TFs), including NAC, bZIP, WRKY, NF-Y, MYB, and AP2/ERF, has been identified and characterized [92]. Recently, a plant stress response system was engineered, which employed the receptor for the plant stress hormone ABA and chemical agonists for initiating a response to drought [93, 94]. In addition, in prokaryotic and eukaryotic systems, many optogenetic switches responsive to green, UV-B, blue, red, and far-red/near-infrared light have been developed and tested to control intracellular signaling pathways with a high spatial and temporal resolution [95]. Furthermore, several optogenetic systems have been applied in plants, such as a CarH-based green light-regulated expression system and a phytochrome-based red light-inducible expression system [96–98]. Light-inducible expression systems with a broad spectrum will be required to optimize a CAM-on-demand system where entrainment of circadian-regulated CAM gene expression patterns will likely be necessary.
4.3. Establishing Gene Activation Systems
Secondly, an activation system that can simultaneously manipulate multiple carboxylation- or decarboxylation-related genes is needed. During the night, the metabolic fluxes from stored carbohydrates towards malate, including the intermediates PEP and OAA, are increased, but the flux towards sucrose is decreased in CAM plants (Figure 2). To achieve this feature in a C3 plant, the key enzymes and transporters, such as β-CA, PEPC, PPCK, NAD(P)-MDH, tDT, and ALMT, will have to be transcriptionally activated to establish the carboxylation module. To date, gene activation can be accomplished with multiple tools (as noted above) in plants, such as by using strong promoter-mediated overexpression, CRISPRa, and a TALE-mediated mTALE-Act system [87]. Among these, multiplex CRISPR-Act2.0 and mTALE-Act, which can manipulate multiple genes simultaneously, appear appropriate for this task. However, neither of these systems can activate more than four genes simultaneously based on the current technology. Therefore, a highly multiplex activation system is required to meet the needs of a fully functional carboxylation or decarboxylation module. Very recently, we developed a de novo multiplex CRISPRa system that can simultaneously perturbate the expression of eight genes in A. thaliana (Yuan et al., unpublished data). Such a system is necessary to simplify the assembly of genetic parts and lower the complexity of the intact model.
The initial target of the gene activation system will be PEP. The synthesis of PEP is indispensable in a fully functional carboxylation module because it is the key substrate for nocturnal CO2 fixation mediated by PEPC, which converts PEP and bicarbonate to OAA (Figure 1). Unlike C3 plants, in which the hydrolytic route mainly degrades starch, typical CAM plants degrade starch to provide a substrate for PEP mainly through the phosphorolytic pathway [24, 61]. Conceptually, C3 plants would benefit from an engineered switch from hydrolytic to phosphorolytic starch breakdown. Specifically, during the day, the metabolic fluxes from accumulated malic acid in the vacuole towards PYR and subsequent storage carbohydrates are increased, but the flux towards sucrose or other soluble storage carbohydrates can be decreased in CAM plants (Figure 2). To achieve this feature in a C3 plant, the key enzymes and regulators, such as NAD(P)-ME, PPDK, PPDK-RP, PEPCK, and others, would have to be transcriptionally activated to establish the decarboxylation module. As discussed above, the multiplex CRISPRa system can also be applied to this task.
In addition to the multiplex CRISPRa system, these engineered systems could be accomplished through a sequence-specific transcriptional activation system (e.g., an adapted GAL4/UAS system or an adapted TALE/RD29A system). The GAL4/UAS system, which was originally developed for studying gene expression and function in Drosophila [99], has become one of the most useful systems for targeted gene expression across different species. For instance, Potri.002G146400-encoded PtrEPSP was identified as a transcriptional repressor using a GAL4/UAS-mediated protoplast transient expression system in Populus [100]. Inspired by this work, an adapted GAL4/UAS system could be used to control carboxylation-related gene expression. This system consists of two individual parts serving for targeting and activation (Figure 4(b)). A Gal4-DNA-binding domain is fused to the transactivator VP16 (GD-VP16) to generate a transcriptional activator that targets the UAS enhancer, and GD-VP16 is driven by a tissue-specific promoter/enhancer. The other component is the carboxylation-related gene driven by a UAS enhancer. Also, the expression of GD-VP16 is regulated by an AND gate (Figure 4(c)). That is, under defined drought and dark conditions, GD-VP16 is bound to the UAS enhancer, thereby activating carboxylation-related gene expression. To provide further precision in expression, the GAL4/UAS system has been further characterized to increase the dynamic range of the system [101]. Simultaneously, an alternate independent activation system can be used to manipulate the decarboxylation-related gene expression. Here, transcriptional activator-like effectors (TALEs), containing a modular DNA-binding domain, can be used to generate chimeric transcriptional activators or repressors. A chimeric TALE-SRDX repressor can be used to repress the transcription of the transgene RD29A::LUC and endogenous gene RD29A in A. thaliana [102]. Again, inspired by this work, an adapted TALE/RD29A system could be used to control decarboxylation-related gene expression (Figure 4(b)). Similar to the GAL4/UAS system, one component is a TALE-DNA-binding domain-fused transactivator VP16 (TALE-VP16) driven by a tissue-specific promoter/enhancer and the other component is the decarboxylation-related gene driven by a RD29A promoter. The expression of TALE-VP16 is regulated by an AND gate (Figure 4(c)).
5. Iterative Design-Build-Test-Learn (DBTL) Cycles of CAM Engineering
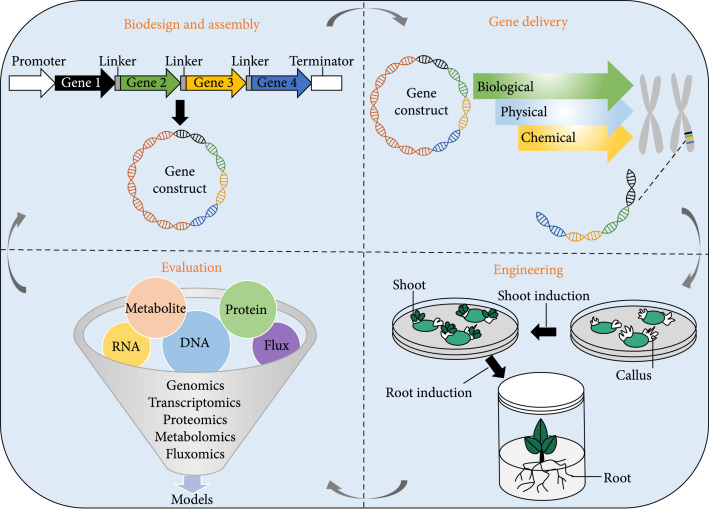
The application of biosystems design to CAM engineering involves DBTL, which has four different phases: (I) biodesigned genetic circuits and assembly of multigene constructs, (II) delivery of biodesigned devices, (III) plant engineering, and (IV) evaluation of engineered plants (Figure 5). In phase I, synthetic devices will likely be essential components of CAM engineering. Although different synthetic switches and biosensors for controlling genome editing, gene regulation, and protein stability have already been utilized in plants, deployment of more complicated genetic circuits for genetic engineering in plants is still a big challenge [90]. This challenge is primarily caused by the experimental bottlenecks (e.g., lack of efficient plant transformation systems) and slow generation times of plants making it difficult to test the genetic circuits in plants. To overcome these limitations, protoplast-based and Agrobacterium-mediated leaf infiltration transient expression assays could be used to provide a rapid and robust analysis of transgene expression and protein subcellular localization and interaction [103]. For the assembly of multigene constructs, there are multiple methods of DNA assembly available, including Gibson assembly, BioBrick assembly, Golden Gate assembly, TOPO cloning, Gateway cloning, TNT cloning, and traditional restriction enzyme cloning [16, 104–106]. Among them, the Golden Gate assembly is capable of assembling up to 24 DNA fragments in a seamless and highly efficient manner [107]. However, unexpected interactions or transcriptional interferences between neighboring transcription units in multigene constructs are commonly found in all eukaryotic organisms including plants [108, 109]. To facilitate modular construction of a CAM gene circuit composed of multiple transcription units, which must have different diel expression patterns, transcriptional interference should be avoided. To overcome this issue, genetic insulators (enhancer blocking or barrier activity) could be deployed in multigene constructs that possibly prevent these unwanted interactions and increase transgene expression in plants [110, 111].
Figure 5.
An overview of synthetic biology-dependent crassulacean acid metabolism (CAM) engineering.
In phase II, the conventional methods to deliver genes to plant cells can be classified into three categories: biological, physical, or chemical approaches, with the most common and preferred method being Agrobacterium-mediated plant transformation. However, to insert large constructs containing multiple genes into the plant genome with high structural and functional stability of the engineered gene modules, new methods need to be developed for multiple rounds of site-specific in planta gene stacking [112]. In phase III, tissue culture-based plant transformation is widely used to create transformed or genome-edited plants. However, creating transgenic plants through tissue culture is a bottleneck of genome editing in plants, because (1) it is only suitable for a limited number of species and genotypes, (2) it is time-consuming with low efficiency, and (3) it might cause unwanted genetic and epigenetic changes [113]. To overcome this bottleneck, two methods were recently developed for the generation of gene-edited dicotyledonous plants via de novo meristem induction by developmental regulators without in vitro culture [114]. In phase IV, robotic high-throughput phenotyping [115], in combination with omics approaches, is needed to advance functional analysis for a quick evaluation of biodesigned devices and circuitry in the transgenic or genome-edited plants. The omics-based system dynamics modeling and diel flux balance analysis [23, 116, 117] will need to be performed for reconstructions of metabolic networks to improve the performance of transgenic plants engineered with CAM. In order to optimize the biosystems design for CAM engineering, multiple iterations of the DBTL cycle will be required and possible adjustments will be made to increase precision and efficiency in each iteration.
6. Conclusion and Perspectives
The engineering of CAM and coadaptive traits, such as tissue succulence, holds a great potential for sustainable production of fiber, food, feed, and biofuels in water-limited areas [9, 15, 19, 60]. Initially, a deep understanding of CAM-related gene function is a key prerequisite for engineering CAM into C3 crops [30]. Many such genes have been identified and organized into separate CAM-related modules (i.e., carboxylation, decarboxylation, and stomatal regulation). The minimum genes that are indispensable to maintain a functional module are proposed based on the knowledge of genomic research and comparative analysis. Genes that play important roles in the CAM pathway are summarized in Table 1, providing a database to guide the user in CAM engineering. Despite decades of notable progress in CAM research, a number of potentially important genes may be yet undiscovered. Comparative analysis of more CAM plant genomes will be needed to accelerate the identification of biological parts (e.g., enzymes, posttranslational modifiers, transporters, and transcription factors) for CAM engineering.
Additional research will be required to characterize the function of candidate genes inferred from the omics and comparative genomics research. Efforts will be needed to reduce the redundancy of CAM-related genes found in different CAM species. To accelerate the C3-to-CAM engineering, wiring of appropriate temporal gene expression and rechanneling of existing metabolic flux will be essential. A CAM-on-demand system that responds to episodic or seasonal drought can be achieved in C3 plants through reversible drought-induced gene expression to increase WUE. Considering the genes mentioned above, using single-cell technologies will enable exploration of photoperiod and cell-type dynamics of CAM-related modules. By integrating with single-cell transcriptome data, the CAM modules can establish the layer of CAM regulation, which is incomplete. The regulatory network of CAM for each module can be explored by various approaches that have not been adapted to CAM research, including ATAC-Seq (Assay for Transposase-Accessible Chromatin followed by high-throughput sequencing), DAP-Seq (DNA affinity purification and sequencing), and DNase-Seq (DNase I hypersensitive site sequencing), which may provide decondensation of accessible chromatin regions that enrich motifs of transcription factors (e.g., NAC, bZIP, WRKY, NF-Y, MYB, and AP2/ERF) mentioned above [118–120].
With the nexus of new technologies like systems genetics, genome editing, synthetic biology, and gene activation systems, we are on the threshold of purposefully accelerating C3-to-CAM progression. The CRISPR toolkit for genome editing and gene regulation provides useful tools required for CAM engineering. The engineering of synthetic circuitry in plant systems has the potential to advance our understanding and ability to manipulate genetic and metabolic networks such as CAM. The strategies for building CAM-on-demand systems are feasible using coordinated systems and synthetic biology. To achieve these goals, synthetic genetic circuits for signaling and tools for manipulating multiple gene expression simultaneously at a large scale need to be developed in plant systems. Meanwhile, some technical challenges need to be overcome. For example, the plant transformation with large-scale multigene stacking that ensures different CAM modules to be properly expressed in transgenic plants will remain a challenge for the foreseeable future. Regardless, with the effective and successful demonstrations already reported in different organisms, synthetic biosystems design holds a great promise to enable C3-to-CAM progression in the near future.
Acknowledgments
This work was supported by the Center for Bioenergy Innovation (CBI), a U.S. Department of Energy Bioenergy Research Center supported by the Office of Science Biological and Environmental Research (BER). The writing of this manuscript was also supported by the Department of Energy (Office of Science, Genomic Science Program) under award number DE-SC0008834. SDL acknowledges support from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2019R1I1A1A01061727). DL acknowledges financial support from the National Science Foundation (NSF) under Award Number 1833402. KM acknowledges support from start-up funding provided by the University of California, Davis. PMS acknowledges support from the Department of Energy (DE-AC02-05CH11231).
Disclosure
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Authors’ Contributions
XY and GY conceived the idea. GY led the writing and revision of the manuscript. MMH, DL, SDL, WCY, JCC, HL, DJW, JGC, KM, PMS, TJT, GAT, and XY contributed to the manuscript revision. All authors accepted the final version of the manuscript.
References
- 1.Division U. N. P.World Population Prospects: 2019 Revision, United Nations New York, NY, 2019 [Google Scholar]
- 2.DeNicola E., Aburizaiza O. S., Siddique A., Siddique A., Khwaja H., and Carpenter D. O., “Climate change and water scarcity: the case of Saudi Arabia,” Annals of Global Health, vol. 81, no. 3, pp. 342–353, 2018 [DOI] [PubMed] [Google Scholar]
- 3.McMichael A. J., Friel S., Nyong A., and Corvalan C., “Global environmental change and health: impacts, inequalities, and the health sector,” BMJ, vol. 336, no. 7637, pp. 191–194, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith P., and Gregory P. J., “Climate change and sustainable food production,” The Proceedings of the Nutrition Society, vol. 72, no. 1, pp. 21–28, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Karp A., and Richter G. M., “Meeting the challenge of food and energy security,” Journal of Experimental Botany, vol. 62, no. 10, pp. 3263–3271, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Kubis A., and Bar-Even A., “Synthetic biology approaches for improving photosynthesis,” Journal of Experimental Botany, vol. 70, no. 5, pp. 1425–1433, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans J. R., “Improving photosynthesis,” Plant Physiology, vol. 162, no. 4, pp. 1780–1793, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducat D. C., and Silver P. A., “Improving carbon fixation pathways,” Current Opinion in Chemical Biology, vol. 16, no. 3-4, pp. 337–344, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borland A. M., Hartwell J., Weston D. J., Schlauch K. A., Tschaplinski T. J., Tuskan G. A., Yang X., and Cushman J. C., “Engineering crassulacean acid metabolism to improve water-use efficiency,” Trends in Plant Science, vol. 19, no. 5, pp. 327–338, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehleringer J. R., and Monson R. K., “Evolutionary and ecological aspects of photosynthetic pathway variation,” Annual Review of Ecology and Systematics, vol. 24, no. 1, pp. 411–439, 1993 [Google Scholar]
- 11.Hatfield J. L., and Dold C., “Water-use efficiency: advances and challenges in a changing climate,” Frontiers in Plant Science, vol. 10, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borland A. M., Griffiths H., Hartwell J., and Smith J. A. C., “Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands,” Journal of Experimental Botany, vol. 60, no. 10, pp. 2879–2896, 2009 [DOI] [PubMed] [Google Scholar]
- 13.White R. P., and Nackoney J., “Drylands, people, and ecosystem goods and services: a web-based geospatial analysis (PDF version),” World Resources Institute, 2003, January 2012, http://pdf.wri.org/drylands.pdf. [Google Scholar]
- 14.Zika M., and Erb K. H., “The global loss of net primary production resulting from human-induced soil degradation in drylands,” Ecological Economics, vol. 69, no. 2, pp. 310–318, 2009 [Google Scholar]
- 15.Yang X., Cushman J. C., Borland A. M., Edwards E. J., Wullschleger S. D., Tuskan G. A., Owen N. A., Griffiths H., Smith J. A. C., de Paoli H. C., Weston D. J., Cottingham R., Hartwell J., Davis S. C., Silvera K., Ming R., Schlauch K., Abraham P., Stewart J. R., Guo H. B., Albion R., Ha J., Lim S. D., Wone B. W. M., Yim W. C., Garcia T., Mayer J. A., Petereit J., Nair S. S., Casey E., Hettich R. L., Ceusters J., Ranjan P., Palla K. J., Yin H., Reyes-García C., Andrade J. L., Freschi L., Beltrán J. D., Dever L. V., Boxall S. F., Waller J., Davies J., Bupphada P., Kadu N., Winter K., Sage R. F., Aguilar C. N., Schmutz J., Jenkins J., and Holtum J. A. M., “A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world,” The New Phytologist, vol. 207, no. 3, pp. 491–504, 2015 [DOI] [PubMed] [Google Scholar]
- 16.DePaoli H. C., Borland A. M., Tuskan G. A., Cushman J. C., and Yang X., “Synthetic biology as it relates to CAM photosynthesis: challenges and opportunities,” Journal of Experimental Botany, vol. 65, no. 13, pp. 3381–3393, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Hultine K. R., Cushman J. C., and Williams D. G., “New perspectives on crassulacean acid metabolism biology,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6489–6493, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X., Liu D., Tschaplinski T. J., and Tuskan G. A., “Comparative genomics can provide new insights into the evolutionary mechanisms and gene function in CAM plants,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6539–6547, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niechayev N. A., Pereira P. N., and Cushman J. C., “Understanding trait diversity associated with crassulacean acid metabolism (CAM),” Current Opinion in Plant Biology, vol. 49, pp. 74–85, 2019 [DOI] [PubMed] [Google Scholar]
- 20.Yang X., Hu R., Yin H., Jenkins J., Shu S., Tang H., Liu D., Weighill D. A., Cheol Yim W., Ha J., Heyduk K., Goodstein D. M., Guo H. B., Moseley R. C., Fitzek E., Jawdy S., Zhang Z., Xie M., Hartwell J., Grimwood J., Abraham P. E., Mewalal R., Beltrán J. D., Boxall S. F., Dever L. V., Palla K. J., Albion R., Garcia T., Mayer J. A., Don Lim S., Man Wai C., Peluso P., van Buren R., de Paoli H. C., Borland A. M., Guo H., Chen J. G., Muchero W., Yin Y., Jacobson D. A., Tschaplinski T. J., Hettich R. L., Ming R., Winter K., Leebens-Mack J. H., Smith J. A. C., Cushman J. C., Schmutz J., and Tuskan G. A., “The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism,” Nature Communications, vol. 8, no. 1, p. 1899, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X., Qi L. S., Jaramillo A., and Cheng Z. M. (. M.)., “Biodesign research to advance the principles and applications of biosystems design,” BioDesign Research, vol. 2019, article 9680853, –4, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurtzel E. T., Vickers C. E., Hanson A. D., Millar A. H., Cooper M., Voss-Fels K. P., Nikel P. I., and Erb T. J., “Revolutionizing agriculture with synthetic biology,” Nature Plants, vol. 5, no. 12, pp. 1207–1210, 2019 [DOI] [PubMed] [Google Scholar]
- 23.Owen N. A., and Griffiths H., “A system dynamics model integrating physiology and biochemical regulation predicts extent of crassulacean acid metabolism (CAM) phases,” The New Phytologist, vol. 200, no. 4, pp. 1116–1131, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Shameer S., Baghalian K., Cheung C. Y. M., Ratcliffe R. G., and Sweetlove L. J., “Computational analysis of the productivity potential of CAM,” Nature Plants, vol. 4, no. 3, pp. 165–171, 2018 [DOI] [PubMed] [Google Scholar]
- 25.Boxall S. F., Dever L. V., Kneřová J., Gould P. D., and Hartwell J., “Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi,” Plant Cell, vol. 29, no. 10, pp. 2519–2536, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medeiros D. B., Barros K. A., Barros J. A. S., Omena-Garcia R. P., Arrivault S., Sanglard L. M. V. P., Detmann K. C., Silva W. B., Daloso D. M., DaMatta F. M., Nunes-Nesi A., Fernie A. R., and Araújo W. L., “Impaired malate and fumarate accumulation due to the mutation of the tonoplast dicarboxylate transporter has little effects on stomatal behavior,” Plant Physiology, vol. 175, no. 3, pp. 1068–1081, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., and Martinoia E., “The Arabidopsis vacuolar malate channel is a member of the ALMT family,” The Plant Journal, vol. 52, no. 6, pp. 1169–1180, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Hurth M. A., Suh S. J., Kretzschmar T., Geis T., Bregante M., Gambale F., Martinoia E., and Neuhaus H. E., “Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast,” Plant Physiology, vol. 137, no. 3, pp. 901–910, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmerlich V., Linka N., Reinhold T., Hurth M. A., Traub M., Martinoia E., and Neuhaus H. E., “The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier,” Proceedings of the National Academy of Sciences of the United States of America, vol. 100, no. 19, pp. 11122–11126, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim S. D., Lee S., Choi W. G., Yim W. C., and Cushman J. C., “Laying the foundation for crassulacean acid metabolism (CAM) biodesign: expression of the C4 metabolism cycle genes of CAM in Arabidopsis,” Frontiers in Plant Science, vol. 10, p. 101, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Astley H. M., Parsley K., Aubry S., Chastain C. J., Burnell J. N., Webb M. E., and Hibberd J. M., “The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis are both bifunctional and interact with the catalytic and nucleotide-binding domains of pyruvate, orthophosphate dikinase,” The Plant Journal, vol. 68, no. 6, pp. 1070–1080, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Males J., and Griffiths H., “Stomatal biology of CAM plants,” Plant Physiology, vol. 174, no. 2, pp. 550–560, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotoh E., Oiwamoto K., Inoue S. I., Shimazaki K. I., and Doi M., “Stomatal response to blue light in crassulacean acid metabolism plants Kalanchoe pinnata and Kalanchoe daigremontiana,” Journal of Experimental Botany, vol. 70, no. 4, pp. 1367–1374, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., Chen M., Mendoza B., Cheng H., Hu R., Li L., Trinh C. T., Tuskan G. A., and Yang X., “CRISPR/Cas9-mediated targeted mutagenesis for functional genomics research of crassulacean acid metabolism plants,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6621–6629, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boxall S. F., Kadu N., Dever L. V., Kneřová J., Waller J. L., Gould P. J. D., and Hartwell J., “Kalanchoë PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes,” Plant Cell, vol. 32, no. 4, pp. 1136–1160, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taybi T., and Cushman J. C., “Abscisic acid signaling and protein synthesis requirements for phosphoenolpyruvate carboxylase transcript induction in the common ice plant,” Journal of Plant Physiology, vol. 159, no. 11, pp. 1235–1243, 2002 [Google Scholar]
- 37.Yin H., Guo H. B., Weston D. J., Borland A. M., Ranjan P., Abraham P. E., Jawdy S. S., Wachira J., Tuskan G. A., Tschaplinski T. J., Wullschleger S. D., Guo H., Hettich R. L., Gross S. M., Wang Z., Visel A., and Yang X., “Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave,” BMC Genomics, vol. 19, no. 1, p. 588, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taybi T., and Cushman J. C., “Signaling events leading to crassulacean acid metabolism induction in the common ice plant,” Plant Physiology, vol. 121, no. 2, pp. 545–556, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dever L. V., Boxall S. F., Kneřová J., and Hartwell J., “Transgenic perturbation of the decarboxylation phase of crassulacean acid metabolism alters physiology and metabolism but has only a small effect on growth,” Plant Physiology, vol. 167, no. 1, pp. 44–59, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyka T. P., Duarte H. M., and Lüttge U. E., “Redundancy of stomatal control for the circadian photosynthetic rhythm inKalanchoë daigremontianaHamet et Perrier,” Plant Biology (Stuttgart, Germany), vol. 7, no. 2, pp. 176–181, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Wai C. M., VanBuren R., Zhang J., Huang L., Miao W., Edger P. P., Yim W. C., Priest H. D., Meyers B. C., Mockler T., Smith J. A. C., Cushman J. C., and Ming R., “Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple,” The Plant Journal, vol. 92, no. 1, pp. 19–30, 2017 [DOI] [PubMed] [Google Scholar]
- 42.Moseley R. C., Tuskan G. A., and Yang X., “Comparative genomics analysis provides new insight into molecular basis of stomatal movement in Kalanchoë fedtschenkoi,” Frontiers in Plant Science, vol. 10, p. 292, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., and Cutler S. R., “Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins,” Science, vol. 324, no. 5930, pp. 1068–1071, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N., Fukuda H., Yamaguchi-Shinozaki K., and Shinozaki K., “A small peptide modulates stomatal control via abscisic acid in long-distance signalling,” Nature, vol. 556, no. 7700, pp. 235–238, 2018 [DOI] [PubMed] [Google Scholar]
- 45.Ziadi A., Uchida N., Kato H., Hisamatsu R., Sato A., Hagihara S., Itami K., and Torii K. U., “Discovery of synthetic small molecules that enhance the number of stomata: C–H functionalization chemistry for plant biology,” Chemical Communications, vol. 53, no. 69, pp. 9632–9635, 2017 [DOI] [PubMed] [Google Scholar]
- 46.Nelson E. A., and Sage R. F., “Functional constraints of CAM leaf anatomy: tight cell packing is associated with increased CAM function across a gradient of CAM expression,” Journal of Experimental Botany, vol. 59, no. 7, pp. 1841–1850, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Nelson E. A., Sage T. L., and Sage R. F., “Functional leaf anatomy of plants with crassulacean acid metabolism,” Functional Plant Biology, vol. 32, no. 5, pp. 409–419, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Borland A. M., Leverett A., Hurtado-Castano N., Hu R., and Yang X., “Functional anatomical traits of the photosynthetic organs of plants with crassulacean acid metabolism,” The Leaf: A Platform for Performing Photosynthesis, Adams W. W. III, and Terashima I., Eds., Springer International Publishing AG, pp. 281–305, 2018 [Google Scholar]
- 49.Lüttge U., and Nobel P. S., “Day-night variations in malate concentration, osmotic pressure, and hydrostatic pressure in Cereus validus,” Plant Physiology, vol. 75, no. 3, pp. 804–807, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith J. A. C., Schulte P. J., and Nobel P. S., “Water flow and water storage in Agave deserti: osmotic implications of crassulacean acid metabolism,” Plant, Cell & Environment, vol. 10, no. 8, pp. 639–648, 1987 [Google Scholar]
- 51.Maxwell K., Caemmerer S. ., and Evans J. R., “Is a low internal conductance to CO2 diffusion a consequence of succulence in plants with crassulacean acid metabolism?,” Australian Journal of Plant Physiology, vol. 24, no. 6, pp. 777–786, 1997 [Google Scholar]
- 52.Griffiths H., Robe W. E., Girnus J., and Maxwell K., “Leaf succulence determines the interplay between carboxylase systems and light use during Crassulacean acid metabolism in Kalanchoë species,” Journal of Experimental Botany, vol. 59, no. 7, pp. 1851–1861, 2008 [DOI] [PubMed] [Google Scholar]
- 53.Kluge M., Brulfert J., Lipp J., Ravelomanana D., and Ziegler H., “A comparative study by δ13C-analysis of crassulacean acid metabolism (CAM) in Kalanchoë (Crassulaceae) species of Africa and Madagascar,” Botanica Acta, vol. 106, no. 4, pp. 320–324, 1993 [Google Scholar]
- 54.Silvera K., Santiago L. S., and Winter K., “Distribution of crassulacean acid metabolism in orchids of Panama: evidence of selection for weak and strong modes,” Functional Plant Biology, vol. 32, no. 5, pp. 397–407, 2005 [DOI] [PubMed] [Google Scholar]
- 55.Barrera Zambrano V. A., Lawson T., Olmos E., Fernandez-Garcia N., and Borland A. M., “Leaf anatomical traits which accommodate the facultative engagement of crassulacean acid metabolism in tropical trees of the genus Clusia,” Journal of Experimental Botany, vol. 65, no. 13, pp. 3513–3523, 2014 [DOI] [PubMed] [Google Scholar]
- 56.Heyduk K., Ray J. N., and Leebens-Mack J., “Leaf anatomy is not correlated to CAM function in a C3+ CAM hybrid species, Yucca gloriosa,” Annals of Botany, 2020 [DOI] [PMC free article] [PubMed]
- 57.Han Y., Wang W., Sun J., Ding M., Zhao R., Deng S., Wang F., Hu Y., Wang Y., Lu Y., du L., Hu Z., Diekmann H., Shen X., Polle A., and Chen S., “Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants,” Journal of Experimental Botany, vol. 64, no. 14, pp. 4225–4238, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicolas P., Lecourieux D., Gomès E., Delrot S., and Lecourieux F., “The grape berry-specific basic helix-loop-helix transcription factor VvCEB1 affects cell size,” Journal of Experimental Botany, vol. 64, no. 4, pp. 991–1003, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim S. D., Yim W. C., Liu D., Hu R., Yang X., and Cushman J. C., “A Vitis vinifera basic helix-loop-helix transcription factor enhances plant cell size, vegetative biomass and reproductive yield,” Plant Biotechnology Journal, vol. 16, no. 9, pp. 1595–1615, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim S. D., Mayer J. A., Yim W. C., and Cushman J. C., “Plant tissue succulence engineering improves water-use efficiency, water-deficit stress attenuation and salinity tolerance in Arabidopsis,” The Plant Journal, vol. 103, no. 3, pp. 1049–1072, 2020 [DOI] [PubMed] [Google Scholar]
- 61.Ceusters N., Frans M., van den Ende W., and Ceusters J., “Maltose processing and not β-amylase activity curtails hydrolytic starch degradation in the CAM orchid Phalaenopsis,” Frontiers in Plant Science, vol. 10, p. 1386, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bianchi T. S., and Canuel E. A.. Chemical Biomarkers in Aquatic Ecosystems, Princeton University Press, 2011 [Google Scholar]
- 63.Sage R. F., Sage T. L., and Kocacinar F., “Photorespiration and the evolution of C4 photosynthesis,” Annual Review of Plant Biology, vol. 63, no. 1, pp. 19–47, 2012 [DOI] [PubMed] [Google Scholar]
- 64.Silvera K., Neubig K. M., Whitten W. M., Williams N. H., Winter K., and Cushman J. C., “Evolution along the crassulacean acid metabolism continuum,” Functional Plant Biology, vol. 37, no. 11, pp. 995–1010, 2010 [Google Scholar]
- 65.Sage R. F., “A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and hall of fame,” Journal of Experimental Botany, vol. 67, no. 14, pp. 4039–4056, 2016 [DOI] [PubMed] [Google Scholar]
- 66.Crayn D. M., Winter K., and Smith J. A. C., “Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae,” Proceedings of the National Academy of Sciences of the United States of America, vol. 101, no. 10, pp. 3703–3708, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bräutigam A., Schlüter U., Eisenhut M., and Gowik U., “On the evolutionary origin of CAM photosynthesis,” Plant Physiology, vol. 174, no. 2, pp. 473–477, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hibberd J. M., and Covshoff S., “The regulation of gene expression required for C4 photosynthesis,” Annual Review of Plant Biology, vol. 61, no. 1, pp. 181–207, 2010 [DOI] [PubMed] [Google Scholar]
- 69.West-Eberhard M. J., Smith J. A. C., and Winter K., “Plant science. Photosynthesis, reorganized,” Science, vol. 332, no. 6027, pp. 311–312, 2011 [DOI] [PubMed] [Google Scholar]
- 70.West-Eberhard M. J.Developmental Plasticity and Evolution, Oxford University Press, 2003 [Google Scholar]
- 71.Brautigam A., and Gowik U., “Photorespiration connects C3 and C4 photosynthesis,” Journal of Experimental Botany, vol. 67, no. 10, pp. 2953–2962, 2016 [DOI] [PubMed] [Google Scholar]
- 72.Winter K., “Ecophysiology of constitutive and facultative CAM photosynthesis,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6495–6508, 2019 [DOI] [PubMed] [Google Scholar]
- 73.Winter K., Holtum J. A. M., and Smith J. A. C., “Crassulacean acid metabolism: a continuous or discrete trait?,” New Phytologist, vol. 208, no. 1, pp. 73–78, 2015 [DOI] [PubMed] [Google Scholar]
- 74.Winter K., and Holtum J. A. M., “Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis,” Journal of Experimental Botany, vol. 65, no. 13, pp. 3425–3441, 2014 [DOI] [PubMed] [Google Scholar]
- 75.Edwards E. J., “Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis,” New Phytologist, vol. 223, no. 4, pp. 1742–1755, 2019 [DOI] [PubMed] [Google Scholar]
- 76.Yang X., Cushman J. C., Borland A. M., and Liu Q., “Editorial: systems biology and synthetic biology in relation to drought tolerance or avoidance in plants,” Frontiers in Plant Science, vol. 11, no. 394, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adli M., “The CRISPR tool kit for genome editing and beyond,” Nature Communications, vol. 9, no. 1, p. 1911, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassan M. M., Yuan G., Chen J. G., Tuskan G. A., and Yang X., “Prime Editing Technology and Its Prospects for Future Applications in Plant Biology Research,” BioDesign Research, vol. 2020, article 9350905, –14, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kampmann M., “CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine,” ACS Chemical Biology, vol. 13, no. 2, pp. 406–416, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Truong V. A., Hsu M. N., Kieu Nguyen N. T., Lin M. W., Shen C. C., Lin C. Y., and Hu Y. C., “CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration,” Nucleic Acids Research, vol. 47, no. 13, article e74, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eamens A., Wang M. B., Smith N. A., and Waterhouse P. M., “RNA silencing in plants: yesterday, today, and tomorrow,” Plant Physiology, vol. 147, no. 2, pp. 456–468, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kamthan A., Chaudhuri A., Kamthan M., and Datta A., “Small RNAs in plants: recent development and application for crop improvement,” Frontiers in Plant Science, vol. 6, p. 208, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., Chen P. J., Wilson C., Newby G. A., Raguram A., and Liu D. R., “Search-and-replace genome editing without double-strand breaks or donor DNA,” Nature, vol. 576, no. 7785, pp. 149–157, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A. V., Raguram A., Doman J. L., Liu D. R., and Gao C., “Prime genome editing in rice and wheat,” Nature Biotechnology, vol. 38, no. 5, pp. 582–585, 2020 [DOI] [PubMed] [Google Scholar]
- 85.Li H., Li J., Chen J., Yan L., and Xia L., “Precise modifications of both exogenous and endogenous genes in rice by prime editing,” Molecular Plant, vol. 13, no. 5, pp. 671–674, 2020 [DOI] [PubMed] [Google Scholar]
- 86.Xie K., Minkenberg B., and Yang Y., “Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system,” Proceedings of the National Academy of Sciences of the United States of America, vol. 112, no. 11, pp. 3570–3575, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lowder L. G., Zhou J., Zhang Y., Malzahn A., Zhong Z., Hsieh T. F., Voytas D. F., Zhang Y., and Qi Y., “Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems,” Molecular Plant, vol. 11, no. 2, pp. 245–256, 2018 [DOI] [PubMed] [Google Scholar]
- 88.Gander M. W., Vrana J. D., Voje W. E., Carothers J. M., and Klavins E., “Digital logic circuits in yeast with CRISPR-dCas9 NOR gates,” Nature Communications, vol. 8, no. 1, article 15459, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lebar T., Bezeljak U., Golob A., Jerala M., Kadunc L., Pirš B., Stražar M., Vučko D., Zupančič U., Benčina M., Forstnerič V., Gaber R., Lonzarić J., Majerle A., Oblak A., Smole A., and Jerala R., “A bistable genetic switch based on designable DNA-binding domains,” Nature Communications, vol. 5, no. 1, article 5007, 2014 [DOI] [PubMed] [Google Scholar]
- 90.Andres J., Blomeier T., and Zurbriggen M. D., “Synthetic switches and regulatory circuits in plants,” Plant Physiology, vol. 179, no. 3, pp. 862–884, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie M., and Fussenegger M., “Designing cell function: assembly of synthetic gene circuits for cell biology applications,” Nature Reviews Molecular Cell Biology, vol. 19, no. 8, pp. 507–525, 2018 [DOI] [PubMed] [Google Scholar]
- 92.Amin A. B., Rathnayake K. N., Yim W. C., Garcia T. M., Wone B., Cushman J. C., and Wone B. W. M., “Crassulacean acid metabolism abiotic stress-responsive transcription factors: a potential genetic engineering approach for improving crop tolerance to abiotic stress,” Frontiers in Plant Science, vol. 10, p. 129, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park S.-Y., Peterson F. C., Mosquna A., Yao J., Volkman B. F., and Cutler S. R., “Agrochemical control of plant water use using engineered abscisic acid receptors,” Nature, vol. 520, no. 7548, pp. 545–548, 2015 [DOI] [PubMed] [Google Scholar]
- 94.Vaidya A. S., Peterson F. C., Yarmolinsky D., Merilo E., Verstraeten I., Park S. Y., Elzinga D., Kaundal A., Helander J., Lozano-Juste J., Otani M., Wu K., Jensen D. R., Kollist H., Volkman B. F., and Cutler S. R., “A rationally designed agonist defines subfamily IIIA abscisic acid receptors as critical targets for manipulating transpiration,” ACS Chemical Biology, vol. 12, no. 11, pp. 2842–2848, 2017 [DOI] [PubMed] [Google Scholar]
- 95.Zhang K., and Cui B., “Optogenetic control of intracellular signaling pathways,” Trends in Biotechnology, vol. 33, no. 2, pp. 92–100, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chatelle C., Ochoa-Fernandez R., Engesser R., Schneider N., Beyer H. M., Jones A. R., Timmer J., Zurbriggen M. D., and Weber W., “A green-light-responsive system for the control of transgene expression in mammalian and plant cells,” ACS Synthetic Biology, vol. 7, no. 5, pp. 1349–1358, 2018 [DOI] [PubMed] [Google Scholar]
- 97.Müller K., Siegel D., Rodriguez Jahnke F., Gerrer K., Wend S., Decker E. L., Reski R., Weber W., and Zurbriggen M. D., “A red light-controlled synthetic gene expression switch for plant systems,” Molecular BioSystems, vol. 10, no. 7, pp. 1679–1688, 2014 [DOI] [PubMed] [Google Scholar]
- 98.Ochoa-Fernandez R., Samodelov S. L., Brandl S. M., Wehinger E., Müller K., Weber W., and Zurbriggen M. D., “Optogenetics in plants: red/far-red light control of gene expression,” Methods in Molecular Biology, vol. 1408, pp. 125–139, 2016 [DOI] [PubMed] [Google Scholar]
- 99.Brand A. H., and Perrimon N., “Targeted gene expression as a means of altering cell fates and generating dominant phenotypes.,” Development, vol. 118, no. 2, pp. 401–415, 1993 [DOI] [PubMed] [Google Scholar]
- 100.Xie M., Muchero W., Bryan A. C., Yee K., Guo H. B., Zhang J., Tschaplinski T. J., Singan V. R., Lindquist E., Payyavula R. S., Barros-Rios J., Dixon R., Engle N., Sykes R. W., Davis M., Jawdy S. S., Gunter L. E., Thompson O., DiFazio S. P., Evans L. M., Winkeler K., Collins C., Schmutz J., Guo H., Kalluri U., Rodriguez M., Feng K., Chen J. G., and Tuskan G. A., “A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus,” Plant Cell, vol. 30, no. 7, pp. 1645–1660, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Belcher M. S., Vuu K. M., Zhou A., Mansoori N., Agosto Ramos A., Thompson M. G., Scheller H. V., Loqué D., and Shih P. M., “Design of orthogonal regulatory systems for modulating gene expression in plants,” Nature Chemical Biology, vol. 16, no. 8, pp. 857–865, 2020 [DOI] [PubMed] [Google Scholar]
- 102.Mahfouz M. M., Li L., Piatek M., Fang X., Mansour H., Bangarusamy D. K., and Zhu J. K., “Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein,” Plant Molecular Biology, vol. 78, no. 3, pp. 311–321, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., and Wang H., “A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes,” Plant Methods, vol. 7, no. 1, p. 30, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Paoli H. C., Tuskan G. A., and Yang X., “An innovative platform for quick and flexible joining of assorted DNA fragments,” Scientific Reports, vol. 6, no. 1, article 19278, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chao R., Yuan Y., and Zhao H., “Recent advances in DNA assembly technologies,” FEMS Yeast Research, vol. 15, no. 1, pp. 1–9, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu R., and Li Q., “Protocol: streamline cloning of genes into binary vectors in agrobacterium via the Gateway® TOPO vector system,” Plant Methods, vol. 4, no. 1, p. 4, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marillonnet S., and Grützner R., “Synthetic DNA assembly using Golden Gate cloning and the hierarchical modular cloning pipeline,” Current Protocols in Molecular Biology, vol. 130, no. 1, 2019 [DOI] [PubMed] [Google Scholar]
- 108.Shearwin K. E., Callen B. P., and Egan J. B., “Transcriptional interference–a crash course,” Trends in Genetics, vol. 21, no. 6, pp. 339–345, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Padidam M., and Cao Y., “Elimination of transcriptional interference between tandem genes in plant cells,” BioTechniques, vol. 31, no. 2, pp. 328–334, 2001 [DOI] [PubMed] [Google Scholar]
- 110.She W., Lin W., Zhu Y., Chen Y., Jin W., Yang Y., Han N., Bian H., Zhu M., and Wang J., “The gypsy insulator of Drosophila melanogaster, together with its binding protein suppressor of hairy-wing, facilitate high and precise expression of transgenes in Arabidopsis thaliana,” Genetics, vol. 185, no. 4, pp. 1141–1150, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang W., Sun L., Yang X., Wang M., Esmaeili N., Pehlivan N., Zhao R., Zhang H., and Zhao Y., “The effects of transcription directions of transgenes and the gypsy insulators on the transcript levels of transgenes in transgenic Arabidopsis,” Scientific Reports, vol. 7, no. 1, article 14757, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCue K. F., Gardner E., Chan R., Thilmony R., and Thomson J., “Transgene stacking in potato using the GAANTRY system,” BMC Research Notes, vol. 12, no. 1, p. 457, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Altpeter F., Springer N. M., Bartley L. E., Blechl A. E., Brutnell T. P., Citovsky V., Conrad L. J., Gelvin S. B., Jackson D. P., Kausch A. P., Lemaux P. G., Medford J. I., Orozco-Cárdenas M. L., Tricoli D. M., van Eck J., Voytas D. F., Walbot V., Wang K., Zhang Z. J., and Stewart CN Jr, “Advancing crop transformation in the era of genome editing,” Plant Cell, vol. 28, no. 7, pp. 1510–1520, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maher M. F., Nasti R. A., Vollbrecht M., Starker C. G., Clark M. D., and Voytas D. F., “Plant gene editing through de novo induction of meristems,” Nature Biotechnology, vol. 38, no. 1, pp. 84–89, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Araus J. L., Kefauver S. C., Zaman-Allah M., Olsen M. S., and Cairns J. E., “Translating high-throughput phenotyping into genetic gain,” Trends in Plant Science, vol. 23, no. 5, pp. 451–466, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheung C. Y. M., Poolman M. G., Fell D. A., Ratcliffe R. G., and Sweetlove L. J., “A diel flux balance model captures interactions between light and dark metabolism during day-night cycles in C3 and crassulacean acid metabolism leaves,” Plant Physiology, vol. 165, no. 2, pp. 917–929, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Borland A. M., and Yang X., “Informing the improvement and biodesign of crassulacean acid metabolism via system dynamics modelling,” The New Phytologist, vol. 200, no. 4, pp. 946–949, 2013 [DOI] [PubMed] [Google Scholar]
- 118.Song L., and Crawford G. E., “DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells,” Cold Spring Harbor Protocols, vol. 2010, no. 2, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Buenrostro J. D., Wu B., Chang H. Y., and Greenleaf W. J., “ATAC-seq: a method for assaying chromatin accessibility genome-wide,” Current Protocols in Molecular Biology, vol. 109, no. 1, pp. 21.29.1–21.29.9, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bartlett A., O'Malley R. C., Huang S. S. C., Galli M., Nery J. R., Gallavotti A., and Ecker J. R., “Mapping genome-wide transcription-factor binding sites using DAP-seq,” Nature Protocols, vol. 12, no. 8, pp. 1659–1672, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]