Abstract
The majority of marine microbes remain uncultured, which hinders the identification and mining of CO2-fixing genes, pathways, and chassis from the oceans. Here, we investigated CO2-fixing microbes in seawater from the euphotic zone of the Yellow Sea of China by detecting and tracking their 13C-bicarbonate (13C-HCO3-) intake via single-cell Raman spectra (SCRS) analysis. The target cells were then isolated by Raman-activated Gravity-driven Encapsulation (RAGE), and their genomes were amplified and sequenced at one-cell resolution. The single-cell metabolism, phenotype and genome are consistent. We identified a not-yet-cultured Pelagibacter spp., which actively assimilates 13C-HCO3-, and also possesses most of the genes encoding enzymes of the Calvin-Benson cycle for CO2 fixation, a complete gene set for a rhodopsin-based light-harvesting system, and the full genes necessary for carotenoid synthesis. The four proteorhodopsin (PR) genes identified in the Pelagibacter spp. were confirmed by heterologous expression in E. coli. These results suggest that hitherto uncultured Pelagibacter spp. uses light-powered metabolism to contribute to global carbon cycling.
1. Introduction
Photosynthesis is a fundamental biological process, which uses solar energy to power the fixation of CO2. This process of photoautotrophy is responsible for nearly all of the Earth’s primary production of 104.9 petagrams of carbon per year (Pg of C/yr), nearly half of which (48.5 Pg of C/yr) is accounted for by photosynthesis in the oceans [1]. Since most marine bacteria are not yet cultured [2, 3], they play an enigmatic yet important role in terms of photosynthesis and CO2 fixation.
Two distinct systems have been discovered so far for harvesting solar energy: chlorophyll-based photochemistry and rhodopsin-based, light-activated proton pumps [4, 5]. Chlorophyll-based systems in cyanobacteria have been studied intensively for decades, and the importance of rhodopsin-based systems has become increasingly clear; for example, a recent survey suggested that microbial rhodopsins are major contributors to solar energy harvesting in the oceans [6]. Although chlorophyll-based photosynthesis involves the assembly of a large, relatively complex network of light-harvesting and charge-separating chlorophyll-protein complexes, there is a greater energetic benefit obtained than that from a rhodopsin-based mechanism [7]. However, the simple and minimal cost of synthesising proteorhodopsin has made it globally abundant in microbes [5, 8–12]. It has also been found that proteorhodopsin (PR) can simulate bacterial growth [13] or support bacteria surviving in poor nutrient conditions [14, 15]. Thus, due to its ubiquitous distribution and high abundance, rhodopsin-based phototrophy plays an important role in nature [16, 17]. Chlorophyll- and rhodopsin-based systems absorb different and complementary wavelengths of the solar spectrum [6] and were thought to rarely coexist in a host. However, recent studies have shown that these two distinct systems can exist in one single bacterium (e.g., Tardiphaga sp.) for enhanced solar energy utilization [18, 19].
Chlorophyll-based photosynthesis is a well-known light-harvesting system for CO2 fixation [1]. PR-based photosynthesis such as Dokdonia sp. MED134 has been reported to fix CO2 in the presence of light [13]. Subsequently, anaplerotic CO2 fixation in marine bacteria has been reported [20–28]. PR-containing bacteria could be the most abundant [29, 30], with microbial rhodopsins making a large contribution to solar energy harvesting in the oceans [6]. However, it is still unknown whether PR-containing bacteria in natural conditions can fix CO2.
Single-cell Raman microspectroscopy, a label-free technique to measure molecular vibrational profiles of single cells, offers a powerful suite of approaches for correlating cellular function and genomics [10, 31–37]. Single-cell Raman spectra (SCRS) display the intrinsic biochemical fingerprints of individual cells, which can be used as a form of metabolic phenotype [38, 39]. When coupled with stable isotope probing (Raman-SIP), SCRS can be used to measure specific functions of cell metabolism, exploiting the shifts in some Raman bands when bacteria incorporate stable isotopes (e.g., 13C, 15N, and 2H) ([31, 39–43]; Hatzenpichler et al.; [34]). Subsequently, individual cells displaying particular bacterial metabolic changes can be selected for genomic analysis using Raman-activated cell sorting (RACS) techniques [32, 44, 45], such as Raman tweezers [38, 46], Raman-activated microfluidic sorting (RAMS) [47–49], Raman-activated droplet sorting (RADS) [50], and Raman-activated cell ejection (RACE) [10, 36, 51, 52].
One challenge of Raman-based single-cell genomics is that genome coverage is usually low (<20%), providing limited genotypic information about the targeted cells. Although minimetagenomics from a small number of sorted cells (rather than one single cell) and binning technology can improve genome coverage [53, 54], they introduce uncertainty in assigning the correlated genomic and functional information to specific cells [10, 36].
In this study, we applied Raman-activated Gravity-driven Encapsulation (RAGE) [55] to precisely isolate CO2-fixing bacteria, one cell at a time. Single bacterial cells from the euphotic zone of the Yellow Sea of China were incubated with 13C-HCO3-, then sorted and whole-genome sequenced based on the Raman shifts in SCRS, which were altered as a result of incorporating 13C. We isolated a strain of Pelagibacter spp. able to fix 13C-HCO3-. It suggests that PR-containing bacteria should contribute to solar energy harvesting in the oceans and could also play an important role in the CO2 global budget.
2. Materials and Methods
2.1. Seawater Sampling and Preparation
Seawater was sampled at the euphotic zone in an inshore site located in a fishery area of Laoshan Bay in the Yellow Sea, China (Figure S1). The euphotic water samples were collected for the subsequent experiments involving incubation with different carbon sources and identification of carotenoid-containing bacteria. The seawater was filtered through a 3 μm membrane to collect microbial biomass.
Five sets of seawater samples were treated with different carbon sources separately, including original seawater (“C-free” for short), 12C-NaHCO3, 13C-NaHCO3, 12C-CO2, and 12C-CO2 plus 13C-NaHCO3 (“C-combined” for short), all in triplicate. Among them, the C-free treatments serve as an incubation control. The final concentration of the total carbon source (12C or 13C bicarbonate) was 2 mM. For the treatment group with CO2, the seawater was bubbled with 0.5% CO2. Seawater microbes were kept in glass bottles in the open air and under constant natural light conditions. Firstly, after pretreatment, the cells were filtered by a 0.22 μm pore-size membrane and then frozen at -80°C until DNA extraction for the following high-throughput 16S rRNA sequencing. Secondly, during incubation, cells in samples with different carbon sources were separately enriched by passing a ~70 ml sample through a Centricon® Plus-70 Ultracel PL-100 centrifugal filter (Merck Millipore, USA). After that, the cells were partly harvested for Raman measurement based on the timeline and partly used for Raman-based single-cell sorting according to the utilization function of 13C-NaHCO3.
As a negative control, dark incubation experiments were carried out for the seawater samples. Specifically, the seawater samples were, respectively, incubated with 12C-NaHCO3 and 13C-NaHCO3, each in triplicates, with the bottles wrapped in tin foil. All incubation conditions and SCRS acquisition setting are identical to the experiments above.
2.2. Chip for Raman-Based Single-Cell Sorting
To decrease Raman background noise, the RAGE chip is made of quartz. The chip consists of two quartz slides: an upper layer with inlet holes and open wells and a bottom layer consisting of a microchannel for cells. The microchannel on the bottom layer is built via the wet-etching method. Specifically, a 2 μm positive photoresist layer is spined on a quartz slide with 100 nm chromium. Then, the slide is covered with a photomask of the microchannel structure and exposed to UV light. The channel is created after etching with chromium etchant (Ce(NH4)2(NO3)6 in HClO4 solution) and quartz etchant (HF-HNO3 solution). The bottom layer with microchannel is then cleaned with acetone and bonded with the upper layer at 100°C. Finally, the quartz chip is treated with dichlorodimethylsilane for hydrophobic decorating, for generating the microdroplets of the proper size.
Single-cell Raman analysis is conducted in the detection window on the chip. The distance from the detection window to the oil well is about 500 μm. A sorting channel connected the detection window and oil well. The scale of the thin sorting channel is adjusted for different sizes of cells. We usually use a ~30 μm width and 10 μm depth microchannel for the sorting of microbial cells and ~50 μm width and 30 μm depth for the sorting of microbial cells.
2.3. Single-Cell Raman Microspectroscopy
All the single-cell Raman spectra were acquired on a RACS-Seq system (Qingdao Single-cell Biotech, China) or a LabRam HR system (Horiba, France). The spectra were analyzed with LabSpec 6 software and customized scripts. The Raman bands in SCRS from carotenoid-containing cells were determined and further analyzed to establish a relationship between the significant Raman shifts and 13C absorption. The Raman bands in SCRS of carotenoid-containing cells from a 12C-labeled sample were used as controls. Cells with a 13C shift in SCRS were isolated using the RAGE chip as described above. After that, the tube which contained the target cells in a one-cell-one-tube manner was then moved into a laminar hood, and lysis buffer (Qiagen, USA) was added for the following cell lysis.
2.4. Proteorhodopsin Overexpression and Purification
Four candidate proteorhodopsin genes identified from RG1 and RG6 were chemically synthesised (Integrated DNA Technologies, USA) with a C-terminal 6xHis tag and flanked by NcoI and PmeI restriction enzyme sites at the N- and C-terminus, respectively. These 4 genes were ligated into the arabinose-inducible pBADMycHisA vector, their integrity confirmed by DNA sequencing (Eurofins Genomics, Germany) and the resulting 4 constructs transformed into chemically competent E. coli C43 cells [56]. Each of the 4 strains was overexpressed by picking a single colony into 10 ml LB containing 100 μg ml-1 ampicillin, growing overnight at 37°C with shaking at 200 rpm, and using this to inoculate 500 ml LB containing 100 μg ml-1 ampicillin in a 2.5 l conical flask. This was grown for 2 hours at 200 rpm and 37°C and then induced with 0.2% () arabinose (Sigma-Aldrich, UK) and 5 μg ml-1 all-trans retinal (Sigma-Aldrich, UK) and grown for a further 4 hours at 37°C. Pelleted cells were resuspended in approximately 10 ml of buffer A (25 ml K2HPO4/KH2PO4 pH 7.4) and lysed by two cycles of French pressing at a pressure of 18 000 psi. The resulting lysate was separated into two halves. Membrane fractions were isolated from one half on a 10-50% continuous sucrose gradient (the sucrose was made up in the cell resuspension buffer and 1.5 ml broken cells loaded per gradient) spun at 30,000 rpm for 2 hours at 4°C. In the second half, the detergent n-dodecyl-β-D-maltopyranoside (β-DDM) was added dropwise to a final concentration of 2% () and left at 4°C for 1 hour. The lysed cells were centrifuged at for 20 minutes and the supernatant decanted into a fresh tube and the pellet discarded. The supernatant was applied to a self-packed anion exchange column containing DEAE-sepharose (GE Healthcare) using buffer A as running buffer. The flow-through, containing the membrane-bound proteorhodopsin complexes, was collected and centrifuged at for 20 minutes at 4°C. The supernatant was discarded and the proteorhodopsin-containing pellet resuspended in buffer A containing 1% () of the detergent octyl-beta-glucoside (OTG) and left overnight at 4°C to solubilise the membrane-bound proteorhodopsin complexes. The solubilised membranes were centrifuged at to remove any unsolubilised material and an absorption spectrum of the supernatant taken using an Agilent Cary 60 UV-Vis spectrophotometer.
3. Results
3.1. A Workflow to Identify and Isolate Individual CO2-Fixing Bacteria from Seawater
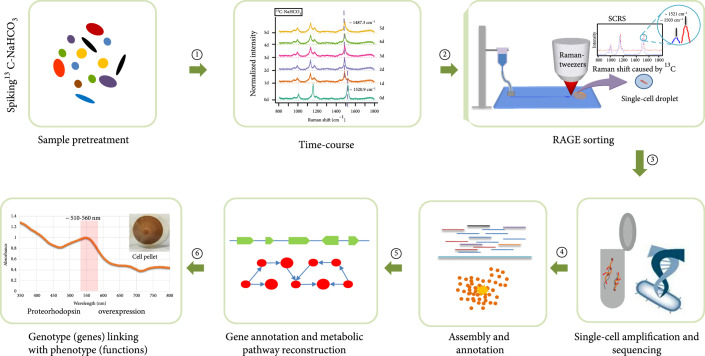
It is important to link the genotype and phenotype of a single cell. This study used Raman spectra to sort cells, establishing a direct link between bacteria function and single-cell genomics (Figure 1). The seawater was sampled from a typical euphotic zone in the Yellow Sea of China (Figure S1; physiochemical parameters in Table S1). A time-course experiment was conducted to identify CO2-fixing bacteria in the presence of 13C-NaHCO3. Raman-activated Gravity-driven Encapsulation (RAGE) [55] was applied to sort single cells. Every single bacterial cell was sorted according to its characteristic SCRS, precisely packaged in a picoliter microdroplet, and exported in an indexed “one-cell-one-tube” manner for downstream single-cell multiple displacement amplification (MDA) and then genome sequencing (Figure 1). The single-cell genomes were reconstructed, and the key functional genes and pathways were identified, according to Raman function analysis. In this case, these include carotenoid and retinal synthesis pathway, PR synthesis, and CO2 fixation pathway. Finally, some novel proteorhodopsin genes were characterized and validated by expressing them in E. coli. Therefore, a holistic picture of the single bacterial cell in terms of function (phenotype) and genomics (genotype) is linked and revealed (Figure 1).
Figure 1.
Workflow for revealing CO2-fixing function of bacteria in the ocean that links genotype to phenotype: ①: feeding of 13C-substrate to probe the metabolism of the seawater sample and then analyzing the time-course Raman spectra of carotenoids containing cells to find the shortest labeling time; ②: target cells were searched by Raman detection according to SCRS which exhibits shifts caused by 13C incorporation and then sorts out by the RAGE device in single-cell-single tube style; ③: the sorted cells were lysed, and its nucleic acid was amplified by MDA; then, the extracted DNA is processed for single-cell genome sequencing by high-throughput sequencing; ④: assembly and annotation for the single-cell genomic data; ⑤: metabolic functions are validated by the reconstructed metabolic pathways and genes; ⑥: proteorhodopsin overexpression was conducted for validating the PR prediction based on genomic analysis, and finally, a link between genotype (genes) and phenotype (functions) is established at the single-cell level.
3.2. Time-Course of 13C Incorporation into CO2-Fixing Bacteria Monitored by Single-Cell Raman Spectra
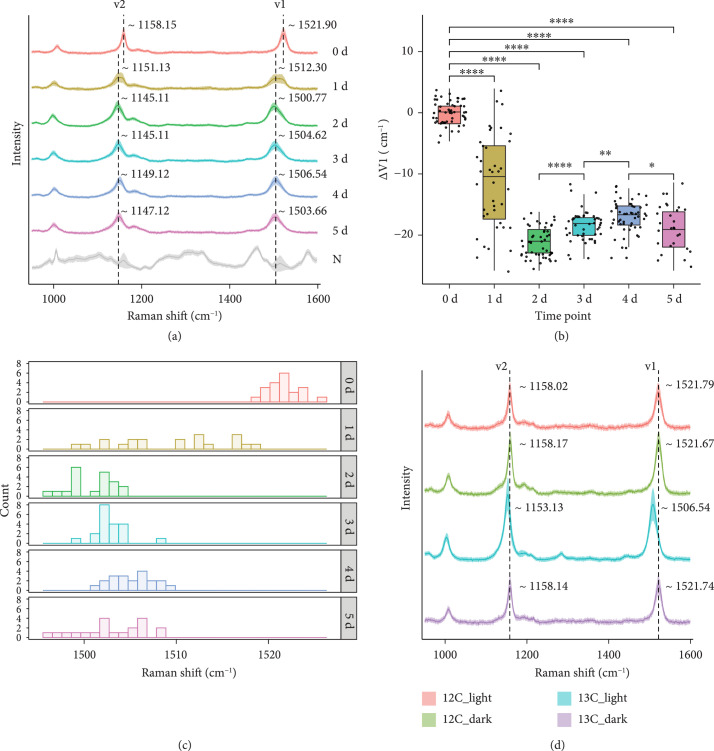
Since the integration of stable isotopes (13C, 15N, 2H, or 18O) into bacterial cells leads to significant shifts of some Raman bands in SCRS, single-cell Raman spectroscopy combined with stable isotope detection (Raman-SIP) has been used to reveal specific metabolic functions of bacteria and to screen active cells in microbial communities [10, 31, 34, 40, 57, 58]. Nearly all photosynthetic bacteria contain either carotenoids or retinoids for light harvesting and structural stabilization [15, 58]. Hence, after uptake and incorporation of 13C-HCO3- or 13CO2 into CO2-fixing bacteria, carotenoid Raman bands shift due to the altered vibrational energy caused by 13C [58]. Raman spectra of carotenoid molecules display three characteristic Raman bands (Figure S2): v1 (in-phase C=C), v2 (C–C stretching vibrations of the polyene chain), and v3 (in-plane rocking mode of CH3 groups attached to the polyene chain) [33, 58–60]. The positions of v1 and v2 in the SCRS of photosynthetic cells show stepwise shifts to lower wavenumbers when cells are incorporated with 13C [58]. In this study, we used the simultaneous shifts in both v1 and v2 as Raman biomarkers, to identify and sort photosynthetic cells that were actively fixing 13C-NaHCO3 in the seawater sample (Figures 2 and 3).
Figure 2.
Raman spectra of carotenoid-containing cells in the seawater which were incubated in closed bottles at room temperature at different times. (a) Dynamic red shift of Raman spectra from carotenoid-containing cells in the seawater which were incubated from day 0 to day 5. Each spectrum represents an average of SCRS from ~45 single cells, and the shadow represents the standard deviation of SCRS. The Raman spectra labeled with N represent an average of SCRS from the non-carotenoid-containing cells. (b) The bar plot showed the intensity ratio of SCRS v1 band in each of the day groups obtained from the marine bacteria cells. (c) Histogram plots for v1 shift in SCRS of carotenoid-containing cells in the seawater which were incubated in closed bottles at room temperature (sampled at time points spanning from day 0 to day 5). (d) Raman spectra from carotenoid-containing cells in the seawater which were incubated with 12C-NaHCO3 or 13C-NaHCO3, under dark or normal light conditions, respectively, on day 5.
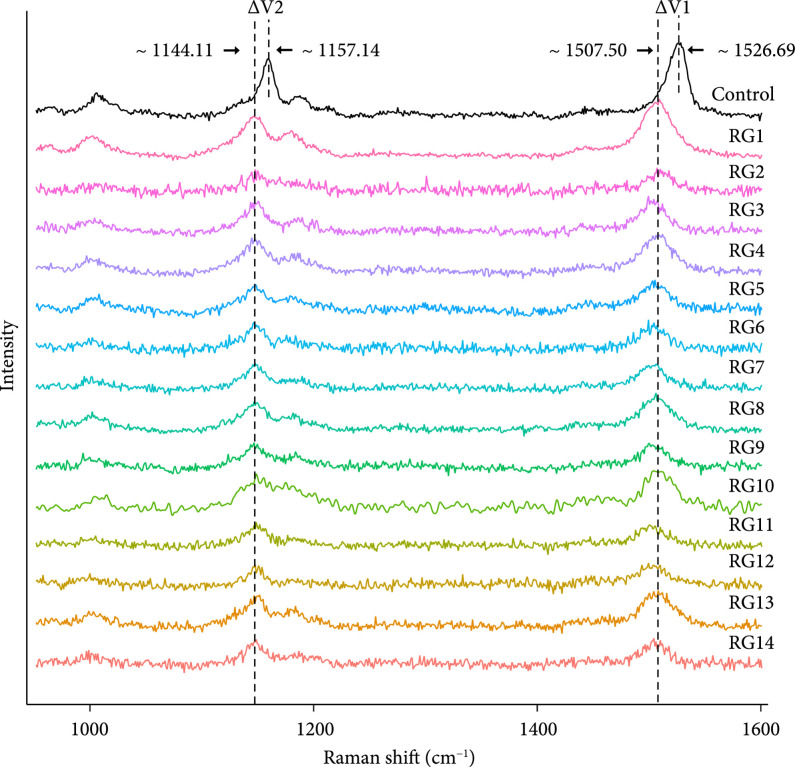
Figure 3.
Single-cell Raman spectra of the CO2-fixing cells sorted by RAGE from the seawater incubated with NaHCO3. “Control”: 12C-NaHCO3-treated cell at 1 day of incubation; “RG1-RG14”: 13C-NaHCO3-treated cell at 1 day of incubation.
The time-course experiment was conducted to estimate the rate of 13C incorporation, and ~45 single-cell Raman spectra were measured at each time point (Figure 2(a)). Within 24 h (day 1) after incubating the seawater with 13C-NaHCO3 at room temperature, Raman shifts (due to 13C incorporation) in marine bacteria were detectable by Raman microspectroscopy (Figures 2(b) and 2(c)). The v2 bands shift from 1158 cm-1 to 1151 cm-1 while v1 bands shift from 1522 cm-1 to 1512 cm-1. In the following days (day 2 to day 5), the v2 and v1 bands downshifted further, to ~1145 cm-1 and ~1502 cm-1, respectively (Figure 2(a)). The data showed that after the first 24 h of incubating the seawater with NaH13CO3, Raman shifts (due to 13C-incorporation) among individual cells were highly heterogeneous, while at 48 h (and beyond), the degree of Raman shifts of the individual cells had all largely reached a steady state (Figures 2(b) and 2(c)).
To investigate the possibility of cross-feeding, we fed NaH13CO3 to a synthetic three-species mock microbiota that consists of Synechococcus elongatus PCC7942 (Se; an autotrophic bacterium), Micrococcus luteus OY14 (Ml; a heterotrophic bacterium), and Saccharomyces cerevisiae BY4742 (Sc; a heterotrophic fungus) mixed in a 1 : 1 : 1 ratio. In this mock community, after NaH13CO3 treatment for 24 h, 13C uptake was detected in Synechococcus elongatus PCC7942 (based on the “red shift” of Raman spectrum), but neither in Micrococcus luteus OY14 nor in Saccharomyces cerevisiae BY4742 (Figure S3). This observation suggests that the possibility of cross-feeding between photoautotrophic and heterotrophic bacteria within 24 h is unlikely in this study.
Based on the above results, the sampling time point for this experiment was set to day 1, and bacteria actively fixing 13C-NaHCO3 were sorted by RAGE, then analyzed by single-cell genomic sequencing (Figure 1). Dark incubation of the seawater samples in the presence of 13C-NaHCO3 was also performed and analyzed (Methods), which showed no Raman shift among the ~45 randomly selected carotenoid-containing cells under such conditions for 5 days (Figure 2(d)). Collectively, the results confirm that the Raman shift in the presence of 13C-NaHCO3 was due to light-responsive CO2 fixation.
3.3. The Impact of Bicarbonate on Microbial Community Structure in the Seawater Sample
To assess the impact of the supplemented carbon source (e.g., 12C-CO2, 12C-NaHCO3, or 13C-NaHCO3) on the microbiota structure, 16S rRNA amplicon sequencing (V3-V4) results were compared for five conditions: day 1 seawater spiked with 13C-NaHCO3 (C13-HCO3), 12C-NaHCO3 (C12-HCO3), 12C- (C12-CO2), 12C-CO2 plus 12C-NaHCO3 (C-combined), and the day 1 seawater control without any treatment (C-free). Three biological replicates were performed. After trimming, screening, and removal of chimeras and singletons, an average of ~64,664 high-quality reads (ranged from 52,931 to 89,432) were obtained (Table S2), showing that in this seawater sample, SAR11 bacteria (~9.6% in the community) were more abundant than cyanobacteria (~3.1%; Figure S4A). Abundance distribution suggests that the supplement of 12C- or 13C-HCO3- had introduced minor impacts on the structure of the microbial community, compared to the original seawater control without any supplement (Figure S4A). Moreover, principal coordinate analysis (PCoA) and UPGMA clustering analysis were performed to evaluate the similarity in the microbial community structure of all samples using the weighted UniFrac distance, which incorporates the degree of divergence in the phylogenetic tree of operational taxonomic units (OTUs). Both analyses show that the C-combined and 12C-CO2 samples were quite similar to each other (probably due to the CO2-caused acidification of seawater), while the HCO3--adding groups were more similar to the C-free group (Figures S4B and S4C). Therefore, since sodium bicarbonate as an external carbon source has a minimal impact on the structure of the seawater microbiota, 13C-HCO3- probing, which was the option we chose here, should more precisely model the in situ functional activity in the seawater sample, than other 13C-labeled carbon substrates tested above.
3.4. Linking CO2 Fixation Functions to the Underlying Genome at Single-Cell Resolution
After one-day incubation of seawater with 13C-NaHCO3, cells were sorted using RAGE (Figure 1; Methods). Cells whose SCRS display both carotenoid bands and 13C shifts were sorted by RAGE (Figures 1 and 3) according to the Raman sorting criteria (Figure 2). The sorted cells trapped in the droplets were transferred into PCR tubes, with each tube containing only one single cell.
Following labeling with 13C-NaHCO3 for 24 hours, 14 individual bacterial cells were sorted independently from the same seawater sample using the sorting criteria (Figures 2 and 3). All sorted cells showed significant Raman shifts in the v1 and v2 bands in SCRS (Figure 3), indicating a likely CO2-fixing function. In order to correlate the observed phenotype (e.g., CO2 fixation) via Raman analysis and the genotype at the single-cell level, the sorted cells were processed to do single-cell genomic analysis. Cells were initially lysed, and then, the lysates were used as templates for MDA. The 16S rRNA sequence of MDA products was examined by PCR to confirm successful amplification (Figure S5; primers shown in Table S3).
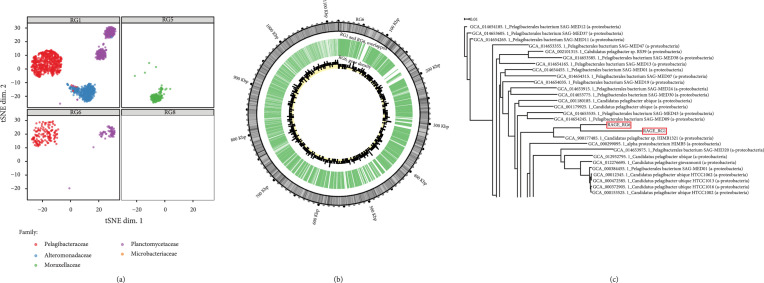
In the same batch of experiments, 6 out of 14 one-cell samples had positive results in the 16S rRNA PCR test. Six positive samples were numbered RG1, RG5, RG6, RG8, RG9, and RG11 (Figure S5). Among these MDA-positive samples, high-throughput sequencing libraries of four (RG1, RG5, RG6, and RG8) were successfully constructed and sequenced (Methods). After quality control, ~17.4 million clean reads for each sample were obtained for data assembly (Table S4). To assign the taxonomy of the target cell, we have employed both GTDB and BLASTN for the RAGE-Seq-derived one-cell assemblies, which revealed essentially identical results (Table S5). According to de novo genome assembly and homology-based annotation, the sorted bacteria belong to five phyla: Pelagibacteraceae, Moraxellaceae, Microbacteriaceae, Planctomycetaceae, and Alteromonadaceae (Table 1). Then, assembled contigs (>2,000 bp), which represented the clusters of genomes in each sample, were visualized using t-SNE (Figure 4(a)).
Table 1.
Predicted genome completeness and 16S rRNA genes of RAGE-sorted bacteria.
| RAGE-sorted samples | Taxonomic classification | Estimated genome completeness by CheckM (%) | Identity score for the identified 16S rRNA in SCGs (%) | Abundance in the community (C-free/13C-HCO3) | Reference supporting presence of carotenoids | Reference evidence of carbon dioxide fixation |
|---|---|---|---|---|---|---|
| RG1 | Pelagibacteraceae | 97.6 | 99.6 | 7.65%/5.84% | [78] | — |
| Planctomycetaceae | 74.5 | — | — | — | — | |
| Alteromonadaceae | 72.9 | — | 0/0 | — | — | |
|
| ||||||
| RG5 | Moraxellaceae | 76.4 | 99.7 | 0/0 | [79] | [80] |
|
| ||||||
| RG6 | Pelagibacteraceae | 80.0 | 99.6 | 7.65%/5.84% | [78] | — |
| Planctomycetaceae | 24.4 | — | — | — | — | |
|
| ||||||
| RG8 | Microbacteriaceae | 10.7 | 100.00 | 6.49%/7.84% | [81] | [82] |
Figure 4.
Binning of SAGs from single-cell genomes derived from one-cell RAGE-Seq from seawater samples. (a) The t-SNE projection of binned contigs from post-RACS single-cell sequencing reveals their taxonomical origin. Contigs are visualized based on 4-mer frequency features. Samples are marked with distinct shapes. Each contig is colored based on its taxonomic annotation at the family level. (b) Circos plot representation of overlaps between recovered Pelagibacter spp. genomes from RG1 and RG6. The RG6 genome is linked from all contigs and represented as the outer track, with coordinates marked on the outermost circle and GC content as an inner heat map. Overlap regions between contigs of RG1 and RG6 are shown as the middle track. The gene numbers per kilobase for RG6 are given as the innermost track. Overall, ~71.25% genomes of RG6 (in length) also have evidence in RG1. (c) Phylogenetic tree based on available Pelagibacteraceae genomes (partial; the complete figure in Figure S5). RG1 and RG6 are compared with genomic sequences of 89 Pelagibacteraceae obtained from NCBI GenBank (GCA) assembly. All genomes were annotated using Prokka. STRIDE was used by OrthoFinder [83] for inferring the root of a species tree by analyzing the distribution of gene duplication events in the genomes of the species in the tree (http://www.stevekellylab.com/stride).
The dominant contigs in RG1 and RG6 were assigned to the SAR11 bacteria of Pelagibacteraceae, although a small portion of contigs was assigned to Planctomycetaceae and Alteromonadaceae (Figure 4(a), Table 1, and File S1). The cross assignment might be due to either the previously unknown Pelagibacteraceae, DNA contaminations, or other symbiotic bacteria. The >1 kb PCR products of 16S rRNA genes (27F and 1492R) from MDA of RG1 and RG6 were purified and sequenced, which showed that both are >99% identical to “Candidatus Pelagibacter ubique” HTCC1062 [61] (File S2).
Based on the RAGE sorting criteria, the targeted cells should contain shifted carotenoid bands in their SCRS due to the incorporation of 13C from 13C-NaHCO3. According to the literature, the sorted bacteria in the three families Pelagibacteraceae, Moraxellaceae, and Microbacteriaceae are able to synthesise carotenoids (Table 1), consistent with the sorting criteria. Although it has been reported that SAR11 can employ PR to harvest sunlight as an energy source [5], it was not known whether SAR11 bacteria could use light to drive the assimilation of HCO3- in the natural environment.
According to the Raman sorting criteria, the sorted Pelagibacter spp. should contain carotenoids and also have the ability to incorporate 13C-NaHCO3. Pelagibacter spp. was in the first group of bacteria (day 1) found to be labeled with 13C in the seawater sample, which was taken from the euphotic zone with a low level of organics (chemical oxygen demand 1.7 mg/l, Table S1). These data suggest that Pelagibacter spp. should have both light-utilizing machinery and CO2 fixation ability. Hence, it is predicted that we should find genes encoding β-carotene biosynthesis, 15,15-dioxygenase, proteorhodopsin (PR), and CO2 fixation pathway in the reconstructed single-cell genome of the sorted Pelagibacter spp. Remarkably, our whole genome analysis indeed found most of these genes in the sorted Pelagibacter spp., as discussed below.
3.5. Reconstruction of the β-Carotene Biosynthesis Pathway from Single-Cell Genomic Data
For the reconstructed Pelagibacter spp. genome of RG1 and RG6, GC content was 29.3% and 29.2%, respectively (Table 2), consistent with the 89 sequenced Pelagibacter spp. strains in the NCBI database (27.8% to 33.4%, with an average of 29.5%). Their completeness was 97.6% and 80.0%, respectively, as estimated by CheckM (based on 695 marker genes from 15 Pelagibacteraceae genomes; Table 1), indicating that RAGE was able to obtain high genome coverage of just one bacteria cell from the microbial community (Figure 1). The reconstructed Pelagibacter spp. genomes of RG1 and RG6 revealed the sharing of 997 kb, i.e., 71.2% genomic content of the latter (Figure 4(b)). A phylogenetic tree of various sequenced SAR11 genomes confirms the identity of the reconstructed genomes from RG1 and RG6 as Pelagibacter spp. (Figure 4(c) and Figure S6).
Table 2.
Data statistics of Pelagibacterales obtained from the functional single-cell genomes of samples RG1 and RG6.
| Sample | Pelagibacter spp. in RG1 | Pelagibacter spp. in RG6 |
|---|---|---|
| Contig number | 5,400 | 862 |
| Bases | 5.036 Mbp | 1.111 Mbp |
| N50 | 1,862 | 3,019 |
| GC% | 29.3 | 29.2 |
| Gene | 4,998 | 1,144 |
The complete gene set for carotenoid biosynthesis was identified in the reconstructed low-GC-content Pelagibacter spp. contigs of RG1 and RG6 but absent in other parts of sequences in the corresponding MDA products (Figure 5 and Table 1). As 13C shift of carotenoid Raman bands was used as criteria for the cell sorting (Figure 3), the SAR11 bacteria of Pelagibacter spp. must be the targeted bacteria that contained carotenoids and incorporated 13C from 13C-HCO3 in the seawater (Table 1 and Figure 4(a)).
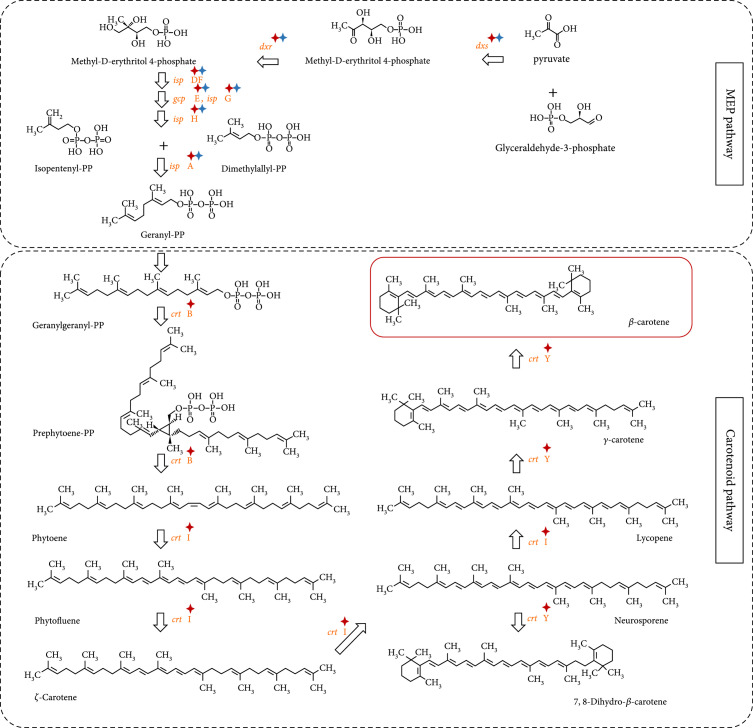
Figure 5.
Reconstructed β-carotene module in the carotenoid synthesis pathway from the single-cell Pelagibacter spp. genomes derived by RAGE-Seq. The known pathways (used as reference) were obtained from the KEGG database. Enzyme genes with red asterisk: found in the Pelagibacter spp. from RG1; with blue asterisk: found in the Pelagibacter spp. from RG6.
To reveal the carbon fixing potential of these functional single-cell genomes, the two single-cell gene sets from the Pelagibacter spp. were mapped to the KEGG pathways, which predict gene function mainly based on sequence homology [62]. Within these gene sets, seven metabolic pathways were examined, including ko00195 (photosynthesis), ko00710 (carbon fixation in photosynthetic organisms), ko00906 (carotenoid biosynthesis), ko00720 (carbon fixation pathways in prokaryotes), ko00860 (porphyrin and chlorophyll metabolism), ko00830 (retinol metabolism), and ko00900 (terpenoid backbone biosynthesis). Most of the seven pathways were detected in these four single-cell genomic samples in our study (details in File S3). Genes encoding the pathway for the synthesis of β-carotene synthesis were identified in the reconstructed Pelagibacter spp. genomes from RG1 and RG6 samples (Figure 5), consistent with the RAGE sorting criteria. Pelagibacter genome obtained from RG1 has all the genes for carotenoid synthesis, and only a partial of these genes were found in RG6 (the specific links between the genomes and the genes are highlighted in Figures 5 and 6).
Figure 6.
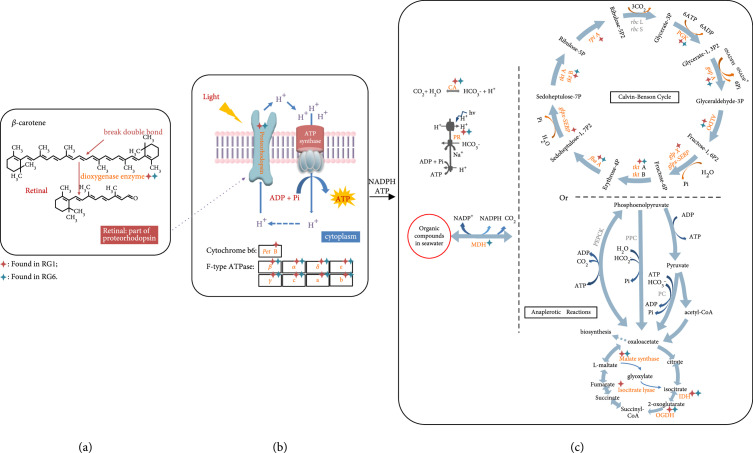
Reconstructed CO2-fixing pathways from the single-cell Pelagibacter spp. genomes derived by RAGE-Seq. (a) β-carotene can be cleaved by dioxygenase into retinal, which is an important element in functional PR; (b) light-harvesting PR; (c) reconstruction of Calvin-Benson cycle pathway and the anaplerotic reactions. The known pathways were obtained from the KEGG database. Enzyme genes with red asterisk: found in the Pelagibacter spp. from RG1; with blue asterisk: found in the Pelagibacter spp. from RG6. CA: carbonic anhydrase; PR: proteorhodopsin; PEPCK: phosphoenolpyruvate carboxykinase; PPC: phosphoenolpyruvate carboxylase; PC: pyruvate carboxylase; MDH: malate dehydrogenase; IDH: isocitrate dehydrogenase; OGDH: 2-oxoglutarate dehydrogenase.
3.6. Establishment of a Light-Utilizing System and a CO2-Fixing Pathway in the RAGE-Seq-Derived SAR11 Bacteria
β-Carotene is the precursor of retinal, a key component of functional PR (Figure 5); the 15,15-dioxygenase cleaves one molecule of β-carotene into two molecules of retinal, which is the cofactor binding within PR to make functional holo-PR (Figure 6(a)) [15, 63]. We not only found the pathway for β-carotene biosynthesis but also identified 15,15-dioxygenase genes from the single-cell Pelagibacter spp. genomes from both RG1 and RG6 samples (Figure 6(a), Figure S7A, and File S4). Notably, we also identified four other proteorhodopsin (PR) genes from single-cell genomes RG1 and RG6 (Figure 6(b), Figures S7B and S8, and File S5). In contrast, the genes required for chlorophyll-based photosynthesis, encoding the biosynthesis of chlorophyll a, the phycobilisome, and the biogenesis of Photosystem I and Photosystem II complexes [64] were all absent from the genome, demonstrating the absence of this chlorophyll mode of photosynthesis from the reconstructed Pelagibacter spp. genomes. Collectively, we have identified the genetic machinery to make holo-PR, and the presence of additional genes encoding the F-type ATPase (Figure 6(b)) suggests that Pelagibacter spp. is able to form a complete “photon to ATP” loop, consistent with the observation in the pure culture of SAR11 strain HTCC1062 [30].
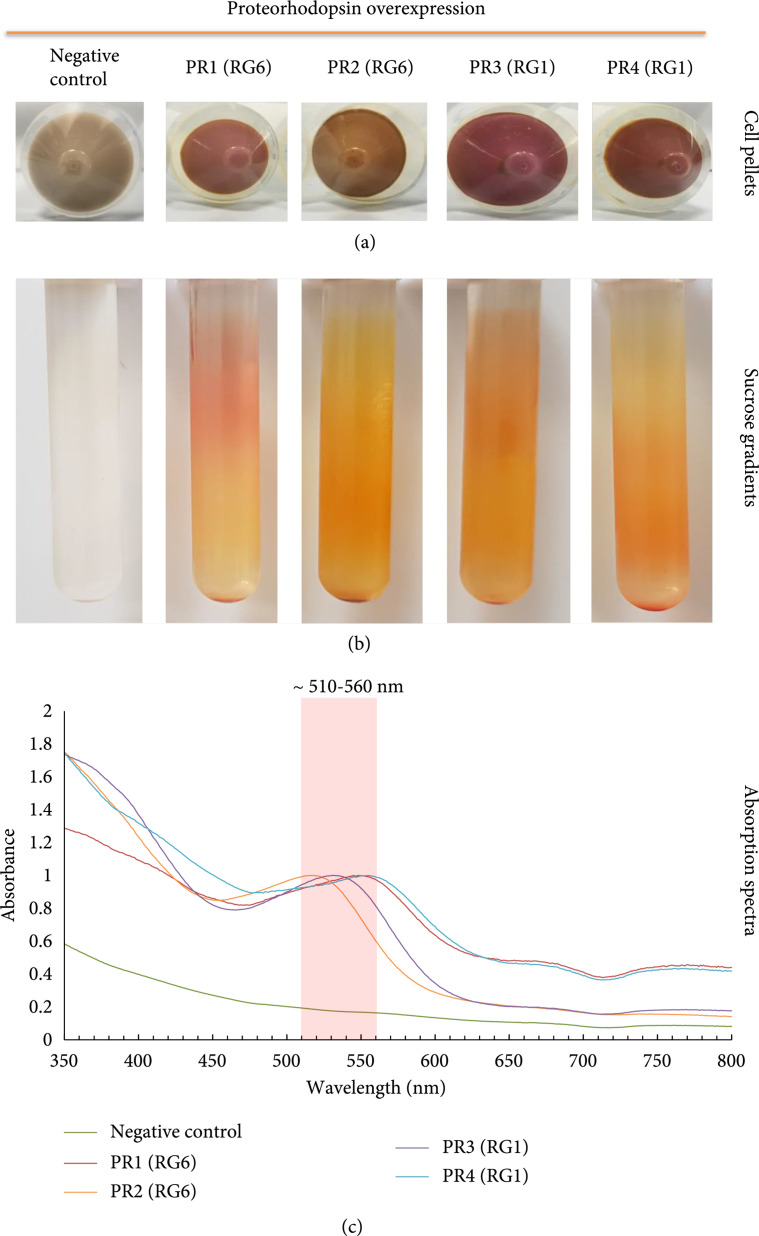
In order to validate the PR prediction based on genomic analysis of RG1 and RG6, we synthesised six PR fragments according to the sequencing data shown in File S5. These putative PR genes were cloned into plasmids and transferred into E. coli C43 strain, which is able to overexpress intracellular membranes for hosting membrane protein PR [56]. The expression of PRs was induced by 0.2% () arabinose in a LB medium supplemented with all-trans retinal. The negative control is E. coli C43 with plasmid backbone that did not produce any pigment, while four of six putative PR genes (PR1-4) cloned into E. coli C43 strains show clear pigment production (Figure 7(a)). The purified membrane of these four clones contains pigments (Figure 7(b)), and the holo-PRs absorb light between 510 and 560 nm (Figure 7(c)), which are the typical absorption wavelength of PR. Among these four PR genes (File S5), PR2 has 98% homology to a PR gene in Candidatus Pelagibacter sp. HIMB1321 (GenBank: LT840186.1), and PR3 has 94% homology to alpha proteobacterium HIMB59, an unclassified Pelagibacteraceae (GenBank: CP003801.1). Interestingly, PR1 and PR4 have no significant homology to any genes in GenBank, NCBI. However, the translated peptide sequence of PR1 is of 90% homology to xanthorhodopsin and 80% homology to bacteriorhodopsin, and PR4 is of 59% homology to rhodopsin in Planctomycetia bacterium. Hence, PR1 and PR4 are novel proteorhodopsin genes and successfully expressed and validated in E. coli.
Figure 7.
Functional characteristics of the PRs mined from the single-cell Pelagibacter spp. genomes derived by RAGE-Seq. (a) Cell pellets obtained after induction with arabinose and retinal for four E. coli C43 strains harbouring pBAD constructs that contain the putative proteorhodopsin (PR) genes from SCG data of RG1 and RG6, respectively. A pellet from the uninduced C43 strain containing pBAD:Hoch PR is shown as the negative control for comparison. (b) Separation of cell membranes from lysed cell pellets following their application to 10-50% continuous sucrose gradients and centrifugation. (c) Absorption spectra of the detergent-solubilised cell membrane pellets that contain the purified PR.
According to the sequencing and binning results, two possible pathways (Calvin-Benson cycle pathway and the anaplerotic reactions) to fix CO2 are shown in Figure 6(c). Most of the genes, except rbcL and rbcS, involved in the Calvin-Benson cycle of CO2 fixation have been identified in the reconstructed single-cell genomes from the sorted RG1 and RG6 Pelagibacter spp. samples (Figure 6(c)). However, some essential genes encoding enzymes catalysing from phosphoenolpyruvate to oxaloacetate in the anaplerotic reactions were absent (Figure 6(c)). These data suggest that the PR-containing Pelagibacter spp. is likely to have the genetic machinery to encode the Calvin-Benson cycle for CO2 fixation (Figure 6(c)) in the sea.
In summary, we used SCRS to identify bacterial cells that contain carotenoids and fix CO2, based on 13C-NaHCO3 labeling (Figure 3), which were then sorted, encapsulated, and whole-genome sequenced at the precisely one-cell resolution, creating a link for each cell between its physiological properties and its genetic composition. The reconstructed single-cell genomes of the SAR11 bacteria in the sample identified individual cells of Pelagibacter spp. harbouring the genes for synthesis of carotenoids (Figure 5), including retinal, proteorhodopsin, enzymes of CO2 fixation, and the F-type ATPase (Figure 6). Thus, each cell has the genetic information for fixing CO2, driven by the solar energy harvested by PR, and therefore, Pelagibacter spp. could contribute to carbon cycling in the oceans.
4. Discussion
4.1. Novel Raman-Activated Cell Sorting to Link Bacterial Functions and Single-Cell Genomics
The marine microbiome constitutes the bulk of the biomass in the ocean, and knowledge of its composition and physiological properties is fundamental for a deep scientific understanding of the global carbon/nitrogen cycle, the solar energy budget, and sustainable exploitation of marine resources. However, most bacteria in the marine microbiome remain uncultured [2, 3]. Thus, there is a need to develop methods for probing the function of environmental microbiomes in situ, which are driven by the following requirements: (i) dissection of the microbiota structure by sequencing and tracking microbiota state, function, and intercellular interaction and (ii) interrogation of a consortium or population of bacteria and the ability to probe individual bacterial cells. Many studies have successfully demonstrated that Raman-SIP is able to precisely pinpoint bacteria with specific metabolic functions at the single-cell level [10, 31, 36, 38, 39, 42, 43, 46, 49, 52, 65]. However, it is important to establish links between metabolism and genomes at the single-cell level using RACS [32, 33].
Although RACE has been developed to sort single cells and sequence their genomes according to bacterial SCRS, genome coverage for single cells was low [36]. Various attempts have been made to improve the genomic quality for single-cell sequencing [66, 67], but high coverage of single-cell genomes remains challenging. Technical limitations, such as high amplification bias with very few DNA templates, lead to lower success rates in single-cell genome amplification and lower target gene hit rates in sequencing data analysis [68]. In previous work, we attempted the identification of CO2-fixing bacteria in the Yellow Sea, but the difficulties in obtaining single-cell genomics required the pooling of over 30 cells for “minimetagenome sequencing” [10].
To tackle these challenges, we applied the RAGE to the marine sample, which sorts and encapsulates precisely one cell on the basis of its SCRS and minimises photo- and thermal damage to single cells by operating in water [55]. More importantly, picoliter encapsulation by RAGE can be readily transformed into an emulsion system, which effectively reduces the MDA reaction volume and achieves high-coverage sequencing of a one-cell genome. We showed that, for marine microbiome samples, SCRS are sufficiently sensitive to identify candidate cells actively fixing CO2 and that for each such cell RAGE allows precisely one-bacterial-cell-per-reaction MDA reaching almost 98% sequence coverage. The ability to profile and correlate bacterial metabolism and high-quality genome sequences at one-cell resolution illustrates the potential for a broad application of such Raman-activated cell sorting and sequencing approaches (RACS-Seq; for a detailed comparison of the various RACS-Seq methods, please refer to Table S2 from [34]).
Notably, the assembled genome RG1 was unusually large (~5 Mbp), much larger than the typical size of Pelagibacter spp. genomes and the ~1.1 Mbp size of Pelagibacter spp. from RG6 (Table 2), although the Pelagibacter spp. from RG1 and RG6, respectively, share 71.2% genomic content. It is possible that RG1 might contain either contaminated DNA or other symbiotic bacteria, although contamination of single-cell sequencing by free floating or cell-surface-attached environmental DNA is usually hard to avoid in field samples [69–74]. However, RG1 and RG6 were identified as SAR11 bacteria with the following evidence: (i) the nearly full-length 16S rRNA sequences (>1.0 kb) of PCR products from MDA of RG1 and RG6 indicated both as Pelagibacter spp. (File S2). (ii) Per CheckM, completeness of the Pelagibacter spp. genome was 97.6% for RG1 and 80.0% for RG6 (Table 1). (iii) The binning result shows that the GC contents of RG1 (~5 Mbp) and RG6 (~1.1 Mbp) were 29.3% and 29.2%, consistent with the typical SAR11 genome feature of low GC content. (iv) Key functional gene sets including carotenoid synthesis, dioxygenase, and PR were found in the reconstructed genomes of Pelagibacter spp. in RG1 and RG6 (Figure 6) but absent in those non-Pelagibacter spp. contigs (e.g., Planctomycetaceae and Alteromonadaceae contigs) in RG1 and RG6.
In the one-cell RAGE-Seq reactions of mock microbiota, without any exception, the top contig bin from each of such precisely one-cell assemblies corresponds to the target cell (i.e., the single-cell 16S rRNA sequencing-based genotype is always consistent with the SCRS-predicted phenotype such as whether the cell harbours pigments), supporting the low possibility of contamination in the RAGE-Seq workflow [34]. The presence of both Pelagibacteraceae and Planctomycetaceae in RG1 and RG6 might implicate the symbiosis hypothesis. However, this hypothesis does not change the key conclusion of this study, i.e., Pelagibacter spp. possesses most of the genes encoding enzymes for CO2 fixation and the full genes necessary for carotenoid synthesis. It is however beyond the scope of this study to validate the symbiosis hypothesis.
4.2. SAR11 in the Oceans Can Fix CO2 in the Presence of Light and Its Implication
In this study, we applied RAGE to obtain single-cell genomes from CO2-fixing bacteria identified by SCRS. We reconstructed genomes of CO2-fixing bacteria originating from the Yellow Sea in China, which were able to assimilate 13C-HCO3- as short as 24 hours at room temperature under natural light, while no CO2-fixating bacterium was observed in the dark experiments. The MDA amplified genomic DNA of the isolated single cells were checked by PCR using 27F and 1492 primers (Table S3), which cover nearly the full 16S rRNA gene length. A typical SAR11 bacterium Pelagibacter spp. was found as the single species in RG1 and RG6 MDA samples. Moreover, GC contents of RG1 and RG6 were 29.3% and 29.2%, respectively, both very similar to those of the other 89 sequenced Pelagibacter spp. strains in the NCBI database (27.8% to 33.4%, with an average of 29.5%). Single-cell genomics revealed that this Pelagibacter spp. contains an entire rhodopsin-based system, including four genes encoding PR and its retinal cofactor. The PR genes were validated by cloning and expression of these genes in E. coli. Thus, we have linked phenotype (the presence of carotenoids), function (the incorporation of 13C-HCO3-), and genotype for a series of Pelagibacter spp. cells, which we suggest as self-sufficient for light-powered energy conversion in the oceans.
The SAR11 clade of marine bacteria has an estimated global population of cells, and conventional metagenomic sequencing indicates that global SAR11 bacteria account for an estimated 25% of all plankton [29, 30]. They are typical oligotrophic microorganisms and are the most abundant bacteria in ocean surface waters, but they reach their maximum number in stratified oligotrophic gyres, which are an expanding habitat in the warming oceans [29]. SAR11 bacteria reportedly play an important role in ocean carbon and nutrient cycling [75, 76]. They are dominant in the ocean and usually contain PR, and a recent survey indicates the significant contribution of microbial rhodopsins to solar energy harvesting in the oceans [6, 29].
Although SAR11 is widely distributed and abundant in the world, our understanding of its ecological effects is limited due to its difficulty in cultivation and the lack of appropriate research tools. We sampled seawater from a euphotic and oligotrophic zone, which has a relatively warm temperature of 16.9°C, plenty of light, and very low chemical oxygen demand (1.7 mg/l) (Table S1), conditions favoring bacteria that are able to harvest light and fix CO2. It is likely that some SAR11 bacteria use organic compounds as electron donors and ATP from PR-light mediated proton pump to fix CO2 (Figure 6). In this model, organic compounds only serve as electron donors, and the energy is provided by PR-based light harvesting, so low energy organics would be sufficient to support cell growth. Indeed, we have shown formate was able to support the growth of PR expressing bacteria (containing genes for the Calvin-Benson cycle) in a separate research [77]. We have used Raman-based imaging, labeling, and sequencing methods to establish a link between PR content and CO2 fixation at the level of single cells, for the SAR11 bacterium Pelagibacter spp. In summary, this study demonstrates that RAGE-mediated analysis of a single-cell genome can establish a reliable link between the phenotype and genotype of uncultured bacteria in the ocean, which solves a basic problem and paves the way for function-based dissection of the “biological dark matter” in the environment.
Acknowledgments
This study was funded by the National Key R&D Program Young Scientists Project of China (No. 2021YFD1900400 to XYJ) and by the National Science Foundation of China (No. 32270109 to XYJ). BM acknowledges the support of the Key Instrument Development Program from Chinese Academy of Sciences (YJKYYQ20170017). WEH acknowledges finance and instrumentation support from the Engineering and Physical Sciences Research Council (EP/M002403/1 and EP/M02833X/1) and Natural Environment Research Council (NE/M002934/1). WEH, PAD, CMC, and CNH acknowledge financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), award number BB/M000265/1. CNH and PAD are supported by the European Research Council Synergy Award 854126. We also thank NSFC (31770112) for the financial support.
Contributor Information
Jian Xu, Email: xujian@qibebt.ac.cn.
Wei E. Huang, Email: wei.huang@eng.ox.ac.uk.
Data Availability
The sequence data reported in this paper have been deposited to the NCBI SRA database with bioproject accession PRJNA609879 and PRJNA611094.
Authors’ Contributions
WEH and XYJ designed the research. XYJ, YM, PAD, and CMC performed the experiments. WEH, TX, BM, and JX introduced and validated the RAGE method and chip. LX provided seawater samples. YHG and XYJ performed the sequencing and data analysis for single-cell genome and 16S rRNA amplicons. WEH, XYJ, YHG, and JX analyzed data and interpreted results. CNH, YTJ, and BM provided critical suggestions. WEH, JX, XYJ, CNH, YHG, and TX wrote the manuscript. Xiaoyan Jing, Yanhai Gong, and Teng Xu contributed equally to this work.
Supplementary Materials
Supplementary file 1: identified 16S rRNA genes from single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 2: identified 16S rRNA genes obtained from MDA products of SAGs from the RAGE-Seq samples. Supplementary file 3: list of carbon metabolism-related genes and their annotations in the single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 4: identified beta-carotene 15,15-dioxygenase genes from the single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 5: identified proteorhodopsin (PR) genes from the single-cell Pelagibacter spp. genomes from RG1 and RG6.
References
- 1.Field C. B., Behrenfeld M. J., Randerson J. T., and Falkowski P., “Primary production of the biosphere: integrating terrestrial and oceanic components,” Science, vol. 281, no. 5374, pp. 237–240, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Amann R. I., Ludwig W., and Schleifer K.-H., “Phylogenetic identification and in situ detection of individual microbial cells without cultivation,” Microbiology and Molecular Biology Reviews, vol. 59, pp. 143–169, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitman W. B., Coleman D. C., and Wiebe W. J., “Prokaryotes: the unseen majority,” Proceedings of the National Academy of Sciences, vol. 95, no. 12, pp. 6578–6583, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant D. A., and Frigaard N.-U., “Prokaryotic photosynthesis and phototrophy illuminated,” Trends in Microbiology, vol. 14, no. 11, pp. 488–496, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Finkel O. M., Beja O., and Belkin S., “Global abundance of microbial rhodopsins,” The ISME Journal, vol. 7, no. 2, pp. 448–451, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Consarnau L., Raven J. A., Levine N. M., Cutter L. S., Wang D., Seegers B., Arístegui J., Fuhrman J. A., Gasol J. M., and Sañudo-Wilhelmy S. A., “Microbial rhodopsins are major contributors to the solar energy captured in the sea,” Science Advances, vol. 5, no. 8, article eaaw8855, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchman D. L., and Hanson T. E., “Bioenergetics of photoheterotrophic bacteria in the oceans,” Environmental Microbiology Reports, vol. 5, no. 2, pp. 188–199, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Campbell B. J., Waidner L. A., Cottrell M. T., and Kirchman D. L., “Abundant proteorhodopsin genes in the North Atlantic Ocean,” Environmental Microbiology, vol. 10, no. 1, pp. 99–109, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman J. A., Schwalbach M. S., and Stingl U., “Proteorhodopsins: an array of physiological roles?,” Nature Reviews Microbiology, vol. 6, no. 6, pp. 488–494, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Jing X., Gou H., Gong Y., Su X., Xu L., Ji Y., Song Y., Thompson I. P., Xu J., and Huang W. E., “Raman-activated cell sorting and metagenomic sequencing revealing carbon- fixing bacteria in the ocean,” Environmental Microbiology, vol. 20, no. 6, pp. 2241–2255, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rusch D. B., Halpern A. L., Sutton G., Heidelberg K. B., Williamson S., Yooseph S., Wu D., Eisen J. A., Hoffman J. M., Remington K., Beeson K., Tran B., Smith H., Baden-Tillson H., Stewart C., Thorpe J., Freeman J., Andrews-Pfannkoch C., Venter J. E., Li K., Kravitz S., Heidelberg J. F., Utterback T., Rogers Y. H., Falcón L. I., Souza V., Bonilla-Rosso G., Eguiarte L. E., Karl D. M., Sathyendranath S., Platt T., Bermingham E., Gallardo V., Tamayo-Castillo G., Ferrari M. R., Strausberg R. L., Nealson K., Friedman R., Frazier M., and Venter J. C., “The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific,” PLoS Biology, vol. 5, no. 3, pp. 398–431, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabehi G., Loy A., Jung K. H., Partha R., Spudich J. L., Isaacson T., Hirschberg J., Wagner M., and Béjà O., “New insights into metabolic properties of marine bacteria encoding proteorhodopsins,” PLoS Biology, vol. 3, no. 8, pp. 1409–1417, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palovaara J., Akram N., Baltar F., Bunse C., Forsberg J., Pedros-Alio C., González J. M., and Pinhassi J., “Stimulation of growth by proteorhodopsin phototrophy involves regulation of central metabolic pathways in marine planktonic bacteria,” Proceedings of the National Academy of Sciences, vol. 111, no. 35, pp. E3650–E3658, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez-Consarnau L., Akram N., Lindell K., Pedersen A., Neutze R., Milton D. L., González J. M., and Pinhassi J., “Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation,” PLoS Biology, vol. 8, no. 4, article e1000358, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y., Cartron M. L., Jackson P. J., Davison P. A., Dickman M. J., Zhu D., Huang W. E., and Hunter C. N., “Proteorhodopsin overproduction enhances the long-term viability of Escherichia coli,” Applied and Environmental Microbiology, vol. 86, article e02087, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beja O., Spudich E. N., Spudich J. L., Leclerc M., and DeLong E. F., “Proteorhodopsin phototrophy in the ocean,” Nature, vol. 411, no. 6839, pp. 786–789, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Béja O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., Jovanovich S. B., Gates C. M., Feldman R. A., Spudich J. L., and Spudich E. N., “Bacterial rhodopsin: evidence for a new type of phototrophy in the sea,” Science, vol. 289, no. 5486, pp. 1902–1906, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa M., Hosaka T., Kojima K., Nishimura Y., Nakajima Y., Kimura-Someya T., Shirouzu M., Sudo Y., and Yoshizawa S., “A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage,” Scientific Reports, vol. 10, no. 1, p. 16752, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y., Chen X., Madsen A. M., Zervas A., Nielsen T. K., Andrei A.-S., Lund-Hansen L. C., Liu Y., and Hansen L. H., “Potential rhodopsin- and bacteriochlorophyll-based dual phototrophy in a high Arctic glacier,” MBio, vol. 11, no. 6, article e02641, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso-Sáez L., Galand P. E., Casamayor E. O., Pedrós-Alió C., and Bertilsson S., “High bicarbonate assimilation in the dark by Arctic bacteria,” The ISME Journal, vol. 4, no. 12, pp. 1581–1590, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Baltar F., and Herndl G. J., “Ideas and perspectives: is dark carbon fixation relevant for oceanic primary production estimates?,” Biogeosciences, vol. 16, no. 19, pp. 3793–3799, 2019 [Google Scholar]
- 22.Beier S., Gálvez M. J., Molina V., Sarthou G., Quéroué F., Blain S., and Obernosterer I., “The transcriptional regulation of the glyoxylate cycle in SAR11 in response to iron fertilization in the Southern Ocean,” Environmental Microbiology Reports, vol. 7, no. 3, pp. 427–434, 2015 [DOI] [PubMed] [Google Scholar]
- 23.Kirchman D. L., Elifantz H., Dittel A. I., Malmstrom R. R., and Cottrell M. T., “Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean,” Limnology and Oceanography, vol. 52, no. 2, pp. 495–507, 2007 [Google Scholar]
- 24.Koedooder C., Guéneuguès A., Van Geersdaële R., Vergé V., Bouget F.-Y., Labreuche Y., Obernosterer I., and Blain S., “The role of the glyoxylate shunt in the acclimation to iron limitation in marine heterotrophic bacteria,” Frontiers in Marine Science, vol. 5, p. 435, 2018 [Google Scholar]
- 25.Moran M. A., and Miller W. L., “Resourceful heterotrophs make the most of light in the coastal ocean,” Nature Reviews Microbiology, vol. 5, no. 10, pp. 792–800, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Pinhassi J., DeLong E. F., Béjà O., González J. M., and Pedrós-Alió C., “Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology, and ecology,” Microbiology and Molecular Biology Reviews, vol. 80, no. 4, pp. 929–954, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quero G. M., Celussi M., Relitti F., Kovačević V., Del Negro P., and Luna G. M., “Inorganic and organic carbon uptake processes and their connection to microbial diversity in meso- and bathypelagic Arctic waters (Eastern Fram Strait),” Microbial Ecology, vol. 79, no. 4, pp. 823–839, 2020 [DOI] [PubMed] [Google Scholar]
- 28.Smith D. P., Kitner J. B., Norbeck A. D., Clauss T. R., Lipton M. S., Schwalbach M. S., Steindler L., Nicora C. D., Smith R. D., and Giovannoni S. J., “Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus pelagibacter ubique,” PLoS One, vol. 5, no. 5, article e10487, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giovannoni S. J., “SAR11 bacteria: the most abundant plankton in the oceans,” Annual Review of Marine Science, vol. 9, no. 1, pp. 231–255, 2017 [DOI] [PubMed] [Google Scholar]
- 30.Giovannoni S. J., Bibbs L., Cho J. C., Stapels M. D., Desiderio R., Vergin K. L., Rappé M. S., Laney S., Wilhelm L. J., Tripp H. J., Mathur E. J., and Barofsky D. F., “Proteorhodopsin in the ubiquitous marine bacterium SAR11,” Nature, vol. 438, no. 7064, pp. 82–85, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Berry D., Mader E., Lee T. K., Woebken D., Wang Y., Zhu D., Palatinszky M., Schintlmeister A., Schmid M. C., Hanson B. T., Shterzer N., Mizrahi I., Rauch I., Decker T., Bocklitz T., Popp J., Gibson C. M., Fowler P. W., Huang W. E., and Wagner M., “Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells,” Proceedings of the National Academy of Sciences, vol. 112, no. 2, pp. 194–203, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Wang X., Ma B., and Xu J., “Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution,” Biotechnology Advances, vol. 37, no. 6, article 107388, 2019 [DOI] [PubMed] [Google Scholar]
- 33.Huang W. E., Ferguson A., Singer A., Lawson K., Thompson I. P., Kalin R. M., Larkin M. J., Bailey M. J., and Whiteley A. S., “Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization,” Applied and Environmental Microbiology, vol. 75, no. 1, pp. 234–241, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing X., Gong Y., Xu T., Meng Y., Han X., Su X., Wang J., Ji Y., Li Y., Jia Z., Ma B., and Xu J., “One-cell metabolic phenotyping and sequencing of soil microbiome by Raman-activated gravity-driven encapsulation (RAGE),” Msystems, vol. 6, no. 3, article e0018121, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y. Z., Cui L., Lopez J. A. S., Xu J. B., Zhu Y. G., Thompson I. P., and Huang W. E., “Raman-deuterium isotope probing for in-situ identification of antimicrobial resistant bacteria in Thames River,” Scientific Reports, vol. 7, no. 1, p. 16648, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y. Z., Kaster A. K., Vollmers J., Song Y. Q., Davison P. A., Frentrup M., Preston G. M., Thompson I. P., Murrell J. C., Yin H., Hunter C. N., and Huang W. E., “Single-cell genomics based on Raman sorting reveals novel carotenoid- containing bacteria in the Red Sea,” Microbial Biotechnology, vol. 10, no. 1, pp. 125–137, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Xu J. B., Kong L. C., Liu T., Yi L. B., Wang H. J., Huang W. E., and Zheng C., “Raman–deuterium isotope probing to study metabolic activities of single bacterial cells in human intestinal microbiota,” Microbial Biotechnology, vol. 13, no. 2, pp. 572–583, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W. E., Ward A. D., and Whiteley A. S., “Raman tweezers sorting of single microbial cells,” Environmental Microbiology Reports, vol. 1, no. 1, pp. 44–49, 2009 [DOI] [PubMed] [Google Scholar]
- 39.Huang W. E., Griffiths R. I., Thompson I. P., Bailey M. J., and Whiteley A. S., “Raman microscopic analysis of single microbial cells,” Analytical Chemistry, vol. 76, no. 15, pp. 4452–4458, 2004 [DOI] [PubMed] [Google Scholar]
- 40.Cui L., Yang K., Li H. Z., Zhang H., Su J. Q., Paraskevaidi M., Martin F. L., Ren B., and Zhu Y. G., “Functional single-cell approach to probing nitrogen-fixing bacteria in soil communities by resonance Raman spectroscopy with15N2labeling,” Analytical Chemistry, vol. 90, no. 8, pp. 5082–5089, 2018 [DOI] [PubMed] [Google Scholar]
- 41.Li M., Ashok P. C., Dholakia K., and Huang W. E., “Raman-activated cell counting for profiling carbon dioxide fixing microorganisms,” The Journal of Physical Chemistry A, vol. 116, no. 25, pp. 6560–6563, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Tao Y. F., Wang Y., Huang S., Zhu P. F., Huang W. E., Ling J. Q., and Xu J., “Metabolic-activity-based assessment of antimicrobial effects by D2O-labeled single-cell Raman microspectroscopy,” Analytical Chemistry, vol. 89, no. 7, pp. 4108–4115, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Huang W. E., Cui L., and Wagner M., “Single cell stable isotope probing in microbiology using Raman microspectroscopy,” Current Opinion in Biotechnology, vol. 41, pp. 34–42, 2016 [DOI] [PubMed] [Google Scholar]
- 44.Song Y. Z., Yin H. B., and Huang W. E., “Raman activated cell sorting,” Current Opinion in Chemical Biology, vol. 33, pp. 1–8, 2016 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q., Zhang P., Gou H., Mou C., Huang W. E., Yang M., Xu J., and Ma B., “Towards high-throughput microfluidic Raman-activated cell sorting,” Analyst, vol. 140, no. 18, pp. 6163–6174, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Lee K. S., Palatinszky M., Pereira F. C., Nguyen J., Fernandez V. I., Mueller A. J., Menolascina F., Daims H., Berry D., Wagner M., and Stocker R., “An automated Raman-based platform for the sorting of live cells by functional properties,” Nature Microbiology, vol. 4, no. 6, pp. 1035–1048, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Crusoe M. R., Alameldin H. F., Awad S., Boucher E., Caldwell A., Cartwright R., Charbonneau A., Constantinides B., Edvenson G., Fay S., Fenton J., Fenzl T., Fish J., Garcia-Gutierrez L., Garland P., Gluck J., González I., Guermond S., Guo J., Gupta A., Herr J. R., Howe A., Hyer A., Härpfer A., Irber L., Kidd R., Lin D., Lippi J., Mansour T., McA'Nulty P., McDonald E., Mizzi J., Murray K. D., Nahum J. R., Nanlohy K., Nederbragt A. J., Ortiz-Zuazaga H., Ory J., Pell J., Pepe-Ranney C., Russ Z. N., Schwarz E., Scott C., Seaman J., Sievert S., Simpson J., Skennerton C. T., Spencer J., Srinivasan R., Standage D., Stapleton J. A., Steinman S. R., Stein J., Taylor B., Trimble W., Wiencko H. L., Wright M., Wyss B., Zhang Q., zyme, and Brown C. T., “The Khmer Software Package: Enabling Efficient Nucleotide Sequence Analysis,” F1000Research, vol. 4, p. 900, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McIlvenna D., Huang W. E., Davison P., Glidle A., Cooper J., and Yin H. B., “Continuous cell sorting in a flow based on single cell resonance Raman spectra,” Lab on a Chip, vol. 16, no. 8, pp. 1420–1429, 2016 [DOI] [PubMed] [Google Scholar]
- 49.Zhang P., Ren L., Zhang X., Shan Y., Wang Y., Ji Y., Yin H., Huang W. E., Xu J., and Ma B., “Raman-activated cell sorting based on dielectrophoretic single-cell trap and release,” Analytical Chemistry, vol. 87, no. 4, pp. 2282–2289, 2015 [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Ren L., Su Y., Ji Y., Liu Y., Li C., Li X., Zhang Y., Wang W., Hu Q., Han D., Xu J., and Ma B., “Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells,” Analytical Chemistry, vol. 89, no. 22, pp. 12569–12577, 2017 [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., Xu J., Kong L., Li B., Li H., Huang W. E., and Zheng C., “Raman-activated sorting of antibiotic-resistant bacteria in human gut microbiota,” Environmental Microbiology, vol. 22, no. 7, pp. 2613–2624, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Ji Y., Wharfe E. S., Meadows R. S., March P., Goodacre R., Xu J., and Huang W. E., “Raman activated cell ejection for isolation of single cells,” Analytical Chemistry, vol. 85, no. 22, pp. 10697–10701, 2013 [DOI] [PubMed] [Google Scholar]
- 53.McLean J. S., Lombardo M.-J., Badger J. H., Edlund A., Novotny M., Yee-Greenbaum J., Vyahhi N., Hall A. P., Yang Y., Dupont C. L., Ziegler M. G., Chitsaz H., Allen A. E., Yooseph S., Tesler G., Pevzner P. A., Friedman R. M., Nealson K. H., Venter J. C., and Lasken R. S., “Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum,” Proceedings of the National Academy of Sciences, vol. 110, no. 26, pp. 2390–2399, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao G., Yu J., Hou Q., Hui W., Liu W., Kwok L.-Y., Menghe B., Sun T., Zhang H., and Zhang W., “A perspective study of koumiss microbiome by metagenomics analysis based on single-cell amplification technique,” Frontiers in Microbiology, vol. 8, p. 165, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu T., Gong Y., Su X., Zhu P., Dai J., Xu J., and Ma B., “Phenome–genome profiling of single bacterial cell by Raman-activated gravity- driven encapsulation and sequencing,” Small, vol. 16, no. 30, article 2001172, 2020 [DOI] [PubMed] [Google Scholar]
- 56.Miroux B., and Walker J. E., “Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels,” Journal of Molecular Biology, vol. 260, no. 3, pp. 289–298, 1996 [DOI] [PubMed] [Google Scholar]
- 57.Li H. Z., Bi Q. F., Yang K., Zheng B. X., Pu Q., and Cui L., “D2O-isotope-labeling approach to probing phosphate-solubilizing bacteria in complex soil communities by single-cell Raman spectroscopy,” Analytical Chemistry, vol. 91, no. 3, pp. 2239–2246, 2019 [DOI] [PubMed] [Google Scholar]
- 58.Li M., Canniffe D. P., Jackson P. J., Davison P. A., FitzGerald S., Dickman M. J., Burgess J. G., Hunter C. N., and Huang W. E., “Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities,” The ISME Journal, vol. 6, no. 4, pp. 875–885, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rimai L., Heyde M., and Gill D., “Vibrational spectra of some carotenoids and related linear polyenes. Raman spectroscopic study,” Journal of the American Chemical Society, vol. 95, no. 14, pp. 4493–4501, 1973 [DOI] [PubMed] [Google Scholar]
- 60.Robert B., “Resonance Raman spectroscopy,” Photosynthesis Research, vol. 101, no. 2-3, pp. 147–155, 2009 [DOI] [PubMed] [Google Scholar]
- 61.Gilbert J. A., Muhling M., and Joint I., “A rare SAR11 fosmid clone confirming genetic variability in the ' Candidatus Pelagibacter ubique ' genome,” The ISME Journal, vol. 2, no. 7, pp. 790–793, 2008 [DOI] [PubMed] [Google Scholar]
- 62.Kanehisa M., Sato Y., and Morishima K., “BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences,” Journal of Molecular Biology, vol. 428, no. 4, pp. 726–731, 2016 [DOI] [PubMed] [Google Scholar]
- 63.Von Lintig J., and Vogt K., “Filling the gap in vitamin A research - molecular identification of an enzyme cleaving beta-carotene to retinal,” Journal of Biological Chemistry, vol. 275, pp. 11915–11920, 2000 [DOI] [PubMed] [Google Scholar]
- 64.Mulkidjanian A. Y., Koonin E. V., Makarova K. S., Mekhedov S. L., Sorokin A., Wolf Y. I., Dufresne A., Partensky F., Burd H., Kaznadzey D., Haselkorn R., and Galperin M. Y., “The cyanobacterial genome core and the origin of photosynthesis,” Proceedings of the National Academy of Sciences, vol. 103, no. 35, pp. 13126–13131, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., and Pevzner P. A., “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing,” Journal of Computational Biology, vol. 19, no. 5, pp. 455–477, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J. F., Darling A., Malfatti S., Swan B. K., Gies E. A., Dodsworth J. A., Hedlund B. P., Tsiamis G., Sievert S. M., Liu W. T., Eisen J. A., Hallam S. J., Kyrpides N. C., Stepanauskas R., Rubin E. M., Hugenholtz P., and Woyke T., “Insights into the phylogeny and coding potential of microbial dark matter,” Nature, vol. 499, no. 7459, pp. 431–437, 2013 [DOI] [PubMed] [Google Scholar]
- 67.Stepanauskas R., Fergusson E. A., Brown J., Poulton N. J., Tupper B., Labonté J. M., Becraft E. D., Brown J. M., Pachiadaki M. G., Povilaitis T., and Thompson B. P., “Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles,” Nature Communications, vol. 8, no. 1, p. 84, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su X., Gong Y., Gou H., Jing X., Xu T., Zheng X., Chen R., Li Y., Ji Y., Ma B., and Xu J., “Rational optimization of Raman-activated cell ejection and sequencing for bacteria,” Analytical Chemistry, vol. 92, no. 12, pp. 8081–8089, 2020 [DOI] [PubMed] [Google Scholar]
- 69.Blainey P. C., and Quake S. R., “Digital MDA for enumeration of total nucleic acid contamination,” Nucleic Acids Research, vol. 39, no. 4, article e19, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan X., Urban A. E., Palejev D., Schulz V., Grubert F., Hu Y., Snyder M., and Weissman S. M., “A procedure for highly specific, sensitive, and unbiased whole-genome amplification,” Proceedings of the National Academy of Sciences, vol. 105, no. 40, pp. 15499–15504, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodrigue S., Malmstrom R. R., Berlin A. M., Birren B. W., Henn M. R., and Chisholm S. W., “Whole genome amplification and de novo assembly of single bacterial cells,” PLoS One, vol. 4, no. 9, article e6864, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woyke T., Sczyrba A., Lee J., Rinke C., Tighe D., Clingenpeel S., Malmstrom R., Stepanauskas R., and Cheng J. F., “Decontamination of MDA reagents for single cell whole genome amplification,” PLoS One, vol. 6, no. 10, article e26161, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woyke T., Tighe D., Mavromatis K., Clum A., Copeland A., Schackwitz W., Lapidus A., Wu D., McCutcheon J. P., McDonald B. R., Moran N. A., Bristow J., and Cheng J. F., “One bacterial cell, one complete genome,” PLoS One, vol. 5, no. 4, article e10314, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., and Arnheim N., “Whole genome amplification from a single cell: implications for genetic analysis,” Proceedings of the National Academy of Sciences, vol. 89, no. 13, pp. 5847–5851, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ottesen E. A., Young C. R., Gifford S. M., Eppley J. M., Marin R., Schuster S. C., Scholin C. A., and DeLong E. F., “Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages,” Science, vol. 345, no. 6193, pp. 207–212, 2014 [DOI] [PubMed] [Google Scholar]
- 76.Sowell S. M., Wilhelm L. J., Norbeck A. D., Lipton M. S., Nicora C. D., Barofsky D. F., Carlson C. A., Smith R. D., and Giovanonni S. J., “Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea,” The ISME Journal, vol. 3, pp. 93–105, 2009 [DOI] [PubMed] [Google Scholar]
- 77.Davison P. A., Tu W., Xu J., Valle S. D., Thompson I. P., Hunter C. N., and Huang W. E., “Engineering a Rhodopsin-based Photo-electrosynthetic System in Bacteria for CO2 Fixation,” ACS Synthetic Biology, 2022, In press. [DOI] [PMC free article] [PubMed]
- 78.Yoshizawa S., Kawanabe A., Ito H., Kandori H., and Kogure K., “Diversity and functional analysis of proteorhodopsin in marine Flavobacteria,” Environmental Microbiology, vol. 14, no. 5, pp. 1240–1248, 2012 [DOI] [PubMed] [Google Scholar]
- 79.Amicucci A., Barbieri E., Sparvoli V., Gioacchini A. M., Calcabrini C., Palma F., Stocchi V., and Zambonelli A., “Microbial and pigment profile of the reddish patch occurring within Tuber magnatum ascomata,” Fungal Biology, vol. 122, no. 12, pp. 1134–1141, 2018 [DOI] [PubMed] [Google Scholar]
- 80.Xia X., Wang Y., Yang Y., Luo T., Van Nostrand J. D., Zhou J., Jiao N., and Zhang R., “Ocean acidification regulates the activity, community structure, and functional potential of heterotrophic bacterioplankton in an oligotrophic gyre,” Journal of Geophysical Research: Biogeosciences, vol. 124, no. 4, pp. 1001–1017, 2019 [Google Scholar]
- 81.Han S.-R., Kim K.-H., Ahn D.-H., Park H., and Oh T.-J., “Complete genome sequence of carotenoid-producing Microbacterium sp. strain PAMC28756 isolated from an Antarctic lichen,” Journal of Biotechnology, vol. 226, pp. 18–19, 2016 [DOI] [PubMed] [Google Scholar]
- 82.Lin G., Sun F., Wang C., Zhang L., and Zhang X., “Assessment of the effect of Enteromorpha prolifera on bacterial community structures in aquaculture environment,” PLoS One, vol. 12, no. 7, article e0179792, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emms D. M., and Kelly S., “OrthoFinder: phylogenetic orthology inference for comparative genomics,” Genome Biology, vol. 20, no. 1, p. 238, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1: identified 16S rRNA genes from single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 2: identified 16S rRNA genes obtained from MDA products of SAGs from the RAGE-Seq samples. Supplementary file 3: list of carbon metabolism-related genes and their annotations in the single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 4: identified beta-carotene 15,15-dioxygenase genes from the single-cell Pelagibacter spp. genomes from RG1 and RG6. Supplementary file 5: identified proteorhodopsin (PR) genes from the single-cell Pelagibacter spp. genomes from RG1 and RG6.
Data Availability Statement
The sequence data reported in this paper have been deposited to the NCBI SRA database with bioproject accession PRJNA609879 and PRJNA611094.