Abstract
Human life intimately depends on plants for food, biomaterials, health, energy, and a sustainable environment. Various plants have been genetically improved mostly through breeding, along with limited modification via genetic engineering, yet they are still not able to meet the ever-increasing needs, in terms of both quantity and quality, resulting from the rapid increase in world population and expected standards of living. A step change that may address these challenges would be to expand the potential of plants using biosystems design approaches. This represents a shift in plant science research from relatively simple trial-and-error approaches to innovative strategies based on predictive models of biological systems. Plant biosystems design seeks to accelerate plant genetic improvement using genome editing and genetic circuit engineering or create novel plant systems through de novo synthesis of plant genomes. From this perspective, we present a comprehensive roadmap of plant biosystems design covering theories, principles, and technical methods, along with potential applications in basic and applied plant biology research. We highlight current challenges, future opportunities, and research priorities, along with a framework for international collaboration, towards rapid advancement of this emerging interdisciplinary area of research. Finally, we discuss the importance of social responsibility in utilizing plant biosystems design and suggest strategies for improving public perception, trust, and acceptance.
1. Introduction
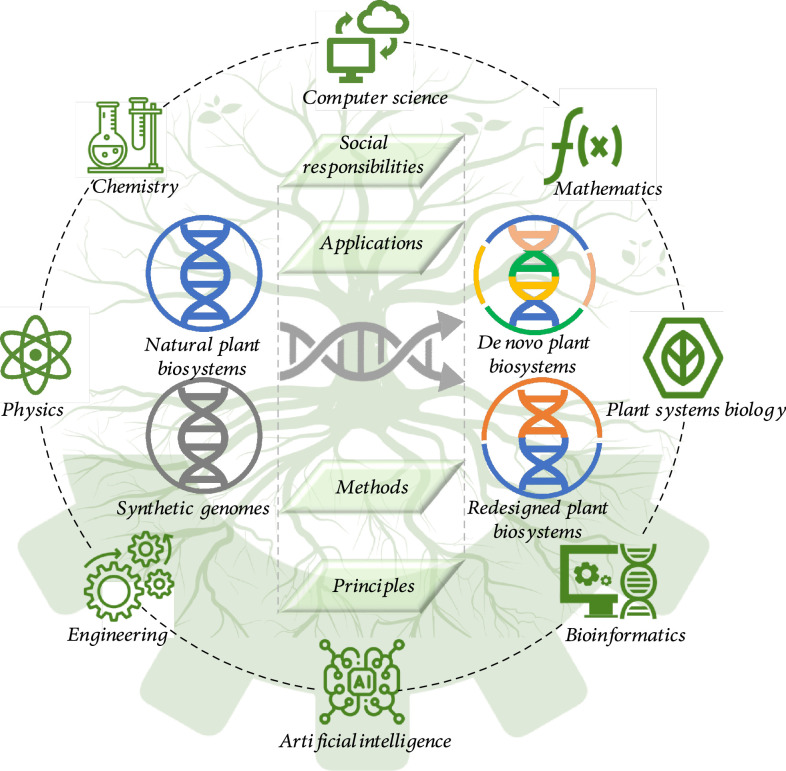
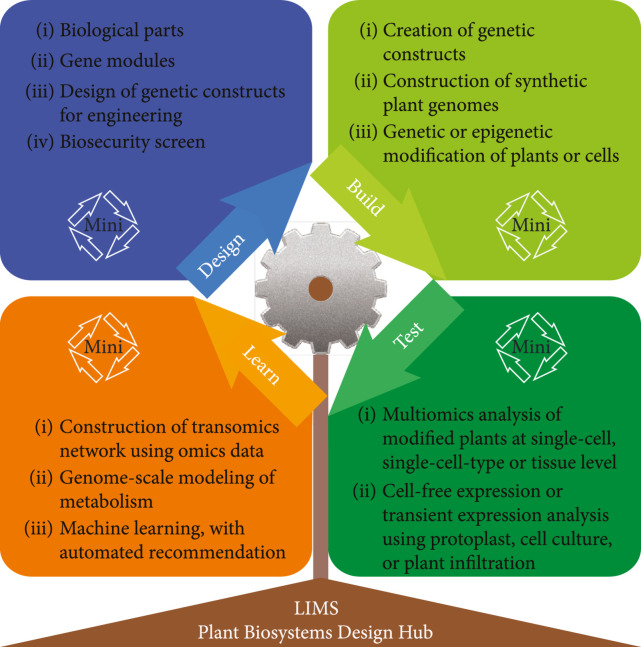
Humans depend on plants for a variety of important resources including sustenance, energy, clothing, bio-based products, and shelter [1–3]. On a global scale, plants play critical roles in biogeochemical cycling and environmental stability [4, 5]. There are currently ~374,000 known plant species on Earth, of which approximately 82% are vascular plants [6], and only a small fraction of these have been domesticated. There still exists vast potential for incorporating useful traits from wild relatives and from natural plant populations to design plants for human and environmental use. It has become increasingly clear that the current trajectories of yield increase for staple crop varieties/cultivars will not be adequate to meet the future demands of the increasing global population [7, 8]. Furthermore, many crop plants may not be sufficiently robust to cope with impending stresses of rapid climate change such as extreme weather, reduced water resources (e.g., reduction in both quantity due to drought and quality due to pollution), and deteriorated soil quality [9, 10]. Therefore, there is an urgent need for new strategies to accelerate crop development and domestication and expand the possibilities for a thriving plant-based bioeconomy (e.g., production of novel bio-based products) to address our projected economic, social, and environmental needs. To this end, a new frontier in plant research called “plant biosystems design” is emerging and quickly evolving. Plant biosystems design is an interdisciplinary field of research that seeks to genetically/epigenetically improve plants or create novel plant traits or organisms through editing, engineering and refactoring of native, heterologous, or synthetic biological parts based on predictive design (Figure 1). To promote this emerging field, we present a roadmap for plant biosystems design that aims to identify knowledge gaps, technical challenges, and opportunities. We review theoretical and technical approaches and propose innovative applications of biosystems design for basic and applied plant science research, along with strategies to enhance social responsibility of scientists and companies in terms of biosafety and ethics (e.g., public beneficence, intellectual freedom and responsibility, and fairness).
Figure 1.
The scope of plant biosystems design research. Plant biosystems design is an interdisciplinary research field integrating plant systems biology, engineering, chemistry, computer science, bioinformatics, artificial intelligence, physics, and mathematics to redesign natural plant systems and construct new plant systems in a predictable and programmable manner. Plant biosystems design research covers four aspects: theories and principles, methods and toolboxes, applications, and social responsibilities. Some icon made by Pixel perfect from http://www.flaticon.com/ and http://www.pngtree.com/.
2. Theoretical Approaches and Principles of Plant Biosystems Design
Plants are complex, multicellular organisms. The predictive design of plant biosystems requires a comprehensive understanding of the principles underlying biological processes across all scales, from molecular interactions to cellular metabolism, cell/tissue/organ growth and development, and environmental responses of plants. Plant biosystems design involves several theoretical approaches: (1) graph theory providing a graphic view of the structure of plant systems, (2) mechanistic models linking genes to phenotypic traits, and (3) evolutionary dynamics theory enabling prediction of the genetic stability and evolvability of genetically modified plants or de novo plant systems. These theoretical approaches enable the design of complex plant systems based on the principles of modular design, dynamic programming, natural and artificial selection (i.e., selective breeding), genetic stability, and upgradability.
2.1. Theoretical Approaches
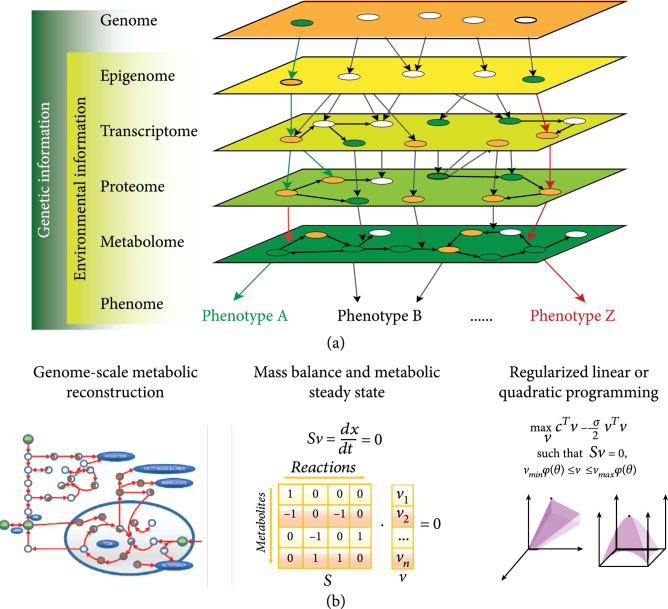
2.1.1. The Graph Theory Approach for Plant Biosystems Design
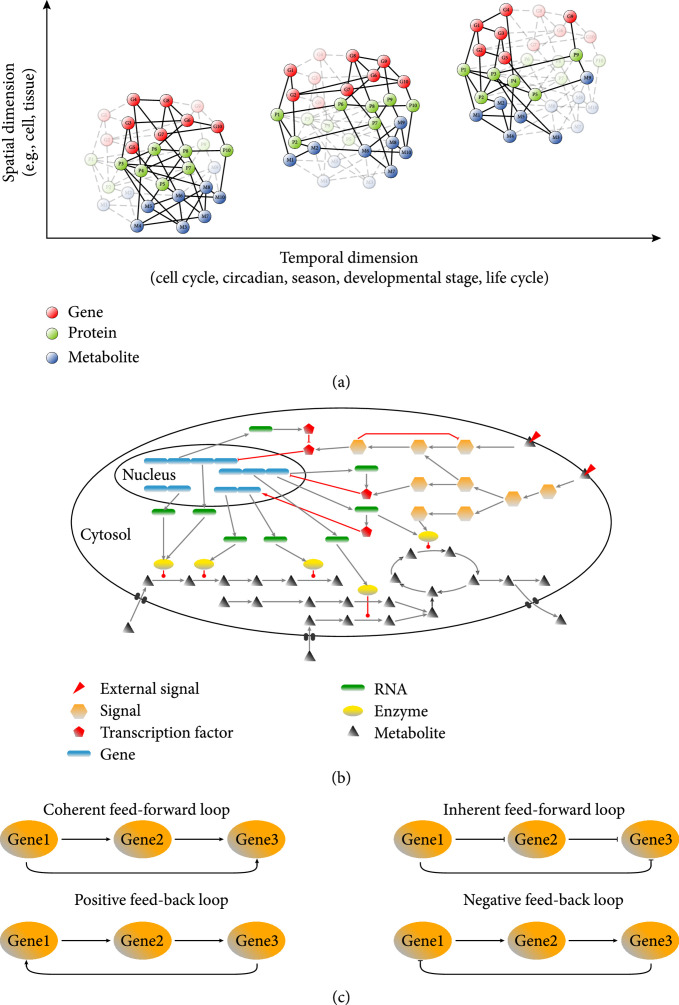
A graph can be used to describe complex biological systems where the components and interactions of the system are represented by thousands of nodes (e.g., genes and metabolites) connected with thousands of edges (e.g., interactions) [11]. Inherent to the graph theoretic approach for describing biological systems is the use of network graphs to represent, for example, the extensive communication between metabolic and gene regulatory networks [12]. Metabolites can regulate protein activity via allosteric regulation and posttranslational modifications [13], and gene expression in plants is subject to epigenetic regulation mediated by metabolic fluxes and cellular redox states [14, 15]. From the perspective of biosystems design, a plant biosystem can be defined as a dynamic network of genes and multiple intermediate molecular phenotypes, such as proteins and metabolites, distributed in a four-dimensional space: three spatial dimensions of structure (e.g., cell and tissue) and one temporal dimension (e.g., cell cycle, circadian time, season, developmental stage, and life cycle) (Figure 2(a)). Along the spatial dimensions, plant tissue/organ growth and development are precisely orchestrated in a distributed fashion through collective interactions of many connected cells [16, 17], and therefore, the subnetworks spatially distributed in individual cells are interconnected as the nodes of tissue/organ-scale networks. Furthermore, tissue/organ-scale subnetworks are interconnected as nodes of whole-plant-scale networks. Along the temporal dimension, genes are turned on and off at various time scales, and their expression profiles vary with changes in cell cycle, circadian clock, growing season, development stage, and life cycle. Also, the products of gene expression (RNAs and proteins) are degraded at various time scales, resulting in variation in their turnover times.
Figure 2.
The network control theory of plant biosystems design. (a) Dynamic networks of genes and metabolites distributed in spatial (e.g., cell and tissue) and temporal (e.g., cell cycle, circadian time, season, developmental stage, and life cycle) dimensions. (b) A plant gene-metabolite network; arrow-shaped edges represent activation; blunt edges represent inhibition, and edges ending with a solid circle indicate enzymatic catalysis; adapted from Gonçalves et al. [370]. (c) The structure of regulatory/signaling network motifs; arrows indicate positive regulation; T-shape arrows indicate negative regulation; adapted from Gupta and Singh [20].
A plant gene-metabolite network contains nodes and edges, where the nodes are genes/RNAs/proteins/metabolites, and the edges represent either promotional or inhibitory relationships in protein-protein, protein-RNA, protein-DNA, protein-metabolite, and RNA-RNA interactions (Figure 2(b)). Moreover, the overall gene-metabolite network of a plant biosystem is complex and can be divided into subnetworks responsible for plant growth, development, and interaction with abiotic and biotic environmental factors. Within the subnetworks, there are network motifs that are statistically overrepresented subgraphs as the simple building blocks of complex systems [18, 19]. The structure of regulatory network motifs can be classified as feed-forward loops or feed-back loops (Figure 2(c)) [20, 21]. So far, plant biosystems design has been limited to subnetworks and basic network motifs (e.g., basic loops). There are significant challenges to be addressed: (1) construction of a genome-scale, metabolic/regulatory network with labelled subnetworks responsible for individual biological processes in relation to plant growth, development, and response to the environment; (2) mathematical modeling of this genome-scale network for accurate prediction of plant phenotypes in response to genetic and environmental perturbations; (3) sharing of this consensus predictive model with the scientific community for plant biosystems design; (4) a lack of knowledge of how the metabolic and genetic networks are linked with each other, such as the binding of metabolites to enzymes and transcription factors, although very good progress has been made in the identification of protein-protein or protein-DNA interactions [22]; and (5) insufficient data about the concentration of metabolites in different compartments and different cell types, as well as the transport between different compartments and cell types, which provides a challenge for modeling. New computational tools like MAGI [23] will be needed to facilitate the integration of metabolic and genetic networks.
2.1.2. The Mechanistic Modeling Theory of Plant Biosystems Design
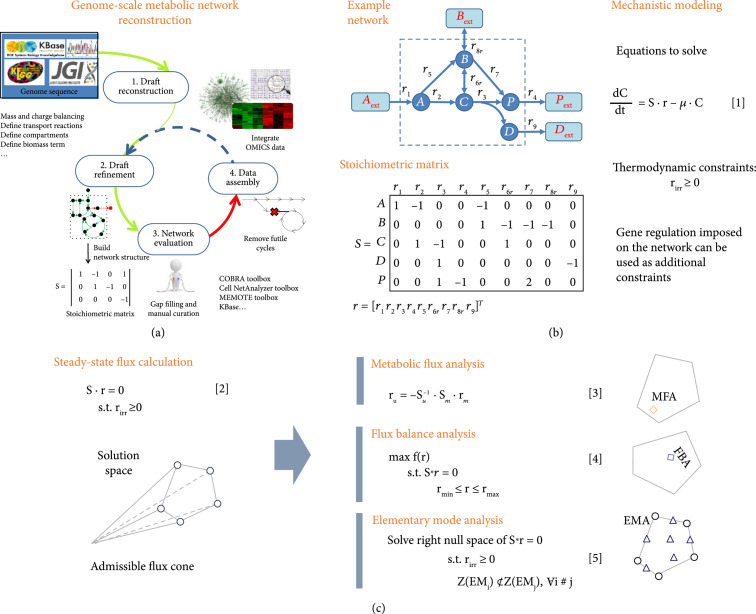
Mechanistic modeling of cellular metabolism, based on the law of mass conservation, is used to interrogate and characterize complex plant biosystems with capabilities of linking genes, enzymes, pathways, cells, tissues, and whole-plant organisms. Starting from the plant genome sequence and omics datasets, a metabolic network can be constructed based on metabolites and reactions representing nodes and edges, respectively [24] (Figure 3(a)). By defining the plant cell as a control system, the mass conservation for each metabolite can be written to decipher the fluxes of chemical elements (e.g., carbon, electron, nitrogen, and phosphate) within the plant system. These fluxes can be used as a basis to quantitatively describe cellular phenotypic characteristics [25–27]. Mathematically, mass conservation can be expressed as a system of ordinary differential equations (ODEs) to delineate the rate of change for each metabolite in the network (Figure 3(b)). The metabolic fluxes are the reaction rates determined by the metabolite concentrations, enzyme activities, enzyme concentrations, and operating conditions (e.g., temperature, pH, and ionic strength) where enzymes are encoded by genes. The first effort to create a genome-scale model (GEM) in plants was achieved for Arabidopsis about a decade ago [28, 29], and this has been improved and expanded since then. Today, there are 35 published GEMs for more than 10 seed plant species [30]. GEMs can be applied to plant biosystems design in the context of metabolic engineering, plant-microbe interactions, evolutionary processes, analysis of biological properties, prediction of cellular phenotypes, and model-driven discovery [31–33]. A plethora of tools can be used to analyze the plant metabolic network and enable the design of new metabolic networks. If the knowledge of reaction kinetics of the network is known, the cell phenotypes can be defined by solving the ODEs directly. Often, this approach is best suited for analyzing small reduced networks due to lack of kinetic information. Alternatively, one can interrogate the steady state solutions of the network (Figure 3(c)). Since the metabolic network is highly underdetermined, the exact phenotypes of the cell can be evaluated by performing extensive flux measurement to make the system being determined via stable isotope-labeling (e.g., 13C-labeled CO2) experiments. Alternatively, constraint-based metabolic analyses can be employed using either flux balance analysis (FBA) or elementary mode analysis (EMA) [30, 34]. FBA can predict a cellular phenotype based on an objective function (e.g., maximization of cell growth or a product synthesis), whereas EMA unbiasedly identifies all possible phenotypes for a given network. Through decades of development, tools for constructing and analyzing metabolic networks are quite mature and useful for plant biosystems design. However, several key challenges still remain: (1) the lack of knowledge of gene functions and their regulation required for accurate and comprehensive network curation and analysis [23]; (2) the lack of experimental data to decipher metabolites, reactions, and pathways that exist in compartments within a cell and among different cell types of a plant; and (3) the hidden underground metabolism due to enzyme promiscuity [22, 35]. Advances in single-cell/single-cell-type omics (see Section 3.6.2) are critically required to address these challenges.
Figure 3.
Mechanistic modeling for plant biosystems design. (a) General steps in reconstructing a genome-scale metabolic network. (b) Mass balance of metabolites in a metabolic network. Equation 1 is a system of ordinary differential equations describing dynamic chemical transformation of metabolites in a metabolic network where is a metabolite concentration vector, is a stoichiometric matrix, is a reaction flux vector, and is the cell growth. (c) Calculation of steady-state flux distributions. Three common methods can be employed to determine metabolic flux distributions including metabolic flux analysis (MFA), flux balance analysis (FBA), and elementary mode analysis (EMA). For a typical metabolic network, a system of homogenous equations [2] is highly underdetermined, resulting in an infinite solution space. MFA determines a physiological state of a cell under a defined condition by calculating based on experimentally measured fluxes that make [2] being determined. Here, , and . FBA also determines a physiological state of a cell by implementing a cellular objective function subject to (s.t.) mass balance and flux bounds. Different from MFA and FBA, EMA unbiasedly seeks to identify all finite admissible fluxes in the solution space by imposing the thermodynamic constraints of reaction direction and pathway nondecomposability. Adapted from [30, 34, 371].
2.1.3. The Evolutionary Dynamics Theory of Plant Biosystems Design
Extant plants are the products of evolution driven by selection and random genetic drift acting on heritable phenotypic variations caused by mutations (e.g., point mutations, insertions, deletions, and gene birth/death), recombination, gene/genome duplication followed by diversification, and transgenerational epigenetic changes [36, 37]. Within an evolutionary context, several important theoretical questions remain to be answered: To what extent are the existing plant biosystems optimized for adaptation vs. production? Which plant genes and metabolites are essential, and which are spandrels: nonadaptive byproducts of evolution [38, 39]? In other words, can we simplify and perhaps improve the gene-metabolite network by removing some optional edges or nodes? Can we rewire/modify natural networks and/or introduce new components into existing networks for genetic improvement of certain traits without negative impact on other traits? The implementation of novel, orthogonal features poses a special challenge, as the interaction with the native network(s) cannot be predicted and require a strong evolutionary adaption of a system.
Plant biosystems design generates either genetically modified or de novo plant genomes, which will likely face evolutionary pressures caused by spontaneous mutations and natural/artificial selection. In Arabidopsis thaliana, the estimated haploid single nucleotide mutation rate and insertion/deletion mutation rate are per site per generation and per site per generation, respectively [40]. Somatic mutations have been reported in various tissues of multiple plant species, with a high proportion of mutations in shoots in perennials being transmissible [41]. In a long-lived woody perennial species Populus trichocarpa, the somatic mutation rate is estimated to be approximately base substitutions per site per generation [42], slightly lower than that in A. thaliana () [43]. Also, the plant phenotype generated from a genome could potentially exert a feed-back loop by either maintaining genome stability or guiding genome variations [44]. Therefore, a key question emerges: What new mechanisms can be implemented to maintain the stability of genetically modified genomes or de novo genomes?
2.2. Principles of Plant Biosystems Design
2.2.1. Modularity of Plant Biosystems Design
Modularity is the fundamental principle of efficient and reproducible construction and maintenance of complex systems [45]. From the perspective of engineering, this has been the driving force for the modern industrialization. A module can be defined as an essential and self-contained functional unit relative to the product of which it is part, with standardized interfaces and interactions that allow composition of products by combination [46]. A modular system can be classified into sessional and chassis-based architectures. The sessional architecture has all components assimilated to modules and shares a common interface, e.g., a piping system, in which pipes are connected with a common interface for fluid transport. The chassis-based architecture can be further subclassified into the bus and slot architectures. The bus architecture (e.g., a USB port) uses the same interface whereas the slot architecture uses different interfaces, e.g., an automobile which is comprised of a chassis with many interfaces with various modules (e.g., wheels and headlights). Plant biological systems exhibit a similar principle of modularity, which has persisted for millions of years under natural selection. The principle of modular design in biological systems has been revealed at the molecular level using network theory in combination with advances in sequencing, omics, and imaging technologies over the past few decades [46–51]. Even though the principles of modular design in both biological and engineered systems are very similar, the former is much more complex, exhibiting all modular design architectures across scales (from genes to enzymes, pathways, cells, and whole organisms), and more importantly, having a unique capability to evolve (e.g., plasticity with a rewiring in response to perturbations).
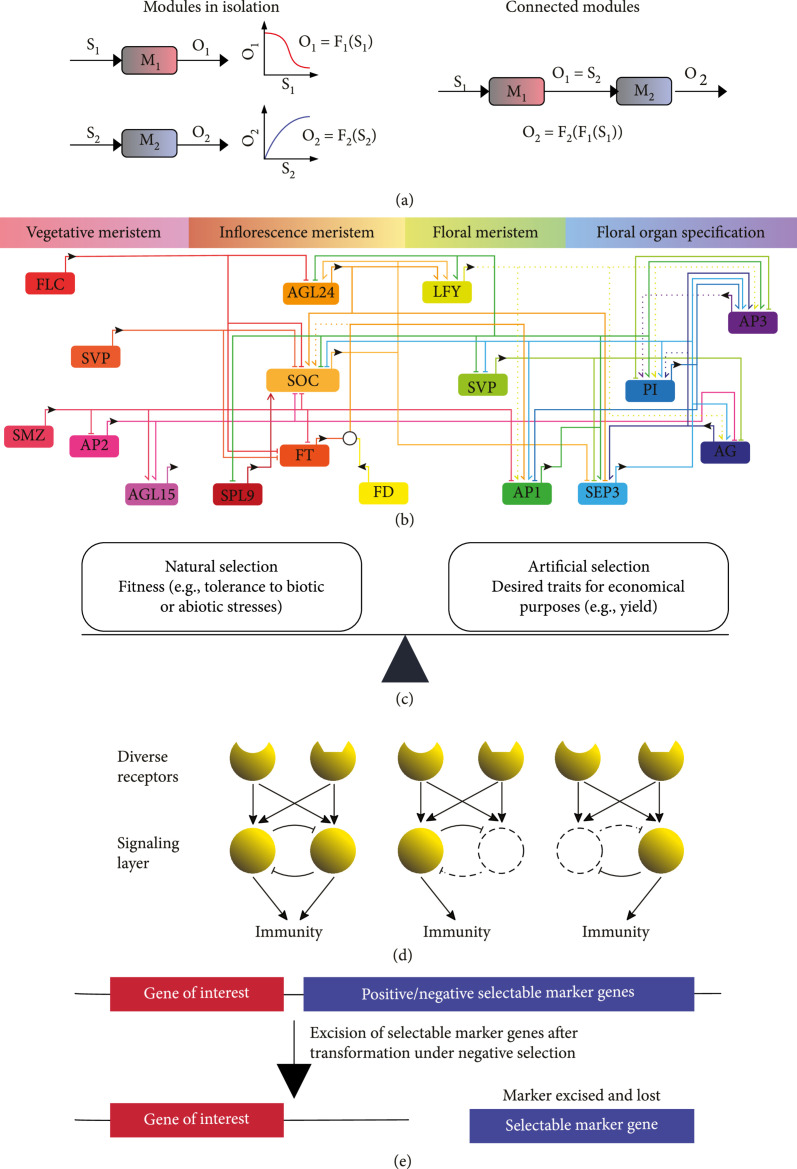
It is critical for plant biosystems design to fundamentally understand the principles of modular design so as to harness them for innovative applications such as challenges related to human health, food, energy, and the environment. For instance, the collective effect of genes within a module or subnetwork should be considered to achieve desirable phenotypic traits. Characterization of input/output properties of the subsystems (or modules) in isolation, and understanding how these are connected to each other, would allow inferring the behavior of complex systems by composing the behaviors of its subsystems (Figure 4(a)). For optimizing existing gene modules in plants, several alternative strategies can be explored, including (1) modifying the protein sequence or changing the gene expression of rate-limiting steps of the signaling or metabolic pathways, (2) manipulating the gene expression of master regulators that control the expression of multiple genes in the target module, (3) engineering enzymes to regulate metabolites that mediate epigenetic control of multiple genes in the target module, and (4) optimizing kinetics of metabolic reaction. These represent “homologous” approaches, whereas “heterologous” approaches, which use cell free systems or a simplified host for reconstruction of modules to evolve and/or identify essential components in the absence of endogenous interference [52, 53], would be useful for optimizing modular design. For installing new modules in plants, all the network components should be configured in gene circuits with the appropriate spatial and temporal expression patterns, with no or minimal negative side effect on the target plants.
Figure 4.
The principles of plant biosystems design. (a) Modularity; M1 and M2 represent two different modules; F1 and F2 represent the processes converting inputs S1 and S2 to outputs O1 and O2, respectively; M1 and M2 are connected, with ; redrawn from Grunberg and Del Vecchio [372]. (b) Dynamic programming as exemplified by the expression of regulators (suppressors or activators) programmed in the sequential developmental stages from a vegetative meristem to a floral meristem; redrawn from Kaufmann et al. [56]. (c) Tradeoff between natural selection and artificial selection. (d) Genetic stability, as exemplified by the pathogen-associated molecular pattern- (PAMP-) triggered immunity signaling network, with the inhibitory loops within the network to provide buffering interference (i.e., loss-of-function of some network components releases associated inhibitory loops allowing other components of the network to compensate for the loss); redrawn from Tyler [373]. (e) Upgradability, as exemplified by marker-free systems, in which the selectable marker gene can be excised from the plant genome after transformation, to allow for unlimited rounds of genetic transformation.
Improved plant biosystems design requires a better understanding of the following questions: What makes up a modular (chassis) cell and exchangeable production modules and their interfaces? How can a modular cell be created as a chassis to effectively couple with exchangeable production modules to achieve desirable phenotypes? How can modular design be implemented to minimize potential tradeoff among robustness, compatibility, and evolution [51, 54]? Critical to addressing these challenges is to reconstruct an accurate plant metabolic network (see Section 2.1.2). Furthermore, recent advances in metabolic network modeling and analyses in combination with Pareto optimization theory, computational algorithms, and high performance computing will help shed light on understanding and harnessing modularity of plants at various levels such as single-cell level (e.g., stem cells and stomata) and tissue level [51, 54, 55]. Application of modular design to plant biosystems needs to consider orthogonal interactions (i.e., if a nonnative/nonnatural metabolite is introduced into the system, how can we test and/or predict if other enzymes and/or transcription factors would react with this unnatural metabolite). This would in principle require an intensive testing of cross-reactivity of metabolites with enzymes and proteins to assess side activities.
2.2.2. Dynamic Programming of Plant Biosystems Design
For biosystems design, it is critical to consider the dynamic genetic programming for plant growth, development, and response to environmental perturbations. Plant biosystems design involves an ability to turn gene networks on or off in the designated tissue, time, and life cycle, while interacting with environmental input. For example, the expression patterns of regulators (suppressors or activators) are genetically programed in the sequential developmental stages from a vegetative meristem to a floral meristem [56], as illustrated in Figure 4(b). The change in gene expression during the transition from apical to floral meristem is governed by various regulators (e.g., ncRNAs, transcription factors, chromatin remodelers, and hormones) in response to environmental signals (e.g., temperature, photoperiod, and nutrient status) and endogenous cues (e.g., plant age) [57, 58]. For dynamic genetic programming in plants, it is necessary to consider not only the abundance of transcripts and proteins but also epigenetic or posttranslational modifications. For example, in Arabidopsis, a novel regulatory mechanism, which depends on cofactor switching mediated by phosphorylation of the photorespiratory enzyme hydroxypyruvate reductase 1, is responsible for the regulation of photorespiratory fluxes in response to the changing environmental conditions [59]. The major challenges in dynamic programming of plant biosystems are: What are sensors and regulators to enable dynamic programming? How can these dynamic regulatory systems be created and controlled, e.g., how can the turnover of mRNAs and proteins be controlled?
2.2.3. Tradeoff between Natural Selection and Artificial Selection
Biosystems design for the industrial purpose of yield maximization or minimal resource utilization may not be orthologous with natural evolution, in which some natural biochemical pathways are presumably optimized for environmental fitness [60]. As most crop plants are grown in open environments, they are still at least partially under natural selection pressure while artificial selection plays an important role in plant domestication. Plant biosystems design might encounter a compromise between the natural selection for fitness and the artificial selection for agricultural and/or industrial purposes (Figure 4(c)). Alternatively, some biosystems design modifications of crops may be selected for under both natural and artificial selection. Engineered photorespiratory bypasses that increase growth rate may form such an example [61, 62]. For example, genetic improvement in yield or quality of crop plants needs to be balanced with stress tolerance enhancement. Alternatively, can these beneficial traits be coupled to ameliorate the tradeoff as part of the biosystems design to increase crop yield and quality under natural environmental inputs and fluctuations?
2.2.4. Genetic Stability of Plant Biosystems Design
As plant genomes are prone to spontaneous mutations (see Section 2.1.3), the capability to maintain the genetic stability of plant biosystems design over a long period of time (e.g., many generations of annual plants and life span of perennial plants) is critical. Also, epigenetic changes may have an impact on the stability of plant biosystems design. Robust traits in multiagent complex systems can be generated through networked buffering mechanism, which features a concurrent, distributed response involving chains of agents with versatility (i.e., agents perform more than one single functional role) and degeneracy (i.e., there exists partial overlap in the functional capabilities of agents) [63]. For example, the pathogen-associated molecular pattern- (PAMP-) triggered immunity signaling network is highly buffered against interference, with the inhibitory loops within the network providing buffering (i.e., loss-of-function of some network components releases associated inhibitory loops to allow other components of the network to compensate for the loss) (Figure 4(d)). Plant biosystems design using long-term buffering strategies such as network buffering or using proteins with multiple functions may produce more robust traits (e.g., disease resistance) that would last for a long period of time (e.g., many generations, tens or hundreds of years).
2.2.5. Upgradability of Plant Biosystems Design
In general, it is important to design a product that can adapt to future required performance and functions via upgrading the components of a biosystem [64]. Since plant biosystems design may require multiple iterations of Design-Build-Test-Learn (DBTL) cycles (for details see Section 3), it is essential that the genetically modified plants or de novo plant systems can be easily upgraded for improving performance or adding new functions. In general, upgrading the plant genome requires consecutive stable plant transformation processes, which is constrained by a limited number of selectable marker genes available for plant transformation, including widely used selectable marker genes conferring antibiotic (e.g., kanamycin and hygromycin) or herbicide (e.g., BASTA) resistance [65], along with some nonantibiotic and nonherbicide markers such as plant phosphomannose isomerase [66], broad-specificity amino acid racemase [67], and fluorescent proteins [68, 69]. For enabling upgradability of plant biosystems design, it would be desirable to consider marker-free plant transformation systems, in which the selectable marker gene can be excised from the plant genome after transformation (Figure 4(e)). Selectable marker genes can be self-excised in plants using various approaches mediated by site-specific recombinase [70–73], zinc finger nuclease [74], and CRISPR [75, 76].
3. Technical Approaches for Plant Biosystems Design
In general, a plant biosystems design approach goes through iterative DBTL cycles. This approach has been widely practiced in the biosystems design of microbial systems [77, 78], but its application to plant biosystems design is still limited, mainly due to much longer time needed to complete DBTL cycles in plants. It would be important to see how recent attempts of accelerating DBTL using cell-free protein synthesis (CFPS) systems [53, 79] would impact cellular approaches in plants. On the other hand, at the organismal level, there is a need for establishing state-of-the-art capabilities for plant biosystems design, including modular cell design, validated biological parts, automated design and build of genetic constructs, generation and testing of plant genetically modified or de novo plant systems, and learning from the test data, and integrating “design (D),” “build (B),” “test (T),” and “learn (L)” as well as executing mini-DBTLs (Figure 5). For effective execution of the DBTL cycles, a laboratory information management system (LIMS) could be used to facilitate local data acquisition and sharing. Also, a Plant Biodesign Hub (PBH) needs to be established as an open access online platform for biological parts registration, genetic circuit design, and predictive modeling based on test data. Although other repositories are already in use and have proven effective, they cannot meet the increasing needs of the growing biosystems design community in terms of data comparability as well as the integration of data curation, submission, biological knowledge, and circuit design.
Figure 5.
Technical approaches for plant biosystems design. In general, plant biosystems design requires iterative cycles of design-build-test-learn. The number of cycles varies with the plant traits to be engineered. LIMS: laboratory information management system.
3.1. Mini-DBTL and Integration
Each component of the Shewhart cycle (D, design; B, build; T, test and L, learn) has their steps for control and continuous improvement forming a DBTL cycle within each D, B, T, and L, named here mini-DBTL. For example, a mini-DBTL within the "D" step could represent formulated DNA sequences that have failed synthetic DNA fragments synthesis and thus have their nucleotide sequence studied, redesigned, resubmitted for synthesis, and reanalyzed to conjunctively inform core adjustments necessary (e.g., maximum local GC content, repeats, homopolymers, and hairpins) for future attempts to succeed [80]. Similarly, researchers may need to (1) attempt different approaches before completing the assemblies (e.g., in the "B" step), (2) evaluate process variation impacting data acquisition (e.g., in the "T" step) [81], (3) develop tools that improve predictive power while making dissimilar suggestions (e.g., in the "L" step) [82], and (4) iterate automation [83]. Such improvements, along with the use of LIMS, robotics, and physical/electronic repositories (e.g., ICE, EDD) [84, 85], will need to be accounted for accelerated and reproducible biosystems design.
3.2. Modular Cell Design
Network modeling has mainly been employed to elucidate complex phenotypes of existing plant biosystems. In principle, the approach can be applied to design plant systems de novo. A recent computational advancement in network modeling using Pareto optimization theory has been described in the ModCell algorithm, which has enabled rational design of a modular (chassis) cell that can be coupled with many exchangeable production modules to achieve various desirable production phenotypes in bacteria [45, 51, 54, 55, 86, 87]. This modular cell engineering approach is aimed at generating production strains rapidly with efficient performance while minimizing the number and cycle time of DBTL cycles. While the ModCell tool may prove very useful to guide the parts, modules, and chassis selection for prokaryotic cells, the compartmentalized endomembrane architecture of eukaryotic cells and the interconnected multicellular nature of complex plant systems featuring specialized cell types remain a considerable challenge for plant biosystems design.
3.3. Curation of Validated Biological Parts
Libraries of validated parts (e.g., protein coding sequences, regulatory elements for gene expression, signaling, and other functional genetic elements) are critical for the engineering of multicomponent biological systems quickly and reliably [88, 89]. Several repositories have been established, such as the iGEM Registry of Standard Biological Parts (http://parts.igem.org) [90, 91], SynBioHub (https://synbiohub.org) [92], and the Addgene repository (https://www.addgene.org) [93, 94]. While these repositories are overwhelmingly dominated by biological parts of prokaryotic origin, they host some DNA parts useful for plant biosystems design, such as the MoClo Toolkit [95, 96] deposited in Addgene. Recently, a library of chloroplast-specific parts was established for plant biosystems design using the plant chloroplast as a chassis [97].
Given the increased complexity of plant genes over those of prokaryotes due to the presence of introns, distal regulatory elements, and posttranscriptional processing signals, a common “Phytobrick” syntax has been developed to enable universal Type IIS assembly with standardized parts (see Section 3.5.1) [89]. Despite these advancements, the conversion of natural DNA sequences into Phytobricks can be laborious, and the removal of “illegal” restriction sites in these sequences to enable Type IIS assembly can introduce unintended alterations to the part’s function. In the recent production of a standardized parts library of 221 Eucalyptus transcription factors and 65 promoters [98], the risk of altering promoter function in their conversion to Phytobricks was minimized by using known single nucleotide polymorphism data in Eucalyptus populations to mutate undesirable restriction sites.
The biological parts for plant biosystems design have been obtained mainly from natural sources (i.e., plant genomes). Recently, a library of synthetic transcriptional regulator systems, which include synthetic activators, synthetic repressors, and synthetic promoters, was established to control plant gene expression in a tissue-specific and environmentally responsive manner [99]. Genome recoding (i.e., rewriting codon meaning through chemical synthesis for new features) has been practiced in microbial systems [100] and could be used to generate novel biological parts for plant biosystems design.
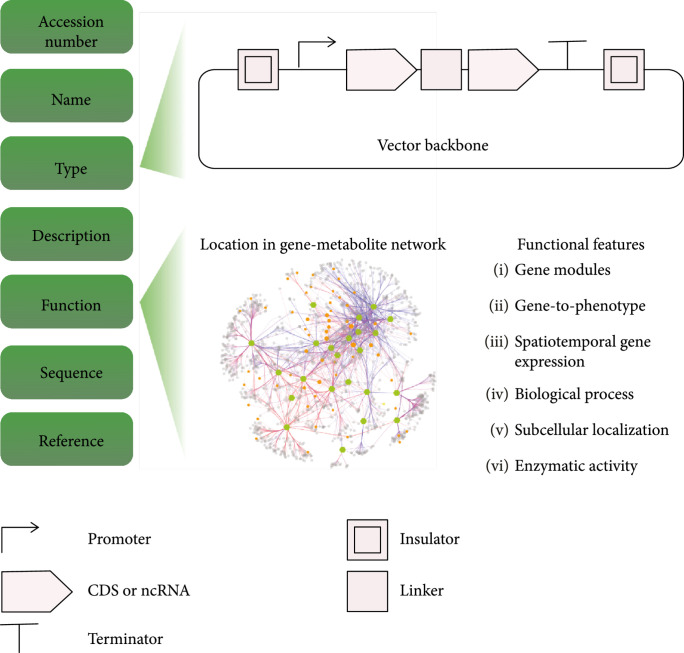
To facilitate international access and reproducibility of data, a centralized knowledgebase of validated biological parts needs to be established, which includes specific experimental context, standardization, and crossed references among different databases (e.g., iGEM, SynBioHub, and Addgene) [85]. The nomenclature of such a knowledgebase would build upon structures already defined by the parts repositories listed above and expand the concept from genomic constructs such as promoters and coding sequences into plant specific structures. Such an initiative could be established through collaboration with KBase [101], with the capability of submission, query, functional mapping onto the gene-metabolite network, as illustrated in Figure 6. There are several challenges to realize the standardization of biological parts across the international plant biodesign research community. For example, (1) how can data be standardized and made comparable between different laboratories? And (2) how can part characterization be rewarded? These challenges may be addressed through international workshops sponsored by a professional society, with participants from academia and industries in the future. For registration of biological parts, it would be important to include negative results, which typically do not get published or reported, to avoid wasteful repetitions in different labs. The negative results would also be very helpful for computational design of synthetic biological parts using machine learning approaches.
Figure 6.
A biological parts registration and curation module for plant biosystems design. The registration form includes accession number, name, type, description, function, sequence (i.e., DNA and protein sequence), and references (e.g., publications associated with the biological parts). For illustration purpose, the functional items are listed for protein-encoding genes only. CDS: protein-encoding sequence; ncRNA: noncoding RNA sequence, including natural noncoding RNAs and guide RNAs for genome editing.
3.4. Genetic Construct Design
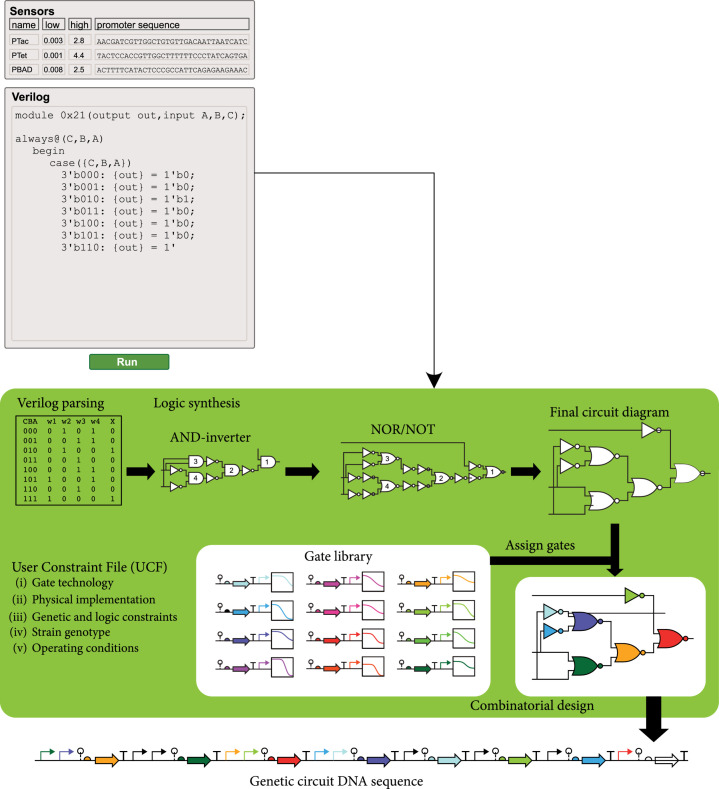
Genetic constructs are designed for nucleic acid sequence modification, gene expression regulation, and metabolic engineering. The genetic design for genomic sequence modification in vivo can be achieved through different methods, including CRISPR/Cas-mediated genome editing, with an emphasis on maximizing on-target efficiency and minimizing off-target effects [102]. Recently, rapid progress has been made in the development of new genome editing technologies, such as high precision prime editing [103], cytosine base editors [104], and adenine base editors [105]. These new technologies have been tested and adapted for genome editing in plants [106–110]. The design for modulating gene expression can be achieved through CRISPR interference (CRISPRi) and activation (CRISPRa) systems, in which nuclease-deactivated Cas9 (dCas9) is tethered to inhibitory and activating domains, respectively, to regulate gene expression [111, 112]. CRISPRi may also be achieved with dCas9 alone acting on promoter or exonic sequences. In addition, RNA editing allows altering splicing or introducing nonheritable changes to protein sequences [113]. It is also possible to multiplex activation, repression, sensing, and emulation of gene expression using homologous CRISPR-sgRNA pairs or different CRISPR-associated RNA scaffolds, which can be potentially used to build complex synthetic programs [102, 114]. For designing predictable gene circuits, genetic design automation (GDA) tools compatible with Synthetic Biology Open Language (SBOL), such as Cello [115] and SBOLDesigner 2 [116], could be adapted to plants and used in an integrated fashion with the knowledgebase of biological parts to streamline the design process, as illustrated in Figure 7.
Figure 7.
An automated gene construct design module for plant biosystems design. Redrawn from Nielsen et al. [115].
3.5. Building Genetic Constructs and Synthetic Plant Genomes
3.5.1. Assembly of DNA Parts into Genetic Constructs
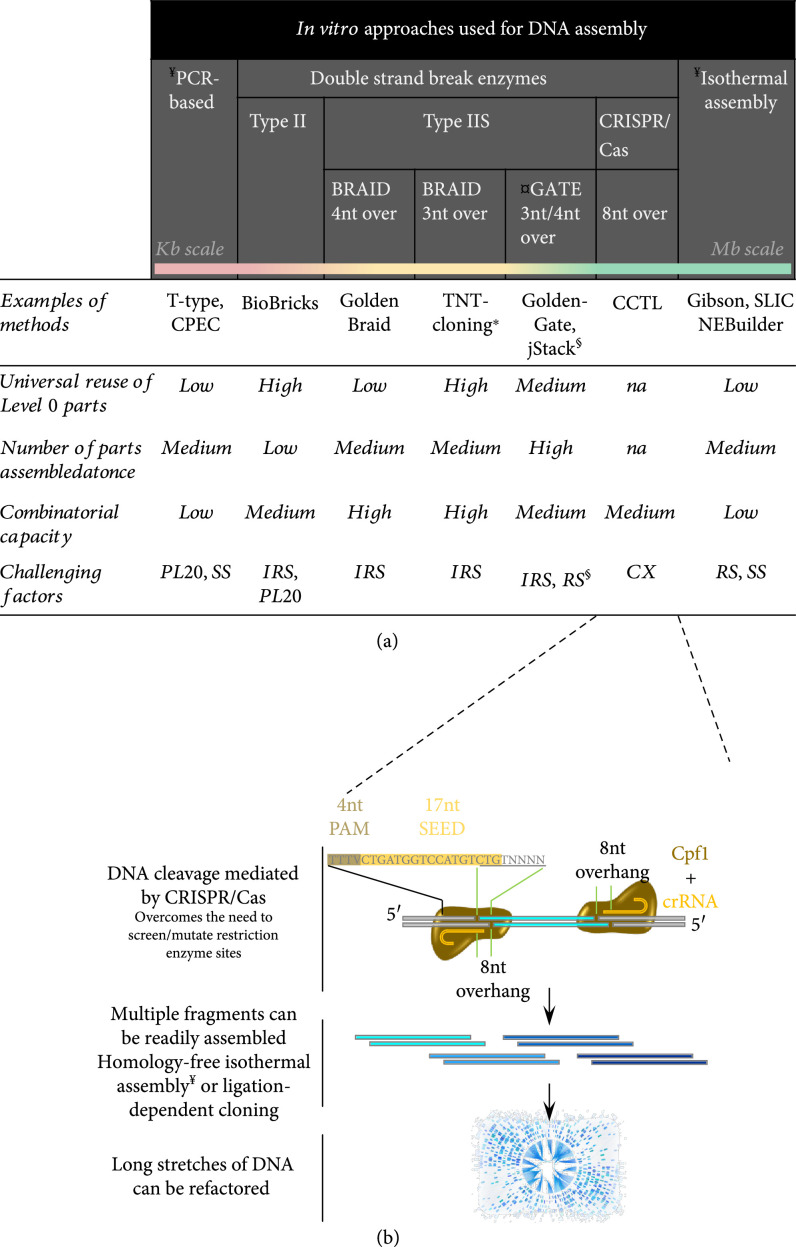
Genetic constructs are built using various DNA assembly methods which can be based solely on PCR reactions (e.g., T-type), sequence-dependent recombinases (e.g., Gateway), enzymes causing specific DNA double strand breaks (e.g., types II and IIS), or an enzyme mix that coordinate nucleotide polymerization (e.g., Gibson assembly) [117–122] (Figure 8(a)). Each of these methods features a unique set of characteristics, and their combinatorial (i.e., shuffling) capacity, hierarchical (i.e., stacking) support, strengths dealing with secondary structure and repetitive sequences will guide their appropriation to different tasks for plant biosystems design. A major challenge in the field has been the editing of specific parts within a large preassembled DNA fragment, which can be addressed by the CCTL method (Cpf1-assisted cutting and Taq DNA ligase-assisted ligation) [123–125] (Figure 8(b)).
Figure 8.
Strategies for DNA assembly across scales relevant for plant biosystems design. (a) Major platforms and key factors considered for executing large DNA assemblies across scales. (b) Detail of the CCTL method. crRNA sequence is from [123]; overhang is underlined with the last 4 nt being programmable [124]. ¥Produce scarless assemblies (NEBuilder allows ssDNA oligos in substitution of homologous overlap).¤May leave scars. Undesired type IIS restriction sites can be partially masked by oligos [122]. For BRAID systems, see [120]. §Hybrid system using golden-gate followed by in vivo homology-based assembly [129]. CX: complex design and execution; PL20: parts size/length beyond 20 kb; RS: repetitive sequences; SS: secondary structures; IRS: internal restriction sites. Recombination-based approaches (e.g., Gateway) were omitted due to limited use for biosystems design.
Type IIS restriction endonuclease-based DNA assembly systems are widely used for hierarchical assembly of DNA fragments into genetic constructs, such as GoldenBraid [120], TNT-cloning [122], and universal Loop assembly (uLoop) [126], which are based on Golden Gate [118]. These approaches have two advantages: (1) the DNA parts can be individually cloned into the entry vectors to establish biological parts libraries, which can be shared in the scientific community, and (2) multiple rounds of binary to hexanary assemblies can be performed to join various numbers of DNA fragments in flexible configurations. However, all type IIS-based approaches suffer from prohibitive internal restriction sites. These sites can be partially masked [122] but not eliminated, and innovative approaches mutating deoxy-adenine within such prohibitive sites to unnatural nucleotides (e.g., deoxy-NaM) during PCR could advance these methods due to E. coli's natural ability to restore the original adenine-rich restriction site in vivo [127].
Multiple DNA fragments with unique short (e.g.,15-20 bp) overlaps between neighboring parts can be assembled using isothermal assembly such as Gibson Assembly, in which a 5 exonuclease removes nucleotides from the 5 ends of double-stranded DNA molecules, complementary single-stranded DNA overhangs are annealed, a DNA polymerase fills the gaps, and a DNA ligase seals the nicks [119]. Recently, another similar approach, called “SureVector,” was developed to assemble multiple DNA fragments with 30 bp overlapping ends, in which DNA parts are denatured and adjacent parts are annealed due to the overlaps followed by DNA polymerase-mediated partial extension of exposed 3-OH ends, resulting in flaps that are digested by an endonuclease and covalently joined by a ligase [128]. These approaches have several key advantages: (1) allowing for assembling multiple blunt-end DNA fragments in a single-tube reaction and (2) no reliance on restriction enzyme digestion of DNA fragments and consequently no requirement for removing or mutating type IIS restriction sites. On the other hand, these methods have a reduced capacity to assemble multiple parts at once (compared to Golden Gate) and have their efficiency strongly impacted by repetitive sequences as well as sequences prone to secondary structure when single stranded. Under such circumstance, using hybrid methods, which combine the advantages of two or more approaches at different levels while being amenable to the drawbacks, will be highly beneficial [129].
3.5.2. Plant Synthetic Genomes
Recently, a 785 kb Caulobacter ethensis-2.0 (C. eth-2.0) genome was constructed in yeast using multiple rounds of homologous gap repair approach [130]. A 4 Mb synthetic Escherichia coli genome was constructed through a high-fidelity convergent total synthesis [131]. However, the construction of plant synthetic chromosomes through DNA synthesis has not been reported yet. A synthetic plant chromosome vector requires a minimum of centromeric and telomeric sequences, origins of replication, and a selectable marker gene [132]. A notable step towards the generation of synthetic plant genomes was the full cloning and yeast-mediated modification of the 204 kb plastid genome of the algae Chlamydomonas reinhardtii [133]. Plastids are among the defining features of plants, and their relatively small and well conserved genomes, along with the potential for high-level expression of desired genes, are currently more tractable candidates for total synthesis than plant nuclear genomes [134].
3.6. Testing Genetic Constructs in Plants
Testing of plant biosystems designs is mainly achieved through stable transformation and transient expression (e.g., agroinfiltration and protoplast transformation) of genetic constructs, sometimes followed by omics (epigenomics, transcriptomics, metabolomics, proteomics, and phenomics) analysis of genetically modified plants or de novo plant systems. Stable transformation is a bottleneck of the DBTL cycle because the main limitation remains to generate many transgenic plants transformed with multigene constructs. What can be improved at a throughput level using protoplasts is at the expense of an understanding of the construct design’s effect on whole-plant performance and fitness, and likewise, protoplast-based systems will not be suitable for all traits being engineered.
3.6.1. Stable Transformation and Transient Expression of Genetic Constructs
Agrobacterium-mediated transformation has been the major approach for plant genetic engineering. However, there is substantial variation in the amenability to Agrobacterium-mediated transformation among plant species and even cultivars of the same species, with high-efficiency transformation protocols available for a limited number of plant species/cultivars. One limitation for using Agrobacterium to transform plants is that not all plant species are Agrobacterium-infectable. Another major bottleneck in Agrobacterium-mediated transformation is in vitro regeneration of shoots or embryos from transformed cells. Ectopic expression of morphogenic or developmental regulator genes (e.g., Baby boom and Wuschel2) can promote somatic cells to form embryos, which develop into whole plants, in monocot species and consequently improve Agrobacterium-mediated transformation efficiencies dramatically [135]. Plant transformation often requires tissue culture by exposure of cells to various hormones, which are inefficient and time-consuming. Recently, a de novo induction of meristem approach based on the use of development regulators was developed in Nicotiana benthamiana, tomato, potato, and grape, avoiding the use of traditional tissue culture [136]. The generation of transgenic roots (hairy roots) through Agrobacterium rhizogenes-mediated transformation, leading to the production of composite plants (i.e., transgenic roots on wild-type shoots), has proven to be a fast and versatile system particularly suited for certain woody plants recalcitrant to transformation such as Eucalyptus [137, 138]. There is an urgent need to extend these methods to other plant species or to develop new capabilities for enabling or improving transformation in a wider range of dicot and monocot species, particularly for the ones that are currently recalcitrant to genetic transformation. In planta transformation methods can be particularly useful because no in vitro regeneration of shoots or embryos is required. It is preferred that morphogenic regulator genes are not used or that the morphogenic regulator genes could be excised out of the genome by inducible recombinase excision system, because expression of these transgenes can affect plant growth and development [139–141].
Besides Agrobacterium-mediated transformation, DNA, RNA, or protein molecules can be directly delivered to target sites via particle bombardment, nanoparticles, or direct injection. For example, Cas9–gRNA ribonucleases (RNPs) were directly injected into plant zygotes for DNA- and selectable-marker-free genome editing in rice [142]. Furthermore, carbon nanotubes were recently used for efficient plasmid DNA delivery into multiple plant species (e.g., arugula, wheat, and cotton) to enable high protein expression levels without transgene integration [143], which has great potential for application to transgene-free genome-modification [144] and may also be useful for functional testing of plant biosystems design.
Multigene transformation is important for plant biosystems design. It can be achieved using binary vectors based on transformation-competent artificial chromosomes, such as pHUGE-Red which is suited for cloning large DNA fragments [145]. Alternatively, multiple binary vectors with compatible replication origins can be hosted in a single Agrobacterium cell for simultaneous delivery of multiple gene constructs into plants [146]. Furthermore, gene stacking based on site-specific recombination and nuclease activity has a potential for in planta stacking of a large number of genes at a single genomic site of the same plant [147]. Also, multiple genes could be stacked using plastid transformation with operon-like and polyprotein expression systems [148–150]. CRISPR/Cas9-mediated targeted T-DNA integration and precise knock-in [102, 151, 152] can potentially be used for in planta stacking.
As an alternative to stable transformation approaches, which are often time-consuming, transient expression techniques via virus-induced gene silencing (VIGS) enable rapid knockdown of a targeted gene in a high-throughput manner, even in plant species that are difficult to transform [153]. However, RNA-directed transcriptional gene silencing cannot induce heritable changes that target the coding region, although targeting of the promoter sequence can cause heritable changes in gene expression mediated by methylation [154]. In combination with CRISPR/Cas systems, viruses can be used to quickly induce heritable changes in plant genomes. Recently, viruses have been used to deliver the guide RNAs or the entire CRISPR–Cas9 cassette for Cas9-mediated gene editing in plants, providing another high-throughput method for testing the function of gene constructs for plant biosystems design [155–157]. Also, multiplexed heritable gene editing was recently achieved through virus-mediated in planta delivery of mobile single guide RNAs (sgRNAs) into Nicotiana benthamiana transgenic plants expressing Cas9 [158], which provides another approach to produce heritable gene editing without tissue culture. Alternatively, a recently developed nanotube-based platform for RNA delivery, which enables stable siRNA delivery and efficient silencing of target genes in intact plant cells [159], might be useful for a broad range of applications including direct delivery of sgRNAs and Cas9 mRNA for DNA-free genome editing.
3.6.2. Omics Analysis of Genetically Modified Plants
Integrative multiomics (e.g., transcriptomics, metabolomics, proteomics, epigenomics, and phenomics) analysis of genetically modified plants could provide rich experimental data for linking genetic design to plant phenotype. So far, plant omics data have been collected at the organ or tissue level, resulting in the molecular, metabolic, and biochemical information averaged over a population of heterogeneous cells [160, 161]. Because plant cellular processes vary spatially, single-cell multiomics is necessary for simultaneous analysis of different biomolecules to achieve accurate assessment of nodes and edges in the gene/metabolite networks operating in an individual cell [162]. Single-cell technologies have evolved intensively in the last decade [163], and transcriptomics and proteomics are viable at the single-cell level [164, 165]. However, most plant species still face major technical hurdles that make it challenging to achieve single-cell resolution, in large part because it is challenging to dissociate cells from the plant tissues [166] and/or collect adequate amounts of the desired biomass when no amplification strategies are available [167, 168]. Progress has been made to solve these challenges. For example, the protoplasts of Arabidopsis root cells have been successfully used for single-cell RNA sequencing using droplet-based microfluidics platform [164, 169–171], and laser capture microdissection has been used to isolate individual cell layers of tomato roots for single-cell-type proteomics [172]. Also, live single-cell mass spectrometry has been used for direct analysis of metabolites in a single live plant cell [173].
3.7. Learning from the Testing of Designed Plant Systems
Integration of multiomics data can provide a multiperspective view of dynamic molecular behavior and interacting networks of genes occurring in plants [174], as demonstrated in an integrative analysis of transcriptomic, proteomic, fluxomic, and phenomic data in relation to lignin biosynthesis in Populus [175]. The omics datasets can be integrated using statistical or advanced machine learning approaches [176] to generate results related to simple pathways or complex networks at cell, tissue, organ, or organism levels, providing insights into the effect of biosystems design on plant phenotype. For example, a machine learning method called Multi-view Factorization AutoEncoder, which uses a deep representation learning approach to simultaneously learn feature and patient embeddings, was recently developed for seamless integration of multiomics data and biological domain knowledge such as molecular interaction networks in humans [177]. This method can be extended to omics data analysis of genetically modified plants or de novo plant systems for improving the design of plant systems in an iterative application to the DBTL cycle, as illustrated in Figure 5. Complementary tools that use probabilistic modeling techniques to converge to the desired specification (e.g., increased expression of a gene cluster) accurately, without requiring full mechanistic understanding of the biological system [82], will need to be recruited to plant biosystems design. An exemplary module for integrative analysis of multiomics data in the “Plant Biodesign Hub” is presented in Figure 9(a). The results from the integrative analysis of multiomics data could be used for metabolic modeling based on the modeling module to be built in the “Plant Biodesign Hub” (Figure 9(b)). Besides learning from engineered plants, it would be important to learn from model species or from cross-species models incorporating prior knowledge through explainable artificial intelligence [178].
Figure 9.
A computational learning module for plant biosystems design. (a) Integrative analysis of multiomics data; redrawn from Yugi et al. [374]. (b) Metabolic modeling; redrawn from Zampieri et al. [375].
4. Applications of Plant Biosystems Design
Biosystems design has great potential for applications in (1) basic plant biology research to gain a deeper understanding of molecular functions and biological processes in plant biosystems (Figure 10) and (2) various aspects of applied plant science research to accelerate the improvement of plant traits or to create new germplasms with improved traits to benefit ecosystem health and human society (Figure 11).
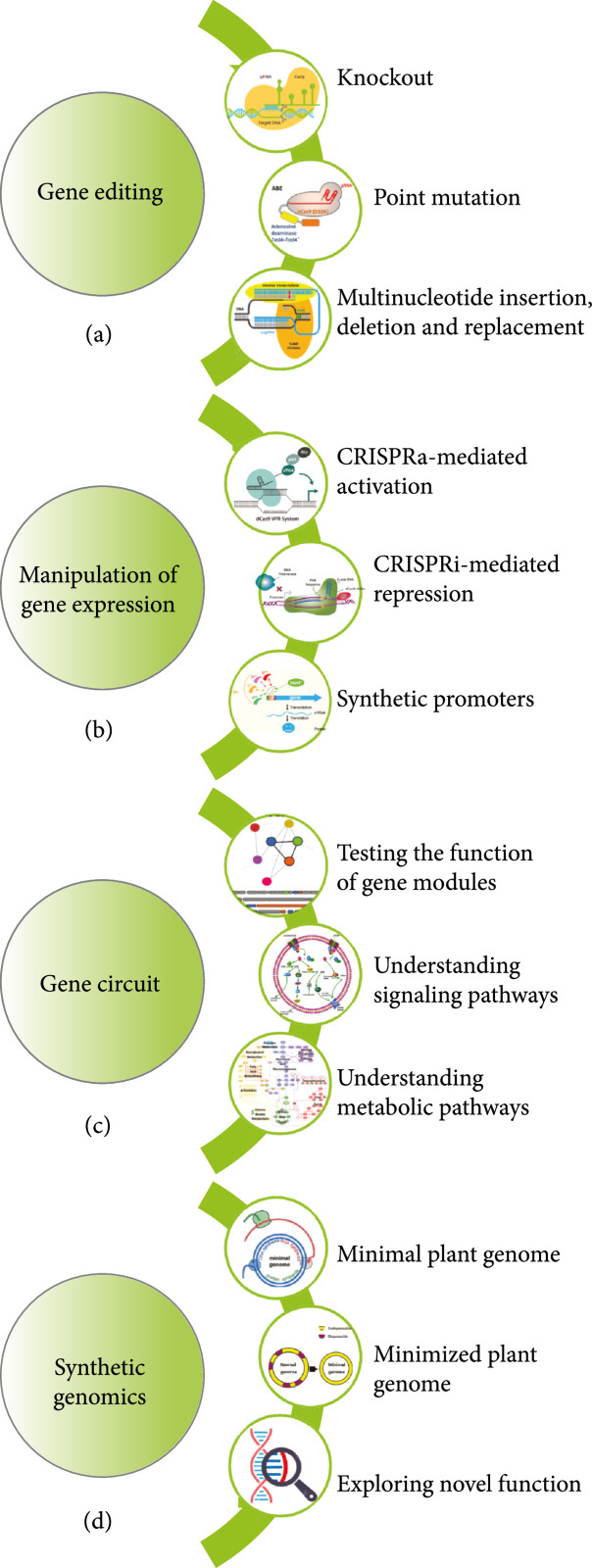
Figure 10.
Applications of biosystems design to basic plant biology research. (a) Elucidating plant gene function through genomic mutations created by using genome editing technologies. (b) Manipulation of gene expression using CRISPR interference (CRISPRi) and activation (CRISPRa) and synthetic promoters. (c) Studying the function of gene modules, signaling, or metabolic pathways using synthetic genetic circuits. (d) Understanding the complexity of plant systems using minimal or minimized plant genomes as well as exploring novel function using synthetic genomics.
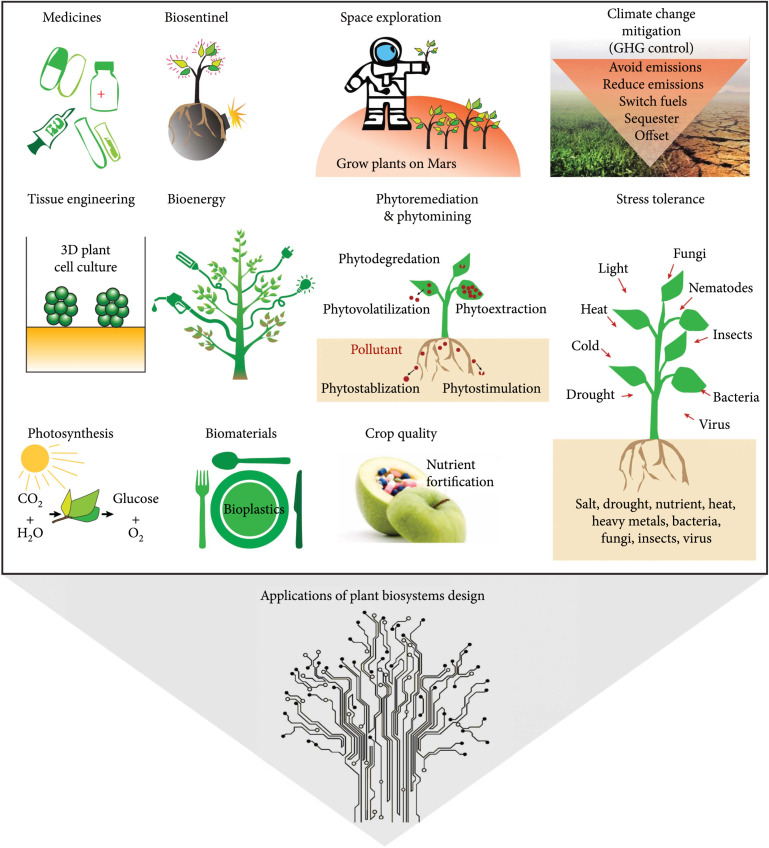
Figure 11.
Applications of biosystems design to applied plant science research. GHG: greenhouse gas. Only representative examples are shown.
4.1. Application of Biosystems Design to Basic Plant Science Research
Biosystems design can be used to further our understanding of molecular mechanisms driving biological processes in plant systems by dissecting the function of individual genes or multigene modules.
4.1.1. Understand Plant Gene Function Using Biosystems Design
Approximately 600 plant species genomes have been sequenced [179], with an increasing number of new plant genome sequences being released every year. However, even in Arabidopsis thaliana, which is one of the best studied model plant species, approximately 60% of predicted enzyme- and transporter-encoding genes do not have credible functional annotations, and this number is even higher in nonmodel plant species [180]. Until recently, only ~5% of genes in the Arabidopsis genome have experimental evidence for their functions (e.g., biochemical activity, subcellular location, and biological role) [181]. Traditionally, experimental characterization of plant gene function depends mainly on (1) knockout mediated by T-DNA insertion or chemical/radiation-induced mutagenesis, (2) knockdown mediated by RNA interference (RNAi) or VIGS, and (3) overexpression of one or a few genes in individual genetically modified plant lines. These traditional approaches suffer from several limitations: (1) the knockout mutations created by T-DNA insertion or chemical/radiation-induction occur as random insertions or deletions and often accompanied by additional unrelated mutations in the genome, (2) it is challenging to obtain homozygous multigene knockout mutants in diploids or single-gene mutants in polyploid species, as it requires multiple generations of self-pollinated plants while being almost impossible in vegetatively propagated plants and perennials with long life cycles, and (3) RNAi works on protein-coding genes only, along with incomplete loss of function and extensive off-target activities [182]. These limitations can be overcome by genome-engineering tools such as CRISPR/Cas-systems, which can be used to generate targeted homozygous knockout mutations without the need for self-fertilization [102, 182]. CRISPR/Cas-mediated gene knockout has one disadvantage for functional genomics research: it is not suitable for studying the function of essential genes due to the lethality of their knockout mutants generated by CRISPR/Cas systems, although it is possible to identify essential genes using CRISPR/Cas targeted mutagenesis in some cases where homozygous knockout mutant seeds can be obtained from heterozygous mutant parents [183]. Still, CRISPRi and CRISPRa offer the opportunities to repress and activate gene expression, respectively, for both coding and noncoding RNAs [112]. However, being similar to the traditional genetic transformation system, the CRISPR/Cas systems have not been established in many plant species. In some cases, low editing efficiency and off-target issues still cannot be fully addressed. Beyond examining the functional roles of a particular gene, more elaborate biosystems design strategies offer a powerful approach for studying the collective function of multigene modules in metabolic pathways, signal transduction cascades, and regulatory networks [184]. It is very challenging to map the protein-DNA interactions in gene regulatory networks using experimental approaches. Biosystems design could enable scalable epitope tagging for high-throughput chromatin immunoprecipitation followed by sequencing (ChIP-Seq) [185], combined with CRISPR/Cas-mediated knockout experiment, to identify accurately the target genes of plant transcriptional factors.
In general, plant functional genomics research is still substantially hindered by labor intensive and time-consuming work and therefore could greatly benefit from biosystems design approaches that provide high-throughput capabilities for determining gene function. For example, multiplex genome editing can generate more than 100 targeting events [186], enabling gain-of-function or loss-of-function screening of a large number of genes. Also, the automated design and high-throughput assembly of gene constructs described in Section 3 would greatly speed up the elucidation of plant gene functions.
4.1.2. Understand the Complexity of Plant Systems Using Biosystems Design
Construction of synthetic genomes (e.g., synthetic minimal genome, massively recoded genome, chimeric genome, and synthetic genome with expanded genetic alphabet) can generate new insights into the basic principles of life and enable valuable applications [187]. Minimal genomes, which are reduced genomes containing only genes essential for life, have been constructed for multiple single-cellular organisms, such as Mycoplasma and Saccharomyces [188–190]. Recent technical advances in DNA synthesis and synthetic genomics may soon allow for the construction of minimal genomes for model plants. A minimal plant genome should enable a complete set of essential features of plants, including plant growth, development, and response to environment. A bottom-up approach based on de novo DNA synthesis could be used to reconstruct minimal plant genomes, but it would require information about the minimal required set of genes for plant viability. Alternatively, minimized genomes can be obtained by deletion of cryptic genes and mobile DNAs in microbes [191]. It is anticipated that creating minimized genomes for plants would be much more challenging than for microbes due to much larger and complex genomes in plants. From a biosystems design perspective, a minimized plant genome could be potentially obtained through reduction via genome-wide gene-knockout using CRISPR/Cas systems. The minimized plant genomes would provide a unique opportunity for dissecting the minimal gene network of a functional plant system and validating modular cell design. Furthermore, it would allow for adding genes or gene modules to study their function.
4.2. Application of Biosystems Design to Applied Plant Science Research
With guidance from the principle of biosystems design, the cutting-edge genome editing and genome-writing/rewriting technologies can be used for modifying or redesigning crop plants for various applications, including genetic improvement of photosynthetic efficiency, plant stress tolerance, crop quality, climate change mitigation, production of biomaterials, bioenergy and medicines, phytoremediation, biosentinel, tissue-engineering, and space exploration.
4.2.1. Plant Biosystems Design for Increasing Photosynthetic Efficiency
The average yields of staple crops currently increase at a rate of about 1% each year, but this will need to increase by two-fold to feed the estimated world population of 9 billion people in 2050 [7]. Improvements in the yield potential of crops could be accelerated through genetic engineering approaches to enhance photosynthetic efficiency. A number of strategies have been employed to achieve this goal (for a detailed discussion see Long et al. [192]), with several recent successes that highlight our capacity to modify both the light-dependent and light-independent reactions of photosynthesis, both of which are important in determining crop yield potential. Here, we discuss some of these strategies and successes that could be further complemented by a plant biosystems design approach.
The photosynthetic reaction center complexes that capture light energy in plants (i.e., photosystem I (PSI) and photosystem II (PSII)) utilize only half of the incident solar energy (i.e., 400 to 700 nm) and work in series, connected by an electron transport chain. Redesign of the photosystems to expand the region of photosynthetic absorption from the visible region of the spectrum to include far-red and infrared regions could improve the efficiency of light capture [193]. For example, introducing novel light-harvesting pigments (e.g., chlorophylls d and f from cyanobacteria Acaryochloris marina and Halomicronema hongdechloris, respectively) could allow for light capture up to 750 nm [194]. In a more ambitious design, it would conceptually be feasible to redesign several components of the photosynthetic electron transport chain to harvest [195]. Thus far, experimental successes to improve light-use efficiency and plant yields have come from overexpressing components of the cytochrome b6f complex, which facilitates the transfer of electrons from PSII to PSI [196, 197]. Furthermore, accelerating the repair of photodamage to PSII, through nuclear overexpression of the core PSII subunit protein D1, can improve photosynthesis and plant productivity and enhance survival under heat stress [198]. Lastly, engineering the photoprotective mechanisms of the photosystems can also facilitate growth enhancements. Photosystems dissipate excess absorbed light energy as heat in full sunlight but do not adapt to fluctuating light conditions rapidly, resulting in suboptimal photosynthetic efficiency and consequently losses of up to 20% of potential yield in field crops. This issue has been addressed by accelerating the induction and recovery from photoprotection in tobacco via bioengineering of an accelerated response to natural shading events, which increased dry biomass yield by ~15% in fluctuating light [199]. In contrast, the same approach in Arabidopsis has led to growth impairments, suggesting that the success of this strategy requires careful balance so as not to interfere with other regulatory processes [200].
For the light-independent reactions of photosynthesis (i.e., CO2 capture and conversion to sugars and starch), ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the key enzyme for CO2 fixation in all plants. Considering its importance, Rubisco is surprisingly inefficient and considered a bottleneck to photosynthetic productivity. Rubisco is a relatively slow enzyme, so to compensate, most plants invest ca. 30% soluble protein in leaves to the Rubisco pool. Furthermore, the dual specificity of Rubisco for CO2 and O2 results in two separate reactions: carboxylation and oxygenation. The latter results in the production of the toxic intermediate phosphoglycolate that is removed by the photorespiratory salvage pathway, causing the loss of carbon and energy. Photorespiration is widely regarded as a necessary but wasteful biochemical pathway [201]. Rubisco is one of the most well-studied enzymes and has been a prime engineering target for decades [202]. However, Rubisco consists of subunits expressed in the nuclear and chloroplast genomes and requires several chaperone proteins for functional assembly. Thus, engineering the catalytic properties of Rubisco has been challenging and progress has been slow. However, recent success in the assembly of Arabidopsis and tobacco Rubisco in E. coli could pave the way for more rapid screens to identify mutants with substantial improvements in function [52]. As an alternative strategy to combat Rubisco oxygenation, several photorespiratory bypasses in the chloroplast have been developed [203, 204] including a synthetic glycolate metabolism pathway that has been shown to increase tobacco biomass yield by ~40% in a field study [205]. Recent work has expanded this approach to rice and highlighted the need for readdressing source-sink flow in plants engineered to have enhanced photosynthetic potential [206]. Several alternative synthetic bypass routes have also been suggested, including a synthetic pathway for converting glyoxylate to hydroxypyruvate in the peroxisomes, which circumvents the mitochondrial reactions, avoiding decarboxylation and deamination [207], and enzyme engineering approaches to perform new-to-nature reactions such as the reduction of glycolate to glycolaldehyde [61]. More ambitious synthetic strategies include the development of alternative carboxylases and synthetic cycling pathways (i.e., not integrated into the Calvin-Benson-Bassham cycle) for CO2 assimilation that completely bypass the shortcomings of Rubisco [208–210]. Computational modeling could be used to design further novel synthetic pathways to assimilate CO2 and bypass photorespiration. Natural evolution in plants has only explored a fraction of the potential metabolic design space to drive photosynthesis [211, 212]; thus, there are likely many opportunities to further redesign and enhance crop performances.
Several photosynthetic organisms have evolved CO2-concentrating mechanisms (CCMs) to overcome the limitations of Rubisco and reduce photorespiration [213]. Plants have evolved two CCM pathways: C4 photosynthesis and crassulacean acid metabolism (CAM) photosynthesis. Although the potential for exploiting CAM photosynthesis for future agricultural production has been highlighted [214], most engineering work has focused on the benefits of C4 photosynthesis. Plants that perform C4 photosynthesis (i.e., C4 plants like maize, sorghum, sugarcane, and switchgrass) typically separate CO2 fixation across two cells types: initial capture of CO2 in mesophyll cells as the C4 acid oxaloacetate and conversion to malate, and then transport to Rubisco-laden bundle sheath cells, where malate is decarboxylated to release CO2. C4 photosynthesis facilitates above atmosphere local concentrations of CO2 around Rubisco, thereby favoring the carboxylation reaction over the oxygenation reaction and reducing photorespiration. As a result, C4 plants generally have higher photosynthetic efficiencies than C3 plants. Nevertheless, several important staple crops (e.g., rice, wheat, and soybean) are C3 plants. International efforts have been undertaken to engineer C4 photosynthesis into rice to increase photosynthetic efficiency and productivity [215]. However, due to the two-cell complexity of C4 photosynthesis, converting C3 into C4 photosynthesis requires considerable reengineering of metabolism and dramatic changes in leaf anatomy, both of which impose a significant challenge. Engineering a single-celled C4 system using biosystems design could be a promising alternative strategy [216]. In addition, introducing the single-celled physical CCMs found in algae and cyanobacteria into plants is predicted to lead to some of the largest improvements in yield potential (>60%) [217, 218]. Promisingly, several of the components required to build such systems have now been successfully introduced into plants [219–221]. Recently, the draft genome sequence of a single-cell C4 (SCC4) plant species, Suaeda aralocaspica, became available [222], providing an excellent genomics resource for engineering SCC4 in C3 plants. C2 photosynthesis, which utilizes glycine decarboxylase activity in the bundle sheath to decarboxylate the photorespiratory glycine produced in the mesophyll and deliver CO2 around Rubisco, is another CCM that operates by capturing, concentrating, and reassimilating CO2 released by photorespiration, and therefore, engineering of C2 photosynthesis has the potential to improve photosynthetic performance under high temperature, bright light, and low CO2 conditions [223, 224].
Roughly 50% of the carbon captured by photosynthesis (net of photorespiration) is subsequently lost, and strategies for cutting this large carbon loss include (1) reducing unnecessary turnover of proteins (e.g., THI4 which is a suicide enzyme with a very high turnover rate) and membranes; (2) replacing, relocating, or rescheduling metabolic activities (e.g., replacing the Phe route to lignin, relocating nitrate assimilation from root to shoot, and rescheduling biosynthetic processes from night to day); (3) suppressing futile cycles (e.g., futile cycles between sucrose synthesis and degradation or between fructose-6P and fructose-1,6BP); and (4) reducing ion transport costs (e.g., reducing efflux of nitrate to the rhizosphere) [225]. These strategies can be implemented through synthetic metabolic engineering approach [226].
4.2.2. Plant Biosystems Design for Increasing Plant Stress Tolerance
Abiotic stresses (e.g., drought, heat, and salt stress) account for more than 60% of the yield loss in some major crops such as maize, wheat, rice, and soybean [227, 228]. Plant resistance to abiotic stresses can be divided into escape, avoidance, and tolerance [229, 230]. Various types of genes have been proposed as candidates for engineering to increase tolerance to abiotic stresses, including genes encoding (1) enzymes for production of protective metabolites (e.g., proline and sugars), (2) enzymes for membrane lipid biosynthesis, (3) enzymes for biosynthesis of antioxidants (e.g., ROS scavenging), (4) protective proteins (e.g., LEAs and molecular chaperones), (5) transporters (e.g., water and ion transport), (6) regulatory proteins, (7) kinases, and (8) proteins regulating transcription (e.g., transcription factors), along with genes involved in posttranscriptional (e.g., microRNAs) and posttranslational (e.g., ubiquitination) regulation of abiotic stress responses [230]. Previous efforts have been focused on engineering of individual genes to enhance tolerance to a specific abiotic stress. Plant biosystems design has the potential of integrating multiple genes to confer resistance to a broad range of abiotic stresses. Also, tissue-specific and stress-inducible expression of genes relevant to abiotic stress tolerance could be implemented via biosystems design to reduce energy costs and avoid pleiotropic effects.
One important example of stress avoidance mechanism is CAM photosynthesis, which is a natural solution to the challenge caused by drought stress. CAM plants close their stomata (the pores on the leaf surface) during the heat of day and open them at night, resulting in lower water loss and higher water use efficiency than C3 or C4 plants, which close their stomata during the nighttime and open them during the daytime [231]. The maximum yield of CAM crops is much higher than that of C3 or C4 crops under water-limited conditions [232]. Engineering of CAM machinery into C3 or C4 plants has great potential for increasing crop yield under drought conditions. CAM engineering requires design of multiple gene modules involved in carboxylation, decarboxylation, and stomatal movement, as well as genes involved in leaf succulence and vacuole size [233–235]. Biosystems design approaches could be used to integrate these CAM-related gene modules into plants, preferably using gene circuits to establish drought-inducible CAM (or CAM-on-demand) systems [236].
Biotic stresses imposed by pathogens and pests can also cause massive losses in crop yield. Engineering synthetic plant immunity would be a promising strategy for increasing or broadening plant resistance to diseases [237]. Immune receptors, such as nucleotide-binding leucine-rich repeat (NLR) receptors, are promising targets for increasing disease resistance using biosystems design approaches [238, 239]. Creating genetically modified crops resistant to insects is a useful approach to reduce the yield loss caused by pests [240], such as transgenic crops overexpressing Bacillus thuringiensis (Bt) insecticidal proteins [241]. It is critical to design gene constructs that specifically target pests without toxic effects on humans or negative impacts on beneficial organisms. Host-induced gene silencing (HIGS), in which double-stranded RNAs (dsRNAs) directed against suitable insect target genes are expressed in transgenic plants, has been used to confer protection against pests [242]. The HIGS approach has two major advantages: (1) dsRNAs can be designed to be highly specific to target insects without negative impact on other organisms, and (2) multiple dsRNAs can be engineered into each individual plant for protection against multiple pests. However, the design of HIGS requires rich genomics resources of target insects and related species.
Beneficial microbes (e.g., bacteria and fungi) can enhance plant resistance to abiotic and biotic sources of stress [243]. Plants can generate molecular and metabolic effectors for promoting beneficial plant-microbe interactions. Synthetic genetic circuits can be engineered into plants to reshape the rhizosphere microbiome to enhance stress tolerance and acquisition of nutrients (e.g., nitrogen). For example, opine biosynthesis pathways have been engineered into plants to reshape rhizosphere populations to increase the population densities of opine-catabolizing bacteria [244]. Similarly, a synthetic pathway has been engineered in Medicago truncatula and barley for the production of the rhizopine scyllo-inosamine to regulate bacterial gene expression in the rhizosphere [245]. There is considerable potential for engineering host plants to promote the beneficial interactions between plants and microbes.
4.2.3. Plant Biosystems Design for Improving Food Crop Quality
The ever-increasing living standard worldwide, combined with limited arable land availability, calls for genetic improvement of food crop quality. Deficiencies in vitamins collectively affect billions of people worldwide and are a cause of substantial morbidity and mortality. Vitamin A deficiency is the global leading cause of preventable blindness [246], iron deficiency delays cognitive development [247], and folate deficiency is especially common among pregnant women and is associated with defects in fetal neural crest development [248]. Eliminating vitamin deficiency is a global public health priority and one of the World Health Organization Millennium Development Goals [249]. Biofortification (i.e., improvement of nutritional quality of food crops during plant growth and development) is a cost-effective strategy to mitigate vitamin deficiencies, particularly in the developing world where other vitamin supplementation programs suffer from logistical problems with transportation. Plant biosystems design is a useful approach to achieve biofortification through the engineering of superior nutritive properties in crop plants. Examples of biofortified crops include those enhanced with beta-carotene (provitamin A) [250, 251]; arachidonic acid [250]; carotenoids associated with eye and cardiovascular health, immunity, and cognitive function [250, 252]; iron [253]; and folate [254], along with efforts underway for α-tocopherol (vitamin E) [255] and zinc [256].
Crop plants generally contain various types of antinutritional factors such as cyanogenic glycosides (e.g., phaseolunatin and dhurrin), enzyme inhibitors (e.g., alkaloids and protease inhibitors), physiological disorganizers (e.g., lectins and saponins), hormone biosynthesis inhibitors (e.g., goitrogens), and antivitamins (e.g., antivitamin E) [257, 258]. For example, cowpea (Vigna unguiculata L. Walp) plants, which are able to grow in semiarid regions with low input requirements, provide a sustainable source of essential nutrients (e.g., high protein and low fat content), but dietary utilization of cowpea has been seriously constrained by its antinutrients (e.g., phytic acid, protease inhibitors, and cyanogenic glucosides) and low protein digestibility [259]. Altogether, biosystems design could simultaneously leverage biofortification, remove antinutritional compounds, and increase protein digestibility, but care needs to be exercised to not consequently increase pest and pathogen susceptibility.
4.2.4. Plant Biosystems Design for Mitigation of Climate Change
Carbon dioxide released into the atmosphere is the primary cause of anthropogenic global warming [260, 261]. Terrestrial plants are a major player of atmospheric CO2 capture and storage [262, 263]. Carbon sustainability and carbon neutrality would be the great benefit that can be achieved with faster growing plants. Biosystems design has great potential in CO2 capture and storage in various aspects, including (1) improving photosynthetic efficiency aboveground and allocation of photosynthates to below ground structures (e.g., source-sink modulation) [264], (2) converting annual crops to perennial plants that have much larger root systems [265], (3) generating more recalcitrant carbon-containing compounds (e.g., lignin and suberin) in the roots [266], (4) establishing deeper root systems [267], (5) enabling animal to plant-sourced protein shifts [268], (6) restoring forests [269], (7) enabling carbon mineralization [270, 271], and (8) increasing biomass accumulation through genetic improvement of photosynthetic efficiency [272] (see also Section 4.2.1). Armed with these biological targets, opportunities to mitigate rising atmospheric CO2 concentrations are many, with promising avenues to pursue through biological carbon capture and storage in soils. This undertaking should be viewed with increasing optimism, especially as the strategies and technologies—aided by plant biosystems design—employed to store carbon in soil pools with long residence times have emerged and will continue to evolve over time. The postgenomics era provides an unprecedented opportunity to identify genes, enzymes, biochemical pathways, and regulatory networks that underlie rate-limiting steps in carbon acquisition, transport, and fate; and thereby yield new approaches to enhance terrestrial carbon sequestration. An investment in plant biosystems design could harness these new approaches to increase biomass production in agricultural crops and fast-growing trees in managed plantations.
4.2.5. Plant Biosystems Design for Bio-Based Materials
Living organisms produce a series of proteins and compounds, i.e., bioproducts, used as building blocks for the manufacture of biomaterials. Such manufacture can take place ex vivo (e.g., through chemical manipulations of extracted bioproducts) or in vivo, being naturally synthetized by the living organism (i.e., biomanufactured). Bio-derived or bio-based products or materials are simply referred to here as “biomaterials”. Importantly, these concepts are independent from bioinspired materials, which are characterized by the application of biological design rules and principles by material scientists during synthetic (nano)material synthesis (Figure 12).
Figure 12.
Diagram showing concepts and examples of bio-based materials from plants. Interdisciplinary gradient connects biological, engineering, medical, physical, and material sciences. Bioproducts are the building blocks of bio-derived materials, which can be manufactured outside (chemically) or inside (biologically) an organism through biomanufacturing. Each class is color coded to its major constituents (organic, inorganic, and/or living cells) with examples in plants (dark green) and elsewhere (light gray). CNC: cellulose nanocrystals; 3HP: 3 hydroxy-propionic acid; PAA: polyacrylic acid; PHA: polyhydroxyalkanoate.
Bioproducts come from organic, inorganic, and/or living cell sources, which can be “mixed and matched” to promote composites with a diverse set of properties and modularity often unavailable to chemists and material scientists [273]. Plants are a prime source for various bioproducts, including (1) biopolymers such as cellulose, lignin, and derivatives; (2) extractives such as latex, starch, and fatty acids (e.g., polyhydroxyalkanoates); (3) small molecules such as phenylacetic acid and muconic acid, which often undergo additional downstream chemical, enzymatic, and/or thermal processing for commercial applications; (4) inorganic biominerals such as phytoliths; and (5) organic-inorganic composites such as calcium oxalate crystals and calcium carbonate. Plant-derived bioproducts can be used for fiber, bioplastics, liquid crystals, energy storage, and insulants [274–283]. Their applications span various fields, including medicine, engineering, and material sciences, representing complementary and replaceable alternatives to bioproducts and biomaterials from animal origin, which can draw environmental and ethical concerns.
One beneficial template example for biomanufacturing is the composite of calcium carbonate (CaCO3), which is a plant crystal equivalent to nacre (mother-of-pearl) in composition. However, their multifactorial architecture is different, with plant crystals being good insulators and nacre having unique mechanical and optical properties. Exploring plant biosystems design with multidisciplinary tools could enhance our predictive power to modify such plant crystals, programming a set of characteristics to solve urgent energy, engineering, and environmental problems [284–289].
Another example, in biomaterials fabrication, is the bioengineering and use of cellulose nanocrystals (CNC) and nanofibrils (CNF). Structural and functional properties of CNFs can be influenced by plant cellulose properties, such as crystallinity and interfacial binding with matrix polysaccharides [290]. The relevance of biologically synthesized 18-(glucan) chain cellulose microfibril structure on CNF properties [290] opens up the prospect of leveraging the biosynthetic processes in plants. Potential biosystems design strategies for varying the chain length and crystallinity of cellulose include altering the distribution and composition of cellulose synthase (CesA) complexes (CSCs), which have been proposed to be composed of 18 CesA proteins [291], to potentially optimize cellulose-derived nanocellulose and nanocellulosic composites. Uses of conventional approaches to enhance cellulose content via overexpression of single secondary wall-associated CesA types have generally not resulted in increased cellulose content in woody plants due to challenges with effective transgene expression in the presence of endogenous copy [292] or varying CesA stoichiometry needed for functional CSCs [293]. Use of biosystems design approaches will aid in understanding the extent to which domain swapping, modified CSC composition, heterologous CesA expression, and use of optimized promoters can impact content, crystallinity, degree of polymerization, and crosslinking properties of cellulose.
In use of biomaterials in clinical contexts, scaffolds of animal origin can lead to variability and environmental and ethical concerns, which can be potentially addressed using animal-free scaffolds such as bioproducts of plant origin [278]. Due to the natural strength of plant and marine algae-derived bioproducts (e.g., nanocellulose and alginate) and their functional roles as scaffolds for growth, plant-derived products, or materials are highly promising as bioinks for printing of novel biomaterials with applications in drug delivery, wound healing, and implantable medical devices [279].
Overall, biosystems design can be a powerful aid in accelerating the research and development of plant-derived bioproducts. Although plants have been the source of biomaterials for a long time, the recent need for petroleum-independent products, associated with the tremendous opportunities and potential for developing renewable and better performing products from biomaterials, has accelerated studies in biomanufacturing [286]. Also, in vivo biomanufacturing provides us with materials having characteristics unable to be reproduced by chemistry alone [273]. However, we still lack understanding of the “material loci” in plants controlling the synthesis, transport, modification, assembly, and storage of biomaterials. In addition, there has been limited exploration of chemical composition, ultrastructure, and bonding within/across interfaces in hybrid biomaterials, which often have appealing physical, optical, and electromagnetic properties. Biosystems design approaches will be required for leveraging such needs as well as recruiting and integrating emerging theoretical, computational, and in situ characterization tools to establish a knowledge toolbox and bridge gaps between disciplines, accelerating the overall biomaterial cycle “design-discover, synthesize, characterize, learn and apply” (DiSCLA). For example, a biosystems design approach has been proposed to reconfigure plant metabolism for cost-effective production of biodegradable plastic [294]. One challenge of this approach is how to minimize the negative impact of biodesign for bio-derived products and materials on plant growth performance both above and below ground [264, 294].
4.2.6. Plant Biosystems Design for Bioenergy Production
The interest in biofuels has largely shifted from bioethanol to drop-in advanced fuels due to the increasing popularity of electric automobiles, which has shifted the major potential for biofuels into aviation, where the use of heavy batteries remains unlikely to become economically feasible. Because energy density is a key consideration for aviation fuels, drop-in fuels are more promising than bioethanol [295]. Current bioethanol production suffers from a number of problems such as relatively small lifecycle reductions in greenhouse gas emissions [296] and a competition with food production that raises the price of staple foodstuffs [297, 298]. In order to overcome these drawbacks, bioenergy crops should be engineered to grow with fewer inputs, on marginal land, or with valuable coproducts such as food, medicine, or industrial chemicals [299]. Biomass feedstocks (e.g., lignocellulose, starches, and lipids) can be converted into jet fuels via several different routes, such as oil to jet fuels (OTJ), syngas to jet fuels (STJ), and alcohol to jet fuels (ATJ) [300]. Currently, the cost of biomass-based jet fuels is relatively high (e.g., 4.4 to 5.1 $/gal from the OTJ route), which can be offset by generating high-value coproducts from the biomass feedstock [300]. Therefore, it is important to design the metabolic pathways in plants for optimizing biomass feedstock for production of both jet fuels and value-added coproducts. Previous efforts have discovered a lot of genes relevant to yield and quality of biomass in multiple bioenergy crops. Biosystems design provides an excellent opportunity for combining the improved traits conferred by individual genes to optimize the performance of bioenergy crops, with simultaneous improvement of biomass quality (e.g., high cellulose content and low recalcitrance to deconstruction) and biomass accumulation under both normal and stress conditions.
Typically, plants have a low photosynthetic efficiency, converting less than 1% of the available sunlight to stored chemical energy [301], which limits the economic feasibility of plant biomass as feedstock for biofuels production [302]. Therefore, it is critical to increase the photosynthetic efficiency of bioenergy crops using biosystems design approaches, as described in Section 4.2.1. In general, the stem of woody bioenergy crops (e.g., poplar) is used for biofuels production while the leaves are discarded as waste. To increase the economic value of woody bioenergy crops, their leaves can be used as bioreactors to produce high-value bio-based products (e.g., biodegradable plastics and specialty or commodity chemicals) using the strategies described in Section 4.2.5 as well as medicine using synthetic biology approaches described in Section 4.2.8. Furthermore, the below-ground tissue of bioenergy crops can be optimized for long-term carbon storage using biosystems design to mitigate climate change (see Section 4.2.4).
4.2.7. Plant Biosystems Design for Phytoremediation and Phytomining
Pollution by heavy metals, which cannot be chemically degraded, poses a serious long-term threat to the environment and human health [303]. Some plants, called hyperaccumulators, can accumulate metal and metalloid trace elements (e.g., nickel, zinc, cadmium, manganese, arsenic, and selenium) to extraordinarily high concentrations in their above-ground living biomass [304]. There are 721 plant species identified as hyperaccumulators [305], and some of the hyperaccumulator plants can take metals up to 1-2% of total dry weight, which may be hundreds or thousands of times greater than commonly grown plants [305, 306]. In comparison with different physical and chemical methods of extracting heavy metals, use of hyperaccumulator plants is perceived as a green, low-cost, and efficient approach [307]. Hyperaccumulator plants can be utilized for phytoremediation to clean up the soils contaminated by heavy metals and/or for phytomining to recover an economic amount of metals (e.g., nickel) from the plants [303, 308].
Comparative analyses of hyperaccumulators and closely related nonhyperaccumulators have improved the understanding of molecular mechanisms of heavy metal uptake, transport, sequestration, and tolerance in hyperaccumulators. Particularly, functional characterization of heavy metal transporters, such as Zinc-regulated transporter Iron-regulated transporter Proteins (ZIP), Heavy Metal transporting ATPases (HMA), Multidrug And Toxin Efflux (MATE), and Metal Transporter Proteins (MTP) gene families, has yielded valuable gene resources for designing and engineering more effective phytoremediation systems [306, 309]. Overexpression of such transporters has been widely successful in enhancing the uptake of heavy metals in model plants or nonhyperaccumulators [306]. However, due to the complexity of plant metal transporting and trafficking systems, a much more sophisticated design of plant biosystems will be required to enhance the capability of phytoremediation.
A nascent area of interest is the role of plants in accumulation of rare earth elements (REEs), which are critical materials with unique light, catalytic, and magnetic activities but do not have reliable supply chains. Plants generally have low concentrations of rare earth elements, which in part reflect diffuse distribution and low concentration of rare elements in soil. However, certain types of plant-like ferns and citrus trees have been reported to have a higher capacity for accumulating rare earth elements [310–312]. Future research on phytomining can focus on the understanding of the molecular mechanism underlying REE accumulation in hyperaccumulating plants and then transfer the REE-accumulating mechanism into the existing crop plants using biosystems design, which may involve engineering of transporters and metabolic pathways.
Biosystems design research for phytoremediation and phytomining can be focused on the following aspects: (1) due to the narrow distribution and low biomass yield of hyperaccumulator plants [305], it is necessary to extend the geographic distribution and biomass yield of existing hyperaccumulators through targeted mutation or gene circuit design; (2) it is important to enhance the uptake and tolerance of heavy metals in nonhyperaccumulators using a biosystems design approach; (3) because hyperaccumulation of lead, copper, cobalt, chromium, and thallium has not been well established in natural plant systems yet [304], it is urgent to either find natural or synthetic genes and pathways for accumulating these heavy metals through systems biology research, in silico modeling, and metal transporter protein engineering; and (4) comparative cross-species studies can unravel the fundamental pathways unique to hyperaccumulators of rare earth elements and present avenues for potential biosystems design approaches in deployable plant species.
4.2.8. Plant Biosystems Design for Medicine Production and Medical Research
Plants have been the primary production chassis for medicine for millennia and continue to play an important role in modern supply chains. Plants are the source of approximately 25% of modern drugs and in many cases remain the most cost effective method for their production [313]. Examples include the antimalarial drug artemisinin [314] and the anesthetic morphine [315]. Many of these high-value medicinal compounds are produced from nonmodel plants, but plant engineering efforts have nonetheless been successful in improving their yield [316], which remains economically competitive with chemical and microbial synthesis [317].
Plants have been used as bioreactors to produce vaccines, such as anticancer or viral vaccines [318]. Genetically engineered plants can produce recombinant proteins at larger scale than conventional platforms and are on track to become cost competitive with conventional production platforms. In fact, plants are being used as a platform to produce a wide range of antibodies in different organs (leaves, roots, seeds, tubers, fruits) of various plant species, such as tobacco, potato, rice, tomato, and pea [319–322]. This has been possible due to rapid improvement in plant genetic engineering and transformation technologies. There are several benefits for the use of plants to produce therapeutic antibodies. First, it reduces the production cost. Plants can be used to produce recombinant proteins at 0.1% and 2–10% of the cost of mammalian cell cultures and microbial fermentation systems, respectively [323]. Second, plants are usually regarded as safe systems for antibody production because they do not harbor mammalian pathogens or produce endotoxin. Finally, production of antibodies in edible tissue will allow convenient, needle-free oral immunization at the gastric mucosal surface [320].
There are several examples of successful use of a plant-based antibody to treat human diseases. One such case is that the secretory antibody “CaroRxTM” derived from tobacco leaves was used to treat Ebola patients during outbreaks of this virus in Africa in 2014 [324]. Other plant-made antibody products that are currently being used to cure human diseases are “DoxoRxTM” for treating drug-induced alopecia, a common side effect of cancer therapy and RhinoRxTM for treating the common cold [320, 325]. Another outstanding example of plant-based molecular pharming to produce biopharmaceuticals is the production of vaccine against the SARS-CoV-2 virus. A biotech company called Medicago Inc. (Quebec City, Canada) is using plants to produce virus-like particles (VLPs), which is the first step in developing a vaccine against COVID-19 before preclinical testing for safety and efficacy. VLPs are the noninfectious viral proteins that lack the key genetic materials for infection. However, they are still recognized by the immune system and therefore can be used to produce antibodies against the SARS-CoV-2. A similar effort is also underway by a US-based company iBio, Inc.
Plants can also be used to produce reagents for detecting human pathogen. For example, plants can be used to generate diagnostic reagents for COVID-19 in multiple ways: (1) generating positive control reagents for RT-PCR detection of SARS-CoV-2 virus by producing artificial RNA containing the target virus genomic regions which are packed inside the VLPs derived from Cowpea mosaic virus (CPMV), (2) generating antibodies for detecting the spike (S) protein of SARS-CoV-2, and (3) generating recombinant proteins for detecting antibodies against SARS-CoV-2 to identify people who are currently infected or recovering from infection [326].
In addition to serving as chassis for producing medicinal compounds and biologics, plants are emerging as a robust discovery and functional validation platform to elucidate the genetic basis of heritable human diseases including cancers and developmental abnormalities. Plants and humans share a common eukaryotic ancestry represented by evolution of the first complex and multicellular organism. Evolution of this complex life form necessitated the emergence of genetic mechanisms to coordinate DNA replication-repair, cell division, signaling between neighboring cells, and their adhesion to facilitate hierarchical assembly into tissues and organs with specialized functions [327]. Given this shared ancestry, proteins underlying basal processes such as DNA replication-repair, cell division, and cell adhesion remain highly conserved across disparate eukaryotes. Additionally, such conservation at the protein level has been shown to result in orthologous phenotypes (or phenologs) across divergent eukaryotes including Arabidopsis, humans, and mice [328], suggesting that comparative studies across eukaryotes have the potential to identify critical amino acid motifs mediating function of these conserved proteins.
Although significant progress has been made in linking mutations to disease outcomes in human populations using genome-wide association studies (GWAS), pin-pointing causal variants that can be used as reliable biomarkers in disease diagnosis remains a major challenge. This is largely due to the fact that heritable diseases are often extremely rare in human populations which limits the ability to acquire sufficient sample sizes for robust statistical associations [270]. In contrast, long-lived perennial plants like poplar (Populus spp.) exhibit wide ranges of phenotypic variation that is underpinned by their ability to maintain diverse sequence variants, including high impact loss-of-function mutations, at surprisingly high levels [329]. As such, high-resolution GWAS in plants often require fewer samples and can precisely identify causal mutations underlying phenotypic expression. For example, using ~300 individual poplar plants, Tuskan et al. [330] demonstrated that shared homology at the protein-level was manifested as an orthologous cell-proliferation phenotype between the poplar and humans. Specifically, they found significant similarities between genes implicated in callus formation in poplar and tumorigenesis in humans. Callus formation, which is the rapid growth of undifferentiated cell masses in plants, is orthologous to the uncontrolled cell proliferation during tumorigenesis in humans. The rate of callus formation was significantly associated with loss-of-function mutations occurring within a poplar SOK1 kinase related to the Mammalian Sterile-20 kinase, which has been shown to function in tumor suppression in humans [331]. In a separate study, Bdeir et al. [332] identified sequence variants in a desmosome protein, which were associated with adhesion of bark tissue in poplar. A functional variant of the protein prevented bark abscission resulting in annual accumulation of bark layers. In humans and mice, orthologous function of the same protein has been implicated in onset of the extremely rare skin cancer, keratoderma, which is manifested as an abnormal accumulation skin layer [333].
Given these unique advantages in precise genetic mapping of causal variants in addition to ease of experimental manipulation as well as fewer ethical issues, plants offer an attractive discovery and validation platform for understanding how mutations modulate the function of proteins that are fundamentally conserved across eukaryotes to provide highly resolved therapeutic targets for treatment of heritable human diseases in the rapidly expanding precision medicine field [334].
4.2.9. Plant Biosystems Design for Biosentinel
Biosensors are defined as molecules, organisms, or devices in a biological context that emit quantifiable signals in response to specific molecules or biological processes [335]. Plants have rapid (within the seconds to minutes time-scale) responses to various biotic and abiotic stimuli [336]. Therefore, inducible expression of reporter genes, such as the genes encoding fluorescent protein (e.g., GFP) and pigments (e.g., anthocyanin and chlorophyll), could be engineered to detect environmental stimuli. For example, a resettable synthetic degreening plant-based biosensor (also called phytosensor) system was successfully created, in which plants lost their chlorophyll from induction of the degreening circuit by a synthetic steroid (4-hydroxytamoxifen, 4-OHT), and regreened after the inducer was removed [337]. Also, a synthetic signal transduction pathway was constructed for detecting the nitroaromatic explosive 2,4,6-trinitrotoluene (TNT) [338].
4.2.10. Plant Biosystems Design for Tissue Engineering
Synthetic morphogenesis enabled by synthetic biology can be achieved for the de novo generation of programmable tissues and organs [339]. Three-dimensional microstructure has recently been created from single plant cells in vitro by mimicking the plant tissue environment and using biocompatible scaffolds similar to those used in mammalian tissue engineering, with the scaffolds providing both developmental cues and structural stability to isolated callus-derived cells grown in liquid culture [340, 341]. Furthermore, one interesting science-fiction-like question is: Can plants be genetically reprogrammed to form new-to-nature structures useful for human life? For example, it has been envisioned that a tree could be potentially reprogrammed to grow into a fully functional house based on the genetic instructions designed by scientists [342].
4.2.11. Plant Biosystems Design for Space Exploration
There are growing interests and ongoing plans for human to travel to Mars in the near future, and plants on Earth need to be redesigned for meeting the needs of humans living in the Martian environment [343]. The targets include increasing drought-resistance to allow for plant growth under water scarcity in an extraterrestrial environment [344], cold tolerance, nutrient-utilization efficiency, and adjusting photosynthetic tuning to optimize for the lower Martian light intensity [343]. For practical reasons, it would be better to engineer plants for survival and yield in a space station and protected facilities on the surface of other celestial bodies (e.g., controlled environment like growth chambers), rather than in the open space on the surfaces of those bodies. The Martian surface temperatures are generally between -60 and 0°C and are not suitable to plant growth without radical engineering out of reach of current technology.
5. Social Responsibility of Plant Biosystems Design
Biosystems design is highly powerful for plant science research with enormous promise and needed caution. While plant biosystems design has great potential for genetic improvement of crop plants or creation of synthetic plant genomes for the benefit of our society, this new research discipline has a huge social responsibility to ensure biosafety and address the potential ethical issues.
5.1. Safety of Plant Biosystems Design
Engineered plants show enormous potential and have been repeatedly shown to be safe by a large number of scientific organizations, including the World Health Organization (https://www.who.int/health-topics/food-genetically-modified#tab=tab_2) and the American Association for the Advancement of Science (https://www.aaas.org/news/statement-aaas-board-directors-labeling-genetically-modified-foods). The Society of American Foresters supports and encourages the use of appropriately regulated Genetically Modified Organisms (GMOs) (https://www.eforester.org/Main/Issues_and_Advocacy/Statements/Regulation_of_Genetically_Modified_Trees.aspx). Nonetheless, plant biotechnology still presents some potential risks that must be considered during plant biosystems design research. These risks can be grouped into six broad categories: nutritional nonequivalence, potential allergenicity, escape of noxious transgenes, creation of resistant weeds and pathogens, unintended changes, and disruption of ecosystem function. Strategies are available to reduce each of these risks but must be implemented through the design, testing, and implementation phases of plant biosystems design in order to effectively manage the risks. While there is a great concern about the potential safety issues caused by transgenes, the risks of not using transgenes have been ignored. It is critical to evaluate the risks of using vs. not using transgenes. For transgenes which are not noxious, their risks can be considered very low if there are no unintended changes (e.g., side effects and off-target effects) caused by the transgenes. Risk analyses should focus on the engineered plant products, not the process through which they are created (e.g., CRISPR/Cas-based gene editing vs. transgenesis). Therefore, genome-edited crops are not regulated as GMOs in more and more countries since genetic variants created through genome editing are indistinguishable from naturally evolved variants [345], although they are still categorized as GMOs in Europe, with concerns on unintended effects (e.g., off-target effects, unintended on-target effects, and other unintended consequences) [346] (see Section 5.1.5 for more details).
5.1.1. Nutritional Nonequivalence
Nutritional nonequivalence is frequently a design feature for transgenic crops, as in the case of biofortified cereal grains [347, 348]. However, there is always the possibility during biosystem engineering of unintended changes to the crop product metabolome, which may result in lower nutritional quality. Careful quantification of compounds metabolically linked to any plant biodesign changes should be carried out under a variety of field conditions to test for changes to nutritional content or production of harmful levels of unintentional off-target metabolites. Flux analysis is a biosystems design tool that can guide the search for relevant metabolites to be analyzed [349].
5.1.2. Potential Allergenicity
Introduced allergenicity is another risk that arises naturally from a design feature. If genes encoding allergenic proteins are introduced into a plant, the resulting engineered plant is likely to act as an allergen. The generation of allergenic plants occurred early in the history of transgenic plant production with soybeans encoding the 2S albumin protein from Brazil nut [350]. Allergies to Brazil nut 2S albumin are relatively common and often severe, so it is likely that introduction of the allergenic soybean to the human food supply would have harmed some consumers, leading to a public relation disaster. Fortunately, the product was never brought to market, but this work acts as a reminder of the seriousness of this issue [351]. All transgenic proteins introduced to crops should be rigorously tested with a library of immune cells and serum samples to ensure no known or unknown allergens are being added to crops.
5.1.3. Potential Escape of Noxious Transgenes
Engineered plants containing transgenes are broadly useful for agriculture, with transgenes intended to remain within the specific cultivars for which they are designed. However, multiple instances have been documented of “transgene escape,” which occurs when transgenes are transferred to other crop cultivars or wild relatives [352, 353]. Transgene escape is widely discussed by opponents of plant biotechnology and could potentially interfere with wild gene pools, generating agronomic problems such as herbicide resistant weeds. Steps should therefore be taken to reduce transgene escape rates. Such methods include the use of transgenesis in the plastid genome rather than the plant nuclear genome, resulting in maternal rather than biparental inheritance [354], self-limiting genes that result in eventual sterilization of the plants [355], male sterility [356], and bisexual sterility [357].
5.1.4. Emergence of Herbicide and Pesticide Resistance
Agricultural fields are usually designed with extremely high densities of genetically similar plants and high levels of nutrients, which are ideal conditions for the spread of disease, parasitism, or opportunistic growth by undesired plants. In the ongoing battle against pathogens, pests, and weeds, new tools tend to be effective for a length of time before evolution of the undesired organisms renders the tool useless [241, 358]. The situation is analogous to the problem of antibiotic resistance in medicine, and the balance will likely continue until the elimination of agriculture or the elimination of the undesired pests, pathogens, and weeds. In some cases, transgene escape could lead to evolution of resistance to existing tools, but even if that case is prevented, evolution will take its course, and organisms will develop resistance to the current agricultural defenses. Agronomic and biotechnological steps can be taken to delay the evolution of resistance including the use of multiple independent forms of defense simultaneously (e.g., several distinct herbicides and herbivory deterrents) and the planting of undefended “refuges” to reduce the selective pressure for undesired organisms to evolve resistance to the crop defense chemical or engineered trait [359].
5.1.5. Unintended Changes
CRISPR/Cas-mediated genome editing has been widely used for genetic improvement of plants. However, there are concerns about potential, unintended genetic modifications caused by gene editing due to the off-target effects. Although off-target mutations in CRISPR/Cas9-edited plants can be negligible and at a level lower than inherent natural variation when highly specific gRNAs are used [360], it would be prudent to carry out risk analysis for practical application or commercialization of genome-edited plants [361, 362]. For example, a recent study assessed, in addition to off-target mutations, potential epigenetic changes attributable to CRISPR-mediated genome editing, reporting no detectable changes on DNA methylation status in edited plants [363].
5.1.6. Disruption of Ecosystem Function
While uncommon, there is a risk that transgenic plants have negative consequences on natural ecosystems. A classic example is the case of plants transformed with the genetic material from the bacterium Bacillus thuringiensis (Bt) and the monarch butterfly, in which a substantial research effort led by the US Department of Agriculture occurred following reports that monarch larvae had a decrease in survival following feedings on milkweed dusted with Bt pollen [364]. Although it was determined that only a very small portion of adjacent milkweed plants accumulate pollen in sufficient concentration to have a negative effect, it is important to recognize that there may be ecological risks associated with the release of transgenic plants.
5.2. Ethics of Plant Biosystems Design
As an emerging cutting-edge discipline, plant biosystems design should meet the following ethic principles: public beneficence, responsible stewardship, intellectual freedom and responsibility, democratic deliberation, and justice and fairness [365].
5.2.1. Ethical Issues of Plant Biosystems Design
Plant biosystems design and plant biotechnology show great promise in improving environmental and human health. Continued funding for applied plant science is critical to ensure this promise is fulfilled, especially for cases without obvious commercial interests such as design of biofortified food for poor consumers in the developing world. Since national research organizations provide a substantial fraction of the funding for plant biosystems design research, there is a moral obligation to ensure that the benefits of the products are shared by as many people as possible.
5.2.2. Solutions to Address the Ethical Issues
A well-known genetically engineered crop is Golden Rice, a widely celebrated biofortified crop that resulted from a major public-private collaboration [366]. The product was designed to benefit the global poor, and the licensing agreements regarding the underlying intellectual property are exemplary: while large scale farmers in Western countries must pay for use of the product, Golden Rice is free for those who need it, specifically breeding programs and smallholder farmers (http://www.goldenrice.org/Content1-Who/who4_IP.php). Such licensing arrangements can simultaneously give companies the required economic incentives to produce products and democratize access to those products to the global poor who stand to benefit most.
6. Conclusion
As an emerging interdisciplinary research field, plant biosystems design shows massive potential for not only increasing our understanding of the mechanisms underlying the biological complexity of plant organisms but also accelerating the domestication of crop plants or creation of novel plant organisms to address challenges related to food and energy security, environmental sustainability, and human health. In this roadmap, we discuss the principles, methods, applications, and social responsibility of plant biosystems design. Currently, the theories and principles of plant biosystems design are only partially understood, and the knowledge gaps are expected to be filled through systems biology research and the DBTL process(es) in the future. Significant progress has already been made in method development for plant biosystems design, such as efficient assembly of DNA parts, high-precision gene editing, enhancement of plant transformation using morphogenic regulators, and virus/nanotube-mediated in planta transformation. Still, new technologies will need to be developed for enabling large-scale genome refactoring or construction of functional synthetic plant genomes. Exciting achievements have been made in the application of biosystems design to pathway engineering for improving agricultural crops, yet the potential of biosystems design needs to be exploited in many other aspects, such as bio-based materials, climate change mitigation, bioenergy production, phytosensor, tissue engineering, and space exploration. The potential of new machine learning capabilities (e.g., explainable artificial intelligence) could be exploited for predictive learning from big genomics and phenomics data. Also, plant biosystems design will need to be deployed for a new type of high precision agriculture and bioeconomy as a contribution to the fourth industrial revolution, in which genetic engineering plays an important role [367–369]. While it will be very important to achieve the scientific and technological advancements in plant biosystems design, we should pay special attention to its social responsibility (i.e., biosafety and ethics) to improve the public perception and acceptance of this new research discipline as benefiting humanity and the environment. In particular, there is a need to engage private and academic stakeholders to bring all these technologies to the poorest populations. Finally, plant biosystems design is much more challenging than traditional genetic engineering and therefore needs extensive interdisciplinary collaborations among many researchers around the world. We hope that the three computational modules in the “Plant Biodesign Hub” (Figures 6, 7, and 9) outlined in this roadmap will serve as a major public platform for national and international collaborations to realize the great promise of plant biosystems design for the future of our global society.
Acknowledgments
The authors are grateful to C.Y. Ralston and R. Evans (Lawrence Berkeley National Laboratory, Berkeley, CA, USA) for their suggestions to the manuscript. The writing of this manuscript was supported by the Center for Bioenergy Innovation, a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Biological and Environmental Research (BER) program, the Laboratory Directed Research and Development program of Oak Ridge National Laboratory, and the U.S. DOE BER Genomic Science Program, as part of the Secure Ecosystem Engineering and Design Scientific Focus Area and the Plant-Microbe Interfaces Scientific Focus Area. YY is supported by NSF Plant Genome Research Project Grant (1740874) and the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project PEN04659 and Accession #1016432. HY is supported by Nonprofit Research Projects (CAFYBB2018ZY001-1) of Chinese Academy of Forestry. CTT acknowledges the financial support from the NSF CAREER award (NSF#1553250) and the DOE BER Genomic Science Program (DE-SC0019412). PMS acknowledges support from the Joint BioEnergy Institute which is supported by the U.S. DOE Office of Science, BER program under Contract No. DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. DL acknowledges financial support through the National Science Foundation (NSF) under Award Number 1833402. AJM acknowledges financial support from the UK Biotechnology and Biological Sciences Research Council (grants BB/M006468/1 and BB/S015531/1) and the Leverhulme Trust (grant RPG-2017-402).
Disclosure
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Authors’ Contributions
XY planned and drafted the manuscript. All authors read and contributed to the content, edited, or reviewed it.
References
- 1.Knapp S., “People and plants: the unbreakable bond,” Plants, People, Planet, vol. 1, no. 1, pp. 20–26, 2019 [Google Scholar]
- 2.Hiscock S. J., Wilkin P., Lennon S., and Young B., “Plants matter: introducing plants, people, planet,” Plants, People, Planet, vol. 1, no. 1, pp. 2–4, 2019 [Google Scholar]
- 3.Chen H.-Z., and Liu Z.-H., “Steam explosion and its combinatorial pretreatment refining technology of plant biomass to bio-based products,” Biotechnology Journal, vol. 10, no. 6, pp. 866–885, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Austin A. T., and Zanne A. E., “Whether in life or in death: fresh perspectives on how plants affect biogeochemical cycling,” Journal of Ecology, vol. 103, no. 6, pp. 1367–1371, 2015 [Google Scholar]
- 5.Dusenge M. E., Duarte A. G., and Way D. A., “Plant carbon metabolism and climate change: elevated CO2 and temperature impacts on photosynthesis, photorespiration and respiration,” New Phytologist, vol. 221, no. 1, pp. 32–49, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Christenhusz M. J. M., and Byng j. W., “The number of known plants species in the world and its annual increase,” Phytotaxa, vol. 261, no. 3, pp. 201–217, 2016 [Google Scholar]
- 7.Long S. P., Marshall-Colon A., and Zhu X.-G., “Meeting the global food demand of the future by engineering crop photosynthesis and yield potential,” Cell, vol. 161, no. 1, pp. 56–66, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Bailey-Serres J., Parker J. E., Ainsworth E. A., Oldroyd G. E. D., and Schroeder J. I., “Genetic strategies for improving crop yields,” Nature, vol. 575, no. 7781, pp. 109–118, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FAOFood and Agriculture Organization of the United Nations, Rome, 2018, p. 60, 2018
- 10.Myers S. S., Smith M. R., Guth S., Golden C. D., Vaitla B., Mueller N. D., Dangour A. D., and Huybers P., “Climate change and global food systems: potential impacts on food security and undernutrition,” Annual Review of Public Health, vol. 38, no. 1, pp. 259–277, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Pavlopoulos G. A., Secrier M., Moschopoulos C. N., Soldatos T. G., Kossida S., Aerts J., Schneider R., and Bagos P. G., “Using graph theory to analyze biological networks,” BioData Mining, vol. 4, no. 1, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson E., MacNeil L. T., Arda H. E., Zhu L. J., and Walhout A. J. M., “Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response,” Cell, vol. 153, no. 1, pp. 253–266, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinschen M. M., Ivanisevic J., Giera M., and Siuzdak G., “Identification of bioactive metabolites using activity metabolomics,” Nature Reviews. Molecular Cell Biology, vol. 20, no. 6, pp. 353–367, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He H., Van Breusegem F., and Mhamdi A., “Redox-dependent control of nuclear transcription in plants,” Journal of Experimental Botany, vol. 69, no. 14, pp. 3359–3372, 2018 [DOI] [PubMed] [Google Scholar]
- 15.Shen Y., Issakidis-Bourguet E., and Zhou D.-X., “Perspectives on the interactions between metabolism, redox, and epigenetics in plants,” Journal of Experimental Botany, vol. 67, no. 18, pp. 5291–5300, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Duran-Nebreda S., and Bassel G. W., “Plant behaviour in response to the environment: information processing in the solid state,” Philosophical Transactions of the Royal Society B: Biological Sciences, vol. 374, no. 1774, article 20180370, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassel G. W., “Information processing and distributed computation in plant organs,” Trends in Plant Science, vol. 23, no. 11, pp. 994–1005, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Wong E., Baur B., Quader S., and Huang C. H., “Biological network motif detection: principles and practice,” Briefings in Bioinformatics, vol. 13, no. 2, pp. 202–215, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorochowski T. E., Grierson C. S., and di Bernardo M., “Organization of feed-forward loop motifs reveals architectural principles in natural and engineered networks,” Science Advances, vol. 4, no. 3, article eaap9751, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta P., and Singh S. K.. Molecular Approaches in Plant Biology and Environmental Challenges, Singh S. P., Upadhyay S. K., Pandey A., and Kumar S., Eds., Springer Singapore, Singapore, 2019 [Google Scholar]
- 21.Jin G.Encyclopedia of Systems Biology, Dubitzky W., Wolkenhauer O., Cho K.-H., and Yokota H., Eds., Springer, New York, NY, 2013 [Google Scholar]
- 22.Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., and Picotti P., “A map of protein-metabolite interactions reveals principles of chemical communication,” Cell, vol. 172, no. 1-2, pp. 358–372.e23, 2018 [DOI] [PubMed] [Google Scholar]
- 23.Erbilgin O., Rübel O., Louie K. B., Trinh M., Raad M. ., Wildish T., Udwary D., Hoover C., Deutsch S., Northen T. R., and Bowen B. P., “MAGI: a method for metabolite annotation and gene integration,” ACS Chemical Biology, vol. 14, no. 4, pp. 704–714, 2019 [DOI] [PubMed] [Google Scholar]
- 24.Thiele I., and Palsson B. Ø., “A protocol for generating a high-quality genome-scale metabolic reconstruction,” Nature Protocols, vol. 5, no. 1, pp. 93–121, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masakapalli S. K., Kruger N. J., and Ratcliffe R. G., “The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a complex response to changes in nitrogen supply,” Plant Journal, vol. 74, no. 4, pp. 569–582, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Williams T. C. R., Sweetlove L. J., and Ratcliffe R. G., “Capturing metabolite channeling in metabolic flux phenotypes,” Plant Physiology, vol. 157, no. 3, pp. 981–984, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen D. K., “Quantifying plant phenotypes with isotopic labeling & metabolic flux analysis,” Current Opinion in Biotechnology, vol. 37, pp. 45–52, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Poolman M. G., Miguet L., Sweetlove L. J., and Fell D. A., “A genome-scale metabolic model of Arabidopsis and some of its properties,” Plant Physiology, vol. 151, no. 3, pp. 1570–1581, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Oliveira Dal'Molin C. G., Quek L.-E., Palfreyman R. W., Brumbley S. M., and Nielsen L. K., “AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis,” Plant Physiology, vol. 152, no. 2, pp. 579–589, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark T. J., Guo L., Morgan J., and Schwender J., “Modeling plant metabolism: from network reconstruction to mechanistic models,” Annual Review of Plant Biology, vol. 71, no. 1, pp. 303–326, 2020 [DOI] [PubMed] [Google Scholar]
- 31.McCloskey D., Palsson B. Ø., and Feist A. M., “Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli,” Molecular Systems Biology, vol. 9, no. 1, p. 661, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberhardt M. A., Palsson B. Ø., and Papin J. A., “Applications of genome-scale metabolic reconstructions,” Molecular Systems Biology, vol. 5, no. 1, p. 320, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price N. D., Reed J. L., and Palsson B. O., “Genome-scale models of microbial cells: evaluating the consequences of constraints,” Nature Reviews Microbiology, vol. 2, no. 11, pp. 886–897, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Trinh C. T., Wlaschin A., and Srienc F., “Elementary mode analysis: a useful metabolic pathway analysis tool for characterizing cellular metabolism,” Applied Microbiology and Biotechnology, vol. 81, no. 5, pp. 813–826, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Notebaart R. A., Szappanos B., Kintses B., Pal F., Gyorkei A., Bogos B., Lazar V., Spohn R., Csorg B., Wagner A., Ruppin E., Pal C., and Papp B., “Network-level architecture and the evolutionary potential of underground metabolism,” Proceedings of the National Academy of Sciences, vol. 111, no. 32, pp. 11762–11767, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Y., Widmer A., and Karrenberg S., “The roles of genetic drift and natural selection in quantitative trait divergence along an altitudinal gradient in Arabidopsis thaliana,” Heredity, vol. 114, no. 2, pp. 220–228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pigliucci M., and Muller G.. Evolution–The Extended Synthesis, The MIT Press, Cambridge, Massachusetts, 2010 [Google Scholar]
- 38.Gould S. J., “The exaptive excellence of spandrels as a term and prototype,” Proceedings of the National Academy of Sciences, vol. 94, no. 20, pp. 10750–10755, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould S. J., and Lewontin R. C., “The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme,” Proceedings of the Royal Society B: Biological Sciences, vol. 205, no. 1161, pp. 581–598, 1979 [DOI] [PubMed] [Google Scholar]
- 40.Weng M.-L., Becker C., Hildebrandt J., Neumann M., Rutter M. T., Shaw R. G., Weigel D., and Fenster C. B., “Fine-grained analysis of spontaneous mutation spectrum and frequency in Arabidopsis thaliana,” Genetics, vol. 211, no. 2, pp. 703–714, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Ji Y., Hu Y., Hu H., Jia X., Jiang M., Zhang X., Zhao L., Zhang Y., Jia Y., Qin C., Yu L., Huang J., Yang S., Hurst L. D., and Tian D., “The architecture of intra-organism mutation rate variation in plants,” PLoS Biology, vol. 17, no. 4, pp. e3000191–e3000191, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmeister B. T., Denkena J., Colomé-Tatché M., Shahryary Y., Hazarika R., Grimwood J., Mamidi S., Jenkins J., Grabowski P. P., Sreedasyam A., Shu S., Barry K., Lail K., Adam C., Lipzen A., Sorek R., Kudrna D., Talag J., Wing R., Hall D. W., Jacobsen D., Tuskan G. A., Schmutz J., Johannes F., and Schmitz R. J., “A genome assembly and the somatic genetic and epigenetic mutation rate in a wild long-lived perennial Populus trichocarpa,” Genome Biology, vol. 21, no. 1, p. 259, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ossowski S., Schneeberger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., Shaw R. G., Weigel D., and Lynch M., “The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana,” Science, vol. 327, no. 5961, pp. 92–94, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auboeuf D., “Physicochemical foundations of life that direct evolution: chance and natural selection are not evolutionary driving forces,” Life, vol. 10, no. 2, p. 7, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia S., and Trinh C. T., “Modular design: implementing proven engineering principles in biotechnology,” Biotechnology Advances, vol. 37, no. 7, article 107403, 2019 [DOI] [PubMed] [Google Scholar]
- 46.Miller T. D., and Elgard P.. Proceedings of the 13th IPS Research Seminar, Fuglsoe, Aalborg Universiy Fuglsoe, 1998 [Google Scholar]
- 47.Han J.-D. J., Bertin N., Hao T., Goldberg D. S., Berriz G. F., Zhang L. V., Dupuy D., Walhout A. J. M., Cusick M. E., Roth F. P., and Vidal M., “Evidence for dynamically organized modularity in the yeast protein–protein interaction network,” Nature, vol. 430, no. 6995, pp. 88–93, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Newman M. E. J., “Modularity and community structure in networks,” Proceedings of the National Academy of Sciences, vol. 103, no. 23, pp. 8577–8582, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravasz E., Somera A. L., Mongru D. A., Oltvai Z. N., and Barabási A.-L., “Hierarchical organization of modularity in metabolic networks,” Science, vol. 297, no. 5586, pp. 1551–1555, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Wagner G. P., Pavlicev M., and Cheverud J. M., “The road to modularity,” Nature Reviews Genetics, vol. 8, no. 12, pp. 921–931, 2007 [DOI] [PubMed] [Google Scholar]
- 51.Garcia S., and Trinh C. T., “Harnessing natural modularity of metabolism with goal attainment optimization to design a modular chassis cell for production of diverse chemicals,” ACS Synthetic Biology, vol. 9, no. 7, pp. 1665–1681, 2020 [DOI] [PubMed] [Google Scholar]
- 52.Aigner H., Wilson R. H., Bracher A., Calisse L., Bhat J. Y., Hartl F. U., and Hayer-Hartl M., “Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2,” Science, vol. 358, no. 6368, pp. 1272–1278, 2017 [DOI] [PubMed] [Google Scholar]
- 53.Guo W., Sheng J., and Feng X., “Mini-review: in vitro metabolic engineering for biomanufacturing of high-value products,” Computational and Structural Biotechnology Journal, vol. 15, pp. 161–167, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia S., and Trinh C. T., “Multiobjective strain design: a framework for modular cell engineering,” Metabolic Engineering, vol. 51, pp. 110–120, 2019 [DOI] [PubMed] [Google Scholar]
- 55.Garcia S., and Trinh C. T., “Comparison of multi-objective evolutionary algorithms to solve the modular cell design problem for novel biocatalysis,” Processes, vol. 7, no. 6, p. 361, 2019 [Google Scholar]
- 56.Kaufmann K., Pajoro A., and Angenent G. C., “Regulation of transcription in plants: mechanisms controlling developmental switches,” Nature Reviews Genetics, vol. 11, no. 12, pp. 830–842, 2010 [DOI] [PubMed] [Google Scholar]
- 57.Xiao J., Jin R., and Wagner D., “Developmental transitions: integrating environmental cues with hormonal signaling in the chromatin landscape in plants,” Genome Biology, vol. 18, no. 1, p. 88, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen D., Yan W., Fu L.-Y., and Kaufmann K., “Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana,” Nature Communications, vol. 9, no. 1, article 4534, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y., Guérard F., Hodges M., and Jossier M., “Phosphomimetic T335D mutation of hydroxypyruvate reductase 1 modifies cofactor specificity and impacts Arabidopsis growth in air,” Plant Physiology, vol. 183, no. 1, pp. 194–205, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson J., Strelkowa N., Stan G. B., Douglas T., Savulescu J., Barahona M., and Papachristodoulou A., “Engineering and ethical perspectives in synthetic biology. Rigorous, robust and predictable designs, public engagement and a modern ethical framework are vital to the continued success of synthetic biology,” EMBO Reports, vol. 13, no. 7, pp. 584–590, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trudeau D. L., Edlich-Muth C., Zarzycki J., Scheffen M., Goldsmith M., Khersonsky O., Avizemer Z., Fleishman S. J., Cotton C. A. R., Erb T. J., Tawfik D. S., and Bar-Even A., “Design and in vitro realization of carbon-conserving photorespiration,” Proceedings of the National Academy of Sciences, vol. 115, no. 49, pp. E11455–E11464, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalal J., Lopez H., Vasani N. B., Hu Z., Swift J. E., Yalamanchili R., Dvora M., Lin X., Xie D., Qu R., and Sederoff H. W., “A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa,” Biotechnology for Biofuels, vol. 8, no. 1, article 175, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitacre J. M., and Bender A., “Networked buffering: a basic mechanism for distributed robustness in complex adaptive systems,” Theoretical Biology & Medical Modelling, vol. 7, no. 1, pp. 20–20, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamada S., Yamada T., Bracke S., and Inoue M., “Upgradable design for reduction of production cost and CO2 emission - case study of a laptop computer,” Applied Mechanics and Materials, vol. 761, pp. 589–593, 2015 [Google Scholar]
- 65.M H., E K., E W., and B R., “Simple and versatile selection of Arabidopsis transformants,” Plant Cell Reports, vol. 21, no. 2, pp. 130–135, 2002 [Google Scholar]
- 66.Hu L., Li H., Qin R., Xu R., Li J., Li L., Wei P., and Yang J., “Plant phosphomannose isomerase as a selectable marker for rice transformation,” Scientific Reports, vol. 6, no. 1, article 25921, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuan Y.-C., Thiruvengadam V., Lin J. S., Liu J. H., Chen T. J., Wu H. M., Wang W. C., and Chen L. J., “Broad-specificity amino acid racemase, a novel non-antibiotic selectable marker for transgenic plants,” Plant Biotechnology Reports, vol. 12, no. 1, pp. 27–38, 2018 [Google Scholar]
- 68.Chin D. P., Shiratori I., Shimizu A., Kato K., Mii M., and Waga I., “Generation of brilliant green fluorescent petunia plants by using a new and potent fluorescent protein transgene,” Scientific Reports, vol. 8, no. 1, article 16556, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang S., Hu Y., Cheng Z., Rice J. H., Miao L., Ma J., Hewezi T., Li Y., and Gai J., “An efficient Agrobacterium-mediated soybean transformation method using green fluorescent protein as a selectable marker,” Plant Signaling & Behavior, vol. 14, no. 7, article 1612682, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polóniová Z., Jopčík M., Matušíková I., Libantová J., and Moravčíková J., “Preparation of plant transformation vector containing “Self-excision” Cre/loxP system,” Journal of Microbiology, Biotechnology and Food Sciences, vol. 2019, pp. 563–572, 2019 [Google Scholar]
- 71.Du D., Jin R., Guo J., and Zhang F., “Construction of marker-free genetically modified maize using a heat-inducible auto-excision vector,” Genes, vol. 10, no. 5, p. 374, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cody J. P., Graham N. D., Zhao C., Swyers N. C., and Birchler J. A., “Site-specific recombinase genome engineering toolkit in maize,” Plant Direct, vol. 4, article e00209, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., Yau Y.-Y., Perkins-Balding D., and Thomson J. G., “Recombinase technology: applications and possibilities,” Plant Cell Reports, vol. 30, no. 3, pp. 267–285, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pathak B. P., Pruett E., Guan H., and Srivastava V., “Utility of I-SceI and CCR5-ZFN nucleases in excising selectable marker genes from transgenic plants,” BMC Research Notes, vol. 12, no. 1, article 272, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh R. K., Sharma L., Bohra N., Anandhan S., Ruiz-May E., and Quiroz-Figueroa F. R.. Advances in Plant Transgenics: Methods and Applications, Sathishkumar R., Kumar S. R., Hema J., and Baskar V., Eds., Springer Singapore, Singapore, 2019 [Google Scholar]
- 76.Srivastava V., Underwood J. L., and Zhao S., “Dual-targeting by CRISPR/Cas9 for precise excision of transgenes from rice genome,” Plant Cell, Tissue and Organ Culture, vol. 129, no. 1, pp. 153–160, 2017 [Google Scholar]
- 77.Galanie S., Thodey K., Trenchard I. J., Filsinger Interrante M., and Smolke C. D., “Complete biosynthesis of opioids in yeast,” Science, vol. 349, no. 6252, pp. 1095–1100, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Opgenorth P., Costello Z., Okada T., Goyal G., Chen Y., Gin J., Benites V., de Raad M., Northen T. R., Deng K., Deutsch S., Baidoo E. E. K., Petzold C. J., Hillson N. J., Garcia Martin H., and Beller H. R., “Lessons from two design–build–test–learn cycles of dodecanol production in Escherichia coli aided by machine learning,” ACS Synthetic Biology, vol. 8, no. 6, pp. 1337–1351, 2019 [DOI] [PubMed] [Google Scholar]
- 79.Perez J. G., Stark J. C., and Jewett M. C., “Cell-free synthetic biology: engineering beyond the cell,” Cold Spring Harbor Perspectives in Biology, vol. 8, p. a023853, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oberortner E., Cheng J.-F., Hillson N. J., and Deutsch S., “Streamlining the design-to-build transition with build-optimization software tools,” ACS Synthetic Biology, vol. 6, pp. 485–496, 2016 [DOI] [PubMed] [Google Scholar]
- 81.Chen Y., Guenther J. M., Gin J. W., Chan L. J. G., Costello Z., Ogorzalek T. L., Tran H. M., Blake-Hedges J. M., Keasling J. D., Adams P. D., García Martín H., Hillson N. J., and Petzold C. J., “Automated “Cells-to-peptides” sample preparation workflow for high-throughput, quantitative proteomic assays of microbes,” Journal of Proteome Research, vol. 18, no. 10, pp. 3752–3761, 2019 [DOI] [PubMed] [Google Scholar]
- 82.Zhang J., Petersen S. D., Radivojevic T., Ramirez A., Pérez-Manríquez A., Abeliuk E., Sánchez B. J., Costello Z., Chen Y., Fero M. J., Martin H. G., Nielsen J., Keasling J. D., and Jensen M. K., “Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism,” Nature Communications, vol. 11, no. 1, pp. 4880–4880, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chao R., Mishra S., Si T., and Zhao H., “Engineering biological systems using automated biofoundries,” Metabolic Engineering, vol. 42, pp. 98–108, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ham T. S., Dmytriv Z., Plahar H., Chen J., Hillson N. J., and Keasling J. D., “Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools,” Nucleic Acids Research, vol. 40, no. 18, p. e141, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morrell W. C., Birkel G. W., Forrer M., Lopez T., Backman T. W. H., Dussault M., Petzold C. J., Baidoo E. E. K., Costello Z., Ando D., Alonso-Gutierrez J., George K. W., Mukhopadhyay A., Vaino I., Keasling J. D., Adams P. D., Hillson N. J., and Garcia Martin H., “The experiment data depot: a web-based software tool for biological experimental data storage, sharing, and visualization,” ACS Synthetic Biology, vol. 6, no. 12, pp. 2248–2259, 2017 [DOI] [PubMed] [Google Scholar]
- 86.Wilbanks B., Layton D. S., Garcia S., and Trinh C. T., “A prototype for modular cell engineering,” ACS Synthetic Biology, vol. 7, pp. 187–199, 2017 [DOI] [PubMed] [Google Scholar]
- 87.Trinh C. T., Liu Y., and Conner D., “Rational design of efficient modular cells,” Metabolic Engineering, vol. 32, pp. 220–231, 2015 [DOI] [PubMed] [Google Scholar]
- 88.Canton B., Labno A., and Endy D., “Refinement and standardization of synthetic biological parts and devices,” Nature Biotechnology, vol. 26, no. 7, pp. 787–793, 2008 [DOI] [PubMed] [Google Scholar]
- 89.Patron N. J., Orzaez D., Marillonnet S., Warzecha H., Matthewman C., Youles M., Raitskin O., Leveau A., Farré G., Rogers C., Smith A., Hibberd J., Webb A. A. R., Locke J., Schornack S., Ajioka J., Baulcombe D. C., Zipfel C., Kamoun S., Jones J. D. G., Kuhn H., Robatzek S., van Esse H. P., Sanders D., Oldroyd G., Martin C., Field R., O'Connor S., Fox S., Wulff B., Miller B., Breakspear A., Radhakrishnan G., Delaux P. M., Loqué D., Granell A., Tissier A., Shih P., Brutnell T. P., Quick W. P., Rischer H., Fraser P. D., Aharoni A., Raines C., South P. F., Ané J. M., Hamberger B. R., Langdale J., Stougaard J., Bouwmeester H., Udvardi M., Murray J. A. H., Ntoukakis V., Schäfer P., Denby K., Edwards K. J., Osbourn A., and Haseloff J., “Standards for plant synthetic biology: a common syntax for exchange of DNA parts,” New Phytologist, vol. 208, no. 1, pp. 13–19, 2015 [DOI] [PubMed] [Google Scholar]
- 90.Smolke C. D., “Building outside of the box: iGEM and the BioBricks Foundation,” Nature Biotechnology, vol. 27, no. 12, pp. 1099–1102, 2009 [DOI] [PubMed] [Google Scholar]
- 91.Goodman C., “Engineering ingenuity at iGEM,” Nature Chemical Biology, vol. 4, no. 1, pp. 13–13, 2008 [DOI] [PubMed] [Google Scholar]
- 92.McLaughlin J. A., Myers C. J., Zundel Z., Mısırlı G., Zhang M., Ofiteru I. D., Goñi-Moreno A., and Wipat A., “SynBioHub: a standards-enabled design repository for synthetic biology,” ACS Synthetic Biology, vol. 7, no. 2, pp. 682–688, 2018 [DOI] [PubMed] [Google Scholar]
- 93.Kamens J., “The Addgene repository: an international nonprofit plasmid and data resource,” Nucleic Acids Research, vol. 43, pp. D1152–D1157, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herscovitch M., Perkins E., Baltus A., and Fan M., “Addgene provides an open forum for plasmid sharing,” Nature Biotechnology, vol. 30, no. 4, pp. 316–317, 2012 [DOI] [PubMed] [Google Scholar]
- 95.Werner S., Engler C., Weber E., Gruetzner R., and Marillonnet S., “Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system,” Bioengineered, vol. 3, pp. 38–43, 2014 [DOI] [PubMed] [Google Scholar]
- 96.Engler C., Youles M., Gruetzner R., Ehnert T. M., Werner S., Jones J. D. G., Patron N. J., and Marillonnet S., “A Golden Gate modular cloning toolbox for plants,” ACS Synthetic Biology, vol. 3, no. 11, pp. 839–843, 2014 [DOI] [PubMed] [Google Scholar]
- 97.Occhialini A., Piatek A. A., Pfotenhauer A. C., Frazier T. P., Stewart C. N. Jr., and Lenaghan S. C., “MoChlo: a versatile, modular cloning toolbox for chloroplast biotechnology,” Plant Physiology, vol. 179, no. 3, pp. 943–957, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hussey S. G., Grima-Pettenati J., Myburg A. A., Mizrachi E., Brady S. M., Yoshikuni Y., and Deutsch S., “A standardized synthetic Eucalyptus transcription factor and promoter panel for re-engineering secondary cell wall regulation in biomass and bioenergy crops,” ACS Synthetic Biology, vol. 8, no. 2, pp. 463–465, 2019 [DOI] [PubMed] [Google Scholar]
- 99.Belcher M. S., Vuu K. M., Zhou A., Mansoori N., Agosto Ramos A., Thompson M. G., Scheller H. V., Loqué D., and Shih P. M., “Design of orthogonal regulatory systems for modulating gene expression in plants,” Nature Chemical Biology, vol. 16, no. 8, pp. 857–865, 2020 [DOI] [PubMed] [Google Scholar]
- 100.Kuo J., Stirling F., Lau Y. H., Shulgina Y., Way J. C., and Silver P. A., “Synthetic genome recoding: new genetic codes for new features,” Current Genetics, vol. 64, no. 2, pp. 327–333, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arkin A. P., Cottingham R. W., Henry C. S., Harris N. L., Stevens R. L., Maslov S., Dehal P., Ware D., Perez F., Canon S., Sneddon M. W., Henderson M. L., Riehl W. J., Murphy-Olson D., Chan S. Y., Kamimura R. T., Kumari S., Drake M. M., Brettin T. S., Glass E. M., Chivian D., Gunter D., Weston D. J., Allen B. H., Baumohl J., Best A. A., Bowen B., Brenner S. E., Bun C. C., Chandonia J. M., Chia J. M., Colasanti R., Conrad N., Davis J. J., Davison B. H., DeJongh M., Devoid S., Dietrich E., Dubchak I., Edirisinghe J. N., Fang G., Faria J. P., Frybarger P. M., Gerlach W., Gerstein M., Greiner A., Gurtowski J., Haun H. L., He F., Jain R., Joachimiak M. P., Keegan K. P., Kondo S., Kumar V., Land M. L., Meyer F., Mills M., Novichkov P. S., Oh T., Olsen G. J., Olson R., Parrello B., Pasternak S., Pearson E., Poon S. S., Price G. A., Ramakrishnan S., Ranjan P., Ronald P. C., Schatz M. C., Seaver S. M. D., Shukla M., Sutormin R. A., Syed M. H., Thomason J., Tintle N. L., Wang D., Xia F., Yoo H., Yoo S., and Yu D., “KBase: The United States Department of Energy Systems Biology Knowledgebase,” Nature Biotechnology, vol. 36, no. 7, pp. 566–569, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu D., Hu R., Palla K. J., Tuskan G. A., and Yang X., “Advances and perspectives on the use of CRISPR/Cas9 systems in plant genomics research,” Current Opinion in Plant Biology, vol. 30, pp. 70–77, 2016 [DOI] [PubMed] [Google Scholar]
- 103.Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., Chen P. J., Wilson C., Newby G. A., Raguram A., and Liu D. R., “Search-and-replace genome editing without double-strand breaks or donor DNA,” Nature, vol. 576, no. 7785, pp. 149–157, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doman J. L., Raguram A., Newby G. A., and Liu D. R., “Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors,” Nature Biotechnology, vol. 38, no. 5, pp. 620–628, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D. I., and Liu D. R., “Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage,” Nature, vol. 551, no. 7681, pp. 464–471, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hassan M. M., Yuan G., Chen J.-G., Tuskan G. A., and Yang X., “Prime editing technology and its prospects for future applications in plant biology research,” BioDesign Research, vol. 2020, article 9350905, pp. 1–14, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bharat S. S., Li S., Li J., Yan L., and Xia L., “Base editing in plants: current status and challenges,” Crop Journal, vol. 8, no. 3, pp. 384–395, 2020 [Google Scholar]
- 108.Mishra R., Joshi R. K., and Zhao K., “Base editing in crops: current advances, limitations and future implications,” Plant Biotechnology Journal, vol. 18, pp. 20–31, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu R., Li J., Liu X., Shan T., Qin R., and Wei P., “Development of plant prime-editing systems for precise genome editing,” Plant Communications, vol. 1, no. 3, article 100043, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A. V., Raguram A., Doman J. L., Liu D. R., and Gao C., “Prime genome editing in rice and wheat,” Nature Biotechnology, vol. 38, no. 5, pp. 582–585, 2020 [DOI] [PubMed] [Google Scholar]
- 111.Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., and Qi L. S., “CRISPR interference (CRISPRi) for sequence-specific control of gene expression,” Nature Protocols, vol. 8, no. 11, pp. 2180–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanson K. R., Hanna R. E., Hegde M., Donovan K. F., Strand C., Sullender M. E., Vaimberg E. W., Goodale A., Root D. E., Piccioni F., and Doench J. G., “Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities,” Nature Communications, vol. 9, no. 1, article 5416, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abudayyeh O. O., Gootenberg J. S., Franklin B., Koob J., Kellner M. J., Ladha A., Joung J., Kirchgatterer P., Cox D. B. T., and Zhang F., “A cytosine deaminase for programmable single-base RNA editing,” Science, vol. 365, no. 6451, pp. 382–386, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., and Charpentier E., “A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity,” Science, vol. 337, no. 6096, pp. 816–821, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nielsen A. A. K., der B. S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E. A., Ross D., Densmore D., and Voigt C. A., “Genetic circuit design automation,” Science, vol. 352, no. 6281, article aac7341, 2016 [DOI] [PubMed] [Google Scholar]
- 116.Zhang M., McLaughlin J. A., Wipat A., and Myers C. J., “SBOLDesigner 2: an intuitive tool for structural genetic design,” ACS Synthetic Biology, vol. 6, no. 7, pp. 1150–1160, 2017 [DOI] [PubMed] [Google Scholar]
- 117.Walhout A. J., “Protein interaction mapping in C. elegans using proteins involved in vulval development,” Science, vol. 287, no. 5450, pp. 116–122, 2000 [DOI] [PubMed] [Google Scholar]
- 118.Engler C., Kandzia R., and Marillonnet S., “A one pot, one step, precision cloning method with high throughput capability,” PLoS One, vol. 3, no. 11, pp. e3647–e3647, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III, and Smith H. O., “Enzymatic assembly of DNA molecules up to several hundred kilobases,” Nature Methods, vol. 6, no. 5, pp. 343–345, 2009 [DOI] [PubMed] [Google Scholar]
- 120.Sarrion-Perdigones A., Vazquez-Vilar M., Palaci J., Castelijns B., Forment J., Ziarsolo P., Blanca J., Granell A., and Orzaez D., “GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology,” Plant Physiology, vol. 162, no. 3, pp. 1618–1631, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang R.-B., Bi L.-J., and Zhang X.-E., “A novel T-type overhangs improve the enzyme-free cloning of PCR products,” Molecular Biotechnology, vol. 55, no. 1, pp. 10–16, 2013 [DOI] [PubMed] [Google Scholar]
- 122.De Paoli H. C., Tuskan G. A., and Yang X., “An innovative platform for quick and flexible joining of assorted DNA fragments,” Scientific Reports, vol. 6, no. 1, article 19278, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lei C., Li S.-Y., Liu J.-K., Zheng X., Zhao G.-P., and Wang J., “The CCTL (Cpf1-assisted cutting and Taq DNA ligase-assisted ligation) method for efficient editing of large DNA constructs in vitro,” Nucleic Acids Research, vol. 45, pp. e74–e74, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moon S. B., Lee J. M., Kang J. G., Lee N.-E., Ha D.-I., Kim D. Y., Kim S. H., Yoo K., Kim D., Ko J.-H., and Kim Y.-S., “Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang,” Nature Communications, vol. 9, no. 1, article 3651, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuchs R. T., Curcuru J., Mabuchi M., Yourik P., and Robb G. B., “Cas12a trans-cleavage can be modulated in vitro and is active on ssDNA, dsDNA, and RNA,” bioRxiv, 2019 [Google Scholar]
- 126.Pollak B., Matute T., Nuñez I., Cerda A., Lopez C., Vargas V., Kan A., Bielinski V., von Dassow P., Dupont C. L., and Federici F., “Universal loop assembly: open, efficient and cross-kingdom DNA fabrication,” Synthetic Biology, vol. 5, no. 1, article ysaa001, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dien V. T., Holcomb M., Feldman A. W., Fischer E. C., Dwyer T. J., and Romesberg F. E., “Progress toward a semi-synthetic organism with an unrestricted expanded genetic alphabet,” Journal of the American Chemical Society, vol. 140, no. 47, pp. 16115–16123, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Braman J. C., and Sheffield P. J., “Seamless assembly of DNA parts into functional devices and higher order multi-device systems,” PLoS One, vol. 14, no. 6, article e0199653, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shih P. M., Vuu K., Mansoori N., Ayad L., Louie K. B., Bowen B. P., Northen T. R., and Loqué D., “A robust gene-stacking method utilizing yeast assembly for plant synthetic biology,” Nature Communications, vol. 7, no. 1, article 13215, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Venetz J. E., del Medico L., Wölfle A., Schächle P., Bucher Y., Appert D., Tschan F., Flores-Tinoco C. E., van Kooten M., Guennoun R., Deutsch S., Christen M., and Christen B., “Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality,” Proceedings of the National Academy of Sciences, vol. 116, no. 16, pp. 8070–8079, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fredens J., Wang K., de la Torre D., Funke L. F. H., Robertson W. E., Christova Y., Chia T., Schmied W. H., Dunkelmann D. L., Beránek V., Uttamapinant C., Llamazares A. G., Elliott T. S., and Chin J. W., “Total synthesis of Escherichia coli with a recoded genome,” Nature, vol. 569, no. 7757, pp. 514–518, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ferrer A., Arró M., Manzano D., and Altabella T.. Advanced Technologies for Protein Complex Production and Characterization, Vega M. C., Ed., Springer International Publishing, Cham, 2016 [Google Scholar]
- 133.O'Neill B. M., Mikkelson K. L., Gutierrez N. M., Cunningham J. L., Wolff K. L., Szyjka S. J., Yohn C. B., Redding K. E., and Mendez M. J., “An exogenous chloroplast genome for complex sequence manipulation in algae,” Nucleic Acids Research, vol. 40, pp. 2782–2792, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schindel H. S., Piatek A. A., Stewart C. N., and Lenaghan S. C., “The plastid genome as a chassis for synthetic biology-enabled metabolic engineering: players in gene expression,” Plant Cell Reports, vol. 37, no. 10, pp. 1419–1429, 2018 [DOI] [PubMed] [Google Scholar]
- 135.Lowe K., Wu E., Wang N., Hoerster G., Hastings C., Cho M. J., Scelonge C., Lenderts B., Chamberlin M., Cushatt J., Wang L., Ryan L., Khan T., Chow-Yiu J., Hua W., Yu M., Banh J., Bao Z., Brink K., Igo E., Rudrappa B., Shamseer P. M., Bruce W., Newman L., Shen B., Zheng P., Bidney D., Falco C., Register J., Zhao Z. Y., Xu D., Jones T., and Gordon-Kamm W., “Morphogenic regulators Baby boom and Wuschel improve monocot transformation,” Plant Cell, vol. 28, no. 9, pp. 1998–2015, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Maher M. F., Nasti R. A., Vollbrecht M., Starker C. G., Clark M. D., and Voytas D. F., “Plant gene editing through de novo induction of meristems,” Nature Biotechnology, vol. 38, no. 1, pp. 84–89, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Plasencia A., Soler M., Dupas A., Ladouce N., Silva-Martins G., Martinez Y., Lapierre C., Franche C., Truchet I., and Grima-Pettenati J., “Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation,” Plant Biotechnology Journal, vol. 14, no. 6, pp. 1381–1393, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dai Y., Hu G., Dupas A., Medina L., Blandels N., San Clemente H., Ladouce N., Badawi M., Hernandez-Raquet G., Mounet F., Grima-Pettenati J., and Cassan-Wang H., “Implementing the CRISPR/Cas9 technology in Eucalyptus hairy roots using wood-related genes,” International Journal of Molecular Sciences, vol. 21, no. 10, p. 3408, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kaur R. P., and Devi S., “In planta transformation in plants: a review,” Agricultural Reviews, vol. 40, pp. 159–174, 2019 [Google Scholar]
- 140.Bahari Z., Sazegari S., Niazi A., and Afsharifar A., “The application of an Agrobacterium-mediated in planta transformation system in a Catharanthus roseus medicinal plant,” Czech Journal of Genetics and Plant Breeding, vol. 56, pp. 34–41, 2020 [Google Scholar]
- 141.Zlobin N. E., Lebedeva M. V., and Taranov V. V., “CRISPR/Cas9 genome editing throughin plantatransformation,” Critical Reviews in Biotechnology, vol. 40, no. 2, pp. 153–168, 2020 [DOI] [PubMed] [Google Scholar]
- 142.Toda E., Koiso N., Takebayashi A., Ichikawa M., Kiba T., Osakabe K., Osakabe Y., Sakakibara H., Kato N., and Okamoto T., “An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice,” Nature Plants, vol. 5, no. 4, pp. 363–368, 2019 [DOI] [PubMed] [Google Scholar]
- 143.Demirer G. S., Zhang H., Goh N. S., González-Grandío E., and Landry M. P., “Carbon nanotube–mediated DNA delivery without transgene integration in intact plants,” Nature Protocols, vol. 14, no. 10, pp. 2954–2971, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He Y., and Zhao Y., “Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants,” aBIOTECH, vol. 1, no. 1, pp. 88–96, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Untergasser A., Bijl G. J. M., Liu W., Bisseling T., Schaart J. G., and Geurts R., “One-step Agrobacterium mediated transformation of eight genes essential for rhizobium symbiotic signaling using the novel binary vector system pHUGE,” PLoS One, vol. 7, no. 10, pp. e47885–e47885, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pasin F., Bedoya L. C., Bernabé-Orts J. M., Gallo A., Simón-Mateo C., Orzaez D., and García J. A., “Multiple T-DNA delivery to plants using novel mini binary vectors with compatible replication origins,” ACS Synthetic Biology, vol. 6, no. 10, pp. 1962–1968, 2017 [DOI] [PubMed] [Google Scholar]
- 147.Srivastava V.Transgenic Plants. Methods in Molecular Biology, Kumar S., Barone P., and Smith M., Eds., Humana Press, New York, NY, vol. vol 1864, 2019 [Google Scholar]
- 148.Jin S., and Daniell H., “The engineered chloroplast genome just got smarter,” Trends in Plant Science, vol. 20, no. 10, pp. 622–640, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang J., Xie X., Xiang N., Tian Z. X., Dixon R., and Wang Y. P., “Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology,” Proceedings of the National Academy of Sciences, vol. 115, no. 36, pp. E8509–E8517, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Márquez-Escobar V. A., González-Ortega O., and Rosales-Mendoza S.. Prospects of Plant-Based Vaccines in Veterinary Medicine, MacDonald J., Ed., Springer International Publishing, Cham, 2018 [Google Scholar]
- 151.Lee K., Eggenberger A. L., Banakar R., McCaw M. E., Zhu H., Main M., Kang M., Gelvin S. B., and Wang K., “CRISPR/Cas9-mediated targeted T-DNA integration in rice,” Plant Molecular Biology, vol. 99, no. 4-5, pp. 317–328, 2019 [DOI] [PubMed] [Google Scholar]
- 152.Miki D., Zhang W., Zeng W., Feng Z., and Zhu J.-K., “CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation,” Nature Communications, vol. 9, no. 1, article 1967, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mellado-Sánchez M., McDiarmid F., Cardoso V., Kanyuka K., and MacGregor D. R., “Virus-mediated transient expression techniques enable gene function studies in black-grass,” Plant Physiology, vol. 183, no. 2, pp. 455–459, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jones L., Ratcliff F., and Baulcombe D. C., “RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance,” Current Biology, vol. 11, no. 10, pp. 747–757, 2001 [DOI] [PubMed] [Google Scholar]
- 155.Ali Z., Abul-faraj A., Li L., Ghosh N., Piatek M., Mahjoub A., Aouida M., Piatek A., Baltes N. J., Voytas D. F., Dinesh-Kumar S., and Mahfouz M. M., “Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system,” Molecular Plant, vol. 8, no. 8, pp. 1288–1291, 2015 [DOI] [PubMed] [Google Scholar]
- 156.Mei Y., Beernink B. M., Ellison E. E., Konečná E., Neelakandan A. K., Voytas D. F., and Whitham S. A., “Protein expression and gene editing in monocots using foxtail mosaic virus vectors,” Plant Direct, vol. 3, article e00181, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma X., Zhang X., Liu H., and Li Z., “Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9,” Nature Plants, vol. 6, no. 7, pp. 773–779, 2020 [DOI] [PubMed] [Google Scholar]
- 158.Ellison E. E., Nagalakshmi U., Gamo M. E., Huang P. J., Dinesh-Kumar S., and Voytas D. F., “Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs,” Nature Plants, vol. 6, no. 6, pp. 620–624, 2020 [DOI] [PubMed] [Google Scholar]
- 159.Demirer G. S., Zhang H., Goh N. S., Pinals R. L., Chang R., and Landry M. P., “Carbon nanocarriers deliver siRNA to intact plant cells for efficient gene knockdown,” Science Advances, vol. 6, no. 26, article eaaz0495, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Talukdar D.Single-Cell Omics, Barh D., and Azevedo V., Eds., Academic Press, San Diego, 2019 [Google Scholar]
- 161.Mergner J., Frejno M., List M., Papacek M., Chen X., Chaudhary A., Samaras P., Richter S., Shikata H., Messerer M., Lang D., Altmann S., Cyprys P., Zolg D. P., Mathieson T., Bantscheff M., Hazarika R. R., Schmidt T., Dawid C., Dunkel A., Hofmann T., Sprunck S., Falter-Braun P., Johannes F., Mayer K. F. X., Jürgens G., Wilhelm M., Baumbach J., Grill E., Schneitz K., Schwechheimer C., and Kuster B., “Mass-spectrometry-based draft of the Arabidopsis proteome,” Nature, vol. 579, no. 7799, pp. 409–414, 2020 [DOI] [PubMed] [Google Scholar]
- 162.Kalia A., and Sharma S. P.. Single-Cell Omics, Barh D., and Azevedo V., Eds., Academic Press, San Diego, 2019 [Google Scholar]
- 163.Svensson V., Vento-Tormo R., and Teichmann S. A., “Exponential scaling of single-cell RNA-seq in the past decade,” Nature Protocols, vol. 13, no. 4, pp. 599–604, 2018 [DOI] [PubMed] [Google Scholar]
- 164.Denyer T., Ma X., Klesen S., Scacchi E., Nieselt K., and Timmermans M. C. P., “Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing,” Developmental Cell, vol. 48, no. 6, pp. 840–852.e5, 2019 [DOI] [PubMed] [Google Scholar]
- 165.Budnik B., Levy E., Harmange G., and Slavov N., “SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation,” Genome Biology, vol. 19, no. 1, p. 161, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Efroni I., and Birnbaum K. D., “The potential of single-cell profiling in plants,” Genome Biology, vol. 17, no. 1, article 65, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zenobi R., “Single-cell metabolomics: analytical and biological perspectives,” Science, vol. 342, no. 6163, article 1243259, 2013 [DOI] [PubMed] [Google Scholar]
- 168.Labib M., and Kelley S. O., “Single-cell analysis targeting the proteome,” Nature Reviews Chemistry, vol. 4, no. 3, pp. 143–158, 2020 [DOI] [PubMed] [Google Scholar]
- 169.Ryu K. H., Huang L., Kang H. M., and Schiefelbein J., “Single-cell RNA sequencing resolves molecular relationships among individual plant cells,” Plant Physiology, vol. 179, no. 4, pp. 1444–1456, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rich-Griffin C., Stechemesser A., Finch J., Lucas E., Ott S., and Schäfer P., “Single-cell transcriptomics: a high-resolution avenue for plant functional genomics,” Trends in Plant Science, vol. 25, no. 2, pp. 186–197, 2020 [DOI] [PubMed] [Google Scholar]
- 171.Shulse C. N., Cole B. J., Ciobanu D., Lin J., Yoshinaga Y., Gouran M., Turco G. M., Zhu Y., O’Malley R. C., Brady S. M., and Dickel D. E., “High-throughput single-cell transcriptome profiling of plant cell types,” Cell Reports, vol. 27, no. 7, pp. 2241–2247.e4, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu Y., Li H., Bhatti S., Zhou S., Yang Y., Fish T., and Thannhauser T. W., “Development of a laser capture microscope-based single-cell-type proteomics tool for studying proteomes of individual cell layers of plant roots,” Horticulture Research, vol. 3, no. 1, article 16026, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fujii T., Matsuda S., Tejedor M. L., Esaki T., Sakane I., Mizuno H., Tsuyama N., and Masujima T., “Direct metabolomics for plant cells by live single-cell mass spectrometry,” Nature Protocols, vol. 10, no. 9, pp. 1445–1456, 2015 [DOI] [PubMed] [Google Scholar]
- 174.Choi H.-K., “Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding,” Genes & Genomics, vol. 41, no. 2, pp. 133–146, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang J. P., Matthews M. L., Williams C. M., Shi R., Yang C., Tunlaya-Anukit S., Chen H. C., Li Q., Liu J., Lin C. Y., Naik P., Sun Y. H., Loziuk P. L., Yeh T. F., Kim H., Gjersing E., Shollenberger T., Shuford C. M., Song J., Miller Z., Huang Y. Y., Edmunds C. W., Liu B., Sun Y., Lin Y. C. J., Li W., Chen H., Peszlen I., Ducoste J. J., Ralph J., Chang H. M., Muddiman D. C., Davis M. F., Smith C., Isik F., Sederoff R., and Chiang V. L., “Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis,” Nature Communications, vol. 9, no. 1, p. 1579, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Misra B. B., Langefeld C., Olivier M., and Cox L. A., “Integrated omics: tools, advances and future approaches,” Journal of Molecular Endocrinology, vol. 62, pp. R21–R45, 2019 [DOI] [PubMed] [Google Scholar]
- 177.Ma T., and Zhang A., “Integrate multi-omics data with biological interaction networks using Multi-view Factorization AutoEncoder (MAE),” BMC Genomics, vol. 20, no. S11, article 944, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Holzinger A., Haibe-Kains B., and Jurisica I., “Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data,” European Journal of Nuclear Medicine and Molecular Imaging, vol. 46, no. 13, pp. 2722–2730, 2019 [DOI] [PubMed] [Google Scholar]
- 179.Kersey P. J., “Plant genome sequences: past, present, future,” Current Opinion in Plant Biology, vol. 48, pp. 1–8, 2019 [DOI] [PubMed] [Google Scholar]
- 180.Niehaus T. D., Thamm A. M. K., de Crécy- V., and Lagard A. D. H., “Proteins of unknown biochemical function: a persistent problem and a roadmap to help overcome it,” Plant Physiology, vol. 169, pp. 1436–1442, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rhee S. Y., and Mutwil M., “Towards revealing the functions of all genes in plants,” Trends in Plant Science, vol. 19, no. 4, pp. 212–221, 2014 [DOI] [PubMed] [Google Scholar]
- 182.Liu D., Chen M., Mendoza B., Cheng H., Hu R., Li L., Trinh C. T., Tuskan G. A., and Yang X., “CRISPR/Cas9-mediated targeted mutagenesis for functional genomics research of crassulacean acid metabolism plants,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6621–6629, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Minkenberg B., Xie K., and Yang Y., “Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes,” Plant Journal, vol. 89, no. 3, pp. 636–648, 2017 [DOI] [PubMed] [Google Scholar]
- 184.Leydon A. R., Gala H. P., Guiziou S., and Nemhauser J. L., “Engineering synthetic signaling in plants,” Annual Review of Plant Biology, vol. 71, no. 1, pp. 767–788, 2020 [DOI] [PubMed] [Google Scholar]
- 185.Xiong X., Zhang Y., Yan J., Jain S., Chee S., Ren B., and Zhao H., “A scalable epitope tagging approach for high throughput ChIP-Seq analysis,” ACS Synthetic Biology, vol. 6, no. 6, pp. 1034–1042, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Armario Najera V., Twyman R. M., Christou P., and Zhu C., “Applications of multiplex genome editing in higher plants,” Current Opinion in Biotechnology, vol. 59, pp. 93–102, 2019 [DOI] [PubMed] [Google Scholar]
- 187.Luo Z., Yang Q., Geng B., Jiang S., Yang S., Li X., Cai Y., and Dai J., “Whole genome engineering by synthesis,” Science China Life Sciences, vol. 61, no. 12, pp. 1515–1527, 2018 [DOI] [PubMed] [Google Scholar]
- 188.Rees-Garbutt J., Chalkley O., Landon S., Purcell O., Marucci L., and Grierson C., “Designing minimal genomes using whole-cell models,” Nature Communications, vol. 11, no. 1, article 836, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou J., Wu R., Xue X., and Qin Z., “CasHRA (Cas9-facilitated Homologous Recombination Assembly) method of constructing megabase-sized DNA,” Nucleic Acids Research, vol. 44, no. 14, pp. e124–e124, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hutchison C. A., Chuang R. Y., Noskov V. N., Assad-Garcia N., Deerinck T. J., Ellisman M. H., Gill J., Kannan K., Karas B. J., Ma L., Pelletier J. F., Qi Z. Q., Richter R. A., Strychalski E. A., Sun L., Suzuki Y., Tsvetanova B., Wise K. S., Smith H. O., Glass J. I., Merryman C., Gibson D. G., and Venter J. C., “Design and synthesis of a minimal bacterial genome,” Science, vol. 351, no. 6280, article aad6253, 2016 [DOI] [PubMed] [Google Scholar]
- 191.Lubrano P., Danchin A., and Acevedo-Rocha C. G.. Minimal Cells: Design, Construction, Biotechnological Applications, Lara A. R., and Gosset G., Eds., Springer, Cham, 2020 [Google Scholar]
- 192.Long S. P., Burgess S., and Causton I.. Sustaining Global Food Security: The Nexus of Science and Policy, Zeigler R. S., Ed., CSIRO Publishing, Clayton South, 2019 [Google Scholar]
- 193.Voloshin R. A., Kreslavski V. D., Zharmukhamedov S. K., Bedbenov V. S., Ramakrishna S., and Allakhverdiev S. I., “Photoelectrochemical cells based on photosynthetic systems: a review,” Biofuel Research Journal, vol. 2, no. 2, pp. 227–235, 2015 [Google Scholar]
- 194.Wolf B. M., and Blankenship R. E., “Far-red light acclimation in diverse oxygenic photosynthetic organisms,” Photosynthesis Research, vol. 142, no. 3, pp. 349–359, 2019 [DOI] [PubMed] [Google Scholar]
- 195.Ort D. R., Merchant S. S., Alric J., Barkan A., Blankenship R. E., Bock R., Croce R., Hanson M. R., Hibberd J. M., Long S. P., Moore T. A., Moroney J., Niyogi K. K., Parry M. A. J., Peralta-Yahya P. P., Prince R. C., Redding K. E., Spalding M. H., van Wijk K. J., Vermaas W. F. J., von Caemmerer S., Weber A. P. M., Yeates T. O., Yuan J. S., and Zhu X. G., “Redesigning photosynthesis to sustainably meet global food and bioenergy demand,” Proceedings of the National Academy of Sciences, vol. 112, no. 28, pp. 8529–8536, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Simkin A. J., McAusland L., Lawson T., and Raines C. A., “Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield,” Plant Physiology, vol. 175, no. 1, pp. 134–145, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ermakova M., Lopez-Calcagno P. E., Raines C. A., Furbank R. T., and von Caemmerer S., “Overexpression of the Rieske FeS protein of the Cytochrome b6f complex increases C4 photosynthesis in Setaria viridis,” Communications Biology, vol. 2, no. 1, article 314, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen J.-H., Chen S. T., He N. Y., Wang Q. L., Zhao Y., Gao W., and Guo F. Q., “Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield,” Nature Plants, vol. 6, no. 5, pp. 570–580, 2020 [DOI] [PubMed] [Google Scholar]
- 199.Kromdijk J., Głowacka K., Leonelli L., Gabilly S. T., Iwai M., Niyogi K. K., and Long S. P., “Improving photosynthesis and crop productivity by accelerating recovery from photoprotection,” Science, vol. 354, no. 6314, pp. 857–861, 2016 [DOI] [PubMed] [Google Scholar]
- 200.Garcia-Molina A., and Leister D., “Accelerated relaxation of photoprotection impairs biomass accumulation in Arabidopsis,” Nature Plants, vol. 6, no. 1, pp. 9–12, 2020 [DOI] [PubMed] [Google Scholar]
- 201.Betti M., Bauwe H., Busch F. A., Fernie A. R., Keech O., Levey M., Ort D. R., Parry M. A. J., Sage R., Timm S., Walker B., and Weber A. P. M., “Manipulating photorespiration to increase plant productivity: recent advances and perspectives for crop improvement,” Journal of Experimental Botany, vol. 67, no. 10, pp. 2977–2988, 2016 [DOI] [PubMed] [Google Scholar]
- 202.Parry M. A. J., Andralojc P. J., Scales J. C., Salvucci M. E., Carmo-Silva A. E., Alonso H., and Whitney S. M., “Rubisco activity and regulation as targets for crop improvement,” Journal of Experimental Botany, vol. 64, no. 3, pp. 717–730, 2013 [DOI] [PubMed] [Google Scholar]
- 203.Kebeish R., Niessen M., Thiruveedhi K., Bari R., Hirsch H. J., Rosenkranz R., Stäbler N., Schönfeld B., Kreuzaler F., and Peterhänsel C., “Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana,” Nature Biotechnology, vol. 25, no. 5, pp. 593–599, 2007 [DOI] [PubMed] [Google Scholar]
- 204.Maier A., Fahnenstich H., von Caemmerer S., Engqvist M. K. M., Weber A. P. M., Flügge U. I., and Maurino V. G., “Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement,” Frontiers in Plant Science, vol. 3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.South P. F., Cavanagh A. P., Liu H. W., and Ort D. R., “Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field,” Science, vol. 363, no. 6422, article eaat9077, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang L.-M., Shen B. R., Li B. D., Zhang C. L., Lin M., Tong P. P., Cui L. L., Zhang Z. S., and Peng X. X., “A synthetic photorespiratory shortcut enhances photosynthesis to boost biomass and grain yield in rice,” Molecular Plant, 2020 [DOI] [PubMed]
- 207.Maurino V. G., “Using energy-efficient synthetic biochemical pathways to bypass photorespiration,” Biochemical Society Transactions, vol. 47, no. 6, pp. 1805–1813, 2019 [DOI] [PubMed] [Google Scholar]
- 208.Bar-Even A., Noor E., Lewis N. E., and Milo R., “Design and analysis of synthetic carbon fixation pathways,” Proceedings of the National Academy of Sciences, vol. 107, no. 19, pp. 8889–8894, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schwander T., Schada von Borzyskowski L., Burgener S., Cortina N. S., and Erb T. J., “A synthetic pathway for the fixation of carbon dioxide in vitro,” Science, vol. 354, no. 6314, pp. 900–904, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Miller T. E., Beneyton T., Schwander T., Diehl C., Girault M., McLean R., Chotel T., Claus P., Cortina N. S., Baret J. C., and Erb T. J., “Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts,” Science, vol. 368, no. 6491, pp. 649–654, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Erb T. J., Jones P. R., and Bar-Even A., “Synthetic metabolism: metabolic engineering meets enzyme design,” Current Opinion in Chemical Biology, vol. 37, pp. 56–62, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wurtzel E. T., Vickers C. E., Hanson A. D., Millar A. H., Cooper M., Voss-Fels K. P., Nikel P. I., and Erb T. J., “Revolutionizing agriculture with synthetic biology,” Nature Plants, vol. 5, no. 12, pp. 1207–1210, 2019 [DOI] [PubMed] [Google Scholar]
- 213.Raven J. A., Beardall J., and Sánchez-Baracaldo P., “The possible evolution and future of CO2-concentrating mechanisms,” Journal of Experimental Botany, vol. 68, no. 14, pp. 3701–3716, 2017 [DOI] [PubMed] [Google Scholar]
- 214.Davis S. C., Simpson J., Gil-Vega K. . C., Niechayev N. A., Tongerlo E. ., Castano N. H., Dever L. V., and Búrquez A., “Undervalued potential of crassulacean acid metabolism for current and future agricultural production,” Journal of Experimental Botany, vol. 70, no. 22, pp. 6521–6537, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ermakova M., Danila F. R., Furbank R. T., and von Caemmerer S., “On the road to C4 rice: advances and perspectives,” Plant Journal, vol. 101, no. 4, pp. 940–950, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jurić I., Hibberd J. M., Blatt M., and Burroughs N. J., “Computational modelling predicts substantial carbon assimilation gains for C3 plants with a single-celled C4 biochemical pump,” PLoS Computational Biology, vol. 15, no. 9, article e1007373, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rae B. D., Long B. M., Förster B., Nguyen N. D., Velanis C. N., Atkinson N., Hee W. Y., Mukherjee B., Price G. D., and McCormick A. J., “Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants,” Journal of Experimental Botany, vol. 68, no. 14, pp. 3717–3737, 2017 [DOI] [PubMed] [Google Scholar]
- 218.Hennacy J. H., and Jonikas M. C., “Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields,” Annual Review of Plant Biology, vol. 71, no. 1, pp. 461–485, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Atkinson N., Feike D., Mackinder L. C. M., Meyer M. T., Griffiths H., Jonikas M. C., Smith A. M., and McCormick A. J., “Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components,” Plant Biotechnology Journal, vol. 14, no. 5, pp. 1302–1315, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Long B. M., Hee W. Y., Sharwood R. E., Rae B. D., Kaines S., Lim Y. L., Nguyen N. D., Massey B., Bala S., von Caemmerer S., Badger M. R., and Price G. D., “Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts,” Nature Communications, vol. 9, no. 1, p. 3570, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Atkinson N., Velanis C. N., Wunder T., Clarke D. J., Mueller-Cajar O., and McCormick A. J., “The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis–Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit,” Journal of Experimental Botany, vol. 70, no. 19, pp. 5271–5285, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang L., Ma G., Wang H., Cheng C., Mu S., Quan W., Jiang L., Zhao Z., Zhang Y., Zhang K., Wang X., Tian C., and Zhang Y., “A draft genome assembly of halophyte Suaeda aralocaspica, a plant that performs C4 photosynthesis within individual cells,” GigaScience, vol. 8, no. 9, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lundgren M. R., “C2 photosynthesis: a promising route towards crop improvement?,” New Phytologist, 2020 [DOI] [PubMed]
- 224.Bellasio C., and Farquhar G. D., “A leaf-level biochemical model simulating the introduction of C2 and C4 photosynthesis in C3 rice: gains, losses and metabolite fluxes,” New Phytologist, vol. 223, no. 1, pp. 150–166, 2019 [DOI] [PubMed] [Google Scholar]
- 225.Amthor J. S., Bar-Even A., Hanson A. D., Millar A. H., Stitt M., Sweetlove L. J., and Tyerman S. D., “Engineering strategies to boost crop productivity by cutting respiratory carbon loss,” Plant Cell, vol. 31, no. 2, pp. 297–314, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhu Q., Wang B., Tan J., Liu T., Li L., and Liu Y. G., “Plant synthetic metabolic engineering for enhancing crop nutritional quality,” Plant Communications, vol. 1, no. 1, article 100017, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fahad S., Bajwa A. A., Nazir U., Anjum S. A., Farooq A., Zohaib A., Sadia S., Nasim W., Adkins S., Saud S., Ihsan M. Z., Alharby H., Wu C., Wang D., and Huang J., “Crop production under drought and heat stress: plant responses and management options,” Frontiers in Plant Science, vol. 8, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ashraf M., Ahmad M. S. A., Öztürk M., and Aksoy A.. Crop Production for Agricultural Improvement, Ashraf M., Öztürk M., Ahmad M. S. A., and Aksoy A., Eds., Springer, Netherlands, Dordrecht, 2012 [Google Scholar]
- 229.Mizoi J., and Yamaguchi-Shinozaki K.. Rice Protocols, Yang Y., Ed., Humana Press, Totowa, NJ, vol. 956, 2013 [Google Scholar]
- 230.Marco F., Bitrián M., Carrasco P., Rajam M. V., Alcázar R., and Tiburcio A. F.. Plant Biology and Biotechnology: Volume II: Plant Genomics and Biotechnology, Bahadur B., Rajam M. V., Sahijram L., and Krishnamurthy K. V., Eds., Springer India, New Delhi, 2015 [Google Scholar]
- 231.Borland A. M., Griffiths H., Hartwell J., and Smith J. A. C., “Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands,” Journal of Experimental Botany, vol. 60, no. 10, pp. 2879–2896, 2009 [DOI] [PubMed] [Google Scholar]
- 232.Davis S. C., LeBauer D. S., and Long S. P., “Light to liquid fuel: theoretical and realized energy conversion efficiency of plants using Crassulacean Acid Metabolism (CAM) in arid conditions,” Journal of Experimental Botany, vol. 65, no. 13, pp. 3471–3478, 2014 [DOI] [PubMed] [Google Scholar]
- 233.Borland A. M., Hartwell J., Weston D. J., Schlauch K. A., Tschaplinski T. J., Tuskan G. A., Yang X., and Cushman J. C., “Engineering crassulacean acid metabolism to improve water-use efficiency,” Trends in Plant Science, vol. 19, no. 5, pp. 327–338, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Liu D., Palla K. J., Hu R., Moseley R. C., Mendoza C., Chen M., Abraham P. E., Labbé J. L., Kalluri U. C., Tschaplinski T. J., Cushman J. C., Borland A. M., Tuskan G. A., and Yang X., “Perspectives on the basic and applied aspects of crassulacean acid metabolism (CAM) research,” Plant Science, vol. 274, pp. 394–401, 2018 [DOI] [PubMed] [Google Scholar]
- 235.Yang X., Cushman J. C., Borland A. M., Edwards E. J., Wullschleger S. D., Tuskan G. A., Owen N. A., Griffiths H., Smith J. A. C., de Paoli H. C., Weston D. J., Cottingham R., Hartwell J., Davis S. C., Silvera K., Ming R., Schlauch K., Abraham P., Stewart J. R., Guo H. B., Albion R., Ha J., Lim S. D., Wone B. W. M., Yim W. C., Garcia T., Mayer J. A., Petereit J., Nair S. S., Casey E., Hettich R. L., Ceusters J., Ranjan P., Palla K. J., Yin H., Reyes-García C., Andrade J. L., Freschi L., Beltrán J. D., Dever L. V., Boxall S. F., Waller J., Davies J., Bupphada P., Kadu N., Winter K., Sage R. F., Aguilar C. N., Schmutz J., Jenkins J., and Holtum J. A. M., “A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world,” New Phytologist, vol. 207, no. 3, pp. 491–504, 2015 [DOI] [PubMed] [Google Scholar]
- 236.Yang X., Cushman J. C., Borland A. M., and Liu Q., “Editorial: systems biology and synthetic biology in relation to drought tolerance or avoidance in plants,” Frontiers in Plant Science, vol. 11, no. 394, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Giannakopoulou A., Bialas A., Kamoun S., and Vleeshouwers V. G. A. A., “Plant immunity switched from bacteria to virus,” Nature Biotechnology, vol. 34, no. 4, pp. 391–392, 2016 [DOI] [PubMed] [Google Scholar]
- 238.Horsefield S., Burdett H., Zhang X., Manik M. K., Shi Y., Chen J., Qi T., Gilley J., Lai J. S., Rank M. X., Casey L. W., Gu W., Ericsson D. J., Foley G., Hughes R. O., Bosanac T., von Itzstein M., Rathjen J. P., Nanson J. D., Boden M., Dry I. B., Williams S. J., Staskawicz B. J., Coleman M. P., Ve T., Dodds P. N., and Kobe B., “NAD+ cleavage activity by animal and plant TIR domains in cell death pathways,” Science, vol. 365, no. 6455, pp. 793–799, 2019 [DOI] [PubMed] [Google Scholar]
- 239.Wan L., Essuman K., Anderson R. G., Sasaki Y., Monteiro F., Chung E. H., Osborne Nishimura E., DiAntonio A., Milbrandt J., Dangl J. L., and Nishimura M. T., “TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death,” Science, vol. 365, no. 6455, pp. 799–803, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Gatehouse A. M. R., Ferry N., Edwards M. G., and Bell H. A., “Insect-resistant biotech crops and their impacts on beneficial arthropods,” Philosophical Transactions of the Royal Society B: Biological Sciences, vol. 366, no. 1569, pp. 1438–1452, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tabashnik B. E., Brévault T., and Carrière Y., “Insect resistance to Bt crops: lessons from the first billion acres,” Nature Biotechnology, vol. 31, no. 6, pp. 510–521, 2013 [DOI] [PubMed] [Google Scholar]
- 242.Yu X.-D., Liu Z. C., Huang S. L., Chen Z. Q., Sun Y. W., Duan P. F., Ma Y. Z., and Xia L. Q., “RNAi-mediated plant protection against aphids,” Pest Management Science, vol. 72, no. 6, pp. 1090–1098, 2016 [DOI] [PubMed] [Google Scholar]
- 243.Porter S. S., Bantay R., Friel C. A., Garoutte A., Gdanetz K., Ibarreta K., Moore B. M., Shetty P., Siler E., and Friesen M. L., “Beneficial microbes ameliorate abiotic and biotic sources of stress on plants,” Functional Ecology, vol. 34, no. 10, pp. 2075–2086, 2020 [Google Scholar]
- 244.Mus F., Crook M. B., Garcia K., Garcia Costas A., Geddes B. A., Kouri E. D., Paramasivan P., Ryu M. H., Oldroyd G. E. D., Poole P. S., Udvardi M. K., Voigt C. A., Ané J. M., and Peters J. W., “Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes,” Applied and Environmental Microbiology, vol. 82, no. 13, pp. 3698–3710, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Geddes B. A., Paramasivan P., Joffrin A., Thompson A. L., Christensen K., Jorrin B., Brett P., Conway S. J., Oldroyd G. E. D., and Poole P. S., “Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria,” Nature Communications, vol. 10, no. 1, article 3430, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wiseman E. M., Bar-El Dadon S., and Reifen R., “The vicious cycle of vitamin a deficiency: a review,” Critical Reviews in Food Science and Nutrition, vol. 57, no. 17, pp. 3703–3714, 2017 [DOI] [PubMed] [Google Scholar]
- 247.Umbreit J., “Iron deficiency: a concise review,” American Journal of Hematology, vol. 78, no. 3, pp. 225–231, 2005 [DOI] [PubMed] [Google Scholar]
- 248.Czeizel A. E., Dudás I., Vereczkey A., and Bánhidy F., “Folate deficiency and folic acid supplementation: the prevention of neural-tube defects and congenital heart defects,” Nutrients, vol. 5, no. 11, pp. 4760–4775, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.United Nations Department of Economic and Social AffairsMillennium Development Goals Report 2009, United Nations Publications, New York, 2009 [Google Scholar]
- 250.Roell M.-S., and Zurbriggen M. D., “The impact of synthetic biology for future agriculture and nutrition,” Current Opinion in Biotechnology, vol. 61, pp. 102–109, 2020 [DOI] [PubMed] [Google Scholar]
- 251.Paul J.-Y., Khanna H., Kleidon J., Hoang P., Geijskes J., Daniells J., Zaplin E., Rosenberg Y., James A., Mlalazi B., Deo P., Arinaitwe G., Namanya P., Becker D., Tindamanyire J., Tushemereirwe W., Harding R., and Dale J., “Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene,” Plant Biotechnology Journal, vol. 15, no. 4, pp. 520–532, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wurtzel E. T., “Changing form and function through carotenoids and synthetic biology,” Plant Physiology, vol. 179, no. 3, pp. 830–843, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Masuda H., Ishimaru Y., Aung M. S., Kobayashi T., Kakei Y., Takahashi M., Higuchi K., Nakanishi H., and Nishizawa N. K., “Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition,” Scientific Reports, vol. 2, no. 1, article 543, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Díaz de la Garza R. I., Gregory J. F., and Hanson A. D., “Folate biofortification of tomato fruit,” Proceedings of the National Academy of Sciences, vol. 104, no. 10, pp. 4218–4222, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mène-Saffrané L., and Pellaud S., “Current strategies for vitamin E biofortification of crops,” Current Opinion in Biotechnology, vol. 44, pp. 189–197, 2017 [DOI] [PubMed] [Google Scholar]
- 256.White P., and Broadley M., “Physiological limits to zinc biofortification of edible crops,” Frontiers in Plant Science, vol. 2, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sinha K., and Khare V., “Review on: antinutritional factors in vegetable crops,” Pharma Innovation Journal, vol. 12, pp. 353–358, 2017 [Google Scholar]
- 258.Sharma R., Bharti S., and Kumar K. V. S., “Diet and thyroid - myths and facts,” Journal of Medical Nutrition and Nutraceuticals, vol. 3, pp. 60–65, 2014 [Google Scholar]
- 259.Gonçalves A., Goufo P., Barros A., Domínguez-Perles R., Trindade H., Rosa E. A. S., Ferreira L., and Rodrigues M., “Cowpea (Vigna unguiculata L. Walp), a renewed multipurpose crop for a more sustainable agri-food system: nutritional advantages and constraints,” Journal of the Science of Food and Agriculture, vol. 96, no. 9, pp. 2941–2951, 2016 [DOI] [PubMed] [Google Scholar]
- 260.IPCCCore Writing Team, Pachauri R., and Meyer L., Eds., IPCC, Geneva, Switzerland, 2014 [Google Scholar]
- 261.Rogelj J., Forster P. M., Kriegler E., Smith C. J., and Séférian R., “Estimating and tracking the remaining carbon budget for stringent climate targets,” Nature, vol. 571, no. 7765, pp. 335–342, 2019 [DOI] [PubMed] [Google Scholar]
- 262.DeLisi C., “The role of synthetic biology in climate change mitigation,” Biology Direct, vol. 14, no. 1, p. 14, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.DeLisi C., Patrinos A., MacCracken M., Drell D., Annas G., Arkin A., Church G., Cook-Deegan R., Jacoby H., Lidstrom M., Melillo J., Milo R., Paustian K., Reilly J., Roberts R. J., Segrè D., Solomon S., Woolf D., Wullschleger S. D., and Yang X., “The role of synthetic biology in atmospheric greenhouse gas reduction: prospects and challenges,” BioDesign Research, vol. 2020, pp. 1–8, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kalluri U. C., Yang X., and Wullschleger S. D., “Plant biosystems design for a carbon-neutral bioeconomy,” BioDesign Research, vol. 2020, pp. 1–5, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Baker B., “Can modern agriculture be sustainable?,” BioScience, vol. 67, no. 4, pp. 325–331, 2017 [Google Scholar]
- 266.Ma T., Dai G., Zhu S., Chen D., Chen L., Lü X., Wang X., Zhu J., Zhang Y., Ma W., He J. S., Bai Y., Han X., and Feng X., “Distribution and preservation of root- and shoot-derived carbon components in soils across the Chinese-Mongolian grasslands,” Journal of Geophysical Research: Biogeosciences, vol. 124, no. 2, pp. 420–431, 2019 [Google Scholar]
- 267.Blaustein R., “Sowing seeds for carbon sequestration,” Bioscience, vol. 69, no. 5, pp. 404–404, 2019 [Google Scholar]
- 268.Harwatt H., “Including animal to plant protein shifts in climate change mitigation policy: a proposed three-step strategy,” Climate Policy, vol. 19, no. 5, pp. 533–541, 2019 [Google Scholar]
- 269.Bastin J.-F., Finegold Y., Garcia C., Mollicone D., Rezende M., Routh D., Zohner C. M., and Crowther T. W., “The global tree restoration potential,” Science, vol. 365, no. 6448, pp. 76–79, 2019 [DOI] [PubMed] [Google Scholar]
- 270.Abbas-Aghababazadeh F., Mo Q., and Fridley B. L., “Statistical genomics in rare cancer,” Seminars in Cancer Biology, vol. 61, pp. 1–10, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.He H., Veneklaas E. J., Kuo J., and Lambers H., “Physiological and ecological significance of biomineralization in plants,” Trends in Plant Science, vol. 19, no. 3, pp. 166–174, 2014 [DOI] [PubMed] [Google Scholar]
- 272.Jansson C., Mockler T., Vogel J., DePaoli H., Hazen S., Srinivasan V., Cousins A., Lemaux P., Dahlberg J., and Brutnell T.. Oxygen Production and Reduction in Artificial and Natural Systems, Barber J., Ruban A. V., and Nixon P. J., Eds., World Scientific Publishing, Singapore, 2019 [Google Scholar]
- 273.U.S. DOEGenome Engineering for Materials Synthesis Workshop Report. DOE/SC-0198, Department of Energy Office of Science, U.S, 2019 [Google Scholar]
- 274.Harfouche A., Meilan R., and Altman A., “Tree genetic engineering and applications to sustainable forestry and biomass production,” Trends in Biotechnology, vol. 29, no. 1, pp. 9–17, 2011 [DOI] [PubMed] [Google Scholar]
- 275.Ramamoorthy S. K., Skrifvars M., and Persson A., “A review of natural fibers used in biocomposites: plant, animal and regenerated cellulose fibers,” Polymer Reviews, vol. 55, no. 1, pp. 107–162, 2015 [Google Scholar]
- 276.Singh P., Kim Y.-J., Zhang D., and Yang D.-C., “Biological synthesis of nanoparticles from plants and microorganisms,” Trends in Biotechnology, vol. 34, no. 7, pp. 588–599, 2016 [DOI] [PubMed] [Google Scholar]
- 277.Fang W., Yang S., Wang X.-L., Yuan T.-Q., and Sun R.-C., “Manufacture and application of lignin-based carbon fibers (LCFs) and lignin-based carbon nanofibers (LCNFs),” Green Chemistry, vol. 19, no. 8, pp. 1794–1827, 2017 [Google Scholar]
- 278.Campuzano S., and Pelling A. E., “Scaffolds for 3D cell culture and cellular agriculture applications derived from non-animal sources,” Frontiers in Sustainable Food Systems, vol. 3, p. 38, 2019 [Google Scholar]
- 279.Jovic T. H., Kungwengwe G., Mills A. C., and Whitaker I. S., “Plant-derived biomaterials: a review of 3D bioprinting and biomedical applications,” Frontiers in Mechanical Engineering, vol. 5, p. 19, 2019 [Google Scholar]
- 280.Karan H., Funk C., Grabert M., Oey M., and Hankamer B., “Green bioplastics as part of a circular bioeconomy,” Trends in Plant Science, vol. 24, no. 3, pp. 237–249, 2019 [DOI] [PubMed] [Google Scholar]
- 281.Mohammadinejad R., Maleki H., Larrañeta E., Fajardo A. R., Nik A. B., Shavandi A., Sheikhi A., Ghorbanpour M., Farokhi M., Govindh P., Cabane E., Azizi S., Aref A. R., Mozafari M., Mehrali M., Thomas S., Mano J. F., Mishra Y. K., and Thakur V. K., “Status and future scope of plant-based green hydrogels in biomedical engineering,” Applied Materials Today, vol. 16, pp. 213–246, 2019 [Google Scholar]
- 282.Bertella S., and Luterbacher J. S., “Lignin functionalization for the production of novel materials,” Trends in Chemistry, vol. 2, no. 5, pp. 440–453, 2020 [Google Scholar]
- 283.Nakata P. A., “Calcium oxalate crystal morphology,” Trends in Plant Science, vol. 7, no. 7, p. 324, 2002 [DOI] [PubMed] [Google Scholar]
- 284.Sun J., and Bhushan B., “Hierarchical structure and mechanical properties of nacre: a review,” RSC Advances, vol. 2, no. 20, pp. 7617–7632, 2012 [Google Scholar]
- 285.Abd A., “Studying the mechanical and electrical properties of epoxy with PVC and calcium carbonate filler,” International Journal of Engineering & Technology, vol. 3, no. 4, pp. 545–553, 2014 [Google Scholar]
- 286.Lertvachirapaiboon C., Pienpinijtham P., Wongravee K., and Ekgasit S., “Optical properties of individual aragonite plates from nacre,” ChemistrySelect, vol. 3, no. 41, pp. 11700–11704, 2018 [Google Scholar]
- 287.Sun W., Jayaraman S., Chen W., Persson K. A., and Ceder G., “Nucleation of metastable aragonite CaCO3 in seawater,” Proceedings of the National Academy of Sciences, vol. 112, no. 11, pp. 3199–3204, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Davies R., Teall O., Pilegis M., Kanellopoulos A., Sharma T., Jefferson A., Gardner D., al-Tabbaa A., Paine K., and Lark R., “Large scale application of self-healing concrete: design, construction, and testing,” Frontiers in Materials, vol. 5, p. 51, 2018 [Google Scholar]
- 289.Chen T., Shi P., Li Y., Duan T., Yu Y., Li X., and Zhu W., “Biomineralization of varied calcium carbonate crystals by the synergistic effect of silk fibroin/magnesium ions in a microbial system,” CrystEngComm, vol. 20, no. 17, pp. 2366–2373, 2018 [Google Scholar]
- 290.Daicho K., Saito T., Fujisawa S., and Isogai A., “The crystallinity of nanocellulose: dispersion-induced disordering of the grain boundary in biologically structured cellulose,” ACS Applied Nano Materials, vol. 1, no. 10, pp. 5774–5785, 2018 [Google Scholar]
- 291.Vandavasi V. G., Putnam D. K., Zhang Q., Petridis L., Heller W. T., Nixon B. T., Haigler C. H., Kalluri U., Coates L., Langan P., Smith J. C., Meiler J., and O’Neill H., “A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers,” Plant Physiology, vol. 170, pp. 123–135, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Joshi C. P., Thammannagowda S., Fujino T., Gou J. Q., Avci U., Haigler C. H., McDonnell L. M., Mansfield S. D., Mengesha B., Carpita N. C., Harris D., DeBolt S., and Peter G. F., “Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen,” Molecular Plant, vol. 4, no. 2, pp. 331–345, 2011 [DOI] [PubMed] [Google Scholar]
- 293.Zhang X., Dominguez P. G., Kumar M., Bygdell J., Miroshnichenko S., Sundberg B., Wingsle G., and Niittylä T., “Cellulose synthase stoichiometry in aspen differs from Arabidopsis and Norway spruce,” Plant Physiology, vol. 177, no. 3, pp. 1096–1107, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lu H., Yuan G., Strauss S. H., Tschaplinski T. J., Tuskan G. A., Chen J. G., and Yang X., “Reconfiguring plant metabolism for biodegradable plastic production,” BioDesign Research, vol. 2020, pp. 1–13, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Savage N., “Fuel options: the ideal biofuel,” Nature, vol. 474, no. 7352, pp. S9–S11, 2011 [DOI] [PubMed] [Google Scholar]
- 296.Rathore D., Nizami A.-S., Singh A., and Pant D., “Key issues in estimating energy and greenhouse gas savings of biofuels: challenges and perspectives,” Biofuel Research Journal, vol. 3, no. 2, pp. 380–393, 2016 [Google Scholar]
- 297.Rosegrant M. W.Biofuels and Grain Prices: Impacts and Policy Responses, International Food Policy Research Institute, Washington, DC, 2008 [Google Scholar]
- 298.German L., Goetz A., Searchinger T., Oliveira G. . L. T., Tomei J., Hunsberger C., and Weigelt J., “Sine Qua Nons of sustainable biofuels: distilling implications of under-performance for national biofuel programs,” Energy Policy, vol. 108, pp. 806–817, 2017 [Google Scholar]
- 299.Markel K., Belcher M. S., and Shih P. M., “Defining and engineering bioenergy plant feedstock ideotypes,” Current Opinion in Biotechnology, vol. 62, pp. 196–201, 2020 [DOI] [PubMed] [Google Scholar]
- 300.Díaz-Pérez M. A., and Serrano-Ruiz J. C., “Catalytic production of jet fuels from biomass,” Molecules, vol. 25, no. 4, p. 802, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.McKendry P., “Energy production from biomass (part 1): overview of biomass,” Bioresource Technology, vol. 83, no. 1, pp. 37–46, 2002 [DOI] [PubMed] [Google Scholar]
- 302.Perin G., and Jones P. R., “Economic feasibility and long-term sustainability criteria on the path to enable a transition from fossil fuels to biofuels,” Current Opinion in Biotechnology, vol. 57, pp. 175–182, 2019 [DOI] [PubMed] [Google Scholar]
- 303.Suman J., Uhlik O., Viktorova J., and Macek T., “Phytoextraction of heavy metals: a promising tool for clean-up of polluted environment?,” Frontiers in Plant Science, vol. 9, p. 1476, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.van der Ent A., Baker A. J. M., Reeves R. D., Pollard A. J., and Schat H., “Hyperaccumulators of metal and metalloid trace elements: facts and fiction,” Plant and Soil, vol. 362, pp. 319–334, 2013 [Google Scholar]
- 305.Reeves R. D., Baker A. J. M., Jaffré T., Erskine P. D., Echevarria G., and van der Ent A., “A global database for plants that hyperaccumulate metal and metalloid trace elements,” New Phytologist, vol. 218, no. 2, pp. 407–411, 2018 [DOI] [PubMed] [Google Scholar]
- 306.Rascio N., and Navari-Izzo F., “Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting?,” Plant Science, vol. 180, no. 2, pp. 169–181, 2011 [DOI] [PubMed] [Google Scholar]
- 307.Ali H., Khan E., and Sajad M. A., “Phytoremediation of heavy metals—concepts and applications,” Chemosphere, vol. 91, no. 7, pp. 869–881, 2013 [DOI] [PubMed] [Google Scholar]
- 308.van der Ent A., Baker A. J., Reeves R. D., Chaney R. L., Anderson C. W., Meech J. A., Erskine P. D., Simonnot M. O., Vaughan J., Morel J. L., and Echevarria G., “Agromining: farming for metals in the future?,” Environmental Science & Technology, vol. 49, no. 8, pp. 4773–4780, 2015 [DOI] [PubMed] [Google Scholar]
- 309.Narender Reddy G., and Prasad M. N. V., “Heavy metal-binding proteins/peptides: occurrence, structure, synthesis and functions. A review,” Environmental and Experimental Botany, vol. 30, no. 3, pp. 251–264, 1990 [Google Scholar]
- 310.Tyler G., “Rare earth elements in soil and plant systems - a review,” Plant and Soil, vol. 267, no. 1-2, pp. 191–206, 2004 [Google Scholar]
- 311.Turra C., De Nadai Fernandes E. A., Bacchi M. A., Sarriés G. A., and Reyes A. E. L., “Uptake of rare earth elements by citrus plants from phosphate fertilizers,” Plant and Soil, vol. 437, no. 1-2, pp. 291–299, 2019 [Google Scholar]
- 312.Mikołajczak P., Borowiak K., and Niedzielski P., “Phytoextraction of rare earth elements in herbaceous plant species growing close to roads,” Environmental Science and Pollution Research International, vol. 24, no. 16, pp. 14091–14103, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Calixto J. B., “The role of natural products in modern drug discovery,” Anais da Academia Brasileira de Ciências, vol. 91, Suppl 3, p. e20190105, 2019 [DOI] [PubMed] [Google Scholar]
- 314.Mortimer J. C., “Plant synthetic biology could drive a revolution in biofuels and medicine,” Experimental Biology and Medicine, vol. 244, no. 4, pp. 323–331, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Brook K., Bennett J., and Desai S. P., “The chemical history of morphine: an 8000-year journey, from resin to de-novo synthesis,” Journal of Anesthesia History, vol. 3, no. 2, pp. 50–55, 2017 [DOI] [PubMed] [Google Scholar]
- 316.Graham I. A., Besser K., Blumer S., Branigan C. A., Czechowski T., Elias L., Guterman I., Harvey D., Isaac P. G., Khan A. M., Larson T. R., Li Y., Pawson T., Penfield T., Rae A. M., Rathbone D. A., Reid S., Ross J., Smallwood M. F., Segura V., Townsend T., Vyas D., Winzer T., and Bowles D., “The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin,” Science, vol. 327, no. 5963, pp. 328–331, 2010 [DOI] [PubMed] [Google Scholar]
- 317.Ikram N. K. B. K., and Simonsen H. T., “A review of biotechnological artemisinin production in plants,” Frontiers in Plant Science, vol. 8, no. 1966, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Mohammadinejad R., Shavandi A., Raie D. S., Sangeetha J., Soleimani M., Shokrian Hajibehzad S., Thangadurai D., Hospet R., Popoola J. O., Arzani A., Gómez-Lim M. A., Iravani S., and Varma R. S., “Plant molecular farming: production of metallic nanoparticles and therapeutic proteins using green factories,” Green Chemistry, vol. 21, no. 8, pp. 1845–1865, 2019 [Google Scholar]
- 319.Lomonossoff G. P., and D'Aoust M. A., “Plant-produced biopharmaceuticals: a case of technical developments driving clinical deployment,” Science, vol. 353, no. 6305, pp. 1237–1240, 2016 [DOI] [PubMed] [Google Scholar]
- 320.Virdi V., and Depicker A., “Role of plant expression systems in antibody production for passive immunization,” International Journal of Developmental Biology, vol. 57, no. 6-7-8, pp. 587–593, 2013 [DOI] [PubMed] [Google Scholar]
- 321.Karg S. R., and Kallio P. T., “The production of biopharmaceuticals in plant systems,” Biotechnology Advances, vol. 27, no. 6, pp. 879–894, 2009 [DOI] [PubMed] [Google Scholar]
- 322.Fischer R., Stoger E., Schillberg S., Christou P., and Twyman R. M., “Plant-based production of biopharmaceuticals,” Current Opinion in Plant Biology, vol. 7, no. 2, pp. 152–158, 2004 [DOI] [PubMed] [Google Scholar]
- 323.Chen M., Liu X., Wang Z., Song J., Qi Q., and Wang P. G., “Modification of plant N-glycans processing: the future of producing therapeutic protein by transgenic plants,” Medicinal Research Reviews, vol. 25, no. 3, pp. 343–360, 2005 [DOI] [PubMed] [Google Scholar]
- 324.Arntzen C., “Plant-made pharmaceuticals: from ‘Edible Vaccines’ to Ebola therapeutics,” Plant Biotechnology Journal, vol. 13, no. 8, pp. 1013–1016, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yao J., Weng Y., Dickey A., and Wang K. Y., “Plants as factories for human pharmaceuticals: applications and challenges,” International Journal of Molecular Sciences, vol. 16, no. 12, pp. 28549–28565, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Capell T., Twyman R. M., Armario-Najera V., Ma J. K. C., Schillberg S., and Christou P., “Potential applications of plant biotechnology against SARS-CoV-2,” Trends in Plant Science, vol. 25, no. 7, pp. 635–643, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.King N., Hittinger C. T., and Carroll S. B., “Evolution of key cell signaling and adhesion protein families predates animal origins,” Science, vol. 301, no. 5631, pp. 361–363, 2003 [DOI] [PubMed] [Google Scholar]
- 328.Woods J. O., Singh-Blom U. M., Laurent J. M., McGary K. L., and Marcotte E. M., “Prediction of gene-phenotype associations in humans, mice, and plants using phenologs,” BMC Bioinformatics, vol. 14, no. 1, p. 203, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Klekowski E. J., “Genetic load and its causes in long-lived plants,” Trees, vol. 2, no. 4, pp. 195–203, 1988 [Google Scholar]
- 330.Tuskan G. A., Mewalal R., Gunter L. E., Palla K. J., Carter K., Jacobson D. A., Jones P. C., Garcia B. J., Weighill D. A., Hyatt P. D., Yang Y., Zhang J., Reis N., Chen J. G., and Muchero W., “Defining the genetic components of callus formation: a GWAS approach,” PLoS One, vol. 13, no. 8, article e0202519, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.O'Neill E. E., Matallanas D., and Kolch W., “Mammalian sterile 20-like kinases in tumor suppression: an emerging pathway:,” Cancer Research, vol. 65, no. 13, pp. 5485–5487, 2005 [DOI] [PubMed] [Google Scholar]
- 332.Bdeir R., Muchero W., Yordanov Y., Tuskan G. A., Busov V., and Gailing O., “Quantitative trait locus mapping of Populus bark features and stem diameter,” BMC Plant Biology, vol. 17, no. 1, article 224, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.McGrath J. A., “Hereditary diseases of desmosomes,” Journal of Dermatological Science, vol. 20, no. 2, pp. 85–91, 1999 [DOI] [PubMed] [Google Scholar]
- 334.Chenoweth M. J., Giacomini K. M., Pirmohamed M., Hill S. L., van Schaik R. H., Schwab M., Shuldiner A. R., Relling M. V., and Tyndale R. F., “Global pharmacogenomics within precision medicine: challenges and opportunities,” Clinical Pharmacology & Therapeutics, vol. 107, pp. 57–61, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Walia A., Waadt R., and Jones A. M., “Genetically encoded biosensors in plants: pathways to discovery,” Annual Review of Plant Biology, vol. 69, no. 1, pp. 497–524, 2018 [DOI] [PubMed] [Google Scholar]
- 336.Kollist H., Zandalinas S. I., Sengupta S., Nuhkat M., Kangasjärvi J., and Mittler R., “Rapid responses to abiotic stress: priming the landscape for the signal transduction network,” Trends in Plant Science, vol. 24, no. 1, pp. 25–37, 2019 [DOI] [PubMed] [Google Scholar]
- 337.Antunes M. S., Ha S. B., Tewari-Singh N., Morey K. J., Trofka A. M., Kugrens P., Deyholos M., and Medford J. I., “A synthetic de-greening gene circuit provides a reporting system that is remotely detectable and has a re-set capacity,” Plant Biotechnology Journal, vol. 4, no. 6, pp. 605–622, 2006 [DOI] [PubMed] [Google Scholar]
- 338.Antunes M. S., Morey K. J., Smith J. J., Albrecht K. D., Bowen T. A., Zdunek J. K., Troupe J. F., Cuneo M. J., Webb C. T., Hellinga H. W., and Medford J. I., “Programmable ligand detection system in plants through a synthetic signal transduction pathway,” PLoS One, vol. 6, no. 1, article e16292, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Teague B. P., Guye P., and Weiss R., “Synthetic morphogenesis,” Cold Spring Harbor Perspectives in Biology, vol. 8, no. 9, article a023929, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Wightman R., and Luo C. J., “From mammalian tissue engineering to 3D plant cell culture,” The Biochemist, vol. 38, no. 4, pp. 32–35, 2016 [Google Scholar]
- 341.Luo C. J., Wightman R., Meyerowitz E., and Smoukov S. K., “A 3-dimensional fibre scaffold as an investigative tool for studying the morphogenesis of isolated plant cells,” BMC Plant Biology, vol. 15, no. 1, article 211, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Bernstein R., Ingram K., and Hart K. M.. BioBuilder: Synthetic Biology in the Lab, O'Reilly Media, Inc., Sebastopol, 2015 [Google Scholar]
- 343.Llorente B., Williams T. C., and Goold H. D., “The multiplanetary future of plant synthetic biology,” Genes, vol. 9, no. 7, p. 348, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Menezes A. A., Montague M. G., Cumbers J., Hogan J. A., and Arkin A. P., “Grand challenges in space synthetic biology,” Journal of The Royal Society Interface, vol. 12, no. 113, pp. 20150803–20150803, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Schmidt S. M., Belisle M., and Frommer W. B., “The evolving landscape around genome editing in agriculture,” EMBO Reports, vol. 21, article e50680, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kawall K., Cotter J., and Then C., “Broadening the GMO risk assessment in the EU for genome editing technologies in agriculture,” Environmental Sciences Europe, vol. 32, no. 1, article 106, 2020 [Google Scholar]
- 347.Paine J. A., Shipton C. A., Chaggar S., Howells R. M., Kennedy M. J., Vernon G., Wright S. Y., Hinchliffe E., Adams J. L., Silverstone A. L., and Drake R., “Improving the nutritional value of Golden Rice through increased pro-vitamin A content,” Nature Biotechnology, vol. 23, no. 4, pp. 482–487, 2005 [DOI] [PubMed] [Google Scholar]
- 348.Wakeel A., Arif S., Bashir M. A., Ahmad Z., Rehman H. ., Kiran A., Ibrahim S., and Khan M. R., “Perspectives of folate biofortification of cereal grains,” Journal of Plant Nutrition, vol. 41, no. 19, pp. 2507–2524, 2018 [Google Scholar]
- 349.Shih M.-L., and Morgan J. A., “Metabolic flux analysis of secondary metabolism in plants,” Metabolic Engineering Communications, vol. 10, article e00123, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Nordlee J. A., Taylor S. L., Townsend J. A., Thomas L. A., and Bush R. K., “Identification of a Brazil-nut allergen in transgenic soybeans,” New England Journal of Medicine, vol. 334, no. 11, pp. 688–692, 1996 [DOI] [PubMed] [Google Scholar]
- 351.Nestle M., “Allergies to transgenic foods--questions of policy,” New England Journal of Medicine, vol. 334, no. 11, pp. 726–728, 1996 [DOI] [PubMed] [Google Scholar]
- 352.Ryffel G. U., “Transgene flow: facts, speculations and possible countermeasures,” GM Crops Food, vol. 5, no. 4, pp. 249–258, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lu B.-R., “Transgene escape from GM crops and potential biosafety consequences: an environmental perspective,” Collection of Biosafety Reviews, vol. 4, pp. 66–141, 2008 [Google Scholar]
- 354.Ruf S., Karcher D., and Bock R., “Determining the transgene containment level provided by chloroplast transformation,” Proceedings of the National Academy of Sciences, vol. 104, no. 17, pp. 6998–7002, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kuvshinov V., Koivu K., Kanerva A., and Pehu E., “Molecular control of transgene escape from genetically modified plants,” Plant Science, vol. 160, no. 3, pp. 517–522, 2001 [DOI] [PubMed] [Google Scholar]
- 356.Daniell H., “Molecular strategies for gene containment in transgenic crops,” Nature Biotechnology, vol. 20, no. 6, pp. 581–586, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Klocko A. L., Lu H., Magnuson A., Brunner A. M., Ma C., and Strauss S. H., “Phenotypic expression and stability in a large-scale field study of genetically engineered poplars containing sexual containment transgenes,” Frontiers in Bioengineering and Biotechnology, vol. 6, p. 100, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Powles S. B., and Yu Q., “Evolution in action: plants resistant to herbicides,” Annual Review of Plant Biology, vol. 61, no. 1, pp. 317–347, 2010 [DOI] [PubMed] [Google Scholar]
- 359.Carrière Y., Fabrick J. A., and Tabashnik B. E., “Can pyramids and seed mixtures delay resistance to Bt crops?,” Trends in Biotechnology, vol. 34, no. 4, pp. 291–302, 2016 [DOI] [PubMed] [Google Scholar]
- 360.Young J., Zastrow-Hayes G., Deschamps S., Svitashev S., Zaremba M., Acharya A., Paulraj S., Peterson-Burch B., Schwartz C., Djukanovic V., Lenderts B., Feigenbutz L., Wang L., Alarcon C., Siksnys V., May G., Chilcoat N. D., and Kumar S., “CRISPR-Cas9 editing in maize: systematic evaluation of off-target activity and its relevance in crop improvement,” Scientific Reports, vol. 9, no. 1, article 6729, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Carroll D., “Collateral damage: benchmarking off-target effects in genome editing,” Genome Biology, vol. 20, no. 1, p. 114, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Abraham P. E., Labbé J. L., and McBride A. A., “Advancing how we learn from biodesign to mitigate risks with next-generation genome engineering,” BioDesign Research, vol. 2020, pp. 1–3, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Lee J. H., Mazarei M., Pfotenhauer A. C., Dorrough A. B., Poindexter M. R., Hewezi T., Lenaghan S. C., Graham D. E., and Stewart C. N. Jr., “Epigenetic footprints of CRISPR/Cas9-mediated genome editing in plants,” Frontiers in Plant Science, vol. 10, p. 1720, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Losey J. E., Rayor L. S., and Carter M. E., “Transgenic pollen harms monarch larvae,” Nature, vol. 399, no. 6733, pp. 214–214, 1999 [DOI] [PubMed] [Google Scholar]
- 365.Gutmann A., “The ethics of synthetic biology: guiding principles for emerging technologies,” Hastings Center Report, vol. 41, no. 4, pp. 17–22, 2011 [DOI] [PubMed] [Google Scholar]
- 366.Beyer P., “Golden Rice and ‘Golden’ crops for human nutrition,” New Biotechnology, vol. 27, no. 5, pp. 478–481, 2010 [DOI] [PubMed] [Google Scholar]
- 367.Maynard A. D., “Navigating the fourth industrial revolution,” Nature Nanotechnology, vol. 10, no. 12, pp. 1005–1006, 2015 [DOI] [PubMed] [Google Scholar]
- 368.Xu M., David J. M., and Kim S. H., “The fourth industrial revolution: opportunities and challenges,” International Journal of Financial Research, vol. 9, no. 2, pp. 90–95, 2018 [Google Scholar]
- 369.Prisecaru P., “Challenges of the fourth industrial revolution,” Knowledge Horizons. Economics, vol. 8, p. 57, 2016 [Google Scholar]
- 370.Gonçalves E., Bucher J., Ryll A., Niklas J., Mauch K., Klamt S., Rocha M., and Saez-Rodriguez J., “Bridging the layers: towards integration of signal transduction, regulation and metabolism into mathematical models,” Molecular BioSystems, vol. 9, no. 7, pp. 1576–1583, 2013 [DOI] [PubMed] [Google Scholar]
- 371.Heirendt L., Arreckx S., Pfau T., Mendoza S. N., Richelle A., Heinken A., Haraldsdóttir H. S., Wachowiak J., Keating S. M., Vlasov V., Magnusdóttir S., Ng C. Y., Preciat G., Žagare A., Chan S. H. J., Aurich M. K., Clancy C. M., Modamio J., Sauls J. T., Noronha A., Bordbar A., Cousins B., el Assal D. C., Valcarcel L. V., Apaolaza I., Ghaderi S., Ahookhosh M., Ben Guebila M., Kostromins A., Sompairac N., le H. M., Ma D., Sun Y., Wang L., Yurkovich J. T., Oliveira M. A. P., Vuong P. T., el Assal L. P., Kuperstein I., Zinovyev A., Hinton H. S., Bryant W. A., Aragón Artacho F. J., Planes F. J., Stalidzans E., Maass A., Vempala S., Hucka M., Saunders M. A., Maranas C. D., Lewis N. E., Sauter T., Palsson B. Ø., Thiele I., and Fleming R. M. T., “Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0,” Nature Protocols, vol. 14, no. 3, pp. 639–702, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Grunberg T. W., and Del Vecchio D., “Modular analysis and design of biological circuits,” Current Opinion in Biotechnology, vol. 63, pp. 41–47, 2020 [DOI] [PubMed] [Google Scholar]
- 373.Tyler B. M., “The fog of war: how network buffering protects plants’ defense secrets from pathogens,” PLoS Genetics, vol. 13, no. 5, article e1006713, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Yugi K., Kubota H., Hatano A., and Kuroda S., “Trans-omics: how to reconstruct biochemical networks across multiple ‘omic’ layers,” Trends in Biotechnology, vol. 34, no. 4, pp. 276–290, 2016 [DOI] [PubMed] [Google Scholar]
- 375.Zampieri G., Vijayakumar S., Yaneske E., and Angione C., “Machine and deep learning meet genome-scale metabolic modeling,” PLoS Computational Biology, vol. 15, no. 7, article e1007084, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]