Dear Editor,
Infertility affects about 15% of couples of childbearing age.1 About half of these cases can be attributed predominantly to a male factor, such as a quantitative or qualitative impairment in spermatogenesis. The etiology of male infertility is related to anatomic, genetic, and environmental causes, being sometimes multifactorial and of unknown or idiopathic origins. At present, genetic screening for nonobstructive azoospermia (NOA) is generally limited to karyotyping for the identification of chromosome abnormalities such as 47,XXY, 46,X,der(X)t(X;Y)(p22.3;p11.2), chromosome rearrangements, and Y chromosome microdeletions. Whole-genome analyses have shown that over 100 gene variants are associated with a male infertility phenotype,2 and the list continues to grow; hence, a large number of genes are involved in spermatogenesis. The continuing identification of genes responsible for male infertility in general and azoospermia, in particular, is a major challenge in both research and the diagnosis of male infertility.
However, gene sequencing has not yet been integrated into the routine clinical management of NOA, even though some researchers have suggested a panel of candidate genes3 or the performance of whole-exome sequencing (WES) before testicular sperm extraction (TESE) or after an initial negative TESE.4 Even though WES might become the “gold standard” genetic analysis in the near future (especially for NOA),5 this step must always be preceded by counseling. Before requesting the patient’s consent for the identification of genetic defects not related to azoospermia (i.e., incidental findings), the counselor must explain the complexity of the data obtained and address the possible outcomes.6
Here, we report a 32-year-old male who consulted in Poissy Saint-Germain en Laye Hospital (Poissy, France) for azoospermia. His personal medical history did not reveal any genital infections, endocrine diseases, genital or spinal trauma, drug abuse, or chronic medication use. His body weight was normal. His pedigree is not remarkable as far as fertility is concerned, except for inbreeding; his parents are first cousins (Supplementary Figure 1a (93.5KB, tif) ). The analysis of two consecutive semen samples revealed complete azoospermia. A physical examination evidenced a small testis volume (6 ml for each testis; normal range: 15–20 ml). The scrotal ultrasound findings were normal. The serum hormone levels were 29.9 IU l−1 for follicle-stimulating hormone (normal range: 1–12 IU l−1), 22 ng l−1 for inhibin B (normal range: 80–400 ng l−1), and 4.5 μg l−1 for total testosterone (normal range: 2.4–8.7 μg l−1). Genetic testing showed a normal karyotype and the absence of Y chromosome microdeletions. After counseling and before TESE, we applied a previously reported WES protocol.4 The study protocol was approved by an independent ethics committee (CPP Ile de France-Ouest, Paris, France; Reference No. 01-132). The patient gave his written informed consent.
We used strict filters to select only rare variants in protein-encoding genes that were involved in spermatogenesis or were testis-enriched, according to the Gene-Tissue Expression (https://gtexportal.org/home/), Human Protein Atlas (https://www.proteinatlas.org), PubMed, and Ensembl databases. Surprisingly (given the known consanguinity), no deleterious homozygous variants were identified. We found a missense hemizygous c.2596G>A, pV866M variant in exon 16 of the zinc-finger, myeloproliferative, and mental retardation-type 3 (ZMYM3) gene on Xq13.1. This variant of uncertain significance and deleterious impact was confirmed by Sanger sequencing (Supplementary Figure 1b (93.5KB, tif) ). Exon 16 is a highly conserved region in humans and mice. The ZMYM3 gene is highly conserved in vertebrates and especially in the mouse. It has been reported that Zmym3 knockout (KO) in the mouse is associated with a low testis volume, spermatogenesis arrest at metaphase I, and infertility in males older than 6 months.7
The ubiquitously expressed ZMYM3 gene is located on the X chromosome and belongs to a family with five other members. It is predominantly expressed in the brain, ovaries, and testis. ZMYM3 protein is expressed in both germ cells (mainly spermatogonia B) and somatic cells (Leydig and Sertoli cells). The protein’s localization in the nucleus is linked to several functions related to DNA protein and zinc ion binding. ZMYM3 protein also has a role in the organization of the cytoskeleton and cell shape regulation.
In view of our present results and according to the American College of Medical Genetics and Genomics classification,8 the c.2596G>A, pV866M mutation is a variant of uncertain significance, even though a deleterious impact is strongly suspected. The patient was given the test result, and TESE was performed. Unfortunately, no spermatozoa were retrieved, with only Sertoli cells being found. A histological analysis highlighted Sertoli cell-only syndrome (SCOS) on 20 seminiferous tubules – the only ones that could be observed. With regard to the discrepancy between the observed phenotype and the KO mouse model, and in order to check for the presence of germ cells inside the patient’s seminiferous tubes, we immunostained testicular tissue samples for stromal antigen 3 (STAG3), a protein that is essential for chromatin cohesion, using a Benchmark Ultra Ventana (Roche, Rotkreuz, Switzerland) automated system and a specific antibody (PA5-63556, Invitrogen, Waltham, MA, USA). The results were compared with a control sample from an individual with obstructive azoospermia and conserved spermatogenesis (Figure 1a). The absence of STAG3 expression in the patient’s samples confirmed the absence of germinal cells and the Sertoli cell-only phenotype (Figure 1b). To screen for the presence of ZMYM3, we immunostained testicular tissue samples with a specific antibody (ab 19165, Abcam, Cambridge, UK). As expected, ZMYM3 was expressed in Sertoli, Leydig and germinal cells in control (Figure 1c), in Sertoli and Leydig cells in the patient (Figure 1d). It was not possible to obtain DNA from the patient’s parents or maternal uncle; we asked the patient to contact them, but he refused. The variant is, therefore, maternally inherited or de novo.
Figure 1.
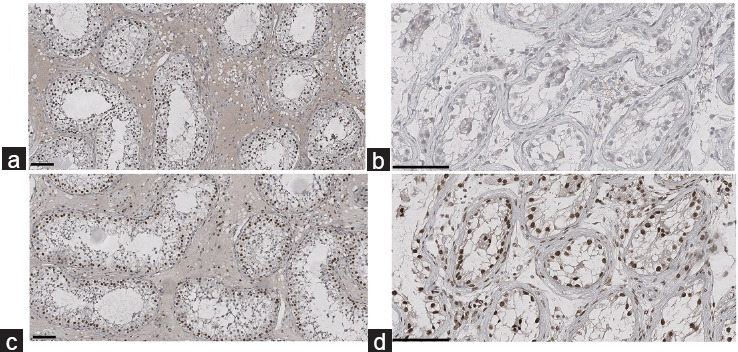
Immunohistochemical staining of the seminiferous tubes. (a) STAG3 expression for the control patient. (b) STAG3 expression for the patient with NOA and a ZMYM3 mutation. (c) ZMYM3 expression for the control patient. (d) ZMYM3 expression for the patient with NOA and a ZMYM3 mutation. Scale bars = 100 μm. NOA: nonobstructive azoospermia; ZMYM3: zinc-finger, myeloproliferative, and mental retardation-type 3; STAG3: stromal antigen 3.
The present case emphasizes the difficulty of evaluating protein expression for a missense variant, especially in germ cells, as no meiotic in vitro model currently exists. Given the very small testis volume, the priority was to find sperm; hence, only a small tissue sample (with just 20 seminiferous tubes analyzed) could be obtained. First, the small size of the sample prevented us from determining whether the testicular phenotype was homogeneous or not; perhaps germ cells would have been observed in another section. Second, it has been reported that the relative decreases in testis sizes, sperm count and fertility in KO mouse model start at the age of 2 months. The mice are infertile by the age of 6 months, after a gradual decrease in the germ cell count. One can hypothesize that germ cell exhaustion occurs in this mouse model. The same might be true in humans. In our present case, TESE might have been attempted too late. Spermatogenesis maturation arrest might constitute a step toward SCOS. Given the absence of a specific phenotype in the Zmym3 KO mouse model (other than infertility), the ZMYM3 variant might be a strong candidate gene for SCOS. However, the variant’s impact must be functionally confirmed in a specific mouse model or an in vitro model using cell lines. At present, there are no in vitro models of meiosis, and it is very complicated (given the 3Rs rule: reduce, replace, and refine) to create an animal model for each variant identified.
In conclusion, we identified a novel ZMYM3 missense hemizygous variant in the X chromosome that, although bioinformatically not predicted to be deleterious, appeared associated with a Sertoli cell-only phenotype. If one day its pathogenicity is proven, it could explain some cases of NOA, as previously suggested for serine peptidase inhibitor Kazal type 2 (SPINK2).9
AUTHOR CONTRIBUTIONS
MLB performed immunohistochemical analyses and Sanger sequencing, and wrote the first draft of the manuscript. NSB and MB enrolled the patient. FG performed whole-exome sequencing interpretation. JF provided technical support for immunohistochemical analyses. CG provided technical support for genetic analyses. VS revised the manuscript. ALS supervised immunohistochemical analyses. FV supervised the study and helped to draft the various versions of the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
(a) Family tree. (b) Sanger sequencing confirmation, using sense primers for control and the patient. The arrow indicates the nucleotide substitution.
ACKNOWLEDGMENT
We would like to thank the staff in the Genetics Department at Poissy St Germain en Laye Hospital for patient management.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, et al. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum Reprod Update. 2021;28:15–29. doi: 10.1093/humupd/dmab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannarella R, Condorelli RA, Paolacci S, Barbagallo F, Guerri G, et al. Next-generation sequencing: toward an increase in the diagnostic yield in patients with apparently idiopathic spermatogenic failure. Asian J Androl. 2021;23:24–9. doi: 10.4103/aja.aja_25_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghieh F, Barbotin AL, Swierkowski-Blanchard N, Leroy C, Fortemps J, et al. Whole-exome sequencing in patients with maturation arrest: a potential additional diagnostic tool for prevention of recurrent negative testicular sperm extraction outcomes. Hum Reprod. 2022;37:1334–50. doi: 10.1093/humrep/deac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghieh F, Barbotin AL, Leroy C, Marcelli F, Swierkowsky-Blanchard N, et al. Will whole-genome sequencing become the first-line genetic analysis for male infertility in the near future? Basic Clin Androl. 2021;31:21. doi: 10.1186/s12610-021-00138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasak L, Lillepea K, Nagirnaja L, Aston KI, Schlegel PN, et al. Actionable secondary findings following exome sequencing of 836 non-obstructive azoospermia cases and their value in patient management. Hum Reprod. 2022;37:1652–63. doi: 10.1093/humrep/deac100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Shen B, Liao S, Ning Y, Ma L. Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death Dis. 2017;8:e2910. doi: 10.1038/cddis.2017.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kherraf ZE, Christou-Kent M, Karaouzene T, Amiri-Yekta A, Martinez G, et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med. 2017;9:1132–49. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Family tree. (b) Sanger sequencing confirmation, using sense primers for control and the patient. The arrow indicates the nucleotide substitution.


