Abstract
Obsessive-compulsive disorder (OCD) is phenotypically heterogeneous and genetically complex. This study aimed to reduce heterogeneity using structural brain imaging to study putative intermediate phenotypes for OCD. We hypothesized that select serotonin gene variants would differ in their relationship with brain volume in specific regions of the cortico-striato-thalamo-cortical (CSTC) circuits between OCD patients and controls. In a total of 200 pediatric subjects, we genotyped candidate serotonin genes (SLC6A4, HTR2A, HTR1B, and HTR2C) and conducted structural magnetic resonance imaging (sMRI) to measure regional brain volumes within CSTC circuits. In males and females separately, we first tested the association between serotonin gene variants and OCD and the effect of serotonin gene variants on brain volume irrespective of diagnosis. We then carried out a series of analyses to assess the effect of genotype-diagnosis interaction on brain volume. In females, but not in males, we identified a statistically significant genotype-diagnosis interaction for two single nucleotide polymorphisms (SNPs) in HTR2C, rs12860460 (interaction term estimate of 5.45 cc and interaction P value of 9.70e-8) and rs12854485 (interaction term estimate of 4.28 cc and interaction P value of 2.07e-6). The tested allele in each SNP was associated with decreased anterior cingulate cortex (ACC) volume in controls and with increased ACC volume in OCD patients. Our findings suggest that, in females, sequence variation in HTR2C influences ACC volume in pediatric OCD. The variants may contribute to differences in ACC volume and to OCD in a sex-specific manner when acting together with other genetic, biological, and/or environmental factors.
Keywords: Obsessive-compulsive disorder, OCD, Serotonin system genes, Neuroimaging, Intermediate phenotype
Introduction
Obsessive-compulsive disorder (OCD) is a neuropsychiatric disorder characterized by intrusive, repetitive thoughts and behaviors that are typically distressing to the patient (National Institutes of Health [NIH] 2008; American Psychiatric Association [APA] 2013). The disorder is heritable, as evidenced by twin studies and direct estimation of heritability from genome-wide common variant data (Davis et al. 2013; Pauls et al. 2014). Furthermore, OCD is genetically complex and phenotypically heterogeneous (Arnold and Richter 2007; Grados 2010). Various approaches have been used to identify genetic variants associated with OCD. These include studies of variants within candidate serotonin system genes. The best-studied serotonin candidate genes for OCD include SLC6A4 (solute carrier family 6 member 4), HTR2A, HTR1B, and HTR2C, which correspond with the serotonin transporter, or SERT, and serotonin receptors 5-HT2A, 5-HT1B, and 5-HT2C respectively (Taylor 2013; Sinopoli et al. 2017). Within SLC6A4, the most extensively studied polymorphism, 5-HTTLPR (serotonin transporter-linked polymorphic region), lies within the promoter region of the gene (Heils et al. 1996; Lesch et al. 1996). The polymorphism exists in two major forms, one long (L) variant which consists of 16 sets of 20–23 bp tandem repeats and one short (S) variant which consists of 14 sets of 20–23 bp tandem repeats. In addition, an intrinsic A/G single nucleotide polymorphism (SNP), rs25531, occurs within the L variant and influences SERT expression. Taken together, the 5-HTTLPR and rs25531 is a triallelic system (LA, LG and S variants) with the LA variant resulting in greater SERT expression compared with the S and LG variants. These variants appear to be additive in their effect (Hu et al. 2006).
Strong evidence supports an association between OCD and this commonly studied polymorphic region of SLC6A4 (Taylor 2013; Walitza et al. 2014; Taylor 2016; Sinopoli et al. 2017); however, findings have been somewhat mixed with respect to which variant of 5-HTTLPR is associated with OCD. These mixed results may be because of heterogeneity in the OCD phenotype that has not been adequately addressed in previous studies. Sources of heterogeneity include multiple overlapping symptom dimensions in OCD (Bloch et al. 2008; Alvarenga et al. 2015), subtypes of OCD that depend on factors such as age of onset or comorbidity with tic disorders (Grados 2010; Leckman et al. 2010; APA 2013; Williams et al. 2013) and sex differences (Alvarenga et al. 2015; Mak et al. 2015).
Biologically salient, quantitative measures that mediate genotype and phenotype, often called “intermediate phenotypes”, are arguably less heterogeneous than clinical diagnostic syndromes like OCD (Meyer-Lindenberg and Weinberger 2006). For example, brain differences measured by neuroimaging techniques like magnetic resonance imaging (MRI) may provide intermediate phenotypes that can be tested for their association with a particular set of candidate genes or used in a genome-wide association study (GWAS). There are two putative advantages of studying intermediate phenotypes compared with psychiatric diagnoses like OCD: 1) Reduced heterogeneity of such measures may lead to increased power to detect genetic association due to increased effect sizes (Leite et al. 2015), although recent imaging genetics studies suggest such effect sizes may be smaller than originally assumed (Carter et al. 2017), and 2) Imaging and other biologically salient measures may help delineate underlying biological mechanisms mediating gene variation and clinical symptoms (Meyer-Lindenberg and Weinberger 2006; Leite et al. 2015). OCD has been repeatedly linked to dysfunction in cortico-striato-thalamo-cortical (CSTC) circuits. Evidence includes volumetric and functional abnormalities in various regions of CSTC circuits including the striatum (caudate and putamen), thalamus, anterior cingulate cortex (ACC), and orbitofrontal cortex (OFC), with some studies reporting differences in the directionality/magnitude of the effects between pediatric and adult OCD patients (MacMaster 2010; Brem et al. 2012; Pauls et al. 2014; Boedhoe et al. 2017). Both pharmacotherapy with a serotonin reuptake inhibitor (SRI) and cognitive behavioral therapy for OCD also alter metabolic activity in CSTC-related brain regions (Pauls et al. 2014).
Only three studies have been published which focus on the association between serotonin system gene variants and brain structure or function as measured by neuroimaging in individuals (predominantly adults) with OCD (Atmaca et al. 2011; Hesse et al. 2011; Honda et al. 2017). Upon examining the ACC, OFC, and thalamus using MRI, Atmaca et al. (2011) reported a significant genotype-diagnosis interaction whereby the effect of the 5-HTTLPR polymorphism on OFC volume was dependent on OCD diagnosis. S variant carriers showed a significantly smaller OFC volume in individuals with OCD, but not in healthy controls. Hesse et al. (2011) used positron emission tomography (PET) with a SERT-selective radiotracer and examined both 5-HTTLPR and STin2 VNTR (variable number of tandem repeats polymorphism in intron 2), another commonly studied polymorphism in SLC6A4. There was no significant effect of the interaction between presence of OCD and 5-HTTLPR genotype on SERT availability. When examining OCD patients alone, however, the authors reported an overall trend towards increased SERT availability in patients with the genotype S/S of 5-HTTLPR (reaching significance in the midbrain), a significant effect of STin2 VNTR genotype on SERT availability in the ACC and putamen, and a significant effect of the interaction between 5-HTTLPR and the STin2 VNTR on SERT availability in the ACC, putamen, and hypothalamus. Neither the study by Atmaca et al. (2011) nor the study by Hesse et al. (2011) corrected for multiple comparisons. The most recent study examined association between 5-HTTLPR and regional brain volumes measured using MRI, while accounting for the differential functionality imparted by rs25531. Authors reported a nominally significant genotype-diagnosis interaction whereby the effect of the 5-HTTLPR polymorphism on the right frontal pole was dependent on OCD diagnosis. They also noted a trend for OCD patients carrying LA to have reduced gray matter volume in the right frontal pole relative to healthy controls carrying LA (Honda et al. 2017). Overall, serotonin system gene variants may be associated with brain changes in OCD but, given the limited number of studies (which used different imaging modalities and different genotyping methods), further and more extensive studies are required.
To date, no studies examining serotonin system gene variants and their relationship to potential neuroimaging phenotypes in OCD have been conducted in pediatric samples. Early-onset OCD (onset typically in childhood or early adolescence) is considered a distinct subtype that differs from late-onset OCD (onset typically in late adolescence or early adulthood) with regard to a number of demographic and clinical features (MacMaster 2010). To reduce heterogeneity, we have focused on children and adolescents with OCD (therefore predominantly capturing individuals with early-onset OCD) (Mattina and Steiner 2016). There is also evidence from a recent meta-analysis suggesting that serotonin system gene variants differ in their association with OCD in males versus in females. Specifically, the S variant may be more associated with OCD in females than in males (Mak et al. 2015). To further reduce heterogeneity in our study, we therefore also chose to stratify by sex (Mattina and Steiner 2016). This decision is further supported by the fact that one of our genes of interest (HTR2C) is located on chromosome X and may therefore be associated with sex differences.
In our study, we examined selected genetic variants in four candidate serotonin system genes, SLC6A4, HTR2A, HTR1B, and HTR2C. Using brain volume in regions previously implicated in OCD, we aimed to identify associations between gene variants and neuroimaging phenotypes, specifically volumes of the ACC and other brain regions that have been implicated in OCD. We focused on children and adolescents, and we examined males and females separately to reduce heterogeneity. If the serotonin system is involved in OCD pathogenesis and if dysfunctional CSTC circuitry is involved in OCD pathogenesis, we hypothesized that we would find serotonin gene variants that differ in their association with brain volume in OCD patients versus in controls. Furthermore, given previously reported sex differences in OCD (Yücel et al. 2008; Mattina and Steiner 2016), we hypothesized that our findings would differ between males and females in our pediatric population.
Materials and methods
Subjects
A total of 411 participants 6–19 years of age were initially recruited at two sites, which included Wayne State University in Detroit, Michigan and the University of Michigan in Ann Arbor, Michigan, as part of a continuing collaborative study (Arnold et al. 2009; Wu et al. 2013). Written and informed consent and assent was obtained for all participants and, when applicable, from their parents. Each site received study approval from its respective Human Investigation Committee prior to recruitment of participants into the study. OCD participants were required to have a lifetime OCD diagnosis and a current, minimum total score of 10 on the Children’s Yale-Brown Obsessive-Compulsive Scale (CY-BOCS) (Scahill et al. 1997). OCD cases and healthy controls were assessed for lifetime diagnosis of Axis I psychiatric disorders, using DSM-IV-TR criteria (APA 2000) based on information obtained through two semi-structured interviews, the Schedule for Affective Disorder and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL) (Kaufman et al. 1997) and the pediatric version of the Schedule for Obsessive-Compulsive and Other Behavioral Syndromes (SOCOBS) (Hanna 2010), administered to both the participant and/or both parents. Composite diagnoses were based on agreement between the two interviews regarding 1) specific diagnoses and 2) level of diagnosis-related interference. OCD cases and healthy controls were also assessed using the Pubertal Development Scale (PDS) (adapted from Petersen et al. 1988).
For both OCD cases and healthy controls, participants were excluded if neurological disorders, such as seizure disorder, were identified. Participants were also excluded if they had a history of head injury with sustained loss of consciousness, chronic medical illness, history of substance abuse/dependence, history of psychosis, bipolar disorder, primary or recurrent major depressive disorder (MDD) (unless onset of MDD was after the onset of OCD), history of conduct disorder, an IQ less than or equal to 80, lifetime diagnosis of autism spectrum disorder (ASD) (including a score of 15 or higher on the Social Communication Questionnaire, lifetime version) (Rutter et al. 2003), lifetime diagnosis of eating disorder, if they were adopted, or if they were living away from both biological parents at the time of the study. In addition to the exclusions above, healthy controls were also screened for lifetime diagnosis of Axis I psychiatric disorders of any kind (APA 2000). For this study, we included only unrelated participants and only participants with both parents of European Caucasian descent to minimize possible population stratification.
Imaging
All participants had T1-weighted structural magnetic resonance imaging (sMRI) data collected at the Children’s Hospital of Michigan Imaging Center. Detailed methods are described elsewhere (Szeszko et al. 2004; Arnold et al. 2009; Wu et al. 2013). Briefly, a Sigma 3.0-Tesla unit (Horizon HDX software, General Electric Medical Systems, Milwaukee, WI) was used to gather four separate contiguous three-dimensional (3D) MRI volumes. A 3D magnetization-prepared 180 degrees radio-frequency pulses and rapid gradient-echo (MP RAGE) sequence was utilized. Acquisition parameters included echo time of 1.6 ms, repetition time of 2200 ms, acquisition matrix of 512 × 512 × 204, pixel dimensions of 0.5 × 0.5 × 0.8 mm3, field of view 256 × 256 mm2, 204 axial slices, flip angle of 13 degrees, and inversion times of 776 ms, 780 ms, 794 ms, and 808 ms for each respective 3D MRI volume. All MRIs were reviewed by a pediatric neuroradiologist to rule out clinically significant abnormalities and magnetic field inhomogeneities. A manual tracing technique was then carried out by trained operators, blind to case-control status, using BRAINS2 image processing software (Magnotta et al. 2002) to measure striatum, caudate, putamen, and thalamus, and using Analyze Direct 11.0 (AnalyzeDirect, Inc., Overland Park, KS) for the ACC and the OFC. This measured regional brain volumes in select structures with a priori evidence of their involvement in OCD, along with the total intracranial volume (ICV) for each subject, in cubic centimeters (cc). Overall, the brain regions we examined were the caudate, the putamen, the striatum (caudate + putamen combined), the thalamus, the ACC, and the OFC. As a result, a total of six regions of interest (ROIs) were examined, with respective parameters for the brain regions outlined further in our past studies (Szeszko et al. 2004; Arnold et al. 2009; Wu et al. 2013). In this study, we combined left and right hemisphere and used the total volume for each ROI and we report intraclass correlation coefficient (ICC) values ranging from 0.970–0.999.
DNA collection and extraction
Either saliva or blood was collected from each participant. Oragene•DNA collection kits were used to extract DNA from saliva using the accompanying, recommended protocol (DNA Genotek). An Autopure LS automated DNA extractor (QIAGEN) was used to extract whole EDTA blood using Puregene chemistry (Gentra). Saliva DNA was quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen) and blood DNA was quantified using an FLx800 Fluorescence Reader (Biotek) and Hoechst 33258 dye (Sigma). DNA quality was verified using gel electrophoresis. Samples with DNA concentrations below 60 ng/μl (if genotyped on the HumanOmni2.5–4 v1.0 or HumanCoreExome-12 v1.0 microarrays (Illumina)) or samples with poor DNA quality were excluded.
Selection of candidate genes
The serotonin system has been the focus of numerous candidate gene studies in OCD, given the efficacy of SRIs in treating the disorder (Sinopoli et al. 2017). We therefore examined variants across serotonin system genes most commonly studied in OCD including SLC6A4, HTR2A, HTR1B, and HTR2C (Taylor 2013) to further understand the relationship of these genes with neuroimaging and OCD. Evidence suggests that the most promising of these genes in OCD are SLC6A4 (and more specifically polymorphism 5-HTTLPR) and HTR2A. There is more limited evidence to suggest the involvement of HTR1B in OCD, with studies indicating that this gene may be specific to certain OCD subgroups (Taylor 2013; Sinopoli et al. 2017). Though there is the least amount of evidence supporting the role of HTR2C in OCD, this gene has been implicated in a number of psychiatric disorders and is of interest to our structural neuroimaging study given that it is almost exclusively expressed in the central nervous system (Werry et al. 2008; Fagerberg et al. 2014).
Genotyping
5-HTTLPR
Given that variation in 5-HTTLPR is not captured using microarrays, we directly genotyped this region of SLC6A4 in a two-step process (Wendland et al. 2006) as previously outlined (Sinopoli et al. under review). This was carried out with The Centre for Applied Genomics (TCAG) at the Hospital for Sick Children (Toronto, Canada). First, we amplified 5-HTTLPR using forward primer 5’-ATGCCAGCACCTAACCCCTAATGT-3′ (5′-labeled with HEX fluorescent dye) and reverse primer 5’-GGACCGCAAGGTGGGCGGGA-3′ (Gelernter et al. 1997). Variants were identified as L or S based on variant length. Second, DNA was digested at the rs25531 SNP location using restriction enzyme MspI (Fermentas, Life Technologies), which only cuts when G is present but not when A is present. Peak Scanner Software v1.0 (Thermo Fisher Scientific) was used to analyze results. Depending on the outcomes of the two steps, variants were identified as LA, LG, or S. Based on their known functionality, we further categorized variants into two allelic groups: LA and [LG + S] which yield increased or decreased SERT expression respectively (Hu et al. 2006).
Quality control (QC) was applied using the standard settings in the Peak Scanner Software v1.0 (Thermo Fisher Scientific). Individuals were genotyped for 5-HTTLPR with a 95% completion rate. Reported siblings were removed. In our final sample, 1 5-HTTLPR-genotyped individual was carrying the previously noted rare SG variant (Wendland et al. 2006; Voyiaziakis et al. 2011). After all QC and removal of ensuing individuals, we had a final N of 160 individuals with the 5-HTTLPR polymorphism genotyped (77 OCD cases and 83 healthy controls).
SNPs across SLC6A4, HTR2A, HTR1B, and HTR2C
Samples were originally genotyped to gather GWAS data for our continuing collaborative study (Arnold et al. 2009; Wu et al. 2013) on the HumanOmni2.5–4 v1.0 or HumanCoreExome-12 v1.0 microarrays (Illumina). We focused on data from SNPs across SLC6A4, HTR2A, HTR1B, and HTR2C (UCSC Genome Browser, hg19).
SNPs with call rate < 0.97 were excluded. Samples were excluded on the basis of sex misspecification and ambiguity and cryptic relatedness of half sibling or above. We verified European ethnicity via principal components analysis (PCA) using EIGENSTRAT version 3.2.10 (Price et al. 2006) and using HapMap populations CEU and TSI to ensure that the genetic data of our participants clustered with that of Europeans, relative to other ethnicities, and then via another PCA to identify and remove outliers within our own population. An additional PCA was run without outliers. A Tracy-Widom test of the resulting principal components (PCs) showed no significant PCs, therefore none were retained to control for population structure in our analyses. After all quality control and removal of ensuing individuals, we had a final N of 154 individuals with SNPs genotyped across our four genes of interest (74 OCD cases and 80 healthy controls).
Chromosomes 6, 13, 17 and X were pre-phased using SHAPEIT2 version 2.790 (Delaneau et al. 2014). SNP data was imputed using IMPUTE2 version 2.3.1 (Howie et al. 2009) for each of the candidate genes, including a 50 kb flanking region on either side of each gene. 1000 Genomes Phase 3 reference data was used for both pre-phasing and imputation. The non-pseudoautosomal region from 1000 Genomes chromosome X was used for chromosome X pre-phasing and imputation. After imputation, SNPs were removed for low quality imputation (information score < 0.8). SNPs with MAF ≥ 0.05 were kept in the sample. Data were kept as dosage calls for all analyses. Table 1 shows SNP counts across SLC6A4, HTR2A, HTR1B, and HTR2C.
Table 1.
SNP counts across 4 candidate genes: All common genotyped and imputed SNPs (MAF ≥ 0.05) are shown for each gene, and its flanking regions, included in the analyses for males and in the analyses for females
| Gene | Number of SNPs in Males | Number of SNPs in Females |
|---|---|---|
| SLC6A4 | 117 | 102 |
| HTR2A | 301 | 313 |
| HTR1B | 241 | 235 |
| HTR2C | 401 | 482 |
SNP, single nucleotide polymorphism; SLC6A4, solute carrier family 6 member 4 (serotonin transporter gene); HTR2A, 5-HT2A receptor gene; HTR1B, 5-HT1B receptor gene; HTR2C, 5-HT2C receptor gene
Statistical analyses
T-tests were used to analyze demographics for statistically significant differences in age, in total ICV, and in PDS for all male cases versus male controls and for all female cases versus female controls. The following analyses were conducted for 5-HTTLPR and for all SNPs across SLC6A4, HTR2A, HTR1B, and HTR2C, using the program, R, Version 3.0.1 (https://www.R-project.org).
Assessing the association of serotonin gene variants with OCD
Logistic regressions were conducted in males and in females separately, using the expected number of tested alleles for each SNP (designated [LG + S] for 5-HTTLPR) against OCD case versus control status, in an additive model with age included as a covariate. The Genetic Type 1 error calculator (GEC) developed to account for dependent SNPs (Li et al. 2012) was used to calculate the P value thresholds for the male and female analyses in the SNPs. The resulting significance P value threshold for SNP analyses was 2.23e-4 for males and 2.15e-4 for females.
Assessing the effect of serotonin gene variants on brain volume
A linear regression was fit for each SNP using the expected number of tested alleles (designated [LG + S] for 5-HTTLPR) against volume for each ROI and across the combined set of cases and controls (i.e., assessing the effect of SNP on brain volume irrespective of OCD diagnosis), in males and in females separately. An additive model was used including age and total ICV as covariates. Analyses involving sMRI data used the GEC-generated values that were then corrected for the number of brain regions under investigation. This resulted in a brain region-adjusted significance P value threshold of 3.72e-5 (2.23e-4 to account for dependent SNPs / 6 ROIs) for SNP analyses in males and of 3.58e-5 (2.15e-4 to account for dependent SNPs / 6 ROIs) for SNP analyses in females.
Assessing the effect of genotype-diagnosis interaction on brain volume
An interaction model was then fit for all individuals (cases and controls) with a main effect for both SNP (tested allele) and OCD (though we only report on SNP main effect, given our gene-driven hypotheses), along with an interaction term of OCD*[tested allele]. This allowed us to assess whether OCD status significantly changed the association of the tested allele with brain region volume (having corrected for age and total ICV by including these variables as covariates in the model). Brain region-adjusted significance P value thresholds were used to assess significance in this model.
Results
Demographic characteristics for all 200 individuals in the study are reported in Table 2. There were no significant differences to report in age, in total ICV, or in PDS between total male cases and total male controls. Between total female cases and total female controls, there was a significant difference in age (absolute t-statistic = 2.3, P value = 0.02) and in PDS (absolute t-statistic = 2.2, P value = 0.03).
Table 2.
Patient and control demographics: Group characteristics for all individuals combined, for OCD cases, and for controls
| All Individuals | ||
|---|---|---|
| Males | Females | |
| Total N | 97 | 103 |
| N with 5-HTTLPR Genotyped | 75 | 85 |
| N with SNPs Genotyped Across SLC6A4, HTR2A, HTR1B, and HTR2C | 75 | 79 |
| OCD Cases | ||
| Males | Females | |
| Total N | 46 | 51 |
| Mean Age (SD) | 13.2 (3.1) | 13.6 (3.8) |
| Age Range | 6.5- <20.0 | 6.2- <20.0 |
| Mean Total ICV (cc) (SD) | 1684.4 (152.7) | 1534.8 (133.3) |
| Mean PDS Score (SD) | 11.6 (3.4) | 12.3 (5.6) |
| PDS Score Range | 5–17 | 1–19 |
| Mean CY-BOCS Score (SD) | 27.0 (6.3) | 28.4 (7.0) |
| CY-BOCS Score Range | 13–37 | 10–39 |
| MDD, N (%) | 2 (4.3) | 6 (11.8) |
| ADHD, N (%) | 9 (19.6) | 11 (21.6) |
| Anxietyi, N (%) | 20 (43.5) | 26 (51.0) |
| Ticsii, N (%) | 16 (34.8) | 8 (15.7) |
| Controls | ||
| Males | Females | |
| Total N | 51 | 52 |
| Mean Age (SD) | 14.2 (3.4) | 15.4 (3.8) |
| Age Range | 6.2–19.9 | 6.8–19.8 |
| Mean Total ICV (cc) (SD) | 1676.6 (135.1) | 1542.7 (120.9) |
| Mean PDS Score (SD) | 12.0 (3.5) | 13.9 (3.4) |
| PDS Score Range | 4–18 | 5–19 |
N, sample size; 5-HTTLPR, serotonin transporter-linked polymorphic region; SNP, single nucleotide polymorphism; SLC6A4, solute carrier family 6 member 4 (serotonin transporter gene); HTR2A, 5-HT2A receptor gene; HTR1B, 5-HT1B receptor gene; HTR2C, 5-HT2C receptor gene; OCD, obsessive-compulsive disorder; SD, standard deviation; ICV, intracranial volume; PDS, Pubertal Development Scale; cc, cubic centimeters; CY-BOCS, Children’s Yale-Brown Obsessive-Compulsive Scale; MDD, major depressive disorder; ADHD, attention-deficit/ hyperactivity disorder
Includes Separation Anxiety Disorder, Panic Disorder (with or without Agoraphobia Specified), Specific Phobia, Social Phobia, Generalized Anxiety Disorder (GAD)
Includes Tourette’s Disorder, Chronic Motor Tic Disorder, Chronic Vocal Tic Disorder, Transient Tic Disorder (Motor and/or Vocal), Lifetime History of Motor/Vocal Tic
Assessing the association of serotonin gene variants with OCD
There was no significant association between 5-HTTLPR and OCD in males or in females. In the SNP analyses, the top 38 SNPs for OCD in males all corresponded with gene HTR1B (odds ratio, OR, ranging from 0.37 to 0.43; P value ranging from 1.27e-2 to 1.98e-2) and 24 of the 25 top SNPs for OCD in females all corresponded with gene SLC6A4 (OR ranging from 0.37 to 3.35; P value ranging from 1.37e-3 to 4.89e-3). The SNP findings did not remain significant after correction for multiple comparisons.
Assessing the effect of serotonin gene variants on brain volume
There were no significant associations between 5-HTTLPR and brain volume for any of the ROIs in males or in females. For the candidate gene SNPs, in males, the top 20 SNPs were all associated with reduced caudate volume, where 17 of the SNPs corresponded with HTR2A (SNP effect ranging from −0.17 to −0.15 cc; P value ranging from 2.68e-4 to 6.53e-4) and 3 of the SNPs corresponded with SLC6A4 (SNP effect of −0.15 cc; P value of 6.23e-4) (Table 3). In females, the top 20 SNPs were all associated with reduced volume of the striatum, where 8 of the SNPs corresponded with HTR1B (SNP effect ranging from −0.18 to −0.15 cc; P value ranging from 2.70e-4 to 6.47e-4) and 8 of the SNPs corresponded with HTR2C (SNP effect ranging from −0.17 to −0.15 cc; P value ranging from 3.64e-4 to 6.49e-4) (Table 4). None of the SNP findings remained significant after correction for multiple comparisons.
Table 3.
Effect of serotonin gene variants on brain volume in males: Top 20 SNP findings
| Brain Region | SNP | Gene | Tested Allele | MAF | SNP Effect (cc) | P value |
|---|---|---|---|---|---|---|
| Caudate | rs7330368 | HTR2A | A | 0.22 | −0.16 | 2.68E-04 |
| Caudate | rs9316235 | HTR2A | A | 0.23 | −0.17 | 2.92E-04 |
| Caudate | rs2770297 | HTR2A | T | 0.26 | −0.16 | 3.87E-04 |
| Caudate | rs61948331 | HTR2A | A | 0.13 | −0.16 | 4.56E-04 |
| Caudate | rs731779 | HTR2A | C | 0.11 | −0.16 | 4.60E-04 |
| Caudate | rs9562687 | HTR2A | A | 0.10 | −0.16 | 4.60E-04 |
| Caudate | rs9562688 | HTR2A | A | 0.10 | −0.16 | 4.60E-04 |
| Caudate | rs61948356 | HTR2A | A | 0.14 | −0.15 | 5.11E-04 |
| Caudate | rs61948354 | HTR2A | C | 0.14 | −0.15 | 5.13E-04 |
| Caudate | rs17287961 | HTR2A | T | 0.07 | −0.16 | 5.49E-04 |
| Caudate | rs61948357 | HTR2A | C | 0.14 | −0.15 | 5.61E-04 |
| Caudate | rs73477746 | HTR2A | G | 0.14 | −0.15 | 5.62E-04 |
| Caudate | rs8078900 | SLC6A4 | A | 0.25 | −0.15 | 6.23E-04 |
| Caudate | rs33980254 | SLC6A4 | T | 0.25 | −0.15 | 6.23E-04 |
| Caudate | rs73268099 | SLC6A4 | G | 0.25 | −0.15 | 6.23E-04 |
| Caudate | rs61647933 | HTR2A | C | 0.17 | −0.15 | 6.38E-04 |
| Caudate | rs61948335 | HTR2A | G | 0.11 | −0.15 | 6.42E-04 |
| Caudate | rs61948318 | HTR2A | T | 0.13 | −0.16 | 6.44E-04 |
| Caudate | rs9534501 | HTR2A | T | 0.13 | −0.16 | 6.53E-04 |
| Caudate | rs9567743 | HTR2A | C | 0.13 | −0.16 | 6.53E-04 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; cc, cubic centimeters; SLC6A4, solute carrier family 6 member 4 (serotonin transporter gene); HTR2A, 5-HT2A receptor gene
Table 4.
Effect of serotonin gene variants on brain volume in females: Top 20 SNP findings
| Brain Region | SNP | Gene | Tested Allele | MAF | SNP Effect (cc) | P value |
|---|---|---|---|---|---|---|
| Striatum | rs9343614 | HTR1B | G | 0.32 | −0.18 | 2.70E-04 |
| Striatum | rs9361228 | HTR1B | A | 0.30 | −0.18 | 2.96E-04 |
| Striatum | rs540285 | HTR2C | T | 0.27 | −0.17 | 3.64E-04 |
| Striatum | rs12836749 | HTR2C | A | 0.13 | −0.16 | 4.07E-04 |
| Striatum | rs1777761 | HTR1B | G | 0.25 | −0.16 | 5.12E-04 |
| Striatum | rs4543330 | HTR1B | C | 0.08 | −0.15 | 5.18E-04 |
| Striatum | rs2798949 | HTR1B | G | 0.24 | −0.16 | 5.22E-04 |
| Striatum | rs1891752 | HTR1B | G | 0.25 | −0.16 | 5.22E-04 |
| Striatum | rs199602838 | HTR2C | T | 0.11 | −0.15 | 5.39E-04 |
| Striatum | rs184155834 | HTR1B | G | 0.08 | −0.15 | 5.48E-04 |
| Striatum | rs9902340 | SLC6A4 | G | 0.44 | −0.16 | 5.72E-04 |
| Striatum | rs12861270 | HTR2C | G | 0.10 | −0.16 | 6.05E-04 |
| Striatum | rs11797988 | HTR2C | A | 0.10 | −0.16 | 6.34E-04 |
| Striatum | rs9567745 | HTR2A | C | 0.06 | −0.15 | 6.41E-04 |
| Striatum | rs3945573 | HTR2A | A | 0.06 | −0.15 | 6.41E-04 |
| Striatum | rs2226183 | HTR1B | A | 0.07 | −0.15 | 6.47E-04 |
| Striatum | rs3764450 | SLC6A4 | A | 0.26 | −0.16 | 6.48E-04 |
| Striatum | rs12855533 | HTR2C | A | 0.18 | −0.16 | 6.49E-04 |
| Striatum | rs12847225 | HTR2C | C | 0.18 | −0.16 | 6.49E-04 |
| Striatum | rs12842363 | HTR2C | A | 0.18 | −0.16 | 6.49E-04 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; cc, cubic centimeters; SLC6A4, solute carrier family 6 member 4 (serotonin transporter gene); HTR2A, 5-HT2A receptor gene; HTR1B, 5-HT1B receptor gene; HTR2C, 5-HT2C receptor gene
Assessing the effect of genotype-diagnosis interaction on brain volume
When we assessed whether OCD diagnosis significantly changed the relationship between 5-HTTLPR and brain volume, there were no significant findings to report for males or for females. When assessing whether OCD diagnosis significantly changed the relationship between SNPs across our four candidate genes and brain volume in males (Table 5), there were no findings that remained significant after correction for multiple comparisons. In females (Table 6), OCD diagnosis significantly changed the relationship between two HTR2C SNPs (rs12860460 and rs12854485) and ACC volume. Both rs12860460 and rs12854485 were associated with a significant SNP main effect in the interaction model (SNP main effect of −4.37 cc and − 3.26 cc respectively; SNP P value of 1.75e-7 and 5.45e-6 respectively). There was a significant genotype-diagnosis interaction for both SNPs (rs12860460, interaction term estimate of 5.45 cc and interaction P value of 9.70e-8; rs12854485, interaction term estimate of 4.28 cc and interaction P value of 2.07e-6). Fig. 1 shows the change in the direction/magnitude of effect of rs12860460 (Fig. 1-a) and rs12854485 (Fig. 1-b) on ACC volume in female OCD patients versus in controls. For both SNPs, the tested allele (versus what we observed for the alternate allele) was associated with reduced ACC volume in female controls and with increased ACC volume in female OCD patients.
Table 5.
Effect of genotype-diagnosis interaction on brain volume in males: Top 20 SNP findings
| Brain Region | SNP | Gene | Tested Allele | MAF | SNP Main Effect (cc) | SNP P value | Interaction Term Estimate (cc) | Interaction P value |
|---|---|---|---|---|---|---|---|---|
| OFC | rs1777764 | HTR1B | G | 0.31 | −0.97 | 3.52E-01 | 5.78 | 1.58E-03 |
| OFC | rs28656158 | HTR1B | C | 0.31 | −0.71 | 4.81E-01 | 5.43 | 2.23E-03 |
| Thalamus | rs17069005 | HTR2A | G | 0.20 | −0.74 | 4.24E-02 | 1.47 | 2.32E-03 |
| Caudate | rs4942577 | HTR2A | T | 0.38 | −0.89 | 3.02E-04 | 1.06 | 3.06E-03 |
| OFC | rs2223832 | HTR1B | C | 0.14 | −2.51 | 1.17E-01 | 7.20 | 3.61E-03 |
| OFC | rs11757592 | HTR1B | C | 0.14 | −2.51 | 1.17E-01 | 7.20 | 3.61E-03 |
| Caudate | rs9534488 | HTR2A | T | 0.46 | −0.69 | 3.04E-03 | 0.97 | 4.44E-03 |
| Caudate | rs9526236 | HTR2A | T | 0.46 | −0.69 | 3.22E-03 | 0.98 | 4.73E-03 |
| OFC | rs9343614 | HTR1B | G | 0.20 | −1.93 | 1.11E-01 | 6.22 | 4.77E-03 |
| Caudate | rs12110491 | HTR1B | T | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.90E-03 |
| Caudate | rs55636038 | HTR1B | T | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.90E-03 |
| Caudate | rs11755194 | HTR1B | C | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.90E-03 |
| Caudate | rs1228798 | HTR1B | G | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.90E-03 |
| Caudate | rs1228797 | HTR1B | T | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.91E-03 |
| Caudate | rs2207056 | HTR1B | T | 0.12 | 0.46 | 2.37E-01 | −1.72 | 4.92E-03 |
| Caudate | rs10806097 | HTR1B | T | 0.12 | 0.48 | 2.34E-01 | −1.74 | 5.06E-03 |
| Caudate | rs1228806 | HTR1B | C | 0.11 | 0.50 | 2.36E-01 | −1.76 | 5.47E-03 |
| Caudate | rs17069005 | HTR2A | G | 0.20 | −0.86 | 7.41E-03 | 1.17 | 5.50E-03 |
| Caudate | rs1228805 | HTR1B | G | 0.11 | 0.50 | 2.36E-01 | −1.77 | 5.56E-03 |
| Caudate | rs1228804 | HTR1B | G | 0.11 | 0.51 | 2.38E-01 | −1.77 | 5.74E-03 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; cc, cubic centimeters; OFC, orbitofrontal cortex; HTR2A, 5-HT2A receptor gene; HTR1B, 5-HT1B receptor gene
Table 6.
Effect of genotype-diagnosis interaction on brain volume in females: Top 20 SNP findings. (Findings significant after correction for multiple comparisons are in bold)
| Brain Region | SNP | Gene | Tested Allele | MAF | SNP Main Effect (cc) | SNP P value | Interaction Term Estimate (cc) | Interaction P value |
|---|---|---|---|---|---|---|---|---|
| ACC | rs12860460 | HTR2C | C | 0.18 | −4.37 | 1.75E-07 | 5.45 | 9.70E-08 |
| ACC | rs12854485 | HTR2C | G | 0.23 | −3.26 | 5.45E-06 | 4.28 | 2.07E-06 |
| Striatum | rs9567737 | HTR2A | T | 0.42 | 0.94 | 8.56E-04 | −1.45 | 3.40E-04 |
| Striatum | rs6561333 | HTR2A | C | 0.42 | 0.95 | 8.77E-04 | −1.45 | 3.42E-04 |
| Striatum | rs1923886 | HTR2A | T | 0.42 | 0.90 | 1.53E-03 | −1.43 | 4.37E-04 |
| Striatum | rs1885884 | HTR2A | G | 0.13 | 1.21 | 1.15E-03 | −1.78 | 6.59E-04 |
| ACC | rs35031982 | HTR2C | G | 0.21 | −2.81 | 2.53E-04 | 3.28 | 6.64E-04 |
| Thalamus | rs1738504 | HTR1B | C | 0.26 | 0.42 | 1.30E-01 | −1.43 | 7.04E-04 |
| Thalamus | rs1777764 | HTR1B | G | 0.34 | 0.50 | 5.10E-02 | −1.34 | 8.82E-04 |
| ACC | rs12861270 | HTR2C | G | 0.10 | −5.46 | 2.19E-03 | 6.20 | 1.01E-03 |
| ACC | rs2192371 | HTR2C | G | 0.26 | −2.36 | 5.44E-04 | 2.86 | 1.06E-03 |
| ACC | rs12862598 | HTR2C | C | 0.10 | −5.40 | 2.57E-03 | 6.17 | 1.14E-03 |
| Thalamus | rs2207055 | HTR1B | A | 0.33 | 0.46 | 7.12E-02 | −1.30 | 1.16E-03 |
| ACC | rs12851998 | HTR2C | T | 0.10 | −5.49 | 1.85E-03 | 6.00 | 1.23E-03 |
| ACC | rs12854729 | HTR2C | G | 0.10 | −5.46 | 2.25E-03 | 6.10 | 1.23E-03 |
| ACC | rs3813928 | HTR2C | A | 0.10 | −5.46 | 2.25E-03 | 6.09 | 1.24E-03 |
| ACC | rs3813929 | HTR2C | T | 0.10 | −5.46 | 2.25E-03 | 6.09 | 1.24E-03 |
| ACC | rs11798441 | HTR2C | G | 0.10 | −5.46 | 2.25E-03 | 6.09 | 1.24E-03 |
| ACC | rs17260565 | HTR2C | G | 0.10 | −5.46 | 2.25E-03 | 6.09 | 1.24E-03 |
| ACC | rs17326429 | HTR2C | A | 0.10 | −5.46 | 2.25E-03 | 6.09 | 1.24E-03 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; cc, cubic centimeters; ACC, anterior cingulate cortex; HTR2A, 5-HT2A receptor gene; HTR1B, 5-HT1B receptor gene; HTR2C, 5-HT2C receptor gene
Fig. 1.
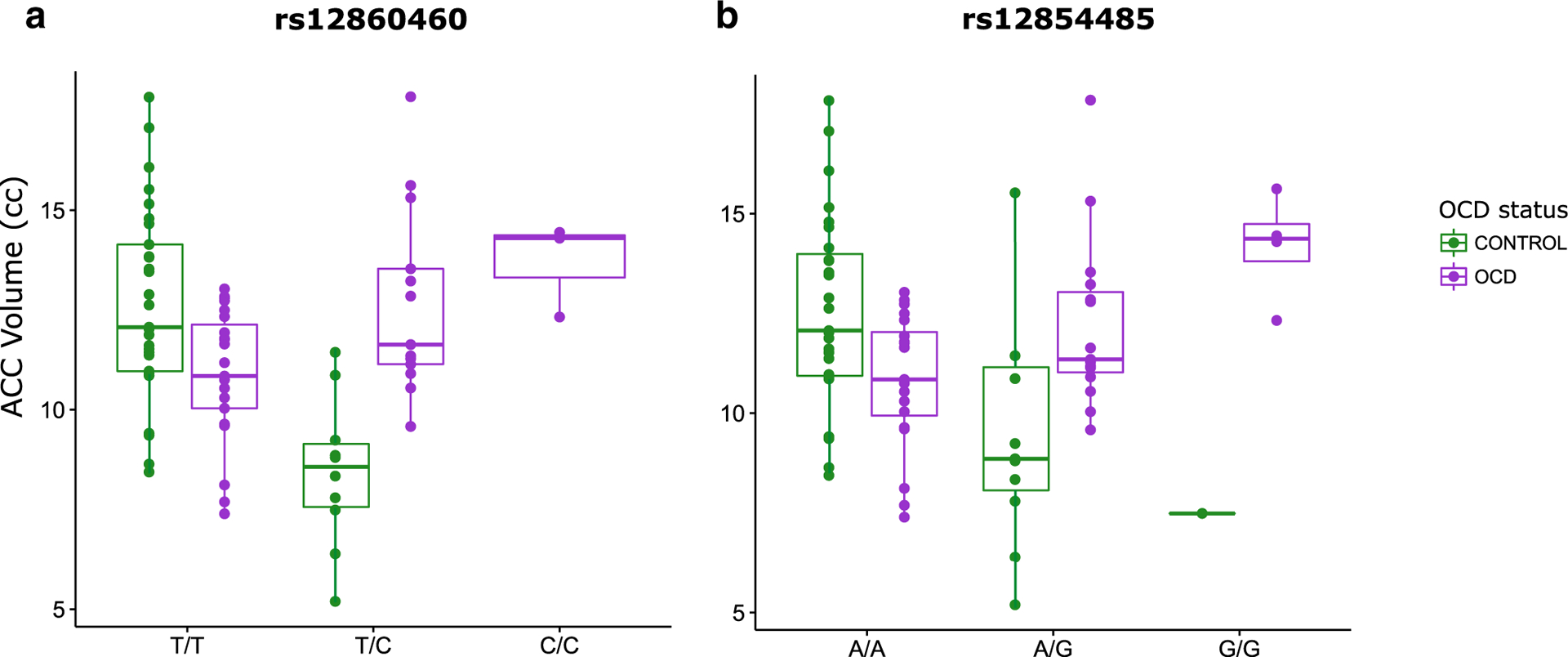
Brain volume versus genotype in females graphed: a) HTR2C SNP rs12860460, showing females with genotype and corresponding ACC volume (cc) for controls (green) and for OCD cases (purple); ACC volume decreases by 4.37 cc in controls (with 0, 1, or 2 tested C alleles) and ACC volume increases by 1.08 cc in OCD cases (with 0, 1, or 2 tested C alleles). b) HTR2C SNP rs12854485, showing females with genotype and corresponding ACC volume (cc) for controls (green) and for OCD cases (purple); ACC volume decreases by 3.26 cc in controls (with 0, 1, or 2 tested G alleles) and ACC volume increases by 1.02 cc in OCD cases (with 0, 1, or 2 tested G alleles). OCD, obsessive-compulsive disorder; ACC, anterior cingulate cortex; cc, cubic centimeters
Discussion
Studies to date have indicated that serotonin system genes may play a role in OCD, but the associated variants reported are inconsistent between studies (Sinopoli et al. 2017). Like other complex disorders, OCD is thought to result from multiple genes of small effect interacting with environmental factors (Pauls et al. 2014). In this study, we adopted a candidate gene approach to examine common potential serotonin system gene polymorphisms hypothesized to be involved in OCD. We aimed to address some of the limitations of previous studies including failure to account for sex or age of onset. To attempt to reduce phenotypic heterogeneity in our study, we used an exclusively pediatric group of individuals and stratified by sex. We also attempted to reduce heterogeneity by using structural brain imaging in order to study putative intermediate phenotypes for OCD, specifically using CSTC circuitry relevant to OCD (MacMaster 2010; Pauls et al. 2014).
In our case-control analyses, the top SNPs nominally associated with OCD in males were located in a region 5′ of HTR1B. These findings did not remain significant after correction for multiple comparisons. In females, the majority of our top SNPs were nominally associated with OCD and corresponded with SLC6A4. Again, these findings did not remain significant after correction for multiple comparisons, but the difference in trends in males and females is in line with previous literature showing that risk variants for OCD may differ by sex (Mattina and Steiner 2016). We did not find any significant association in males or in females between OCD and the most commonly studied candidate gene polymorphism, 5-HTTLPR, which has been associated with the disorder in previous literature (Taylor 2013). Though our case-control analyses were underpowered, our findings suggest a need to analyze candidate serotonin system genes in males and in females separately.
For our brain region analyses, we first examined the effect of candidate serotonin gene variants on brain volume in cases and controls combined (irrespective of diagnosis). None of the candidate variants were significantly associated with regional brain volumes in males or in females after correction for multiple comparisons. The results from our analyses assessing the effect of genotype-diagnosis interaction on brain volume supported our hypothesis that the relationship between some serotonin system gene variants and brain volume differs between OCD patients and controls and that these differences vary between males and females. Our findings in males did not survive correction for multiple comparisons. In females, we identified a significant genotype-diagnosis interaction for two SNPs in HTR2C, rs12860460 and rs12854485. We also note that both significant SNPs were genotyped and not imputed, further supporting the reliability of our findings. Specifically, these HTR2C SNPs were differentially associated with ACC volume in female children/adolescents in such a way that the tested allele (versus what we observed for the alternate allele) in each SNP was associated with decreased ACC volume in healthy individuals and with increased ACC volume in OCD patients. The effect of gene variant on ACC volume was also smaller in OCD cases than it was in controls. We cannot refer to one of the variants in either SNP as a genetic risk factor, however, since we did not identify any to be significantly associated with OCD in our first set of analyses (though underpowered).
Our study implicated the ACC, consistent with prior findings of increased ACC volume in pediatric OCD (Rosenberg and Keshavan 1998; Szeszko et al. 2004) and findings of ACC abnormalities being more pronounced in female OCD patients than in male OCD patients (Yücel et al. 2008). Our study highlights a possible mechanism in OCD where the allelic variants in the implicated HTR2C SNPs act differently in cases versus in controls with regard to the direction and magnitude of effect on ACC volume.
Our diagnosis-specific findings are similar to what has previously been reported. Two prior studies that examined 5-HTTLPR in OCD showed a nominally significant genotype-diagnosis interaction effect on brain volume (Atmaca et al. 2011; Honda et al. 2017). Atmaca et al., (2011) noted a stronger effect of 5-HTTLPR genotype on OFC in OCD patients versus in controls, with reduced volume in the OFC of S carriers with OCD (versus individuals of genotype L/L with OCD). Honda et al., (2017) noted a stronger effect of 5-HTTLPR genotype on the right frontal pole in OCD patients versus in controls, with reduced gray matter volume in the right frontal pole in LA carriers with OCD versus LA carriers without OCD (Honda et al. 2017). We similarly identified significant genotype-diagnosis interaction effects on brain volume for serotonin system gene variants and our findings survived correction for multiple comparisons. Unlike these previous studies, though, we did not observe a significant genotype-diagnosis effect for 5-HTTLPR. Instead, our strongest findings were within HTR2C. Furthermore, the effects of the significant HTR2C SNPs were more marked in controls as opposed to in OCD cases, opposite to what was observed for 5-HTTLPR in previous studies.
The 5-HT2C receptor is widely distributed throughout the central nervous system, plays a number of roles in cell signalling, and is important in mood, sex, and appetite regulation (Molineaux et al. 1989). Studies in psychiatry have also suggested the receptor’s involvement in anxiety, schizophrenia, depression, and suicide (Werry et al. 2008), with a group of studies reporting significant differences in RNA editing for HTR2C in post-mortem brain tissue of suicide victims (Niswender et al. 2001; Gurevich et al. 2002; Iwamoto and Kato 2003; Dracheva et al. 2008; Lyddon et al. 2013; Di Narzo et al. 2014; Weissmann et al. 2016).
Rs12854485 and rs12860460 are adjacent to one another (~4.67 kb apart) in intron 2 of HTR2C. Nearby the SNPs, and overlapping the same region as HTR2C intron 2, are 4 microRNA (miRNA) genes (MIR764, MIR1912, MIR1264, MIR1298) which yield non-coding RNAs involved in gene expression and post-transcriptional regulation, and an H/ACA box small nucleolar RNA (snoRNA) gene (SNORA35), part of a class yielding non-coding RNAs which facilitate rRNA or spliceosomal RNA modification (Lestrade and Weber 2006; UCSC Genome Browser, hg19). SNORA35 is predominantly expressed in the brain (UCSC Genome Browser, hg19; GTEx, Release V6). It could be the case that our significant SNPs lie in potential regulatory regions of HTR2C that affect gene expression, or more specifically RNA editing or alternative splicing, depending on additional risk factors (Werry et al. 2008; Lu et al. 2012; Wang et al. 2015). The SNPs may also be tagging variants in another region of the gene(s) that are implicated in HTR2C expression or functionality or in the expression or functionality of the small regulatory genes overlapping HTR2C. Our significant genotype-diagnosis interaction shows us that the effect of each SNP variant on female ACC volume differs in OCD cases versus in controls. HTR2C variation is likely to be only one contributing factor in the complex etiology of OCD, which interacts with additional genetic, epigenetic, biological, and/or environmental risk factors to produce disease-relevant changes in ACC volume in female children and adolescents.
Though our study provides evidence of the value of studying the genetics of neural mechanisms for OCD in homogenous subgroups reflective of sex and age (children/adolescents versus adults), it had a number of limitations. First, subgrouping by sex resulted in reduced sample size for each analysis, which could have reduced our power to detect associations in the absence of strong sex effects. To address this issue, we performed post hoc analyses for all autosomal variants (excluding HTR2C which is on chromosome X) combining males and females. There were no significant findings for these combined analyses after correction for multiple comparisons. The second limitation of our study was that our sample was too small to consider genome-wide data and all potential brain regions, both of which would have conferred a stricter significance threshold to account for multiple comparisons. We therefore elected to focus on identifying associations of interest confined to select candidate serotonin system genes and a priori brain regions pertinent to OCD.
Future studies are warranted to replicate our findings in a larger sample and, ideally, using a more comprehensive, genome-wide approach. Should our findings be replicated, more extensive studies would be necessary to identify additional pathological factors driving the development of OCD and that are specific to the relationship between ACC and HTR2C in pediatric females. Future genetic studies of OCD should similarly stratify by sex and age group, given that we have evidence supporting genetic association in a pediatric sample that would not have been identified without stratifying based on sex. Age has been shown to have an effect on brain volume, particularly in subcortical brain regions, with puberty status describing such developmental changes even better than age (Wierenga et al. 2018). Given that the age range of our sample traverses adolescence, we included age as a covariate in our study to control for its effects on brain volume. Future studies in larger samples may benefit from more detailed analyses of the influence of age and pubertal development (i.e., by stratifying based on these variables) given the effect of puberty on brain structure. Our sample size was insufficient to account for symptom dimensions in OCD, but future studies with larger samples should test for genetic associations based on symptom dimensions. This will help us to better understand the phenotypic heterogeneity of OCD and the complexity of the disorder at the level of gene systems and brain morphology.
In summary, although limited by a small sample size and by its focus on single candidate genes, our study is important given the novelty of our approach. Ours is one of the first studies of its kind (and the largest to date) to look across several candidate serotonin genes in OCD, using brain imaging as a potential mediating biological factor in OCD and sex-based analyses to reduce phenotypic heterogeneity. This approach has not yet been implemented on a genome-wide scale in OCD. Our findings, therefore, help lay methodological groundwork and will help guide future genome-wide studies aimed at understanding the genetic and neural mechanisms driving OCD. Consistent with a precision medicine approach, identification of potential genetic and neurobiological mechanisms in OCD, combined with information regarding key demographic features (sex, age of onset), may allow us to target OCD treatment to specific subgroups of patients.
Highlights.
Significant effect of genotype-diagnosis interaction on ACC volume in females.
Effect of HTR2C SNPs on ACC volume differs between OCD patients and controls.
No significant genotype-diagnosis interaction found in males
Funding
This work was supported by the National Institutes of Health (NIH) grants R01-MH101493, R01-MH085300, R01-MH085321, and R01-MH59299. Dr. Arnold receives support from the Alberta Innovates Health Solutions (AIHS) Translational Research Chair in Child and Youth Mental Health. Dr. Rosenberg receives support from the Lycaki-Young Fund, State of Michigan, Miriam L. Hamburger Endowed Chair of Child Psychiatry, Paul and Anita Strauss Endowment, Children’s Hospital of Michigan Foundation, and Donald and Mary Kosch Foundation. Vanessa Sinopoli received funding from the Canadian Institutes of Health Research (CIHR) Master’s Award: Frederick Banting and Charles Best Canada Graduate Scholarships, Ontario Graduate Scholarship (OGS), and the Hospital for Sick Children Restracomp Studentship.
Footnotes
Conflict of interest The authors have no conflicts of interest to disclose.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent Written and informed consent and assent was obtained for all participants. Each site received study approval from its Human Investigation Committee prior to recruitment of participants.
References
- Alvarenga PG, Cesar RC, Leckman JF, Moriyama TS, Torres AR, Bloch MH, Coughlin CG, Hoexter MQ, Manfro GG, Polanczyk GV, Miguel EC, & do Rosario MC (2015). Obsessive-compulsive symptom dimensions in a population-based, cross-sectional sample of school-aged children. Journal of Psychiatric Research, 62, 108–114. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. (2000). Diagnostic and statistical manual of mental disorders (4th ed., text rev.). Washington, DC. [Google Scholar]
- American Psychiatric Association (2013). Diagnostic and statistical manual of mental disorders (5th ed.). Arlington, VA. [Google Scholar]
- Arnold P, & Richter MA (2007). Genetics of obsessive-compulsive disorder: Evidence from pediatric and adult studies. In Storch EA & Greffken GR (Eds.), Handbook of child and adolescent obsessive-compulsive disorder. New Jersey: Lawrence Erlbaum Associates Inc.. [Google Scholar]
- Arnold PD, MacMaster FP, Hanna GL, Richter MA, Sicard T, Burroughs E, Mirza Y, Easter PC, Rose M, Kennedy JL, & Rosenberg DR (2009). Glutamate system genes associated with ventral prefrontal and thalamic volume in pediatric obsessive-compulsive disorder. Brain Imaging and Behavior, 3(1), 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmaca M, Onalan E, Yildirim H, Yuce H, Koc M, Korkmaz S, & Mermi O (2011). Serotonin transporter gene polymorphism implicates reduced orbito-frontal cortex in obsessive-compulsive disorder. Journal of Anxiety Disorders, 25(5), 680–685. [DOI] [PubMed] [Google Scholar]
- Bloch MH, Landeros-Weisenberger A, Rosario MC, Pittenger C, & Leckman JF (2008). Meta-analysis of the symptom structure of obsessive-compulsive disorder. The American Journal of Psychiatry, 165(12), 1532–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe PS, Schmaal L, Abe Y, Ameis SH, Arnold PD, Batistuzzo MC, Benedetti F, Beucke JC, Bollettini I, Bose A, Brem S, Calvo A, Cheng Y, Cho KI, Dallaspezia S, Denys D, Fitzgerald KD, Fouche JP, Giménez M, Gruner P, Hanna GL, Hibar DP, Hoexter MQ, Hu H, Huyser C, Ikari K, Jahanshad N, Kathmann N, Kaufmann C, Koch K, Kwon JS, Lazaro L, Liu Y, Lochner C, Marsh R, Martínez-Zalacaín I, Mataix-Cols D, Menchón JM, Minuzzi L, Nakamae T, Nakao T, Narayanaswamy JC, Piras F, Piras F, Pittenger C, Reddy YC, Sato JR, Simpson HB, Soreni N, Soriano-Mas C, Spalletta G, Stevens MC, Szeszko PR, Tolin DF, Venkatasubramanian G, Walitza S, Wang Z, van Wingen GA, Xu J, Xu X, Yun JY, Zhao Q, ENIGMA OCD Working Group, Thompson PM, Stein DJ, & van den Heuvel OA (2017). Distinct subcortical volume alterations in pediatric and adult OCD: A worldwide Meta- and mega-analysis. American Journal of Psychiatry, 174(1), 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem S, Hauser TU, Iannaccone R, Brandeis D, Drechsler R, & Walitza S (2012). Neuroimaging of cognitive brain function in paediatric obsessive compulsive disorder: A review of literature and preliminary meta-analysis. Journal of Neural Transmission (Vienna), 119(11), 1425–1448. [DOI] [PubMed] [Google Scholar]
- Carter CS, Bearden CE, Bullmore ET, Geschwind DH, Glahn DC, Gur RE, Meyer-Lindenberg A, & Weinberger DR (2017). Enhancing the informativeness and replicability of imaging genomics studies. Biological Psychiatry, 82(3), 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, Neale BM, Yang J, Lee SH, Evans P, Barr CL, Bellodi L, Benarroch F, Berrio GB, Bienvenu OJ, Bloch MH, Blom RM, Bruun RD, Budman CL, Camarena B, Campbell D, Cappi C, Cardona, Silgado JC, Cath DC, Cavallini MC, Chavira DA, Chouinard S, Conti DV, Cook EH, Coric V, Cullen BA, Deforce D, Delorme R, Dion Y, Edlund CK, Egberts K, Falkai P, Fernandez TV, Gallagher PJ, Garrido H, Geller D, Girard SL, Grabe HJ, Grados MA, Greenberg BD, Gross-Tsur V, Haddad S, Heiman GA, Hemmings SM, Hounie AG, Illmann C, Jankovic J, Jenike MA, Kennedy JL, King RA, Kremeyer B, Kurlan R, Lanzagorta N, Leboyer M, Leckman JF, Lennertz L, Liu C, Lochner C, Lowe TL, Macciardi F, McCracken JT, McGrath LM, Mesa Restrepo SC, Moessner R, Morgan J, Muller H, Murphy DL, Naarden AL, Ochoa WC, Ophoff RA, Osiecki L, Pakstis AJ, Pato MT, Pato CN, Piacentini J, Pittenger C, Pollak Y, Rauch SL, Renner TJ, Reus VI, Richter MA, Riddle MA, Robertson MM, Romero R, Rosàrio MC, Rosenberg D, Rouleau GA, Ruhrmann S, Ruiz-Linares A, Sampaio AS, Samuels J, Sandor P, Sheppard B, Singer HS, Smit JH, Stein DJ, Strengman E, Tischfield JA, Valencia Duarte AV, Vallada H, Van Nieuwerburgh F, Veenstra-Vanderweele J, Walitza S, Wang Y, Wendland JR, Westenberg HG, Shugart YY, Miguel EC, McMahon W, Wagner M, Nicolini H, Posthuma D, Hanna GL, Heutink P, Denys D, Arnold PD, Oostra BA, Nestadt G, Freimer NB, Pauls DL, Wray NR, Stewart SE, Mathews CA, Knowles JA, Cox NJ, & Scharf JM (2013). Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genetics, 9(10), e1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, & 1000 Genomes Project Consortium. (2014). Integrating sequence and array data to create an improved 1000 genomes project haplotype reference panel. Nature Communications, 5, 3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Narzo AF, Kozlenkov A, Roussos P, Hao K, Hurd Y, Lewis DA, Sibille E, Siever LJ, Koonin E, & Dracheva S (2014). A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. Human Molecular Genetics, 23(18), 4801–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, Patel N, Woo DA, Marcus SM, Siever LJ, & Haroutunian V (2008). Increased serotonin 2C receptor mRNA editing: A possible risk factor for suicide. Molecular Psychiatry, 13(11), 1001–1010. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, & Uhlen M (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics : MCP, 13(2), 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler H, & Cubells JF (1997). Serotonin transporter protein (SLC6A4) allele and haplotype frequencies and linkage disequilibria in african- and european-american and japanese populations and in alcohol-dependent subjects. Human Genetics, 101(2), 243–246. [DOI] [PubMed] [Google Scholar]
- Grados MA (2010). The genetics of obsessive-compulsive disorder and tourette syndrome: An epidemiological and pathway-based approach for gene discovery. Journal of the American Academy of Child and Adolescent Psychiatry, 49(8), 810–9, 819.e1–2. [DOI] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, & Schmauss C (2002). Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron, 34(3), 349–356. [DOI] [PubMed] [Google Scholar]
- Hanna GL (2010). Schedule for obsessive-compulsive and other behavioral syndromes (SOCOBS). Ann Arbor, MI: University of Michigan. [Google Scholar]
- Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, & Lesch KP (1996). Allelic variation of human serotonin transporter gene expression. Journal of Neurochemistry, 66(6), 2621–2624. [DOI] [PubMed] [Google Scholar]
- Hesse S, Stengler K, Regenthal R, Patt M, Becker GA, Franke A, Knüpfer H, Meyer PM, Luthardt J, Jahn I, Lobsien D, Heinke W, Brust P, Hegerl U, & Sabri O (2011). The serotonin transporter availability in untreated early-onset and late-onset patients with obsessive-compulsive disorder. The International Journal of Neuropsychopharmacology / Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP), 14(5), 606–617. [DOI] [PubMed] [Google Scholar]
- Honda S, Nakao T, Mitsuyasu H, Okada K, Gotoh L, Tomita M, Sanematsu H, Murayama K, Ikari K, Kuwano M, Yoshiura T, Kawasaki H, & Kanba S (2017). A pilot study exploring the association of morphological changes with 5-HTTLPR polymorphism in OCD patients. Annals of General Psychiatry, 16, 2-017-0126-6. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, & Marchini J (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics, 5(6), e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, & Goldman D (2006). Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. American Journal of Human Genetics, 78(5), 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, & Kato T (2003). RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neuroscience Letters, 346(3), 169–172. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, & Ryan N (1997). Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): Initial reliability and validity data. Journal of the American Academy of Child and Adolescent Psychiatry, 36(7), 980–988. [DOI] [PubMed] [Google Scholar]
- Leckman JF, Denys D, Simpson HB, Mataix-Cols D, Hollander E, Saxena S, Miguel EC, Rauch SL, Goodman WK, Phillips KA, & Stein DJ (2010). Obsessive-compulsive disorder: A review of the diagnostic criteria and possible subtypes and dimensional specifiers for DSM-V. Depression and Anxiety, 27(6), 507–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite J, Hespanhol R, & Buchpiguel CA (2015). Molecular imaging in genetics. Neuroimaging Clinics of North America, 25(1), 17–29. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Müller CR, Hamer DH, & Murphy DL (1996). Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science (New York, N.Y.), 274(5292), 1527–1531. [DOI] [PubMed] [Google Scholar]
- Lestrade L, & Weber MJ (2006). snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Research, 34(Database issue), D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Yeung JM, Cherny SS, & Sham PC (2012). Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics, 131(5), 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZX, Jiang P, & Xing Y (2012). Genetic variation of pre-mRNA alternative splicing in human populations. Wiley Interdisciplinary Reviews RNA, 3(4), 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyddon R, Dwork AJ, Keddache M, Siever LJ, & Dracheva S (2013). Serotonin 2c receptor RNA editing in major depression and suicide. The World Journal of Biological Psychiatry : The Official Journal of the World Federation of Societies of Biological Psychiatry, 14(8), 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMaster FP (2010). Translational neuroimaging research in pediatric obsessive-compulsive disorder. Dialogues in Clinical Neuroscience, 12(2), 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O’Leary DS, Yuh WT, & Heckel D (2002). Structural MR image processing using the BRAINS2 toolbox. Computerized Medical Imaging and Graphics : The Official Journal of the Computerized Medical Imaging Society, 26(4), 251–264. [DOI] [PubMed] [Google Scholar]
- Mak L, Streiner DL, & Steiner M (2015). Is serotonin transporter polymorphism (5-HTTLPR) allele status a predictor for obsessive-compulsive disorder? A meta-analysis. Archives of Women’s Mental Health, 18(3), 435–445. [DOI] [PubMed] [Google Scholar]
- Mattina GF, & Steiner M (2016). The need for inclusion of sex and age of onset variables in genetic association studies of obsessive-compulsive disorder: Overview. Progress in NeuroPsychopharmacology & Biological Psychiatry, 67, 107–116. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, & Weinberger DR (2006). Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nature Reviews. Neuroscience, 7(10), 818–827. [DOI] [PubMed] [Google Scholar]
- Molineaux SM, Jessell TM, Axel R, & Julius D (1989). 5-HT1c receptor is a prominent serotonin receptor subtype in the central nervous system. Proceedings of the National Academy of Sciences of the United States of America, 86(17), 6793–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health. (2008). Novel interventions for neurodevelopmental disorders (R21/R33). Retrieved from http://grants.nih.gov/grants/guide/rfa-files/RFA-MH-09-021.html [Google Scholar]
- Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, & Sanders-Bush E (2001). RNA editing of the human serotonin 5-HT2C receptor. Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 24(5), 478–491. [DOI] [PubMed] [Google Scholar]
- Pauls DL, Abramovitch A, Rauch SL, & Geller DA (2014). Obsessive-compulsive disorder: An integrative genetic and neurobiological perspective. Nature Reviews. Neuroscience, 15(6), 410–424. [DOI] [PubMed] [Google Scholar]
- Petersen AC, Crockett L, Richards M, & Boxer A (1988). A self-report measure of pubertal status: Reliability, validity and initial norms. Journal of Youth and Adolescence, 7(2), 117–133. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, & Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38(8), 904–909. [DOI] [PubMed] [Google Scholar]
- Rosenberg DR, & Keshavan MS (1998). A.E. Bennett research award. Toward a neurodevelopmental model of obsessive-compulsive disorder. Biological Psychiatry, 43(9), 623–640. [DOI] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Berument SK, Lord C, & Pickles A (2003). Social communication questionnaire. Los Angeles, CA: Western Psychological Services. [Google Scholar]
- Scahill L, Riddle MA, McSwiggin-Hardin M, Ort SI, King RA, Goodman WK, Cicchetti D, & Leckman JF (1997). Children’s Yale-Brown obsessive compulsive scale: Reliability and validity. Journal of the American Academy of Child and Adolescent Psychiatry, 36(6), 844–852. [DOI] [PubMed] [Google Scholar]
- Sinopoli VM, Erdman L, Burton CL, Park LS, Dupuis A, Shan J, Goodale T, Li B, Shaheen S-M, Crosbie J, Schachar RJ, Arnold PD (under review). Serotonin system genes and obsessive-compulsive trait dimensions in a population-based, pediatric sample: A genetic association study. Journal of Child Psychology and Psychiatry. [DOI] [PubMed] [Google Scholar]
- Sinopoli VM, Burton CL, Kronenberg S, & Arnold PD (2017). A review of the role of serotonin system genes in obsessive-compulsive disorder. Neuroscience and Biobehavioral Reviews, 80, 372–381. [DOI] [PubMed] [Google Scholar]
- Szeszko PR, MacMillan S, McMeniman M, Chen S, Baribault K, Lim KO, Ivey J, Rose M, Banerjee SP, Bhandari R, Moore GJ, & Rosenberg DR (2004). Brain structural abnormalities in psychotropic drug-naive pediatric patients with obsessive-compulsive disorder. The American Journal of Psychiatry, 161(6), 1049–1056. [DOI] [PubMed] [Google Scholar]
- Taylor S (2013). Molecular genetics of obsessive-compulsive disorder: A comprehensive meta-analysis of genetic association studies. Molecular Psychiatry, 18(7), 799–805. [DOI] [PubMed] [Google Scholar]
- Taylor S (2016). Disorder-specific genetic factors in obsessive-compulsive disorder: A comprehensive meta-analysis. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of Psychiatric Genetics, 171B(3), 325–332. [DOI] [PubMed] [Google Scholar]
- Voyiaziakis E, Evgrafov O, Li D, Yoon HJ, Tabares P, Samuels J, Wang Y, Riddle MA, Grados MA, Bienvenu OJ, Shugart YY, Liang KY, Greenberg BD, Rasmussen SA, Murphy DL, Wendland JR, McCracken JT, Piacentini J, Rauch SL, Pauls DL, Nestadt G, Fyer AJ, & Knowles JA (2011). Association of SLC6A4 variants with obsessive-compulsive disorder in a large multicenter US family study. Molecular Psychiatry, 16(1), 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walitza S, Marinova Z, Grunblatt E, Lazic SE, Remschmidt H, Vloet TD, & Wendland JR (2014). Trio study and meta-analysis support the association of genetic variation at the serotonin transporter with early-onset obsessive-compulsive disorder. Neuroscience Letters, 580, 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu J, Huang BO, Xu YM, Li J, Huang LF, Lin J, Zhang J, Min QH, Yang WM, & Wang XZ (2015). Mechanism of alternative splicing and its regulation. Biomedical Reports, 3(2), 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann D, van der Laan S, Underwood MD, Salvetat N, Cavarec L, Vincent L, Molina F, Mann JJ, Arango V, & Pujol JF (2016). Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Translational Psychiatry, 6(8), e878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland JR, Martin BJ, Kruse MR, Lesch KP, & Murphy DL (2006). Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Molecular Psychiatry, 11(3), 224–226. [DOI] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, & Christopoulos A (2008). RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacology & Therapeutics, 119(1), 7–23. [DOI] [PubMed] [Google Scholar]
- Wierenga LM, Bos MGN, Schreuders E, Kamp F.v., Peper JS, Tamnes CK, & Crone EA (2018). Unraveling age, puberty and testosterone effects on subcortical brain development across adolescence. Psychoneuroendocrinology, 91, 105–114. [DOI] [PubMed] [Google Scholar]
- Williams MT, Mugno B, Franklin M, & Faber S (2013). Symptom dimensions in obsessive-compulsive disorder: Phenomenology and treatment outcomes with exposure and ritual prevention. Psychopathology, 46(6), 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Hanna GL, Easter P, Kennedy JL, Rosenberg DR, & Arnold PD (2013). Glutamate system genes and brain volume alterations in pediatric obsessive-compulsive disorder: A preliminary study. Psychiatry Research, 211(3), 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel M, Wood SJ, Wellard RM, Harrison BJ, Fornito A, Pujol J, Velakoulis D, & Pantelis C (2008). Anterior cingulate glutamate-glutamine levels predict symptom severity in women with obsessive-compulsive disorder. The Australian and New Zealand Journal of Psychiatry, 42(6), 467–477. [DOI] [PubMed] [Google Scholar]


