Abstract
Interleukin-17F (IL-17F), considered a pro-inflammatory cytokine, has been shown to contribute to skeletal tissue degradation and hence chronic inflammation in rheumatoid arthritis (RA). In this study we utilized bioinformatics tools to analyze the effect of three exonic SNPs (rs2397084, rs11465553, and rs763780) on the structure and function of the IL-17F gene, and evaluated their association with RA in Pakistani patients. The predicted deleterious and damaging effects of identified genetic variants were assessed through the utilization of multiple bioinformatics tools including PROVEAN, SNP&GO, SIFT, and PolyPhen2. Structural and functional effects of these variants on protein structures were evaluated through the use of additional tools such as I-Mutant, MutPred, and ConSurf. Three-dimensional (3D) models of both the wild-type and mutant proteins were constructed through the utilization of I-TASSER software, with subsequent structural comparisons between the models conducted through the use of the TM-align score. A total of 500 individuals, 250 cases and 250 controls, were genotyped through Tri-ARMS-PCR method and the resultant data was statistically analyzed using various inheritance models. Our bioinformatics analysis showed significant structural differences for wild type and mutant protein (TM-scores and RMSD values were 0.85934 and 2.34 for rs2397084 (E126G), 0.87388 and 2.49 for rs11465553 (V155I), and 0.86572 and 0.86572 for rs763780 (H161R) with decrease stability for the later. Overall, these tools enabled us to predict that these variants are crucial in causing disease phenotypes. We further tested each of these single nucleotide variants for their association with RA. Our analysis revealed a strong positive association between the genetic variant rs763780 and the risk of developing rheumatoid arthritis (RA) at both the genotypic and allelic levels. The genotypic association was statistically significant[χ2 = 111.8; P value <0.0001], as was the allelic level [OR 3.444 (2.539–4.672); P value 0.0008]. These findings suggest that the presence of this genetic variant may increase the susceptibility to RA. Similarly, we observed a significant distribution of the genetic variant rs11465553 at the genotypic level [χ2 = 25.24; P value = 0.0001]. However, this variant did not show a significant association with RA at the allelic level [OR = 1.194 (0.930–1.531); P value = 0.183]. However, the distribution of variant rs2397084 was more or less random across our sample with no significant association either at genotypic and or allelic level. Put together, our association study and in silico prediction of decreasing of IL17-F protein stabilty confirmed that two SNPs, rs11465553 and rs763780 are crucial to the suscetibility of and showed that these RA in Pakistani patients.
Introduction
Rheumatoid Arthritis is an inflammatory autoimmune disorder which mainly affects the joints, resulting in high levels of rheumatoid factor (RF) and anti-citrullinated protein antibody (ACPA) [1]. RA is a devastating disease, causing bone and cartilage deformities, and systemic injuries, if left untreated and unmanaged [2]. Its complications can be mild joints swelling or severe polyarthritis related with erosion of bones or cartilages [3]. The prevalence of RA remains relatively consistent across various populations worldwide, with a global incidence of 0.5 to 1.0%. [4]. Although its pathogenesis is still unclear, many genetic, environmental, hormonal, and infectious factors have significant roles in RA [5]. Approximately 50–60% of RA cases can be attributed to genetic factors [6]. The genetic factors include polymorphisms in cytokine receptors as well as in other functional pathways genes. However, several association studies and meta-analyses have linked about 150 genes/loci with RA in various populations [7–11]. Genetic variations in Th17 cytokines could affect the transcriptional regulation of these genes, which in response; increase the susceptibility of patients to diseases. For example, the polymorphisms in Th17 regimens are related with different complex diseases, including RA, Crohn’s disease, multiple sclerosis, and juvenile idiopathic arthritis [12]. Nearly 20 genes are known to be closely associated with the development and activity of Th17 cells [13].
The IL-17 family of proinflammatory cytokines is made up of six distinct members: IL-17A, IL-17B, IL-17C, IL-17D, IL-17E, and IL-17F [14]. Despite the co-localization of the genes encoding IL-17A and IL-17F on the chromosome 6p12, other members of the IL-17 family are distributed across different chromosomes [15–17]. The expression of IL-17A and IL-17F stimulate the generation of additional cytokines, chemokines, and antimicrobial peptides, leading to the degradation of skeletal tissues and chronic inflammation in affected individuals [15]. The expression of both IL-17A and IL-17F is detectable within synovial tissue, and their presence has been implicated in the pathogenesis of rheumatoid arthritis (R [18]. Based on in vitro evidence demonstrated that IL-17F may be involved in the regulation of angiogenesis, as well as the production of specific cytokines from both endothelial cells (including CXCL1, ICAM1, IL-6, and IL-8) and epithelial cells (such as G-CSF) [19–22].
Recent evidence has revealed that the H161R variant functions as a natural antagonist to wild-type IL17F. This is achieved through its ability to bind to the IL17F receptor without inducing downstream signaling pathways, effectively blocking the induction of IL8 [23, 24]. Several inflammatory diseases, including rheumatoid arthritis, inflammatory bowel disease, asthma, Graves’ disease, ulcerative colitis, and cancer, have identified IL-17F as a promising candidate gene. These findings highlight the potential involvement of IL-17F in the pathogenesis of these conditions and suggest that targeting this cytokine may hold therapeutic promise [25–32]. Numerous studies have demonstrated a clear association between levels of IL-17 in both serum and synovial fluid and markers of inflammation, including rheumatoid factor (RF), C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), and the Disease Activity Score 28 (DAS-28) in patients with rheumatoid arthritis (RA) [33].
Being an important component of inflammatory pathway, the single nucleotide variants of IL-17F have been subjected to association studies and the link with RA has been reported for rs2397084, rs11465553 and rs763780 variants in various populations [14–19]. Each of these variants lies in exonic regions of IL-17F gene and result in non-synonymous changes in the encoded protein.
The epigenome encompasses a variety of mechanisms that modulate gene expression patterns and phenotypic outcomes in response to environmental factors, such as nutrition, pathogens, and climate. These mechanisms include DNA methylation, chromatin remodeling, histone tail modifications, microRNAs, and long noncoding RNAs. The complex interplay between the epigenome and environmental cues highlights the crucial role of epigenetic regulation in shaping biological processes and underscores its potential as a therapeutic target for a broad range of diseases. [20, 21]. Environment, genome and epigenome might be involved in multi-level interaction [22]. Additionally, epigenome variation appears to have an impact on health and productivity [23–25]. In eukaryotes, gene expression is regulated through complex, temporal, and multidimensional mechanisms [26]. In each type of tissue, only a small fraction of the genome is expressed, and gene expression varies with developmental stage; therefore, eukaryotes express genes differently based on their tissues [27, 28]. The expression of a gene is influenced by the amount of its gene products in the tissue where it is expressed, as well as in other tissues that contribute to the formation of the gene product [29]. The analysis of genes and proteins associated with diseases and important traits entails studying at the cellular or chromosomal level 1 [30].
In this study we applied various bioinformatics tools to determine the effect of, rs11465553 (V155I), rs2397084 (H161R) and rs763780 (E126G) on the structure, stability and hence function of IL-17 F protein. Apart from in silico analysis, we also performed a case control study to determine the association between these three SNPs and RA in Pakistani population.
Methodology
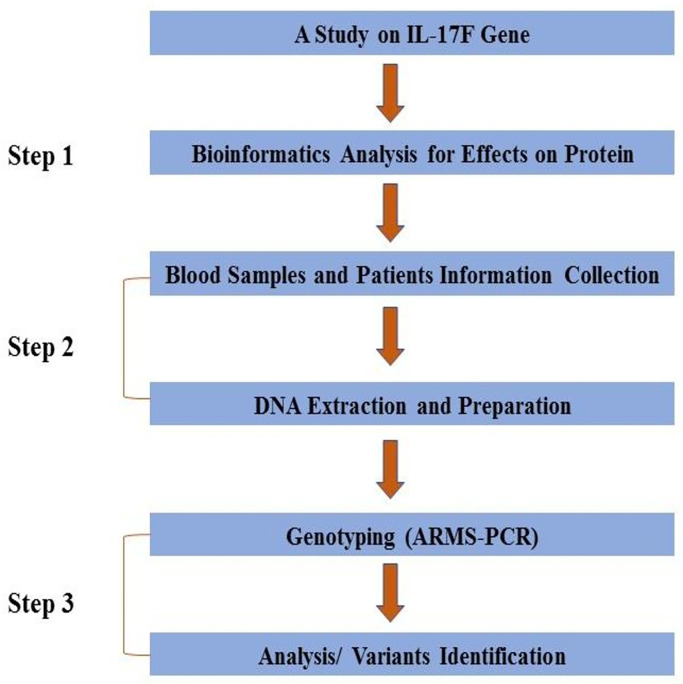
A workflow for the complete methodology is given in (Fig 1).
Fig 1. Workflow of the study.
Bioinformatics analysis
Different bioinformatics tools, including PhD-SNP (Predictor of human Deleterious SNP) (https://snps.biofold.org/phd-snp/phd-snp.html) [31], SNPs&GO (https://snps.biofold.org/snps-and-go/snps-and-go.html) [32], SIFT (Sorting Intolerant from Tolerant) (https://sift.bii.a-star.edu.sg/) [33], PROVEAN (Protein Variation Effect Analyzer) (http://provean.jcvi.org/seq_submit.php) [34], and PolyPhen2 (Polymorphism Phenotyping 2) (http://genetics.bwh.harvard.edu/pph2/) [35] were applied to evaluate deleterious effect of these variants. Structural and functional effect of the variants was predicted by MutPred (http://mutpred.mutdb.org/) [36] and their effects on the stability of the targeted protein were analyzed using I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html) [37]. Furthermore, the ConSurf server (http://consurf.tau.ac.il/2016/) helped to predict the evolutionary conserved residues [38]. For 3D modeling of these proteins, the Protein Data Bank (PDB) file was produced by I-TASSER (https://seq2fun.dcmb.med.umich.edu//I-TASSER/) [39]. Chimera 1.11 was used for the molecular properties and visualization of resultant proteins [40]. We compared the structures of wild type and mutant proteins using TM-align (https://seq2fun.dcmb.med.umich.edu//TM-align/example/873772.html), a computational tool that calculates TM scores and RMSD values. This helped us understand how the mutation impacted the protein’s shape and function. [41].
Genetic analysis
For genetic analysis, 250 seropositive RA patients (187 females, 63 males) with the mean age of 43.5 (±14.5) years were recruited from Department of Rheumatology, Lady Reading Hospital (LRH) Peshawar, Pakistan. The patients’ diagnoses were made by experienced rheumatologists who followed the rigorous guidelines set out by the American College of Rheumatology (ACR) [42]. Each patient was examined for the number of joints involved, the extra-articular manifestations as well as for the clinical features, such as rheumatoid factor (RF), erythrocyte sedimentation rate (ESR) and ACPA. Similarly, a control group of 250 healthy individuals (181 female, 69 males) was included for comparison. The mean age of the control group was 42 (±12.6) years with a standard deviation. Individuals with any other autoimmune complex disease or family history of such illnesses were excluded. Prior to the study, written informed consent was obtained from all participants, and the study was conducted in accordance with ethical guidelines set by both the Abdul Wali Khan University Mardan and LRH Peshawar in Pakistan. The clinical characteristics of the rheumatoid arthritis (RA) patients are detailed in Table 1.
Table 1. Characteristics of both rheumatoid arthritis patients and healthy controls.
| Variable | Cases (n = 250) | Controls (n = 250) | P value |
|---|---|---|---|
| Gender (male/female) | 63/187 | 69/181 | 0.6121 |
| Age in years Mean (±SD) | 43.5 (±14.5) | 42 (±12.6) | 0.217 |
| Disease duration in years Mean (±SD) | 4.1 (±3.7) | - | - |
| Sero-positive antibody Mean (±SD) | 100% (RF positive) | - | - |
| ESR Mean (±SD) | 40.60 (±15.8) | - | - |
Blood sample (5 mL) was taken from each participant and DNA was isolated via phenol-chloroform method [43]. Three IL-17F gene SNPs rs2397084, rs11465553, and rs763780 were genotyped using Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS-PCR)with allele-specific primers (two forward and one reverse) and details of the nucleotide has been provided in Table 2. The primer sets included the following primers; 1) for IL-17F-rs2397084 (T/C), F1: 5’-TCCGGACGACCAGGGTCC-3’; F2: 5’-CTCCGGACGACCAGGGTCT R: 5’-CCAGGCTGTGTGGCTCCAGAA-3’. 2) for IL-17F-rs11465553 (A/G), F1: 5’-TGACTGTTGGCTGCACCTGCA-3’; F2: 5’-ACTGTTGGCTGCACCTGCG-3’; R: 5’-CTGTTTCCATCCGTGCAGGTC-3’; and 3) for IL-17F-rs763780 (C/T), F1: 5’-ATATGCACCTCTTACTGCACAC-3’; F2: 5’-GATATGCACCTCTTACTGCACAT-3’; R: 5’-TACCCCTCGGAAGTTGTACAG-3’. To amplify our DNA samples and study the IL-17F gene SNPs, we used a carefully crafted set of PCR conditions. First, we began with an initial denaturation step at 94°C for 5 minutes, to help prepare the DNA strands for amplification. Next, we embarked on a series of 35 cycles, each of which included denaturation at 94°C for 30 seconds, annealing at a temperature range of 57–60°C for 30 seconds, and extension at 72°C for 1 minute. These cycles were designed to efficiently amplify the specific regions of interest in the DNA samples. Finally, we performed a final extension step at 72°C for 7 minutes to ensure that all of the amplified DNA fragments were completely synthesized. With these carefully controlled PCR conditions, we were able to generate high-quality DNA amplification products for our research. We used a 2% agarose gel to separate the amplified products and looked for distinct band patterns. By comparing these patterns to known standards, we were able to accurately determine the genotype calls for each sample.
Table 2. Details of amino acid changes in the IL17-F gene.
| Gene | SNP ID | Reference Allele | Alter Allele | Amino acid Changes |
|---|---|---|---|---|
| IL-17F | rs2397084 | T | C | Glu126Gly (E126G) |
| rs11465553 | G | A | Val155Ile (V155I) | |
| rs763780 | C | T | His161Arg (H161R) |
The data was subjected to quality controls check using Hardy-Weinberg Equilibrium (HWE), and association of IL-17F rs2397084, rs11465553, and rs763780 with RA was tested using the statistical models. We performed statistical analyses using both the Chi-square (χ2) and Fisher’s exact tests, with a confidence interval of 95% (95% CI). The Chi-square test was utilized for larger sample sizes to compare observed and expected frequencies, while Fisher’s exact test was used for smaller sample sizes to calculate the probability of obtaining a frequency distribution under the assumption of independence. We only considered a P-value less than 0.05 to be statistically significant in all of our analyses.
Results
Bioinformatics analysis
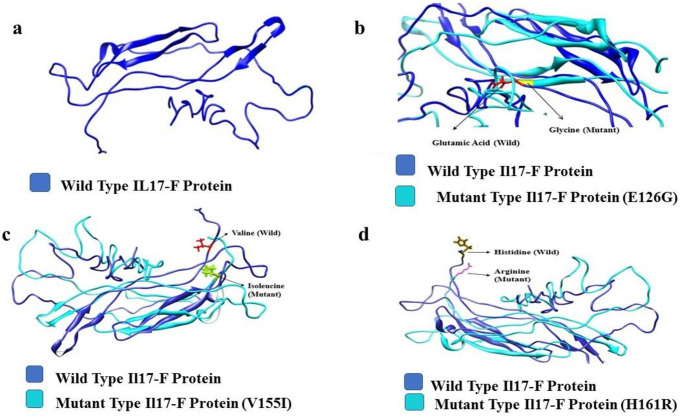
The results of bioinformatics analyses are listed in Table 3. In PhD-SNP and SNPs&GO tools, the SNPs with a prediction score >0.5 were evaluated deleterious. The scores for each variant (E126G, V155I, and H161R) were less than 0.5, which were evaluated to be neutral. In SIFT the cut-off value for tolerance index (TI) was set to 0.05. The variant V155I (0.12) was found to be tolerated, while the E126G (0.02) and H161R (0.05) were evaluated as deleterious through SIFT. The PROVEAN (threshold value -2.5) predicted V155I (-0.926) as neutral and E126G (-5.786) and H161R (-2.620) variants as deleterious. PolyPhen2 (score range 0–1) predicted V155I (1) and E126G (0.999) as probably damaging and H161R (0.048) as a benign variant. Furthermore, I-Mutant predicted that all the three variants decreased the stability of IL-17F protein [E126G (RI = 9); V155I (RI = 4); H161R (RI = 5)]. MutPred tool predicted structural and functional consequences including gain of intrinsic disorder, loss of allosteric site, creation of glycosylation and catalytic site, and altered membrane protein etc. Only H161R showed loss of strand, altered transmembrane protein, and altered protein stability. ConSurf predicted that the variants E126G and V155I were highly conserved, exposed and functional residues while H161R was highly conserved and exposed but not functionally active residue. The 3-D proteins structures were produced using I-TASSER and were subjected to TM-align analysis. Thus, significant structural changes were determined through TM-scores and RMSD values of the variants. TM-scores and RMSD values were 0.85934 and 2.34 for E126G, 0.87388 and 2.49 for V155I, and 0.86572 and 0.86572 for H161R, respectively. Finally, protein structures were viewed and characterized through Chimera 1.11and are shown in (Fig 2).
Table 3. Bioinformatic analyses of IL-17F gene.
| Bioinformatics tools | rs2397084 (E126G) | rs11465553 (V155I) | rs763780 (H161R) | |
|---|---|---|---|---|
| PHD SNP (Threshold 0.5) | Prediction | Neutral | Neutral | Neutral |
| Score | 0.145 | 0.381 | 0.138 | |
| SNP& GO (Threshold 0.5) | Prediction | Neutral | Neutral | Neutral |
| Score | 0.145 | 0.159 | 0.050 | |
| SIFT | Prediction | Not-tolerated | Tolerated | Not-tolerated |
| Score | 0.02 | 0.12 | 0.05 | |
| PROVEAN | Prediction | Disease | Neutral | Disease |
| Score | -5.786 | -0.926 | -2.620 | |
| POLYPHEN-2 | Prediction | Probably Damaging | Probably Damaging | Benign |
| Score | 1 | 0.999 | 0.048 | |
| I-Mutant | Prediction | Decrease | Decrease | Decrease |
| Score | 9 | 4 | 5 | |
| ConSurf | Prediction | Highly Conserved and Exposed (F) | Highly Conserved and Exposed (F) | Highly Conserved and Exposed |
| Conservation score | 9 | 8 | 5 | |
| MutPred | Top Features | - | - | LOSS Of Strand (P = 0.05) Altered Transmembrane protein (P = 1.3e-03) Altered Stability (P = 0.03) |
0.0< TM-score < 0.30, random structural similarity 0±0.3 and 0.5 < TM-score < 1.00, in about the same fold 0.5±1.
Fig 2.
3-D structural models visualized in Chimera 1.11 (a). wild type IL-17F protein (b). IL-17F superimposed with E126G mutant (c). IL-17F superimposed with V155I mutant (d). IL-17F superimposed with H161R mutant.
Association analysis
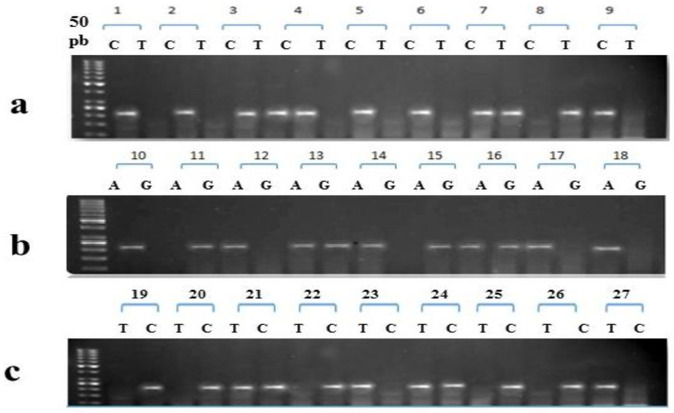
The findings of association analyses are given in Table 4 and gel electrophoresis pattern are shown in (Fig 3). An insignificant distribution of rs2397084 was observed at both the genotypic [χ2 = 3.202; P value 0.201] and allelic levels [OR 0.858 (0.668–1.101); P value 0.253]. Whereas, in case of rs11465553 a significant distribution of genotypes was observed in co-dominant [χ2 = 25.24; P value 0.0001] and homozygous models [OR 0.415 (0.268–0.641); P value 0.0001], but the distribution of its genotypes was insignificant at allelic level [OR 1.194(0.930–1.531); P value 0.183]. Similarly, the distribution of rs763780 in cases and controls was significant at genotypic [χ2 = 111.8; P value 0.0001] as well as allelic level [OR 3.444(2.539–4.672); P value 0.0008].
Table 4. Statistical models used in association analysis.
| Genes | Models | Genotype | Cases 250 | Controls 250 | OR 95% Cl | χ2 -value | P-value |
|---|---|---|---|---|---|---|---|
| IL17F rs2397084 | Co- | n (% age) | n (% age) | ||||
| Dominant | CC | 32 (13%) | 35 (14%) | - | 3.202 | 0.201 | |
| Model | CT | 160 (64%) | 173 (70%) | ||||
| TT | 58 (23%) | 42 (16%) | |||||
| Homozygous | TT | 58 (23%) | 42 (16%) | 1.496 | |||
| Dominant | CT + CC | 192 (77%) | 208 (84%) | (0.960–2.330) | - | 0.093 | |
| Homozygous | CC | 32 (13%) | 35 (14%) | 0.901 | |||
| Recessive | CT + TT | 218 (87%) | 215 (86%) | (0.538–1.509) | - | 0.793 | |
| Heterozygou | CT | 160 (64%) | 173 (70%) | 0.791 | |||
| s | CC+TT | 90 (36%) | 77 (30%) | (0.549–1.155) | - | 0.255 | |
| Additive | C | 224 (44.8%) | 243 (48.6%) | 0.858 | |||
| T | 276 (45.2%) | 257 (51.4%) | (0.668–1.101) | 0.253 | |||
| IL17F rs11465553 | Co- | AA | 32 (13%) | 48 (18%) | |||
| Dominant | AG | 179 (72%) | 125 (50%) | - | 25.24 | 0.0001 | |
| Model | GG | 39 (16%) | 77 (32%) | ||||
| Homozygous | GG | 39 (16%) | 77 (32%) | 0.415 | |||
| Dominant | AG +AA | 211 (84%) | 173 (68%) | (0.268–0.641) | — | 0.0001 | |
| Homozygous | AA | 32 (13%) | 48 (18%) | 0.617 | |||
| Recessive | GG +AG | 218 (87%) | 202 (82%) | (0.379–1.005) | — | 0.066 | |
| Heterozygou | AG | 179 (72%) | 125(50%) | 1.19 | |||
| s | AA+GG | 71 (28%) | 125 (50%) | (0.790–1.791) | — | 0.466 | |
| Additive | A | 243 (48.6%) | 221 (44.2%) | 1.194 | |||
| G | 257 (51.4%) | 279 (54.8%) | (0.930–1.531) | — | 0.183 | ||
| IL17F Irs763780 | Co- | TT | 79 (31.6%) | 192 (76.8%) | |||
| Dominant | CC | 18 (7.2%) | 17 (6.8%) | - | 111.8 | <0.0001 | |
| Model | CT | 153 (61.2%) | 41 (16.4%) | ||||
| Homozygous | TT | 79 (39%.76) | 192(28.23%) | 0.139 | |||
| Dominant | CT + CC | 171(60.24%) | 58(61.67%) | (0.093–0.027) | - | 0.001 | |
| Homozygous | CC | 18 (16.20%) | 17 (21.08%) | 1.063 | |||
| Recessive | CT + TT | 232 (83.80%) | 233 (78.92%) | (0.534–2.115) | - | 1.00 | |
| Heterozygou | CT | 153 (61.2%) | 41 (16.4%) | 4.193 | |||
| s | CC+TT | 97 38.8%) | 109(83.6%) | (2.70–6.51) | - | 0.0001 | |
| Additive | C | 189(37.8%) | 75 (15%) | 3.444 | |||
| T | 311 (62.2%) | 425 (85%) | (2.539–4.672) | 0.0001 |
Fig 3. Tri-Primer ARMS-PCR Gel Pattern of IL17-F Gene, a) IL-17F rs2397084 Genotypes, b) IL-17F rs11465553 Genotypes, and c) IL-17F rs763780 Genotypes.
Discussion
IL-17 plays a crucial role in rheumatoid arthritis (RA) by promoting inflammation and angiogenesis. In the early stages of RA, IL-17 activates fibroblast-like synoviocytes (FLS) to produce vascular endothelial growth factor (VEGF), which leads to increased angiogenesis and inflammation in the joint [44]. IL-17 also boosts the production of inflammatory mediators such as IL-6, IL-8, prostaglandin E2 (PGE2), and granulocyte colony-stimulating factor (G-CSF) from synovial fibroblasts [45–47]. Besides RA, many studies have elucidated relationship of IL-17F rs2397084 (E126G) and rs763780 (H161R) polymorphisms to osteoarthritis [48], IBD and UC [49]. Similarly, The IL-17F H161R genetic variation has also been studied in relation to various other disorders., including functional dyspepsia (FD), BD, chronic fatigue syndrome (CFS) or gastric cancer [50] FD and H. pylori-infected patients [51].
The in-silico and computational methods have identified several variants that significantly affect the structure and function of certain proteins [52–55]. Therefore, we utilized multiple in-silico tools to assess the deleterious impacts of the variants on the structure and function of the IL17F protein. Bioinformatics tools predicted deleterious effect of variants and the findings were cross-checked with CADD, REVEL, Mutation Assessors and MetalR in Ensemble genome browser 96 (accessed: 25th January, 2022). When a variant receives a CADD score of 30, it falls within the top 0.1% of the most damaging single nucleotide polymorphisms (SNPs) known to science. Similarly, a CADD score of 20 signifies that the variant is among the top 1% of the most harmful SNP variants found throughout the entirety of the human genome. The CADD score of E126G variant was 27 followed by V155I and H161R that were 23 and 21, respectively. MutPred1.2 server predicted that E126G variant had highest P-value of 0.677, followed by V155I and H161R (0.252 and 0.088, respectively). These results suggested that these variants may structurally and functionally affect IL17F protein. Furthermore, I-Mutant predicted that these variants decreased the protein stability, as listed in Table 3. To ensure the reliability of our findings, we double-checked the results from I-Mutant by comparing them with those generated by the CUPSAT server (http://cupsat.tu-bs.de/), which ultimately provided further support for our initail findings. ConSurf analysis facilitated the assessment of protein evolutionary conservation, with the identification of the most highly conserved amino acids providing key insights into their fundamental role in protein structure and function [38]. Furthermore, the highly conserved residues are very important for protein-protein interactions. According to Miller and Kumar, highly conserved nsSNPs are most damaging ones [33].
In this study, the variants E126G and V155I were identified within a region of high evolutionary conservation, and were predicted to be critical for protein function., while H161R was functionally inactive. The protein structure was modelled using I-TASSER with protein FASTA sequences. We utilized the RAMPAGE server to perform a Ramachandran Plot Analysis and compute RAMPAGE values for the modeled protein structures [56]. Protein structures with RAMPAGE values > 80% were considered as a better structure [57]. The wild type IL17-F protein achieved a RAMPAGE value of 84.8% (favored), 12.5% (allowed), and (2.7%) outlier. Similarly, for the variant E126G; 83.4% (favored), 11.5 (allowed), 5.1 (outlier), and for V155I; 81.2% (favored), 11.6% (allowed), 7.2% (outlier), and for H161R 85.4% (favored), 11.5% (allowed), 3.1% (outlier).
In this study, we analyzed three variants (rs763780, rs11465553 and rs2397084) of IL-17F genes for their association with RA and found significant association for two of them (rs763780 and rs11465553) in Pakistani population. Contrary to our findings, a case-control study on Polish population reported no association of IL-17F rs763780 and rs2397084 with RA. However, a significant association was observed, for rs763780, upon stratification of individuals, based on joints tenderness, HAQ score or DAS-28-CRP level. Furthermore, rs2397084 was associated with longer disease activity in RA cases [14]. Similarly, another Polish study reported IL-17F rs763780 as risk factor of RA [15]. A group of investigators, genotyped IL-17F (rs763780, rs11465553, rs2397084) using RFLP method and established an insignificant distribution in Turkish population [16]. There were different observations even in the same population, when the literature was reviewed. A comparatively larger case-controls study on individuals of Polish decent (422 RA cases and 337 control) determined an insignificant association of IL-17F (rs763780, rs11465553, rs2397084) through TaqMan assays [17]. Marwa et al. performed a case-control study in Tunisian population using RFLP method and showed a significant association of IL-17F rs2397084 and rs763780 with RA [19]. In a meta-analysis of 7,474 patients and 10,628 controls, obtained from 25 different studies, established that IL-17F rs763780 was significantly involved with increasing the RA risk [18]. To our knowledge, genetic factors have rarely been studied in Pakistanis. Few studies have replicated the known SNPs/variants in Pakistani RA patients [11, 58–61]. Recently, IL-17F gene was sequenced in 50 RA cases and 50 controls of Pakistani origin and established a significant association of rs763780 [62]. Furthermore, there is no newly established and authentic data available on the prevalence of RA in Pakistani population. However, the trend in disease progression with respect to gender differences have been shown [63], with the prevalence rate of 0.14% [64].
There is a limitation of the study. We did not perform haplotype analyses of the three SNPs and the information of linkage disequilibrium is not available. Although our study provides valuable insights, it is important to note that ethnic variability may exist, highlighting the need for large-scale, multi-ethnic population studies to fully elucidate the pathogenic mechanisms and evolutionary background of the genetic factors implicated.
Conclusions
It is concluded that IL-17F rs763780 (H161R) and rs11465553 (V155I) are the risk factors for RA in Pakistani patients. Furthermore, these are important coding variants, encoding highly conserved amino acids. Thus, these variants decrease the stability of IL-17F protein, thereby causing structural changes. Thus, on the basis of these findings IL-17F can be considered to be one of the potential therapeutic targets.
Acknowledgments
We are grateful to all the participants and their families. Thanks, are also due to the Consultant Rheumatologist, Lady Ready Hospital-MTI Peshawar, Pakistan for providing RA samples and Department of Biotechnology, Abdul Wali Khan University Mardan, Pakistan for providing lab facilities.
Data Availability
All relevant data are within the paper.
Funding Statement
Dr. Fazal Jalil received funding from HEC-NRPU (The Higher Education of Pakistan (HEC) - National Research Programs for Universities (NRPU)) Project # 5203, under whose supervision the study was designed. The project funding provided resources for data collection, analysis, and manuscript preparation, which were carried out by the research team under his guidance.
References
- 1.Li G, Xia Z, Liu Y, Meng F, Wu X, Fang Y, et al. SIRT1 inhibits rheumatoid arthritis fibroblast-like synoviocyte aggressiveness and inflammatory response via suppressing NF-κB pathway. Bioscience reports. 2018;38(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McInnes I, Schett G. A patogênese da artrite reumatóide. N Engl J Med. 2011;365:2205–19. [DOI] [PubMed] [Google Scholar]
- 3.Dörr S, Lechtenböhmer N, Rau R, Herborn G, Wagner U, Müller-Myhsok B, et al. Association of a specific haplotype across the genes MMP1 and MMP3 with radiographic joint destruction in rheumatoid arthritis. Arthritis Res Ther. 2004;6(3):R199. doi: 10.1186/ar1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silman AJ, Pearson JE. Epidemiology and genetics of rheumatoid arthritis. Arthritis research & therapy. 2002;4(S3):S265. doi: 10.1186/ar578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowes J, Barton A. Recent advances in the genetics of RA susceptibility. Rheumatology. 2008;47(4):399–402. doi: 10.1093/rheumatology/ken005 [DOI] [PubMed] [Google Scholar]
- 6.Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nature Reviews Rheumatology. 2013;9(3):141. doi: 10.1038/nrrheum.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nature genetics. 2010;42(6):515. doi: 10.1038/ng.583 [DOI] [PubMed] [Google Scholar]
- 8.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature genetics. 2010;42(6):508. doi: 10.1038/ng.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalil SF, Bhatti A, Demirci FY, Wang X, Ahmed I, Ahmed M, et al. Replication of european rheumatoid arthritis loci in a Pakistani population. The Journal of Rheumatology. 2013;40(4):401–7. doi: 10.3899/jrheum.121050 [DOI] [PubMed] [Google Scholar]
- 10.Akhtar M, Ali Y, Islam Z-u, Arshad M, Rauf M, Ali M, et al. Characterization of Rheumatoid Arthritis Risk-Associated SNPs and Identification of Novel Therapeutic Sites Using an In-Silico Approach. Biology. 2021;10(6):501. doi: 10.3390/biology10060501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali Y, Khan S, Chen Y, Farooqi N, Islam Z-U, Akhtar M, et al. Association of AFF3 Gene Polymorphism rs10865035 with Rheumatoid Arthritis: A Population-Based Case-Control Study on a Pakistani Cohort. Genetics Research. 2021;2021. doi: 10.1155/2021/5544198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roeleveld DM, Koenders MI. The role of the Th17 cytokines IL-17 and IL-22 in Rheumatoid Arthritis pathogenesis and developments in cytokine immunotherapy. Cytokine. 2015;74(1):101–7. doi: 10.1016/j.cyto.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 13.van Hamburg JP, Tas SW. Molecular mechanisms underpinning T helper 17 cell heterogeneity and functions in rheumatoid arthritis. Journal of autoimmunity. 2018;87:69–81. doi: 10.1016/j.jaut.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 14.Paradowska‐Gorycka A, Wojtecka‐Lukasik E, Trefler J, Wojciechowska B, Lacki J, Maslinski S. Association between IL‐17F gene polymorphisms and susceptibility to and severity of Rheumatoid Arthritis (RA). Scandinavian journal of immunology. 2010;72(2):134–41. doi: 10.1111/j.1365-3083.2010.02411.x [DOI] [PubMed] [Google Scholar]
- 15.Bogunia-Kubik K, Świerkot J, Malak A, Wysoczańska B, Nowak B, Białowąs K, et al. IL-17A, IL-17F and IL-23R gene polymorphisms in Polish patients with rheumatoid arthritis. Archivum immunologiae et therapiae experimentalis. 2015;63(3):215–21. doi: 10.1007/s00005-014-0319-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkol İnal E, Görükmez O, Dündar Ü, Görükmez Ö, Yener M, Özemri Sağ Ş, et al. The influence of polymorphisms of interleukin-17A and-17F genes on susceptibility and activity of rheumatoid arthritis. Genetic testing and molecular biomarkers. 2015;19(8):461–4. doi: 10.1089/gtmb.2015.0064 [DOI] [PubMed] [Google Scholar]
- 17.Pawlik A, Kotrych D, Malinowski D, Dziedziejko V, Czerewaty M, Safranow K. IL17A and IL17F gene polymorphisms in patients with rheumatoid arthritis. BMC musculoskeletal disorders. 2016;17(1):208. doi: 10.1186/s12891-016-1064-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eskandari-Nasab E, Moghadampour M, Tahmasebi A. Meta-analysis of risk association between interleukin-17A and F gene polymorphisms and inflammatory diseases. Journal of Interferon & Cytokine Research. 2017;37(4):165–74. doi: 10.1089/jir.2016.0088 [DOI] [PubMed] [Google Scholar]
- 19.Marwa OS, Kalthoum T, Wajih K, Kamel H. Association of IL17A and IL17F genes with rheumatoid arthritis disease and the impact of genetic polymorphisms on response to treatment. Immunology letters. 2017;183:24–36. doi: 10.1016/j.imlet.2017.01.013 [DOI] [PubMed] [Google Scholar]
- 20.Barazandeh A, Mohammadabadi M, Ghaderi-Zefrehei M, Nezamabadipour HJIJoAAS. Predicting CpG islands and their relationship with genomic feature in cattle by hidden markov model algorithm. 2016;6(3):571–9. [Google Scholar]
- 21.Masoudzadeh S, Mohammadabadi M, Khezri A, Kochuk-Yashchenko O, Kucher D, Babenko O, et al. Dlk1 gene expression in different Tissues of lamb. 2020;10(4):669–77. [Google Scholar]
- 22.Mohamadipoor Saadatabadi L, Mohammadabadi M, Amiri Ghanatsaman Z, Babenko O, Stavetska R, Kalashnik O, et al. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. 2021;17(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barazandeh A, Mohammadabadi M, Ghaderi-Zefrehei M, Nezamabadi-Pour HJCJoAS. Genome-wide analysis of CpG islands in some livestock genomes and their relationship with genomic features. 2016;61(11):487–95. [Google Scholar]
- 24.Safaei SMH, Dadpasand M, Mohammadabadi M, Atashi H, Stavetska R, Klopenko N, et al. An Origanum majorana Leaf Diet Influences Myogenin Gene Expression, Performance, and Carcass Characteristics in Lambs. 2022;13(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordbar F, Mohammadabadi M, Jensen J, Xu L, Li J, Zhang LJA. Identification of candidate genes regulating carcass depth and hind leg circumference in simmental beef cattle using Illumina Bovine Beadchip and next-generation sequencing analyses. 2022;12(9):1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahsavari M, Mohammadabadi M, Khezri A, Asadi Fozi M, Babenko O, Kalashnyk O, et al. Correlation between insulin-like growth factor 1 gene expression and fennel (Foeniculum vulgare) seed powder consumption in muscle of sheep. 2021:1–11. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadabadi M, Masoudzadeh SH, Khezri A, Kalashnyk O, Stavetska RV, Klopenko NI, et al. Fennel (Foeniculum vulgare) seed powder increases Delta-Like Non-Canonical Notch Ligand 1 gene expression in testis, liver, and humeral muscle tissues of growing lambs. 2021;7(12):e08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadinejad F, Mohammadabadi M, Roudbari Z, Sadkowski TJA. Identification of Key Genes and Biological Pathways Associated with Skeletal Muscle Maturation and Hypertrophy in Bos taurus, Ovis aries, and Sus scrofa. 2022;12(24):3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammadabadi MJABJ. Expression of calpastatin gene in Raini Cashmere goat using Real Time PCR. 2020;11(4):219–35. [Google Scholar]
- 30.Mohammadabadi M, Asadollahpour Nanaei HJABJ. Leptin gene expression in Raini Cashmere goat using Real Time PCR. 2021;13(1):197–214. [Google Scholar]
- 31.Capriotti E, Altman RB. Improving the prediction of disease-related variants using protein three-dimensional structure. BMC bioinformatics. 2011;12(4):1–11. doi: 10.1186/1471-2105-12-S4-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calabrese R, Capriotti E, Casadio R, editors. Predicting the Insurgence of Human Genetic Diseases Due to Single Point Protein Mutation using Machine Learning Approach. Communications to SIMAI Congress; 2006. [DOI] [PubMed] [Google Scholar]
- 33.Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Human molecular genetics. 2001;10(21):2319–28. doi: 10.1093/hmg/10.21.2319 [DOI] [PubMed] [Google Scholar]
- 34.Cho J-H, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, et al. Systems biology of interstitial lung diseases: integration of mRNA and microRNA expression changes. BMC medical genomics. 2011;4(1):1–20. doi: 10.1186/1755-8794-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 35.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25(21):2744–50. doi: 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capriotti E, Fariselli P, Casadio R. I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic acids research. 2005;33(suppl_2):W306–W10. doi: 10.1093/nar/gki375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, et al. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20(8):1322–4. doi: 10.1093/bioinformatics/bth070 [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25(13):1605–12. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research. 2005;33(7):2302–9. doi: 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO III, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis & rheumatism. 2010;62(9):2569–81. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook P, Cohen M, Eisman J, Pocock N, Champion G, Yeates M. Effects of low dose corticosteroids on bone mass in rheumatoid arthritis: a longitudinal study. Annals of the rheumatic diseases. 1989;48(7):535–8. doi: 10.1136/ard.48.7.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu S, Lee J, Kim S. IL-17 increased the production of vascular endothelial growth factor in rheumatoid arthritis synoviocytes. Clinical rheumatology. 2006;25(1):16–20. doi: 10.1007/s10067-005-1081-1 [DOI] [PubMed] [Google Scholar]
- 45.Inoue H, Takamori M, Shimoyama Y, Ishibashi H, Yamamoto S, Koshihara Y. Regulation by PGE2 of the production of interleukin‐6, macrophage colony stimulating factor, and vascular endothelial growth factor in human synovial fibroblasts. British journal of pharmacology. 2002;136(2):287–95. doi: 10.1038/sj.bjp.0704705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onishi RM, Gaffen SL. Interleukin‐17 and its target genes: mechanisms of interleukin‐17 function in disease. Immunology. 2010;129(3):311–21. doi: 10.1111/j.1365-2567.2009.03240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benedetti G, Miossec P. Interleukin 17 contributes to the chronicity of inflammatory diseases such as rheumatoid arthritis. European journal of immunology. 2014;44(2):339–47. doi: 10.1002/eji.201344184 [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi M, Takahashi D, Hizawa N, Suzuki S, Matsukura S, Kokubu F, et al. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. Journal of Allergy and Clinical Immunology. 2006;117(4):795–801. doi: 10.1016/j.jaci.2005.12.1346 [DOI] [PubMed] [Google Scholar]
- 49.Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p. His161Arg polymorphism in IBD. Inflammatory bowel diseases. 2008;14(4):437–45. doi: 10.1002/ibd.20339 [DOI] [PubMed] [Google Scholar]
- 50.Jang W-C, Nam Y-H, Ahn Y-C, Lee S-H, Park S-H, Choe J-Y, et al. Interleukin-17F gene polymorphisms in Korean patients with Behçet’s disease. Rheumatology international. 2008;29(2):173. [DOI] [PubMed] [Google Scholar]
- 51.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. Journal of clinical immunology. 2008;28(1):44–9. doi: 10.1007/s10875-007-9125-8 [DOI] [PubMed] [Google Scholar]
- 52.Anwer F, Yun S, Nair A, Ahmad Y, Krishnadashan R, Deeg HJ. Severe refractory immune thrombocytopenia successfully treated with high-dose pulse cyclophosphamide and eltrombopag. Case reports in hematology. 2015;2015. doi: 10.1155/2015/583451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sneha P, Doss CGP. Molecular dynamics: new frontier in personalized medicine. Advances in protein chemistry and structural biology. 2016;102:181–224. doi: 10.1016/bs.apcsb.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 54.Mohajer FS, Parvizpour S, Razmara J, Khoshkhooy Yazdi M, Shamsir MS. Structural, functional and molecular dynamics analysis of the native and mutated actin to study its effect on congenital myopathy. Journal of Biomolecular Structure and Dynamics. 2017;35(7):1608–14. doi: 10.1080/07391102.2016.1190299 [DOI] [PubMed] [Google Scholar]
- 55.Akhtar M, Jamal T, Jamal H, Din JU, Jamal M, Arif M, et al. Identification of most damaging nsSNPs in human CCR6 gene: In silico analyses. International journal of immunogenetics. 2019;46(6):459–71. doi: 10.1111/iji.12449 [DOI] [PubMed] [Google Scholar]
- 56.Lovell SC, Davis IW, Arendall WB III, De Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics. 2003;50(3):437–50. [DOI] [PubMed] [Google Scholar]
- 57.Morris AL, MacArthur MW, Hutchinson EG, Thornton JMJPS, Function,, Bioinformatics. Stereochemical quality of protein structure coordinates. 1992;12(4):345–64. [DOI] [PubMed]
- 58.Jalil SF, Ahmed I, Gauhar Z, Ahmed M, Malik JM, John P, et al. Association of Pro12Ala (rs1801282) variant of PPAR gamma with Rheumatoid Arthritis in a Pakistani population. Rheumatology international. 2014;34(5):699–703. doi: 10.1007/s00296-013-2768-2 [DOI] [PubMed] [Google Scholar]
- 59.Arshad M, Bhatti A, John P. Identification and in silico analysis of functional SNPs of human TAGAP protein: A comprehensive study. PloS one. 2018;13(1):e0188143. doi: 10.1371/journal.pone.0188143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akhtar M, Khan S, Ali Y, Haider S, ud Din J, Islam Z-u, et al. Association study of CCR6 rs3093024 with Rheumatoid Arthritis in a Pakistani cohort. Saudi Journal of Biological Sciences. 2020;27(12):3354–8. doi: 10.1016/j.sjbs.2020.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aslam MM, John P, Fan K-H, Bhatti A, Aziz W, Ahmed B, et al. Investigating the GWAS-Implicated Loci for Rheumatoid Arthritis in the Pakistani Population. Disease markers. 2020;2020. doi: 10.1155/2020/1910215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amin A, Sheikh N, Mukhtar M, Saleem T, Akhtar T, Fatima N, et al. Association of interleukin-17 gene polymorphisms with the onset of Rheumatoid Arthritis. Immunobiology. 2021;226(1):152045. doi: 10.1016/j.imbio.2020.152045 [DOI] [PubMed] [Google Scholar]
- 63.Jalil F, Arshad M, Bhatti A, Jamal M, Ahmed M, Malik JM, et al. Progression pattern of rheumatoid arthritis: A study of 500 Pakistani patients. Pakistan journal of pharmaceutical sciences. 2017;30(4):1219–23. [PubMed] [Google Scholar]
- 64.Dai S-M, Han X-H, Zhao D-B, Shi Y-Q, Liu Y, Meng J-M. Prevalence of rheumatic symptoms, rheumatoid arthritis, ankylosing spondylitis, and gout in Shanghai, China: a COPCORD study. The Journal of rheumatology. 2003;30(10):2245–51. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.