Abstract
Identifying isocitrate dehydrogenase (IDH)-mutation and glioma subtype during surgery instead of days later can aid in modifying tumor resection strategies for better survival outcomes. We report intraoperative identification of IDH-mutant glioma (N = 12 patients) with a clinically compatible fluorescence lifetime imaging (FLIm) device (excitation: 355 nm; emission spectral bands: 390/40 nm, 470/28 nm, 542/50 nm). The fluorescence-derived parameters were analyzed to study the optical contrast between IDH-mutant tumors and surrounding brain tissue. IDH-mutant oligodendrogliomas exhibited shorter lifetimes (3.3 ± 0.1 ns) than IDH-mutant astrocytomas (4.1 ± 0.1 ns). Both IDH-mutant glioma subtypes had shorter lifetimes than white matter (4.6 ± 0.4 ns) but had comparable lifetimes to cortex. Lifetimes also increased with malignancy grade within IDH-mutant oligodendrogliomas (grade 2: 2.96 ± 0.08 ns, grade 3: 3.4 ± 0.3 ns) but not within IDH-mutant astrocytomas. The current results support the feasibility of FLIm as a surgical adjuvant for identifying IDH-mutant glioma tissue.
Keywords: astrocytoma, fluorescence lifetime, IDH-mutant glioma, imaging, neurosurgery, oligodendroglioma
1 |. INTRODUCTION
Adult-type diffuse gliomas are classified as astrocytoma, oligodendroglioma, and glioblastoma according to the World Health Organization (WHO) 2021 Classification of Tumors of the Central Nervous System (CNS) [1]. This updated classification, referred to as WHO CNS5, preferentially uses key molecular alterations to define tumor subgroups, compared to traditional histopathological markers used in the past. In the current classification scheme, the main differentiating factor among diffuse glioma subgroups is the mutational status of isocitrate dehydrogenase (IDH) genes, which are critical to tumor metabolism. Gliomas with IDH mutations have a better prognosis than those that do not have the mutations. Glioblastoma is now defined exclusively as IDH-wildtype, while astrocytoma and oligodendroglioma have mutations in the IDH1/2 genes. Within the IDH-mutant tumors, 1p/19q codeletion distinguishes oligodendroglioma (1p/19q co-deleted) from astrocytoma (1p/19q non-co-deleted). Tumor grades are defined within every subtype. Astrocytomas are graded as CNS WHO grade 2, 3, or 4, whereas oligodendrogliomas include CNS WHO grades 2 and 3. Within IDH mutant tumors, only tumor cells harbor the mutation, whereas normal surrounding glia are IDH-wildtype, allowing histopathologic differentiation between tumor and brain parenchyma.
Surgical resection remains the gold standard for the initial treatment of adult-type diffuse glioma [2, 3]. However, different glioma subtypes have different prognoses and respond differently to post-surgical therapy (chemotherapy and radiotherapy) [3, 4]. Identifying the type of diffuse glioma before or during surgery instead of several days post-surgery (current standard methods) will allow personalized optimal surgical strategies.
Oligodendrogliomas are known to respond to therapy more favorably than astrocytomas and are associated with significantly longer overall survival [1, 2, 5, 6]. While the extent of surgical resection has been associated with improved survival for gliomas in general [7], the improved response of oligodendrogliomas to adjuvant therapy reduces the importance of gross total resection in these tumors. More aggressive resections, while beneficial to survival, are more likely to damage eloquent parts of the brain [8]. Therefore, aggressive surgery with the goal of complete tumor removal is more important and appropriate for patients with astrocytomas than oligodendrogliomas. However, there remains a standing need for an intraoperative tool to identify the type of diffuse glioma in real-time to allow surgeons to modify the approach.
Radiomics and radiogenomics have been employed in classifying gliomas according to their malignancy grades and IDH mutational status, but they do not provide information to differentiate glioma subtypes within IDH-mutant tumors [9]. Currently, the standard techniques to identify IDH-mutant glioma subtype are based on molecular analyses (1p/19q assessment by fluorescence in-situ hybridization [FISH] or sequencing) that provide results weeks after tissue resection [1]. Intraoperative frozen section histology is often performed during surgery with an unsatisfactory agreement with the final diagnosis [10, 11].
Label-free fluorescence lifetime imaging (FLIm) leverages changes in tissue metabolism associated with the different glioma subtypes to address this challenge. The most prominent endogenous fluorophores in the brain are nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) and flavin adenine dinucleotide (FAD), which are directly involved in the tissue’s metabolic activity. Other compounds, including collagen, pyridoxal phosphate (PLP), pyridoxamine phosphate (PMP) and glutamate decarboxylase (GAD) [12–14], as well as lipopigments and porphyrins [15], also contribute to the intrinsic fluorescence of brain tissue.
The use of fluorescence lifetime to identify brain malignancies in human patients has evolved from the earlier studies on ex vivo freshly excised samples [15–19] to in vivo spectroscopy and imaging during neurosurgery [20–23]. These studies have shown the ability of FLIm to resolve non-pathologic tissue types (i.e., cortex from white matter) [16, 17, 22] and brain lesions (i.e., tumors and necrotic tissue) [15–17, 19–22]. Building on the previous work, this study aims to examine the potential of FLIm to identify the glioma subtype within IDH-mutant tumors intraoperatively. Such knowledge will aid the neurosurgeon’s real-time decision-making in modifying tumor resection strategies, avoiding unnecessarily aggressive resection, and thereby preserving eloquent tissue and neurological function. Intraoperative FLIm could personalize the treatment strategy leading to improved survival outcomes.
2 |. MATERIALS AND METHODS
2.1 |. Study workflow
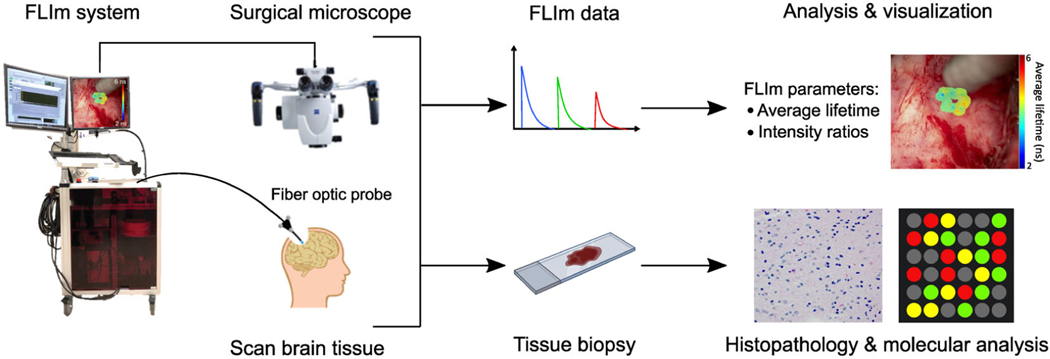
In the operating room (OR), a clinically compatible mesoscopic FLIm device [24, 25] developed by our group was interfaced with a conventional surgical microscope (OPMI Pentero 900; Zeiss, Germany) routinely used in open craniotomy procedures. During tumor resection surgery, clinically relevant regions at the resection margins where the neurosurgeon suspected tumor infiltration were FLIm-interrogated using a hand-held fiber optic probe and subsequently biopsied. Typically, 5–10 scans were obtained from the tumor cavity of each patient. The FLIm measurements obtained from each scan were labeled based on tumor subtype, tumor cellular density, and tissue type according to the histopathological evaluation and molecular analysis performed on the collected specimen. The workflow of FLIm data acquisition and biopsy collection for pathological validation is shown in Figure 1. This study was approved by the University of California, Davis Institutional Review Board (IRB), and all patients provided informed consent before surgery.
FIGURE 1.
Workflow of data collection using FLIm. The FLIm system was connected with the surgical microscope in the OR. The neurosurgeon used the distal end of a sterile fiber optic probe to scan brain tissue of interest. FLIm parameters were extracted and analyzed in real time and augmented on the white-light images of the surgical field-of-view obtained from the surgical microscope. Biopsies were collected from the scanned regions for histopathology (tissue type, tumor cellular density) and molecular analysis (IDH-mutation, 1p/19q codeletion).
2.2 |. Human patients
The study cohort comprised 12 patients with IDH-mutant gliomas confirmed by histopathology post-surgery. Table 1 summarizes the patient diagnosis and demographic information, the device used for collecting FLIm data, and the number of samples collected from each patient. Table 2 shows the number of scans/samples acquired for each glioma subtype and tumor malignancy grade.
TABLE 1.
Demographic information of IDH-mutant glioma patients recruited for the study
| Subject | Diagnosis | Sex | Age | Race | Ethnicity | Device | No. of samples |
|---|---|---|---|---|---|---|---|
| 1 | Astrocytoma, grade 3 | Male | 38 | White | Not Hispanic/Latino | S1 | 2 |
| 2 | Oligodendroglioma, grade 3 | Male | 54 | White | Not Hispanic/Latino | S1 | 7 |
| 3 | Astrocytoma, grade 4 | Male | 30 | White | Hispanic/Latino | S1 | 4 |
| 4 | Astrocytoma, grade 2 | Transgender female | 30 | White | Not Hispanic/Latino | S1 and S2 | 5 (S1) and 3 (S2) |
| 5 | Oligodendroglioma, grade 2 | Male | 36 | White | Not Hispanic/Latino | S1 and S2 | 4 (S1) and 4 (S2) |
| 6 | Astrocytoma, grade 3 | Male | 26 | White | Not Hispanic/Latino | S1 and S2 | 3 (S1) and 3 (S2) |
| 7 | Oligodendroglioma, grade 2 | Female | 29 | White | Not Hispanic/Latino | S2 | 3 |
| 8 | Oligodendroglioma, grade 3 | Female | 41 | White | Not Hispanic/Latino | S2 | 8 |
| 9 | Astrocytoma, grade 3 | Female | 39 | Unknown | Not Hispanic/Latino | S2 | 4 |
| 10 | Oligodendroglioma, grade 3 | Female | 40 | Unknown | Hispanic/Latino | S2 | 6 |
| 11 | Astrocytoma, grade 4 | Female | 41 | White | Not Hispanic/Latino | S2 | 5 |
| 12 | Astrocytoma, grade 2 | Male | 26 | Black | Hispanic/Latino | S3 | 5 |
TABLE 2.
Number of samples categorized by glioma subtype and malignancy grade
| Number of samples |
||||||
|---|---|---|---|---|---|---|
| Glioma subtype | Malignancy grade | Number of patients | Tumor | White matter | Cortex | Total |
| Oligodendroglioma | Grade 2 | 2 | 4 | 5 | 2 | 11 |
| Grade 3 | 3 | 10 | 6 | 5 | 21 | |
| Subtotal | 5 | 14 | 11 | 7 | 32 | |
| Astrocytoma | Grade 2 | 2 | 2 | 7 | 4 | 13 |
| Grade 3 | 3 | 5 | 7 | 0 | 12 | |
| Grade 4 | 2 | 5 | 2 | 2 | 4 | |
| Subtotal | 7 | 12 | 16 | 6 | 34 | |
| Total | 12 | 26 | 27 | 13 | 66 | |
2.3 |. FLIm instrumentation
FLIm data was collected using two custom-built, clinically compatible mesoscopic FLIm devices developed in our laboratory and described in detail elsewhere [24–26] Both devices had all components for signal generation, acquisition, data processing, and display integrated into a custom cart. Tissue autofluorescence was excited using a 355 nm pulsed Nd-YAG microchip laser (STV-02 E-1×0, TEEM photonics, France) (0.25 μJ pulse energy, 600 ps pulse duration, 120 Hz repetition rate). The light was delivered to the tissue using a 3-m long, 365 μm core diameter, 0.22 NA fiber optic sterilized hand-held probe. The neurosurgeon operated the distal end of the fiber optic probe to perform freehand scans of regions of interest in the brain. Each scan took between 2 and 5 s. The fluorescent light emitted from the tissue was collected by the same probe for multispectral detection in three spectral bands, 390/40 nm, 470/28 nm, and 542/50 nm. These spectral bands typically capture fluorescence from collagen/GAD/PMP, NAD(P)H, and FAD, respectively, whose emission maximums fall within the respective spectral bands.
The two FLIm devices differed in the detection system. To sample the fluorescence decays, the first system (S1) used a single microchannel plate photomultiplier tube (MCP-PMT, R3809U-50; Hamamatsu, Japan) connected to an amplifier (AM-1607–3000; Miteq Inc.) and a high-speed digitizer (12.5 GS/s, PXIe-5185; National Instruments, TX) [24]. The second system (S2) used three avalanche photodetector (APD) modules with an integrated transimpedance amplifier and a digitizer (2.5 GS/s, NI PXIe-5162, National Instruments, TX) [25].
S2 represents a technical upgrade from S1 with independent gain adjustment in each spectral band, 4-times faster imaging speed, and a 5-fold reduction of lifetime measurement variability [25]. The standard deviation of lifetime measurements acquired with S2 is typically smaller than those acquired with S1 (Figure S3). The FLIm parameters used in this study from both devices were comparable. The average lifetimes of both devices can be directly compared based on standard dye measurements [24, 25]. A correction factor was applied to the intensities in S1 to account for differences in detector sensitivity in each spectral band. The results obtained with either device independently were comparable to those reported here combining both datasets.
To ensure precise tracking and visualization of the point measurements on the tissue, an aiming beam from a 440 nm continuous-wave solid-state laser (TECBL-50G-440-USB-TTL, Worldstartech, Canada) was delivered to the tissue through the hand-held fiber optic probe. Precise motion tracking ensures that the tissue exposure to laser light falls within the maximum permissible exposure (MPE) limits in compliance with the American Standard for Safe Use of Lasers ANSI Z136.1 (2014) [27].
2.4 |. FLIm data acquisition, analysis, and visualization
FLIm scans (~10 mm2) were acquired from the surgical resection margins. The data acquisition, processing, and display sequentially occurred in real-time (33 ms/point measurement) to provide an augmented reality view of the surgical field-of-view with the FLIm parameters (example in Figure 2A) [22]. The FLIm data were also processed and analyzed independently from the real-time processes, and these results are reported here.
FIGURE 2.
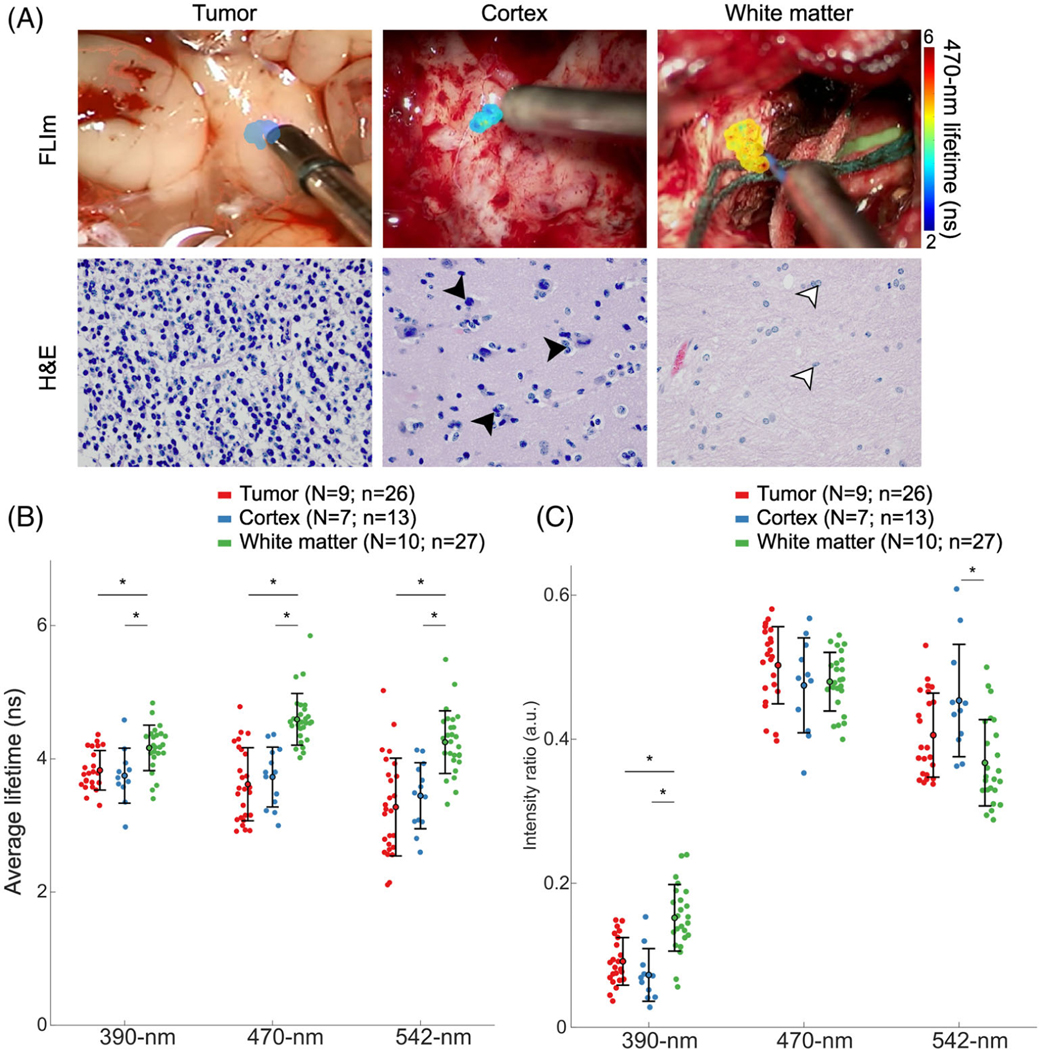
FLIm provides optical contrast between tumor and white matter. (A) Surgical field-of-view with the FLIm scanned area overlaid with the 470-nm average lifetime for a region of tumor, normal cortex, and normal white matter, and corresponding H&E images (×40 magnification). Tumor tissue is histologically characterized by high cellular density. Cortex tissue is composed of neuron bodies (black arrowheads) and appears darker than white matter. Glial cells are present in the white matter (white arrowheads). (B) Average lifetimes and (C) spectral intensity ratios of tumor (red), cortex (blue) and white matter (green) across the three spectral channels. Each point corresponds to the median of each scan. The mean and standard deviation of each tissue group are indicated by the error bars. *p-value < 0.01; non-significant statistics are left unmarked. N indicates the number of patients; n indicates the number of scans
The fluorescence decays, the average fluorescence lifetime, and the intensity ratio in each spectral channel were extracted by a constrained least-squares deconvolution with the Laguerre expansion method described elsewhere [28]. The medians of the fluorescence lifetimes and intensity ratios of each scan (n) were used as the basic unit of analysis (individual points displayed), unless noted otherwise. The means and standard deviations for each group are reported, and the Wilcoxon Rank Sum test was used to determine statistical differences between groups. Differences were considered significant at p < 0.01 and are indicated by * in the figures. Sensitivity, specificity, and ROC-AUC curves are also reported using lifetimes for distinguishing astrocytoma from oligodendroglioma tumors. These parameters were computed by setting threshold values from 2 to 5 ns, in increments of 0.5 ns, for all three lifetimes (Figure 3C and Table 5).
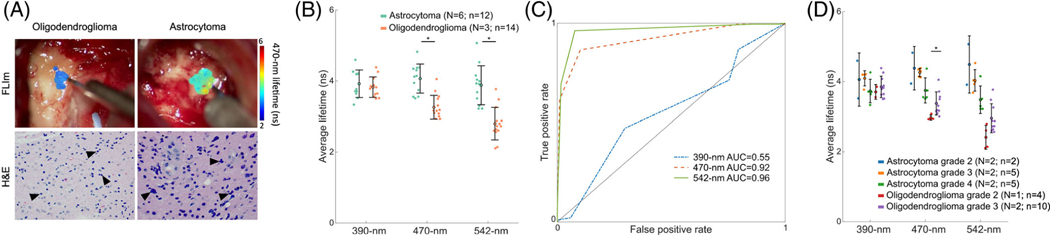
FIGURE 3.
FLIm provides optical contrast between astrocytoma and oligodendroglioma. (A) Surgical field-of-view of representatives of oligodendroglioma (left) and astrocytoma (right) tumors with 470-nm lifetime overlaid in the scanned regions and the corresponding H&E scan (×40 magnification). (B) Average lifetimes of tumor tissue from astrocytoma (teal) and oligodendroglioma (orange) across the three spectral channels. (C) ROC curves computed for the data point-level identification of astrocytoma from oligodendroglioma tumors for the 390-nm, 470-nm, and 542-nm lifetimes. (D) Average lifetimes of tumor tissue from different grades of astrocytoma and oligodendroglioma samples across the three spectral channels. Each point corresponds to the median of each scan. The mean and standard deviation of each tissue group are indicated by the error bars. *p-value <0.01; non-significant statistics are left unmarked. N indicates the number of patients; n indicates the number of scans.
TABLE 5.
Sensitivity, specificity, and AUC statistics for predicting astrocytoma from oligodendroglioma tumor
| Sensitivity | Specificity | AUC | |
|---|---|---|---|
| 390/40 nm | 0.47 | 0.70 | 0.55 |
| 470/28 nm | 0.87 | 0.90 | 0.92 |
| 542/50 nm | 0.96 | 0.92 | 0.96 |
2.5 |. Histopathological and molecular assessment
Tissue biopsies (1–2 mm3) were collected from the scanned regions. Biopsied tissue was analyzed by the neuropathologist (H.S.L. or L.-W.J.) following standard procedures. The tissue type (white matter/cortex) and the tumor cellular density for each sample were assessed from hematoxylin and eosin (H&E) stained tissue slices and confirmed with IDH immunohistochemistry stain. A semi-quantitative scale was used to define four classes of tumor cellular density as absent, low, moderate, and high, according to the following criteria. Absent: the collected tissue from the resection margin showed no evidence of tumor cells. Low: <200 cells per high power field (HPF; 0.5 mm field diameter using a ×40 objective) or tumor nuclei covering <10% of surface area. Moderate: 200–1000 cells per HPF or tumor nuclei covering 10%–25% of surface area. High: >1000 tumor cells per HPF or tumor nuclei covering >25% of surface area. Figure S1 shows representative H&E images and corresponding IDH stains for the different classes of tumor cellular density.
For this study, we designate as “normal” all FLIm data from scans of regions of interest where biopsies showed absent and low tumor cellular density, and as “tumor” all FLIm data from scans where biopsies showed moderate and high tumor cellular density. The evaluation of molecular markers (i.e., IDH1/2 mutation and 1p/19q codeletion mutation) served as the ground truth to validate the IDH-mutation status of the samples and their tumor subtype (astrocytoma or oligodendroglioma), and it was used to label the FLIm scans accordingly.
3 |. RESULTS
3.1 |. Fluorescence lifetimes of IDH-mutant tumors are shorter compared to white matter, but comparable to cortex
To determine if the FLIm parameters can distinguish IDH-mutant tumor from surrounding brain parenchyma (IDH-wildtype), we compared the average lifetimes and intensity ratios of the three tissue types labeled according to the biopsy results (tumor, cortex, and white matter) (Table 3 and Figure 2).
TABLE 3.
Average lifetimes and intensity ratios of the different tissue categories. Gray cells indicate comparisons with statistical significance (p < 0.01).
| Average lifetime (ns) |
Intensity ratios (a.u.) |
|||||
|---|---|---|---|---|---|---|
| Tissue | 390-nm | 470-nm | 542-nm | 390-nm | 470-nm | 542-nm |
| Tumor | 3.83 ± 0.30 | 3.62 ± 0.55 | 3.28 ± 0.73 | 0.09 ± 0.03 | 0.50 ± 0.05 | 0.41 ± 0.06 |
| Cortex | 3.75 ± 0.41 | 3.73 ± 0.45 | 3.45 ± 0.50 | 0.07 ± 0.04 | 0.47 ± 0.07 | 0.45 ± 0.08 |
| White matter | 4.16 ± 0.34 | 4.59 ± 0.39 | 4.25 ± 0.47 | 0.15 ± 0.05 | 0.48 ± 0.04 | 0.37 ± 0.06 |
| P (t-c) | 0.659 | 0.465 | 0.348 | 0.084 | 0.254 | 0.105 |
| P (t-w) | 0.007 | 5.06 ×10−6 | 1.09 ×10−4 | 2.05 ×10−4 | 0.929 | 0.002 |
| P (c-w) | 0.002 | 1.20 ×10−7 | 5.03 ×10−6 | 4.59 ×10−5 | 0.083 | 0.019 |
t – tumor; c – cortex; w – white matter
Figure 2A shows a representative FLIm scan overlaid onto the surgical field-of-view acquired in each tissue type and the corresponding H&E images. The average lifetime values for each tissue type in all three spectral channels are shown in Figure 2B and in Table 3. Tumor tissue, comprising both IDH-mutant oligodendroglioma and IDH-mutant astrocytoma, had shorter lifetime values when compared to white matter in all the channels (390-nm: 3.8 ± 0.3 ns vs. 4.2 ± 0.3 ns, p < 0.01; 470-nm: 3.6 ± 0.6 ns vs. 4.6 ± 0.4 ns, p < 0.01; 542 nm: 3.3 ± 0.7 ns vs. 4.3 ± 0.5 ns, p < 0.01). However, tumor lifetime values were comparable to cortex values (390-nm: 3.8 ± 0.4 ns, p = 0.66; 470-nm: 3.7 ± 0.5 ns, p = 0.47; 542-nm: 3.5 ± 0.5 ns, p = 0.35). Note that white matter samples had longer lifetime values than cortex samples in all three channels. We found that whereas lifetime is not enough to identify tumors infiltrating cortex tissue, there is a clear distinction in lifetime values between IDH-mutant tumors and white matter.
Figure 2C and Table 3 show the intensity ratios from each scanned area. The strongest contribution in intensity was at the 470 nm band for all tissue types, consistent with the predominant fluorescence from NAD(P) H. Despite all tissue types having a similar spectral profile, small differences were captured in the 390-nm channel. In this spectral band, white matter exhibited a relatively higher intensity (0.15 ± 0.05 a.u.) when compared to cortex (0.07 ± 0.04 a.u., p < 0.01) and tumor (0.09 ± 0.03 a.u., p < 0.01). On the metabolic channels (470-nm and 542-nm), the intensity ratio did not provide discrimination among the tissue types.
3.2 |. Fluorescence lifetime values differ between astrocytoma and oligodendroglioma
To determine whether FLIm can provide optical contrast between IDH-mutant astrocytoma and IDH-mutant oligodendroglioma, the tumor scans were analyzed according to their tumor type (Table 4). Figure 3A shows the fluorescence lifetimes in the 470-nm channel overlaid on the surgical field-of-view of representative oligodendroglioma and astrocytoma tumors and their corresponding H&E images. The average lifetimes of tumor tissue from astrocytoma and oligodendroglioma are shown in Figure 3B. Oligodendroglioma tumors had shorter lifetimes in the metabolic channels (470-nm: 3.3 ± 0.1 ns; 542-nm: 2.8 ± 0.2 ns) compared to astrocytoma (470-nm: 4.1 ± 0.1 ns; 542-nm: 3.9 ± 0.2 ns; p < 0.01 for each channel pair). The average lifetimes of the two IDH-mutant tumor types were comparable in the 390-nm band.
TABLE 4.
Average lifetimes of the different tumor types and malignancy grades
| Average lifetime (ns) |
|||||
|---|---|---|---|---|---|
| Tissue | Number of patients | Number of samples | 390-nm | 470-nm | 542-nm |
| Astrocytoma | 6 | 12 | 3.83 ± 0.60 | 4.05 ± 0.14 | 3.86 ± 0.22 |
| Oligodendroglioma | 3 | 14 | 3.82 ± 0.13 | 3.25 ± 0.13 | 2.78 ± 0.24 |
| P (A-O) | 0.93 | 1.57 × 10−4 | 1.92 × 10−4 | ||
| Astrocytoma grade 2 | 2 | 2 | 3.64 ± 0.32 | 4.30 ± 0.68 | 4.38 ± 0.91 |
| Astrocytoma grade 3 | 2 | 5 | 4.10 ± 0.22 | 4.25 ± 0.10 | 4.05 ± 0.31 |
| Astrocytoma grade 4 | 2 | 5 | 3.70 ± 0.32 | 3.75 ± 0.37 | 3.45 ± 0.38 |
| P (A2-A3) | 0.267 | 1.000 | 0.857 | ||
| P (A2-A4) | 1.000 | 0.191 | 0.191 | ||
| P (A3-A4) | 0.064 | 0.151 | 0.056 | ||
| Oligodendroglioma grade 2 | 1 | 4 | 3.69 ± 0.17 | 2.96 ± 0.08 | 2.40 ± 0.33 |
| Oligodendroglioma grade 3 | 2 | 10 | 3.85 ± 0.29 | 3.36 ± 0.34 | 2.93 ± 0.41 |
| P (O2-O3) | 0.606 | 0.004 | 0.054 | ||
Note: Gray cells indicate comparisons with statistical significance (p < 0.01).
Abbreviation: A, astrocytoma; O, oligodendroglioma; A2, astrocytoma grade 2; A3, astrocytoma grade 3; A4, astrocytoma grade 4; O2, oligodendroglioma grade 2; O3, oligodendroglioma grade 3.
The average lifetime values of the IDH-mutant glioma subtypes differ from those of the surrounding infiltrating tissue in different ways (Figure S2). In the plane defined by the 470-nm and 542-nm average lifetimes, oligodendroglioma separated from white matter but completely overlapped with cortex tissue (Figure S2C,D). For IDH-mutant astrocytoma, while the white matter has longer lifetimes on average than the tumor tissue, there is an overlapping region between the two distributions of average lifetime (Figure S2A). Unlike oligodendrogliomas, the lifetime of astrocytoma seemed separated from that of cortex tissue (Figure S2B). However, the astrocytoma tumor points correspond to a patient who underwent a laser interstitial thermal therapy (LITT) procedure before surgery. It is plausible that the high lifetime values observed in this case indicate changes due to LITT.
In this study, tissue specimens without tumor cells and those with low tumor cellular density were assigned as “normal” tissue and specimens with a moderate or high degree of tumor cellular density were assigned as “tumor.” For white matter tissue, the average fluorescence lifetime values in channels 470 nm and 542 nm, and to a minor degree in channel 390 nm, exhibited shorter values in specimens with moderate and high tumor cellularity than in specimens without tumor cells or with low tumor cellular density (Figure S3). But there was no difference in lifetime values between the moderate and high cellular density groups, nor between the absent and low cellular density groups in either glioma subtype. Importantly, despite the interpatient variability in lifetime values in each tumor cellular density group, these trends were observed in all patients for which data from the “normal” and “tumor” groups were available.
3.3 |. Fluorescence lifetime values may provide further contrast between grades of IDH-mutant tumors
Within each IDH-mutant glioma subtype, the mean fluorescence lifetime also differed between grades (Figure 3D and Table 4). With the current limited number of samples (Table 2), the differences between grades in IDH-mutant astrocytoma were non-significant across all channels (p > 0.01). However, oligodendroglioma grade 2 showed shorter lifetimes than grade 3 in the channel centered at 470 nm (2.96 ± 0.08 ns vs. 3.4 ± 0.3, p < 0.01). Fluorescence lifetime values at 390 nm (p = 0.31) and 542 nm (p = 0.05) were comparable between the two grades.
3.4 |. Diagnostic potential of fluorescence lifetimes
The distinct average fluorescence lifetimes of IDH-mutant oligodendroglioma and IDH-mutant astrocytoma in channels 470-nm and 542-nm (Figure 3B) indicate FLIm’s potential to diagnose between the two glioma subtypes. The sensitivity and specificity were computed for a range of thresholds between 2 and 5 ns to generate ROC-AUC curves in each spectral band (Figure 3C and Table 5). While the performance of the 390-nm lifetime was marginal (AUC = 0.55) as expected due to the overlapping lifetime values (Figure 3B), the 470-nm and 542-nm lifetimes provided high diagnostic potential with AUC470-nm = 0.92 and AUC542-nm = 0.96, respectively.
4 |. DISCUSSION
Current results indicate that FLIm-derived parameters (i.e., average fluorescence lifetime) differ between IDH-mutant astrocytoma and oligodendroglioma tumors, underscoring the potential of mesoscopic FLIm as an intraoperative tool for glioma subtype identification. Moreover, FLIm can identify IDH-mutant gliomas from surrounding white matter.
The differences in fluorescence lifetimes between brain parenchyma and the distinct tumor subtypes evaluated in this study are most likely the result of alterations in metabolic activity [29] and microenvironmental conditions (pH, tissue oxygenation) [30] that occur in gliomas. These components affect the optical properties of metabolic cofactors NAD(P)H and FAD, the primary sources of tissue autofluorescence in the brain, which can be detected by the mesoscopic clinical FLIm device without additional contrast agents.
4.1 |. Fluorescence lifetime contrast between white matter and cortex tissue
To address whether FLIm provides optical contrast between tumors and the surrounding brain in IDH-mutant gliomas, we independently compared tumor FLIm parameters with those from white matter and cortex. Cortex and white matter have different biochemical compositions and structural arrangements that result in different fluorescence lifetimes (Figures 2 and S2). This finding is in line with previous studies, which report longer average lifetime values in white matter than in cortex in human tissue [16, 17, 22], and in rat models [15]. The lifetime differences between the tissue types of the brain parenchyma must be considered when discriminating IDH-mutant tumors from surrounding healthy tissue.
4.2 |. FLIm can identify IDH-mutant tumors from white matter, but not from cortex
The current results also confirm that IDH-mutant tumors have significantly lower lifetimes than surrounding white matter, which is consistent with previous time-resolved fluorescence studies [14, 17, 21, 22]. The shorter lifetime values of IDH-mutant tumors in the 470-nm and 542-nm channels compared to surrounding white matter (Figures 2, S2, and S3) can be explained by the reduction of the NAD(P)H and FAD levels in tumor cells due to the combined effect of cancer energetics [31, 32] and IDH mutations [33]. Typically, cancer cells adopt a glycolytic phenotype, even in the presence of oxygen (aerobic glycolysis), as opposed to the oxidative metabolism of nonpathogenic cells (Warburg effect) [31]. Aerobic glycolysis leads to increased free NADH in cells [34, 35], which has shorter lifetimes compared to protein-bound NADH, which is more abundant in non-cancerous cells [30, 36]. Moreover, the Warburg effect indicates that elevated levels of glucose metabolism lead to acidification of the tumor microenvironment [31], which is another factor known to affect the fluorescence lifetime [37]. In addition to changes in energetic metabolism, IDH mutations lead to reduced NADPH levels in the tumor cells, which also result in a decreased lifetime [33, 35]. NADH and NADPH have overlapping spectral profiles, with an emission peak at 470 nm [30], and are thus indistinguishable through the FLIm approach used in this study. FAD is another key coenzyme involved in redox reactions [32, 38] and has correlated alterations with NADH in the energetic metabolism [32]. These metabolic changes are expected to contribute to the observed lifetime variations. In the 390-nm channel, there is a considerable amount of signal, possibly from GAD and PMP or remnants of collagen in the brain parenchyma [14], even though the intensity collected in this band is the lowest of the three channels (Figure 2C). The lifetime values at 390-nm are relatively constant (~4 ns) across tissue types, indicating that environmental factors or changes in their metabolism are not strong enough to alter their lifetime or that the current approach is not sensitive enough to detect it.
Tumor tissue, and particularly IDH-mutant oligodendroglioma, however, had comparable fluorescence lifetime values to cortex (Figure S2D), consistent with previous studies [15, 17, 22]. The differences in cellular and extracellular environment makeup between the cortex and white matter likely contribute to their differing baseline in lifetime values that result in a lack of contrast between tumor tissue and cortex.
IDH-mutant tumor and cortex had smaller intensity ratios than white matter in the 390-nm band. The mean intensity ratios were comparable between all three tissue types in the rest of the spectral channels (Figure 2C). Spectral shifts were not prominent between the three tissue types, indicating that intensity ratios do not contribute to identifying IDH-mutant tumors from normal brain tissue as much as lifetimes do.
4.3 |. Fluorescence lifetime decreases with increasing tumor cellular density
Despite a non-negligible degree of interpatient variability, for each patient, the lifetime values of tumor-infiltrating white matter tissue decreased with increasing tumor cellular density (Figure S3). Notably, distinct lifetime values were observed between specimens of the low and moderate tumor cellular density regions within each IDH-mutant oligodendroglioma. The same pattern was observed in IDH-mutant astrocytoma, albeit less pronounced. These findings may indicate that the current sensitivity threshold of FLIm for detecting IDH-mutant tumors lies between low and moderate tumor cellular density. It is worth noting that specimens with high tumor cellular density in our dataset belong to IDH-mutant oligodendroglioma. More data are necessary to evaluate the influence of tumor cellular density on the tissue lifetime.
4.4 |. Optical contrast between two types of IDH-mutant brain tumors: Astrocytoma and oligodendroglioma
FLIm shows promising results in differentiating oligodendroglioma from astrocytoma (Figure 3). The histologic features and molecular mechanisms of gliomagenesis are different between these tumor subtypes [2]. On histopathology, oligodendrogliomas typically show higher tumor cellular density than grade-matched astrocytomas. The higher proportion of IDH-mutant tumor cells with altered NAD(P)H and FAD pathways could elicit higher lifetime variations in oligodendrogliomas over grade-matched astrocytomas. Also contributing to changes in fluorescence lifetime are the different expression levels of glycolysis-related proteins and metabolite yields between IDH-mutant astrocytoma and oligodendroglioma. For example, IDH-mutant astrocytomas have an increased lactate production that leads to a more acidic microenvironment [39]. In fact, tumor acidity has been correlated with tumor malignancy [40]. The fluorescence lifetime is sensitive to the pH of the fluorophore microenvironment [41], hence differences in pH between the tumor types are also expected to contribute to the overall fluorescence response.
A difference in average lifetime was also found in the 470-nm and 542-nm channels between oligodendroglioma grade 2 and oligodendroglioma grade 3 (Figure 3D), albeit with a more limited sample size. Such differences in lifetime values were not observed among IDH-mutant astrocytoma grades. A recent study comparing the molecular landscape of IDH-mutant astrocytoma and oligodendroglioma grade 2 showed that oligodendroglioma are more homogenous and have increased tumor purity than astrocytoma [42]. Homogeneity and increased tumor purity, which is the ratio of tumor cells to non-tumor cells (i.e., stromal cells and immune cells) [43], may be associated with significant differences observed in the optical properties of low-grade (grade 2) vs. high-grade (grade 3) oligodendroglioma. The heterogeneity in the tumor cells could lead to non-significant differences in lifetimes in astrocytoma grades.
The ROC curves depicted in Figure 3C show promising results toward building classification algorithms with high accuracies to differentiate IDH-mutant astrocytomas from IDH-mutant oligodendrogliomas in real-time with FLIm. With an expanded dataset, improved performance could be expected for detecting the malignancy grade and the glioma subtype with high precision.
Overall, the results of this study suggest that FLIm can be used to differentiate between IDH-mutant oligodendrogliomas and astrocytomas in real-time during surgical resection. An available diagnosis during the surgical procedure may have a role in optimizing the extent of surgical resection for each glioma subtype. Such application of this technology has real implications for patient outcome, based on the known impact of extent of resection on survival. Increased extent of resection has been shown to improve overall survival for low and high-grade astrocytomas [44–46]. In contrast, oligodendrogliomas, which have increased sensitivity to adjuvant radiation and chemotherapy given routinely after surgery, are not as dependent on the surgical resection to achieve maximal overall survival [4, 47]. As the extent of resection is increased, the risk for neurologic injury and significant impact on quality of life increases as well. This suggests that for oligodendrogliomas, where greater resection does not necessarily improve survival, surgery should be less aggressive. Currently, the distinction between oligodendrogliomas and astrocytomas cannot be made intraoperatively, requiring surgeons to be maximally aggressive as a default. Using FLIm for real-time tumor subtype identification could better guide surgeons and modify the goal of the surgical procedure accordingly.
4.5 |. Challenges and future directions
While FLIm contrast was observed between IDH-mutant tumors and surrounding brain parenchyma (i.e., white matter) and between IDH-mutant astrocytoma and oligodendroglioma tumors, the number of patients recruited for this study was small. To implement intraoperative diagnosis of IDH-mutant glioma subtype using FLIm, a larger dataset needs to be collected and analyzed from a range of IDH-mutant glioma subtypes and grades. With enough data, classification algorithms could be developed for automated prediction of tumor diagnostics based on intraoperative FLIm data in real-time. Nevertheless, the current study shows, for the first time to our knowledge, that FLIm has the potential to identify IDH-mutant tumor subtypes (astrocytoma vs. oligodendroglioma) intraoperatively.
Of note, one of the oligodendroglioma grade 3 patients recruited for the study had been administered 5-aminolevulinic acid (5-ALA) before surgery. 5-ALA fluorescence in low-grade patients is weak [48–50] and we did not see any significant differences in the fluorescence data singularly attributed to 5-ALA administration in the spectral channels used for this study. Another patient (astrocytoma grade 2) underwent a LITT procedure for tumor ablation prior to the open craniotomy for tumor resection. In this case, the average lifetimes were relatively higher in the tumor tissue compared to all other patients, which could be a direct effect of the thermal ablation of the tissue (Figures 3B and S2B).
Another challenge lies in verifying the FLIm-scanned region with biopsied tissue. The scans were collected from a localized region that the neurosurgeon confirmed was representative of the biopsy collected. However, the FLIm scans (~10 mm2) may cover a portion larger than the biopsy (1–2 mm3). Future analysis will address this challenge by precisely identifying the scan region that was biopsied.
5 |. CONCLUSION
This study examines the ability of FLIm to provide endogenous contrast between IDH-mutant tumor types and surrounding normal tissue intraoperatively. IDH-mutant tumor tissue had shorter lifetime values compared to surrounding white matter. Within the IDH-mutant tumors, oligodendrogliomas had shorter lifetime values than astrocytoma. Also, lifetimes in the metabolic channels provided optical contrast between different grades of oligodendroglioma. Together, these results support the feasibility of FLIm as an intraoperative tool for real-time surgical guidance, and the potential to identify IDH-mutant glioma subtype in situ. The successful implementation of such a surgical guidance tool in the operating room could lead to improving treatment for glioma patients.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (Grant No. R21CA252510, R01CA250512) to L.M. and O.B. and the University of California, Davis Comprehensive Cancer Center’s Brain Malignancies Innovation Group funded through the Comprehensive Cancer Center Support Grant (P30CA093373). The authors would like to thank Lisanne Kraft, Roberto P. Frusciante, and Sukhkaran Singh Aulakh for their assistance in data collection. The authors would also like to acknowledge the use of Biorender and gramm library [51] in MATLAB for generating the figures.
Funding information
National Institutes of Health, Grant/Award Numbers: R01CA250512, R21CA252510; University of California, Davis, Comprehensive Cancer Center, Grant/Award Number: P30CA093373
Abbreviations:
- APD
avalanche photodetector
- CNS
central nervous system
- FAD
flavin adenine dinucleotide
- FISH
fluorescence in-situ hybridization
- FLIm
fluorescence lifetime imaging
- GAD
glutamate decarboxylase
- H&E
hematoxylin and eosin
- HPF
high power field
- IDH
isocitrate dehydrogenase
- IRB
institutional review board
- LITT
laser interstitial thermal therapy
- MPE
maximum permissible exposure
- NAD(P)H
nicotinamide adenine dinucleotide (phosphate)
- OR
operating room
- PLP
pyridoxal phosphate
- PMP
pyridoxamine phosphate
- WHO
world health organization
Footnotes
CONFLICT OF INTEREST
The authors declare no financial or commercial conflict of interest.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- [1].Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW, Neuro Oncol 2021, 23, 1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].van den Bent MJ, Smits M, Kros JM, Chang SM, J Clin Oncol 2017, 35, 2394. [DOI] [PubMed] [Google Scholar]
- [3].Choi J, Kim SH, Ahn SS, Choi HJ, Yoon HI, Cho JH, Roh TH, Kang S, Chang JH, Suh C, Sci. Rep. 2020, 10, 2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patel SH, Bansal AG, Young EB, Batchala PP, Patrie JT, Lopes MB, Jain R, Fadul CE, Schiff D, Am J Neuroradiol 2019, 40, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wesseling P, van den Bent M, Perry A, Acta Neuropathol 2015, 129, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Craig Tork A, Atkinson C, StatPearls 2022. [Online]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK559184/?report=printable. [Google Scholar]
- [7].Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, Gilbert MR, Yang C, Br J Cancer 2020, 122, 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abdullah A, el Shitany H, Abbass W, Safwat A, Elsamman AK, el Refaee E, J Neurosci Rural Pract 2016, 7, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Singh G, Manjila S, Sakla N, True A, Wardeh AH, Beig N, Vaysberg A, Matthews J, Prasanna P, Spektor V, Br. J. Cancer 2021, 125, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tofte K, Berger C, Torp SH, Solheim O, Surg Neurol Int 2014, 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ogrinc N, Saudemont P, Takats Z, Salzet M, Fournier I, Trends Mol Med 2021, 27, 602. [DOI] [PubMed] [Google Scholar]
- [12].Moreira Franco YE, Alves MJ, Uno M, Moretti IF, Trombetta-Lima M, de Siqueira Santos S, dos Santos AF, Arini GS, Baptista MS, Lerario AM, Oba-Shinjo SM, Marie SKN, Cancer Metab 2021, 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Z, Liu F, Fan N, Zhou C, Li D, Macvicar T, Dong Q, Bruns CJ, Zhao Y, Front. Oncol. 2020, 10, 589508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Butte PV, Fang Q, Jo JA, Yong WH, Pikul BK, Black KL, Marcu L, J Biomed Opt 2010, 15, 027008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lukina M, Yashin K, Kiseleva EE, Alekseeva A, Dudenkova V, Zagaynova EV, Bederina E, Medyanic I, Becker W, Mishra D, Berezin M, Shcheslavskiy VI, Shirmanova M, Front. Oncol. 2021, 11, 666059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marcu L, Jo JA, Butte PV, Yong WH, Pikul BK, Black KL, Thompson RC, Photochem. Photobiol. 2004, 80, 3. [DOI] [PubMed] [Google Scholar]
- [17].Yong WH, Butte PV, Pikul BK, Jo JA, Fang Q, Papaioannou T, Black KL, Marcu L, Front Biosci 2006, 11, 1199-, pp. 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Butte PV, Pikul BK, Hever A, Yong WH, Black KL, Marcu L, J Biomed Opt 2005, 10, 064026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zanello M, Poulon F, Pallud J, Varlet P, Hamzeh H, Lahoud GA, Andreiuolo F, Ibrahim A, Pages M, Chretien F, Rocco FD, Dezamis E, Nataf F, Turak B, Devaux B, Haidar DA, Sci. Rep. 2017, 7, 41724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun Y, Hatami N, Yee M, Phipps J, Elson DS, Gorin F, Schrot RJ, Marcu L, J Biomed Opt 2010, 15, 056022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Butte PV, Mamelak AN, Nuno M, Bannykh SI, Black KL, Marcu L, Neuroimage 2011, 54, S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Alfonso-Garcia A, Bec J, Weaver SS, Hartl B, Unger J, Bobinski M, Lechpammer M, Girgis F, Boggan J, Marcu L, J Biophotonics 2019, 13, e201900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kantelhardt SR, Kalasauskas D, Konig K, Kim E, Weinigel M, Uchugonova A, Giese A, J Neurooncol 2016, 127, 473. [DOI] [PubMed] [Google Scholar]
- [24].Yankelevich DR, Ma D, Liu J, Sun Y, Sun Y, Bec J, Elson DS, Marcu L, Rev. Sci. Instrum. 2014, 85, 034303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou X, Bec J, Yankelevich D, Marcu L, Opt. Express 2021, 29, 20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ma D, Bec J, Gorpas D, Yankelevich D, Marcu L, Biomed Opt Express 2015, 6, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].“ANSI Z136.1 Safe Use of Lasers,” 2014. [Google Scholar]
- [28].Liu J, Sun Y, Qi J, Marcu L, Phys Med Biol 2012, 57, 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Alfonso-Garcia A, Bec J, Weyers BW, Marsden M, Zhou X, Lai C, Marcu L, J Biophoton 2021, 14, e202000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Datta R, Heaster TM, Sharick JT, Gillette AA, Skala MC, J Biomed Opt 2020, 25, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liberti MV, Locasale JW, Trends Biochem Sci 2016, 41, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Croce AC, Bottiroli G, Eur J Histochem 2014, 58, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huang J, Yu J, Tu L, Huang N, Li H, Luo Y, Front Oncol 2019, 9, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kennedy BE, Sharifa T, Martella E, Daia C, Kimb Y, Lee PWK, Gujar SA, Pharmacol Res 2016, 114, 274. [DOI] [PubMed] [Google Scholar]
- [35].Ranjit S, Malacrida L, Stakic M, Gratton E, J Biophotonics 2019, 12, e201900156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schroeder AB, Pointer KB, Clark PA, Datta R, Kuo JS, Eliceiri KW, J Biomed Opt 2020, 25, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Feiner-Gracia N, Pujals S, Delcanale P, Albertazzi L, Smart Nanoparticles for Biomedicine 2018, p. 219. Amsterdam, Netherlands: Elsevier. 10.1016/B978-0-12-814156-4.00015-X [DOI] [Google Scholar]
- [38].Georgakoudi I, Quinn KP, Annu. Rev. Biomed. Eng. 2012, 14, 351. [DOI] [PubMed] [Google Scholar]
- [39].Yao J, Hagiwara A, Raymond C, Shabani S, Pope WB, Salamon N, Lai A, Ji M, Nghiemphu PL, Liau LM, Cloughesy TF, Ellingson BM, Sci. Rep. 2020, 10, 11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Corbet C, Feron O, Nat Rev Cancer 2017, 17, 577. [DOI] [PubMed] [Google Scholar]
- [41].Schmitz R, Tweed K, Walsh C, Walsh AJ, Skala MC, J Biomed Opt 2021, 26, 056502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhao B, Xia Y, Yang F, Wang Y, Wang Y, Wang Y, Dai C, Wang Y, Ma W, Mol Med 2022, 28, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang C, Cheng W, Ren X, Wang Z, Liu X, Li G, Han S, Jiang T, Wu A, Clin Cancer Res 2017, 23, 6279. [DOI] [PubMed] [Google Scholar]
- [44].Sanai N, Polley M-Y, McDermott MW, Parsa AT, Berger MS, J Neurosurg 2011, 115, 3. [DOI] [PubMed] [Google Scholar]
- [45].Aghi MK, Nahed BV, Sloan AE, Ryken TC, Kalkanis SN, Olson JJ, J Neurooncol 2015, 125, 503. [DOI] [PubMed] [Google Scholar]
- [46].Kavouridis VK, Boarro A, Dorr J, Cho EY, Iorgulescu JB, Reardon DA, Arnaout O, Smith TR, J Neurosurg 2019, 133, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Alattar AA, Brandel MG, Hirshman BR, Dong X, Carroll KT, Ali MA, Carter BS, Chen CC, J Neurosurg 2018, 128, 1076. [DOI] [PubMed] [Google Scholar]
- [48].Kiesel B, Freund J, Reichert D, Wadiura L, Erkkilae MT, Woehrer A, Hervey-Jumper S, Berger MS, Widhalm G, Front Oncol 2021, 11, 699301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Díez Valle R, Hadjipanayis CG, Stummer W, J Neuro-Oncol 2019, 141, 487. [DOI] [PubMed] [Google Scholar]
- [50].Suero Molina E, Black D, Kaneko S, Müther M, Stummer W, J Neurosurg 2022, 137, 1. [DOI] [PubMed] [Google Scholar]
- [51].Morel J Open Source Softw 2018, 3, 568. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.