Abstract
Objective
The global burden of diabetes mellitus will impact strongly American countries in the coming decades. Type 2 diabetes mellitus (T2DM) is a multifactorial disease and the basis for its genetic susceptibility remains not fully understood. Different population studies have demonstrated that variants of the TCF7L2 gene are strongly associated with an increased risk of T2DM. Moreover, institutions or countries with limited budget to conduct genetic research need cost effective methods for detecting DNA variants.
Subjects and methods
We standardized a rapid and simple allele-specific PCR method for genotyping the rs12255372 single nucleotide polymorphism (SNP) in a pilot study exploring the association of three TCF7L2 polymorphisms (rs7903146, rs12255372 and DG10S478) with T2DM in 70 patients and 73 controls from Venezuela.
Results
The performance of the designed allele-specific PCR reaction for rs12255372 genotyping was reliable and accurate. Patients carrying the TCF7L2 rs7903146 T allele (CT + TT genotypes) and heterozygous CT genotype had a significantly higher risk for T2DM (OR = 2.9 and 2.3, respectively). Although rs12255372 and DG10S478 risk alleles predominated in T2DM group no statistical significance was found.
Conclusions
We developed a novel allele-specific PCR method for easier and rapid detection of rs12255372 polymorphism without the use of expensive instrumentation and reagents. Our study in a relatively small sample of the Venezuelan population replicated the association of the rs7903146 SNP with T2DM. Further studies with larger sample size and more biochemical data should be conducted to explore the genetic basis of T2DM susceptibility in Venezuela.
Keywords: Type 2 diabetes mellitus, TCF7L2, allele-specific PCR, gene polymorphism, Venezuela
INTRODUCTION
Diabetes mellitus is an endocrine disorder characterized by dysregulation of insulin production, insulin sensitivity, and glucose control. Diabetes is generally classified in type 1 diabetes mellitus (T1DM), which consists of an autoimmune etiology, and T2DM, which includes a heterogeneous group of metabolic disorders characterized by the inability of pancreatic beta cells to increase insulin secretion in order to compensate for insulin resistance (1). T2DM patients are typically diagnosed in adulthood and usually after the age of 40. T2DM is the most common form of diabetes, accounting for 90% of cases in the world (1). The burden of T2DM is growing worldwide with near 80% of patients living in low- and middle-income countries (2). By 2035 the number of people with diabetes in South and Central America will increase by 60% (2). Moreover, in Venezuela, T2DM was ranked as the fifth leading cause of mortality in 2011 (3).
The development of T2DM results from interaction between environmental factors and a robust genetic component (4). These environmental factors, including obesity, sedentary lifestyle, small or large birth weight and stress (5), do not influence everyone in the same way, which highlights the role that genetic susceptibility plays in the etiology and manifestation of T2DM. Multiple T2DM risk genes were initially identified using candidate gene and linkage-based studies, and more recently by genome-wide association studies (GWAS) (reviewed at 6,7).
TCF7L2 (transcription factor 7-like 2) is among the highly replicated genes showing robust association with T2DM (8,9). The TCF7L2 gene spans 215.9 kb on chromosome 10q25.3, and its product is a high-mobility box-containing transcription factor involved in the Wnt signaling pathway (10). Wnt pathway activity plays an important role for lipid and glucose metabolism, pancreatic islets proliferation, regeneration and function, and for production of the insulinotropic hormone glucagon-like peptide 1 (GLP-1) (11-13). Therefore, alteration in this pathway could cause the reduction of GLP-1 secretion, then affecting insulin secretion, and subsequently, among other effects, impairing blood glucose homeostasis.
After the initial report on the association of a common microsatellite within intron 3 of TCF7L2 (DG10S478) with T2DM (14), successive replication studies conducted in populations of different ethnic origin confirmed the strong association between a number of TCF7L2 polymorphisms, especially rs7903146 and rs12255372, and T2DM (15-22). Moreover, among Latin American countries there are only reports on the association of TCF7L2 polymorphisms and T2DM from Colombia (23), Brazil (24-28) and Mexico (9,29-32). Latin American populations are characterized by high socioeconomic and genetic heterogeneity with varying degrees of ancestral population proportions from country to country, thus more studies are needed exploring the basis of T2DM susceptibility from these mixed populations.
Most commonly used methods for detecting DNA polymorphisms are restriction enzyme digestion based. However, they generally require 2 days to complete entire procedure. Sequencing or real-time PCR systems are the alternative, but they are costly. Therefore, we used PCR assays for rapid and simple genotyping in a pilot study to explore the role of TCF7L2 gene polymorphisms (rs7903146, rs12255372 and DG10S478) in conferring susceptibility to T2DM in the Venezuelan population. For this purpose, we designed an allelic-specific PCR reaction for genotyping the rs12255372 (G/T) SNP, thus providing a simple, reproducible and low-cost assay.
SUBJECTS AND METHODS
Subjects
The patient population consists of 70 subjects with T2DM (31 men and 39 women), while the control group included 73 non-diabetic subjects (25 men and 48 women) randomly selected from the general population. We collected all blood samples at the Hospital Antonio María Pineda (HAMP), Barquisimeto, Venezuela.
Patient group consisted of subjects with diagnosis of Diabetes (defined according to the American Diabetes Association criteria) made at least two years before entering the study, who were on drug treatment for the disease. Subjects who met inclusion criteria were enrolled into the control group: age at examination over 40 years, absence of family history of diabetes in the previous two generations and fasting plasma glucose of less than 110 mg/dL (6.1 mmol/l). Anthropometric measurements including weight, height, and waist circumference (WC) were obtained using standardized techniques. The body mass index (BMI) was calculated as the weight in kilograms divided by the square of height in meters (kg/m2).
The study was approved by the Bioethics Committee of the School of Health Sciences, Universidad Centroccidental Lisandro Alvarado (UCLA), and performed in accordance with the Declaration of Helsinki. Informed consent was obtained from each patient before participation in the study.
Genotyping
Peripheral blood samples of patient and control groups were collected in EDTA-anticoagulated tubes, and then DNA was extracted using Wizard® Genomic DNA Purification kit (Promega). DNA concentration was checked by UV absorption at 260 and 280 nm. All amplification reactions were carried out on a GeneAmp® PCR System 9700 (Applied Biosystems) thermo cycler. Specific PCR reactions were conducted in a final volume of 25 µl containing 1X Green GoTaq® Flexi Buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U of GoTaq DNA Polymerase (Promega) and 20-50 ng of genomic DNA.
Primers used in PCR reactions carried out in this work are shown in table 1. We designed an allele-specific PCR reaction for genotyping the rs12255372 SNP. For this reaction we designed two doubly mismatched reverse primers with mismatches in their last 3’ nucleotide and at the third nucleotide from the 3’ end such that allow to specifically amplify one of the two variants (G/T) of the rs12255372 polymorphism. Briefly, for each sample, two PCR reactions were run in parallel, combining a common forward primer (CFw) and one of the two reverse primers in order to detect the presence of the polymorphic base (Rev_G and Rev_T). The observation of a 160-bp band in PCR reaction G or T indicated the presence of the respective allele. PCR reactions contained 0.3 µM of each primer and the cycling conditions were as follows: initial denaturation for 3 min at 95 °C; followed by 35 cycles of 20 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C; then a final extension for 5 min at 72 °C. PCR products were electrophoresed on 3% agarose gels.
Table 1. Primers used in this work for TCF7L2 polymorphisms genotyping.
| Polimorfism | Primer Sequence (5’ → 3’) | Detection | Origin |
|---|---|---|---|
| rs7903146 C/T | rs7903146 C: GAACAATTAGAGAGCTAAGCACTTTTTAGAAAC | Allele-specific PCR | Dutra and cols. (33) |
| rs7903146 T: GAACAATTAGAGAGCTAAGCACTTTTTAGAGAT | |||
| rs7903146 R: AGATGAAATGTAGCAGTGAAGTGC | |||
| rs12255372 G/T | CFw: CTGGAAACTAAGGCGTGAGGGA | Allele-specific PCR | This work |
| Rev_G: CAGAGGCCTGAGTAATTATCAGAATATGATC | |||
| Rev_T: CAGAGGCCTGAGTAATTATCAGAATATGCTA | |||
| DG10S478 STR | Fw: TTCAGGCCATTGGTGTTGTA | PCR | Grant and cols. (14) |
| Rev: AAAGTTCCCACCATCCCACT |
Bold underlined bases indicate a mismatch for increasing PCR specificity. Nucleotides at 3’ end (indicated as bold cursive case) are specific for each one of the rs12255372 alleles (G/T).
We conducted the rs7903146 genotype assays by an allele-specific PCR reaction using primers and cycling conditions previously reported by Dutra and cols. (33). We used primers described by Grant and cols. (14) at an annealing temperature of 62 °C to amplify microsatellite DG10S478, then the PCR products were separated by electrophoresis on a 3% w/v agarose gel for allele detection using the nomenclature proposed by these authors: allele 0 (384-bp) is the smallest and all the other are included in a composite allele which is referred to as allele X.
We used the Kodak 1D Molecular Imaging Software (Eastman Kodak, Rochester, NY) for image analysis of ethidium bromide-stained agarose gels and for allelic assignment of the PCR products and restrictions fragments.
Statistical analysis
Genotype frequencies were examined for deviations from their corresponding Hardy-Weinberg equilibrium using Arlequin ver. 2.000 software. SPSS software, version 14.0, was used for the rest of statistical analyses. Mean age of cases and controls, fasting plasma glucose, BMI and WC were compared by t test. The significance of the differences in the distribution of the polymorphisms was calculated using the χ2 or Fisher’s exact test. Association between genotypes and T2D was tested by logistic regression. A P value of < 0.05 was considered to be significant.
RESULTS
Table 2 shows clinical and biochemical characteristics of the study subjects. Comparison between groups showed that the fasting plasma glucose levels were significantly higher in the diabetic subjects, while mean age, BMI and WC were similar in both groups (P > 0.05). The genotypic distribution in both the control and the T2DM groups was in Hardy-Weinberg equilibrium.
Table 2. Characteristics of the normal and diabetic study subjects.
| Controls n = 73 | T2D cases n = 70 | P value | |
|---|---|---|---|
| Age (years) | 50.8 ± 8.5.5 | 53.9 ± 10.5 | 0.055 |
| WC (cm) | 91.8 ± 12.6 | 91.9 ± 10.9 | 0.740 |
| BMI (kg/m2) | 27.0 ± 4.9 | 26.8 ± 4.8 | 0.867 |
| Fasting plasma glucose (mg/dL) | 97.0 ± 7.7 | 179.7 ± 87.1 | 0.000 |
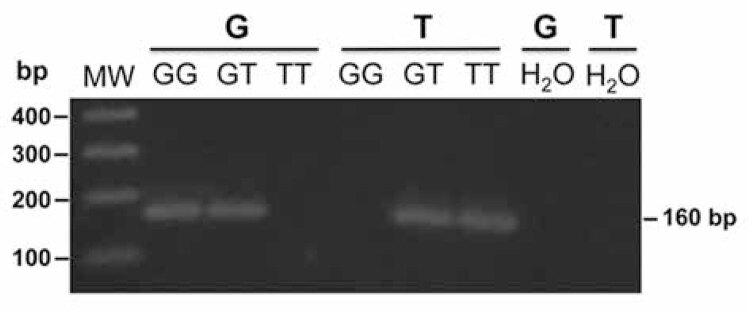
We were able to genotype all the samples included in the study with the performed PCR reactions. We used the PCR-RFLP protocol described by Bodhini and cols. (18) to demonstrate the accuracy of our allele-specific PCR reaction for the genotype discrimination of rs12255372 polymorphism. As expected, the combination of the two reverse primers with a single forward primer in separated reactions, allowed the specific amplification of each one of the alleles (G/T) of the rs12255372 polymorphism (Figure 1).
Figure 1. Genotyping of the TCF7L2 rs12255372 (G/T) polymorphism by an allele-specific PCR assay. For each sample, two PCR reactions were performed, each including a common forward primer and rs12255372 G (reaction G) or rs12255372 T (reaction T) reverse primer, according to the allele to be amplified. A 160-bp DNA fragment indicated the presence of the allele, while amplification failure indicated the absence of the allele in the sample. In a 3% w/v agarose gel, MW indicates molecular size standard, G and T indicate the product from reaction G and reaction T, respectively, amplified from selected DNA sample of each possible genotypes (GG, GT and TT) and H2O (negative control).
Table 3 shows allele and genotype frequencies of the TCF7L2 polymorphisms. The presence of the TCF7L2 rs7903146 T allele (CT + TT genotypes) and heterozygous CT genotype was associated with higher diabetes risk (OR = 2.9 and 2.3, respectively). The distribution of rs12255372 and DG10S478 polymorphisms showed no significant difference between the two groups, despite the homozygote genotypes and the carriers for the mutant alleles (T for rs12255372 and X for DG10S478) predominated in T2DM patients (Table 3). None of the polymorphisms showed significant correlation with fasting plasma glucose, BMI or WC in either the control or the diabetic subjects. Moreover, haplotypes constructed using the three polymorphisms were not significantly associated with T2DM risk.
Table 3. Genotype distribution of the TCF7L2 rs7903146, rs12255372 and DG10S478 polymorphisms in controls and T2DM patients from Venezuela.
| Marker | Controls n = 73 |
T2D cases n = 70 |
P value | OR (%95 CI) | |||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | ||||
| rs7903146 | |||||||
| CC | 46 | (63.0) | 26 | (37.1) | 0.002 | 0.3 (0.18–0.68) | |
| CT | 22 | (30.1) | 35 | (50.0) | 0.015 | 2.3 (1.17–4.60) | |
| TT | 5 | (6.8) | 9 | (12.9) | 0.227 | 2.0 (0.64–6.32) | |
| TT + CT | 27 | (37.0) | 44 | (62.9) | 0.002 | 2.9 (1.46–5.69) | |
| rs12255372 | |||||||
| GG | 46 | (63.0) | 33 | (47.1) | 0.056 | 0.5 (0.27–1.02) | |
| GT | 24 | (32.9) | 29 | (41.4) | 0.290 | 1.4 (0.73–2.86) | |
| TT | 3 | (4.1) | 8 | (11.4) | 0.101 | 3.0 (0.77–11.85) | |
| TT + GT | 27 | (37.0) | 37 | (52.9) | 0.056 | 1.9 (0.98–3.73) | |
| DG10S478 | |||||||
| 0/0 | 47 | (64.4) | 39 | (55.7) | 0.290 | 0.7 (0.34–1.36) | |
| 0/X | 23 | (31.5) | 24 | (34.3) | 0.724 | 1.1 (0.56–2.28) | |
| X/X | 3 | (4.2) | 7 | (10.0) | 0.174 | 2.6 (0.63–10.31) | |
| X/X + 0/X | 26 | (35.6) | 31 | (44.3) | 0.290 | 1.4 (0.73–2.81) | |
DISCUSSION
According to the Global Burden of Disease report, the prevalence of T2DM will have an increasing and the highest impact on the health systems and society in Latin America countries (34). Moreover, years of potential life lost rates due to diabetes among adults are more concentrated in Oceania, Latin America, North Africa, and the Middle East (35). Therefore, among other efforts necessary for the prevention and treatment of diabetes in Latin American countries, identifying of specific genetic markers and/or validates those examined in multiple ethnicities for better prediction of T2DM risk for our mixed populations is essential.
Grant and cols. (14), when reported the association of the STR DG10S478 of TCF7L2 with T2DM in white patients from several populations, found that the rs12255372 and rs7903146 SNPs were in strong linkage disequilibrium with the microsatellite and showing similarly robust associations with disease. These and other authors have recommended that these two SNPs should be genotyped in all attempts at replication (14,36). Moreover, numerous studies and meta-analyses examining various ethnic groups demonstrated an association between TCF7L2 SNPs and T2DM and, although with some inconsistent results, the two most highly correlated are rs7903146 and rs12255372 (36-38).
Herein, we observed that subjects harboring CT genotype and carriers of the allele T of TCF7L2 rs7903146 SNP had a higher T2DM risk (OR = 2.9 and 2.3, respectively). Similarly, this polymorphism was found associated with diabetes in several works carried out in admixed populations from other Latin American countries (9,23-25,28,29,31,32). However, Barros and cols. (27) failed to find an association of this variant with T2DM in northeastern populations from Brazil; while, Parra and cols. (30) also reported no association between the rs7903146 SNPs and T2DM in a replicate case-control study conducted in Mexico City.
In our study, although the frequency of known risk alleles of rs12255372 and DG10S478 polymorphisms was higher in T2DM patients, the association with disease was not strong. Similarly, Furgeri and cols. (26) and Barros and cols. (27) did not find association of the TCF7L2 rs12255372 SNP with T2DM in Brazilian populations. Moreover, Franco and cols. (39) found that TCF7L2 rs7903146 and rs12255372 variants did not make major contributions to the incidence of glucose tolerance abnormalities in Japanese-Brazilians, a population with a high prevalence of T2DM. Finally, recent large meta-analyses confirmed that the rs12255372 SNP is significantly associated with susceptibility to T2DM in the global population, although in some ethnic populations, no significant associations were found (36,38,40).
Although the precise role of TCF7L2 with regard to T2DM risk is still under investigation, evidence suggests that TCF7L2 gene variants may affect the susceptibility to T2DM by indirectly altering GLP-1 levels, whose gene is transcriptionally regulated by this transcription factor (11). T allele of the rs7903146 SNP is associated with enhanced expression of TCF7L2 in human islets, as well as with impaired insulin secretion, incretin effects, and enhanced rate of hepatic glucose production (41,42). Moreover, Gaulton and cols. (43) suggested that the rs7903146 SNP affects T2DM susceptibility by altering cis regulation and local chromatin structure in islet cells.
Genetic analysis in developing countries can often be limited by lack of funding and expertise. Therefore, we also standardized a reliable and time-efficient allele-specific PCR method for genotyping the rs12255372 SNP. Although, a typical PCR-RFLP assay is comparable in cost, it requires additional steps of incubation and electrophoresis. Moreover, this method does not require sequence analysis of PCR products or the costly and time consuming high-tech genotyping protocols used in previous TCF7L2 association studies. Finally, we consider necessary to recommend for the allele-specific PCR assays used for genotyping the rs12255372 (this work) and rs7903146 (33) SNPs, that users must optimize the PCR conditions, especially template starting amount and the number of cycles.
In summary, the designed method will enable researchers to carry out genetic polymorphism studies for rs12255372 risk factor associated with diverse diseases without the use of expensive instrumentation and reagents. Our results replicated the association of the rs7903146 polymorphism of TCF7L2 gene with T2DM in the Venezuelan population. Although the genotypic and allelic frequencies for the TCF7L2 rs12255372 and DG10S478 polymorphisms did not differ significantly between the patients and controls, the predominance of genotypes carrying mutant alleles in T2DM subjects cannot be dismissed. Further studies with larger sample size and biochemical data for glucose tolerance and measurements on insulin secretion should be conducted in Venezuelan population to confirm the putative role of this gene in insulin secretion.
Acknowledgments
Research supported by CDCHT-UCLA grant N° 001-CS-2013.
REFERENCES
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365(9467):1333-46. [DOI] [PubMed]
- 2.International Diabetes Federation (IDF). Diabetes Atlas. 6th ed. Brussels, Belgium: IDF; 2013. Available from: <http://www.eatlas.idf.org>.
- 3.Ministerio del Poder popular para la Salud. Anuario de mortalidad. 2011. Available from: <http://www.mpps.gob.ve/>.
- 4.Lange EM, Sun J, Lange LA, Zheng SL, Duggan D, Carpten JD, et al. Family-based samples can play an important role in genetic association studies. Cancer Epidemiol. Biomarkers Prev. 2008;17(9):2208-14. [DOI] [PMC free article] [PubMed]
- 5.Adeghate E, Schattner P, Dunn E. An update on the etiology and epidemiology of diabetes mellitus. Ann N Y Acad Sci. 2006;1084:1-29. [DOI] [PubMed]
- 6.Imamura M, Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives. Endocr J. 2011;58(9):723-39. [DOI] [PubMed]
- 7.Ntzani EE, Kavvoura FK. Genetic risk factors for type 2 diabetes: insights from the emerging genomic evidence. Curr Vasc Pharmacol. 2012;10(2):147-55. [DOI] [PubMed]
- 8.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881-5. [DOI] [PubMed]
- 9.Cruz M, Valladares-Salgado A, Garcia-Mena J, Ross K, Edwards M, Angeles-Martinez J, et al. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev. 2010;26(4):261-70. [DOI] [PubMed]
- 10.Duval A, Busson-Leconiat M, Berger R, Hamelin R. Assignment of the TCF-4 gene (TCF7L2) to human chromosome band 10q25.3. Cytogenet Cell Genet. 2000;88(3-4):264-5. [DOI] [PubMed]
- 11.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005;280(2):1457-64. [DOI] [PubMed]
- 12.Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57(3):645-53. [DOI] [PubMed]
- 13.Shu L, Zien K, Gutjahr G, Oberholzer J, Pattou F, Kerr-Conte J, et al. TCF7L2 promotes beta cell regeneration in human and mouse pancreas. Diabetologia. 2012;55(12):3296-307. [DOI] [PubMed]
- 14.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320-3. [DOI] [PubMed]
- 15.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55(10):2903-8. [DOI] [PubMed]
- 16.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, et al. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55(9):2640-4. [DOI] [PubMed]
- 17.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55(9):2645-8. [DOI] [PubMed]
- 18.Bodhini D, Radha V, Dhar M, Narayani N, Mohan V. The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism. 2007;56(9):1174-8. [DOI] [PubMed]
- 19.Chang YC, Chang TJ, Jiang YD, Kuo SS, Lee KC, Chiu KC, et al. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes. 2007;56(10):2631-7. [DOI] [PubMed]
- 20.Hayashi T, Iwamoto Y, Kaku K, Hirose H, Maeda S. Replication study for the association of TCF7L2 with susceptibility to type 2 diabetes in a Japanese population. Diabetologia. 2007;50(5):980-4. [DOI] [PubMed]
- 21.Guinan KJ. Worldwide distribution of type II diabetes-associated TCF7L2 SNPs: evidence for stratification in Europe. Biochem Genet. 2012;50(3-4):159-79. [DOI] [PubMed]
- 22.Danquah I, Othmer T, Frank LK, Bedu-Addo G, Schulze MB, Mockenhaupt FP. The TCF7L2 rs7903146 (T) allele is associated with type 2 diabetes in urban Ghana: a hospital-based case–control study. BMC Med Genet. 2013;14:96. [DOI] [PMC free article] [PubMed]
- 23.Campbell DD, Parra MV, Duque C, Gallego N, Franco L, Tandon A, et al. Amerind ancestry, socioeconomic status and the genetics of type 2 diabetes in a Colombian population. PLoS One. 2012;7(4):e33570. [DOI] [PMC free article] [PubMed]
- 24.Marquezine GF, Pereira AC, Sousa AG, Mill JG, Hueb WA, Krieger JE. TCF7L2 variant genotypes and type 2 diabetes risk in Brazil: significant association, but not a significant tool for risk stratification in the general population. BMC Med Genet. 2008;9:106. [DOI] [PMC free article] [PubMed]
- 25.Barra GB, Dutra LAS, Watanabe SC, Costa PGG, Cruz PSM, Azevedo MF, Amato AA, et al. Association of the rs7903146 single nucleotide polymorphism at the Transcription Factor 7-like 2 (TCF7L2) locus with type 2 diabetes in Brazilian subjects. Arq Bras Endocrinol Metabol. 2012;56(8):479-84. [DOI] [PubMed]
- 26.Furgeri DT, Marson FA, Ribeiro AF, Bertuzzo CS. Association between the IVS4G>T mutation in the TCF7L2 gene and susceptibility to diabetes in cystic fibrosis patients. BMC Res Notes. 2012;5:561. [DOI] [PMC free article] [PubMed]
- 27.Barros CM, Araujo-Neto AP, Lopes TR, Barros MA, Motta FJ, Canalle R, et al. Association of the rs7903146 and rs12255372 polymorphisms in the TCF7L2 gene with type 2 diabetes in a population from northeastern Brazil. Genet Mol Res. 2014;13(3):7889-98. [DOI] [PubMed]
- 28.Assmann TS, Duarte GC, Rheinheimer J, Cruz LA, Canani LH, Crispim D. The TCF7L2 rs7903146 (C/T) polymorphism is associated with risk to type 2 diabetes mellitus in Southern-Brazil. Arq Bras Endocrinol Metabol. 2014;58(9):918-25. [DOI] [PubMed]
- 29.Lehman DM, Hunt KJ, Leach RJ, Hamlington J, Arya R, Abboud HE, et al. Haplotypes of transcription factor 7-like 2 (TCF7L2) gene and its upstream region are associated with type 2 diabetes and age of onset in Mexican Americans. Diabetes. 2007;56(2):389-93. [DOI] [PubMed]
- 30.Parra EJ, Cameron E, Simmonds L, Valladares A, McKeigue P, Shriver M, et al. Association of TCF7L2 polymorphisms with type 2 diabetes in Mexico City. Clin Genet. 2007;71(4):359-66. [DOI] [PubMed]
- 31.Martínez-Gómez LE, Cruz M, Martínez-Nava GA, Madrid-Marina V, Parra E, García-Mena J, et al. A replication study of the IRS1, CAPN10, TCF7L2, and PPARG gene polymorphisms associated with type 2 diabetes in two different populations of Mexico. Ann Hum Genet. 2011;75(5):612-20. [DOI] [PubMed]
- 32.Gamboa-Meléndez MA, Huerta-Chagoya A, Moreno-Macías H, Vázquez-Cárdenas P, Ordóñez-Sánchez ML, Rodríguez-Guillén R, et al. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61(12):3314-21. [DOI] [PMC free article] [PubMed]
- 33.Dutra LAS, Costa PGG, Velasco LFR, Amato AA, Barra GB. Allele-specific PCR assay to genotype SNP rs7903146 in TCF7L2 gene for rapid screening of diabetes susceptibility. Arq Bras Endocrinol Metabol. 2008;52(8):1362-6. [DOI] [PubMed]
- 34.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095-128. [DOI] [PMC free article] [PubMed]
- 35.Sepúlveda J, Murray C. The state of global health in 2014. Science. 2014;345(6202):1275-8. [DOI] [PubMed]
- 36.Tong Y, Lin Y, Zhang Y, Yang J, Zhang Y, Liu H, Zhang B. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15. [DOI] [PMC free article] [PubMed]
- 37.Wang J, Hu F, Feng T, Zhao J, Yin L, Li L, et al. Meta-analysis of associations between TCF7L2 polymorphisms and risk of type 2 diabetes mellitus in the Chinese population BMC Med Genet. 2013;14:8. [DOI] [PMC free article] [PubMed]
- 38.Wang J, Zhang J, Li L, Wang Y, Wang Q, Zhai Y, et al. Association of rs12255372 in the TCF7L2 gene with type 2 diabetes mellitus: a meta-analysis. Braz J Med Biol Res. 2013;46(4):382-93 [DOI] [PMC free article] [PubMed]
- 39.Franco LF, Crispim F, Pereira AC, Moisés RS; Japanese-Brazilian Diabetes Study Group. Variants of transcription factor 7-like 2 (TCF7L2) gene and incident glucose intolerance in Japanese-Brazilians. Braz J Med Biol Res. 2011;44(3):240-4. [DOI] [PubMed]
- 40.Peng S, Zhu Y, Lü B, Xu F, Li X, Lai M. TCF7L2 gene polymorphisms and type 2 diabetes risk: a comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis. 2013;28(1):25-37. [DOI] [PubMed]
- 41.Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117(8):2155-63. [DOI] [PMC free article] [PubMed]
- 42.Wagner R, Li J, Kenar E, Kohlbacher O, Machicao F, Häring HU, et al. Clinical and non-targeted metabolomic profiling of homozygous carriers of Transcription Factor 7-like 2 variant rs7903146. Sci Rep. 2014;4(5296). [DOI] [PMC free article] [PubMed]
- 43.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255-9. [DOI] [PMC free article] [PubMed]